Abstract
The ability of 17β-estradiol (E2) to regulate the proliferation of prostate cancer (PCa) cells in the absence of androgen is poorly understood. Here, we show the predominant estrogen receptor (ER) isoform expressed in PCa specimens and cell lines is ERβ. Our data indicate that E2 induces the formation of a complex between androgen receptor (AR), ERβ, and a proline-, glutamic acid-, and leucine-rich cofactor protein 1 (PELP1) in PCa cells. This protein complex is formed on AR's cognate DNA-responsive elements on the promoter in response to E2. Formation of this complex enables the transcription of AR-responsive genes in response to E2. Knockdown of PELP1, AR, or ERβ blocks the assembly of this complex, blocks E2-induced genomic activation of AR-regulated genes, and blocks E2-stimulated proliferation of PCa cells. Overall, this study shows that PELP1 may enable E2-induced AR signaling by forming a protein complex between AR, ERβ, and PELP1 on the DNA, leading to the proliferation of PCa cells in the absence of androgen. PELP1 may bridge the signal between E2 bound to ERβ and AR and thus allow for cross talk between these steroid receptors. These data suggest a novel mechanism of AR activation in the absence of androgens in PCa cells. Our data indicate that disruption of the complex between AR and PELP1 may be a viable therapeutic strategy in advanced PCa.
The first line of therapy for advanced prostate cancer (PCa) is androgen deprivation. However, castration-resistant PCa (CRPC) invariably develops in patients with metastatic PCa (1). In CRPC, the androgen receptor (AR) is functionally active, resulting from either 1) AR amplification, 2) gain-of-function in AR (largely occurring in the ligand-binding domain and conferring ligand promiscuity), 3) intracrine androgen production (thus providing tumor-produced ligand to AR), or 4) indirect or nonandrogenic AR activation (2, 3).
Nonandrogenic AR activation in CRPC may occur through indirect AR activation by growth factors or other steroid receptors (4). Estrogens have been reported to stimulate proliferation of cultured PCa cells (5). The first estrogen receptor (ER) expressed in fetal prostate, and the predominant form in its epithelium, is ERβ, which, together with AR, appears to mediate the initial stages of gland development. ERα gene appears to be transcriptionally inactivated by promoter DNA methylation in most PCa cell lines and specimens (6–8). ERβ expression in metastastic lesions has been noted (9, 10). However, the expression ligand binding, heterodimerization, transactivation, and subcellular localization of the two major ER subtypes, ERα and ERβ, have been variously reported in both normal and diseased human prostate (11–14). Discrepancies in the literature make it difficult to define the relative biological roles of the predominant ER subtypes, ERβ and ERα, in PCa (11–16). Specifically, the role of ERβ in PCa is unclear, with evidence for both an association between loss of ERβ expression in CRPC and progression, (6, 12, 17–19) and between retained expression of ERβ in metastatic lesions and increased mortality (10, 18, 19).
In this report, we examined the ability of estrogens to mediate nonandrogenic proliferation of PCa cells via AR and its cofactor, the proline-, glutamic acid-, and leucine-rich protein 1 (PELP1) (20–23). PELP1 is known to interact with several nuclear receptors (NR), including ER and AR (24, 25). PELP1 appeared to be predominantly localized in the nuclear compartment, interacted with histones and histone-modifying enzymes, and has been shown to be involved in chromatin remodeling for ligand-bound NR (26–28). PELP1 thus represented a protooncogene involved in cellular proliferation (29, 30). PELP1 uniquely has been shown to be important for both the genomic and nongenomic action of NR (24, 28). Because PELP1 has been shown to interact with AR in PCa cells (31), we examined whether PELP1 could couple ER and AR and facilitate 17β-estradiol (E2)-induced activation of AR signaling in PCa cells.
We found that E2 stimulates PCa proliferation in an ERβ-, PELP1-, and AR-dependent manner, suggesting a novel mechanism by which CRPC can be stimulated to grow in the absence of androgen.
Results
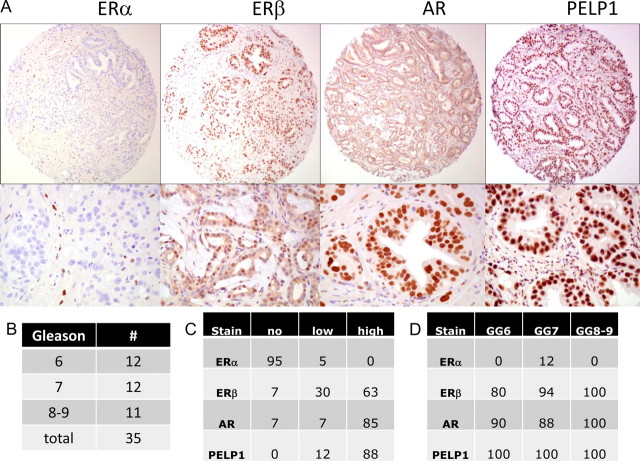
Because the expression of the ER isoforms in PCa was not clearly established, we evaluated, using immunohistochemistry, clinical prostate specimens from 35 consecutive patients who had undergone a radical prostatectomy for clinically localized PCa (Fig. 1, A and B). We noted low or no expression of ERα in the vast majority of primary prostate tumors (Fig. 1, A and C). In contrast, high levels of expression of ERβ, AR, and PELP1 expression were noted in prostate tumors (Fig. 1, A and C). Although the expression of AR and PELP1 were noted to be predominantly nuclear, ERβ expression was detected in both the nucleus and cytoplasm of the prostate tumors. The expression level and subcellular localization of AR, PELP1, and ERβ was not influenced by the grade of the tumor (Fig. 1D). Further evaluation revealed that ERβ, but not ERα, was expressed in a vast majority of PCa cell lines (Supplemental Fig. 1, published on The Endocrine Society's Journals Online web site at http://mend.endojournals.org). These data together suggested that ERβ is the predominant isoform in primary PCa and PCa cell lines.
Fig. 1.
A, Immunohistochemical analysis of ERα, ERβ, AR, and PELP1 in prostatic cancer specimens is shown at magnification ×100 (top row) and ×400 (bottom row). Tables show the distribution of the Gleason sum (B), the staining pattern for different markers by percentage of specimens with no staining, low staining, and high staining (C), and the percentage of specimens with any degree of positivity as stratified by Gleason sum (D).
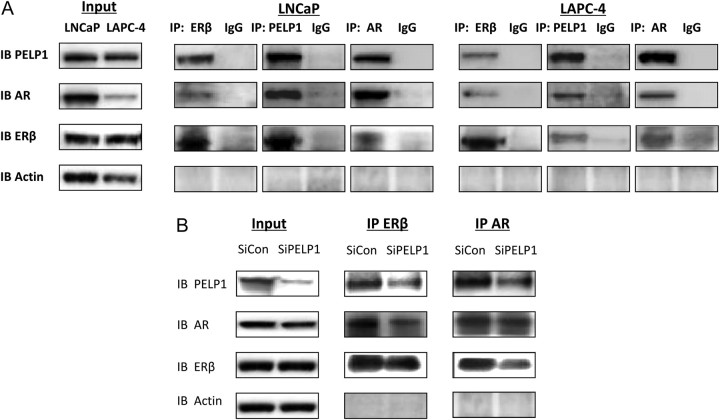
Because ER and AR both interact with the scaffolding protein PELP1, and PELP1 has a documented role in both genomic and nongenomic signaling mediated by AR, we postulated that PELP1 may be involved in E2-mediated activation of AR. Using coimmunoprecipitation experiments, a complex between ERβ, AR, and PELP1 was noted in LNCaP and LAPC4 cells (Fig. 2A). Similar complexes were noted in C4-2, CWR22v1, and PC-3(AR)2 cell lines (data not shown).
Fig. 2.
ERβ, AR, and PELP1 can form a complex. A, One hundred micrograms of protein extracts from LNCaP and LAPC4 cells under normal growing conditions were evaluated by immunoprecipitation by AR, ERβ, PELP1, or IgG control and immunoblotted with corresponding antibody. Input lysates are shown on the left. B, Effect of siRNA to PELP1 on AR, PELP1, and ERβ complex formation. LAPC4 cells were transfected with siRNA for 48 h, and their lysates were subjected to IP with AR or with ER-β and then immunoblotted with antibodies to AR, PELP1, or ERβ Input lysates are shown on the left. IB, Immunoblot; IP, immunoprecipitation.
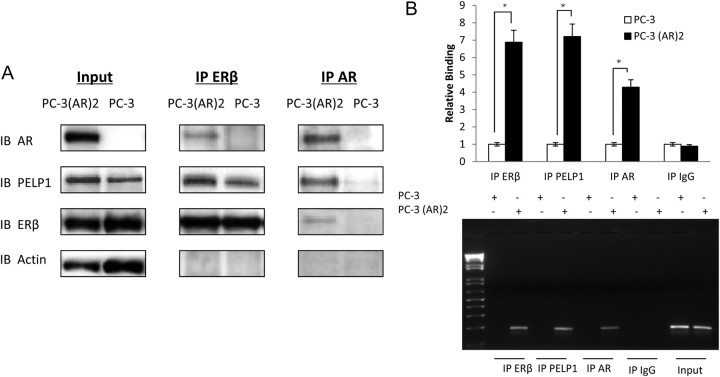
Knockdown of PELP1 decreased the interactions between PELP1 and AR, PELP1 and ERβ, and AR and ERβ (Fig. 2B and Supplemental Fig. 2). In contrast, knockdown of AR or ERβ did not affect the interaction between ERβ and PELP1 and AR and PELP1, respectively (data not shown). Indeed, in PC-3 cells, which lack AR expression, an interaction between ERβ and PELP1 was noted: upon introduction of AR in PC-3(AR)2 cells, the complex between AR, ERβ, and PELP1 was restored, without significant increase in the interaction between ERβ and PELP1 (Fig. 3A). These data suggest that PELP1 acts a bridge between AR and ERβ.
Fig. 3.
ERβ, AR, and PELP1 can form a complex on DNA. A, Fifty micrograms of protein extracts from PC-3 cells or PC-3(AR)2 (PC-3 cells stably transfected with AR under the control of its native promoter) were evaluated by immunoprecipitation by AR, ERβ, and control and immunoblotted with corresponding antibody. Input lysates are shown on the left. B, One hundred microliters of sheared DNA lysates from PC-3 cells or PC-3-AR2 were subject to ChIP assays with the PSA promoter after immunoprecipitation with ERβ, AR, PELP1, or IgG control. A representative PCR gel is shown below. IB, Immunoblot; IP, immunoprecipitation; *, statistically significant.
Because the subcellcular localization in clinical specimens indicate that AR, ERβ, and PELP1 are predominantly nuclear, chromatin immunoprecipitation (ChIP) assays were used to determine whether this complex was formed on androgen-responsive elements (ARE) on the prostate-specific antigen (PSA) promoter. In PC-3 cells, in the absence of AR, neither ERβ nor PELP1 could bind the PSA promoter. Thus, even though ERβ and PELP1 can be detected in a complex, they do not bind the PSA promoter. In the presence of AR, in PC-3(AR)2 cells, AR, ERβ, and PELP1 form a complex associated with the ARE region of the gene (Fig. 3B). These data suggest that the transcriptional machinery on the ARE may include the protein complex between AR, ERβ, and PELP1, with AR binding to its cognate ARE sequence, PELP1 binding AR, and ERβ binding PELP1.
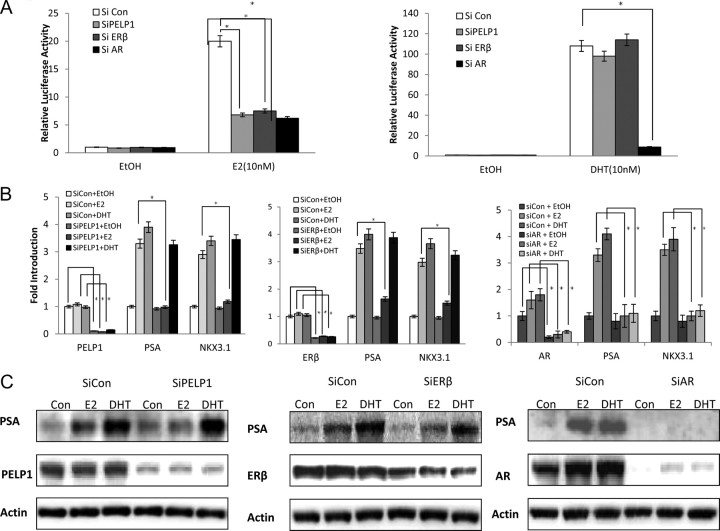
The functional import of the protein complex between AR, ERβ, and PELP1 was revealed by increased transcription from and ARE in response to E2 in knockdown experiments (Fig. 4 and Supplemental Fig. 3). As previously reported, E2 was less potent than dihydrotestosterone (DHT) in inducing transcription from ARE (5). Further, E2-induced gene expression required each member of the complex, as evidenced by the effect of the knockdown of either AR, ERβ, or PELP1 on the transcription from an ARE luciferase (Fig. 4A), transcription of AR-responsive genes, such as NKx3.1 and PSA in quantitative RT-PCR assays (Fig. 4B) or protein expression of PSA in Western blottings (Fig. 4C and Supplemental Fig. 4). Interestingly, knockdown of ERβ or PELP1 did not affect the ability of DHT to induce PSA gene expression: only knockdown of AR affected DHT-induced PSA gene expression (Fig. 4, A–C). These data suggest that the protein complex between AR, ERβ, and PELP1 is important for E2-mediated regulation of androgen-responsive genes in PCa cells.
Fig. 4.
Role of protein complex between ERβ, AR, and PELP1 on expression of androgen-responsive genes. A, 500,000 C4-2 cells were transfected with siRNA and with 4 μg of ARE-luciferase reporter in P-60 dishes, then plated into six-well plates and subjected to androgen-free media for 48 h. Cells were then treated with DHT (10 nm) or E2 (10 nm) and harvested for luciferase 48 h later. B, mRNA level: 100,000 LNCaP cells in six-well plates were transfected with siRNA and subjected to androgen-free media for 2 d. Ethanol (EtOH), E2 (10 nm), or DHT (10 nm) was added, and mRNA was harvested 24 h later for QPCR analyses. C, Protein level 100,000 LNCaP cells in six-well plates were transfected with siRNA and subjected to androgen-free media for 2 d. E2 (10 nm) and DHT (10 nm) were added, and lysates were harvested 24 h later for Western blot analyses. siRNA, Small interfering RNA; si con, control siRNA; *, statistically significant.
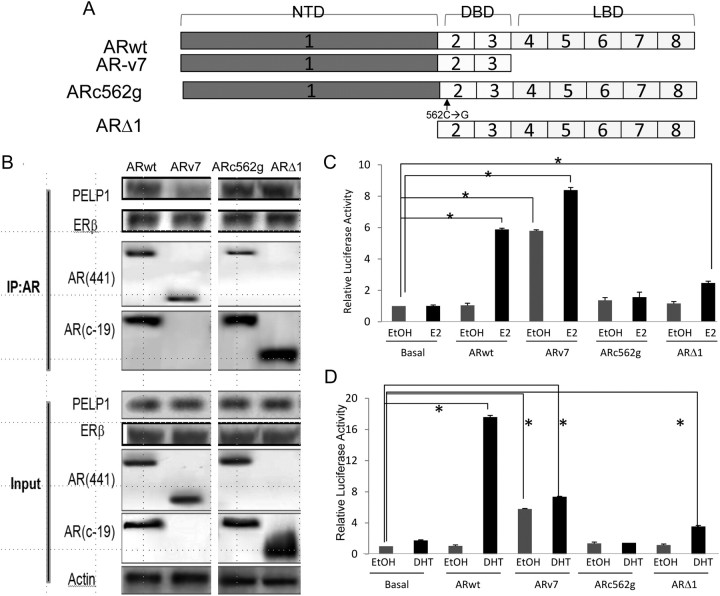
Further confirmation of the role of this complex in gene transcription is derived from transient transfection experiments with AR splice variants in PC-3 cells (Fig. 5A). Because PELP1 has 10 LXXLL motifs, PELP1 most likely binds to the ligand-binding domain of AR. Full-length AR is able to bind PELP1 and ERβ (Fig. 5B) and activate transcription from an ARE in response to either E2 (Fig. 5C) or DHT (Fig. 5D). The AR splice variant, ARv7, which lacks the ligand-binding domain, does not bind PELP1 well (Fig. 5B) and is associated with a significantly higher basal level of transcription from the ARE luciferase but does not significantly further induce transcription from an ARE in response to either E2 (Fig. 5C) or DHT (Fig. 5D). ARc562g binds to PELP1 and ERβ (Fig. 5B) but cannot activate transcription to either E2 (Fig. 5C) or DHT (Fig. 5D), because it cannot bind DNA [the single amino acid change (C to G) disrupts the zinc finger DNA-binding domain of AR]. These data indicate that AR needs to bind PELP1, ERβ, and DNA to modulate E2-mediated transcription from ARE. Finally, a synthetic AR construct, ARΔ1 (which lacks exon 1 and its powerful transactivation domain), binds PELP1, ERβ, and DNA but is able to weakly transactivate in response to both E2, as a consequence of its weakened transactivational ability (Fig. 5C) or DHT (Fig. 5D). These data strongly support the central role for PELP1 and its interaction with both ERβ and AR on the ARE promoter sequence in modulating E2-mediated transcriptional activation.
Fig. 5.
Effect of AR mutants on AR-PELP1 binding and E2-mediated transactivation. A, Structures of five AR constructs used: ARwt is the full-length AR, ARv7 is an AR splice variant that lacks the C-terminal ligand-binding domain, ARc562g is a point mutant AR construct that does not translocate to the nucleus upon DHT treatment, and ARΔ1 is a synthetic construct lacking AR amino terminus and the transactivation domain. NTD, Amino-terminal domain; DBD, DNA-binding domain; LBD, ligand-binding domain. B, PC-3 cells were transiently transfected with these AR constructs and then evaluated for complex formation between AR and PELP1 and AR and ERβ by immunoprecipitation with the AR441 antibody (directed against an epitope on the amino terminus of AR) or the AR c-19 antibody (directed against an epitope on the carboxy terminus of AR). Immunoprecipitation of extracts from PC3 cells transfected with ARwt, ARv7, and ARc562g was performed with the AR 441 antibody (against the N terminus), whereas those from cells transfected with ARΔ1 were perfomed with the AR c-19 antibody (against the C terminus). The immunoblotting was performed with antibodies to PELP1 and ERβ and both the AR-441 antibody and the AR c-19 antibody. Input lanes reveal the expression and immunoreactivity of PELP1, ERβ, and AR in these transiently PC-3-transfected cells (bottom four rows). As expected, both the ARwt and ARc562g are recognized by both AR antibodies. The ARΔ1 splice variant is recognized by only by the C-terminal antibody, whereas the ARv7 splice variant is only recognized by the N-terminal antibody. Transcription in PC-3 cells transiently transfected with AR splice variants, along with an ARE luciferase promoter, was evaluated in the absence and presence of E2 (10 nm) (C) or DHT (10 nm) for 24 h (D). IP, Immunoprecipitation; *, statistically significant.
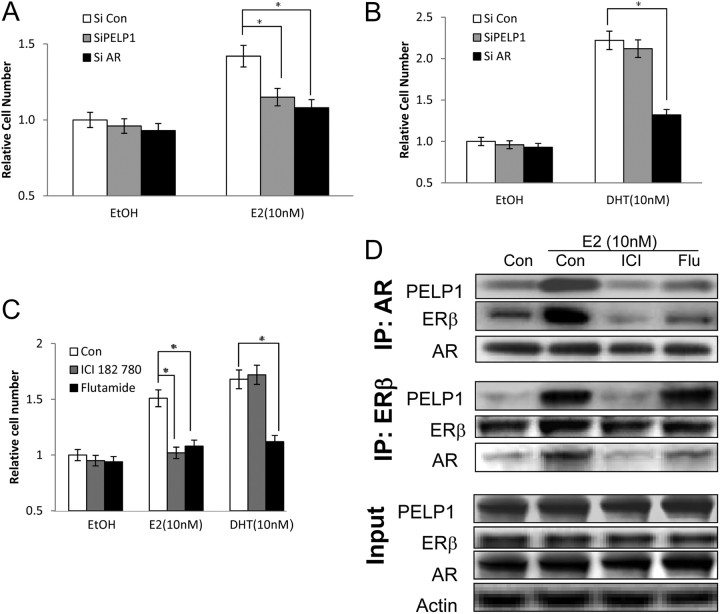
Because E2 is known to regulate proliferation of PCa cells, we then examined the importance of this protein complex in modulating cell proliferation. C4-2 cells transfected with small interfering RNA (siRNA) were subject to androgenic deprivation for 48 h and then treated with E2 or DHT (Fig. 6). E2-induced proliferation of PCa cells was blocked by knockdown of either PELP1 or AR on 3-(4,5-dimethylthiazol-2-yl)-2,5-diphenyltetrazolium bromide (MTT) assays (Fig. 6A) and verified by green fluorescent CyQUANT (Invitrogen, Carlsbad, CA) cell proliferation assays in C4-2 cells (data not shown). In contrast, DHT-induced proliferation of PCa cells was blocked by knockdown of AR but not of PELP1 (Fig. 6B). E2-induced proliferation was dependent on a functional ERβ and AR, as confirmed by chemical blockers of ER (ICI 182,780) or AR (flutamide) in LAPC4 cells (Fig. 6C). Both ICI 182,780 and flutamide block the ability of E2-induced protein complex formation between ERβ, AR, and PELP1 (Fig. 6D). Flutamide has no effect E2-induced protein complex between ERβ and PELP1 but appears to decrease the ability of AR to bind this protein complex. Taken together, these data strongly support the central role for AR in E2-induced and DHT-induced proliferation in PCa cells.
Fig. 6.
Role of PELP1 and AR in E2 and DHT-mediated proliferation of PCa cells. A and B, 50,000 C4-2 cells in 24-well plates were transfected with siRNA and subjected to androgen-free media for 48 h. E2 (10 nm) (A) or DHT(10 nm) (B) were added and proliferation evaluated at 48 h. C and D, 50,000 LAPC-4 cells were subjected to androgen-free media for 48 h, incubated with either ICI 182,780 (10 μm) or flutamide (25 μm) for 2 h before the addition of either E2 (10 nm) or DHT (10 nm). Proliferation was assessed by the MTT assay at 48 h (C). From a parallel batch of LAPC4 cells, protein extracts were prepared and subjected to immunoprecipitation by AR or ERβ and immunoblotted with corresponding antibody. Input lysates are shown on the bottom. EtOH, Ethanol; IP, immunoprecipitation; siRNA, small interfering RNA; si con, control siRNA; ICI, ICI182780; Flu, flutamide; *, statistically significant.
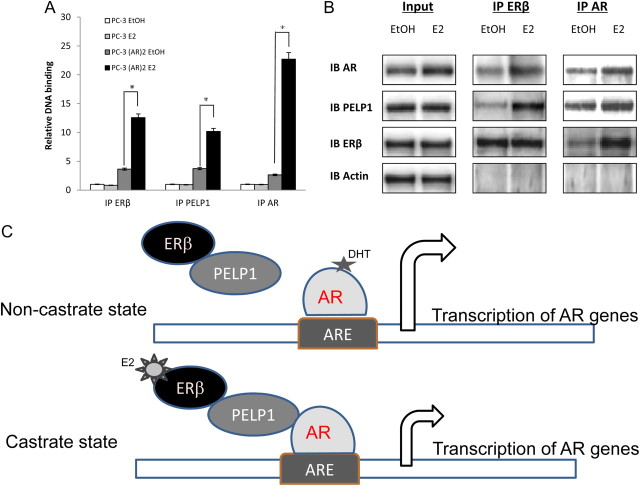
Mechanistically, the treatment with E2 resulted in the increased recruitment of AR, ERβ, and PELP1 to the complex formation on the PSA promoter on ChIP assays (Fig. 7A). E2 treatment increased the interaction between ERβ and PELP1, between ERβ and AR, and between AR and PELP1 (Fig. 7B). Further validation of these findings was obtained from ChIP-re-ChIP experiments, which revealed an E2-mediated increase in AR, ERβ, and PELP1 complex formation on the PSA promoter (Supplemental Fig. 5). Critically, the complex on the PSA promoter induced by E2 appears to be distinct from that induced by DHT, which further supports this alternative pathway for activation of androgen-responsive genes. Taken together, these data strongly suggest that in the presence of DHT (noncastrate state), that the transcription from androgen-responsive genes is driven by AR. ERβ and PELP1 may be present in a protein complex with AR but are not required for DHT-mediated transcription. In contrast, in the castrate state, DHT may be absent, and exposure of PCa cells to E2 may recruit ERβ and PELP1 to the DNA complex with AR and activate transcription of genes regulated by ARE (Fig. 7C). Thus, this model indicates a role for PELP1 in regulating nonandrogenic activation of AR signaling in PCa cells.
Fig. 7.
Mechanism of E2-regulated gene expression. A, PC-3 cells or PC-3(AR)2 were subject to androgen-free media for 2 d and then treated with 10 nm E2 for 24 h; 100 μl of sheared DNA lysates were subject to ChIP assays with the PSA promoter after immunoprecipitation with ERβ, AR, or PELP1. B, C4-2 cells were subject to androgen-free media for 2 d and then treated with 10 nm E2 for 24 h; 50 μg of protein extracts were evaluated by immunoprecipitation by AR or ERβ and immunoblotted with corresponding antibody. Input lysates are shown on the left. C, Model showing the activation of AR-responsive genes in the noncastrate state in the presence of androgen and in the castrate state in the absence of androgen. EtOH, Ethanol; IB, immunoblot; IP, immunoprecipitation; *, statistically significant.
Discussion
The role of estrogens in PCa initiation and development is complex and poorly understood. In men, serum testosterone levels drop by about 35% between the ages of 21 and 85, whereas E2 levels remain constant or increase (19). These levels are thought to be maintained by increased aromatization of adrenal androgens within peripheral adipose tissue, which also tends to increase in older males (19, 32–34) This alteration of the E2/testosterone ratio temporally mirrors the onset of prostate disease and in particular PCa (19, 35, 36). In addition, serum levels of estrogens in African American men (who have the highest incidence of PCa) are higher than in Caucasian and Japanese men (lowest incidence of PCa); serum testosterone levels did not significantly vary among these men (19, 32). Data from aromatase-knockout mice indicate that PCa develops more rapidly when estrogen was administered in addition to testosterone (37). Expression array analyses indicate a central role for ER-related pathways in PCa (19, 38, 39). These data strongly suggest a role for estrogens in PCa.
The intraprostatic concentration of E2 in patients with PCa after castration is not known. Although a 10 nm concentration of E2 is physiologic, the relevance to the prostatic milieu is not known (19). Clearly, E2 is not as potent as DHT: the concentrations required for maximal activation of AR (10 nm E2) may not be reached. However, we have shown that even at 0.1–1 nm concentrations, E2 is able to activate AR signaling. This activation of AR signaling may be sufficient in CRPC and represents an alternative pathway for AR activation. E2 may directly bind AR. However, the ability of ERβ and PELP1 knockdowns to diminish AR activation strongly suggests that direct binding of E2 to AR is not the primary driver of E2-induced AR signaling.
Our data suggest a possible mechanism for studies that show a poorer outcome with higher ERβ expression in metastatic PCa. Elevated expression of ERβ may allow for the nonandrogenic activation of AR signaling and proliferation of CRPC cells potentially in response to lower E2 concentrations. Activation of ERβ leads to increased binding of the protein complex of AR, ERβ, and PELP1 to the DNA and subsequent activation of transcription from these promoters. Disruption of this complex blocks the ability of E2 to activate AR genomic signaling.
In this manuscript, we have described a novel mechanism for AR activation in the absence of androgens via the scaffolding protein PELP1. PELP1 expression is deregulated in a wide variety of hormone-driven tumors, including breast, ovarian, and PCa (23, 24, 29, 40). PELP1 contains several motifs that may be involved in its myriad of protein-protein interactions, including 10 NR-interacting boxes (LXXLL motifs), a zinc finger motif, and proline-rich domains (41). PELP1 likely interacts with NR such as AR, ER, glucocorticoid, and progesterone receptor (PR) via these LXXLL motifs and modulates the function of these NR (24). PELP1 directly also interacts with kinases such as c-Src and the p85 subunit of phosphatidylinositol 3 kinase via its proline-rich domains (42). This ability of PELP1 to interact with multiple proteins via distinct motifs allows it to function as a “scaffolding” protein bringing disparate NR and kinases together. Our data indicate that PELP1 bridges the interaction between AR and ERβ in PCa cells and enables the activation of AR by the activation of ERβ. The physical interaction of PELP1 with both AR and ERβ likely facilitates the cross talk between these two NR.
Our data indicate that PELP1 forms a complex with ERβ in the absence of AR in PC-3 PCa cells. However, this complex is unable to bind ARE or direct transcription from an ARE. Thus, although both PELP1 and ERβ can both bind DNA, their effect on transcription from an ARE requires AR binding to the ARE. These data are further confirmed by the AR mutant experiments, which demonstrate that the transactivation of AR-responsive genes by E2 requires both AR DNA-binding domain as well as AR interaction with PELP1. Further, the fold of transcriptional activation of AR-responsive genes is driven by AR transactivation domain, because deletion of this domain significantly attenuates AR transactivation. E2 treatment strongly induces the formation of the AR-PELP1-ERβ complex on the DNA. Our data indicate that PELP1 enables the bridging of the E2 signal from ERβ to AR and thus allowing AR to activate transcription of AR-regulated genes.
Interestingly, the AR-PELP1-ERβ complex does not appear to be critical for DHT-induced transcription from an ARE. DHT-induced AR gene expression does require AR, as evidenced by knockdown experiments. DHT and E2 treatment of PCa cells are known to induce transcription of many similar genes, as evidenced by microarray analyses (43). These data establish a novel mechanism for E2-induced transcription of AR-responsive genes in PCa cells. E2 appears to be less potent than DHT in inducing transcription from an ARE. However, the similar fold induction of PSA and Nkx3.1 gene expression by E2 and DHT suggests that additional mechanisms may also be involved in E2-regulated gene expression. Our data suggest that other proposed mechanisms, including possible direct E2-ERβ binding to ARE or direct binding of E2 to AR, are unlikely to play a major role in E2-mediated transcription of AR-responsive genes. Previous studies have indicated that E2 may bind weakly to a mutant AR T887A that is seen in LNCaP cells. However, binding of E2 to unmutated AR such as that found in LAPC4 cells has not been reported (44). The activity of E2 in LAPC4 cells as well as in PC3 cells with stably transfected wild-type AR cannot be attributed to E2 binding to AR.
Knockdown of PELP1 expression by conventional siRNA and short hairpin RNA techniques appears to be of limited value. In our hands, we can routinely achieve only a 70% knockdown of PELP1 protein expression. Despite multiple approaches to achieve higher levels of PELP1 knockdown, PCa cells appear to require some expression of PELP1. Similar findings have been noted in ovarian cancer cell lines, where a 70–80% knockdown of PELP1 could be achieved (30). Consequently, although the knockdown experiments strongly indicate a role for PELP1 in the nonandrogenic activation of AR, they likely underestimate the true measure of the role of PELP1 in nonandrogenic activation of AR. Novel approaches may be required to block PELP1 function in PCa cells.
The ability of E2 to activate AR in PCa cells raises several additional mechanistic questions. The effect of E2, PELP1, or ERβ on AR nuclear translocation has not been reported. Several factors are postulated to be involved in AR nuclear translocation, including heat shock proteins (hsp), hsp27 and hsp90, importin, and others. The role of AR posttranslational modifications such as sumolyation or phosphorylation on AR nuclear translocation is not known. PELP1 may serve as a scaffolding protein involved in alternative mechanisms of AR nuclear translocation and AR activation. PELP1 may couple the E2 signal to AR activation, by enabling AR posttranslational modifications or by disrupting critical protein-protein interactions involved in AR cytoplasmic tethering. These putative mechanisms are under active investigation in our laboratory.
The key findings in this manuscript, that PELP1 to couple ERβ with AR and to activate AR in the absence of androgenic ligand, are both novel and interesting. Because PELP1 may directly interact with ERα, glucocorticoid, and PR (45), PELP1 may couple the activation of AR signaling to other steroid receptors as well. Moderate to strong PR expression is identified in 60% of metastatic lesions and in 54% of recurrent tumors after androgen deprivation therapy (46). Thus, this cross talk between steroid receptors resulting in nonandrogenic activation of AR may be an escape mechanism for AR activation in PCa cells despite blockade of androgen production. We believe that this cross talk pathway via PELP1 may represent a viable therapeutic target for CRPC.
Materials and Methods
Case selection
Formalin-fixed paraffin-embedded PCa tissues were retrieved from March 2004 to October 2004 at our institution, using pathology archives. Tissue samples from 35 consecutive patients undergoing a radical prostatectomy for PCa were included in the construction of tissue microarray (TMA). We used consecutive patients to minimize the potential biases in selection of specific patients. The Institutional Review Board reviewed and approved the study. For each case, comprehensive clinicopathologic data were obtained from patients' medical files and entered into an institutional review board-approved database.
Pathologic evaluation
All hematoxylin and eosin (H&E)-stained section from each specimen were reviewed by a staff pathologist (Dr. Payal Kapur) to select representative areas of the tumor from which to acquire cores for microarray analysis. Two to three samples, for each patient, two from the tumor and one from high-grade prostatic intraepithelial neoplasm if present, were identified and circled on the H&E-stained slides. Only cases with large tumor volume (more than two sections of tumor with at least 0.5 cm of the tumor dimension on each slide) were included in the study.
Construction of TMA blocks
TMA were built using a semiautomatic arraying instrument (Beecher Instruments, Silver Spring, MD) that uses two separate core needles for punching the donor and recipient blocks and a micrometer-precise coordinate system for assembling tissue samples on a block. For each case, two to three 1.0-mm core diameter sample punches were taken, each of which corresponded to the earlier selected areas on the H&E section. These punches from each “donor” block and placed on separate “recipient” TMA block and spaced 0.5 mm apart. Multiple sausage internal controls of normal prostate were also placed in the TMA block. Serial 3- to 4-μm sections were obtained from the microarray for immunohistochemical staining, and the first slide was stained with H&E to confirm the presence and grade of tumor. Tumor samples were randomly arranged on the blocks. Sample tracking was based on coordinate positions for each tissue spot in the TMA block; the spots were transferred onto TMA slides for staining. This sample tracking system was linked to a Microsoft Excel database containing demographic, clinical, pathologic, and survival data on each patient. The array was read according to the given TMA map, and each core was scored individually. The cores that contained very little tumor were not evaluated. Each case had at least one core punches with sufficient tumor (at least three to four high-power fields of viable tumor) for immunohistochemical evaluation.
Immunohistochemical staining
We performed immunohistochemical staining for PELP1, ERα, ERβ, and AR using serial sections from the paraffin-embedded TMA block. Immunostaining was performed on either Ventana Benchmark XT (ER and PELP) automatic immunostainer (Ventana, Tucson, AZ) or DAKO (AR) Autostainer (DAKO, Carpinteria, CA). Heat-induced epitope retrieval was performed using EDTA buffer and a modified pressure cooker. Optimum primary antibody dilutions were predetermined, using a PCa as positive control sample for PELP1 (monoclonal mouse, SX53G8, dilution 1:150; DAKO), ERα (monoclonal rabbit, SP1, prediluted; Ventana), ERβ (monoclonal mouse, 14C8, dilution 1:50; Abcam, Cambridge, MA), and AR (polyclonal rabbit, N-20, dilution 1:50; Santa Cruz Biotechnology, Inc., Santa Cruz, CA). Negative controls were run by omitting the primary antibody.
Immunohistochemical analysis and scoring
Number of positive tumor cells in relation to the total number of cells encountered and the intensity of nuclear staining (weak or strong) for each of the markers were quantified manually per tissue core by a staff pathologist (Dr. Payal Kapur), who was blinded from the clinical data. The percentage of positively staining tumor cells was assessed. The mean of the duplicate scores was calculated for data analysis.
Cell lines and culture
LNCaP and DU145 were obtained from American Type Culture Collection (Manassas, VA). LAPC4 was kindly provided by C. Sawyers (Memorial Sloan Cancer Center, New York, NY), CWR22v1 was kindly provided by T. G. Pretlow (Case Western Reserve University, Cleveland, OH), and PC-3(AR)2 was kindly provided by T. J. Brown (University of Toronto, Toronto, Canada). LNCaP, C4-2, PC-3, PC-3(AR)2, and DU145 were maintained in T medium (Invitrogen) supplemented with 5% fetal bovine serum (FBS); CWR22v1 in DMEM (Mediatech, Manassas, VA) containing 10% FBS; and LAPC-4 cell in Iscove's DMEM (Mediatech) containing 10% FBS. All growth media were supplement with penicillin (100 IU/ml) and streptomycin (100 μg/ml).
Androgen-free media
Cells were carefully washed with PBS, and growth medium was changed to phenol red-free RPMI 1640 with 1% charcoal strip FBS for 48 h before treatment.
Antibodies and reagents
AR, β-actin antibody was purchased from Sigma (St. Louis, MO); PELP1 antibody from Bethyl Laboratories (Montgomery, TX); ERβ antibody (MCA1974S) from AbDSerotec (Raleigh, NC); ERβ, AR (AR441 and AR C-19) PSA antibodies from Santa Cruz Biotechnology, Inc.; and ER-α from Cell Signaling Technology (Boston, MA). E2, DHT, ICI 182,780, and flutamide were purchased from Sigma-Aldrich (St. Louis, MO). E2, DHT, and flutamide were solubilized in ethanol; ICI 182,780 was solubilized in dimethylsulfoxide.
RNA interference
Twenty micromolar human PELP1 SMARTpool siRNA or nonspecific control siRNA from Dharmacon (Chicago, IL), or siRNA against human AR, ERβ from Invitrogen (20 μm), was transfected using LipofectAMINE 2000 (Invitrogen) according to manufacturer's protocol.
Proliferation assays
Cells in 96-well plates were subjected to androgen deprivation for 48 h. Cells were then treated with either ethanol, E2, or DHT for 48 h. Chemical inhibitors were added 2 h before treatment. Cell proliferation was measured using MTT colorimetric assay. All experiments were performed in triplicate.
Western blot analysis
After treating with ethanol, E2, and DHT for 24 h, cells were washed twice with PBS and total cellular protein extracted from cell pellets by protein lysis buffer [50 mmol/liter HEPES (pH 7.5), 150 nmol/liter NaCl, 10% glycerol, 1% Triton X-100, and 1.5 mmol/liter MgCl2] containing protease inhibitor cocktail. Protein samples (15–30 μg) were loaded on polyacrylamide gels (NuPAGE 10% bis-Tris gel; Invitrogen) and subjected to electrophoretic analysis and subsequent blocking. Membranes were incubated with primary antibody (overnight at 4 C) and relevant secondary antibodies (1 h at room temperature). After extensive washing, membranes were developed using ECL Plus (Amersham, Piscataway, NJ) or SuperSignal West Dura Extended Duration Substrate (Pierce, Rockford, IL).
RNA isolation and real-time RT-PCR
Total cellular RNA was extracted with RNeasy mini kit (QIAGEN, Valencia, CA) according to manufacturer's instructions. One microgram of RNA was subjected to cDNA synthesis (Bio-Rad, Hercules, CA); 2.5 μl of cDNA were subjected to a 25-μl PCR carried out in iCycler thermal cycler (Bio-Rad) using iQ SYBR Green Supermix (Bio-Rad) with denaturing at 95 C for 3 min followed by 36 cycles of amplification at 95 C for 30 sec, 60 C for 30 sec, and 72 C for 60 sec. Primers to homeobox protein NK-3 homolog A and PSA were obtained from Invitrogen. 18S rRNA was used as internal control. All experiments were repeated in triplicate. Fold induction of homeobox protein NK-3 homolog A, PSA mRNA was determined by normalizing threshold of cycle value of these cDNA with Δ threshold of cycle value of 18S rRNA cDNA of each sample.
Immunoprecipitation
AR, ERβ, and PELP1 antibodies were bound with Dynabeads Protein G (Invitrogen), and target antigens in whole cell lysates (200 μg) were immunoprecipitated according to manufacturer's protocol.
Luciferase assay
Appropriately transfected cells were divided into 12-well plates and culture media replaced after 24 h with androgen-free media. Identical cell populations were stimulated with E2, DHT, or vehicle for additional 24 h. All experiments were performed in triplicate, luciferase activity in cell lysates were measured using luciferase assay system (Promega, Madison, WI) and were normalized to sample protein concentration. Results are presented as fold change from baseline by dividing the relative luciferase activity of treated cells over that obtained for untreated cells.
Chromatin immunoprecipitation
Transfected cells were subject to androgen deprivation for 48 h and then treated with equal volumes of 0.1% ethanol, E2, or DHT. Twenty-four hours later, cells were fixed in 1% formaldehyde solution in PBS for cross-linking, which was stopped using 125 mmol/liter glycine. Cells were treated with 750-μl cell lysis buffer [5 mmol/liter PIPES (pH 8.0), 85 mmol/liter KCl, 0.5% Nonidet P-40, 12 ng of benzamidine, 100 ng of 1,10-phenanthroline, 100 ng of aprotinin, and 100 ng/ml leupeptine] for 5 min on ice. Nuclear lysis buffer [400 μl; 50 mmol/liter Tris-Cl (pH 8.1), 10 mmol/liter EDTA, and 1% sodium dodecyl sulfate (SDS)] was added to the pellet for 10 min on ice, and samples were sonicated to shear DNA to 200- to 1000-bp fragments. Lysates were precleared with rotating incubation with 20 μl of Sepharose beads for 2 h at 4 C. As a positive control for DNA fragmentation, 10 μl (10% volume of the immunoprecipitates) of input samples were collected at this stage. For immunoprecipitation, 100 μl of lysate were incubated with 5 μg of AR antibody. Immunoprecipitation was performed in 400 μl of IP buffer [150 mmol/liter NaCl, 1.0% Nonidet P-40, 0.5% deoxycholate, 0.1% SDS, and 50 mmol/liter Tris (pH 8.0)] for 2 h at 4 C. Then protein A-Sepharose (Amersham) was added for 2 h at 4 C with rotation. Protein A beads were washed with Super-IP (IP buffer plus 150 mmol/liter NaCl), thrice with IP buffer and once with TE buffer [10 mmol/liter Tris (pH 8.0) and 1 mmol/liter EDTA], with 5-min rotations between each wash at room temperature. To extract DNA, all samples (including inputs) were incubated with 150 μl of ChIP extraction buffer (1% SDS and 0.1 mol/liter NaHCO3) along with 10 μl of 5 mol/liter NaCl and 0.3 μl of ribonuclease A. This solution was incubated at 65 C overnight. DNA was purified using QIAquick PCR Purification kit (QIAGEN). PCR amplification was done on DNA recovered from immunoprecipitated and input chromatin samples. PCR conditions were: 94 C for 3 min followed by 36 cycles of 94 C for 30 sec, 60 C for 30 sec, and 72 C for 60 sec. Quantification by real-time PCR was done as indicated.
ChIP-re-ChIP assay
Ten micrograms of sheared chromatin from variously treated LNCAP cells were extracted as described (ChIP-IT Express Enzymatic Magnetic Cromatin Re-immunopreciptation kit and Enzymatic Shearing kit; catalog nos. 53009 and 53035; Active Motif, Carlsbad, CA) and added to 25 μl of progtein G magnetic beads, 20 μl of ChIP buffer 1, and protease inhibitor cocktail (Re-ChIP-IT Magnetic Chromatin Re-immunopreciptation kit, catalog no. 53016; Active Motif) in a siliconized 1.7-ml microcentrifuge tube. Three micrograms of either AR or PELP1 antibody were added last and the mixture incubated for 4 h at 4C. The tubes were then placed on a magnetic stand to pellet the beads on the side of the tubes and the supernatant discarded. The beads were washed with the ChIP buffers provided and then the chromatin eluted using the Re-ChIP-IT Elution buffers. The chromatin was then desalted using a prepared desalted column.
A portion of the chromatin (10 μl) from this stage was processed and used to evaluate the efficacy of the immunoprecipitation using quantitative polymerase chain reaction (QPCR). Most of the chromatin (90 μl) from this stage was then used for the re-ChIP evaluation and was mixed with 25 μl of protein G magnetic beads, 10 μl of ChIP buffer 1, 1 μl of protease inhibitor cocktail, and 11 μl of distilled water. 3 μg of either AR or PELP1 antibody were added last and the mixture incubated for 4 h at 4C. The tubes were then placed on a magnetic stand to pellet the beads on the side of the tubes and the supernatant discarded. The beads were washed with the ChIP buffers provided and then the chromatin eluted using the Re-ChIP-IT Elution buffers. The cross-links were reversed using reverse cross-linking buffer. Processed DNA samples then used for QPCR evaluation.
Statistical analysis
All error bars in graphical data represent mean ± sd. Student's two-tailed t test was used for the determination of statistical relevance between groups with P < 0.05 considered as significant.
Acknowledgments
This work was supported by a Durden Foundation-Prostate Cancer Foundation Young Investigator grant and by the Dorothy and James Cleo Thompson Foundation (G.V.R.).
Disclosure Summary: The authors have nothing to disclose.
Footnotes
- AR
- Androgen receptor
- ARE
- androgen-responsive element
- ChIP
- chromatin immunoprecipitation
- CRPC
- castration-resistant PCa
- DHT
- dihydrotestosterone
- E2
- 17β-estradiol
- ER
- estrogen receptor
- FBS
- fetal bovine serum
- H&E
- hematoxylin and eosin
- hsp
- heat shock protein
- MTT
- 3-(4,5-dimethylthiazol-2-yl)-2,5-diphenyltetrazolium bromide
- NR
- nuclear receptor
- PCa
- prostate cancer
- PELP1
- proline-, glutamic acid-, and leucine-rich protein 1
- PR
- progesterone receptor
- PSA
- prostate-specific antigen
- QPCR
- quantitative polymerase chain reaction
- SDS
- sodium dodecyl sulfate
- siRNA
- small interfering RNA
- TMA
- tissue microarray.
References
- 1. Knudsen KE , Scher HI. 2009. Starving the addiction: new opportunities for durable suppression of AR signaling in prostate cancer. Clin Cancer Res 15:4792–4798 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2. Chen CD , Welsbie DS , Tran C , Baek SH , Chen R , Vessella R , Rosenfeld MG , Sawyers CL. 2004. Molecular determinants of resistance to antiandrogen therapy. Nat Med 10:33–39 [DOI] [PubMed] [Google Scholar]
- 3. Chen Y , Sawyers CL , Scher HI. 2008. Targeting the androgen receptor pathway in prostate cancer. Curr Opin Pharmacol 8:440–448 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Edwards J , Bartlett JM. 2005. The androgen receptor and signal-transduction pathways in hormone-refractory prostate cancer. Part 1: modifications to the androgen receptor. BJU Int 95:1320–1326 [DOI] [PubMed] [Google Scholar]
- 5. Takahashi Y , Perkins SN , Hursting SD , Wang TT. 2007. 17β-estradiol differentially regulates androgen-responsive genes through estrogen receptor-β- and extracellular-signal regulated kinase-dependent pathways in LNCaP human prostate cancer cells. Mol Carcinog 46:117–129 [DOI] [PubMed] [Google Scholar]
- 6. Risbridger GP , Ellem SJ , McPherson SJ. 2007. Estrogen action on the prostate gland: a critical mix of endocrine and paracrine signaling. J Mol Endocrinol 39:183–188 [DOI] [PubMed] [Google Scholar]
- 7. Li LC , Carroll PR , Dahiya R. 2005. Epigenetic changes in prostate cancer: implication for diagnosis and treatment. J Natl Cancer Inst 97:103–115 [DOI] [PubMed] [Google Scholar]
- 8. Li LC , Chui R , Nakajima K , Oh BR , Au HC , Dahiya R. 2000. Frequent methylation of estrogen receptor in prostate cancer: correlation with tumor progression. Cancer Res 60:702–706 [PubMed] [Google Scholar]
- 9. Leav I , Lau KM , Adams JY , McNeal JE , Taplin ME , Wang J , Singh H , Ho SM. 2001. Comparative studies of the estrogen receptors β and α and the androgen receptor in normal human prostate glands, dysplasia, and in primary and metastatic carcinoma. Am J Pathol 159:79–92 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Lai JS , Brown LG , True LD , Hawley SJ , Etzioni RB , Higano CS , Ho SM , Vessella RL , Corey E. 2004. Metastases of prostate cancer express estrogen receptor-β. Urology 64:814–820 [DOI] [PubMed] [Google Scholar]
- 11. Ito T , Tachibana M , Yamamoto S , Nakashima J , Murai M. 2001. Expression of estrogen receptor (ER-α and ER-β) mRNA in human prostate cancer. Eur Urol 40:557–563 [DOI] [PubMed] [Google Scholar]
- 12. Horvath LG , Henshall SM , Lee CS , Head DR , Quinn DI , Makela S , Delprado W , Golovsky D , Brenner PC , O'Neill G , Kooner R , Stricker PD , Grygiel JJ , Gustafsson JA , Sutherland RL. 2001. Frequent loss of estrogen receptor-β expression in prostate cancer. Cancer Res 61:5331–5335 [PubMed] [Google Scholar]
- 13. Setlur SR , Mertz KD , Hoshida Y , Demichelis F , Lupien M , Perner S , Sboner A , Pawitan Y , Andrén O , Johnson LA , Tang J , Adami HO , Calza S , Chinnaiyan AM , Rhodes D , Tomlins S , Fall K , Mucci LA , Kantoff PW , Stampfer MJ , Andersson SO , Varenhorst E , Johansson JE , Brown M , Golub TR , Rubin MA. 2008. Estrogen-dependent signaling in a molecularly distinct subclass of aggressive prostate cancer. J Natl Cancer Inst 100:815–825 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Signoretti S , Loda M. 2001. Estrogen receptor β in prostate cancer: brake pedal or accelerator? Am J Pathol 159:13–16 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Pasquali D , Rossi V , Esposito D , Abbondanza C , Puca GA , Bellastella A , Sinisi AA. 2001. Loss of estrogen receptor β expression in malignant human prostate cells in primary cultures and in prostate cancer tissues. J Clin Endocrinol Metab 86:2051–2055 [DOI] [PubMed] [Google Scholar]
- 16. Pravettoni A , Mornati O , Martini PG , Marino M , Colciago A , Celotti F , Motta M , Negri-Cesi P. 2007. Estrogen receptor β (ERβ) and inhibition of prostate cancer cell proliferation: studies on the possible mechanism of action in DU145 cells. Mol Cell Endocrinol 263:46–54 [DOI] [PubMed] [Google Scholar]
- 17. McPherson SJ , Hussain S , Balanathan P , Hedwards SL , Niranjan B , Grant M , Chandrasiri UP , Toivanen R , Wang Y , Taylor RA , Risbridger GP. 2010. Estrogen receptor-β activated apoptosis in benign hyperplasia and cancer of the prostate is androgen independent and TNFα mediated. Proc Natl Acad Sci USA 107:3123–3128 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Benbrahim-Tallaa L , Liu J , Webber MM , Waalkes MP. 2007. Estrogen signaling and disruption of androgen metabolism in acquired androgen-independence during cadmium carcinogenesis in human prostate epithelial cells. Prostate 67:135–145 [DOI] [PubMed] [Google Scholar]
- 19. Carruba G. 2006. Estrogens and mechanisms of prostate cancer progression. Ann NY Acad Sci 1089:201–217 [DOI] [PubMed] [Google Scholar]
- 20. Heemers HV , Regan KM , Schmidt LJ , Anderson SK , Ballman KV , Tindall DJ. 2009. Androgen modulation of coregulator expression in prostate cancer cells. Mol Endocrinol 23:572–583 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Edwards J , Bartlett JM. 2005. The androgen receptor and signal-transduction pathways in hormone-refractory prostate cancer. Part 2: androgen-receptor cofactors and bypass pathways. BJU Int 95:1327–1335 [DOI] [PubMed] [Google Scholar]
- 22. Brooke GN , Parker MG , Bevan CL. 2008. Mechanisms of androgen receptor activation in advanced prostate cancer: differential co-activator recruitment and gene expression. Oncogene 27:2941–2950 [DOI] [PubMed] [Google Scholar]
- 23. Dimple C , Nair SS , Rajhans R , Pitcheswara PR , Liu J , Balasenthil S , Le XF , Burow ME , Auersperg N , Tekmal RR , Broaddus RR , Vadlamudi RK. 2008. Role of PELP1/MNAR signaling in ovarian tumorigenesis. Cancer Res 68:4902–4909 [DOI] [PubMed] [Google Scholar]
- 24. Chakravarty D , Tekmal RR , Vadlamudi RK. 2010. PELP1: a novel therapeutic target for hormonal cancers. IUBMB Life 62:162–169 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Haas D , White SN , Lutz LB , Rasar M , Hammes SR. 2005. The modulator of nongenomic actions of the estrogen receptor (MNAR) regulates transcription-independent androgen receptor-mediated signaling: evidence that MNAR participates in G protein-regulated meiosis in Xenopus laevis oocytes. Mol Endocrinol 19:2035–2046 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Nair SS , Nair BC , Cortez V , Chakravarty D , Metzger E , Schüle R , Brann DW , Tekmal RR , Vadlamudi RK. 2010. PELP1 is a reader of histone H3 methylation that facilitates oestrogen receptor-α target gene activation by regulating lysine demethylase 1 specificity. EMBO Rep 11:438–444 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Choi YB , Ko JK , Shin J. 2004. The transcriptional corepressor, PELP1, recruits HDAC2 and masks histones using two separate domains. J Biol Chem 279:50930–50941 [DOI] [PubMed] [Google Scholar]
- 28. Brann DW , Zhang QG , Wang RM , Mahesh VB , Vadlamudi RK. 2008. PELP1—a novel estrogen receptor-interacting protein. Mol Cell Endocrinol 290:2–7 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Habashy HO , Powe DG , Rakha EA , Ball G , Macmillan RD , Green AR , Ellis IO. 2010. The prognostic significance of PELP1 expression in invasive breast cancer with emphasis on the ER-positive luminal-like subtype. Breast Cancer Res Treat 120:603–612 [DOI] [PubMed] [Google Scholar]
- 30. Chakravarty D , Roy SS , Babu CR , Dandamudi R , Curiel TJ , Vivas-Mejia P , Lopez-Berestein G , Sood AK , Vadlamudi RK. 2011. Therapeutic targeting of PELP1 prevents ovarian cancer growth and metastasis. Clin Cancer Res 17:2250–2259 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Cheskis BJ , Greger J , Cooch N , McNally C , Mclarney S , Lam HS , Rutledge S , Mekonnen B , Hauze D , Nagpal S , Freedman LP. 2008. MNAR plays an important role in ERa activation of Src/MAPK and PI3K/Akt signaling pathways. Steroids 73:901–905 [DOI] [PubMed] [Google Scholar]
- 32. Prins GS. 2008. Endocrine disruptors and prostate cancer risk. Endocr Relat Cancer 15:649–656 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Singh PB , Matanhelia SS , Martin FL. 2008. A potential paradox in prostate adenocarcinoma progression: oestrogen as the initiating driver. Eur J Cancer 44:928–936 [DOI] [PubMed] [Google Scholar]
- 34. Prins GS , Korach KS. 2008. The role of estrogens and estrogen receptors in normal prostate growth and disease. Steroids 73:233–244 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. Ellem SJ , Risbridger GP. 2006. Aromatase and prostate cancer. Minerva Endocrinol 31:1–12 [PubMed] [Google Scholar]
- 36. Prins GS , Putz O. 2008. Molecular signaling pathways that regulate prostate gland development. Differentiation 76:641–659 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37. McPherson SJ , Wang H , Jones ME , Pedersen J , Iismaa TP , Wreford N , Simpson ER , Risbridger GP. 2001. Elevated androgens and prolactin in aromatase-deficient mice cause enlargement, but not malignancy, of the prostate gland. Endocrinology 142:2458–2467 [DOI] [PubMed] [Google Scholar]
- 38. Fujimoto N , Suzuki T , Honda H , Kitamura S. 2004. Estrogen enhancement of androgen-responsive gene expression in hormone-induced hyperplasia in the ventral prostate of F344 rats. Cancer Sci 95:711–715 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39. Härkönen PL , Mäkelä SI. 2004. Role of estrogens in development of prostate cancer. J Steroid Biochem Mol Biol 92:297–305 [DOI] [PubMed] [Google Scholar]
- 40. Nair S , Vadlamudi RK. 2007. Emerging significance of ER-coregulator PELP1/MNAR in cancer. Histol Histopathol 22:91–96 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41. Vadlamudi RK , Wang RA , Mazumdar A , Kim Y , Shin J , Sahin A , Kumar R. 2001. Molecular cloning and characterization of PELP1, a novel human coregulator of estrogen receptor α. J Biol Chem 276:38272–38279 [DOI] [PubMed] [Google Scholar]
- 42. Nair SS , Guo Z , Mueller JM , Koochekpour S , Qiu Y , Tekmal RR , Schüle R , Kung HJ , Kumar R , Vadlamudi RK. 2007. Proline-, glutamic acid-, and leucine-rich protein-1/modulator of nongenomic activity of estrogen receptor enhances androgen receptor functions through LIM-only coactivator, four-and-a-half LIM-only protein 2. Mol Endocrinol 21:613–624 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43. Ngan S , Stronach EA , Photiou A , Waxman J , Ali S , Buluwela L. 2009. Microarray coupled to quantitative RT-PCR analysis of androgen-regulated genes in human LNCaP prostate cancer cells. Oncogene 28:2051–2063 [DOI] [PubMed] [Google Scholar]
- 44. Veldscholte J , Ris-Stalpers C , Kuiper GG , Jenster G , Berrevoets C , Claassen E , van Rooij HC , Trapman J , Brinkmann AO , Mulder E. 1990. A mutation in the ligand binding domain of the androgen receptor of human LNCaP cells affects steroid binding characteristics and response to anti-androgens. Biochem Biophys Res Commun. 173:534–540 [DOI] [PubMed] [Google Scholar]
- 45. Vadlamudi RK , Kumar R. 2007. Functional and biological properties of the nuclear receptor coregulator PELP1/MNAR. Nucl Recept Signal 5:e004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46. Bonkhoff H , Fixemer T , Hunsicker I , Remberger K. 2001. Progesterone receptor expression in human prostate cancer: correlation with tumor progression. Prostate 48:285–291 [DOI] [PubMed] [Google Scholar]