Abstract
Copy number variants have been associated with intellectual disability, multiple congenital anomalies and craniofacial disorders. It has been reported that microduplication of 15q13.3 is associated with autism, cognitive impairment, seizures, and attention-deficit hyperactivity disorder. Here, the author identified microduplications in the 15q13.3 region in 4 cases from 3 Chinese families using chromosomal microarray analysis–single nucleotide polymorphism array (CMA-SNP). These 4 cases include 2 fetuses from 2 unrelated families and a father and a daughter from a third family. The identified microduplications of 15q13.3 are approximately 400 kb in size, encompassing just 1 gene, cholinergic receptor, neuronal nicotinic, alpha polypeptide 7 (CHRNA7). Three-fourths of the probands exhibit oral clefts, which has not been previously reported in cases with this duplication genotype. Therefore, in this study, the author describes for the first time the common feature of oral clefts in patients carrying a microduplication of 15q13.3 encompassing the CHRNA7 gene, which sheds light on the correlation between CHRNA7 and cleft palate.
Keywords: autism, 15q13.3 microduplication, CMA-SNP array, CHRNA7, oral clefts
Microduplications of 15q13.3 involving cholinergic receptor, neuronal nicotinic, alpha polypeptide 7 (CHRNA7) have been previously described, with duplication sizes ranging from 350 kb to 1.6 Mb and with various breakpoints within the 15q13.3 region.1–5 Although the pathogenicity of 15q13.3 duplications encompassing CHRNA7 have a lower estimation of penetrance (0.87) than most other genomic disorders,6 patients with such microduplications exhibit various clinical symptoms such as developmental delay/mental retardation, autism, muscular hypotonia, and a variety of neuropsychiatric disorders.1–5 However, the relationship between birth defects such as oral clefts and microduplication of 15q13.3 involving CHRNA7 has not been previously explored.
Oral clefts are among the most common birth defects and include 3 anatomical defects: cleft lip, cleft lip and palate, and cleft palate. They can occur as a singular phenotype or as one feature of a specific syndrome.7 It has been demonstrated that the etiology of oral clefts is complex and that both genetic and environmental factors contribute to its pathology. Indeed, multiple genes or their regulatory genetic elements have been implicated in the etiology of oral clefts.8–14 Furthermore, high rates of Mendelian inconsistencies were observed in 11 different genes, suggesting the existence of additional microdeletions/microduplications among cases with oral cleft.15 For instance, 4q35qter deletions have recently been associated with oral clefts.16
Here, the author reports 3 patients, including 2 fetuses and 1 nonconsanguineous Chinese family member, carrying microduplications of 15q13.3 encompassing only the CHRNA7 gene. These probands all had a common phenotype of cleft palate, which has not been previously associated with microduplications of 15q13.3 involving CHRNA7. Therefore, in this study, the author provides the first evidence for the involvement of microduplications at 15q13.3 encompassing CHRNA7 in the etiology of oral clefts.
Materials and Methods
Clinical Description
Clinical features of the 3 patients were assessed by experienced clinical geneticists. Informed consent was obtained from all patients involved in this study, and this was approved by the Ethics Committee.
Patient 1 was a female and the first child of healthy and nonconsanguineous parents (mother 42 years and father 45 years). She was delivered by cesarean section because of fetal distress at 42 +3 weeks of gestation. At birth, her weight was 5300 g, and her length was 50 cm. Cleft palate was observed at birth without surgery. Brain computed tomography (CT) of this patient at 6 months showed mild hydrocephalus which had disappeared upon reexamination at 2 years of age. Further clinical evaluation was performed because of mental retardation and cleft palate at the age of 16 years. She displayed dysmorphologic features including macrocephaly, short stature, short limbs, and black tongue (Figure 1D). She also exhibited mental retardation, hearing impairment, and an inability to speak.
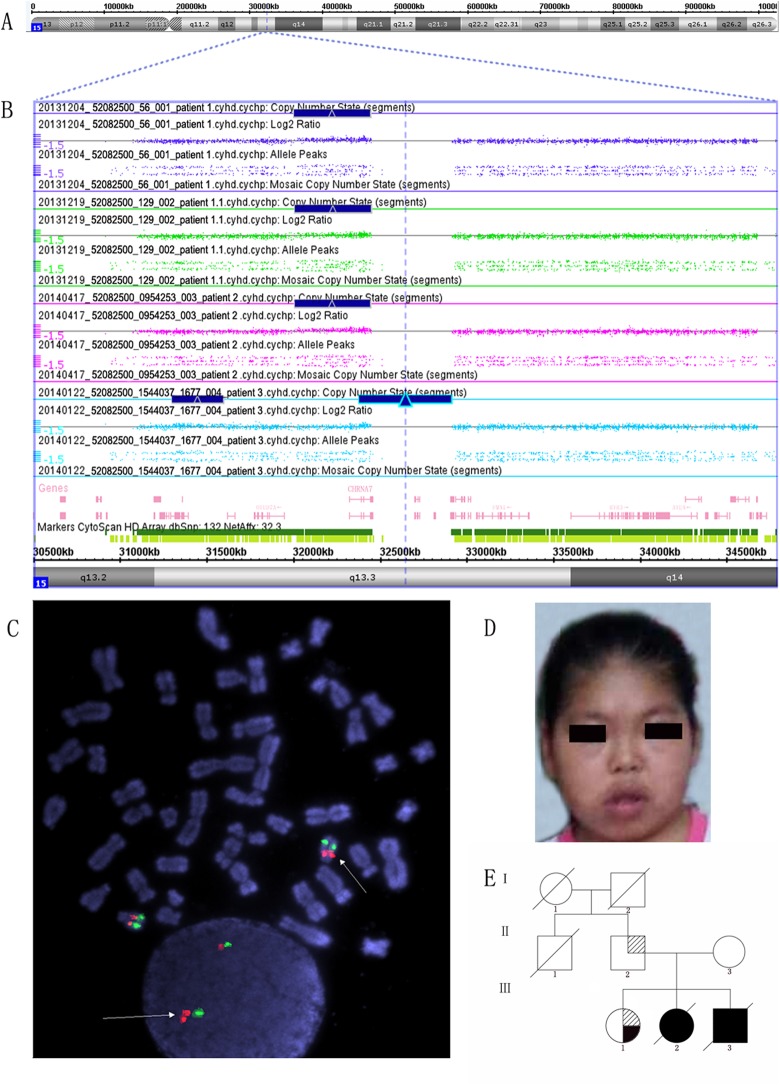
Figure 1.
A, Chromosome view. B, Zoom-in view of the 15q13.3 microduplication from the CMA-SNP array analysis. The microduplicated region of 15q13.3 is represented by the blue color box. C, Fluorescent in situ hybridization results of patient 1. Fluorescent in situ hybridization analysis was performed using the RP11-115G22 probe (red) located within the microduplicated region of 15q13.3 and the control probe RP11-2M12 (green) in patient 1. Interphase fluorescent in situ hybridization analysis performed on cultured lymphocytes derived from patient 1 showed 3 red signals, with 2 signals clustered together and well separated from the third signal, suggesting a homozygous duplication. No translocation or insertion of the duplicated region was detected in metaphase cells. D, Facial appearance of patient 1 at 16 years of age showing macrocephaly, short limbs, and black tongue. E, Pedigree of the family of Patient 1. III-1 is the proband. Solid black indicates cleft palate.102 × 141 mm (300 × 300 DPI).
The parents of patient 1 had an adverse reproductive history, with 2 fetuses both showing cleft lip and palate. Induction of labor was performed without any genetic testing. The brother of patient 1’s father also had a cleft lip and palate and was known to be dead without any further information (Pedigree of the family is shown in Figure 1E). Cytogenetic analysis and CMA (array SNP) were performed for patient 1 and her parents.
Patients 2 and 3 were both fetuses. One was from a 26-year-old, gravida para 0 woman and the other was from a 32-year-old, gravida para 1 woman. The 26-year-old and 32-year-old pregnant women were first referred to the hospital for a genetic consultation at 25 and 21 weeks of gestation, respectively, because cleft lip was detected by prenatal ultrasound. The fathers were 26 and 34 years old, respectively. The 2 couples were nonconsanguineous and had no family history of congenital malformations on either side. After genetic counseling, cordocentesis was performed for the 2 fetuses under ultrasound guidance for cytogenetic analysis and CMA (array SNP) at 25 weeks and 21 weeks of gestation, respectively.
Karyotype
Standard karyotypes were obtained by high-resolution G-banding techniques in all patients.
CMA-SNP Array Analysis
A 2 mL sample of peripheral blood or umbilical cord blood was collected from patients and their parents. Genomic DNA was extracted from the uncultured amniocytes using a QIAamp DNA Blood Mini Kit (Qiagen, Valencia, CA). A total of 250 ng DNA was amplified, labeled, and hybridized to the CytoScan HD array platform (Affymetrix, Santa Clara, California) according to the manufacturer’s instructions. This array is designed specifically for cytogenetic analysis and offers more than 2 700 000 markers across the whole genome, including 750 000 SNP probes and 1 950 000 probes for the detection of copy number variations (Cyto-arrays). The data were analyzed with Chromosome Analysis Suite software (Affymetrix) and interpreted based on annotations of the human genome version GRCH37/hg19.
Fluorescent In Situ Hybridization
Fluorescent in situ hybridization was performed using standard protocols with a probe derived from the BAC clone RP11-458P22 (BlueGnome, United Kingdom) located within the duplication region on 15q13.3. Probe RP11-2M12 (BlueGnome) was used as a control probe.
Quantitative Polymerase Chain Reaction
Quantitative polymerase chain reaction (qPCR) was used to confirm the SNP array results and analyze the affected patients. Primer sequences are available upon request. The qPCR was performed on an ABI Prism 7900HT Sequence Detection System.
Results
A whole-genome CMA-SNP array was performed on all patients and their parents to identify possible microdeletions or microduplications. Our data revealed a microduplication at 15q13.3 of approximately 400 kb in all patients and in the father of patient 1 (Table 1). Only 1 gene, CHRNA7, was identified within this region (Figure 1A and B). The existence of this microduplication was further confirmed using qPCR (data not shown) and fluorescent in situ hybridization analysis (Figure 1C).
Table 1.
SNP Array Revealed a Microduplication 15q13.3 in the Patients.
| ID | Sample Type | sex | Clinical Data | Duplication Boundaries(hg19) | Size, kb | Inherited | Gene | |
|---|---|---|---|---|---|---|---|---|
| Start | End | |||||||
| P1 | pb | female | MR and CP | 32003537 | 32444044 | 441 | p | CHRNA7 |
| P1.1a | pb | male | Normal | 32011475 | 32446830 | 435 | _ | CHRNA7 |
| P2 | ub | male | Fetal oral clefts | 32011475 | 32444044 | 433 | dn | CHRNA7 |
| P3 | ub | male | Fetal oral clefts | 32379160 | 32914240 | 535 | dn | CHRNA7 b |
Abbreviations: pb, peripheral blood; ub, umbilical cord blood; MR, mental retardation; CP, cleft palate; _: data not access.
aP1.1 is the father of P1.
b CHRNA7, overlap with part of CHRNA7.
Discussion
The first patient carrying the 15q13.3 microduplication was identified by Van Bon et al3 in 2009. Since then, over 58 cases of smaller 15q13.3 microduplications encompassing CHRNA7 (350-680 kb in size) have been reported.2,4 The duplication lies distal to the Prader-Willi/Angelman Syndrome critical region between breakpoints 4 and 5. Indeed, there are 2 different sizes of recurrent nonallelic homologous recombination-mediated deletions or duplications at this locus: The larger copy number variants utilize breakpoint 4 and breakpoint 5 as substrate pairs and typically span a 1.6 Mb genomic region, whereas the smaller copy number variants arise between 2 low copy repeats and are 350 to 680 kb in size.2
Copy number variants of chromosome 15q13.3 involving CHRNA7 have been associated with multiple neurological and neuropsychiatric phenotypes.17–21 For example, studies have reported that 15q13.3 duplications were not associated with schizophrenia22 but were significantly associated with developmental delay and intellectual disability.6 Furthermore, a small CHRNA7 duplication was found as a risk factor for attention-deficit hyperactivity disorder.5 In addition to neurological disorders, a deletion with a size of 5.3 Mb distal to the Prader-Willi/Angelman region including CHRNA7 has been identified.23 The affected patient had a heart defect, cleft palate, recurrent infections, and developmental delay, which suggested that CHRNA7 might also be involved in body development.
The CHRNA7 gene (OMIM #118511) contains 10 exons and spans approximately 75 kb on chromosome 15q13.3.24 It is a member of the nicotinic acetylcholine receptor superfamily of ligand-gated ion channels that mediate signal transmission at synapses. The CHRNA7 protein forms a homopentameric synaptic ion channel that is highly expressed in the brain,25 which is consistent with the neurological dysfunctions identified in patients carrying a microduplication at 15q13.3. In addition to neurodevelopmental functions, the dosage sensitivity of the CHRNA7 gene has been associated with developmental delays and other phenotypes.26
It has been noted that the pathogenicity of microdeletions or microduplications of 15q13.3 encompassing CHRNA7 had a lower penetrance and led to a variation in phenotypes from normal to severe clinical symptoms.3 It was speculated that different mosaic states of microdeletion or microduplication of 15q13.3 encompassing CHRNA7 can be responsible for this variability and studies in monozygotic twins carrying mosaic 15q13.3 encompassing CHRNA7 confirmed the impact of mosaicism on phenotypic variability.17
Here, the author reported 3 nonconsanguineous families with 2 affected fetuses and 2 adults carrying a 15q13.3 breakpoints 4-breakpoint 5 duplication of ∼535 kb including only the CHRNA7 gene. Oral clefts were identified in 3 of 4 of these patients carrying the microduplication at 15q13.3. The author cannot exclude the possibility that there can be some small brain deformations that could not be detected by prenatal ultrasound. Indeed, the expression of the duplicate alone yielded protein expression but no functional receptor or coexpression with alpha 7.27 Salih et al28 reported 2 patients with high-arched palates with clear expression of integrin alpha 7 at the muscle fiber surface and in the blood vessels. Previous studies of palatal shelves from embryonic mice cultured in serum-free media and treated with nicotine inhibited palatal fusion in vitro in a dose-dependent manner, and the alpha 7 subunit of the nicotinic receptor was expressed in the medial edge epithelia during palate fusion and increased in nicotine-treated tissues.29
These 3 patients provide suggestive but insufficient evidence to conclude that the duplication of CHRNA7 alone might be responsible for oral clefts. Although the family of patient 1 included 2 cleft lip fetuses that were not tested for the 15q13.3 microduplication, it is highly suspected that they carried the microduplication because the same phenotype was observed in the prenatal ultrasound and because the father carries the same 15q13.3 microduplication.
Indeed, our study is the first to establish the relationship between 15q13.3 microduplication involving CHRNA7 and oral clefts. In the future, additional cases need to be described and compared to evaluate and confirm the association of the 15q13.3 microduplication with oral clefts.
Acknowledgments
The author would like to thank the Clinical Cytogenetics laboratory for helping the collection of data presented here. The author is also grateful to the individuals included in this study as well as their families.
Footnotes
Declaration of Conflicting Interests: The author(s) declared no potential conflicts of interest with respect to the research, authorship, and/or publication of this article.
Funding: The author(s) disclosed receipt of the following financial support for the research, authorship, and/or publication of this article: This study was supported by the National Natural Science Foundation Committee of China (NSFC-81500974 to Xie Yingjun).
Ethical Approval: Ethical approval was obtained for this study from the Ethics Committee of the First Affiliated Hospital of Sun Yat-sen University (IRB 201301)
References
- 1. Beal JC. Case report: neuronal migration disorder associated with chromosome 15q13.3 duplication in a boy with autism and seizures. J Child Neurol. 2014;29(12):NP186–NP188. [DOI] [PubMed] [Google Scholar]
- 2. Szafranski P, Schaaf CP, Person RE, et al. Structures and molecular mechanisms for common 15q13.3 microduplications involving CHRNA7: benign or pathological? Hum Mutat. 2010;31(7):840–850. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. van Bon BW, Mefford HC, Menten B, et al. Further delineation of the 15q13 microdeletion and duplication syndromes: a clinical spectrum varying from non-pathogenic to a severe outcome. J Med Genet. 2009;46(8):511–523. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Miller DT, Shen Y, Weiss LA, et al. Microdeletion/duplication at 15q13.2q13.3 among individuals with features of autism and other neuropsychiatric disorders. J Med Genet. 2009;46(4):242–248. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Williams NM, Franke B, Mick E, et al. Genome-wide analysis of copy number variants in attention deficit hyperactivity disorder: the role of rare variants and duplications at 15q13.3. Am J Psychiatry. 2012;169(2):195–204. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Cooper GM, Coe BP, Girirajan S, et al. A copy number variation morbidity map of developmental delay. Nature Genet. 2011;43(9):838–846. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Dixon MJ, Marazita ML, Beaty TH, Murray JC. Cleft lip and palate: understanding genetic and environmental influences. Nat Rev Genet. 2011;12(3):167–178. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Jugessur A, Shi M, Gjessing HK, et al. Genetic determinants of facial clefting: analysis of 357 candidate genes using two national cleft studies from Scandinavia. PloS One. 2009;4(4):e5385. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Wang Y, Li X, Zhu WL, et al. Genome-wide and interaction linkage scan for nonsyndromic cleft lip with or without cleft palate in two multiplex families in Shenyang, China. Biomed Environ Sci. 2010;23(5):363–370. [DOI] [PubMed] [Google Scholar]
- 10. Pan Y, Ma J, Zhang W, et al. Replication of two novel susceptibility loci for non-syndromic orofacial clefts in a Chinese population. Oral Dis. 2011;17(3):304–308. [DOI] [PubMed] [Google Scholar]
- 11. Beaty TH, Murray JC, Marazita ML, et al. A genome-wide association study of cleft lip with and without cleft palate identifies risk variants near MAFB and ABCA4. Nat Genet. 2010;42(6):525–529. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Blanton SH, Burt A, Stal S, Mulliken JB, Garcia E, Hecht JT. Family-based study shows heterogeneity of a susceptibility locus on chromosome 8q24 for nonsyndromic cleft lip and palate. Birth Defects Res A Clin Mol Teratol. 2010;88(4):256–259. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Ingersoll RG, Hetmanski J, Park JW, et al. Association between genes on chromosome 4p16 and non-syndromic oral clefts in four populations. Eur J Hum Genet. 2010;18(6):726–732. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Shi M, Mostowska A, Jugessur A, et al. Identification of microdeletions in candidate genes for cleft lip and/or palate. Birth Defects Res A Clin Mol Teratol. 2009;85(1):42–51. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Younkin SG, Scharpf RB, Schwender H, et al. A genome-wide study of de novo deletions identifies a candidate locus for non-syndromic isolated cleft lip/palate risk. BMC Genet. 2014;15:24. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Strehle EM, Yu L, Rosenfeld JA, et al. Genotype-phenotype analysis of 4q deletion syndrome: proposal of a critical region. Am J Med Genet Part A. 2012;158A(9):2139–2151. [DOI] [PubMed] [Google Scholar]
- 17. Popovici C, Busa T, Missirian C, Milh M, Moncla A, Philip N. Mosaic 15q13.3 deletion including CHRNA7 gene in monozygotic twins. Eur J Med Genet. 2013;56(5):274–277. [DOI] [PubMed] [Google Scholar]
- 18. Liao J, DeWard SJ, Madan-Khetarpal S, Surti U, Hu J. A small homozygous microdeletion of 15q13.3 including the CHRNA7 gene in a girl with a spectrum of severe neurodevelopmental features. Am J Med Genet A. 2011;155A(11):2795–800. [DOI] [PubMed] [Google Scholar]
- 19. Ancin I, Cabranes JA, Santos JL, et al. CHRNA7 haplotypes are associated with impaired attention in euthymic bipolar disorder. J Affect Disord. 2011;133(1-2):340–345. [DOI] [PubMed] [Google Scholar]
- 20. Joo EJ, Lee KY, Kim HS, Kim SH, Ahn YM, Kim YS. Genetic Association Study of the Alpha 7 Nicotinic Receptor (CHRNA7) with the development of schizophrenia and bipolar disorder in Korean population. Psychiatry Invest. 2010;7(3):196–201. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Ben-Shachar S, Lanpher B, German JR, et al. Microdeletion 15q13.3: a locus with incomplete penetrance for autism, mental retardation, and psychiatric disorders. J Med Genet. 2009;46(6):382–388. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Sahoo T, Theisen A, Rosenfeld JA, et al. Copy number variants of schizophrenia susceptibility loci are associated with a spectrum of speech and developmental delays and behavior problems. Genet Med. 2011;13(10):868–880. [DOI] [PubMed] [Google Scholar]
- 23. Erdogan F, Ullmann R, Chen W, et al. Characterization of a 5.3 Mb deletion in 15q14 by comparative genomic hybridization using a whole genome “tiling path” BAC array in a girl with heart defect, cleft palate, and developmental delay. Am J Med Genet A. 2007;143(2):172–178. [DOI] [PubMed] [Google Scholar]
- 24. Gault J, Robinson M, Berger R, et al. Genomic organization and partial duplication of the human alpha7 neuronal nicotinic acetylcholine receptor gene (CHRNA7). Genomics. 1998;52(2):173–185. [DOI] [PubMed] [Google Scholar]
- 25. Weng PH, Chen JH, Chen TF, et al. CHRNA7 polymorphisms and response to cholinesterase inhibitors in Alzheimer’s disease. PloS One. 2013;8(12):e84059. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Urraca N, Cleary J, Brewer V, et al. The interstitial duplication 15q11.2-q13 syndrome includes autism, mild facial anomalies and a characteristic EEG signature. Autism Res. 2013;6(4):268–279. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Araud T, Graw S, Berger R, et al. The chimeric gene CHRFAM7A, a partial duplication of the CHRNA7 gene, is a dominant negative regulator of alpha7*nAChR function. Biochem Pharmacol. 2011;82(8):904–914. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Salih MA, Al Rayess M, Cutshall S, et al. A novel form of familial congenital muscular dystrophy in two adolescents. Neuropediatrics. 1998;29(6):289–293. [DOI] [PubMed] [Google Scholar]
- 29. Kang P, Svoboda KK. Nicotine inhibits palatal fusion and modulates nicotinic receptors and the PI-3 kinase pathway in medial edge epithelia. Orthod Craniofac Res. 2003;6(3):129–142. [DOI] [PMC free article] [PubMed] [Google Scholar]