Abstract
Androgen receptor (AR) is a major therapeutic target that plays pivotal roles in prostate cancer (PCa) and androgen insensitivity syndromes. We previously proposed that compounds recruited to ligand-binding domain (LBD) surfaces could regulate AR activity in hormone-refractory PCa and discovered several surface modulators of AR function. Surprisingly, the most effective compounds bound preferentially to a surface of unknown function [binding function 3 (BF-3)] instead of the coactivator-binding site [activation function 2 (AF-2)]. Different BF-3 mutations have been identified in PCa or androgen insensitivity syndrome patients, and they can strongly affect AR activity. Further, comparison of AR x-ray structures with and without bound ligands at BF-3 and AF-2 showed structural coupling between both pockets. Here, we combine experimental evidence and molecular dynamic simulations to investigate whether BF-3 mutations affect AR LBD function and dynamics possibly via allosteric conversation between surface sites. Our data indicate that AF-2 conformation is indeed closely coupled to BF-3 and provide mechanistic proof of their structural interconnection. BF-3 mutations may function as allosteric elicitors, probably shifting the AR LBD conformational ensemble toward conformations that alter AF-2 propensity to reorganize into subpockets that accommodate N-terminal domain and coactivator peptides. The induced conformation may result in either increased or decreased AR activity. Activating BF-3 mutations also favor the formation of another pocket (BF-4) in the vicinity of AF-2 and BF-3, which we also previously identified as a hot spot for a small compound. We discuss the possibility that BF-3 may be a protein-docking site that binds to the N-terminal domain and corepressors. AR surface sites are attractive pharmacological targets to develop allosteric modulators that might be alternative lead compounds for drug design.
Androgen receptor (AR, NR3C4) is a ligand-activated transcription factor (1) that belongs to the nuclear receptor (NR) superfamily (2). AR plays specific roles in male development, prostate cancer (PCa), androgen insensitivity syndromes (AIS), and the rare neurodegenerative spinal and bulbar muscular atrophy (3–13). Like other NR, AR displays a modular architecture, composed of an N-terminal domain (NTD), a DNA-binding domain (DBD), a hinge region, and a C-terminal ligand-binding domain (LBD) (14). AR LBD adopts the canonical NR LBD fold (15): a three-layered α-helical sandwich with the ligand buried inside the hydrophobic ligand-binding pocket (LBP) [Fig. 1, supporting information (SI); and Table 1]. The LBD harbors a major coactivator binding surface [activation function-2 (AF-2)], which acts as a docking site for short hydrophobic peptide motifs (NR boxes) featured in AR coactivators and in the AR NTD and mediates AR functional amino/carboxy (N/C)-terminal interaction (16–20).
Fig. 1.
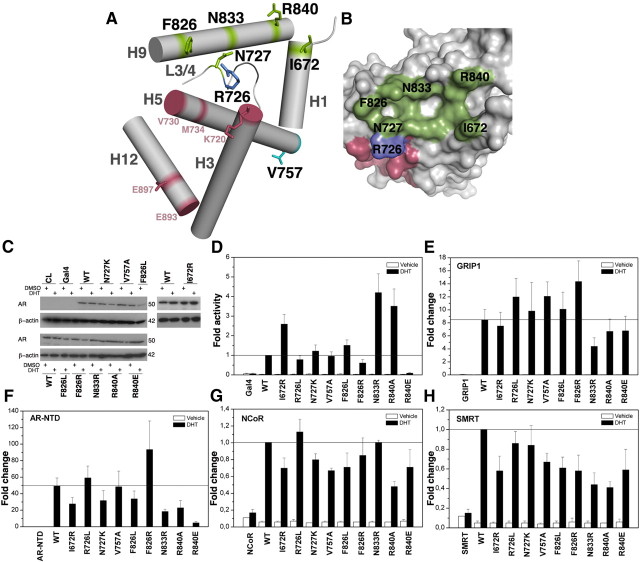
Mutations in AR LBD, AF-2 transcriptional activation, and mammalian two-hybrid assays. A, Simplified model representation of AR LBD structure. Helices H1, H3, H4–5, H9, and H12 are depicted as gray cylinders. The AF-2 coactivator binding pocket is lined by H3, H5, and H12, whereas the BF-3 pocket is formed by H1, H9, and the loop linking H3 with H4–5 (L3/4), which is shown as a thin gray wire. BF-3 residues studied herein are shown as green sticks. Key AF-2 residues implicated in engaging in important contacts with coactivator peptides as shown in previous crystal structures are highlighted in raspberry, and the charge clamp residues K720 and E897 are shown as rasperry sticks. The residues R726 and N727 belonging to the boundary between AF-2 and BF-3 pockets are shown as dark blue and green sticks, respectively. The residue V757 the mutation of which, V757A, has been associated with PCa is depicted as a blue stick located at the end of H5 and is surface exposed. B, Solid-surface representation of AR LBD in gray showing the residues lining BF-3 in green, the residues lining AF-2 in raspberry and residue R726 in blue. C, Western blot showing the protein expression level of all AR LBDs. D and E, AR AF-2 activity in the absence (D) or presence (E) of GRIP1 coactivator. WT AR LBD AF-2 activity is 1%. F–H, Mammalian two-hybrid assays with (F) AR NTD domain, and corepressors (G) N-CoR or (H) SMRT. HeLa cells were transfected with 100 ng DNA and treated with (D–F) 100 nm or 10 nm (G–H) DHT. Results are the mean of at least five independent experiments performed in triplicate.
Table 1.
Summary of the experimental results obtained in vitro using AF-2 transcriptional activation and mammalian two-hybrid assays with AR LBD WT and mutant variants, and computational parameters from their corresponding MD simulations.
| AR LBD | Site | Luciferase Activity | Fold Increase Activity with GRIP1 | Fold Increase NTD Interaction | Fold Increase NCoR Interaction | Fold Increase SMRT Interaction | rel mean all b-factor | rel mean BF-3 b-factor | rel mean AF-2 b-factor | BF-3-AF-2 corr (r2) | BF-3-LBP corr (r2) | avgeASA DHT |
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| WT | WT | WT | WT | WT | WT | 1.00 | 1.00 | 1.00 | 0.34 | 0.13 | 0.33 | |
| Super-Activating | ||||||||||||
| N833R | BF-3 | VERY HIGH | LOWER | VERY LOW | WT | LOWEST | 0.90 | 1.36 | 1.07 | 0.12 | 0.05 | 1.90 |
| R840A | BF-3 | VERY HIGH | WT | VERY LOW | LOWEST | LOWEST | 0.87 | 0.94 | 1.26 | 0.50 | 0.17 | 1.22 |
| I672R | H1-BF-3 | VERY HIGH | WT | VERY LOW | LOWER | LOWER | 1.14 | 1.53 | 0.92 | 0.87 | 0.03 | 0.19 |
| PCa | ||||||||||||
| V757A | H5 | WT | HIGHER | HIGHER-MILD | LOWER | LOWER | 1.38 | 1.64 | 1.14 | 0.89 | 0.10 | 0.14 |
| R726L | BF-3 | WT | HIGHER | HIGHER-MILD | WT | WT | 1.13 | 1.34 | 1.14 | 0.72 | 0.36 | 2.48 |
| AF-2 | ||||||||||||
| AIS | ||||||||||||
| N727K (MAIS) | BF-3 | HIGHER-MILD | HIGHER-MILD | LOWER-MILD | LOWER | WT | 0.83 | 1.14 | 1.33 | 0.76 | 0.16 | 3.73 |
| AF-2 | ||||||||||||
| F826L (PAIS) | BF-3 | HIGHER-MILD | HIGHER-MILD | LOWER-MILD | LOWER | LOWER | 0.83 | 1.04 | 0.91 | 0.61 | 0.01 | 1.70 |
| Inhibiting | ||||||||||||
| F826R | BF-3 | LOWER | HIGHEST | HIGHEST | WT | LOWER | 0.92 | 0.81 | 1.12 | 0.49 | 0.16 | 0.33 |
| R840E | H1-BF-3 | DEAD | WT | LOWEST | LOWER | LOWER | 0.86 | 0.87 | 1.03 | 0.50 | 0.01 | 0.25 |
Rel, Relative.
Androgen binding is known to trigger widespread structural and dynamic alterations within the AR LBD, although detailed structural data are missing. By analogy with other NR LBDs, unliganded (apo-) AR LBD may feature a dislodged helix 12 (H12) adopting an unstructured molten globule organization. Upon admission of the hormone into the core of the LBD, the overall stability of the domain is increased, achieving a more defined structure (21). The best-described allosteric rearrangement in NRs upon ligand binding takes place with the conformational change of H12 that completes AF-2 (22).
AR LBD is subject to mutations in advanced PCa and AIS (Androgen Receptor Gene Mutations Database: http://androgendb.mcgill.ca). PCa mutations often result in increased transactivation or expanded ligand binding preference (5, 7, 8, 10, 23, 24). Conversely, AIS mutations usually reduce AR activity and cause varying degrees of fertility problems and undervirilization (4, 6, 9, 13, 25). Whereas AR mutations that arise in both diseases commonly affect known functional regions of the protein, including the ligand-binding pocket and AF-2 surface, many others affect regions of the AR surface with no assigned function, implying that they disrupt as yet undefined aspects of AR activity.
AR is the pharmacological target for antiandrogens used in PCa treatment. Current PCa clinical treatments involve combinations of androgen-deprivation therapy and antiandrogens (e.g. bicalutamide) that inhibit AR action by competing for androgen binding and displacing H12 to prevent formation of a productive AF-2 pocket. Unfortunately, prolonged antiandrogen treatment results in emergence of hormone-refractory PCa with poor prognosis; incompletely defined mechanisms result in reactivation of AR in the absence of androgens and presence of antiandrogens (12, 26–29).
We have proposed that the AR surface may harbor attractive sites for intervention with small molecules (30–33). AR AF-2 undergoes subtle induced fit rearrangements upon coactivator binding and several residue side chains (e.g., K720, M734, and M894) move to create hydrophobic subpockets that bind apolar side chains of coactivator NR boxes and can deepen further to accommodate bulky hydrophobic side chains of W/FxxLF motifs that characterize AR LBD binding peptides within the NTD- and AR-specific coactivators (19, 20). These surface cavities are attractive targets for small molecules and, because AR is reactivated in recurrent PCa, such small molecules could inhibit growth of both early-stage PCa and late-stage hormone-refractory forms of the disease (30–33). In a previous study, we screened chemical libraries for small molecules that inhibit coactivator binding to AF-2 (30). Surprisingly, several compounds, including Triac and members of the fenamic acid series of antiinflammatories, inhibited AR/LxxLL peptide interactions but preferentially were localized by x-ray crystallography to a distinct surface, binding function 3 (BF-3) (30); BF-3 which is topologically adjacent to, but distinct from, AF-2, displays characteristics of a protein-binding site, and is target for PCa and AIS mutations (30–32). Site-directed mutagenesis of BF-3-lining residues confirmed its modulating role in AR activity (30). Furthermore, comparisons of AR LBD crystal structures with and without Triac at BF-3 suggested that compound binding triggers allosteric alterations that propagate to AF-2 and inhibit coregulator binding (30). Thus, x-ray structures suggest that BF-3 and AF-2 pockets are structurally coupled and that allosteric communication between them exists, but the succession of conformational changes and function of these effects are not clear.
Here, we employed a combination of in vitro transactivation assays, mammalian two-hybrid assays with AR LBD, and computational molecular dynamics (MD) simulations to understand how mutations in residues in or near BF-3 may influence AR function and dynamics and how allosteric communication between BF-3 by AF-2 may take place. Our data show that mutations in BF-3 act as allosteric elicitors of conformational changes that are transmitted towards AF-2, and that this allosteric communication affects AR LBD function as experimentally shown in vitro. Furthermore, a series of residues from BF-3, the boundary of BF-3/AF-2, and AF-2 are structurally interconnected and allosterically coupled. Moreover, our data indicate that several residues belonging to BF-3 and AF-2 surface pockets are key players of an allosteric network that may influence multiple aspects of AR LBD function.
Materials and Methods
Cell culture and transfection assays with AR LBD
Transcriptional activities of wild-type (WT) and mutant human AR LBD GAL4-DBD constructs (GAL4-AR LBD) were determined in transient cotransfection assays using human cervix adenocarcinoma epithelial HeLa cells. Vectors and assay procedures were previously described (20, 30). The GK1-Luciferase (LUC) reporter plasmid used contained five GAL4 response elements upstream of a minimal promoter. GAL4-AR LBD WT and constructs of mutants I672R, R726L, F826R, N833R, and R840E have been previously described (30). Mutants N727K, V757A, F826L, and R840A were made by using the QuikChange Site-Directed Mutagenesis Kit (Stratagene, La Jolla, CA). HeLa cells were maintained in Dulbecco's modified Eagle, 4.5 g/liter d-glucose medium (GIBCO) containing 10% fetal bovine serum, 0.58 g/liter l-glutamine, 0.11 g/liter sodium pyruvate, 100 u/ml penicillin, and 100 mg/ml streptomycin. Cells were collected 24 h before transfection in fresh medium containing 10% charcoal-stripped fetal bovine serum and seeded in 24-well culture plates (Corning, Inc., Corning, NY) at a density of 1.5 × 105 cells per well. They were transfected using FuGENE HD reagent (Promega Corp., Madison, WI) as described by the manufacturer. The DNA mixture was composed of 300 ng/well of GK1-LUC; 2.5 ng/well of Renilla-LUC; 100 ng/well of WT or mutant GAL4-AR LBD or empty control vector; and 100 ng/well of pSG5-GR-interacting protein (GRIP)1, VP16-AR NTD (1-504), and VP16-NCoR (1925–2440) or VP16-SMRT (2025–2525) plasmids. The cells were incubated 5 h after transfection with vehicle (1%, vol/vol, dimethylsulfoxide) or hormone [dihydrotestosterone (DHT), dissolved in dimethylsulfoxide], which was purchased from Sigma-Aldrich (St. Louis, MO). Finally, cells were washed twice with PBS and lysed in 100 μl of passive lysis buffer (Promega). LUC and Renilla-LUC activities were measured on 25 μl of the extracts in a GloMax 96 Microplate Luminometer (Promega) using the Dual-Luciferase Reporter Assay System (Promega), according to the manufacturer's instructions.
Western blot analysis
For AR Western blot analysis, HeLa cells were transfected with 1 μg of AR LBD expression constructs as described above. Cells were washed with cold PBS 24 h after hormone treatment, lysed with cold RIPA buffer (150 mm NaCl, 1% Triton X-100, 0.1% sodium dodecyl sulfate, 0.5% sodium deoxycholate and 50 mm Tris, pH 7.5) supplemented with complete protease inhibitors (Roche Diagnostics, Indianapolis, IN) and centrifuged for 10 min at 14,000 rpm. Lysates were boiled and loaded onto a NuPAGE 4–12% Bis-Tris gel (Invitrogen, Carlsbad, CA), after which proteins were transferred to a polyvinylidene difluoride membrane. Western immunoblotting was performed using a polyclonal antibody against the C-terminus of AR (C-19, Santa Cruz Biotechnology, Inc., Santa Cruz, CA), followed by incubation with a horseradish peroxidase-conjugated mouse antirabbit antibody (Abcam, Cambridge, MA). AR LBDs were visualized by a reaction with Luminata Forte Western HRP substrate (Millipore).
Molecular dynamic simulations
Preparation of AR LBD input structures
We used the x-ray structure of AR LBD in complex with the hormone dihydrotestosterone (DHT) (PDB code 1T7T; 1.7 Å resolution) (19) as starting template to model all the mutants described herein, using the AMBER module LEAP. The PDB file was converted into an AMBER structure file using LEAP. The preparation of the files for the hormone DHT was done with the AMBER modules ANTECHAMBER and LEAP.
As a first preparation step, the crystallographic water molecules were removed from the structure. Then a quick minimization step was performed to optimize the structure within the force field in vacuum. Later on, each AR LBD structure to study was placed in a periodic truncated octahedron box of TIP3P water molecules (the minimum distance between protein and walls of the box was set at 12 Å). Na+ and Cl− counterions were then added to the solvent bulk of protein/water with LEAP to neutralize the system and achieve 150 mm NaCl concentration.
System setup and simulation protocol
We performed the MD simulations using the force field AMBER parm99 of the AMBER10 package (34, 35). Our equilibration protocol for each MD simulation started by 1 psec minimization with a 50 kcal/(mol · Å2) harmonic potential to restrain the protein atoms to minimize the solvent molecules. Then, we applied 40 psec of MD simulation at constant pressure (1 atm) with a 12 Å nonbonding cut-off distance during which the temperature was raised from 50 to 300K using Langevin dynamics, applying restraints of 25 kcal/(mol · Å2) to all the protein atoms. After this, we ran 40 psec MD simulation with restraints of 10 kcal/(mol · Å2), and another 40 psec MD simulation with restraints of 5 kcal/(mol · Å2) on all the protein atoms. Then, we applied 20 psec MD simulation with 5 kcal/(mol · Å2) restraints only to the protein backbone atoms, and another 20 psec MD simulation with 1 kcal/(mol · Å2) restraints on the protein backbone atoms as well. Finally, we ran 60 psec MD simulations in which all the atoms of the system were able to move freely. After the above-mentioned system equilibration step, 40 nsec of MD simulation were run for each mutant/complex at constant pressure (1 atm) and a temperature of 300 K. The particle mesh Ewald method was used, with a collision frequency of 0.2 psec−1 excluding bonds involving hydrogen atoms.
B-Factors derived from MD simulation
We calculated the temperature factors (B-factors) from the MD simulation, using the ptraj AMBER tool (35). B-Factors were computed as atomic positional fluctuations multiplied by 8/3π2 and then mass weighted and averaged for each residue. For the sake of comparison, the original B-factors derived from x-ray diffraction data in the AR LBD PDB 1T7T were also mass weighted and averaged by residue.
Analysis of cross-correlation matrices
Correlation matrices, representing all the residue-residue pair-wise root mean square deviation correlations along the complete MD trajectory, were computed using the ptraj AMBER tool (35). The standard convention was used with a positive value between 0 and 1 reflecting correlated motion and a negative coefficient between 0 and −1 reflecting anticorrelated motion. Atoms with correlated motions move in phase whereas atoms with perfect anticorrelated motion move in antiphase.
Accessible surface area calculations
The hormone DHT accessible surface area (ASA) along the simulations was computed with the ICM-Browser program (www.molsoft.com), based on the center of a spherical probe of 1.4 Å radius rolling over the structure of AR bound to the hormone (36). This parameter gives a measure of the solvent accessibility of DHT, which is buried within the LBP.
Analysis of surface cavities along MD simulations
To identify the evolution of surface cavities or ligand pockets during the MD simulations, we used the fpocket program and its module MDpocket developed to track the persistence of pockets within MD trajectories (37, 38). MDpocket ran fpocket iteratively on 100 trajectory snapshots (extracted every 400 psec) to compute α-spheres (defined as those in contact with four atoms without containing any internal atom inside). Then the density of the conserved positions of the α-spheres during the trajectory was calculated. High-density regions corresponded to stable and well-defined cavities whereas lower densities indicated transient pockets. To visualize all volumes detected by MDpocket for each trajectory, we used an isovalue of 2.19, which selects the top 1% of the detected volume for the WT density data.
Contacting pairs and distance calculations
We calculated all contact pairs (residue-residue minimal interatomic distance <4 Å) formed by residues in the peptide-binding region (i.e. those at a distance <4 Å from the ARA70 peptide in the structure 1T5Z after superimposing AR coordinates) (20), in the snapshots generated every 400 psec from the MD trajectories. We compared the frequency of such contact pairs in the mutants with respect to the WT.
Results
BF-3 mutations alter AR AF-2 activity
To address functional effects of different BF-3 mutations, we tested in vitro the transactivation activity of agonist-bound WT AR LBD (WT AR LBD) and nine AR LBD mutants (Fig. 1) and performed in parallel an exhaustive comparison of dynamics of WT AR and mutant ARs using MD simulation (see Figs. 2–5 and SI). The chosen mutated residues are located either lining the BF-3 pocket (I672, F826, N833, R840) or at the boundary between AF-2 and BF-3, hence part of both pockets (R726, N727) (Fig. 1, A and B). Additionally, we chose for control purposes a mutation (V757A) located at the end of H5, hence distant from both studied pockets (Fig. 1A). The selected dataset of mutations studied herein includes: I672R (http://androgendb.mcgill.ca), V757A (39–40), and R726L (41–46), associated with PCa; F826L (47) and N727K (48), found in AIS patients, as well as mutations that have not been associated with pathology (F826R, R840A, R840E, and N833R) (Fig. 1, A and B, and Table 1). We have not studied mutations F826L, N727K, and R840A before (Fig. 1 and Table 1). WT and all mutant AR LBDs investigated exhibit comparable levels of expressed protein as assessed by Western blot (Fig. 1C).
Fig. 2.
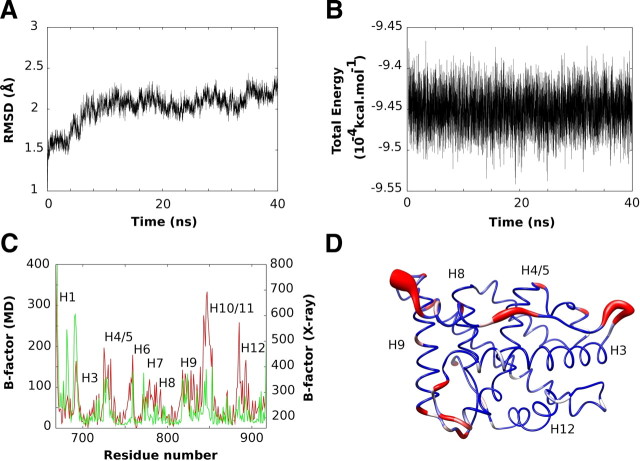
Simulation of AR LBD WT and mutants. A, Evolution of root mean square deviation (RMSD) with respect to the initial structure along the MD trajectory. B, Evolution of global energy along MD trajectory. C, Comparison between experimental (in red) and AMBER-based simulated B factors (in green). For consistency, experimental B factors from PDB 1T7T were transformed to be comparable to simulated values (see Materials and Methods for details). AMBER B-factors per residue were computed from an ensemble of 200 frames along the MD trajectory selected every 200 psec. D, Simulated B-factors mapping on AR. The AR receptor B-factors are shown as worms with variable thickness and color according to their corresponding value (B-factors < 46 in blue; 46 < B-factors < 77 in white; B-factors > 119 in red). ns, Nanoseconds.
Fig. 3.
Evolution of pair-wise minimal contact distance for selected residue pairs in WT AR (blue), I672R mutant (green), F826L mutant (red), and R840E mutant (magenta) along the MD trajectory for selected residue pairs. ns, Nanoseconds.
Fig. 4.
Surface residues of the AF-2 pocket. A, WT AR x-ray structure, PDB 1T7T. B, AR bound to ARA70 peptide x-ray structure, PDB 1T5Z. C, AR bound to ARA70 peptide, with peptide removed for the sake of clarity. D, WT AR during MD simulation (snapshot taken at 10.4 nsec, representative of the first third of the trajectory). E, I672R mutant during MD simulation (snapshot taken at 29.6 nsec, representative of the last half of the trajectory). F, R840E mutant during MD simulation (snapshot taken at 4 nsec, representative of the first half of the trajectory).
Fig. 5.

Conformational changes in WT AR and mutants during MD simulation. A, Detail of the H3–H4/5 loop in WT AR during MD simulation (in gray, snapshot taken at 10.4 nsec, representative of the first third of the trajectory) with respect to the x-ray structure in yellow. B, I672R mutant after MD simulation (in gray, snapshot taken at 29.6 nsec, representative of the last half of the trajectory) compared with WT AR x-ray structure (in yellow). C, Same loop detail in R840E mutant after MD simulation (in gray, snapshot taken at 4 nsec, representative of the first half of the trajectory) compared with WT AR x-ray structure (in yellow). D, Residue pairs in which contact frequency during MD simulation in I672R mutant significantly change with respect to the AR WT simulation; residues are represented as spheres, and the link is colored in red or green depending whether the contact frequency in the corresponding AR mutant MD simulation is significantly smaller or larger, respectively, than in WT AR MD simulation. E and F, Similar analysis for F826L mutant (E), and for R840E mutant (F).
We observe that mutants I672R and N833R behave as super-AR variants, as earlier reported (30), as well as the new mutant R840A. Mutants N727K and F826L also moderately enhance AR AF-2 activity although without exhibiting super-AR behavior. Also along previous observations, F826R and R726L marginally reduce, and R840E totally abolishes AR LBD activity in vitro (Fig. 1D and Table 1).
BF-3 mutations affect AR LBD activation by GRIP1
We additionally investigated the functionality of the studied AR LBDs by measuring AR AF-2 transactivation activity in vitro in the presence of coactivator and by addressing AR LBD interaction either with its N-terminal domain or corepressors using a mammalian two-hybrid experiment.
We first determined effects of the coactivator GRIP1 on WT and mutant AR activity (Fig. 1E). GRIP1 is the mouse orthologue of the human protein transcription intermediary factor 2, which is known to interact with the AR AF-2 pocket in a hormone-dependent manner (49). GRIP1 enhances activity of all the mutants, but there are variations in the extent of GRIP1 potentiation (Fig. 1E and Table 1). GRIP1 enhances activity of super-mutants I672R and R840A similarly to WT, but fold increase in activity of supermutant N833R by GRIP1 is lower. GRIP1 also enhances activity of PCa mutants V757A and R726L, AIS-associated mutations F826L and N727K, although to a lesser degree and rescues activity of the medium-inhibiting mutant F826R, resulting in the highest fold increase among all the studied AR LBD mutants. Even the very weak activity of the R840E mutant is enhanced by GRIP1. Thus, some BF-3 mutations (notably N833R and F826R) alter the extent of GRIP1 coactivation, but none abolish GRIP1 interaction.
BF-3 mutations affect the N/C interdomain interaction
To assess whether mutations at the BF-3 pocket affect AR LBD/NTD (N/C) interaction, a mammalian two-hybrid was performed (Fig. 1F and Table 1). Again, some of the BF-3 mutants altered AR N/C interaction, but the pattern of effects was different from that with GRIP1. All three supermutant AR LBDs, I672R, N833R, and R840A, show a decreased interaction with the NTD as compared with WT. Mutants R726L and V757A, associated with PCa, display a WT or moderately higher increase in their interaction with the NTD. Medium-inhibiting mutant F826R, which exhibited the highest increase in activity in the presence of coactivator, also exhibits the largest fold induction with the NTD. Mutants N727K and F826L, associated with AIS, show mild impairment in NTD interaction, superactivating mutant N833R exhibits the lowest capacity for N/C interaction, and activity of the severely impaired mutant R840E is not enhanced by NTD overexpression, unlike the case with GRIP1.
BF-3 mutations alter AR interaction with N-CoR and SMRT
Because DHT-liganded AR also interacts weakly with NR corepressors (50), we used a mammalian two-hybrid assay to assess how BF-3 mutants affect this interaction. The chosen corepressors were the silencing mediator of retinoid and thyroid receptors (SMRT) and the nuclear receptor corepressor (N-CoR) (Fig. 1, G and H).
As seen with GRIP1 and the AR NTD, BF-3 mutants broadly affect the capacity of AR to bind corepressors (Fig. 1, G and H, and Table 1). Both supermutants I672R and R840A exhibit a significant decrease in their ability to interact with the receptor-interacting domains of corepressors SMRT and N-CoR, being R840 the one that disrupts such interactions the most among all the AR LBD herein studied. V757A, F826L, and the severe mutant R840E also reduce corepressor interaction. Supermutant N833R and mutant F826R show impaired interaction with SMRT, but maintain a WT-like interaction with N-CoR, whereas N727K decreases interaction with N-CoR but leaves interaction with SMRT unaffected. Finally, R726L has little effect on corepressor interaction in these assays.
AR LBD MD simulations
To understand the structural and dynamic effects of the above-described mutations, we first analyzed WT AR LBD by running MD simulation for 40 nsec in explicit solvent, after an initial step to allow equilibration of AR LBD. The WT AR LBD structure appeared stable along the dynamics, and there was no evidence of large-scale reorganization or denaturation processes (Fig. 2, A and B).
The overall apparent mobility of WT AR LBD amino acids along the MD trajectory resembled that of the AR LBD x-ray structure PDB 1T7T (19). We estimated residue B-factor values by computing fluctuations of each residue along the MD simulation. Figure 2C shows these values against B-factor values derived from electron density uncertainties in the AR LBD structure (19). In general, B-factors derived from MD simulation were similar to the crystallographic ones, implying that our simulation produces a reasonably accurate representation of the molecular motions that are detected in the crystal although the most N-terminal helix of AR LBD (H1) and amino acids 687–695 (H1-H3 loop) appear more flexible in the simulation whereas two highly flexible regions [amino acids 844–850 (H9-H10) and 880–886 (H11 and H11/12 loop)] appear more rigid than equivalent regions in the x-ray structure.
Interestingly, the B-factor measurements flag the H3-H4/5 loop and the S3-H9 loop as relatively flexible compared with other regions of the AR LBD; this was seen both in MD simulations and in B-factors derived from the x-ray structure (Fig. 2C and Supplemental Fig.1 for mutant B-factors fluctuations). Both regions are located in the boundary of AF-2 and BF-3 pockets, suggesting a possible dynamic link between these binding surfaces (Fig. 2D).
Next, we studied the effects of mutations on AR LBD dynamics, by obtaining 40 nsec MD trajectories for each of the mutants (Supplemental Fig. S2). Interestingly, consideration of mobility of the entire LBD reveals that mutations I672R, F826L, and N727K, which enhance AR LBD activity, exhibit larger average B-factors (Table 1) indicative of higher mobility. This is not true for supermutants N833R and R840A. On the contrary, inhibitory AR mutations generally display smaller average B-factors, indicative of reduced mobility (Table 1). Correlation of B-factors with activity is even more striking when mobility of only the BF-3 residues is considered (again, excluding N833R and R840A). There is no similar correlation between mobility of the AF-2 residues and activity (Table 1).
AR mutations reveal allosteric coupling between BF-3 and AF-2
To analyze how specific AR mutations affect dynamics of BF-3 and other regions of the LBD and pinpoint possible allosteric effects, we computed the motion correlation of all AR residues against each other along the MD trajectories, shown as correlation matrices (see Supplemental Fig. 3 and Materials and Methods). The cross-correlation plots for WT activating (I672R, F826L) and inhibiting (R840E) mutations are shown in Supplemental Fig. 3. The values for WT AR LBD (Supplemental Fig. 3) show regions that are correlated (in red) or anticorrelated (in blue) along the MD trajectory. There is little obvious coupling between different regions of the WT AR LBD. Interestingly, the correlation matrix for AR I672R shows significant differences: there is stronger correlation between motion of residues 672–673 (H1, the mutation site), residues 710–740 (H3 and loop 3/4, mostly AF-2 and the boundary region of AF-2/BF-3) and residues 820–840 (the BF-3 lining loop S3/H9 and H9). In the case of F826L, we observe, in general, less motion correlation between regions than with I672R, but there is significant correlation between the region of the mutated residue (H9, close to BF-3) and the adjacent AF-2 pocket and this is stronger than WT. Increased correlation of mobility of BF-3 and AF-2 is not seen with the N833R and R840A mutants. In the case of R840E mutant, there is less motion correlation and no clearly correlated regions that can be highlighted. Reductions in correlation between mobility of the BF-3 and AF-2 pockets are also seen with other BF-3 mutants that reduce AR LBD activity.
Calculation of correlation coefficients (r2) along the MD trajectories between the residues forming BF-3, AF-2, and LBP (where DHT is bound) sites underscores impressions from the mobility correlation matrices (Table 1). Activating mutants (with the exception of N833R and R840A) have BF-3/AF-2 correlation r2 more than 0.7 (significantly higher than WT), whereas inhibiting mutants have r2 around 0.5–0.6, smaller than that of the activating mutants, but still higher than WT. Thus, there is increased allosteric coupling between BF-3 and AF-2 when the transcriptional output is enhanced activation.
We do not observe any correlation between AF-2 or BF-3 and residues that line the LBP for WT AR or any of the analyzed AR mutants. Curiously, if we exclude the most activating mutants (N833R, R840A, I672R, and F826L), there is some correlation between the average DHT ASA relative to WT and activity (Supplemental Fig. 4). This raises the possibility that enhanced plasticity of LBP observed during MD simulations may favor AR activity.
BF-3 mutations change dynamics of AF-2
Because the results obtained from our MD simulations suggest close structural communication between BF-3 and AF-2 surface pockets, we analyzed effects of BF-3 mutations on AF-2 conformation in more detail. To do this, we computed all possible contact pairs of AF-2 residues that exhibited at least a 15% reduction or increase in their pair-wise interaction frequency relative to WT AR during simulations (see Materials and Methods for details). Figure. 3 and Supplemental Figs. 5 and 6 show evolution of contacts between selected residue pairs along the simulations.
BF-3 mutations result in significant changes in pair-wise interactions within AF-2 residues over time. For activating mutants I672R and F826L, the proximity between the AF-2 key residues K720-M734 is greatly reduced relative to WT AR or R840E (Fig. 3). In contrast, K720 (one of the charge clamp AF-2 residues) forms more extensive contacts with H729, N727, L728, and V730. Among other AF-2 residues, M734 establishes frequent contacts with L712, M894, and I898, whereas V730 forms more frequent contacts with V716 in the context of both activating mutants. Effects of the AR R840E (inhibitor) mutation are opposite to that of the activating mutants. Specifically, pair-wise interactions between K720 and M734 are enhanced, whereas interactions between R726, K720, Q733, and V730 appear less frequently, V730 remains further away from K720 and V716, and contacts between K720 and Q733 are less prevalent (whereas contacts between K720 and H729, N727, or L728 are never formed, as in WT).
The net effect of these changes in pair-wise interactions is that there are alterations in the frequency of formation of AF-2 subpockets that accommodate bulky side chains of F1xxLF5 peptides present in AR-specific coactivators (Supplemental Fig. 7). In the x-ray WT AR structure (PDB 1T7T) (19), K720 and M734 are in contact, closing the entrance for the second F side chain (at position 5) of the FxxLF motif (Fig. 4A). During the WT MD simulation, these residues open frequently to create an organization that resembles that of AR in complex with the ARA70 FxxLF motif (PDB 1T5Z) (20) (Fig. 4B). In the I672R mutant simulations, K720 and M734 separate even more widely and frequently, opening a larger hydrophobic pocket (Fig. 4E), an effect that can also be seen in the other activating mutant F826L (data not shown). By contrast, in the AR R840E (inhibitor) mutant, the arrangement of K720 and M734 along the simulation resembles that of the original AR x-ray structure PDB 1T7T (19), and the deep AF-2 subpocket fails to open (Fig. 4F), unlike the dynamic opening and closing of the subpocket seen in simulations with WT AR (Fig. 4D). Thus, MD simulations seem to suggest that BF-3 mutations alter the propensity of AF-2 to form subpockets that accommodate FxxLF peptides; activating mutations enhance subpocket formation, whereas the inhibitory mutation reduces subpocket formation.
Allosteric paths on AF-2
We examined snapshots of the MD simulations to search for dynamic structural rearrangements that could be responsible for the allosteric transmission of information from BF-3 to AF-2 (Fig. 5). Analysis of the simulations of activating AR mutants I672R and F826L revealed a large conformational change in the H3-H4/5 loop (residues 723–734) toward the end of the simulation (see a conformational snapshot of I672R in Fig. 5B). Movements of this loop relative to the initial position (Supplemental Fig. 8) were more extensive than the ones seen in WT AR or any other mutant simulation [interestingly, the mutants with higher fold increase in activity in the presence of GRIP1 (V757A, R726L and, to a lower extent, F826R), have these loops more extensively moved than WT along the dynamics]. Within the H3–H4/5 loop, there are striking changes in the spatial location of residues H729 and V730 (Fig. 3). A possible mechanism for this effect is that the I672R side chain interacts with E837 (located in H9 and forming a salt bridge with R840 in WT AR). This effect would alter H9 position and free the H3–H4/5 loop so that K720 (H3) can interact with H729 (H3-H4/5 loop). H729 moves from being in contact with a lysine residue (K822, loop S3-H9) to contacting K720 backbone as a result of the new loop conformation.
Similar analysis for F826L (Fig. 5E) also reveals opening of the H3–H4/5 loop. Early in the MD simulation, L826-N823 interaction breaks leading to distortion of the S3-H9 loop and breakage of K822-H729 contact. This effect, which is not seen in WT AR, leaves H729 free to interact with K720 and results in a H729-K720 conformation that is exactly the same as the one seen in the I672R simulation and is not found in any other mutant. By contrast, the K822-H729 contact is more stable in the simulations with the R840E inhibitory mutant relative to WT-AR and certainly much more stable than in activating mutants (Fig. 3), underscoring the potential importance of this interaction in BF-3/AF-2 communication.
These new conformations of the H3-H4/5 and S3-H9 loops (especially in the activating mutants) open a small hydrophobic cavity around Y739 (H4) that is adjacent to AF-2 and close to BF-3. Comparison of the organization of this cavity reveals that it is closed throughout the simulations with the inhibitor mutation R840E.
Discussion
We have previously discovered a small molecule-binding surface on the AR LBD (BF-3), and our mutational analysis has revealed that it is highly important for AR function (30). AR mutants that affect BF-3 and have been documented in PCa and AIS variously influence androgen binding and dissociation, coregulator recruitment, N/C-interaction, and transactivation (6, 7, 30, 42, 47, 51–69). We envision two possible physiological roles for BF-3, which are not mutually exclusive. First, our combined functional and computational assays support our previous hypothesis that BF-3 is an allosteric modulator of the adjacent AF-2 pocket, affecting its function. Second, BF-3 may be a protein-protein interaction site for coregulator proteins.
Our studies confirm and extend our previous data, which show that BF-3 mutants alter overall AR activity in different, and hard to predict, ways. We show that mutants in BF-3 and residues that lie between BF-3 and AF-2 produce a range of effects from superactivators (I672R, N833R, R840A), moderate enhancers (F826L, N727K), weak inhibitors (F826R, R726L), and very potent inhibitors (R840E). We have also shown a wide range of effects upon coregulator binding; BF-3 mutations do not completely inhibit functional interactions with GRIP1 or NTD but do affect coactivation differentially by both proteins. This stands in marked contrast to effects of mutations in the AF-2 surface that consistently block AR interactions with GRIP1, the AR NTD, and other coregulators. Finally, BF-3 mutations moderately inhibit corepressor interactions, but, here again, there are diverse effects with some mutations strongly inhibiting N-CoR and SMRT binding, others exhibiting milder effects or no effects, and some distinguishing between N-CoR and SMRT.
Our MD simulations do not systematically explain effects of all AR BF-3 mutations but do suggest a plausible general explanation for their diverse effects on AR activity; BF-3 is coupled to AF-2, and BF-3 mutations alter the propensity of the AF-2 surface to form deep subpockets that accommodate bulky side chains of coregulator motifs. MD simulation on AR LBD shows a dynamic link between BF-3 and AF-2, and two regions that form the boundary between AF-2 and BF-3 pockets play a key role in allosteric communication: the H3-H4/5 loop, where R726 and N727 are located, and the S3-H9 loop, where F826 resides. Interestingly, consideration of mobility of the entire LBD reveals that three mutations that enhance AR LBD in vitro activity (I672R, F826L, N727K), exhibit a greater mobility than WT. On the contrary, AR inhibitory mutations generally feature smaller mobility with respect to WT. Analysis of the flexibility per pocket indicates that BF-3 flexibility degree is inversely correlated to the AR LBD in vitro function, but there is no correlation with the AF-2 mobility.
BF-3 mutations induce conformational changes in several side chains of the adjacent AF-2 pocket. For activating mutants I672R and F826L, residues K720 and M734 stand out as already observed in the x-ray crystal structures of coactivator peptides bound to AF-2 (20). Pair-wise residue contacts with several key AF-2 residues are manifested, specially implicating L712, H729, N727, L728, V730, and H12 M894 and I898. Effects of the AR R840E (inhibitor) mutation on the observed pair-wise contact formation are opposite to that of the activating mutants. The net effect of these changes in pair-wise interactions is alterations in the frequency of formation of AF-2 subpockets that accommodate NR boxes present in coactivator peptides and the NTD domain. There is thus a shift in the conformational ensemble of the AF-2 groove. Residues K720 and M734, which are in a closed conformation in the x-ray WT AR structure without coactivator peptide (PDB 1T7T) (19), remain more frequently along the MD trajectory in the AF-2 open conformation, similar to the one observed in the crystal structure of AR bound to the ARA70 FxxLF motif (PDB 1T5Z) (20). In the I672R and F826L mutant MD simulation, K720 and M734 separate even more widely, frequently opening a larger hydrophobic pocket. However, K720 and M734 are found in the closed conformation along the MD trajectory in the inhibitory AR mutant R840E. Thus, MD simulation suggests that BF-3 mutations alter the propensity of AF-2 to reorganize its subpockets to accept the entering coactivator peptides. Whereas several activating mutations enhance the formation of subpockets, inhibitory ones reduce them.
Other paths may also be involved in the allosteric transmission of information from BF-3 to AF-2, and from/to other areas of the receptor. Our studies have evidenced possible roles for identified residues located in the H3-H4/5 and S3-H9 loops that are responsible for conformational changes and allosteric cross talk among AR LBD surfaces. These new conformations of the H3-H4/5 and S3-H9 loops (especially in the activating mutants) open a small hydrophobic cavity around Y739 (H4) that is adjacent to AF-2 and close to BF-3. Comparison of the organizations of this cavity reveals that it is closed throughout the simulations in the inhibitor mutation R840E. Interestingly, we observed weak binding of an apolar small molecule (salycylaldehyde) at this cavity adjacent to AF-2 in our initial screening of surface inhibitor compounds (PDB 2PIR) (30). Y739 is one of the residues that interacted with this surface binder drug, and, most importantly, K822 (loop S3-H9) was at a distance that allowed electrostatic stabilization of the drug (30). We call this groove “binding function 4” (BF-4). We did not observe correlation between AF-2 or BF-3 and the residues that line the LBP for WT AR or any of the AR analyzed mutants. However, it is interesting to point out that if the most activating mutants are excluded (N833R, R840A, I672R, and F826L), there seems to be a correlation between the average ASA of the LBP, where DHT is cocooned, indicating that an increase in LBP adaptability during the MD simulation may influence AR activity.
Our data indicate that two superactivating BF-3 mutations may work by a distinct mechanism. Our MD simulations evidence that N833R and R840A do not exhibit larger overall flexibility than WT AR, unlike I672R and other activating mutations, suggesting that they enhance AR activity via a mechanism that differs from allosteric communication. Because BF-3 exhibits characteristics of a protein interaction surface, it is attractive to speculate that both mutations could alter direct contacts with an unknown protein(s) that could either potentiate or silence AR function. Several lines of evidence suggest that BF-3 could be involved in protein contact. Equivalent regions of the thyroid hormone receptor (70) and the nuclear receptor LRH-1 bind to corepressor and the wnt-signaling dependent coactivator β-catenin (71). Furthermore, functional evidence links this region of AR to contacts with the chaperone FKBP52 (72). It is even possible that BF-3 might contact GRIP1 coactivator, the NTD, and corepressors and that these proteins have an interaction surface on AR LBD that extends beyond the AF-2 pocket toward BF-3 as we have previously suggested (30, 33).
Acknowledgments
We dedicate this work to the memory of Professor John D. Baxter, who sadly passed away just a few days before the submission of this manuscript. Professor J. D. Baxter's passion for endocrinology and leadership has inspired many young scientists like us during these years.
We thank Drs. A. T. Hagler (University of Massachusetts) and M. Orozco (Institute for Research in Biomedicine (IRB), Barcelona) for fruitful discussions. We thank Drs. P. Fuentes-Prior (Hospital de Sant Pau, Barcelona), J. Lüders at the Institute for Research in Biomedicine (IRB, Barcelona), and M. Vegetti for useful comments on the manuscript. We thank E. Hondares (IBUB-University of Barcelona) and A. Cvoro (Methodist Hospital) for useful technical advice.
This work was supported by Plan Nacional I+D+i grants SAF-2008-03562, SAF-2011-29681 and BIO2010-22324 [MICINN (Ministerio de Ciencia e Innovación), Gobierno de España], and Marie-Curie International Reintegration Grant IRG-code 224812 (EU). E.E.P. is supported by a RyC (Programa Ramón y Cajal)-2007 fellowship. L.R.C. is recipient of a FPI (Formación de Personal Investigador) fellowship from the MICINN. Obtained funding: E.E.P., J.F.R.; Overall responsibility: E.E.P., J.F.R. G.B. is the recipient of the Martin Harris Research Fellowship.
Disclosure Summary: Conception and design: E.E.P., J.F.R.; Analysis and interpretation: S.G., L.R.C., V.B., G.B., C.B., P.W., E.E.P., J.F.R.; Data collection: S.G., L.R.C., V.B., P.N.; Writing the article: E.E.P., J.F.R., P.W. with contributions of C.B., S.G., L.R.C., V.B., J.D.B.; Critical revision of the article: E.E.P., J.F.R., C.B., P.W., J.D.B.; Final approval of the article: E.E.P., J.F.R., C.B., P.W.
NURSA Molecule Pages†:
Nuclear Receptors: AR;
Ligands: Dihydrotestosterone.
Annotations provided by Nuclear Receptor Signaling Atlas (NURSA) Bioinformatics Resource. Molecule Pages can be accessed on the NURSA website at www.nursa.org.
- AF-2
- Activation function 2
- AIS
- androgen insensitivity syndromes
- AR
- androgen receptor
- BF-3 and -4
- binding functions 3 and 4
- DBD
- DNA-binding domain
- DHT
- dihydrotestosterone
- FxxLF
- phenylalanine-rich motif
- GRIP1
- GR-interacting protein1
- H12
- helix 12
- LBD
- ligand-binding domain
- LBP
- ligand-binding pocket
- LxxLL
- Leucine-rich motif
- MD
- molecular dynamics
- N-CoR
- nuclear receptor corepressor
- NH2/COOH interaction
- amino-terminal/carboxy-terminal interaction
- NTD
- N-terminal domain
- NR
- nuclear receptor
- PCa
- prostate cancer
- SMRT
- silencing mediator of retinoid and thyroid receptors
- SI
- supporting information
- WT
- wild-type.
References
- 1. Lubahn DB , Joseph DR , Sar M , Tan J , Higgs HN , Larson RE , French FS , Wilson EM. 1988. The human androgen receptor: complementary deoxyribonucleic acid cloning, sequence analysis and gene expression in prostate. Mol Endocrinol 2:1265–1275 [DOI] [PubMed] [Google Scholar]
- 2. Mangelsdorf DJ , Thummel C , Beato M , Herrlich P , Schütz G , Umesono K , Blumberg B , Kastner P , Mark M , Chambon P , Evans RM. 1995. The nuclear receptor superfamily: the second decade. Cell 83:835–839 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. La Spada AR , Wilson EM , Lubahn DB , Harding AE , Fischbeck KH. 1991. Androgen receptor gene mutations in X-linked spinal and bulbar muscular atrophy. Nature 352:77–79 [DOI] [PubMed] [Google Scholar]
- 4. Quigley CA , French FS. 1994. Androgen insensitivity syndromes. Curr Ther Endocrinol Metab 5:342–351 [PubMed] [Google Scholar]
- 5. Brinkmann A , Trapman J. 2000. Genetic analysis of androgen receptors in development and disease. Adv Pharmacol:317–341. [DOI] [PubMed] [Google Scholar]
- 6. Brinkmann AO. 2001. Molecular basis of androgen insensitivity. Mol Cell Endocrinol 179:105–109 [DOI] [PubMed] [Google Scholar]
- 7. Buchanan G , Greenberg NM , Scher HI , Harris JM , Marshall VR , Tilley WD. 2001. Collocation of androgen receptor gene mutations in prostate cancer. Clin Cancer Res 7:1273–1281 [PubMed] [Google Scholar]
- 8. Gottlieb B , Beitel LK , Wu J , Elhaji YA , Trifiro M. 2004. Nuclear receptors and disease: androgen receptor. Essays Biochem 40:121–136 [DOI] [PubMed] [Google Scholar]
- 9. Hughes IA , Deeb A. 2006. Androgen resistance. Best Pract Res Clin Endocrinol Metab 20:577–598 [DOI] [PubMed] [Google Scholar]
- 10. Brooke GN , Bevan CL. 2009. The role of androgen receptor mutations in prostate cancer progression. Curr Genomics 10:18–25 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Tindall DaM , Mohler J, eds. 2009. Androgen action in prostate cancer. ISBN 978-0-387-69177-0 [Google Scholar]
- 12. Knudsen KE , Penning TM. 2010. Partners in crime: deregulation of AR activity and androgen synthesis in prostate cancer. Trends Endocrinol Metab 21:315–324 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Szafran AT , Sun H , Hartig S , Shen Y , Mediwala SN , Bell J , McPhaul MJ , Mancini MA , Marcelli M. 2011. Androgen receptor mutations associated with androgen insensitivity syndrome: a high content analysis approach leading to personalized medicine. Adv Exp Med Biol 707:63–65 [DOI] [PubMed] [Google Scholar]
- 14. Jenster G , van der Korput HA , van Vroonhoven C , van der Kwast TH , Trapman J , Brinkmann AO. 1991. Domains of the human androgen receptor involved in steroid binding, transcriptional activation, and subcellular localization. Mol Endocrinol 5:1396–1404 [DOI] [PubMed] [Google Scholar]
- 15. Matias PM , Donner P , Coelho R , Thomaz M , Peixoto C , Macedo S , Otto N , Joschko S , Scholz P , Wegg A , Bäsler S , Schäfer M , Egner U , Carrondo MA. 2000. Structural evidence for ligand specificity in the binding domain of the human androgen receptor. Implications for pathogenic gene mutations. J Biol Chem 275:26164–26171 [DOI] [PubMed] [Google Scholar]
- 16. Heery DM , Kalkhoven E , Hoare S , Parker MG. 1997. A signature motif in transcriptional co-activators mediates binding to nuclear receptors. Nature 387:733–736 [DOI] [PubMed] [Google Scholar]
- 17. He B , Kemppainen JA , Wilson EM. 2000. FXXLF and WXXLF sequences mediate the NH2-terminal interaction with the ligand binding domain of the androgen receptor. J Biol Chem 275:22986–22994 [DOI] [PubMed] [Google Scholar]
- 18. Christiaens V , Bevan CL , Callewaert L , Haelens A , Verrijdt G , Rombauts W , Claessens F. 2002. Characterization of the two coactivator-interacting surfaces of the androgen receptor and their relative role in transcriptional control. J Biol Chem 277:49230–49237 [DOI] [PubMed] [Google Scholar]
- 19. Hur E , Pfaff SJ , Sturgis PE , Hanne G , Buehrer BM , Fletterick RJ. 2004. Recognition and accommodation at the androgen receptor coactivator binding interface. PLoS Biol 2:E274. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Estébanez-Perpiñá E , Moore JM , Mar E , Delgado-Rodrigues E , Nguyen P , Baxter JD , Buehrer BM , Webb P , Fletterick RJ , Guy RK. 2005. The molecular mechanisms of coactivator utilization in ligand-dependent transactivation by the androgen receptor. J Biol Chem 280:8060–8068 [DOI] [PubMed] [Google Scholar]
- 21. Jasuja R , Ulloor J , Yengo CM , Choong K , Istomin AY , Livesay DR , Jacobs DJ , Swerdloff RS , Miksovská J , Larsen RW , Bhasin S. 2009. Kinetic and thermodynamic characterization of dihydrotestosterone-induced conformational perturbations in androgen receptor ligand-binding domain. Mol Endocrinol 23:1231–1241 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Watanabe C , Watanabe H , Tanaka S. 2010. An interpretation of positional displacement of the helix12 in nuclear receptors: preexistent swing-up motion triggered by ligand binding. Biochim Biophys Acta 1804:1832–1840 [DOI] [PubMed] [Google Scholar]
- 23. McPhaul MJ. 2008. Mechanisms of prostate cancer progression to androgen independence. Best Pract Res Clin Endocrinol Metab 22:373–388 [DOI] [PubMed] [Google Scholar]
- 24. Nacusi LP , Tindall DJ. 2009. Androgen receptor abnormalities in castration-recurrent prostate cancer. Expert Rev Endocrinol Metab 4:417–422 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Gottlieb B , Pinsky L , Beitel LK , Trifiro M. 1999. Androgen insensitivity. Am J Med Genet 89:210–217 [DOI] [PubMed] [Google Scholar]
- 26. Huggins C. 1967. Endocrine-induced regression of cancers. Science 156:1050–1054 [DOI] [PubMed] [Google Scholar]
- 27. Culig Z , Hoffmann J , Erdel M , Eder IE , Hobisch A , Hittmair A , Bartsch G , Utermann G , Schneider MR , Parczyk K , Klocker H. 1999. Switch from antagonist to agonist of the androgen receptor bicalutamide is associated with prostate tumour progression in a new model system. Br J Cancer 81:242–251 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Bohl CE , Gao W , Miller DD , Bell CE , Dalton JT. 2005. Structural basis for antagonism and resistance of bicalutamide in prostate cancer. Proc Natl Acad Sci USA 102:6201–6206 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Yuan X , Balk SP. 2009. Mechanisms mediating androgen receptor reactivation after castration. Urol Oncol 27:36–41 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Estébanez-Perpiñá E , Arnold LA , Arnold AA , Nguyen P , Rodrigues ED , Mar E , Bateman R , Pallai P , Shokat KM , Baxter JD , Guy RK , Webb P , Fletterick RJ. 2007. A surface on the androgen receptor that allosterically regulates coactivator binding. Proc Natl Acad Sci USA 104:16074–16079 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Axerio-Cilies P , Lack NA , Nayana RS , Chan KH , Yeung A , Leblanc E , Guns E , Rennie P , Cherkasov A. 2011. Inhibitors of androgen receptor activation function-2 (AF2) site indentified through virtual screening. J Med Chem 54:6197–6205 [DOI] [PubMed] [Google Scholar]
- 32. Estébanez-Perpiñá E , Jouravel N , Fletterick RJ. 2007. Perspectives on designs of antiandrogens for prostate cancer. Expert Opin Drug Discov 2:1341–1355 [DOI] [PubMed] [Google Scholar]
- 33. Buzón V , Carbó LR , Estruch SB , Fletterick RJ , Estébanez-Perpiñá E. 2012. A conserved surface on the ligand binding domain of nuclear receptors for allosteric control. Mol Cell Endocrinol 348:394–402 [DOI] [PubMed] [Google Scholar]
- 34. Wang P , Cieplak P , Kollman PA. 2000. How well does a restrained electrographic potential (RESP) model perform in calculating conformational energies of organic and biological molecules? J Comput Chem 21:1049–1074 [Google Scholar]
- 35. Case DA , Cheatham TE , Darden T , Gohlke H , Luo R , Merz KM , Onufriev A , Simmerling C , Wang B , Woods RJ. 2005. The Amber biomolecular simulation programs. J Comput Chem 26:1668–1688 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36. Lee B , Richards FM. 1971. The interpretation of protein structures: estimation of static accessibility. J Mol Biol 55:379–400 [DOI] [PubMed] [Google Scholar]
- 37. Le Guilloux V , Schmidtke P , Tuffery P. 2009. Fpocket: an open source platform for ligand pocket detection. BMC Bioinformatics 10:168. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38. Schmidtke P , Souaille C , Estienne F , Baurin N , Kroemer RT. 2010. Large-scale comparison of four binding site detection algorithms. J Chem Inf Model 50:2191–2200 [DOI] [PubMed] [Google Scholar]
- 39. Marcelli M , Ittmann M , Mariani S , Sutherland R , Nigam R , Murthy L , Zhao Y , DiConcini D , Puxeddu E , Esen A , Eastham J , Weigel NL , Lamb DJ. 2000. Androgen receptor mutations in prostate cancer. Cancer Res 60:944–949 [PubMed] [Google Scholar]
- 40. Duff J , Davies P , Watt K , McEwan IJ. 2006. Structural dynamics of the human androgen receptor: implications for prostate cancer and neurodegenerative disease. Biochem Soc Trans 34:1098–1102 [DOI] [PubMed] [Google Scholar]
- 41. Elo JP , Kvist L , Leinonen K , Isomaa V , Henttu P , Lukkarinen O , Vihko P. 1995. Mutated human androgen receptor gene detected in a prostatic cancer patient is also activated by estradiol. J Clin Endocrinol Metab 80:3494–3500 [DOI] [PubMed] [Google Scholar]
- 42. Koivisto PA , Schleutker J , Helin H , Ehren-van Eekelen C , Kallioniemi O-P , Trapman J. 1999. Androgen receptor gene alterations and chromosomal gains and losses in prostate carcinomas appearing during finasteride treatment for benign prostatic hyperplasia. Clin Cancer Res 5:3578–3582 [PubMed] [Google Scholar]
- 43. Mononen N , Syrjäkoski K , Matikainen M , Tammela TL , Schleutker J , Kallioniemi OP , Trapman J , Koivisto PA. 2000. Two percent of Finnish prostate cancer patients have a germ-line mutation in the hormone-binding domain of the androgen receptor gene. Cancer Res 60:6479–6481 [PubMed] [Google Scholar]
- 44. Hyytinen ER , Haapala K , Thompson J , Lappalainen I , Roiha M , Rantala I , Helin HJ , Jänne OA , Vihinen M , Palvimo JJ , Koivisto PA. 2002. Pattern of somatic androgen receptor gene mutations in patients with hormone-refractory prostate cancer. Lab Invest 82:1591–1598 [DOI] [PubMed] [Google Scholar]
- 45. Koivisto PA , Hyytinen ER , Matikainen M , Tammela TL , Ikonen T , Schleutker J. 2004. Germline mutation analysis of the androgen receptor gene in Finnish patients with prostate cancer. J Urol 171:431–433 [DOI] [PubMed] [Google Scholar]
- 46. Yan J , Feng J , Goldman D , Cook EH , Craddock N , Jones IR , Heston LL , Sommer SS. 2004. Mutation scanning of the androgen receptor gene in patients with psychiatric disorders reveals highly conserved variants in alcoholic and phobia patients. Psychiatr Genet 14:57–60 [DOI] [PubMed] [Google Scholar]
- 47. Wong HY , Hoogerbrugge JW , Pang KL , van Leeuwen M , van Royen ME , Molier M , Berrevoets CA , Dooijes D , Dubbink HJ , van de Wijngaart DJ , Wolffenbuttel KP , Trapman J , Kleijer WJ , Drop SL , Grootegoed JA , Brinkmann AO. 2008. A novel mutation F826L in the human androgen receptor in partial androgen insensitivity syndrome; increased NH2-/COOH-terminal domain interaction and TIF2 co-activation. Mol Cell Endocrinol 292:69–78 [DOI] [PubMed] [Google Scholar]
- 48. Yong EL , Ng SC , Roy AC , Yun G , Ratnam SS. 1994. Pregnancy after hormonal correction of severe spermatogenic defect due to mutation in androgen receptor gene. Lancet 344:826–827 [PubMed] [Google Scholar]
- 49. Lim J , Ghadessy FJ , Abdullah AA , Pinsky L , Trifiro M , Yong EL. 2000. Human androgen receptor mutation disrupts ternary interactions between ligand, receptor domains, and the coactivator TIF2 (transcription intermediary factor 2). Mol Endocrinol 14:1187–1197 [DOI] [PubMed] [Google Scholar]
- 50. Dotzlaw H , Moehren U , Mink S , Cato AC , Iñiguez Lluhí JA , Baniahmad A. 2002. The amino terminus of the human AR is target for corepressor action and antihormone agonism. Mol Endocrinol 16:661–673 [DOI] [PubMed] [Google Scholar]
- 51. Imasaki K , Hasegawa T , Okabe T , Sakai Y , Haji M , Takayanagi R , Nawata H. 1994. Single amino acid substitution (840Arg→His) in the hormone-binding domain of the androgen receptor leads to incomplete androgen insensitivity syndrome associated with a thermolabile androgen receptor. Eur J Endocrinol 1:569–574 [DOI] [PubMed] [Google Scholar]
- 52. Beitel LK , Kazemi-Esfarjani P , Kaufman M , Lumbroso R , DiGeorge AM , Killinger DW , Trifiro MA , Pinsky L. 1994. Substitution of arginine-839 by cysteine or histidine in the androgen receptor causes different receptor phenotypes in cultured cells and coordinate degrees of clinical androgen resistance. J Clin Invest 94:546–554 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53. Melo KF , Latronico AC , Costa EM , Billerbeck AE , Mendonca BB , Arnhold IJ. 1999. A novel point mutation (R840S) in the androgen receptor in a Brazilian family with partial androgen insensitivity syndrome. Hum Mutat 14:353. [DOI] [PubMed] [Google Scholar]
- 54. Nazareth LV , Stenoien DL , Bingman WE , James AJ , Wu C , Zhang Y , Edwards DP , Mancini M , Marcelli M , Lamb DJ , Weigel NL. 1999. A C619Y mutation in the human androgen receptor causes inactivation and mislocalization of the receptor with concomitant sequestration of SRC-1 (steroid receptor coactivator 1). Mol Endocrinol 13:2065–2075 [DOI] [PubMed] [Google Scholar]
- 55. Buchanan G , Yang M , Harris JM , Nahm HS , Han G , Moore N , Bentel JM , Matusik RJ , Horsfall DJ , Marshall VR , Greenberg NM , Tilley WD. 2001. Mutations at the boundary of the hinge and ligand binding domain of the androgen receptor confer increased transactivation function. Mol Endocrinol 15:46–56 [DOI] [PubMed] [Google Scholar]
- 56. Wang Q , Lu J , Yong EL. 2001. Ligand- and coactivator-mediated transactivation function (AF2) of the androgen receptor ligand-binding domain is inhibited by the cognate hinge region. J Biol Chem 276:7493–7499 [DOI] [PubMed] [Google Scholar]
- 57. Tahiri B , Auzou G , Nicolas JC , Sultan C , Lupo B. 2001. Participation of critical residues from the extreme C-terminal end of the human androgen receptor in the ligand binding function. Biochemistry 40:8431–8437 [DOI] [PubMed] [Google Scholar]
- 58. Gelmann EP. 2002. Molecular biology of the androgen receptor. J Clin Oncol 20:3001–3015 [DOI] [PubMed] [Google Scholar]
- 59. Ong YC , Kolatkar PR , Yong EL. 2002. Androgen receptor mutations causing human androgen insensitivity syndromes show a key role of residue M807 in helix 8-helix 10 interactions and in receptor ligand-binding domain stability. Mol Hum Reprod 8:101–108 [DOI] [PubMed] [Google Scholar]
- 60. McPhaul MJ. 2002. Molecular defects of the androgen receptor. Recent Prog Horm Res 57:181–194 [DOI] [PubMed] [Google Scholar]
- 61. Shi XB , Ma AH , Xia L , Kung HJ , de Vere White RW. 2002. Functional analysis of 44 mutant androgen receptors from human prostate cancer. Cancer Res 62:1496–1502 [PubMed] [Google Scholar]
- 62. Gruber SB , Chen H , Tomsho LP , Lee N , Perrone EE , Cooney KA. 2003. R726L androgen receptor mutation is uncommon in prostate cancer families in the United States. Prostate 54:306–309 [DOI] [PubMed] [Google Scholar]
- 63. Mazen I , Lumbroso S , Abdel Ghaffar S , Salah N , Sultan C. 2004. Mutation of the androgen receptor (R840S) in an Egyptian patient with partial androgen insensitivity syndrome: review of the literature on the clinical expression of different R840 substitutions. J Endocrinol Invest 27:57–60 [DOI] [PubMed] [Google Scholar]
- 64. Quigley CA , Tan JA , He B , Zhou ZX , Mebarki F , Morel Y , Forest MG , Chatelain P , Ritzén EM , French FS , Wilson EM. 2004. Partial androgen insensitivity with phenotypic variation caused by androgen receptor mutations that disrupt activation function 2 and the NH(2)- and carboxyl-terminal interaction. Mech Ageing Dev 125:683–695 [DOI] [PubMed] [Google Scholar]
- 65. Ledig S , Jakubiczka S , Neulen J , Aulepp U , Burck-Lehmann U , Mohnike K , Thiele H , Zierler H , Brewer C , Wieacker P. 2005. Novel and recurrent mutations in patients with androgen insensitivity syndromes. Horm Res 63:263–269 [DOI] [PubMed] [Google Scholar]
- 66. Yen JL , Chang KH , Sheu JC , Lee YJ , Tsai LP. 2005. Partial androgen insensitivity syndrome with R840H mutation in androgen receptor: report of one case. Acta Paediatr Taiwan 46:101–105 [PubMed] [Google Scholar]
- 67. Haelens A , Tanner T , Denayer S , Callewaert L , Claessens F. 2007. The hinge region regulates DNA binding, nuclear translocation, and transactivation of the androgen receptor. Cancer Res 67:4514–4523 [DOI] [PubMed] [Google Scholar]
- 68. Deeb A , Jääskeläinen J , Dattani M , Whitaker HC , Costigan C , Hughes IA. 2008. A novel mutation in the human androgen receptor suggests a regulatory role for the hinge region in amino-terminal and carboxy-terminal interactions. J Clin Endocrinol Metab 93:3691–3696 [DOI] [PubMed] [Google Scholar]
- 69. Tanner TM , Denayer S , Geverts B , Van Tilborgh N , Kerkhofs S , Helsen C , Spans L , Dubois V , Houtsmuller AB , Claessens F , Haelens A. 2010. A 629RKLKK633 motif in the hinge region controls the androgen receptor at multiple levels. Cell Mol Life Sci 67:1919–1927 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70. Marimuthu A , Feng W , Tagami T , Nguyen H , Jameson JL , Fletterick RJ , Baxter JD , West BL. 2002. TR surfaces and conformations required to bind nuclear receptor corepressor. Mol Endocrinol 16:271–286 [DOI] [PubMed] [Google Scholar]
- 71. Yumoto F , Nguyen P , Sablin EP , Baxter JD , Webb P , Fletterick RJ. 2012. 2012 Structural basis of coactivation of liver receptor homolog-1 by β-catenin. Proc Natl Acad Sci USA 109:143–148 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72. De Leon JT , Iwai A , Feau C , Garcia Y , Balsiger HA , Storer CL , Suro RM , Garza KM , Lee S , Kim YS , Chen Y , Ning YM , Riggs DL , Fletterick RJ , Guy RK , Trepel JB , Neckers LM , Cox MB. 2011. Targeting the regulation of androgen receptor signaling by the heat shock protein 90 cochaperone FKBP52 in prostate cancer cells. Proc Natl Acad Sci USA 108:11878–11883 [DOI] [PMC free article] [PubMed] [Google Scholar]