Abstract
A national workgroup convened by the Centers for Disease Control and Prevention identified principles and made recommendations for standardizing the description of sequence data contained within the variant file generated during the course of clinical next-generation sequence analysis for diagnosing human heritable conditions. The specifications for variant files were initially developed to be flexible with regard to content representation to support a variety of research applications. This flexibility permits variation with regard to how sequence findings are described and this depends, in part, on the conventions used. For clinical laboratory testing, this poses a problem because these differences can compromise the capability to compare sequence findings among laboratories to confirm results and to query databases to identify clinically relevant variants. To provide for a more consistent representation of sequence findings described within variant files, the workgroup made several recommendations that considered alignment to a common reference sequence, variant caller settings, use of genomic coordinates, and gene and variant naming conventions. These recommendations were considered with regard to the existing variant file specifications presently used in the clinical setting. Adoption of these recommendations is anticipated to reduce the potential for ambiguity in describing sequence findings and facilitate the sharing of genomic data among clinical laboratories and other entities.
Next-generation sequencing (NGS) has revolutionized the analysis of the human genome. NGS has been widely adopted in the clinical environment. Recent publications have documented the utility of NGS for the diagnosis of rare diseases and cancer and to inform decisions pertaining to drug selection and dosing.1, 2 Clinical laboratories are also using NGS for human leukocyte antigen typing, pharmacogenetics, and infectious and chronic disease testing.3, 4, 5, 6, 7, 8 NGS fundamentally differs from Sanger sequencing in both method and description of findings.9 Sanger sequencing is most effective for analysis of limited regions of the genome, often targeted to specific genes or transcripts. As a consequence, variant types and positions are reported within the context of the targeted genes or transcripts. The advent of NGS permits analysis at the genomic level and as a consequence fostered the need to represent sequence findings based on a genomic reference. The shift from a gene/transcript to a genomic reference required modification of existing methods for describing sequence findings.
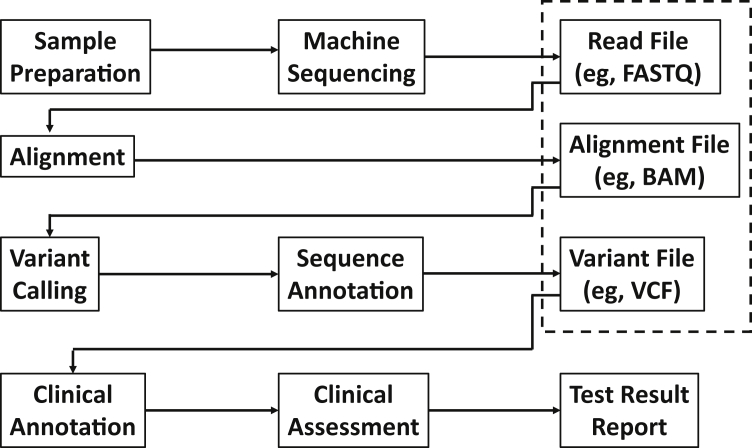
NGS depends on a number of file types to store data at various stages of the analysis (Figure 1). The variant file stores the calls made from the alignment of the patient's sequence to a reference. The identified variants are subsequently analyzed to determine which are clinically relevant to the patient. The research community has developed variant file specifications to support a broad range of research applications. These file specifications have been adopted by the clinical laboratory community, but their inherent flexibility has resulted in variation among laboratories with respect to how content is represented. Such flexibility is essential for research to accommodate different types of studies. On the other hand, such flexibility generates challenges for clinical laboratories, especially when standardized conventions for data descriptions have not been uniformly adopted. For example, unless the reference sequence used in assigning base positions is explicitly described, the usefulness of the data returned from a database query made to identify clinically relevant variants may be problematic because the base coordinate systems may be different.
Figure 1.
Next-generation sequencing workflow and associated data files (designated with a dashed-line box). Machine sequencing of the patient sample produces a large number of short reads deposited in a file with associated quality scores (eg, FASTQ). These reads are aligned to a reference assembly or sequence and the results are deposited in an alignment file (eg, BAM). Variants are called and their properties relevant to the sequence (eg, type of variant) are annotated and deposited in the variant file [eg, variant call format (VCF)]. The data in the variant file are further analyzed to determine what findings are clinically relevant and reportable to the physician to inform medical decision making.
Principles and recommendations are presented, based on the deliberations of a nonfederal, independent workgroup, convened by the Centers for Disease Control and Prevention, to promote standardization for the data content of variant files that are initially generated after variant calling (that are quality checked to remove entries deemed to be artifacts and make other corrections). The intent is to promote consistency in the representation of sequence findings to facilitate meaningful interlaboratory comparisons and to provide a common format for data contained within the variant file to facilitate downstream processing to ultimately identify disease-associated variants, when present.
Materials and Methods
A 2012 national workgroup that developed guidance for the design and optimization of a clinical NGS informatics pipeline articulated the need for standardizing the content of NGS variant files.10 As a consequence, a new workgroup was formed and tasked to identify principles and make recommendations directed to improving the uniformity of content contained within the variant call format (VCF) and similar file formats. This new workgroup, the Clinical Grade Variant-File Workgroup, was convened and facilitated by the Centers for Disease Control and Prevention and other federal partners (the National Center for Biotechnology Information, The National Institute of Standards and Technology, and the Food and Drug Administration). Members of this workgroup included informaticians, research and clinical laboratory directors, and representatives from industry, accrediting bodies, the HL7 Clinical Genomics workgroup, and federal agencies noted above and the National Human Genome Research Institute. Participants were chosen for their roles as leaders and contributors to the advancement of the integration of NGS into clinical applications. The workgroup was formed in 2014 and completed work in 2016. Discussions were had once or twice a month by telephone and web conference. The workgroup initially reviewed existing variant files, their uses and limitations, before more focused discussions that led to the principles and recommendations presented herein.
The workgroup considered both laboratory processes (eg, selection and alignment to a reference sequence) and the data presentation within variant files, both considered important to arriving at recommendations targeted to standardizing content. This article provides the outcomes of the workgroup's discussions and recommendations. In some instances, suggestions are given for consideration in lieu of recommendations because workgroup members could not arrive at consensus that certain practices were sufficiently mature to warrant a formal recommendation. Recommendations were based on workgroup member agreement without dissension.
Results
The workgroup derived the following recommendations from their discussions. The rationale for deriving these is described after this listing.
Recommendations for Laboratory Processes before Generation of the Variant File
Laboratory-selected reference sequences not available from publically accessible databases (eg, RefSeq, LRG) should be submitted to one or more of these databases for publication to allow for comparable cross mapping against the human genome reference assembly.
Variant callers should be configured to output reference, variant, and no-calls, together with local phasing information at least for those regions likely to harbor clinically important variants. As a caveat, no-calls associated with low-confidence sequence findings should be output for all sequences targeted for analysis.
Recommendations for Standardizing the Content within the Variant File
The variant file should include a description of both the specification and the version used. The human genome reference assembly should be used as the standard for assignment of genomic coordinates derived from NGS testing. The accession.version numbers of the sequences and assembly used for alignment and position assignment should be specified within the variant file to describe an unambiguous reference from which the genomic coordinates are derived. Variants should be described using Human Genome Variation Society (HGVS) descriptions that follow the published rules and that the use of abbreviated HGVS descriptions be linked back to the full HGVS description. When data sources are specified, their origin, build, version number, or other relevant parameters should be included to uniquely identify the source of the data elements. The Human Genome Nomenclature Committee (HGNC) descriptions should be used for specifying the targeted genes.
Variant File Formats
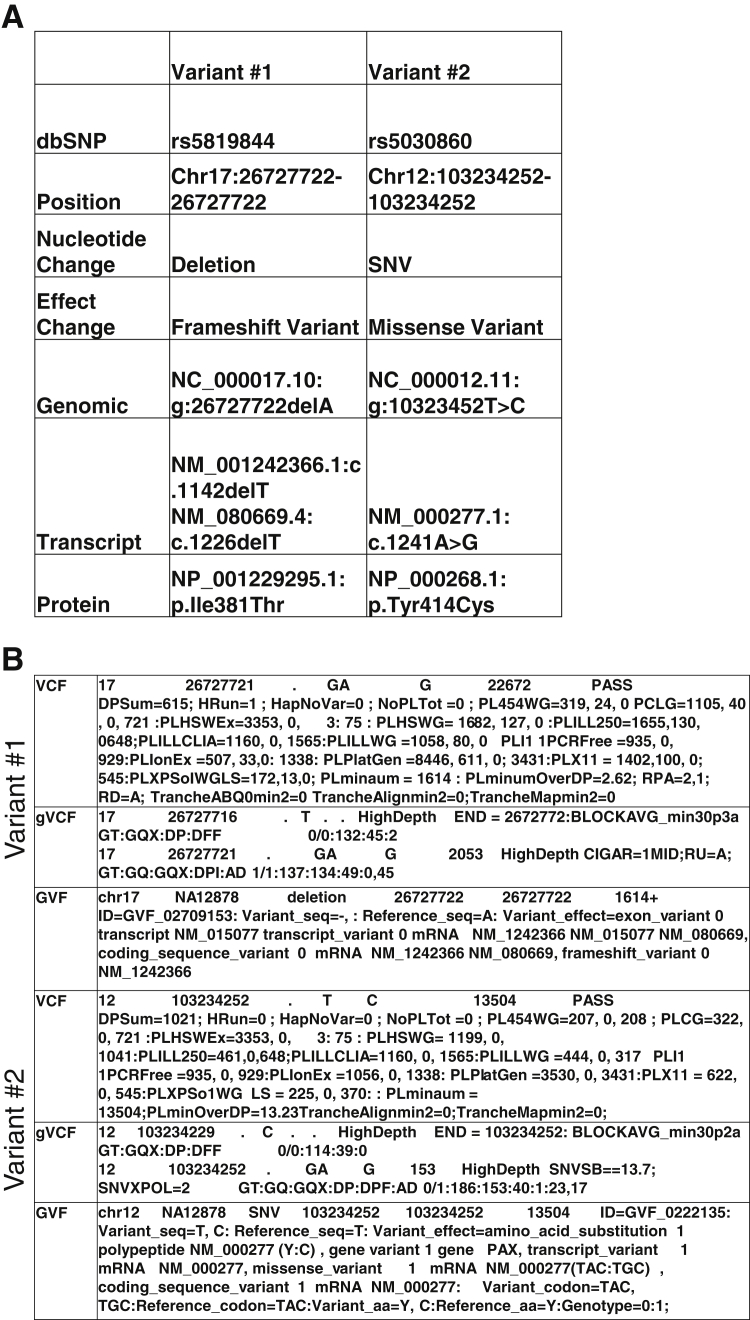
The workgroup discussions considered what could be learned from existing variant files in use. Several variant file formats have been established. These include the VCF (https://github.com/samtools/hts-specs, last accessed December 5, 2016), genomeVCF (https://sites.google.com/site/gvcftools/home/about-gvcf, last accessed December 5, 2016), and the genome variation format (https://github.com/The-Sequence-Ontology/Specifications/blob/master/gvf.md, last accessed December 5, 2016) (Figure 2).11, 12 Since the completion of the 1000 Genomes Project, stewardship of the VCF specification has been transferred to the Global Alliance for Genomics and Health under the file formats task team of the data working group. The genomeVCF specification builds on the VCF with additional features. The genomeVCF allows for the representation of both variant and nonvariant positions by default. The genome variation format is another specification and is based on the GFF3 format, a tool initially developed to permit comparison of gene annotations among different organisms.12 The evolution of the genome variation format provides for a detailed annotation of a genome, and uses terms from ontologies to describe the sequence alterations and the expected effect on the gene product.12
Figure 2.
Variant representation in three common variant file specifications. A: The two variants are listed from sample NA12878 from the 1000Genomes database. Variant 1 is a deletion of A, and variant 2 is a substitution of A to G. The dbSNP identifiers, chromosome number, nucleotide change, and the predicted effect are shown. The Human Genome Variation Society nomenclature for the change is shown relative to the genomic DNA, the mRNA, and protein RefSeq sequences. B: Contrast the differences among the variant file specifications for each of the two variants. The genome variant call format (gVCF) includes the invariant regions, not typically reported by the VCF. The genome variation format (GVF) includes additional annotation of the effect of the variant on the reference annotated features. SNV, single-nucleotide variants.
Specifications for these file formats were primarily generated to support research rather than clinical applications, and were designed to either catalog population variations (eg, VCF) or capture deep annotation of a single personal genome (eg, genome variation format). These formats were designed to provide a level of consistency with respect to the data elements while allowing for flexibility in content presentation to accommodate a broad range of research applications. Variant files typically include annotated sequence information and metadata. The annotated sequence information describes the sequence and related data important for understanding specific context such as the position and type of variant. Metadata provides information that typically refers to the complete data set.
The workgroup did not endorse a single file format recognizing that specifications continue to evolve and existing software enables translation among file specifications. However, the workgroup recognized that as of 2016 the VCF specification has the greatest adoption into clinical practice as a consequence of its long history, evolving specification, and the availability of a rich set of tools for manipulating these types of files. Irrespective of the variant file specification used in practice, changes are made as the specification advances. As such, the workgroup recommends that the variant file includes a description of both the specification and the version used. Ideally, the most current version will be adopted, but variation of versions used in practice will likely continue to exist into the foreseeable future.
Using the Human Genome Reference Assembly
The workgroup endorses the recommendations made by other standard/guideline setting bodies for use of the human genome reference assembly as the standard for assignment of genomic coordinates derived from NGS testing (http://www.ncbi.nlm.nih.gov/projects/genome/assembly/grc/human, last accessed December 5, 2016).10, 13, 14
The reference assembly is meant to model genome sequences across the global population. Initial models developed by the Human Genome Project used a single haploid assembly model.15 Subsequent analysis uncovered highly polymorphic regions of the human genome that made it impossible to describe a single sequence for that region.16, 17 The Genome Reference Consortium developed an improved assembly model that allows for representation of various sequences (termed alternate loci) at regions with high diversity.18, 19 Many of these regions contain medically relevant genes, such as the human leukocyte antigen, KIR, and CYP gene families. A genome build refers to a specific version of the reference assembly. The most recent human genome build GRCh38p9 was released in September 2016 by the Genome Reference Consortium. This assembly represents a significant advance, with one of the most important additions relevant for clinical testing being the inclusion of 261 alternate loci across 178 regions. Only 72 alternate loci were available in the previous assembly. Although this represents a significant advance in describing the human genome, it has generated a challenge for clinical laboratories because of the absence of validated tools that are able to incorporate alternate assemblies into their analytic algorithm. The development of tools that can take advantage of these new sequences will be critical for improved genome analysis.19
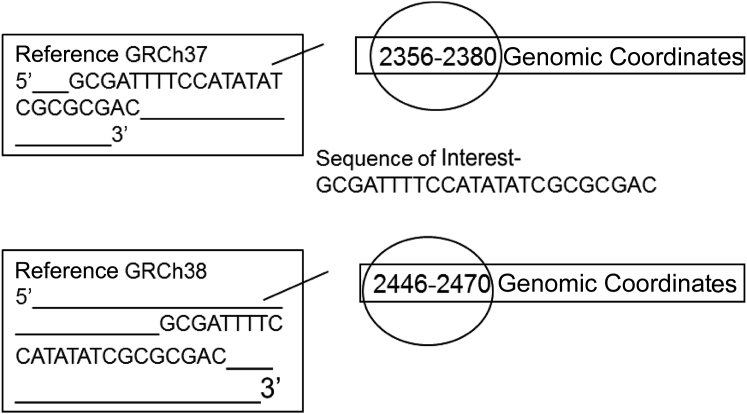
The human genome reference assembly is regularly updated by the Genome Reference Consortium and submitted to GenBank to obtain stable, traceable identifiers, accession, and versions, for both the sequences contained in the assembly as well as the assembly itself. Major updates are released infrequently and include corrections and additions to fill in existing gaps. Genomic coordinate assignments are extensively revised in major updates and are designated by different accession numbers (Figure 3). Minor updates, called patch releases, occur quarterly and do not affect the sequences released as part of the major release; as a consequence, the genomic coordinate assignments do not change. The workgroup recommends that the accession.version numbers of the sequences and assembly used for alignment and position assignment should be specified within the variant file to describe an unambiguous reference from which the genomic coordinates are derived.
Figure 3.
Origin of genomic coordinates. Genomic coordinates of sequence contained within the variant file are made in reference to a genomic build/reference and assigned based on a 5′ to 3′ numbering of the positive strand. Genomic coordinates can change during major updates to the reference assembly, as illustrated in comparing GRCh37 to GRCh38.
Clinical laboratories access databases that use one or a combination of historical data (before the use of the genomic reference assembly) and data based on the reference assembly in designating base coordinates. Attention to the origin of the coordinate system used and mapping to the appropriate reference assembly is required when querying databases. The National Center for Biotechnology Information and others have generated tools for remapping different versions of the reference assembly to permit comparisons of findings derived using a different accession.version of the reference assembly (http://www.ncbi.nlm.nih.gov/genome/tools/remap, last accessed December 5, 2016).
Alignment against the full genome during exome and genome sequencing has been recommended as a means to reduce forced alignments that can result in miscalls because of the presence of homologous sequences.10 Other practices, such as the use of decoy sequences, can minimize misalignment of reads to homologous off-target sequences.
Many laboratories align reads from analysis of multigene panels against laboratory-selected sequences that correspond to genes included in the panel. This practice requires mapping of sequence findings back to the human genome reference sequence to establish the corresponding genomic coordinates. Laboratory-selected sequences may be taken from databases available through the National Center for Biotechnology Information or European Informatics Institute [eg, RefSeq (http://www.ncbi.nlm.nih.gov/refseq, last accessed December 5, 2016) and LRG (http://www.lrg-sequence.org, last accessed December 5, 2016)] in which cross mapping against the human reference assembly using standardized methods is performed and results posted with the database entry. This provides an unambiguous description and assignment of genomic coordinates for all entries. The workgroup recommends that laboratory-selected reference sequences not available from publically accessible databases (eg, RefSeq, LRG) should be submitted to one or more of these databases for publication to allow for comparable cross mapping against the human genome reference assembly. The workgroup discourages cross mapping outside these methods because differences in software and settings among laboratories raise the risk for incorrect assignment of genomic coordinates.
The description and assignment of position for insertions and deletions within a repeated sequence requires consideration of the file specification and this is influenced by whether genomic DNA or a transcript is being described. The VCF and other variant file specifications designate the starting position of an insertion or deletion variant using the genomic coordinate associated with the left most (5′) base associated with the insertion or deletion (left justification). Genomic coordinates are always assigned relative to the + strand of the genome irrespective of transcript direction. Position assignment within transcripts typically use the HGVS conventions. HGVS assigns coordinates with respect to the 5′ to 3′ direction of the transcript. This is irrespective of its genomic directionality (the + or − strand of the genome). HGVS specifies the transcript position assignment for an insertion or duplication be the most 3′ (right) base possible.20, 21, 22 The differences associated with position assignments between genomic DNA and transcripts using the conventions described require an understanding of how variant file formats and HGVS assignments are made. This requires laboratory professionals to be aware of when and how these conventions are applied and take these issues into account when making sequence comparisons and mapping between transcript and the genomic sequences.
HGVS remains an evolving standard, and ambiguous descriptions can result because of changing conventions. There are also reports of incorrect application of the nomenclature that has resulted in additional ambiguity.20, 23 As such, the workgroup recommended that variants should be described using HGVS descriptions that follow the published rules and that the use of abbreviated HGVS descriptions be linked back to the full HGVS description.10, 13, 14, 20, 24 To minimize the possibility for position ambiguity, sequences should be reported in conjunction with unambiguous genomic coordinates. In 2013, HGVS adopted versioning that is helpful for resolving differences that can result from modification of the rules used to describe sequence findings.
Some current software programs are designed to promote consistent HGVS assignment (eg, https://mutalyzer.nl, last accessed December 5, 2016).25, 26 The workgroup emphasized that there remains variability in how these programs develop HGVS descriptions and encouraged clinical laboratories to be cautious in their use and to perform a comprehensive validation targeted for the intended clinical applications. Nonetheless, adoption of such tools may be helpful to ensure consistent HGVS descriptions, including position assignments.
Variant files often contain a data field for each sequence entry that permits reference to a data source (eg, the ID field corresponding to a sequence entry within the VCF data section). For variant detection, corresponding dbSNP (http://www.ncbi.nlm.nih.gov/SNP, last accessed September 17, 2016) entries are commonly cited and more recently the ClinVar Variation ID (http://www.ncbi.nlm.nih.gov/clinvar/docs/variation_report, last accessed December 5, 2016). The ClinVar database continues to develop and is designed to report the relationships between variants and their clinical relevance. For NGS somatic analysis, the COSMIC database (http://cancer.sanger.ac.uk/cosmic, last accessed December 5, 2016) is an important resource developed to contain a listing of somatic variants found in human cancer. For pharmacogenomic haplotypes, the Pharmacogenomics knowledgebase can be referenced (https://www.pharmgkb.org, last accessed December 5, 2016). For human leukocyte antigen, the International ImMunoGeneTics Project/human leukocyte antigen database can be used.27 The workgroup recommends that when data sources are specified, their origin, build, version number, or other relevant parameters should be included to uniquely identify the source of the data elements.
Considerations for Variant Calling and Sequence Representation within the VCF and Similar Variant Files
We use the term local phasing to refer to the short-range phasing information that is output by some variant callers using information from the reads to determine whether multiple (two or greater) variants are located on the same chromosome (cis) or on different homologous chromosomes (trans). Local phasing data allow for assembly of individual and short sequences typically reported from variant callers to fully define a haplotype. Early versions of the VCF specification did not permit inclusion of reference calls, but recent versions permit this, an adaption from the genomeVCF specification (http://www.1000genomes.org/wiki/Analysis/vcf4.0, last accessed December 5, 2016; the most recent update can be found at https://samtools.github.io/hts-specs, last accessed December 5, 2016). The challenge with inclusion of reference calls is the generation of large files that many of the existing software tools are not suited to manage. This limitation has been handled by targeting hot spots, areas of the targeted regions likely to contain clinically relevant variants, for the collection of the full data set that includes reference calls.10, 28 This greatly decreases the file size, especially when exome or genome analysis is performed. The workgroup recommends that variant callers should be configured to output reference, variant, and no-calls, together with local phasing information at least for those regions likely to harbor clinically important variants. As a caveat, no-calls associated with low-confidence sequence findings should be output for all sequences targeted for analysis.
No-calls refers to one or more base positions that cannot be precisely determined during the course of sequence analysis.29 This can include one or more base positions in which the local sequence quality was not sufficient to permit a confident diploid call, or in which there was a complex variant with one or more unresolved bases. For the latter example, in not identifying a no-call, the complex variant may not be properly described. The VCF specification permits the description of these types of findings. Failure to identify no-calls can result in mistakes associated with position or zygosity assignment. A no-call can also occur as a consequence of an insertion of indeterminate length into a known location. This occurs when sequence reads and/or mate-paired reads are too short to span a repetitive region of the genome. Failure to identify these can also result in incorrect position assignment and misattribution of a repetitive element. Describing no-call insertions presents a challenge because it can be difficult to determine whether the number of copies of the repeated segment has changed because of an insertion or deletion. Another example of a no-call is a variant that occurs within a duplicated region but its precise position cannot be determined. Additional esoteric situations involving low complexity or repetitive regions, or other undetectable balanced structural variants, can also lead to no-calls. No-calls can influence the clinical interpretation of test results. For example, an insertion within a trinucleotide repeat region that is missed during sequence analysis can result in the reporting of fewer repeats than are actually present. Manual review of the data is typically necessary to identify no-calls. Eventually, these problems will be mitigated as read lengths increase and better library preparation and bioinformatics methods become available, but, as of 2016, they are inherent in NGS data sets and should be considered when interpreting sequence results.30, 31 The recommendation to include no-calls associated with low-confidence reads is useful for identifying regions that may require alternative methods for sequence determination and understanding the limitations of the analysis.
The VCF and other variant files are designed to capture one or a few variants within a minimal read length on each sequence data line. The designation of complex variants as a haplotype often requires the assembly of sequence from multiple short sequences using local phasing (Figure 4). This is important for knowing the phase (cis or trans) of two or greater closely spaced variants in deriving a haplotype.32 Correctly describing complex variants has implications for establishing or negating heterozygosity for two disease-associated variants or making correlations with disease severity.33, 34 Findings also have implications for other family members with respect to risk for or severity of disease.
Figure 4.
Phasing data are required to establish the cis or trans relationship of two nonadjacent variants (bold and underlined). A: The true diplotype describing an individual having one allele with two nonadjacent variants that are not found in the same allele of the homologous chromosome. B: Output from a variant caller. C: Interpretation of the output of the variant caller. In the presence of phasing data, the correct haplotypes are established. In the absence of phasing data, the cis or trans association of the two variants cannot be distinguished.
As of 2016, there is no consensus in the community as to whether variant callers should be set up to output longer sequences requiring minimal phasing information or shorter phased sequences (referred to as primitives). For the latter, one proposal is for variant calls to span a genomic window that is appropriate given the local distribution of variants and the sequencing read length, which would typically result in window sizes in the order of 50 bp.35 The workgroup recognized the validity of both approaches, suggesting that complex variants be output either as phased primitive single-nucleotide variants and insertions/deletions or as a single complex event. Nearby heterozygous variants should always be phased when possible in deriving an accurate haplotype description.
Describing Genes within the Variant File
The workgroup recommends that the HGNC descriptions should be used for specifying the targeted genes. This is consistent with recommendations from other professional groups.13, 14, 36 HGNC assigns an identification number, symbol, and name to each gene. For some genes, the symbol and/or name will change over time, but the identification number remains unique. HGNC tracks use of these identifiers over time. The workgroup decided not to advocate for any one HGNC gene representation given that old and new names and symbols are routinely used by many laboratories, with the identification number used the least. However, the benefit of using identification numbers was recognized and the workgroup suggested that this parameter be included to minimize the potential for ambiguity. Historically, human genes have acquired more than one name. Many oncologists, physicians, molecular biologists, and genetic counselors are most familiar with common or older gene names and, as such, laboratories often include these in addition to the HGNC descriptors, and they may be included as metadata within the variant file, but not to the exclusion of HGNC designations.
Assessing the Quality of the Data
The VCF primarily contains quality data applicable to the confidence of the sequence represented that often includes the variant quality score. These scores are often used as a metric to include or exclude variants in the final variant set that is reviewed for clinical interpretation. Generation of these measures is method specific, negating direct comparison among different platforms.13, 14 For methods developed within the laboratory, validation established the performance specifications of the method used.13 The absence of standardized metrics applicable across platforms can lead to variability of sequence calls among laboratories that may influence what is called and the false-positive and false-negative rates for the assay. As an area still in development, the workgroup opted not to offer specific recommendations related to this topic.
Additional Considerations for Downstream Analysis and File Formats
As of 2016, both manual and automated processes are used to analyze data within the variant file to derive clinical assertions. Further automation of this process should greatly reduce the analysis time and provide a high level of consistency and quality among laboratories. The obstacles to achieving such automation are the variation in the representation of content within the variant files used and the absence of curated databases designed to serve medical applications. Efforts are underway to address these gaps that include the development of methods for querying the National Center for Biotechnology Information ClinVar database with a VCF file.37, 38 One approach may be the adoption of a standard to store variants within databases using genomic coordinates, canonicalized using a common scheme that recognizes when identical variants are defined differently. This is being addressed through software development, but standard methods have not yet been implemented.39 Linking to ClinVar in this way advances the prospects for automating the extraction of structured and standardized data useful to support downstream analyses used to make clinical assertions. These assertions can then be described and reported using accepted nomenclature, such as HGVS for variants, American College of Medical Genetics–approved terms for clinical significance,21 and VariO (http://variationontology.org, last accessed December 5, 2016) and Sequence Ontology terms for variation type, molecular consequence, and functional consequence.40
Discussion
Properties of a Prototypic Clinical-Grade Variant File
Standardizing variant files is proposed to expedite a number of functions important to advancing clinical laboratory practice. These include data sharing among laboratories, querying of databases, and streamlined processes for downstream data analysis to identify the clinically relevant variants. The workgroup recognized that certain steps, such as uniform assignment of genomic coordinates, need to be taken before generation of the variant file to promote content uniformity and enable effective data sharing and management.
Future Directions
Use of a uniform representation of sequence data within variant files derived from genome, exome, or panel analysis will be needed to advance the exchange of data among laboratories, databases, and, in time, patient records. This is envisioned as the primary incentive to begin the process of adopting standards for variant file content among clinical laboratories and software developers and implementers. Uniformity in data representation is important for several critical functions. These include the comparison of derived data among laboratories as a quality assurance process and for proficiency testing. Uniform representation of sequence data also provides for a common input to downstream algorithms that may minimize variation in downstream results that can occur as a consequence of how sequence data are presented within the variant file. This is particularly relevant for the detection of variants that were not targeted during the test validation.
The principles and recommendations presented are not comprehensive but provide a context for what the workgroup believed could be addressed from a clinical testing perspective as of 2016. For example, the workgroup discussed the potential need to include more robust identifiers within the variant file that links to a patient or sample. The workgroup was not able to agree to a specific recommendation regarding this topic. From a regulatory perspective, the Food and Drug Administration published a draft guidance document addressing the use of NGS in vitro diagnostics for germline analysis (http://www.fda.gov/downloads/MedicalDevices/DeviceRegulationandGuidance/GuidanceDocuments/UCM509838.pdf, last accessed December 5, 2016). Although the final guidance has yet to be published as of September 2016, the draft document does emphasize the need to describe and document data processes and analyses relevant to the validation of the bioinformatics pipeline.
Although specifications will continue to advance, variant files are expected to remain a standard tool in the coming years. Nonetheless, new technologies and techniques, such as work underway to advance the use of graph theory that provides a statistical and visual representation of sequence findings, may fundamentally displace use of variant files to provide a more useful means for describing genomic sequences.19
Ultimately, the expectation is that an electronic health record will contain structured sequence data. The focus today is to work toward inclusion of clinically relevant findings that would typically be reported in the patient's test result report. How these data are captured, displayed, and used is the subject of use cases and guidance that are being developed by several groups that include the HL7 Clinical Genomics Workgroup (http://www.hl7.org/special/committees/clingenomics, last accessed December 5, 2016) and the Health and Medicine Division of the Academies (http://iom.nationalacademies.org/Activities/Research/GenomicBasedResearch/Innovation-Collaboratives/EHR.aspx?page=1#sthash.Bta487hi.dpuf, last accessed December 5, 2016). The capability to provide patient exome or genome sequence data in a structured format able to support clinical decision making is an exciting prospect for the future. This will support the capability to query larger data sets, such as ClinVar, using automated processes to expedite the identification of clinically important variants.
Footnotes
Supported by an appointment of E.R.L. to the Research Participation Program at the Centers for Disease Control and Prevention (CDC) administered by the Oak Ridge Institute for Science and Education through an interagency agreement between the US Department of Energy and CDC; National Human Genome Research Institute grants R01HG008628 (K.E.) and U41HG006834 (H.L.R.); and the National Institute of General Medical Sciences grant GM109737 (A.W.Z.).
Disclosures: L.J.B. is an employee at Sunquest, Inc.; D.B. is an employee of Complete Genomics; D.M.C. is an employee and shareholder at 10× Genomics and Personalis, Inc.; S.C. is an employee at Fluidigm and was previously employed at Complete Genomics; K.E. is a consultant to Omicia, Inc.; F.H. is an employee at Life Technologies; D.M. holds shares in Pfizer, Inc. and Merck, Inc.; J.D.P. is the cofounder of and reports employment at PierianDx and P&V Licensing LLC; H.L.R. is an employee at Partners Laboratory for Molecular Medicine and BWH and Harvard Medical School; R.T. is an employee and shareholder at Invitae, and was previously used at Complete Genomics; E.W. is an employee at the HudsonAlpha Institute for Biotechnology, is the Founder and Chief Product Development Officer and shareholder at Envision Genomics, holds a patent that describes methods and apparatus for identification of disease-associated mutations, and has a patent pending that addresses genomic data conversion; A.W.Z. is an employee, shareholder, and board member of Curoverse, Inc.
The findings and conclusions in this report are those of the authors and do not necessarily represent the views of the Centers for Disease Control and Prevention or the US Agency for Toxic Substances and Disease Registry, the Food and Drug Administration, the National Institute of Standards and Technology, or the NIH.
Use of trade names and commercial sources is for identification only and does not imply endorsement by the Centers for Disease Control and Prevention, the Public Health Service, the US Department of Health and Human Services, or the US National Institute of Standards and Technology, nor does it imply that the products identified are necessarily the best available for the purpose.
All authors are members of the Clinical-Grade Variant File Workgroup, established and facilitated by the Centers for Disease Control and Prevention, tasked to consider principles and make laboratory practice recommendations that are presented within this article.
Current address of S.C., Fluidigm, South San Francisco, CA; of E.W., The HudsonAlpha Institute, Huntsville, AL. D.B. is currently unaffiliated.
A guest editor acted as the Editor-in-Chief for this manuscript. No person at the Centers for Disease Control and Prevention was involved in the peer review process or final disposition of this article.
References
- 1.Boycott K.M., Vanstone M.R., Bulman D.E., MacKenzie A.E. Rare-disease genetics in the era of next-generation sequencing: discovery to translation. Nat Rev Genet. 2013;14:681–691. doi: 10.1038/nrg3555. [DOI] [PubMed] [Google Scholar]
- 2.Chang F., Li M.M. Clinical application of amplicon-based next-generation sequencing in cancer. Cancer Genet. 2013;206:413–419. doi: 10.1016/j.cancergen.2013.10.003. [DOI] [PubMed] [Google Scholar]
- 3.Liu C., Yang X., Duffy B., Mohanakumar T., Mitra R.D., Zody M.C., Pfeifer J.D. ATHLATES: accurate typing of human leukocyte antigen through exome sequencing. Nucleic Acids Res. 2013;41:e142. doi: 10.1093/nar/gkt481. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Mori A., Deola S., Xumerle L., Mijatovic V., Malerba G., Monsurro V. Next generation sequencing: new tools in immunology and hematology. Blood Res. 2013;48:242–249. doi: 10.5045/br.2013.48.4.242. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Gillis N.K., Patel J.N., Innocenti F. Clinical implementation of germ line cancer pharmacogenetic variants during the next-generation sequencing era. Clin Pharmacol Ther. 2014;95:269–280. doi: 10.1038/clpt.2013.214. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Bertelli C., Greub G. Rapid bacterial genome sequencing: methods and applications in clinical microbiology. Clin Microbiol Infect. 2013;19:803–813. doi: 10.1111/1469-0691.12217. [DOI] [PubMed] [Google Scholar]
- 7.Barzon L., Lavezzo E., Costanzi G., Franchin E., Toppo S., Palu G. Next-generation sequencing technologies in diagnostic virology. J Clin Virol. 2013;58:346–350. doi: 10.1016/j.jcv.2013.03.003. [DOI] [PubMed] [Google Scholar]
- 8.Churko J.M., Mantalas G.L., Snyder M.P., Wu J.C. Overview of high throughput sequencing technologies to elucidate molecular pathways in cardiovascular diseases. Circ Res. 2013;112:1613–1623. doi: 10.1161/CIRCRESAHA.113.300939. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Beck T.F., Mullikin J.C., Program N.C.S., Biesecker L.G. Systematic evaluation of Sanger validation of next-generation sequencing variants. Clin Chem. 2016;62:647–654. doi: 10.1373/clinchem.2015.249623. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Gargis A.S., Kalman L., Bick D.P., da Silva C., Dimmock D.P., Funke B.H. Good laboratory practice for clinical next-generation sequencing informatics pipelines. Nat Biotechnol. 2015;33:689–693. doi: 10.1038/nbt.3237. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Danecek P., Auton A., Abecasis G., Albers C.A., Banks E., DePristo M.A., Handsaker R.E., Lunter G., Marth G.T., Sherry S.T., McVean G., Durbin R., 1000 Genomes Project Analysis Group The variant call format and VCFtools. Bioinformatics. 2011;27:2156–2158. doi: 10.1093/bioinformatics/btr330. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Reese M.G., Moore B., Batchelor C., Salas F., Cunningham F., Marth G.T., Stein L., Flicek P., Yandell M., Eilbeck K. A standard variation file format for human genome sequences. Genome Biol. 2010;11:R88. doi: 10.1186/gb-2010-11-8-r88. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Rehm H.L., Bale S.J., Bayrak-Toydemir P., Berg J.S., Brown K.K., Deignan J.L., Friez M.J., Funke B.H., Hegde M.R., Lyon E., Working Group of the American College of Medical Genetics and Genomics Laboratory Quality Assurance Committee ACMG clinical laboratory standards for next-generation sequencing. Genet Med. 2013;15:733–747. doi: 10.1038/gim.2013.92. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.CLSI . Clinical and Laboratory Standards Institute; Wayne, PA: 2014. Nucleic Acid Sequencing Methods in Diagnostic Laboratory Medicine. CLSI document MM09-A2 February 2014 ed. [Google Scholar]
- 15.Green E.D., Watson J.D., Collins F.S. Human Genome Project: twenty-five years of big biology. Nature. 2015;526:29–31. doi: 10.1038/526029a. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Zody M.C., Jiang Z., Fung H.C., Antonacci F., Hillier L.W., Cardone M.F., Graves T.A., Kidd J.M., Cheng Z., Abouelleil A., Chen L., Wallis J., Glasscock J., Wilson R.K., Reily A.D., Duckworth J., Ventura M., Hardy J., Warren W.C., Eichler E.E. Evolutionary toggling of the MAPT 17q21.31 inversion region. Nat Genet. 2008;40:1076–1083. doi: 10.1038/ng.193. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Sudmant P.H., Kitzman J.O., Antonacci F., Alkan C., Malig M., Tsalenko A., Sampas N., Bruhn L., Shendure J., Genomes P., Eichler E.E. Diversity of human copy number variation and multicopy genes. Science. 2010;330:641–646. doi: 10.1126/science.1197005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Church D.M., Schneider V.A., Graves T., Auger K., Cunningham F., Bouk N. Modernizing reference genome assemblies. PLoS Biol. 2011;9:e1001091. doi: 10.1371/journal.pbio.1001091. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Church D.M., Schneider V.A., Steinberg K.M., Schatz M.C., Quinlan A.R., Chin C.S., Kitts P.A., Aken B., Marth G.T., Hoffman M.M., Herrero J., Mendoza M.L., Durbin R., Flicek P. Extending reference assembly models. Genome Biol. 2015;16:13. doi: 10.1186/s13059-015-0587-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Berwouts S., Morris M.A., Girodon E., Schwarz M., Stuhrmann M., Dequeker E. Mutation nomenclature in practice: findings and recommendations from the cystic fibrosis external quality assessment scheme. Hum Mutat. 2011;32:1197–1203. doi: 10.1002/humu.21569. [DOI] [PubMed] [Google Scholar]
- 21.Richards S., Aziz N., Bale S., Bick D., Das S., Gastier-Foster J., Grody W.W., Hegde M., Lyon E., Spector E., Voelkerding K., Rehm H.L., ACMG Laboratory Quality Assurance Committee Standards and guidelines for the interpretation of sequence variants: a joint consensus recommendation of the American College of Medical Genetics and Genomics and the Association for Molecular Pathology. Genet Med. 2015;17:405–424. doi: 10.1038/gim.2015.30. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.den Dunnen J.T., Dalgleish R., Maglott D.R., Hart R.K., Greenblatt M.S., McGowan-Jordan J., Roux A.F., Smith T., Antonarakis S.E., Taschner P.E. HGVS recommendations for the description of sequence variants: 2016 update. Hum Mutat. 2016;37:564–569. doi: 10.1002/humu.22981. [DOI] [PubMed] [Google Scholar]
- 23.Tack V., Deans Z.C., Wolstenholme N., Patton S., Dequeker E.M. What's in a name? a coordinated approach toward the correct use of a uniform nomenclature to improve patient reports and databases. Hum Mutat. 2016;37:570–575. doi: 10.1002/humu.22975. [DOI] [PubMed] [Google Scholar]
- 24.Kalman L.V., Agundez J., Appell M.L., Black J.L., Bell G.C., Boukouvala S. Pharmacogenetic allele nomenclature: international workgroup recommendations for test result reporting. Clin Pharmacol Ther. 2016;99:172–185. doi: 10.1002/cpt.280. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Wildeman M., van Ophuizen E., den Dunnen J.T., Taschner P.E. Improving sequence variant descriptions in mutation databases and literature using the Mutalyzer sequence variation nomenclature checker. Hum Mutat. 2008;29:6–13. doi: 10.1002/humu.20654. [DOI] [PubMed] [Google Scholar]
- 26.Hart R.K., Rico R., Hare E., Garcia J., Westbrook J., Fusaro V.A. A Python package for parsing, validating, mapping and formatting sequence variants using HGVS nomenclature. Bioinformatics. 2015;31:268–270. doi: 10.1093/bioinformatics/btu630. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Marsh S.G., Albert E.D., Bodmer W.F., Bontrop R.E., Dupont B., Erlich H.A., Fernandez-Vina M., Geraghty D.E., Holdsworth R., Hurley C.K., Lau M., Lee K.W., Mach B., Maiers M., Mayr W.R., Muller C.R., Parham P., Petersdorf E.W., Sasazuki T., Strominger J.L., Svejgaard A., Terasaki P.I., Tiercy J.M., Trowsdale J. Nomenclature for factors of the HLA system, 2010. Tissue Antigens. 2010;75:291–455. doi: 10.1111/j.1399-0039.2010.01466.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Singh R.R., Patel K.P., Routbort M.J., Reddy N.G., Barkoh B.A., Handal B., Kanagal-Shamanna R., Greaves W.O., Medeiros L.J., Aldape K.D., Luthra R. Clinical validation of a next-generation sequencing screen for mutational hotspots in 46 cancer-related genes. J Mol Diagn. 2013;15:607–622. doi: 10.1016/j.jmoldx.2013.05.003. [DOI] [PubMed] [Google Scholar]
- 29.Carnevali P., Baccash J., Halpern A.L., Nazarenko I., Nilsen G.B., Pant K.P., Ebert J.C., Brownley A., Morenzoni M., Karpinchyk V., Martin B., Ballinger D.G., Drmanac R. Computational techniques for human genome resequencing using mated gapped reads. J Comput Biol. 2012;19:279–292. doi: 10.1089/cmb.2011.0201. [DOI] [PubMed] [Google Scholar]
- 30.Loomis E.W., Eid J.S., Peluso P., Yin J., Hickey L., Rank D., McCalmon S., Hagerman R.J., Tassone F., Hagerman P.J. Sequencing the unsequenceable: expanded CGG-repeat alleles of the fragile X gene. Genome Res. 2013;23:121–128. doi: 10.1101/gr.141705.112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Peters B.A., Kermani B.G., Sparks A.B., Alferov O., Hong P., Alexeev A., Jiang Y., Dahl F., Tang Y.T., Haas J., Robasky K., Zaranek A.W., Lee J.H., Ball M.P., Peterson J.E., Perazich H., Yeung G., Liu J., Chen L., Kennemer M.I., Pothuraju K., Konvicka K., Tsoupko-Sitnikov M., Pant K.P., Ebert J.C., Nilsen G.B., Baccash J., Halpern A.L., Church G.M., Drmanac R. Accurate whole-genome sequencing and haplotyping from 10 to 20 human cells. Nature. 2012;487:190–195. doi: 10.1038/nature11236. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Tewhey R., Bansal V., Torkamani A., Topol E.J., Schork N.J. The importance of phase information for human genomics. Nat Rev Genet. 2011;12:215–223. doi: 10.1038/nrg2950. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Chen N., Schrijver I. Allelic discrimination of cis-trans relationships by digital polymerase chain reaction: GJB2 (p.V27I/p.E114G) and CFTR (p.R117H/5T) Genet Med. 2011;13:1025–1031. doi: 10.1097/GIM.0b013e3182272e0b. [DOI] [PubMed] [Google Scholar]
- 34.Lucarelli M., Narzi L., Pierandrei S., Bruno S.M., Stamato A., d'Avanzo M., Strom R., Quattrucci S. A new complex allele of the CFTR gene partially explains the variable phenotype of the L997F mutation. Genet Med. 2010;12:548–555. doi: 10.1097/GIM.0b013e3181ead634. [DOI] [PubMed] [Google Scholar]
- 35.Zook J.M., Chapman B., Wang J., Mittelman D., Hofmann O., Hide W., Salit M. Integrating human sequence data sets provides a resource of benchmark SNP and indel genotype calls. Nat Biotechnol. 2014;32:246–251. doi: 10.1038/nbt.2835. [DOI] [PubMed] [Google Scholar]
- 36.Gargis A.S., Kalman L., Berry M.W., Bick D.P., Dimmock D.P., Hambuch T. Assuring the quality of next-generation sequencing in clinical laboratory practice. Nat Biotechnol. 2012;30:1033–1036. doi: 10.1038/nbt.2403. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.NCBI Resource Coordinators Database resources of the National Center for Biotechnology Information. Nucleic Acids Res. 2016;44:D7–D19. doi: 10.1093/nar/gkv1290. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Lappalainen I., Lopez J., Skipper L., Hefferon T., Spalding J.D., Garner J., Chen C., Maguire M., Corbett M., Zhou G., Paschall J., Ananiev V., Flicek P., Church D.M. DbVar and DGVa: public archives for genomic structural variation. Nucleic Acids Res. 2013;41:D936–D941. doi: 10.1093/nar/gks1213. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Sun C., Medvedev P. VarMatch: robust matching of small variant datasets using flexible scoring schemes. Bioinformatics. 2016 doi: 10.1093/bioinformatics/btw797. [Epub ahead of print] doi: 10.1093/bioinformatics/btw797. [DOI] [PubMed] [Google Scholar]
- 40.Mungall C.J., Batchelor C., Eilbeck K. Evolution of the Sequence Ontology terms and relationships. J Biomed Inform. 2011;44:87–93. doi: 10.1016/j.jbi.2010.03.002. [DOI] [PMC free article] [PubMed] [Google Scholar]