Abstract
Previous studies shows that connexins appear very early during murine embryo development, the gap junctional intercellular communication found in the inner cell mass of early embryo is also maintained in embryonic stem cells (ESC), and expression of oxytocin receptor (OTR) is developmentally regulated at early embryonic development. However, effect of oxytocin (OT) on the regulation of the connexin43 (Cx43) and maintenance of undifferentiation is not fully understood in stem cells. Therefore, we investigated the effect of OT on Cx43 expression and related signaling cascades in mouse ESC. OT increased Cx43 expression that was inhibited by the OTR inhibitor atosiban. In experiments to examine whether the effect of OT depends on lipid rafts, caveolin-1 (cav-1), cav-2, and flotillin-2, but not OTR, were detected in lipid raft fractions. Also, colocalization of OTR, cav-1, and cav-2 was not detected. Moreover, the lipid raft disruptor methyl-β-cyclodextrin did not attenuate OT-induced Cx43 expression. In experiments to examine related signaling pathways, OT activated cAMP/protein kinase A (PKA) which was inhibited by adenylyl cyclase inhibitor SQ 22536 and PKA inhibitor PKI. OT increased nuclear factor κ-light-chain-enhancer of activated B cells (NF-κB) phosphorylation which was inhibited by PKI. OT also increased cAMP response element-binding (CREB)/CREB-binding protein (CBP) expression in the nucleus and induced the formation of CREB1/NF-κB/CBP complexes, which was blocked by the NF-κB-specific small interfering RNA, NF-κB inhibitors, SN50, and bay11–7082. Complex disruption by NF-κB inhibitors decreased OT-induced Cx43 expression. In conclusion, OT stimulates Cx43 expression through the NF-κB/CREB/CBP complex via the lipid raft-independent OTR-mediated cAMP/PKA in mouse ESC.
Embryonic stem cells (ESC) are cells with self-renewal capacity and the ability to differentiate into multiple cell types (1, 2). The remarkable self-renewal and differentiation properties of ESC have been targeted for research and use in therapeutic applications (1, 2). Over the last few years, gap junctions, generally thought of as specialized intercellular connections, have become recognized as key players in the regulation of ESC self-renewal and differentiation (3–6). Various connexins (Cx) or gap junction proteins have been found to be expressed in ESC including Cx31, Cx43, and Cx45 (3). Cx43 is a highly enriched and prominent subtype in undifferentiated ESC compared with their differentiated counterparts and is identified as an ”undifferentiated mouse ESC marker” (3). Interactions between stem cells and hormones especially are important to regulate stem cell function including proliferation or differentiation through alter gene expression and regulated signaling molecules such as matrix metalloproteinase 2 (7). Despite the amazing role of gap junctions in ESC development, regulatory mechanisms of Cx43 and its roles remain unknown in maintenance of mouse ESC undifferentiated state.
There are few reports that oxytocin (OT) has been shown to play various roles in stem cells. In human umbilical cord blood-derived mesenchymal stem cells, OT stimulates migration and proliferation through regulating matrix metalloproteinase 2 expression (7). Also, OT promotes the differentiation via down-regulation of Nanog and Oct4, and activated Erk and Wnt are found at very low levels in human bone marrow-derived mesenchymal stem cells (8). Recently, new functions have also been reported for OT as a positive and negative regulator of gap junctions in uterine myocytes and myoepithelial cells (9–11), and gap junctions are overly sensitive to OT in embryos (12). These different effects seem to be mediated by different signaling pathways elicited by differences in OT receptor (OTR) locations in myoepithelial and cancer cells (10). In turn, these different OTR locations ultimately lead to different temporal patterns of cellular functions through various signaling mechanisms: 1) The coupling of OTR located outside lipid rafts to OT leads to cAMP, protein kinase A (PKA), and calcium ion (Ca2+) signaling (10, 11, 13) whereas 2) the coupling of OTR located inside lipid rafts leads to phosphatidylinositol 3-kinase/serine/threonine protein kinase (PI3K/Akt), MAPK, and epidermal growth factor receptor signaling (14, 15).
Previous studies shows that connexins appear very early during murine embryo development, the gap junctional intercellular communication (GJIC) found in the inner cell mass of the early embryo is also maintained in ESC, and expression of oxytocin receptor is developmentally regulated at early embryonic development (5, 12, 16, 17). These results suggested that interaction between oxytocin and GJIC might be involved in the maintenance of the self-renewal and pluripotent state of ESC. In addition, a better knowledge of these processes is crucial for the management of these cells in medical applications and may improve the understanding of the mechanisms involved in the regulation of early development. Therefore, we examined the effect of OT on Cx43 expression and its related signaling cascades in the maintenance of undifferentiated state of ESC.
Materials and Methods
Materials
Mouse ESC were obtained from the American Type Culture Collection (ES-E14TG2a; Manassas, VA). Fetal bovine serum (FBS) was purchased from BioWhittaker, Inc. (Walkersville, MO). Oxytocin (OT), atosiban, SQ 22356, 8-bromoadenosine (8-Br)-cAMP, forskolin (FSK), methyl-β-cyclodextrin (MβCD), propidium iodide (PI), Lucifer yellow (LY), and mouse leukemia inhibitory factor (LIF) were obtained from Sigma Chemical Co. (St. Louis, MO). Protein kinase inhibitor (PKI) and SN 50 were purchased from Calbiochem (La Jolla, CA). Bay11–7082 was purchased from Biomol International, LP (Plymouth Meeting, PA). Anti-Nanog, Oct4, SSEA-1, Cx43, OTR, caveolin (cav)-1, cav-2, phospho-nuclear factor κB (NF-κB) p60, NF-κB, CREB, CREB-binding protein (CBP), β-actin, and lamin A/C were purchased from Santa Cruz Biotechnology, Inc. (Santa Cruz, CA). Horseradish peroxidase-conjugated goat antirabbit and goat antimouse IgG were purchased from Jackson ImmunoResearch Laboratories (West Grove, PA). Liquiscint was obtained from National Diagnostics (Parsippany, NJ). All other reagents were of the highest purity commercially available and were used as received.
ESC culture
Mouse ESC were cultured for 5 d in DMEM (Life Technologies, Inc., Gaithersburg, MD) supplemented with 3.7 g/liter sodium bicarbonate, 1% penicillin and streptomycin, 1.7 mm l-glutamine, 0.1 mm β-mercaptoethanol, 5 ng/ml mouse LIF, and 15% FBS, without a feeder layer. The cells were grown on gelatinized 12-well plates or 60-mm culture dishes in an incubator maintained at 37 C in an atmosphere containing 5% CO2 and air. After 2–3 d, the cells were washed twice with PBS and maintained in serum-free DMEM with all supplements. After a 24-h incubation period, the cells were washed twice with PBS and incubated with fresh serum-free media including all supplements and designated agents for the indicated period.
Scrape loading/dye transfer assay
The gap junction intercellular communication level was measured using a slightly modified scrape loading/dye transfer (SL/DT) method (18). The SL/DT assay was carried out using a noncytotoxic dose of the samples, as determined by Thiazolyl Blue Tetrazolium Blue (MTT) assay. Mouse ESC were treated in the presence or absence of OT (10−7 m) for 3 h after which cells on 35-mm dishes were rinsed twice with 2 ml of PBS. After rinsing with PBS, a mixture of 1% LY (Sigma Chemical Co., St. Louis, MO) and 1% of Rhodamine-dextran (RD, Invitrogen, Carsbad, CA) in PBS was added. The cell layer was then scratched by applying a 26-gauge needle. After exactly 3 min incubation in the dye mixture, the cells are rinsed three times with PBS. Cells are fixed for 20 min at room temperature with 4% paraformaldehyde/PBS (pH 7.5) and washed three times with PBS. LY and RD were observed by optical microscopy (FluoView 300, Olympus; Tokyo, Japan).
RNA isolation and RT-PCR
Total RNA was extracted from mouse ESC using STAT-60, which is a monophasic solution of phenol and guanidine isothiocyanate purchased from Tel-Test, Inc. (Friendswood, TX; http://www.bioresearchonline.com). Reverse transcription (RT) was carried out using 3 μg of RNA using a RT system kit (AccuPower RT PreMix; Bioneer, Daejeon, Korea; http://www.bioneer.com) with oligo(dT)18 primers. A PCR kit (AccuPower PCR PreMix; Bioneer) was used to amplify 5 μl of the RT product under the following conditions: denaturation at 94 C for 5 min, 30 cycles at 94 C for 45 sec, Annealing melting temperature (Tm) (Table 1) for 30 sec, and 72 C for 30 sec, followed by 5 min of extension at 72 C. Amplifications of connexin family members (26, 30.3, 31, 32, 37, 40, 43, 45, 46, 47) cDNA were performed in mouse ESC using the primers described in Table 1. PCR of β-actin was also performed as a control for RNA quantity.
Table 1.
Primers used for PCR
| Gene | Identification | Primer sequence, 5′-3′ | Annealing Tm. (° C) | Size (bp) |
|---|---|---|---|---|
| Cx26 | Sense | CGGAAGTTCATGAAGGGAGAGAT | 55 | 365 |
| Antisense | ACGAGTCCTTTCAGGTTTTCTGG | |||
| Cx30.3 | Sense | TCAAACATGGGCCCAATG | 50 | 182 |
| Antisense | CGAACGAGACACTGAGGG | |||
| Cx31 | Sense | AGAAGCACGGGGAGCAAT | 55 | 182 |
| Antisense | GTCACGCGGTCGTATCAT | |||
| Cx32 | Sense | CTGCTCTACCCCGGCTATGC | 60 | 386 |
| Antisense | TTCTCGCTGGCTACGAGTCGGAC | |||
| Cx37 | Sense | GGCTGGACCATGGAGCCGGT | 60 | 422 |
| Antisense | CGGGGGGTCCCACCGGCTTT | |||
| Cx40 | Sense | CTGTCCCCACCCAGTCAACT | 56 | 460 |
| Antisense | CGATGGTATCACTGTTTGCC | |||
| Cx43 | Sense | TACCACGCCACCACTGGCCCA | 57 | 294 |
| Antisense | ATTCTGGTTGTCGTCGGGGAAATC | |||
| Cx45 | Sense | TTCCAAGTCCACCCATTTTAT | 55 | 444 |
| Antisense | AGTCTTACCGAGTCCTTGCTA | |||
| Cx46 | Sense | GGAAAGGCCACAGGGTTTCCTGG | 60 | 332 |
| Antisense | GGCAACCAGGAGGACCTGGG | |||
| Cx47 | Sense | TAGCCCCACAGTATGCCCTTAG | 64 | 530 |
| Antisense | CGTCTGCGCTCCTGTTCC | |||
| Nanog | Sense | CACCCACCCATGCTAGTCTT | 54 | 1071 |
| Antisense | ACCCTCAAACTCCTG GTCCT | |||
| Oct4 | Sense | CGTGAGACTTTGCAGCCTGA | 50 | 519 |
| Antisense | GGGATGTAAGTGATCTGCTG | |||
| FoxD3 | Sense | TCTCTGGGGCAATCACACTC | 50 | 550 |
| Antisense | GTACATTTGTTGATAAAGGG | |||
| Sox2 | Sense | GGCAGCTACAGCATGATGCAGGAGC | 50 | 171 |
| Antisense | CTGGTCATGGAGTTGTACTGCAGG | |||
| β-Actin | Sense | AGCCATGTACGTAGCCATCC | 55 | 350 |
| Antisense | CTCTCAGCTGTGGTGGTGAA |
FoxD3, Forkhead box D3; Oct4, octamer-binding transcription factor 4; Sox2, sex determining region Y box 2.
Real-time PCR
Total RNA was extracted from mouse ESC using STAT-60, which is a monophasic solution of phenol and guanidine isothiocyanate purchased from Tel-Test, Inc. (Friendswood, TX; http://www.bioresearchonline.com). RT was carried out with 3 μg of RNA using a RT system kit (AccuPower RT PreMix; Bioneer, Daejeon, Korea; http://www.bioneer.com) with oligo(dT)18 primers. The real-time quantification of RNA targets was performed using a Rotor-Gene 6000 real-time thermal cycling system (Corbett Research, New South Wales, Australia) with a QuantiTect SYBR Green RT-PCR Kit (QIAGEN, Chatsworth, CA), 20 μl reaction mixture contained 200 ng cDNA, and 0.5 μm of each primer, enzymes, and fluorescent dyes. The primers used are described in Table 1. The data were collected during the extension step and analyzed using the manufacturer's software. To verify the specificity and identity of PCR products, the amplification cycles were followed by a high-resolution melting cycle from 65 C to 99 C at a rate of 0.1 C/2 sec. When the Tm was reached, double-stranded DNA was denatured and the SYBR was released, which caused a dramatic decrease in fluorescence intensity. The rate of this change was determined by plotting the derivative of the fluorescence relative to the temperature (dF/dT) vs. temperature by data analysis software of the real-time PCR instrument. The temperature at which a peak occurred on the plot corresponded to the Tm of the DNA duplex. β-Actin was used as an endogenous control, and a normalization control was used as a defined calibrator.
Detergent-free purification of caveolin-rich membrane fraction
Caveolin-enriched membrane fractions were prepared as described previously (19). Cells were washed twice with ice-cold PBS, scraped into 2 ml of 500 mm sodium carbonate (pH 11.0), transferred to a plastic tube, and homogenized with a Sonicator 250 apparatus (Branson Ultrasonic, Danbury, CT) using three 20-sec bursts. The homogenate was adjusted to 45% sucrose by the addition of 2 ml 90% sucrose prepared in 2-(N-morpholino)ethanesulfonic acid (MES)-buffered solution consisting of 25 mm MES-buffer solution (pH 6.5) and 0.15 m NaCl and placed at the bottom of an ultracentrifuge tube. A 5%–35% discontinuous sucrose gradient was formed (4 ml each of 5% and 35% sucrose, both in MES-buffer solution containing 250 mm sodium carbonate) and centrifuged at 40,000 rpm for 20 h in a SW 41 Rotor (Beckman Coulter, Fullerton, CA). Twelve fractions (1 ml each) were collected and analyzed by 8–12% SDS-PAGE.
Nuclear and nonnuclear protein fractionation
Cells were lysed in hypotonic buffer [20 mm HEPES (pH 7), 10 mm KCl, 2 mm MgCl2, 0.5% Nonidet P-40, 1 mm sodium orthovanadate] containing a cocktail of protease inhibitors. Lysates were homogenized gently by 30 passes in a syringe with a 19-gauge needle and 30 passes with a 25-gauge needle and then centrifuged at 1500 × g for 5 min at 4 C. The supernatants containing the nonnuclear fraction were cleared by centrifugation at 12,000 × g for 5 min and preserved at −70 C. The pellets containing the nuclear fraction were washed three times in hypotonic buffer. After homogenization in hypertonic buffer containing 0.5 m NaCl and a cocktail of protease inhibitors, pellets were centrifuged at 12,000 × g for 10 min and the supernatants preserved at −70 C.
Immunofluorescence microscopy
Cells were plated onto coverslips, serum starved for 24 h, and then stimulated for 1 or 3 h with 50 μm OT. Cells were fixed with 3.5% paraformaldehyde in PBS, permeabilized for 10 min with 0.1% (vol/vol) Triton X-100, and washed three times for 10 min each with PBS. Cells, preincubated with 10% BSA (Sigma-Aldrich) in PBS for 20 min to decrease nonspecific antibody binding, were incubated for 60 min with a 1:100 dilution of primary antibody (anti-Cx43 and anti-phospho-NF-κB antibody) in a solution containing 1% (vol/vol) BSA in PBS, and washed three times for 10 min each with PBS. Cells were then incubated with 1% (vol/vol) BSA for 5 min, incubated for 60 min with fluorescein isothiocyanate-conjugated secondary antibody antimouse or antirabbit IgM-fluorescein isothiocyanate (green) antibody, counterstained with PI in PBS containing 1% (vol/vol) BSA, and washed three times for 10 min each with PBS. Samples were mounted on slides and visualized with an Olympus FluoView 300 confocal microscope with ×400 objective.
Immunoassay for cAMP
Cells were cultured until they reached 70% confluence and then treated with OT or AH 6809 for various times and concentrations at 37 C. The cells were washed with cold PBS and then harvested in ice-cold ethanol. The collected samples were vacuum dried and resuspended in assay buffer as suggested in the kit instructions. The amount of intracellular cAMP was determined using a cAMP enzyme immunoassay kit according to the manufacturer's instructions (R&D, Minneapolis, MN).
Analysis of PKA activity
The activity of PKA was determined as described previously (20). Briefly, cells were washed with cold PBS, lysed in HP buffer [10 mm potassium phosphate (pH 6.8), 1 mm β-mercaptoethanol, 10 μg/ml leupeptin, 10 mm magnesium acetate, 10 μm ATP, and 300 μg/ml Kemptide substrate], and then homogenized by passing 15 times through a 26-gauge needle. The homogenates were centrifuged at 8000 rpm at 4 C for 5 min, and the supernatant was collected. The activity of PKA was measured in a total volume of 100 μl containing 5 × 105 cpm [γ-32P]ATP (3000 Ci/mmol). The reaction mixtures were incubated at 30 C for 5 min, after which 30 μl of reaction mixture were spotted onto Whatman P-81 paper. The paper was washed in 75 mm phosphoric acid twice and acetone once and then subjected to scintillation counting for measurement of Kemptide phosphorylation levels.
Small interfering RNA (siRNA) transfection
Mouse ESC were grown until 75% of the plate surface was covered after which they were transfected for 24 h with either siRNA specific for NF-κB, CREB, CBP (100 nmol/liter; Bioneer, Alameda, CA) or nontargeting siRNA (as a negative control; 100 nmol/liter; Dharmacon, Inc., Lafayette, CO; http://www.dharmacon.com) using Hyperfectamine (QIAGEN) according to the manufacturer's instructions. After 24 h, transfection mixtures were replaced with regular medium, and cells were maintained in normal culture conditions (DMEM supplemented with LIF and FBS) before the experiments. The sequences used are described in Table 2. Next, we determined each siRNA efficacy, respectively (Supplemental Fig. 1).
Table 2.
siRNA sequence for transfection
| Gene | Sequence, 5′-3′ |
|---|---|
| OTR | GACACACACACCUAUGCAU |
| AUGCAUACCUCUGUGUGUC | |
| GACACACACACCUAUGCAU | |
| AUGCAUAGGUGUGUGUGUC | |
| NF-κB | CUGCAAAGGUUAUCGUUCA |
| UGAACGAUAACCUUUGCAG | |
| GAAGAAAAUGGCGGAGUUU | |
| AAACUCCGCCAUUUUCUUC | |
| CREB1 | CUGUACAUAUGCUACUGAU |
| AUCAGUAGCAUAUGUACAG | |
| GAGUGUGUGCUAUGGUACA | |
| UGUACCAUAGCACACACUC | |
| CBP | UCAUCACAGCAGCAACCAA |
| UUGGUUGCUGCUGUGAUGA | |
| GUGACAAGCGAAACCAACA | |
| UGUUGGUUUCGCUUGUCAC | |
| Cx43 | GACAAGGUCCAAGCCUACU |
| AGUAGGCUUGGACCUUGUC | |
| CUCACAGAUUUGAAUCGAA | |
| UUCGAUUCAAAUCUGUGAG | |
| Nt | CCUACGCCACCAAUUUCGU |
| ACGAAAUUGGUGGCGUAGG | |
| UGGUUUACAUGUCGACUAA | |
| UUAGUCGACAUGUAAACCA |
Nt, Nontargeting.
Western blot analysis
Cells were harvested, washed twice with PBS, and lysed with buffer [20 mm Tris (pH 7.5), 1 mm EDTA, 1 mm EGTA, 1% Triton X-100, 1 mg/ml aprotinin, and 1 mm phenylmethylsulfonylfluoride (PMSF)] for 30 min on ice. The lysates were then cleared by centrifugation (15,000 rpm at 4 C for 10 min). Protein concentration was determined by the Bradford method (21). Equal amounts of protein (20 μg) were resolved by 10% SDS-PAGE and transferred to polyvinylidene fluoride membranes. After the blots were washed with TBST [10 mm Tris-HCl (pH 7.6), 150 mm NaCl, and 0.05% Tween-20] the membranes were blocked with 5% skim milk for 1 h and incubated with an appropriate primary antibody at the dilution recommended by the supplier. The membrane was then washed, and primary antibodies were detected with a horseradish peroxidase-conjugated secondary antibody. The bands were visualized by enhanced chemiluminescence (Amersham Pharmacia Biotech, Inc., Buckinghamshire, UK). Densitometric analysis was performed using the TINA version 2.09 program package. The ratios between each treated and control sample were calculated for each individual experiment and expressed as a percentage of control.
Proximity ligation assay (PLA)
Cells were fixed in 4% paraformaldehyde/PBS for 20 min, permeabilized with 0.25% Triton X-100/PBS for 5 min, and blocked in 10% FBS in 0.1% Triton X-100/PBS for 20 min. Primary antibody (anti-phospho-NF-κB, anti-CREB, or anti-CBP) diluted in 0.1% Triton X-100/PBS/1% FBS was added for 2 h. The Duolink (Olink Biosciences, Uppsala, Sweden) in situ PLA was performed according to the manufacturer's protocol. PLA probes were diluted in 0.1% Triton X-100/PBS/1% FBS and incubated in a preheated humidity chamber for 1 h at 37 C, followed by hybridization, ligation, amplification, and detection. The distance between the two primary antibodies must be less than 40 nm to generate a signal in this assay, making the methodology highly specific for physically interacting protein-protein complexes. Samples were mounted on slides and visualized with an Olympus FluoView 300 confocal microscope with a ×400 objective.
Alkaline phosphatase (AP) staining
Cells were washed twice with PBS and fixed with 4% formaldehyde for approximately 15 min at room temperature. After washing the cells with PBS, they were incubated with AP substrate solution [200 mg/ml naphthol AS-MX phosphate, 2% N, N dimethylformamide, 0.1 m Tris (pH 8.2), and 1 mg/ml Fast RedTR salt] for approximately 10–15 min at room temperature. After washing with PBS, the cells were photographed.
Statistical analysis
Results are expressed as means ± se. All experiments were analyzed by ANOVA, followed in some cases by a comparison of treatment means to the control using the Bonferroni-Dunn test. Differences were considered statistically significant at P < 0.05.
Results
Effect of OT on Cx43 expression via lipid raft-independent OTR
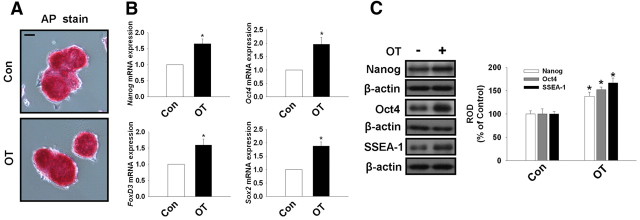
The undifferentiated state of the mouse ESC used in this experiment was confirmed by examining the expression of undifferentiated stem cell markers, including alkaline phosphatase (AP) activity, Oct4, Sox2, FoxD3, and Nanog expression. As shown in Fig. 1A, mouse ESC, in both the presence and absence of OT, maintained AP enzyme activity, which was detected by immunofluorescent staining. Mouse ESC treated with OT (10−7 m) for 72 h increased Oct4, Sox2, FoxD3, and Nanog mRNA expression (Fig. 1B). In addition, we observed Oct4, SSEA-1, and Nanog protein expression (Fig. 1C) was increased after treatment with OT (10−7 m) for 72 h. Therefore, these results suggest that mouse ESC were maintained in an undifferentiated state under the experimental condition used in this study.
Fig. 1.
Effect of OT on undifferentiated marker genes of mouse ESC. A, AP enzyme activity was assessed in mouse ESC treated in the presence or absence of OT (10−7 m) for 72 h as described in Materials and Methods. Scale bars, 20 μm (magnification ×400). B, The cells were treated with OT (10−7 m) for 72 h. Total RNA from mouse ESC was reverse transcribed, and Nanog, Oct4, FoxD3, Sox2, and β-actin cDNA were amplified by real-time PCR as described in Materials and Methods. The data are reported as the mean ± se of four independent experiments, each conducted in triplicate. *, P < 0.05 vs. control. C, The cells were treated with OT (10−7 m) for 72 h. Total protein was extracted and blotted with Nanog, Oct4, SSEA-1, and β-actin antibodies. Each example shown is representative of four independent experiments. The right part (C) depicting the bars denotes the mean ± se of four independent experiments for each condition determined from densitometry relative to β-actin. *, P < 0.05 vs. control. Con, Control; ROD, Relative Optical Density.
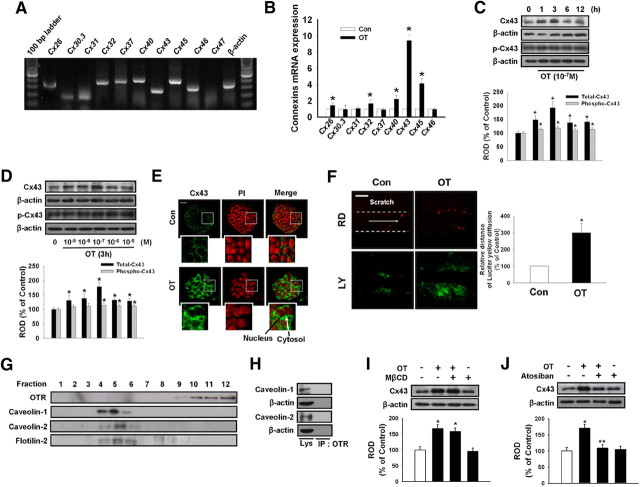
In experiments to determine the existence of connexin family members in ESC, we detected signals for amplicons of Cx26, Cx30.3, Cx31, Cx32, Cx37, Cx40, Cx43, Cx45, and Cx46 mRNA, but not Cx47 mRNA (Fig. 2A). Among them, OT maximally stimulated the Cx43 mRNA expression level (Fig. 2B). Thus, we examined the effect of OT on Cx43 expression and analyzed the underlying mechanisms involved in regulatory activities related to OTR in mouse ESC. The effect of OT on Cx43 protein expression levels was examined by treating mouse ESC with 10−7 m OT for various time periods (0–12 h) or with various doses (0 to 10−5 m) for 3 h. As shown in Fig. 2C and D, the maximum increase in total Cx43 protein expression was observed after 3 h incubation with an OT dose of 10−7 m. Although OT slightly increased Cx43 phosphorylation (14% increased compared with the control), OT more increased total Cx43 protein expression level (81% increased compared with the control) than phosphorylated form of Cx43. Cx43 protein expression levels assessed using immunofluorescence staining showed a complimentary pattern (Fig. 2E) Next, we examined GJIC in mouse ESC using the SL/DT assay with LY and RD. When cells treated with OT (10−7 m) were scraped and incubated in the presence of LY and RD, extensive LY diffusion through the colonies was observed, whereas LY and RD remained at the site of the scrape in the control group. Mean ± sd (inch in photographs) of maximal distance of lateral spreading of LY on mouse ESC treated with OT was significantly increased than in control group (Fig. 2F). To assess whether OTR localized without the lipid raft fraction, the latter fractions were prepared by detergent-free purification using discontinuous sucrose density gradient centrifugation. The plasma membrane lipid raft fraction was found to reside mainly in the light buoyant membranes (Fig. 2G, fractions 4, 5, and 6). Western blot analysis for OTR, cav-1, cav-2, and flotilin-2 demonstrated only localization of cav-1 and cav-2 in the lipid raft fraction, but not OTR (Fig. 2G). Also, OTR did not colocalize with cav-1 and −2 as shown in immunoprecipitation experiments (Fig. 2H). The lipid raft disruptor MβCD (10−4 m) and OTR inhibitor atosiban (10−5 m) were also used to further confirm lipid raft-independent OTR signaling in OT-induced Cx43 expression. Pretreatment with atosiban significantly blocked OT-induced Cx43 protein expression, but no effect was observed with MβCD pretreatment (Fig. 2, I and J).
Fig. 2.
OT increases Cx43 expression via lipid raft-independent OTR. A, Total RNA from mouse ESC was reverse transcribed, and connexin family members cDNA was amplified by PCR as described in Materials and Methods. Each example shown is representative of five independent experiments. B, Cells were treated with 10−7 m OT for 3 h. The mRNA expression of connexin family members was measured in cells using real-time PCR as described in Materials and Methods. The data is reported as the mean ± se of four independent experiments, each conducted in triplicate. *, P < 0.05 vs. control. Time (C) and dose (D) responses of OT on Cx43 protein expression levels: Mouse ESC were treated with OT for various times (0–12 h) or different doses of OT (0–10−5 m) for 3 h and then harvested. Total protein was extracted and blotted with antibody against total Cx43 and phospho-Cx43. Each of the examples shown is representative of five independent experiments. The lower part (C and D) depicting the bars denotes the mean ± se of five independent experiments for each condition determined from densitometry relative to β-actin. *, P < 0.05 vs. control. E, Mouse ESC were incubated with OT for 3 h, and Cx43 was detected by immunostaining with anti-Cx43 antibody counterstained with PI. Insets show magnified versions of the sections in the white boxes. Scale bars, 20 μm (magnification, ×400). F, Mouse ESC treated with OT (10−7 m) for 3 h and GJIC analysis was carried out using a SL/DT assay, as described in Materials and Methods. The right panel depicts the mean ± sd of five independent experiments for each condition as determined from quantification of GJIC (the relative distance of LY diffusion compared with control). *, P < 0.05 vs. control. Scale bars, 20 μm (magnification, ×400). G, Control lysate was subjected to discontinuous sucrose density gradient fractionation, after which OTR, caveolin (cav)-1, cav-2, and flotilin-2 were detected. Each fraction was assessed by Western blot. H, Normal mouse ESC lysates were analyzed by Western blotting with antibodies that recognize OTR, cav-1, and cav-2. Immunoprecipitation of anti-OTR was analyzed by Western blotting with antibodies that recognize cav-1 and cav-2. Each of the examples is representative of five independent experiments. I, Cells were pretreated with lipid raft disruptor MβCD (10−4 m) for 30 min before OT (10−7 m) treatment for 3 h. Total protein was extracted and blotted with Cx43 or β-actin antibodies. Each example shown is representative of five independent experiments. J, Cells were pretreated with the OTR inhibitor atosiban (10−5 m) for 30 min before OT treatment for 3 h. Total protein was extracted and blotted with Cx43 or β-actin antibodies. Each example shown is a representative of five independent experiments. The lower part (H–J) depicting the bars denotes the mean ± se of five independent experiments for each condition determined from densitometry relative to β-actin. *, P < 0.05 vs. control, **, P < 0.05 vs. OT alone. Con, Control; IP, immunoprecipitation; ROD, Relative Optical Density.
Effect of OT on cAMP production and PKA activity
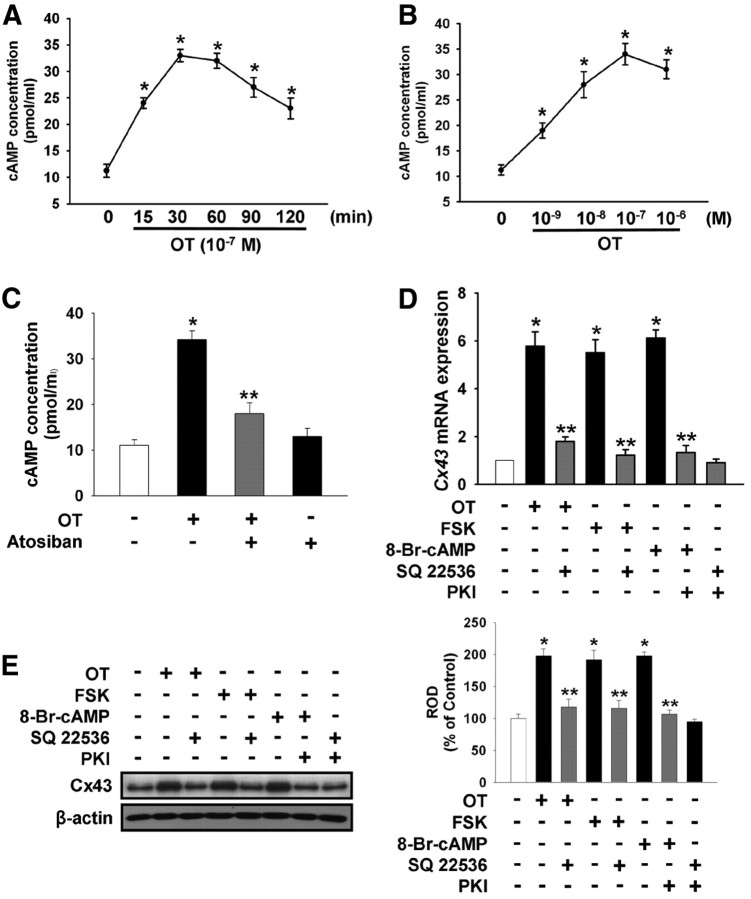
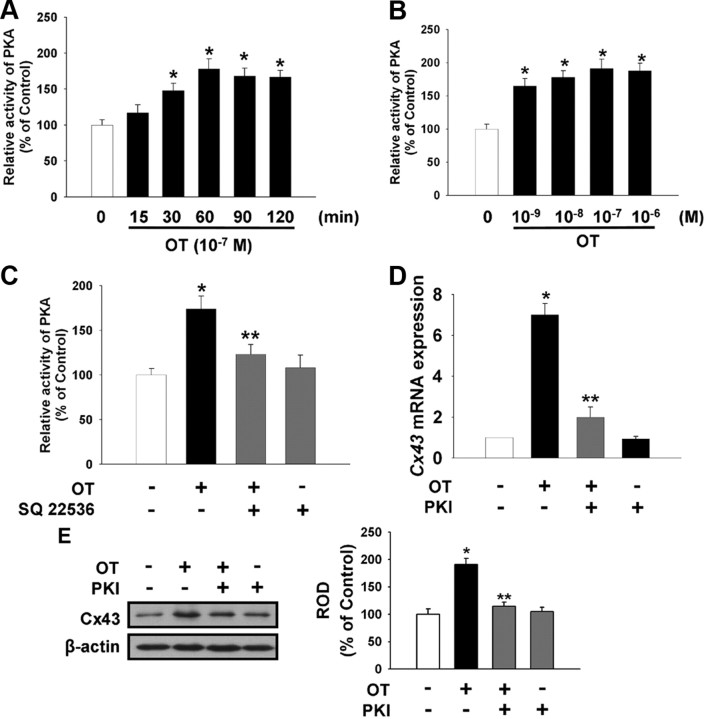
The effect of OT on intracellular cAMP production was also examined. As shown in Fig. 3A, there was increased production of intracellular cAMP after a 15-min incubation with 10−7 m OT. Also, an increasing concentration of OT from 10−9 m to 10−6 m induced intracellular cAMP production in a dose-dependent manner (Fig. 3B). As shown in Fig. 3C, atosiban (10−5 m) reduced the OT-induced increase in intracellular cAMP. A significant increase in Cx43 mRNA and protein expression levels were observed in cells treated with the adenylate cyclase activator FSK (10−5 m) and the membrane-permeable cAMP analog 8-Br-cAMP (2 × 10−3 m) as well as OT, which were blocked by adenylyl cyclase inhibitor SQ 22536 (10−5 m) or protein kinase A inhibitor PKI (4 × 10−5 m) (Fig. 3, D and E). To examine the involvement of PKA activity in OT-induced Cx43 expression, we assessed whether OT induced PKA activity in mouse ESC. As shown in Fig. 4A, PKA activity increased after a 30-min incubation with 10−7 m OT. Also, increasing the concentration of OT from 10−9 m to 10−6 m induced increased PKA activity (Fig. 4B). SQ 22536 reduced the OT-induced increase in PKA activity (Fig. 4C). Pretreatment with PKI (4 × 10−5 m) significantly blocked the OT-induced increase in Cx43 mRNA and protein expression level (Fig. 4, D and E).
Fig. 3.
OT elevate intracellular cAMP levels. Time (A) and dose (B) response of OT on intracellular cAMP levels. Mouse ESC were treated with OT for various times (0–120 min) or with different doses of OT (0–10−6 m) for 30 min. The maximum increase in intracellular cAMP levels was observed after a 30-min incubation with 10−7 m OT. The values represent the mean ± se of four independent experiments with triplicate dishes. *, P < 0.05 vs. control. C, Cells were pretreated with atosiban (10−5 m) for 30 min before OT treatment for 30 min. The values represent the mean ± se of four independent experiments with triplicate dishes. *, P < 0.05 vs. control; **, P < 0.05 vs. OT alone. D, Real-time RT-PCR quantification of the Cx43 gene. Cells were pretreated with adenylyl cyclase inhibitor SQ 22536 (10−5 m) or/and PKA inhibitor PKI (4 × 10−5 m) for 30 min before the adenylate cyclase activator FSK (10−5 m), the membrane-permeable cAMP analog 8-Br-cAMP (2x10−3 m), or OT (10−7 m) treatment for 3 h. Total RNA from mouse ESC was reverse-transcribed, and Cx43 cDNA was amplified by real-time PCR as described in Materials and Methods. The data are reported as the mean ± se of four independent experiments, each conducted in triplicate. *, P < 0.05 vs. control; **, P < 0.05 vs. OT alone. E, Cells were pretreated with SQ 22536 or/and PKI (4 × 10−5 m) for 30 min before FSK (10−5 m), 8-Br-cAMP (2 × 10−3 m), or OT (10−7 m) treatment for 3 h. Total protein was extracted and blotted with Cx43 or β-actin antibodies. Each example shown is representative of five independent experiments. The right part (E) depicting the bars denotes the mean ± se of five independent experiments for each condition determined from densitometry relative to β-actin. *, P < 0.05 vs. control; **, P < 0.05 vs. OT alone; ROD, Relative Optical Density.
Fig. 4.
Effect of OT on PKA activity. Time (A) and dose (B) response of OT on PKA activity. Mouse ESC were treated with OT for various times (0–120 min) or with different doses of OT (0–10−6 m) for 60 min. The maximum increase in PKA activity was observed after 60 min incubation with 10−7 m OT. The values represent the mean ± se of four independent experiments with triplicate dishes. *, P < 0.05 vs. control. C, Cells were pretreated with SQ 22536 (10−5 m) for 30 min before OT treatment for 60 min. The values represent the mean ± se of four independent experiments with triplicate dishes. *, P < 0.05 vs. control; **, P < 0.05 vs. OT alone. D, Real-time RT-PCR quantification of the Cx43 gene. Cells were pretreated with PKA inhibitor PKI (4 × 10−5 m) for 30 min before OT treatment for 3 h. Total RNA from mouse ESC was reverse transcribed, and Cx43 cDNA was amplified by real-time PCR as described in Materials and Methods. The data are reported as the mean ± se of five independent experiments, each conducted in triplicate. *, P < 0.05 vs. control; **, P < 0.05 vs. OT alone. E, Cells were pretreated with PKI for 30 min before OT treatment for 3 h. Total protein was extracted and blotted with Cx43 or β-actin antibodies. Each example shown is a representative of five independent experiments. The right part (E) depicting the bars denotes the mean ± se of five independent experiments for each condition determined from densitometry relative to β-actin. *, P < 0.05 vs. control; **, P < 0.05 vs. OT alone; ROD, Relative Optical Density.
Role of NF-κB in OT-induced Cx43 protein expression
To examine the role of NF-κB in OT-induced Cx43 protein expression, we determined whether OT (10−7 m) induced the phosphorylation of NF-κB in the nuclear fraction of mouse ESC. As shown in Fig. 5A, Western blot analysis revealed an increase in phosphorylated NF-κB 30 to 120 min after OT treatment. In addition, a nuclear and nonnuclear protein fractionation assay showed that OT induced phosphorylated-NF-κB significantly in nuclear fractions. Enrichment of the nuclear fraction was validated by the nuclear marker lamin A/C (Fig. 5B). The increased accumulation of phosphorylated-NF-κB in the nucleus induced by OT treatment was further confirmed by immunofluorescent nuclear staining and counterlabeling with PI (Fig. 5C). Also, PKI inhibited OT-induced phosphorylated NF-κB in the nuclear fraction (Fig. 5D). As shown in Fig. 5E, NF-κB inhibitors SN 50 (500 ng/ml) and bay 11–7082 (10−5 m) inhibited OT-induced Cx43 protein expression levels. In addition, NF-κB-specific siRNA reduced the basal levels of NF-κB and OT-induced Cx43 protein expression (Fig. 5, F and G).
Fig. 5.
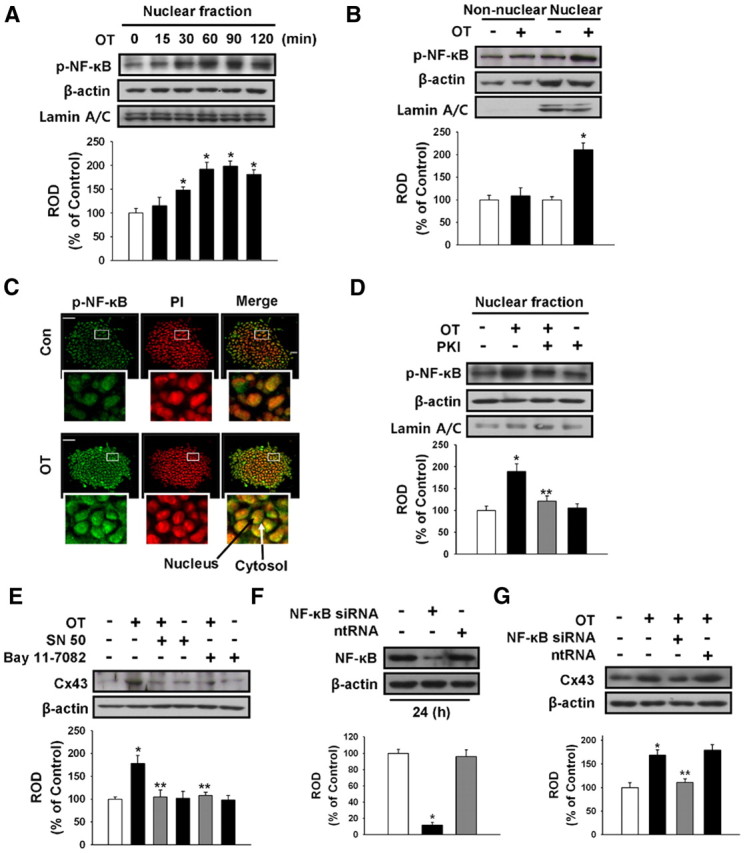
Involvement of NF-κB in OT-induced Cx43 expression. A, Mouse ESC were incubated with OT(10−7 m) for 0–120 min and then harvested. Cell lysates were immunoblotted with anti-phospho-NF-κB p65, anti-β-actin, or antilamin A/C antibody. Each example shown is representative of five independent experiments. B, Mouse ESC were incubated with OT (10−7 m) for 90 min, and phospho-NF-κB levels in the nuclear or nonnuclear fractions were detected. Cell lysates were immunoblotted with anti-phospho-NF-κB, anti-β-actin, or antilamin A/C antibody. Each example shown is representative of five independent experiments. The lower part (A and B) depicting the bars denotes the mean ± se of five independent experiments for each condition determined from densitometry relative to β-actin. *, P < 0.05 vs. control. C, Immunofluorescence shows phospho-NF-κB redistribution in the nucleus induced by OT. Mouse ESC were treated with vehicle or OT (10−7 m) for 90 min. Fixed cells were immunolabeled with anti-phospho-NF-κB antibody and counterstained with PI. Insets show magnified versions of sections in the white boxes. Scale bars, 20 μm (magnification, ×400). D, Mouse ESC were pretreated with PKI (4 × 10−5 m) for 30 min before OT(10−7 m) for 90 min. Total protein was extracted and blotted with anti-phospho-NF-κB, anti-β-actin, or antilamin A/C antibody. Each example shown is representative of five independent experiments. E, Mouse ESC were treated with OT (10−7 m) in the presence or absence of NF-κB inhibitors SN 50 (500 ng/ml) and bay 11–7082 (10−5 m), resulting in significant inhibition of OT-induced Cx43 protein expression. Each example shown is representative of five independent experiments. F, Mouse ESC were transfected for 24 h with either NF-κB-specific siRNA (100 nmol/liter) or nontargeting control siRNA (100 nmol/liter) using Hyperfectamine before 1 h OT treatment. NF-κB expression was analyzed using Western blot. Each of the examples shown is representative of five independent experiments. G, Mouse ESC were transfected for 24 h with either NF-κB-specific siRNA or nontargeting control siRNA before OT(10−7 m) treatment for 3 h, after which Cx43 protein expression levels were detected. Each example shown is representative of five experiments. The lower part (D–G) depicting the bars denotes the mean ± se of five independent experiments for each condition determined from densitometry relative to β-actin. *, P < 0.05 vs. control; **, P < 0.05 vs. OT alone. Con, Control; ntRNA, nontargeting RNA; ROD, Relative Optical Density.
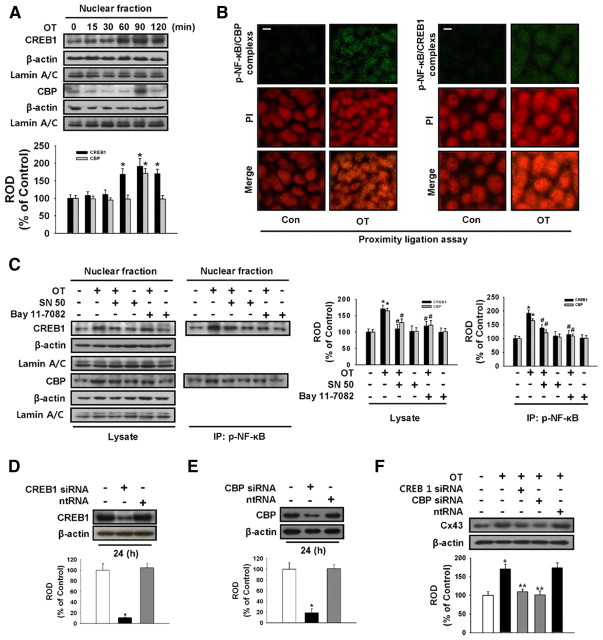
Effect of OT on formation of NF-κB/CREB1/CBP complex in nuclear fraction
To determine whether or not OT plays a role in regulating NF-κB/CREB1/CBP complex formation, NF-κB, CREB1, and CBP were analyzed with Western blotting, PLA, or immunoprecipitation. As shown in Fig. 6A, OT increased CREB1 and CBP expression in the nuclear fraction part. In situ PLA (22) is a powerful tool to rather easily screen for protein-protein interactions. Confocal micrographs collected at 0.38-μm intervals and merged together show high numbers of phospho-NF-κB/CREB1 interactions with 10−7 m OT (Fig. 6B, each green dot represents one detected interaction). Phospho-NF-κB/CBP interaction was also observed with treatment of OT (Fig. 6B, green dots). In addition, OT increased the formation of NF-κB and CREB1, and NF-κB and CBP complexes as shown in immunoprecipitation experiments (Fig. 6C), which was blocked by NF-κB inhibitors SN 50 (500 ng/ml) and bay 11–7082 (10−5 m). The efficiency of CREB1 and CBP-specific siRNA transfection was determined using CREB1 and CBP Western blots (Fig. 6, D and E). To elucidate the involvement of CREB1 and CBP on OT-induced Cx43 protein expression levels, the cells were transfected with CREB1 and CBP-specific siRNA (100 nmol/liter) or nontargeting siRNA (100 nmol/liter) before OT treatment. CREB1 and CBP-specific siRNA reduced OT-induced Cx43 protein expression levels.
Fig. 6.
OT-mediated expression and clustering of NF-κB/CREB1/CBP complex promotes Cx43 expression. A, Mouse ESC were treated with OT(10−7 m) for different time periods (0–120 min). Cell lysates were immunoblotted with anti-CREB1, anti-CBP, anti-β-actin, or antilamin A/C antibody. Each example shown is representative of five independent experiments. The lower part (A) depicting the bars denotes the mean ± se of five independent experiments for each condition determined from densitometry relative to β-actin. *, P < 0.05 vs. control. B, Immunofluorescence confocal microscopy in combination with in situ PLA, which detects protein-protein complexes, was used to explore interactions between phospho-NF-κB, CREB1, and CBP. Each detected complex is represented by a green dot. Nuclei were counterstained by PI (red). Scale bars, 20 μm (magnification, ×400). C, Mouse ESC were pretreated in the absence and presence of SN 50 (500 ng/ml) or bay 11–7082 (10−5 m) for 30 min before 90 min OT treatment. Cell lysates were analyzed by Western blotting with antibodies that recognize phospho-NF-κB, CREB1, and CBP. Immunoprecipitation of phospho-NF-κB was analyzed by Western blotting with antibodies that recognize CREB1 and CBP. Each of the examples is representative of five independent experiments. The right part (A) depicting the bars denotes the mean ± se of five independent experiments for each condition determined from densitometry relative to β-actin. *, P < 0.05 vs. control. D, Mouse ESC were transfected for 24 h with either CREB1-specific siRNA (100 nmol/liter) or nontargeting control siRNA (100 nmol/liter) using Hyperfectamine before 1 h OT treatment. CREB1 expression was analyzed by Western blot. Each of the examples shown is representative of four independent experiments. E, Mouse ESC were transfected for 24 h with either CBP-specific siRNA (100 nmol/liter) or nontargeting control siRNA (100 nmol/liter) using Hyperfectamine before 1 h OT treatment. CBP expression was analyzed by Western blot. Each of the examples shown is representative of four independent experiments. F, Mouse ESC were transfected for 24 h with either CREB1-specific siRNA, CBP-specific siRNA, or nontargeting control siRNA before OT (10−7 m) treatment for 3 h, after which Cx43 protein expression levels were detected. Each example shown is representative of four experiments. The lower part (D–F) depicting the bars denotes the mean ± se of four independent experiments for each condition determined from densitometry relative to β-actin. *, P < 0.05 vs. control; **, P < 0.05 vs. OT alone. Con, Control; ntRNA, nontargeting RNA; ROD, Relative Optical Density.
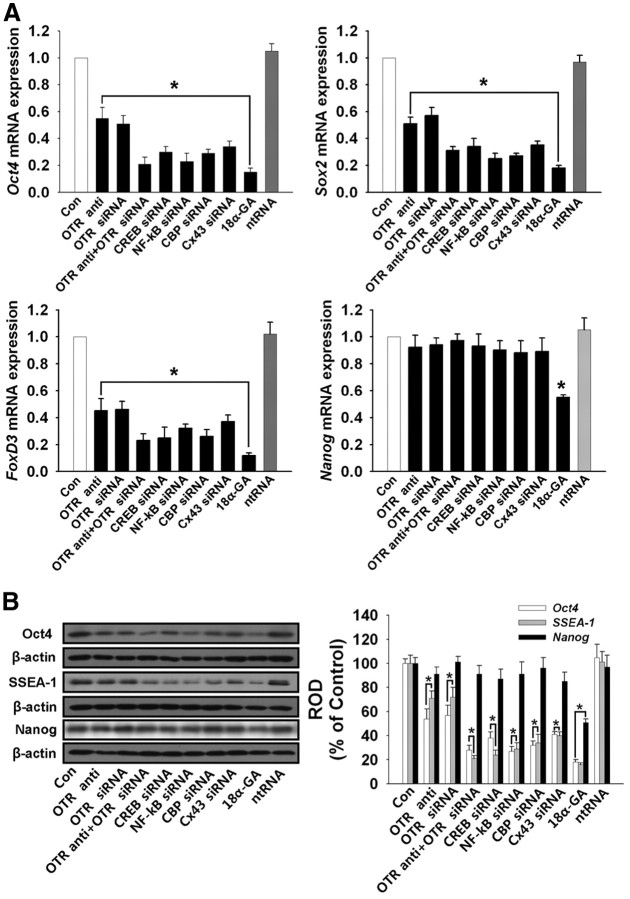
Role of OT in maintaining ESC in an undifferentiated state
We examined the roles of OT on level of undifferentiated marker genes and proteins expression in mouse ESC. We treated the OTR-neutralizing antibody, OTR-specific siRNA, combination of OTR neutralizing antibody and OTR-specific siRNA, CREB-, NF-κB-, CBP-, and CX43 → Cx43 specific siRNA in normal ESC culture conditions to determine the role of gap junction. Each treatment down-regulated the mRNA and protein expression level of undifferentiated markers, Oct4, Sox2, FoxD3, and SSEA-1 but not Nanog (Fig. 7, A and B). However, treatment of gap junction inhibitor 18α-glycyrrhetinic acid (18α-GA, 10 μg/ml) down-regulated mRNA and protein expression level of all undifferentiated markers including Nanog (Fig. 7, A and B). These results suggested that OT may contribute to maintain undifferentiated state of mouse ESC through gap junction regulation.
Fig. 7.
Role of OT in maintaining ESC in an undifferentiated state. A, Real-time RT-PCR quantification of undifferentiated marker genes. Cells were treated with OTR-neutralizing antibody (0.1 μg/ml), OTR-specific siRNA, combination of OTR-neutralizing antibody and OTR-specific siRNA, CREB-, NF-κB-, CBP-, CX43-specific siRNA, 18α-GA (10 μg/ml), and ntRNA for 72 h. Total RNA from mouse ESC was reverse transcribed, and Oct4, Sox2, FoxD3, Nanog, and β-actin cDNA were amplified by real-time PCR as described in Materials and Methods. The data are reported as the mean ± se of five independent experiments, each conducted in triplicate. *, P < 0.05 vs. control. B, Cells were treated with OTR-neutralizing antibody, OTR-specific siRNA, combination of OTR-neutralizing antibody and OTR-specific siRNA, CREB-, NF-κB-, CBP-, CX43-specific siRNA, 18α-GA, and ntRNA for 72 h. Total protein was extracted and blotted with Oct-4, SSEA-1, Nanog, and β-actin antibodies. Each example shown is representative of five independent experiments. The right part of panel B depicting the bars denotes the mean ± se of five independent experiments for each condition determined from densitometry relative to β-actin. *, P < 0.05 vs. control. Con, Control; ntRNA, nontargeting RNA; ROD, Relative Optical Density.
Discussion
The present data demonstrated that OT stimulates expression of the gap junctional protein Cx43, which is mediated by the NF-κB/CREB/CBP complex via lipid raft-independent OTR-mediated cAMP/PKA in mouse ESC. Of the various functions of OT, evidence has accumulated for a role as a cellular function transducer whereby multiple signals are initiated (14, 23). Signaling initiation and biological behaviors of stem cell commonly have relevance to specific binding to membrane-bound receptors that belong to the rhodopsin-like G protein-coupled receptor family, a large group of receptors that use G proteins to transduce signals across the cell membrane (24). Previous reports show that OTR exists in cell membranes and the OT/OTR system regulates early embryonic development (12). As such, OT/OTR may also initiate various effects on other cell functions. These different temporal patterns of cellular function, such as regulation of cell growth and gap junction, probably are due to different signaling pathways related to the location of OTR (11, 23). For example, the OT stimulates cAMP and PKA signaling via coupling of OTR located outside lipid rafts (10–11, 13) whereas OT stimulates PI3K/Akt, MAPK through coupling of OTR located inside lipid rafts (14–15). In addition, previous studies have reported on embryonic kidney cells stably expressing OTR localized within lipid rafts (10), and a location of OTR outside lipid rafts has been described on human myometrial cells and uterine myocytes (10–11, 13). Although the existence of OTR was also detected in undifferentiated ESC and derived differentiated cells (12, 25), the exact location of OTR is yet to be determined and OT/OTR-related mechanism effects on Cx43 protein expression are not clear in mouse ESC. In this study, we showed that OT promotes Cx43 expression via lipid raft (cav)-independent OTR in mouse ESC. We then examined detergent-resistant membrane fractions (DRM, lipid raft region) to further confirm the exclusive localization of OTR in mouse ESC. In addition, Cx43 protein expression levels were increased upon application of OT and attenuated by atosiban, but not MβCD. Therefore, our data strongly suggest that lipid raft-independent OTR are an important mediator in OT-induced Cx43 protein expression in mouse ESC.
It has also been found that the cAMP/PKA signaling pathway is an important regulator of various cellular processes and may be mediated effects of OT/OTR in various cell types such as myoepithelial cell, cancer cell, and uterine myocytes (10, 11). Therefore, we hypothesized that OT is linked to the lipid raft-independent OTR-mediated cAMP/PKA pathway in mouse ESC. The cAMP second messenger stimulates transcription regulation of a broad variety of genes for many biological processes including cell-to-cell junction regulation (26). PKA activation may regulate gap junctions at the level of junction assembly by increasing the number of junctional plaques on the cell surface that are essential for Cx43 expression (27, 28). Although a previous study showed that OT regulates the cAMP/PKA pathway in a human myometrial cell line (13), interaction of lipid raft-independent OTR-mediated cAMP/PKA and OT-induced Cx43 protein expression in the mouse is still unclear. In the present study, we determined that intracellular cAMP/PKA levels increased upon application of OT. Also, SQ 22536 and PKI inhibited OT-induced Cx43 protein expression. On the other hand, the Gαi inhibitor pertussis toxin, or Ca2+ mobilization blockers had no effect to inhibit OT induction of Cx43 expression (Supplemental Fig. 2), although OT effects on uterine smooth muscle are known to be mediated via Gi and Gq, resulting in cAMP inhibition and increased calcium signaling (10, 11). Therefore, the present results suggest that lipid raft-independent OTR-mediated cAMP/PKA is an important mediator in OT-induced Cx43 protein expression in mouse ESC.
In addition, our study sheds light on the potential role of NF-κB/CREB/CBP complex formation in OT-induced Cx43 protein expression. It is well known that the free catalytic subunits of PKA can affect IκB-NF-κB complexes in the cytoplasm, and that IκB sequesters PKA by masking its catalytic domain in somatic cells (29). As a consequence, PKA increases NF-κB phosphorylation at Ser273 sites (30). This PKA-dependent phosphorylation of NF-κB facilitates recruitment of the transcription coactivator CBP (31, 32). Moreover, activated PKA enhances activation of CREB (33), which then initiates transcription by recruiting CBP/p300 (34). In this context, we investigated the role of NF-κB/CREB/CBP complex formation on OT effects in mouse ESC. We found that OT significantly increased NF-κB phosphorylation and formation of complexes with NF-κB and CREB, and NF-κB and CBP in the nucleus. These results are supported by previous studies showing that NF-κB-dependent signaling recruited CBP and CBP-activated CREB (31, 34). In addition, the importance of NF-κB and CREB signals in the regulation of Cx43 protein expression levels in stem cells has been previously suggested (7, 35). Therefore, it is possible that PKA-dependent NF-κB, -CREB, and -CBP are key players involved in the regulation of Cx43 protein expression. The present study also showed that OT-induced stimulation of Cx43 protein expression was blocked by NF-κB, CREB, and CBP-specific siRNA transfection. These results suggest that formation of NF-κB/CREB/CBP complexes in the nucleus play an important role in OT-induced Cx43 protein expression (Fig. 8). In addition, our results show that OT maintains expression of undifferentiated marker genes and proteins that were down-regulated by inhibition of OTR as well as OT-related signal molecules. These data suggest that OT may maintain an undifferentiated state through gap junctions in mouse ESC. In previous reports, endogenous OT was produced or secreted naturally in neural progenitor cells and mouse embryo (12, 36). Based upon these results as well as our results, we suggest the possibility that the mouse ESC produce endogenous OT which is acting in an autoregulatory manner to maintain the undifferentiated state, although OT induced differentiation of mesenchymal stem cells (8). This discrepancy could be due to a difference in animal species, cell types, or experimental conditions used for study. Taken together, defining the role and mechanisms of OT in stimulating gap junction protein expression in mouse ESC represents a significant advancement in knowledge related to how mouse embryonic cell-to-cell junctions are regulated and can provide a clue for maintenance of the undifferentiated state of mouse ESC by OT. In conclusion, OT stimulates Cx43 expression through the NF-κB/CREB/CBP complex via lipid raft-independent OTR-mediated cAMP/PKA in mouse ESC, which contribute to maintain an undifferentiated state of mouse ESC.
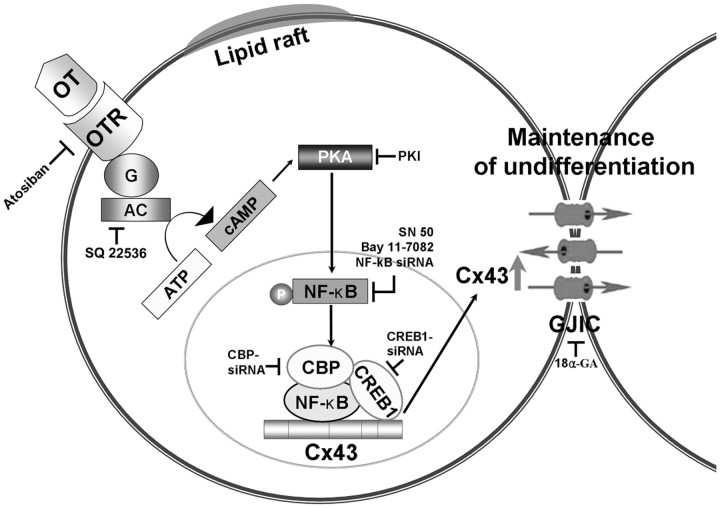
Fig. 8.
A model for OT-induced Cx43 expression and its related signal pathways. OT activates cAMP/PKA signal pathways via lipid raft-independent OTR. This signal increased NF-κB phosphorylation after which NF-κB nuclear localization and accumulation and subsequently increased formation of CREB1/NF-κB/CBP complex. Finally, OT stimulated Cx43 expression, thereby contributing to maintaining ESC in an undifferentiated state. AC, Adenylyl cyclase.
Acknowledgments
This study was supported by the Basic Science Research Program through the National Research Foundation of Korea (NRF) (2012-0000184; 2010-0020268), and graduate fellowship of the Brain Korea 21 project provided by the Ministry of Education, Science and Technology, Republic of Korea.
Disclosure Summary: S.P.Y., S.P., J.M.R., J.H.P., M.O.K., J.-H.L., and H.J.L. have nothing to declare.
Footnotes
- AP
- Alkaline phosphatase
- 8-Br
- 8-bromoadenosine
- CBP
- CREB-binding protein
- CREB
- cAMP response element-binding
- Cx43
- connexin43
- ESC
- embryonic stem cells
- FBS
- fetal bovine serum
- FSK
- forskolin
- 18α-GA
- 18α-glycyrrhetinic acid
- GJIC
- gap junctional intercellular communication
- LIF
- leukemia inhibitory factor
- LY
- lucifer yellow
- NF-κB
- nuclear factor κB
- MβCD
- methyl-β-cyclodextrin
- MES
- 2-(N-morpholino)ethanesulfonic acid
- OT
- oxytocin
- OTR
- OT receptor
- PI
- propidium iodide
- PKA
- protein kinase A
- PKI
- protein kinase inhibitor
- PLA
- proximity ligation assay
- RD
- rhodamine-dextran
- RT
- reverse transcription
- siRNA
- small interfering RNA
- SL/DT
- scrape loading/dye transfer
- Tm
- melting temperature.
References
- 1. Griffiths DS , Li J , Dawson MA , Trotter MW , Cheng YH , Smith AM , Mansfield W , Liu P , Kouzarides T , Nichols J , Bannister AJ , Green AR , Göttgens B. 2011. LIF-independent JAK signalling to chromatin in embryonic stem cells uncovered from an adult stem cell disease. Nat Cell Biol 13:13–21 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2. Kamiya D , Banno S , Sasai N , Ohgushi M , Inomata H , Watanabe K , Kawada M , Yakura R , Kiyonari H , Nakao K , Jakt LM , Nishikawa S , Sasai Y. 2011. Intrinsic transition of embryonic stem-cell differentiation into neural progenitors. Nature 470:503–509 [DOI] [PubMed] [Google Scholar]
- 3. Wörsdörfer P , Maxeiner S , Markopoulos C , Kirfel G , Wulf V , Auth T , Urschel S , von Maltzahn J , Willecke K. 2008. Connexin expression and functional analysis of gap junctional communication in mouse embryonic stem cells. Stem Cells 26:431–439 [DOI] [PubMed] [Google Scholar]
- 4. Oyamada Y , Komatsu K , Kimura H , Mori M , Oyamada M. 1996. Differential regulation of gap junction protein (connexin) genes during cardiomyocytic differentiation of mouse embryonic stem cells in vitro. Exp Cell Res 229:318–326 [DOI] [PubMed] [Google Scholar]
- 5. Todorova MG , Soria B , Quesada I. 2008. Gap junctional intercellular communication is required to maintain embryonic stem cells in a non-differentiated and proliferative state. J Cell Physiol 214:354–362 [DOI] [PubMed] [Google Scholar]
- 6. Jäderstad J , Jäderstad LM , Li J , Chintawar S , Salto C , Pandolfo M , Ourednik V , Teng YD , Sidman RL , Arenas E , Snyder EY , Herlenius E. 2010. Communication via gap junctions underlies early functional and beneficial interactions between grafted neural stem cells and the host. Proc Natl Acad Sci USA 107:5184–5189 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Kim YS , Kwon JS , Hong MH , Kim J , Song CH , Jeong MH , Cho JG , Park JC , Kang JC , Ahn Y. 2010. Promigratory activity of oxytocin on umbilical cord blood-derived mesenchymal stem cells. Artif Organs 34:453–461 [DOI] [PubMed] [Google Scholar]
- 8. Elabd C , Basillais A , Beaupied H , Breuil V , Wagner N , Scheideler M , Zaragosi LE , Massiéra F , Lemichez E , Trajanoski Z , Carle G , Euller-Ziegler L , Ailhaud G , Benhamou CL , Dani C , Amri EZ. 2008. Oxytocin controls differentiation of human mesenchymal stem cells and reverses osteoporosis. Stem Cells 26:2399–2407 [DOI] [PubMed] [Google Scholar]
- 9. Gassanov N , Devost D , Danalache B , Noiseux N , Jankowski M , Zingg HH , Gutkowska J. 2008. Functional activity of the carboxyl-terminally extended oxytocin precursor peptide during cardiac differentiation of embryonic stem cells. Stem Cells 26:45–54 [DOI] [PubMed] [Google Scholar]
- 10. Reversi A , Cassoni P , Chini B. 2005. Oxytocin receptor signaling in myoepithelial and cancer cells. J Mammary Gland Biol Neoplasia 10:221–229 [DOI] [PubMed] [Google Scholar]
- 11. Zhou XB , Lutz S , Steffens F , Korth M , Wieland T. 2007. Oxytocin receptors differentially signal via Gq and Gi proteins in pregnant and nonpregnant rat uterine myocytes: implications for myometrial contractility. Mol Endocrinol 21:740–752 [DOI] [PubMed] [Google Scholar]
- 12. Beretsos P , Loutradis D , Koussoulakos S , Margaritis LH , Kiapekou E , Mastorakos G , Papaspirou I , Makris N , Makrigiannakis A , Antsaklis A. 2006. Oxytocin receptor is differentially expressed in mouse endometrium and embryo during blastocyst implantation. Ann NY Acad Sci 1092:466–479 [DOI] [PubMed] [Google Scholar]
- 13. Burghardt RC , Barhoumi R , Stickney M , Monga M , Ku CY , Sanborn BM. 1996. Correlation between connexin43 expression, cell-cell communication, and oxytocin-induced Ca2+ responses in an immortalized human myometrial cell line. Biol Reprod 55:433–438 [DOI] [PubMed] [Google Scholar]
- 14. Reversi A , Rimoldi V , Brambillasca S , Chini B. 2006. Effects of cholesterol manipulation on the signaling of the human oxytocin receptor. Am J Physiol Regul Integr Comp Physiol 291:R861–R869 [DOI] [PubMed] [Google Scholar]
- 15. Rimoldi V , Reversi A , Taverna E , Rosa P , Francolini M , Cassoni P , Parenti M , Chini B. 2003. Oxytocin receptor elicits different EGFR/MAPK activation patterns depending on its localization in caveolin-1 enriched domains. Oncogene 22:6054–6060 [DOI] [PubMed] [Google Scholar]
- 16. Lo CW , Chung QY. 1979. The sedative effect of acupuncture. Am J Chin Med 7:253–258 [DOI] [PubMed] [Google Scholar]
- 17. Lo CW , Gilula NB. 1979. Gap junctional communication in the post-implantation mouse embryo. Cell 18:411–422 [DOI] [PubMed] [Google Scholar]
- 18. Zhang Y , Kakinuma Y , Ando M , Katare RG , Yamasaki F , Sugiura T , Sato T. 2006. Acetylcholine inhibits the hypoxia-induced reduction of connexin43 protein in rat cardiomyocytes. J Pharmacol Sci 101:214–222 [DOI] [PubMed] [Google Scholar]
- 19. Song KS , Li Shengwen , Okamoto T , Quilliam LA , Sargiacomo M , Lisanti MP. 1996. Co-purification and direct interaction of Ras with caveolin, an integral membrane protein of caveolae microdomains. Detergent-free purification of caveolae microdomains. J Biol Chem 271:9690–9697 [DOI] [PubMed] [Google Scholar]
- 20. Michael LF , Asahara H , Shulman AI , Kraus WL , Montminy M. 2000. The phosphorylation status of a cyclic AMP-responsive activator is modulated via a chromatin-dependent mechanism. Mol Cell Biol 20:1596–1603 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Bradford MM. 1976. A rapid and sensitive method for the quantitation of microgram quantities of protein utilizing the principle of protein-dye binding. Anal Biochem 72:248–254 [DOI] [PubMed] [Google Scholar]
- 22. Söderberg O , Gullberg M , Jarvius M , Ridderstråle K , Leuchowius KJ , Jarvius J , Wester K , Hydbring P , Bahram F , Larsson LG , Landegren U. 2006. Direct observation of individual endogenous protein complexes in situ by proximity ligation. Nat Methods 3:995–1000 [DOI] [PubMed] [Google Scholar]
- 23. Herbert Z , Bötticher G , Aschoff A , Sendemir E , Zermann DH , Arnold R , Mall G , Jirikowski GF. 2007. Changing caveolin-1 and oxytocin receptor distribution in the ageing human prostate. Anat Histol Embryol 36:361–365 [DOI] [PubMed] [Google Scholar]
- 24. Gimpl G , Fahrenholz F. 2001. The oxytocin receptor system: structure, function, and regulation. Physiol Rev 81:629–683 [DOI] [PubMed] [Google Scholar]
- 25. Hatami L , Valojerdi MR , Mowla SJ. 2007. Effects of oxytocin on cardiomyocyte differentiation from mouse embryonic stem cells. Int J Cardiol 117:80–89 [DOI] [PubMed] [Google Scholar]
- 26. Schajnovitz A , Itkin T , D'Uva G , Kalinkovich A , Golan K , Ludin A , Cohen D , Shulman Z , Avigdor A , Nagler A , Kollet O , Seger R , Lapidot T. 2011. CXCL12 secretion by bone marrow stromal cells is dependent on cell contact and mediated by connexin-43 and connexin-45 gap junctions. Nat Immunol 12:391–398 [DOI] [PubMed] [Google Scholar]
- 27. Paulson AF , Lampe PD , Meyer RA , TenBroek E , Atkinson MM , Walseth TF , Johnson RG. 2000. Cyclic AMP and LDL trigger a rapid enhancement in gap junction assembly through a stimulation of connexin trafficking. J Cell Sci 113:3037–3049 [DOI] [PubMed] [Google Scholar]
- 28. Wang Z , Do CW , Valiunas V , Leung CT , Cheng AK , Clark AF , Wax MB , Chatterton JE , Civan MM. 2010. Regulation of gap junction coupling in bovine ciliary epithelium. Am J Physiol Cell Physiol 298:C798–C806 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Gambaryan S , Kobsar A , Rukoyatkina N , Herterich S , Geiger J , Smolenski A , Lohmann SM , Walter U. 2010. Thrombin and collagen induce a feedback inhibitory signaling pathway in platelets involving dissociation of the catalytic subunit of protein kinase A from an NFκB-IκB complex. J Biol Chem 285:18352–18363 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Zhong H , SuYang H , Erdjument-Bromage H , Tempst P , Ghosh S. 1997. The transcriptional activity of NF-κB is regulated by the IκB-associated PKAc subunit through a cyclic AMP-independent mechanism. Cell 89:413–424 [DOI] [PubMed] [Google Scholar]
- 31. Gao X , Wang Q , Li W , Yang B , Song H , Ju W , Liu S , Cheng J. 2011. Identification of nucleolar and coiled-body phosphoprotein 1 (NOLC1) minimal promoter regulated by NF-κB and CREB. BMB Rep 44:70–75 [DOI] [PubMed] [Google Scholar]
- 32. Hadad N , Tuval L , Elgazar-Carmom V , Levy R , Levy R. 2011. Endothelial ICAM-1 protein induction is regulated by cytosolic phospholipase A2α via both NF-κB and CREB transcription factors. J Immunol 186:1816–1827 [DOI] [PubMed] [Google Scholar]
- 33. Chen YQ , Xie X. 2010. Podophyllotoxin induces CREB phosphorylation and CRE-driven gene expression via PKA but not MAPKs. Mol Cells 29:41–50 [DOI] [PubMed] [Google Scholar]
- 34. Leahy P , Crawford DR , Grossman G , Gronostajski RM , Hanson RW. 1999. CREB binding protein coordinates the function of multiple transcription factors including nuclear factor I to regulate phosphoenolpyruvate carboxykinase (GTP) gene transcription. J Biol Chem 274:8813–8822 [DOI] [PubMed] [Google Scholar]
- 35. Schubert SW , Abendroth A , Kilian K , Vogler T , Mayr B , Knerr I , Hashemolhosseini S. 2008. bZIP-Type transcription factors CREB and OASIS bind and stimulate the promoter of the mammalian transcription factor GCMa/Gcm1 in trophoblast cells. Nucleic Acids Res 36:3834–3846 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36. Markakis EA , Palmer TD , Randolph-Moore L , Rakic P , Gage FH. 2004. Novel neuronal phenotypes from neural progenitor cells. J Neurosci 24:2886–2897 [DOI] [PMC free article] [PubMed] [Google Scholar]