Abstract
Central and peripheral mechanisms modulate food intake and energy balance in mammals and the precise role of the type 1 cannabinoid receptor (CB1) in these processes is still being explored. Using the zebrafish, Danio rerio, we show that rimonabant, a CB1-specific antagonist with an EC50 of 5.15 × 10−8 m, decreases embryonic yolk sac reserve use. We reveal a developmental overlap between CART genes and CB1 expression in the hypothalamus and medulla oblongata, two brain structures that play crucial roles in appetite regulation in mammals. We show that morpholino knockdown of CB1 or fasting decreases cocaine- and amphetamine-related transcript (CART)-3 expression. Strikingly, this down-regulation occurs only in regions coexpressing CB1 and CART3, reinforcing the link between CB1, CART, and appetite regulation. We show that rimonabant treatment impairs the fasting-induced down-regulation of CART expression in specific brain regions, whereas vehicle alone-treated embryos do not display this rescue of CART expression. Our data reveal that CB1 lies upstream of CART and signals the appetite through the down-regulation of CART expression. Thus, our results establish the zebrafish as a promising system to study appetite regulation.
A combination of central and peripheral mechanisms modulate food intake and energy balance in mammals and the constant progression of metabolic disorders creates the need for a better understanding of these mechanisms (1–5). Appetite and food intake are regulated within the central nervous system by a complex circuitry involving several specific regions of the hypothalamus as well as the amygdala, the limbic forebrain, and the brainstem (5, 6). In these structures, many neuropeptides as well as different classes of molecules (biogenic amines, fatty acid derivatives, etc.) regulate appetite, either positively (orexigenic) or negatively (anorexigenic) (3, 4, 6). The receptors for these various classes of molecules are promising drug targets because regulation of their activities could reduce food intake and thus shift the tightly regulated balance that adjusts energy homeostasis over time.
Evidence has accumulated recently to suggest that endocannabinoids act as orexigenic signals via the type 1 cannabinoid receptor (CB1), a G protein-coupled receptor expressed in the central nervous system (3, 7). Blocking the CB1 receptor inhibits food intake, resulting in weight loss in rodents (3). Endocannabinoids such as anandamide and 2-arachidonoyl glycerol stimulate appetite and food intake in both rodents and humans, and CB1 receptor knockout mice are resistant to diet-induced obesity (8–11). In accordance with this notion, CB1 antagonists such as rimonabant (SR141716) have been studied as drugs to reduce human obesity (12, 13).
Nevertheless, the mechanisms by which endocannabinoids or rimonabant regulate food intake via CB1 are still unclear. It would be desirable to better understand these mechanisms to reduce side effects associated with the use of CB1 antagonists (14). The unique synaptic organization of the endocannabinoid/CB1 system and the fact that endocannabinoids are generated in postsynaptic neurons and retrogradely diffuse to presynaptic axon terminals to stimulate the CB1 receptor have led to the suggestion that the CB1 receptors can act via anorexigenic peptide mediators (3, 15). Cocaine- and amphetamine-related transcript (CART) was proposed to be such a factor, and it was found that administration of rimonabant did not affect feeding behavior in CART-deficient mice (15, 16). Reduced CART expression was observed in mice deficient for fatty acid amide hydrolase (FAAH; the enzyme responsible for the degradation of anandamine) and treatment with rimonabant showed an increase in CART levels in these mice (15–17). This suggests that CART inhibition might play a crucial role in the hypothalamic mediation of orexigenic endocannabinoid action. Nevertheless, it has been shown that CART expression is reduced in CB1 knockout mice, an observation that does not fit with the above model (17). In addition, several conflicting reports have been published regarding the effects (either orexigenic or anorexigenic) of CART on appetite regulation (18–21). Of note, CART-deficient mice did not show an increase in body weight until after the 14th wk of a high-fat diet, suggesting that a CART role in obesity is context dependent (22). Thus, the precise relationship linking the cannabinoid receptor system and CART may be more complex than anticipated and requires further evaluation, as do their roles in regulating food intake.
In this paper, we used the zebrafish, an already relevant and established model to study obesity (23). We used all the advantages that the zebrafish offers to study the regulation of appetite genes during embryonic development and larval stages to characterize the cannabinoid and CART signaling pathways in vivo. Two forms of the CART gene were previously reported in goldfish, another excellent teleost model to study metabolism and appetite regulation (24). In this study we found that rimonabant decreased yolk reserve utilization in a CB1-dependent manner and reduced fat levels in developing embryos. We characterized four CART genes whose expression overlapped with that of CB1 in two major regions of the brain known to be implicated in appetite regulation in mammals: the hypothalamus and the medulla oblongata. In particular, we noted early coexpression of CART3 and CB1, and we found that CART3 expression was decreased when we knocked down CB1 expression through morpholino injection. We observed that fasting decreased the expression of CART, including CART3. This decrease occurred in the specific areas of the central nervous system that showed coexpression between CB1 and CART3, whereas rimonabant treatment specifically suppressed this CART down-regulation. Taken together, our results establish that CB1 lies upstream of CART and during fasting signals to the CART pathway the need for food. Our results also highlight zebrafish as a promising system to study appetite regulation and further emphasize the context dependency of CB1/CART regulation.
Results
Rimonabant decreases yolk sac use in a CB1-dependent manner
To study the activity of rimonabant in zebrafish, we first characterized the effect of rimonabant on zebrafish CB1. We found that the agonist ligand HU-210 stimulated cAMP production in mammalian cells transfected by a zebrafish CB1 expression vector and that this effect was inhibited when increasing doses of rimonabant were added (apparent affinity ∼10 nm; Supplemental Fig. 1A, published on The Endocrine Society's Journals Online web site at http://mend.endojournals.org). This indicates that, as in mammals, rimonabant is a CB1 antagonist. In addition, using radiolabeled rimonabant, we observed that this compound accumulated in embryos as well as in larvae, suggesting that, when added to the water, it effectively enters the fish (Supplemental Fig. 1A).
Then we treated zebrafish embryos at 10 h post fertilization (hpf), just after gastrulation to avoid possible early morphogenetic effects, with either 10 or 5 μm of rimonabant. Under careful examination, we noticed that the yolk sac of the rimonabant-treated fish was consistently bigger, at an equivalent age, than the one of mock-treated fish (Fig. 1, A and B). Morphometric measurements of yolk sac at 48 hpf (not shown) or at 3 d post fertilization (dpf; Fig. 1C) show that rimonabant treated embryos consistently exhibited an increase of ca. 27% ± 2.8 in their yolk sac volume excluding the yolk sac extension, whereas other measures, such as head width, were not modified (Fig. 1A). This may indicate that rimonabant induces a malabsorption syndrome during development (23). To substantiate this finding, we performed Oil Red O staining and showed that rimonabant decreased fat accumulation in the embryos. Conversely, treatment with the CB1 agonist WIN 55,212–2 (that did not induce smaller yolk sac in zebrafish embryos) induced more fat accumulation (Fig. 1D). These data thus suggest that rimonabant regulates yolk sac reserve use, an effect that, strikingly, can be compared with a decrease in appetite at the embryonic level (see Discussion).
Fig. 1.
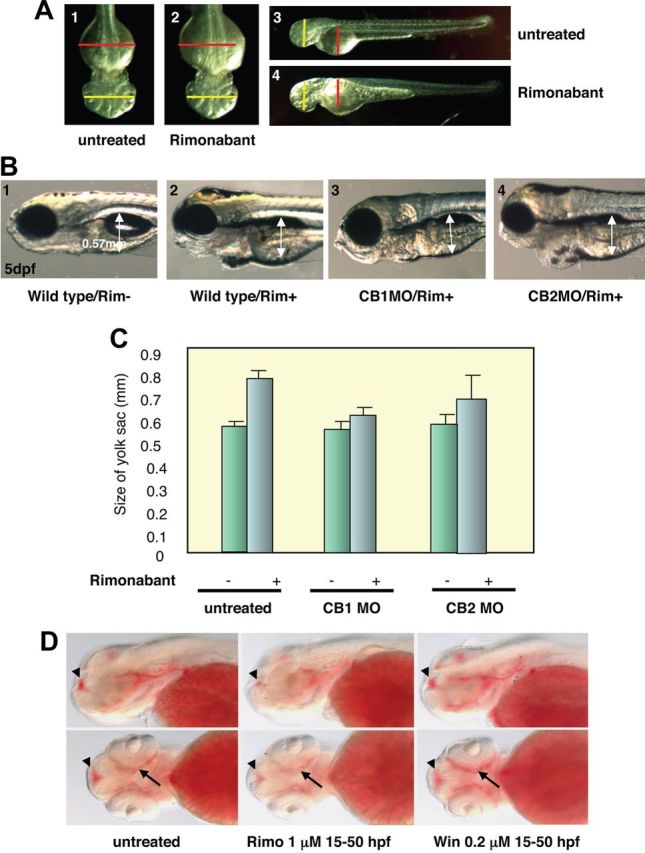
Rimonabant exposure decreases yolk size use in a CB1-dependant manner. A, Embryos treated with 5 μm of rimonabant from 10 to 48 hpf display a bigger yolk sac (2, 4) compared with untreated wild type (1, 3). The red lines represent the maximum length of the yolk sac in wild types, whereas the yellow lines represent the maximum length of the head in wild types. Notes that rimonabant-treated embryos have a bigger yolk sac but have the same head size compared with wild type. B, 1, Wild-type embryos at 5 dpf with an average yolk sac size of 0.57 mm (2). Rimonabant-treated embryos, 1 μm from 10 hpf onward display a bigger yolk sac (3). Injection of CB1 morpholino abolishes the effect of rimonabant exposure, whereas injection of CB2 morpholino (4) does not. See Supplemental Fig. 2 for the efficacy of the CB1 morpholino. C, Rimonabant exposure from 5 to 72 hpf led to ca. 20% increase of the yolk sac size. This effect is blocked by CB1 morpholino injection but not by CB2 morpholino injection. D, Fat accumulation is decreased in rimonabant-treated embryos (middle panel) compared with the wild type (left panel). Note the decrease of Oil Red O staining in the telencephalon (arrowhead) and around the eye (arrow). Exposure to the CB1 agonist WIN 55,212-2 increases fat accumulation (right panel).
We next determined the developmental expression pattern of the zebrafish CB1 receptor by in situ hybridization (Supplemental Fig. 1B). We noticed that, in addition to a complex central nervous system expression, CB1 was transiently expressed at the pharyngeal level (with a peak at 36 hpf and a decreased level at 48 hpf onward), a location that can be hypothesized to play a role in controlling yolk sac reserve use (see also Ref. 24). We therefore tested the effect of knocking down CB1 expression on rimonabant action. Interestingly, in presence of rimonabant, CB1 morphants exhibited the same yolk sac size as mock-treated fish (Fig. 1, B and C), whereas embryos treated with a random morpholino did exhibit a bigger yolk sac when treated with rimonabant. Of note, this effect has been observed with two different CB1 morpholinos targeting different regions of the CB1 mRNA (data not shown), and only a marginal effect was observed in presence of morpholinos targeting type 2 cannabinoid receptor (CB2) (Fig. 1C). The observed effect on yolk sac size was statistically relevant under rimonabant treatments (Wilcoxon test; P = 0.00015). However, when we exposed CB1 morphants to rimonabant, the effect on the yolk sac size was attenuated (Wilcoxon test; P = 0.0073), whereas effects due to the CB2 receptor remained marginal (P = 0.062). These results suggest that the rimonabant effects are not linked to a toxicological action of the compound, which would have slowed down the development but are specific CB1-dependent effects.
CB1 and CART genes are coexpressed in the hypothalamus and the medulla oblongata
Because CART has been proposed as a possible effector of CB1 action, we cloned the zebrafish CART gene. To our surprise, bioinformatic mining of the zebrafish genome, confirmed by experimental isolation of the relevant cDNA, demonstrated that no fewer than four CART genes (CART1, CART2a, CART2b, and CART3) are present in zebrafish (Supplemental Fig. 3). This is consistent with recent reports highlighting the multiplicity of CART genes in other fish, such as goldfish or pufferfish (25, 26). These four genes can be phylogenetically mapped into three groups, one of which, CART1, is clearly orthologous to the mammalian CART gene (Supplemental Fig. 4A). The other groups, CART2a, CART2b, and CART3, have no orthologs in mammals. This multiplicity is consistent with an early duplication of the gene [probably at the base of vertebrates during the so-called two rounds of genome duplications (27)] followed by a further duplication of CART2 in several fish including zebrafish (28). This scenario would imply the loss of CART2 and CART3 genes in mammals. Several other cases of gene loss are known to have occurred at the base of the mammalian tree (29, 30).
We then compared the expression pattern of the four CART genes with that of CB1. The expression patterns of the zebrafish CART genes are presented in Supplemental Fig. 4. Their most salient features are shown in Fig. 2 in which the neuroanatomical structures are indicated (from Ref. 31). CART1 was detected at 72 hpf in the diencephalon with no earlier expression being observed. CART2a was first observed at 30 and 36 hpf in the diencephalon and later detected at 48 hpf in the dorsal thalamus and ventral part of posterior tuberculum. A dynamic expression pattern was also observed in the medulla oblongata region at 48 hpf (Fig. 2D) and 72 hpf (Fig. 2G). CART2b was first detected at 36 hpf in the hindbrain. At 48 hpf a new stronger expression was detected in the medulla oblongata (Fig. 2E). Later, at 72 hpf, the expression diversified and reached the pretectal area: the lens, the pharynx, and the medulla oblongata (Fig. 2G). CART3 exhibited the earliest and highest expression levels and was found specifically at 24 and 36 hpf in precise regions in the hypothalamus and hindbrain. At 48 hpf, the same expression was reiterated (see Fig. 2, B and F), but two new expression domains were visible within the olfactory bulb and midbrain tegmentum. At 72 hpf the expression further diversified in a more posterior area in the medulla oblongata.
Fig. 2.
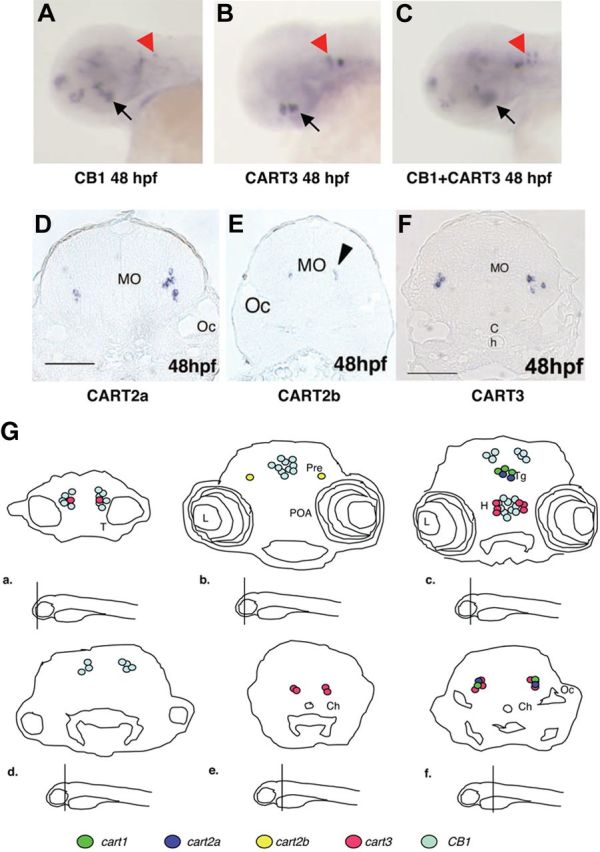
CB1 and CART genes are coexpressed in the hypothalamus and the medulla oblongata during embryogenesis. A, At 48 hpf, CB1 transcripts are detected in the telencephalon, the tegmentum, the preoptic area, the hypothalamus (black arrow), and the medulla oblongata (anterior hindbrain, red arrowhead). B, At 48 hpf, CART3 transcripts are detected in the hypothalamus (black arrow) and in the medulla oblongata (red arrowhead). C, Double in situ hybridization of CB1 and CART3 showing their coexpression in the hypothalamus (black arrow) and medulla oblongata (red arrowhead). D–F, Sagittal sections of 48 hpf embryos showing expression of CART2a, CART2b, and CART3, respectively, in the medulla oblongata (see black arrowhead in E). Note that these genes are expressed in a similar domain as CB1 in the medulla oblongata (red arrowhead in A). G, Schematic representation of different sagittal section of 72 hpf embryos showing the expression of CB1, CART1, CART2a, CART2b, and CART3. The level of the section is shown under each panel. L, Lens; H, hypothalamus; Oc, otic capsule, Ch, optic chiasma; POA, preoptic area; Pre, pretectum; T, tectum; Tg, tegmentum.
Taken together, these expression patterns show two areas of interest in which coexpression occurs: 1) in the hypothalamus at 48 hpf, which exhibited extensive overlap between CART3 and CB1 (see Fig. 2C), and no other CART were expressed and 2) in the medulla oblongata in which, starting at 48 hpf (Fig. 2, D–F) and then at 72 hpf (Fig. 2G), extensive overlap between CART2a, CART2b, CART3, and CB1 were observed (see Refs. 32 and 33 for an extensive description of CB1 expression).
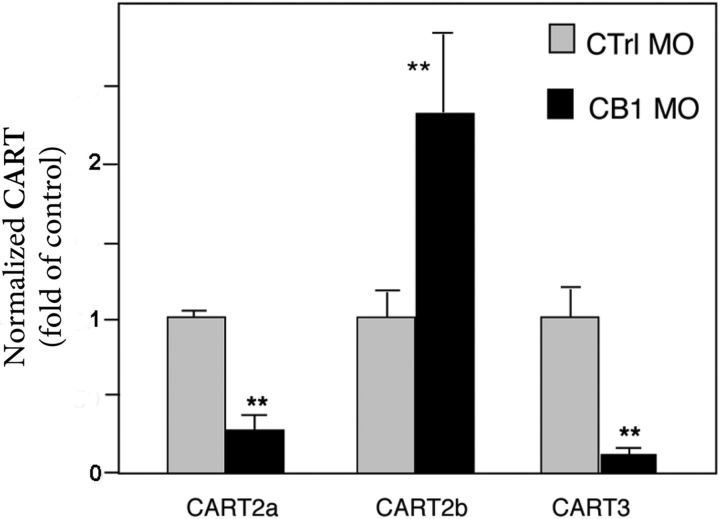
The expression of CB1 and CART genes in the same area suggests that CB1 may play a role in controlling CART expression. To test this hypothesis, we injected CB1 morpholinos and monitored CART expression. By studying CART mRNA levels at 48 hpf by quantitative PCR (Q-PCR) experiments in CB1 morphants, we found a reproducible effect of the knockdown of CB1 on CART2a, CART2b, and CART3 expression (Fig. 3). CART1, which is not expressed at 48 hpf, was not monitored. These data revealed that CART2a and CART3 expression was strongly reduced when CB1 activity was diminished, indicating that their expression is dependent on the integrity of the CB1 pathway. On the other hand, we observed a strong up-regulation of CART2b in CB1 morphants (Fig. 3). These results showed that the expression of all CART genes detected at 48 hpf was dependent on CB1, although the effect was the opposite on CART2b compared with the two others, CART2a and CART3.
Fig. 3.
CB1 regulates CART expression. Quantitative RT-PCR of CART2a, CART2b, and CART3 at 48 hpf in CB1 morphants showing that CART2b and CART3 expression is strongly decreased, whereas CART2a expression is up-regulated. Values for CART mRNA were subtracted from the α-actin mRNA and converted from log linear to linear term. The data represent mean ± sem values. CART1 not being expressed at 48 hpf was not monitored. **, P < 0.01.
Fasting regulates CART3 expression in specific brain areas
Several recent reports link CART expression and appetite regulation in mammals and fish (26, 34). In goldfish, intracerebroventricular administration of human CART decreases food intake. In both the mammal and the fish brain, CART mRNA levels decrease during fasting (16, 26). We thus tested the effect of fasting on CART expression in the brain of adult zebrafish. As observed previously in goldfish (26), Q-PCR analysis of CART mRNA levels in the brain showed a strong decrease after 3 d of fasting (Fig. 4A). This effect was more pronounced for CART3 and CART1 than for the two CART2 genes.
Fig. 4.
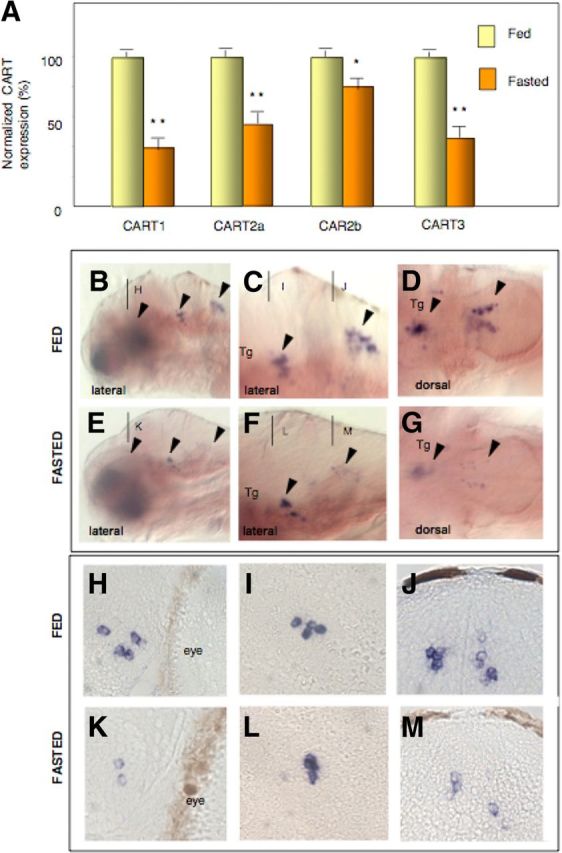
Fasting decreases CART genes expression. A, Effects of 3 d fasting on CART expression in adult zebrafish brain showing a decrease of all CART genes. The data represent mean ± sem values from three zebrafish brains. Fasting decreases CART3 expression specifically in the hypothalamus (K) and the medulla oblongata (E–G, M), whereas expression in the tegmentum remains unaffected by fasting (F, G, and L). B, C, E, and F, Lateral views; D, and G, dorsal views; H–M, transverse sections. *, P < 0.05; **, P < 0.01.
To relate this effect to the expression patterns of CART genes, we performed the same fasting experiment on young larvae, which are still permeable enough to allow whole-mount in situ hybridization. In our experimental conditions, larvae started to feed normally after complete yolk sac resorbtion at 6 dpf. We let these larvae feed for 3 d. Then we either fed or fasted them over 4 d and monitored gene expression in 12 dpf larvae (see experimental scheme on Supplemental Fig. 5). We decided to focus on Cart3 because Cart3 and CB1 showed extensive overlap in their expression patterns. CART3 transcripts were also well detected during larval stages and CB1 morphants exhibited a decrease in CART3 expression. As shown in Fig. 4, B–D, in 12 dpf larvae, CART3 was expressed in the hypothalamus, tegmentum, and medulla oblongata (see Fig. 4, B–D, arrowheads in the general and close-up views). Because the hypothalamic labeling is not visible in Fig. 4B, we performed sections that revealed clearly the three main areas: the hypothalamus (Fig. 4H), tegmentum (Fig. 4I), and medulla oblongata (Fig. 4J). In fasted larvae, we observed a specific decrease of CART3 labeling in the hypothalamus (visible only in sections; compare Fig. 4K with Fig. 4H) as well as in the medulla oblongata (see Fig. 4, F and G, compared with Fig. 4, C and D, and the 4sections in Fig. 4M compared with Fig. 4J). In contrast, no effect was seen in the tegmentum area (compare Fig. 4L with Fig. 4I). These data suggest that CART3 is down-regulated by fasting only in regions in which coexpression with CB1 is observed. This supports the notion that CART3 expression depends in part on CB1 signaling.
The effect of fasting on CART3 expression is CB1 dependent
In our experimental setting, in which an effect is detected at 12 dpf, morpholino inhibition cannot be used to directly test whether the CART3 decrease we observed is linked to CB1 signaling. To test the possible link between CB1 and the starvation-induced inhibition of CART3 expression, we therefore used rimonabant as a pharmacological agent that specifically inhibits the CB1 pathway.
We used either fasted or fed fish that we did or did not treat with rimonabant at 1 μm (see Supplemental Fig. 5). We first performed the experiments in adults (Fig. 5, A–D) and showed by Q-PCR that fasting decreased CART3 expression. Interestingly, rimonabant treatment in fasted fish specifically inhibited this decrease in CART3 expression (Fig. 5D). These data suggest that the fasting-dependent inhibition of CART3 expression is CB1 dependent. This effect was even more pronounced with CART2b, which displayed an increase of expression after rimonabant exposure. However, CART1 and CART2a were not affected by rimonabant treatment (Fig. 5, A–C).
Fig. 5.
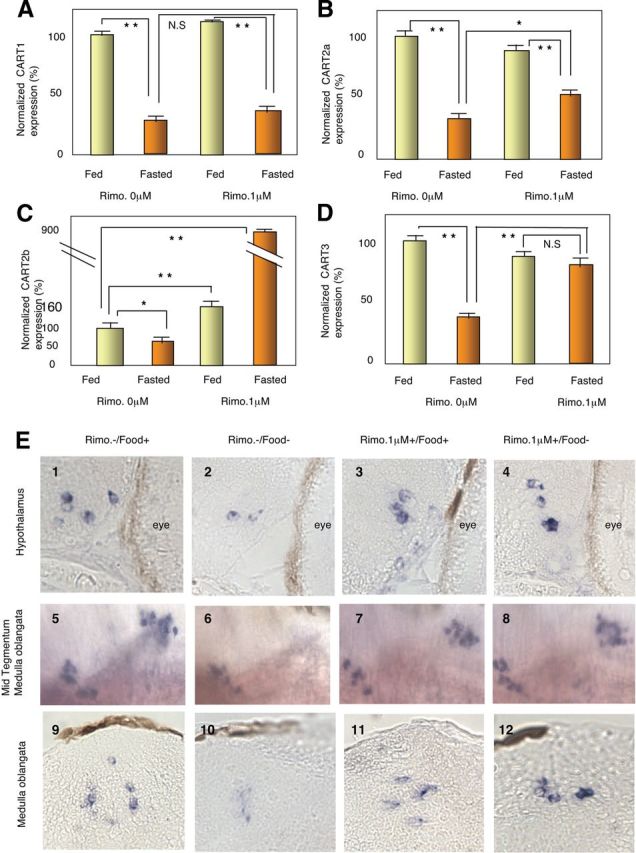
Rimonabant exposure restores CART3 expression in fasted larvae. A–D, Effects of rimonabant exposure conjugated with 4 d fasting on CART expression in 12 dpf larvae monitored by Q-PCR. Note that CART2b (C) and CART3 (D) expression is restored in fasted larvae exposed to rimonabant. The data represent mean ± sem values from six zebrafish larvae. E, Expression of CART3 is restored in cell coexpressing CB1 in fasted 12 dpf larvae exposed to rimonabant. CART3 expression in the hypothalamus (4) and in the medulla oblongata (8, 12) is restored in fasted larvae treated with rimonabant. Expression in the tegmentum remains unaffected (7, 11). Panels 1–4 and 9–12, transverse sections; panels 5–8, lateral views. NS, Not significant; *, P < 0.05; **, P < 0.01.
Because we saw earlier (see Fig. 1) that rimonabant induces a decrease in yolk sac reserve use, we performed the same experiments in 12 dpf larvae to see whether, at an early stage, rimonabant treatment can antagonize the CART3 expression decrease induced by fasting (Fig. 5E). Interestingly, we observed that in the hypothalamus (Fig. 5E, upper panels) and in the medulla oblongata (Fig. 5E, lower panels) but not in the tegmentum region (Fig. 5E, middle panels), rimonabant impairs the CART3 expression decrease observed after fasting. For example, in the hypothalamic region, it is obvious that CART3 levels were decreased after fasting (compare Fig. 5E2 with Fig. 5E1) and that rimonabant suppressed this effect (compare Fig. 5E4 with Fig. 5E2). Two further observations should be made concerning these experiments: 1) rimonabant treatment by itself has only marginal effects in fed fish, suggesting that the inhibition of CB1 signaling is important only in a certain physiological state and 2) even if this is not seen in Q-PCR analysis of total brain mRNA, we consistently observed that the whole-mount in situ signal, after rimonabant treatment, in the hypothalamus and the medulla oblongata was higher than the signal observed in fed fish without rimonabant treatment (compare Fig. 5E4 with Fig. 5E1 and Fig. 5E12 with Fig. 5E9).
Taken together, these data suggest that the CB1 signaling pathway regulates CART3 expression in specific brain regions, and this occurs only when the embryos are in a specific physiological state induced by fasting. These results thus refine our current vision of the intricate relationship existing between the cannabinoid pathway, CART, and appetite regulation and suggest that these relationships are extremely context specific.
Discussion
Our results showed that rimonabant decreased yolk sac use and inhibited the fasting-induced decrease of CART3 expression in specific brain areas implicated in food intake. It is tempting to link these two observations and to suggest that, by acting on CART3 (and possibly other CART genes, see below), rimonabant decreases the speed at which the embryo uses its limited nutrient reserve for its development. Recently it has been shown that treatment of zebrafish embryos with clofibrate induces a malabsorption syndrome that is reminiscent of the effect we observed in our study (23). This reinforces the notion that rimonabant treatment effectively impairs the use of the vitelline reserve of the embryo, linking, at a metabolic level, yolk sac use with appetite regulation. It is important to stress that the effect of rimonabant treatment is not a general toxic effect and is CB1 specific because it is not observed in the absence of the CB1 receptor, after morpholino knockdown of CB1. The fact that rimonabant, which is a CB1 antagonist, inhibits yolk sac uses whereas CB1 knockdown does not produce the same effect is puzzling and may suggest that either rimonabant induces a specific inhibitory action at the receptor level or, alternatively, that the apoolipoprotein-CB1 receptor has a specific action. A detailed genetic comparison of the CB1 knockout models with animals in which the production of endogeneous ligand is impaired could bring interesting data with that regard.
CB1 action on fasting on specific territories of CART in the brain
Recent data, obtained in mammals, converge toward the notion that the brain endocannabinoid system controls food intake at two levels: 1) it reinforces the motivation for food searching and consumption, most likely by interacting with the mesolimbic reward system (3), and 2) it is activated in the hypothalamus after short-term food deprivation and then transiently regulates the levels and/or action of other orexigenic and anorexigenic mediators to induce appetite. It is at this second level that the regulation observed in our analysis is taking place.
After fasting, the endocannabinoid system in the hypothalamus is transiently activated, which increases energy uptake, because we have shown that, in zebrafish, CB1 lies upstream of CART. This may explain why CART3 expression is down-regulated by fasting in only some brain areas in which CB1 is expressed, including the hypothalamus. This is consistent with data obtained in mice showing that mice deficient in FAAH, which is responsible for the degradation of anandamide, have reduced levels of CART in brain regions implicated in appetite control (15). Of note, the CB1 inhibition by morpholino, which led to a decrease of CART3 (and CART2a) expression, suggests that CB1 normally activates CART3 expression. This contrasts to the effect observed in fasted animals, in which CB1 was activated, and we observed a decrease in CART expression, reversed by rimonabant. However, it has to be mentioned that these experimental paradigms represent different physiological situations that are difficult to compare. Indeed, the effect of morpholinos was studied in normal embryos that had access to their yolk reserve, which was comparable with being fed. In these later conditions, the fact that CART2b behaved oppositely may be explained by a different signal transduction pathway that translates the agonist action of endocannabinoids into negative gene regulation. Alternatively, it is possible that in certain brain areas, in which CART2b was expressed, CB1 was not occupied by agonists.
Interestingly, rimonabant treatment of FAAH knockout mice induces an increase in CART levels, an observation similar to the effect on CART3 that we observed in zebrafish. Despite the striking difference in gene number, this highlights the very similar regulatory inputs controlling CART expression between teleost fish and mammals, specifically concerning the cannabinoid system-CART relationship. This suggests that the regulation of CART by endocannabinoids is an ancient, conserved, and essential node in appetite control in vertebrates.
If it has been clearly shown that CART is a downstream mediator of the orexigenic effect of endocannabinoids, the mechanisms behind this effect are not well understood (3). Because of the predominantly presynaptic localization of CB1 receptors and their capacity to decrease neurotransmitter release via inhibition of presynaptic Ca2+ channels (35), it is possible, based on our expression data in zebrafish (Fig. 5), one scenario of the CB1 effect on anorexic signals is that CB1-mediated inhibition of the release of an anorexigenic mediator, such as CART, could be one of the main factors responsible for the appetite-stimulating effect of cannabinoids, although this remains to be directly tested. Indeed, endogenous CART may actively inhibit food intake, as indicated by the ability of human CART to act as a potent satiety factor when centrally injected in goldfish (36). This is further supported by the fasting-induced decrease in CART expression in brain regions involved in appetite control, including the hypothalamus, an effect that we also observed in zebrafish 12 dpf larvae (34, 37).
Of note, the effect we describe in this paper is mechanistically very different from the regulation proposed in the above studies. In our study, we investigated CART mRNA levels and not the amount of CART peptide present, a task that would be more complex, given that at least four different gene products exist in zebrafish. This means that, in our study, CB1 inhibition by rimonabant led to CART3 gene transcription through a classical signal transduction pathway that, in the nucleus, modifies CART3 promoter activity. This mechanism is different, and not mutually exclusive, from the more direct effect of CB1 on the release of CART peptide. Interestingly, the transcriptional effect we describe can last longer than a direct effect of CART peptide release. It will be important, in the future, to delineate the roles of these two mechanisms in food intake and in the regulation of energy metabolism.
We have to point out that our study does not yet show a direct effect of this regulation in food intake in the 12-d larvae. In addition, our conclusion of an effect on appetite relies on the observation of rimonabant effects on yolk use in embryos. It will be important in the future, using video monitoring, to directly survey the food intake of the zebrafish larvae. Finally, it is also important to mention that leptin has been identified in mammals as a third important player in this regulation (38). Interestingly, in zebrafish, the leptin receptor is expressed in the medulla oblongata and the hypothalamus, suggesting that it can play a role in the regulatory hierarchy (39). CART levels in the hypothalamus are reduced in leptin-deficient obese mice and leptin can increase CART levels in mammals (20, 40). Murine leptin has been shown to induce a significant decrease in food intake in goldfish and is able to modulate orexigenic neuropeptides (41, 42). Therefore, it will be important to study the effect of leptin on CART gene levels in zebrafish in the presence or absence of rimonabant.
Zebrafish as a model system for appetite regulation
In addition to providing a better understanding of the complex regulations linking CB1 and CART, our work also reveals that several aspects of the pathways regulating appetite are conserved between zebrafish and mammals. Zebrafish could be a powerful genetic system to better understand the mechanisms controlling appetite and obesity in an in vivo context (43). In addition, because zebrafish development is entirely external, it is an ideal model to performed pharmacological studies. Previous reports have indicated that key players in appetite regulation are present in other fish species, such as goldfish, which was previously the best known nonmammalian model of appetite regulation. For example, in goldfish, several appetite-regulated neuropeptides, such as neuropeptide Y, corticotropin-releasing factor, cholecystokinin, and gastrin-releasing peptide, have been isolated (26). Several CART genes are known to be present in this species, and their expression has been shown to be induced by food intake and decreased by fasting. In addition, because it is relatively easy to monitor food intake in a slow fish like the goldfish, it has been shown that injection of human CART induced a significant decrease in food intake. This suggests that CART has an anorexigenic effect in goldfish (36). Measuring food intake in zebrafish may prove to be difficult because of their high mobility compared with goldfish. However, zebrafish appear to be a promising model for these studies. Given the transparency of the embryo and larvae (after 1-phenyl-2-thiourea treatment) for more than 10 dpf, it is possible to combine pharmacological treatments and gene expression studies to determine the precise relationship between a given signaling pathway (e.g. the cannabinoid pathway) and its putative targets (e.g. CART genes). In addition, because gene expression knockdown by morpholino injection is easy to perform, zebrafish allow us to directly test the epistatic relationship between several genes during embryogenesis. Finally, zebrafish allow us to perform genetic screens to isolate genes implicated in various and complex developmental processes. It would be very interesting, for example, to isolate mutants for which rimonabant does not induce a decrease in the yolk sac use because this could lead to the identification of new important players in appetite regulation.
Materials and Methods
Bioinformatics
Basic local alignment search tool analysis of the National Center for Biotechnology Information NCBI expressed sequence tag database (Bethesda, MD) was used to identify expressed sequence tags with sequence similarity to mammalian CART and CB genes. From the resulting sequences, a new alignment was constructed using Muscle, and we performed a PhyML analysis using the default parameters (1000 bootstrap replicates).
Isolation of zebrafish CART genes
CART and CB1 cDNA were cloned from a zebrafish 48 or 72 hpf total RNA by using TRIzol Reagent (Invitrogen, Carlsbad, CA). RT-PCR was done to detect and amplify CART cDNA using primers designed from the zebrafish cDNA sequence. The cDNA was prepared by reverse transcription of total RNA with random primers (Promega, Madison, WI) and Moloney murine leukemia virus reverse transcriptase (Invitrogen, Carlsbad, CA). Primers for each gene are indicated in Supplemental Fig. 2. The complete sequences of the four CART cDNA have been deposited in Genbank under the following accession numbers: GU057833-GU057836.
cAMP response and incorporation of labeled rimonabant
The construction of the p658-derived vector expressing the zebrafish CB1 receptor was adapted from Shire et al. (44), and transfected using Lipofectamine 2000 (Invitrogen) into Chines hamster ovary cells. cAMP response was determined as previously reported (44).
For the calculations, each curve was fitted using SAS (SAS Institute, Cary, NC) to calculate the respective EC50. The empirical pA2 value as an affinity parameter was calculated from the IC50 according to the formula of Furchgott: pA2 = log (CR-1) − log ([antag]).
Embryos and larvae, 30 or 60 individuals, were incubated in 20-ml dishes (E3 buffer) in the presence of 0.5 μm rimonabant supplemented with 0.1 μCi/ml [3H]rimonabant (TRK1028; Amersham, Piscataway, NJ) for 96 h at 28 C. Then embryos and larvae were collected and washed in 20 ml PBS before their transfer and drying on a Whatman filter (Middlesex, UK). Embryos and larvae were vortexed and dissolved in 500 μl NaOH 1 n. Scintillation cocktail (10 ml) was added, and the radioactivity associated for each embryo and larvae groups was quantified using a liquid scintillation analyzer, Tri-Carb 2900 TR (PerkinElmer, Norwalk, CT).
Quantitative RT-PCR
For all experiments, reverse transcription was performed with Moloney murine leukemia virus-reverse transcriptase (Invitrogen) using 1 μg of total RNA mixed with 500 ng of hexamer oligonucleotides according to the manufacturer's instructions. One 100th of cDNA were used for Q-PCR using specific primers (as listed below) and SYBR-Green PCR kit (QIAGEN, Valencia, CA). Q-PCR were performed at least three times in duplicate on a DNA engine. Each couple of oligonucleotides hybridizes on different exons. The sequences of the oligonucleotides used are indicated on Supplemental Fig. 3. Specificity of the amplification was optimized by determining melting curves of the amplicons. In all experiments α-actin was used for the normalization of the amplified products.
Animals and injections
Zebrafish (Danio rerio) were kept at 28 C in a 14-h light, 10-h dark cycle with light on at 0900 h [Zeitgeber time (ZT) 0] and light turned off at 2300 h (ZT14). Adults were crossed overnight, resulting in spawning and fertilization around ZT0 the next morning. Embryos were collected after spawning to perform the morpholino injection and then raised at 28 C in petri dishes. To prevent pigmentation, 0.2 mm 1-phenyl-2-thiourea (Sigma, St. Louis, MO) was added to the water at 12 hpf. Parafin sections from embryos were cut at 10 μm and processed according to current protocols (45). Antisense morpholinos (Gene Tools, Philomath, OR) were injected at the one- to four-cell stage. The targeted sequences of the morpholinos used are as follows: CB1 untranslated region MO1, 5′-GAATGACTACGCTTACATGGACATC-3′; CB1 AUG MO2, 5′-AACAGCATGGTCAGAGATGCTCTAG-3′; CB2 AUG MO1, 5′-CTGCTCTTGTGTGGTCAAAACCATG-3′; and standard control MO, 5′-CCTCTTACCTCAGTTACAATTTAT-3′.
Whole-mount in situ hybridization and immunohistochemistry
Embryos at 48 hpf stage were fixed in 4% paraformaldehyde in PBS overnight at 4 C and then stored in methanol. Whole-mount in situ hybridization using digoxigenin- and FLU-labeled riboprobes was performed as described elsewhere (46). The zebrafish CART probes for in situ hybridization were PCR amplified using the same primers we used for RT-PCR and subcloned into pCRII-TOPO vector (Invitrogen). Control experiments performed in parallel with the CART sense RNA probe did not reveal any staining. Nomenclature of brain areas was based on the work of Mueller and Wullimann (32).
Starving experiments
For treatments on larvae, 60 individuals in 30-ml dishes were fed with paramecia from 6 to 10 dpf and from 10 dpf supplemented with brine shrimp. For the fasting group, embryos from 6 dpf were transferred to 30-ml dishes and kept there until the animals were killed. We performed in situ hybridization of the two groups in the same tube.
For the adults, animals (n = 4) were either fed (once a day) or fasted for 96 h and were collected on d 5. Whole brains were collected, frozen in liquid nitrogen, and stored at −80 C until analyzed, and total RNA was extracted using Trizol (Invitrogen). Deoxyribonuclease I-treated total RNA (2 μg) from each tissue was used for Q-PCR analysis to quantitate mRNA using SYBR Green dye. Oil Red O staining was performed as previously described (47).
Acknowledgments
We thank Dr. Warren G. Hill (Harvard Medical School/Beth Israel Deaconess Medical Center), Frederic Flamant (Institut de Génomique Fonctionelle de Lyon), and Daniel Fraher (Deakin University) for their critical reading of the manuscript. We are extremely grateful to Kei Yamamoto, Philippe Vernier, and Mario Wulliman for their advice and help in neurobiology and Marie Semon for her help in statistics.
This work was supported by Sanofi-Aventis.
This paper is part of the GenObCB1 research program funded by the French Ministry of Industry and Ministry of Research. We thank the Centre National de la Recherche Scientifique, Ministere de l'Education Nationale, de la Recherche et de la Technologie for financial support. Y.G. received a grant from the Association pour la Recherche Contre le Cancer.
Disclosure Summary: The authors declare they have no conflict of interest and nothing to disclose.
Footnotes
- CART
- Cocaine- and amphetamine-related transcript
- CB1
- type 1 cannabinoid receptor
- CB2
- type 2 cannabinoid receptor
- dpf
- d post fertilization
- FAAH
- fatty acid amide hydrolase
- hpf
- h post fertilization
- ZT
- Zeitgeber time.
References
- 1. Ahima RS , Lazar MA. 2008. Adipokines and the peripheral and neural control of energy balance. Mol Endocrinol 22:1023–1031 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2. Arora S , Anubhuti. 2006. Role of neuropeptides in appetite regulation and obesity—a review. Neuropeptides 40:375–401 [DOI] [PubMed] [Google Scholar]
- 3. Di Marzo V , Matias I. 2005. Endocannabinoid control of food intake and energy balance. Nat Neurosci 8:585–589 [DOI] [PubMed] [Google Scholar]
- 4. Kalra SP , Kalra PS. 2010. Neuroendocrine control of energy homeostasis: update on new insights. Prog Brain Res 181:17–33 [DOI] [PubMed] [Google Scholar]
- 5. Morton GJ , Cummings DE , Baskin DG , Barsh GS , Schwartz MW. 2006. Central nervous system control of food intake and body weight. Nature 443:289–295 [DOI] [PubMed] [Google Scholar]
- 6. Horvath TL. 2005. The hardship of obesity: a soft-wired hypothalamus. Nat Neurosci 8:561–565 [DOI] [PubMed] [Google Scholar]
- 7. Zimmer A , Zimmer AM , Hohmann AG , Herkenham M , Bonner TI. 1999. Increased mortality, hypoactivity, and hypoalgesia in cannabinoid CB1 receptor knockout mice. Proc Natl Acad Sci USA 96:5780–5785 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Di Marzo V , Goparaju SK , Wang L , Liu J , Bátkai S , Járai Z , Fezza F , Miura GI , Palmiter RD , Sugiura T , Kunos G. 2001. Leptin-regulated endocannabinoids are involved in maintaining food intake. Nature 410:822–825 [DOI] [PubMed] [Google Scholar]
- 9. Jamshidi N , Taylor DA. 2001. Anandamide administration into the ventromedial hypothalamus stimulates appetite in rats. Br J Pharmacol 134:1151–1154 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Kirkham TC. 2005. Endocannabinoids in the regulation of appetite and body weight. Behav Pharmacol 16:297–313 [DOI] [PubMed] [Google Scholar]
- 11. Ravinet Trillou C , Delgorge C , Menet C , Arnone M , Soubrié P. 2004. CB1 cannabinoid receptor knockout in mice leads to leanness, resistance to diet-induced obesity and enhanced leptin sensitivity. Int J Obes Relat Metab Disord 28:640–648 [DOI] [PubMed] [Google Scholar]
- 12. Bermudez-Silva FJ , Viveros MP , McPartland JM , Rodriguez de Fonseca F. 2010. The endocannabinoid system, eating behavior and energy homeostasis: the end or a new beginning? Pharmacol Biochem Behav 95:375–382 [DOI] [PubMed] [Google Scholar]
- 13. Mackie K. 2006. Mechanisms of CB1 receptor signaling: endocannabinoid modulation of synaptic strength. Int J Obes (Lond) 30(Suppl 1):S19–S23 [DOI] [PubMed] [Google Scholar]
- 14. Bifulco M. 2009. The endocannabinoid system: from biology to therapy. Pharmacol Res 60:75–76 [DOI] [PubMed] [Google Scholar]
- 15. Osei-Hyiaman D , Depetrillo M , Harvey-White J , Bannon AW , Cravatt BF , Kuhar MJ , Mackie K , Palkovits M , Kunos G. 2005. Cocaine- and amphetamine-related transcript is involved in the orexigenic effect of endogenous anandamide. Neuroendocrinology 81:273–282 [DOI] [PubMed] [Google Scholar]
- 16. Hunter RG , Philpot K , Vicentic A , Dominguez G , Hubert GW , Kuhar MJ. 2004. CART in feeding and obesity. Trends Endocrinol Metab 15:454–459 [DOI] [PubMed] [Google Scholar]
- 17. Cota D , Marsicano G , Tschöp M , Grübler Y , Flachskamm C , Schubert M , Auer D , Yassouridis A , Thöne-Reineke C , Ortmann S , Tomassoni F , Cervino C , Nisoli E , Linthorst AC , Pasquali R , Lutz B , Stalla GK , Pagotto U. 2003. The endogenous cannabinoid system affects energy balance via central orexigenic drive and peripheral lipogenesis. J Clin Invest 112:423–431 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Abbott CR , Rossi M , Wren AM , Murphy KG , Kennedy AR , Stanley SA , Zollner AN , Morgan DG , Morgan I , Ghatei MA , Small CJ , Bloom SR. 2001. Evidence of an orexigenic role for cocaine- and amphetamine-regulated transcript after administration into discrete hypothalamic nuclei. Endocrinology 142:3457–3463 [DOI] [PubMed] [Google Scholar]
- 19. Kong WM , Stanley S , Gardiner J , Abbott C , Murphy K , Seth A , Connoley I , Ghatei M , Stephens D , Bloom S. 2003. A role for arcuate cocaine and amphetamine-regulated transcript in hyperphagia, thermogenesis, and cold adaptation. FASEB J 17:1688–1690 [DOI] [PubMed] [Google Scholar]
- 20. Kristensen P , Judge ME , Thim L , Ribel U , Christjansen KN , Wulff BS , Clausen JT , Jensen PB , Madsen OD , Vrang N , Larsen PJ , Hastrup S. 1998. Hypothalamic CART is a new anorectic peptide regulated by leptin. Nature 393:72–76 [DOI] [PubMed] [Google Scholar]
- 21. Wang C , Billington CJ , Levine AS , Kotz CM. 2000. Effect of CART in the hypothalamic paraventricular nucleus on feeding and uncoupling protein gene expression. Neuroreport 11:3251–3255 [DOI] [PubMed] [Google Scholar]
- 22. Asnicar MA , Smith DP , Yang DD , Heiman ML , Fox N , Chen YF , Hsiung HM , Köster A. 2001. Absence of cocaine- and amphetamine-regulated transcript results in obesity in mice fed a high caloric diet. Endocrinology 142:4394–4400 [DOI] [PubMed] [Google Scholar]
- 23. Song Y , Cone RD. 2007. Creation of a genetic model of obesity in a teleost. FASEB J 21:2042–2049 [DOI] [PubMed] [Google Scholar]
- 24. Volkoff H , Peter RE. 2001. Characterization of two forms of cocaine- and amphetamine-regulated transcript (CART) peptide precursors in goldfish: molecular cloning and distribution, modulation of expression by nutritional status, and interactions with leptin. Endocrinology 142:5076–5088 [DOI] [PubMed] [Google Scholar]
- 25. Raldúa D , André M , Babin PJ. 2008. Clofibrate and gemfibrozil induce an embryonic malabsorption syndrome in zebrafish. Toxicol Appl Pharmacol 228:301–314 [DOI] [PubMed] [Google Scholar]
- 26. Nishio S , Gibert Y , Bernard L , Brunet F , Triqueneaux G , Laudet V. 2008. Adiponectin and adiponectin receptor genes are coexpressed during zebrafish embryogenesis and regulated by food deprivation. Dev Dyn 237:1682–1690 [DOI] [PubMed] [Google Scholar]
- 27. Kehoe AS , Volkoff H. 2007. Cloning and characterization of neuropeptide Y (NPY) and cocaine and amphetamine regulated transcript (CART) in Atlantic cod (Gadus morhua). Comp Biochem Physiol A Mol Integr Physiol 146:451–461 [DOI] [PubMed] [Google Scholar]
- 28. Dehal P , Boore JL. 2005. Two rounds of whole genome duplication in the ancestral vertebrate. PLoS Biol 3:e314. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Jaillon O , Aury JM , Brunet F , Petit JL , Stange-Thomann N , Mauceli E , Bouneau L , Fischer C , Ozouf-Costaz C , Bernot A , Nicaud S , Jaffe D , Fisher S , Lutfalla G , Dossat C , Segurens B , Dasilva C , Salanoubat M , Levy M , Boudet N , Castellano S , Anthouard V , Jubin C , Castelli V , Katinka M , Vacherie B , et al. 2004. Genome duplication in the teleost fish, Tetraodon nigroviridis, reveals the early vertebrate proto-karyotype. Nature 431:946–957 [DOI] [PubMed] [Google Scholar]
- 30. Bertrand S , Brunet FG , Escriva H , Parmentier G , Laudet V , Robinson-Rechavi M. 2004. Evolutionary genomics of nuclear receptors: from twenty-five ancestral genes to derived endocrine systems. Mol Biol Evol 21:1923–1937 [DOI] [PubMed] [Google Scholar]
- 31. Danchin EG , Gouret P , Pontarotti P. 2006. Eleven ancestral gene families lost in mammals and vertebrates while otherwise universally conserved in animals. BMC Evol Biol 6:5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Mueller T , Wulliman MF. 2005. Atlas of the early zebrafish brain development. Amsterdam: Elsevier [Google Scholar]
- 33. Lam CS , Rastegar S , Strähle U. 2006. Distribution of cannabinoid receptor 1 in the CNS of zebrafish. Neuroscience 138:83–95 [DOI] [PubMed] [Google Scholar]
- 34. Watson S , Chambers D , Hobbs C , Doherty P , Graham A. 2008. The endocannabinoid receptor, CB1, is required for normal axonal growth and fasciculation. Mol Cell Neurosci 38:89–97 [DOI] [PubMed] [Google Scholar]
- 35. Vicentic A , Jones DC. 2007. The CART (cocaine- and amphetamine-regulated transcript) system in appetite and drug addiction. J Pharmacol Exp Ther 320:499–506 [DOI] [PubMed] [Google Scholar]
- 36. Wilson RI , Kunos G , Nicoll RA. 2001. Presynaptic specificity of endocannabinoid signaling in the hippocampus. Neuron 31:453–462 [DOI] [PubMed] [Google Scholar]
- 37. Volkoff H , Peter RE. 2000. Effects of CART peptides on food consumption, feeding and associated behaviors in the goldfish, Carassius auratus: actions on neuropeptide Y- and orexin A-induced feeding. Brain Res 887:125–133 [DOI] [PubMed] [Google Scholar]
- 38. McAlister ED , Van Vugt DA. 2004. Effect of leptin administration versus re-feeding on hypothalamic neuropeptide gene expression in fasted male rats. Can J Physiol Pharmacol 82:1128–1134 [DOI] [PubMed] [Google Scholar]
- 39. Jelsing J , Larsen PJ , Vrang N. 2009. The effect of leptin receptor deficiency and fasting on cannabinoid receptor 1 mRNA expression in the rat hypothalamus, brainstem and nodose ganglion. Neurosci Lett 463:125–129 [DOI] [PubMed] [Google Scholar]
- 40. Liu Q , Chen Y , Copeland D , Ball H , Duff RJ , Rockich B , Londraville RL. 2010. Expression of leptin receptor gene in developing and adult zebrafish. Gen Comp Endocrinol 166:346–355 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41. Elias CF , Lee C , Kelly J , Aschkenasi C , Ahima RS , Couceyro PR , Kuhar MJ , Saper CB , Elmquist JK. 1998. Leptin activates hypothalamic CART neurons projecting to the spinal cord. Neuron 21:1375–1385 [DOI] [PubMed] [Google Scholar]
- 42. Volkoff H , Canosa LF , Unniappan S , Cerdá-Reverter JM , Bernier NJ , Kelly SP , Peter RE. 2005. Neuropeptides and the control of food intake in fish. Gen Comp Endocrinol 142:3–19 [DOI] [PubMed] [Google Scholar]
- 43. Volkoff H , Eykelbosh AJ , Peter RE. 2003. Role of leptin in the control of feeding of goldfish, Carassius auratus: interactions with cholecystokinin, neuropeptide Y and orexin A, and modulation by fasting. Brain Res 972:90–109 [DOI] [PubMed] [Google Scholar]
- 44. Calandra B , Portier M , Kernéis A , Delpech M , Carillon C , Le Fur G , Ferrara P , Shire D. 1999. Dual intracellular signaling pathways mediated by the human cannabinoid CB1 receptor. Eur J Pharmacol 374:445–455 [DOI] [PubMed] [Google Scholar]
- 45. Nusslein-Volhard C , Dahm R. 2002. Zebrafish: a practical approach. United States ed. Oxford: Oxford University Press [Google Scholar]
- 46. Thisse C , Thisse B. 2008. High-resolution in situ hybridization to whole-mount zebrafish embryos. Nat Protoc 3:59–69 [DOI] [PubMed] [Google Scholar]
- 47. Schlombs K , Wagner T , Scheel J. 2003. Site-1 protease is required for cartilage development in zebrafish. Proc Natl Acad Sci USA 100:14024–14029 [DOI] [PMC free article] [PubMed] [Google Scholar]