Abstract
Cancer cells use alternate energetic pathways; however, cancer stem cell (CSC) metabolic energetic pathways are unknown. The purpose of this study was to define the metabolic characteristics of head and neck cancer at different points of its pathogenesis with a focus on its CSC compartment. UPLC-MS/MS-profiling and GC-MS-validation studies of human head and neck cancer tissue, saliva, and plasma were used in conjunction with in vitro and in vivo models to carry out this investigation. We identified metabolite biomarker panels that distinguish head and neck cancer from healthy controls, and confirmed involvement of glutamate and glutaminolysis. Glutaminase, which catalyzes glutamate formation from glutamine, and aldehyde dehydrogenase (ALDH), a stemness marker, were highly expressed in primary and metastatic head and neck cancer tissues, tumorspheres, and CSC versus controls. Exogenous glutamine induced stemness via glutaminase, whereas inhibiting glutaminase suppressed stemness in vitro and tumorigenesis in vivo. Head and neck CSC (CD44hi/ALDHhi) exhibited higher glutaminase, glutamate, and sphere levels than CD44lo/ALDHlo cells. Glutaminase drove transcriptional and translational ALDH expression, and glutamine directed even CD44lo/ALDHlo cells toward stemness. Glutaminolysis regulates tumorigenesis and CSC metabolism via ALDH. These findings indicate that glutamate is an important marker of cancer metabolism whose regulation via glutaminase works in concert with ALDH to mediate cancer stemness. Future analyses of glutaminolytic-ALDH driven mechanisms underlying tumorigenic transitions may help in the development of targeted therapies for head and neck cancer and its CSC compartment.
Keywords: metabolomics, head and neck squamous cell carcinoma, glutaminolysis, glutamic acid, stemness, cancer stem cells, tumorigenesis
Graphical Abstract

INTRODUCTION
A hallmark of cancer cells is an ability to shift to alternate energetic pathways. Cancer stem cells are thought to contribute to cancer development and progression; however, their metabolic energetic pathways are not known. Two major metabolic pathways by which mammalian cells obtain energy are lactate fermentation and aerobic respiration. Cancer cells shift from aerobic respiration (TCA cycle) to lactic acid fermentation (glucose to pyruvate to lactate) for energy production: the Warburg Effect.1,2 Cancer cells can also shift to alternate energy sources, including shifting from glycolysis or lactic acid fermentation to glutaminolysis.3–5 Glutaminolysis, or the conversion of glutamine to lactate via glutamate, α-ketoglutarate, and the TCA cycle, complements glucose metabolism/the Warburg effect by providing high-energy cofactors.6 In fact, glutaminolysis dependence may supersede that of glucose-dependence for the synthesis of certain metabolic components. Glutaminolysis leads to a glutamine addiction in some cancer cell systems, and the oncogene Myc can coordinate the expression of genes necessary for glutamine catabolism.3 Thus, cancer cells adapt to alternate energy sources depending on their genetic profile and substrate availability.5,7 These unique cancer cell properties underscore the important concept that targeting one metabolic pathway may not be sufficient. Furthermore, resistance emerges in aggressive cancers, enabling them to elude targeted therapies.8 This resistance profile may include an altered metabolic signature9–12 Thus, knowing how regulators of metabolism are altered during carcinogenesis, including stemness, is important for developing novel therapies.
Cancer stem cells (CSCs) contribute to cancer development and progression, yet their metabolic energetic pathways are not completely known.13 The fact that many stem cells rely heavily on anaerobic glycolysis suggests that these bioenergetic pathways may be central to CSC properties.14 Indeed, knowledge about the processes that govern CSC metabolism and energetics is emerging.7,15–21
Smoking, alcohol consumption, and HPV are risk factors for head and neck cancer. Like other cancers, evidence suggests that CSCs play a role in the tumorigenicity of these malignancies. Oral squamous cell carcinoma, the most common malignant oral neoplasm, accounts for 90% of all oral malignancies and has a poor 5-year survival rate that has not changed in decades, underscoring the need to identify novel therapeutic targets.22 Understanding processes that govern tumorigenesis of CSC may contribute to identifying new avenues for therapeutics.
In addition to the expressed genome and proteome, scientists can now study a substantial component of the tumor metabolome. Such an analysis can provide an in-depth understanding of how metabolic pathways are involved in the pathogenesis of cancer. Mass spectrometry and NMR-based metabolomics offer a novel platform for the development of marker panels that are characteristic of disease phenotypes or cellular processes that are readily measured in biofluids and tissues. Metabolomic profiling yields a signature set of functional metabolites of disease phenotypes. Using NMR-based metabolomic profiling of human primary and metastatic head and neck squamous cell carcinoma (HNSCC) tissue specimens, we previously determined that elevated glutamate was a distinctive feature of primary and metastatic HNSCC.23 Elevated levels of glutamate suggested that HNSCC depends on highly active glutaminolysis for carbons that contribute to anaplerosis of the TCA cycle. Indeed, others have shown that glutamine addiction/glutamate metabolism is important in multiple cancer types, including HNSCC.3,24–27 Glutamic acid is a nonessential amino acid whose carboxylate anions and salts are known as glutamates. Glutamate, a key compound in cellular metabolism, is generated from glutamine by the enzyme glutaminase, whereas glutamine is synthesized by the enzyme glutamine synthetase from glutamate and ammonia. Glutaminolysis is a series of biochemical reactions in which the amino acid glutamine is degraded to glutamate, aspartate, CO2, pyruvate, lactate, alanine, and citrate. The purpose of our study was to define the metabolic characteristics of HNSCC at different points of its pathogenesis with a focus on its CSC compartment.
MATERIALS AND METHODS
Ethical Review Board
This study was conducted under Ethical Review Board approved protocols, including those reviewed by the University of Michigan and affiliates of ProteoGenex.
Tissue Sample Collection and Histopathological Evaluation
Frozen HNSCC tissue specimens were obtained from ProteoGenex (ProteoGenex Inc., Culver City, CA, USA). Inclusion criteria consisted of histological confirmation of HNSCC, and tissue specimens were histologically confirmed to contain over 90% tumor tissue prior to analysis. Confounding variables included age, sex, race/ethnicity, tumor site, cancer stage, and grade. All tissues were snap-frozen in liquid nitrogen immediately after surgery and preserved in −80 °C. In total, 103 tissue samples (normal, tumor, and metastatic) originating from the tongue, oral cavity, lip, larynx, and lymph-nodes of 47 HNSCC patients (41 males and 6 females with an age range of 39–88 years and mean age of 58.1 ± 5.7 years) were investigated in the present study. Among these tissues, matched normal, tumor, and metastatic tissues were obtained from 19 patients, matched normal and tumor tissues were obtained from 18 patients, and metastatic only tissues were obtained from 10 patients. The patients’ information (race, ethnicity, gender, and age), clinical diagnosis, TNM [extent of tumor (T), extent of spread to lymph nodes (N), and the presence of distant metastasis (M)] staging, and tumor grade, established by histopathological evaluation, are included in Table 1 and Supporting Information; Tables S1, S3, S6, and S7.
Table 1.
Demographic and Clinical Characteristics of Healthy and HNSCC Patients
| Tissues | Saliva | Plasma | |
|---|---|---|---|
| Number of samples | 103 | 75 | 14 |
| Male | 41 | 57 | 10 |
| Female | 6 | 18 | 4 |
| Age | 39–88 | 34–84 | 40–63 |
| Race/Ethnicity | Caucasian/European/American/White | Caucasian/European/American/White | Caucasian/European/American/White |
| Primary disease site | tongue, oral cavity, lip, larynx, hypopharynx, lymp node | tongue, oral cavity, lip, larynx, hypopharynx, oropharynx, nasopharynx | oral cavity, mouth floor mucosa, mandible |
Saliva Sample Collection
Subjects were instructed to refrain from eating, drinking, smoking, and oral hygiene for a minimum of 2 h before saliva collection. All saliva was collected in the AM hours to control for circadian variation in salivary gland function. There was no exclusion on the basis of age, sex, and ethnicity. Subjects were instructed to rinse out with warm water and avoid use of mouthwashes or fluoride rinses. After saliva collection, the volumes of all saliva samples were determined gravimetrically on an analytical balance, assuming a specific gravity of 1.0. Saliva samples were aliquoted and stored at −80 °C. In total, 75 saliva samples from 55 HNSCC patients (mean age 61.0 ± 13.0 years) and 18 saliva samples from healthy controls (mean age 55.1 ± 5.7 years) were obtained from the UM Head and Neck SPORE and ProteoGenex for the metabolomic profiling and validation studies. Whole saliva samples were processed by centrifugation and then aliquoted and stored at −80 °C until analyzed. Clinical information for the patients is summarized in Table 1 and Supporting Information; Tables S2 and S4.
Plasma Sample Collection
Blood samples were collected into EDTA vacutainer tubes and immediately mixed gently after blood collection. Plasma was separated from the entire blood by centrifugation at 1500g for 10 min at 4 °C. Plasma samples were aliquoted in 2.0 mL cryovials and stored at −80 °C. In total, 14 plasma samples from 7 HNSCC patients and 7 plasma samples from healthy controls (mean age 52.4 ± 8.25) were obtained from ProteoGenex for validation studies (Table 1 and Supporting Information; Table S5).
Metabolomic Profiling
Sample Preparation
Samples were sent to Metabolon, Inc. (Durham, NC) for metabolomic profiling studies. Briefly, samples were prepared using the automated MicroLab STAR system from the Hamilton Company (Reno, NV). A recovery standard was added prior to the first step in the extraction process for Quality Control (QC) purposes. To remove protein, dissociate small molecules bound to protein or trapped in the precipitated protein matrix, and to recover chemically diverse metabolites, proteins were precipitated with methanol under vigorous shaking for 2 min (Glen Mills GenoGrinder 2000) followed by centrifugation. The resulting extract was divided into five fractions: one for analysis by Ultrahigh Performance Liquid Chromatography-Tandem Mass Spectroscopy (UPLC-MS/MS) with positive ion mode electrospray ionization, one for analysis by UPLC-MS/MS with negative ion mode electrospray ionization, one for analysis by UPLC-MS/MS polar platform (negative ionization), one for analysis by Gas Chromatography–Mass Spectroscopy (GC-MS), and one sample was reserved for backup. Samples were placed briefly on a TurboVap (Zymark) to remove the organic solvent. For LC, the samples were stored overnight under nitrogen before preparation for analysis. For GC, each sample was dried under vacuum overnight before preparation for analysis. For a complete description of quality assurance and quality control measures please review Supporting Information reports (tissue and saliva); Report S1.
Validation of Glutamate and Glutamine by GC-MS
The levels of glutamate and glutamine were assessed in tissues, cell lines, saliva, and plasma samples using isotype dilution GC-MS as described previously.28 In brief, frozen tissues, saliva, plasma, or cell lysate pellets were homogenized in methanol after spiking with labeled internal standards (d5-glutamic acid and d4-glutamine) and extracted overnight at 4 °C, with constant shaking. The extraction was carried out using a 1:1 molar ratio of water/chloroform at room temperature. The aqueous methanolic layer was collected and dried completely. The dried methanolic extract was azeotroped twice by adding 100 μL of dimethylformamide, mixed by vortexing, and dried using a Speed Vac for 30 min. One hundred microliters of dimethylformamide and N-methyl-N-tert-butylmethylsilyltri-fluoroacetamide + 1% tert-butyl-dimethylchlorosilane were then added to the dried sample, capped, and incubated at 60 °C for 1 h. Selective ion monitoring was used for quantification. The amount of glutamate and glutamine were calculated by measuring the peak area of the native glutamate (m/z = 432) and glutamine (m/z = 431) to that corresponding to spiked isotope-labeled glutamate (m/z = 437) and glutamine (m/z = 436), respectively. The levels of glutamate and glutamine were normalized to the tissue weight.
Cell Lines and Culture
The human HNSCC cell lines were as follows: UM-SCC-17B (supraglottis/soft tissue-neck) and UM-SCC-14A (floor of mouth; both provided by Thomas Carey, University of Michigan, Ann Arbor, MI);29 HSC-3 (tongue; provided by Randall Kramer, University of California, San Francisco, CA);30 OSCC-3 (tongue; provided by Mark Lingen, University of Chicago, Chicago, IL). HNSCC cell line authentication and origin were provided by their sources. HNSCC cells were maintained in Dulbecco’s modified Eagle’s medium containing 10% fetal bovine serum, 1% penicillin, and 1% streptomycin. Primary human oral keratinocytes (ScienCell, Carlsbad, CA) were maintained in oral keratinocyte medium (OKM) (ScienCell, Carlsbad, CA).
Immunohistochemical Staining
Standard immunohistochemical analyses were used to evaluate glutaminase expression in normal and tumor tissue sections using a glutaminase primary antibody (AP8809b, Abgent, San Diego, CA). Staining intensities [1 (weak), 2 (moderate), and 3 (strong)] for glutaminase were graded and analyzed in a blinded manner by a pathologist. Low expression was defined as intensity 1, and high expression was defined as intensity 2 or 3.
Immunoblot Analysis
To evaluate the protein expression levels of glutaminase and ALDH1, standard Western blot analyses were performed using an antiglutaminase primary antibody (ab607709, Abcam, San Francisco, CA) or an ALDH1 primary antibody (61195, BD Transduction, Franklin Lakes, NJ). β-Actin (SC-1615, Santa Cruz Biotechnology) served as a loading control.
Tumorspheres and Control Adherent HNSCC Cells
HNSCC sphere assays, designated tumorspheres, were used to assess stemness, a property thought to contribute to metastatic potential. HNSCC tumorspheres (UM-SCC-14A) were prepared as previously reported.31 In other experiments, cells were treated with either glutamine (10–30 μg/mL) (49419, Sigma-Aldrich, St. Louis, MO) or 6-diazo-5-oxo-L-norleucine (DON; 1 mM) (D2141, Sigma-Aldrich, St. Louis, MO) and cultured under adherent or suspension conditions for 72 h.
Development of Stable Glutaminase Suppression in HNSCC Cell Lines
UM-SCC-14A cells were transduced with glutaminase-shRNA (SC-105592-V, Santa Cruz Biotecnology, Santa Cruz, CA) or scrambled-shRNA (SC-108080; Santa Cruz Biotechnology) lentiviral particles in 0.5 mL of serum-free media, and then selected in 10 μg/mL puromycin (sc-108071; Santa Cruz Biotechnology, Santa Cruz, CA) for an additional 10 days. Surviving cell colonies were picked and propagated before testing for glutaminase expression using Western blot analysis.
Oral Cancer Mouse Model
To validate the significance of glutaminolysis/glutaminase in regulating tumorsphere formation or stemness and tumorigenesis in vivo, an oral cancer mouse model was used. All protocols were approved by the UM University Committee on Use and Care of Animals. HNSCC cells grown as tumorspheres or under control adherent conditions were prepared as described above and used in the mouse model as described previously.31,32 For these experiments, 3-week old male mice (NCr-nu/nu strain, NCI, Frederick, MD) were purchased and used for all experimental protocols. In another set of experiments, HNSCC cells (1 × 106 cells/0.05) that exhibited stably suppressed levels of glutaminase (described in detail above) and control transduced cells (1 × 106 cells/0.05) were also tested in the mouse model.
Isolation of ALDHhi CD44hi and ALDHlo CD44lo HNSCC Cells and Flow Cytometry
The ALDEFLUOR assay kit (01700, StemCell, Vancouver, British Columbia, Canada) was used to isolate the ALDHhi CD44hi and ALDHlo CD44lo HNSCC cells according to the manufacturer’s instructions. Briefly, HNSCC cells (UM-SCC-14A) were trypsinized and washed with PBS, and then the Aldefluor substrate was added to 1 × 106 cells/mL suspended in Aldefluor assay buffer and incubated at 37 °C for 45 min. For each sample, 5 μL of diethylaminobenzaldehyde (DEAB), a specific ALDH inhibitor, was added to 0.5 mL of Aldefluor-stained cells as a negative control. Cells were costained with an APC antihuman CD44 antibody (559942, BD Pharmingen, San Diego, CA). The expression of CD44 and ALDH was analyzed by flow cytometry using a FACS Aria Cell Sorter (BD Biosciences, San Jose, CA).
For tumorsphere assays, ALDHhi CD44hi and ALDHlo CD44lo cells, derived from FACS, were plated on poly-HEMA coated plates at a density of 5000 viable cells/well. Tumorsphere growth was observed under a microscope after 10 days.
Quantitative PCR
Total RNA was isolated from cells and tissue samples using the RNeasy mini, RNA isolation kit (74104, Qiagen, Vaencia, CA) according to manufacturer’s instructions. The cDNA was synthesized using total RNA, oligo-dt, primers, and reverse transcriptase (4331182, Applied Biosystems, city). Real-time qPCR was performed with gene-specific primers (Glutaminase, ALDH1A1, or c-myc; Supporting Information; Table S8) and the SYBR Green PCR Master Mix (4369016, Qiagen, Vaencia, CA) using a thermal cycler (Applied Biosystems 7700, Foster city, CA). The gene of interest was normalized against the reference gene glyceraldehyde-3-phosphate dehydrogenase (GAPDH). The expression level of each gene was calculated using the 2−ΔΔCt method.
Statistical Analyses and Data Management Systems
Metabolic Profiling and Validation
Following log transformation and imputation of missing values, with the minimum observed value for each compound, ANOVA contrasts (tissues) and Welch’s two-sample t test (saliva) were used to identify metabolites that differed significantly between experimental groups (p ≤ 0.05 and 0.05 < p < 0.10). Analysis by two-way ANOVA with repeated measures identified features exhibiting the main effects of the group experimental parameter. An estimate of the false discovery rate (q-value) is calculated to take into account the multiple comparisons that normally occur in metabolomic-based studies. Q-values are provided for further evaluation of metabolomic trends within the data set; a typical q-value cutoff used with metabolomic data sets is q ≤ 0.05 (a potential for 5% false observations). However, where metabolites move in a consistent direction across a pathway, a lower q-value threshold may be used. Refer to the Supporting Information for general definitions, further descriptions of false discovery rate, and other statistical tests and analyses or classification and clustering systems used for data management, including the Random Forest Classification, Hierarchical Clustering, and Principal Component Analysis (Supporting Information (tissue and saliva); Report S1.
For all other analyses and functional studies, values were expressed as means ± SD. Intergroup differences were determined using ANOVA and a Tukey–Kramer HSD posthoc test. Statistical significance was defined as p ≤ 0.05. For the in vivo studies, independent t tests with unequal variances were used. All experiments were repeated at least three times.
RESULTS
Metabolomic Profiling Reveals Significant Numbers of Altered Metabolites in Matched Primary and Metastatic HNSCC and Normal Adjacent Control Tissues
Global metabolic profiling of 19 (n = 57) matched pairs of primary and metastatic HNSCC tissues and normal adjacent control tissues revealed that 405 (p < 0.05) metabolites were statistically different in multiple comparisons out of 569 named features (Supporting Information; Report S1, (tissue)). When comparing primary HNSCC to control tissues, 301 metabolites were elevated and 84 were downregulated or a total of 385 metabolites were altered (p < 0.05). When comparing metastatic HNSCC to control tissues, 288 metabolites were elevated and 95 were downregulated or a total of 383 metabolites were altered (p < 0.05). When comparing metastatic to primary HNSCC tissues, 22 metabolites were elevated and 41 were downregulated or a total of 63 metabolites were altered (p < 0.05) (Supporting Information (tissue); Reports S2 and S3, and Supporting Information; Figure S1 (tissue)).
Metabolomic Profiling Reveals Significant Numbers of Altered Metabolites in HNSCC Saliva Compared to Healthy Controls
Global metabolic profiling of 60 HNSCC (n = 47) and normal (n = 13) saliva samples revealed that 234 (p < 0.05) metabolites were statistically different in comparisons out of 481 named biochemicals (Supporting Information; Report S1, (saliva)). Of these, 120 were upregulated and 114 were downregulated (p < 0.05). (Supporting Information; Reports S4 and S5 (saliva)) and Supporting Information; Figure S1 (saliva)).
Principal Component Analyses of Metabolic Variables Show Separation of HNSCC Tumors from Normal Tissues and by Grade, and Separation of HNSCC from Normal Saliva
Using principal component analysis (PCA) to transform the large number of metabolic variables into a smaller number of orthogonal variables in order to analyze variation between groups and to provide a high-level overview of the data set, we found that samples derived from normal adjacent tissue showed good separation from tumor samples (Figure 1A). Primary and metastatic tumor samples formed an overlapping population in the PCA (Figure 1A). When analyzed by tumor grade, tumor samples again formed overlapping populations; however, low-grade tumors appeared to pull away from high-grade tumors on the PCA (Figure 1B). Nevertheless, a larger sample size is needed to examine this further. In the hierarchical clustering analysis (HCA), control samples tended to cluster separately from the tumor samples (Figure 1C). Tumor samples tended to form subclusters with their matched tumor pair rather than by tumor status (primary or metastatic), indicating subtle differences in metabolism in the individual tumors.
Figure 1.
Metabolomic profiling of altered metabolites in matched primary and metastatic HNSCC and normal adjacent control tissues, and saliva samples. (A) PCA score plot showing differential grouping among normal, primary, and metatastic tissues and (B) by tumor grade. (C) HCA of normal, primary, and metastatic tissues. (D) PCA score plot showing differential grouping among normal and HNSCC saliva samples.
PCA of saliva metabolites derived from normal and HNSCC samples tended to form partially overlapping populations, with HNSCC samples showing a wider spread across the PCA (Figure 1D). In hierarchical clustering, control and HNSCC saliva samples tended to cluster by disease status (data not shown).
Random Forest Analyses of Metabolic Variables for HNSCC Tumor Tissue and Saliva Yield a High Predictive Accuracy
Random forest analyses (RFA) showed good efficiency at separating control, primary, and metastatic samples, with a predictive accuracy of 67% (Figure 2A). Random chance would be expected to yield a predictive accuracy of 33% in this analysis. The top 30 metabolites for predicting control, primary, and metastatic groups included metabolites related to lipid metabolism (caprate, docosatrienoate, CDP-choline, adrenate), the urea cycle (pro-hydroxy-pro, putrescine, N-acetylglutamate), inflammation or niacin biosynthesis (kynurenine), and glutaminolysis/glutamate metabolism (glutamate) (Figure 2A). RFA of saliva was very effective at separating normal controls from HNSCC samples, with a predictive accuracy of 100% (a 50% predictive accuracy would be expected by random chance) (Figure 2B). The top 30 metabolites for predicting groups included biochemicals related to xenobiotics (benzamide, levulinate, dihydroferulic acid, 5-hydroxymethylfurfural, benzoate), carbohydrates (3-phosphoglycerate, an isobar of fructose/glucose 1,6-diphosphate or myo-inositol 1,4 or 1,3-diphosphate, glucose-6-phosphate, and maltose), lipids (maleate, caproate, heptanoate, carnitine), nucleotides (2′,3′-cGMP, 3′-AMP, allantoin), and energetics/glutaminolysis/TCA metabolism (α-ketoglutarate).
Figure 2.

RFA of metabolic variables for HNSCC tumor tissue and saliva yield a high predictive accuracy. (A) RFA of metabolic variables for normal, primary, and metastatic tumor tissue; (B) RFA of metabolic variables for normal and HNSCC saliva samples showing the top 30 metabolites.
Validation Studies Confirm Highly Active Glutaminolysis in Primary and Metastatic HNSCC Tissues and Cells, Marked by High Glutamate Levels, Low Glutamine Levels, and High Glutaminase Expression
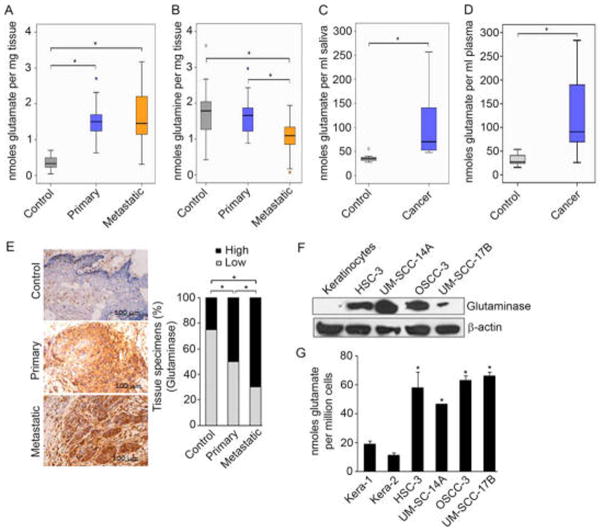
Given our previous NMR-based metabolomic studies of HNSCC cells and tissues,23,33 and the current data underscoring the important role of altered energetics in HNSCC tissues and saliva, we performed additional experiments to validate the role of glutaminolysis in HNSCC tumorigenesis. For these studies, we examined additional sets of HNSCC tissues and found that glutamate was differentially elevated in primary and metastatic HNSCC tissues compared to normal adjacent tissues (Figure 3A). Glutamine, the precursor to glutamate, was lowest in the metastatic tissues, suggesting an active level of glutaminolysis in metastatic tissues (Figure 3B). Parallel to changes in HNSCC tissues, studies of unmatched saliva and plasma samples from patients with HNSCC showed that glutamate levels were also significantly elevated in samples derived from HNSCC patients compared to healthy controls (Figure 3C and D).
Figure 3.
Validation studies in HNSCC revealed highly active glutaminolysis in primary and metastatic HNSCC tissues and cells, marked by high glutamate levels, low glutamine levels, and high glutaminase expression. (A and B) Box plots showing the levels of glutamate and glutamine in metastatic HNSCC (Mets; n = 26), primary HNSCC (n = 26), and normal control (n = 24) tissues based on isotope dilution GC-MS. (C and D) Box plots showing the levels of salivary glutamate in HNSCC patients (cancer; n = 10) and normal healthy controls (control; n = 8), and (D) plasma glutamate levels in HNSCC patients (cancer; n = 7) and normal healthy controls (control; n = 7) based on isotope dilution GC-MS. Intergroup differences were determined by Kruskal–Wallis tests for three-way comparisons between groups. Statistical significance was defined as *p ≤ 0.05. (E) (left) Representative normal control, primary HNSCC, or metastatic HNSCC tissue sections immunostained with antibodies against glutaminase; (right) percentage of tissue specimens expressing glutaminase determined by immunohistochemical staining. Staining intensity was graded as High or Low. McNemar’s test was used to compare the two proportions with significant differences set at *p ≤ 0.05. (F) Immunoblots show glutaminase levels in keratinocytes and HNSCC cells. β-Actin served as a loading control. (G) Glutamate levels in keratinocytes (Kera 1 and 2) and HNSCC cells (HSC-3, UM-SCC-14A, OSCC-3, and UM-SCC-17B), based on isotope dilution GC-MS.
The enzyme glutaminase catalyzes the formation of glutamate from glutamine; therefore, glutaminase expression was also examined in these HNSCC tissues. Immunohistochemical staining analyses revealed that glutaminase expression was high in primary and metastatic HNSCC tissues compared to normal tissues (Figure 3E). These findings support that glutaminase and glutaminolysis are important markers for the transition to tumorigenesis and to a more aggressive metastatic phenotype in HNSCC.
At the cellular level, HNSCC cells also exhibited significantly higher levels of glutaminase expression compared to normal human oral keratinocytes (Figure 3F). Corresponding validation studies confirmed that these HNSCC cells had higher metabolic levels of glutamate than primary human oral keratinocytes (Figure 3G). In summary, these validation studies showed that primary and metastatic HNSCC tissues and HNSCC cells exhibit highly active glutaminolysis marked by high glutamate levels, low glutamine levels, and high glutaminase expression. These data support the concept that as keratinocytes undergo transformative events toward a tumorigenic and metastatic phenotype, their metabolism becomes progressively more dependent on glutaminolysis.
Glutaminolysis Regulates HNSCC Stemness Properties
Cancer stemness is thought to contribute to tumorigenesis, and it may contribute to aggressive or metastatic tumor potential.13 To test the hypothesis that glutaminolysis promotes cancer stemness, glutaminase expression and glutamate levels were examined in the context of sphere assays. HNSCC spheres, designated tumorspheres, showed elevated levels of glutaminase expression compared to their counterpart adherent controls (Figure 4A). Metabolic validation studies revealed that these HNSCC tumorspheres exhibited elevated levels of glutamate, compared to adherent controls (Figure 4B). Using an oral cancer floor-of-mouth xenograft model, we found that mouse tumors derived from these tumorsphere cells exhibited a larger mean tumor volume and incidence and higher increases in the levels of glutamate compared to those derived from adherent control cells (Figure 4C and D). Exogenous addition of glutamine promoted tumorsphere formation and coincident glutaminase expression, whereas depriving cells of glutamine suppressed tumorsphere formation (Figure 4E and F).
Figure 4.
HNSCC stemness properties are defined by elevated glutamate and glutaminase expression in vitro and in vivo and regulated by glutaminolysis. (A) (left) Phase contrast images; (right) levels of glutamate in HNSCC cells (UM-SCC-14A) cultured under adherent or suspension conditions for 3 days. (B) Immunoblots showing the levels of glutaminase expression in HNSCC cells (UM-SCC-14A) cultured under adherent or suspension conditions for the indicated time points. β-Actin served as a loading control. (C) Representative images of tumors dissected from mice 6 weeks after injection with HNSCC cells cultured under adherent or tumorsphere conditions (n = 6/8 adherent and n = 9/9 tumorsphere developed tumors). (D) Bar/box plot showing levels of glutamate in mouse tumor tissues derived from adherent HNSCC cells and tumorspheres, based on isotope dilution GC-MS. Intergroup differences were determined by Kruskal–Wallis tests for two-way comparisons between groups. Statistical significance was defined as *p ≤ 0.05. (E) (left) Phase contrast images; (right) graph showing fold tumorsphere induction (total area) of UM-SCC-14A cells cultured in the presence or absence of glutamine (30 μg/mL) under suspension conditions in glutamine free media for the indicated time points. (F) Immunoblots showing the levels of glutaminase expression in HNSCC cells cultured in the presence or absence of glutamine, as indicated under suspension conditions in glutamine free media for 72 h. β-Actin served as a loading control. Scale bars represent 50 μm. (Photograph courtesy of Pachiyappan Kamarajan, copyright 2016.)
Inhibiting Glutaminolysis Abrogates HNSCC Stemness Properties in Vitro and Tumorigenesis in Vivo
Chemically inhibiting glutaminase activity with 6-diazo-5-oxo-L-norleucine (DON), a glutamine antagonist, led to suppression of tumorsphere formation and decreased glutamate levels in HNSCC cells in vitro (Figure 5A and B). Similarly, stably suppressing glutaminase expression in these cells with lentiviral shRNA also abrogated HNSCC tumorsphere formation and glutamate levels in HNSCC cells in vitro (Figure 5C and D). Mouse tumors derived from HNSCC cells stably transduced with glutaminase shRNA exhibited significantly decreased tumor volumes. Consistently, metabolic validation studies confirmed decreased levels of glutamate levels in these tumors (Figure 5E and F).
Figure 5.
Glutaminase suppression inhibits HNSCC tumorsphere formation in vitro and tumorigenesis in vivo via glutaminase. (A) (left) Phase contrast images; (right) graph showing fold tumorsphere inhibition (total area). (B) Levels of glutamate in UM-SCC-14A cells cultured in the presence or absence of the glutaminase inhibitor (DON) under adherent or suspension conditions for 3 days, based on isotope dilution GC-MS. (C) (left) Phase-contrast images; (right) graph showing fold tumorsphere inhibition (total area) of cells stably transduced with control scrambled-shRNA or glutaminase-shRNA and then cultured under adherent or suspension conditions (tumorsphere) for 3 days. (inset) Immunoblots showing the levels of glutaminase expression in cells transduced with control shRNA or glutaminase-shRNA. β-Actin served as a loading control. (D) Levels of glutamate in cells stably transduced with control shRNA or glutaminase-shRNA and then cultured under adherent or suspension conditions (tumor-spheres) for 3 days. (E) Images show the dissected tumors. (F) Levels of glutamate in mouse tumors derived from mice injected with UM-SCC-14A cells that were transduced with control shRNA or glutaminase-shRNA and grown under adherent conditions (n = 4/4 control shRNA; n = 4/4 glutaminase shRNA). Scale bars represent 50 μm. (Photograph courtesy of Pachiyappan Kamarajan, copyright 2016.)
HNSCC CSC (CD44hi/ALDHhi) and Their ALDH Expression Are Regulated by Glutaminolysis
HNSCC cancer stem cells (CSCs) have been defined by high expression of the stem cell surface markers CD44 and aldehyde dehydrogenase (ALDH).34,35 Using flow cytometry to isolate HNSCC CSCs (CD44hi/ALDHhi), we determined that these cells express high levels of glutaminase expression, and metabolic validation studies showed that they also exhibited high levels of glutamate compared to control cells (CD44lo/ALDHlo) (Figure 6A–C). These HNSCC CSC (CD44hi/ALDHhi) also exhibited a greater potential for tumorsphere formation (Figure 6D). Glutamine enrichment drove even CD44lo/ALDHlo HNSCC cells toward stemness. CD44lo/ALDHlo cells cultured in glutamine-enriched media formed more tumorspheres and exhibited higher ALDH expression compared to cells cultured in nonenriched media (Figure 6E and F). Suppressing glutaminase expression in HNSCC cells resulted in transcriptional and translational suppression of ALDH expression (Figure 6G and H). Consistently, we noted that chemical inhibition of glutaminase inhibits CD44 expression (Supporting Information; Figure S3). Thus, glutaminolysis regulates HNSCC and HNSCC CSC metabolic energetic pathways via ALDH.
Figure 6.
HNSCC cancer stem cells (CD44hi/ALDHhi) and ALDH expression are regulated by glutaminolysis. (A) Representative FACS-based sorting of ALDHhi CD44hi and ALDHlo CD44lo fractions from UM-SCC-14A cells stained with the Aldefluor reagent. The nonviable cells were excluded using 7-aminoactinomycin D (P1 = viable cells). The ALDH inhibitor N,N-diethylaminobenzaldehyde was used as a negative control. ALDHhi and ALDHlo fractions were gated against CD44 to select ALDHhiCD44hi (P7 = 1.4%) and ALDHloCD44lo (P6 = 1.9%) cells. (B) Immunoblots showing the levels of glutaminase expression in ALDHhiCD44hi and ALDHloCD44lo cells. β-Actin served as a loading control. (C) Graph showing the levels of glutamate in ALDHhiCD44hi and ALDHloCD44lo cells. (D) (left) Phase contrast images; (right) graph showing the number of tumorspheres formed in ALDHhiCD44hi and ALDHloCD44lo cells cultured under suspension conditions for 10 days. (E) (left), Phase contrast images; (right) graph showing the number of tumorspheres formed in ALDHhiCD44hi and ALDHloCD44lo cells cultured in the presence or absence of glutamine (30 μg/mL) under suspension conditions for 10 days. (F) Immunoblots showing the levels of ALDH1A1 in ALDHloCD44lo cells cultured in the presence and absence of glutamine (30 μg/mL) under suspension conditions for 10 days. β-Actin served as a loading control. (G) Graphs showing the levels of glutaminase and ALDH1A1 mRNA in cells transduced with control shRNA or glutaminase-shRNA. (H) Immunoblots showing the levels of ALDH1A1 and glutaminase expression in cells transduced with control shRNA or glutaminase-shRNA and then cultured under adherent or suspension conditions (tumorspheres) for 3 days. Graphs showing the levels of (I) glutaminase and (J) ALDH1A1 mRNA in metastatic HNSCC (Mets; n = 24), primary HNSCC (n = 24), and normal adjacent (n = 24) tissues measured by qPCR. Scale bars represent 50 μm. (Photograph courtesy of Pachiyappan Kamarajan, copyright 2016.)
In human HNSCC tissues, glutaminase RNA expression levels increased, as did those of ALDH expression (Figure 6I and J). Taken in aggregate, these data indicate that glutaminolytic metabolic pathways regulate the expression of the CSC marker ALDH.
DISCUSSION
Compared to other “-omes” (genome/proteome/transcriptome), the metabolome is relatively small (~3,200 compounds), highly conserved, and highly defined. Metabolomic data sets are more computationally approachable, more amenable to interpretation, and more precise in characterization. The metabolome can be integrated into systems biology or with other -omics data sets. Since detection of metabolites is less complex than proteomic/genomic alternatives and routinely used in clinical settings, target metabolites identified in association with primary or metastatic HNSCC could be used as prognostic markers, for identification of disease progression, staging, and treatment monitoring in a clinical setting.
Overall, the results from this global metabolomic study comparing control adjacent, primary and metastatic tumor samples and saliva samples from control or tumor-bearing subjects differed in a number of metabolic readouts, including changes in metabolites related to energetics, lipid metabolism, inflammation, markers of oxidative stress, and xenobiotics. In PCA, tumor samples (primary and metastatic) formed an overlapping population that was well-separated from control samples. The overall changes in metabolites in saliva and tissue matrices showed good correlation, suggesting altered biochemicals may be useful prognostic biomarkers of disease.
Glutaminolysis and its associated metabolites, including glutamate, have diagnostic potential for HNSCC. Our data have identified a highly accurate (100% predictive accuracy) signature metabolome of 30 metabolites that distinguishes normal saliva from HNSCC-derived saliva. Our data have also identified a signature metabolome of 30 metabolites that distinguishes normal tissues from primary and metastatic HNSCC tissues. The glutaminolytic pathway and its component metabolites ranked among these signature metabolites that distinguished normal from disease states across both tissues and saliva samples. Previously, using NMR spectroscopy combined with PCA plus partial least-squares discriminant analyses, we similarly found that glutamate and other metabolites showed a clear distinction between tumors and normal tissues.23 Others have also shown that glutamate is an important metabolite in tumorigenesis, including HNSCC, and resonances from glutamate, taurine, choline, lactic acid, and lipids were found to have diagnostic potential for HNSCC; however, our data reveal the mechanistic significance of glutaminolysis in tumorigenesis and its regulatory role in modulating stemness.25,27
Our earlier metabolomic analyses of HNSCC cells showed that several metabolites, including glutamate and those involved with glycolysis, are important in distinguishing HNSCC cells from normal oral keratinocytes.33 Specifically, unsupervised PCA performed on NMR data of HNSCC cell lines revealed a clear separation between HNSCC cells and normal human oral keratinocytes. HNSCC cells exhibited significantly altered levels of lactate, acetate, aspartate, isoleucine, taurine, tyrosine, NAD+, valine, creatine, myo-inositol, alanine, glutamine, uridine diphosphate coupled sugars, glutathione, adenosine mono-/diphosphate, phenylalanine, glycine, fumarate, glycerophosphocholine, phosphocholine, arachidonic acid, lysophosphatidylcholine, and sphingomyelin. These altered metabolites clearly revealed dysregulation of multiple metabolic events, including aerobic glycolysis (Warburg effect), oxidative phosphorylation (Pasteur’s effect), energy metabolism, TCA cycle anaplerotic flux, hexosamine pathway, osmoregulatory, and antioxidant mechanisms. It is noteworthy that an increased glutamate/glutamine ratio was observed in HNSCC cells, suggesting altered regulation of glutaminolysis during cell proliferation. Interestingly, mass spectrometry/liquid chromatography analyses of HNSCC cell lines by another group indicated that glucose rather than glutamine is the dominant energy source required for proliferation and survival of HNSCC cells.36 Thus, these studies suggest that both glycolysis and glutaminolysis are important energy determinants in HNSCC; however, glucose metabolism may be more important for survival and proliferation, whereas glutamine/glutamate metabolism may be critical for subsequent aggressive transitions, such as acquiring stemness properties and metastatic potential. Also, although analyses of cellular metabolism in a cell culture setting are helpful, the complex metabolism of tissues, which incorporate rich multicellular and extracellular matrix interactions, provides a more holistic overview of the overall metabolic processes that define tumorigenesis.
Metabolomic analyses of biofluids from head and neck cancer or oral cancer patients have also demonstrated the ability to discriminate between cancer patients and normal controls using NMR or mass spectroscopy-based approaches. Analysis of blood samples from 15 oral cancer patients using NMR spectroscopy combined with a multivariate chemometric analysis successfully discriminated between serum samples from cancer patients and from a control group.37 NMR-based metabolomics of plasma from patients with oral SCC (OSCC) or oral leukoplakia, and healthy controls, also successfully differentiated OSCC patients from oral leukoplakia patients and controls.38 Gas chromatography–mass spectrometry-based serum analyses recently identified glutamic acid, lactic acid, and aspartic acid as potential biomarkers for the efficacy of induction chemotherapy in OSCC.39 Mass spectrometry-based metabolomic approaches have similarly been useful in distinguishing oral cancer from other cancers, healthy controls, or patients with periodontal disease using saliva or other biofluid analyses.40,41 The current data expand these findings and underscore the strength of saliva and plasma in accurately discriminating between health and HNSCC with a 100% level of accuracy in the case of saliva. Thus, combined saliva- and biofluid-based metabolomic studies have identified a variety of useful discriminating metabolite biomarkers, including those involved in energetics, such as glutamate. Differences in the metabolite biomarkers identified and their levels may reflect differences in the NMR or mass spectroscopy methods used, statistical analyses employed, or heterogeneity of the samples.
Multiple mechanisms have been ascribed to aldehyde dehydrogenases (ALDHs) in promoting cancer metabolism, cancer chemoresistance, and CSC survival. ALDHs are a family of enzymes that help detoxify a variety of aldehydes, including those in drugs, xenobiotics, and intracellular components, such as alcohol. ALDH can assist with the biosynthesis of retinoic acid, which is thought to be important in determining cell fate, cancer cell signaling, and gene expression. ALDHs are also thought to play a role as antioxidants via their NADPH recycling function. In terms of chemoresistance, ALDHs are capable of direct enzymatic inactivation of alkylating agents, such as cyclophosphamide.42,43 ALDHs can also confer resistance to other chemotherapeutic drugs, such as cisplatin and doxorubicin, although the mechanisms have not been fully delineated.44,45 Other evidence reveals that ALDH is often coexpressed with antioxidant factors.46 For a more comprehensive discussion about the role of ALDH in promoting cancer cell processes, the reader is directed to a recent review on this subject.47 Thus, ALDH seems to functionally promote multiple cancer cell processes via direct drug inactivation, promoting oxidative stress resistance, and mediating retinoic acid signaling mechanisms that favor cell survival and other tumorigenic processes. The current study has identified a new mechanism whereby ALDH can influence CSC properties. Specifically, this study has identified a novel relationship between the stemness marker ALDH and glutamate/glutaminase/glutaminolysis. The current data indicate a causal relationship between glutamate/glutaminase and ALDH levels. It is known that the active site of the ALDH enzyme requires priming by glutamate to catalyze the oxidation of aldehydes to carboxylic acids. Thus, it may be that glutaminase, by upregulating glutamate levels, regulates the priming of ALDH activity and ultimately ALDH levels. In aggregate, these findings indicate that stemness energetics are regulated, in part, by glutaminolysis, which regulates ALDH levels/functionality.
Driving these elevated levels of glutaminolysis and ALDH in tumorigenesis may be mutations in the tumor promoter c-Myc or ALDH itself. c-Myc mutations can lead to increased expression of glutamate transporters that results in elevated glutamate in tumor cells.3 Also, deficiency in glutamine but not glucose induces MYC-dependent apoptosis in human cancer cells.48 Thus, Myc-mediated elevated glutamate levels may in turn drive the increase in ALDH levels that promote HNSCC stemness. Our data support this premise as the basis for the current findings, since c-Myc levels were elevated in tandem with glutaminase and ALDH levels in the human HNSCC tissue samples (Figure 6I and J and Supporting Information; Figure S2). Alternatively, mutations or polymorphisms in ALDH genes, several of which have been described, could confer a hyper-responsiveness to glutamate levels that then drive the stemness phenotype.49,50 For example, the glutamate to lysine single nucleotide polymorphism (Glu504Lys) was reported to have decreased activity of the ALDH enzyme and caused much higher blood levels of acetaldehyde; in a meta-analysis this showed an increased risk for cancer, especially esophageal cancer.50
CONCLUSIONS
In summary, the results from this global metabolomic study of tissues, saliva, plasma, and cell and animal models of HNSCC led to the discovery of a number of important metabolic readouts. The data helped identify biomarker panels with high predictive accuracy. Profiling and validation studies further fueled cell and animal model experiments that uncovered the mechanistic role of the glutaminolytic–aldehyde dehydrogenase relationship in regulating cancer cell stemness. This discovery helped unite two facets of cancer biology that heretofore were not well understood, namely cancer cell metabolic energetics and their relationship to cancer stemness, especially as relates to the well-known cancer stem cell marker ALDH. Future analyses of glutaminolytic-ALDH driven mechanisms underlying tumorigenic transitions may help in the development of targeted therapies for HNSCC and its CSC compartment.
Supplementary Material
Acknowledgments
This research was supported by funding from NIH (R56DE023333 to Y.L.K. and the UM Metabolomics core grant DK097153). We thank the patients, head and neck surgeons, and staff and faculty at the University of Michigan (UM) Head and Neck Specialized Program of Research Excellence (SPORE), who helped provide saliva samples. The SPORE was supported by grant funding from NIH (NCI P50-CA-097248). We thank Charles Burant and Arul M. Chinnaiyan for their helpful comments and Adam Banda for his technical assistance.
Footnotes
The authors declare no competing financial interest.
ORCID
Yvonne L. Kapila: 0000-0003-1330-0654
The Supporting Information is available free of charge on the ACS Publications website at DOI: 10.1021/acs.jproteome.6b00936.
Metabolic profiling and immunoblots (PDF)
Tables of clinical information and qPCR reagents (PDF)
Descriptions of two supplementary experiments and their results and conclusions (PDF)
Report S2. (Tissue-multiple worksheets) Original scaled data, pathway heat map (statistically significant biochemicals), box plots alphaphetical and box plots patways(XLSX)
Report S3. (Tissue) Statistics report (XLSX)
Report S4. (Saliva-multiple worksheets) Original scaled data, pathway heat map (statistically significant biochemicals), box plots alphaphetical and box plots patways(XLSX)
Report S5. (Saliva) Statistics report (XLSX)
References
- 1.Warburg O. On the origin of cancer cells. Science. 1956;123:309–314. doi: 10.1126/science.123.3191.309. [DOI] [PubMed] [Google Scholar]
- 2.Ferreira LM. Cancer metabolism: the Warburg effect today. Exp Mol Pathol. 2010;89:372–380. doi: 10.1016/j.yexmp.2010.08.006. [DOI] [PubMed] [Google Scholar]
- 3.Wise DR, DeBerardinis RJ, Mancuso A, Sayed N, Zhang XY, Pfeiffer HK, Nissim I, Daikhin E, Yudkoff M, McMahon SB, Thompson CB. Myc regulates a transcriptional program that stimulates mitochondrial glutaminolysis and leads to glutamine addiction. Proc Natl Acad Sci U S A. 2008;105:18782–18787. doi: 10.1073/pnas.0810199105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Yang C, Sudderth J, Dang T, Bachoo RM, McDonald JG, DeBerardinis RJ. Glioblastoma cells require glutamate dehydrogenase to survive impairments of glucose metabolism or Akt signaling. Cancer Res. 2009;69:7986–7993. doi: 10.1158/0008-5472.CAN-09-2266. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Cairns RA, Harris IS, Mak TW. Regulation of cancer cell metabolism. Nat Rev Cancer. 2011;11:85–95. doi: 10.1038/nrc2981. [DOI] [PubMed] [Google Scholar]
- 6.DeBerardinis RJ, Mancuso A, Daikhin E, Nissim I, Yudkoff M, Wehrli S, Thompson CB. Beyond aerobic glycolysis: transformed cells can engage in glutamine metabolism that exceeds the requirement for protein and nucleotide synthesis. Proc Natl Acad Sci U S A. 2007;104:19345–19350. doi: 10.1073/pnas.0709747104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Hanahan D, Weinberg RA. Hallmarks of cancer: the next generation. Cell. 2011;144:646–674. doi: 10.1016/j.cell.2011.02.013. [DOI] [PubMed] [Google Scholar]
- 8.Holohan C, Van Schaeybroeck S, Longley DB, Johnston PG. Cancer drug resistance: an evolving paradigm. Nat Rev Cancer. 2013;13:714–726. doi: 10.1038/nrc3599. [DOI] [PubMed] [Google Scholar]
- 9.Butler EB, Zhao Y, Munoz-Pinedo C, Lu J, Tan M. Stalling the engine of resistance: targeting cancer metabolism to overcome therapeutic resistance. Cancer Res. 2013;73:2709–2717. doi: 10.1158/0008-5472.CAN-12-3009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Wen YA, Stevens PD, Gasser ML, Andrei R, Gao T. Downregulation of PHLPP expression contributes to hypoxia-induced resistance to chemotherapy in colon cancer cells. Mol Cell Biol. 2013;33:4594–4605. doi: 10.1128/MCB.00695-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Cioce M, Valerio M, Casadei L, Pulito C, Sacconi A, Mori F, Biagioni F, Manetti C, Muti P, Strano S, Blandino G. Metformin-induced metabolic reprogramming of chemoresistant ALDHbright breast cancer cells. Oncotarget. 2014;5:4129–4143. doi: 10.18632/oncotarget.1864. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Hernandez-Davies JE, Tran TQ, Reid MA, Rosales KR, Lowman XH, Pan M, Moriceau G, Yang Y, Wu J, Lo RS, Kong M. Vemurafenib resistance reprograms melanoma cells towards glutamine dependence. J Transl Med. 2015;13:210. doi: 10.1186/s12967-015-0581-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Kreso A, Dick JE. Evolution of the cancer stem cell model. Cell Stem Cell. 2014;14:275–291. doi: 10.1016/j.stem.2014.02.006. [DOI] [PubMed] [Google Scholar]
- 14.Ito K, Suda T. Metabolic requirements for the maintenance of self-renewing stem cells. Nat Rev Mol Cell Biol. 2014;15:243–256. doi: 10.1038/nrm3772. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Janiszewska M, Suva ML, Riggi N, Houtkooper RH, Auwerx J, Clement-Schatlo V, Radovanovic I, Rheinbay E, Provero P, Stamenkovic I. Imp2 controls oxidative phosphorylation and is crucial for preserving glioblastoma cancer stem cells. Genes Dev. 2012;26:1926–1944. doi: 10.1101/gad.188292.112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Dong C, Yuan T, Wu Y, Wang Y, Fan TW, Miriyala S, Lin Y, Yao J, Shi J, Kang T, Lorkiewicz P, St Clair D, Hung MC, Evers BM, Zhou BP. Loss of FBP1 by Snail-mediated repression provides metabolic advantages in basal-like breast cancer. Cancer Cell. 2013;23:316–331. doi: 10.1016/j.ccr.2013.01.022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Hirsch HA, Iliopoulos D, Struhl K. Metformin inhibits the inflammatory response associated with cellular transformation and cancer stem cell growth. Proc Natl Acad Sci U S A. 2013;110:972–977. doi: 10.1073/pnas.1221055110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Liu PP, Liao J, Tang ZJ, Wu WJ, Yang J, Zeng ZL, Hu Y, Wang P, Ju HQ, Xu RH, Huang P. Metabolic regulation of cancer cell side population by glucose through activation of the Akt pathway. Cell Death Differ. 2014;21:124–135. doi: 10.1038/cdd.2013.131. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Lagadinou ED, Sach A, Callahan K, Rossi RM, Neering SJ, Minhajuddin M, Ashton JM, Pei S, Grose V, O’Dwyer KM, Liesveld JL, Brookes PS, Becker MW, Jordan CT. BCL-2 inhibition targets oxidative phosphorylation and selectively eradicates quiescent human leukemia stem cells. Cell Stem Cell. 2013;12:329–341. doi: 10.1016/j.stem.2012.12.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Flavahan WA, Wu Q, Hitomi M, Rahim N, Kim Y, Sloan AE, Weil RJ, Nakano I, Sarkaria JN, Stringer BW, Day BW, Li M, Lathia JD, Rich JN, Hjelmeland AB. Brain tumor initiating cells adapt to restricted nutrition through preferential glucose uptake. Nat Neurosci. 2013;16:1373–1382. doi: 10.1038/nn.3510. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Shen YA, Wang CY, Hsieh YT, Chen YJ, Wei YH. Metabolic reprogramming orchestrates cancer stem cell properties in nasopharyngeal carcinoma. Cell Cycle. 2015;14:86–98. doi: 10.4161/15384101.2014.974419. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. [accessed 26 Feb, 2016];Annual Report to the Nation on the Status of Cancer, 1975–2012. Available from URL: https://seer.cancer.gov/report_to_nation/
- 23.Somashekar BS, Kamarajan P, Danciu T, Kapila YL, Chinnaiyan AM, Rajendiran TM, Ramamoorthy A. Magic angle spinning NMR-based metabolic profiling of head and neck squamous cell carcinoma tissues. J Proteome Res. 2011;10:5232–5241. doi: 10.1021/pr200800w. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Bhutia YD, Babu E, Ramachandran S, Ganapathy V. Amino Acid transporters in cancer and their relevance to “glutamine addiction”: novel targets for the design of a new class of anticancer drugs. Cancer Res. 2015;75:1782–1788. doi: 10.1158/0008-5472.CAN-14-3745. [DOI] [PubMed] [Google Scholar]
- 25.El-Sayed S, Bezabeh T, Odlum O, Patel R, Ahing S, MacDonald K, Somorjai RL, Smith IC. An ex vivo study exploring the diagnostic potential of 1H magnetic resonance spectroscopy in squamous cell carcinoma of the head and neck region. Head Neck. 2002;24:766–772. doi: 10.1002/hed.10125. [DOI] [PubMed] [Google Scholar]
- 26.Jacque N, Ronchetti AM, Larrue C, Meunier G, Birsen R, Willems L, Saland E, Decroocq J, Thiago TT, Lambert M, Poulain L, Hospital MA, Sujobert P, Joseph L, Chapuis N, Lacombe C, Moura IC, Demo S, Sarry JE, Recher C, Mayeux P, Tamburini J, Bouscary D. Targeting glutaminolysis has antileukemic activity in acute myeloid leukemia and synergizes with BCL-2 inhibition. Blood. 2015;126:1346–1356. doi: 10.1182/blood-2015-01-621870. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Okada A, Takehara H, Yoshida K, Nishi M, Miyake H, Kita Y, Komi N. Increased aspartate and glutamate levels in both gastric and colon cancer tissues. Tokushima J Exp Med. 1993;40:19–25. [PubMed] [Google Scholar]
- 28.Sreekumar A, Poisson LM, Rajendiran TM, Khan AP, Cao Q, Yu J, Laxman B, Mehra R, Lonigro RJ, Li Y, Nyati MK, Ahsan A, Kalyana-Sundaram S, Han B, Cao X, Byun J, Omenn GS, Ghosh D, Pennathur S, Alexander DC, Berger A, Shuster JR, Wei JT, Varambally S, Beecher C, Chinnaiyan AM. Metabolomic profiles delineate potential role for sarcosine in prostate cancer progression. Nature. 2009;457:910–914. doi: 10.1038/nature07762. [DOI] [PMC free article] [PubMed] [Google Scholar] [Research Misconduct Found]
- 29.Brenner JC, Graham MP, Kumar B, Saunders LM, Kupfer R, Lyons RH, Bradford CR, Carey TE. Genotyping of 73 UM-SCC head and neck squamous cell carcinoma cell lines. Head Neck. 2010;32:417–426. doi: 10.1002/hed.21198. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Matsumoto K, Horikoshi M, Rikimaru K, Enomoto S. A study of an in vitro model for invasion of oral squamous cell carcinoma. J Oral Pathol Med. 1989;18:498–501. doi: 10.1111/j.1600-0714.1989.tb01350.x. [DOI] [PubMed] [Google Scholar]
- 31.Kamarajan P, Hayami T, Matte B, Liu Y, Danciu T, Ramamoorthy A, Worden F, Kapila S, Kapila Y, Nisin ZP. a Bacteriocin and Food Preservative, Inhibits Head and Neck Cancer Tumorigenesis and Prolongs Survival. PLoS One. 2015;10:e0131008. doi: 10.1371/journal.pone.0131008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Alhazzazi TY, Kamarajan P, Joo N, Huang JY, Verdin E, D’Silva NJ, Kapila YL. Sirtuin-3 (SIRT3), a novel potential therapeutic target for oral cancer. Cancer. 2011;117:1670–1678. doi: 10.1002/cncr.25676. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Tripathi P, Kamarajan P, Somashekar BS, MacKinnon N, Chinnaiyan AM, Kapila YL, Rajendiran TM, Ramamoorthy A. Delineating metabolic signatures of head and neck squamous cell carcinoma: phospholipase A2, a potential therapeutic target. Int J Biochem Cell Biol. 2012;44:1852–1861. doi: 10.1016/j.biocel.2012.06.025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Ginestier C, Hur MH, Charafe-Jauffret E, Monville F, Dutcher J, Brown M, Jacquemier J, Viens P, Kleer CG, Liu S, Schott A, Hayes D, Birnbaum D, Wicha MS, Dontu G. ALDH1 is a marker of normal and malignant human mammary stem cells and a predictor of poor clinical outcome. Cell Stem Cell. 2007;1:555–567. doi: 10.1016/j.stem.2007.08.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Prince ME, Sivanandan R, Kaczorowski A, Wolf GT, Kaplan MJ, Dalerba P, Weissman IL, Clarke MF, Ailles LE. Identification of a subpopulation of cells with cancer stem cell properties in head and neck squamous cell carcinoma. Proc Natl Acad Sci U S A. 2007;104:973–978. doi: 10.1073/pnas.0610117104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Sandulache VC, Ow TJ, Pickering CR, Frederick MJ, Zhou G, Fokt I, Davis-Malesevich M, Priebe W, Myers JN. Glucose, not glutamine, is the dominant energy source required for proliferation and survival of head and neck squamous carcinoma cells. Cancer. 2011;117:2926–2938. doi: 10.1002/cncr.25868. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Tiziani S, Lopes V, Gunther UL. Early stage diagnosis of oral cancer using 1H NMR-based metabolomics. Neoplasia. 2009;11:269–276. 264–269. doi: 10.1593/neo.81396. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Zhou J, Xu B, Huang J, Jia X, Xue J, Shi X, Xiao L, Li W. 1H NMR-based metabonomic and pattern recognition analysis for detection of oral squamous cell carcinoma. Clin Chim Acta. 2009;401:8–13. doi: 10.1016/j.cca.2008.10.030. [DOI] [PubMed] [Google Scholar]
- 39.Ye G, Liu Y, Yin P, Zeng Z, Huang Q, Kong H, Lu X, Zhong L, Zhang Z, Xu G. Study of induction chemotherapy efficacy in oral squamous cell carcinoma using pseudotargeted metabolomics. J Proteome Res. 2014;13:1994–2004. doi: 10.1021/pr4011298. [DOI] [PubMed] [Google Scholar]
- 40.Sugimoto M, Wong DT, Hirayama A, Soga T, Tomita M. Capillary electrophoresis mass spectrometry-based saliva metabolomics identified oral, breast and pancreatic cancer-specific profiles. Metabolomics. 2010;6:78–95. doi: 10.1007/s11306-009-0178-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Wang Q, Gao P, Wang X, Duan Y. The early diagnosis and monitoring of squamous cell carcinoma via saliva metabolomics. Sci Rep. 2014;4:6802. doi: 10.1038/srep06802. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Russo JE, Hilton J. Characterization of cytosolic aldehyde dehydrogenase from cyclophosphamide resistant L1210 cells. Cancer Res. 1988;48:2963–2968. [PubMed] [Google Scholar]
- 43.Sladek NE, Kollander R, Sreerama L, Kiang DT. Cellular levels of aldehyde dehydrogenases (ALDH1A1 and ALDH3A1) as predictors of therapeutic responses to cyclophosphamide-based chemotherapy of breast cancer: a retrospective study. Rational individualization of oxazaphosphorine-based cancer chemotherapeutic regimens. Cancer Chemother Pharmacol. 2002;49:309–321. doi: 10.1007/s00280-001-0412-4. [DOI] [PubMed] [Google Scholar]
- 44.Canino C, Luo Y, Marcato P, Blandino G, Pass HI, Cioce M. A STAT3-NFkB/DDIT3/CEBPbeta axis modulates ALDH1A3 expression in chemoresistant cell subpopulations. Oncotarget. 2015;6:12637–12653. doi: 10.18632/oncotarget.3703. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Croker AK, Allan AL. Inhibition of aldehyde dehydrogenase (ALDH) activity reduces chemotherapy and radiation resistance of stem-like ALDHhiCD44(+) human breast cancer cells. Breast Cancer Res Treat. 2012;133:75–87. doi: 10.1007/s10549-011-1692-y. [DOI] [PubMed] [Google Scholar]
- 46.Kim RJ, Park JR, Roh KJ, Choi AR, Kim SR, Kim PH, Yu JH, Lee JW, Ahn SH, Gong G, Hwang JW, Kang KS, Kong G, Sheen YY, Nam JS. High aldehyde dehydrogenase activity enhances stem cell features in breast cancer cells by activating hypoxia-inducible factor-2alpha. Cancer Lett. 2013;333:18–31. doi: 10.1016/j.canlet.2012.11.026. [DOI] [PubMed] [Google Scholar]
- 47.Xu X, Chai S, Wang P, Zhang C, Yang Y, Yang Y, Wang K. Aldehyde dehydrogenases and cancer stem cells. Cancer Lett. 2015;369:50–57. doi: 10.1016/j.canlet.2015.08.018. [DOI] [PubMed] [Google Scholar]
- 48.Yuneva M, Zamboni N, Oefner P, Sachidanandam R, Lazebnik Y. Deficiency in glutamine but not glucose induces MYC-dependent apoptosis in human cells. J Cell Biol. 2007;178:93–105. doi: 10.1083/jcb.200703099. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Vasiliou V, Thompson DC, Smith C, Fujita M, Chen Y. Aldehyde dehydrogenases: from eye crystallins to metabolic disease and cancer stem cells. Chem-Biol Interact. 2013;202:2–10. doi: 10.1016/j.cbi.2012.10.026. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Cai Q, Wu J, Cai Q, Chen EZ, Jiang ZY. Association between Glu504Lys polymorphism of ALDH2 gene and cancer risk: a meta-analysis. PLoS One. 2015;10:e0117173. doi: 10.1371/journal.pone.0117173. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.