Abstract
Chromosomal microarray (CMA) testing to detect copy number aberrations among individuals with multiple congenital anomalies and/or developmental delay is typically performed on peripheral blood DNA. However, the use of saliva DNA may be preferred for some patients, which prompted our validation study using six saliva DNA samples with a range of bacterial content (approximately 3% to 21%) and 20 paired blood and saliva specimens on the Agilent Technologies, Illumina, and Affymetrix CMA platforms. Ten of the 20 paired specimens were previously determined to carry clinically significant copy number aberrations by clinical CMA testing on blood DNA (100 kb to 2.56 Mb; five deletions, eight duplications). Notably, the quality of saliva DNA (DNA Genotek) was equivalent to blood DNA regardless of bacterial content, as was CMA quality and single-nucleotide polymorphism genotyping quality with all CMA platforms. The number of copy number variants and absence of heterozygosity regions detected by CMA were comparable between paired blood and saliva DNA and, more important, all 13 clinically significant copy number aberrations were detected in saliva DNA by all CMA platforms. These data confirm that the quality of saliva DNA is comparable to blood DNA regardless of bacterial content, including important CMA and single-nucleotide polymorphism quality metrics, and that saliva DNA is a reliable alternative for the detection of clinically significant copy number aberrations by clinical CMA testing.
Chromosomal microarray (CMA) is currently considered a first-tier test for the detection of constitutional copy number aberrations among individuals with idiopathic multiple congenital anomalies and/or developmental delay,1, 2, 3, 4, 5 which is supported by CMA validation and interpretation practice guidelines by both the American College of Medical Genetics and Genomics6, 7 and the European Society of Human Genetics.8 The rapid adoption of CMA testing in clinical cytogenetics laboratories was driven by its improved diagnostic yield (approximately 10% to 20%)3, 5, 9 over G-banded karyotype testing (approximately 3%)10, 11, 12, 13 for the detection of clinically significant copy number aberrations among patients with multiple congenital anomalies/developmental delay. The recent availability of higher-resolution CMA platforms that incorporate single-nucleotide polymorphism (SNP) genotyping enables even greater diagnostic yields.14 In addition to copy number aberrations, CMA platforms with SNP genotyping also detect absence of heterozygosity (AOH), which is indicative of uniparental disomy or identity by descent, as well as atypical genome-wide genotyping patterns that facilitate the detection of polyploidy.
Most postnatal clinical molecular assays in human genetics, including CMA testing, are performed using DNA isolated from peripheral blood. Although the DNA yield from peripheral blood is high, the use of saliva for DNA isolation and subsequent molecular testing offers some potential advantages over peripheral blood, including a noninvasive collection procedure and the capacity for sample stability at room temperature. In addition, improved detection of mosaic abnormalities may be possible with saliva DNA because of the presence of multiple cell types that comprise the oral microenvironment.15 Despite these advantages of saliva DNA, the variable presence of microbial DNA in human saliva could potentially interfere with hybridization-based molecular assays, such as CMA, indicating that thorough validation is warranted when incorporating saliva DNA into clinical laboratory protocols.
Although saliva DNA has been reported to have concordant genotyping call rates to blood DNA for research genome-wide association studies using SNP genotyping microarrays,16, 17 the potential impact of bacterial DNA on CMA quality and constitutional copy number variant (CNV) detection has not been adequately studied16, 17, 18, 19, 20, 21 and no CMA study has previously tested the ability to detect clinically significant copy number aberrations using saliva DNA. The paucity of available validation data on saliva DNA for clinical CMA testing prompted our study with paired blood and saliva DNA specimens using the three major CMA platforms commonly adopted by clinical cytogenetics laboratories (Agilent Technologies, Illumina, Inc., and Affymetrix, Inc.) (Table 1).
Table 1.
CMA Platform Probe and Resolution Characteristics
| Variable | Agilent G3 ISCA CGH + SNP 4 × 180K |
Illumina CytoSNP-850K BeadChip |
Affymetrix CytoScan HD Array |
|---|---|---|---|
| CGH/copy number probes | 110,712 | NA | 1.95 × 106 |
| SNP probes | 59,647 | 850,000 | 750,000 |
| Total probes | 179,080∗ | 850,000 | 2.7 × 106 |
| Median probe spacing | 25.3 kb (∼5 kb in ISCA regions) | 1.8 kb | 1.15 kb (∼880 bp in ISCA regions) |
CGH, comparative genomic hybridization; CMA, chromosomal microarray; HD, high density; ISCA, International Standards for Cytogenomic Arrays; NA, not applicable; SNP, single-nucleotide polymorphism.
Includes replicates and internal quality control probes.
Materials and Methods
Study Subjects and Specimens
Six independent saliva samples with quantified bacterial DNA content were obtained from anonymized healthy adult volunteers. Bacterial DNA content was previously determined by real-time quantitative PCR using universal primers targeted to the bacterial 16S rRNA gene, as per manufacturer's instructions (Bacterial DNA assay, PD-PR-065; DNA Genotek, Ottawa, ON, Canada).22 In addition, 20 paired blood and saliva specimens were obtained from 10 anonymized healthy adult volunteers and 10 deidentified subjects with clinically significant copy number aberrations that were identified through clinical CMA testing at the Mount Sinai Genetic Testing Laboratory. Clinically significant was defined as any copy number aberration that was reported by the Mount Sinai Genetic Testing Laboratory on the basis of American College of Medical Genetics and Genomics guidelines6, 7 as a variant of uncertain clinical significance (n = 10), a variant of uncertain clinical significance–likely pathogenic (n = 2), or pathogenic (n = 1). For prenatal specimens (n = 1) and probands (n = 6) that were unable to provide a saliva specimen (as determined by referring physician), paired blood and saliva specimens were collected from the transmitting parent when available.
DNA Isolation
Peripheral blood was collected in EDTA vacutainer tubes using standard practices, and DNA was isolated from 500 μL of whole blood using the QiaSymphony (Qiagen, Valencia, CA) or Chemagic (Perkin Elmer, Baesweiler, Germany), according to the manufacturer's instructions. Saliva samples were collected using the Oragene Dx kit (OGD-500; DNA Genotek), and DNA was manually isolated using the prepIT•L2P protocol, as per the manufacturer's instructions (DNA Genotek). Total DNA samples from blood and saliva were quantified using the Nanodrop 1000 Spectrophotometer (Thermo Scientific, Waltham, MA). The fraction of high-molecular-weight human DNA within saliva DNA samples was inferred using standard curves generated from densitometry measurements following agarose electrophoresis [ImageJ software version 1.48 (NIH, Bethesda, MD; http://imagej.nih.gov/ij) bundled with Java version 1.6.0 (Oracle, Redwood Shores, CA)], and these human DNA concentrations were used to calculate input DNA for CMA testing.
CMA Analysis
The number of comparative genomic hybridization/copy number and SNP probes of the three CMA platforms used in this study (Agilent, Illumina, Affymetrix), including their resolution characteristics, are summarized in Table 1.
Agilent SurePrint G3 International Standards for Cytogenomic Arrays Comparative Genomic Hybridization + SNP 4 × 180K Array
Agilent CMA testing was performed as previously reported23 and as per the manufacturer's instructions (Agilent Technologies, Santa Clara, CA). Images were processed with Feature Extraction software version 9.5.1 and the data analyzed with Agilent Genomic Workbench software version 7.0 (both from Agilent Technologies). Copy number aberrations were identified using the ADM1 algorithm (or ADM2 where noted) at a threshold of 6.0 and a four-probe aberration filter. Identified CNVs were filtered to exclude those <100 kb, nested aberrations, Y chromosome calls in females, and reference DNA CNVs, and AOH was defined by a minimum of 10 probes and >2 Mb.
Illumina Infinium 850K BeadChip Array
Processing of the Infinium 850K BeadChip arrays (Illumina, San Diego, CA) was performed at the Mount Sinai Genomics Core facility according to the Illumina HD Assay Super protocol. Array data files (.idat) were converted to .gtc files using Beeline version 1.0.37 (Illumina), which were then imported into BlueFuse Multi version 4.0 (Illumina) and analyzed using the default BlueFuse algorithm and settings (10 contiguous markers for CNVs and 500 contiguous markers for AOH). The manifest and cluster files used for CytoSNP-850K BeadChip processing were cytosnp-850k_bpm and CytoSNP-850K_APCAs cluster file4.egt, respectively. CNV and AOH calls <100 kb and <2 Mb, respectively, were manually excluded.
Affymetrix Cytoscan HD Array
Processing of the Cytoscan HD array was performed by Affymetrix, Inc. (Santa Clara, CA), and raw data (.cel) files were uploaded into Affymetrix's Chromosome Analysis Suite version 3.1 for variant calling. Variant calling was performed using the default settings in Chromosome Analysis Suite, coupled with a size filter to exclude CNV and AOH calls <100 kb and <2 Mb, respectively.
Concordance and Statistical Analysis
Concordance between paired blood and saliva samples was analyzed using raw CNV and AOH calls derived from Genomic Workbench, BlueFuse Multi, and Chromosome Analysis Suite. Concordance was calculated for each sample as the fraction of overlapping variants in blood and saliva divided by the sum of variants in blood and saliva. Descriptive statistics were performed using the Descriptive Statistics function within the Microsoft Excel Analysis Toolkit (Microsoft Corporation, Redmond, WA). Linear regression modeling to examine a correlation between CMA quality metrics and the number of copy number variants in blood and saliva was also performed using the Analysis Toolkit. All unpaired and paired t-tests were performed using GraphPad Prism 5 software (GraphPad Software, La Jolla, CA).
Results
Saliva DNA Quality
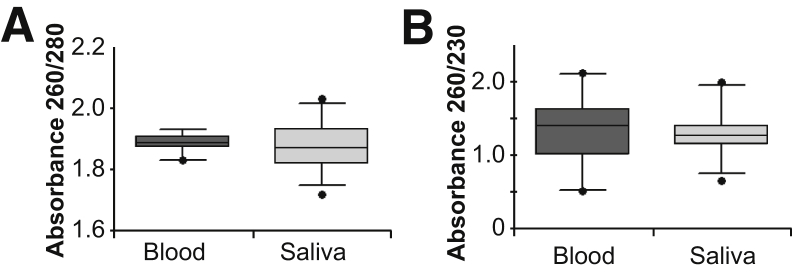
Assessing DNA quality is integral for CMA quality assurance as poor DNA quality can lead to suboptimal array performance. To determine whether the quality of saliva DNA is acceptable for CMA testing, the six saliva samples with known bacterial content and 20 paired blood and saliva samples were analyzed by UV spectrophotometry. No significant differences in the A260/280 and A260/230 ratios were detected between the blood and saliva DNA samples (Figure 1), indicating that the overall quality of saliva DNA is comparable to blood DNA. The concentrations of DNA extracted from blood and saliva ranged from approximately 100 to 500 ng/μL and approximately 80 to 700 ng/μL, respectively.
Figure 1.
Evaluation of overall saliva DNA quality. All 20 blood and saliva DNA samples were assessed for quality by measuring UV spectrophotometry absorbance. Illustrated are box-and-whisker plots of the A260/280 (A) and A260/230 (B) values for blood and saliva DNA, with medians denoted by horizontal lines and 95th percentiles denoted by vertical lines. Data are expressed as median box-and-whisker plots.
Saliva Microbial DNA and CMA Quality
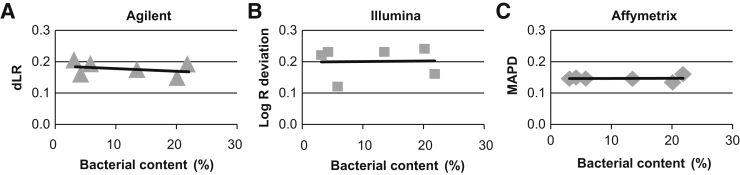
Although the quantity of microbial DNA in human saliva can vary widely depending on source and collection method,16, 18, 19, 20 the median percentage of bacterial DNA in human saliva collected using the Oragene kit has been reported to be approximately 12%.22 To determine whether higher bacterial content results in lower CMA quality, six saliva samples with a wide range of bacterial DNA content (approximately 3% to 21%) were tested on the three CMA platforms. General CMA quality was assessed using the following platform-specific quality metrics: derivative log ratio (Agilent), log R deviation (Illumina), and median absolute pairwise difference (Affymetrix). More important, bacterial content did not affect CMA quality for any platform (Figure 2), and the SNP genotyping quality metrics [SNP call rate (Agilent), median B allele frequency (Illumina), SNP quality control (Affymetrix)] were also comparable across all samples (data not shown). These data indicate that DNA isolated from saliva with higher bacterial content can produce CMA results with quality metrics that are comparable to those derived from blood DNA.
Figure 2.
Effect of bacterial content on chromosomal microarray (CMA) quality. The effect of bacterial content on CMA quality was assessed using the following platform-specific quality metrics: derivative log ratio (dLR; Agilent; A), log R deviation (Illumina; B), and median absolute pairwise difference (MAPD; Affymetrix; C). The scatter plots indicate that increasing bacterial content does not influence CMA quality metrics.
Comparison of Blood and Saliva CMA Performance
Blood and Saliva CMA Quality
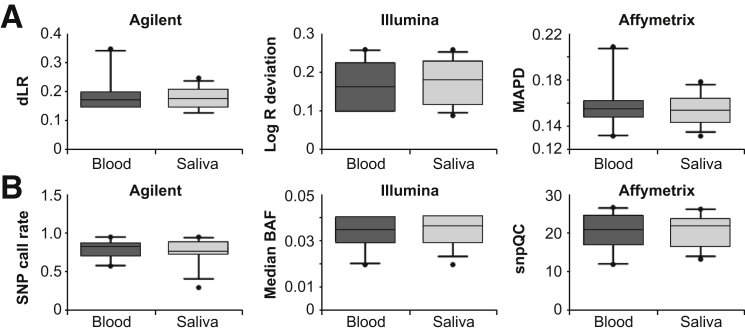
Given that whole blood is the most commonly used specimen to isolate DNA for postnatal constitutional CMA, the CMA and SNP quality metrics of the three CMA platforms were analyzed in the 20 blood and saliva DNA samples. No significant differences were detected between the CMA quality metric values (derivative log ratio, log R deviation, and median absolute pairwise difference) between the blood and saliva DNA samples (Figure 3A). Similarly, the SNP quality metrics (SNP call rate, median B allele frequency, SNP quality control) for blood and saliva specimens were also not significantly different for any of the three CMA platforms (Figure 3B).
Figure 3.
Comparison of chromosomal microarray (CMA) and single-nucleotide polymorphism (SNP) quality metrics between blood and saliva DNA. CMA quality was assessed using the derivative log ratio (dLR; Agilent), log R deviation (Illumina), and median absolute pairwise difference (MAPD; Affymetrix), and SNP genotyping quality metrics were assessed using the SNP call rate (Agilent), median B allele frequency (BAF; Illumina), and SNP quality control (snpQC; Affymetrix). Illustrated are box-and-whisker plots of the CMA (A) and SNP (B) quality values for blood and saliva DNA, with medians denoted by horizontal lines and 95th percentiles denoted by vertical lines.
Blood and Saliva CMA Detection of CNVs
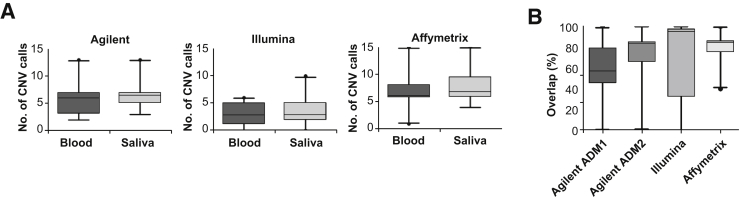
In addition to assessing CMA and SNP quality metrics across the three CMA platforms, identified CNV calls (>100 kb) were evaluated in the 20 paired blood and saliva DNA. Comparable numbers of raw CNV calls were observed in paired blood and saliva DNA by all CMA platforms (Figure 4A). In addition to numbers of variants, direct concordance of detected raw CNV calls was evaluated (Figure 4B). Median percentage concordance was variable and ranged from 57% to 85% across the three CMA platforms, which was not correlated with CMA quality metrics (derivative log ratio, Agilent; log R deviation, Illumina; median absolute pairwise difference, Affymetrix). Variability in direct overlap of raw CNV calls was most likely the result of false-positive CNV calls that were not manually filtered or curated and differences in array chemistry and resolution, as previously reported.24 Variability would also likely be influenced by differences in CNV detection algorithm thresholds, as evidenced by the increased percentage concordance detected (57% to 67%) using the Agilent ADM2 algorithm, which incorporates probe quality, compared to the ADM1 algorithm (Figure 4B).
Figure 4.
Copy number variant (CNV) concordance between paired blood and saliva DNA. Illustrated are box-and-whisker plots for raw CNV calls (>100 kb; A) and percentage overlap of CNV calls for the three chromosomal microarray (CMA) platforms (B). Medians are denoted by horizontal lines, and 95th percentiles denoted by vertical lines. Although comparable numbers of CNVs were detected in paired blood and saliva DNA, the percentage overlap of identified CNVs was variable across all CMA platforms.
Blood and Saliva CMA Detection of Clinically Significant Copy Number Aberrations
To determine whether clinically significant copy number aberrations can be detected using saliva DNA, 10 cases with previously reported copy number aberrations identified by clinical CMA testing of blood DNA on the Agilent platform were selected for CMA testing using paired blood and saliva DNA on all three platforms. A total of 13 copy number aberrations (100 kb to 2.56 Mb; five deletions, eight duplications) were reported in these 10 cases (Table 2), and all variant of uncertain clinical significance–likely pathogenic (n = 2) and pathogenic (n = 1) aberrations were independently confirmed by fluorescence in situ hybridization testing as per Mount Sinai Genetic Testing Laboratory protocol (data not shown). More important, all clinically significant aberrations identified in blood DNA were detected in saliva DNA by all three CMA platforms (Table 2), indicating that saliva DNA is a reliable alternative to blood DNA to detect clinically significant copy number aberrations using the Agilent, Illumina, and Affymetrix CMA platforms.
Table 2.
Clinically Significant Copy Number Aberrations Tested by CMA on Paired Blood and Saliva DNA
| Agilent (Genomic Workbench) |
Illumina (Bluefuse) |
Affymetrix (ChAS) |
||||
|---|---|---|---|---|---|---|
| Case | ISCN 2016 aberration nomenclature (blood) | Saliva | Blood | Saliva | Blood | Saliva |
| Proband | arr[hg19] 16p11.2(29656684_30190568)x1 | Detected | Detected | Detected | Detected | Detected |
| Proband | arr[hg19] 21q22.11q22.12(35734654_35905168)x3 | Detected | Detected | Detected | Detected | Detected |
| Parent | arr[hg19] 15q13.2q13.3(30819465_32509926)x3 | Detected | Detected | Detected | Detected | Detected |
| Parent | arr[hg19] 3p26.3(2354154_2967372)x3 | Detected | Detected | Detected | Detected | Detected |
| Parent | arr[hg19] 11p14.3(22261179_22361577)x3 | Detected | Detected | Detected | Detected | Detected |
| Parent | arr[hg19] Xp22.31(6260533_7196120)x1 | Detected | Detected | Detected | Detected | Detected |
| Parent | arr[hg19] 2p14(69038958_69172533)x1 | Detected | Detected | Detected | Detected | Detected |
| Proband | arr[hg19] 6q16.1(92505173_95065711)x3 | Detected | Detected | Detected | Detected | Detected |
| Parent | arr[hg19] 12q14.1(60338750_62223708)x1 | Detected | Detected | Detected | Detected | Detected |
| Proband | arr[hg19] 10q26.13(125544062_125726005)x3 | Detected | Detected | Detected | Detected | Detected |
| Proband | arr[hg19] 2p13.3(71584516_71712891)x3 | Detected | Detected | Detected | Detected | Detected |
| Proband | arr[hg19] 8p23.2(3235094_3602403)x3 | Detected | Detected | Detected | Detected | Detected |
| Proband | arr[hg19] 15q21.1(47613566_47939687)x1 | Detected | Detected | Detected | Detected | Detected |
ChAS, Chromosome Analysis Suite; CMA, chromosomal microarray; ISCN, International Standards for Human Cytogenetic Nomenclature.
Blood and Saliva CMA Detection of AOH
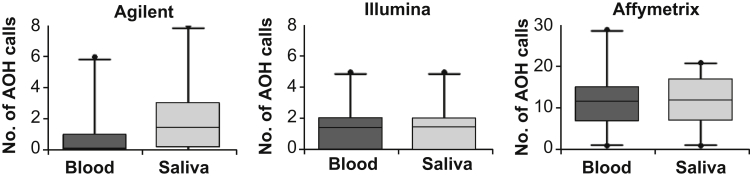
Although clinically significant AOH was not reported among the cases subjected to clinical CMA testing, the total number of AOH calls (>2 Mb) were assessed in all paired blood and saliva samples. Comparable numbers of AOH regions were detected between the paired blood and saliva DNA by all CMA platforms; however, as expected, CMA platforms based on SNP genotyping and with higher resolution (ie, Illumina and Affymetrix) were more consistent between the paired specimens (Figure 5 and Table 1). Moreover, the limited number of AOH regions detected in these specimens (approximately 0 to 2 AOH calls/specimen by Agilent and Illumina) resulted in an inability to accurately assess direct AOH concordance between the paired blood and saliva DNA.
Figure 5.
Absence of heterozygosity (AOH) concordance between blood and saliva DNA. Illustrated are median box-and-whisker plots for the number of detected AOH calls (>2 Mb), medians denoted by horizontal lines and 95th percentiles denoted by vertical lines. The numbers of AOH calls detected in paired blood and saliva DNA were comparable by chromosomal microarray (CMA) testing; however, CMA platforms based on single-nucleotide polymorphism genotyping chemistry were most consistent.
Discussion
Our validation study on the use of saliva as a clinical specimen for CMA testing confirms that saliva DNA has overall quality that is comparable to blood DNA regardless of microbial content, including important CMA and SNP quality metrics, and that saliva DNA is a reliable alternative to blood DNA for the detection of clinically significant copy number aberrations using the Agilent, Illumina, and Affymetrix CMA platforms. The detection of AOH by saliva DNA was also evaluated; however, the limited number of AOH regions detected indicates that additional validation of saliva DNA with clinically significant AOH cases (eg, probands from consanguineous families, heterodisomic uniparental disomy) is still warranted.
Validation of new specimen types for clinical genetic testing requires adequate demonstration of sample integrity and equivocal performance relative to the current gold standard. Previous studies that used saliva DNA on SNP genotyping arrays have reported variable outcomes with respect to SNP genotyping call rate, which were most likely because of the use of different saliva sample collection methods, the quality and percentage of isolated human DNA, and the wide variety of genotyping platforms used.16, 17, 18, 19, 20, 21 Moreover, all published saliva DNA reports were research studies that did not typically measure saliva DNA bacterial content, and these studies were also not specifically designed to interrogate CNVs or clinically significant copy number aberrations. However, our analysis of human DNA isolated from saliva collected with the Oragene kit indicates that higher bacterial concentrations do not have an appreciable effect on overall DNA or CMA quality.
Irrespective of specimen type, false-positive CNV calls are not uncommon by CMA analysis, which often can be platform specific and typically require manual curation by an experienced cytogeneticist for proper filtering and interpretation. Although numerous analytical and algorithm factors can affect CNV calling, spurious calls in saliva or blood DNA would affect CNV concordance between the two specimens. Although CMA analysis of paired blood and saliva DNA in our study resulted in equivalent CMA quality metrics, SNP call rates, and total numbers of raw CNV calls, the direct overlap of raw CNV calls was variable. Our study was not designed to directly compare CMA platforms; however, greater concordance of raw CNV calls between specimens correlated with higher probe density arrays. Moreover, greater CNV concordance for each CMA platform would likely have been achieved after manually filtering false-positive CNV calls, which is evidenced by the greater concordance observed when the more stringent Agilent ADM2 algorithm was used compared to the ADM1 results.
Taken together, our results strongly support the use of saliva DNA as a reliable alternative for clinical copy number aberration detection. Although our preliminary data suggest that saliva DNA can also adequately detect constitutional AOH by CMA testing, further validation using additional samples with clinically significant copy-neutral AOH is still warranted.
Acknowledgments
We thank Drs. Ethylin Wang Jabs, Bryn Webb, and Amy Yang for their assistance with sample collection; and Drs. Melanie Babcock, Lisong Shi, and Hui Mei for their assistance with clinical chromosomal microarray testing.
Footnotes
Supported by the Mount Sinai Genetic Testing Laboratory at the Icahn School of Medicine at Mount Sinai and the National Institute of General Medical Sciences of the NIH grant K23 GM104401 (S.A.S.). Chromosomal microarrays and reagents were supplied by Agilent Technologies, Illumina Inc., and Affymetrix Inc. DNA Genotek supplied the Oragene saliva collection kits and provided support for processing the Illumina microarrays. Affymetrix provided support for processing the Affymetrix arrays.
Disclosures: None declared.
References
- 1.Aradhya S., Manning M.A., Splendore A., Cherry A.M. Whole-genome array-CGH identifies novel contiguous gene deletions and duplications associated with developmental delay, mental retardation, and dysmorphic features. Am J Med Genet A. 2007;143A:1431–1441. doi: 10.1002/ajmg.a.31773. [DOI] [PubMed] [Google Scholar]
- 2.Battaglia A., Doccini V., Bernardini L., Novelli A., Loddo S., Capalbo A., Filippi T., Carey J.C. Confirmation of chromosomal microarray as a first-tier clinical diagnostic test for individuals with developmental delay, intellectual disability, autism spectrum disorders and dysmorphic features. Eur J Paediatr Neurol. 2013;17:589–599. doi: 10.1016/j.ejpn.2013.04.010. [DOI] [PubMed] [Google Scholar]
- 3.Gijsbers A.C., Lew J.Y., Bosch C.A., Schuurs-Hoeijmakers J.H., van Haeringen A., den Hollander N.S., Kant S.G., Bijlsma E.K., Breuning M.H., Bakker E., Ruivenkamp C.A. A new diagnostic workflow for patients with mental retardation and/or multiple congenital abnormalities: test arrays first. Eur J Hum Genet. 2009;17:1394–1402. doi: 10.1038/ejhg.2009.74. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Manning M., Hudgins L., Professional Practice and Guidelines Committee Array-based technology and recommendations for utilization in medical genetics practice for detection of chromosomal abnormalities. Genet Med. 2010;12:742–745. doi: 10.1097/GIM.0b013e3181f8baad. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Miller D.T., Adam M.P., Aradhya S., Biesecker L.G., Brothman A.R., Carter N.P., Church D.M., Crolla J.A., Eichler E.E., Epstein C.J., Faucett W.A., Feuk L., Friedman J.M., Hamosh A., Jackson L., Kaminsky E.B., Kok K., Krantz I.D., Kuhn R.M., Lee C., Ostell J.M., Rosenberg C., Scherer S.W., Spinner N.B., Stavropoulos D.J., Tepperberg J.H., Thorland E.C., Vermeesch J.R., Waggoner D.J., Watson M.S., Martin C.L., Ledbetter D.H. Consensus statement: chromosomal microarray is a first-tier clinical diagnostic test for individuals with developmental disabilities or congenital anomalies. Am J Hum Genet. 2010;86:749–764. doi: 10.1016/j.ajhg.2010.04.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Kearney H.M., South S.T., Wolff D.J., Lamb A., Hamosh A., Rao K.W., Working Group of the American College of Medical Genetics American College of Medical Genetics recommendations for the design and performance expectations for clinical genomic copy number microarrays intended for use in the postnatal setting for detection of constitutional abnormalities. Genet Med. 2011;13:676–679. doi: 10.1097/GIM.0b013e31822272ac. [DOI] [PubMed] [Google Scholar]
- 7.South S.T., Lee C., Lamb A.N., Higgins A.W., Kearney H.M., Working Group for the American College of Medical Genetics and Genomics Laboratory Quality Assurance Committee ACMG Standards and Guidelines for constitutional cytogenomic microarray analysis, including postnatal and prenatal applications: revision 2013. Genet Med. 2013;15:901–909. doi: 10.1038/gim.2013.129. [DOI] [PubMed] [Google Scholar]
- 8.Hanemaaijer N.M., Sikkema-Raddatz B., van der Vries G., Dijkhuizen T., Hordijk R., van Essen A.J., Veenstra-Knol H.E., Kerstjens-Frederikse W.S., Herkert J.C., Gerkes E.H., Leegte L.K., Kok K., Sinke R.J., van Ravenswaaij-Arts C.M. Practical guidelines for interpreting copy number gains detected by high-resolution array in routine diagnostics. Eur J Hum Genet. 2012;20:161–165. doi: 10.1038/ejhg.2011.174. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Sagoo G.S., Butterworth A.S., Sanderson S., Shaw-Smith C., Higgins J.P., Burton H. Array CGH in patients with learning disability (mental retardation) and congenital anomalies: updated systematic review and meta-analysis of 19 studies and 13,926 subjects. Genet Med. 2009;11:139–146. doi: 10.1097/GIM.0b013e318194ee8f. [DOI] [PubMed] [Google Scholar]
- 10.Faed M.J., Robertson J., Field M.A., Mellon J.P. A chromosome survey of a hospital for the mentally subnormal. Clin Genet. 1979;16:191–204. doi: 10.1111/j.1399-0004.1979.tb00990.x. [DOI] [PubMed] [Google Scholar]
- 11.Jacobs P.A., Matsuura J.S., Mayer M., Newlands I.M. A cytogenetic survey of an institution for the mentally retarded, I: chromosome abnormalities. Clin Genet. 1978;13:37–60. doi: 10.1111/j.1399-0004.1978.tb04127.x. [DOI] [PubMed] [Google Scholar]
- 12.Kondo I., Hamaguchi H., Nakajima S., Haneda T. A cytogenetic survey of 449 patients in a Japanese institution for the mentally retarded. Clin Genet. 1980;17:177–182. doi: 10.1111/j.1399-0004.1980.tb00130.x. [DOI] [PubMed] [Google Scholar]
- 13.Sutherland G.R., Murch A.R., Gardiner A.J., Carter R.F., Wiseman C. Cytogenetic survey of a hospital for the mentally retarded. Hum Genet. 1976;34:231–245. doi: 10.1007/BF00295286. [DOI] [PubMed] [Google Scholar]
- 14.D'Amours G., Langlois M., Mathonnet G., Fetni R., Nizard S., Srour M., Tihy F., Phillips M.S., Michaud J.L., Lemyre E. SNP arrays: comparing diagnostic yields for four platforms in children with developmental delay. BMC Med Genomics. 2014;7:70. doi: 10.1186/s12920-014-0070-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Shehab M.I., Mazen I., Bint S. Tissue-specific mosaicism for tetrasomy 9p uncovered by array CGH. Am J Med Genet A. 2011;155A:2496–2500. doi: 10.1002/ajmg.a.34176. [DOI] [PubMed] [Google Scholar]
- 16.Bahlo M., Stankovich J., Danoy P., Hickey P.F., Taylor B.V., Browning S.R., Australian and New Zealand Multiple Sclerosis Genetics Consortium (ANZgene) Brown M.A., Rubio J.P. Saliva-derived DNA performs well in large-scale, high-density single-nucleotide polymorphism microarray studies. Cancer Epidemiol Biomarkers Prev. 2010;19:794–798. doi: 10.1158/1055-9965.EPI-09-0812. [DOI] [PubMed] [Google Scholar]
- 17.Hoffmann T.J., Kvale M.N., Hesselson S.E., Zhan Y., Aquino C., Cao Y. Next generation genome-wide association tool: design and coverage of a high-throughput European-optimized SNP array. Genomics. 2011;98:79–89. doi: 10.1016/j.ygeno.2011.04.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Herraez D.L., Stoneking M. High fractions of exogenous DNA in human buccal samples reduce the quality of large-scale genotyping. Anal Biochem. 2008;383:329–331. doi: 10.1016/j.ab.2008.08.015. [DOI] [PubMed] [Google Scholar]
- 19.Hu Y., Ehli E.A., Nelson K., Bohlen K., Lynch C., Huizenga P., Kittlelsrud J., Soundy T.J., Davies G.E. Genotyping performance between saliva and blood-derived genomic DNAs on the DMET array: a comparison. PLoS One. 2012;7:e33968. doi: 10.1371/journal.pone.0033968. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Marenne G., Rodriguez-Santiago B., Closas M.G., Perez-Jurado L., Rothman N., Rico D., Pita G., Pisano D.G., Kogevinas M., Silverman D.T., Valencia A., Real F.X., Chanock S.J., Genin E., Malats N. Assessment of copy number variation using the Illumina Infinium 1M SNP-array: a comparison of methodological approaches in the Spanish Bladder Cancer/EPICURO study. Hum Mutat. 2011;32:240–248. doi: 10.1002/humu.21398. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Rasi S., Bruscaggin A., Rinaldi A., Cresta S., Fangazio M., De Paoli L., Monti S., Gargiulo E., Kwee I., Foa R., Bertoni F., Gaidano G., Rossi D. Saliva is a reliable and practical source of germline DNA for genome-wide studies in chronic lymphocytic leukemia. Leuk Res. 2011;35:1419–1422. doi: 10.1016/j.leukres.2011.05.024. [DOI] [PubMed] [Google Scholar]
- 22.James C., Iwasiow R.M., Birnboim H.C. DNA Genotek Inc.; Ottawa, ON, Canada: 2011. Human genomic DNA content of saliva samples collected with the Oragene self-collection kit, Technical Manual, PD-WP-011 Issue 4/2011-11. [Google Scholar]
- 23.Scott S.A., Cohen N., Brandt T., Toruner G., Desnick R.J., Edelmann L. Detection of low-level mosaicism and placental mosaicism by oligonucleotide array comparative genomic hybridization. Genet Med. 2010;12:85–92. doi: 10.1097/GIM.0b013e3181cc75d0. [DOI] [PubMed] [Google Scholar]
- 24.Tucker T., Montpetit A., Chai D., Chan S., Chenier S., Coe B.P., Delaney A., Eydoux P., Lam W.L., Langlois S., Lemyre E., Marra M., Qian H., Rouleau G.A., Vincent D., Michaud J.L., Friedman J.M. Comparison of genome-wide array genomic hybridization platforms for the detection of copy number variants in idiopathic mental retardation. BMC Med Genomics. 2011;4:25. doi: 10.1186/1755-8794-4-25. [DOI] [PMC free article] [PubMed] [Google Scholar]