Abstract
RNA interference (RNAi) is a functional genomics tool to correlate genotype and phenotype by delivering targeted, gene-specific, and complementary dsRNA into a host via injection, feeding, or other means in order to reduce gene expression. In the red flour beetle, Tribolium castaneum, RNAi has been successful via injected dsRNA at all life stages. Traditionally, successful transcript knockdown has been quantified by qPCR on a gene-by-gene basis, where only expression of the target gene and normalization genes are evaluated. In this study, RNA-Seq was used to quantify transcript expression in larvae injected with dsRNA for aspartate 1-decarboxylase (ADC), which gives a reliable phenotype of an adult with a black cuticle instead of the wild-type red-brown. ANOVA of control, mock-injected, and ADC-dsRNA injected larvae indicated that target gene expression was significantly (P = 0.002) reduced 4-fold, and the black phenotype was achieved in all adults injected with ADC-dsRNA as larvae. In a pairwise analysis, significant (P < 0.05) differential expression of other genes in ADC-injected larvae suggested connections between gene pathways. One gene, dopamine receptor 2, was increased in expression 227-fold (P = 0.025), presumably connected to previous data that showed a reduction in expression of ADC results in increased levels of dopamine. To evaluate the hypothesis that increased dopamine levels can affect mobility, T. castaneum adults injected with ADC-dsRNA as larvae were significantly impaired in movement tests compared to controls, similar to black mutants in Drosophila melanogaster. The data demonstrate that RNA-Seq can reveal gene connectivity and provide more complete data validation and analysis compared to qPCR.
Keywords: Tribolium castaneum, RNAi, ADC, RNA-Seq, gene expression
RNA interference (RNAi) is a popular tool in functional genomics and, in pest insects like Tribolium castaneum, a potential strategy for molecular pest control (Baum et al. 2007, Aronstein et al. 2011, Ulrich et al. 2015). RNAi is an evolutionarily conserved mechanism that relies on natural cellular pathways to target and degrade double stranded RNA (dsRNA) of viruses, as described in nematodes (Fire et al. 1998). This natural process can be mimicked by the introduction of dsRNA into an organism (e.g. injection). The endoribonuclease, Dicer, cleaves dsRNA into 21–23 nucleotide short interfering RNAs (siRNAs), which are incorporated into an RNA-induced silencing complex (RISC) that targets and degrades RNA with complementary sequence.
RNAi has been used successfully to increase insect mortality or developmental abnormalities via microinjection or feeding in vitro or in planta (Baum et al. 2007, Huvenne and Smagghe 2010, Aronstein et al. 2011). In T. castaneum, injection of 520 base pair dsRNA fragments was sufficient to elicit a response at concentrations ranging from 0.0001 to 0.001 μg/μl (Miller et al. 2012). Other studies have demonstrated that injection can be successful in any T. castaneum life stage (reviewed in Aronstein et al. 2011).
The most common downstream validation of knockdown after RNAi is quantitative polymerase chain reaction (qPCR). This type of analysis is dependent on gene-specific primers, including onerous optimization that is often circumvented, and generally only used to measure the target gene and a normalizer gene as a standard. We suggest this method limits the information gained from RNAi, and that RNA transcriptome-sequencing and quantitation (RNA-Seq) is more informative. RNA-Seq can monitor target gene knockdown and off-target effects, and provides results similar to qPCR (Morris et al. 2009, Chen et al. 2016). In our hands, RNA-Seq has been beneficial in cases where RNAi does not show a visible phenotype, and attempts at validation by qPCR are ambiguous. For example, qPCR validation of knockdown of a major gut cysteine peptidase gene in T. castaneum was inconclusive by qPCR, and there was no difference in phenotype (unpublished data). However, using RNA-Seq, we demonstrated that knockdown was in fact successful, but closely related cysteine peptidase as well as serine peptidase genes, were compensating for the loss of target gene expression (Perkin et al. 2017). Thus, sequence similarity and redundancy in gene function, as well as compensation responses embedded in the transcriptome response, can mask the effects of RNAi, making it difficult to verify with qPCR alone.
To illustrate that RNA-Seq can be a superior method to validate the results of RNAi, we report here the result of knockdown of aspartate 1-decarboxylase (ADC) in T. castaneum larvae. ADC is one of two decarboxylases (the other is DDC, TC013480) needed for proper insect cuticle tanning in T. castaneum (sclerotization and pigmentation). The process is complex and requires the conjugation and cross-linking of cuticle proteins, leading to an insoluble, hard, and darkened red-brown exoskeleton (Roseland et al. 1987). ADC catalyzes the synthesis of β-alanine, which plays a critical role in cuticle tanning because of its conjugation with dopamine to produce Ν-β-alanyldopamine (NBAD), a substrate for the phenol oxidase laccase that catalyzes the synthesis of the cuticle protein cross-linking agents and pigment precursors (Kramer et al. 1984). Knockdown of ADC leads to a black cuticle phenotype and the accumulation of dopamine (Arakane et al. 2009).
We demonstrate that RNAi targeting ADC, as verified by RNA-Seq and black phenotype, changes the expression of other genes that may be interconnected. One affected gene, dopamine receptor 2, led us to test whether movement was impaired in T. castaneum adults injected with ADC-dsRNA as larvae, uncovering a previously unknown interaction between ADC and genes controlling movement in T. castaneum.
Materials and Methods
Insects
The T. castaneum lab strain was originally collected from a grain bin in Kansas and has been reared at the Center for Grain and Animal Health Research (CGAHR, Agricultural Research Service, United States Department of Agriculture, Manhattan, KS) for over 20 yr. Insects are maintained on a diet of 95% wheat flour and 5% Brewer’s yeast at 28 °C, 75% relative humidity, 0:24 (L:D) h.
Primers and dsRNA
Primers were designed via Primer-BLAST (http://www.ncbi.nlm.nih.gov/tools/primer-blast/) to the ADC gene in T. castaneum using default parameters. All primers were specific, and selected primer sets were unique to the ADC gene. The first round of PCR amplified the entire gene using the primary primer sets: ADC forward 5′-AAGGCGAAGGGAACATCAGG-3′ and reverse 5′-CTCCCCAACCGCTCAATCTC-3′. PCR reactions were 25 μl total volume using genomic DNA template from the lab strain and standard thermal cycle conditions (Perkin et al. 2017). The product was assessed on a 1% agarose E-gel (ThermoFisher, Waltham, MA) to ensure the amplified region was the correct size.
A second set of primers was designed to amplify within the first amplification region, and the products were used for dsRNA amplification. Primers were designed to target each sequence towards the 3′ end, according to previous data that suggested this region was best for specific and maximal knockdown (Whyard et al. 2009, Perkin et al. 2017). ADC-dsRNA primers were as in Arakane et al. (2009), forward 5′-AAGGCGAAGGGAACATCAGG-3′ and reverse 5′-TCCCCAACCGCTCAATCTC-3′. All secondary PCR primers had a T7 construct attached to the 5′ end (TAATACGACTCACTATAGGG). Another round of PCR was performed with each secondary primer set and using the template obtained for each respective gene in the primary reaction as the template, using similar PCR conditions (100 μl reaction; Perkin et al. 2017). Secondary PCR products were evaluated via 1% agarose E-gel for appropriate length and sufficient amplification.
The PCR product from the secondary amplification was used to make dsRNA via a MEGAscript T7 kit (Invitrogen, Life Technologies, Carlsbad, CA), and was purified via a MEGAclear kit (ThermoFisher, Waltham, MA; Perkin et al. 2017). Size and quantity of products were verified on a TapeStation (Agilent, Santa Clara, CA) and quantity also was verified by a digital nanophotometer (Implen, Westlake Village, CA). Negative controls included noninjected larvae (Control) and mock-injected with water and dye (Mock).
Micro-injected dsRNA
There were three groups in this study: T. castaneum larvae injected with water/dye (Mock); larvae injected with ADC-dsRNA construct (ADC) mixed with dye; and a noninjected (Control) group. Immediately before injection, treatments (water or dsRNA) were mixed with blue dye (FD&C Blue 1, Kroger Food Colors, Cincinnati, OH; diluted 1:20) to aid in visualization of the injected liquid. Actively feeding, third instar larvae were briefly placed on ice and transferred to double-sided tape on a microscope slide placed on a small tissue culture flask ice block (Posnien et al. 2009). Briefly, a Drummond Nanoject II (Drummond Scientific Co., Broomall, PA) with a “bee-stinger” needle was set at 69 nl, and dsRNA was diluted to provide 200 ng of dsRNA per larva (Perkin et al. 2017) and loaded into the needle. Needles were made with 3.5 Drummond glass capillary tubes (3-000-203-G) and a micropipette puller (Sutter Instrument Co. Model P-97, Novato, CA). After injection, each group was allowed to recover for 2 h at room temperature, and then were covered with diet (95% wheat flour, 5% Brewer’s yeast) and kept at 28 °C, 75% relative humidity, 0:24 (L:D) h. All injections in a single replicate were done on the same day, with a total of three independent replicates per treatment.
mRNA Extraction
From each treatment group, eight T. castaneum larvae were randomly selected at 7-d postinjection, ground in liquid nitrogen with a disposable pellet pestle, and total RNA was obtained using a Qiagen RNeasy Plus kit (Qiagen, Hilden, Germany). The Plus version of this kit utilizes a “gDNA Eliminator” spin column as a pretreatment to further diminish any contamination of the RNA with DNA, and an optional step of on-column DNase digestion also was used. Quantity and quality at each step of mRNA collection were evaluated by TapeStation, and quantity was verified by a nanophotometer (Implen, Westlake Village, CA). Aliquots were stored at −80 °C.
RNAseq
A stranded mRNA-Seq Kit (KAPA Biosystems, Wilmington, MA) with appropriate Ion Torrent primers, adapters, and bar codes (Integrated DNA Technologies, Coralville, IA) was used for library preparation of 4 µg of total RNA from each biological replicate from each treatment. A final concentration of adapter and barcodes of 100 nM was used without optimization of the concentration. Eight equimolar barcoded libraries from all treatment groups of a biological replicate were pooled based on concentration estimates from a KAPA Library Quantification Kit for Ion Torrent platform (KK4827), and were placed in an Ion Chef (ThermoFisher, Grand Island, NY) for template preparation and loading onto an Ion PI Chip v2 for sequencing on the Ion Proton sequencer (Perkin et al. 2017).
Data Analysis
Transcripts were mapped to the Tcas5.2 genome build (NCBI) in SeqManNGen (DNAStar, Madison, WI), and differential gene expression was analyzed in ArrayStar (DNAStar version 14) using default parameters. Genes were normalized using Reads Per Kilobase per Million (RPKM; Mortazavi et al. 2008) and filtered to transcripts >8-fold change between groups in Student’s t-test comparisons, and later were filtered to a 90% confidence interval after correction for multiple comparisons with False Discovery Rate (FDR) analysis (Benjamini and Hochberg 1995), with exceptions as noted. We also used gene expression data from different developmental stages (egg, larvae, pupae, and adult) of T. castaneum, as detailed in Perkin et al. (2016).
Nonannotated differentially expressed genes were submitted to BLAST2GO PRO (Valencia, Spain) to elucidate possible function. From this analysis, additional functions were identified in Rfam (Nawrocki et al. 2015).
Each replicate had >7 million reads with a total of >31 million reads per sample (Supp Table 1 [online only]). Sequences from this study were deposited into NCBI Sequence Read Archive PRJNA302304.
Behavior Assays and Analysis
To assess movement and behavioral patterns in ADC-injected larvae, 25 late instar larvae were collected from ADC, Mock, and Control groups and placed into 1 oz solo cups with holes in the lid. Media composed of 90% stabilized wheat germ and 10% flour was added. Each group was monitored daily, and pupae were collected and placed in individual wells of a 24-well plate and allowed to emerge as adults.
At 3- to 12-d posteclosion, individual beetles were placed in the center of a 90-mm diameter plastic petri dish using vacuum suction, facing the same direction at the start of each assay. After the lid was placed on the dish, individuals were allowed to acclimate for 10 s before a video recording (Sony Handycam HDR-XR520V) was taken of each beetle. After 5 min, the recording was terminated and beetles were removed from the petri dish and discarded. There were 18 individuals from each treatment group (ADC-injected and Mock) and control that were recorded for movement behavior.
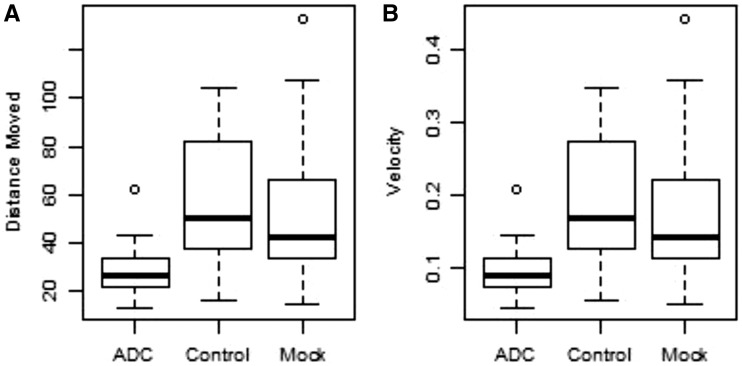
Videos were analyzed using Ethovision XT (Version 8.0, Noldus Information Technology, Wageningen, The Netherlands). Video tracks were analyzed for Distance Moved (cm); Mobile, Immobile, or Highly Mobile states (defined as the changes in body position between frames at 10 frames per second, where 0%, 20%, and 60% of the body changes position) in sec; and velocity (distance traveled per time between frames, measured as the mean in cm/s). Distance, Velocity, and the three mobility states were analyzed using individual PROC GLM statements in SAS (SAS Institute, Cary NC, version 9.4), where strain was the main effect in the model and pairwise differences were Tukey HSD adjusted.
Results
In this study, we reduced the expression of the T. castaneum aspartate 1-decarboxylase (ADC, LOC100124592, TC034596) via RNAi in larvae, and we used RNA-Seq to validate knockdown. ADC is a gene commonly used as a positive RNAi phenotypic control. Most previous validation experiments for RNAi in T. castaneum have been by qPCR, but we chose RNA-Seq because we wanted to examine other potential effects on reduced transcript levels of ADC.
ANOVA of All Treatment Groups
All T. castaneum larvae injected with ADC-dsRNA in all replicates developed into adults with a black cuticle compared to the wild-type red/brown cuticle, presumably due to manipulation of the pigmentation and sclerotization pathways (Arakane et al. 2009). In T. castaneum larvae injected with ADC-dsRNA, ADC expression was reduced 4.25-fold compared to a water-injected control (Mock; Fig. 1). An ANOVA comparing Control, Mock and ADC treatments indicated that ADC expression was significantly different between treatments (P = 0.002; Table 1). Other genes were also differentially expressed (>2-fold) in ADC-dsRNA injected larvae, many related to gene regulation. The most highly up-regulated gene (7.39-fold) encodes a leucine-rich repeat-containing protein (LOC103314786), which is annotated in insects, mollusks, and chordates (Supp Fig. 2 [online only]). In UniProt, the gene is annotated in Caenorhabditis elegans and Trichinella spp. (parasitic roundworms), but most functional data is from C. elegans where it was expressed in multiple tissues in embryo through larval development, and was important in maintaining apical extracellular matrix integrity (Mancuso et al. 2012, Finn et al. 2016). However, the relationship of this gene to ADC is not apparent. Additionally, a gene encoding a hypothetical noncoding RNA (LOC107397781) was dramatically down-regulated (974-fold).
Fig. 1.
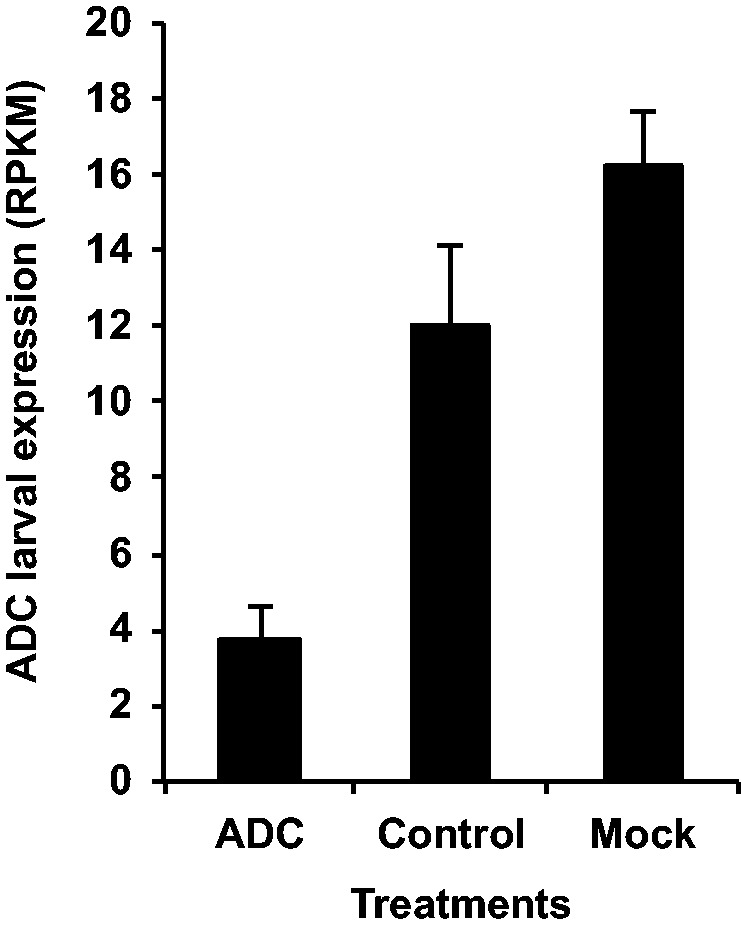
ADC expression (average total RPKM) differences between larvae injected with dsRNA targeting ADC (ADC), the noninjected control (Control), and the mock-injected (Mock) group. Error bars denote SE.
Table 1.
ANOVA of control, mock-injected, and ADC-dsRNA injected T. castaneum larvae, filtered to linear RPKM > 1 in at least one of the groups (values to right for each group), >2-fold change in Mock versus ADC, and P < 0.05 (FDR Benjamini and Hochberga)
| Name | Description | Fold changeb | P value | ADC RPKM | Control RPKM | Mock RPKM |
|---|---|---|---|---|---|---|
| LOC103314786 | Leucine-rich repeat-containing protein egg-6-like | 7.385 | 0.044600 | 7.067 | 0.802 | 0.957 |
| LOC107397729 | Pfamc: Retrotransposon gag protein, Reverse transcriptase | 6.253 | 0.014700 | 2.695 | 0.559 | 0.431 |
| LOC103315154 | Leucine-rich repeat-containing protein let-4-like | 3.267 | 0.035500 | 4.513 | 1.958 | 1.381 |
| LOC658512 | Carboxylesterase 1E | 2.383 | 0.001280 | 52.77 | 22.45 | 22.15 |
| LOC656320 | Trypsin I-P1 isoform X1 | 2.180 | 0.000184 | 47.91 | 25.74 | 21.97 |
| LOC103314471 | Ribulose-phosphate 3-epimerase | −2.424 | 0.012800 | 33.22 | 66.39 | 80.52 |
| LOC664050 | Serine/threonine-protein kinase RIO3 | −2.771 | 0.001390 | 9.336 | 25.40 | 25.88 |
| LOC107398585 | CPDFd: protein kinase | −3.815 | 0.031300 | 0.380 | 0.696 | 1.449 |
| LOC100124592 | Aspartate 1-decarboxylase | −4.245 | 0.001800 | 0.563 | 2.123 | 2.390 |
| LOC107397781 | pfam: ncRNA | −974.2 | 0.000001 | 0.001 | 0.001 | 1.106 |
Target gene (ADC) is shaded.
Fold change in ADC-injected larvae.
pfam: protein families database (Finn et al. 2016).
CPDF: Conserved Protein Domain Family, NCBI.
Pairwise Analysis of ADC-injected Versus Mock-injected
We compared gene expression of ADC-injected T. castaneum larvae to the mock-injected control (Table 2 andSupp Fig. 1 [online only]). The expression of dopamine receptor 2 (LOC661535) was increased 227-fold in ADC compared to Mock (P = 0.025), but the expression of dopamine receptor 1 was unchanged (data not shown). The expression of two other genes also increased, an uncharacterized gene (245-fold; P = 0.029) and a protein with unknown function with an alkaline phosphatase motif (141-fold; P = 0.004).
Table 2.
Significant differentially expressed genes in a pairwise comparison of ADC-dsRNA injected T. castaneum larvae to mock-injected larvae (> 8-fold change, >90% confidence interval, Student’s t-test P values with Benjamini and Hochberga FDR correction, with linear total RPKM values on the right)
| Name | Descriptionb | Fold changeb | P value | Mock RPKM | ADC RPKM |
|---|---|---|---|---|---|
| LOC107398333 | Uncharacterized | 245.2 | 0.0290 | 0.001 | 0.278 |
| LOC661535 | Dopamine receptor 2 | 227.0 | 0.0250 | 0.001 | 0.258 |
| LOC659605 | Pfamc: Protein of unknown function (DUF229), alkaline phosphatase | 141.4 | 0.0044 | 0.001 | 0.161 |
| LOC103313283 | Pfam: Domain of unknown function (DUF4550) | −66.93 | 0.0338 | 0.076 | 0.001 |
| LOC107398197 | Uncharacterized | −81.96 | 0.0301 | 0.093 | 0.001 |
| LOC657226 | Glutamate dehydrogenase, mitochondrial-like | −90.06 | 0.0290 | 0.112 | 0.001 |
| LOC107398565 | Odorant receptor 85a-like | −136.7 | 0.0250 | 0.155 | 0.001 |
| LOC107398092 | CPDFd: 7tm chemosensory receptor | −146.3 | 0.0250 | 0.166 | 0.001 |
| LOC107398783 | Uncharacterized | −176.0 | 0.0250 | 0.200 | 0.001 |
| LOC663271 | Pfam: haemolymph juvenile hormone binding protein (JHBP) | −257.7 | 0.0003 | 0.293 | 0.001 |
| LOC100142608 | Pfam: 7tm odorant receptor (OR139) | −240.6 | 0.0145 | 0.273 | 0.001 |
| LOC107398253 | Pfam: (ncRNA/snoRNA U3) | −593.3 | 0.0081 | 0.674 | 0.001 |
| LOC657178 | odorant binding protein 18 (OBP18/OBP4A) | −789.3 | 0.0250 | 0.896 | 0.001 |
| LOC107397781 | Pfam: ncRNA | −974.2 | 0.0244 | 1.106 | 0.001 |
| LOC100216358 | Allatotropin I preprohormone isoform X1 | −1,411 | 0.0011 | 1.603 | 0.001 |
Fold change in ADC-injected larvae.
Pfam: protein families database (Finn et al. 2016).
CPDF: Conserved Protein Domain Family, (Marchler-Bauer et al. 2015).
Injection of ADC-dsRNA also significantly decreased the expression of 12 additional genes compared to the mock-injected control (P< 0.03; Table 2). The most severely repressed gene was LOC100216358, a gene encoding allatotropin I preprohormone, that was decreased 1,411-fold (P = 0.001). Several chemosensory genes (LOC107398565, LOC107398092, LOC100142608, LOC657178), three of which have a conserved odorant receptor domain, were significantly decreased 137- to 240-fold (P < 0.03). There were two other uncharacterized genes (LOC107397781 and LOC107398253) that decreased 974- and 593-fold, respectively, and both had motifs similar to ncRNAs. Further analysis with Rfam found the latter is part of the small nucleolar RNA (snoRNA) U3 family. This family is predicted to guide site-specific cleavage of ribosomal RNA (rRNA) during pre-rRNA processing (Clery et al. 2007).
Movement Assays
The black cuticle phenotype of beetles injected with ADC-dsRNA has been demonstrated to associated with an accumulation of dopamine (Arakane et al. 2009). However, this is the first demonstration that a reduction in ADC leads to an increase in expression of a dopamine receptor gene. Previous work in D. melanogaster indicated that body color mutants often have neurological phenotypes (Wittkopp et al. 2003), including black mutants that show reduced activity at the larval/pupal boundary or “wandering stage” (Phillips et al. 2005). Therefore, we conducted a movement behavior assay on T. castaneum adults injected with ADC-dsRNA as larvae to evaluate if increased expression of dopamine receptor 2 results in a similar neurological phenotype.
Tribolium castaneum larvae injected with dsRNA targeting ADC were significantly slower and moved less often as adults than both Mock beetles and Control beetles (Table 3 and Supp Video 1 [online only]). Slowed movement was demonstrated by a reduction in mean velocity (cm/s) and total distance traveled in the ADC-injected beetles compared to Mock and Control (Fig. 2). Over the 5-min period of uninhibited movement, ADC-injected beetles traveled on an average 23.11 cm less than Mock beetles (P = 0.018) and had an average velocity 0.076 cm/s slower than Mock beetles (P = 0.018). Overall, neither ADC-injected nor mock beetles had long bouts of highly mobile activity (where 60% of body position changes per frame) and the durations were not significantly different (ADC-injected = 0.32 s; Mock = 0.82 s; P = 0.21). However, ADC-injected beetles spent significantly more time in an immobile state (where 0% of body changes position per frame at 10 frames per second). ADC-injected beetles spent 297.4 s in an immobile state, while Mock spent 289.9 s immobile (P = 0.011; Table 3).
Table 3.
Statistical data of movement patterns of T. castaneum adults injected as larvae with ADC-dsRNA compared to Control and Mock
| Trait | Type III SS | Mean square | F value | Pr > F | Tukey’s HSD P value |
|---|---|---|---|---|---|
| Distance moved | 8,042 | 4,020 | 6.68 | 0.003 | 0.018 |
| Velocity | 0.089 | 0.045 | 6.68 | 0.003 | 0.018 |
| High mobility | 2.580 | 1.290 | 1.74 | 0.190 | 0.210 |
| Immobile | 609.4 | 304.7 | 5.44 | 0.007 | 0.011 |
| Mobile | 532.7 | 266.3 | 5.47 | 0.007 | 0.011 |
Values are ANOVA for each movement metric; source of ANOVA variation is from the main effect of strain (DF = 2 for all comparisons). Distance moved is the total distance moved in cm for a 5-min assay; Velocity is the mean velocity in cm/s as measured by distance moved between frames; High Mobility refers to when 60% or more of the body changes location between frames, at 10 frames per second; Immobile means 0% of the body has moved between frames, whereas Mobile means 20% of the body is changing per frame. The far right column includes the Tukey HSD adjusted P values for the ADC/Mock comparison.
Fig. 2.
Box plots showing the distance moved (A) and velocity (B) of ADC-, Control-, and Mock-injected beetles. Solid black lines indicate the median distance and velocity. The vertical edges of the boxes represent 25% and 75% of the data distribution (Interquartile Range or IQR) with whiskers representing 1.5 × (IQR), a measure of the maximum and minimum values in the dataset.
Discussion
Our approach demonstrates that RNA-Seq documents target knockdown by RNAi of ADC in T. castaneum through normalized gene expression data, even in small differences of expression. All ADC-injected larvae resulted in the expected phenotype, adults with a black cuticle and a knockdown of 4.25-fold decrease. The black cuticle phenotype was the result of decreased ADC leading to an accumulation of dopamine upstream in the cuticle-tanning pathway in epidermal cells, which may be the predominant site of expression of this gene (Arakane et al. 2009).
RNA-Seq as an RNAi validation method also revealed differences in expression of genes not previously associated with the target gene, ADC. Most significant was the increased expression of dopamine receptor 2 (LOC661535) in the ADC RNAi treatment that led us to evaluate possible behavioral differences. Dopamine is a neurotransmitter important for a variety of life functions in vertebrates and invertebrates, and many human disorders have been attributed to disruptions in dopamine pathways (reviewed in Beaulieu and Gainetdinov 2011). Dopamine receptors are G protein-coupled receptors separated into D1 and D2 classes. In vertebrates, D1-class dopamine receptors include D1 and D5, and they stimulate cyclic-AMP production by adenylate cyclase, whereas the D2-class receptors include D2, D3, and D4, and they inhibit adenylate cyclase and thus lead to a decrease in cyclic AMP (reviewed in Civelli et al. 1993). In humans, the D2 receptor is associated with different structures in the brain, including olfaction. Insects have orthologs of dopamine receptors 1 and 2, and there are genes encoding dopamine receptors 1 and 2 in T. castaneum (Hauser et al. 2008, Tribolium Genome Sequencing Consortium 2008). Our data suggests that expression of ADC and dopamine receptor 2 are diametrically opposed in T. castaneum.
To understand how dopamine and cuticle pigmentation pathways may interact, we turned to the abundant literature on D. melanogaster pigmentation. D.melanogaster pigmentation is more complicated than T. castaneum due to multiple color patterns and sexual dimorphism. However, most D. melanogaster pigment genes have orthologs in T. castaneum. For example, the D. melanogaster black gene is orthologous to the T. castaneum ADC gene, both genes encoding aspartate 1-decarboxylase, and contributing to the black phenotype in mutants for this gene. Many studies have shown that pigmentation gene mutants in D. melanogaster, such as black, ebony, and tan not only have body color phenotypes, but also neurological phenotypes (Wittkopp et al. 2003). For example, D. melanogaster black mutants have reduced activity at the larval/pupal boundary or “wandering stage”. In the adult stage, black null mutants had significantly reduced number of initiated walks compared to wild-type (Phillips et al. 2005). In T. castaneum black mutants, the loss of ADC was attributed to lower transcript levels (Arakane et al. 2009), similar to what we achieved through RNAi of wild-type insects. We examined the movement patterns of ADC-dsRNA-injected T. castaneum and T. castaneum mutants with black phenotype (data not shown) and observed reduction in distance moved, overall velocity, and mobility compared to Mock and Control. Thus, T. castaneum adults with a disrupted ADC gene function similarly to D. melanogaster black mutants, suggesting that T. castaneum mutants with black phenotype may also have a mutation in the ADC gene and/or reduction in ADC expression.
Expression of two predicted ncRNAs was significantly altered in the ADC RNAi treatment. These RNAs are transcribed from DNA but are not translated to proteins, hence noncoding. They are proposed to serve as epigenetic regulators and include miRNA, siRNA, piRNA, and lncRNA. ADC-dsRNA induced a 974-fold decrease of ncRNA LOC107397781; this sequence is upstream of the gene white (TC007047), an eye pigment gene. However, the expression of white was not significantly changed (data not shown). The other differentially expressed ncRNA was LOC107398253, which is part of the snoRNA U3 family. These ncRNAs are found in the nucleolus, and instead of functioning as an epigenetic regulator, are proposed to guide site-specific cleavage of rRNA during the pre-processing of rRNA (Clery et al. 2007). The data suggest that ADC RNAi not only reduced targeted transcript expression, but also may have affected the expression of other genes controlling the processing of RNA through changes in expression of ncRNA.
Other genes identified in the study include four chemosensory-related genes that were significantly decreased in the ADC-dsRNA treatment group, which also may indicate that expression of these genes is affected by increased dopamine. Expression of LOC657178, odorant binding protein 18 (OBP18), was the most significantly reduced, 789-fold in ADC-injected larvae. OBP18 (also known as OBP4A) was highly expressed in the T. castaneum larval head and mouthparts compared to the body (Dippel et al. 2014). Three chemosensory receptor genes belong to the 7tm receptor gene class, LOC100142608, LOC107398092, and LOC107398565. Engsontia et al. (2008) found that LOC100142608, also referred to as OR139, is part of a T. castaneum-specific clade in an odorant receptor gene tree consisting of sequences from Tribolium, Drosophila, Apis, and Heliothis, and expression was isolated to the adult and larval head. While less is known about the latter two 7tm receptor genes, the data suggest that these chemoreceptors may have functional similarity, and that they may be responsive to dopamine levels in the head.
ADC-dsRNA injection also decreased the expression of a gene encoding allatotropin I preprohormone, 1,411-fold. The product of this gene is a precursor to the neuropeptides that regulate biosynthesis of juvenile hormone (JH) in insects (Abdel-Latief and Hoffmann 2014). In holometabolous insects such as T. castaneum, JH decreases between the pupal and adult molt, and adult structures begin to form in the presence of ecdysteroids (Hiruma and Kaneko 2013). The biosynthesis of JH is stimulated or inhibited by allatotropin or allatostatin, respectively, two neuropeptides in the corpora allata. Other roles may incude inhibition of active ion transport, photic entrainment of the circadian clock, and also may be myoactive (Andinet and Sweedler 2007). The data suggest that ADC and allatotropin genes may be interconnected in regulatory pathways.
We propose that, based on our data, a decrease in ADC results in the accumulation of dopamine and an increased expression of dopamine receptor 2, and consequently decreased adenylate cyclase and production of cAMP. As both dopamine receptor 2 and allatotropin I are expressed in the T. castaneum brain, we speculate that decreased expression of allatotropin I was due to decreased cAMP. While it is known that allatotropin I expression results in increased cAMP in T. castaneum (Vuerinckx et al. 2011), our data suggests that expression of allatotropin I may also be sensitive to decreased cAMP levels in a feedback loop mechanism; this hypothesis remains to be tested.
Finally, we note that in the examination of RNA-Seq data, we began our analyses with all differentially expressed genes, to examine those that may be functionally related even if genes do not meet the criteria for inclusion (i.e., statistical or minimal expression values). In the pairwise analysis, the RPKM values of both ADC-injected or Mock-injected were mostly below the threshold of 1 (Table 1), but the data was still useful in making relevant biological predictions, as was the case for increased dopamine receptor 2 and decreased mobility. Our data analysis and statistical significance may be improved with additional coverage. However, while we understand the need to carefully examine RNA-Seq data, significant information may be missed in situations where the data is less than optimal (i.e., low RPKM values) but with statistically significant, large changes in expression with adequate biological replicates.
In summary, we propose that RNA-Seq is superior to qPCR to quantify RNAi knockdown, because differential expression of other genes may provide new information on gene interconnectivity. Our results demonstrated a connection between ADC and dopamine receptor 2 genes in T. castaneum with additional evidence via functional tests, as well as other possible gene connections that remain to be tested.
Supplementary Data
Supplementary data are available at Journal of Insect Science online.
Supplementary Material
Acknowledgments
We would like to thank technicians Ken Friesen and Tom Morgan for their contribution, injecting larvae and RNA extraction and library preparation, respectively, Sierra Upton for video editing, and Dr Subbaratnam Muthukrishnan for his comments on an earlier version of the manuscript. Mention of trade names or commercial products in this publication is solely for the purpose of providing specific information and does not imply recommendation or endorsement by the U.S. Department of Agriculture. USDA is an equal opportunity provider and employer.
References Cited
- Abdel-Latief M., Hoffmann H. K. 2014. Functional activity of allatotropin and allatostatin in the pupal stage of a holometablous insect, Tribolium castaneum (Coleoptera, Tenebrionidae). Peptides 53: 172–184. [DOI] [PubMed] [Google Scholar]
- Andinet A., Sweedler J. V. 2007. Neuropeptide precursors in Tribolium castaneum. Peptides 28: 1282–1291. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Arakane Y., Lomakin J., Beeman R. W., Muthukrishnan S., Gehrke S. H., Kanost M. R., Kramer K. J. 2009. Molecular and functional analyses of amino acid decarboxylases involved in cuticle tanning in Tribolium castaneum. J. Biol. Chem. 284: 16584–16594. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Aronstein K., Oppert B., Lorenzen M. 2011. RNAi in agriculturally-important arthropods, pp. 157–180. In (eds.), RNA processing. INTECH Open access Publisher. [Google Scholar]
- Baum J. A., Bogaert T., Clinton W., Heck G. R., Feldmann P., Ilagan O., Johnson S., Plaetinck G., Munyikwa T., Pleau M. et al. 2007. Control of coleopteran insect pests through RNA interference. Nat. Biotechnol. 25: 1322–1326. [DOI] [PubMed] [Google Scholar]
- Beaulieu J. M., Gainetdinov R. R. 2011. The physiology, signaling, and pharmacology of dopamine receptors. Pharm. Rev. 63: 182–217. [DOI] [PubMed] [Google Scholar]
- Benjamini Y., Hochberg Y. 1995. Controlling the false discovery rate: a practical and powerful approach to multiple testing. J. R. Stat. Soc. B (Methodological) 57: 289–300. [Google Scholar]
- Chen X., Xiong W., Li C., Gao S., Song X., We W., Bin L. 2016. Comparative RNA-sequencing profiling reveals novel Delta-class glutathione S-transferases relative genes expression patterns in Tribolium castaneum. Gene 593: 13–20. [DOI] [PubMed] [Google Scholar]
- Civelli O., Bunzow J. R., Grandy D. K. 1993. Molecular diversity of the dopamine receptors. Annu. Rev. Pharmacol. Toxicol. 32: 281–307. [DOI] [PubMed] [Google Scholar]
- Clery A., Senty-Segault V., Leclerc F., Raue H. A., Branlant C. 2007. Analysis of sequence and structural features that identify the B/C motif of U3 small nucleolar RNA as the recognition site for the Snu13p-Rrp9p protein pair. Mol. Cell Biol. 27: 1191–1206. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dippel S., Oberhofer G., Kahnt J., Gerischer L., Opitz L., Schachtner J., Stanke M., Schutz S., Wimmer E. A., Angeli S. 2014. Tissue-specific transcrtipomics, chromosomal localization, and phylogeny of chemosensory and odorant binding proteins from the red flour beetle Tribolium castaneum reveal subgroup specificities for olfaction or more generalized functions. BMC Genomics 15: 1141. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Engsontia P., Sanderson A. P., Cobb M., Walden K. K., Robertson H. M., Brown S. 2008. The red flour beetle’s large nose: an expanded odorant receptor gene family in Tribolium castaneum. Insect Biochem. Mol. Biol. 38: 387–397. [DOI] [PubMed] [Google Scholar]
- Finn R. D., Coggill P., Eberhardt R. Y., Eddy S. R., Mistry J., Mitchell A. L., Potter S. C., Punta M., Qureshi M., Sangrador-Vegas A. et al. 2016. The Pfam protein families database: towards a more sustainable future. Nucleic Acids Res. 44: D279–D285. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fire A., Xu S., Montgomery M. K., Kostas S. A., Driver S. E., Mello C. C. 1998. Potent and specific genetic interference by double-stranded RNA in Caenorhabditis elegans. Nature 391: 806–811. [DOI] [PubMed] [Google Scholar]
- Hauser F., Cazzamali G., Williamson M., Park Y., Li B., Tanaka Y., Predel R., Neupert S., Schachtner J., Verleyen P. et al. 2008. A genome-wide inventory of neurohormone GPCRs in the red flour beetle Tribolium castaneum. Front. Neuroendocrinol. 29: 142–165. [DOI] [PubMed] [Google Scholar]
- Hiruma K., Kaneko Y. 2013. Hormonal regulation of insect metamorphosis with special reference to juvenile hormone biosynthesis, pp. 73–78. InShi Y.-B. (ed.), Animal metamorphosis. Elevier, Oxford. [DOI] [PubMed] [Google Scholar]
- Huvenne H., Smagghe G. 2010. Mechanisms of dsRNA uptake in insects and potential of RNAi for pest control: a review. J. Insect Physiol. 56: 227–235. [DOI] [PubMed] [Google Scholar]
- Kramer K. J., Morgan . D., Hopkins T. L., Roseland C. R., Aso Y., Beeman R. W., Lookhart G. L. 1984. Catecholamines and β-Alanine in the red flour beetle, Tribolium castaneum. Roles in cuticle sclerotization and melanization. Insect Biochem. 14: 293–298. [Google Scholar]
- Mancuso V. P., Parry J. M., Storer L., Poggioli C., Nguyen K. C., Hall D. H., Sundaram M. V. 2012. Extracellular leucine-rich repeat proteins are required to organize the apical extracellular matrix and maintain epithelial junction integrity in C. elegans. Development 139: 979–990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Marchler-Bauer A., Derbyshire M. K., Gonzales N. R., Lu S., Chitsaz F., Geer L. Y., Geer R. C., He J., Gwadz M., Hurwitz D. I. et al. 2015. CDD: NCBI’s conserved domain database. Nucleic Acids Res. 43: D222–D226. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Miller S. C., Miyata K., Brown S. J., Tomoyasu Y. 2012. Dissecting systemic RNA interference in the red flour beetle Tribolium castaneum: parameters affecting the efficiency of RNAi. PLoS One 7: e47431.. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mortazavi A., Williams B. A., McCue K., Schaeffer L., Wold B. 2008. Mapping and quantifying mammalian transcriptomes by RNA-seq. Nat. Methods 5: 621–628. [DOI] [PubMed] [Google Scholar]
- Morris K., Lorenzen M. D., Hiromasa Y., Tomich J. M., Oppert C., Elpidina E. N., Vinokurov K., Jurat-Fuentes J. L., Fabrick J., Oppert B. 2009. Tribolium castaneum Larval gut transcriptome and proteome: A resource for the study of the Coleopteran gut. Journal of Proteome Research. 8: 3889–3898. [DOI] [PubMed] [Google Scholar]
- Nawrocki E. P., Burge S. W., Bateman A., Daub J., Eberhardt R. Y., Eddy S. R., Floden E. W., Gardner P. P., Jones T. A., Tate J. et al. 2015. Rfam 12.0: updates to the RNA families database. Nucleic Acids Res. 43: D130–D137. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Perkin L. C., Elpidina E. N., Oppert B. 2016. Expression patterns of cysteine peptidase genes across thetribolium castaneum life cycle provide clues to biological function. PeerJ 4: e1581.. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Perkin L. C., Elpidina E. N., Oppert B. 2017. RNA interference and dietary inhibitors induce a similar compensation response in Tribolium castaneum larvae. Insect Mol. Biol. 26: 35–45. [DOI] [PubMed] [Google Scholar]
- Phillips A. M., Smart R., Strauss R., Brembs B., Kelly L. E. 2005. The Drosophila black enigma: the molecular and behavioural characterization of the Black1 mutant allele. Gene 351: 131–142. [DOI] [PubMed] [Google Scholar]
- Posnien N., Schinko J., Grossmann D., Shippy T. D., Konopova B., Bucher G. 2009. RNAi in the red flour beetle (Tribolium), in: Harbor C.S. (Ed.), Cold Spring Harbor Protocols, p. doi:10.1101/pdb.prot5256. [DOI] [PubMed] [Google Scholar]
- Roseland C. R., Kramer K. J., Hopkins T. L. 1987. Cuticular strength and pigmentation of red-rust and black strains of Tribolium castaneum. Insect Biochem. 17: 21–28. [Google Scholar]
- Tribolium Genome Sequencing Consortium. 2008. The genome of the model beetle and pest Tribolium castaneum. Nature 452: 949–955. [DOI] [PubMed] [Google Scholar]
- Ulrich J., Dao V. A., Majumdar U., Schmitt-Engel C., Schwirz J., Schultheis D., Strohlein N., Troelenberg N., Grossmann D., Richter T. et al. 2015. Large scale RNAi screen in Tribolium reveals novel target genes for pest control and the proteasome as prime target. BMC Genomics 16: 674.. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vuerinckx K., Verlinden H., Lindemans M., Vanden Broeck J., Huybrechts R. 2011. Characterization of an allatotropin-like peptide receptor in the red flour beetle, Tribolium castaneum. Insect Biochem. Mol. Biol. 41: 815–822. [DOI] [PubMed] [Google Scholar]
- Wittkopp P. J., Carroll S. B., Kopp A. 2003. Evolution in black and white: genetic control of pigment patterns in Drosophila. Trends Genet. 19: 495–504. [DOI] [PubMed] [Google Scholar]
- Whyard S., Singh A. D., Wong S. 2009. Ingested double-stranded RNAs can act as species-specific insecticides. Insect Biochem. and Mol. Biol. 39: 824–832. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.