Abstract
ETS family gene fusions are common in prostate cancer and molecularly define a tumor subset. ERG is the most commonly rearranged, leading to its overexpression, followed by ETV1, ETV4, and ETV5, and these alterations are generally mutually exclusive. We validated the Decipher prostate cancer assay to detect ETS alterations in a Clinical Laboratory Improvement Amendments–accredited laboratory. Benchmarking against ERG immunohistochemistry and ETV1/4/5 RNA in situ hybridization, we examined the accuracy, precision, and reproducibility of gene expression ETS models using formalin-fixed, paraffin-embedded samples. The m-ERG model achieved an area under curve of 95%, with 93% sensitivity and 98% specificity to predict ERG immunohistochemistry status. The m-ETV1, -ETV4, and -ETV5 models achieved areas under curve of 98%, 88%, and 99%, respectively. The models had 100% robustness for ETS status, and scores were highly correlated across sample replicates. Models predicted 41.5% of a prospective radical prostatectomy cohort (n = 4036) to be ERG+, 6.3% ETV1+, 1% ETV4+, and 0.4% ETV5+. Of prostate tumor biopsy samples (n = 509), 41.2% were ERG+, 8.6% ETV1+, 0.4% ETV4+, and none ETV5+. Higher Decipher risk status tumors were more likely to be ETS+ (ERG or ETV1/4/5) in the radical prostatectomy and the biopsy cohorts (P < 0.05). These results support the utility of microarray-based ETS status prediction models for molecular classification of prostate tumors.
The landscape of somatic genomic alterations in primary prostate cancer has largely been elucidated.1 The most common alterations are gene rearrangements resulting in overexpression of ETS gene family transcription factors, including ERG most commonly, and ETV1, ETV4, ETV5, and FLI1.1, 2, 3, 4, 5 ETS gene alterations occur in approximately 50% of primary prostate tumors arising in patients of European descent,6 and they are somewhat less common in patients with other ancestry.7, 8, 9 ETS rearrangement status is not associated with altered oncologic outcomes in patients treated with radical prostatectomy6; however, ETS status may modify the association of other alterations and lifestyle factors with prognosis.10, 11 Although many prostate tumors demonstrate marked genomic heterogeneity, because ETS rearrangements are among the earliest genomic alterations to occur,12 tumor ETS status could also be exploited to track tumor clonality and recurrence.13, 14, 15, 16, 17 Finally, tumor ETS gene rearrangement status remains an attractive target for novel therapeutic and imaging methods under development.18, 19, 20 Thus, routine determination of tumor ETS status is useful for accurate molecular classification of prostate tumors and could be useful as a predictive biomarker if targeted therapies are developed in the future.
ETS gene rearrangements result in the overexpression of the ETS family member, generally because of fusion with an active, androgen-regulated promoter from the 5′ partner.2 With rare exceptions, rearrangements in one ETS gene are mutually exclusive with rearrangements of any other ETS family members,1, 21 suggesting functional redundancy among the various ETS alterations.1 In addition, some primary tumors overexpress full-length ETS transcripts (perhaps because of cryptic translocations and/or epigenetic regulation), and these tumors also lack ETS fusions.1 ETS fusion status can be determined by DNA sequencing or RT-PCR in fresh-frozen material.2, 22 In clinical samples, which are generally formalin-fixed and paraffin-embedded (FFPE), ERG status has historically been determined by DNA fluorescence in situ hybridization (DNA-FISH)23 or by immunohistochemistry (IHC) to detect ERG protein overexpression,6, 24, 25 with a very high concordance (>95%) between the two methods in most studies.6, 24 However, the lack of reliable antibodies for ETV1, ETV4, and ETV5 has meant that RNA in situ hybridization (RISH) is the only reliable method to detect fusions involving these genes in clinical specimens.26 Thus, multiple assays and different methods are required to fully characterize ETS status in FFPE clinical material, and this presents a challenge for many diagnostic pathology laboratories.
To address this issue, we validated a method for comprehensive determination of prostate tumor ETS status in FFPE tissue using a clinical high-resolution gene expression microarray assay, the Decipher prostate cancer classifier.27, 28, 29 Decipher is run on the Affymetrix Human Exon 1.0 ST microarray, and beyond determination and reporting of a validated metastasis signature, genome-wide gene expression data are automatically collected on all cases, enabling simultaneous comprehensive assessment of ETS status along with tumor risk score. We previously developed and validated a model to predict ERG DNA-FISH status from Decipher Genomics Resource Information Database (GRID) gene expression microarray data in a set of primary prostate tumors from the Mayo Clinic.9 Here, we expand to include models for ETV1, ETV4, and ETV5 status and validate the four models against gold standard IHC and RISH in a total of 456 prostate tumor cases from the Johns Hopkins Hospitals. Finally, we interrogate ETS status in >4500 clinical prostate tumor samples run on the Decipher GRID transcriptome database.
Materials and Methods
Patient and Tissue Selection
After institutional review board approval, a total of 456 unique patients were selected from two overlapping and previously published radical prostatectomy (RP) cohorts at Johns Hopkins.28, 30, 31 These cohorts were originally designed in Decipher validation studies to test for prognostic markers and were highly enriched for adverse oncologic outcomes. Tumor tissue from the dominant tumor nodule and benign tissue were sampled in quadruplicate on 16 individual tissue microarrays (TMAs) using 0.6-mm cores. Tissues were simultaneously punched with a 1-mm punch for Decipher assay. Expression profiles from 536 benign prostate tissues at RP from Decipher GRID and 65 LNCaP expression profiles were used as controls. LNCaP cells are negative for ERG rearrangement, but they harbor a rearrangement inserting the entire ETV1 locus into a transcriptionally active locus on chromosome 14.4 For model precision evaluation, 110 FFPE tumor samples from 11 patients (10 each) were profiled. These samples were run in different batches, with different operators and using different reagent lots. For model evaluation in prospective samples, we evaluated de-identified expression profiles from 4036 prospective RP tumor samples and 509 prospective biopsy samples available for research in the Decipher GRID.
Preprocessing and Expression Profiling Using Human Exon 1.0 ST Arrays for Decipher Assay
RNA extraction from FFPE tissues, amplification, labeling, and hybridization to Affymetrix Human Exon 1.0 ST microarrays was performed in a Clinical Laboratory Improvement Amendments (CLIA)-certified clinical laboratory using the Decipher prostate cancer classifier (GenomeDx Biosciences, San Diego, CA) as described previously.27, 28, 30 The SCAN algorithm was used for individual patient profile pre-processing and normalization.
Detection of ERG Overexpression by IHC
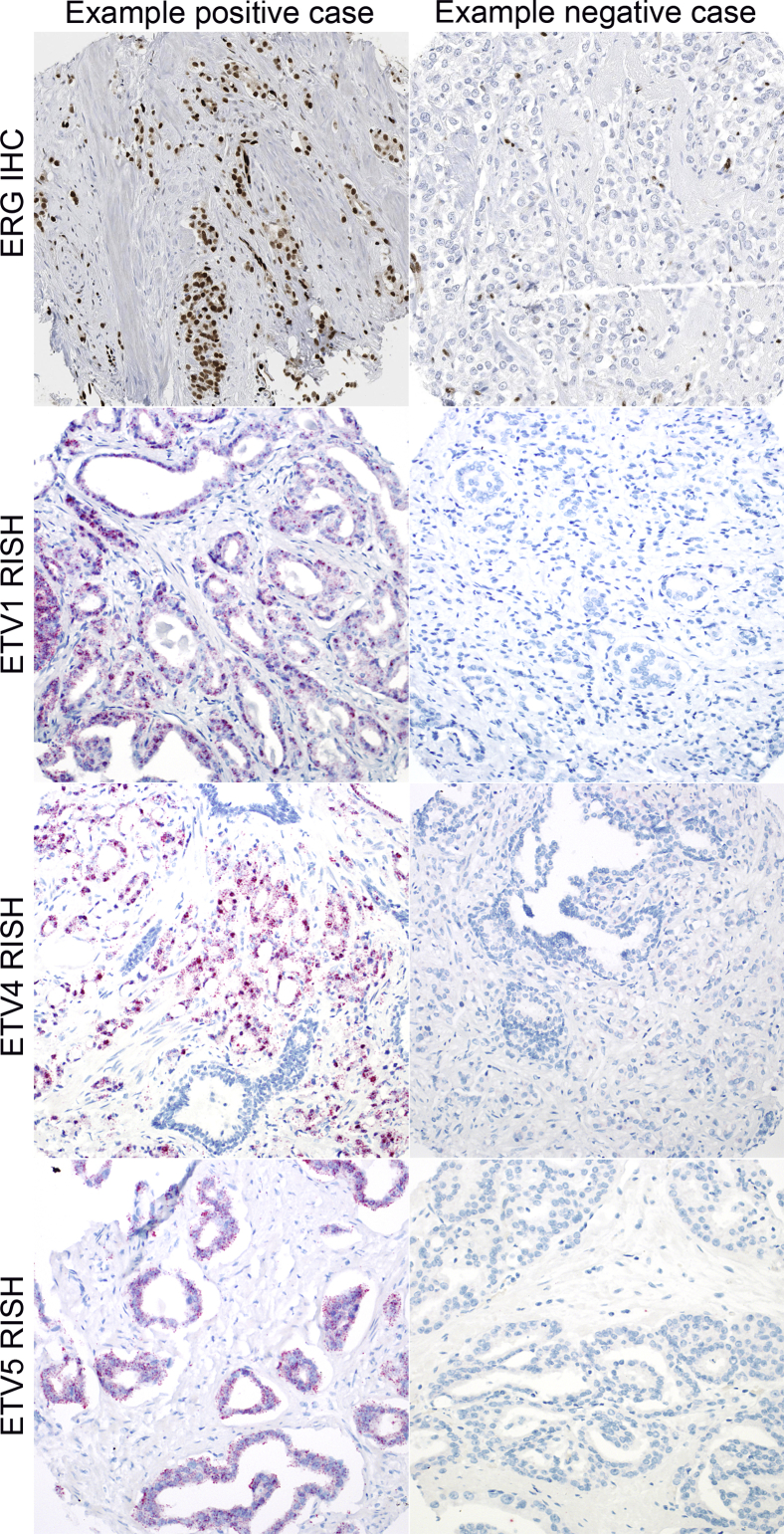
ERG IHC was performed on the Ventana Benchmark autostaining system using a rabbit monoclonal antibody (EPR3864, C-terminal) after antigen retrieval in CC1 buffer followed by detection with the Optiview HRP system (Roche/Ventana Medical Systems, Tucson, AZ). One important caveat to note for this antibody is that it reacts with the C-terminal of the protein and can cross-react with FLI1,32 another ETS gene documented to be rearranged in a small number of prostate cancer cases.5 However, on comparison of ERG immunostaining with EPR3864 and a mouse monoclonal anti-ERG antibody reacting to the N-terminus (9FY; Biocare Medical, Concord CA) which does not cross-react with FLI1, we found that only 1 of 120 cases showed discordance between the two antibodies, suggesting that cross-reactivity of EPR3864 with FLI1 may rarely be a problem. Each tissue microarray spot containing tumor was visually dichotomously scored for the presence or absence of nuclear ERG signal by a urologic pathologist blinded to the gene expression data (T.L.L.). A spot was considered to be ERG+ if any tumor nuclei showed ERG positivity, using endothelial cells as an internal positive control in all cases. A tumor was considered ERG+ if all sampled spots were scored as ERG+, and as ERG− if all sampled spots were scored as ERG− (Figure 1). In the case of mixed scoring in a given tumor (some spots negative and some spots positive, 3.5% of cases, 16 of 456 cases), the case was excluded from the validation because of heterogeneity. This low percentage of cases that are heterogeneous for ERG status is consistent with what has been reported in other studies, and these cases may represent collision tumors.5, 12, 13, 33 To evaluate reproducibility of the ERG IHC, a total of 132 prostate tumors from the Hopkins cohort were independently sampled in duplicate on more than one TMA. Of these, 127 had interpretable ERG IHC results, with 95.3% (121 of 127) showing agreement in ERG status across the two TMA sets.
Figure 1.
Examples of ERG IHC and ETV1/4/5 RISH results across the Johns Hopkins retrospective radical prostatectomy cohort. All depicted cases are concordant with m-ERG and ETV1/4/5 model calls by gene expression microarray. Original magnification, ×200. IHC, immunohistochemistry; RISH, RNA in situ hybridization.
Detection of ETV1/4/5 Overexpression by in Situ Hybridization
Chromogenic in situ hybridization for ETV1/4/5 RNA was performed with the RNAscope FFPE kit 2.5 from Advanced Cell Diagnostics (Hayward, CA) according to the manufacturer's recommendations. ETV1 (NM_004956), ETV4 (NM_001986.2), and ETV5 (NM_004454.2) probes, validated in a recent study, were used.26 Probes for PPIB (NM_000942.4) were used as positive control. We revalidated the assay in our own laboratory, using cases known by sequencing to be positive for ETV1, ETV4, or ETV5 fusions,34 and the LNCaP cell line (ETV1+).4 Normal prostate tissues from RP specimens were used as negative control tissue. All cases were qualitatively scored by a blinded surgical pathologist (T.L.L.), using a dichotomous scoring system to assess for positive cases (cases with any distinct red punctae present in any tumor cells in any punch) (Figure 1). Cases that were scored as ETV1/4/5+ in punches from one TMA set but scored as negative in the duplicate punches on another TMA set were given a final score of ETV1/4/5 heterogeneous (1.1% or 5 of 456 for ETV1; 1.1% or 5 of 456 for ETV4; and 0.2% or 1 of 456 for ETV5).
ETS Model Development
The m-ERG model was previously developed and described by Tomlins et al.9 Briefly, the ERG status (by FISH) and whole-genome expression using the Human Exon 1.0 ST Arrays was available for 407 of 580 patients as previously reported in the RP cohort from the Mayo Clinic.27 To build a model for predicting ERG status, we trained a Random Forest model using random 252 samples (of the 407 samples) with FISH status using 65 probe sets of 132 spanning the ERG locus. The 65 probe sets were summarized into three features for the Random Forest. The model was tested on 155 patients from the Mayo Clinic demonstrating significant prediction (AUC, 0.94) with 91% sensitivity and 98% specificity.
The other m-ETS models were developed from the expression of probe selection regions falling within each gene. Fifty-two, 22, and 58 probe selection regions were used for ETV1, ETV4, ETV5 models, respectively, to generate scores for the three genes by averaging the expression of probe selection regions. With the use of outlier analysis method based on extremevalues R package (R Project), a threshold was defined using the 252 samples from the Mayo Clinic samples, with expression values above threshold annotated fusion positive. Although FLI1 is an additional ETS family member documented to be rearranged in prostate cancer,5 the m-FLI1 model predicted only one FLI1+ case in the 456 tumors in the current cohort (data not shown). Given this, we did not include FLI1 in the validation set.
Statistical Analysis
Statistical analyses were performed in R version 3.2.2 (R Project), and all tests were two-sided using a 5% significance level. Fisher's exact test was used to study the association between categorical variables. Area under curve (AUC), specificity, sensitivity, positive predictive value (PPV), and negative predictive value (NPV) were used for model performance evaluation.
Results
Analytical Validation of ETS Gene Expression Models in Cell Lines and Control Tissues
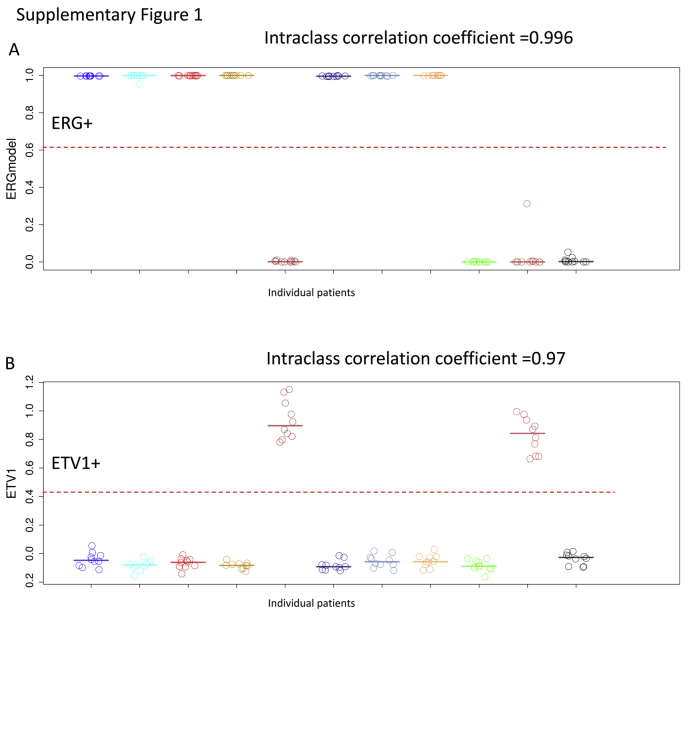
To evaluate the reproducibility and precision of the gene expression models, we used 11 different patients each with 10 FFPE samples. Across the 11 patients, the 10 replicates had 100% agreement on the four model calls with intraclass correlation coefficient of 0.99, 0.97, 0.98, and 0.95 for m-ERG, ETV1, ETV4, and ETV5, respectively (Supplemental Figure S1).
As a control, 65 LNCaP gene expression profiles, known to be ETV1+, from the Decipher GRID were used to evaluate the specificity and robustness of the models. The ETV1 model called all 65 samples as ETV1+ with 100% sensitivity, whereas other models were negative in the LNCaP cell line. To further demonstrate specificity, we tested 536 benign prostate tissues, either adjacent or spatially distant from the tumor, from RP samples. In total, 11 of 536 samples (2%) were ERG+ according to the m-ERG model score. Of interest, eight of them showed high PCA3 expression and 10 showed high α-methylacyl-coenzyme A racemase expression, suggesting that a small percentage of the samples might have been inadvertently contaminated with tumor or high-grade prostatic intraepithelial neoplasia which may be positive for ERG.35
Analytical Validation of ETS Gene Expression Models in the Retrospective RP Cohort
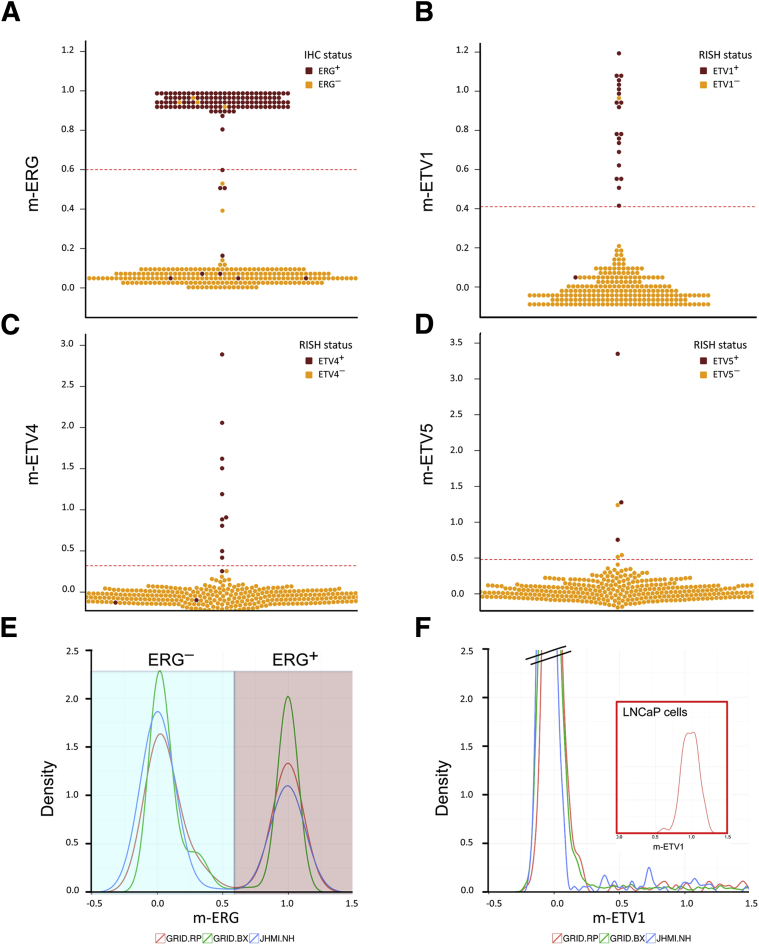
The ETS gene expression models were then applied to 456 RP samples with known ETS status by IHC or chromogenic RISH. ERG status was determined in the RP cohort by IHC, using a previously genetically validated IHC protocol.24 From the RP cohort, a total of 98% (446 of 456) of samples had interpretable ERG results by IHC (Figure 1, Table 1), with 2% (10 of 456) having absent tumor tissue on the TMAs or ambiguous staining results. Of the samples with interpretable ERG results by IHC, 37% (166 of 446) were ERG+, 59% (264 of 446) were ERG−, and 4% (16 of 446) showed heterogeneous ERG expression, with some tumor spots scored as ERG+ and some scored as ERG−. Only three (2%) of the ERG+ cases were also found to be ETV1+ (n = 2) or ETV4+ (n = 1) by RISH (see below), and all cases were heterogeneously positive for ERG. Of the 446 samples with interpretable ERG IHC results, 78% (349 of 446) had available RNA expression profiling from the Decipher assay, whereas 21% (97 of 446) did not have available expression profiling because of failure to isolate high-quality RNA. Next, we evaluated the concordance between the ERG IHC results and m-ERG model results (Figure 2A). We excluded a total of 10 samples with heterogeneous ERG status by IHC. Benchmarked against IHC, the m-ERG model achieved an AUC of 95.5 (93%–98%), with 93% sensitivity and 98% specificity. The PPV of the m-ERG model was 97% and the NPV was 96% compared with IHC.
Table 1.
Concordance between m-ERG Model Results and ERG IHC Results in Retrospective Radical Prostatectomy Cohort
IHC, immunohistochemistry.
Data were used for calculation of performance characteristics in Table 5.
Figure 2.
Validation of the m-ERG and ETV1/4/5 gene expression models by comparison with IHC and RISH. A–D: Beeswarm plots of ERG, ETV1, ETV4, and ETV5 models in JHMI RP cohort showing model scores are highly concordant with IHC and RISH calls. The red dotted lines indicate the threshold of each model for calling ETS positivity. E and F: Density plots of m-ERG and ETV1 models in a subset of the JHMI RP cohort, prospective Decipher GRID RP, and biopsy cohorts and LNCaP cells with ETV1 fusion, showing distribution of models are highly similar across different data sets. ERG−, blue background; ERG+, mauve background. n = 358 JHMI RP cohorts (E and F); n = 4036 prospective Decipher GRID RP cohorts (E and F); n = 509 biopsy cohorts (E and F). GRID, Genomics Resource Information Database; IHC, immunohistochemistry; JHMI, Johns Hopkins Medical Institute; RISH, RNA in situ hybridization; RP, radical prostatectomy.
ETV1, ETV4, and ETV5 status for each case was determined in the RP cohort by RISH. From the RP cohort, a total of 98% (448 of 456) of samples had interpretable ETV1 RISH results (Figure 1, Table 2), with 2% (8 of 456) having absent tumor tissue on the TMA. Of the samples with interpretable ETV1 results, 6% (29 of 448) were ETV1+, 92% (414 of 448) were ETV1−, and 1% (5 of 448) showed heterogeneous ETV1 expression, with some tumor spots scored as ETV1+ and some scored as ETV1−. Of the 448 samples with interpretable ETV1 RISH results, 78% (351 of 448) had available RNA expression profiling from the Decipher assay, whereas 22% (97 of 448) did not have available expression profiling because of failure to isolate high-quality RNA. Next, we examined the concordance between the ETV1 RISH results and ETV1 model results (Figure 2B). We excluded the five samples with heterogeneous ETV1 status by RISH. Benchmarked against RISH, the ETV1 model achieved an AUC of 98% (94%–100%), with 93% sensitivity and 99% specificity. The PPV of the ETV1 model was 93% and the NPV was 99% compared with IHC.
Table 2.
Concordance between m-ETV1 Model Results and ETV1 RISH Results in Retrospective Radical Prostatectomy Cohort
RISH, RNA in situ hybridization.
Data were used for calculation of performance characteristics in Table 5.
From the RP cohort, a total of 99% (451 of 456) of samples had interpretable ETV4 RISH results (Figure 1, Table 3), with 1% (5 of 456) having absent tumor tissue on the TMA. Of the samples with interpretable ETV4 results, 3% (13 of 451) were ETV4+, 96% (433 of 451) were ETV4−, and 1% (5 of 451) showed heterogeneous ETV4 expression, with some tumor spots scored as ETV4+ and some scored as ETV4−. Of the 451 samples with interpretable ETV4 RISH results, 78% (354 of 451) had available RNA expression profiling from the Decipher assay, whereas 22% (97 of 451) did not have available expression profiling because of failure to isolate high-quality RNA. Next, we examined the concordance between the ETV4 RISH results and ETV4 model results (Figure 2C). We excluded the five samples with heterogeneous ETV4 status by RISH. Benchmarked against RISH, the ETV4 model achieved an AUC of 88% (76%–99%), with 77% sensitivity and 100% specificity. The PPV of the ETV4 model was 100% and the NPV was 99% compared with IHC.
Table 3.
Concordance between m-ETV4 Model Results and ETV4 RISH Results in Retrospective Radical Prostatectomy Cohort
RISH, RNA in situ hybridization.
Data were used for calculation of performance characteristics in Table 5.
From the RP cohort, a total of 99% (450 of 456) of samples had interpretable ETV5 RISH results (Figure 1, Table 4), with 1% (6 of 456) having absent tumor tissue on the TMA. Of the samples with interpretable ETV5 results, 1% (4 of 450) were ETV5+, 99% (445 of 450) were ETV5−, and 0.2% (1 of 450) showed heterogeneous ETV5 expression, with some tumor spots scored as ETV5+ and some scored as ETV5−. Of the 450 samples with interpretable ETV5 RISH results, 78% (352 of 450) had available RNA expression profiling from the Decipher assay, whereas 22% (98 of 450) did not have available expression profiling because of failure to isolate high-quality RNA. Next, we evaluated concordance between the ETV5 RISH results and ETV5 model results (Figure 2D). We excluded the single sample with heterogeneous ETV5 status by RISH. Benchmarked against RISH, the ETV5 model achieved an AUC of 99.6% (99%–100%), with 100% sensitivity and 99% specificity. The PPV of the ETV5 model was 50% and the NPV was 100% compared with IHC. A comparison of the performance characteristics of mERG, ETV1, ETV4, and ETV5 models is included in Table 5.
Table 4.
Concordance between m-ETV5 Model Results and ETV5 RISH Results in Retrospective Radical Prostatectomy Cohort
RISH, RNA in situ hybridization.
Data were used for calculation of performance characteristics in Table 5.
Table 5.
Performance Metrics of m-ETS Models Compared with Genetically Validated Immunohistochemistry and Chromogenic RNA in Situ Hybridization
| m-ETS model | Metric |
|||
|---|---|---|---|---|
| Sensitivity | Specificity | PPV | NPV | |
| ERG | 93 | 98 | 97 | 96 |
| ETV1 | 93 | 99 | 93 | 99 |
| ETV4 | 77 | 100 | 100 | 99 |
| ETV5 | 100 | 99 | 50 | 100 |
All values are percentages.
NPV, negative predictive value; PPV, positive predictive value.
Implementing and Evaluating the ETS Gene Expression Models in Prospective RP and Biopsy Samples
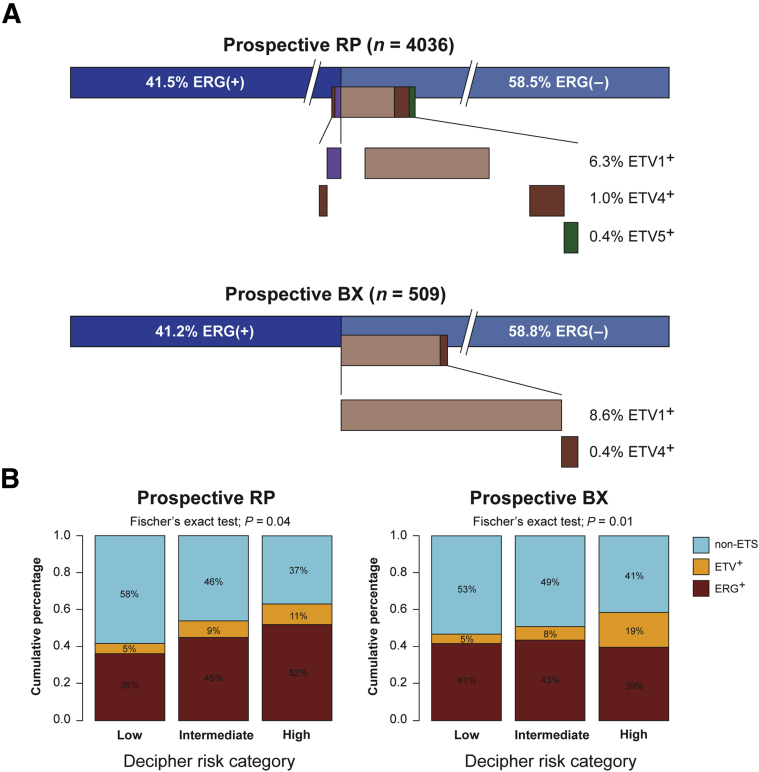
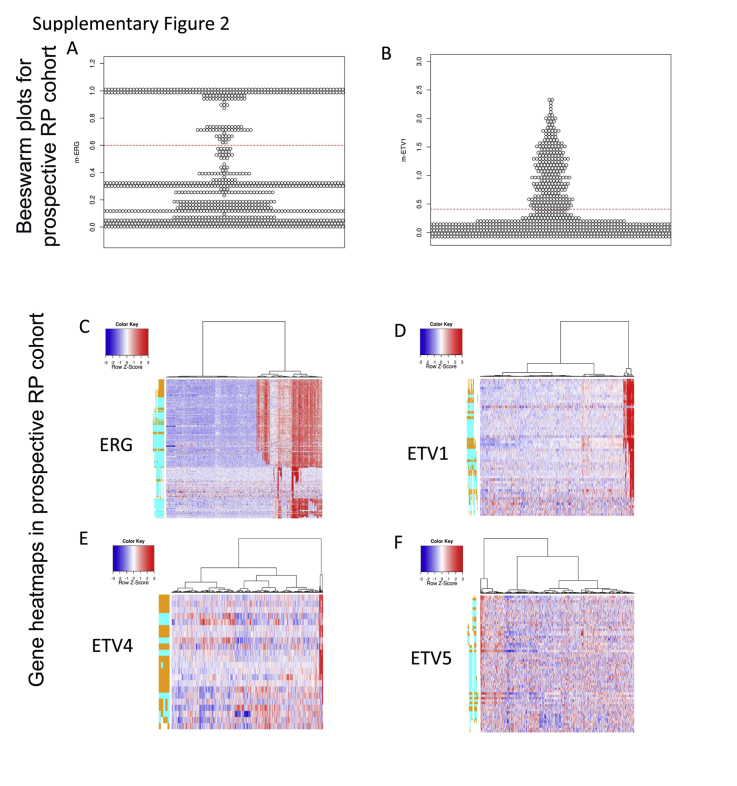
We implemented the ETS models prospectively as part of the Decipher assay performed on 4036 RP and 509 biopsy samples in a CLIA-accredited laboratory (Supplemental Figure S2). The ERG and ETV1 models had a similar distribution of scores in the retrospective RP cohort as in the prospective RP and biopsy cohorts (Figure 2, E and F). In both the prospective RP and biopsy cohorts, 41% of the patients were found to be m-ERG+, in agreement with previous large-scale studies.6 In total, 6.3% and 8.6% of the RP and biopsy samples, respectively, were ETV1+. ETV4 and ETV5 were detected at low rate >1% in both the RP and biopsy samples. Overall, 11 samples were found to be both ERG+ and ETV1 and one case was ERG+/ETV4+ in the RP samples, similar to the low frequency of collision tumor described in other reports.26 No double-positive cases were identified in the biopsy cohort (Figure 3A).
Figure 3.
Examination of m-ERG and ETV1/4/5 gene expression models in prospective Decipher GRID RP and BX specimens. A: In GRID RP and BX data, ERG and ETV1/4/5 status are highly, although not entirely, mutually exclusive. B: Among cases in the RP cohort with high Decipher risk score compared with low Decipher risk score, a higher proportion of cases are m-ERG+. Among cases in the BX cohort with high Decipher risk score compared with low Decipher risk score, a high proportion of cases are ETV1/4/5+ (ETV+). BX, biopsy; GRID, Genomics Resource Information Database; RP, radical prostatectomy.
Because definitive outcome data are not available on the prospective samples, we correlated ETS gene expression classification with the Decipher risk category in these samples, because the Decipher risk category correlates with risk of metastasis in multiple retrospective RP cohorts.28, 36 Overall, 52% of samples classified as high risk by Decipher were ERG+, whereas only 36% of samples classified as low risk were ERG+ in the prospective RP cohort (Fisher's test, P = 0.04). In the prospective biopsy cohort, ERG frequency was not significantly different among Decipher risk categories, but ETV1/4/5+ cases comprised 19% of the high-risk Decipher group compared with 5% in low-risk Decipher (Fisher's test, P = 0.01) (Figure 3B). When combining ERG and ETV1/4/5+ cases into one ETS+ group, the associations between ETS+ and metastatic risk was more pronounced. In the high-risk Decipher group, 63% and 59% of RP and biopsy samples, respectively, were ETS+, compared with 42% and 46% in the low-risk Decipher RP and biopsy samples, respectively.
Discussion
Fusions involving the ETS family genes define the largest molecularly distinct subclass of prostate tumors and are among the earliest events in the clonal evolution of the disease. Although the presence of ETS rearrangements by themselves is not associated with adverse oncologic outcomes in surgically treated patients,6 ERG status has been prognostic in tumor samples from transurethral resections in a conservatively managed population37 and in biopsies in some contexts. Most notably, assessing a cohort of 217 active surveillance patients, Berg et al38 reported that patients with any ERG+ cores at diagnosis (by IHC) were more than twice as likely to progress compared with ERG− patients; ERG+ status was the most significant predictor of active surveillance progression in multivariable Cox regression analysis. In addition, there is increasing evidence that ERG fusions may modify the association of other molecular alterations and lifestyle factors with prognosis. For example, loss of the PTEN tumor suppressor gene is more strongly associated with death from prostate cancer among ERG− prostate tumors compared with those that are ERG+.10 Obesity is also associated with a significantly elevated risk of death from prostate cancer, specifically among ERG+ but not ERG− tumors.11
The high prevalence of ETS alterations and their relatively early occurrence in tumor progression makes them an attractive diagnostic and potentially therapeutic target. Prostate cancer is commonly multifocal, with multiple genetically distinct tumor foci that may be indistinguishable by routine histology. In contrast to other alterations involving genes such as PTEN, ERG and ETS status are almost invariably clonal and homogeneous within a given prostate tumor and accordingly are considered to be truncal alterations in most primary tumors.2, 12, 33 Thus, discerning ETS status in diagnostic biopsies and/or metastatic biopsies may have utility in evaluating tumor multifocality, tracking individual tumor clones, and assessing for disease recurrence in liquid biopsies.13, 14, 15, 16 Finally, therapies that exploit potential vulnerabilities unique to ETS-positive cells have the potential for dramatic efficacy in prostate tumors that are otherwise clonally heterogeneous.17, 18, 19, 20 Although more targeted therapies are still under development, there is early evidence that ETS status may be predictive for response to standard therapies as well.39 Thus, assays to comprehensively assess ETS status could be potentially predictive.
The current gold standard for assessing ETS status in FFPE clinical prostate cancer tissues involves multiple IHC and DNA/RNA in situ hybridization assays. Assessment for ERG rearrangement by IHC is used clinically in a number of laboratories. However, performing this stand-alone assay misses as many as 15% or so of prostate tumors that harbor fusions in alternative ETS family members, such as ETV1, ETV4, and ETV5, and are molecularly similar to ERG-rearranged tumors. Previously, we reported on the development of a model to predict ERG DNA-FISH status from Decipher gene expression microarray data.27, 28 Here, we have further evaluated the Decipher clinical-grade assay as a streamlined approach to comprehensively assess ETS status in the Decipher GRID, which has been made available to the urologic oncology community for research. The m-ERG model was developed in a set of 252 primary prostate tumors at RP from the Mayo Clinic and validated in an independent set of 155 RP samples from the same institute, achieving 91% sensitivity and 98% specificity.9 In the present study, we expanded this validation to include ETV1, ETV4, and ETV5 (compared with RISH) and ERG (compared with IHC rather than FISH) using a retrospective RP cohort from Johns Hopkins. We show that the m-ERG models was highly specific (100%) and the ETV1 model was highly sensitive (100%) in the LNCaP cells (known to be ETV1+). ETS models were highly reproducible across 11 patients with 10 technical replicate samples each run in different batches, operators, or reagent lots. In benign prostate tissue samples, m-ERG was 98% specific, whereas only 11 samples of 536 were called ERG+. Of interest, a small fraction of histologically benign-appearing glands have previously been reported to be ERG+.40 In addition, a deeper look at these 11 samples revealed that 8 expressed high levels of PCA3 and 10 expressed α-methylacyl-coenzyme A racemase, raising the possibility of tumor or high-grade prostatic intraepithelial neoplasia contamination.
When applied to a cohort of prostate tumors from RP samples at Johns Hopkins, the m-ERG model was 93% sensitive for ERG expression as measured by IHC and 98% specific for lack of ERG expression. Because ERG fusion status is heterogeneous in a minority of cases (perhaps because of collisions of two independent clones of tumor), it is likely that some of the discordance between the m-ERG model and IHC may be due to sampling of two separate tumor clones.13 Similarly, the ETV1/4/5 gene expression models showed high sensitivity for detecting cases with underlying RNA overexpression by RISH, with sensitivity for ETV1, the second most commonly rearranged ETS gene, at 93%. Notably, lower sensitivity was seen for the more uncommon cases overexpressing ETV4. Importantly, however, specificity at or exceeding 99% was seen for ETV1/4/5. In part, the low rate of positive events may have adversely affected the performance characteristics for ETV4 and ETV5, because any discordant cases have a disproportionally large effect on sensitivity when the number of positive cases is small. Reasons for the discordance between the RISH and microarray-based expression assays are multiple, including potential for tumor heterogeneity and RNA degradation in older material that may have more severely affected one assay compared with the other. We strongly suspect that the later explanation may have been dominant here. Of note, >20% of cases in the validation cohort failed RNA quality controls for Decipher, and almost all of these cases had FFPE tumor blocks that were substantially older than 10 years,28 a potential limitation for the Decipher assay in older material. However, despite failure for Decipher, at least a few of the same cases had clearly positive RISH results (Table 2, Table 3, and 4), indicating potentially differing tolerance of each assay to RNA degradation occurring in FFPE over time. Ultimately, additional validation in prospective cohorts from our institution with newer FFPE material could be useful to test whether tissue block age may have been a factor in the relatively low concordance between the two methods.
When applied to prospective cohorts of RP and biopsy samples, ERG- and ETV-positive cases were identified at the expected frequencies, with distribution of the m-ERG scores similar to that seen in the Hopkins archival RP cases. Interestingly, there was a correlation of m-ERG/ETS model frequency with Decipher risk category, with a relatively higher frequency of m-ERG+ and m-ETV1/4/5+ cases seen among cases with high Decipher risk category compared with low Decipher risk category in the RP and biopsy cohorts, respectively. Because high Decipher risk category is strongly associated with poor oncologic outcomes in prostate cancer, these findings might suggest that m-ERG+ and/or ETV1/4/5+ cases are associated with a worse prognosis. Importantly, however, the presence of ETS rearrangements was not associated with adverse outcomes in the current cohorts as previously reported by our group,30 similar to what has been reported previously using Decipher in other cohorts9 and similar to most studies querying ERG status alone by other methods in surgical cohorts.6 Thus, the significance of this finding is unclear.
In conclusion, we anticipate that incorporating these comprehensive and validated models to predict tumor ETS status into the clinically available Decipher prostate cancer classifier assay will have potential near term clinical utility for the molecular classification of prostate tumors. ETS status may be useful as a prognostic biomarker in active surveillance, either alone or in combination with other markers, as a tool to evaluate tumor clonality and to track disease recurrence or during clinical trial design for targeted therapies. Importantly, IHC and RISH for ETS classification are less expensive assays that can be performed in any pathology laboratory; thus, these will still be useful for tumors that require only limited characterization. However, in the setting of high-risk tumors that may benefit from Decipher classification, ETS classification can be additionally determined simultaneously in a CLIA-accredited laboratory using the Decipher gene expression platform.
Acknowledgments
We thank Angelo Michael De Marzo and Qizhi Zheng for their help with RNA in situ hybridization protocols.
Footnotes
Supported in part by the Congressionally Directed Medical Research Programs Transformative Impact Award (T.L.L.) and NIH/National Cancer Institute Prostate Specialized Programs of Research Excellence grant P50CA58236.
A.T. and M.A. contributed equally to this work.
Disclosures: M.A., N.E., J.C., L.L., L.L.C.L., E.D., E.A.G., and K.Y. are employees of GenomeDx Biosciences with stock options, the company that provided the funding for the microarray-based expression analysis in this study. A.E.R. is a consultant for GenomeDx. T.L.L. has received research funding from Ventana Medical Systems and GenomeDx Biosciences. S.A.T. is a co-inventor on a patent issued to the University of Michigan on ETS gene fusions in prostate cancer. The diagnostic field of use has been licensed to Hologic/Gen-Probe, Inc., which has sublicensed rights to Roche/Ventana Medical Systems. S.A.T. has served as a consultant for and received honoraria from Roche/Ventana Medical Systems, Janssen, AbbVie, and Astellas/Medivation. S.A.T. had a sponsored research agreement with and received travel support from Compendia Biosciences/Life Technologies/Thermo Fisher. S.A.T. has received research funding from GenomeDX Biosciences and Astellas. S.A.T. is a co-founder, equity holder in, and consultant for Strata Oncology.
Supplemental material for this article can be found at http://dx.doi.org/10.1016/j.jmoldx.2017.01.007.
Supplemental Data
Supplemental Figure S1.
m-ERG (A) and ETV1 (B) model scores across 11 patients, each with 10 replicate samples, showing reproducibility and precision of models. The red dotted lines indicate the threshold of each model for calling ETS positivity.
Supplemental Figure S2.
Examination of m-ERG and ETV1 gene expression models in prospective Decipher GRID RP specimens. A and B: Beeswarm plots of m-ERG and m-ETV1 model scores in the GRID RP cohort. The red dotted lines indicate the threshold of each model for calling ETS positivity. C–F: Heatmaps of ERG, ETV1, ETV4, ETV5 PSRs across GRID RP cohort showing the feasibility of predicting fusion using the Human Exon 1.0 ST arrays. GRID, Genomics Resource Information Database; PSR, probe selection region; RP, radical prostatectomy.
References
- 1.Cancer Genome Atlas Research Network The molecular taxonomy of primary prostate cancer. Cell. 2015;163:1011–1025. doi: 10.1016/j.cell.2015.10.025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Tomlins S.A., Rhodes D.R., Perner S., Dhanasekaran S.M., Mehra R., Sun X.W., Varambally S., Cao X., Tchinda J., Kuefer R., Lee C., Montie J.E., Shah R.B., Pienta K.J., Rubin M.A., Chinnaiyan A.M. Recurrent fusion of TMPRSS2 and ETS transcription factor genes in prostate cancer. Science. 2005;310:644–648. doi: 10.1126/science.1117679. [DOI] [PubMed] [Google Scholar]
- 3.Tomlins S.A., Mehra R., Rhodes D.R., Smith L.R., Roulston D., Helgeson B.E., Cao X., Wei J.T., Rubin M.A., Shah R.B., Chinnaiyan A.M. TMPRSS2:ETV4 gene fusions define a third molecular subtype of prostate cancer. Cancer Res. 2006;66:3396–3400. doi: 10.1158/0008-5472.CAN-06-0168. [DOI] [PubMed] [Google Scholar]
- 4.Tomlins S.A., Laxman B., Dhanasekaran S.M., Helgeson B.E., Cao X., Morris D.S., Menon A., Jing X., Cao Q., Han B., Yu J., Wang L., Montie J.E., Rubin M.A., Pienta K.J., Roulston D., Shah R.B., Varambally S., Mehra R., Chinnaiyan A.M. Distinct classes of chromosomal rearrangements create oncogenic ETS gene fusions in prostate cancer. Nature. 2007;448:595–599. doi: 10.1038/nature06024. [DOI] [PubMed] [Google Scholar]
- 5.Paulo P., Barros-Silva J.D., Ribeiro F.R., Ramalho-Carvalho J., Jeronimo C., Henrique R., Lind G.E., Skotheim R.I., Lothe R.A., Teixeira M.R. FLI1 is a novel ETS transcription factor involved in gene fusions in prostate cancer. Genes Chromosomes Cancer. 2012;51:240–249. doi: 10.1002/gcc.20948. [DOI] [PubMed] [Google Scholar]
- 6.Pettersson A., Graff R.E., Bauer S.R., Pitt M.J., Lis R.T., Stack E.C., Martin N.E., Kunz L., Penney K.L., Ligon A.H., Suppan C., Flavin R., Sesso H.D., Rider J.R., Sweeney C., Stampfer M.J., Fiorentino M., Kantoff P.W., Sanda M.G., Giovannucci E.L., Ding E.L., Loda M., Mucci L.A. The TMPRSS2:ERG rearrangement, ERG expression, and prostate cancer outcomes: a cohort study and meta-analysis. Cancer Epidemiol Biomarkers Prev. 2012;21:1497–1509. doi: 10.1158/1055-9965.EPI-12-0042. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Magi-Galluzzi C., Tsusuki T., Elson P., Simmerman K., LaFargue C., Esgueva R., Klein E., Rubin M.A., Zhou M. TMPRSS2-ERG gene fusion prevalence and class are significantly different in prostate cancer of Caucasian, African-American and Japanese patients. Prostate. 2011;71:489–497. doi: 10.1002/pros.21265. [DOI] [PubMed] [Google Scholar]
- 8.Khani F., Mosquera J.M., Park K., Blattner M., O'Reilly C., MacDonald T.Y., Chen Z., Srivastava A., Tewari A.K., Barbieri C.E., Rubin M.A., Robinson B.D. Evidence for molecular differences in prostate cancer between African American and Caucasian men. Clin Cancer Res. 2014;20:4925–4934. doi: 10.1158/1078-0432.CCR-13-2265. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Tomlins S.A., Alshalalfa M., Davicioni E., Erho N., Yousefi K., Zhao S., Haddad Z., Den R.B., Dicker A.P., Trock B.J., DeMarzo A.M., Ross A.E., Schaeffer E.M., Klein E.A., Magi-Galluzzi C., Karnes R.J., Jenkins R.B., Feng F.Y. Characterization of 1577 primary prostate cancers reveals novel biological and clinicopathologic insights into molecular subtypes. Eur Urol. 2015;68:555–567. doi: 10.1016/j.eururo.2015.04.033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Ahearn T.U., Pettersson A., Ebot E.M., Gerke T., Graff R.E., Morais C.L., Hicks J.L., Wilson K.M., Rider J.R., Sesso H.D., Fiorentino M., Flavin R., Finn S., Giovannucci E.L., Loda M., Stampfer M.J., De Marzo A.M., Mucci L.A., Lotan T.L. A prospective investigation of PTEN loss and ERG expression in lethal prostate cancer. J Natl Cancer Inst. 2016;108:djv346. doi: 10.1093/jnci/djv346. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Pettersson A., Lis R.T., Meisner A., Flavin R., Stack E.C., Fiorentino M., Finn S., Graff R.E., Penney K.L., Rider J.R., Nuttall E.J., Martin N.E., Sesso H.D., Pollak M., Stampfer M.J., Kantoff P.W., Giovannucci E.L., Loda M., Mucci L.A. Modification of the association between obesity and lethal prostate cancer by TMPRSS2:ERG. J Natl Cancer Inst. 2013;105:1881–1890. doi: 10.1093/jnci/djt332. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Gumuskaya B., Gurel B., Fedor H., Tan H.L., Weier C.A., Hicks J.L., Haffner M.C., Lotan T.L., De Marzo A.M. Assessing the order of critical alterations in prostate cancer development and progression by IHC: further evidence that PTEN loss occurs subsequent to ERG gene fusion. Prostate Cancer Prostatic Dis. 2013;16:209–215. doi: 10.1038/pcan.2013.8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Fontugne J., Davis K., Palanisamy N., Udager A., Mehra R., McDaniel A.S., Siddiqui J., Rubin M.A., Mosquera J.M., Tomlins S.A. Clonal evaluation of prostate cancer foci in biopsies with discontinuous tumor involvement by dual ERG/SPINK1 immunohistochemistry. Mod Pathol. 2016;29:157–165. doi: 10.1038/modpathol.2015.148. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Perner S., Svensson M.A., Hossain R.R., Day J.R., Groskopf J., Slaughter R.C., Jarleborn A.R., Hofer M.D., Kuefer R., Demichelis F., Rickman D.S., Rubin M.A. ERG rearrangement metastasis patterns in locally advanced prostate cancer. Urology. 2010;75:762–767. doi: 10.1016/j.urology.2009.10.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Ulz P., Belic J., Graf R., Auer M., Lafer I., Fischereder K., Webersinke G., Pummer K., Augustin H., Pichler M., Hoefler G., Bauernhofer T., Geigl J.B., Heitzer E., Speicher M.R. Whole-genome plasma sequencing reveals focal amplifications as a driving force in metastatic prostate cancer. Nat Commun. 2016;7:12008. doi: 10.1038/ncomms12008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Attard G., Swennenhuis J.F., Olmos D., Reid A.H., Vickers E., A'Hern R., Levink R., Coumans F., Moreira J., Riisnaes R., Oommen N.B., Hawche G., Jameson C., Thompson E., Sipkema R., Carden C.P., Parker C., Dearnaley D., Kaye S.B., Cooper C.S., Molina A., Cox M.E., Terstappen L.W., de Bono J.S. Characterization of ERG, AR and PTEN gene status in circulating tumor cells from patients with castration-resistant prostate cancer. Cancer Res. 2009;69:2912–2918. doi: 10.1158/0008-5472.CAN-08-3667. [DOI] [PubMed] [Google Scholar]
- 17.Reig O., Marin-Aguilera M., Carrera G., Jimenez N., Pare L., Garcia-Recio S., Gaba L., Pereira M.V., Fernandez P., Prat A., Mellado B. TMPRSS2-ERG in blood and docetaxel resistance in metastatic castration-resistant prostate cancer. Eur Urol. 2016;70:709–713. doi: 10.1016/j.eururo.2016.02.034. [DOI] [PubMed] [Google Scholar]
- 18.Roychowdhury S., Chinnaiyan A.M. Advancing precision medicine for prostate cancer through genomics. J Clin Oncol. 2013;31:1866–1873. doi: 10.1200/JCO.2012.45.3662. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Brenner J.C., Ateeq B., Li Y., Yocum A.K., Cao Q., Asangani I.A., Patel S., Wang X., Liang H., Yu J., Palanisamy N., Siddiqui J., Yan W., Cao X., Mehra R., Sabolch A., Basrur V., Lonigro R.J., Yang J., Tomlins S.A., Maher C.A., Elenitoba-Johnson K.S., Hussain M., Navone N.M., Pienta K.J., Varambally S., Feng F.Y., Chinnaiyan A.M. Mechanistic rationale for inhibition of poly(ADP-ribose) polymerase in ETS gene fusion-positive prostate cancer. Cancer Cell. 2011;19:664–678. doi: 10.1016/j.ccr.2011.04.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Han S., Brenner J.C., Sabolch A., Jackson W., Speers C., Wilder-Romans K., Knudsen K.E., Lawrence T.S., Chinnaiyan A.M., Feng F.Y. Targeted radiosensitization of ETS fusion-positive prostate cancer through PARP1 inhibition. Neoplasia. 2013;15:1207–1217. doi: 10.1593/neo.131604. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Svensson M.A., LaFargue C.J., MacDonald T.Y., Pflueger D., Kitabayashi N., Santa-Cruz A.M., Garsha K.E., Sathyanarayana U.G., Riley J.P., Yun C.S., Nagy D., Kosmeder J.W., Pestano G.A., Tewari A.K., Demichelis F., Rubin M.A. Testing mutual exclusivity of ETS rearranged prostate cancer. Lab Invest. 2011;91:404–412. doi: 10.1038/labinvest.2010.179. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Tu J.J., Rohan S., Kao J., Kitabayashi N., Mathew S., Chen Y.T. Gene fusions between TMPRSS2 and ETS family genes in prostate cancer: frequency and transcript variant analysis by RT-PCR and FISH on paraffin-embedded tissues. Mod Pathol. 2007;20:921–928. doi: 10.1038/modpathol.3800903. [DOI] [PubMed] [Google Scholar]
- 23.Perner S., Demichelis F., Beroukhim R., Schmidt F.H., Mosquera J.M., Setlur S., Tchinda J., Tomlins S.A., Hofer M.D., Pienta K.G., Kuefer R., Vessella R., Sun X.W., Meyerson M., Lee C., Sellers W.R., Chinnaiyan A.M., Rubin M.A. TMPRSS2:ERG fusion-associated deletions provide insight into the heterogeneity of prostate cancer. Cancer Res. 2006;66:8337–8341. doi: 10.1158/0008-5472.CAN-06-1482. [DOI] [PubMed] [Google Scholar]
- 24.Chaux A., Albadine R., Toubaji A., Hicks J., Meeker A., Platz E.A., De Marzo A.M., Netto G.J. Immunohistochemistry for ERG expression as a surrogate for TMPRSS2-ERG fusion detection in prostatic adenocarcinomas. Am J Surg Pathol. 2011;35:1014–1020. doi: 10.1097/PAS.0b013e31821e8761. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Park K., Tomlins S.A., Mudaliar K.M., Chiu Y.L., Esgueva R., Mehra R., Suleman K., Varambally S., Brenner J.C., MacDonald T., Srivastava A., Tewari A.K., Sathyanarayana U., Nagy D., Pestano G., Kunju L.P., Demichelis F., Chinnaiyan A.M., Rubin M.A. Antibody-based detection of ERG rearrangement-positive prostate cancer. Neoplasia. 2010;12:590–598. doi: 10.1593/neo.10726. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Kunju L.P., Carskadon S., Siddiqui J., Tomlins S.A., Chinnaiyan A.M., Palanisamy N. Novel RNA hybridization method for the in situ detection of ETV1, ETV4, and ETV5 gene fusions in prostate cancer. Appl Immunohistochem Mol Morphol. 2014;22:e32–e40. doi: 10.1097/PAI.0000000000000095. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Erho N., Crisan A., Vergara I.A., Mitra A.P., Ghadessi M., Buerki C., Bergstralh E.J., Kollmeyer T., Fink S., Haddad Z., Zimmermann B., Sierocinski T., Ballman K.V., Triche T.J., Black P.C., Karnes R.J., Klee G., Davicioni E., Jenkins R.B. Discovery and validation of a prostate cancer genomic classifier that predicts early metastasis following radical prostatectomy. PLoS One. 2013;8:e66855. doi: 10.1371/journal.pone.0066855. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Ross A.E., Johnson M.H., Yousefi K., Davicioni E., Netto G.J., Marchionni L., Fedor H.L., Glavaris S., Choeurng V., Buerki C., Erho N., Lam L.L., Humphreys E.B., Faraj S., Bezerra S.M., Han M., Partin A.W., Trock B.J., Schaeffer E.M. Tissue-based genomics augments post-prostatectomy risk stratification in a natural history cohort of intermediate- and high-risk men. Eur Urol. 2016;69:157–165. doi: 10.1016/j.eururo.2015.05.042. [DOI] [PubMed] [Google Scholar]
- 29.Knudsen B.S., Kim H.L., Erho N., Shin H., Alshalalfa M., Lam L.L., Tenggara I., Chadwich K., Van Der Kwast T., Fleshner N., Davicioni E., Carroll P.R., Cooperberg M.R., Chan J.M., Simko J.P. Application of a clinical whole-transcriptome assay for staging and prognosis of prostate cancer diagnosed in needle core biopsy specimens. J Mol Diagn. 2016;18:395–406. doi: 10.1016/j.jmoldx.2015.12.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Johnson M.H., Ross A.E., Alshalalfa M., Erho N., Yousefi K., Glavaris S., Fedor H., Han M., Faraj S.F., Bezerra S.M., Netto G., Partin A.W., Trock B.J., Davicioni E., Schaeffer E.M. SPINK1 defines a molecular subtype of prostate cancer in men with more rapid progression in an at risk, natural history radical prostatectomy cohort. J Urol. 2016;196:1436–1444. doi: 10.1016/j.juro.2016.05.092. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Faraj S.F., Bezerra S.M., Yousefi K., Fedor H., Glavaris S., Han M., Partin A.W., Humphreys E., Tosoian J., Johnson M.H., Davicioni E., Trock B.J., Schaeffer E.M., Ross A.E., Netto G.J. Clinical validation of the 2005 ISUP Gleason grading system in a cohort of intermediate and high risk men undergoing radical prostatectomy. PLoS One. 2016;11:e0146189. doi: 10.1371/journal.pone.0146189. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Tomlins S.A., Palanisamy N., Brenner J.C., Stall J.N., Siddiqui J., Thomas D.G., Lucas D.R., Chinnaiyan A.M., Kunju L.P. Usefulness of a monoclonal ERG/FLI1 antibody for immunohistochemical discrimination of Ewing family tumors. Am J Clin Pathol. 2013;139:771–779. doi: 10.1309/AJCPN4L1BMRQPEIT. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Krohn A., Freudenthaler F., Harasimowicz S., Kluth M., Fuchs S., Burkhardt L., Stahl P., C Tsourlakis M., Bauer M., Tennstedt P., Graefen M., Steurer S., Sirma H., Sauter G., Schlomm T., Simon R., Minner S. Heterogeneity and chronology of PTEN deletion and ERG fusion in prostate cancer. Mod Pathol. 2014;27:1612–1620. doi: 10.1038/modpathol.2014.70. [DOI] [PubMed] [Google Scholar]
- 34.Weier C., Haffner M.C., Mosbruger T., Esopi D.M., Hicks J., Zheng Q., Fedor H., Isaacs W.B., De Marzo A.M., Nelson W.G., Yegnasubramanian S. Nucleotide resolution analysis of TMPRSS2 and ERG rearrangements in prostate cancer. J Pathol. 2013;230:174–183. doi: 10.1002/path.4186. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Morais C.L., Guedes L.B., Hicks J., Baras A.S., De Marzo A.M., Lotan T.L. ERG and PTEN status of isolated high-grade PIN occurring in cystoprostatectomy specimens without invasive prostatic adenocarcinoma. Hum Pathol. 2016;55:117–125. doi: 10.1016/j.humpath.2016.04.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Karnes R.J., Bergstralh E.J., Davicioni E., Ghadessi M., Buerki C., Mitra A.P., Crisan A., Erho N., Vergara I.A., Lam L.L., Carlson R., Thompson D.J., Haddad Z., Zimmermann B., Sierocinski T., Triche T.J., Kollmeyer T., Ballman K.V., Black P.C., Klee G.G., Jenkins R.B. Validation of a genomic classifier that predicts metastasis following radical prostatectomy in an at risk patient population. J Urol. 2013;190:2047–2053. doi: 10.1016/j.juro.2013.06.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Demichelis F., Fall K., Perner S., Andren O., Schmidt F., Setlur S.R., Hoshida Y., Mosquera J.M., Pawitan Y., Lee C., Adami H.O., Mucci L.A., Kantoff P.W., Andersson S.O., Chinnaiyan A.M., Johansson J.E., Rubin M.A. TMPRSS2:ERG gene fusion associated with lethal prostate cancer in a watchful waiting cohort. Oncogene. 2007;26:4596–4599. doi: 10.1038/sj.onc.1210237. [DOI] [PubMed] [Google Scholar]
- 38.Berg K.D., Vainer B., Thomsen F.B., Roder M.A., Gerds T.A., Toft B.G., Brasso K., Iversen P. ERG protein expression in diagnostic specimens is associated with increased risk of progression during active surveillance for prostate cancer. Eur Urol. 2014;66:851–860. doi: 10.1016/j.eururo.2014.02.058. [DOI] [PubMed] [Google Scholar]
- 39.Galletti G., Matov A., Beltran H., Fontugne J., Miguel Mosquera J., Cheung C., MacDonald T.Y., Sung M., O'Toole S., Kench J.G., Suk Chae S., Kimovski D., Tagawa S.T., Nanus D.M., Rubin M.A., Horvath L.G., Giannakakou P., Rickman D.S. ERG induces taxane resistance in castration-resistant prostate cancer. Nat Commun. 2014;5:5548. doi: 10.1038/ncomms6548. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Tomlins S.A., Palanisamy N., Siddiqui J., Chinnaiyan A.M., Kunju L.P. Antibody-based detection of ERG rearrangements in prostate core biopsies, including diagnostically challenging cases: ERG staining in prostate core biopsies. Arch Pathol Lab Med. 2012;136:935–946. doi: 10.5858/arpa.2011-0424-OA. [DOI] [PMC free article] [PubMed] [Google Scholar]