Abstract
Steroid hormones are critical for organismal development and health. The rate-limiting step in steroidogenesis is the transport of cholesterol from the outer mitochondrial membrane (OMM) to the cytochrome P450 enzyme CYP11A1 in the inner mitochondrial membrane (IMM). Cholesterol transfer occurs through a complex termed the “transduceosome,” in which cytosolic steroidogenic acute regulatory protein interacts with OMM proteins translocator protein and voltage-dependent anion channel (VDAC) to assist with the transfer of cholesterol to OMM. It has been proposed that cholesterol transfer from OMM to IMM occurs at specialized contact sites bridging the two membranes composed of VDAC and IMM adenine nucleotide translocase (ANT). Blue native PAGE of Leydig cell mitochondria identified two protein complexes that were able to bind cholesterol at 66- and 800-kDa. Immunoblot and mass spectrometry analyses revealed that the 800-kDa complex contained the OMM translocator protein (18-kDa) and VDAC along with IMM CYP11A1, ATPase family AAA domain-containing protein 3A (ATAD3A), and optic atrophy type 1 proteins, but not ANT. Knockdown of ATAD3A, but not ANT or optic atrophy type 1, in Leydig cells resulted in a significant decrease in hormone-induced, but not 22R-hydroxycholesterol-supported, steroid production. Using a 22-phenoxazonoxy-5-cholene-3-beta-ol CYP11A1-specific probe, we further demonstrated that the 800-kDa complex offers the microenvironment needed for CYP11A1 activity. Addition of steroidogenic acute regulatory protein to the complex mobilized the cholesterol bound at the 800-kDa complex, leading to increased steroid formation. These results identify a bioactive, multimeric protein complex spanning the OMM and IMM unit that is responsible for the hormone-induced import, segregation, targeting, and metabolism of cholesterol.
Traditionally viewed as the powerhouse of the cell, mitochondria in recent years have demonstrated increasing roles in the number of cellular processes that are critical for the survival of the cell. Due to the large number of biochemical reactions that occur in the mitochondria, the architecture of the mitochondria must be tightly regulated to ensure the correct structural organization of the enzymatic/protein complexes. This organization is necessary both for the channeled flow of intermediates and for the optimal protein-protein interactions that regulate the flow of intermediates (1).
Mammalian gonadal and adrenal tissues synthesize steroids from cholesterol in response to stimulation by circulating pituitary hormones. The rate-limiting step in steroid production is the transport of cholesterol across the outer mitochondrial membrane (OMM) to the inner mitochondrial membrane (IMM) where cytochrome P450 enzyme CYP11A1 converts cholesterol to pregnenolone, the precursor of all steroids (2). The transfer of cholesterol into the mitochondria has been demonstrated to occur through a protein complex termed the “transduceosome” which is composed of OMM proteins voltage-dependent anion channel (VDAC) and translocator protein (TSPO, 18-kDa) and cytosolic proteins, acyl-coenzyme A binding domain containing 3 and the cAMP-dependent protein kinase-regulatory subunit Iα (PKA-RIα) (3). Steroid production is initiated at this complex by the hormone-responsive, mitochondrial-targeted steroidogenic acute regulatory (STAR) protein (4). The mitochondrial proteins present in this complex anchor the cytosolic proteins, allowing PKA-RIα to induce local, cAMP-dependent, phosphorylation of STAR required for maximal steroidogenic activity (2). STAR acts at the OMM to facilitate cholesterol transfer to the IMM (5) through the assistance of TSPO (4). TSPO is a high-affinity cholesterol- and drug-ligand binding protein that undergoes polymerization upon hormonal stimulation; this act increases TSPO ligand binding affinity and stimulates cholesterol transfer to the IMM (6). The role of the transduceosome in regulating cholesterol import into the OMM is clear. However, how the OMM proteins channel the flow of cholesterol into IMM, where CYP11A1 is located, remains unknown. Currently this process is proposed to occur through an OMM/IMM contact site composed of transduceosome proteins VDAC and TSPO with the IMM protein adenine nucleotide transporter (ANT) (7, 8).
In this study we set about to identify the mechanism upon which cholesterol is transferred from the OMM to the IMM where CYP11A1 is located. Utilizing blue-native polyacrylamide gel electrophoresis (BN-PAGE) coupled to mass spectrometry (MS), we identified a novel bioactive mitochondrial protein complex composed of both OMM and IMM proteins, specifically OMM proteins TSPO and VDAC and IMM proteins ATPase family AAA domain-containing protein 3A (ATAD3A) and CYP11A1, forming a complex that does not contain ANT. These proteins form a dynamic complex mediating cholesterol trafficking for steroid production.
Materials and Methods
Materials
Antibodies for VDAC, cytochrome c oxidase subunit IV (CoxIV), CYP11A1, optic atrophy type 1 (OPA1), and ATAD3A were obtained from Abcam (Cambridge, MA), ANT antibody was obtained from Santa Cruz Biotechnology, Inc. (Santa Cruz, CA). The TSPO polyclonal antibody was developed as previously described (9). MA-10 Leydig cells were maintained in DMEM/Ham's F12 (50:50) supplemented with 5% fetal bovine serum and 2.5% horse serum.
Steroid biosynthesis
MA-10 cells were plated into 96-well plates at a density of 2.5 × 104 cells per well. After 24 h, cells were stimulated with 50 ng/mg human chorionic gonadotropin (hCG) for the stated time points or stated concentration of hCG for 2 h. RIA analysis was performed as previously described (4) with antiprogesterone and antipregnenolone serum purchased from MP-Biomedicals (Irvine, CA). For measurement of isolated mitochondria steroid production, MA-10 cells were cultured and treated as previously described (10). In brief, cells were treated with 1 nm hCG in the presence of 0.76 mm aminoglutethimide, an inhibitor of CYP11A1. At the indicated time points, cell media were removed, and cells were frozen in liquid nitrogen. Frozen cells were resuspended in PBS, and mitochondria were isolated and incubated in 125 mm sucrose, 1 mm ATP, 1 mm reduced nicotinamide adenine dinucleotide, 50 mm KCl, 0.05 mm ADP, 2 mm dithiothreitol, 5 mm Na-succinate, 2 mm Mg(OA)2, 2 mm KH2PO4, and 10 mm HEPES buffer at pH 7.5 supplemented with 5 μm trilostane (4a,5-epoxy-17β-hydroxy-3-keto-5α-androstan-2α-carbonitrile to inhibit metabolism of pregnenolone to progesterone) at 300 μg/ml. The rate of pregnenolone formation, reflecting the amount of cholesterol available to CYP11A1, was determined by a RIA.
Cholesterol cross-linking assay
Mitochondria isolation, BN-PAGE, and two-dimensional SDS-PAGE were performed as previously described (11). Mitochondria were isolated from MA-10 cells cultured for 2 h in serum-free media and treated with 1 mm dibutyryl-cAMP (dbcAMP; BIOMOL Research Laboratories,) or 50 ng/mg hCG for 2 h. Mitochondria (50 μg) were incubated with 5 μCi (83.5 pmol) 7-azi-5α-[3,5,6-3H]cholestan-3β-ol ([3H]Photocholesterol) (American Radiolabeled Chemicals, Inc., St. Louis, MO) for 30 min. Half of the sample was cross-linked with a UVP 3UV Ultraviolet Lamp (Pierce Chemical Co., Rockford, IL) at 365 nm, 8 Watts for 16 min, with a 30-mm distance from the source to the sample. Wild-type (WT) and C258 STAR constructs were prepared, and their sequence was confirmed by sequencing (4). STAR in vitro transcription and translation was performed as previously stated (4). Briefly, 1 μl synthesized WT STAR or mutant C258 STAR was incubated with 50 μg isolated mitochondria preloaded with [3H]Photocholesterol. Samples were removed after 5 and 15 min and cross-linked as described above. One-way ANOVA was performed to determine significance.
Two-dimensional (2D)-SDS-PAGE
A lane from the one-dimensional BNP gel was removed and incubated with reducing solution (50 mm dithiothreitol and 1× NuPAGE LDS Sample Buffer; Invitrogen, Carlsbad, CA) for 30 min. The solution was removed and replaced with Alkylating Solution (Invitrogen) for 30 min before the addition of Quenching Solution (Invitrogen) for 15 min. The gel strip was placed on the top of a new gel for 4–20% Tris-Glycine 2D SDS-PAGE (Invitrogen), overlaid with NuPAGE LDS Sample Buffer, and run at 120 V. The gel was transferred and analyzed as described above.
Immuno (Western) blot analysis
Proteins separated by BN-PAGE or SDS-PAGE were electrophoretically transferred to a nitrocellulose membrane as previously described (9). Membranes were incubated with primary antibodies against TSPO (1:2500), VDAC (1:2000), CoxIV (1:1000), CYP11A1 (1:1000), OPA1 (1:1000), ATAD3A (1:1000), and ANT (1:500). Immunoreactive proteins were visualized using an enhanced chemiluminescence kit (Amersham Biosciences, Arlington Heights, IL) and horseradish peroxidase-goat antirabbit and horseradish peroxidase-rabbit antidonkey secondary antisera used at 1:5000 and 1:3000 dilutions, respectively.
CYP11A1 probe synthesis
The fluorogenic probe 22-phenoxazonoxy-5-cholene-3-beta-ol (cholesterol-resorufin) was synthesized in a four-step chemical synthesis (U.S. Patent 5,208,332 with modifications) (12). 3-β-acetoxy-23, 24-bisnor-5-cholenic acid (Steraloids, Inc., Newport, RI) (1.0 g) was dissolved in 5 ml methylene chloride cooled to 4 C. Next, 900 mg tert-butoxy thionyl chloride were added and stirred for 20 h. After a liquid/liquid extraction using NaCl solution, the organic phase was dried and 2.3 ml of 1.0 m tri-tert-butoxyaluminhydride in tetrahydrofuran was added under argon and cooled to −65 C. After 1 h, the mixture was extracted with NaCl/HCl solution. The dry organic phase contained compound I (3-β-acetoxy-5-cholene-22-ol) purified on a silica chromatography column. Next, 175 mg p-toluenesulfonyl chloride was added to compound I dissolved in pyridine. The mixture was stirred for 20 h at room temperature. 3-β-acetoxy-22-p-toluenesulfonyl-5-cholene was obtained after a liquid/liquid extraction with diethyl ether/HCl/NaCl. Next, 200 mg resorufin was added to the compound in DMSO and stirred under argon at 55 C for 10 d. Compound II (3-β-acetoxy-22-phenoxazonoxy-5-cholene) was obtained by purification using a silica chromatography column. Compound II was suspended in 5% KOH/methanol containing 1% water and refluxed for 30 min under argon. After washing the mixture with HCl/water, fluorogenic probe 22-phenoxazonoxy-5-cholene-3-β-ol was obtained as an orange solid crystallized in methylene chloride/hexane. After synthesis, the sample was dissolved in 10 mg/ml ethanol, sonicated, and filtered through a 0.22-μm filter.
Steroidogenic CYP11A1 probe assay
MA-10 cells were incubated with 5 μm CYP11A1 probe for 24 h and plated into 96-well plates at a density of 2.5 × 104 cells per well for another 24 h. Half of the wells were stimulated with dbcAMP, and the other half was left unstimulated as controls. Fluorescence was measured at 530ex/595em at 5-min intervals using the Victor multilabel plate reader (PerkinElmer Life Sciences, Boston, MA). Isolated mitochondria were resuspended in steroidogenic buffer (250 mm sucrose, 10 mm potassium phosphate buffer, 15 mm triethanolamine-HCl, 20 mm KCl, 5 mm MgCl2, 5 μm trilostane, pH 7.0), and 5 μm CYP11A1 probe was added. Steroidogenesis was initiated by addition of one tenth volume of buffer C (150 mm isocitrate, 5 mm NADP), and fluorescence was measured at 530ex/595em. When indicated, aminoglutethimide (0.67 mm, Sigma Chemical Co., St. Louis, MO) was added before addition of CYP11A1 probe and buffer C. BN-PAGE gel slices were incubated with 50 μm CYP11A1 probe and steroidogenic buffer in a 96-well plate. Steroidogenesis was initiated by adding one tenth volume of buffer C, and after 30 min, fluorescence was measured at 530ex/595em.
In-gel digestion and MS analysis
The BN-PAGE gel was divided into 18 horizontal slices starting at 20 kDa, cut into 3-mm slices, increasing in molecular mass until 1200 kDa and subjected to in-gel digestion and MS as previously described (11). In brief, resulting in-gel digests were desalted using C18 Zip Tips (Millipore Corp., Medford, MA) before analysis by nanoflow reversed-phase liquid chromatography using an Agilent 1100 LC system (Agilent Technologies, Inc., Palo Alto, CA) coupled online to a linear ion trap mass spectrometer (LTQ, Thermo Scientific, San Jose, CA). Reversed-phase separations were performed using 75 μm inner diameter × 360 μm outer diameter × 10 cm long capillary columns (Polymicro Technologies, Inc., Phoenix, AZ) that were slurry packed in house with a 5-μm, 300 Å pore size Jupiter C-18 silica-bonded stationary phase (Phenomenex, Torrance, CA). After being injected with 5 μl of sample, the column was washed for 20 min with 98% solvent A (0.1% formic acid in water, vol/vol), and peptides were eluted using a linear gradient of 2% solvent B (0.1% formic acid in 100% acetonitrile, vol/vol) to 85% solvent B for 110 min at a constant flow rate of 250 nl/min. The linear ion trap-MS was operated in a data-dependent mode in which each full MS scan was followed by seven MS/MS scans in which the most abundant peptide molecular ions were dynamically selected for collision-induced dissociation using a normalized collision energy of 36%. The temperature of the heated capillary and electrospray voltage (applied on column base) was 180 C and 1.7 kV, respectively. The collision-induced dissociation spectra were searched against a nonredundant mouse protein database using SEQUEST (Thermo Scientific, San Jose, CA), and results were tabulated for each identified peptide/protein. Control and hCG-treated BN-PAGE mitochondrial protein separations were performed in triplicate, and each gel slice was analyzed by MS in triplicate. A two-tailed t test was performed to determine significance between control and hCG-treated gel slices.
Forster resonance energy transfer (FRET) analysis
The CYP11A1 and ANT1 cDNA was obtained from Open Biosystems and inserted into eCFP-N1. The VDAC1 cDNA (Open Biosystems, Huntsville, AL) was inserted into DsRED-N1. All constructs were verified by sequencing at the McGill University Genome Quebec Innovation Centre. MA-10 cells were plated on a 35-mm FluoroDish Sterile Culture Dish (World Precision Instruments, Inc., Sarasota, FL) and transiently transfected with Lipofectamine 2000 (Invitrogen). Cells were evaluated using a scanning laser confocal microscope (FluoViewTM FV1000; Olympus Corp., Lake Success, NY; software version 1.70.16) at 100× with an oil immersion objective (UPLSAP). Images were processed using Adobe Photoshop and Image-Pro Plus (version 6.3) and analyzed using the pFRET plug-in for Image-J. A Mann-Whitney U test was performed to determine the statistical difference in E% between two sets of independent samples (before and after 2-h dbcAMP treatment).
Small interfering RNA (siRNA) treatment and RIA
MA-10 cells were transfected with siRNA Trifecta kit (Integrated DNA Technologies, Coralville, IA) per manufacturer's instructions. RNA was obtained using RNAmax (Invitrogen). After 2 d of treatment, cells were plated into 96-well plates at a density of 2.5 × 104 cells per well. After 24 h, cells were stimulated with 1 mm dbcAMP, 50 ng/mg hCG, or 20 μm 22R-hydroxycholesterol for RIA. RIA analysis was performed as previously described (4) with antiprogesterone and antipregnenolone serum purchased from MP Biomedicals. Erastin (Sigma) was incubated at the indicated concentrations with hCG, and after 2 h the samples were harvested. RIA data were analyzed using Graphpad Software (San Diego, CA). Cells were harvested for immunoblot and qPCR analyses 3 d after treatment, as previously described (11).
ATAD3A: NM_179203, used at 150 nm; Duplex 1, 5′-CCAUCGCAACAAGAAAUACCAAGAA-3′; Duplex 2, 5′ -CCAGUUUGACUAUGGAAAGAAAUGC-3′; Duplex 3, 5′-AGGACAAAUGGAGCAACUUCGACCC-3′
OPA1: NM_133752, used at 50 nm; Duplex 1, 5′- GGAAGUCCAUGCGCCAUUGGGAGGG-3′; Duplex 2, 5′-GUCAUUGUCGGAGCAGGAAUCGGAC-3′; Duplex 3, 5′- GGAAUACAAAGAAACGUACCGCUCC-3′
VDAC1: NM_011964, used at 50 nm; Duplex 1, 5′-GCUAUAAGACGGAUGAAUUCCACT-3′; Duplex 2, 5′-CGAAGUCAGAGAAUGGAUUGGAATT-3′; Duplex 3, 5′-GGAAUUUCAAGCAUAAAUGAAUATT
ANT1: NM_007450, used at 25 nm; Duplex 1, 5′- CGCCUUCAAAGACAAGUACAAGCAG-3′;
Duplex 2, 5′-AGCUCACAAGUUCACAGAUCCAUTG-3′; Duplex 3, 5′-GGCGUAGGCAAGAGCAAACGAGCGG-3′
Scrambled: 5′-CGUUAAUCGCGUAUAAUACGCGUAT-3′
Mitochondria morphology
For confocal microscopy imaging, cells were treated with siRNA as previously stated; on the third day cells were plated into a 24-well glass bottom dish previously treated with poly-l-lysine. Cells were grown for an additional 24 h, treated with 100 nm mitotraker and Hoechst (Invitrogen). Confocal microscopy was performed as previously stated. Counts are representative of two independent experiments with 200 cells scored per experiment. For electron microscopy (EM) MA-10 cells were treated with siRNA as previously stated. At the end of the treatments cells were harvested and washed in sodium cacodylate buffer (0.1 m, pH 7.4; MECALAB, Montreal, Quebec, Canada). After centrifugation (1500 × g for 5 min), the pellet was fixed with 2.5% glutaraldehyde (Sigma) overnight, at 4 C. After three washes with dH2O, cells were stained with OsO4 1% Osmium (MECALAB) for 2 h at 4 C. Cells were subsequently dehydrated by incubation in increasing acetone concentration from 30 to 100%. After a step of infiltration with different mixtures of acetone-epon (1:1, 1:1:3, vol/vol), the samples were embedded in pure Epon (MECALAB). Polymerization was performed by incubation at 60 C for 48 h. Blocks were cut in slices of 100 nm, collected on 200 mesh copper grids (Electron Microscopy Sciences, Fort Washington, PA), and poststained with 4% uranyl acetate for 5 min followed by Reynold's lead for 5 min. Cells on grids were observed with a transmission electron microscope FEI TECNAI 12 operated at 120 kV at the Facility for Electron Microscopy Research, McGill University. Images were collected on a charge-coupled device camera (AMT XR 80 C). Quantitative evaluation of altered mitochondrial morphology of cells treated with the various siRNA was obtained by observing an average of 400 mitochondria in 20 cells and counting altered mitochondria compared with control. Values are reported as a percentage of mitochondria normalized to control.
Mitochondrial integrity
To measure the cytotoxicity of the siRNA used, cells plated into 96-well plates were treated with the various siRNA as indicated earlier. At the end of the treatment cells were incubated with 3-(4,5-dimethylthiazol-2-yl)-2,5-diphenyltetrazolium bromide as described in the CellTiter 96 Non-radioactive Cell Proliferation Assay (Promega) based on the conversion of water-soluble 3-(4,5-dimethylthiazol-2-yl)-2,5-diphenyltetrazolium bromide (MTT) to insoluble formazan. Formazan blue formation was quantified at 600 and 690 nm using the Victor multilabel plate reader, and the results were expressed as (OD600 − OD690). ATP levels were determined using the ATP Determination Kit (PerkinElmer Corp., Wellesley, MA). Cells were treated with siRNA as previously stated, plated into six-well plates and harvested after 24 h. Cells plated into six-well plates were treated with the various siRNA, and, at the end of the treatment, cells were washed in phosphate buffer saline followed by ATP determination as specified in the manufacturer's instructions. Samples were analyzed for luminescence using the Victor multilabel plate reader. For all experiments, samples were analyzed in replicates of three, and ATP levels were determined according to ATP standard curves (in the range of 0.5 to 10 μm) of fresh ATP solution.
Statistical analysis
Statistical analysis was performed using Student's t test and one-way ANOVA followed by the Dunnett's Multiple Comparison Test, using the Prism 4.02 software package from GraphPad (San Diego, CA).
Results
Identification of cholesterol-binding protein complexes
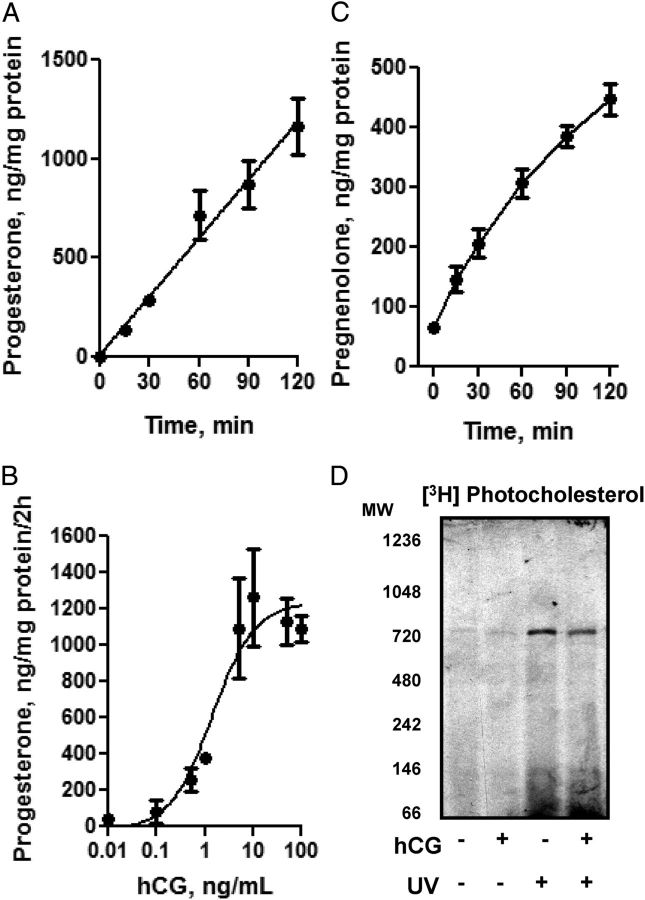
Stimulation of mouse Leydig cells, MA-10, by hCG results in a rapid and dose-dependent rise in steroid production (Fig. 1, A and B). To confirm that steroid production was due to cholesterol transferred to the mitochondrial CYP11A1, MA-10 cells were treated with hCG together with the CYP11A1 inhibitor aminoglutethimide. The inhibitor was removed from the cells, and the mitochondria were isolated in the presence of the 3β-hydroxysteroid dehydrogenase inhibitor trilostane, allowing pregnenolone formation to occur again (Fig. 1C). Because no additional cholesterol was added to the mitochondria, this allowed confirmation that pregnenolone formation was due to the transfer and accumulation of cholesterol at the CYP11A1 in the IMM. We next set about to identify the mechanism by which cholesterol enters in the MA-10 mitochondria. To accomplish this we used a photoactivatable cholesterol derivative, [3H]Photocholesterol, that covalently cross-links to neighboring proteins (13). After incubation of [3H]Photocholesterol with isolated mitochondria and subsequent cross-linking by UV light, we were able to identify protein complexes by BN-PAGE; a technique in which digitonin-solubilized membranes are run on a native gel, ensuring that protein complexes remain intact and protein complexes can then be identified via immunoblot analysis (14). Through this process we identified two protein complexes at 50–80 kDa, named 66 kDa because it migrated at the same place as the 66-kDa molecular mass marker did, and 800 kDa that bound [3H]Photocholesterol (Fig. 1D). No significant changes were observed upon hCG treatment before mitochondrial isolation, suggesting that, in both basal and hormone stimulation conditions, cholesterol enters the mitochondria through one conserved pathway.
Fig. 1.
hCG-induced mitochondrial steroid formation. A, Time-dependent effect of 50 ng/ml hCG incubation on MA-10 Leydig cell progesterone formation. B, Dose-dependent production of progesterone after a 2-h incubation of stated concentration of hCG on MA-10 Leydig cells. Results shown in A and B are means ± sem (n = 2). C, Cultured MA-10 cells were washed with serum-free medium and treated with hCG (1 nm) and aminoglutethimide (0.76 mm). At the indicated time points mitochondrial were prepared, and the rate of pregnenolone formation was measured, as described in Material and Methods. The results shown are the means ± sem (n = 3). D. Mitochondria were isolated from control or hCG-treated MA-10 cells and incubated with [3H]Photocholesterol for 30 min. Upon completion, half of the sample was cross-linked with a UV light, solubilized with 1% digitonin buffer, and run on a BN-PAGE gel. The gel was transferred to polyvinylidene difluoride and exposed to film, and the [3H]Photocholesterol was identified at 66-kDa and 800-kDa molecular masses. MW, Molecular weight (molecular mass).
Identification of novel mitochondrial contact site formation
To further characterize the identified 66- and 800-kDa protein complexes, where [3H]Photocholesterol bound, the BN-PAGE gel was divided into 18 horizontal gel slices starting at 20 kDa and increasing to 1200 kDa and analyzed using Mass Spectrometry (MS). The identified proteins from the average of three slices of control and hCG-treated mitochondria were organized according to their molecular mass on the BN-PAGE gel. Focusing on the results from the 66-kDa and 800-kDa complex gel slices, where [3H]Photocholesterol bound, we identified several proteins critical for steroid production that were up-regulated upon hormonal stimulation, suggesting that this complex possesses specific steroidogenic activity (Table 1). These proteins included, CYP11A1, ferredoxin reductase, VDAC1, and LONP1.
Table 1.
Major proteins present in the 66- and 800-kDa mitochondrial complexes identified by MS
| Protein complex | Accession no. | Peptides control (n) | Peptides hCG (n) | P |
|---|---|---|---|---|
| 66-kDa | ||||
| MDH2 | P08249 | 7 | 30 | 0.004 |
| CYP11A1 | Q9QZ82 | 24 | 34 | 0.06 |
| FDXR | Q3UZ58 | 56 | 64 | 0.493 |
| IDH2 | P54071 | 13 | 16 | 0.587 |
| 800-kDa | ||||
| VDAC1 | Q60932 | 11 | 21 | 0.0005 |
| ACAT1 | Q8QZT1 | 14 | 20 | 0.013 |
| LONP1 | Q3V2D0 | 31 | 48 | 0.043 |
| CYP11A1 | Q9QZ82 | 23 | 31 | 0.396 |
| ATAD3A | Q925I1 | 6 | 8 | 0.643 |
| OPA1 | P58281 | 6 | 8 | 0.643 |
| IDH2 | P54071 | 8 | 9 | 0.767 |
| MDH2 | P08249 | 8 | 7 | 0.677 |
Mitochondrial samples from three separate experiments were isolated from control and hCG-treated MA-10 cells; statistical analysis was performed via a two-tailed t test on control vs. hCG-treated samples (see Fig. 2). FDXR, Ferredoxin reductase
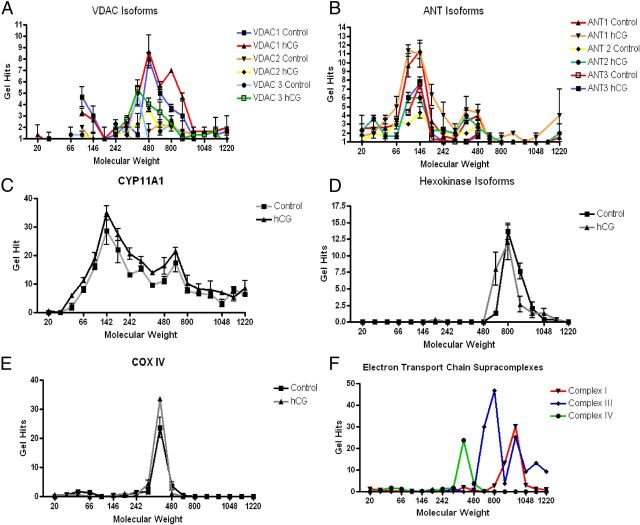
Further analysis of the MS results of the BN-PAGE slices identified that whereas the 800-kDa cholesterol cross-linking complex contains VDAC isoforms, which significantly increase upon hormonal stimulation (Fig. 2A), very little ANT isoforms were identified (Fig. 2B). ANT isoforms were primarily identified at the 146- and 480-kDa molecular mass complexes whereas VDAC isoforms hits were mainly seen at the 480- and 800-kDa complexes. These data suggest that VDAC and ANT, which are proposed to form the mitochondrial contact site for cholesterol transfer, do not interact in the identified 800-kDa complex although they might interact in other protein complexes, specifically at the 146- or 480-kDa molecular masses. CYP11A1 was identified at all molecular masses (Fig. 2C) with an increased, although not significant, manner upon hCG treatment, suggesting an improved association with the IMM or homodimerization (15). Hexokinase isoforms were identified at the 800-kDa complex (Fig. 2D), implying that the 800-kDa complex could be a functional mitochondrial contact site, possibly participating in cholesterol transport for steroid production (16).
Fig. 2.
MS Analysis of BN-PAGE gel identifies protein complex at 800 kDa. Mitochondria from control and hCG-treated cells were isolated, and protein complexes were isolated by BN-PAGE. The gel was divided into 18 slices (3 mm in width) and subjected to MS analysis. The resulting protein identification was classified according to gel slice/molecular mass of the BN-PAGE and the average samples identified of the three gel slices from control and hCG treatment. VDAC isoforms (panel A), ANT isoforms (panel B), CYP11A1 (panel C), hexokinase isoforms (panel D), Cox IV (panel E), and electron transport chain supracomplexes (panel F) were each identified at the stated molecular mass with VDAC and CYP11A1 present at 800 kDa.
From these results, we then proposed that the 800-kDa complex is a multiprotein complex capable of steroidogenic activity. To test this hypothesis, we set out to identify other known multiprotein complexes present in the MS results. Because the electron transport chain complexes have previously been demonstrated to organize into supramolecular complexes, we used this complex as our control. We identified the individual subunits of each of the electron chain complexes I, III, and IV from the BN-PAGE MS from the three gel slices from the control isolated mitochondria. CoxIV, a subunit of electron transport chain complex IV, was identified at the 440-kDa complex (Fig. 2E), which also associated with the rest of complex IV at 440 kDa (Fig. 2F), confirming our hypothesis that multiprotein complexes could be identified by MS analysis of the separated BN-PAGE protein complexes. In these studies we also observed that the electron transport chain complexes further interacted to form larger complexes, as demonstrated by the formation of electron transport chain subunit complexes I and III into supramolecular complexes (Fig. 2F) in agreement with previous data (17). These results further validate the utilization of this methodology for the identification of novel mitochondrial protein complexes, specifically in the identification of a functional steroidogenic complex.
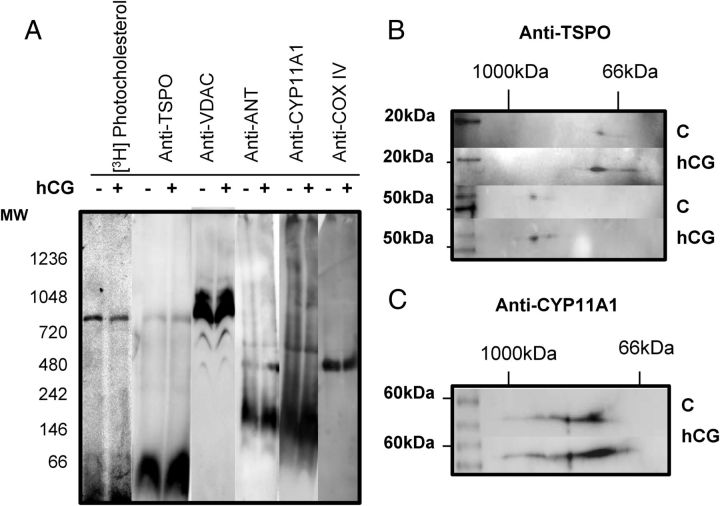
To further identify whether other proteins from the transduceosome complex are present in the 800-kDa complex and further validate the MS results, we then used immunoblot analysis. As expected, we were unable to detect TSPO via MS, due to the large amount of recombinant TSPO needed to obtain a MS profile of the protein (data not shown); therefore, we initiated the immunoblot analysis with anti-TSPO antibodies. The results obtained revealed the presence of TSPO both in the 66-kDa and 800-kDa protein complexes, corresponding to the [3H]Photocholesterol-labeled complexes (Fig. 3A). VDAC1 was detected at multiple molecular masses as was observed in the MS results, confirming its localization in the 800-kDa complex (Fig. 3A). ANT was only detected at 146-kDa and CYP11A1 was present mainly at 146-kDa although immunoreactive bands at higher molecular sizes were seen (Fig. 3A). Cox IV was identified at 440-kDa (Fig. 3A), demonstrating that the immunoblot analysis of BN-PAGE is in agreement with results seen by MS.
Fig. 3.
A, Identification of cholesterol-binding mitochondrial protein complexes. Mitochondria were isolated and treated as previously stated. Immunoblot analysis of BN-PAGE identified TSPO at 66 and 800 kDa, same as the [3H]Photocholesterol, CYP11A1 at 146 kDa and higher molecular masses, VDAC laddering at 440, 600, and 800 kDa and higher complexes, and CoxIV was located at 440 kDa. B, 2D-SDS-PAGE was performed on BN-PAGE gel strip, identifying TSPO as an 18-kDa monomer at the 66-kDa complex and a 54-kDa polymer at 800-kDa complex. C, 2D SDS-PAGE gel identified CYP11A1 forming a band at 55 kDa. MW, Molecular weight (molecular mass).
Because BN-PAGE preserves intact protein complexes, the site of antigen binding might not be available to the antibody in the protein complex; therefore further analysis of the BN-PAGE complexes by 2D SDS-PAGE, in which the first dimension was the BN-PAGE gel, was used to further determine the composition of the protein complexes. Through this method it was identified that TSPO was present as an 18-kDa monomer in the 66-kDa complex and as a 56-kDa trimer in the 800-kDa complex (Fig. 3B). Because TSPO is known to form polymers upon hormonal stimulation (6), this process would increase the concentration of cholesterol found specifically at the 800-kDa complex. 2D-gel immunoblot analysis revealed that CYP11A1 is present at higher molecular masses from 140 to 1000 kDa (Fig. 3C) as was seen with the MS results, suggesting it is in complex with other mitochondrial proteins and unable to be detected via immunoblot in BN-PAGE.
Steroidogenic bioactivity identified in BN-PAGE
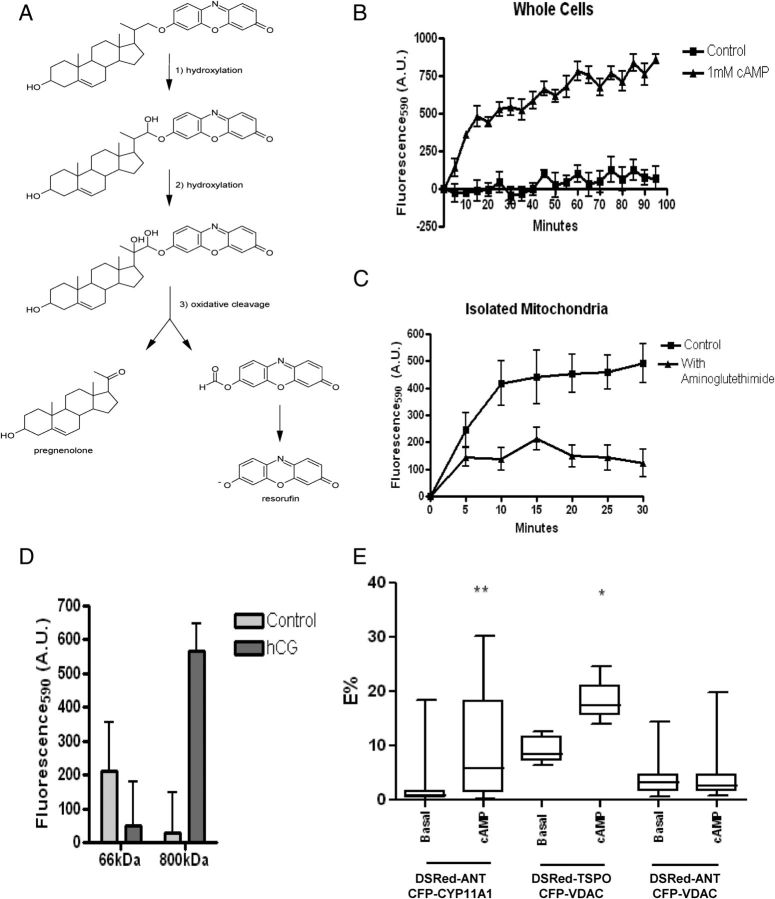
Because cholesterol bound to the 800-kDa complex could be mobilized for transport to the IMM, we then investigated whether the cholesterol transferred through this complex is used for pregnenolone synthesis. We synthesized a CYP11A1 probe in which a resorufin dye was conjugated to the side chain of cholesterol and upon cleavage of the cholesterol side chain by CYP11A1, resorufin levels can be measured at 590 nm (Fig. 4A). Cholesterol-resorufin has been used to assess CYP11A1 activity in gonadal cells (12) and in isolated CYP11A1 reconstituted in proteoliposomes (18). MA-10 cells incubated with the CYP11A1 probe showed a time-dependent increase in fluorescence after being stimulated with 1 mm dbcAMP (Fig. 4B). Mitochondrial specificity was confirmed by inhibiting the increase in fluorescence by addition of the CYP11A1 inhibitor aminoglutethimide to isolated mitochondria (Fig. 4C).
Fig. 4.
Cholesterol-Resorufin probe detects CYP11A1 activity. A, The enzymatic activity of CYP11A1 on the cholesterol-resorufin probe results in the production of fluorescent resorufin and pregnenolone. B, MA-10 cells incubated with 5 μm of the cholesterol resorufin probe for 24 h, after which the addition of cAMP results in a time-dependent increase in fluorescence; measurement was at 530ex/595em at the stated time points. C, Isolated mitochondria were incubated with 5 μm cholesterol-resorufin probe or cholesterol-resorufin probe and CYP11A1 inhibitor aminoglutethimide (AG). Measurement at 530ex/595em at stated time points demonstrate an increase in fluorescence in mitochondria not incubated with AG. D, Cholesterol-resorufin probe (50 μm) was incubated in control and hCG-stimulated gel slices. After addition of isocitrate to stimulate steroid production, CYP11A1 activity was measured at 530ex/595em, demonstrating increased activity at the 800-kDa complexes upon hCG treatment. E, Transiently transfected MA-10 cells were imaged 48 h after transfection of DsRed-ANT, CFP-CYP11A1 pair; DsRed-ANT, CFP-VDAC pair; or DsRed-TSPO, CFP-VDAC pair. Cells were treated for 2 h with dbcAMP and imaged. Mann-Whitney U test was performed to determine statistical differences of the E% between samples obtained before and after cAMP treatment. Results shown are means ± sem (n = 3); *, P < 0.05 **, P < 0.01.
BN-PAGE gel slices containing the 66-, or 800-kDa complexes were incubated with the CYP11A1 probe, and steroidogenesis was initiated by the addition of isocitrate. The activity of the hCG-treated 66-kDa complex was decreased compared with control, but the hCG-treated 800-kDa complexes showed an increase in resorufin emission (Fig. 4D). Because all complexes studied demonstrate minimal CYP11A1 activity, these data confirmed that CYP11A1 is constitutively active in the IMM but lacks the cholesterol substrate (19). Because we propose only the 800-kDa complex possesses the capability for cholesterol transfer across the intermembrane space, this mitochondrial complex could allow for both the rapid induction and termination of steroid biosynthesis by factors acting upon it, such as the transduceosome protein STAR. hCG-induced changes in the architecture of the mitochondria, as demonstrated by the increased resorufin fluorescence in the 800-kDa mitochondrial complex isolated from hCG-treated cells, suggests that the 800-kDa complex offers the microenvironment needed for CYP11A1 activity (20).
Mitochondrial contact site formation upon hormonal stimulation
It has been hypothesized that cholesterol transfer from the OMM to IMM occurs primarily at mitochondrial contact sites (7) where VDAC and ANT are major components (8). TSPO is enriched in steroidogenic mitochondria contact sites (21) and is known to interact with VDAC and ANT (22). To determine whether hormone treatment induces reorganization of the mitochondrial contact sites, FRET was used to study the interactions between TSPO, VDAC, ANT, and CYP11A1. Our results demonstrate that the tagged protein pairs ANT-CYP11A1 and TSPO-VDAC, but not VDAC-ANT, showed significantly increased FRET upon dbcAMP stimulation (Fig. 4E). The lack of increase in FRET between ANT and VDAC upon dbcAMP stimulation suggests that these proteins interact but they are in a separate protein complex from the 800-kDa complex, as was also observed by BN-PAGE.
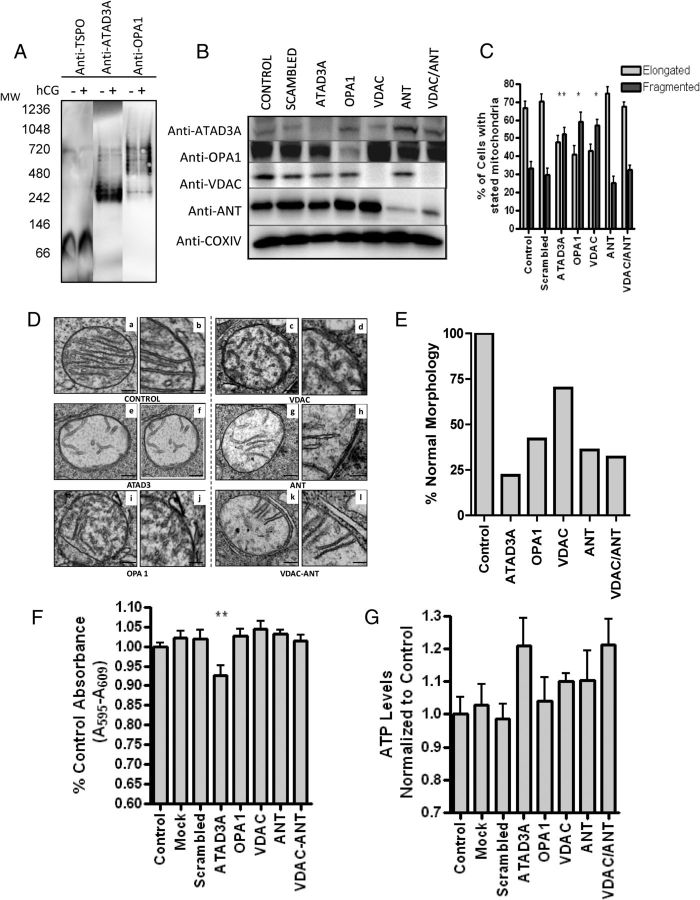
To determine the mechanism for cholesterol transfer to the IMM, we analyzed the MS data for proteins located in the 800-kDa complex that could play a role in cholesterol transfer to the IMM, identifying both ATAD3A and Dynamin-like 120-kDa protein OPA1. ATAD3A is an ATPase that is located in the mitochondrial matrix with its N terminus anchored in the OMM, where it may bridge the two mitochondrial membranes, controlling contact site formation in an ATP-dependent manner (23). Decreased ATAD3A expression results in reduced hormone-stimulated steroidogenesis, suggesting that altering the contact site formation alters cholesterol transfer across the intermembrane space (23). OPA1 is an IMM protein involved in mitochondrial fusion and shaping that could function in the formation of mitochondrial contact sites, although its role in steroid production has not been demonstrated (24). To confirm their presence in the 800-kDa complex, we probed BN-PAGE membrane with anti-ATAD3A and OPA1, verifying their presence in this complex (Fig. 5A). To examine which protein plays a role in cholesterol transfer, MA-10 cells were transfected with siRNA against ATAD3A, OPA1, VDAC, ANT, or VDAC/ANT together. Efficient knockdown of protein expression was confirmed by both Western blot (Fig. 5B) and altered mitochondrial morphology (Fig. 5, C and D). An increase in fragmented mitochondria over WT was observed through confocal microscopy upon the treatment of siRNA toward OPA1, ATAD3A, and VDAC. Because these proteins have been previously demonstrated to alter mitochondrial fusion ability, this further confirmed efficient protein knockdown (Fig. 5C). EM analysis was subsequently performed to further study changes in mitochondrial morphology and, more specifically, the mitochondrial cristae; because the initial step in steroidogenesis occurs at the matrix side of the IMM, alteration of cristae morphology could alter steroid production. Quantification of altered mitochondria morphology further confirmed the confocal imaging data showing significant alteration of mitochondria cristae formation in response to siRNA treatments targeting ATAD3A and OPA1 but also toward VDAC/ANT-treated cells (Fig. 5, D and E). Because of the observed changes in mitochondrial morphology, we investigated the state of mitochondrial integrity using the MTT assay, which assesses the metabolism of MTT into formazan by the mitochondrial succinic dehydrogenase. A slight but significant reduction was observed only in cells in which the ATAD3A protein was knocked down (Fig. 5F). However, this effect was not detrimental to the cells as demonstrated by the measurement of cellular ATP production (Fig. 5G). Interestingly, the various siRNA treatments used to knock down the indicated mitochondrial proteins had no significant effect on ATP production; in contrast, a slight increase was observed in cells in which VDAC/ANT and ATAD3A proteins were knocked down (Fig. 5G). This could be due to an increase in cytosolic glycolysis ATP production (25).
Fig. 5.
Effect of ATAD3A, VDAC, OPA1, and ANT knockdown on MA-10 mitochondrial morphology. A, BN-PAGE immunoblot identifies TSPO, OPA1, and ATAD3A at the 800-kDa molecular mass complex. B, MA-10 cells were transfected with the siRNA targeted for the indicated gene products. After 3 d of treatment, specific protein expression was assessed by immunoblot analysis. Immunoblots shown are representative of three independent experiments. C, The percent elongated vs. tubular mitochondria of the mitochondria treated with stated siRNA as indicated in panel B. One-way ANOVA with Dunnett's Multiple Comparison Test on the difference in percent elongated was performed. Results shown are means ± sem (n = 2; 200 cells scored each); *, P < 0.05, **, P < 0.01. D, Transmission EM images of MA-10 cells treated with or without mitochondrial protein-specific siRNAs. a, c, e, g, I, and k are low magnification images (200 nm) whereas b, d, f, h, j, and i are higher magnification images (100 nm) of representative mitochondria from cells treated as indicated under each panel. E, Quantitative evaluation of altered mitochondrial morphology in response to the various siRNA treatments obtained as described under Materials and Methods. Values are reported as percentage of normal mitochondria relative to control; control, scrambled, and mock have the same percentage of transfection. F, MTT analysis compared with control (%) and (G) ATP levels normalized to control in siRNA-treated cells. Samples are ± sem (n = 2; each point in triplicate); **, P < 0.01.
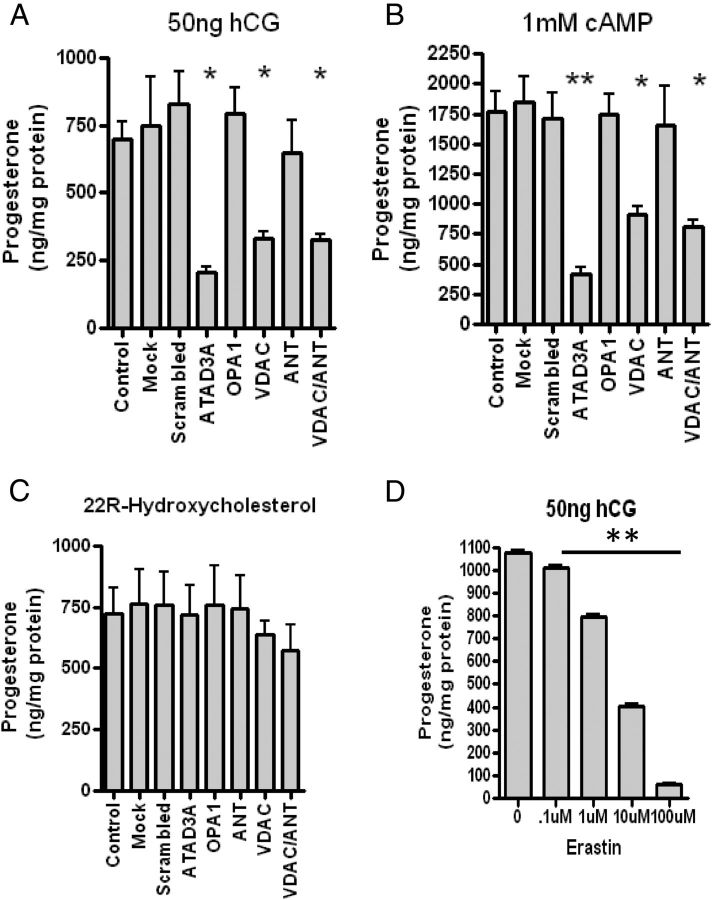
To assess the steroidogenic ability of the MA-10 cells depleted of the indicated mitochondrial proteins, cells were stimulated with either hCG (Fig. 6A) or dbcAMP (Fig. 6B), and steroid synthesis was determined. A significant decrease in progesterone production upon treatment with hCG or dbcAMP was observed in cells transfected with siRNA targeting ATAD3A, VDAC1, the main VDAC present in MA-10 cells, and VDAC/ANT. However, treatment of the cells with siRNA targeting ANT alone or OPA1 did not affect the hormone- or cAMP-induced steroid formation (Fig. 6, A and B). To determine that there was no alteration in CYP11A1-mediated mitochondrial steroid formation, the siRNA-treated cells were incubated with 22R-hydroxycholesterol (22R), a membrane-permeable CYP11A1 intermediate, which resulted in no change in steroid production (Fig. 6C), confirming that CYP11A1 activity and steroidogenic pathway were not affected by the treatments. To further assess the role of VDAC, we examined the impact of altering its permeability using erastin, a drug targeting VDAC permeability resulting in altered ROS production (26). This alteration resulted in reduced steroidogenesis (Fig. 6D), confirming that VDAC is important for contact site formation and steroid biosynthesis. In contrast, because knocking down ANT had no effect on steroidogenesis, its role in cholesterol transport and contact site formation is questionable.
Fig. 6.
Role of ATAD3A, VDAC, OPA1, and ANT in MA-10 steroidogenesis. MA-10 cells treated with various siRNA were stimulated with 50 ng/ml hCG (A) 1 mm dbcAMP (B) or 20 μm 22R-hydroxycholesterol (C). One-way ANOVA with Dunnett's Multiple Comparison Test was performed with the results shown as means ± sem (n = 3); *, P < 0.05, **, P < 0.01. D, Effect of the VDAC permeability inhibitor erastin on MA-10 cell steroid formation. Cells were incubated with the indicated concentrations of erastin and hCG for 2 h before harvesting. Results shown are means ± sem (n = 2; each point in triplicate); **, P < 0.01.
Mobilization of cholesterol bound at the OMM
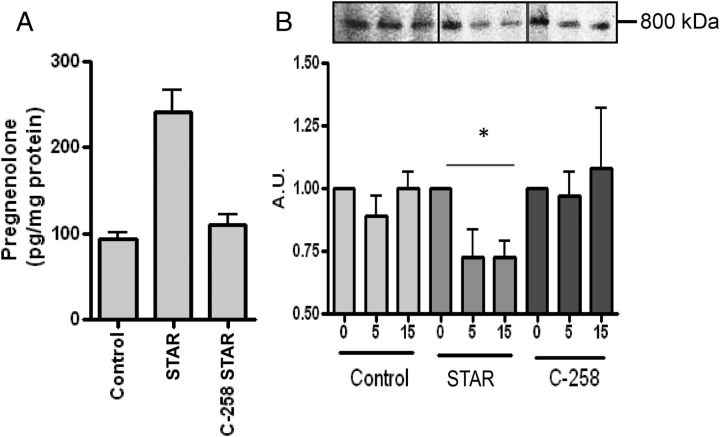
Because ATAD3A knockdown resulted in a significant decrease in steroid production, a protein identified only at the 800-kDa complex, we next set out to examine whether this complex was the elusive site of action of STAR. To accomplish this we used an in vitro transcription/translation system to generate WT STAR and the deletion mutant C258 STAR. C258 STAR lacks the STAR-related lipid transfer domain and is thus devoid of its ability to bind cholesterol (27) or stimulate mitochondrial steroidogenesis (Fig. 7A). We incubated the STAR constructs with mitochondria preloaded with [3H]Photocholesterol, removing an aliquot of the mitochondria reaction after a 5- or 15-min incubation, due to the short 7.5-min half-life of STAR upon incubation with isolated mitochondria (28). After photoactivation, samples were analyzed using BN-PAGE. A significant decrease of [3H]Photocholesterol cross-linking at the 800-kDa complex was observed after incubation with STAR (Fig. 7B); however. with C258 STAR incubation the [3H]Photocholesterol cross-linking levels were not altered, confirming its inability to bind cholesterol for steroidogenesis (Fig. 7B). This result further confirms that the 800-kDa complex functions as the primary site of cholesterol transfer to the IMM and that cholesterol bound at TSPO polymers at the 800-kDa complex can be mobilized by STAR and directly transferred to CYP11A1 at the 800-kDa complex for pregnenolone formation.
Fig. 7.
STAR mobilizes cholesterol bound at the 800-kDa complex. A, Steroid production with isolated mitochondria incubated with in vitro transcription/translation-generated WT or mutant C258 STAR demonstrated that mutant STAR and control samples are unable to stimulate steroid production as compared with WT. B, Isolated mitochondria preloaded with [3H]Photocholesterol were incubated with WT and mutant STAR. Aliquots were removed after 5 and 15 min, cross-linked, and run on BN-PAGE. Decrease in cholesterol binding was observed at 800 kDa in WT STAR but not control or mutant STAR. Results shown are means ± sem (n = 3) or representative blots from at least three experiments. *, P < 0.05.
Discussion
Multiprotein enzyme complexes that carry out sequential reactions on a substrate have been referred to as supramolecular complexes, protein machines, or metabolons (1). Metabolons pass substrates from one active site to another without equilibration and diffusion with the surrounding environment. Proteins in metabolons are held together by noncovalent interactions and are often stabilized by membrane anchoring. Their formation prevents unwanted cross talk with other pathways, including unwanted interference with general mitochondrial function and metabolism, and optimizes substrate concentration and subsequent targeting to an enzyme (29). With the lack of identifiable intermediates in the steroid biosynthesis pathway, the existence of such complexes in steroidogenic mitochondria was proposed by Lieberman and Prasad (30); these complexes were referred to as “hormonads” because of their sensitivity to hormones. Our study provides the first evidence of this complex, in which an 800-kDa mitochondrial protein complex containing the OMM components of the transduceosome TSPO, VDAC, the matrix protein ATAD3A, and the IMM protein CYP11A1 that is required for steroidogenesis allows this complex to be capable of binding and segregating cholesterol from structural OMM lipids and has the ability to synthesize steroids. Therefore, upon hormonal stimulation, this multiprotein mitochondrial complex could function in the pooling of cholesterol at the OMM, an activity that would then allow STAR to mobilize the cholesterol for transfer into the adjacent IMM and its subsequent conversion to pregnenolone.
The initial awareness of an intracellular metabolon for steroidogenesis was demonstrated with the identification of a multiprotein complex associated with the OMM, the transduceosome (3). By initially functioning in propagating the cAMP signaling and initiating cholesterol transfer to the mitochondria, the transduceosome functioned as a sieve at the OMM by separating and restricting the cholesterol for steroidogenic activity. This regulated, controlled flow of cholesterol into the mitochondria is accomplished by anchoring of the cytosolic components of the transduceosome; coenzyme A binding domain containing 3, PKA-RIα, and the hormone-induced mitochondria targeted STAR to the OMM proteins TSPO and VDAC. Although the initial components of the proposed hormonad allow cholesterol transfer to the OMM, the transduceosome does not identify the key mechanism of this metabolon of cholesterol transfer to the IMM and CYP11A1. Therefore, to further understand this process and identify the proposed hormonad regulating the rate-limiting step in steroid production, we used BN-PAGE, which allows us to observe the architecture of the mitochondrial protein complexes that play a role in this process. [3H]Photocholesterol labeling allowed us to identify 66- and 800-kDa protein cholesterol-binding complexes. Interestingly, upon hormonal stimulation, these complexes do not change or are newly identified cholesterol cross-linking complexes, suggesting that cholesterol enters the mitochondria through an established mechanism used both for normal mitochondrial import of cholesterol and for hormone-induced steroidogenesis.
Through immunoblot analysis it was established that TSPO, VDAC, and CYP11A1 are located at the 800-kDa complex, although the lack of identification of ANT that would assist with cholesterol transport to the IMM at the 800-kDa complex led us to wonder about its role in steroidogenesis. We confirmed the ability of the 800-kDa complex to produce steroids utilizing cholesterol-resorufin. Interestingly, both the cholesterol-binding complexes, 66- and the 800-kDa in control cells, were shown to produce steroids. Because CYP11A1 has been demonstrated to be constitutively active in the IMM but lacking in substrate cholesterol (19), this activity was expected. The ability of the 800-kDa mitochondrial complex to form steroids was dramatically induced by the treatment of the cells with hCG. This could be due either to the modest increase in CYP11A1, as identified via BN-PAGE and MS, or more likely to the increased efficiency of cholesterol transfer to CYP11A1 and therefore steroidogenesis. The reasons for the latter are unknown and may involve either hormone-dependent events such as an increase in free fatty acids and reorganization of cardiolipin in the IMM (32, 33) or altered regulation of electron transfer to CYP11A1 affecting its enzymatic activity (15, 34).
The detection of the 800-kDa complex led us to discover the presence of AAA+ ATPase ATAD3A (36), a protein recently shown to play a role in adrenal steroid synthesis (23). The N terminus of ATAD3A is located in the OMM with the C terminus located in the matrix, effectively bridging the two membranes together to form a contact site (23). Knockdown of this protein in our cell model significantly decreased steroid production, possibly due to the fact that cholesterol is not able to bypass the hydrophilic inner membrane space and insert into the IMM. There are several isoforms of ATAD3A, with the long isoform of ATAD3A containing an additional 50 amino acids at the N terminus, which are proposed to form an α-helix that are proposed to then insert either back into the OMM or even insert into other organelles, such as the endoplasmic reticulum (ER) (36). Mitochondria-ER contact sites, termed “mitochondria associated membranes”, contain as much as 5–20% of the OMM (37). These structures play an import role in the transfer of lipids, such as cholesterol and specific phospholipids, along with calcium into the mitochondria (38, 39). It is likely that the long isoform of ATAD3A could also provide a link between CYP11A1 and the site of cholesterol biosynthesis in the ER. Such a link could ensure that cholesterol, a hydrophobic molecule, would pass between two hydrophilic spaces formed by membrane contact sites, thus continuing the metabolon into the ER.
VDAC1, in addition to its role in the mitochondrial contact site formation, is also proposed to play a role in the mitochondria-ER contact site formation, interacting with the inositol 1,4,5-triphosphate receptor, which functions in mitochondrial Ca2+ transfer. Increasing the calcium concentration in the matrix regulates several enzymes of the citric acid cycle, specifically isocitrate dehydrogenase, that we found in both the 66- and 800-kDa mitochondrial complexes (Table 1), providing the NADPH used by ferredoxin reductase for the electron transport chain for CYP11A1 activity (31). This increased activity further augments ATP production, necessary for ATAD3A function and overall cellular health. Considering that despite the substantial knocking down of VDAC1 in our studies steroid production was not completely abolished, it is likely that the ER might be a source of cholesterol but not the sole source. Interestingly, knocking down the two other isoforms of VDAC, VDAC2 and VDAC3, did not decrease steroid production (data not shown), suggesting a specific role for VDAC1.
Previous data suggested that TSPO, VDAC, and ANT interact to form the OMM/IMM mitochondrial contact site (22). Although VDAC and ANT have been found in mitochondrial contact sites (8) and TSPO has been shown to be concentrated in mitochondrial contract sites in steroidogenic cells (21), the presence and role of the IMM protein ANT remained elusive. The MS and FRET data presented herein, together with our previous cross-linking studies (3), clearly indicate that ANT is not part of the protein complex mediating cholesterol transport and steroid formation. These results were further corroborated by the ANT knockdown experiments in MA-10 cells, in which the hormone-induced steroid formation was not affected by the absence of ANT. This observation, however, should not lead to the conclusion that the formation of VDAC- and ANT-containing contact sites is not favorable for other energetic demands. As with ANT, knocking down OPA1, VDAC1, and ATAD3A did alter mitochondrial cristae morphology as observed through EM, but it did not affect steroid production in response to the membrane-permeable 22R-hydroxycholesterol, suggesting the presence of an intact CYP11A1 activity. VDAC1 and ATAD3A knockdowns also drastically inhibited the hormone- and cAMP-induced steroid synthesis, indicating an effect on cholesterol availability to CYP11A1. In contrast, despite the effect of knocking down the mitochondrial-shaping protein OPA1 on MA-10 mitochondrial cristae morphology, the cells maintained their full response to hormone- and cAMP-stimulated progesterone formation, suggesting that OPA1 is not critical for hormone-induced steroidogenesis.
Our results demonstrated that VDAC1 and ATAD3A are both required for steroid production. Because both proteins were present at the 800-kDa complex in MA-10 control mitochondria, the hormone-induced initiation of cholesterol transfer to the CYP11A1 in this complex is likely to be accomplished by STAR. The STAR-induced decrease of the bound cholesterol present at the 800-kDa protein complex, further resulting in an increase in steroid production, demonstrated that this was the case. It is unknown at present whether this is the same mitochondrial contact site used to import STAR into the mitochondria. However, the finding that the protease demonstrated to be responsible for STAR's degradation, LONP1 (35), is present at the 800-kDa complex (Table 1) suggests that STAR associated with this particular complex when imported into mitochondria.
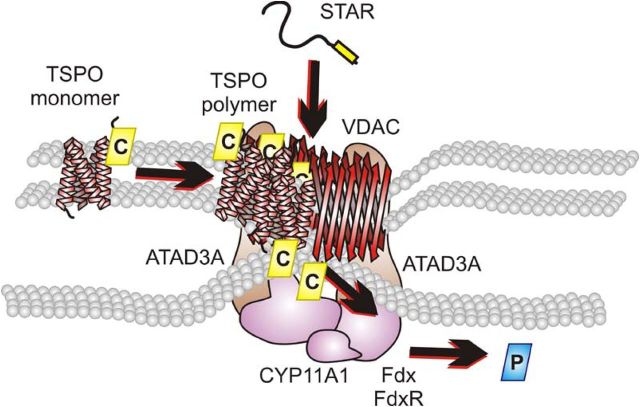
In conclusion, several cytosolic and mitochondrial proteins are required to facilitate the transport of cholesterol to the IMM (Fig. 8). The OMM protein TSPO is necessary to bind, pool, and segregate the cholesterol. The mitochondrial-targeted STAR is required to initiate transport into the IMM. The OMM protein VDAC is present to anchor TSPO and STAR. Finally, ATAD3A forms the bridge of the OMM and IMM, thus allowing access of cholesterol to CYP11A1. Because each of these proteins has been shown to be required for efficient steroid production, our results herein further identify the mechanism through which they interact. We documented a novel, hormone-dependent mitochondrial metabolon (hormonad) that facilitates import, segregation, and transfer of the steroidogenic pool of cholesterol to the IMM protein CYP11A1, the rate-determining steps in steroid formation.
Fig. 8.
Schematic representation of the hormone-responsive steroidogenic metabolon (hormonad). The TSPO monomer was demonstrated to bind cholesterol at 66 kDa. Upon hormonal stimulation, TSPO undergoes polymerization and associates with the 800-kDa complex where it is associated with VDAC. The pooling of cholesterol bound to TSPO is acted upon by STAR where it is able to be translocated to the IMM through the formation of a contact site by VDAC and the mitochondrial matrix protein ATAD3A present in the 800-kDa complex. Cholesterol is then metabolized by CYP11A1, also present in the 800-kDa complex, supported by the electron transfer proteins, ferredoxin and ferredoxin reductase, to form pregnenolone. The proteins function together to form a hormone-dependent mitochondrial metabolon, the hormonad.
Acknowledgments
We thank Dr. Mario Ascoli (University of Iowa) for the MA-10 cells; the National Hormone and Pituitary Program (NIH) for supplying the hCG; Stegram Pharmaceuticals (Sussex, UK) for supplying the trilostane; the staff of the Facility for Electron Microscopy Research at McGill University for their assistance with the electron microscopy studies; and Dr. John Bergeron (McGill University) for his critical reading of the manuscript.
This work was supported by a grant from the Canadian Institutes of Health Research (MOP102647) and a Canada Research Chair in Biochemical Pharmacology (to V.P.). M.R. was supported in part by a postdoctoral fellowship from The Research Institute of McGill University Health Center. A.M. was supported by postdoctoral fellowships from the Canadian Institutes of Health Research (TGF36110) and Le Fonds de la recherche du Québec-santé. The Research Institute of McGill University Health Center was supported by a Center grant from Le Fonds de la recherche du Québec-santé.
Disclosure Summary: The authors have nothing to disclose.
Footnotes
- ANT
- Adenine nucleotide transporter
- ATAD3A
- ATPase family AAA domain-containing protein 3A
- BN-PAGE
- blue native polyacrylamide gel electrophoresis
- CoxIV
- cytochrome c oxidase subunit IV
- 2D
- two-dimensional
- dbcAMP
- dibutyryl-cAMP
- cholesterol-resorufin
- 22-phenoxazonoxy-5-cholene-3-β-ol
- CYP11A1
- cytochrome P450 side-chain cleavage enzyme
- EM
- electron microscopy
- ER
- endoplasmic reticulum
- FRET
- Forster resonance energy transfer
- hCG
- human chorionic gonadotropin
- IMM
- inner mitochondrial membrane
- MS
- mass spectrometry
- MTT
- 3-(4,5-dimethylthiazol-2-yl)-2,5-diphenyltetrazolium bromide
- OMM
- outer mitochondrial membrane
- OP1
- optic atrophy type 1
- [3H]Photocholesterol
- 7-azi-5α-[3,5,6-3H] cholestan-3β-ol
- PKA
- cAMP-dependent protein kinase A
- PKA-RIα
- cAMP-dependent protein kinase-regulatory subunit Iα
- STAR
- steroidogenic acute regulatory protein
- TSPO
- translocator protein (18 kDa)
- VDAC
- voltage-dependent anion channel
- WT
- wild-type
References
- 1. Srere PA. 1987. Complexes of sequential metabolic enzymes. Annu Rev Biochem 56:89–124 [DOI] [PubMed] [Google Scholar]
- 2. Rone MB , Fan J , Papadopoulos V. 2009. Cholesterol transport in steroid biosynthesis: role of protein-protein interactions and implications in disease states. Biochim Biophys Acta 1791:646–658 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Liu J , Rone MB , Papadopoulos V. 2006. Protein-protein interactions mediate mitochondrial cholesterol transport and steroid biosynthesis. J Biol Chem 281:38879–38893 [DOI] [PubMed] [Google Scholar]
- 4. Hauet T , Yao ZX , Bose HS , Wall CT , Han Z , Li W , Hales DB , Miller WL , Culty M , Papadopoulos V. 2005. Peripheral-type benzodiazepine receptor-mediated action of steroidogenic acute regulatory protein on cholesterol entry into Leydig cell mitochondria. Mol Endocrinol 19:540–554 [DOI] [PubMed] [Google Scholar]
- 5. Bose HS , Lingappa VR , Miller WL. 2002. Rapid regulation of steroidogenesis by mitochondrial protein import. Nature 417:87–91 [DOI] [PubMed] [Google Scholar]
- 6. Delavoie F , Li H , Hardwick M , Robert JC , Giatzakis C , Péranzi G , Yao ZX , Maccario J , Lacapère JJ , Papadopoulos V. 2003. In vivo and in vitro peripheral-type benzodiazepine receptor polymerization: functional significance in drug ligand and cholesterol binding. Biochemistry 42:4506–4519 [DOI] [PubMed] [Google Scholar]
- 7. Thomson M. 2003. Does cholesterol use the mitochondrial contact site as a conduit to the steroidogenic pathway? Bioessays 25:252–258 [DOI] [PubMed] [Google Scholar]
- 8. Crompton M , Barksby E , Johnson N , Capano M. 2002. Mitochondrial intermembrane junctional complexes and their involvement in cell death. Biochimie 84:143–152 [DOI] [PubMed] [Google Scholar]
- 9. Li H , Yao Z , Degenhardt B , Teper G , Papadopoulos V. 2001. Cholesterol binding at the cholesterol recognition/ interaction amino acid consensus (CRAC) of the peripheral-type benzodiazepine receptor and inhibition of steroidogenesis by an HIV TAT-CRAC peptide. Proc Natl Acad Sci USA 98:1267–1272 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Boujrad N , Gaillard JL , Garnier M , Papadopoulos V. 1994. Acute action of choriogonadotropin on Leydig tumor cells: induction of a higher affinity benzodiazepine-binding site related to steroid biosynthesis. Endocrinology 135:1576–1583 [DOI] [PubMed] [Google Scholar]
- 11. Rone MB , Liu J , Blonder J , Ye X , Veenstra TD , Young JC , Papadopoulos V. 2009. Targeting and insertion of the cholesterol-binding translocator protein into the outer mitochondrial membrane. Biochemistry 48:6909–6920 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Marrone BL , Simpson DJ , Yoshida TM , Unkefer CJ , Whaley TW , Buican TN. 1991. Single cell endocrinology: analysis of P-450scc activity by fluorescence detection methods. Endocrinology 128:2654–2656 [DOI] [PubMed] [Google Scholar]
- 13. Mintzer EA , Waarts BL , Wilschut J , Bittman R. 2002. Behavior of a photoactivatable analog of cholesterol, 6-photocholesterol, in model membranes. FEBS Lett 510:181–184 [DOI] [PubMed] [Google Scholar]
- 14. Simpson RJ. 2003. Proteins and proteomics: a laboratory manual. Cold Spring Harbor, NY: Cold Spring Harbor Laboratory Press [Google Scholar]
- 15. Praporski S , Ng SM , Nguyen AD , Corbin CJ , Mechler A , Zheng J , Conley AJ , Martin LL. 2009. Organization of cytochrome P450 enzymes involved in sex steroid synthesis: Protein-protein interactions in lipid membranes. J Biol Chem 284:33224–33232 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Wilson JE. 2003. Isozymes of mammalian hexokinase: structure, subcellular localization and metabolic function. J Exp Biol 206:2049–2057 [DOI] [PubMed] [Google Scholar]
- 17. Acin-Pérez R , Fernández-Silva P , Peleato ML , Pérez-Martos A , Enriquez JA. 2008. Respiratory active mitochondrial supercomplexes. Mol Cell 32:529–539 [DOI] [PubMed] [Google Scholar]
- 18. Yamada M , Homma R , Ohta Y , Kawato S. 1977. Electron transfer interactions of mitochondrial cytochrome P450 in membranes. J Inorg Biochem 67:115 [Google Scholar]
- 19. Jefcoate C. 2002. High-flux mitochondrial cholesterol trafficking, a specialized function of the adrenal cortex. J Clin Invest 110:881–890 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Tanaka T , Strauss JF. 1982. Stimulation of luteal mitochondrial cholesterol side-chain cleavage by cardiolipin. Endocrinology 110:1592–1598 [DOI] [PubMed] [Google Scholar]
- 21. Culty M , Li H , Boujrad N , Amri H , Vidic B , Bernassau JM , Reversat JL , Papadopoulos V. 1999. In vitro studies on the role of the peripheral-type benzodiazepine receptor in steroidogenesis. J Steroid Biochem Mol Biol 69:123–130 [DOI] [PubMed] [Google Scholar]
- 22. McEnery MW , Snowman AM , Trifiletti RR , Snyder SH. 1992. Isolation of the mitochondrial benzodiazepine receptor: association with the voltage-dependent anion channel and the adenine nucleotide carrier. Proc Natl Acad Sci USA 89:3170–3174 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Gilquin B , Taillebourg E , Cherradi N , Hubstenberger A , Gay O , Merle N , Assard N , Fauvarque MO , Tomohiro S , Kuge O , Baudier J. 2010. The AAA+ ATPase ATAD3A controls mitochondrial dynamics at the interface of the inner and outer membranes. Mol Cell Biol 30:1984–1996 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Griffin EE , Detmer SA , Chan DC. 2006. Molecular mechanism of mitochondrial membrane fusion. Biochim Biophys Acta 1763:482–489 [DOI] [PubMed] [Google Scholar]
- 25. Midzak AS , Chen H , Aon MA , Papadopoulos V , Zirkin BR. 2011. ATP synthesis, mitochondrial function, and steroid biosynthesis in rodent primary and tumor Leydig cells. Biol Reprod 84:976–985 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Yagoda N , von Rechenberg M , Zaganjor E , Bauer AJ , Yang WS , Fridman DJ , Wolpaw AJ , Smukste I , Peltier JM , Boniface JJ , Smith R , Lessnick SL , Sahasrabudhe S , Stockwell BR. 2007. RAS-RAF-MEK-dependent oxidative cell death involving voltage-dependent anion channels. Nature 447:864–868 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Arakane F , Sugawara T , Nishino H , Liu Z , Holt JA , Pain D , Stocco DM , Miller WL , Strauss JF. 1996. Steroidogenic acute regulatory protein (StAR) retains activity in the absence of its mitochondrial import sequence: implications for the mechanism of StAR action. Proc Natl Acad Sci USA 93:13731–13736 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Artemenko IP , Zhao D , Hales DB , Hales KH , Jefcoate CR. 2001. Mitochondrial processing of newly synthesized steroidogenic acute regulatory protein (StAR), but not total StAR, mediates cholesterol transfer to cytochrome P450 side chain cleavage enzyme in adrenal cells. J Biol Chem 276:46583–46596 [DOI] [PubMed] [Google Scholar]
- 29. Møller BL. 2010. Plant science. Dynamic metabolons. Science 330:1328–1329 [DOI] [PubMed] [Google Scholar]
- 30. Lieberman S , Prasad VV. 1990. Heterodox notions on pathways of steroidogenesis. Endocr Rev 11:469–493 [DOI] [PubMed] [Google Scholar]
- 31. Bernardi P. 1999. Mitochondrial transport of cations: channels, exchangers, and permeability transition. Physiol Rev 79:1127–1155 [DOI] [PubMed] [Google Scholar]
- 32. Dhariwal MS , Jefcoate CR. 1989. Cholesterol metabolism by purified cytochrome P-450scc is highly stimulated by octyl glucoside and stearic acid exclusively in large unilamellar phospholipid vesicles. Biochemistry 28:8397–8402 [DOI] [PubMed] [Google Scholar]
- 33. Lambeth JD , Kitchen SE , Farooqui AA , Tuckey R , Kamin H. 1982. Cytochrome P-450scc-substrate interactions. Studies of binding and catalytic activity using hydroxycholesterols. J Biol Chem 257:1876–1884 [PubMed] [Google Scholar]
- 34. Miller WL. 2005. Minireview: regulation of steroidogenesis by electron transfer. Endocrinology 146:2544–2550 [DOI] [PubMed] [Google Scholar]
- 35. Granot Z , Kobiler O , Melamed-Book N , Eimerl S , Bahat A , Lu B , Braun S , Maurizi MR , Suzuki CK , Oppenheim AB , Orly J. 2007. Turnover of mitochondrial steroidogenic acute regulatory (StAR) protein by Lon protease: the unexpected effect of proteasome inhibitors. Mol Endocrinol 21:2164–2177 [DOI] [PubMed] [Google Scholar]
- 36. Hubstenberger A , Labourdette G , Baudier J , Rousseau D. 2008. ATAD 3A and ATAD 3B are distal 1p-located genes differentially expressed in human glioma cell lines and present in vitro anti-oncogenic and chemoresistant properties. Exp Cell Res 314:2870–2883 [DOI] [PubMed] [Google Scholar]
- 37. Rizzuto R , Pinton P , Carrington W , Fay FS , Fogarty KE , Lifshitz LM , Tuft RA , Pozzan T. 1998. Close contacts with the endoplasmic reticulum as determinants of mitochondrial Ca2+ responses. Science 280:1763–1766 [DOI] [PubMed] [Google Scholar]
- 38. Hayashi T , Fujimoto M. 2010. Detergent-resistant microdomains determine the localization of σ-1 receptors to the endoplasmic reticulum-mitochondria junction. Mol Pharmacol 77:517–528 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39. de Brito OM , Scorrano L. 2010. An intimate liaison: spatial organization of the endoplasmic reticulum-mitochondria relationship. EMBO J 29:2715–2723 [DOI] [PMC free article] [PubMed] [Google Scholar]