Abstract
We previously demonstrated that long-term pretreatment of rat FRTL-5 thyroid cells with TSH or cAMP-generating reagents potentiated IGF-I-dependent DNA synthesis. Under these conditions, cAMP treatment increased tyrosine phosphorylation of a 125-kDa protein (p125) and its association with a p85 regulatory subunit of phosphatidylinositol 3-kinase (p85 PI3K), which were suggested to mediate potentiation of DNA synthesis. This study was undertaken to identify p125 and to elucidate its roles in potentiation of DNA synthesis induced by IGF-I. Matrix-assisted laser desorption ionization-time of flight mass spectrometry (MALDI-TOF MS) analysis revealed p125 to be a rat ortholog of human XB130, which we named PI3K-associated protein (PI3KAP). cAMP treatment elevated PI3KAP/XB130 mRNA and protein levels as well as tyrosine phosphorylation and interaction with p85 PI3K leading to increased PI3K activities associated with PI3KAP/XB130, supporting the role of PI3KAP/XB130 in DNA synthesis potentiation. Importantly, PI3KAP/XB130 knockdown attenuated cAMP-dependent potentiation of IGF-I-induced DNA synthesis. Furthermore, c-Src was associated with PI3KAP/XB130 and was activated in response to cAMP. Addition of Src family kinase inhibitors, PP1 or PP2, during cAMP treatment abolished tyrosine phosphorylation of PI3KAP/XB130 and its interaction with p85 PI3K. Finally, introduction of PI3KAP/XB130 into NIH3T3 fibroblasts lacking endogenous PI3KAP/XB130 enhanced IGF-I-induced DNA synthesis; however, a mutant Y72F incapable of binding to p85 PI3K did not show this response. Together, these data indicate that cAMP-dependent induction of PI3KAP/XB130, which is associated with PI3K, is required for enhancement of IGF mitogenic activities.
The IGF play important roles in normal mammalian development and growth (1–3). In many cell types, IGF exert a wide variety of bioactivities such as cell proliferation, differentiation, survival, and maintenance of differentiated cell functions (4). However, bioactivities of IGF by themselves are generally weak and often potentiated by other bioactive factors including growth factors (5, 6), steroid hormones (7, 8), and tropic hormones (9–11). Therefore, to understand how the various bioactivities of IGF are induced, it is important to elucidate the molecular mechanisms underlying the potentiation of IGF bioactivities by other factors.
In cultured thyroid cells, we and others have shown that TSH synergistically potentiates the mitogenic activity of IGF-I (10, 11). This potentiating effect of TSH on the activity of IGF-I is also observed in vivo. For example, in patients with hypopituitarism, GH therapy increased IGF-I levels but did not restore their small thyroids to normal size when TSH levels were low (12). Other studies have indicated that high levels of IGF-I increased thyroid volume/weight in patients with acromegaly or in mice overexpressing IGF-I/IGF-I receptor in the thyroid gland (13, 14). All of these findings suggest that TSH and IGF-I collaborate to promote thyroid cell proliferation in vitro and in vivo.
FRTL-5 cells are a nontransformed line of rat thyroid follicular cells widely used as a model to study aspects of thyroid physiology such as cell proliferation, iodide uptake, and thyroglobulin biosynthesis (15). We have investigated the molecular mechanisms of synergistic cell proliferation induced by TSH and IGF-I using this cell line. In our previous research, we reported that prolonged pretreatment with TSH or other agents that increased intracellular cAMP potentiated IGF-I-dependent DNA synthesis synergistically compared with each single treatment in FRTL-5 cells (11). Additional studies showed that long-term TSH or cAMP treatment produced a time- and concentration-dependent increase in tyrosine phosphorylation of intracellular proteins including a 125-kDa protein (p125) (16). Moreover, we found that a prolonged cAMP stimulus induced association of phosphotyrosyl p125 with p85 regulatory subunit of phosphatidylinositol 3-kinase (PI3K) as well as activation of PI3K. Chronic activation of PI3K in response to cAMP was necessary for potentiation of DNA synthesis induced by IGF-I (17). All of these results suggest that p125 plays an important role in cAMP-dependent potentiation of DNA synthesis induced by IGF-I.
The aim of the present study was to identify p125 and to elucidate its function in potentiation of IGF-I-induced DNA synthesis. Here we demonstrate that p125, named PI3K-associated protein (PI3KAP) is an ortholog of human XB130. Moreover, we show that prolonged cAMP stimulus increases PI3KAP levels, its tyrosine phosphorylation by c-Src, and its binding to p85 PI3K. Moreover, we demonstrate that this protein is required for potentiation of IGF-I-dependent DNA synthesis.
Materials and Methods
Materials
Coon's modified Ham's F-12 (Coon's F-12), bovine insulin, and bovine TSH (1.23 U/mg) for culture were purchased from Sigma-Aldrich (St. Louis, MO). DMEM, PBS, and Hanks' balanced salt solution were obtained from Nissui (Tokyo, Japan). Newborn bovine serum and fetal bovine serum (FBS) were obtained from JRH Bioscience (Tokyo, Japan). Transferrin and dibutyryl cAMP (Bt2cAMP) was purchased from Nacalai Tesque, Inc. (Kyoto, Japan). Recombinant human IGF-I was kindly donated by Dr. Toshiaki Ohkuma (Fujisawa Pharmaceutical Co., Osaka, Japan). Purified bovine TSH (30 U/mg) for biological studies was generously donated by the National Hormone and Pituitary Program (National Institute of Diabetes and Digestive and Kidney Diseases, Bethesda, MD). PP1 (4-amino-5-(4-methylphenyl)-7-(t-butyl)pyrazolo[3,4-d]-pyrimidine) and PP2 (4-amino-5-(4-chlorolphenyl)-7-(t-butyl)pyrazolo[3,4-d]-pyrimidine) were purchased from Merck (Darmstadt, Germany). Anti-PI3KAP/XB130 antibody was raised in rabbits using a PI3KAP/XB130 carboxyl-terminal peptide NH2-CVTGKGTVLQKAKEWEKKGAS-COOH as an antigen. Anti-p85 PI3K regulatory subunit antibody and anti-Src GD11 antibody were purchased from Upstate Biotechnology, Inc. (Lake Placid, NY). Anti-phospho-Src family (Tyr-416) antibody was purchased from Cell Signaling Technology, Inc. (Beverly, MA). Anti-cyclin D1 antibody (72-13G) and anti-GFP antibody (FL) were purchased from Santa Cruz Biotechnology, Inc. (Santa Cruz, CA). Anti-FLAG M2 antibody, anti-phosphotyrosine antibody, anti-β-actin antibody, and nonimmune mouse IgG were obtained from Sigma-Aldrich. Horseradish peroxidase (HRP)-linked antimouse IgG antibody and HRP-linked antirabbit IgG antibody were purchased from GE Healthcare UK Ltd. (Buckinghamshire, UK). Random control, PI3KAP-specific, and Src-specific small interfering RNA (siRNA) were purchased from RNAi Corp. (Tokyo, Japan). The sequences of the PI3KAP siRNA used were 5′-GUCACGCUGGUGCUUUGUUAG-3′ (no. 1, nucleotides (nts) 1064–1086 in AB568258), 5′-CUUUUGUGCUACAAAUCGUCC-3′ (no. 2, nts 564–586), and 5′-UCCGGUCAAGUCUUCCAUAAAAC-3′ (no. 3, nts 2545–2567 in XM_217643). The Src siRNA sequences were 5′-CACCGGACAGACCGGUUACAUCC-3′ (no. 1, nts 384–406 in AF130457.1) and 5′-AGCAGCUUGUGGCUUACUACUCC-3′ (no. 2, nts 680–702). PI3KAP siRNA no. 3 was targeted to the 3′-untranslated region of PI3KAP, and the others were targeted to the open reading frame. The control siRNA sequence was 5′-GUACCGCACGUCAUUCGUAUC-3′. Other chemicals were of reagent grade available commercially.
Cell culture
Cells of a line of rat thyroid follicular FRTL-5 (15) (no. CRL8305; American Type Culture Collection, Manassas, VA) were kindly provided by Dr. Leonard Kohn (Ohio University and Edison Biotechnology Institute, Athens, OH) and the Interthyr Research Foundation (Baltimore, MD). FRTL-5 cells were routinely cultured at 37 C in a humidified 5% CO2-controlled atmosphere in Coon's F-12 medium containing 2.785 mg/ml NaHCO3, 0.33 mg/ml glutamine, MEM nonessential amino acids (ICN Biochemicals, Costa Mesa, CA), 50 IU/ml penicillin, 50 μg/ml streptomycin, 0.5 μg/ml amphotericin B, and 100 μg/ml kanamycin supplemented with 5% newborn bovine serum and a mixture of three hormones, including TSH (1 mU/ml), insulin (10 μg/ml), and transferrin (5 μg/ml). NIH3T3 cells were purchased from Health Science Research Resources Bank (Osaka, Japan). NIH3T3 cells were cultured in DMEM containing 10% FBS and 1 mg/ml NaHCO3 (10% FBS/DMEM). For cells to become quiescent, FRTL-5 cells and NIH3T3 cells were cultured for 24 h in Coon's F-12 medium containing 0.1% BSA or DMEM containing 0.1% BSA, respectively. PLAT-E cells for retrovirus production were a kind gift from Dr. Toshio Kitamura (The Institute of Medical Science, The University of Tokyo, Tokyo, Japan). PLAT-E cells were cultured in 10% FBS/DMEM supplemented with 1 μg/ml puromycin and 10 μg/ml blasticidin.
Animal treatment and hormone assay
Male Wistar rats (Nippon Clea Inc., Shizuoka, Japan) initially weighing 260 g (8 wk of age) were divided into two groups, each comprising seven animals: a methimazole (MMI)-treated group or a control group. The rats were supplied with either 0.03% MMI (Sigma-Aldrich) in drinking water (MMI-treated group) or water alone (control) ad libitum for 4 wk. After 4 wk of MMI treatment, the rats were fasted overnight, and tissue samples of thyroid glands were collected under anesthesia and frozen in liquid nitrogen. Blood samples were collected from carotid artery, mixed with EDTA at final concentrations of 1 mg/ml, and then centrifuged at 1000 × g. The obtained plasma samples were stored at −80 C until TSH measurement. Plasma TSH levels were measured using ELISA kits for rat TSH (Shibayagi Co., Ltd., Gunma, Japan). All procedures for animal research in this study were approved by the Committee on Laboratory Animal Care, Graduate School of Agriculture and Life Sciences, The University of Tokyo.
Plasmid construction
IMAGE cDNA clone encoding the mouse ortholog of PI3KAP/XB130 (GenBank accession no. BC031515) was obtained from Invitrogen (Carlsbad, CA). The mammalian expression vector expressing N-terminally FLAG-tagged PI3KAP/XB130 (pFLAG-PI3KAP/XB130) was constructed as follows. Mouse PI3KAP/XB130 cDNA containing the full-length open reading frame was amplified by PCR using two primers, 5′-GGGATGGAGCGCTACAAAGCA-3′ and 5′-CTAACTGGCTCCTTTCT-TC-3′. The PCR product was digested by DraI and XbaI and was cloned into the DraI-XbaI-digested pShuttle2 vector (Clontech, Mountain View, CA). Site-directed mutagenesis was performed using overlap-extension PCR mutagenesis (18) to introduce a Y72F mutation in PI3KAP/XB130. We used the following primers in this procedure: 5′-TTCTGATGAGGAGTTTATTTACATGAAACA-3′, 5′-TGTTCATGTAAATAAACTCCTCATCAGA-3′, and the aforementioned two primers. The resulting DNA fragment encoding FLAG-Y72F was cloned into the DraI-XbaI-digested pShuttle2 vector. A retroviral vector pMXs-neo was a kind gift from Dr. Toshio Kitamura and was used for retrovirus production. The DNA fragments encoding FLAG-PI3KAP/XB130 or FLAG-Y72F were amplified by PCR using two primers, 5′-AAATTAATTAACGTCAGAATTAACCATGGACTAC-3′ and 5′-TGTGAAATTTGTGATGCTATTGCT-3′. The amplified fragments were digested by PacI and NotI and cloned into the PacI-NotI-cut pMXs-neo vector. The mammalian expression vector expressing EGFP (pEGFP-N1) was purchased from Clontech.
Transfection of siRNA
Transfection of siRNA into FRTL-5 cells was performed using Lipofectamine 2000 according to the manufacturer's protocol (Invitrogen) with some modifications. Briefly, cells were plated into six-well plates at 5 × 105 cells per well. Cells were cultured for 2–3 d, and then 500 μl OptiMEM (Invitrogen) containing the siRNA-Lipofectamine 2000 mixture (250 pmol siRNA, 5 μl Lipofectamine 2000) were added to the cells cultured in 1.5 ml growth medium. For transfection of Src siRNA, 5 × 105 cells were stripped and resuspended in 1.5 ml growth medium and mixed with 500 μl of the aforementioned OptiMEM/siRNA/Lipofectamine mixture. After 4 h incubation, the medium was replaced by fresh growth medium, and cell culture was continued for 24 h. Thereafter, cells were serum starved for 24 h, followed by treatment with indicated hormones/chemicals. Transfection efficiency of siRNA into FRTL-5 cells was over 90%.
Retrovirus production and infection
PLAT-E cells were transfected with pMXs-neo, pMXs-FLAG-PI3KAP/XB130, or pMXs-FLAG-Y72F using Lipofectamine 2000 according to the manufacturer's protocol and were incubated overnight in 10% FBS/DMEM. After replacement of the medium with fresh 10% FBS/DMEM, cells were incubated for an additional 24 h, until the conditioned medium containing retrovirus was collected. NIH3T3 cells were infected with retroviruses by overnight incubation in the medium containing 3 μg/ml polybrene (Sigma-Aldrich). Infected cells were selected by 500 μg/ml G418 (Nacalai Tesque) for 1 wk.
DNA synthesis assay
Quiescent FRTL-5 cells on 48-well plates were cultured for 24 h in Coon's F-12 medium with 0.1% BSA with or without 1 mm Bt2cAMP. After this pretreatment, the cells were washed three times with Hanks' balanced salt solution and then treated with or without 100 ng/ml IGF-I for 24 h. [Methyl-3H]thymidine (0.3 μCi/well, 1 μCi/ml; GE Healthcare UK) was added to each well 4 h before the termination of each experiment. The labeling was stopped by adding 1 m ascorbic acid. The cells were washed twice with ice-cold PBS and twice with ice-cold 10% trichloroacetic acid. Trichloroacetic acid-precipitated materials were solubilized with 250 μl 0.2 n NaOH and 0.1% sodium dodecyl sulfate (SDS), mixed into 5 ml clear-sol II (Nacalai Tesque), and the radioactivity was measured using a liquid scintillation counter (Aloka, Tokyo, Japan).
Immunoblotting
Quiescent cells in 100-mm dishes were pretreated and treated with or without test agents as indicated. Cells were lysed at 0 C in 400 μl lysis buffer containing 50 mm Tris-HCl (pH 7.4), 150 mm NaCl, 1 mm NaF, 1 mm EDTA, 1 mm EGTA, 1% Triton X-100, 10% glycerol, 500 μm Na3VO4, 100 kallikrein-inactivating U/ml aprotinin, 20 μg/ml phenylmethylsulfonyl fluoride, 10 μg/ml leupeptin, and 5 μg/ml pepstatin. The lysates were centrifuged at 15,000 × g for 10 min at 4 C. The protein assay of the supernatant was performed using a protein assay kit (Bio-Rad Laboratories, Inc., Hercules, CA). Equal amounts of proteins (25 μg protein) of each sample were mixed with a half-volume of 3× Laemmli's buffer [30 mm Tris-HCl (pH 7.8), 9% SDS, 15% glycerol, 6% 2-mercaptoethanol, 0.05% bromophenol blue]. The mixtures were boiled for 5 min, subjected to SDS-PAGE, and transferred onto nitrocellulose membrane (BA-85; Schleicher & Schuell Bio-Science, Keene, NH). The indicated first antibodies and HRP-conjugated second anti-IgG antibodies were hybridized according to standard immunoblotting protocols. Chemiluminescence reactions were performed using the enhanced chemiluminescence kit (ECL kit; PerkinElmer Life Science, Inc. Boston, MA), and the luminescence was exposed onto x-ray film (XOmat; Kodak, Tokyo, Japan). Densitometric analysis was performed using the ImageJ version 1.37 program (http://rsb.info.nih.gov/ij/; National Institutes of Health, Bethesda, MD).
Immunoprecipitation
One milligram of total protein of cell lysates was mixed with indicated antibodies (at concentrations recommended by the manufacturer) and made up to 1 ml with the lysis buffer described above. Samples were incubated at 4 C for 1–2 h, and then 10 μl protein A-Sepharose or protein G-Sepharose (GE Healthcare UK) was added, and incubation was continued for 1 h. Precipitates were washed with the lysis buffer three times. Samples to be analyzed by immunoblotting were diluted with 1× Laemmli's buffer, boiled for 5 min, and subjected to SDS-PAGE.
Purification of p125
Reduction and N-ethylmaleimide treatment of cell lysates were performed as previously described (19). Eighteen dishes (100 mm in diameter) of cells per sample were washed with ice-cold PBS and then lysed and reduced on ice in SDS buffer [100 mm HEPES (pH 7.4), 4% SDS, 300 mm NaCl, 10 mm dithiothreitol, 10 μg/ml leupeptin, 5 μg/ml pepstatin, 20 μg/ml phenylmethylsulfonyl fluoride, 100 kallikrein-inactivating U/ml aprotinin]. The lysates were homogenized with a Dounce homogenizer and then centrifuged at 15,000 × g for 10 min at 4 C. The supernatants (6.5 ml) were boiled for 5 min to denature proteins, treated with 30 mm N-ethylmaleimide to cap the thiol groups, and then diluted with 91 ml ice-cold 1.7% Thesit buffer [50 mm HEPES (pH 7.4), 1.7% Thesit (Sigma), 150 mm NaCl]. The mixtures were centrifuged at 3000 × g for 10 min, and the supernatants were filtered with a 0.45-μm syringe filter. The samples were then mixed with nonimmune rabbit IgG-conjugated protein G-Sepharose for 3 h at 4 C, followed by the centrifugation at 3000 × g for 10 min at 4 C to precipitate proteins that nonspecifically bound to IgG-beads. The supernatants were mixed with 300 μg anti-phosphotyrosine antibody. After overnight incubation at 4 C, 30 μl protein G-Sepharose was added, and the incubation was continued for 1 h at 4 C. The precipitates were washed seven times with 1 ml 0.5% Thesit buffer [50 mm HEPES (pH 7.4), 0.5% Thesit, 150 mm NaCl], mixed with 75 μl 1× Laemmli's buffer, and incubated for 1 h at 60 C. After the centrifugation at 15,000 × g, the supernatants were subjected to SDS-PAGE, followed by gel staining using SilverQuest (Invitrogen) according to the manufacturer's recommended protocols.
Identification of p125
Gel slices were destained according to the protocols in the gel staining kits and then equilibrated with 50 mm NH4NCO3 for 15 min. After the reagent was removed, gel slices were mixed with dehydration buffer (50 mm NH4NCO3, 50% CH3CN) and vortexed for 15 min. After the dehydration procedure was repeated again, gels were dried in rotary evaporator. Gels were then reswollen in reducing buffer (10 mm DTT, 25 mm NH4NCO3) and incubated for 1 h at 56 C. After the equal volume of alkylation buffer (55 mm iodoacetamide, 25 mm NH4NCO3) was added, the mixtures were shaded and vortexed for 30 min at room temperature. Gels were then equilibrated with 50 mm NH4NCO3, dehydrated with 50% CH3CN, and dried in a centrifugal concentrator as described above. After reswelling on ice in digestion buffer [10 μg/ml Trypsin Gold, MA Spectrometry Grade (Promega, Madison, WI), 50 mm NH4NCO3, 5 mm CaCl2], gels were incubated for 16 h at 37 C. To extract the tryptic peptides, 5% trifluoroacetic acid (TFA) was added, and the supernatant was recovered. Gels were then vortexed in 5% TFA/30% CH3CN for 5 min and sonicated for 20 min, and the supernatant was recovered. The same procedure was repeated using 5% TFA/50% CH3CN, and 5% TFA/70% CH3CN, sequentially. All supernatants were collected and concentrated to 10 μl in a centrifugal concentrator, and TFA (final concentration, 0.1%) was added to the samples. To desalt the sample, ZipTip μC18 (Millipore) was swollen in 0.1% TFA/50% CH3CN and equilibrated with 0.1% TFA/2% CH3CN, and the samples were then adsorbed and washed with 0.1% TFA/2% CH3CN. The tryptic peptides were eluted with 1 μl of 0.1% TFA/66% CH3CN onto a 100-well gold sample plate (Applied Biosystems, Foster City, CA), mixed with 1 μl of matrix solution (10 mg/ml α-cyano-4-hydroxy cinnamic acid in 0.2% TFA/60% CH3CN), and dried at room temperature. The samples were analyzed by matrix-assisted laser desorption ionization-time of flight mass spectrometry (MALDI-TOF MS) Voyager-DE STR (Applied Biosystems) in peptide-sensitivity reflector mode. The spectra were obtained by the accumulation of 200-1000 consecutive laser shots and calibrated by mass values of auto-digested trypsin peptides. The mass data were subjected to MASCOT PMF search tools (Matrix Science, London, UK; http://www.matrixscience.com/search_form_select.html). The maximum tolerance level for the peptide masses was 100 parts per million, and the modifications accepted were carbamidomethyl cysteines and artifactual oxidation of methionines. The NCBI nonredundant databases were used for the searches.
Cloning of rat PI3KAP
Total RNA was isolated from cAMP-treated FRTL-5 cells using the TRIzol reagent according to the manufacturer's protocol (Invitrogen). To amplify rat p125 by RT-PCR, we designed sets of reverse-transcription primers and PCR primers shown in Table 1 using Genetyx Mac software version 11.1 (Software Development Co., Ltd., Tokyo, Japan) based on the predicted nucleotide sequence of similar to AU041783 (XM_217643.2). First-strand cDNA was synthesized using SuperScript2 RT-PCR kit (Invitrogen) and then subjected to PCR to amplify rat p125 fragments. Six amplified products were subcloned into pBluescript KS (+) vector and sequenced. We assembled the six sequences to obtain the 2523-bp sequence, which was submitted to DDBJ/EMBL/GenBank under the accession number AB568258.
Table 1.
Primers used for amplification of rat p125 by RT-PCR
| Reverse transcription primer (5′–3′) | PCR primer (forward) (5′–3′) | PCR primer (reverse) (5′–3′) |
|---|---|---|
| TCATTGGCATGATCTTGAGCTT | CGACTTCCTCAAGGTTCTAGAC | TCATTGGCATGATCTTGAGCTT |
| CAGTCTGGAAGACATATATGGG | GAGGCTGAGCCATTTGACAC | TCACCTCCTGGATGACCAAC |
| CAGTCTGGAAGACATATATGGG | GAGGCTGAGCCATTTGACAC | CAGTCTGGAAGACATATATGGG |
| CAGTCTGGAAGACATATATGGG | GGAAGAAATCTACCTCACTGGA | CAGTCTGGAAGACATATATGGG |
| ACTTGACCGGAGCAGGATTG | CTCTCAGAGTCCGGCTCCAA | CTCTGTCCGGTTCTTCCCAA |
| ACTTGACCGGAGCAGGATTG | AGCTGAGACCCTCACAGTAG | ACTTGACCGGAGCAGGATTG |
| ACTTGACCGGAGCAGGATTG | CTCTCAGAGTCCGGCTCCAA | ACTTGACCGGAGCAGGATTG |
RT-PCR and real-time PCR analysis
For analysis of tissue distribution of PI3KAP/XB130, 3-month-old male rats were killed, and total RNA was isolated from tissues using the TRIzol reagent. For analysis of cyclin D1 mRNA levels, quiescent FRTL-5 cells were treated with 1 mm Bt2cAMP followed by 100 ng/ml IGF-I for the indicated hours, and then total RNA was isolated using the TRIzol reagent. First-strand cDNA was synthesized from 2 μg total RNA with oligo(dT) primers using the SuperScript2 RT-PCR kit. First-strand cDNA was then subjected to PCR. PI3KAP/XB130-specific primers, 5′-GGAAGAAATCTACCTCACTGGA-3′ and 5′-CAGTCTGGAAGACATATATGGG-3′, and cyclin D1-specific primers, 5′-GCCACCATGCTGCTGGACCCG-3′ and 5′-TCCTGCCTGGTTGGGACGCCT-3′, were used for PCR. Ribosomal protein S29 (RPS29, primers were 5′-TGAAGGCAAGATGGGTCACCAGCAGC-3′ and 5′-CAGGGTAGACAGTTGGTTTCATTGGG-3′) was used as the internal control. The PCR products were separated by electrophoresis in 2% agarose gel and stained with ethidium bromide.
For quantitative analysis of PI3KAP/XB130 mRNA in rat thyroid glands, real-time PCR was performed using ABI Prism 7900HT Sequence Detection System (Applied Biosystems) with SYBR green dye. PCR was performed using QuantiTect SYBR Green PCR kit (QIAGEN, Valencia, CA) in a final volume of 20 μl containing 0.4 μm of each primer and 2 μl of first-strand cDNA (equivalent to 3 ng total RNA). The settings for the PCR amplification were as follows: initial denaturation (15 min at 95 C) followed by 40 amplification cycles (15 sec at 94 C, 30 sec at 60 C, and 30 sec at 72 C). Fluorescence was measured after extension of each cycle. One of the cDNA samples was serially diluted and analyzed in parallel to obtain standard curves. The threshold cycle values and the relative amounts of mRNA were calculated using the Sequence Detector Software SDS version 2.3 (Applied Biosystems). The relative mRNA amounts were normalized to an internal control gene RPS29. The PI3KAP/XB130 primers used were 5′-CCAGTCAACTCTGCATCCGT-3′ and 5′-TGTCAGCAAGATCCATGACCT-3′. The RPS29 primers are described above.
Northern blotting
Quiescent FRTL-5 cells in three 100-mm dishes for each sample were treated with 1 mm Bt2cAMP for the indicated hours. Total cellular RNA was isolated using the TRIzol reagent. RNA (30 μg) was fractionated by electrophoresis through a 1.5% agarose gel containing formaldehyde, transferred to nylon membranes (GeneScreen Plus; PerkinElmer Life Science), and cross-linked by UV light. The membranes were hybridized at 42 C overnight with random primed 32P-labeled cDNA produced by the Megaprime DNA labeling Kit (GE Healthcare UK) and then washed at 65 C and subjected to autoradiography.
PI3K assay
Immunoprecipitates with anti-PI3KAP antibody were washed with the lysis buffer, LiCl buffer [100 mm Tris-HCl (pH 7.5), 500 mm LiCl], distilled water, and Tris-NaCl-EDTA buffer [10 mm Tris-HCl (pH 7.5), 150 mm NaCl, 1 mm EDTA] and finally resuspended in 45 μl of the PI3K reaction buffer [20 mm Tris-HCl (pH 7.5), 100 mm NaCl, 0.5 mm EGTA]. PI3K assay was initiated by the addition of 5 μl [γ-32P]ATP/MgCl2/phosphatidylinositol to give a final concentration of 20 mm MgCl2, 1 mm DTT, 30 μm ATP, 10 μCi [γ-32P]ATP (3000 Ci/mmol; GE Healthcare UK), and 20 μg bovine liver phosphatidylinositol (Avanti Polar Lipids, Inc., Alabaster, AL). After the incubation at 25 C for 15 min, 100 μl chloroform-methanol-HCl (10:20:1) was added to the reaction mixture to stop the reaction. Each lipid product was extracted, spotted onto a silica gel plate, and developed with a solvent containing chloroform-methanol-25% ammonia water-water (43:38:6:6). 32P radioactivity incorporated into phosphatidylinositol was measured by autoradiography.
Statistical analyses
Statistical analyses of data were performed by one-way factorial ANOVA using StatView software (Abacus Concepts, Inc., Berkeley, CA). Fisher's projected least significant difference was performed to study the significance between different conditions. The results shown are the mean ± sem. P < 0.05 was considered statistically significant.
Results
Identification of p125
To identify p125, we developed a multistep procedure based on immunoprecipitation from p125-enriched cAMP-treated FRTL-5 cells (see Materials and Methods). Tyrosine-phosphorylated p125 was readily detected in total cell lysates (Fig. 1A, left) and in immunoprecipitates (Fig. 1A, middle). In larger-scale cell lysates, p125 shown in SDS-PAGE of immunoprecipitates by silver staining was excised and analyzed by MALDI-TOF MS. A gel slice of similar electrophoretic mobility in the cAMP-untreated sample was similarly treated and analyzed as a negative control. Using peptide mass finger printing, multiple peptides detected only in samples prepared from cAMP-treated cells (Fig. 1B) were assigned to a predicted protein similar to expressed sequence AU041783. Twenty of the 36 mass data used for the search were assigned to this protein with sequence coverage of 24%. Nucleotide sequence encoding this protein was predicted by an annotation program based on genomic sequences. We confirmed the sequence of 2523 bp by RT-PCR covering almost the full length of this mRNA, except for a short 5′ region containing a predicted start codon. The sequence was submitted to DDBJ/EMBL/GenBank databases, and it was assigned the accession number AB568258. We named this protein PI3KAP. In addition, we found that this protein is an ortholog of human XB130 (20) and a homolog of actin filament-associated protein (AFAP)-110 (21).
Fig. 1.
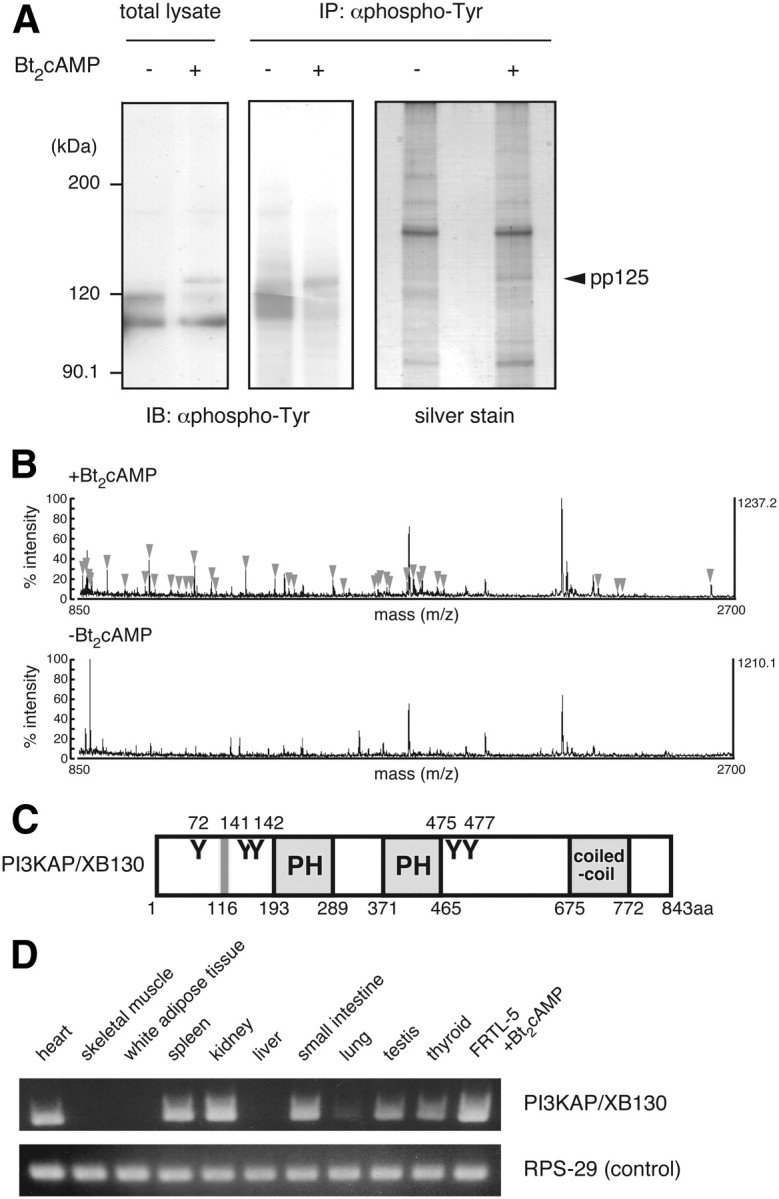
Identification of the 125-kDa tyrosine-phosphorylated protein (p125). A, Quiescent FRTL-5 cells were treated with 1 mm Bt2cAMP for 24 h. Cell lysates were prepared and subjected to immunoprecipitation with anti-phosphotyrosine (αphospho-Tyr) antibody, as described in Materials and Methods. Samples were analyzed by immunoblotting with anti-phosphotyrosine antibody or silver staining. B, The band corresponding to p125 in the cAMP-treated sample (indicated as pp125) was subjected to MALDI-TOF MS analysis. The gel region with a similar electrophoretic mobility in the untreated sample was used as a negative control. MS peaks detected only in the Bt2cAMP-treated sample (arrowheads) were analyzed by peptide mass finger printing. C, Schematic diagram of mouse PI3KAP/XB130. Y represents putative tyrosine phosphorylation sites for Src family kinases, PH is the PH domain, and coiled-coil is the coiled-coil domain. Gray line represents the SH3 domain-binding motif. Amino acid numbers of putative tyrosine phosphorylation sites, and ends of each domain/motif are indicated. D, PI3KAP/XB130 mRNA levels in rat tissues or FRTL-5 cells treated with 1 mm Bt2cAMP for 24 h were analyzed by RT-PCR. RPS-29 was analyzed as a loading control.
The schematic structure of PI3KAP/XB130 is shown in Fig. 1C, based on amino acid sequence analysis using Pfam (http://pfam.sanger.ac.uk/) and Scansite (http://scansite.mit.edu/). PI3KAP/XB130 contains two pleckstrin homology (PH) domains in the middle region and one coiled-coil domain in the C-terminal region. In addition, this protein possesses five tyrosine residues possibly phosphorylated, one of which could serve as a docking site for p85 PI3K.
We next examined tissue distribution of PI3KAP/XB130 in rats (Fig. 1D). PI3KAP/XB130 mRNA was detected in thyroid, testis, heart, spleen, kidney, and small intestine, and a weaker signal was detected in lung. By contrast, PI3KAP/XB130 mRNA was not detected in white adipose tissue, skeletal muscle, or liver.
TSH or cAMP treatment increased expression and tyrosine phosphorylation of PI3KAP/XB130, leading to the formation of a PI3KAP/XB130-PI3K complex
We then analyzed whether TSH regulates PI3KAP/XB130 gene expression in vivo to show physiological relevance of this gene. It is well known that treatment of rats with MMI, a potent inhibitor of thyroid hormone synthesis leads to hypothyroidism and increases TSH secretion, and this model is often used to study gene expression in response to TSH (22). Therefore, we treated rats with MMI at 0.03% in drinking water for 4 wk and then analyzed plasma TSH levels and PI3KAP/XB130 gene expression in thyroid. MMI treatment increased plasma TSH levels compared with control rats (Fig. 2A) and enlarged thyroid glands (Fig. 2B). Importantly, the expression of PI3KAP/XB130 mRNA was up-regulated in the thyroid of MMI-treated rats (Fig. 2C), suggesting that TSH does regulate PI3KAP/XB130 gene expression in vivo.
Fig. 2.
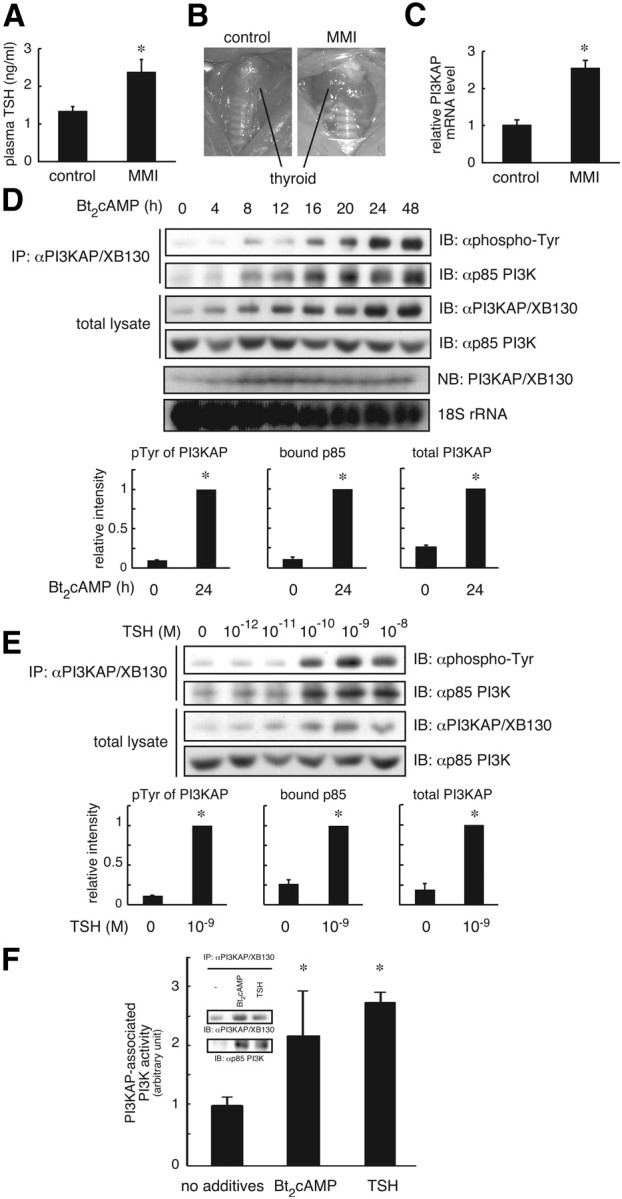
TSH or cAMP treatment increases gene expression and tyrosine phosphorylation of PI3KAP/XB130, leading to the increased formation of PI3KAP/XB130-PI3K complex. A–C, The 8-wk-old male Wistar rats were fed 0.03% MMI in drinking water for 4 wk; A, plasma TSH levels were evaluated by ELISA; B, thyroid glands were photographed before removal of any thyroid tissue, with the enlarged thyroid glands in MMI-treated rats and control thyroid glands indicated and representative photographs shown; C, relative mRNA levels of PI3KAP/XB130 in the thyroid glands were analyzed by real-time PCR. The means ± sem from seven rats are shown. D and E, Quiescent FRTL-5 cells were treated with 1 mm Bt2cAMP for the indicated hours or with indicated concentrations of TSH for 24 h. PI3KAP/XB130 protein levels, tyrosine phosphorylation, and its interaction with p85 PI3K were examined by immunoprecipitation (IP) and immunoblotting (IB) analysis. PI3KAP/XB130 mRNA levels were analyzed by Northern blotting (NB). The results of ethidium bromide staining of 18S rRNA are shown for a loading control. The means ± sem of the densitometric analysis results of three independent experiments are shown in the lower graphs. F, Quiescent FRTL-5 cells were treated with 1 nm TSH or 1 mm Bt2cAMP for 24 h. The cell lysates were subjected to immunoprecipitation followed by immunoblotting with indicated antibodies (inset), or PI3K activity was measured in the immunoprecipitates. The means ± sem of three replicate dishes are shown. Similar results were obtained in two independent experiments. *, P < 0.05.
We next studied whether cAMP treatment increases PI3KAP/XB130 tyrosine phosphorylation in FRTL-5 cells, which is an important functional modification of p125 as previously reported (16). PI3KAP/XB130 was immunoprecipitated with anti-PI3KAP/XB130 antibody, and then its tyrosine phosphorylation was analyzed by immunoblotting with an anti-phosphotyrosine antibody. As shown in Fig. 2D, PI3KAP/XB130 tyrosine phosphorylation was increased by cAMP treatment in a time-dependent manner. The increases in PI3KAP/XB130 tyrosine phosphorylation in response to cAMP treatment were parallel to increases in protein and mRNA levels of PI3KAP/XB130 (Fig. 2D). We confirmed TSH treatment also elevated tyrosine phosphorylation and protein expression of PI3KAP/XB130 in a concentration-dependent manner (Fig. 2E).
To examine effects of cAMP treatment on binding of PI3KAP/XB130 to PI3K, immunoprecipitated PI3KAP/XB130 complexes were analyzed for the presence of p85 PI3K. In agreement with elevation of tyrosine phosphorylation and protein levels, PI3KAP/XB130 interaction with p85 PI3K was also increased in response to cAMP or TSH treatment (Fig. 2, D and E). In addition, PI3K activities bound to PI3KAP/XB130 were 2- to 3-fold higher in TSH- or Bt2cAMP-treated cells (Fig. 2F).
We further explored intracellular localization of PI3KAP/XB130. Immunofluorescence analysis showed that PI3KAP/XB130 was colocalized with filamentous actin (F-actin) in the peripheral region in cAMP-treated cells as well as untreated cells (Supplemental Fig. 1, published on The Endocrine Society's Journals Online web site at http://mend.endojournals.org). These results are consistent with a recent report by Lodyga et al. (23) showing PI3KAP/XB130 localized to the lamellipodial F-actin meshwork.
PI3KAP/XB130 was necessary for cAMP-dependent potentiation of the mitogenic activity of IGF-I
To investigate roles of PI3KAP/XB130 in cell cycle progression in response to cAMP and IGF-I, we examined effects of PI3KAP/XB130 depletion on PI3K signaling and DNA synthesis in FRTL-5 cells using siRNA against PI3KAP/XB130. After siRNA transfection and serum starvation, cells were pretreated with Bt2cAMP for 24 h. In cells treated with Bt2cAMP, PI3KAP/XB130 protein levels were repressed 80% by PI3KAP/XB130 siRNA transfection (Fig. 3A). Knockdown of PI3KAP/XB130 was also determined at a single-cell level by immunofluorescence analysis (Supplemental Fig. 2A). In PI3KAP/XB130-depleted cells, tyrosine phosphorylation of PI3KAP/XB130 and its PI3K binding were decreased (Fig. 3B). Reduction in PI3K activities bound to PI3KAP/XB130 was also observed in these cells (Fig. 3C). After cAMP pretreatment, cells were treated with IGF-I for additional 24 h, and DNA synthesis was measured. In control siRNA-transfected cells, IGF-I-induced DNA synthesis was potentiated by cAMP pretreatment (Fig. 3D), as previously reported (11). By contrast, transfection of PI3KAP/XB130 siRNA attenuated this potentiating effect of cAMP (Fig. 3D). Similarly, bromodeoxyuridine (BrdU) uptake assay showed that BrdU-positive cells were increased by cAMP pretreatment and transfection of PI3KAP/XB130 siRNA inhibited this potentiation (Supplemental Fig. 2B). These results suggest that PI3KAP/XB130 mediated the potentiating effect of cAMP on IGF-I-induced DNA synthesis. Furthermore, reintroduction of PI3KAP/XB130 into siRNA-transfected cells showed a trend toward recovery of potentiation in two independent experiments (Supplemental Fig. 2B).
Fig. 3.
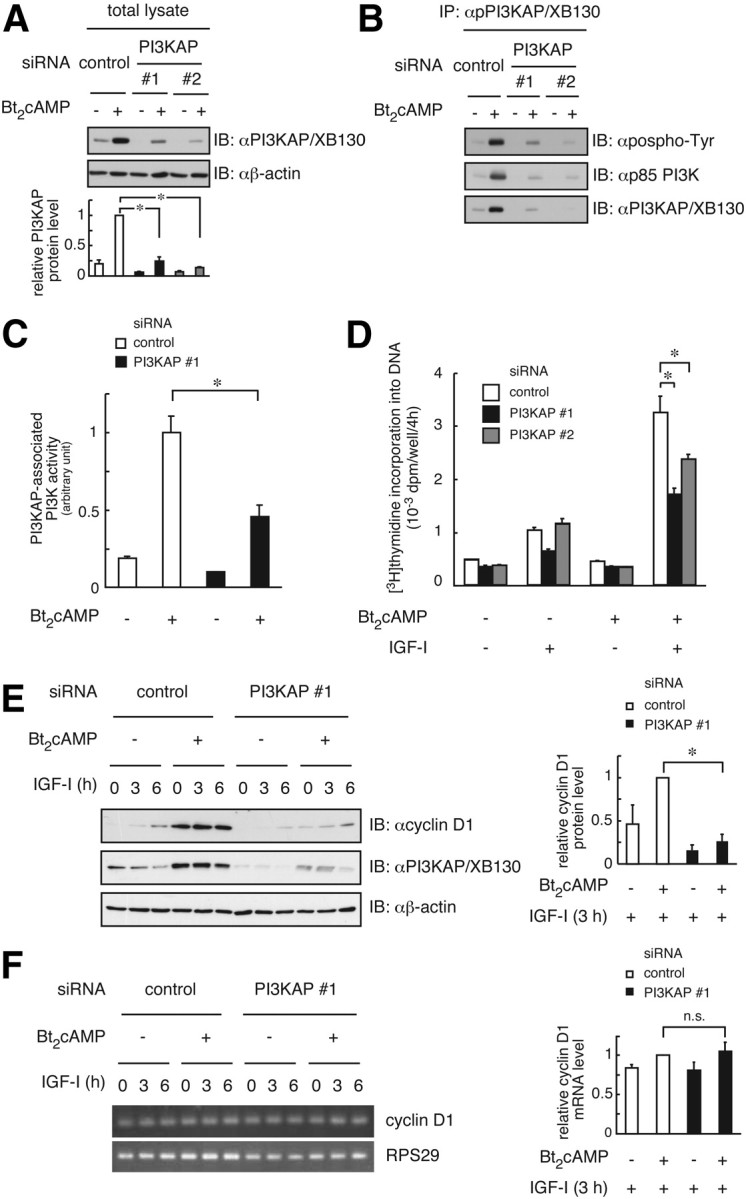
Knockdown of PI3KAP/XB130 suppresses cAMP-dependent potentiation of IGF-I-induced DNA synthesis in FRTL-5 cells. FRTL-5 cells were transfected with control siRNA or siRNA against PI3KAP/XB130. After serum starvation, cells were pretreated with or without 1 mm Bt2cAMP for 24 h. A, The knockdown efficiency was evaluated by immunoblotting (IB). The means ± sem of the densitometric analysis results of three independent experiments are shown in the lower graphs. B, PI3KAP/XB130 tyrosine phosphorylation and its interaction with p85 PI3K were examined by immunoprecipitation (IP) and immunoblotting analysis. C, PI3K activity was measured in the immunoprecipitates with anti-PI3KAP/XB130 antibody. The means ± sem of three replicate dishes are shown. Similar results were obtained in two independent experiments. D–F, After washing to remove reagents, cells were treated with 100 ng/ml IGF-I for 24 h (D) or 3–6 h (E and F). [Methyl-3H]thymidine incorporation into DNA was measured during the last 4 h (D). The means ± sem of three replicate wells are shown. Similar results were obtained in three independent experiments. Cyclin D1 protein levels were analyzed by immunoblotting (E), and the cyclin D1 mRNA levels were analyzed by RT-PCR (F). RPS29 was a loading control. The means ± sem of the densitometric analysis results of three independent experiments are shown in the right graphs (E and F). *, P < 0.05. n.s., Not significant.
Next, we tested the roles of PI3KAP/XB130 in changes in mRNA and protein levels of cyclin D1, which is important for cell cycle progression from G1 phase to S phase. cAMP treatment increased cyclin D1 protein levels, and knockdown of PI3KAP/XB130 reduced them (Fig. 3E), indicating that PI3KAP/XB130 mediates cAMP-dependent enhancement of IGF-I mitogenic activity partly via elevating cyclin D1 levels. We also found that knockdown of PI3KAP/XB130 did not affect cyclin D1 mRNA levels (Fig. 3F). These data raised the possibility that PI3KAP/XB130 mediates up-regulation of cyclin D1 protein levels through increasing its translation, which is consistent with our previous report that cAMP-dependent activation of PI3K enhanced cyclin D1 translation (24).
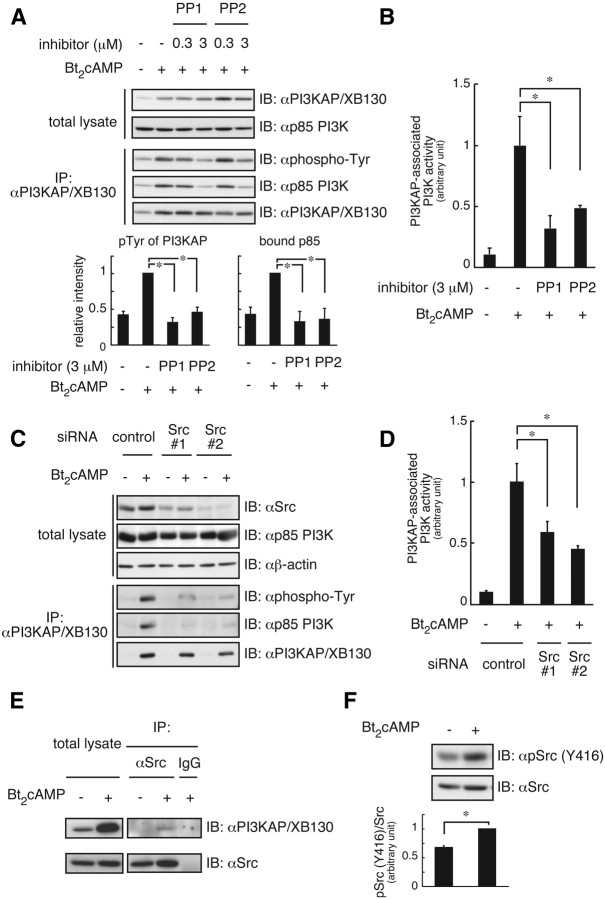
PI3KAP/XB130 was tyrosine phosphorylated by Src family tyrosine kinases followed by binding to PI3K
We further determined the involvement of c-Src in tyrosine phosphorylation of PI3KAP/XB130 in cAMP-treated FRTL-5 cells using PP1 or PP2, specific inhibitors of Src family kinases. Figure 4A demonstrates cAMP-dependent increases in tyrosine phosphorylation of PI3KAP/XB130 and its interaction with PI3K were inhibited by addition of PP1 or PP2. PI3K activities bound to PI3KAP/XB130 were also decreased by treatment with PP1 or PP2 (Fig. 4B). Moreover c-Src knockdown attenuated tyrosine phosphorylation of PI3KAP/XB130, its PI3K binding (Fig. 4C), and PI3KAP/XB130-associated PI3K activities (Fig. 4D). We further tested whether cAMP treatment could increase interaction between PI3KAP/XB130 and c-Src. As shown in Fig. 4E, c-Src interacted with PI3KAP/XB130 in response to cAMP treatment. In addition, Western blotting analysis using a phospho-specific Src antibody, which recognizes the activated form of this kinase, revealed that cAMP treatment resulted in an increase in c-Src phosphorylation at tyrosine 416, suggesting its activation (Fig. 4F). We also investigated whether tyrosine phosphorylation of PI3KAP/XB130 regulated intracellular localization of this protein and found that PP1 or PP2 treatment did not affect colocalization of PI3KAP/XB130 with F-actin (Supplemental Fig. 3). These results indicate that activation of Src family tyrosine kinases in response to cAMP may be required for the tyrosine phosphorylation of PI3KAP/XB130 that leads to its interaction with p85 PI3K.
Fig. 4.
PI3KAP/XB130 is phosphorylated by Src family tyrosine kinases and forms a complex with PI3K. Quiescent FRTL-5 cells were treated with or without 1 mm Bt2cAMP for 24 h. A and B, During cAMP treatment, the indicated concentrations of PP1 or PP2 were added. PI3KAP/XB130 protein levels, tyrosine phosphorylation, and its interaction with p85 PI3K were examined by immunoprecipitation (IP) and immunoblotting (IB) analysis. The means ± sem of the densitometric analysis results of three independent experiments are shown in the lower graphs (A). PI3K activity was measured in the immunoprecipitates with anti-PI3KAP/XB130 antibody. The means ± sem of three independent experiments are shown (B). C and D, Before serum starvation and cAMP treatment, FRTL-5 cells were transfected with control siRNA or siRNA against Src. PI3KAP/XB130 protein levels, tyrosine phosphorylation, and its interaction with p85 PI3K were examined by immunoprecipitation and immunoblotting analysis (C). PI3K activity was measured in the immunoprecipitates with anti-PI3KAP/XB130 antibody. The means ± sem of three replicate dishes are shown (D). E, The interaction of PI3KAP/XB130 with Src was analyzed by immunoprecipitation followed by immunoblotting with the indicated antibodies. Nonimmune IgG was used as a negative control. Similar results were obtained in two independent experiments. F, Src protein and the Y416 phosphorylation levels were analyzed by immunoblotting. The means ± sem of the densitometric analysis results of three independent experiments are shown as pSrc to total Src ratio in the graphs. *, P < 0.05.
The binding of PI3K to PI3KAP/XB130 was necessary for potentiation of the mitogenic activity of IGF-I
To determine the roles of PI3KAP/XB130 in association with PI3K in IGF-I-dependent DNA synthesis, we generated a mutant of PI3KAP/XB130 that does not bind to PI3K and examined effects of overexpression of wild-type or mutant PI3KAP/XB130 on IGF-I-dependent DNA synthesis. Because PI3KAP/XB130 contains one tyrosine (tyrosine 72) in the YXXM motif, a putative binding site for p85 Src homology 2 (SH2) domains (25), this tyrosine was mutated to phenylalanine (Y72F). First we tried to obtain FRTL-5 clones in which PI3KAP/XB130 is stably knocked down followed by expression of wild-type or mutated PI3KAP/XB130 to evaluate the effects of these proteins for mitogenic activity of IGF-I. However, because PI3KAP/XB130 is required for proliferation of FRTL-5 cells treated with TSH and high concentrations of insulin in the presence of serum, we could not establish PI3KAP/XB130 knocked-down FRTL-5 cells. Instead, wild-type PI3KAP/XB130 or the Y72F mutant was expressed in NIH3T3 cells, which lack endogenous protein expression of PI3KAP/XB130 (Fig. 5A). Both wild-type PI3KAP and the Y72F mutant were tyrosine phosphorylated (Fig. 5A), indicating that tyrosine residues other than Y72 are phosphorylated. Wild-type PI3KAP/XB130 was bound to p85 PI3K in these cells, whereas binding of the Y72F mutant to p85 PI3K was not observed (Fig. 5A), confirming that Y72 is required for PI3K binding. Wild-type or mutant PI3KAP/XB130-expressing NIH3T3 cells were stimulated with IGF-I, and then DNA synthesis was measured. As shown in Fig. 5B, expression of PI3KAP/XB130 enhanced IGF-I-dependent DNA synthesis, but this enhancement was not observed in cells expressing the Y72F mutant (Fig. 5B). These results suggested that binding of PI3K to PI3KAP/XB130 potentiated IGF-I-induced DNA synthesis.
Fig. 5.
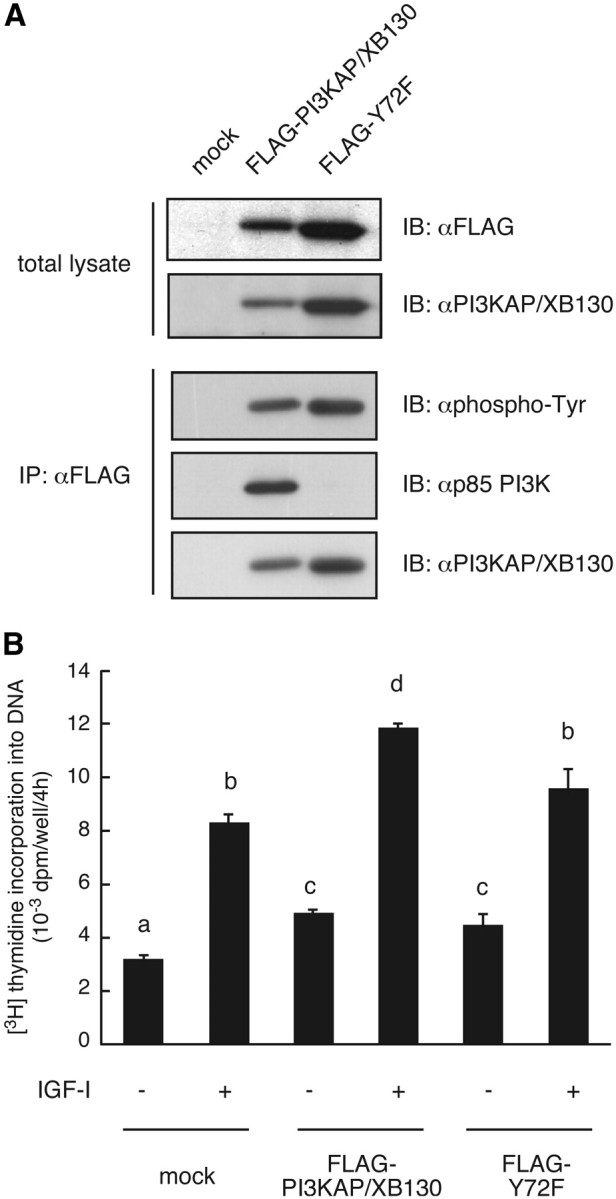
Interaction of PI3KAP/XB130 with PI3K is required for potentiation of DNA synthesis induced by IGF-I. NIH3T3 cells were infected with control retrovirus pMXs-neo (mock) or retrovirus encoding FLAG-PI3KAP/XB130 or FLAG-Y72F. A, The cell lysates were subjected to immunoprecipitation (IP) with anti-FLAG antibody. The immunoprecipitates and the total cell lysates were subjected to immunoblotting (IB) with indicated antibodies. B, The retrovirus-infected cells were serum starved for 24 h and then treated with 100 ng/ml IGF-I for 16 h. [Methyl-3H]thymidine incorporation into DNA was measured during the last 4 h. The means ± sem of three replicate wells are shown. There are significant differences between values with different letters (P < 0.05). Similar results were obtained in three independent experiments.
Discussion
In general, short-term activation of the cAMP-dependent pathway is shown to inhibit growth factor-dependent signaling and bioactivities through repression of activation of downstream serine/threonine kinases such as Raf for example. In endocrine cells, the cAMP-dependent pathway has been thought to be important for cell differentiation and maintenance of cell functions (40). However, we and others found that long-term activation of the cAMP-dependent pathway by tropic hormones promoted growth factor-dependent cell cycle progression in endocrine cells including thyroid follicular cells. Specifically, our previous studies have shown that the PI3K activated by long-term cAMP stimulus plays an important role in enhancement of IGF-I-induced DNA synthesis in FRTL-5 cells and have suggested involvement of a 125-kDa phosphotyrosyl protein p125 in this process (17). In the present study, we identified p125 as PI3KAP/XB130 using MALDI-TOF MS. In addition, our study demonstrated that cAMP-dependent increases in PI3KAP/XB130 mRNA and protein levels were associated with increased tyrosine phosphorylation of PI3KAP/XB130 followed by its interaction with PI3K and with increased activity of PI3K bound to PI3KAP/XB130 (Fig. 2). These results suggest that PI3KAP/XB130 can mediate the long-term activation of the cAMP-dependent pathway via PI3K.
PI3KAP/XB130 has 39% homology with AFAP-110 in amino acid sequences, which has been reported to alter actin filament integrity and to be involved in podosome formation and conversion of mechanical force into biochemical signaling (26–28). AFAP-110 also has two PH domains, a potential SH3 domain-binding motif, and several tyrosine phosphorylation sites containing SH2-domain-binding motifs, a structure similar to PI3KAP/XB130 (Fig. 1). These structural similarities suggest that PI3KAP/XB130 and AFAP-110 belong to a new family of proteins, as proposed by Xu et al. (20). Colocalization of PI3KAP/XB130 with F-actin, and disruption of cortical actin structures in PI3KAP/XB130-depleted cells were observed in our results (Supplemental Fig. 2). Similar disruption reported by Lodyga et al. (23) was attributed to conserved characteristics of this family in the aspects of intracellular localization and actin cytoskeleton organization. However, there are interesting structural differences between these proteins. For example, AFAP-110 seems to have no tyrosine phosphorylation site recognized by p85 PI3K, suggesting a characteristic role of PI3KAP/XB130 in the activation of the PI3K signaling pathway.
We found that PI3KAP/XB130 is expressed in various tissues (Fig. 1D). Lodyga et al. (29) reported that PI3KAP/XB130 was detected in human thyroid tissues and papillary thyroid cancer and in TPC1 papillary thyroid cancer cells. These results suggest that PI3KAP/XB130 functions in a variety of tissues and cell types. In this study, we for the first time showed the marked increases in PI3KAP/XB130 mRNA in thyroid cells stimulated with TSH or Bt2cAMP (Fig. 2), which are consistent with cAMP-dependent transcriptional regulation of PI3KAP/XB130. Moreover, our results showing that PI3KAP/XB130 gene expression is up-regulated in thyroid of MMI-treated rats (Fig. 2) suggest physiological significance of regulation of PI3KAP/XB130 gene expression in vivo. It has been reported that TSH stimulation induces protein kinase A (PKA) activation followed by cAMP-response element binding protein (CREB) phosphorylation within an hour in thyroid cells (30), and PKA/CREB is a candidate for signaling molecules involved in the early steps that regulate PI3KAP/XB130 gene expression. Therefore, we searched for a cAMP-responsive element using the 5′-flanking region of mouse and human PI3KAP/XB130 genes. However, we did not find a typical cAMP-responsive element within this region. We speculate that CREB does not directly regulate PI3KAP/XB130 gene expression but instead regulates through induction or activation of intermediate transcription factors. Additional investigation is necessary to elucidate mechanisms for transcriptional regulation of PI3KAP/XB130.
We showed that cAMP treatment increased interaction between PI3KAP/XB130 and c-Src (Fig. 4). PI3KAP/XB130 possesses SH2 or SH3 domain-binding motifs (Fig. 1), and it has been reported that Src SH2 domain and SH3 domain bind to PI3KAP/XB130 (20). Thus, these binding motifs may be binding sites for c-Src. In addition, the weak interaction between PI3KAP/XB130 and c-Src suggests an enzyme-substrate relationship between these two proteins. We also demonstrated that cAMP treatment induced c-Src activation in FRTL-5 cells (Fig. 4). Xu et al. (20) reported that c-Src could be activated by overexpression of PI3KAP/XB130 in COS7 cells (20), raising the possibility that PI3KAP/XB130 itself contributed to cAMP-induced c-Src activation in FRTL-5 cells. To evaluate this possibility, we measured the c-Src activity in FRTL-5 cells transfected with PI3KAP/XB130 siRNA. Our results showed that PI3KAP/XB130 knockdown did not affect c-Src autophosphorylation significantly (data not shown). These results suggest that cAMP-dependent c-Src activation may occur through mechanisms independent of PI3KAP/XB130. Because PKA phosphorylates and activates c-Src in other cell lines (31, 32), PKA may be involved in cAMP-induced c-Src activation in FRTL-5 cells. Because up-regulation of PI3KAP/XB130 and c-Src activation were required for binding of PI3KAP/XB130 to PI3K (Figs. 3 and 4), at least two pathways appear to be required for PI3K binding: 1) cAMP stimulation increases PI3KAP/XB130 gene expression, and 2) cAMP stimulation increases Src activity. These two pathways may converge to markedly increase tyrosine-phosphorylated PI3KAP/XB130 and its binding to PI3K. Our results showing reduced PI3K activities associated with PI3KAP/XB130 in PI3KAP/XB130-depleted cell or PP1/PP2-treated cells (Figs. 3 and 4) support our conclusion that these pathways are required for cAMP-dependent activation of PI3K pathways.
It has been reported that PI3KAP/XB130 is also phosphorylated by Rearranged in Transformation/Papillary Thyroid Carcinomas (RET/PTC), a constitutively active oncogenic tyrosine kinase, resulting in increased binding of PI3K (29). Our results indicated that even in nontransformed cells, PI3KAP/XB130 was phosphorylated by c-Src in response to a physiological stimulus, such as cAMP. These results suggest that under different physiological conditions, different tyrosine kinases phosphorylate PI3KAP/XB130 resulting in activation of the PI3K pathways.
We demonstrated that tyrosine 72 in the YXXM motif was required for its p85 binding (Fig. 5). Given the importance of the YIYM motif and its conservation in rat and human, we searched the NCBI BLAST database for orthologs of rat PI3KAP. We found putative orthologs of PI3KAP/XB130 in cattle (NP_001069843), dog (XP_544028), chicken (XP_421770), frog (NP_001079952), and zebrafish (XP_695754). The YIYM sequence was conserved in all of them, suggesting that the PI3KAP/XB130-dependent PI3K pathway may be evolutionarily conserved, at least in vertebrates.
Two sets of experiments, one showing suppression of PI3KAP/XB130 reduced potentiation of IGF-I-induced DNA synthesis (Fig. 3) and the other showing overexpression of wild-type but not Y72F mutant PI3KAP/XB130 enhanced potentiation (Fig. 5), demonstrate the key role of PI3KAP/XB130, specifically Y72 in this potentiation. The tyrosine phosphorylation level of the Y72F mutant comparable to wild type (Fig. 5) indicates existence of tyrosine phosphorylation sites other than Y72. Tyrosine residues phosphorylated in AFAP-110 by c-Src (33) are conserved in PI3KAP/XB130 (Y142, 474, and 477), and these sites are candidates for tyrosine phosphorylation sites. Our finding that the knockdown of PI3KAP/XB130 was associated with reduced cyclin D1 protein levels and unchanged mRNA levels (Fig. 3) is consistent with our previous studies showing that cAMP-dependent PI3K activation is required for increased cyclin D1 translation and enhanced IGF-I-dependent DNA synthesis (17, 24). Taken together with a report that down-regulation of PI3KAP/XB130 inhibited cell cycle progression in TPC1 papillary thyroid cancer cells (29), these results suggest PI3KAP/XB130 plays an important role in cell cycle progression of nontransformed thyroid cells as well as thyroid tumor cells.
The gradual increase in PI3KAP/XB130 in response to cAMP and concomitant formation of complexes with PI3K (Fig. 2) may lead to conversion of short-term cAMP signaling to the prolonged activation of PI3K we previously reported (17). As described above, cAMP-dependent prolonged activation of PI3K appears to mediate enhancement of translation of cell cycle regulatory genes including cyclin D1. Analysis of mechanisms for the regulation of translation mediated by prolonged activation of PI3K pathways is in progress in our laboratory. Although many studies have focused on transient PI3K activation triggered by extracellular stimuli (34–36), it is becoming clear that prolonged activation of PI3K is also important for cell proliferation of fibroblasts and thyrocytes (17, 37) and for differentiation of ovarian follicular cells (38) and preadipocytes (39). Therefore, it is necessary to shed light on molecular mechanisms of biological effects mediated by the long-term PI3K activation. We propose that PI3KAP/XB130 may play important roles in the long-term activation of the PI3K pathway in a novel manner.
In summary, our data show that long-term cAMP stimulus increases mRNA and protein expression of PI3KAP/XB130. Concomitantly, cAMP stimulus leads to c-Src activation, which is required for PI3KAP/XB130 tyrosine phosphorylation followed by its binding to PI3K. These processes are necessary for cAMP-dependent amplification of IGF-I-induced cell proliferation, and PI3KAP/XB130 plays a key role in the augmentation of bioactivities of IGF-I.
Acknowledgments
We thank Dr. Leonard Kohn (Edison Biotechnology Institute, Ohio University, Athens, OH) and Interthyr Research Foundation (Baltimore, MD) for the gift of FRTL-5 cells, the National Hormone and Pituitary Program (National Institute of Diabetes and Digestive and Kidney Diseases) for providing bovine TSH, and Dr. Toshio Kitamura (the Institute of Medical Science, the University of Tokyo, Tokyo, Japan) for the kind gift of PLAT-E cells and pMXs-neo vector. We also thank Dr. Toshiaki Ohkuma and Kitajima Mitsuhiro (Fujisawa Pharmaceutical Co., Osaka, Japan, currently Astellas Pharma Inc., Tokyo, Japan) for the donation of recombinant human IGF-I and Dr. Shinji Nagata (Department of Agriculture and Life Sciences, the University of Tokyo, Tokyo, Japan) for the helpful advice on mass spectrometry analysis. We appreciate helpful discussions during our experiments and the writing of our manuscript with Dr. Asako Takenaka (Faculty of Agriculture, Meiji University, Kanagawa, Japan) and Dr. Susan H. Hall (The University of North Carolina at Chapel Hill, Chapel Hill, NC).
This work was supported by a Grant-in-Aid for the Scientific Research Fund of the Ministry of Education, Science, Culture, and Sports, Japan, to S.-I.T. [(A)16208028 and (A)22248030] and to F.H. [(C)21580345].
Present address for T.F.: Department of Medical Science, Graduate School of Medicine, University of Hiroshima, 1-2-3 Kasumi, Minami-ku, Hiroshima City, Hiroshima 734-8553, Japan.
Present address for T.N.: Department of Applied Biosciences, Graduate School of Life Sciences, Toyo University, 1-1-1 Izumino, Itakura-machi, Ora-gun, Gunma 374-0193, Japan.
D.Y., T.A., K.C., S.M., K.S., F.H., and S.-I.T. designed research; D.Y., T.A., T.F., T.N., and C.K. conducted research and analyzed data; D.Y., T.A., F.H., and S.-I.T. wrote paper. S.-I.T. had primary responsibility for final content. All authors read and approved the final manuscript.
Disclosure Summary: The authors have nothing to disclose.
Footnotes
- AFAP
- Actin filament-associated protein
- BrdU
- bromodeoxyuridine
- Bt2cAMP
- dibutyryl cAMP
- CREB
- cAMP-response element binding protein
- DTT
- dithiothreitol
- F-actin
- filamentous actin
- FBS
- fetal bovine serum
- HRP
- horseradish peroxidase
- MALDI-TOF MS
- matrix-assisted laser desorption ionization-time of flight mass spectrometry
- MMI
- methimazole
- nts
- nucleotides
- p125
- 125-kDa protein
- PH
- pleckstrin homology
- PI3K
- phosphatidylinositol 3-kinase
- PI3KAP
- PI3K-associated protein
- PKA
- protein kinase A
- SDS
- sodium dodecyl sulfate
- SH2
- Src homology 2
- siRNA
- small interfering RNA
- TFA
- trifluoroacetic acid.
References
- 1. Powell-Braxton L , Hollingshead P , Warburton C , Dowd M , Pitts-Meek S , Dalton D , Gillett N , Stewart TA. 1993. IGF-I is required for normal embryonic growth in mice. Genes Dev 7:2609–2617 [DOI] [PubMed] [Google Scholar]
- 2. Baker J , Liu JP , Robertson EJ , Efstratiadis A. 1993. Role of insulin-like growth factors in embryonic and postnatal growth. Cell 75:73–82 [PubMed] [Google Scholar]
- 3. Liu JP , Baker J , Perkins AS , Robertson EJ , Efstratiadis A. 1993. Mice carrying null mutations of the genes encoding insulin-like growth factor I (Igf-1) and type 1 IGF receptor (Igf1r). Cell 75:59–72 [PubMed] [Google Scholar]
- 4. Jones JI , Clemmons DR. 1995. Insulin-like growth factors and their binding proteins: biological actions. Endocr Rev 16:3–34 [DOI] [PubMed] [Google Scholar]
- 5. Qureshi FG , Tchorzewski MT , Duncan MD , Harmon JW. 1997. EGF and IGF-I synergistically stimulate proliferation of human esophageal epithelial cells. J Surg Res 69:354–358 [DOI] [PubMed] [Google Scholar]
- 6. Stiles CD , Capone GT , Scher CD , Antoniades HN , Van Wyk JJ , Pledger WJ. 1979. Dual control of cell growth by somatomedins and platelet-derived growth factor. Proc Natl Acad Sci USA 76:1279–1283 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Stewart AJ , Johnson MD , May FE , Westley BR. 1990. Role of insulin-like growth factors and the type I insulin-like growth factor receptor in the estrogen-stimulated proliferation of human breast cancer cells. J Biol Chem 265:21172–21178 [PubMed] [Google Scholar]
- 8. Itagane Y , Inada H , Fujita K , Isshiki G. 1991. Interactions between steroid hormones and insulin-like growth factor-I in rabbit chondrocytes. Endocrinology 128:1419–1424 [DOI] [PubMed] [Google Scholar]
- 9. Meroni SB , Riera MF , Pellizzari EH , Galardo MN , Cigorraga SB. 2004. FSH activates phosphatidylinositol 3-kinase/protein kinase B signaling pathway in 20-day-old Sertoli cells independently of IGF-I. J Endocrinol 180:257–265 [DOI] [PubMed] [Google Scholar]
- 10. Tramontano D , Moses AC , Veneziani BM , Ingbar SH. 1988. Adenosine 3′,5′-monophosphate mediates both the mitogenic effect of thyrotropin and its ability to amplify the response to insulin-like growth factor I in FRTL5 cells. Endocrinology 122:127–132 [DOI] [PubMed] [Google Scholar]
- 11. Takahashi S , Conti M , Van Wyk JJ. 1990. Thyrotropin potentiation of insulin-like growth factor-I dependent deoxribonucleic acid synthesis in FRTL-5 cells: mediation by an autocrine amplification factor(s). Endocrinology 126:736–745 [DOI] [PubMed] [Google Scholar]
- 12. Cheung NW , Lou JC , Boyages SC. 1996. Growth hormone does not increase thyroid size in the absence of thyrotropin: a study in adults with hypopituitarism. J Clin Endocrinol Metab 81:1179–1183 [DOI] [PubMed] [Google Scholar]
- 13. Clément S , Refetoff S , Robaye B , Dumont JE , Schurmans S. 2001. Low TSH requirement and goiter in transgenic mice overexpressing IGF-I and IGF-Ir receptor in the thyroid gland. Endocrinology 142:5131–5139 [DOI] [PubMed] [Google Scholar]
- 14. Cheung NW , Boyages SC. 1997. The thyroid gland in acromegaly: an ultrasonographic study. Clin Endocrinol (Oxf) 46:545–549 [DOI] [PubMed] [Google Scholar]
- 15. Ambesi-Impiombato FS , Parks LA , Coon HG. 1980. Culture of hormone-dependent functional epithelial cells from rat thyroids. Proc Natl Acad Sci USA 77:3455–3459 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Takahashi S , Conti M , Prokop C , Van Wyk JJ , Earp HS. 1991. Thyrotropin and insulin-like growth factor I regulation of tyrosine phosphorylation in FRTL-5 cells. Interaction between cAMP-dependent and growth factor-dependent signal transduction. J Biol Chem 266:7834–7841 [PubMed] [Google Scholar]
- 17. Nedachi T , Akahori M , Ariga M , Sakamoto H , Suzuki N , Umesaki K , Hakuno F , Takahashi SI. 2000. Tyrosine kinase and phosphatidylinositol 3-kinase activation are required for cyclic adenosine 3′,5′-monophosphate-dependent potentiation of deoxyribonucleic acid synthesis induced by insulin-like growth factor-I in FRTL-5 cells. Endocrinology 141:2429–2438 [DOI] [PubMed] [Google Scholar]
- 18. Ho SN , Hunt HD , Horton RM , Pullen JK , Pease LR. 1989. Site-directed mutagenesis by overlap extension using the polymerase chain reaction. Gene 77:51–59 [DOI] [PubMed] [Google Scholar]
- 19. Kane S , Sano H , Liu SC , Asara JM , Lane WS , Garner CC , Lienhard GE. 2002. A method to identify serine kinase substrates. Akt phosphorylates a novel adipocyte protein with a Rab GTPase-activating protein (GAP) domain. J Biol Chem 277:22115–22118 [DOI] [PubMed] [Google Scholar]
- 20. Xu J , Bai XH , Lodyga M , Han B , Xiao H , Keshavjee S , Hu J , Zhang H , Yang BB , Liu M. 2007. XB130, a novel adaptor protein for signal transduction. J Biol Chem 282:16401–16412 [DOI] [PubMed] [Google Scholar]
- 21. Lodyga M , Bai XH , Mourgeon E , Han B , Keshavjee S , Liu M. 2002. Molecular cloning of actin filament-associated protein: a putative adaptor in stretch-induced Src activation. Am J Physiol Lung Cell Mol Physiol 283:L265–L274 [DOI] [PubMed] [Google Scholar]
- 22. Grozovsky R , Morales MM , Carvalho DP. 2007. Biphasic modulation of insulin receptor substrate-1 during goitrogenesis. Braz J Med Biol Res 40:679–686 [DOI] [PubMed] [Google Scholar]
- 23. Lodyga M , Bai XH , Kapus A , Liu M. 2010. Adaptor protein XB130 is a Rac-controlled component of lamellipodia that regulates cell motility and invasion. J Cell Sci 123:4156–4169 [DOI] [PubMed] [Google Scholar]
- 24. Fukushima T , Nedachi T , Akizawa H , Akahori M , Hakuno F , Takahashi S. 2008. Distinct modes of activation of phosphatidylinositol 3-kinase in response to cyclic adenosine 3′, 5′-monophosphate or insulin-like growth factor I play different roles in regulation of cyclin D1 and p27Kip1 in FRTL-5 cells. Endocrinology 149:3729–3742 [DOI] [PubMed] [Google Scholar]
- 25. Songyang Z , Shoelson SE , Chaudhuri M , Gish G , Pawson T , Haser WG , King F , Roberts T , Ratnofsky S , Lechleider RJ , Neel BG , Birge RB , Fajardo JE , Chou MM , Hanafusa H , Schaffhausen B , Cantley LC. 1993. SH2 domains recognize specific phosphopeptide sequences. Cell 72:767–778 [DOI] [PubMed] [Google Scholar]
- 26. Baisden JM , Qian Y , Zot HM , Flynn DC. 2001. The actin filament-associated protein AFAP-110 is an adaptor protein that modulates changes in actin filament integrity. Oncogene 20:6435–6447 [DOI] [PubMed] [Google Scholar]
- 27. Gatesman A , Walker VG , Baisden JM , Weed SA , Flynn DC. 2004. Protein kinase Cα activates c-Src and induces podosome formation via AFAP-110. Mol Cell Biol 24:7578–7597 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Han B , Bai XH , Lodyga M , Xu J , Yang BB , Keshavjee S , Post M , Liu M. 2004. Conversion of mechanical force into biochemical signaling. J Biol Chem 279:54793–55801 [DOI] [PubMed] [Google Scholar]
- 29. Lodyga M , De Falco V , Bai XH , Kapus A , Melillo RM , Santoro M , Liu M. 2009. XB130, a tissue-specific adaptor protein that couples the RET/PTC oncogenic kinase to PI 3-kinase pathway. Oncogene 28:937–949 [DOI] [PubMed] [Google Scholar]
- 30. Uyttersprot N , Costagliola S , Dumont JE , Miot F. 1999. Requirement for cAMP-response element (CRE) binding protein/CRE modulator transcription factors in thyrotropin-induced proliferation of dog thyroid cells in primary culture. Eur J Biochem 259:370–378 [DOI] [PubMed] [Google Scholar]
- 31. Obara Y , Labudda K , Dillon TJ , Stork PJ. 2004. PKA phosphorylation of Src mediates Rap1 activation in NGF and cAMP signaling in PC12 cells. J Cell Sci 117:6085–6094 [DOI] [PubMed] [Google Scholar]
- 32. Schmitt JM , Stork PJ. 2002. PKA phosphorylation of Src mediates cAMP's inhibition of cell growth via Rap1. Mol Cell 9:85–94 [DOI] [PubMed] [Google Scholar]
- 33. Guappone AC , Weimer T , Flynn DC. 1998. Formation of a stable src-AFAP-110 complex through either an amino-terminal or a carboxy-terminal SH2-binding motif. Mol Carcinog 22:110–119 [DOI] [PubMed] [Google Scholar]
- 34. Kong M , Mounier C , Wu J , Posner BI. 2000. Epidermal growth factor-induced phosphatidylinositol 3-kinase activation and DNA synthesis. Identification of Grb2-associated binder 2 as the major mediator in rat hepatocytes. J Biol Chem 275:36035–36042 [DOI] [PubMed] [Google Scholar]
- 35. Lawlor MA , Feng X , Everding DR , Sieger K , Stewart CE , Rotwein P. 2000. Dual control of muscle cell survival by distinct growth factor-regulated signaling pathways. Mol Cell Biol 20:3256–3265 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36. Cheatham B , Vlahos CJ , Cheatham L , Wang L , Blenis J , Kahn CR. 1994. Phosphatidylinositol 3-kinase activation is required for insulin stimulation of pp70 S6 kinase, DNA synthesis, and glucose transporter translocation. Mol Cell Biol 14:4902–4911 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37. Jones SM , Kazlauskas A. 2001. Growth-factor-dependent mitogenesis requires two distinct phases of signalling. Nat Cell Biol 3:165–172 [DOI] [PubMed] [Google Scholar]
- 38. Alam H , Maizels ET , Park Y , Ghaey S , Feiger ZJ , Chandel NS , Hunzicker-Dunn M. 2004. Follicle-stimulating hormone activation of hypoxia-inducible factor-1 by the phosphatidylinositol 3-kinase/AKT/Ras homolog enriched in brain (Rheb)/mammalian target of rapamycin (mTOR) pathway is necessary for induction of select protein markers of follicular differentiation. J Biol Chem 279:19431–19440 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39. Xia X , Serrero G. 1999. Inhibition of adipose differentiation by phosphatidylinositol 3-kinase inhibitors. J Cell Physiol 178:9–16 [DOI] [PubMed] [Google Scholar]
- 40. Adashi EY , Resnick CE. 1984. Forskolin-induced differentiation of cultured rat granulosa cells: new evidence for an intermediary role of adenosine 3′,5′-monophosphate in the mechanism of action of follicle-stimulating hormone. Endocrinology 115:183–190 [DOI] [PubMed] [Google Scholar]