Abstract
Recent studies indicate that neural EGFL-like 1 (Nell-1), a secretive extracellular matrix molecule, is involved in chondrogenic differentiation. Herein, we demonstrated that Nell-1 serves as a key downstream target of runt-related transcription factor 2 (Runx2), a central regulator of chondrogenesis. Unlike in osteoblast lineage cells where Nell-1 and Runx2 demonstrate mutual regulation, further studies in chondrocytes revealed that Runx2 tightly regulates the expression of Nell-1; however, Nell-1 does not alter the expression of Runx2. More important, Nell-1 administration partially restored Runx2 deficiency–induced impairment of chondrocyte differentiation and maturation in vitro, ex vivo, and in vivo. Mechanistically, although the expression of Nell-1 is highly reliant on Runx2, the prochondrogenic function of Nell-1 persisted in Runx2−/− scenarios. The biopotency of Nell-1 is independent of the nuclear import and DNA binding functions of Runx2 during chondrogenesis. Nell-1 is a key functional mediator of chondrogenesis, thus opening up new possibilities for the application of Nell-1 in cartilage regeneration.
Chondrogenesis is an obligatory step for skeletogenesis, which results in the construction of the primary skeleton of the vertebrate embryo.1 Chondrogenesis begins as the mesenchymal cells migrate and tightly pack at the presumptive skeletogenic sites to form cell mass condensations.1, 2, 3 Next, the condensed cells undergoing earlier chondrogenic differentiation proliferate and produce cartilage extracellular matrix, such as collagen II and aggrecan (alias cartilage-specific proteoglycan core protein or chondroitin sulfate proteoglycan 1).1, 2, 3, 4 For long bone development, the proliferating chondrocytes go through prehypertrophy, hypertrophy, and terminal maturation with matrix-mineralization that serves as a template for the subsequent deposition of bone matrix.1, 2, 3, 4 A great number of molecules regulate chondrogenic differentiation, an essential process for bone and cartilage formation.1, 2, 3, 4, 5, 6, 7, 8, 9, 10, 11 In particular, SRY-Box 9 and runt-related transcription factor 2 (Runx2; alias core-binding factor subunit α1 or Cbfa1) are two master transcriptional factors for chondrogenesis initiation and hypertrophic maturation, respectively.1, 2, 3, 4, 7, 8, 9
Our previous study showed that neural EGFL-like 1 (Nell-1), an extracellular matrix molecule distributed in human uncalcified articular cartilage,12 enhances chondrogenic marker expression and cartilage nodule formation of rabbit chondrocytes.13 In addition, application of Nell-1 induces hyaline cartilage regeneration, as demonstrated in a rabbit knee subchondral defect model,14 and administration of Nell-1–overexpressed bone marrow mesenchymal stem cells promotes articular cartilage reestablishment in critical-sized goat mandibular condyle osteochondral defects.15 Developmentally, in comparison with wild-type (WT) littermates, newborn Nell-1 overexpression transgenic mice exhibit premature hypertrophy and apoptosis in the chondrocranium region,16 whereas neonatal Nell-1–deficient mice have shorter and deformed rib cages and vertebral bodies with compressed intervertebral spaces accompanied with reduced cartilage extracellular matrix.17 Moreover, Nell-1 deficiency also results in reduced expression of multiple cartilage-related genes.17 Taken together, these findings indicate that Nell-1 may also play a role in chondrogenic differentiation and maturation. To better understand the potential regulatory roles of Nell-1 in chondrogenesis, the current study focuses on Runx2−/− mice to eliminate the pivotal contribution of Runx2.
Materials and Methods
Animal Maintenance and Skeletal Analysis
Mice were bred and maintained, as previously described,7, 17, 18, 19 under institutional approval by the Chancellor's Animal Research Committee at UCLA (protocol number 2012-041). Runx2 heterozygous deficient mice (Runx2+/−; generated by embryonic stem cells derived from the 129 mouse strain backcrossed with the C57BL/6 strain7) were mated with Nell-1–overexpressing mice (CMV-Nell-1; derivative of the B6C3 strain19) to obtain Runx2−/−/CMV-Nell-1 mice.18 Because severe reduction of Nell-1 expression in homozygotes [Nell-16R/6R; Nell-16R: an N-ethyl-N-nitrosourea–induced point mutation that truncates an 810 amino acid Nell-1 protein at residue 50220, 21] induces neonatal death,17 Nell-16R heterozygous mice (Nell-1+/6R) were used to produce Nell-16R/6R fetuses. Mouse genotypes were determined by PCR, and mRNA expression levels of Nell-1 and Runx2 were monitored using quantitative real-time PCR.
The animals in this study were euthanized with an overdose of phenobarbital (Piramal Healthcare, Maharashtra, India). After euthanization, three neonatal Runx2−/−/CMV-Nell-1 mice and five Runx2−/− littermates were skinned, dissected, and fixed in 95% ethanol for 16 hours before standard skeletal staining with Alcian Blue and Alizarin Red to provide gross distinction between cartilage and mineralized tissues. For paraffin embedding, hind limbs isolated from mouse embryos at different stages and from neonatal mice were fixed in 4% paraformaldehyde (Sigma-Aldrich, St. Louis, MO) at 4°C overnight. For Runx2-deficient mice and N-ethyl-N-nitrosourea–induced Nell-1–deficient mice, embryo pairs from six litters were used for each utero stage. Four neonatal Runx2−/−/CMV-Nell-1 mice and three Runx2−/− littermates were also used for histological analysis. Hematoxylin and eosin staining was performed on paraffin sections (5 μm thick) for histological analysis, whereas Safranin O staining, Alcian Blue staining, and immunohistochemical and immunofluorescence staining were performed following standard protocols.13, 14 Primary antibodies against collagen II (Developmental Studies Hybdridoma Bank, Iowa City, IA), collagen X (Developmental Studies Hybdridoma Bank), Nell-1 (Allele Biotechnology, San Diego, CA), osteocalcin (Abcam, Cambridge, UK), and Runx2 (Santa Cruz Biotechnology, Santa Cruz, CA) were used for immunohistochemical and/or immunofluorescence staining, respectively. DAPI (Sigma-Aldrich) was used for nuclear counterstaining in immunofluorescence staining.
Mouse Primary Chondrocyte Isolation and Cultivation
After removing soft tissues by 2 mg/mL protease (Roche, Nutley, NJ) and 3 mg/mL collagenase II (Roche), the rib cages of neonatal mouse embryos were digested in 1 mg/mL collagenase II for 3 hours to achieve single-cell suspension. Chondrocytes were cultured in a basal culture medium (Dulbecco's modified Eagle's medium with 10% fetal bovine serum, 100 U/mL penicillin, and 100 μg/mL streptomycin). Medium was changed every 3 days and cells were passaged at 70% to 90% confluence. All cell culture reagents were purchased from Invitrogen (Carlsbad, CA).
A total of 5 × 104 cells/well passage 2 (P2) chondrocytes were seeded in 6-well plates with basal culture medium for 6 hours. Recombinant human Nell-1 protein was synthesized by Aragen Bioscience Inc. (Morgan Hill, CA) with a purity of 92%. Before treatment, cells were synchronized by culture in starvation medium [Dulbecco's modified Eagle's medium + 1% insulin–transferrin-sodium selenite media supplement (BD Biosciences, San Jose, CA)] for 18 hours.
For adenoviral infection, P2 mouse chondrocytes at 80% confluence were infected with AdLacZ, AdNell-1, or AdRunx2 at a multiplicity of infection of 50, 250, or 500 plaque-forming units per cell, as described previously.22
Mouse Primary Mesenchymal Progenitor Cell Isolation and Cultivation
Primary embryonic mesenchymal progenitor cells were isolated from limb buds of E11.5 embryonic mice, as previously described.23 Briefly, limbs were dissected from the embryos and then digested with 1 mg/mL Dispase (Roche) for 1.5 hours at 37°C. Cells were filtered through a prewashed 40-μm cell strainer to generate single-cell suspension. After wash with Puck's Saline A solution, cells were suspended in culture medium [Dulbecco's modified Eagle's medium:F12 = 2:3 (Invitrogen) containing 10% fetal bovine serum, 100 U/mL penicillin, 100 μg/mL streptomycin, 0.5 mmol/L glutamine, 50 μg/mL vitamin C (Sigma-Aldrich), 10 nmol/L β-glycerophosphate (Sigma-Aldrich)] with or without 2 μg/mL recombinant human Nell-1 protein, and were plated at the density of 2 × 105 cells per tube, centrifuged at 500 × g for 5 minutes, then incubated at 37°C in a 5% CO2 humidified incubator. Medium was changed every 48 hours. Pellets were cultured at 7 or 21 days for further analysis.
Mouse Limb Bud Isolation and Explant Cultivation
Mouse hind limbs were isolated from embryonic day (E)14.5 mouse embryos. Isolated limbs were cultured in BGJb medium (Invitrogen) for 24 hours on polycarbonate tissue culture inserts (pore size, 0.1 μm; EMD Millipore, Billerica, MA). For the treatment, α-modified Eagle's medium containing 5% fetal bovine serum, 100 U/mL penicillin, 100 μg/mL streptomycin, and 0.5 mmol/L glutamine (Invitrogen) with or without 2 μg/mL recombinant human Nell-1 protein was changed every 48 hours for 5 days to simulate the environment in neonatal mice (term, 19.5 days). Limb explants were fixed in 4% paraformaldehyde at 4°C overnight, embedded in paraffin, and divided into sections (5 μm thick).
Proliferation Assay
5-Bromo-2′-deoxyuridine (Invitrogen) was added 6 hours before sample collection, and pellets were fixed in 4% paraformaldehyde at 4°C overnight, embedded in paraffin, and divided into sections (5 μm thick). DAPI was used for nuclear counterstaining.
Quantitative Real-Time PCR
Total RNA was isolated by TRIzol Reagent (Invitrogen) followed by DNase (Invitrogen) treatment. RNA (1 μg) was injected for reverse transcription (RT) with the SuperScript II Reverse Transcriptase Kit (Invitrogen), as per the manufacturer's instructions. Real-time PCR was performed on the 7300 Real-Time PCR system with SYBR Green Mastermix (Invitrogen). The primer pair sequences were as follows: Acan, 5′-CCAGGCTCCACCAGATACTC-3′ (forward) and 5′-TGCTCATAGCCTGCCTCATA-3′ (reverse); Adamts4, 5′-ATGGCCTCAATCCATCCCAG-3′ (forward) and 5′-AAGCAGGGTTGGAATCTTTGC-3′ (reverse); Col2α1, 5′-GTCCTGAAGGTGCTCAAGGT-3′ (forward) and 5′-TTTGGCTCCAGGAATACCAT-3′ (reverse); Mmp13, 5′-TGTTTGCAGAGCACTACTTGAA-3′ (forward) and 5′-CAGTCACCTCTAAGCCAAAGAAA-3′ (reverse); Nell-1, 5′-TCCTGGGTAGATGGTGACAA-3′ (forward) and 5′-CATTGGCCAGAAATATGCAC-3′ (reverse); Runx2, 5′-AACGATCTGAGATTTGTGGGC-3′ (forward) and 5′-CCTGCGTGGGATTTCTTGGTT-3′ (reverse); and SRY-Box 9, 5′-ACGGCTCCAGCAAGAACAAG-3′ (forward) and 5′-TTGTGCAGATGCGGGTACTG-3′ (reverse). Concomitant glyceraldehyde 3-phosphate dehydrogenase was also evaluated in separate tubes for each RT reaction as a housekeeping standard [5′-ATTCAACGGCACAGTCAAGG-3′ (forward) and 5′-GATGTTAGTGGGGTCTCGCTC-3′ (reverse)]. Relative gene expression was analyzed by the ΔΔCT method.24
Image Processing
Images were acquired at room temperature with the CellSens Standard 1.9 software (Olympus, America Inc., Center Valley, PA) on a microscope (Olympus) using 4× (dry HC Plan Apochromat; numerical aperture, 0.13), 10× (dry HC Plan Apochromat; numerical aperture, 0.30), and 20× (dry HC Plan Apochromat; numerical aperture, 0.17) objective lenses. Images were processed in Photoshop CS2 and Illustrator CS4 (Adobe Systems Computer Software Company, San Jose, CA) for merging.
Statistical Analysis
Statistical analysis was performed by OriginPro 8 (Origin Lab Corp., Northampton, MA) and included the one-way analysis of variance and two-sample t-test. The Mann-Whitney test was used for nonparametric data. Statistical significance was determined at the P < 0.05 level.
Results
Nell-1 and Runx2 Exhibit Similar Spatiotemporal Expression Patterns during Late Limb Development of Mouse Femurs
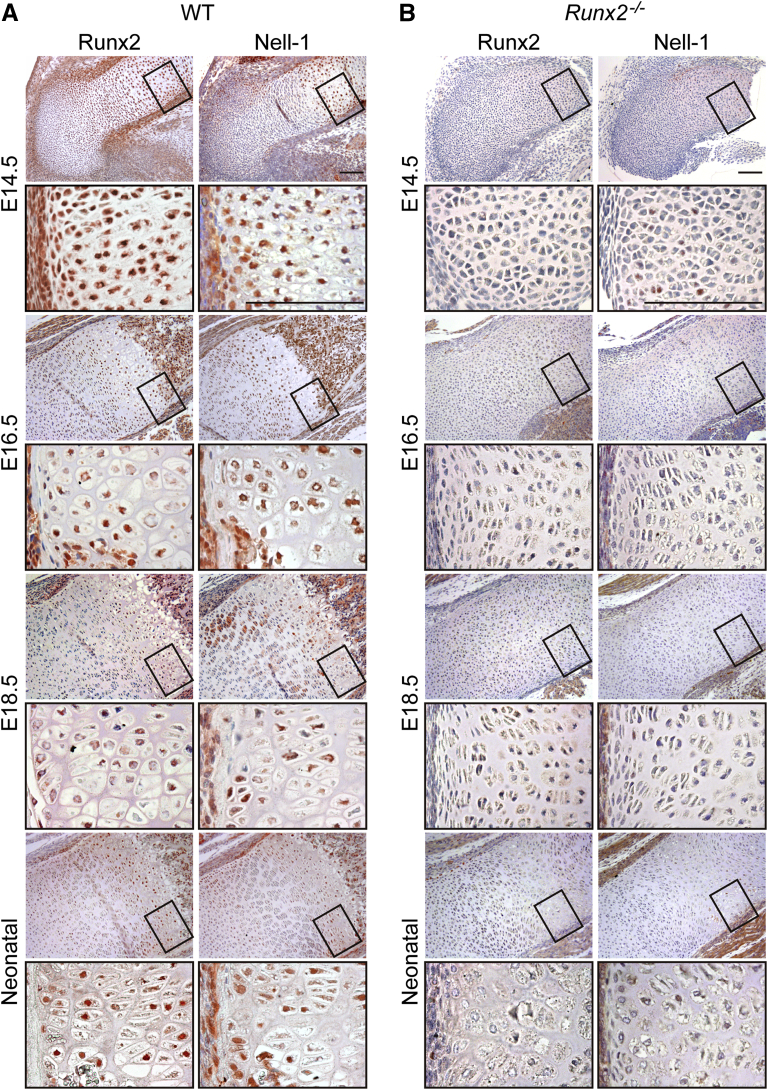
To reveal the spatiotemporal expression pattern of Nell-1 and Runx2 during long bone development, the hind limbs of fetal and neonatal WT (Runx2+/+) and Runx2−/− mice were harvested for immunohistochemical examination. In WT animals, from E16.5 onward, both Nell-1 and Runx2 staining were observed in the superficial osteoblasts of the newly formed trabeculae (Figure 1A). Runx2 and Nell-1 were also coexpressed in the perichondrium and prehypertrophic chondrocytes of the WT E14.5 femurs (Figure 1A). Beginning at E16.5, Runx2 was preferentially expressed in hypertrophic chondrocytes of WT mice, where intense Nell-1 expression was also observed (Figure 1A and Supplemental Figure S1). This spatiotemporal overlap in endogenous Nell-1 and Runx2 expression at different chondrogenic differentiation zones during long bone development suggested a possible regulatory relationship between Runx2 and Nell-1 in chondrogenesis, at least during the late stages of chondrogenic differentiation.
Figure 1.
Spatiotemporal expression of Nell-1 and Runx2 in the femurs of mouse embryos during chondrogenesis. Nell-1 and Runx2 expression in the femoral bone of WT (A) and Runx2−/− (B) embryos at embryonic day (E)14.5, E16.5, E18.5, and neonatal stage as examined by immunohistochemical staining. Boxed areas are shown below at higher magnification. Scale bars = 100 μm (A and B).
Unlike WT mice with strong Nell-1 staining in the resting, proliferating, and prehypertrophic chondrocytes of femurs from E16.5 to newborn (Figure 1A), Runx2−/− mice barely exhibited Nell-1 staining throughout the whole femurs during the entire examining period, accompanied with impaired endochondral ossification (Figure 1B and Supplemental Figure S1). Therefore, at least during late limb development, Nell-1 expression in chondrocytes is highly reliant on Runx2.
Nell-1 Is a Downstream Target of Runx2 in Mouse Chondrocytes
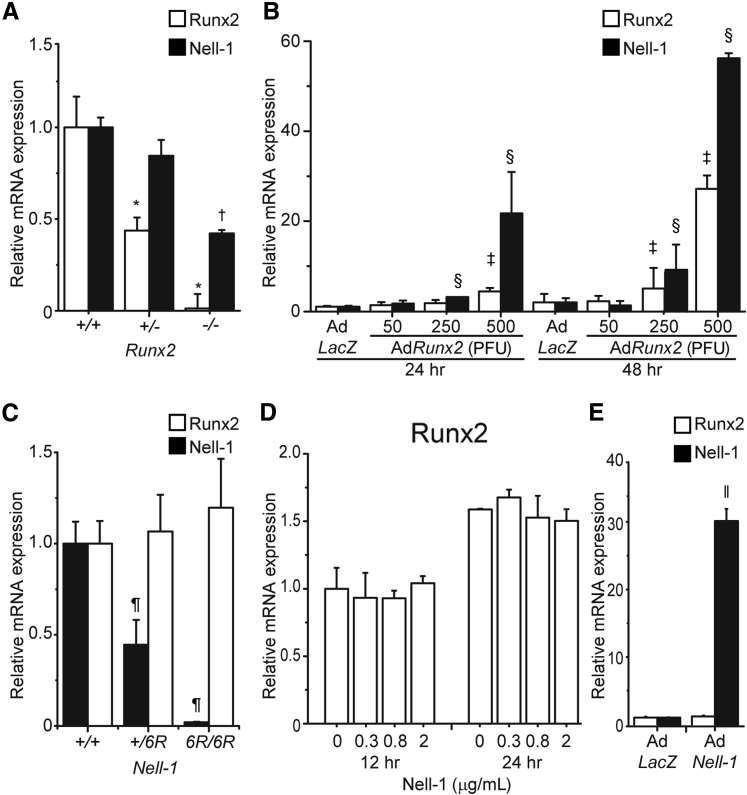
To examine the regulatory relationship between Nell-1 and Runx2 in chondrocytes, we next evaluated the expression of Runx2 mRNA and Nell-1 mRNA in chondrocytes of neonatal mice with different genotypes. The expression level of Nell-1 mRNA in Runx2−/− mouse chondrocytes was only half of that in WT control (Figure 2A). In addition, forcing Runx2 overexpression in WT mouse chondrocytes by adenovirus AdRunx2 significantly stimulated Nell-1 mRNA expression in a viral dose-dependent manner (Figure 2B). In contrast, expression of Runx2 mRNA was retained at similar levels in mouse chondrocytes with different Nell-1 genotypes (Figure 2C). Moreover, neither Nell-1 protein nor AdNell-1 adenovirus had a significant effect on Runx2 mRNA expression in the Nell-1+/+ chondrocytes (Figure 2, D and E). These results clearly demonstrated that, in chondrocytes, Runx2 regulated Nell-1 expression but that Nell-1 did not alter Runx2 expression.
Figure 2.
Nell-1 is a downstream target of Runx2 in mouse chondrocytes. A: mRNA levels of Nell-1 in mouse chondrocytes with different genotypes of Runx2. B: Transcriptional levels of Nell-1 and Runx2 in AdRunx2-transduced Runx2+/+ mouse chondrocytes. C: mRNA levels of Runx2 in mouse chondrocytes with different genotypes of Nell-1. D: Transcriptional levels of Runx2 in Nell-1 protein stimulated Nell-1+/+ mouse chondrocytes. E: Transcriptional levels of Nell-1 and Runx2 in AdNell-1–transduced Nell-1+/+ chondrocytes. Two sample t-tests were used to compare the data from two groups. Data are expressed as means + SD of three independent experiments performed in triplicate (A–E). ∗P < 0.05 versus Runx2 mRNA expression in Runx2+/+ cells; †P < 0.05 versus Nell-1 mRNA expression in Runx2+/+ cells; ‡P < 0.05 versus Runx2 mRNA expression in AdLacz-transduced chondrocytes for 24 hours; §P < 0.05 versus Nell-1 mRNA expression in AdLacz-transduced chondrocytes for 24 hours; ¶P < 0.05 versus Nell-1 expression in Nell-1+/+ cells; ‖P < 0.05 versus Nell-1 expression in AdLacz-transduced Nell-1+/+ chondrocytes for 24 hours. PFU, plaque-forming unit.
Nell-1 Plays Critical Roles in Chondrogenic Differentiation and Maturation
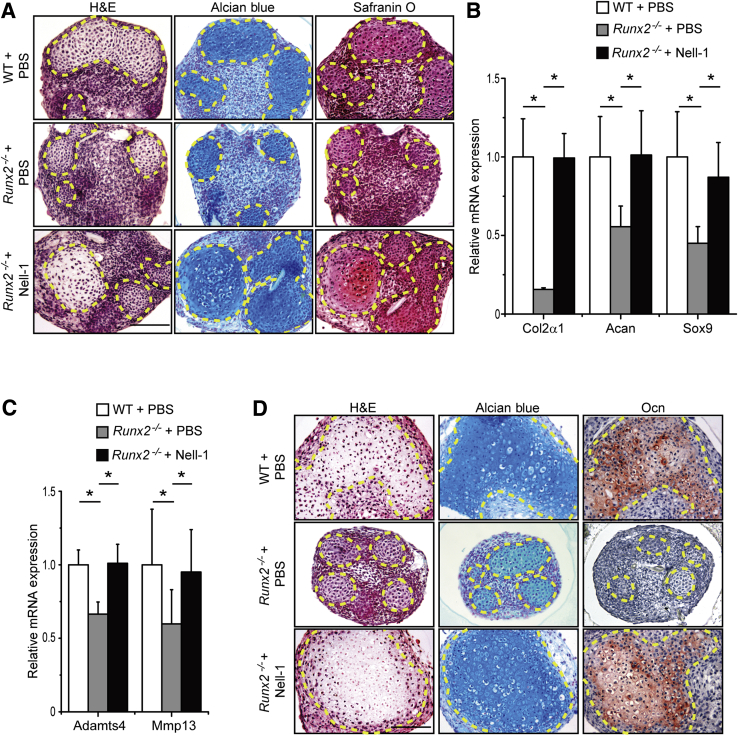
After 7 days of chondrogenic differentiation, in vitro pellet cultures of primary mesenchymal progenitor cells isolated from Runx2−/− mouse embryonic limb buds exhibited a larger undifferentiated area with less intense Alcian Blue and Safranin O staining in cartilaginous components in comparison to the pellet controls formed by cells isolated from WT littermates (Figure 3A). Interestingly, administration of Nell-1 protein not only promoted cell proliferation in Runx2−/− pellets (Supplemental Figure S2), but also increased cartilaginous nodule formation in the Runx2−/− pellets, which contained rich cartilage matrix characterized by high staining intensity of Alcian Blue and Safranin O (Figure 3A). Transcriptionally, expression of genes encoding chondrogenic markers, such as Col2α1 (encoding collagen II, α 1, an abundant and specific protein in cartilage), Acan (encoding aggrecan core protein, one of the major structural components in cartilage matrix), and SRY-Box 9,1, 2, 3, 4 in Runx2−/− pellets was significantly lower than that in WT pellets (Figure 3B). However, disruption of chondrogenic differentiation in Runx2−/− pellets was rescued by Nell-1 protein administration (Figure 3, A and B).
Figure 3.
Exogenous Nell-1 protein promotes cartilaginous nodule formation and maturation of primary Runx2−/− mesenchymal progenitor cells. A: Nell-1 treatment promotes the chondrogenic differentiation of Runx2−/− cells at day 7 shown by hematoxylin and eosin (H&E), Alcian Blue, and Safranin O staining. The cartilage nodules are circled by the dotted yellow lines. B: Nell-1 stimulation rescues chondrogenic marker expression in Runx2−/− limb bud pellets at day 7. C: Nell-1 treatment promotes the terminal chondrogenic maturation markers of Runx2−/− cells at day 21 by gene expression. D: Nell-1 treatment promotes the chondrogenic maturation of Runx2−/− cells at day 21 shown by H&E, Alcian Blue, and osteocalcin (Ocn) staining. The cartilage nodules are circled by the dashed yellow lines. Two sample t-tests were used to compare the data from two groups. Data are expressed as means + SD (B and C). n = 6 (B and C). ∗P < 0.05. Scale bars = 100 μm (A and D). PBS, phosphate-buffered saline; WT, wild-type.
Meanwhile, after 21 days of cultivation, Runx2−/− pellets exhibited significantly decreased expression of terminal chondrogenic differentiation markers [Adamts4 (encoding aggrecanase-1)25 and Mmp13 (encoding matrix metallopeptidase 13)1, 2, 3, 4] when compared to WT pellets (Figure 3C). Mature chondrogenic hypertrophy was limited in Runx2−/− pellets with reduced proteoglycan deposition (Figure 3D). In addition, matrix mineralization observed in WT pellets after 21 days of cultivation was also absent in Runx2−/− pellets, as characterized by the lack of osteogenic marker osteocalcin1, 26 (Figure 3D). However, Nell-1 administration also restored chondrogenic maturation and ossification in Runx2−/− pellets (Figure 3, C and D).
Nell-1 Partially Restores Delayed Chondrocyte Hypertrophy in Runx2−/− Mouse Hind Limbs
To avoid systemic influences and initial differentiation stage differences,27 we took advantage of an established ex vivo approach28 to further evaluate the influence of Nell-1 on chondrocyte/cartilage maturation. Not only was collagen II observed, but also collagen X, a marker for chondrogenic maturity and hypertrophy,1, 2, 3, 4 in the hypertrophic zone of E14.5 WT mouse limb explants after 5 days of cultivation (Supplemental Figure S3). In contrast, absence of chondrocyte hypertrophy was observed in the age-matched Runx2−/− explants, as characterized by negligible collagen X expression but excessive collagen II expression (Supplemental Figure S3). Administration of Nell-1 protein partially reestablished collagen X expression and reduced collagen II level in Runx2−/− explants (Supplemental Figure S3), indicating the restoration of the hypertrophic chondrocyte phenotype and marker expression pattern of WT explants.
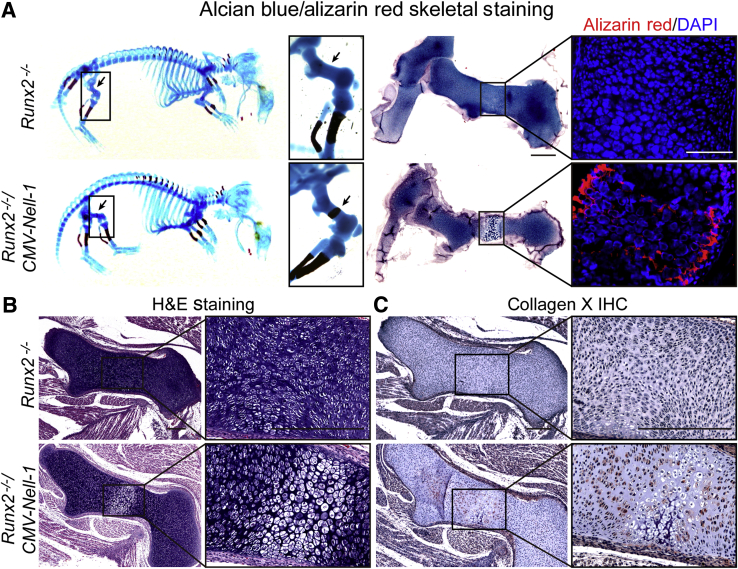
Furthermore, only Alcian Blue staining was observed throughout the whole femur of neonatal Runx2−/− mice (Figure 4A), which reflected the diminished endochondral calcification due to the Runx2 deficiency. Histological examination further confirmed the lack of chondrogenic maturation by the complete absence of the hypertrophic zone, the disorganized chondrocyte architecture without an apparent proliferating or prehypertrophic zone, and the lack of collagen X expression in the neonatal Runx2−/− mouse femurs (Figure 4, B and C). By crossing Nell-1–overexpressing transgenic mice with Runx2+/− mice,18 we obtained Runx2−/−/CMV-Nell-1 mice that overexpress Nell-1 in a Runx2−/− background. Overexpression of Nell-1 was unable to keep the neonatal Runx2−/− mice alive, as we previously described,18 suggesting that Nell-1 cannot completely overcome the global influence of Runx2 deficiency on fetal mouse development. However, the mid-shaft of the neonatal Runx2−/−/CMV-Nell-1 mouse femurs exhibited obvious signs of chondrocyte hypertrophy and calcification characterized by positive staining with collagen X and Alizarin Red (Figure 4).
Figure 4.
Partial rescue of chondrogenic differentiation in neonatal Runx2−/− by Nell-1 overexpression. A: Alcian Blue/Alizarin Red skeletal staining of neonatal Runx2−/− and Runx2−/−/CMV-Nell-1 mice. The mid-shaft of the femur and lower thoracic vertebrae in neonatal Runx2−/−/CMV-Nell-1 mice clearly stain positively with Alizarin Red (arrows). Endochondral ossification is clearly absent in the vertebrae and proximal limbs of Runx2−/− mice. Calcified cartilage, which stains positively with Alizarin Red, is apparent in Runx2−/−/CMV-Nell-1 femurs. B: Hematoxylin and eosin (H&E) staining shows the cartilage morphology of Runx2−/− and Runx2−/−/CMV-Nell-1 femurs. C: Restoration of a distinct hypertrophic chondrocyte zone is clearly shown by immunostaining of collagen X in restricted regions of the limbs in Runx2−/−/CMV-Nell-1 mice. Boxed areas are shown at higher magnification to the right (A–C). Scale bars: 500 μm (A, left column); 50 μm (A, right column); 200 μm (B and C). IHC, immunohistochemistry.
Discussion
Nell-1 was originally identified as a craniosynostosis-associated molecule from resected human fusing and fused coronal sutures.29 Functionally, Nell-1 overexpression in mice leads to premature suture closure simulating nonsyndromic craniosynostosis,19, 22 whereas N-ethyl-N-nitrosourea–induced Nell-1–deficient mice exhibit a cleidocraniodysplasia-like defect.30 Mechanistically, overexpression of the critical osteogenesis regulator, Runx2,1, 31, 32 in osteoblast lineage cells results in up-regulated Nell-1 expression. Runx2 deficiency, however, leads to significantly reduced Nell-1 expression,33 which manifested the regulation of Runx2 on Nell-1 expression during osteogenesis. Further investigation demonstrated that, as a downstream regulator of Runx2 in osteogenesis, Nell-1 reciprocally induces the expression and activation of Runx2 in osteoblast lineage cells.18, 33 In fact, it is clear that Nell-1 exhibits its pro-osteogenic functions by activating the mitogen-activated protein kinase signaling pathway in osteoblast lineage cells.18, 33, 34, 35 The importance of the Runx2→Nell-1→mitogen-activated protein kinase axis in osteogenic differentiation has been extensively studied by multiple independent research groups,18, 33, 36, 37 and the osteogenic activity of Nell-1 has been proven in various small and large animal models.17, 38, 39, 40, 41, 42, 43, 44, 45
Herein, we noticed that Nell-1 expression was devoid in neonatal mouse femurs because of the Runx2 deficiency. Further investigation revealed that, similar to the preosteoblasts, osteoblasts, and osteosacoma cell line,33, 34 Nell-1 is a downstream target of Runx2 in chondrocytes. This similarity implies that Runx2→Nell-1 regulation may be a general characteristic shared by osteochondral lineage cells. However, unlike the mutual regulation of Nell-1 and Runx2 observed in osteoblast lineage cells,9, 13 our findings demonstrated that Nell-1 could not induce Runx2 expression in chondrocytes. This indicates a cell-specific mode of regulation between Nell-1 and Runx2.
Functionally, Nell-1 deficiency results in abnormal cartilage phenotypes in the rib cage, vertebral and long bones, and chondrocranial tissues.17, 30 Nell-1 protein also exhibits prochondrogenic effects in cartilage regeneration both in vitro12, 13 and in vivo.14, 15 To better explore the possible functional role of Nell-1 in chondrogenesis from its upstream regulator, Runx2, that is a predominant transcription factor associated with chondrocyte hypertrophy differentiation,1, 2, 3, 4 we selected mouse subjects with Runx2−/− and Nell-1 overexpression (Runx2−/−/CMV-Nell-1) after crossing Nell-1 overexpression transgenic mice and Runx2 haploinsufficient mice. Although overexpression of Nell-1 was unable to rescue Runx2−/− mice from perinatal lethality,6, 7 CMV-driven Nell-1 overexpression resulted in partial rescue of the Runx2 deficiency–induced impairments of chondrogenic differentiation and maturation. This was evidenced by mineralized cartilage in the mid-shaft of the femurs of Runx2−/−/CMV-Nell-1 mice.
Runx2 not only regulates the late stage of chondrogenic maturation, but is also involved in chondrogenic initiation.46, 47, 48 Interestingly, although Runx2 is able to bind directly to the Indian Hedgehog (Ihh) promoter9 to stimulate the Ihh-Gli pathway, the most plausible signaling pathway that induces chondrogenic differentiation,11, 48 recent studies revealed that the nuclear import and DNA binding functions of Runx2 are insufficient for chondrocyte differentiation.31 To avoid systemic influences and initial differentiation stage differences,27 in vitro and ex vivo approaches were applied to verify the prochondrogenic bioactivities of Nell-1 in a Runx2−/− background. In agreement with the in vivo results, Nell-1 protein was able to restore the chondrocyte hypertrophy of Runx2−/− limb explants and primary mesenchymal progenitor cell pellets. In addition, we also found that Nell-1 protein enhanced primary mesenchymal progenitor cell proliferation and early chondrogenic differentiation in limb bud pellet cultivation. Taken together, in agreement with previous publications,12, 14, 15, 17, 30 our current findings clearly demonstrated the essential roles of Nell-1 in chondrogenic commitment, differentiation, and maturation independent from Runx2. Although the expression of Nell-1 is highly dependent on Runx2, the prochondrogenic function of Nell-1 persists in Runx2−/− scenarios. This inconsistency demonstrates the complexity of the Runx2/Nell-1 regulatory network that governs chondrogenesis. Particularly, Nell-1 promotes chondrogenesis, at least partially, via an alternative pathway that is independent of the nuclear import and DNA binding functions of Runx2.
In summary, the identification of Nell-1, a secretory molecule, as a key downstream mediator of Runx2-mediated chondrogenesis opens up new possibilities for the application of Nell-1 in cartilage regeneration. This is especially important when considering the fact that cartilage regeneration continues to pose a big challenge in healing critical-sized defects.49 Because Nell-1 has the ability to induce mesenchymal stem cell commitment to chondrogenic differentiation,12 it could have the potential advantage to enhance stem cell–based therapies for cartilage regeneration.14, 15 In particular, we have found that the combination of Nell-1, bone morphogenetic protein-6, and transforming growth factor β-3 significantly promotes human perivascular stem cell chondrogenic differentiation while inhibiting their hypertrophic, fibrotic, osteogenic, and apoptotic differentiation.12 Further investigation is therefore necessary not only to dissect the details on how Nell-1 orchestrates chondrogenic initiation and maturation, but also to explore the broader applications of Nell-1 as a therapeutic agent for bone and chondrogenic regeneration. The overexpression of Nell-1 was observed in multiple bone and cartilage tumors.50, 51 Moreover, our previous studies showed that the teratocarcinoma-derived chondrogenic cell line ATDC5 has distinguished responses to Nell-1 stimulation than do primary mouse chondrocytes.52 Thus, the involvement of Nell-1 in bone and cartilage-related tumor formation also warrants further investigation.
Acknowledgments
We thank Drs. Wenfang Wang and Bjorn R. Olsen (Harvard University) for providing the Runx2+/− mice and Dr. Renny T. Franceschi (University of Michigan) for providing the Runx2 adenovirus and related plasmids.
Footnotes
Supported by NIH–National Institute of Arthritis and Musculoskeletal and Skin Diseases grants R01AR066782 (K.T.), R01AR068835 (C.S.), and R01AR061399 (C.S.); UCLA/NIH Clinical and Translational Science Institute (CTSI) grant UL1TR000124 (C.S.); National Aeronautical and Space AdministrationGA-2014-154 (K.T.); and International S&T Cooperation Program of China grant 2013DFB30360 (K.T. and Y.Z.).
Disclosures: K.T., X.Z., and C.S. are inventors of Nell-1–related patents. C.T.C. is an inventor of Nell-1–related patents filed from ORNL. K.T., X.Z., and C.S. are founders and/or board members of Bone Biologics Inc./Bone Biologics Corp. that sublicenses Nell-1 patents from the UC Regents, which also holds equity in the company. C.T.C. is a founder of NellOne Therapeutics, Inc., which licensed Nell-1–related patent applications from ORNL.
Supplemental material for this article can be found at http://dx.doi.org/10.1016/j.ajpath.2016.12.026.
Contributor Information
Xinli Zhang, Email: xzhang@dentistry.ucla.edu.
Chia Soo, Email: bsoo@ucla.edu.
Supplemental Data
Supplemental Figure S1.
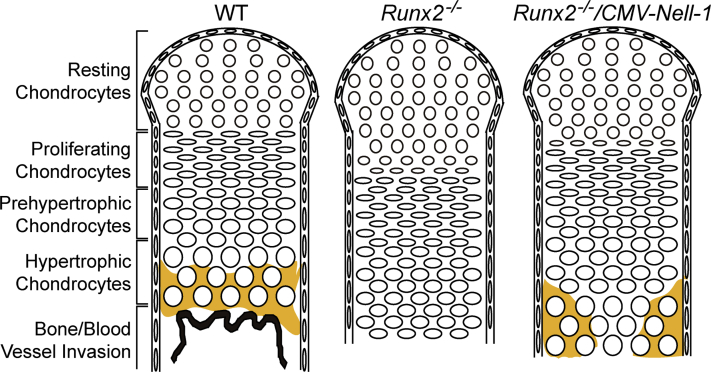
Proposed model for the compensation of chondrogenic deficit by Nell-1 in Runx2 deficiency. Transgenic overexpression of Nell-1 results in a partial rescue of the Runx2−/− phenotype by presence of hypertrophic chondrocytes and mineralized cartilage in the mid-shaft of the femur. The region of calcified cartilage is shown in yellow. WT, wild-type.
Supplemental Figure S2.
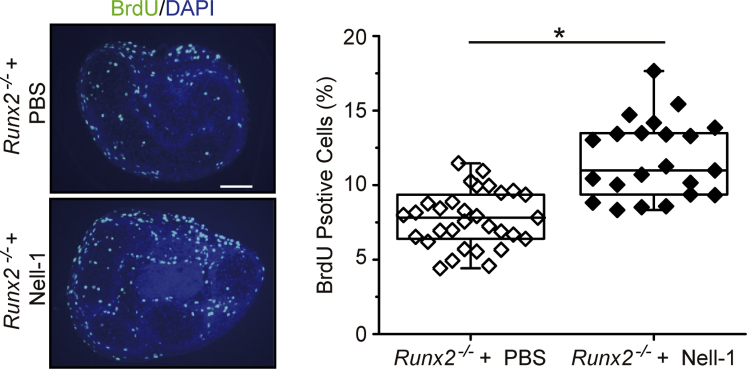
Nell-1 promotes the proliferation of Runx2−/− chondrocytes in vitro. Proliferation analysis of primary Runx2−/− mesenchymal progenitor cell pellets at day 7 by 5-bromo-2′-deoxyuridine (BrdU) quantification. The cell number in each pellet was counted on BrdU/DAPI-stained sections and expressed as a ratio of BrdU-positive cells/total counted cells. The raw data were presented with box plot. Minimum and maximum values are depicted by short black lines, the box signifies the upper and lower quartiles, and the median is represented by a black line within the box for each group. Nell-1 stimulation significantly increases the proliferation of primary Runx2−/− mesenchymal progenitor cells. ∗P < 0.05. Scale bar = 50 μm. PBS, phosphate-buffered saline.
Supplemental Figure S3.
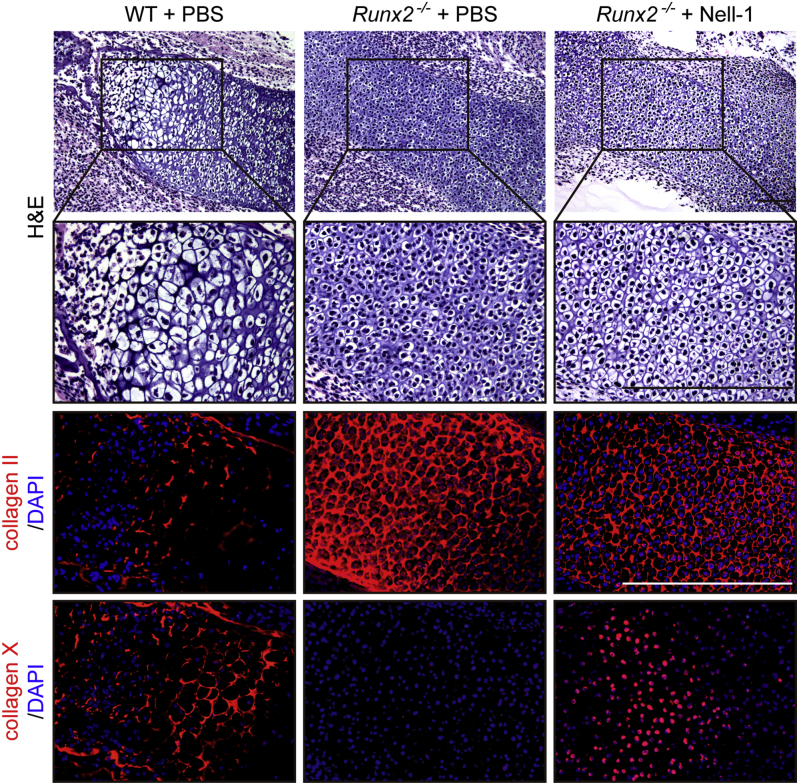
Nell-1 promotes chondrogenic differentiation and maturation of ex vivo cultured Runx2−/− E14.5 hind limbs. Hind limb explants were isolated from WT and Runx2−/− E14.5 mouse embryos, cultured in media with or without 2.0 μg/mL Nell-1 protein for 5 days, fixed in 4% paraformaldehyde, divided into sections, and stained with hematoxylin and eosin (H&E) for histological evaluation and immunohistochemistry of collagen II and collagen X to assess chondrogenic differentiation and maturation. Boxed areas are shown at higher magnification below. Scale bars = 100 μm. PBS, phosphate-buffered saline; WT, wild-type.
References
- 1.Lefebvre V., Bhattaram P. Vertebrate skeletogenesis. Curr Top Dev Biol. 2010;90:291–317. doi: 10.1016/S0070-2153(10)90008-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Lefebvre P., Martin P.J., Flajollet S., Dedieu S., Billaut X., Lefebvre B. Transcriptional activities of retinoic acid receptors. Vitam Horm. 2005;70:199–264. doi: 10.1016/S0083-6729(05)70007-8. [DOI] [PubMed] [Google Scholar]
- 3.Wuelling M., Vortkamp A. Chondrocyte proliferation and differentiation. Endocr Dev. 2011;21:1–11. doi: 10.1159/000328081. [DOI] [PubMed] [Google Scholar]
- 4.Goldring M.B. Chondrogenesis, chondrocyte differentiation, and articular cartilage metabolism in health and osteoarthritis. Ther Adv Musculoskelet Dis. 2012;4:269–285. doi: 10.1177/1759720X12448454. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Olsen B.R., Reginato A.M., Wang W. Bone development. Annu Rev Cell Dev Biol. 2000;16:191–220. doi: 10.1146/annurev.cellbio.16.1.191. [DOI] [PubMed] [Google Scholar]
- 6.Komori T., Yagi H., Nomura S., Yamaguchi A., Sasaki K., Deguchi K., Shimizu Y., Bronson R.T., Gao Y.H., Inada M., Sato M., Okamoto R., Kitamura Y., Yoshiki S., Kishimoto T. Targeted disruption of Cbfa1 results in a complete lack of bone formation owing to maturational arrest of osteoblasts. Cell. 1997;89:755–764. doi: 10.1016/s0092-8674(00)80258-5. [DOI] [PubMed] [Google Scholar]
- 7.Otto F., Thornell A.P., Crompton T., Denzel A., Gilmour K.C., Rosewell I.R., Stamp G.W., Beddington R.S., Mundlos S., Olsen B.R., Selby P.B., Owen M.J. Cbfa1, a candidate gene for cleidocranial dysplasia syndrome, is essential for osteoblast differentiation and bone development. Cell. 1997;89:765–771. doi: 10.1016/s0092-8674(00)80259-7. [DOI] [PubMed] [Google Scholar]
- 8.Inada M., Yasui T., Nomura S., Miyake S., Deguchi K., Himeno M., Sato M., Yamagiwa H., Kimura T., Yasui N., Ochi T., Endo N., Kitamura Y., Kishimoto T., Komori T. Maturational disturbance of chondrocytes in Cbfa1-deficient mice. Dev Dyn. 1999;214:279–290. doi: 10.1002/(SICI)1097-0177(199904)214:4<279::AID-AJA1>3.0.CO;2-W. [DOI] [PubMed] [Google Scholar]
- 9.Kim I.S., Otto F., Zabel B., Mundlos S. Regulation of chondrocyte differentiation by Cbfa1. Mech Dev. 1999;80:159–170. doi: 10.1016/s0925-4773(98)00210-x. [DOI] [PubMed] [Google Scholar]
- 10.Lian J.B., Stein G.S. Runx2/Cbfa1: a multifunctional regulator of bone formation. Curr Pharm Des. 2003;9:2677–2685. doi: 10.2174/1381612033453659. [DOI] [PubMed] [Google Scholar]
- 11.Yoshida C.A., Yamamoto H., Fujita T., Furuichi T., Ito K., Inoue K., Yamana K., Zanma A., Takada K., Ito Y., Komori T. Runx2 and Runx3 are essential for chondrocyte maturation, and Runx2 regulates limb growth through induction of Indian hedgehog. Genes Dev. 2004;18:952–963. doi: 10.1101/gad.1174704. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Li C.S., Zhang X., Peault B., Jiang J., Ting K., Soo C., Zhou Y.H. Accelerated chondrogenic differentiation of human perivascular stem cells with NELL-1. Tissue Eng Part A. 2016;22:272–285. doi: 10.1089/ten.tea.2015.0250. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Lee M., Siu R.K., Ting K., Wu B.M. Effect of Nell-1 delivery on chondrocyte proliferation and cartilaginous extracellular matrix deposition. Tissue Eng Part A. 2010;16:1791–1800. doi: 10.1089/ten.TEA.2009.0384. [DOI] [PubMed] [Google Scholar]
- 14.Siu R.K., Zara J.N., Hou Y., James A.W., Kwak J., Zhang X., Ting K., Wu B.M., Soo C., Lee M. NELL-1 promotes cartilage regeneration in an in vivo rabbit model. Tissue Eng Part A. 2012;18:252–261. doi: 10.1089/ten.tea.2011.0142. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Zhu S., Zhang B., Man C., Ma Y., Hu J. NEL-like molecule-1-modified bone marrow mesenchymal stem cells/poly lactic-co-glycolic acid composite improves repair of large osteochondral defects in mandibular condyle. Osteoarthritis Cartilage. 2011;19:743–750. doi: 10.1016/j.joca.2011.02.015. [DOI] [PubMed] [Google Scholar]
- 16.Zhang X., Cowan C.M., Jiang X., Soo C., Miao S., Carpenter D., Wu B., Kuroda S., Ting K. Nell-1 induces acrania-like cranioskeletal deformities during mouse embryonic development. Lab Invest. 2006;86:633–644. doi: 10.1038/labinvest.3700430. [DOI] [PubMed] [Google Scholar]
- 17.Desai J., Shannon M.E., Johnson M.D., Ruff D.W., Hughes L.A., Kerley M.K., Carpenter D.A., Johnson D.K., Rinchik E.M., Culiat C.T. Nell1-deficient mice have reduced expression of extracellular matrix proteins causing cranial and vertebral defects. Hum Mol Genet. 2006;15:1329–1341. doi: 10.1093/hmg/ddl053. [DOI] [PubMed] [Google Scholar]
- 18.Zhang X., Ting K., Bessette C.M., Culiat C.T., Sung S.J., Lee H., Chen F., Shen J., Wang J.J., Kuroda S., Soo C. Nell-1, a key functional mediator of Runx2, partially rescues calvarial defects in Runx2(+/−) mice. J Bone Miner Res. 2011;26:777–791. doi: 10.1002/jbmr.267. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Zhang X., Kuroda S., Carpenter D., Nishimura I., Soo C., Moats R., Iida K., Wisner E., Hu F.Y., Miao S., Beanes S., Dang C., Vastardis H., Longaker M., Tanizawa K., Kanayama N., Saito N., Ting K. Craniosynostosis in transgenic mice overexpressing Nell-1. J Clin Invest. 2002;110:861–870. doi: 10.1172/JCI15375. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Rinchik E.M., Carpenter D.A., Selby P.B. A strategy for fine-structure functional analysis of a 6- to 11-centimorgan region of mouse chromosome 7 by high-efficiency mutagenesis. Proc Natl Acad Sci U S A. 1990;87:896–900. doi: 10.1073/pnas.87.3.896. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Rinchik E.M., Carpenter D.A., Johnson D.K. Functional annotation of mammalian genomic DNA sequence by chemical mutagenesis: a fine-structure genetic mutation map of a 1- to 2-cM segment of mouse chromosome 7 corresponding to human chromosome 11p14-p15. Proc Natl Acad Sci U S A. 2002;99:844–849. doi: 10.1073/pnas.022628199. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Zhang X., Carpenter D., Bokui N., Soo C., Miao S., Truong T., Wu B., Chen I., Vastardis H., Tanizawa K., Kuroda S., Ting K. Overexpression of Nell-1, a craniosynostosis-associated gene, induces apoptosis in osteoblasts during craniofacial development. J Bone Miner Res. 2003;18:2126–2134. doi: 10.1359/jbmr.2003.18.12.2126. [DOI] [PubMed] [Google Scholar]
- 23.Aydelotte M.B., Kochhar D.M. Development of mouse limb buds in organ culture: chondrogenesis in the presence of a proline analog, L-azetidine-2-carboxylic acid. Dev Biol. 1972;28:191–201. doi: 10.1016/0012-1606(72)90137-6. [DOI] [PubMed] [Google Scholar]
- 24.Livak K.J., Schmittgen T.D. Analysis of relative gene expression data using real-time quantitative PCR and the 2(-Delta Delta C(T)) Method. Methods. 2001;25:402–408. doi: 10.1006/meth.2001.1262. [DOI] [PubMed] [Google Scholar]
- 25.Djouad F., Delorme B., Maurice M., Bony C., Apparailly F., Louis-Plence P., Canovas F., Charbord P., Noel D., Jorgensen C. Microenvironmental changes during differentiation of mesenchymal stem cells towards chondrocytes. Arthritis Res Ther. 2007;9:R33. doi: 10.1186/ar2153. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Gouttenoire J., Valcourt U., Ronziere M.C., Aubert-Foucher E., Mallein-Gerin F., Herbage D. Modulation of collagen synthesis in normal and osteoarthritic cartilage. Biorheology. 2004;41:535–542. [PubMed] [Google Scholar]
- 27.Takarada T., Hinoi E., Nakazato R., Ochi H., Xu C., Tsuchikane A., Takeda S., Karsenty G., Abe T., Kiyonari H., Yoneda Y. An analysis of skeletal development in osteoblast-specific and chondrocyte-specific runt-related transcription factor-2 (Runx2) knockout mice. J Bone Miner Res. 2013;28:2064–2069. doi: 10.1002/jbmr.1945. [DOI] [PubMed] [Google Scholar]
- 28.Wuelling M., Vortkamp A. Cartilage explant cultures. Methods Mol Biol. 2014;1130:89–97. doi: 10.1007/978-1-62703-989-5_7. [DOI] [PubMed] [Google Scholar]
- 29.Ting K., Vastardis H., Mulliken J.B., Soo C., Tieu A., Do H., Kwong E., Bertolami C.N., Kawamoto H., Kuroda S., Longaker M.T. Human NELL-1 expressed in unilateral coronal synostosis. J Bone Miner Res. 1999;14:80–89. doi: 10.1359/jbmr.1999.14.1.80. [DOI] [PubMed] [Google Scholar]
- 30.Zhang X., Ting K., Pathmanathan D., Ko T., Chen W., Chen F., Lee H., James A.W., Siu R.K., Shen J., Culiat C.T., Soo C. Calvarial cleidocraniodysplasia-like defects with ENU-induced Nell-1 deficiency. J Craniofac Surg. 2012;23:61–66. doi: 10.1097/SCS.0b013e318240c8c4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Chen H., Ghori-Javed F.Y., Rashid H., Adhami M.D., Serra R., Gutierrez S.E., Javed A. Runx2 regulates endochondral ossification through control of chondrocyte proliferation and differentiation. J Bone Miner Res. 2014;29:2653–2665. doi: 10.1002/jbmr.2287. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Bialek P., Kern B., Yang X., Schrock M., Sosic D., Hong N., Wu H., Yu K., Ornitz D.M., Olson E.N., Justice M.J., Karsenty G. A twist code determines the onset of osteoblast differentiation. Dev Cell. 2004;6:423–435. doi: 10.1016/s1534-5807(04)00058-9. [DOI] [PubMed] [Google Scholar]
- 33.Truong T., Zhang X., Pathmanathan D., Soo C., Ting K. Craniosynostosis-associated gene nell-1 is regulated by runx2. J Bone Miner Res. 2007;22:7–18. doi: 10.1359/jbmr.061012. [DOI] [PubMed] [Google Scholar]
- 34.James A., Shen J., Velasco O., Asatrian G., Chung C.G., Khadarian K., Zhang Y., Chang L., Goyal R., Zhang X., Adams J., Ting K., Soo C. Systemic administration of NELL-1, a Wnt/β-Catenin regulator, induces bone formation in osteoporotic mice via Integrinβ1. J Bone Miner Res. 2013;28(Suppl 1):S1. [Google Scholar]
- 35.Cowan C.M., Jiang X., Hsu T., Soo C., Zhang B., Wang J.Z., Kuroda S., Wu B., Zhang Z., Zhang X., Ting K. Synergistic effects of Nell-1 and BMP-2 on the osteogenic differentiation of myoblasts. J Bone Miner Res. 2007;22:918–930. doi: 10.1359/jbmr.070312. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Bokui N., Otani T., Igarashi K., Kaku J., Oda M., Nagaoka T., Seno M., Tatematsu K., Okajima T., Matsuzaki T., Ting K., Tanizawa K., Kuroda S. Involvement of MAPK signaling molecules and Runx2 in the NELL1-induced osteoblastic differentiation. FEBS Lett. 2008;582:365–371. doi: 10.1016/j.febslet.2007.12.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Chen F., Walder B., James A.W., Soofer D.E., Soo C., Ting K., Zhang X. NELL-1-dependent mineralisation of Saos-2 human osteosarcoma cells is mediated via c-Jun N-terminal kinase pathway activation. Int Orthop. 2012;36:2181–2187. doi: 10.1007/s00264-012-1590-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Cowan C.M., Cheng S., Ting K., Soo C., Walder B., Wu B., Kuroda S., Zhang X. Nell-1 induced bone formation within the distracted intermaxillary suture. Bone. 2006;38:48–58. doi: 10.1016/j.bone.2005.06.023. [DOI] [PubMed] [Google Scholar]
- 39.Aghaloo T., Cowan C.M., Zhang X., Freymiller E., Soo C., Wu B., Ting K., Zhang Z. The effect of NELL1 and bone morphogenetic protein-2 on calvarial bone regeneration. J Oral Maxillofac Surg. 2010;68:300–308. doi: 10.1016/j.joms.2009.03.066. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Zhang X., Zara J., Siu R.K., Ting K., Soo C. The role of NELL-1, a growth factor associated with craniosynostosis, in promoting bone regeneration. J Dent Res. 2010;89:865–878. doi: 10.1177/0022034510376401. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Turner N.J., Londono R., Dearth C.L., Culiat C.T., Badylak S.F. Human NELL1 protein augments constructive tissue remodeling with biologic scaffolds. Cells Tissues Organs. 2013;198:249–265. doi: 10.1159/000356491. [DOI] [PubMed] [Google Scholar]
- 42.Lee S., Zhang X., Shen J., James A.W., Chung C.G., Hardy R., Li C., Girgius C., Zhang Y., Stoker D., Wang H., Wu B.M., Peault B., Ting K., Soo C. Brief report: human perivascular stem cells and Nel-like protein-1 synergistically enhance spinal fusion in osteoporotic rats. Stem Cells. 2015;33:3158–3163. doi: 10.1002/stem.2103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.James A.W., Shen J., Zhang X., Asatrian G., Goyal R., Kwak J.H., Jiang L., Bengs B., Culiat C.T., Turner A.S., Seim Iii H.B., Wu B.M., Lyons K., Adams J.S., Ting K., Soo C. NELL-1 in the treatment of osteoporotic bone loss. Nat Commun. 2015;6:7362. doi: 10.1038/ncomms8362. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Aghaloo T., Jiang X., Soo C., Zhang Z., Zhang X., Hu J., Pan H., Hsu T., Wu B., Ting K., Zhang X. A study of the role of nell-1 gene modified goat bone marrow stromal cells in promoting new bone formation. Mol Ther. 2007;15:1872–1880. doi: 10.1038/sj.mt.6300270. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Lu S.S., Zhang X., Soo C., Hsu T., Napoli A., Aghaloo T., Wu B.M., Tsou P., Ting K., Wang J.C. The osteoinductive properties of Nell-1 in a rat spinal fusion model. Spine J. 2007;7:50–60. doi: 10.1016/j.spinee.2006.04.020. [DOI] [PubMed] [Google Scholar]
- 46.Lengner C.J., Drissi H., Choi J.Y., van Wijnen A.J., Stein J.L., Stein G.S., Lian J.B. Activation of the bone-related Runx2/Cbfa1 promoter in mesenchymal condensations and developing chondrocytes of the axial skeleton. Mech Dev. 2002;114:167–170. doi: 10.1016/s0925-4773(02)00050-3. [DOI] [PubMed] [Google Scholar]
- 47.Stricker S., Fundele R., Vortkamp A., Mundlos S. Role of Runx genes in chondrocyte differentiation. Dev Biol. 2002;245:95–108. doi: 10.1006/dbio.2002.0640. [DOI] [PubMed] [Google Scholar]
- 48.Kim E.J., Cho S.W., Shin J.O., Lee M.J., Kim K.S., Jung H.S. Ihh and Runx2/Runx3 signaling interact to coordinate early chondrogenesis: a mouse model. PLoS One. 2013;8:e55296. doi: 10.1371/journal.pone.0055296. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Huey D.J., Hu J.C., Athanasiou K.A. Unlike bone, cartilage regeneration remains elusive. Science. 2012;338:917–921. doi: 10.1126/science.1222454. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Shen J., LaChaud G., Khadarian K., Shrestha S., Zhang X.L., Soo C., Ting K., Dry S.M., James A.W. NELL-1 expression in benign and malignant bone tumors. Biochem Biophys Res Commun. 2015;460:368–374. doi: 10.1016/j.bbrc.2015.03.040. [DOI] [PubMed] [Google Scholar]
- 51.Shen J., LaChaud G., Shrestha S., Asatrian G., Zhang X.L., Dry S.M., Soo C., Ting K., James A.W. NELL-1 expression in tumors of cartilage. J Orthop. 2015;12:S223–S229. doi: 10.1016/j.jor.2015.10.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Chen W., Zhang X., Siu R.K., Chen F., Shen J., Zara J.N., Culiat C.T., Tetradis S., Ting K., Soo C. Nfatc2 is a primary response gene of Nell-1 regulating chondrogenesis in ATDC5 cells. J Bone Miner Res. 2011;26:1230–1241. doi: 10.1002/jbmr.314. [DOI] [PMC free article] [PubMed] [Google Scholar]