Abstract
Type I toxin-antitoxin (TA) systems have been identified in a wide range of bacterial genomes. Here, we report the characterization of a new type I TA system present on the chromosome of the major human gastric pathogen, Helicobacter pylori. We show that the aapA1 gene encodes a 30 amino acid peptide whose artificial expression in H. pylori induces cell death. The synthesis of this toxin is prevented by the transcription of an antitoxin RNA, named IsoA1, expressed on the opposite strand of the toxin gene. We further reveal additional layers of post-transcriptional regulation that control toxin expression: (i) transcription of the aapA1 gene generates a full-length transcript whose folding impedes translation (ii) a 3΄ end processing of this message generates a shorter transcript that, after a structural rearrangement, becomes translatable (iii) but this rearrangement also leads to the formation of two stem-loop structures allowing formation of an extended duplex with IsoA1 via kissing-loop interactions. This interaction ensures both the translation inhibition of the AapA1 active message and its rapid degradation by RNase III, thus preventing toxin synthesis under normal growth conditions. Finally, a search for homologous mRNA structures identifies similar TA systems in a large number of Helicobacter and Campylobacter genomes.
INTRODUCTION
The bacterial pathogen Helicobacter pylori is the etiologic agent of chronic gastritis and peptic ulcers and plays a major role in the genesis of gastric cancer (1). About half of the human population is infected by this bacterium, which is responsible for about 700 000 deaths worldwide every year (2). To chronically survive and multiply in the human stomach, H. pylori has developed original strategies to modulate its gene expression in response to various stresses. Riboregulation, which has emerged as a major level of regulation in bacteria, was also proposed to play an important role in the adaptive response of H. pylori (3). However apart from housekeeping RNAs, transfer-messenger RNA, signal recognition particle RNA, 6S RNA and M1 RNA (RNase P), none of the enterobacterial small non-coding RNAs (sRNAs) are conserved in this bacterium. A combination of bioinformatics and genome wide RNA-seq analysis allowed us to characterize the H. pylori transcriptome and to reveal the existence of more than 60 new sRNAs in H. pylori strain 26695 (4). Regulator of polymeric G repeats (RepG) was identified as the first example of a trans-acting sRNA in H. pylori, repressing the expression of TlpB, a chemotaxis receptor (5). Although several of the RNA-seq-identified sRNAs in H. pylori are putative regulators, their mechanisms and functions are still unknown.
Among the sRNA with the highest level of expression in H. pylori strain 26695 was an intriguing family of six homologous cis-encoded antisense RNAs (named IsoA1 to IsoA6) that are expressed on the opposite strand of a novel class of small mRNAs (4). Using in vitro translation, we previously showed that each of the small mRNA of the A family expresses a short peptide (30 amino acids), designated AapA (Antisense-associated peptide family A). We also showed that the AapA and IsoA transcripts are both constitutively expressed in vivo during exponential growth, defining a small expression module, repeated many times at six different chromosomal loci (I–VI) (4). In vitro translation of AapA1 and AapA3 mRNAs was specifically inhibited by their cognate IsoA1 and IsoA3 antisense RNA, respectively. Due to the gene organization of these loci, it was hypothesized that these expression modules might constitute a new family of chromosomally encoded type I toxin-antitoxin (TA) systems. The TAs systems are categorized into six types based on their genetic organization and the nature of the antitoxin (6,7). In the type I, the toxin is downregulated by base-pairing of the antitoxin sRNA with the stable mRNA of the toxin (8). These systems were initially discovered on plasmids, where they play a key role in their stabilization during bacterial cell division, a phenomenon also known as post-segregational killing (9). When present on the chromosome, the identification of their function is less intuitive. A few of them have been reported to play important roles in adaptive responses to stress, including phenomena such as bacterial persistence (10).
In the present study, we characterize the aapA1/IsoA1 locus of H. pylori and demonstrate that it belongs to a new family of type I TA system. By using an artificial expression system, we show that the aapA1 gene encodes a small peptide whose expression leads to toxicity. The synthesis of the toxin is prevented by IsoA1 sRNA which thus acts as an antitoxin. Surprisingly, the use of rifampicin during RNA decay measurements reveals the existence of a transcript generated from a 3΄ processing of the highly stable AapA1 full-length (FL) mRNA. By using in vitro translation assays and footprinting experiments, we further demonstrate that, in contrast to the FL mRNA, the processed AapA1 mRNA can be translated due to a structural rearrangement of the 5΄ untranslated region (UTR). This truncated transcript binds IsoA1, creating an extended duplex that prevents ribosome binding and that is targeted for degradation by RNase III. This degradation prevents the accumulation of the active message, and, together with the particular folding of the FL mRNA allow H. pylori growth despite the presence of a toxic gene in its genome. Finally, we take advantage of the strong conservation of the mRNA folding properties of this new TA system to identify many homologs in other Helicobacter and Campylobacter species. Interestingly, they are not only present on the chromosome but also associated with mobile genetic elements (MGE) such as plasmids, prophages and integrative and conjugative elements (ICE).
MATERIALS AND METHODS
Molecular techniques
Molecular biology experiments were performed according to standard procedures and the supplier (NEB) recommendations. QIAprep Spin Miniprep Kit (Qiagen), PureLink® HiPure Plasmid Maxiprep Kit (Thermo Fisher Scientific) and QIAamp DNA Mini Kit (Qiagen) were used for plasmids preparations and H. pylori genomic DNA extractions, respectively. PCR were performed either with Taq Core DNA polymerase (MP Biomedicals), or with Phusion Hot Start DNA polymerase (Finnzymes) when the product required high fidelity polymerase.
H. pylori strains and culture conditions
The H. pylori strains used in this study (Supplementary Table S1) were 26 695 (11) B128 (12,13) and X47-2AL (14). Strains were grown on Columbia agar plates supplemented with 7% horse blood and Dent selective supplement (Oxoid, Basingstoke, UK) for 24–48 h depending on the strain. Liquid cultures were performed in brain-heart infusion medium (Oxoid) supplemented with 10% fetal bovine serum and Dent. H. pylori plates and liquid cultures were incubated at 37°C under microaerobic conditions (10% CO2, 6% O2, 84% N2) using an Anoxomat (MART microbiology) atmosphere generator. For liquid cultures, bacteria harvested from plates were inoculated at an optical density at 600 nm of 0.05 (OD600 = 0.05) into 5 ml (tubes, shaking at 175 rpm) brain-heart infusion medium supplemented with 10% fetal bovine serum and Dent supplement. After 12–24 h, pre-cultures were diluted to an OD600 of 0.05 into 25 ml (flasks, shaking at 125 rpm). Plasmids used for cloning were amplified in Escherichia coli strain JM109, which was grown in Luria–Bertani medium, supplemented either with kanamycin (50 μg.ml−1) or chloramphenicol (30 μg.ml−1). For H. pylori mutant selection and culture, antibiotics were used at the following final concentrations 20 μg.ml−1 kanamycine (Sigma) and 8 μg.ml−1 chloramphenicol (Sigma).
Construction of H. pylori mutant strains
Chromosomal mutants of H. pylori (Supplementary Table S1) were obtained by natural transformation as previously described (15). The ΔaapA1/IsoA1, Δrnc, aapA1Δ-10 box mutants and X47-2AL aapA126695 or aapA1B128 complementation strains were constructed by homologous recombination using a PCR cassette carrying an antibiotic resistance gene (aphA-3 or catGC gene conferring kanamycin or chloramphenicol resistance, respectively), flanked by approximately 500 base-pairs (bp) regions upstream and downstream of the gene of interest, as previously described (16). For the aapA1/IsoA1 deletion, the region between nt 1 245 653 to 1 245 866 (encompassing the Shine–Dalgarno (SD) sequence and the ATG and TAG codons) was removed, placing aphA-3 (from its own SD to its own stop codon) under the control of the aapA1 promoter and upstream of a transcriptional terminator (Supplementary Figure S8). The B128 and X47-2AL Δrnc strains were constructed by replacing the rnc Open Reading Frame (ORF) (from the ATG to 24 nt before its stop codon) by the aphA-3 gene amplified from its own ATG to its own TAG (Supplementary Figure S11). The aapA1Δ-10 box mutant was constructed by removing the two last nt of the aapA1 -10 box sequence (TAAAAT) (Supplementary Figure S9). To construct the X47-2AL aapA126695/ aapA1B128 complementation strains, the aapA1 locus (nt 1 245 624 to 1 245 986 for 26695 strain and 287 085 to 286 691 for B128 strain) with its own promoter (either from strain 26695 or B128) was fused to the catGC resistance gene and inserted into the rdxA locus of the X47-2AL strain. RdxA is a non-essential gene routinely used for complementation in H. pylori (17). The X47-2AL has a non-homologous copy of the aapA1/IsoA1 module (Supplementary Figure S10). The genomic DNA of H. pylori strain 26695, B128, P12 repG::aphA-3 (5) and the vectors pUC18K2 (kanamycin resistance gene) and pILL2150 (chloramphenicol resistance gene) were used as template for all PCR amplifications (see plasmids and primers list, Supplementary Table S2 and S3, respectively).
Plasmids constructions
Three plasmids carrying different isoforms of the aapA1/IsoA1 locus (from the start codon to the 3΄ UTR) were generated for this study. The pA1-IsoA1 plasmid contains the wild-type aapA1/IsoA1 sequence; the pA1 plasmid carries two mutations that inactivate the isoA1 promoter without changing the coding sequence of the peptide and pA1* is a derivative of pA1 containing an additional mutation in the start codon of AapA1 (ATG → ATT). The pA1-IsoA1 and pA1 plasmids were obtained by PCR amplification of genomic DNA from the 26695 strain, with the primer pairs FD213/FD180 and FD212/FD180, respectively. These products were cloned into pILL2157bis (18) between the NdeI and BamHI restriction sites (Supplementary Table S2). The pA1* plasmid was obtained by site-directed mutagenesis of the pA1 plasmid using primers FD608/FD609. The resulting plasmids were introduced into H. pylori strain B128 by mobilization, as described in Backert et al. (19).
Total RNA extraction
For RNA extraction, bacterial growth was stopped at the desired OD600 by adding 1.25 ml cold stop solution (95% ethanol, 5% acidic phenol) to 10 ml of culture, which was placed on ice. Cells were then centrifuged for 10 min at 3500 rpm at 4°C, and the pellets were stored at −80°C. Cell pellets were resuspended in 600 μl lysis solution (20 mM NaAc pH 5.2, 0.5% sodium dodecyl sulphate (SDS), 1 mM ethylenediaminetetraacetics acid (EDTA)) and added to 600 μl hot acidic phenol. After 6–10 min incubation at 65°C, the mixture was centrifuged for 10 min at 13 000 rpm at room temperature. Next the aqueous phase was transferred to a phase locked gel tube (Eppendorf) with an equal volume of chloroform and centrifuged 10 min at 13 000 rpm at room temperature. Total RNA was then precipitated from the aqueous phase by adding 2.5 volumes of EtOH 100% and 10% 3 M NaOAc pH 5.2. For RNA half-life determinations, rifampicin (Sigma, prepared at 34 mg.ml−1 in methanol) was added to the culture at a final concentration of 80 μg.ml−1 and cells were harvested after 0, 2, 5, 10, 20, 30, 60, 120 and 240 min of incubation. A culture where rifampicin was replaced by the same volume of methanol served as a non-treated control.
Northern blot
For Northern blot analysis, 5–20 μg of total RNA were separated on an 8% polyacrylamide (PAA), 7 M urea, 1X Tris Borate EDTA (TBE) gel. RNA was transferred to a nylon membrane (HybondTM-N, GE Healthcare Life Science) by electroblotting in TBE 1X at 8V overnight. Then RNA was cross-linked to the membrane by UV irradiation and hybridized with 5΄-labeled (γ32P) oligodeoxynucleotides (see Supplementary Table S3) in a modified Church buffer (1 mM EDTA, 0.5 M NaPO4 pH 7.2, 7% SDS) overnight at 42°C. Membranes were washed two times for 5 min in 2X SSC, 0.1% SDS and revealed using a Pharos FX phosphorimager (Biorad).
For riboprobe synthesis, a DNA template containing a T7 promoter sequence was amplified by PCR with primers FD671/FD672 from the 26695 strain genomic DNA as template. The AapA1 riboprobe was prepared as described in the Maxiscript Kit (Ambion) and purified on a Sephadex G50 column. Hybridization was performed in the modified Church buffer at 64°C and the membrane was washed 2 times for 5 min in 2X SSC, 0.1% SDS at 64°C.
In vitro transcription and translation assays
For in vitro synthesis of the AapA and IsoA RNAs, DNA templates were amplified from H. pylori 26695 genomic DNA using the primer pairs FD54/FD452 (AapA1_FL), FD54/FD55 (AapA1_Tr1), FD9 /FD15 (IsoA1 wt), FD205/FD234 (IsoA1L1L2), each forward primer carrying a T7 promoter sequence (see Supplementary Table S3). FD205/FD234 primers carry both three mutations resulting in the synthesis of the IsoA1L1L2 RNA that contains three mutations in each loop. In vitro transcription was carried out using the MEGAscript® T7 Transcription Kit (Ambion #AM1334) according to the manufacturer's protocol. After phenol:chloroform extraction followed by isopropanol precipitation, the RNA samples were desalted by gel filtration using a sephadex G-50 or G-25 (GE Healthcare) column, depending on the RNA size. For in vitro translation of the AapA1_FL and AapA1_Tr1 mRNAs, 0.05 to 1 μg of RNA was added to the E. coli S30 kit (Promega, #L1030) as previously described (4).
In vitro structure probing
AapA1 transcripts were first dephosphorylated by the CIP alkaline phosphatase (NEB) and then labeled with 10 pmol of γ32P-ATP and T4 PNK enzyme (NEB). Labeled RNA was purified on an 8% PAA containing 7 M urea 1X TBE and eluted overnight at 4°C under shaking in 750 μl elution buffer (0.1 M NaOAc pH 5.2, 0.1% SDS). Then, RNA was extracted by phenol:chloroform, desalted and concentrated by ethanol precipitation, pellets were resuspended in 50 μl H2O and stored at −20°C. Before use, each in vitro transcribed RNA was denatured by incubation at 90°C for 2 min in the absence of magnesium and salt, then chilled on ice for 1 min, followed by a renaturation step at room temperature for 15 min in 1X Structure Buffer (10 mM Tris-HCl pH 7.0, 10 mM MgCl2, 100 mM KCl).
Structures probing analyses were performed as described previously (4,20), using 0.1 pmol of labeled AapA1 RNA. To determine the secondary structure of RNA in native conditions (N), 1 μl RNase T1 (0.01 U.μl−1; Ambion, #AM2283), 1 μl RNase TA (0.005 μg.ml−1; Ambion, #AM2275) and 1 μl of RNase V1 (0.0005 U.μl−1; Ambion, #AM2275)) were added to the labeled RNA and incubated in 1X Structure Buffer for 1 to 2 min at 37°C. For the denaturing conditions (D), 1X Sequencing Buffer (20 mM Sodium Citrate, pH 5.0, 1 mM EDTA, 7M Urea) was used, with the same RNase T1 concentration and 1 μl of RNase TA (0.01U.μl-1) and incubation was performed at 37°C for 5 min. We performed lead acetate (5 mM final concentration) digestions on both aapA1_Tr1 and aapA1_FL in the absence or in the presence of a 2- or 10-fold excess of wild-type or mutated IsoA1 RNAs. All the reactions were stopped by adding 10 μl of 2X Loading Buffer (95% formamide, 18 mM EDTA, Xylene Blue and Bromophenol Blue) and stored at −20°C. Cleaved fragments were then analyzed on an 8% denaturing PAA gel containing 7 M urea and 1X TBE. Gels were dried 45 min at 80°C, and revealed using a Pharos FX phosphorimager (Biorad).
Sequence conservation analysis
A sequence similarity search for AapA mRNA homologs was carried out on the Helicobacter and Campylobacter genomes using the Geneious program on an iterative process (Geneious version 8.1.3 http://www.geneious.com (21)). Genome sequences used are listed in Supplementary Table S4. A sequence similarity with a threshold of 60% was used as a first restraint with the H. pylori 26695 aapA/IsoA nucleotide (nt) sequence (see Supplementary Figure S12 for more details). Once identified, the secondary structure of the AapA and IsoA RNAs were analyzed via the RNA fold plugin (22) implemented in Geneious. Finally, each locus was manually inspected for the presence of a promoter for both aapA and IsoA RNAs, as well as the presence of a SD sequence upstream of the AapA ORF. Every new result was added to the query database and the process was completed when no new result was found (Supplementary Figure S13).
RESULTS
The expression of AapA1 peptide is toxic to H. pylori
In the present study, our first goal was to establish whether the aapA1/IsoA1 module located at the locus I of the H. pylori strain 26695 encodes a type I TA system (Figure 1). Indeed, we previously reported that in vitro translation of the AapA1 mRNA leads to the synthesis of a small peptide of 30 amino acids whose expression is repressed in vitro by a small antisense RNA named IsoA1 (4). To this end, we cloned the coding sequence of this peptide into an E. coli/H. pylori shuttle vector (18) under the control of an isopropyl-β-D-thiogalactopyranoside (IPTG)-inducible promoter to generate plasmid pA1-IsoA1 (Figure 2A). Two mutated variants of this plasmid were also constructed. The first one carries two mutations in the IsoA1 antisense promoter (TATAAT → TACAAG) that prevents the expression of the antisense RNA without changing the peptide sequence (pA1 plasmid). The second one contains an additional mutation in the AapA1 mRNA start codon (ATG → ATT), which prevents the expression of the AapA1 peptide (plasmid pA1*) (Figure 2A). Each plasmid was transformed into H. pylori B128 strain deleted for the chromosomal copy of aapA1/IsoA1. The transcripts expressed from the different constructs were analyzed by Northern blot (Supplementary Figure S1A). We confirmed the inducible expression of the AapA1 transcripts in presence of IPTG despite some leakiness of the promoter in absence of IPTG. We also showed that the mutated isoA1 promoter is inactive, as shown by the absence of IsoA1 (Supplementary Figure S1A).
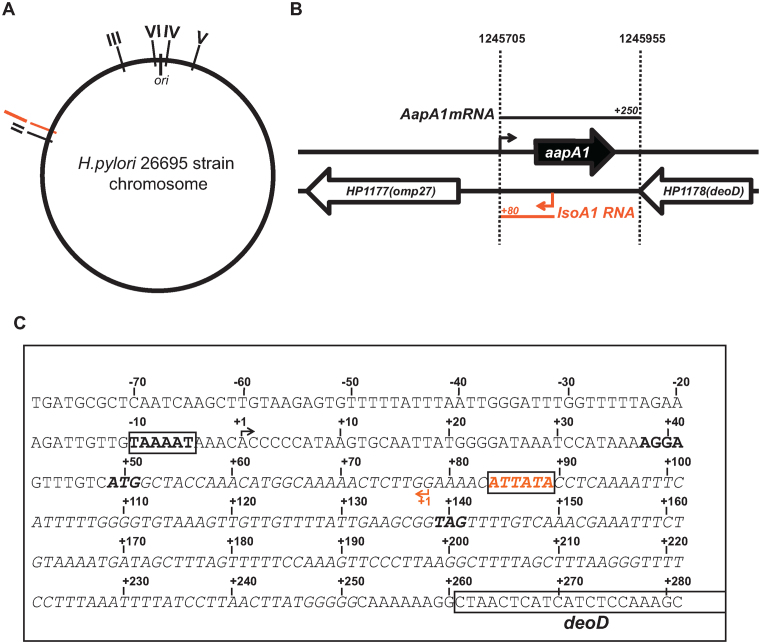
Figure 1.
Genetic organization of the aapA1/IsoA1 locus in the H. pylori 26695 strain. (A) Localization of the six different loci (I–VI) containing an homolog of the IsoA sRNA on the chromosome of H. pylori 26695 strain. The locus I containing the AapA1/IsoA1 module is shown in red. All loci are in the same orientation on the chromosome, the aapA gene being always on the positive strand (forward). (B) Genomic organization of the locus I. The open reading frame encoding the 30 amino acids peptide AapA1 (in black) and IsoA1 small RNA (in red) are transcribed from the intergenic region between the deoD gene (HP1178, purine nucleotide phosphorylase) and the omp27 gene (HP1177, outer membrane protein). Arrows indicate the respective transcriptional start sites of each transcript and black and red bars their approximate length determined by RNA-seq analysis (4). (C) The sequence of the intergenic region containing the aapA1/IsoA1 module in 26695 H. pylori strain is shown. The AapA1 fragment cloned into the pILL2157 vector is shown in italic. Promoter sequences (boxes) of aapA1 and isoA1 genes are shown in black and red, respectively. The Shine–Dalgarno sequence (SD), the start and stop codons are indicated by bold letters.
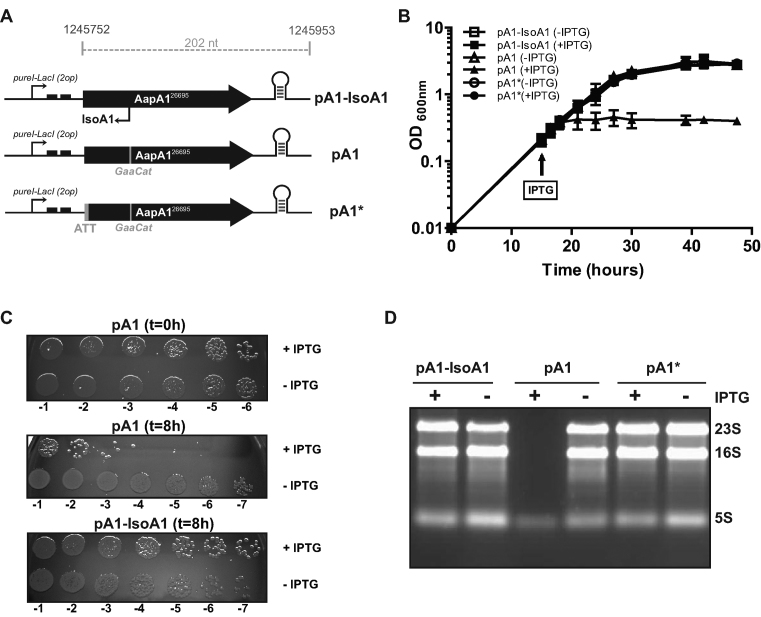
Figure 2.
Expression of the AapA1 peptide in H. pylori induces cell growth arrest, loss of viability and ribosomal RNA degradation. (A) Schematic representation of the aapA1 gene cloned into the pILL2157-bis plasmid (Supplementary Table S2). The fragment encompassing the region from the start codon to the 3΄ untranslated end of the aapA1 gene (202 nucleotides, the precise coordinates of the genomic DNA cloned in the vector are indicated, see the sequence in Figure 1C) was cloned downstream a ureI-derived promoter and two LacI repressor sequences (black boxes) (18). Thus the transcription of this construct leads to the production of a recombinant form of AapA1 mRNA and of an IsoA1 RNA that only shares 30 nucleotides with the original sequence and was therefore renamed IsoA1-rec (see expression in Supplementary Figure S1). The pA1 vector carries the same module except that two point mutations have been introduced to inactivate the isoA1 promoter without changing the AapA1 amino acids sequence. In pA1*, both the isoA1 promoter and the AapA1 start codon are mutated. Each vector has been introduced into H. pylori B128 strain deleted for the endogenous aapA1/IsoA1 module (B) Growth curves of the strains carrying either pA1, pA1* or pA1-IsoA1 vectors, in presence (black) or absence (white) of IPTG. IPTG was added at 16 h of culture. Data shown are the mean values ± standard deviations of three biological replicates. (C) Cell viability of the strains collected at 0 or 8 h after IPTG addition. Serial dilutions of the culture were plated on CAB-chloramphenicol agar plates. (D) Expression of AapA1 in B128 ΔaapA1/IsoA1 strain results in the degradation of rRNA. Ethidium-bromide 1% agarose gel was used to analyze rRNA (RNA samples were extracted after 8 h induction as shown in B and C). One microgram of total RNA was loaded in each lane. The positions of 23S, 16S and 5S rRNAs are indicated.
We next analyzed the effects of the AapA1 peptide expression on both growth rate (Figure 2B) and cell viability (Figure 2C). In the strain carrying plasmid pA1, induction of AapA1 expression caused an immediate growth arrest and a 104-fold decrease in cell number (Figure 2B and C). When IPTG was washed out from the cultures after 8 h of induction, cells expressing AapA1 were not able to recover growth (data not shown). Thus, expression of AapA1 is strongly toxic and bactericidal, and the onset of toxicity is fast relative to the generation time (3.8 h for pA1 expressing strain compared to 3.6 h for pA1-IsoA1 and pA1*). Of note, no cell lysis was observed upon expression of the toxin (Figure 2B). Interestingly, this lethality was correlated with a dramatic degradation of the 23S and 16S ribosomal RNAs while the toxin-encoding mRNA was still present (Figure 2D and Supplementary Figure S1B). The toxicity was not observed in the strain carrying the pA1-IsoA1 plasmid, demonstrating that transcription of IsoA1 prevents the toxicity of AapA1 and thus indeed acts as an RNA antitoxin. In addition, the absence of toxicity of the pA1* plasmid (in which the translation of the AapA1 mRNA is abolished) demonstrates that the toxicity is specifically due to the AapA1 peptide expression and not to the AapA1-encoding mRNA. The same results were obtained when using the wild-type background instead of the ΔaapA1 B128 strain, indicating that the chromosomal copy of IsoA1 cannot neutralize the ectopic expression of AapA1 (Supplementary Figure S1B).
Altogether these experiments demonstrate that the aapA1/IsoA1 module functions like a type I TA system in which the expression of a toxic peptide is prevented by the expression of an RNA antitoxin.
3΄-processing of AapA1 mRNA triggers translation activation
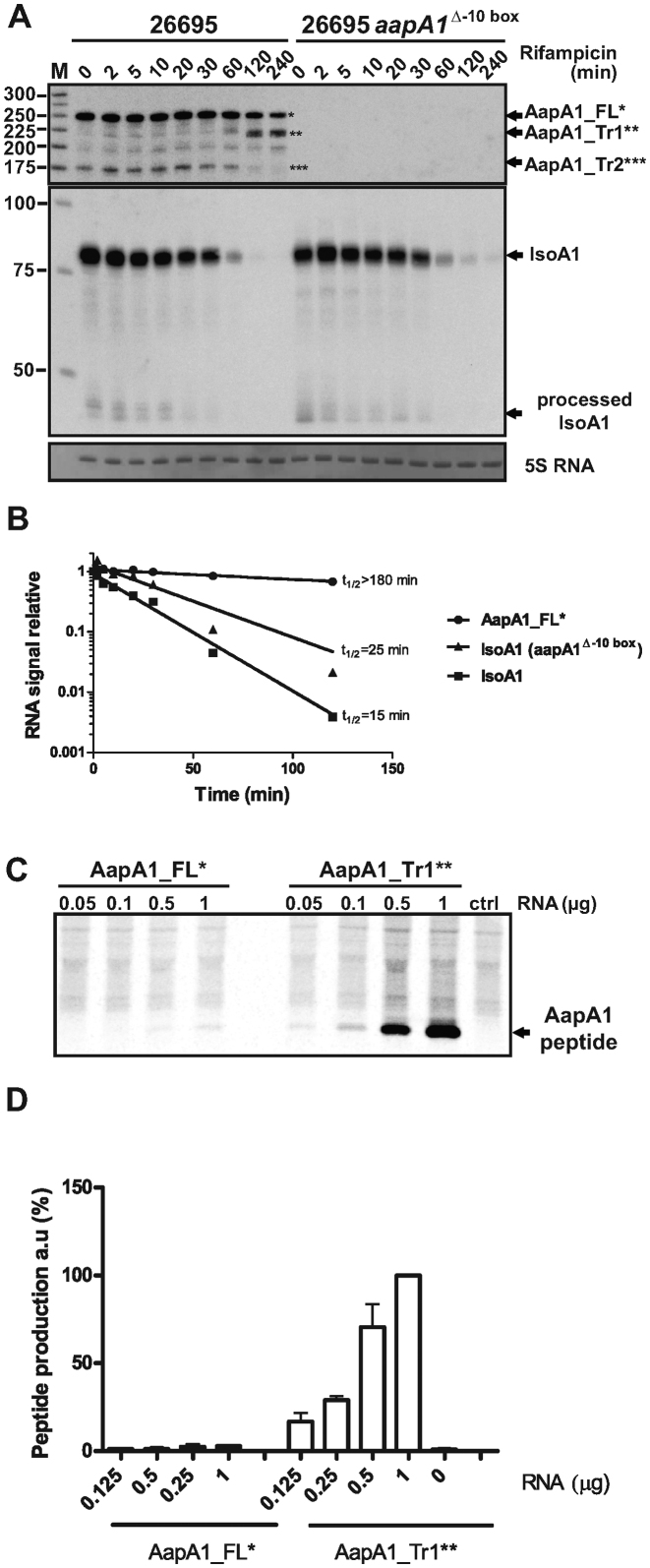
Type I TA systems are often characterized by a differential stability between the toxin-encoding mRNA and the antitoxin. To analyze the stability of both AapA1 and IsoA1 transcripts, cultures of H. pylori 26695 strain were treated with rifampicin to block transcription, and total RNA was extracted at different time points and analyzed by Northern Blot (Figure 3A). An oligonucleotide probe (FD47) targeting the AapA1 coding region detected several species from 175 to 250 nucleotides (nt) (Figure 3A, left panel). A similar pattern of expression was confirmed with a riboprobe directed against the first 225 nt of the mRNA (Supplementary Figure S2). None of these transcripts were detected in a strain deleted for the aapA1 promoter (Figure 3A right panel) showing that they are all produced from the same transcription start site at the position 1245705 (4). The strongest signal corresponds to the FL transcript of 250 nt (designated AapA1_FL), which displays a very slow decay with a half-life longer than 3 h. In contrast, the IsoA1 RNA has a relative short half-life of 15 min (Figure 3B). Most interestingly, 120 min after rifampicin addition, a transcript of around 225 nt appears. Since transcription is blocked, this transcript is the result of AapA1_FL processing and was consequently designated as AapA1_Tr1 (for truncated) (Figure 3A). Different probes targeting either the 5΄ or 3΄ end of the aapA1_FL revealed that the truncation occurs around 25 nt upstream of the 3΄ end of the FL message (Supplementary Figure S3). Another transcript of 172 nt, named AapA1_Tr2 was detected with FD47 but not with FA115 and FA116 probes (corresponding to the 5΄ and 3΄ ends of AapA1) indicating that this transcript is the result of cleavages of the FL message at both ends. Another faint band corresponding to a transcript of ∼200 nt was detected. However, since it was not observed in other H. pylori strains tested with the riboprobe (Supplementary Figure S2), we did not explore it in the present study. During the course of the rifampicin experiment we observed that AapA1_Tr1 appears at the same time that IsoA1 disappears. Since AapA1 mRNA expression leads to toxicity when the antitoxin is absent (Figure 2), we propose that AapA1_Tr1 is the active message. To test this hypothesis, the translatability of AapA1_FL and AapA1_Tr1 transcripts was assessed by in vitro translation assays (Figure 3C). Whereas a very faint signal corresponding to AapA1 peptide was observed with the FL transcript, a strong signal was detected with the Tr1 species (Figure 3C and 3D). These results demonstrate that a 3΄-end processing is required for a fully efficient in vitro translation of the AapA1 peptide.
Figure 3.
A truncated AapA1 mRNA species revealed by rifampicin treatment is efficiently translated in vitro (A) AapA1 and IsoA1 transcripts half-lives were determined in the 26695 H. pylori strain (left panel) or in an isogenic mutant deleted for aapA1 promoter (Δ-10 box, right panel). After rifampicin addition (at OD600 = 1.7), aliquots of cultures were collected at several time points (as indicated on top of the gel), RNA extracted and subjected to Northern blot analysis. The same membrane was successively probed with FD47 and FD198 labeled oligonucleotides to detect either AapA1 or IsoA1 transcripts, respectively. Arrows indicate the different transcripts: AapA1_FL (one star), AapA1_Tr1 (two stars), AapA1_Tr2 (three stars) and IsoA1 full-length and processed transcripts. A labeled DNA marker (lane M) was used for size estimation. Proper loading was assessed by staining the membrane with methylene blue (level of 5S rRNA is shown). The Northern blot shown here is a representative of more than three independent experiments using several biological replicates. (B) RNA decay was determined by plotting normalized intensities (RNA signal relative to time 0) of band corresponding to AapA1_FL* and IsoA1 transcripts, either in the wild-type or Δ-10 box mutated strains, as function of time after rifampicin addition. Approximate half-lives (t1/2, in min) are indicated for each transcript analyzed. (C) In vitro translations assays of AapA1_FL (left) and AapA1_Tr1 (right). Increasing amounts of in vitro synthesized mRNAs (μg) were added to E. coli S30 extracts in presence of 35S methionine. Control lane (Ctrl) shows the translation background obtained without exogenous mRNA. Here is a representative experiment out of several independent assays. (D) The amount of AapA1 peptide produced from both transcripts (AapA1_FL and Tr1) was quantified and the intensity of the band at the highest AapA_Tr1 quantity (1 μg) was set to 100%. Data are the average and standard deviations of three independent experiments.
The SD sequence is accessible in the truncated mRNA and sequestered in the full-length transcript
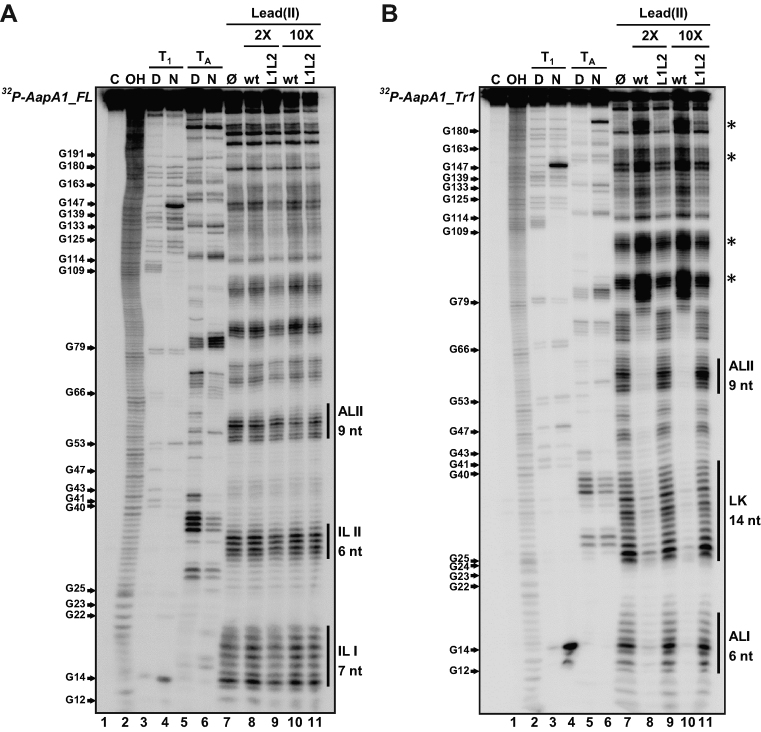
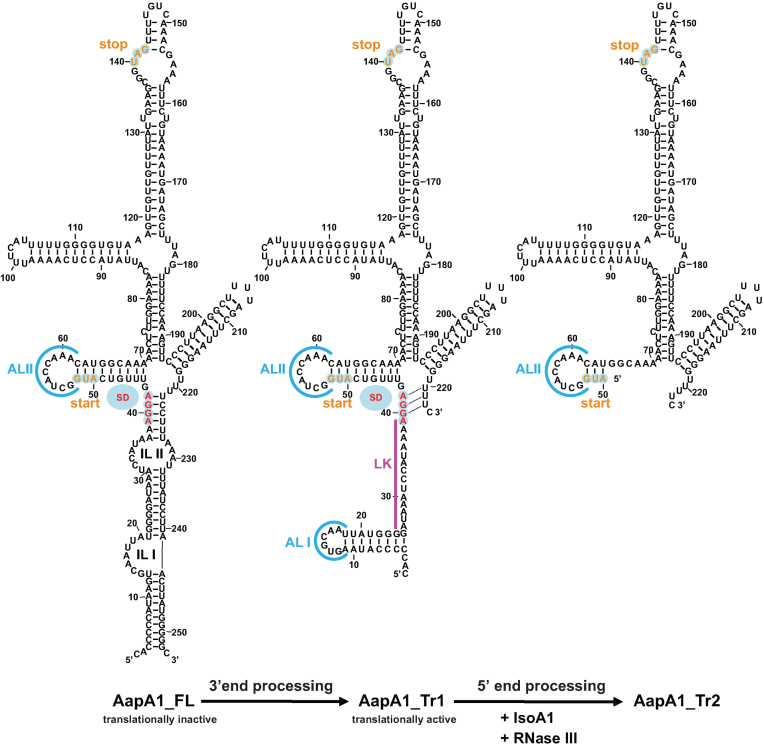
To investigate the reasons for the differential translation efficiency of the two AapA1 transcripts (FL and Tr1), we performed in vitro structure probing by combining chemical and enzymatic footprinting (Figure 4). RNAstructure software 5.2 (22) was used to generate secondary structure predictions of both transcripts according to the experimental data obtained (see the structures Figure 5). Lead cleavage showed a strong structural rearrangement of the 5΄ UTR between the two transcripts. Indeed while the cleavage pattern of AapA1_FL (Figure 4A, lane 7) fits very well with the formation of two internal loops (IL I and IL II, Figure 5), this region folds as two apical loops (AL I and AL II, Figure 5) connected by a 14 nt linker in AapA1_Tr1 (Figure 4B, lane 7). AapA1_FL folding involves a long distance interaction between the 5΄ and 3΄ ends of the transcript where the SD sequence is sequestered (Figure 5). This structure prediction explains the high stability of the transcript (few single stranded nucleotides accessible to RNases) but also why it is poorly translated. On the contrary, the 3΄-end truncation of AapA1_FL leads to an important structural rearrangement in the 5΄ UTR in which the SD sequence becomes accessible to ribosomes (Figure 5). In addition we also observed increased lead cleavages in the residues 66–75 in Tr1 versus the FL transcript, indicating that the second stem-loop might be more accessible to ribosomes in the truncated transcript than in the full length mRNA.
Figure 4.
Structure probing of (A) AapA1_FL and (B) AapA1_Tr1 RNAs in presence or absence of IsoA1. The secondary structure of each in vitro transcribed RNA was probed by submitting ∼0.1 pmol 32P- labeled RNA to partial enzymatic digestion either under native (N) or denaturing conditions (D) (RNase T1 cleaving single stranded G residues, lanes 3 and 4 ; RNase TA cleaving single stranded A residues, lanes 5 and 6). The interaction between AapA1 and IsoA1 was mapped using lead probing (lanes 7–11) in the absence (lane 7) or presence of either 2 or 10 times excess of wild-type IsoA1 (wt, lanes 8 and 10) or IsoA1 mutated in its apical loop (L1L2, lanes 9 and 11). Structure mapping, as well as secondary structures of IsoA1 wt and L1L2, are shown in Supplementary Figure S4. Untreated RNA (lane 1, denoted C) and partially alkali digested RNA (lane 2, denoted OH) served as control and ladder, respectively. Cleaved fragments were analyzed on an 8% denaturing polyacrylamide gel. Positions of all G residues are indicated relative to the transcription start site of the aapA1 gene (left of the gel). Single stranded regions involved in the 5΄ structural rearrangement are indicated by vertical black bars (right of the gel). The internal loops I and II (IL I, IL II) present in AapA1_FL are replaced by an apical loop (AL II) and a 14-nucleotides linker (LK) in AapA1_Tr1. Black stars on the right of the gel indicate other structural rearrangements observed following IsoA1 RNA binding.
Figure 5.
mRNA folding predictions of the different AapA1 transcripts. The sequences of the three different AapA1 mRNA species were inferred from the RNA-seq data (4) and from Northern Blot analyzed with a combination of various probes (Supplementary Figure S3). Each secondary structure was predicted using RNAstructure 5.2 software (22) in agreement with the experimental data obtained by enzymatic and chemical footprinting (Figure 4 and Supplementary Figure S5). The secondary structures were visualized and designed with the VARNA applet (46). The AapA1-FL transcript corresponds to a 253 nt transcript in which most of the 5΄ UTR is engaged in a long distance interaction with the 3΄ end of the message. A 3΄ processing event removing the last 30 nt leads to the formation of a truncated transcript (AapA1_Tr1) that undergoes a structural rearrangement, rendering the SD accessible to the ribosome. The first 76 nt of this translationally active message interact with IsoA1 to form an extended duplex (not shown here, see Supplementary Figure S5). This long duplex is then cleaved by RNase III to generate the AapA1_Tr2 transcript. The start and stop codons of the AapA1 ORF are indicated in orange and the SD sequence is in red. Apical loops (AL) are shown in blue. Two internal loops (IL) are shown in the AapA1_FL transcript.
This structural analysis provides the basis for the much higher translation efficiency observed with the truncated form of the AapA1 transcript as compared to the full-length message.
The truncated AapA1 mRNA and IsoA1 RNAs interact via loop–loop interactions to form a full duplex
To characterize the interaction between AapA_Tr1 and IsoA1 RNA, we performed lead probing on 5΄end-labeled AapA_Tr1 transcript in absence or presence of IsoA1 RNA (Figure 4B, lanes 7, 8 and 10). A comparison of the cleavage patterns showed that a 2-fold excess of IsoA1 resulted in a complete protection of the mRNA region that is complementary to IsoA1 (Figure 4B, lane 8). Indeed, lead cleavage was strongly reduced in the 5΄ UTR encompassing AL I, the 14 nt linker and AL II and this was even more pronounced when IsoA1 was added in a 10-fold excess (Figure 4B, lane 10). Moreover, other structural rearrangements (highlighted with a star) were observed indicating a refolding of the AapA1_Tr1 transcript upon the formation of an extended duplex with IsoA1 (Figure 4B). Under the same conditions, no interaction could be detected between AapA1_FL and IsoA1 (Figure 4A, lanes 8 and 10), indicating that the antisense RNA specifically targets the 3΄end-processed mRNA. Additionally, structure mapping and secondary structure predictions performed on IsoA1 RNA (Supplementary Figure S4) showed that the AapA1_Tr1 5΄ UTR and IsoA1 both fold into two complementary stem-loops that could favor the formation of loop–loop interactions.
To determine whether such kissing-loop complexes could represent the initial step of the AapA1/IsoA1 hybrid formation, we performed in vitro footprinting experiments in presence of neomycin. This antibiotic of the aminoglycoside family is known to stabilize kissing complexes and to prevent the formation of extended duplexes (23). In presence of increasing concentration of neomycin, the LK region of AapA1_Tr1 was no longer base-paired whereas the ALI and ALII loops were still protected (Supplementary Figure S5A), confirming that loop–loop interactions are promoting the interaction between the two transcripts. In addition, we could map neomycin's binding regions at the boundaries of the helices (Supplementary Figure S5A, lanes 6 and 7), explaining why this antibiotic prevents the AapaA_Tr1/IsoA1 hybrid to extend into a full duplex (Supplementary Figure S5A lanes 10–11). No interaction was observed with IsoA3 which displays a similar fold than IsoA1 but a different sequence in the loop (Supplementary Figure S4B).
To further confirm that the initial interaction is mediated via loop–loop interaction, we constructed an IsoA1 variant mutated in its two apical loops L1 and L2 (named IsoA1 L1L2) and tested its ability to bind the AapA1_Tr1 transcript (Figure 4 and Supplementary Figure S6). We first verified that the three nt mutations introduced in each loop of IsoA1 did not alter its folding (Supplementary Figure S6). In contrast to the wild-type (wt) IsoA1, the addition of IsoA1 L1L2 did not change the digestion profile of AapA1_Tr1 (Figure 4B, lanes 9 and 11) indicating that this mutant cannot bind despite a strong sequence complementarity. This result confirms that the interaction between AapA1_Tr1 and IsoA1 is mediated via a loop–loop interaction. Finally, we performed the reverse experiment, i.e lead cleavage of 5΄ end-labeled IsoA1 upon AapA1_Tr1 addition. The results confirmed the duplex formation between IsoA1 and the first 76 nt of AapA1_Tr1 mRNA (Supplementary Figure S6A). This very long hybrid is masking the ribosome binding site (RBS) of AapA1_Tr1 (Supplementary Figure S5B) suggesting that the primary role of the antitoxin is to inhibit the translation of the toxin.
Taken together these data indicate that IsoA1 antisense RNA binds specifically to the AapA1_Tr1 active transcript via kissing-loop interactions (Supplementary Figure S5B). These short interactions are key determinants in the specificity of the interaction and explain why each IsoA RNA only represses the translation of its cognate mRNA, as shown previously (4).
RNase III cleavage ensures rapid turnover of the translationally active message
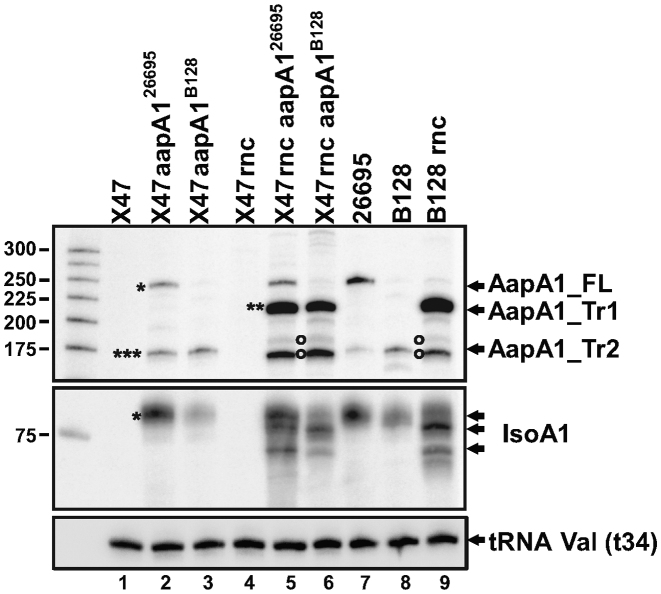
Base-pairing between AapA1_Tr1 and IsoA1 creates a long duplex of 76 bp (Supplementary Figure S5B), which could be a good substrate for the double-stranded specific ribonuclease RNase III. To investigate the role of this ribonuclease in aapA1/IsoA1 regulation in vivo, we compared both transcripts in wt and RNase III-deficient H. pylori strains. Since we could not delete the rnc (HP0662) gene encoding RNase III in the 26695 strain, we used two other strains, B128 and X47-2AL, for which the deletion of rnc could be obtained. We inserted the complete aapA1/IsoA1 module of either the 26695 or the B128 strain into the rdxA gene of the wt and Δrnc X47-2AL strains (see Supplementary Figure S10 for details). We then analyzed the expression of AapA1 and IsoA1 RNAs by Northern blot (Figure 6). The introduction of either aapA1/IsoA1 module in the X47-2AL background resulted in the expression pattern observed for the parental strain (Figure 6, compare lanes 2 and 3 with lanes 7 and 8). Thus, transcription of the aapA1/IsoA1 module is not influenced by the recipient strain under these conditions. In both the X47-2AL and B128 backgrounds, the rnc deletion led to a strong accumulation of the AapA1_Tr1 truncated form (two stars) while the levels of AapA1_FL (one star) were unaffected (Figure 6, lanes 5, 6 and 9). Same samples were run longer for a better separation of the different species (Supplementary Figure S7). In addition AapA1_Tr2 (three stars) was absent in the Δrnc strains, indicating that this transcript results from RNase III cleavage of AapA1_Tr1 (Supplementary Figures S2 and S7). In contrast, the amount of the full-length IsoA1 RNA seemed unaffected in absence of RNase III. We have shown that, in wild-type cells, IsoA1 RNA is in large excess over AapA1_Tr1. Therefore, the proportion of IsoA1 transcripts base-paired with AapA_Tr1 mRNA is negligible, explaining why the global level of IsoA1 full-length RNA appears unchanged in wt and Δrnc backgrounds (Figure 6, compare lanes 2, 3 and 8 with lanes 5, 6 and 9). Interestingly, in absence of RNase III, two other transcripts (highlighted by open circles) of a size similar to AapA1_Tr2 accumulate (Figure 6, lanes 5, 6 and 9) and are probably the result of an alternative degradation of AapA1_Tr1 mRNA by unknown ribonucleases.
Figure 6.
RNase III prevents accumulation of the active AapA1_Tr1 mRNA. Expression of AapA1 and IsoA1 RNAs in wild type and isogenic Δrnc H. pylori strains (X47-2AL, 26695 and B128) were analyzed by Northern blot. The aapA1/IsoA1 locus of the 26695 strain (AapA126695) or of the B128 strain (AapA1B128) was inserted in the X47-2AL strain and its Δrnc derivative. Samples from each strain were collected when culture reached OD600 ≈ 1. tRNA Val (t34) served as a loading control and was probed with the FD499 radiolabeled oligonucleotide. Upper panel: AapA1 transcripts revealed with FD47 probe; middle panel: IsoA1 transcripts revealed with FD198 probe.
Overall these data indicate that RNase III is the major enzyme responsible for the AapA1_Tr1/IsoA1 duplex degradation thereby ensuring the rapid turnover of the AapA1 active message.
Identification of new aapA/IsoA systems in other Helicobacter and Campylobacter species through structural conservation
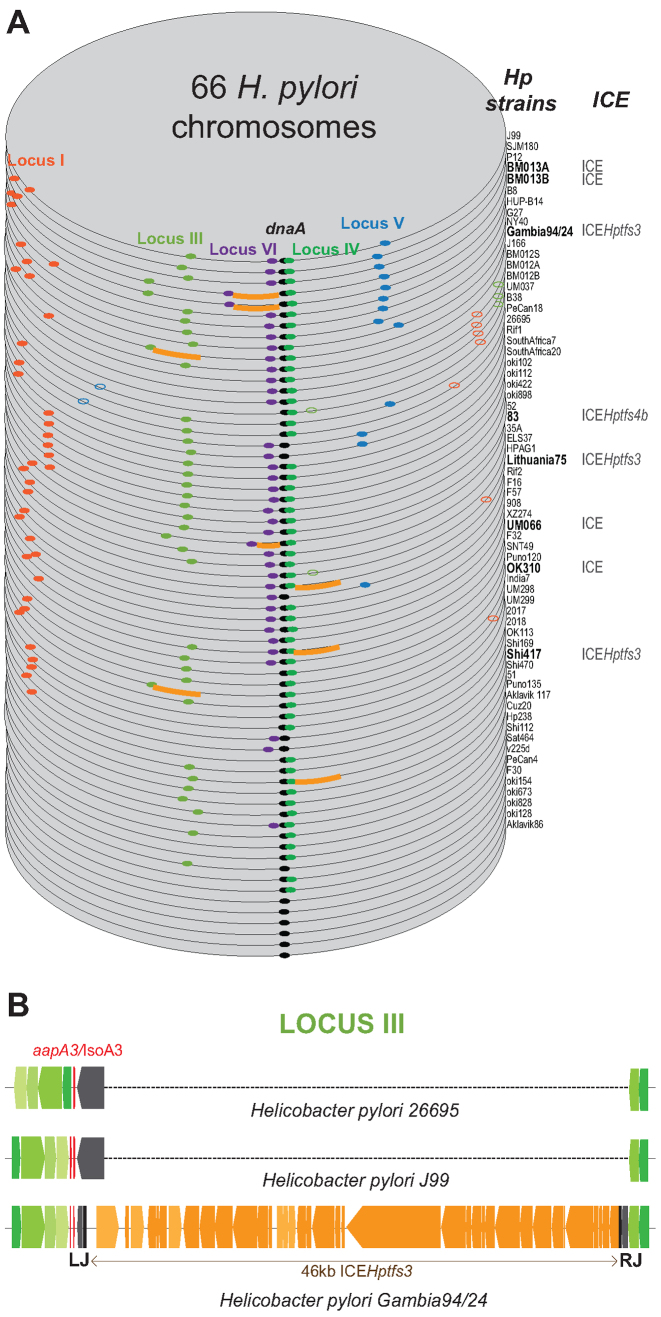
We analyzed the conservation of the newly characterized type I aapA/IsoA TA system among the H. pylori strains. A PSI-BLAST search to detect small peptides (as described in (24)) resulted in very few positive hits because few ORFs encoding small proteins have been annotated. We next conducted a TBLASTN search against nucleotide database. This search revealed the presence of aapA/IsoA homologs in almost every H. pylori genome and in the closely related genomes of Helicobacter acinonychis and Helicobacter cetorum (Supplementary Table S4). Moreover, the synteny associated with the different loci was strongly conserved between the various strains (Supplementary Figure S12). The aapA/IsoA loci were thus classified according to their position at the 5 loci named I, III, IV, V and VI (Figure 7A). Although IsoA2 at locus II is conserved, no functional promoter upstream of a putative AapA2 mRNA nor a conserved small ORF longer than four amino acids were detected. Thus, this locus seems not functional and therefore was no longer considered in this study. Among the 66 H. pylori complete genomes analyzed, the aapA/IsoA modules were present in one or multiple copies at the 5 conserved loci in all but 6 genomes that are free of intact aapA/IsoA systems (Figure 7A and Supplementary Table S4). We then hypothesized that, given the importance of mRNA folding in the control of the AapA1 toxin expression, this folding should be conserved. We thus carried out a search for structural mRNA homologs against all available Helicobacter complete genomes and other closely related species such as Campylobacter (Supplementary Figure S13). This search identified many new loci that were all manually inspected for features such as mRNA folding, presence of a conserved SD sequence, start and stop codons, as well as a promoter for the antisense RNA (Supplementary Figure S13). This manual inspection allowed us to dismiss a considerable fraction of detected loci. Each new positive locus was implemented for the next search in an iterative manner (see the detail of this search in Supplementary Figure S13). Interestingly, this new search identified many AapA mRNA homologs in other Helicobacter species that were not found with the TBLASTN or PSI-BLAST searches. Since the synteny was not conserved in these Helicobacter species, these systems were named aapA/IsoA without any specific number associated (Supplementary Table S4). Surprisingly, this search revealed the presence of this system near prophage encoding genes in H. felis ATC 49179 and H. bizzozeronii CIII-1 or near a transposon in H. cetorum MIT-99 5656 (Supplementary Figure S14). A closer look at the H. pylori loci revealed that some of them were associated with another type of MGE such as the ICE recently characterized in H. pylori genomes (25,26). For instance in Gambia94/24, a complete ICE of 46 kb named ICEHptfs3 was present in the intergenic region corresponding to the locus III, creating a much larger genomic region (Figure 7B). Finally, although no aapA/IsoA system was detected in Helicobacter plasmids, one was identified on a Campylobacter jejuni plasmid as well on the chromosome of some C. jejuni strains in a region involved in plasmid stabilization (Supplementary Figure S15).
Figure 7.
Conservation of aapA/isoA TA systems in 66 Helicobacter pylori strains. (A) Each genome is stacked according to the decreasing number of systems present at each locus and aligned to the origin of replication (next to the dnaA gene). Each locus containing at least one copy of a functional aapA/IsoA module (presence of both promoters and of a small peptide with the consensus length of 30 amino acids) is represented according to the following color code: locus I (red), III (light green), IV (dark green), V (blue) and VI (purple). Most systems have the mRNA on the positive strand; the few ones on the minus strand are shown as open dots. A thick orange line shows the presence of an integrative and conjugative element (ICE) next to an aapA/isoA module. (B) Schematic representation of the locus III alignment in H. pylori strains 26695, J99 and Gambia94/24. Blue and green colored genes represent the 5΄ and 3΄ side of the locus III, respectively. The aapA gene is in red. All the ORF contained in the ICE Hptfs3 are shown in orange. Left (LJ) and right (RJ) junctions of the ICE are indicated.
Altogether these results indicate that while these TA systems have been discovered initially on the chromosome of H. pylori, their localization near MGEs such as ICE, transposon and prophage suggests that they might have been acquired via horizontal gene transfer and played a role in stabilizing these mobile elements.
DISCUSSION
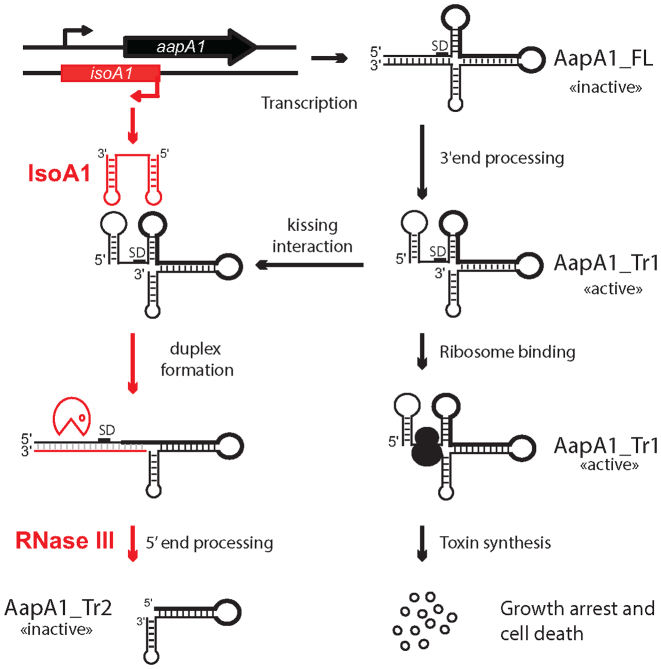
In this article we characterize for the first time a type I toxin–antitoxin system in H. pylori. We have shown that the aapA1 gene encodes a small peptide whose expression is toxic to H. pylori and found that several layers of post-transcriptional regulation are preventing the expression of this toxin, as described in our working model in Figure 8. The transcription of the aapA1 gene generates a full-length mRNA of 250 nt, denoted AapA1_FL that is translationally inactive. This inert form is constitutively processed at its 3΄ end to generate a truncated transcript of 225 nt which is translatable. This active form denoted AapA1_Tr1, base pairs with the IsoA1 antisense RNA to form an extended duplex that is rapidly degraded by RNase III to generate AapA1_Tr2. In absence of IsoA1, the synthesis of AapA1 toxin can occur leading to a growth arrest and cell death.
Figure 8.
Model of the aapA1/IsoA1 TA system regulation. This model is based on all experimental data presented and is described in the text (see discussion). Simplified structures of AapA1 transcripts (FL, Tr1 and Tr2) are based on Figure 5. The region encoding the AapA1 ORF in the transcript is shown with a thick black line. The Shine-Dalgarno sequence is indicated as SD. The translationally active and inactive states of the different mRNA species are also indicated.
Metastable structures prevent premature toxin expression during mRNA synthesis
While most TA systems including type II, III, IV and V, have adopted a specific operon organization allowing the production of the antitoxin before that of the toxin, type I TA systems are using an alternative strategy to prevent toxin production. Most type I toxin encoding mRNAs have their SD sequence sequestered in a stable double-stranded region that prevents expression of the toxin while the RNA is transcribed (20,27–31). In some cases, this sequestration involves a long distance base-pairing between the 5΄- and 3΄- UTRs (31,32). However due to transcription–translation coupling that occurs in bacteria, the RBS could be, in principle, available before the 3΄ region is synthesized. This particular mode of regulation implies that metastable structures sequestering the RBS are formed during transcription in order to prevent translation initiation on the nascent mRNA (31,32). To predict such transient secondary structures that are formed during transcription, we used the Kinefold stochastic simulations that predict co-transcriptional folding paths of functional RNAs (33). This simulation showed that three successive structures masking the RBS can potentially form at various stages of mRNA transcription (Supplementary Figure S16 and Video SM1 for AapA1_FL mRNA, data not shown for the other loci). A more stable structure replaces the first two metastable structures when transcription reaches the end of the message. Besides sequestration of the RBS, the 5΄ to 3΄ end-pairing of the full-length mRNA forms an 11 bp helix including 6 GC pairs (Figure 5) that induces an efficient transcription termination at this position. Another putative terminator stem-loop was previously annotated at the position +225 (4) but our work shows that it is not able to induce transcription termination very efficiently. In addition, the IsoA1 could, in principle, bind to the nascent AapA1 mRNA, before the RNA polymerase reaches the end of the message. This interaction could then promote the production of the Tr1 transcript. However, we believe this scenario quite unlikely because the AapA1_Tr1 is only visible after rifampicin treatment at late stages when IsoA1 is completely degraded. In addition we would never observed accumulation of the full length mRNA. Altogether our results clearly indicate that IsoA1 is required for the degradation of Tr1 but not for its production.
AapA1 mRNA requires a 3΄ end processing to be translated
For type I toxin mRNAs having their SD sequence sequestered, the mechanism by which the toxin mRNA gains activation is still poorly documented. Our results show that for AapA1, a 3΄ end-processing event is required to refold the mRNA and to unmask the SD (Figure 5). Until now, two examples of type I TA systems for which the toxin is produced after the cleavage of the mRNA have been described (either in 5΄ for TisB or 3΄ end for Hok (20,34)). In contrast to the 3΄ end processing involved in the maturation of the Hok mRNA, the 3΄ to 5΄ exonucleolytic activity of the polynucleotide phosphorylase and of the ribonuclease II is not sufficient to process the 3΄ end of the AapA1_FL mRNA (H. Arnion, unpublished results). As for TisB (20), the enzymatic activity responsible for the processing remains to be identified.
IsoA1 inhibits toxin expression primarily at the translational level
In order to avoid toxin expression, IsoA1 is constitutively synthesized in a large excess over AapA1_Tr1. The double stem-loop structure in the 5΄UTR of AapA1-Tr1 mRNA mediates a specific recognition by the IsoA1 RNA via loop–loop interactions. In vitro this hybrid is then extended into a full duplex that is cleaved by the double-stranded specific RNase III. In vivo this degradation is very rapid since the AapA1_Tr1 does not accumulate. However, RNase III is not essential in the B128 strain despite the presence of several aapA/IsoA systems in its genome, suggesting that, at least in this strain, RNase III cleavage is not mandatory for the repression of the toxin. The binding of IsoA1 to AapA1_Tr1 is completely masking the RBS and in absence of RNase III, the stability of the long duplex formed (76 bp) is probably sufficient to prevent ribosome binding. Thus, the repression of AapA1 toxin expression by IsoA1 RNA may primarily occur at the translational level, as previously shown for other type I toxins (i.e. Hok (35), TisB (20), Fst (29) and BsrG (28)). In contrast, in B. subtilis, the degradation of the TxpA and YonT toxin mRNAs by RNase III is essential to prevent toxin synthesis (27,36).
Analogy with other type I TA systems
This new system identified in epsilon proteobacter species shares many similarities with other type I TA previously identified in enterobacteria. For instance the aapA1/IsoA1 system share common features with the overlapping cis-encoded antisense RNA regulating TA systems characterized in E. coli (37). Similar to hok/Sok and ldr/Rdl TA systems, this system encodes a small toxic protein (size < 50 aa) that is synthesized from a highly stable mRNA and which production is repressed by a small unstable antisense RNA. In addition, as for hok/Sok, the aapA1/IsoA1 requires a 3΄ processing to get activated (34). However, in contrast to these two TA systems, its translation activation does not seem to require the translation of an overlapping small ORF (like the mok and ldrX peptides of the hok/Sok and ldr/Rdl systems, respectively) (38,39). Another major difference with other TA systems relies on the fact that the IsoA1 antitoxin is directly targeting the translation initiation region of the toxin gene. This specificity is due to the genomic organization of this TA system, where the antitoxin is fully complementary to the 5΄ UTR of the toxin-encoding mRNA, as also shown for the symE/SymR TA system in E. coli (40).
Conservation of aapA/IsoA TA systems in Helicobacter and Campylobacter
Few reports have highlighted the difficulties in predicting the presence of type I TA system in bacterial genomes (24,41). Indeed the large diversity of toxin sequences as well as their small nucleotide size complicates the finding of new type I TA systems. In this report we present a new strategy taking advantage of the specific folding adopted by the AapA mRNA to identify more representatives of these systems. Our search revealed the presence of a large number of these systems in Helicobacter genomes, larger than anticipated (4). With the exception of a few strains that only have ghost copies of aapA/IsoA (i.e. with either no functional promoter and/or no SD and/or no ORF longer than 5–6 amino acids), most H. pylori strains contain one or multiple copies of these systems at conserved loci on the chromosome. At a given locus, they can be present in single or multiple copies, depending on the strains. The locus IV, located close to the replication origin is the most conserved one, as it is found in all the strains analyzed. Our data show that these TA systems are sometimes inactivated by point mutations, genomic rearrangements or insertion of an IS (insertion sequence) element. This is similar to what was observed for the hok/Sok systems present on the chromosome of E. coli (41). However, we also found that in some cases, the aapA/IsoA systems are associated with MGE. For instance our search reveals their presence on Campylobacter plasmids and within genomic islands coding either for an ICE element (strains shown in bold in Figure 7A) or prophage genes (Helicobacter felis ATCC49179 and Helicobacter bizozzeroni MIT 99–5656). In these cases, these newly identified TA systems may be part of a plasmid stabilization and/or addiction system as was reported for several TA systems (42). This phenomenon, also known as post-segregational killing, was first described for hok/Sok system (43). If a plasmid bearing a TA module is not transmitted to a daughter cell, the unstable antitoxin is degraded while the stable toxin acts on cellular targets and kill the plasmid-free cells. For instance type II systems have already been shown to promote the maintenance of ICE element and conjugative plasmids (44,45). Whether a similar role could be played by the aapA/IsoA systems remains to be explored.
CONCLUSION
To conclude, the characterization of a new TA system in H. pylori revealed the importance of mRNA folding in type I TA regulation. We propose that the AapA mRNA requires two structural RNA switches to gain activation. The first one consists of a spontaneous conformational rearrangement, in which metastable 5΄-end structures are disrupted in favor of a more stable 5΄ end/3΄ end interaction. The second switch is induced after a processing event done by a yet unknown RNase, which renders the mRNA translationally active. This sophisticated mechanism may be conserved for other TA systems. Another interesting characteristic of the aapA/IsoA system, is the high stability of the mRNA versus the instability of the antitoxin that makes it a particularly efficient addiction module. Indeed, the cell should not be able to get rid of such TA system unless a non-sense mutation is present in the toxin gene. Finally our bioinformatic analysis revealed that the aapA/IsoA system belongs to a novel large family of type I TA system that is not only present on the chromosome but also associated with MGEs. Although our data provide some hints for a role in stabilizing MGEs, these systems might have diverse functions depending on their genetic localization or on the organisms that host them.
Supplementary Material
ACKNOWLEDGEMENTS
The authors thank Denis Dupuy, Axel Innis and Cathy Staedel for critical reading of the manuscript and all present and past members of the ARNA laboratory for helpful discussions.
SUPPLEMENTARY DATA
Supplementary Data are available at NAR Online.
FUNDING
INSERM ; Université de Bordeaux; Agence Nationale de la Recherche (http://www.agence-nationale-recherche.fr/) [ANR-12-BSV5-0025-Bactox1, ANR-12-BSV6-007-asSUPYCO]; European Union's Horizon 2020 research and innovation programme under the Marie Sklodowska-Curie grant agreement [642738]. Funding for open access charge: INSERM.
Conflict of interest statement. None declared.
REFERENCES
- 1. Cover T.L., Blaser M.J.. Helicobacter pylori in health and disease. Gastroenterology. 2009; 136:1863–1873. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2. Ferlay J., Shin H.-R., Bray F., Forman D., Mathers C., Parkin D.M.. Estimates of worldwide burden of cancer in 2008: GLOBOCAN 2008. Int. J. Cancer. 2010; 127:2893–2917. [DOI] [PubMed] [Google Scholar]
- 3. Pernitzsch S.R., Sharma C.M.. Transcriptome complexity and riboregulation in the human pathogen Helicobacter pylori. Front. Cell. Infect. Microbiol. 2012; 2, 14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Sharma C.M., Hoffmann S., Darfeuille F., Reignier J., Findeiß S., Sittka A., Chabas S., Reiche K., Hackermüller J., Reinhardt R. et al. The primary transcriptome of the major human pathogen Helicobacter pylori. Nature. 2010; 464:250–255. [DOI] [PubMed] [Google Scholar]
- 5. Pernitzsch S.R., Tirier S.M., Beier D., Sharma C.M.. A variable homopolymeric G-repeat defines small RNA-mediated posttranscriptional regulation of a chemotaxis receptor in Helicobacter pylori. Proc. Natl. Acad. Sci. U.S.A. 2014; 111:E501–510. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Goeders N., Van Melderen L.. Toxin-antitoxin systems as multilevel interaction systems. Toxins. 2014; 6:304–324. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Wen J., Fozo E.M.. sRNA antitoxins: more than one way to repress a toxin. Toxins. 2014; 6:2310–2335. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Brantl S., Jahn N.. sRNAs in bacterial type I and type III toxin-antitoxin systems. FEMS Microbiol. Rev. 2015; 39:413–427. [DOI] [PubMed] [Google Scholar]
- 9. Gerdes K., Gultyaev A.P., Franch T., Pedersen K., Mikkelsen N.D.. Antisense RNA-regulated programmed cell death. Annu. Rev. Genet. 1997; 31:1–31. [DOI] [PubMed] [Google Scholar]
- 10. Gerdes K., Maisonneuve E.. Bacterial persistence and toxin-antitoxin loci. Annu. Rev. Microbiol. 2012; 66:103–123. [DOI] [PubMed] [Google Scholar]
- 11. Tomb J.F., White O., Kerlavage A.R., Clayton R.A., Sutton G.G., Fleischmann R.D., Ketchum K.A., Klenk H.P., Gill S., Dougherty B.A. et al. The complete genome sequence of the gastric pathogen Helicobacter pylori. Nature. 1997; 388:539–547. [DOI] [PubMed] [Google Scholar]
- 12. Farnbacher M., Jahns T., Willrodt D., Daniel R., Haas R., Goesmann A., Kurtz S., Rieder G.. Sequencing, annotation, and comparative genome analysis of the gerbil-adapted Helicobacter pylori strain B8. BMC Genomics. 2010; 11, 335. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. McClain M.S., Shaffer C.L., Israel D.A., Peek R.M., Cover T.L.. Genome sequence analysis of Helicobacter pylori strains associated with gastric ulceration and gastric cancer. BMC Genomics. 2009; 10, 3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Ermak T.H., Giannasca P.J., Nichols R., Myers G.A., Nedrud J., Weltzin R., Lee C.K., Kleanthous H., Monath T.P.. Immunization of mice with urease vaccine affords protection against Helicobacter pylori infection in the absence of antibodies and is mediated by MHC class II-restricted responses. J. Exp. Med. 1998; 188:2277–2288. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Skouloubris S., Thiberge J.M., Labigne A., De Reuse H.. The Helicobacter pylori UreI protein is not involved in urease activity but is essential for bacterial survival in vivo. Infect. Immun. 1998; 66:4517–4521. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Stingl K., Brandt S., Uhlemann E.-M., Schmid R., Altendorf K., Zeilinger C., Ecobichon C., Labigne A., Bakker E.P., de Reuse H.. Channel-mediated potassium uptake in Helicobacter pylori is essential for gastric colonization. EMBO J. 2007; 26:232–241. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Goodwin A., Kersulyte D., Sisson G., Veldhuyzen van Zanten S.J., Berg D.E., Hoffman P.S.. Metronidazole resistance in Helicobacter pylori is due to null mutations in a gene (rdxA) that encodes an oxygen-insensitive NADPH nitroreductase. Mol. Microbiol. 1998; 28:383–393. [DOI] [PubMed] [Google Scholar]
- 18. Boneca I.G., Ecobichon C., Chaput C., Mathieu A., Guadagnini S., Prévost M.-C., Colland F., Labigne A., de Reuse H.. Development of inducible systems to engineer conditional mutants of essential genes of Helicobacter pylori. Appl. Environ. Microbiol. 2008; 74:2095–2102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Backert S., Kwok T., König W.. Conjugative plasmid DNA transfer in Helicobacter pylori mediated by chromosomally encoded relaxase and TraG-like proteins. Microbiol. Read. Engl. 2005; 151:3493–3503. [DOI] [PubMed] [Google Scholar]
- 20. Darfeuille F., Unoson C., Vogel J., Wagner E.G.H.. An antisense RNA inhibits translation by competing with standby ribosomes. Mol. Cell. 2007; 26:381–392. [DOI] [PubMed] [Google Scholar]
- 21. Kearse M., Moir R., Wilson A., Stones-Havas S., Cheung M., Sturrock S., Buxton S., Cooper A., Markowitz S., Duran C. et al. Geneious Basic: an integrated and extendable desktop software platform for the organization and analysis of sequence data. Bioinformatics. 2012; 28:1647–1649. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Reuter J.S., Mathews D.H.. RNAstructure: software for RNA secondary structure prediction and analysis. BMC Bioinformatics. 2010; 11, 129. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Bernacchi S., Freisz S., Maechling C., Spiess B., Marquet R., Dumas P., Ennifar E.. Aminoglycoside binding to the HIV-1 RNA dimerization initiation site: thermodynamics and effect on the kissing-loop to duplex conversion. Nucleic Acids Res. 2007; 35:7128–7139. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Fozo E.M., Makarova K.S., Shabalina S.A., Yutin N., Koonin E.V., Storz G.. Abundance of type I toxin-antitoxin systems in bacteria: searches for new candidates and discovery of novel families. Nucleic Acids Res. 2010; 38:3743–3759. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Fischer W., Breithaupt U., Kern B., Smith S.I., Spicher C., Haas R.. A comprehensive analysis of Helicobacter pylori plasticity zones reveals that they are integrating conjugative elements with intermediate integration specificity. BMC Genomics. 2014; 15, 310. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Kersulyte D., Lee W., Subramaniam D., Anant S., Herrera P., Cabrera L., Balqui J., Barabas O., Kalia A., Gilman R.H. et al. Helicobacter Pylori's plasticity zones are novel transposable elements. PloS One. 2009; 4:e6859. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Durand S., Jahn N., Condon C., Brantl S.. Type I toxin-antitoxin systems in Bacillus subtilis. RNA Biol. 2012; 9:1491–1497. [DOI] [PubMed] [Google Scholar]
- 28. Jahn N., Brantl S.. One antitoxin–two functions: SR4 controls toxin mRNA decay and translation. Nucleic Acids Res. 2013; 41:9870–9880. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Shokeen S., Patel S., Greenfield T.J., Brinkman C., Weaver K.E.. Translational regulation by an intramolecular stem-loop is required for intermolecular RNA regulation of the par addiction module. J. Bacteriol. 2008; 190:6076–6083. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Wen J., Won D., Fozo E.M.. The ZorO-OrzO type I toxin-antitoxin locus: repression by the OrzO antitoxin. Nucleic Acids Res. 2014; 42:1930–1946. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Møller-Jensen J., Franch T., Gerdes K.. Temporal translational control by a metastable RNA structure. J. Biol. Chem. 2001; 276:35707–35713. [DOI] [PubMed] [Google Scholar]
- 32. Nagel J.H., Gultyaev A.P., Gerdes K., Pleij C.W.. Metastable structures and refolding kinetics in hok mRNA of plasmid R1. RNA. 1999; 5:1408–1418. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Xayaphoummine A., Bucher T., Isambert H.. Kinefold web server for RNA/DNA folding path and structure prediction including pseudoknots and knots. Nucleic Acids Res. 2005; 33:W605–W610. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Thisted T., Nielsen A.K., Gerdes K.. Mechanism of post-segregational killing: translation of Hok, SrnB and Pnd mRNAs of plasmids R1, F and R483 is activated by 3΄-end processing. EMBO J. 1994; 13:1950–1959. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. Gerdes K., Nielsen A., Thorsted P., Wagner E.G.. Mechanism of killer gene activation. Antisense RNA-dependent RNase III cleavage ensures rapid turn-over of the stable hok, srnB and pndA effector messenger RNAs. J. Mol. Biol. 1992; 226:637–649. [DOI] [PubMed] [Google Scholar]
- 36. Durand S., Gilet L., Condon C.. The essential function of B. subtilis RNase III is to silence foreign toxin genes. PLoS Genet. 2012; 8:e1003181. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37. Kawano M. Divergently overlapping cis-encoded antisense RNA regulating toxin-antitoxin systems from E. coli: hok/sok, ldr/rdl, symE/symR. RNA Biol. 2012; 9:1520–1527. [DOI] [PubMed] [Google Scholar]
- 38. Thisted T., Gerdes K.. Mechanism of post-segregational killing by the hok/sok system of plasmid R1. Sok antisense RNA regulates hok gene expression indirectly through the overlapping mok gene. J. Mol. Biol. 1992; 223:41–54. [DOI] [PubMed] [Google Scholar]
- 39. Kawano M., Oshima T., Kasai H., Mori H.. Molecular characterization of long direct repeat (LDR) sequences expressing a stable mRNA encoding for a 35-amino-acid cell-killing peptide and a cis-encoded small antisense RNA in Escherichia coli. Mol. Microbiol. 2002; 45:333–349. [DOI] [PubMed] [Google Scholar]
- 40. Kawano M., Aravind L., Storz G.. An antisense RNA controls synthesis of an SOS-induced toxin evolved from an antitoxin. Mol. Microbiol. 2007; 64:738–754. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41. Pedersen K., Gerdes K.. Multiple hok genes on the chromosome of Escherichia coli. Mol. Microbiol. 1999; 32:1090–1102. [DOI] [PubMed] [Google Scholar]
- 42. Van Melderen L., Saavedra De Bast M.. Bacterial toxin-antitoxin systems: more than selfish entities. PLoS Genet. 2009; 5:e1000437. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43. Gerdes K., Thisted T., Martinussen J.. Mechanism of post-segregational killing by the hok/sok system of plasmid R1: sok antisense RNA regulates formation of a hok mRNA species correlated with killing of plasmid-free cells. Mol. Microbiol. 1990; 4:1807–1818. [DOI] [PubMed] [Google Scholar]
- 44. Carraro N., Poulin D., Burrus V.. Replication and active partition of integrative and conjugative elements (ICEs) of the SXT/R391 family: the line between ICEs and conjugative plasmids is getting thinner. PLoS Genet. 2015; 11:e1005298. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45. Wozniak R.A.F., Waldor M.K.. A toxin-antitoxin system promotes the maintenance of an integrative conjugative element. PLoS Genet. 2009; 5:e1000439. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46. Darty K., Denise A., Ponty Y.. VARNA: interactive drawing and editing of the RNA secondary structure. Bioinformatics. 2009; 25:1974–1975. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.