Abstract
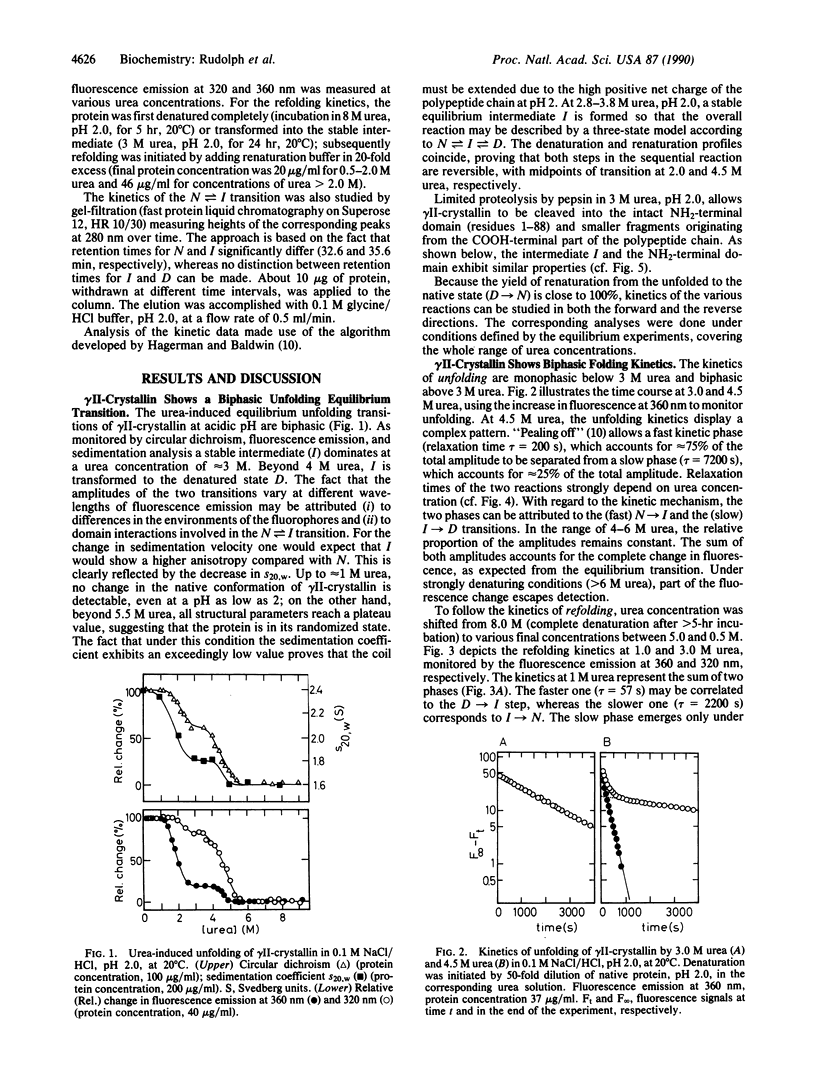
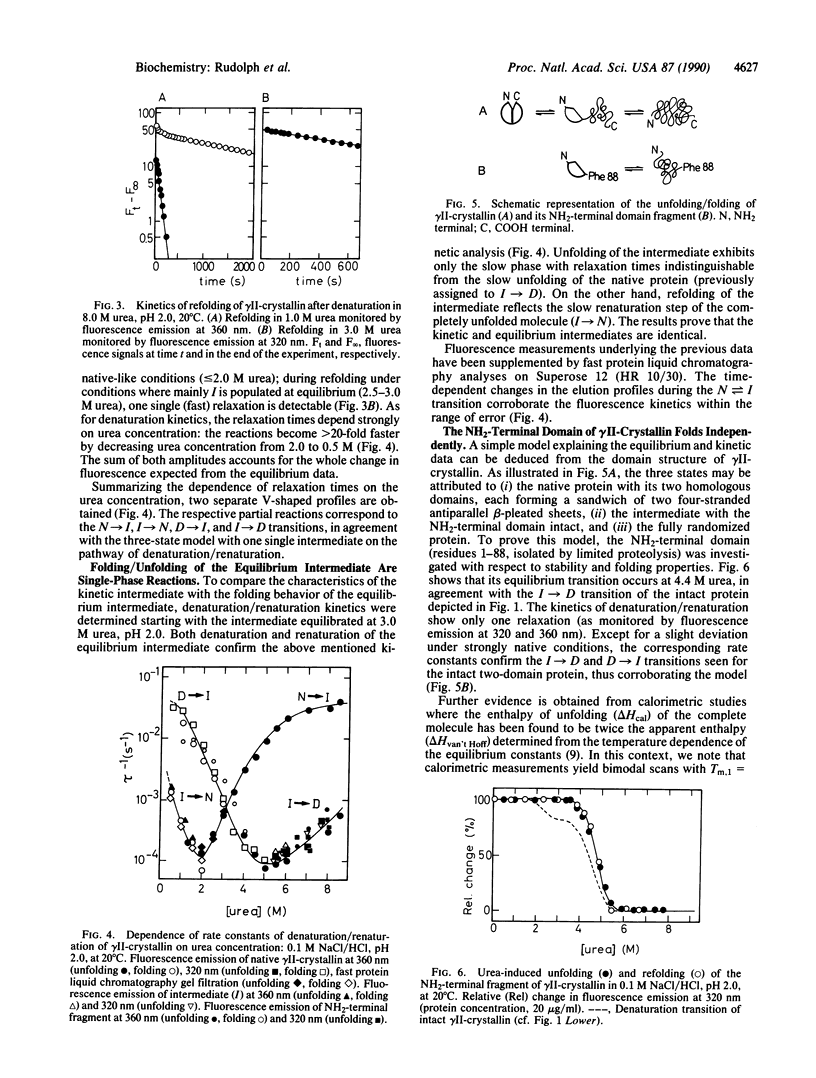
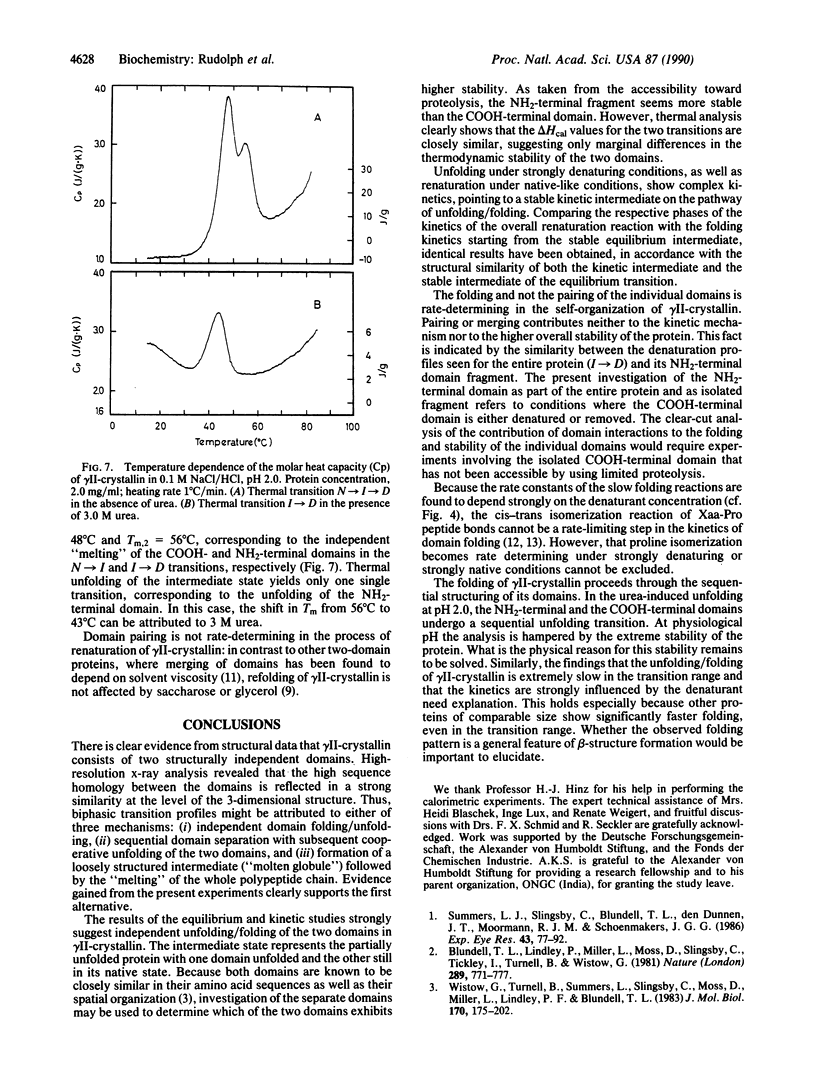
gamma II-crystallin from calf eye lens consists of two homologous domains, each composed of two similar "Greek key" motifs. As a consequence of the bilobal structure, a biphasic transition is seen upon unfolding by urea at low pH (monitored by circular dichroism, fluorescence emission, and ultracentrifugal analysis). In 3.3 +/- 0.5 M urea, a stable intermediate is formed at equilibrium, whereas 5.5 M urea causes maximum denaturation. Unfolding/folding kinetics display a complex pattern characterized by two kinetic phases. Both reactions exhibit strong dependence on the urea concentration; in the range of the respective transition, their rates are extremely slow (k approximately 1 x 10(-4)s-1). The kinetic mechanism of unfolding and refolding may be described by a three-state model: native in equilibrium intermediate in equilibrium denatured. The rate-determining steps are domain folding rather than domain pairing or proline isomerization. Kinetic analysis of the unfolding/folding of the intermediate populated in 3.0 M urea, pH 2.0, reveals that the kinetic and the equilibrium intermediates have similar structures. Limited proteolysis of gamma II-crystallin by pepsin in 3 M urea, pH 2, allows the NH2-terminal domain of the protein to be isolated. Unfolding/refolding of the fragment parallels the second transition in the above scheme, thus proving that the intermediate contains the COOH-terminal domain in its random state, whereas the NH2-terminal domain is still in its native conformation. In conclusion, folding of gamma II-crystallin proceeds through the independent sequential structuring of the domains.
Full text
PDF

Selected References
These references are in PubMed. This may not be the complete list of references from this article.
- Blundell T., Lindley P., Miller L., Moss D., Slingsby C., Tickle I., Turnell B., Wistow G. The molecular structure and stability of the eye lens: x-ray analysis of gamma-crystallin II. Nature. 1981 Feb 26;289(5800):771–777. doi: 10.1038/289771a0. [DOI] [PubMed] [Google Scholar]
- Hagerman P. J., Baldwin R. L. A quantitative treatment of the kinetics of the folding transition of ribonuclease A. Biochemistry. 1976 Apr 6;15(7):1462–1473. doi: 10.1021/bi00652a017. [DOI] [PubMed] [Google Scholar]
- Jaenicke R. Folding and association of proteins. Prog Biophys Mol Biol. 1987;49(2-3):117–237. doi: 10.1016/0079-6107(87)90011-3. [DOI] [PubMed] [Google Scholar]
- Kim P. S., Baldwin R. L. Specific intermediates in the folding reactions of small proteins and the mechanism of protein folding. Annu Rev Biochem. 1982;51:459–489. doi: 10.1146/annurev.bi.51.070182.002331. [DOI] [PubMed] [Google Scholar]
- Summers L. J., Slingsby C., Blundell T. L., den Dunnen J. T., Moormann R. J., Schoenmakers J. G. Structural variation in mammalian gamma-crystallins based on computer graphics analyses of human, rat and calf sequences. 1. Core packing and surface properties. Exp Eye Res. 1986 Jul;43(1):77–92. doi: 10.1016/s0014-4835(86)80047-1. [DOI] [PubMed] [Google Scholar]
- Wistow G., Turnell B., Summers L., Slingsby C., Moss D., Miller L., Lindley P., Blundell T. X-ray analysis of the eye lens protein gamma-II crystallin at 1.9 A resolution. J Mol Biol. 1983 Oct 15;170(1):175–202. doi: 10.1016/s0022-2836(83)80232-0. [DOI] [PubMed] [Google Scholar]
- van Dam A. F. Purification and composition of beta-s-crystallin. Exp Eye Res. 1966 Oct;5(4):255–266. doi: 10.1016/s0014-4835(66)80035-0. [DOI] [PubMed] [Google Scholar]



