Abstract
To gain insight into the cellular and molecular interactions mediating the desmoplastic reaction and aggressive malignancy of mass-forming intrahepatic cholangiocarcinoma (ICC), we modeled ICC desmoplasia and progression in vitro. A unique three-dimensional (3D) organotypic culture model was established; within a dilute collagen–type I hydrogel, a novel clonal strain of rat cancer-associated myofibroblasts (TDFSM) was co-cultured with a pure rat cholangiocarcinoma cell strain (TDECC) derived from the same ICC type as TDFSM. This 3D organotypic culture model reproduced key features of desmoplastic reaction that closely mimicked those of the in situ tumor, as well as promoted cholangiocarcinoma cell growth and progression. Our results supported a resident liver mesenchymal cell origin of the TDFSM cells, which were not neoplastically transformed. Notably, 3D co-culturing of TDECC cells with TDFSM cells provoked the formation of a dense fibrocollagenous stroma in vitro that was associated with significant increases in both proliferative TDFSM myofibroblastic cells and TDECC cholangiocarcinoma cells accumulating within the gel matrix. This dramatic desmoplastic ICC-like phenotype, which was not observed in the TDECC or TDFSM controls, was highly dependent on transforming growth factor (TGF)-β, but not promoted by TGF-α. However, TGF-α was determined to be a key factor for promoting cholangiocarcinoma cell anaplasia, hyperproliferation, and higher malignant grading in this 3D culture model of desmoplastic ICC.
Next to hepatocellular carcinoma (HCC), intrahepatic cholangiocarcinoma (ICC) is the second most common type of primary liver cancer. Both HCC and ICC are aggressive and most often fatal malignant neoplasms that usually present at advanced stages with limited treatment options for achieving appreciably improved patient survival outcomes. Unlike conventional HCC, mass-forming ICC typically exhibits a prominent desmoplastic reaction characterized by the formation of a dense collagen-enriched tumor stroma containing α-smooth muscle actin–positive (α-SMA+) cancer-associated myofibroblasts (CAFs) that are often abundant in number1, 2 and reported to correlate with poorer survival outcomes in ICC patients after surgical resection.3, 4 However, our current understanding of the complex cellular and molecular events provoking the desmoplastic response in ICC, as well as its clinical significance as a modifier of cholangiocarcinoma cell behavior and as a potential therapeutic target, are only just beginning to be elucidated.1, 2 Furthermore, despite increasing evidence suggesting that the desmoplastic stroma of ICC does not simply function as a static host-derived connective tissue barrier to malignant progression, but rather represents an evolving and collaborative microenvironment fostering cancer cell-stromal interactions that promote malignant cell growth, invasiveness, and chemoresistance,1, 2, 5 there have been limited efforts to specifically investigate the cellular and molecular interactions between stromal cells and cancer cells that induce and regulate the desmoplastic microenvironment of ICC.
The isolation and comprehensive biological, cellular, and molecular characterization of purified and stable CAF populations from ICC are important prerequisites for establishing relevant cellular models useful for investigating the complex stromal cell–cancer cell interactions provoking the desmoplastic reaction, promoting malignant cell growth and progression, and potentially identifying novel molecular therapeutic targets in ICC. Currently, there have been only few published studies on the isolation and culturing of CAFs from either human3, 6, 7, 8 or rat cholangiocarcinoma tissue.9, 10 These studies in large part involved only limited characterizations of the CAF isolates in terms of their phenotypic, genotypic, molecular, and functional properties, and to our knowledge published results from comprehensive gene expression profiling of fibroblastic cells derived from cholangiocarcinomas were only described by Utispan et al6 for human and by our laboratory for rat.9, 10
Previous studies aimed at investigating mesenchymal cell–cholangiocarcinoma cell interactions were also largely performed using two-dimensional (2D) co-culture systems of long-standing human or rat cholangiocarcinoma cell lines in vitro with either immortalized activated LX-2 hepatic stellate cells,11 and more recently immortalized portal fibroblast cell lines,12 myofibroblastic human primary hepatic stellate cells,11 primary human cholangiocarcinoma derived CAFs,3 conditioned medium from cultured CAFs from human cholangiocarcinoma,3 or cultured medium from primary hepatic myofibroblasts derived from liver samples obtained from patients undergoing partial hepatectomy for metastatic colon cancer.13 Although these 2D co-culture systems served as useful experimental models for addressing the specific functional aims of the studies in which they were used, they do not reproduce in vitro the dense collagen-enriched matrix characteristic of desmoplastic ICC tissue in situ. Even more limiting is the fact that they were not conducted with cholangiocarcinoma cells and CAFs derived from the same tumor type, but as noted above, typically used established long-standing cholangiocarcinoma cell lines of various biliary tumor origins, which were combined with stromal fibroblastic cells of different tumor origins as the cholangiocarcinoma cells or obtained from non-neoplastic liver.
Unlike 2D culture systems, three-dimensional (3D) organotypic culture models have the potential to more accurately reproduce the complex stromal cell–cancer cell and cancer cell–extracellular matrix interactions of native cancer tissue, thereby mimicking in a more realistic manner the features of solid tumors in situ.14, 15 In this context, we have previously reported on the development of a 3D organotypic culture model of rat cholangiocarcinoma that was established by co-culturing in a dilute collagen type I hydrogel a cholangiocarcinoma cell strain with that of CAFs, each of which were derived from an orthotopic well-differentiated desmoplastic cholangiocarcinoma formed in syngeneic rat liver after bile duct inoculation of spontaneously transformed tumorigenic rat cholangiocytes.10 In this initial study, we first demonstrated that when co-cultured over a 6- to 10-day period in medium containing 10% fetal bovine serum (FBS), increased numbers of plated CAFs interacted proportionally with cholangiocarcinoma cells initially plated at a fixed cell density to significantly enhance the formation of cholangiocarcinoma cell spheroid/ductal-like structures within the gel matrix, as well as to promote a more progressed cholangiocarcinoma cell phenotype when compared with mono-cell cultures of cholangiocarcinoma cells only. We further showed that 3D culturing of the cholangiocarcinoma cells in the presence of the CAFs (or conditioned medium) induced significantly up-regulated expressions of genes (eg, Muc1, Cxcr4, Hgf) known to be associated with ICC progression and/or invasion. Moreover, we provided precursory histochemical evidence of prominently enhanced fibrous collagen overproduction and secretion within the hydrogel matrix of the co-cultures over that produced by the cholangiocarcinoma cells alone in 3D mono-cell culture, suggesting an in vitro model of desmoplastic cholangiocarcinoma.10
Herein, we have greatly expanded on our initial efforts to establish an in vitro model of desmoplastic cholangiocarcinoma that closely mimics key cellular and stromal features of mass-forming ICC in situ. Specifically, we have rigorously characterized a novel CAF cell strain, which we designated as TDFSM based on its strong constitutive expression of α-SMA, as well as a pure cholangiocarcinoma cell strain (TDECC) in terms of their unique biological, karyotypic, phenotypic, molecular, and functional properties. Both the TDFSM CAF and TDECC cancer cell strains were derived as single-cell clones from the same parent organotypic desmoplastic cholangiocarcinoma (BDEspT) described previously in our initial article.10 Quantitative methods were used to assess TDFSM cells for their ability to provoke a strong desmoplastic-like reaction when interacted in 3D co-culture with TDECC cells. We further used global gene expression profiling together with immunohistochemistry and/or Western blot validation for select gene products in an effort to delineate the cellular origins (cholangiocarcinoma cell or CAF) of overexpressed genes characterizing the parent tumor from pair-matched normal liver. The tumor-derived CAF and cholangiocarcinoma cell strains were also evaluated for their differential expression of the matricellular protein periostin (Postn) and the tumor differentiation antigen mesothelin (Msln), both of which have been reported to be overexpressed in human ICC and each having been suggested as being potential prognostic indicators and/or therapeutic targets for this hepatobiliary cancer.6, 16, 17, 18 In addition, we analyzed several clinically relevant targeted agents, including the dual ErbB1/ErbB2 tyrosine kinase inhibitor lapatinib,19 the Sonic Hedgehog antagonist vismodegib (GDC-0449),20 the multikinase inhibitor sorafenib,21 the transforming growth factor-β (TGF-β) receptor 1 kinase inhibitor LY2157299 (galunisertib),22 and the TGF-β signaling inhibitor halofuginone23 for their ability to selectively attenuate the desmoplastic-like reaction prominently induced in vitro under defined conditions in our 3D co-culture model. Moreover, we now describe findings aimed at showing that 3D co-culturing of TDECC cholangiocarcinoma cells with TDFSM CAFs generates an interactive milieu augmenting the overproduction of TGF-β, as well as significantly increasing proliferating cell nuclear antigen (PCNA) expression. Last, using gene set enrichment analysis (GSEA), we have demonstrated components of the TGF-β signaling pathway to be markedly expressed in the parent rat cholangiocarcinoma from which the TDECC and TDFSM cell strains were derived, as well as provided novel experimental evidence supporting TGF-β as being a key mediator of the desmoplastic-like reaction induced in our 3D cholangiocarcinoma culture model, and furthermore, that TGF-α is critically important for provoking cholangiocarcinoma cell anaplasia and hyperproliferation in this 3D desmoplastic cholangiocarcinoma culture model.
Materials and Methods
Cell Strains and 3D Co-Culturing
All of the animal experiments described in this study were performed in accordance with and approved by Virginia Commonwealth University Institutional Animal Care and Use Committee. The method used to establish the TDFSM and TDECC cell strains from orthotopic rat desmoplastic cholangiocarcinoma tissue (BDEspT),24 together with the establishment of 3D organotypic co-cultures of the TDECC cholangiocarcinoma cells with TDFSM fibroblastic cells, was as previously described.10 Briefly, 3D organotypic cultures of our cholangiocarcinoma cell strain with clonal TDFSM from the same tumor type were, unless otherwise indicated, developed by co-culturing TDECC cells at an initial plating density of 2 × 105 cells and at cell viabilities of ≥90% with TDFSM cells at initial plating densities that ranged from 0 to 8 × 105 cells and with cell viabilities of ≥90% in a 2-mL gel matrix of rat tail type I collagen (BD Biosciences, Bedford, MA) at a concentration of 1 mg/mL. The cells were initially delivered aseptically and gently mixed within collagen solution (pH approximately 7.4) on ice and then allowed to gel in plastic culture dishes at 37°C for 1 hour, according to a slight modification of the manufacturer's protocol. The cultures were maintained in our standard medium composed of Dulbecco's modified Eagle's medium supplemented with 100 U/mL penicillin, 100 μg/mL streptomycin, 0.1 μmol/L insulin, and 5 μg/mL transferrin. Unless otherwise specified, the culture medium also contained 10% FBS for the first 4 or 24 hours of culturing, which was then typically reduced to 1.0% for specific experiments. Different cell strains were passaged four to eight times. Each of the cell strains described in this study tested negative for Mycoplasma using the MycoProbe Mycoplasma detection kit from R&D Systems, Inc. (Minneapolis, MN).
Bioassays and Cytogenetic Analysis
Anchorage-dependent growth curves were determined from viable cell counts of trypsinized TDECC and TDFSM cells individually cultured in standard medium with 10% FBS over a 14-day period on rat tail collagen type I–coated plastic culture dishes using the Countess automated cell counter for cell counting with trypan blue stain (Life Technologies Corp., Grand Island, NY). The respective cell strains were assayed for anchorage-independent growth in soft agar using the CytoSelect 96-well cell transformation assay (Cell Biolabs, Inc., San Diego, CA), according to a slight variation of the manufacturer's protocol. Cytogenetic analysis was performed on a total of 10 G-banded metaphase cells per cell strain by Cell Line Genetics (Madison, WI). The Promega CytoTox Non-Radioactive Cytotoxicity Assay Kit (Promega Corp., Madison, WI) was used to test for LY2157299-induced acute cytotoxicity against viable TDECC and TDFSM cells individually cultured in 96-well plastic tissue culture plates coated with rat tail collagen type I.
Tumorigenicity assays were performed by bile duct inoculation of either 10 × 106 TDECC cells or 15 × 106 TDFSM cells in 0.1 mL Hank's balanced salt solution (≥90% cell viability) into the livers of syngeneic Fischer rats, according to our original method.24 Specifically, rats under deep ketamine/xylazine anesthesia first underwent a laparotomy, permitting exposure of the left hepatic bile duct at the liver hilas. The cells were then slowly inoculated into the duct through a 26-gauge, 3/8-inch needle directed toward the left hepatic lobe. At 5 seconds after completion of the cell inoculation, the needle is carefully removed under cover of sterile gauze placed with gentle compression over the injection site for at least 30 seconds. Performed properly, no bleeding is observed and there is no evidence on the removed gauze of leakage of cell inoculum. The whole surgical procedure from the initial laparotomy to closure of the incision is typically accomplished within a 10- to 15-minute period.
Gene Expression Microarray and GSEAs
Comprehensive gene expression profiling using our previously described Affymetrix genechip analysis method9, 10 was performed on comparably passaged TDECC and TDFSM cells individually cultured to 60% to 70% confluence on collagen-coated plastic (https://www.ncbi.nlm.nih.gov/geo; accession number GSE70587) and compared to transcriptomes of parent orthotopic rat desmoplastic BDEspT (https://www.ncbi.nlm.nih.gov/geo; accession number GSE38889) versus pair-matched right hepatic lobe without cancer from the same rat liver as the tumor. To test for sets of related genes that might be systematically altered in BDEspT cholangiocarcinoma compared with pair-matched normal liver, we used the GSEA approach, as described by Subramanian et al.25 GSE data were generated using the GSEA software version 2.1.0 obtained from the Broad Institute of Massachusetts Institute of Technology and Harvard (Cambridge, MA). The gene sets database was compiled from the KEGG database. P values for the gene sets were computed by permuting the genes 1000 times in this study, and the q value for selecting significantly enriched gene sets was set at 0.1.
Targeted Agent Treatments
Targeted agents tested in our 3D cholangiocarcinoma co-culture model included the following: lapatinib (LC Laboratories, Woburn, MA), GDC-0449 (vismodegib) (Selleckchem, Houston, TX), sorafenib (LC Laboratories), LY2157299 (galunisertib) (Selleckchem), and halofuginone (Sigma-Aldrich, St. Louis, MO). Stock solutions of each of the targeted test agents listed above, with the exception of halofuginone, were prepared in dimethyl sulfoxide, and were then diluted with culture medium to achieve the desired concentration levels used to treat the cultures. Halofuginone was dissolved in 0.44 mol/L lactic acid buffer, pH 7.4, for its stock solution.
Collagen Staining, Immunofluorescence, Immunohistochemistry, and Microscopic Imaging
The picrosirius red staining kit from Polysciences, Inc. (Warrington, PA) was used, as previously described,10 to stain for fibrous collagen in 10% neutral buffered formalin-fixed, paraffin-embedded sections from the 3D gel cultures and from rat TDECC liver tumors (n = 9) and pair-matched noncancerous right liver lobes from the same adult tumor-bearing rats (n = 3), as well as in comparably fixed and processed human specimens of desmoplastic small duct ICC (n = 8) and normal adult donor liver transplant tissue (n = 3). The human specimens were from A.E.S.'s human cholangiocarcinoma archive and contained no identifiers or codes that would classify them as human subject research. Picrosirius red staining was viewed under an Olympus BX41 light microscope (OPELCO, Dulles, VA) equipped with a BX-POL polarizer.10 With this staining method under polarized light microscopy, thick fibrillar collagen stains yellow-orange and thin collagen fibers green, which are useful parameters for assessing collagen structural content, spatial orientation, and amounts.26, 27, 28
Immunofluorescence biomarker profile staining of the individual cholangiocarcinoma and fibroblastic cell strains cultured on collagen type I–coated coverslips and fixed for approximately 10 minutes with ice-cold 4% paraformaldehyde in phosphate-buffered saline, pH 7.4, was performed using the following primary antibodies: CK19 mouse monoclonal (VP-C415, b170) (Vector Laboratories, Inc., Burlingame, CA), α-SMA mouse monoclonal (M0851, 1A4) (Dako Cytomation, Inc., Carpinteria, CA), desmin polyclonal goat IgG (sc-7559, Y-20), vimentin (Vim) mouse monoclonal (sc-6260, V9), thymocyte differentiation antigen-1 (Thy-1) mouse monoclonal (sc-53116, OX7), CD34 mouse monoclonal (sc-7324, ICO115), and CD45 mouse monoclonal (sc-53047, OX30) (Santa Cruz Biotechnology, Inc., Paso Robles, CA), anti-rat elastin rabbit polyclonal IgG (CL55041AP, Lot 333K) (Cedarlane Labs, Burlington, NC), and anti-Postn rabbit polyclonal IgG (ab14041) (Abcam, Inc., Cambridge, MA). Fluorochrome conjugated secondary antibodies that were used included goat anti-rabbit IgG H&L (Alexa Fluor 488) preadsorbed (ab150081), goat anti-mouse IgG H&L (Alexa Fluor 488) preadsorbed (ab150117), and goat anti-mouse IgG H&L (Alexa Fluor 647) preadsorbed (ab150119) from Abcam, Inc. and rabbit anti-goat IgG H&L (Alexa Fluor 546) preabsorbed (A-21085) from Life Technologies Corp. Vectashield mounting medium with DAPI (Vector Laboratories, Inc.) was used to mount the coverslips with cells onto glass slides. Immunofluorescence staining was viewed under a Zeiss Axio Imager A1 microscope equipped with an Axiocam MRc color charge-coupled device camera located in the Virginia Commonwealth University Microscopy Facility. Zeiss Axiovision software version 4.8.2.0 was used to capture grayscale images at ×20, and cellSens Dimension software version 1.11 (Olympus America, Inc., Center Valley, PA) in A.E.S.'s laboratory used to pseudo-color the images. Confocal microscopic immunofluorescence imaging of the ectonucleoside triphosphate diphosphohydrolase nucleoside triphosphate diphosphohydrolase 2 (NTPDase2) expressed in mono-cell cultures of TDECC and TDFSM, respectively, was performed as previously described12 using rabbit polyclonal anti-NTPDase2 (rN2-6L, Ectonucleotidases-Ab) (1:200) as the primary antibody and Alexa 488–conjugated goat anti-rabbit IgG (1:1000) as the secondary antibody.
Immunohistochemistry for CK19, α-SMA, desmin, Postn (anti-periostin rabbit polyclonal antibody ab92460; Abcam, Inc.), Msln [anti-rat C-ERC/mesothelin (306) rabbit IgG affinity purify (code number 28001; lot number 0G-718); Clontech Laboratories, Inc., Mountain View, CA], PCNA [anti-PCNA mouse monoclonal (PC10) ab29], as well as anti–TGF-β/TGF-β1 (anti–TGF-β and anti–TGF-β1 rabbit polyclonal antibodies ab66043 and ab92486, respectively; Abcam, Inc.), was performed on formalin-fixed, paraffin-embedded sections prepared from the gel cultures, and/or from the rat and/or human cholangiocarcinoma and liver tissue specimens as previously described10, 29 using Vectastain Elite ABC immunoperoxidase kits, antigen retrieval using either heat-induced epitope retrieval in citrate buffer (pH 6.0 or 9.0) or proteolytic-induced epitope retrieval with either proteinase K or trypsin pretreatment, and diaminobenzidine staining.
Computational image analysis using CellSens Dimension 1.11 image analysis software was used to quantify picrosirius red staining intensity, reflecting fibrous collagen content, in digital images of multiple nonserial histological sections prepared from formalin-fixed, paraffin-embedded gel cultures. Image analysis was also used as previously described10 to facilitate quantifying numbers of CK19-positive cholangiocarcinoma cell spheroids/ductal-like structures and of α-SMA–positive CAFs in randomized microscopic fields of sections (10 μm thick) prepared from the gel cultures.
Western Blotting
Western blot analysis of total protein in lysates prepared in radioimmunoprecipitation assay buffer supplemented with the protease inhibitor cocktail cOmplete, Mini (catalog number 11836153001ROCHE) and Phosphatase Inhibitor Cocktail 2 (catalog number P5726) from Sigma-Aldrich, Inc., was performed as previously described16, 24, 29 on either 2D monocultures of TDECC or TDFSM cells, respectively, plated on collagen type I–coated plastic substratum, or from specified 3D organotypic gel mono- and co-cultures, using the same primary antibodies described in the preceding section of Materials and Methods for CK19, α-SMA, desmin, Thy-1, elastin, Postn, Msln, TGF-β (ab66043; Abcam, Inc.), and PCNA. In addition, caspase-3 antibody (9662), and cyclin D1 rabbit monoclonal antibody (2978) were purchased from Cell Signaling Technology (Beverly, MA). Epidermal growth factor receptor (EGFR; A10): sc-373746 and phosphorylated EGFR (p-EGFR; Tyr 1173) were from Santa Cruz Biotechnology, Inc. Differential expression of the NTPDase2 in cultured TDFSM and TDECC cells was also determined by Western blotting using rabbit polyclonal antibody BZ3-4F raised against rat NTPDase2.30 Unless otherwise indicated, the initial viable cell plating densities for the 3D gel cultures were 4 × 105 TDECC cells/gel and 1.6 × 106 TDFSM cells/gel. Protein bands were detected by chemiluminescence using species-appropriate horseradish peroxidase–conjugated secondary antibodies (catalog numbers 170-6515, 170-6516, and 172-1034) and Clarity Western ECL Substrate (170-5061) from Bio-Rad (Hercules, CA). Protein bands normalized to vinculin or actin were imaged with the Bio-Rad ChemiDoc XRS imaging system using Quantity One 1-D analysis software version 4.6.9 (Bio-Rad). Relative protein band densities from a minimum of three independent sample measurements were quantified using Bio-Rad Image Lab software version 5.2.1.
Statistical Analysis
For the microarray data analyses, background correction, normalization, and estimation of probe set expression summaries were performed, as previously described,10 using the log-scale robust multiarray analysis method of Irizarry et al.31 Statistical significance for multivariate analysis to assess probe set–specific false discovery rates was performed by estimating the q values, using the Bioconductor q-value package.10 Student's two-tailed t-test was used to determine P values (P ≤ 0.05, P ≤ 0.01, P ≤ 0.001, and P ≤ 0.0001), with P ≤ 0.05 considered significant. The minimum number of cultures analyzed per data set was three, with image analysis data generated from counts made between six and nine histological sections (3 to 10 random fields/slide) for each analyzed group of control and experimental gel cultures. On average, individual cell culture experiments were repeated two to four times. Graphic data are presented as means ± SD. The synergy ratio R, between the observed (Cobs) and expected (Cexp) was calculated as follows: R = Cobs/Cexp, with a value >1 indicating the interaction to be synergistic.32
Results
Anchorage-Dependent and Anchorage-Independent Cell Growth, Cytogenetic and Tumorigenic Properties Differentiating BDEspT Cholangiocarcinoma-Derived TDFSM and TDECC Cell Strains
As an important first step toward classifying the TDFSM and TDECC strains in terms of their unique identifying characteristics, we comparatively investigated each of these respective cholangiocarcinoma-derived cell strains for their distinctive cytogenetic features, anchorage-dependent and anchorage-independent growth rates in 2D culture, and their tumorigenic potential in vivo. When maintained in 2D monolayer culture on collagen type I–coated plastic substratum, TDECC cells exhibited an epithelial cell morphology (Supplemental Figure S1). In contrast, under these same culture conditions, TDFSM cells showed a mesenchymal cell–like morphology.
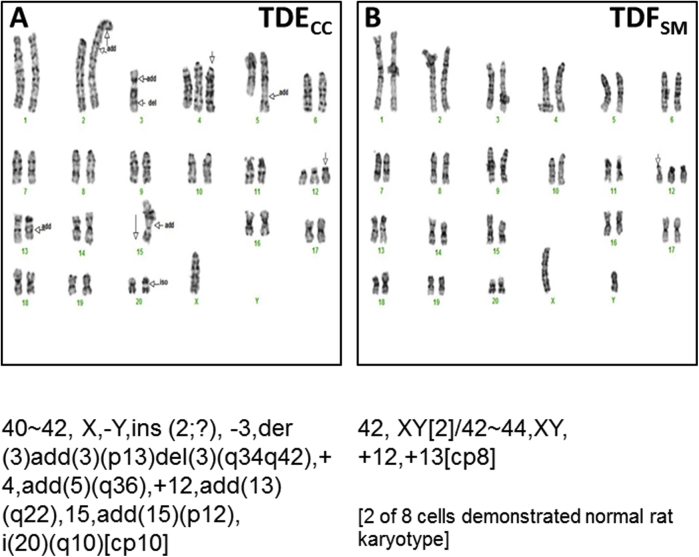
Both cell strains demonstrated abnormal, but nevertheless distinct karyotypes, and as expected, like the TDFSM cell strain, the cholangiocarcinoma cell strain had many more chromosome aberrations. The common chromosomal abnormalities characterizing each of the cholangiocarcinoma-derived cell strains are shown in Supplemental Figure S2. Interestingly, both the TDECC and TDFSM cell strains exhibited an extra chromosome 12. However, this abnormality could possibly represent a culturing effect (see Discussion), because the trisomy 12 patterns were different for each of these two distinct single cell–derived cell strains. Moreover, not all of the analyzed metaphase spreads exhibited this defect, as exemplified by our finding that two of the TDFSM spreads showed a normal rat karyotype.
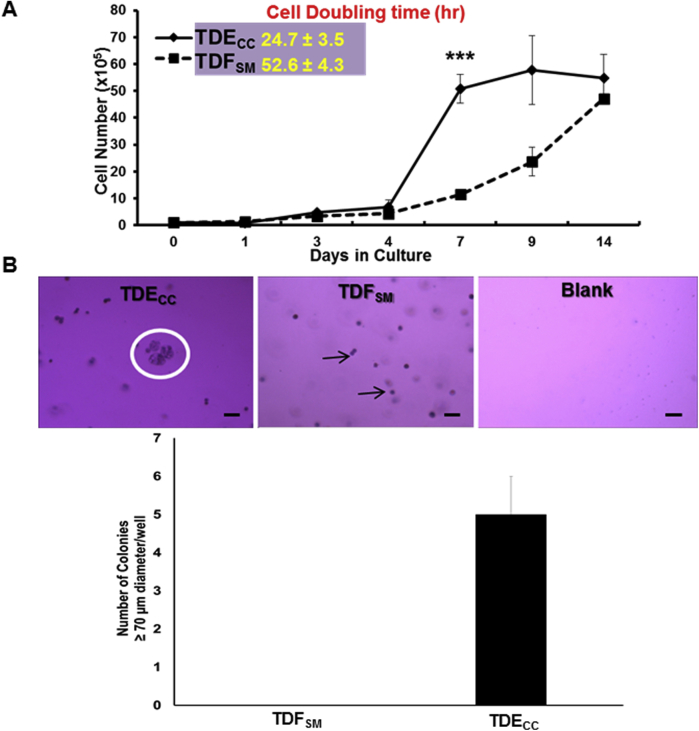
The TDECC and TDFSM cell strains each exhibited anchorage-dependent growth when cultured on collagen type I–coated plastic. However, under identical 2D culture conditions, the TDECC strain exhibited an anchorage-dependent growth rate that was significantly greater than that of the TDFSM cell strain, having a mean cell doubling time calculated to be approximately half that determined for the TDFSM cell strain (Supplemental Figure S3A). Only TDECC cells exhibited anchorage-independent growth, a hallmark of cell transformation, as monitored by soft agar colony formation (Supplemental Figure S3B).
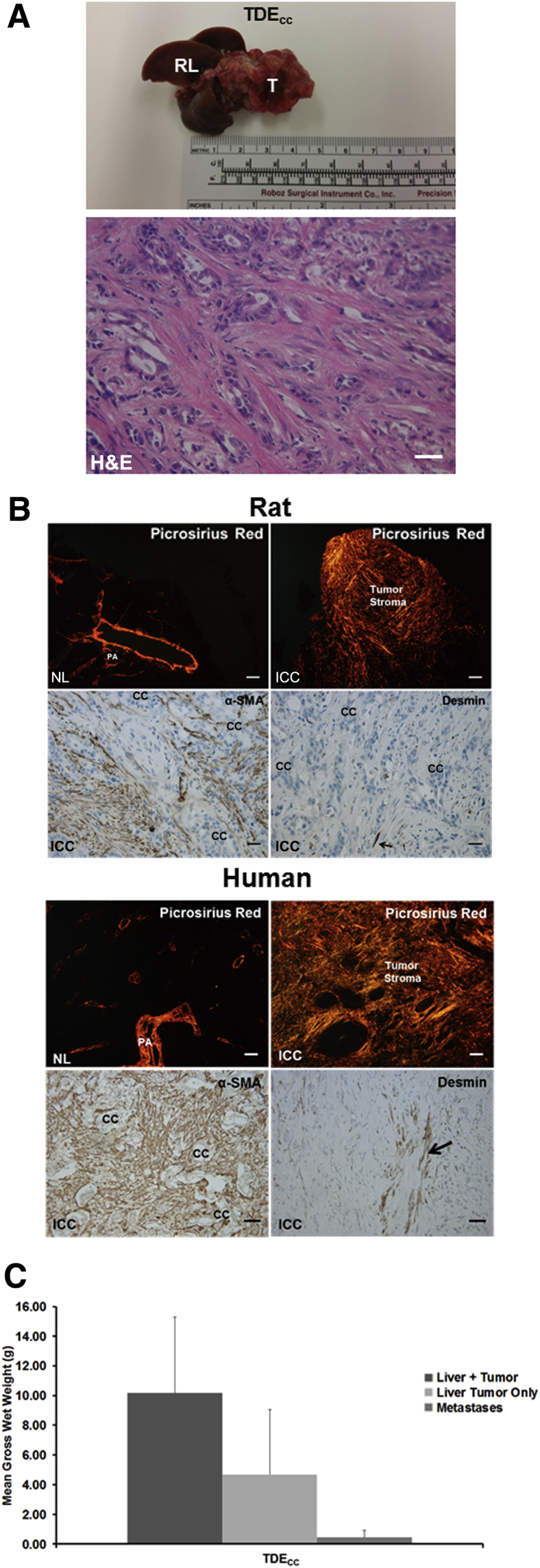
Consistent with the soft agar data, TDECC cells were highly tumorigenic, giving rise at a 100% incidence in liver to well-to-moderately differentiated desmoplastic small duct cholangiocarcinomas, together with associated gross peritoneal metastases by 53 days after their bile duct inoculation into the livers of syngeneic rats (Figure 1, A and C). No tumors formed in rats at 53 days after bile duct inoculation of TDFSM cells. Furthermore, cholangiocarcinomas that formed in rat liver after bile duct inoculation of TDECC cells—like human mass-forming desmoplastic small duct ICC—were characterized by a dense fibrous collagenous stroma intensely stained with picrosirius red, which was accompanied in both the rat and human tumors by an abundant increase in α-SMA–positive CAFs accumulated within the tumor microenvironment (Figure 1B). In comparison, positive immunostaining for desmin, commonly reported as a marker of quiescent/activated hepatic stellate cells,33, 34 was observed in only a minority of fibroblastic cells in the desmoplastic stroma of both the rat and human tumors (Figure 1B). In corresponding sections of normal liver tissue, picrosirius red staining was confined largely to the collagen of the portal areas and intrahepatic small vessel walls, with α-SMA immunostaining not detected in the hepatic parenchyma or sinusoidal areas (data not shown).
Figure 1.
A: Representative gross and microscopic pathology of a tumor formed in the liver of a young adult male Fischer 344 rat at 53 days after bile duct inoculation of 10 × 106 TDECC cholangiocarcinoma cells [hematoxylin and eosin (H&E)]. Note that the left lateral and median liver lobes have been totally replaced by tumor tissue (T) and that in this model, the right (RL), as well as caudate liver lobes are typically without evidence of gross tumor. The accompanying representative photomicrograph shows that the tumor that forms in syngeneic rat liver after bile duct inoculation of TDECC cells exhibits the histopathological features of a well-to-moderately differentiated desmoplastic cholangiocarcinoma. B: Representative photomicrographs of picrosirius red staining under polarized light for dense collagen (orange staining) and under bright-field microscopy for α-SMA versus desmin immunoreactivity (brown staining) in histological sections from rat and human ICC tissue samples. Arrows point to desmin-positive cells. C: Mean gross wet weights of cholangiocarcinomas and associated pooled peritoneal metastases formed in rats over a 23- to 53-day period after TDECC cell transplantation into liver via bile duct inoculation. Data are expressed as means ± SD. n = 5 (C). Scale bars: 50 μm (A, and B, human tumor sections immunostained for α-SMA and desmin); 200 μm (B, picrosirius red–stained sections); 20 μm (B, rat tumor α-SMA and desmin immunostaining). cc, cholangiocarcinoma cells; H&E, hematoxylin and eosin; NL, normal adult rat and human liver; PA, portal area.
TDECC and TDFSM Cell Strains Are Immunophenotypically Distinct
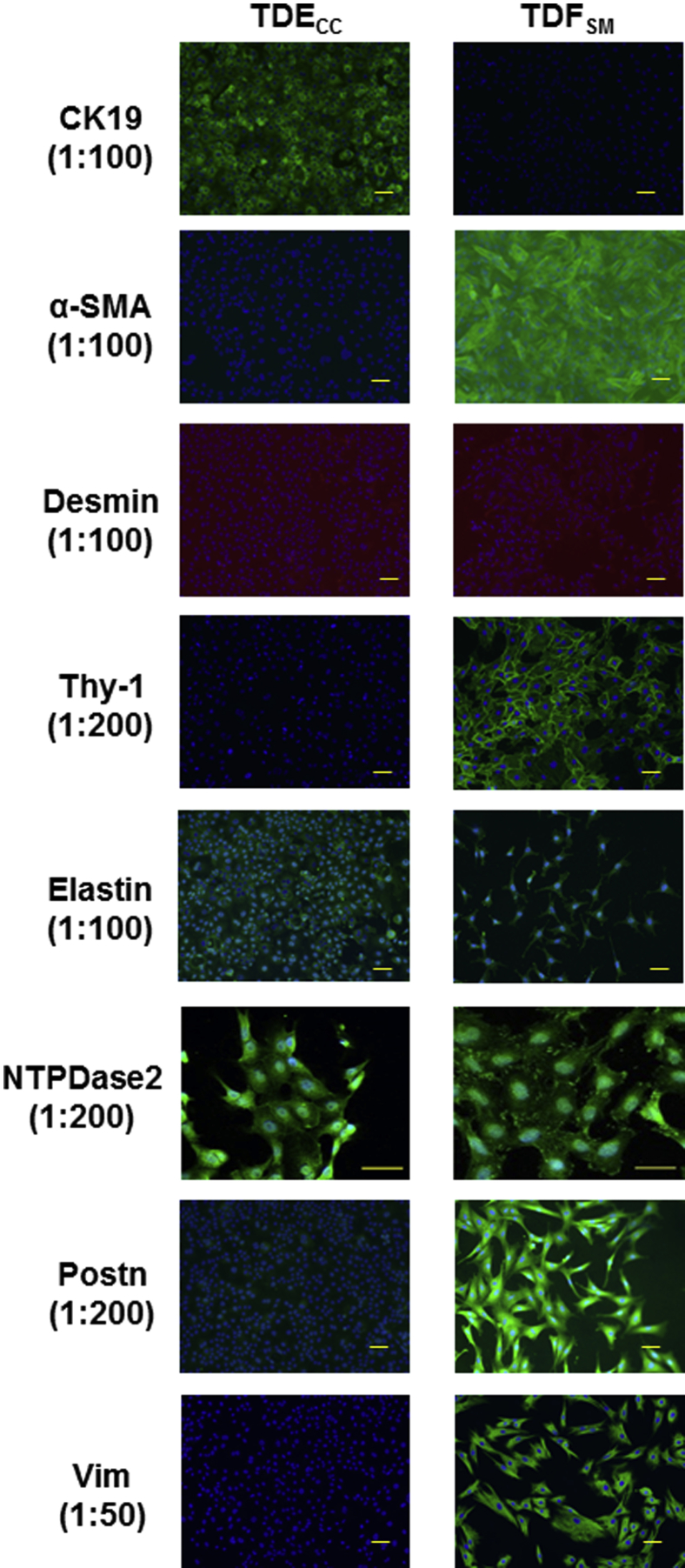
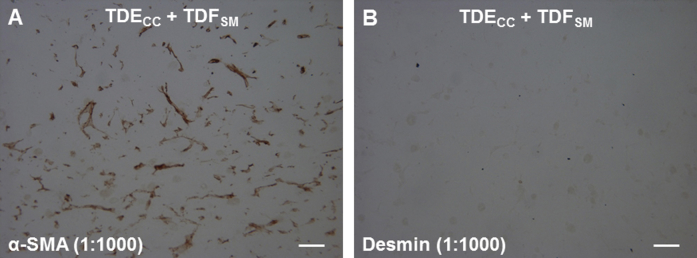
Immunofluorescence microscopy combined with quantitative Western blotting were used to profile the TDECC and TDFSM cell strains in short-term 2D culture for their respective expression of select biliary epithelial cell and fibroblastic/myofibroblastic cell markers. Only the TDECC cell strain expressed CK19, a widely used biomarker of normal cholangiocytes and cholangiocarcinoma cells in mammalian liver (Figure 2 and Figure 3A). In comparison, prominent constitutive expression of α-SMA, a hallmark feature of myofibroblasts, was a characteristic feature of the TDFSM cell strain and not detected in the cholangiocarcinoma cells comparably maintained in 2D monoculture. Neither the TDECC nor TDFSM cells were immunoreactive for desmin by either immunocytofluorescence (Figure 2) or Western blotting (data not shown) when reacted with an anti-desmin antibody, which we validated by both methods for positive immunoreactivity against a rat fibroblastic-like cell strain expressing desmin (data not shown).
Figure 2.
Differential immunophenotyping by biomarker immunofluorescence staining of TDECC cholangiocarcinoma versus TDFSM cell strains individually cultured for 24 to 48 hours on rat tail collagen type I–coated glass coverslips in standard medium containing 10% FBS. Primary antibody dilutions are shown in parentheses. In each case, corresponding controls with secondary antibody alone, omitting the primary antibody, were negative for immunofluorescence staining. Green staining: secondary antibody conjugated with Alexa Fluor 488; orange staining: secondary antibody conjugated with Alexa Fluor 546; blue nuclei: DAPI staining. Scale bars: 50 μm (except for NTPDase 2); 40 μm (NTPDase 2). α-SMA, α-smooth muscle actin; CK19, cytokeratin-19; NTPDase 2, nucleoside triphosphate diphosphohydrolase 2; Postn, periostin; Thy-1, thymocyte differentiation antigen-1; Vim, vimentin.
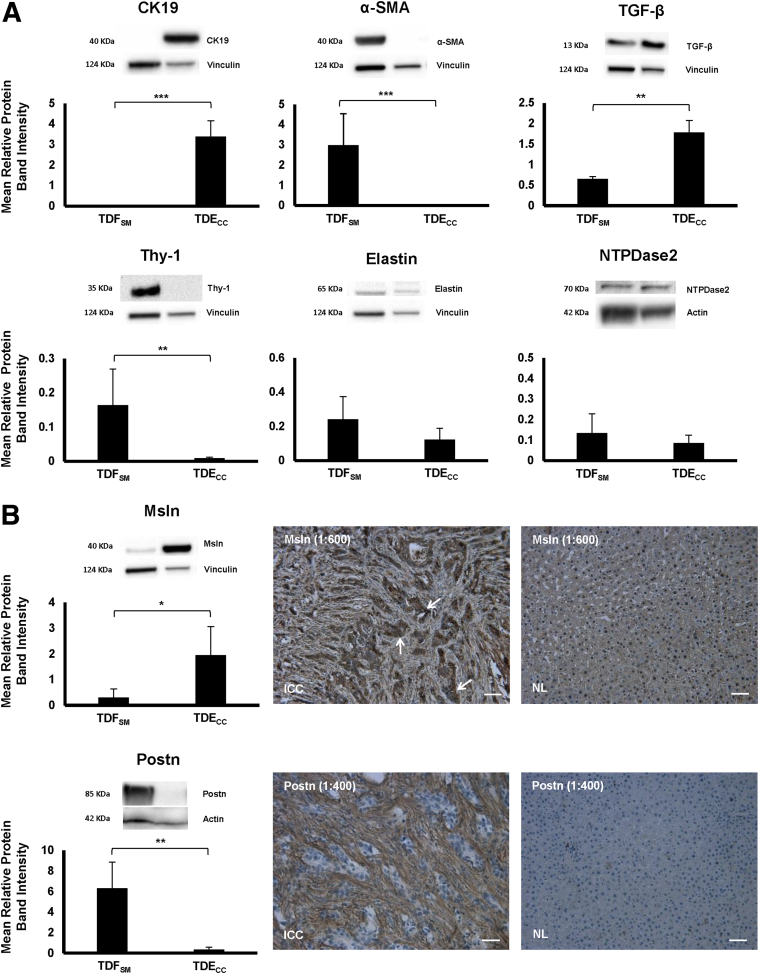
Figure 3.
A: Quantitative Western blot analysis of select protein biomarkers differentially expressed in cultured TDFSM and TDECC cell strains individually grown to 95% to 100% confluency under standard culture conditions (10% FBS supplemented medium) in 10-cm-diameter plastic culture dishes coated with rat tail collagen type I. B: In the case of the secreted protein Postn, Western blotting was performed on concentrated medium samples prepared using an Amicon Ultra-2 Centrifugal Filter Device with Ultracel-50 membrane (catalog number UFC205024; EMD Millipore Corp., Darmstadt, Germany) from monolayer cells initially grown on collagen type I–coated plastic to 60% confluence in standard medium containing 10% FBS and then maintained for an additional 48 hours in medium containing 1% FBS. In the case of transforming growth factor (TGF)-β, cell lysates were from cultures maintained for 48 hours in medium with 1.0% FBS. Blotting conditions were optimized for each protein biomarker, with primary/secondary antibody dilutions indicated as follows: cytokeratin-19 (CK19), 1:1000/1:3000; α-smooth muscle actin (α-SMA), 1:750/1:5000; desmin, 1:500/1:3000; thymocyte differentiation antigen-1 (Thy-1), 1:100/1:3000; elastin, 1:750/1:5000; nucleoside triphosphate diphosphohydrolase 2 (NTPDase2), 1:200/1:5000; mesothelin (Msln), 1:1000/1:3000; periostin (Postn), 1:1000/1:3000; TGF-β, 1:500/1:3000. Protein band intensity values normalized to actin or to vinculin are from measurements made on three separate Western blots per biomarker protein. B: Representative photomicrographs demonstrate strong immunoreactivity for Msln in the cholangiocarcinoma cells (arrows) and for Postn specifically in the stroma of desmoplastic cholangiocarcinoma (ICC) formed in rat liver orthotopically transplanted with TDECC cells. Corresponding pair-matched right liver lobe tissue exhibit a normal liver histology (NL) sampled from the same tumor bearing rats and do not immunostain for either Msln or Postn. Histological sections were counterstained with hematoxylin. Primary antibody dilutions shown in parentheses. Data are expressed as means ± SD (A and B). ∗P < 0.05, ∗∗P < 0.01, and ∗∗∗P < 0.001. Scale bars = 50 μm.
Cell surface Thy-1, which has been reported to be expressed in myofibroblasts in periportal areas of rat liver, but not found in hepatic stellate cells after carbon tetrachloride–induced liver injury,35 was most strongly expressed in the cultured TDFSM cells and essentially not detected in the TDECC cells. Elastin and NTPDase2, whose positive expression has been shown to be a phenotypic feature of rat portal fibroblasts/myofibroblasts,12, 30, 36 were detected in both BDEspT-derived cell strains, being detected at similarly expressed levels in each of the cultured CAF and cholangiocarcinoma cell types.
Msln, which has been demonstrated to be overexpressed in the malignant cholangiocytes of human ICCs,17, 18 and further reported to be expressed in activated murine portal fibroblasts,36 was found by us to be most strongly expressed in the TDECC cholangiocarcinoma cell strain, but also detected by Western blotting at appreciably lower levels in the TDFSM cell strain than in the TDECC strain (Figure 3B). Conversely, Postn, which is highly expressed in α-SMA–positive CAFs and hypersecreted into the desmoplastic extracellular matrix of human and rat desmoplastic ICCs, but either not or only weakly expressed in the cholangiocarcinoma cells in these tumors6, 9, 10, 37 (Figures 2 and 3B), was determined to be significantly increased in amount in the culture medium from 2D cultures of TDFSM cells over that of comparably cultured TDECC cell cultures, where this matricellular protein was barely detected by Western blotting in the medium from the cholangiocarcinoma cell cultures (Figure 3B). Although both tumor-derived cell strains exhibited protein band densities for the cleaved 13-kDa form of TGF-β, the cultured TDECC cholangiocarcinoma cell strain expressed this mature form of TGF-β at a significantly higher mean level that was more than twice the mean level of that determined by Western blot analysis for the TDFSM cell strain (Figure 3A).
This orthotopic rat cholangiocarcinoma model closely reproduces the immunostaining patterns of Msln overexpression in the cholangiocarcinoma cells, and conversely, Postn immunoreactivity is a predominant feature of the desmoplastic stroma and not detected in the cholangiocarcinoma cells, as also previously described for human ICCs (Figure 3B).6
Unlike the TDFSM cell strain characterized in this study, the TDECC cholangiocarcinoma cell strain was not immunoreactive for the mesenchymal cell intermediate filament protein Vim (Figure 2), which is often used as a biomarker for epithelial-mesenchymal transition. Neither the TDECC nor TDFSM cell strains were immunoreactive for CD45, characteristically expressed in bone marrow–derived fibrocytes,38 or for CD34, a hematopoeitic stem cell marker,39 also expressed in bone marrow–derived fibrocytes40 (Supplemental Figure S4).
Gene Expression Profiles of TDECC and TDFSM Cells Compared with Those of Parent BDEspT and Pair-Matched Noncancerous Liver
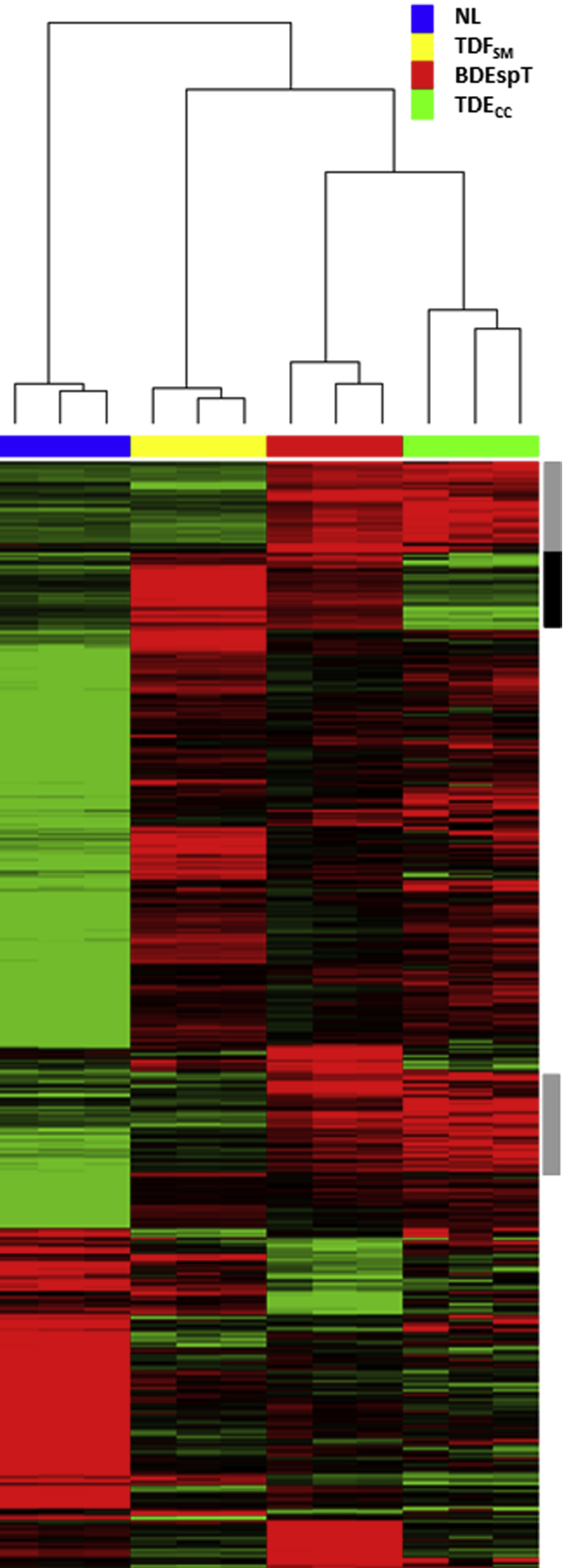
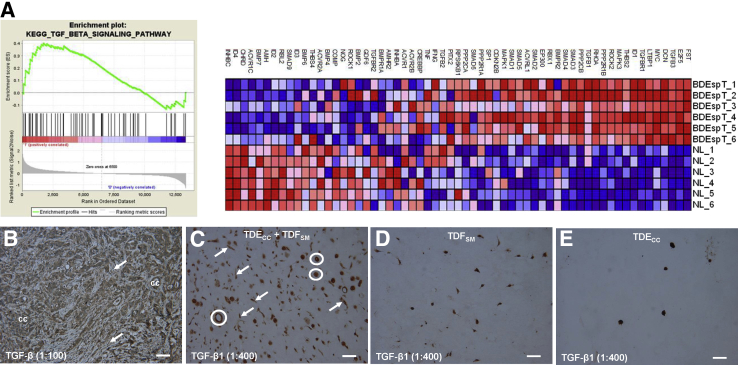
Because the BDEspT-derived cell strains analyzed in this study were each established from single-cell clones, it was important to determine whether the signature gene expression patterns, notably those characterizing the TDECC and α-SMA–positive TDFSM cell strains, reflected more generally the overall pattern of genes overexpressed in the parent tumor when compared with the gene expression profile of pair-matched histologically normal rat liver samples obtained from the same rats as the tumors. The heat map shown in Figure 4, based on an analysis of 1252 significantly altered probe sets, demonstrates a close agreement between defined regions of overexpressed genes exhibited by the TDECC cells when matched against genes overexpressed in BDEspT cholangiocarcinoma relative to normal liver. TDFSM cells also contributed to the overall pattern of overexpressed genes exhibited by the parent tumor.
Figure 4.
Two-dimensional hierarchical clustering of cultured cell and tissue samples using Pearson's (centered) correlation and average linkage, based on 1252 probe sets that were significantly different (q ≤ 0.05) between TDECC and TDFSM cell strains used to cluster all of the individual samples indicated by the blue, yellow, red, and green bars. Analyzed cell samples were from 2D monolayer cultures of comparably passaged TDECC and TDFSM cells, respectively, harvested when at 60% and 70% confluence on collagen type I–coated plastic. BDEsp-T refers to parent tumor tissue from which the TDECC cholangiocarcinoma and TDFSM myofibroblastic cell strains were derived, whereas NL refers to corresponding pair-matched right liver lobe tissue samples without histological evidence of cholangiocarcinoma. Each of the colored rows in the heat map shows the relative expression for that specific gene in the separate specimen samples (columns), where the relative extent of gene overexpression is indicated in red and that of gene underexpression is shown in green when compared to the median intensity across all samples. The gray sidebars reflect common probe sets overexpressed in both the TDECC cell strain and BDEsp-T tissue samples relative to TDFSM and NL; the black side bar emphasizes genes commonly overexpressed in the TDFSM cells and BDEsp-T tumor compared with TDECC and NL.
Results from our comparative transcriptomic analysis of signature genes differentiating the TDFSM myofibroblasts from the TDECC cholangiocarcinoma cells are shown in Table 1. From this analysis, which also included BDEspT and pair-matched normal liver, it is evident that the pattern of overexpressed genes in the TDFSM cells is consistent with their having a mesenchymal (CAF) tumor stromal mRNA expression signature (eg, Col1a2, Col1a1, Postn, Tnc, Vim, fibronectin1, Acta2), whereas that of the TDECC cell strain reflects a cholangiocarcinoma cell mRNA expression signature (eg, Krt19, Cdh1, Cd24, Tacstd1, Muc1). The mRNA expression patterns for Krt19, Acta2 (α-SMA), Thy-1, and Msln also closely agreed with our immunostaining and Western blot findings for these specific markers (Figures 2 and 3). Of further note, a number of the signature genes overexpressed in the TDFSM cell strain relative to TDECC cells, including those transcribing gremlin 1, Thy-1, fibronectin 1, fibulin 2, cofilin 1 (nonmuscle), and collagen type XV, α1, have also been shown to be overexpressed in rodent portal myofibroblasts relative to hepatic stellate cell myofibroblasts.35, 36, 41, 42
Table 1.
Differential Expression of Select Biomarker Genes Distinguishing TDFSM from TDECC Cholangiocarcinoma Cells
| Gene name | Gene symbol | NL∗ | BDEspT† | TDECC | TDFSM | Fold change‡ | q Value |
|---|---|---|---|---|---|---|---|
| Collagen, type I, α 2 | Col1a2 | 6.88 ± 0.63 | 12.63 ± 0.40 | 3.15 ± 0.13 | 14.25 ± 0.02 | 2200.5 | 1.73×10-5 |
| Periostin, osteoblast specific factor | Postn | 4.04 ± 0.17 | 12.66 ± 0.57 | 3.18 ± 0.24 | 13.59 ± 0.25 | 1358.7 | 4.48×10-5 |
| Protease, serine, 23 | Prss23 | 7.97 ± 0.13 | 8.71 ± 0.20 | 3.69 ± 0.28 | 13.46 ± 0.10 | 875.4 | 3.93×10-5 |
| Fibronectin 1 | Fn1 | 13.50 ± 0.04 | 12.69 ± 0.23 | 4.01 ± 0.47 | 13.68 ± 0.08 | 814.3 | 8.47×10-5 |
| Serine (or cysteine) peptidase inhibitor, clade H, member 1 | Serpinh1 | 8.39 ± 0.14 | 11.33 ± 0.10 | 4.25 ± 0.24 | 13.52 ± 0.10 | 618.0 | 3.25×10-5 |
| Collagen, type I, α 1 | Col1a1 | 6.05 ± 0.33 | 12.80 ± 0.19 | 4.74 ± 0.49 | 13.83 ± 0.10 | 541.3 | 1.06×10-4 |
| Gremlin 1 | Grem1 | 3.75 ± 0.08 | 6.25 ± 1.82 | 3.34 ± 0.26 | 12.25 ± 0.11 | 482.1 | 4.33×10-5 |
| Tenascin C | Tnc | 3.81 ± 0.21 | 9.95 ± 0.64 | 3.38 ± 0.47 | 12.09 ± 0.17 | 419.6 | 1.12×10-4 |
| Insulin-like growth factor binding protein 7 | Igfbp7 | 10.90 ± 0.17 | 11.98 ± 0.21 | 4.07 ± 0.30 | 12.66 ± 0.07 | 384.6 | 4.62×10-5 |
| Secreted acidic cysteine-rich glycoprotein | Sparc | 8.11 ± 0.11 | 11.02 ± 0.12 | 4.53 ± 0.41 | 12.47 ± 0.06 | 245.3 | 9.03×10-5 |
| Chemokine (C-X-C motif) ligand 12 | Cxcl12 | 11.72 ± 0.16 | 8.28 ± 1.33 | 3.82 ± 0.08 | 11.55 ± 0.06 | 212.0 | 1.73×10-5 |
| Thymus cell antigen 1, θ | Thy1 | 5.06 ± 0.09 | 7.86 ± 0.24 | 4.18 ± 0.26 | 11.75 ± 0.11 | 189.8 | 4.83×10-5 |
| Vimentin | Vim | 8.91 ± 0.30 | 12.63 ± 0.06 | 7.94 ± 0.54 | 14.75 ± 0.01 | 112.1 | 2.05×10-4 |
| Smooth muscle α-actin | Acta2 (α-SMA) | 7.40 ± 0.64 | 10.67 ± 0.39 | 6.61 ± 0.95 | 11.60 ± 0.07 | 31.8 | 1.54×10-3 |
| Matrix metallopeptidase 2 | Mmp2 | 6.42 ± 0.21 | 10.17 ± 0.51 | 4.57 ± 0.18 | 9.31 ± 0.02 | 26.7 | 5.39×10-5 |
| Fibulin 2 | Fbln2 | 6.18 ± 0.24 | 7.64 ± 0.63 | 6.05 ± 0.27 | 10.07 ± 0.10 | 16.2 | 1.63×10-4 |
| Cofilin 1 | Cfl1 | 7.57 ± 0.22 | 8.75 ± 0.17 | 8.65 ± 0.06 | 11.17 ± 0.18 | 5.7 | 1.72×10-4 |
| Collagen, type XV, α 1 | Col15a1 | 7.14 ± 0.22 | 10.34 ± 0.83 | 6.90 ± 0.22 | 8.63 ± 0.05 | 3.3 | 6.07×10-4 |
| Desmin | Des | 6.71 ± 0.04 | 8.11 ± 0.40 | 5.72 ± 0.09 | 6.35 ± 0.05 | 1.5 | 1.06×10-3 |
| Oncomodulin | Ocm | 4.39 ± 0.28 | 12.95 ± 0.17 | 13.12 ± 0.22 | 9.28 ± 0.10 | −14.4 | 1.35×10-4 |
| Mucin 1, transmembrane | Muc1 | 5.96 ± 0.25 | 9.55 ± 0.95 | 9.62 ± 0.95 | 5.42 ± 0.11 | −18.4 | 2.30×10-3 |
| Mesothelin | Msln | 6.19 ± 0.26 | 11.22 ± 0.53 | 12.84 ± 0.31 | 5.83 ± 0.17 | −129.2 | 8.56×10-5 |
| Tumor-associated calcium signal transducer 1 (Epcam) | Tacstd1 | 6.96 ± 0.27 | 13.43 ± 0.30 | 12.88 ± 0.31 | 3.20 ± 0.12 | −821.4 | 4.50×10-5 |
| CD24 antigen | Cd24 | 5.08 ± 0.07 | 12.28 ± 0.53 | 12.56 ± 0.60 | 3.58 ± 0.03 | −503.5 | 1.47×10-4 |
| Cadherin 1 | Cdh1 | 10.00 ± 0.33 | 12.08 ± 0.34 | 12.77 ± 0.35 | 3.48 ± 0.12 | −623.2 | 5.55×10-5 |
| Keratin 19 | Krt19 | 5.45 ± 0.22 | 13.88 ± 0.36 | 14.34 ± 0.19 | 3.58 ± 0.18 | −1731.6 | 2.71×10-5 |
Data are expressed as means ± SEM unless otherwise indicated. n = 3 samples analyzed per individual specimen group.
TDECC, cholangiocarcinoma cells; TDFSM, cancer-associated myofibroblasts.
Normal liver (NL) is represented by cholangiocarcinoma-free right liver lobe tissue pair-matched with BDEspT from the same animal.
BDEspT, orthotopic rat cholangiocarcinoma from which TDFSM and TDECC cell strains were derived.
Fold change of gene expression in TDFSM versus TDECC.
Modeling the Desmoplastic Reaction of ICC in 3D Organotypic Culture
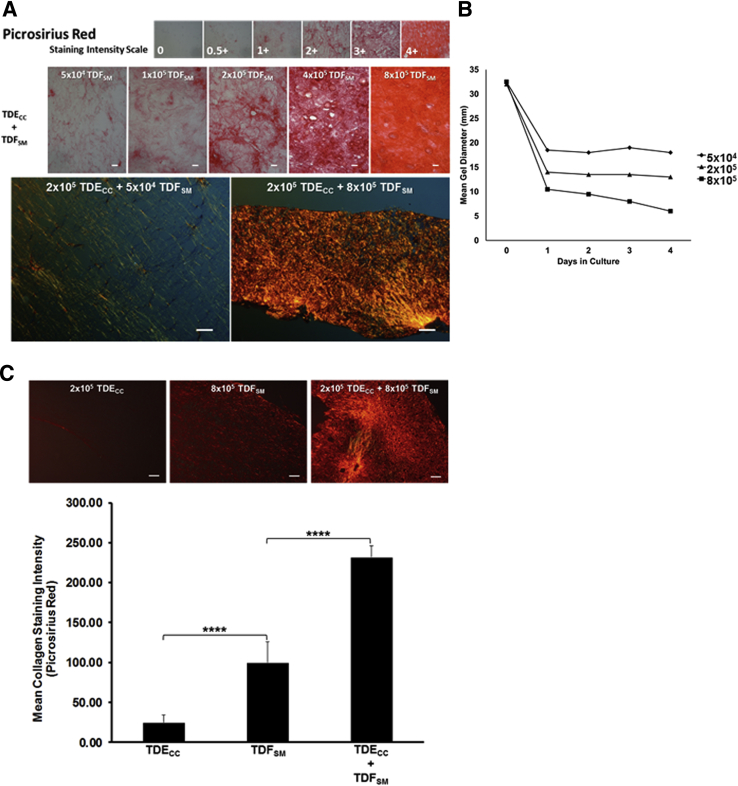
We were able to consistently reproduce key features of the desmoplastic stroma of ICC in vitro (namely, the induction of a dense fibrous collagen stroma in association with a marked increase in α-SMA–positive myofibroblasts within the cancer cell microenvironment) by co-culturing within a dilute rat tail type I collagen gel matrix TDECC cholangiocarcinoma cells at an initial fixed plating density with increasing numbers of TDFSM cells (Figures 5 and 6). Herein, we could demonstrate a progressive increase in picrosirius red staining intensity (Figure 5A) together with an accompanying proportional and relatively rapid decrease in gel shrinkage, indicative of matrix contraction (Figure 5B), as being a function of an increasing number of viable TDFSM cells when initially mixed with 2 × 105 viable TDECC cells within the 3D co-culture gel matrix.
Figure 5.
A: Photomicrographs depicting the graded increase in picrosirius red staining for collagen seen under bright-field microscopy (red staining) in representative histological sections from 3D organotypic gel cultures in which TDECC cholangiocarcinoma cells (initially plated at 2 × 105 viable cells) were co-cultured with TDFSM myofibroblastic cells at increasing initial viable cell plating densities ranging from 5 × 104 to 8 × 105 cells/gel. Note that the picrosirius staining reaction is proportionally enhanced in sections prepared from gel cultures in which the TDECC cells were co-cultured with TDFSM cells as a function of increasing initial cell plating densities. When viewed microscopically under polarized light, the obvious differences in the intensities of the picrosirius red staining reaction for dense collagen fiber production and deposition into the gel matrix that distinguishes TDECC + TDFSM co-cultures at the low versus the high TDFSM proportions is clearly visualized. B: Graphic representation further showing gel matrix shrinkage (contraction) to also be a function of increasing initial TDFSM myofibroblastic cell proportion when co-cultured with a constant number of TDECC cholangiocarcinoma cells at an initial cell plating density of 2 × 105 cells/gel. Each value indicates the mean gel diameter determined from individual measurements made on two separate gel cultures per time point. C: Quantitative imaging data demonstrating picrosirius red staining intensity under polarized light for fibrous collagen to be most prominently increased in 3D gel co-cultures of TDECC + TDFSM cells over staining intensity values measured in histological sections from TDFSM, and TDECC mono-cell cultures, respectively. n = 3 cultures analyzed per data point, with staining intensity measurements made on 3 random sections/culture. ∗∗∗∗P < 0.0001. Scale bars: 20 μm (A, top row); 100 μm (A, bottom row, and C).
Figure 6.
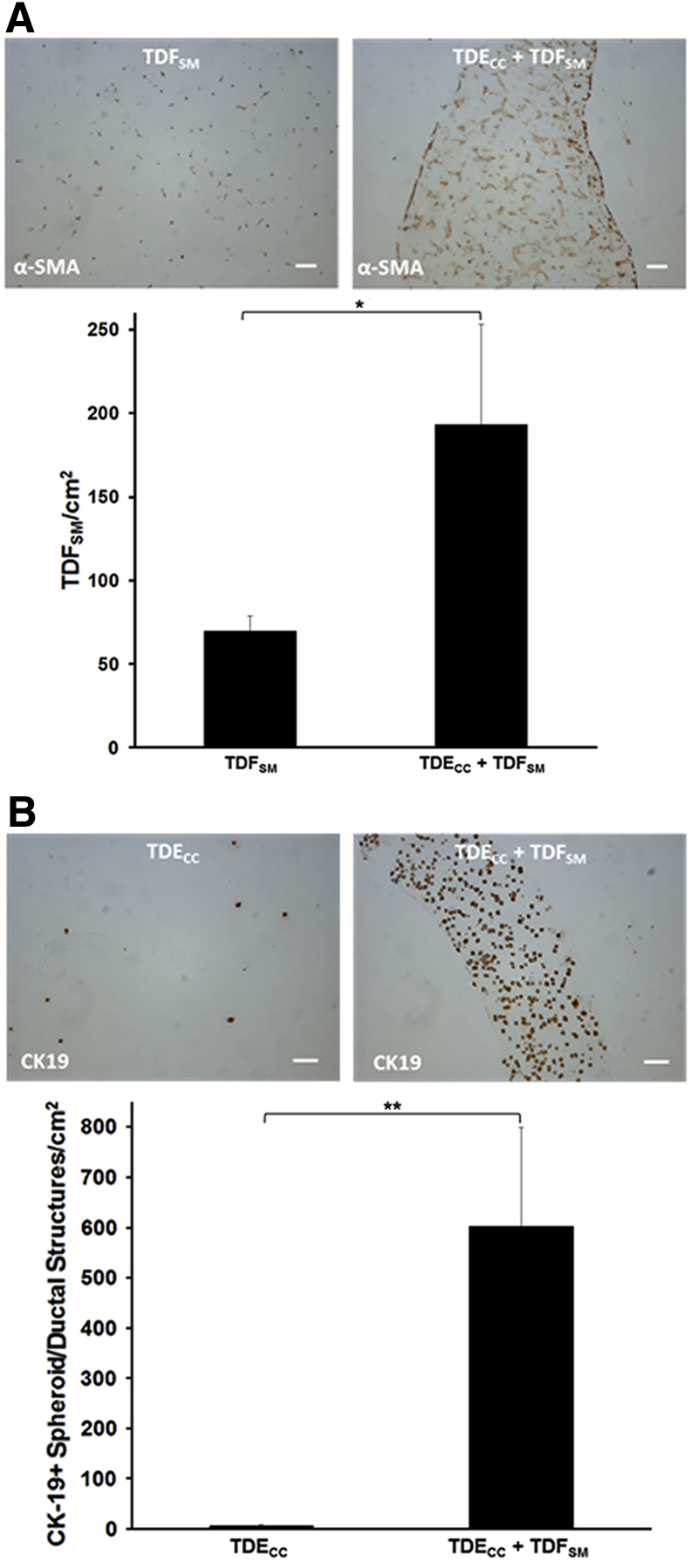
A: 3D gel co-culturing of TDECC with TDFSM cells over a 4-day period significantly increases the accumulation of α-SMA–positive fibroblastic cells within the gel matrix over those determined over the same time period for TDFSM mono-cell cultures. B: 3D gel co-culturing of TDFSM cells with TDECC cells significantly increases the number of CK19-positive cholangiocarcinoma structures/cm2 section area detected in random histological sections from these gel cultures over those determined for TDECC cells cultured alone. The initial viable cell plating density/gel for TDECC cells was 2 × 105 and for TDFSM, 8 × 105. The FBS concentration in each culture was reduced from 10% to 1.0% at 24 hours after the initial cell plating and the cultures analyzed 72 hours later. Data are expressed as means ± SD (A and B). n = 3 cultures analyzed per data point, with measurements made on random fields of 6 sections/culture (A and B). ∗P < 0.05, ∗∗P < 0.01. Scale bars = 100 μm (A and B).
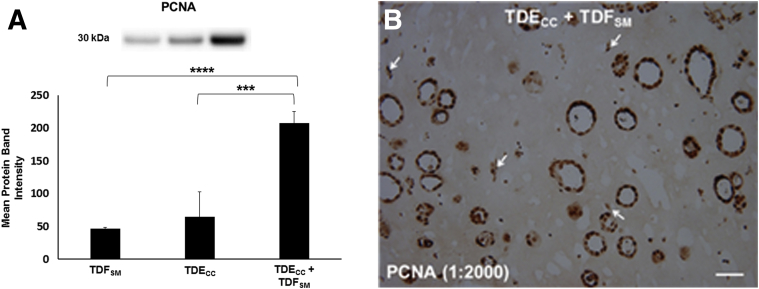
Using the picrosirius red staining-polarization method combined with quantitative image analysis, we could further show a significant potentiation of the production and deposition of dense fibrous collagen within the gel matrix of the TDECC + TDFSM co-cultures when compared with TDECC and TDFSM mono-cell cultures, respectively (Figure 5C). Furthermore, co-culturing TDECC cells with TDFSM cells within 3D gels markedly enhanced the numbers of both α-SMA–positive, desmin-negative fibroblastic cells (Figure 6A and Supplemental Figure S5), as well as CK19-positive spheroid/ductal-like cholangiocarcinoma structures (Figure 6B) formed within the collagen gel matrix over those measured in corresponding control mono-cell cultures. In this context, it is of significance that PCNA protein, a marker of cell mitogenic activity, was found to be synergistically increased in Western blots of protein lysates from whole TDECC + TDFSM gel co-cultures over that of respective TDFSM and TDECC mono-cell cultures (Figure 7). Compatible with the quantitative Western blot results, both the cholangiocarcinoma cells and fibroblastic cells within the gel co-culture exhibited strongly positive immunoreactivity for nuclear PCNA, with the cholangiocarcinoma cells showing a noticeably diffuse nuclear labeling pattern.
Figure 7.
A: Quantitative Western blot demonstrating PCNA protein to be significantly increased in 3D gel co-cultures of TDECC + TDFSM cells over levels determined under comparable conditions for TDECC and TDFSM mono-cell cultures, respectively. Initial cell plating for the TDECC cell strain was 4 × 105 cells/gel and for TDFSM cells, 1.6 × 106 cells/gel, with cell viabilities at initial cell plating ranging from 93% to 98%. After 24 hours, FBS was omitted from the culture medium, and after washings with phosphate-buffered saline, the cultures were maintained without serum for an additional 48 hours, followed by total protein extraction from individual gels for analysis by Western blotting. Bar values are from measurements made on triplicate cultures for each condition. A synergy ratio R value of >1 was calculated based on the ratio between the observed measured relative PCNA band density values for TDECC + TDFSM co-culture over the added individual mean values determined for each TDECC and TDFSM mono-cell culture. B: A representative photomicrograph demonstrating extensive nuclear PCNA immunostaining in abundant spheroid/ductal-like cholangiocarcinoma structures formed when TDECC cells were organotypically co-cultured with TDFSM cells. Note also in this histological section evidence for nuclear PCNA labeling in ostensible fibroblastic cells (arrows) within the gel matrix. Primary antibody dilution shown in parentheses. Initial viable cell plating density for TDECC cells was 2 × 105 cells/gel and for TDFSM cells, 8 × 105 cells/gel. At 24 hours after initial cell plating, the gel co-cultures were washed with phosphate-buffered saline and then maintained for an additional 7 days with fresh daily changes of standard medium containing 1.0% FBS before being processed for PCNA immunohistochemistry. Data are expressed as means ± SD. ∗∗∗P < 0.001, ∗∗∗∗P < 0.0001. Scale bar = 50 μm.
TGF-β Is an Important Driver of the Desmoplastic-Like Reaction Induced in Organotypic Cholangiocarcinoma
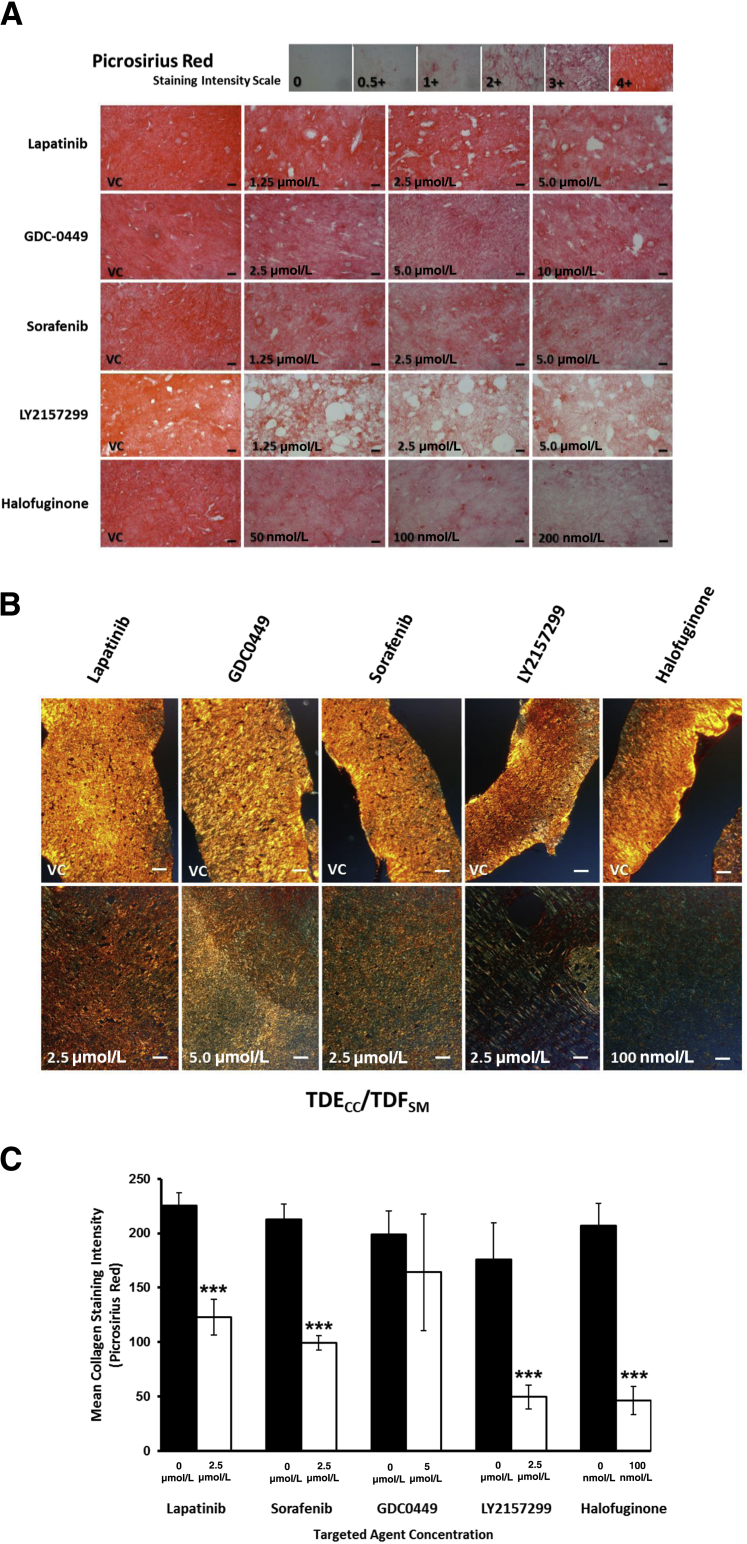
In an effort to identify molecular pathways driving the desmoplastic-like reaction induced in the TDECC + TDFSM co-cultures, we investigated several select and clinically relevant signaling pathway inhibitors for their ability to significantly attenuate the dense fibrous collagen overproduction and deposition, as well as associated increase in TDFSM CAFs accumulated within the 3D gel matrix in our organotypic culture model. Under the described in vitro conditions used for this experiment, the specific TGF-β receptor I kinase inhibitor LY2157299 and the TGF-β–mediated collagen type I synthesis inhibitor halofuginone were found to be the most potent of the various signaling pathway inhibitors tested for their ability to elicit a significant concentration-dependent attenuation of dense fibrous collagen formed within the gel matrix (Figure 8).
Figure 8.
A: Photomicrographic composite demonstrating differential concentration-dependent effects of select clinically relevant anti-cancer targeted agents to attenuate dense collagen matrix (red picrosirius red staining under bright-field microscopy) produced by 3D organotypic co-culturing of TDECC cells with TDFSM cells. Initial viable cell plating density for TDECC cells = 2 × 105 cells/gel; for TDFSM cells = 8 × 105 cells/gel. Targeted agents added daily beginning at 24 hours after initial cell plating at the indicated concentrations to cultures maintained in standard culture medium containing 1.0% FBS. Photomicrographs taken of stained sections prepared from gels processed at 96 hours after initial cell plating. B: Representative photomicrographs of picrosirius red stained histological sections (20 μm thick) under polarized light of vehicle control (VC) cultures versus corresponding targeted agent-treated 3D co-cultures of TDECC + TDFSM cells depicted in A. Each preparation was photomicrographed under identical polarized light settings and magnification. C: Quantitative imaging results demonstrating differing degrees of reduction in picrosirius red staining intensity levels produced by treatments with the select targeted agents compared with VC values, and as reflected in the photomicrographs shown in B. n = 3 cultures analyzed per data point. ∗∗∗P < 0.001 versus control. Scale bars: 20 μm (A); 100 μm (B).
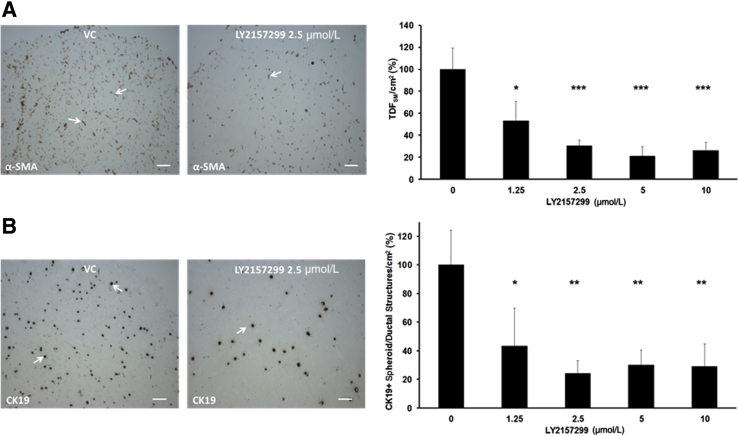
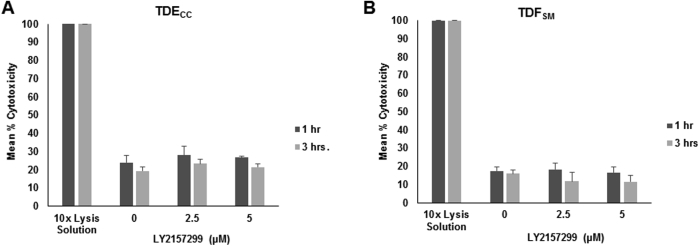
Focusing on LY2157299, we further observed that treatment of the gel co-cultures with this TGF-β pathway inhibitor also concomitantly produced significant concentration-dependent decreases in both α-SMA–positive myofibroblastic cells and CK19-positive cholangiocarcinoma structures accumulated within the gel matrix when compared with vehicle control (VC)–treated cultures (Figure 9). LY2157299 at both the effective 2.5 and 5.0 μmol/L concentration levels was not acutely cytotoxic to either TDECC cells or TDFSM cells in 2D culture on collagen type I–coated plastic, as determined by measured levels of lactic acid dehydrogenase released into the medium of the treated cultures relative to those of VC (0) control cultures (Supplemental Figure S6).
Figure 9.
A: Treatment of TDECC + TDFSM 3D co-cultures with LY2157299 significantly inhibits the accumulation of α-SMA–positive fibroblastic cells/cm2 section area in a concentration-dependent manner. B: Likewise, the LY2157299 treatment also significantly reduces the number of CK19-positive cholangiocarcinoma cell spheroid/ductal structures/cm2 section area formed within the TDECC + TDFSM co-cultures. Arrows in A and B exemplify each cell type/structure within the gel matrix over those of the vehicle control (VC) cultures. Initial viable cell plating density for TDECC = 2 × 105/gel and for TDFSM = 8 × 105/gel. Beginning at 24 hours after initial cell plating, cultures containing 1% FBS were treated daily with LY2157299 in dimethyl sulfoxide over a 72-hour period and analyzed on day 4. Data are expressed as means ± SD. n = 3 cultures analyzed for each indicated concentration level, with α-SMA–positive cell counts made on three random histological sections per culture and those of CK19-positive cell structures made on 2 random sections per culture. ∗P < 0.05, ∗∗P < 0.01, and ∗∗∗P < 0.001 versus control. Scale bars = 100 μm.
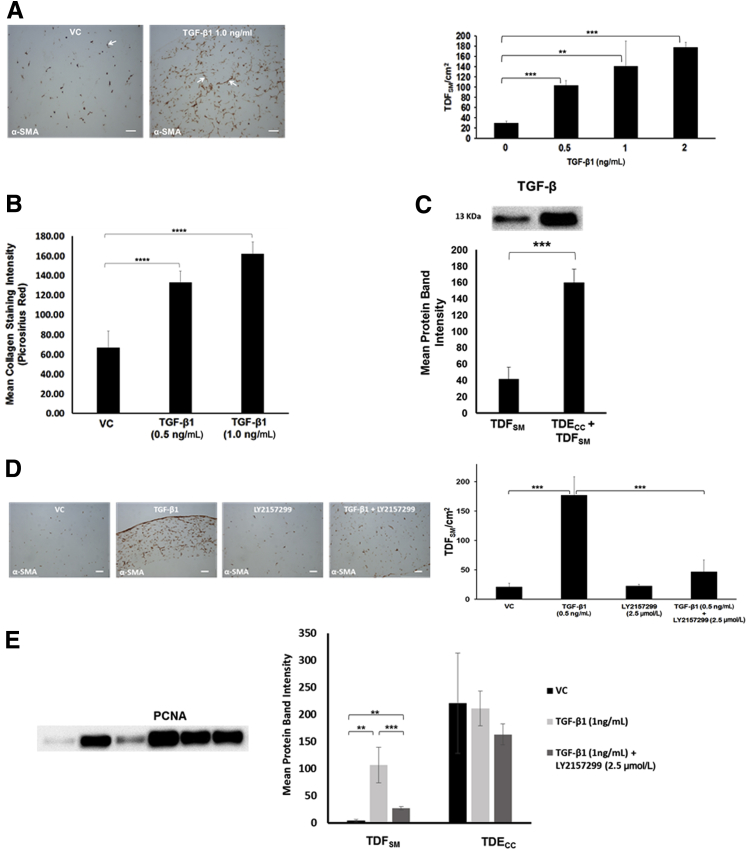
We were able to reproduce the desmoplastic-like reaction induced in the TDECC + TDFSM gel co-cultures by culturing TDFSM CAFs alone within dilute collagen type I gel in the presence of increasing concentrations of recombinant human TGF-β1. These results demonstrate significant concentration-dependent increases in the mean number of α-SMA–positive fibroblastic cells accumulated within the gel matrix as a function of increasing concentrations of TGF-β1 exogenously added over a 4- to 96-hour period in the presence of 1.0% FBS in the culture medium over those determined for the VC control cultures (Figure 10A). This increase in mean TDFSM cell number per cm2 gel section area positively correlated with an overproduction of dense fibrous collagen within the gel matrix determined by quantitative imaging of picrosirius red staining intensity under polarized light (Figure 10B). More important, co-culturing TDECC cells with TDFSM cells in the 3D gel cultures dramatically increased the mean level of mature TGF-β protein (13 kDa) determined in whole gel culture lysates over that determined for TDFSM cells cultured under comparable conditions without TDECC cells (Figure 10C).
Figure 10.
A: Recombinant human TGF-β1 (R&D Systems, Inc., Minneapolis, MN) induces significant concentration-dependent increases in α-SMA–positive fibroblastic cell number/cm2 gel section area when exogenously added to 3D monocultures of TDFSM cells. Initial viable cell plating density of TDFSM cells = 8 × 105/gel. TGF-β1 at the indicated concentration levels was added daily over a 72-hour period beginning at 4 hours after initial cell plating to cultures maintained in standard medium containing 1.0% FBS. Arrows point to representative α-SMA–positive TDFSM myofibroblastic cells. B: Corresponding concentration-dependent effect in 3D mono-cell gel cultures of TDFSM cells of exogenously added TGF-β1 on significantly enhancing dense fibrous collagen production measured as picrosirius red staining intensity under polarized light over that produced in vehicle control (VC) cultures. C: Comparative quantitative Western blot analysis of mature TGF-β protein expression in 3D gel cultures of TDFSM myofibroblastic cells alone and in co-culture with TDECC cholangiocarcinoma cells. Initial viable plating densities for TDFSM cells = 1.6 × 106 cells per gel; for TDECC cells = 4 × 105 cells per gel. Western blotting was performed on total gel protein lysates prepared from cultures that were maintained in serum-free medium for 48 hours, beginning 1 day after initial cell plating. Primary antibody: rabbit polyclonal TGF-β (ab66043, Abcam Inc.) at 1:500 followed by secondary antibody conjugated with horseradish peroxidase (1:3000). D: Concomitantly added LY2157299 significantly blocks the TGF-β1–induced increase in TDFSM cells/cm2 area of histological sections prepared within the 3D collagen gels. E: Effect of exogenously added recombinant human TGF-β1 alone versus in the presence of LY2157299 on PCNA protein expression in 3D gel monocultures of TDFSM cells compared with those of TDECC cells. Initial viable plating densities were 1.6 × 106 TDFSM cells/gel and 4 × 105 TDECC cells/gel, respectively. Cultures were treated at the indicated concentrations with either recombinant human TGF-β1 alone, TGF-β1 + LY2157299, or VC, each of which was added daily in the presence of standard medium containing 1.0% FBS beginning at 4 hours after initial cell plating and then continued daily at 24, 48, and 72 hours after the start of treatment. The cultures were then processed for Western blotting at 96 hours after the initial cell plating. Data are expressed as means ± SD from measurements made on 3 separate cultures per data point (A–E). ∗∗P < 0.01, ∗∗∗P < 0.001, and ∗∗∗∗P < 0.0001. Scale bars = 100 μm (A and D).
As was the case in the TDECC + TDFSM co-cultures, the increased number of TDFSM cells observed in the gel matrix after exposure to exogenously added TGF-β1 was almost completely suppressed by LY2157299 treatment (Figure 10D). Moreover, consistent with the quantitative immunohistochemical imaging results shown in Figure 10, A and D, PCNA protein expression in the TDFSM cell cultures treated with exogenously added recombinant TGF-β1 was determined by quantitative Western blotting to be significantly enhanced over that of TDFSM cells cultured without TGF-β1 (Figure 10E), as well as significantly suppressed by LY2157299. In contrast, as depicted in the same graph (Figure 10E), comparable treatment of cultured TDECC cells with exogenous TGF-β1 alone or in the presence of LY2157299 had no significant effect on PCNA levels expressed in these cholangiocarcinoma cells over vehicle control levels. Under each of these respective conditions, including those cultures that were treated with LY2157299, Western blotting detected only the full length inactivated form of caspase-3 (35 kDa) without evidence of its cleaved 17-kDa peptide fragment, a product of caspase-3 activation in cells undergoing apoptosis (data not shown).
Extending our findings for TGF-β, we determined using GSEA of parent BDEspT desmoplastic cholangiocarcinoma from which the TDECC and TDFSM cell strains were derived that genes involved in the TGF-β signaling pathway, including TGF-β3 and TGF-β1, as well as TGFBR1 (the molecular target of LY2157299), were significantly overexpressed in the tumor tissue samples when compared with pair-matched right liver tissue samples without histologically discernable cholangiocarcinoma involvement (Figure 11A). Furthermore, both the cholangiocarcinoma cells and stromal fibroblastic cells of orthotopic tumors formed in syngeneic rat liver after bile duct inoculation of TDECC cells were each observed to be positively immunostained when separately reacted with two different commercially available anti–TGF-β antibodies, one of which was generated against a synthetic peptide derived from human TGF-β3 protein (ab66043), but also predicted to react with TGF-β1 (Figure 11B), and the other (ab92486) against a synthetic peptide corresponding to human TGF-β1 (data not shown). Using ab92486, we also detected positive cytoplasmic immunoreactivity for TGF-β1 in both cholangiocarcinoma spheroid/ductal-like structures and fibroblastic cells, each of which were much more numerously abundant in random histological sections prepared from TDECC + TDFSM gel co-cultures (Figure 11C) than in those from gel mono-cell cultures of either TDFSM cells (Figure 11D) or of TDECC cells (Figure 11E).
Figure 11.
A: GSEA data demonstrating parent BDEsp-T cholangiocarcinoma tissue from which the TDECC and TDFSM cell strains were derived to be significantly enriched in expressed TGF-β signaling pathway genes relative to corresponding pair-matched right liver lobe tissue without histological evidence of cholangiocarcinoma. Individual liver tumors and corresponding right liver lobe samples from six different rats were analyzed. Red indicates overexpressed genes; blue, underexpressed genes. B: Photomicrograph demonstrating positive immunohistochemical staining for TGF-β in both cholangiocarcinoma cells (cc) and CAFs (arrows) within a representative histological section from a desmoplastic cholangiocarcinoma formed in rat liver after bile duct inoculation of TDECC cholangiocarcinoma cells. C: Photomicrograph of histological section from a 3D gel co-culture of TDECC + TDFSM cells showing strong positive immunoreactivity for TGF-β1 in both the cholangiocarcinoma spheroid/ductal-like structures (circles) and in the TDFSM fibroblastic cells (arrows). D and E: Photomicrographs of sections of 3D gel mono-cell cultures of TDFSM fibroblastic cells (D) and TDECC spheroids (E), respectively, demonstrating each cell type to be positively immunostained for TGF-β1 under comparable experimental conditions as in B. Note that the combined TGF-β1–positive cell structures in C are consistently seen to be more numerous than those observed in D or E. The tumor tissue section in B was reacted with primary anti–TGF-β antibody (ab66043), whereas those in C–E were reacted with anti–TGF-β1 antibody (ab92486). Primary antibody dilutions are shown in parentheses. In each case, corresponding negative controls, which omitted the primary antibody, did not exhibit TGF-β or TGF-β1 immunostaining. Scale bars = 50 μm.
Effect of TGF-α on the Desmoplastic Response and Malignant Progression in 3D Co-Cultures of TDECC + TDFSM
We had previously shown that ligands (TGF-α and amphiregulin) for the EGFR are predominantly expressed in rat cholangiocarcinoma cells relative to CAFs derived from the same orthotopic liver tumor.10 TGF-α together with collagen type I has also been shown to facilitate the activation of hepatic stellate cells,43 and bile acids have been demonstrated to transactivate the epidermal growth factor receptor of cholangiocarcinoma cells via a TGF-α mechanism to promote cellular growth.44 More recently, Clapéron et al13 have reported that hepatic myofibroblasts promote the progression of human cholangiocarcinoma cells through EGFR activation. Thus, we were interested in determining the effect of a potent activator of the EGFR-like TGF-α on the desmoplastic-like reaction in our 3D organotypic cholangiocarcinoma culture model, and of even more interest, on whether TGF-α could induce a prominent increase in malignant cholangiocarcinoma cell proliferation and progression in this 3D cholangiocarcinoma culture model.
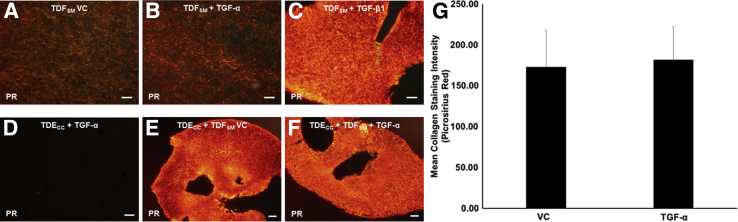
Exogenously added recombinant TGF-α was found not to significantly contribute to the already intense dense fibrocollagenous matrix response induced under basal conditions in 3D TDECC + TDFSM gel cultures (Figure 12, E–G). Moreover, by strictly comparing mono-cell gel cultures of TDFSM cells treated with recombinant TGF-β1 (Figure 12C) versus those treated with recombinant TGF-α (Figure 12B), we further determined that unlike the profound fibrocollagenous response elicited by TGF-β1, TGF-α was without discernable effect in increasing the production of dense collagen fibers into the gel matrix, as detected by changes in picrosirius red staining over that observed in histological sections from control cultures maintained without TGF-α (Figure 12A). Furthermore, exogenously added TGF-α to mono-cell gel cultures of TDECC cells was also without effect on enhancing picrosirius red staining in sections from these cholangiocarcinoma cell gel cultures (Figure 12D) over the low gel matrix background staining determined for 3D TDECC mono-cell gel cultures maintained without added TGF-α (Figure 5C).
Figure 12.
A–C: Photomicrographs of representative picrosirius red (PR)–stained sections viewed under polarized light demonstrating the differential effects of exogenously added recombinant TGF-β1 alone versus that of recombinant TGF-α (R&D Systems, Inc.) alone on promoting dense fibrocollagen overproduction by TDFSM CAFs in 3D gel culture. Note that TGF-β1 elicited an intense PR histochemical staining reaction for dense fibrocollagen in the TDFSM gel cultures (C) compared with the notably weaker PR staining reactions exhibited in sections from TDFSM cultures treated with TGF-α alone (B), or with vehicle control (VC; A), respectively. D–F: Representative photomicrographs of PR-stained sections under polarized light showing comparably intense histochemical staining reactions for dense fibrocollagen in TDECC + TDFSM gel co-cultures either treated with TGF-α (F) or VC (E), relative to the negligible collagen content exhibited in PR-stained sections from mono-cell gel cultures of TDECC cholangiocarcinoma cells treated with TGF-α (D). G: Graphic demonstration of mean collagen fiber staining intensities measured under polarized light for PR stained sections from TDECC + TDFSM gel co-cultures treated with TGF-α versus VC. Each bar value is determined from individual measurements made on seven separate gel cultures per treatment group. In A–C, TDFSM cells were plated at an initial cell density of 8 × 105 viable cells/gel. After 4 hours in culture, the FBS content of the medium was reduced from 10% to 1.0% and the cultures were then maintained until 96 hours, at which time they were processed for PR histochemistry. For the cultures represented in D–G, the initial cell plating densities for the TDECC cells = 2 × 105 viable cells/gel and for TDFSM cells = 8 × 105 viable cells/gel, with the experimental culture conditions being the same as those described for A–C. For this experiment, recombinant human TGF-α at 10 ng/mL or TGF-β1 at 2 ng/mL was added to 3D cultures maintained with standard media containing 1.0% FBS, beginning at 4 hours after initial cell plating and then daily for an additional 72 hours. The cultures were then processed for PR histochemical staining at 96 hours after initial cell plating. Data are expressed as means ± SD. Scale bars: 100 μm (A–C); 50 μm (D); 200 μm (E and F).
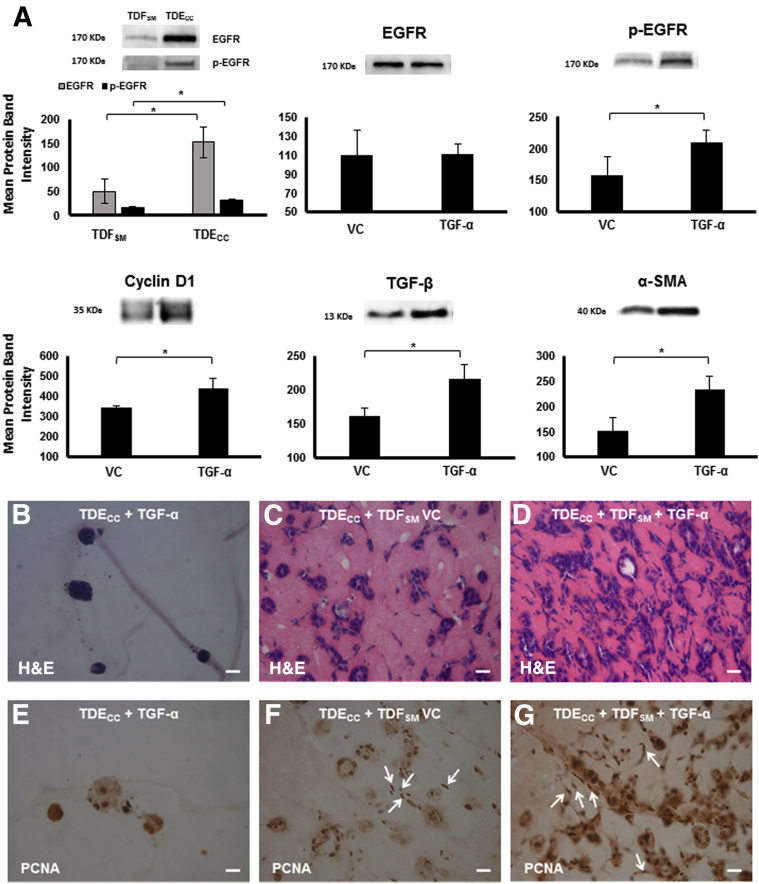
Using quantitative Western blotting, we determined the mean protein band intensity levels for both EGFR and its constitutively activated form (tyrosine -1173 p-EGFR) to be significantly increased by threefold and twofold, respectively, in mono-cell gel cultures of TDECC cells over those of comparably cultured TDFSM cells (Figure 13A). Treatment of 3D gel co-cultures of TDECC + TDFSM with exogenously added recombinant TGF-α under low serum conditions significantly increased the mean protein band intensity of p-EGFR in these co-cultures over those without TGF-α treatment but had no significant effect on the mean EGFR protein band intensity from that of the VC co-cultures (Figure 13A). This further increase in activated EGFR was, in turn, accompanied by significant increases in the mean protein band intensities for the cell cycle regulatory protein, cyclin D1, as well as for TGF-β and α-SMA when compared with those determined for the VC co-cultures.
Figure 13.
A: Quantitative Western blot analysis demonstrating the effects of exogenously added TGF-α versus vehicle control (VC) on EGFR, tyrosine (1173) phosphorylated EGFR (p-EGFR), cyclin-D1, TGF-β, and α-SMA expression in 3D TDECC + TDFSM gel co-cultures. The top left graph shows both EGFR and p-EGFR to each be expressed at significantly higher levels in TDECC cholangiocarcinoma cells in mono-cell gel cultures than in TDFSM CAFs in mono-cell gel cultures. In the TDECC + TDFSM gel co-cultures, EGFR was expressed at comparable protein band intensity levels in both the TGF-α–treated and VC cultures. In contrast, the TGF-α treatment significantly increased the protein band intensities of p-EGFR, cyclin D1, TGF-β, and α-SMA, respectively, in the gel co-cultures over corresponding VC levels. B–D: Representative photomicrographs of hematoxylin and eosin (H&E)–stained sections demonstrating TGF-α to accelerate cholangiocarcinoma cell progression as manifested by increased malignant cell grade in 3D gel co-cultures of TDECC cholangiocarcinoma cells + TDFSM cells (D) when compared with co-cultures without TGF-α (C) and even more notably when contrasted with TDECC mono-cell cultures that were comparably treated with TGF-α in the absence of TDFSM cells (B). E–G: Representative photomicrographs demonstrating differences in nuclear PCNA labeling patterns distinguishing high-grade cholangiocarcinoma cell aggregates formed in TDECC + TDFSM gel co-culture maintained in the presence of exogenously added TGF-α (G) versus the less frequent PCNA nuclear labeling pattern exhibited by more differentiated cholangiocarcinoma ductal-like structures formed in co-cultures without TGF-α treatment (F) and that of the much lower total number of well-circumscribed low-grade cholangiocarcinoma cell spheroids that formed in mono-cell gel culture without TDFSM cells, but in the presence of added TGF-α (E). Note that strongly positive nuclear PCNA immunoreactivity is most prominently detected in the epithelial TDECC cells within the TGF-α–treated gel co-culture, but also localized to TDFSM cell nuclei (arrows) within the collagen gel matrix of co-cultures maintained with or without TGF-α treatment. Initial viable cell plating density of TDECC = 2 × 105 cells/gel; for TDFSM = 8 × 105 cells/gel. PCNA antibody dilution = 1:4000. The histological results shown herein were repeated in three separate experiments, with a minimum of triplicate cultures analyzed per TGF-α and VC treatment groups. TGF-α treatment conditions for this experiment were essentially the same as those described for Figure 12. In the case of the mono-cell cultures used for Western blotting, the initial cell plating density for TDECC and TDFSM cells was 8 × 105 cells/gel. For the gel co-cultures, the initial cell plating density for the TDECC cells was 2 × 105 cells/gel and that of the TDFSM cells was 8 × 105 cell/gel. Protein extraction for EGFR, p-EGFR, and TGF-β was performed at 48 hours after initial cell plating, whereas that for cyclin D1 and α-SMA was performed at 96 hours after initial cell plating. Primary antibody dilutions: EGFR (1:100), p-EGFR (1:100), cyclin-D1 (1:1000), TGF-β (1:500), α-SMA (1:1000). ∗P < 0.05. Scale bars = 20 μm.
TGF-α treatment of the TDECC + TDFSM 3D gel co-cultures maintained in medium with 1.0% FBS resulted in an increased anaplastic and histologically higher grade malignant cell phenotype when compared with that of VC co-cultures (Figure 13C), and even more profoundly manifested when contrasted with TDECC gel mono-cell cultures treated under comparable conditions with recombinant TGF-α (Figure 13B). Particularly note the tissue-like appearance of histological sections prepared from the TDECC + TDFSM gel co-cultures, which in hematoxylin and eosin–stained sections of the TGF-α–treated co-cultures show a strong eosinophilic staining of the gel matrix (Figure 13D) that is visibly somewhat more pronounced than that of the TDECC + TDFSM co-cultures without TGF-α treatment (Figure 13C) and only weakly exhibited by the gel matrix of TGF-α–treated TDECC cells in gel mono-cell culture (Figure 13B). This increased eosinophilia of the matrix is likely because of increased acidophilic protein production and secretion in the TGF-α–treated co-cultures; however, we were not able to discern differences between the TGF-α–treated and VC co-cultures in terms of our measurements of picrosirius red staining intensity under polarized light (Figure 12).
The morphologically disorganized aggregates of cholangiocarcinoma cells intermingled with fibroblastic cells, which formed in TDECC + TDFSM 3D gel co-cultures treated with exogenously added TGF-α, exhibited a significantly enhanced nuclear PCNA immunoreactivity that was prominently localized to the cancer cells, but also detected in the fibroblastic cells, thereby indicating an active cell proliferative activity for both cell types (Figure 13G). In comparison, nuclear PCNA labeling of cholangiocarcinoma cells was noticeably less expansive in sections prepared from co-cultures maintained without TGF-α (Figure 13F), and even much more so when contrasted with that of well-differentiated cholangiocarcinoma cell spheroids/ductal-like structures formed in TDECC mono-cell gel cultures maintained in the presence of TGF-α (Figure 13E).
Discussion
The 3D co-culture model described herein has the potential to serve as a highly reproducible preclinical in vitro platform that is particularly well suited to directly investigating critical cancer cell–tumor stromal cell interactions specifically mediating ICC desmoplasia and malignant tumor growth and progression, and for rapidly identifying agents that target the desmoplastic stroma of advanced ICC. As an important step toward the development of this model, we were able to establish and rigorously characterize a novel CAF cell strain designated TDFSM, which was derived from orthotopic rat desmoplastic cholangiocarcinoma formed in syngeneic rat liver from a spontaneously transformed rat cholangiocyte line, in comparison with a transplantable cholangiocarcinoma cell strain, TDECC, derived from the same tumor type.
Tumors formed in liver from orthotopically transplanted TDECC cholangiocarcinoma cells closely mimicked the classic histopathological characteristics and clinical features (eg, peritoneal metastases) of human well-to-moderately differentiated, mass-forming desmoplastic small duct ICC. Not surprisingly, the TDECC cholangiocarcinomas were observed to be intermediate in their degree of malignant aggressiveness when contrasted with previously determined liver tumor growth rates and metastatic potential of parent BDEspT from which the TDECC cell strain was derived,24 as well as with the more highly malignant cholangiocarcinomas that formed in syngeneic rat liver after bile duct inoculation of our highly tumorigenic rat BDEneu cholangiocyte cell line genetically engineered to overexpress activated rat neu oncogene.24 We have further shown that TDECC cholangiocarcinomas closely recapitulate key features of the desmoplastic stroma of human ICC (namely, by exhibiting a dense fibrocollagenous matrix highly enriched in α-SMA–positive CAFs and also by being strongly immunoreactive for Postn, which was first reported by Utispan et al6 to be localized to the CAFs and not cancer cells of human ICCs). Furthermore, as has also been reported for human ICCs,17, 18 Msln was seen to be strongly immunoreactive in the cancer cells of the TDECC cholangiocarcinomas, but only weakly expressed in the stroma of these rat orthotopic tumors.
Using immunohistochemistry, we further identified a minor population of desmin-positive fibroblastic-like cells within the desmoplastic stroma in both orthotopic rat TDECC and in a small sampling of human desmoplastic ICC. This latter finding is somewhat consistent with that of Okabe et al,4 who had detected desmin immunoreactivity in the cancer stroma of 6 of 18 human ICC samples, but also observed that the number of stromal cells in their analyzed cases of human ICC coexpressing α-SMA and desmin was lower than expected and that the degree of desmin expression in the stromal cells was small. In the context of our study, the extent, if any, to which the desmin-positive stromal cells observed infrequently by us in both histological sections from our rat orthotopic cholangiocarcinoma model and in select analyzed samples of human desmoplastic small duct ICCs may be contributing to the desmoplastic reaction characterizing these respective hepatobiliary tumors remains to be established. What seems apparent, however, is that the TDFSM cell strain exhibited phenotypic features, as well as a biomarker gene expression profile (eg, Table 1), that identify this cell type as being a constitutively activated myofibroblast, mirroring the α-SMA–positive, desmin-negative immunohistochemical staining pattern characterizing the vast majority of CAFs observed within the desmoplastic stroma of our orthotopic rat cholangiocarcinoma model.
It is beyond the scope of our current analysis to use genetic fate tracing45, 46, 47 as a means of attempting to definitively identify the origins of the TDFSM cell strains derived from orthotopic rat desmoplastic BDEspT cholangiocarcinoma. Nevertheless, our overall comprehensive characterization of this CAF cell strain would strongly suggest that it was derived from resident liver fibroblasts and not likely through cholangiocarcinoma cell epithelial-mesenchymal transition, as evidenced in part by the absence of known cholangiocarcinoma cell markers (eg, CK19) being expressed in the TDFSM cells or of established mesenchymal/epithelial-mesenchymal transition cell markers (eg, Vim) being expressed in the TDECC cells. Moreover, the results of our comprehensive microarray analysis further support this conclusion (Figure 4 and Table 1). Neither the myofibroblastic cell strain nor the cholangiocarcinoma cell strain were immunoreactive for either CD45 or for CD34, suggesting that hematopoietic stem cells or bone marrow–derived fibrocytes do not function as progenitors of the TDFSM or TDECC cell types. Primary CAFs from human HCC are negative for CD34 and CD45, but like the TDFSM cell strain, are immunoreactive for α-SMA and Vim.48
Our finding that the clonal TDECC cholangiocarcinoma cell strain exhibited more numerous, as well as distinctive chromosomal alterations when compared with the karyotype of the TDFSM myofibroblastic cell strain, and that some of the TDFSM cells were found to have a normal rat karyotype, further argues against epithelial-mesenchymal transition as a likely mechanism for the origin of the TDFSM cell strain. It is of interest, though, that duplication of chromosome 12 was seen to be a common feature of both the cultured TDECC and TDFSM cell strains. A possible explanation for this unique chromosomal alteration might relate to the fact that cultured cells can acquire specific chromosomal aberrations even within a few passages in culture, as exemplified coincidentally by a high incidence of chromosomal 12 duplications reported to occur fairly rapidly in propagated human stem cell populations as a result of culture adaptation.49, 50, 51 However, further studies on the influence of cell culturing on chromosomal instability and specific chromosome duplication are needed to validate this possibility.
CAFs are not normal fibroblastic cells, and their differentiation and phenotypic biomarker profiles can be influenced by the tumor microenvironment and cross talk with cancer cells. This makes the use of various biomarkers that have been commonly reported to help establish the hepatic stellate cell or portal fibroblast origins of myofibroblastic cell types in experimental liver fibrosis induced by carbon tetrachloride or bile duct ligation35, 36, 42 unreliable for use as biomarkers to identify stromal fibroblast/myofibroblast cell origins in desmoplastic ICC. However, such biomarkers can have value for delineating specific CAF phenotypes. In this context, we have shown by transcriptomic analysis and/or by immunophenotyping that TDFSM myofibroblasts prominently overexpress gene products (eg, gremlin 1, cell surface Thy-1, fibulin 2, cofilin 1) relative to TDECC cells and/or normal adult rat liver, which were previously described by others as being overexpressed in rat portal myofibroblasts relative to activated hepatic stellate cells.35, 42 CAFs from colorectal metastases in human liver specimens have also been previously shown by Mueller et al52 to display a phenotype (α-SMA+, Thy-1+, Vim+, desmin-negative) similar to our rat cholangiocarcinoma-derived TDFSM myofibroblastic cell strain. NTPdase2 and elastin, which are also commonly used as biomarkers to distinguish portal fibroblasts from hepatic stellate cells,12, 30, 36, 53, 54 were further determined by Western blot analysis and immunofluorescence cytochemistry to be expressed in TDFSM myofibroblasts. Of added interest, we were also able to detect NTPdase2 and elastin protein in the TDECC cholangiocarcinoma cells, albeit at a nonsignificantly lower mean expression level than that of TDFSM cells. However, these findings may not be without precedent, because mRNA transcripts for NTPDase2 have been previously reported to be expressed in human Mz-ChA-1 cholangiocarcinoma cells,55 and furthermore, that malignant epithelium, along with fibroblastic cells, was shown to be a source of elastin in desmoplastic breast cancers.56
Msln, which has also been proposed to be a novel biomarker of portal myofibroblasts36 and shown to be expressed in mesothelial cells of the liver capsule,53 was only weakly detected by Western blotting in protein lysates from cultured TDFSM cells, but significantly overexpressed in the cultured TDECC cholangiocarcinoma cells. The comparably low levels of Msln protein assessed by quantitative Western blotting in the TDFSM cells, as well as the relatively low level of expression of Msln mRNA transcripts detected by our microarray analysis in TDFSM cells relative to TDECC cells would also appear to rule out capsular mesothelial cells as being a major cellular source of the TDFSM cell strain. In addition, the significantly high levels of secreted Postn protein, as well as Postn mRNA transcripts overexpressed in the TDFSM cells when compared with the TDECC cells, as well as those of Msln mRNA transcripts and protein overexpressed in the TDECC cells relative to TDFSM cells is highly compatible with our microarray data showing significantly high levels of Msln and Postn mRNA transcripts in BDEspT tissue when compared with the levels expressed in pair-matched normal liver (Table 1). Our immunohistochemical demonstration of strongly positive immunoreactivity for Msln in the cholangiocarcinoma cells compared with the stroma and of Postn in the desmoplastic stroma compared with cholangiocarcinoma cells in tissue sections from orthotopic TDECC cholangiocarcinoma (Figure 3B) also supports the TDECC and TDFSM cell strains as being representative of the cancer and stromal cellular composition of the in situ tumor. That both Msln and Postn have been proposed to be potential indicators of ICC progression, and possibly may serve as promising targets for ICC therapy6, 16, 17, 18 provides further support to our rat organotypic culture and orthotopic cholangiocarcinoma models as having added translational research relevance for human ICC.
Our ability to reproduce hallmark features of the desmoplastic reaction in culture of mass-forming ICC in situ was dependent on combining TDECC cells with increasing numbers of TDFSM myofibroblastic-like cells in a dilute collagen type I hydrogel. To our knowledge, this unique 3D organotypic co-culture model is the first of its kind to demonstrably reproduce key histological features (dense fibrocollagenous matrix in association with a significant accumulation of α-SMA–positive myofibroblastic-like cells) of desmoplastic cancer stroma in situ.
Kadaba et al,57 who by co-culturing different proportions of a strain of h-TERT immortalized human pancreatic stellate cells (expressing both α-SMA and desmin) and Capan1 or AsPc1 human pancreatic adenocarcinoma cells also demonstrated that increasing proportions of stellate cells to cancer cells yielded maximum effects on gel contraction, cancer cell proliferation, and invasion. We too had earlier determined with our organotypic rat cholangiocarcinoma model that gel shrinkage and cholangiocarcinoma cell spheroid/ductal-like growth were both significantly enhanced as a function of increasingly higher initial CAF plating densities relative to a fixed initial cell plating density for the cholangiocarcinoma cells.10 In the current study, we have further shown that the intense desmoplastic-like reaction induced by 3D co-culturing of TDFSM cells with TDECC cells in dilute type I collagen gel is maximized by proportionally increasing the numbers of the CAFs initially mixed into the gel culture with a fixed number of cholangiocarcinoma cells, with the greatest response in dense fibrous collagen production and deposition within the gel matrix as measured by picrosirius red staining intensity being produced at an initial CAF to cancer cell plating density ratio of 4:1. It is also noteworthy that co-culturing the TDFSM with the TDECC cells significantly enhanced the accumulation of the cholangiocarcinoma spheroid/ductal-like structures within the gel matrix over those measured in mono-cell culture controls for each cell strain, which we demonstrated to be associated with a significant increase in PCNA protein expression within total gel lysates from the TDECC + TDFSM co-cultures when compared with TDECC or TDFSM mono-cell cultures, as well as reflected by extensive nuclear PCNA labeling exhibited by the cholangiocarcinoma cells within histological sections from the TDECC + TDFSM co-cultures.
Using picrosirius red staining intensity under circularly polarized light to assess dense collagen content, we were readily able to differentiate between different clinically relevant targeted agents for their ability to attenuate the desmoplastic-like reaction promoted by co-culturing TDFSM cells with TDECC cells in organotypic culture. With this simple and highly reproducible histological detection assay, we convincingly identified the TGF-β pathway inhibitors LY2157299 and halofuginone as being the most potent of the clinically relevant targeted agents tested in this 3D culture model. Our microarray analysis further revealed Col1a2 and Col1a1 to be highly expressed in TDFSM cells, consistent with the intense yellow-orange picrosirius red staining observed in 3D untreated or vehicle control TDECC + TDFSM co-cultures and in 3D TDFSM mono-cell cultures treated with recombinant TGF-β1 as being largely because of the induced overproduction of collagen type I being organized into dense collagenous fibers within the gel matrix. Collagen, type XV, α1 mRNA transcripts, recently demonstrated to be overexpressed in rat liver portal myofibroblasts relative to hepatic stellate cell myofibroblasts,42 were also determined by our microarray analysis to be more highly expressed in TDFSM cells relative to TDECC cells, as well as in BDEspT relative to pair-matched normal liver, but were at dramatically lower levels of expression than those determined for Col1a2 and Col1a1 in these respective sampled specimens (Table 1).
That treatment of the 3D TDECC + TDFSM cell co-cultures with the selective TGF-β receptor 1 serine/threonine kinase inhibitor LY2157299 led to a prominent dose-dependent attenuation of the dense fibrocollagenous matrix accompanied by significant decreases in the numbers of α-SMA–positive TDFSM myofibroblastic-like cells within the gel matrix most clearly suggests a model in which the TGF-β pathway functions as a major contributor to the development of desmoplastic stroma in mass-forming ICC. Similar results as those obtained with LY2157299 were also demonstrated by us in co-cultures comparably treated with the specific TGF-β receptor tyrosine kinase inhibitor SB432542 (data not shown). As added proof, we showed that the addition of recombinant TGF-β1 to 3D monocultures of TDFSM cells also significantly increased in a concentration-dependent manner the numbers of α-SMA–positive myofibroblastic cells accumulated within the gel matrix, which correlated with a significant overproduction of dense collagen fibers elaborated into the matrix, and that this desmoplastic-like response could also be suppressed by treatment with LY2157299, similar to what we observed in the TDECC + TDFSM co-cultures. Herein, it is of particular importance that the active mature form of TGF-β (TGF-β1) protein is significantly increased in 3D co-cultures of TDECC cholangiocarcinoma cells + TDFSM CAFs exhibiting enhanced dense fibrocollagen production when contrasted with comparably cultured TDFSM cells. This latter finding indicates that while the mature 13-kDa form of TGF-β is expressed in cultured TDECC and TDFSM cells, respectively (Figures 3 and 11), the amount of this cytokine generated in TDFSM mono-cell cultures is insufficient to support a robust desmoplastic-like response as that characterizing the TDECC + TDFSM co-culture. Further studies are needed, however, to determine whether the significantly higher mean level of TGF-β (TGF-β1) protein elaborated in the TDECC + TDFSM gel co-cultures reflects the prominent increases in both cholangiocarcinoma spheroid/ductal-like structures and α-SMA–positive myofibroblastic cells formed under this 3D co-culture condition or if it also involves an increase in cancer cell and/or CAF TGF-β transcription and processing. In this context, it is of interest that Sukowati et al48 found TGF-β1 gene expression to be up-regulated in HuH7 and JHH6 HCC cell lines co-cultured with CAFs derived from human HCC nodules, but not when co-cultured with fibroblasts from distant nontumoral liver tissues.
TGF-β (eg, TGF-β1) treatment induces the transformation of both quiescent rodent hepatic stellate cells and portal fibroblasts into myofibroblasts in 2D cell culture,53, 54, 58 as well as being an important autocrine/paracrine regulator of collagen type I production by activated hepatic stellate cells and portal fibroblasts.59 On the other hand, TGF-β1 treatment has been reported to either inhibit or be without effect on cell proliferation in 2D cultures of rodent hepatic stellate cells or portal fibroblasts.53, 60, 61 In comparison, our findings (Figure 10) demonstrate TGF-β1 to stimulate cell growth, reflected by enhanced PCNA protein expression together with a significantly increased cell number, in pure 3D gel monocultures of TDFSM CAFs constitutively immunoreactive for α-SMA. Our data further showed that TDECC cholangiocarcinoma cell growth in mono-cell gel culture was not affected by treatments with either TGF-β1 alone or in combination with LY2157299, whereas LY2157299 significantly suppressed the cell growth of TDFSM cells induced by TGF-β1. Moreover, taking into account that LY2157299 was found not to be acutely cytotoxic to either TDECC or TDFSM cells in 2D monolayer culture nor to activate caspase-3 in 3D gel cultures of these respective cell types, provides support for a mechanism whereby selective growth inhibition of TDFSM CAFs mediated by a specific TGF-β receptor 1 kinase inhibitor significantly diminishes their capacity through myofibroblastic cell attrition and a concomitant decrease in dense fibrocollagen production to promote TDECC cholangiocarcinoma spheroid/ductal-like growth within the 3D co-culture microenvironment.
Of particular relevance to our study, PANC1 pancreatic tumor cells (known neither to induce desmoplasia nor to express high amounts of TGF-β1) when transfected to overexpress TGF-β1 have been shown after transplantion into nude mouse pancreas to induce desmoplasia with increased production of collagen type I and fibronectin similar to that seen in human pancreatic adenocarcinoma.62 Also, as we have now demonstrated for TDECC cholangiocarcinoma cells, TGF-β1 was previously observed not to influence the proliferation of human ICC cholangiocarcinoma cell lines in cell culture.63, 64 Human ICCs, like desmoplastic liver tumors formed from orthotopically transplanted rat TDECC cholangiocarcinoma cells, were further seen to express histochemically detectable TGF-β1 in the cytoplasm of the cholangiocarcinoma cells and of surrounding mesenchymal cells, but it is not expressed in normal bile ducts.63 Moreover, GSEA data for rat BDEspT and for more aggressive BDEneu cholangiocarcinomas (data not shown), together with our TGF-β (TGF-β1) immunostaining results for the TDECC cholangiocarcinomas, demonstrate these experimental hepatobiliary cancers, like human ICCs,65 to each be highly enriched in TGF-β signaling pathway genes when compared with pair-matched liver without cancer. Overall, these results strongly support the TGF-β signaling pathway as being particularly active in desmoplastic ICC, suggesting its potential as a target for ICC therapy.
Some limited studies have suggested that targeting the TGF-β pathway and hepatic stellate cell activation may prevent cholangiocarcinoma development. For example, halofuginone, when administered orally to rats before thioacetamide-induced liver fibrosis, has been reported to prevent hepatic stellate cell activation, and more relevant to our current results, when given to rats with established fibrosis, halofuginone was found to cause complete resolution of the fibrotic condition.66 Halofuginone was also determined to prevent pancreatic xenograft tumor development in nude mice by inhibiting pancreatic stellate cell activation.67 In addition, murine neutralizing TGF-β monoclonal antibody 1D11, when administered to rats with established hepatic fibrosis induced by thioacetamide, was shown to result in a regression of tissue injury and fibrosis when compared with livers from vehicle control-treated animals. This antifibrotic effect was further found to be concomitantly associated with a reduction in the development of cholangiocarcinomas formed in the thioacetamide-treated rats.68 More recently, and in contrast to results obtained with the rat thioacetamide model of hepatic fibrosis and cholangiocarcinogenesis, it was recently shown that hepatic fibrosis induced by treatments such as carbon tetrachloride or bile duct ligation was not altered in mice in which TGF-β signaling was inhibited by multiple cell Cre deleters to delete floxed TGF-β receptor II gene (Tgfbr2) in hepatocytes, cholangiocytes, or in both populations, and furthermore, that concomitant deletion of both Tgfbr2 and of Pten favored the development of cholangiocarcinoma in mice with liver-specific ablation of both of these genes by a mechanism linked to enhanced cholangiocyte proliferation.69 This latter finding would suggest that TGF-β signaling in liver may actually protect against cholangiocarcinoma development by restricting cholangiocyte proliferation. However, likely differences in the origins of cholangiocarcinomas formed in rats treated with thioacetamide, which progress from preneoplastic cholangiofibrotic lesions, versus those developed in Tgfbr2/Pten-depleted mouse livers, which have been suggested to develop from hepatocyte-derived cholangiocytes, could explain differences in divergent responses to TGF-β inhibition. Moreover, the mouse model does not take into account that human ICCs are well known to form in livers with underlying cholangitis, periductal fibrosis, biliary ductal dysplasia, and partially impaired bile flow, together with the complexity of the desmoplastic microenvironment and molecular cross talk involving TGF-β signaling between cholangiocarcinoma cells with CAFs within the fully developed cancer.
We cannot yet say if our in vitro outcomes with LY2157299, which is currently in phase I/II clinical trials against various solid tumor types (eg, pancreatic cancer, HCC), or halofuginone when tested in our 3D TDECC + TDFSM co-culture model will translate into a significant therapeutic response when comparatively tested against early and advanced desmoplastic cholangiocarcinomas formed in rat liver after bile duct inoculation of TDECC cholangiocarcinoma cells or with our even more highly malignant tumorigenic rat BDEneu malignant cholangiocyte cell line. These studies, which involve the use of our orthotopic patient-like rat model of cholangiocarcinoma progression24 (Figure 1), are currently being developed and are focused on desmoplastic cholangiocarcinoma therapy rather than prevention. A potentially attractive translational feature of this orthotopic cancer model is that it is amenable to surgical resection of the median/left-lateral liver lobes containing progressive desmoplastic cholangiocarcinoma, which can then be combined with adjuvant targeted treatments aimed at repressing peritoneal metastases originated from advanced liver tumor.
The compelling data of Clapéron et al13 demonstrating that hepatic myofibroblasts interacting with cholangiocarcinoma cells through the heparin-binding EGF/EGFR axis contributes to human cholangiocarcinoma progression, and furthermore, that TGF-β1 produced by cholangiocarcinoma cells induces HB-EGF in human liver myofibroblasts is highly relevant to our overall findings. In this context, our results in Figures 12 and 13 would appear to suggest that although the EGFR ligand TGF-α, unlike TGF-β1, is not directly in itself a major contributor to the desmoplastic-like matrix reaction characterizing the 3D organotypic TDECC + TDFSM co-culture model, TGF-α can potentially act in concert with TDFSM CAFs to provide a potent and essential stimulus for accelerating TDECC cholangiocarcinoma cell proliferation and progression. Ostensibly, this can occur by augmenting EGFR activation predominantly expressed in the cholangiocarcinoma cells, thereby leading to an increase in the cancer cell population and an associated enhancement of active TGF-β production, which in turn is mitogenic for TDFSM CAFs, but not mito-inhibitory for TDECC cholangiocarcinoma cells (Figure 10E). Histologically, this TGF-α effect is manifested in our 3D organotypic cholangiocarcinoma model via a higher malignant cell grade and hyperproliferative cancer cell response. Further studies, however, are now needed to identify which specific factors produced by CAFs are most critical in cross talking with the EGF family signaling pathway to enhance the malignant potential of cholangiocarcinoma cells, as well as to assess the therapeutic potential of combinational targeting of the TGF-β and EGFR signaling pathways in desmoplastic ICC.
Footnotes
Supported by NIH grants R01 CA083650 (A.E.S.) and R56 DK076735 (J.A.D.) and Gilead Sciences Research Scholars Program in Liver Disease Award UAM#271G141616-01 (M.F.). Some of the microscopy was performed at the Virginia Commonwealth University Department of Anatomy and Neurobiology Microscopy Facility supported, in part, by NIH–National Institute of Neurological Diseases and Stroke Center Core grant 5P30 NS047463 and, in part, by NIH–National Cancer Institute Cancer Center Support grant P30 CA016059.
M.A.M., A.U., and D.J.C. contributed equally to this work.
Disclosures: None declared.
Supplemental material for this article can be found at http://dx.doi.org/10.1016/j.ajpath.2017.01.013.
Supplemental Data
Supplemental Figure S1.
Characteristic morphological features of TDECC and TDFSM cell strains in two-dimensional culture. A: Representative photomicrograph shows that TDECC cells when cultured on rat tail collagen type I–coated plastic exhibit an epithelial pattern in their morphology. B: Representative photomicrograph demonstrates that TDFSM cells, cultured under identical conditions as A, exhibit a fibroblastic stellate-like shape. Compare also with TDFSM images in Figure 2. Scale bars = 50 μm. H&E, hematoxylin and eosin.
Supplemental Figure S2.
Chromosomal analysis of TDECC (A) and TDFSM (B) cell strains cultured on rat tail collagen type I–coated plastic. Representative karyotypes are shown for each cell strain. n = 10 separate karyotypes analyzed per cell strain.
Supplemental Figure S3.
Anchorage-dependent and anchorage-independent growth properties of orthotopic rat BDEsp cholangiocarcinoma-derived cancer and stromal fibroblastic cell strains. A: Comparative anchorage-dependent growth curves for TDECC cholangiocarcinoma cells versus TDFSM fibroblastic cells, respectively, cultured on rat tail collagen type I–coated plastic wells. Each value is from individual cell counts made on triplicate cultures per time point. Cell doubling times were calculated from cell counts plotted over the 4- to 7-day period (linear portion of the curves). B: Representative photomicrographs showing anchorage-independent colony cell growth formed in soft agar by the TDECC cells, but not exhibited by TDFSM cells cultured under comparable conditions. White circle indicates a TDECC cell colony formed in soft agar culture; arrows, single TDFSM cells that do not form colonies in soft agar. The accompanying graph quantifies the number of anchorage-independent colonies ≥70 μm in diameter detected only in the TDECC cultures, with no cell colonies having been formed in soft agar from the cultured TDFSM cells. Initial number of viable cells plated in soft agar for each of the cell strains analyzed was 5 × 103 per well in a 96-well sterile flat bottom microplate. The samples were analyzed in triplicate. Data are expressed as means ± SD (A and B). ∗∗∗P ≤ 0.001 versus TDFSM. Scale bars = 50 μm.
Supplemental Figure S4.
Photomicrographs demonstrating the lack of immunofluorescence staining for both CD45 and CD34 in TDECC and TDFSM cells, respectively, maintained for up to 3 days in 2D monolayer culture on collagen type I–coated coverslips. Secondary antibody for CD45 conjugated with Alexa Fluor 488 (1:200) and for CD34 conjugated with Alexa Fluor 647 (1:200). Blue fluorescence nuclei reflect nuclear DNA counterstaining with DAPI. Scale bars = 50 μm.
Supplemental Figure S5.
Representative photomicrographs showing numerous α-SMA+ fibroblastic cells accumulated within the gel matrix of a 3D TDECC + TDFSM co-culture (A) and absence of desmin-immunoreactive cells in TDECC + TDFSM gel co-cultures (B). Cholangiocarcinoma cell spheroids/ductal-like structures did not immunostain for either α-SMA or for desmin in these co-cultures. Initial viable cell plating densities: TDECC = 2 × 105 cells/gel; TDFSM = 8 × 105 cells/gel. Cultures were maintained in standard medium containing 10% FBS until 48 hours after initial cell plating. At 48 hours, FBS was reduced to 1.0%, with the fresh culture medium changes then made daily until the gel cultures were processed at 48 hours later. Primary antibody dilution shown in parentheses. Scale bars = 50 μm.
Supplemental Figure S6.
Evaluation of LY2157299 for acute cytotoxicity against individually cultured TDECC and TDFSM cells. Cytotoxicity was assessed using the CytoTox96 Non-Radioactive Cytotoxicity Assay (Promega Corp., Madison, WI) as a function of lactic acid dehydrogenase (LDH) released into the culture medium measured at 1 and 3 hours after their initial exposure to LY2157299 in dimethyl sulfoxide (DMSO) at the indicated concentrations. Each well was assayed in triplicate with an initial cell plating density of 15 × 103 cells/well in 96-well culture plates coated with collagen type I. Blank wells did not contain cells, and vehicle control cultures were treated with 10 μL DMSO in culture medium without LY2157299 (0 concentration). Indicated values represent the relative to maximum LDH release produced by treatment with lysis buffer provided with the assay kit. Note under these conditions that the LY2157299 treatment is not acutely toxic to either the cultures of TDECC (A) or TDFSM (B) cells. Data are expressed as means ± SD.
References
- 1.Sirica A.E. The role of cancer-associated myofibroblasts in intrahepatic cholangiocarcinoma. Nat Rev Gastroenterol Hepatol. 2012;9:44–54. doi: 10.1038/nrgastro.2011.222. [DOI] [PubMed] [Google Scholar]
- 2.Sirica A.E., Gores G.J. Desmoplastic stroma and cholangiocarcinoma: clinical implications and therapeutic targeting. Hepatology. 2014;59:2397–2402. doi: 10.1002/hep.26762. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Chuaysri C., Thuwajit P., Paupairoj A., Chau-in S., Suthiphongchai T., Thuwajit C. Alpha-smooth muscle actin-positive fibroblasts promote biliary cell proliferation and correlate with poor survival in cholangiocarcinoma. Oncol Rep. 2009;21:957–969. doi: 10.3892/or_00000309. [DOI] [PubMed] [Google Scholar]
- 4.Okabe H., Beppu T., Hayashi H., Horino K., Masuda T., Komori H., Ishikawa S., Watanabe M., Takamori H., Iyama K., Baba H. Hepatic stellate cells may relate to progression of intrahepatic cholangiocarcinoma. Ann Surg Oncol. 2009;16:2555–2564. doi: 10.1245/s10434-009-0568-4. [DOI] [PubMed] [Google Scholar]
- 5.Cadamuro M., Morton S.D., Strazzabosco M., Fabris L. Unveiling the role of tumor reactive stroma in cholangiocarcinoma: an opportunity for new therapeutic strategies. Transl Gastrointest Cancer. 2013;2:130–144. doi: 10.3978/j.issn.2224-4778.2013.04.02. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Utispan K., Thuwajit P., Abiko Y., Charngkaew K., Paupairoj A., Chau-in S., Thuwajit C. Gene expression profiling of cholangiocarcinoma-derived fibroblast reveals alterations related to tumor progression and indicates periostin as a poor prognostic marker. Mol Cancer. 2010;9:13. doi: 10.1186/1476-4598-9-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Mertens J.C., Fingas C., Christensen J.D., Smoot R.L., Bronk S.F., Werneburg N.W., Gustafson M.P., Dietz A.B., Roberts L.R., Sirica A.E., Gores G.J. Therapeutic effects of deleting cancer-associated fibroblasts in cholangiocarcinoma. Cancer Res. 2012;73:897–907. doi: 10.1158/0008-5472.CAN-12-2130. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Massani M., Stecca T., Fabris L., Caratozzolo E., Ruffolo C., Furlanetto A., Morton S., Cadamuro M., Strazzabosco M., Bassi N. Isolation and characterization of biliary epithelial and stromal cells from resected human cholangiocarcinoma: a novel in vitro model to study tumor-stroma interactions. Oncol Rep. 2013;30:1143–1148. doi: 10.3892/or.2013.2568. [DOI] [PubMed] [Google Scholar]
- 9.Dumur C.I., Campbell D.J.W., DeWitt J.L., Oyesanya R.A., Sirica A.E. Differential gene expression profiling of cultured neu-transformed versus spontaneously-transformed rat cholangiocytes and of corresponding cholangiocarcinomas. Exp Mol Pathol. 2010;89:227–235. doi: 10.1016/j.yexmp.2010.08.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Campbell D.J.W., Dumur C.I., Lamour N.F., DeWitt J.L., Sirica A.E. Novel organotypic culture model of cholangiocarcinoma progression. Hepatol Res. 2012;42:1119–1130. doi: 10.1111/j.1872-034X.2012.01026.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Fingas C.D., Bronk S.F., Werneburg N.W., Mott J.L., Guicciardi M.E., Cazanave S.C., Mertens J.C., Sirica A.E., Gores G.J. Myofibroblast-derived PDGF-BB promotes hedgehog survival signaling in cholangiocarcinoma cells. Hepatology. 2011;54:2076–2088. doi: 10.1002/hep.24588. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Fausther M., Goree J.R., Lavoie É.G., Graham A.L., Sévigny J., Dranoff J.A. Establishment and characterization of rat portal myofibroblast cell lines. PLoS One. 2015;10:e0121161. doi: 10.1371/journal.pone.0121161. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Clapéron A., Mergey M., Aoudjehane L., Ho-Bouldoires T.H.N., Wendum D., Prignon A., Merabtene F., Firrincieli D., Desbois-Mouthon C., Scatton O., Conti F., Housset C., Fouassier L. Hepatic myofibroblasts promote the progression of human cholangiocarcinoma through activation of epidermal growth factor receptor. Hepatology. 2013;58:2001–2011. doi: 10.1002/hep.26585. [DOI] [PubMed] [Google Scholar]
- 14.Shamir E.R., Ewald A.J. Three-dimensional organotypic culture: experimental models of mammalian biology and disease. Nat Rev Mol Cell Biol. 2014;15:647–664. doi: 10.1038/nrm3873. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Weigelt B., Ghajar C.M., Bissell M.J. The need for complex 3-D culture models to unravel novel pathways and identify accurate biomarkers in breast cancer. Adv Drug Deliv Rev. 2014;69-70:42–51. doi: 10.1016/j.addr.2014.01.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Sirica A.E., Almenara J.A., Li C. Periostin in intrahepatic cholangiocarcinoma: pathological insights and clinical implications. Exp Mol Pathol. 2014;97:515–524. doi: 10.1016/j.yexmp.2014.10.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Yu L., Feng M., Kim H., Phung Y., Kleiner D.E., Gores G.J., Qian M., Wang X.W., Ho M. Mesothelin as a potential therapeutic target in human cholangiocarcinoma. J Cancer. 2010;1:141–149. doi: 10.7150/jca.1.141. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Nomura R., Fujii H., Abe M., Sugo H., Ishizaki Y., Kawasaki S., Hino O. Mesothelin expression is a prognostic factor in cholangiocellular carcinoma. Int Surg. 2013;98:164–169. doi: 10.9738/INTSURG-D-13-00001.1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Zhang Z., Oyesanya O., Campbell D.J.W., Almenara J.A., DeWitt J.L., Sirica A.E. Preclinical assessment of simultaneous targeting of epidermal growth factor receptor (ERBB1) and ERBB2 as a strategy for cholangiocarcinoma therapy. Hepatology. 2010;52:975–986. doi: 10.1002/hep.23773. [DOI] [PubMed] [Google Scholar]
- 20.Singh B.N., Fu J., Srivastava R.K. Shankar S Hedgehog signaling antagonist GDC-0449 (Vismodegib) inhibits pancreatic cancer stem cell characteristics: molecular mechanisms. PLoS One. 2011;6:e27306. doi: 10.1371/journal.pone.0027306. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Kuczynski E.A., Lee C.R., Man S., Chen E., Kerbel R.S. Effect of sorafenib dose on acquired reversible resistance and toxicity in hepatocellular carcinoma. Cancer Res. 2015;75:2510–2519. doi: 10.1158/0008-5472.CAN-14-3687. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Rodón J., Carducci M., Sepulveda-Sánchez J.M., Azaro A., Calvo E., Seoane J., Braña I., Sicart E., Gueorguieva I., Cleverly A., Sokalingum Pillay N., Desaiah D., Estrem S.T., Paz-Ares L., Holdhoff M., Blakeley J., Lahn M.M., Baselga J. Pharmacokinetic, pharmacodynamics and biomarker evaluation of transforming growth factor-β receptor I kinase inhibitor, galunisertib, in phase 1 study in patients with advanced cancer. Invest New Drugs. 2015;33:357–370. doi: 10.1007/s10637-014-0192-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Gnainsky Y., Spira G., Paizi M., Bruck R., Nagler A., Abu-Amara S.N., Geiger B., Genina O., Monsonego-Ornan E., Pines M. Halofuginone, an inhibitor of collagen synthesis by rat stellate cells, stimulates insulin-like growth factor binding protein-1 synthesis by hepatocytes. J Hepatol. 2004;40:269–277. doi: 10.1016/j.jhep.2003.10.020. [DOI] [PubMed] [Google Scholar]
- 24.Sirica A.E., Zhang Z., Lai G.H., Asano T., Shen X.N., Ward D.J., Mahatme A., DeWitt J.L. A novel “patient-like” model of cholangiocarcinoma progression based on bile duct inoculation of tumorigenic rat cholangiocyte cell lines. Hepatology. 2008;47:1178–1190. doi: 10.1002/hep.22088. [DOI] [PubMed] [Google Scholar]
- 25.Subramanian A., Tamayo P., Mootha V.K., Mukherjee S., Ebert B.L., Gillette M.A., Paulovich A., Pomeroy S.L., Golub T.R., Lander E.S., Mesirov J.P. Gene set enrichment analysis: a knowledge-based approach for interpreting genome-wide expression profiles. Proc Natl Acad Sci U S A. 2005;102:15545–15550. doi: 10.1073/pnas.0506580102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Rich L., Whittaker P. Collagen and picrosirius red staining: a polarized light assessment of fibrillar hue and special distribution. Braz J Morphl Sci. 2005;22:97–104. [Google Scholar]
- 27.Lattouf R., Younes R., Lutomski D., Naaman N., Godeau G., Senni K., Changotade S. Picrosirius red staining: a useful tool to appraise collagen networks in normal and pathological tissues. J Histochem Cytochem. 2014;62:751–758. doi: 10.1369/0022155414545787. [DOI] [PubMed] [Google Scholar]
- 28.Sharma R., Rehani S., Mehendiratta M., Kardam P., Kumra M., Mathias Y., Yadav J., Sahay K. Architectural analysis of picrosirius red stained collagen in oral epithelial dysplasia and oral squamous cell carcinoma using polarization microscopy. J Clin Diagn Res. 2015;9:EC13–EC16. doi: 10.7860/JCDR/2015/13476.6872. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Lai G.H., Zhang Z., Shen X.N., Ward D.J., DeWitt J.L., Holt S.E., Rozich R.A., Hixson D.C., Sirica A.E. erbB2/neu transformed rat cholangiocytes recapitulate key cellular and molecular features of human bile duct cancer. Gastroenterology. 2005;129:2047–2057. doi: 10.1053/j.gastro.2005.10.010. [DOI] [PubMed] [Google Scholar]
- 30.Dranoff J.A., Kruglov E.A., Robson S.C., Braun N., Zimmermann H., Sévigny J. The ecto-nucleoside triphosphate diphosphohydrolase NTPDase2/CD39L1 is expressed in a novel functional compartment within the liver. Hepatology. 2002;36:1135–1144. doi: 10.1053/jhep.2002.36823. [DOI] [PubMed] [Google Scholar]
- 31.Irizarry R.A., Bolstad B.M., Collin F., Cope L.M., Hobbs B., Speed T.P. Summaries of Affymetrix GeneChip probe level data. Nucleic Acids Res. 2003;31:e15. doi: 10.1093/nar/gng015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Kosman E., Cohen Y. Procedures for calculating and differentiating synergism and antagonism in action of fungicide mixtures. Phytopathology. 1996;86:1263–1272. [Google Scholar]
- 33.Geerts A., Eliasson C., Niki T., Wielant A., Vaeyens F., Pekny M. Formation of normal desmin intermediate filaments in mouse hepatic stellate cells requires vimentin. Hepatology. 2001;33:177–188. doi: 10.1053/jhep.2001.21045. [DOI] [PubMed] [Google Scholar]
- 34.Iwaisako K., Brenner D.A., Kisseleva T. What's new in liver fibrosis? the origins of myofibroblasts in liver fibrosis. J Gastroenterol Hepatol. 2012;27:65–68. doi: 10.1111/j.1440-1746.2011.07002.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Dudas J., Mansuroglu T., Batusic D., Ramadori G. Thy-1 is expressed in myofibroblasts but not found in hepatic stellate cells following liver injury. Histochem Cell Biol. 2009;131:115–127. doi: 10.1007/s00418-008-0503-y. [DOI] [PubMed] [Google Scholar]
- 36.Iwaisako K., Jiang C., Zhang M., Cong M., Moore-Morris T.J., Park T.J., Liu X., Xu J., Wang P., Paik Y.H., Meng F., Asagiri M., Murray L.A., Hofmann A.F., Iida T., Glass C.K., Brenner D.A., Kisseleva T. Origin of myofibroblasts in the fibrotic liver in mice. Proc Natl Acad Sci U S A. 2014;111:E3297–E3305. doi: 10.1073/pnas.1400062111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Sirica A.E., Dumur C.I., Campbell D.J.W., Almenara J.A., Ogunwobi O.O., DeWitt J.L. Intrahepatic cholangiocarcinoma progression: prognostic factors and basic mechanisms. Clin Gastroenterol Hepatol. 2009;7:S68–S78. doi: 10.1016/j.cgh.2009.08.023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Xu J., Kisseleva T. Bone marrow-derived fibrocytes contribute to liver fibrosis. Exp Biol Med (Maywood) 2015;240:691–700. doi: 10.1177/1535370215584933. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Yovchev M.I., Grozdanov P.N., Zhou H., Racherla H., Guha C., Dabeva M.D. Identification of adult hepatic progenitor cells capable of repopulating injured rat liver. Hepatology. 2008;47:636–647. doi: 10.1002/hep.22047. [DOI] [PubMed] [Google Scholar]
- 40.Roderfeld M., Rath T., Voswinckel R., Dierkes C., Dietrich H., Zahner D., Graf J., Roeb E. Bone marrow transplantation demonstrates medullar origin of CD34+ fibrocytes and ameliorates hepatic fibrosis in Abcb4−/− mice. Hepatology. 2010;51:267–276. doi: 10.1002/hep.23274. [DOI] [PubMed] [Google Scholar]
- 41.Bosselut N., Housset C., Marcelo P., Rey C., Burmester T., Vinh J., Vaubourdolle M., Cadoret A., Baudin B. Distinct proteomic features of two fibrogenic liver cell populations: hepatic stellate cells and portal myofibroblasts. Proteomics. 2010;10:1017–1028. doi: 10.1002/pmic.200900257. [DOI] [PubMed] [Google Scholar]
- 42.Lemoinne S., Cadoret A., Rauton P.E., El Mourabit H., Ratziu V., Corpechot C., Rey C., Bosselut N., Barbu V., Wendum D., Feldmann G., Boulanger C., Henegar C., Housset C., Thabut D. Portal myofibroblasts promote vascular remodeling underlying cirrhosis formation through the release of microparticles. Hepatology. 2015;61:1041–1055. doi: 10.1002/hep.27318. [DOI] [PubMed] [Google Scholar]
- 43.Lee K.S., Buck M., Houglum K., Chojkier M. Activation of hepatic stellate cells by TGFα and collagen type I is mediated by oxidative stress through c-myb expression. J Clin Invest. 1995;96:2461–2468. doi: 10.1172/JCI118304. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Werneburg N.W., Yoon J.H., Higuchi H., Gores G.J. Bile acids activate EGF receptor via TGF-α-dependent mechanism in human cholangiocyte cell lines. Am J Physiol Gastrointest Liver Physiol. 2003;285:G31–G36. doi: 10.1152/ajpgi.00536.2002. [DOI] [PubMed] [Google Scholar]
- 45.Lua I., James D., Wang J., Wang K.S., Asahina K. Mesodermal mesenchymal cells give rise to myofibroblasts, but not epithelial cells, in mouse liver injury. Hepatology. 2014;60:311–322. doi: 10.1002/hep.27035. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Chu A.S., Diaz R., Hui J.J., Yanger K., Zong Y., Alpini G., Stanger B.Z., Wells G. Lineage tracing demonstrates no evidence of cholangiocyte epithelial-to-mesenchymal transition in murine models of hepatic fibrosis. Hepatology. 2011;53:1685–1695. doi: 10.1002/hep.24206. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Mederacke I., Hsu C.C., Troeger J.S., Huebener P., Mu X., Dapito D.H., Pradere J.P., Schwabe R.F. Fate-tracing reveals hepatic stellate cells as dominant contributors to liver fibrosis independent of its etiology. Nat Commun. 2013;4:2823. doi: 10.1038/ncomms3823. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Sukowati C.H.C., Anfuso B., Crocé L.S., Tiribelli C. The role of multipotent cancer associated fibroblasts in hepatocarcinogenesis. BMC Cancer. 2015;15:188. doi: 10.1186/s12885-015-1196-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Baker D.E., Harrison N.J., Maltby E., Smith K., Moore H.D., Shaw P.J., Heath P.R., Holden H., Andrews P.W. Adaptation to culture of human embryonic stem cells and oncogenesis in vivo. Nat Biotechnol. 2007;25:207–215. doi: 10.1038/nbt1285. [DOI] [PubMed] [Google Scholar]
- 50.Mayshar Y., Ben-David U., Lavon N., Biancotti J.C., Yakir B., Clark A.T., Plath K., Lowry W.E., Benvenisty N. Identification and classification of chromosomal aberrations in human induced pluripotent stem cells. Cell Stem Cell. 2010;7:521–531. doi: 10.1016/j.stem.2010.07.017. [DOI] [PubMed] [Google Scholar]
- 51.Ben-David U., Mayshar Y., Benvevisty N. Large-scale analysis reveals acquisition of lineage-specific chromosomal aberrations in human adult stem cells. Cell Stem Cell. 2011;9:97–102. doi: 10.1016/j.stem.2011.06.013. [DOI] [PubMed] [Google Scholar]
- 52.Mueller L., Goumas F.A., Affeldt M., Sandtner S., Gehling U.M., Brilloff S., Walter J., Karnatz N., Lamszus K., Rogiers X., Broering D.C. Stromal fibroblasts in colorectal liver metastases originate from resident fibroblasts and generate an inflammatory microenvironment. Am J Pathol. 2007;171:1608–1618. doi: 10.2353/ajpath.2007.060661. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Lua I., Yuchang Y., Zagory J.A., Wang K.S., French S.W., Sévigny J., Asahina K. Characterization of hepatic stellate cells, portal fibroblasts, and mesothelial cells in normal and fibrotic livers. J Hepatol. 2016;64:1137–1146. doi: 10.1016/j.jhep.2016.01.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Li Z., Dranoff J.A., Chan E.P., Uemura M., Sévigny J., Wells R.G. Transforming growth factor-β and substrate stiffness regulate portal fibroblast activation in culture. Hepatology. 2007;46:1246–1256. doi: 10.1002/hep.21792. [DOI] [PubMed] [Google Scholar]
- 55.Goree J.R., Lavoie E.G., Fausther M., Dranoff J.A. Expression mediators of purinergic signaling in human liver cell lines. Purinergic Signal. 2014;10:631–638. doi: 10.1007/s11302-014-9425-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Krishnan R., Cleary E.G. Elastin gene expression in elastotic human breast cancers and epithelial cell lines. Cancer Res. 1990;50:2164–2171. [PubMed] [Google Scholar]
- 57.Kadaba R., Birke H., Wang J., Hooper S., Andl C.D., Di Maggio F., Soylu E., Ghallab M., Bor D., Froeling F.E.M., Bhattacharya S., Rustgi A.K., Sahai E., Chelala C., Sasieni P., Kocher H.M. Imbalance of desmoplastic stromal cell numbers drives aggressive cancer processes. J Pathol. 2013;230:107–117. doi: 10.1002/path.4172. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Olsen A.L., Bloomer S.A., Chan E.P., Gaca M.D.A., Georges P.C., Sackey B., Sackey B., Uemura M., Janmey P.A., Wells R.G. Hepatic stellate cells require a stiff environment for myofibroblastic differentiation. Am J Physiol Gastrointest Liver Physiol. 2011;301:G110–G118. doi: 10.1152/ajpgi.00412.2010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Puche J.E., Saiman Y., Friedman S.L. Hepatic stellate cells and liver fibrosis. Compr Physiol. 2013;3:1473–1492. doi: 10.1002/cphy.c120035. [DOI] [PubMed] [Google Scholar]
- 60.Shen H., Huang G.J., Gong Y.W. Effect of transforming growth factor beta and bone morphogenetic proteins on rat hepatic stellate cell proliferation and trans-differentiation. World J Gastroenterol. 2003;9:784–787. doi: 10.3748/wjg.v9.i4.784. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Wells R.G., Kruglov E., Dranoff J.A. Autocrine release of TGF-β by portal fibroblasts regulates cell growth. FEBS Lett. 2004;559:107–110. doi: 10.1016/S0014-5793(04)00037-7. [DOI] [PubMed] [Google Scholar]
- 62.Löhr M., Schmidt C., Ringel J., Kluth M., Müller P., Nizze H., Jesnowski R. Transforming growth factor-β1 induces desmoplasia in an experimental model of human pancreatic carcinoma. Cancer Res. 2001;61:550–555. [PubMed] [Google Scholar]
- 63.Zen Y., Harada K., Sasaki M., Chen T.C., Chen M.F., Yeh T.S., Jan Y.Y., Huang S.F., Nimura Y., Nakanuma Y. Intrahepatic cholangiocarcinoma escapes from growth inhibitory effect of transforming growth factor-β1 by overexpression of cyclin D1. Lab Invest. 2005;85:572–581. doi: 10.1038/labinvest.3700236. [DOI] [PubMed] [Google Scholar]
- 64.Shimizu T., Yokomura S., Mizuguchi Y., Kawahigashi Y., Arima Y., Taniai N., Mamada Y., Yoshida H., Akimara K., Tajiri T. Effect of transforming growth factor-β1 on human intrahepatic cholangiocarcinoma cell growth. World J Gastroenterol. 2006;12:6316–6324. doi: 10.3748/wjg.v12.i39.6316. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Xue T.C., Zhang B.H., Ye S.L., Ren Z.G. Differentially expressed gene profiles of intrahepatic cholangiocarcinoma, hepatocellular carcinoma, and combined hepatocellular-cholangiocarcinoma by integrated microarray analysis. Tumour Biol. 2015;36:5891–5899. doi: 10.1007/s13277-015-3261-1. [DOI] [PubMed] [Google Scholar]
- 66.Bruck R., Genina O., Aeed H., Alexiev R., Nagler A., Avni Y., Pines M. Halofuginone to prevent and treat thioacetamide-induced liver fibrosis in rats. Hepatology. 2001;33:379–386. doi: 10.1053/jhep.2001.21408. [DOI] [PubMed] [Google Scholar]
- 67.Spector I., Honig H., Kawada N., Nagler A., Genin O., Pines M. Inhibition of pancreatic stellate cell activation by halofuginone prevents pancreatic xenograft tumor development. Pancreas. 2010;39:1008–1015. doi: 10.1097/MPA.0b013e3181da8aa3. [DOI] [PubMed] [Google Scholar]
- 68.Ling H., Roux E., Hempel D., Tao J., Smith M., Lonning S., Zuk A., Arbeeny C., Ledbetter S. Transforming growth factor β neutralization ameliorates pre-existing hepatic fibrosis and reduces cholangiocarcinoma in thioacetamide-treated rats. PLoS One. 2013;8:e54499. doi: 10.1371/journal.pone.0054499. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Mu X., Pradere J.P., Affò S., Dapito D.H., Friedman R., Lefkovich J.H., Schwabe R.F. Epithelial transforming growth factor-β signaling does not contribute to liver fibrosis but protects mice from cholangiocarcinoma. Gastroenterology. 2016;150:720–733. doi: 10.1053/j.gastro.2015.11.039. [DOI] [PMC free article] [PubMed] [Google Scholar]