Abstract
FSH is a key hormonal regulator of Sertoli cell secretory activity, required to optimize sperm production. To fulfil its biological function, FSH binds a G protein-coupled receptor, the FSH-R. The FSH-R-transduced signaling network ultimately leads to the transcription or down-regulation of numerous genes. In addition, recent evidence has suggested that FSH might also regulate protein translation. However, this point has never been demonstrated conclusively yet. Here we have addressed this issue in primary rat Sertoli cells endogenously expressing physiological levels of FSH-R. We observed that, within 90 min of stimulation, FSH not only enhanced overall protein synthesis in a mammalian target of rapamycin-dependent manner but also increased the recruitment of mRNA to polysomes. m7GTP pull-down experiments revealed the functional recruitment of mammalian target of rapamycin and p70 S6 kinase to the 5′cap, further supported by the enhanced phosphorylation of one of p70 S6 kinase targets, the eukaryotic initiation factor 4B. Importantly, the scaffolding eukaryotic initiation factor 4G was also recruited, whereas eukaryotic initiation factor 4E-binding protein, the eukaryotic initiation factor 4E generic inhibitor, appeared to play a minor role in translational regulations induced by FSH, in contrast to what is generally observed in response to anabolic factors. This particular regulation of the translational machinery by FSH stimulation might support mRNA-selective translation, as shown here by quantitative RT-PCR amplification of the c-fos and vascular endothelial growth factor mRNA but not of all FSH target mRNA, in polysomal fractions. These findings add a new level of complexity to FSH biological roles in its natural target cells, which has been underappreciated so far.
In the male gonad, FSH is a critical factor controlling the environment that leads to the expansion of spermatogenesis. Its receptor, the FSH-R, is expressed at the surface of Sertoli cells, which secrete nutrients for each individual germ cell stage to which they intimately associate. It has been known for several decades that Sertoli cell stimulation by peritoneal injection of FSH for 1 h leads to enhanced overall protein synthesis (1). In vitro, similar results were obtained after 6 h of FSH stimulation (2), but no data are available with shorter stimulation time to alleviate mRNA transcriptional up-regulations. FSH likely acts at multiple regulation levels to enhance the protein content of the cell, as suggested for the androgen receptor (3). More recently microarray analyses of in vitro-stimulated rat Sertoli cells (4) and Sertoli cells from hypogonadal mouse stimulated in vivo (5) have shown that FSH impacts on the steady-state level of several mRNA. Although the whole FSH-induced proteome is not available yet, it is striking to observe that pioneering works on the subject had identify only a limited number of neosynthesized proteins, which does not match the several hundreds of genes up-regulated or down-regulated in response to FSH, as evidenced from transcriptomic analyses. This suggests that FSH proteome and transcriptome do not match and that regulations take place in between. Importantly, a recent work has quantified the part of gene regulations occurring at the level of transcription vs. translation in the same cell type and in a unique set of experiments. This work proposed a mathematical model that demonstrates that the protein content of a cell is predominantly controlled at the level of translation (6).
Previous reports have suggested that FSH could regulate the translation efficacy of preexisting mRNA. The signaling mechanisms that control the translational machinery involve changes in the phosphorylation of translation initiation and elongation factors as well as protein/protein interactions/dissociations. One important signaling pathway controlling mRNA translation involves the mammalian target of rapamycin (mTOR) within the mTORC1 and mTORC2 complexes, which triggers key regulatory steps of the protein synthesis machinery. One of them involves the ribosomal protein S6 kinases, which phosphorylate ribosomal protein S6 and eukaryotic initiation factor 4B (eIF4B) (7–10), among other proteins. Second, mTOR controls the ability of the 5′ methylguanosine triphosphate (m7-GTP) cap-binding protein, the eukaryotic initiation factor 4E (eIF4E), to initiate a molecular assembly that recruits the 40S ribosomal subunit to the mRNA, leading to formation of the 43S preinitiation complex. Activation of mTOR leads to sequential hyperphosphorylation of the eukaryotic initiation factor 4E-binding protein (4E-BP1) and its release from eIF4E, allowing eIF4E to bind the scaffolding protein eukaryotic initiation factor 4G (eIF4G) and form active preinitiation complexes (11). eIF4G scaffolds other proteins such as eukaryotic initiation factor 4A, an RNA helicase that unwinds secondary structure in mRNA 5′-untranslated regions that may otherwise impede scanning of the initiator codon by the 43S preinitiation complex (12).
FSH stimulates Sertoli cell division in the neonate and differentiation in prepubertal animals, and these biological responses are correlated to qualitative as well as quantitative differences in FSH signaling. The FSH-R is a G protein-coupled receptor (GPCR), which activates G protein- and β-arrestin-dependent signaling. Coupling of the FSH-R to Gs enables adenylyl cyclase to produce cAMP, whereas β-arrestins have been shown to stimulate the late phase of ERK activation (13, 14). In addition, a subtle interplay between cAMP- and phosphoinositide-dependent signaling is required for p70 S6 kinase 1 (p70S6K) activation by FSH in Sertoli cells: both pathways are costimulatory in differentiating cells (15), but they exert opposite effects in proliferating cells (16). This is presumably due to up-regulation of phosphatase and tensin homolog deleted on chromosome 10 (PTEN) expression and activity by FSH in differentiating Sertoli cells, which prevents the level of phosphatidyl-inositide (3, 4, 5) trisphosphate to raise upon phosphoinositide-3 kinase activation (17). cAMP and phosphoinositides are also involved in p70S6K regulation in granulosa cells, where tuberous sclerosis 2 is a critical regulatory checkpoint triggered both by protein kinase A-stimulated ERK (18) and by Akt (19). Furthermore, in granulosa cells, FSH exposure also enhances the phosphorylation of p70S6K and of 4E-BP1 on Ser65 (19, 20). Likewise, FSH stimulation has been shown to inhibit AMP-activated protein kinase and consequently induces granulosa cell proliferation (21) by reducing the cell cycle inhibitor p27Kip level (22). AMP-activated protein kinase is a regulator of cell energy consumption. As such, it sustains tuberin inhibitory effect on the mTOR pathway when the ATP level drops intracellularly to limit energy-consuming anabolic processes (23).
To summarize, several studies have identified that signaling mechanisms classically involved in translational control can be activated in response to FSH in vitro. However, although the anabolic role of FSH in Sertoli cells is well established, it is still unknown whether FSH directly impacts on the translational machinery to enhance protein neosynthesis. Here we addressed this question in differentiating Sertoli cells from prepubertal rats, which naturally express the FSH-R at physiological levels.
Materials and Methods
Primary Sertoli cells
Sertoli cells were isolated from testes of 19-d-old Wistar rats (Iffa-Credo, Lyon, France). Animals were treated following the current ethical guidelines of the European Community 86/609/CEE. Following purification as reported previously (44), Sertoli cells were seeded on CellBind plates (Corning Life Sciences, Wilkes Barre, PA) in DMEM (Sigma Aldrich, St. Louis, MO) complemented with penicillin (100 IU/ml), streptomycin (100 μg/ml), glutamine (2 mm), retinol (50 ng/ml), vitamin E (200 ng/ml), and human transferrin (5 μg/ml), all purchased from Sigma. On average, our Sertoli cell cultures were 90% pure, as previously quantified (44). Twenty-four or 48 h after initial seeding, cells were exposed to 100 ng/ml porcine FSH, obtained from Dr. George R. Bousfield (Wichita State University, Wichita KS).
Measurement of protein synthesis by 35S-labeling
Sertoli cells were seeded at various densities the day before labeling and FSH stimulation. Each condition was assayed in triplicate. Then cells were preincubated in DMEM without methionine and without cysteine (Sigma Aldrich) for 30 min before the addition of 25 μCi/ml L-[35S]methionine/L-[35S]cysteine (PerkinElmer, Waltham, MA) and 100 ng/ml FSH for 1.5 h. In some experiments, PP242 (Sigma Aldrich), a potent mTOR inhibitor, was added 10 min before FSH stimulation. Cells were then washed thrice with ice-cold PBS and were lysed with TNET extraction buffer [20 mm Tris, pH 7.8; 50 mm NaCl; 5 mm EGTA; 1% (vol/vol) Triton X-100; 1 mm phenylmethylsulfonyl fluoride; 4 mm Na3VO4; 5 μg/ml leupeptin; 5 μg/ml pepstatin; and 5 μg/ml aprotinin]. One aliquot of each sample was precipitated with 10% (wt/vol) trichloroacetic acid on 3MM GF/C glass microfiber filters (Whatman, GE Healthcare UK Ltd., Buckinghamshire, UK), and precipitated radiolabeled proteins were quantified by scintillation counting with a Packard Tri-Carb 1600 counter (Perkin Elmer, MA\). An equivalent aliquot of each sample was directly placed into the scintillation liquid to quantify the total incorporation of radiolabeled amino acids within cells.
Polysomal fractionation
Forty-eight hours after their initial seeding, Sertoli cells (15 × 106 cells per condition) were treated with FSH for 90 min at 34 C. Fifteen minutes before harvesting, 100 μg/ml of cycloheximide (CHX) was added. Then cells were washed with ice-cold PBS/CHX and resuspended in polysome lysis buffer [10 mm Tris-HCl, pH 7.8; 10 mm MgCl2; 1.5 mm KCl; 1% Nonidet P-40; 1% deoxycholate; 2.5 mm dithiothreitol (DTT); 100 μg/ml CHX] and 500 U/ml RNasin (Promega, Madison, WI). Cellular debris was removed by centrifugation at 10,000 rpm for 5 min, and supernatants were layered onto a 12-ml linear sucrose gradient (10–45% sucrose in 10 mm Tris-HCl, pH 7.8; 10 mm MgCl2; 80 mm KCI; and 1 mm DTT) (Beckman Coulter, Palo Alto, CA) and centrifuged at 38,000 rpm in an SW41-Ti rotor at 4 C for 150 min. Gradients were collected while absorbance was monitored at 254 nm using a density gradient fractionation system (ISCO, Inc., Columbus, OH). Fractions were treated with 200 μg/ml proteinase K (Ambion, Austin, TX) for 25 min at 50 C in 10 mm Tris-HCl (pH 8.0), 1 mm EDTA, and 0.5% sodium dodecyl sulfate, and then deproteinized with an equal volume of phenol-chloroform (1:1) and precipitated with isopropanol. The RNA from each fraction was dissolved in 30 μl diethyl pyrocarbonate-treated H2O, and an aliquot was subjected to electrophoresis through a 1% agarose gel and stained with GelRed (Interchim, Montluçon, France). A control experiment was done as above, except that the samples were pretreated with 50 mm EDTA for 10 min before loading and that the linear sucrose gradient was supplemented with 10 mm EDTA.
Real-time RT-PCR analysis
One microgram of total RNA was treated with deoxyribonuclease I before reverse transcription using Superscript II and random primers (all from Invitrogen, Life Technologies, Carlsbad, CA) according to the manufacturer's instructions. Single-step, real-time PCR was performed in a LightCycler 480 II (Roche Diagnostics, Basel, Switzerland) in a final volume of 10 μl containing 25 ng of reverse transcribed RNA, 8 μl Platinum SYBR Green qPCR Super-Mix UDG (Invitrogen), and 1 μm of each primer. Primers, designed using the Primer3 software (http://frodo.wi.mit.edu/primer3/) have been synthesized by Eurogentec (Liège Science Park, Seraing, Belgium) and are listed in Supplemental Table 1, published on The Endocrine Society's Journals Online web site at http://mend.endojournals.org. Conditions were determined to avoid primer dimer formation and obtain optimal dynamic range for quantification. The relative expression of each target gene was normalized to glyceraldehyde-3-phosphate dehydrogenase (GAPDH) expression, and the Cp (crossing point) values were compared by calculating the 2−ΔCp (45). Then 2−ΔCp values were compared between the stimulated and unstimulated cells.
m7GTP pull-down assay
Twenty million cells at 60% confluency were rinsed once with PBS and then resuspended in 300 μl of 2× lysis buffer (40 mm HEPES, pH 7.4; 0.2 mm Na3VO4; 100 mm NaCl; 0.4 mm EDTA; 20 mg/ml aprotinin; 20 mg/ml leupeptin; 1 mm Pefablock 4-(2-aminoethyl)benzene sulfonyl fluoride hydrochloride; 2 mm DTT; 100 mm β-glycerophosphate; 20 mm pyrophosphate; and 100 mm NaF, all from Sigma Aldrich) and gently sonicated on ice. Cell debris were discarded by centrifugation for 15 min at 13,000 × g at 4 C, and proteins were quantified in the supernatants by a Bradford assay (Bio-Rad, Hercules, CA). For each sample, 50 μl of m7GTP-coupled Sepharose beads (GE Healthcare UK Ltd.) was equilibrated twice in 1× lysis buffer and were saturated with 10% BSA. Then 100 μg of cell lysate were added to the beads in 500 μl 2× buffer, and the samples were tumbled for 1.5 h at 4 C. After three washes in 1× lysis buffer complemented to 100 mm NaCl, samples were boiled in 50 μl of 2× Laemmli sample buffer and analyzed by Western blot.
Western blot analysis
Cell lysates corresponding to an equal cell number were resolved by 10 or 15% acrylamide SDS-PAGE, electrophoretically transferred to a nitrocellulose membrane (Whatman, Dassel, Germany), and probed with the appropriate primary antibody, generally diluted 1:1000. Antibodies raised against phospho-eIF4B [S422], phospho-4E-BP1 [S65], [T70], mTOR, eIF4G, and 4E-BP1 were purchased from Cell Signaling Technology Inc. (Beverly, MA). Antibodies to p70S6K and to c-fos were purchased from Santa Cruz Biotechnology Inc. (Santa Cruz, CA), and anti-eIF4E was from BD Biosciences Pharmingen (San Jose, CA).
Horseradish peroxidase-coupled antirabbit and antimouse antibodies were purchased from GE Healthcare UK Ltd. and were diluted 1:3000 or 1:5000, respectively, to detect antigen-antibody interactions by West Pico chemioluminescence (Thermo Scientific Life Science, Rockford, IL). Membranes were reprobed with an anti-GAPDH antibody (Santa Cruz Biotechnology) to monitor gel loading between lanes. All the films were scanned and the OD of the signals was measured with the ImageMaster 1D Elite version 4 software (Amersham Biosciences, Arlington Heights, IL).
Data analysis
Experiments shown are representative of at least three independent experiments and are expressed as mean values ± sem. One-way ANOVA was used to compare different samples or two-way ANOVA was used for comparison of different groups. Both analyses were followed by a Bonferroni's posttest. Differences between two groups were assessed by a two-tailed unpaired t test. Differences among means were considered as significantly different at P < 0.05. Data were analyzed by using the GraphPad PRISM software (San Diego, CA).
Results
FSH enhances protein neosynthesis in Sertoli cells cultured at high density
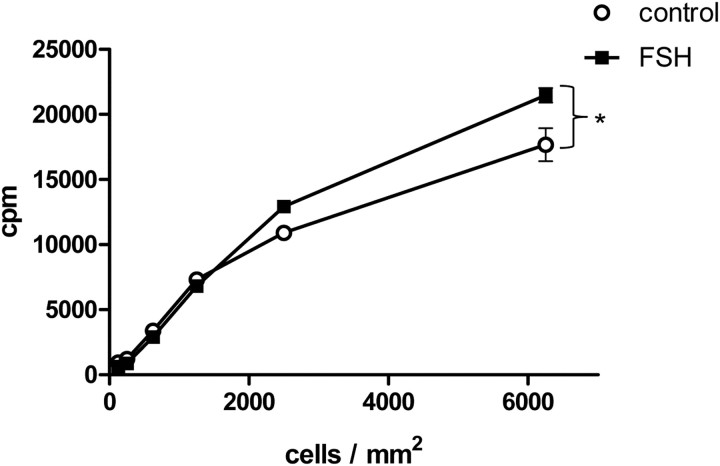
We first sought to determine whether FSH enhanced protein neosynthesis in our culture conditions. Experiments were performed with short duration of FSH stimulation, i.e. 90 min to minimize transcriptional regulations. This procedure was chosen over actinomycin D treatment because this drug inhibits the transcription of translation factors themselves (24). Supplemental Fig. 1 indicates that, except for immediate-early genes such as tissue-type plasminogen activator (tPA) or c-fos, well-known FSH target genes such as transferrin or anti-Mullerian hormone (AMH) are not or modestly up-regulated after 90 min. This observation is consistent with previous works indicating that in mouse, the peak of transcriptional regulations by FSH occurs at 4 h (5). Incorporation of 35S-labeled Met/Cys into neosynthesized proteins was quantified in Sertoli cells (Fig. 1) seeded at various densities and stimulated with FSH. In these conditions, a significant stimulatory impact of FSH on protein neosynthesis was visible. Incorporation of the radiolabeled aminoacids was moderately enhanced by FSH in the nonprecipitable fraction (Supplemental Fig. 2) but to a lesser extent than in the trichloroacetic acid precipitates. Previous evidence had been obtained that amino acid transport into testicular cells played only a minor role in the increased protein biosynthesis induced by FSH in vivo (1). Surprisingly, the hormone stimulatory effect was observed only when cells were seeded at high density. Hence, in the following, experiments were carried out at a cell density higher than 6250 Sertoli cells/mm2.
Fig. 1.
Short FSH stimulation enhances protein neosynthesis as a function of Sertoli cell density. Cells were seeded at increasing density, as indicated, before stimulation with 100 ng/ml FSH along with 35S-labeled methionine and cysteine. Ninety minutes later, cells were harvested and radiolabeled proteins were quantitated as trichloroacetic precipitable counts per minute. Results were expressed as means ± sem of triplicates. Statistical difference was calculated using two-way ANOVA with a Bonferroni posttest to compare control vs. FSH-stimulated cells. *, P < 0.05.
FSH enhances the recruitment of mRNA to the polysomes
Because we ruled out an important transcriptional component to regulate protein neosynthesis in response to short FSH stimulation, except for immediate-early genes, we next investigated at which posttranscriptional level FSH stimulation occurred. To explore whether mRNA translation could be impacted by FSH-mediated signaling, mRNA from control or FSH-stimulated cells were fractionated on a sucrose gradient, and the OD at 254 nm of the collected fractions led to discriminate between free RNA, 40 S and 60 S ribosomal subunits, 80 S monosomes, and polysomes, which were resolved as clear peaks (Fig. 2, A and B). Densitometry analysis of agarose gels loaded with aliquots of each of these fractions indicates that FSH stimulation enhanced the recruitment of mRNA to the polysomal fractions (Fig. 2C). EDTA treatment before loading the samples on the sucrose gradient supplemented with EDTA induced a shift toward the light, nonpolysomal fractions, indicative of the dissociation of monosomes and polysomes (Supplemental Fig. 3).
Fig. 2.
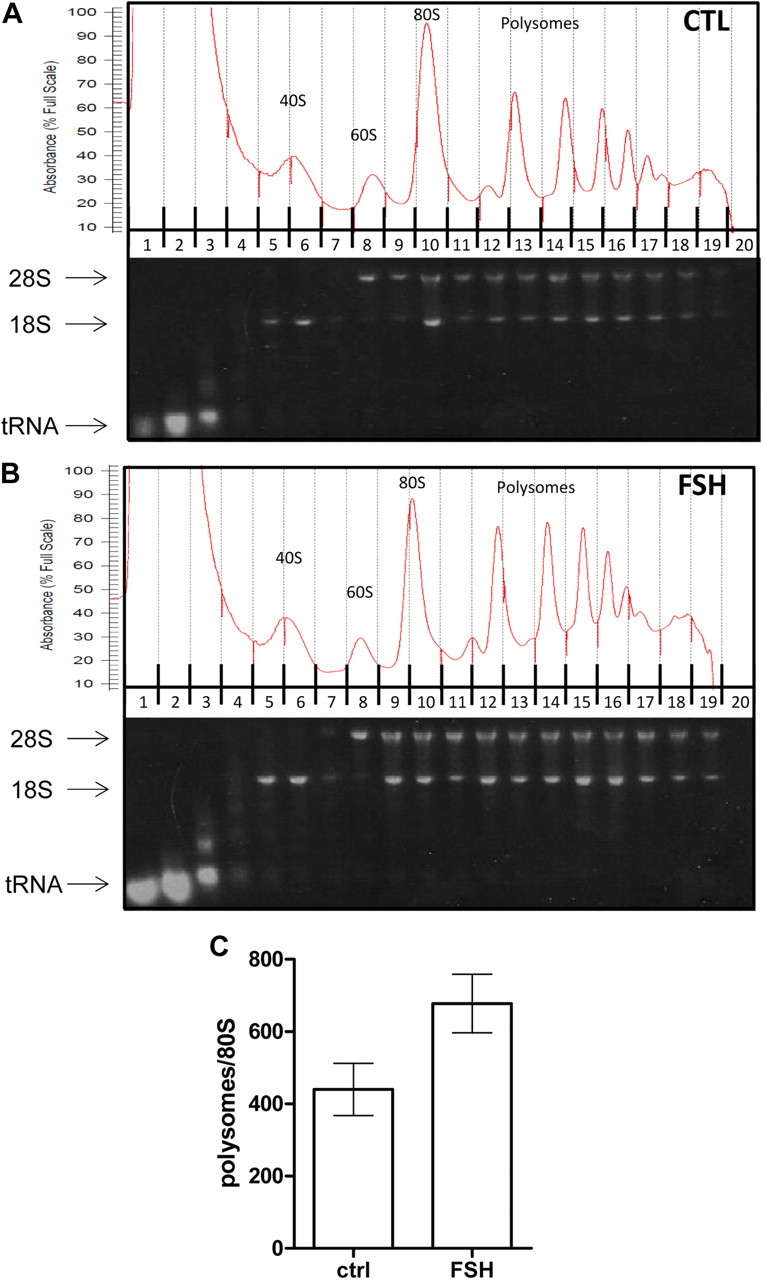
FSH stimulation enhances the assembly of polysomes in Sertoli cells. Sertoli cells were treated with FSH for 90 min and cell lysates were separated on a sucrose gradient. A, An experiment done with control Sertoli cells. B, FSH-stimulated cells have been fractionated. Upper panel, Representative absorbance profile at OD = 254 nm, of each collected fraction of RNA separated by velocity sedimentation through a 10–45% sucrose gradient. The positions of the 40 S and 60 S ribosomal subunits, 80 S single ribosomes and polysomes containing two to nine ribosomes are indicated. The nonpolysomal fractions also contain free RNA. Lower panel, Electrophoresis in a 1% agarose gel of an aliquot of RNA isolated from each of the 20 fractions. The 28 S and 18 S rRNA, and the tRNA are visible upon staining and UV illumination. C, Quantification of the band intensity in each fraction by scanning densitometry of the agarose gels. The histogram represents the ratio of intensity in the polysome vs. 80 S fraction, and the results are expressed as the means ± sem of five independent experiments. Statistical difference was calculated using a two-tailed unpaired t test to compare control vs. FSH-stimulated cells. P = 0.0503.
Signaling events at the mRNA 5′cap in response to FSH
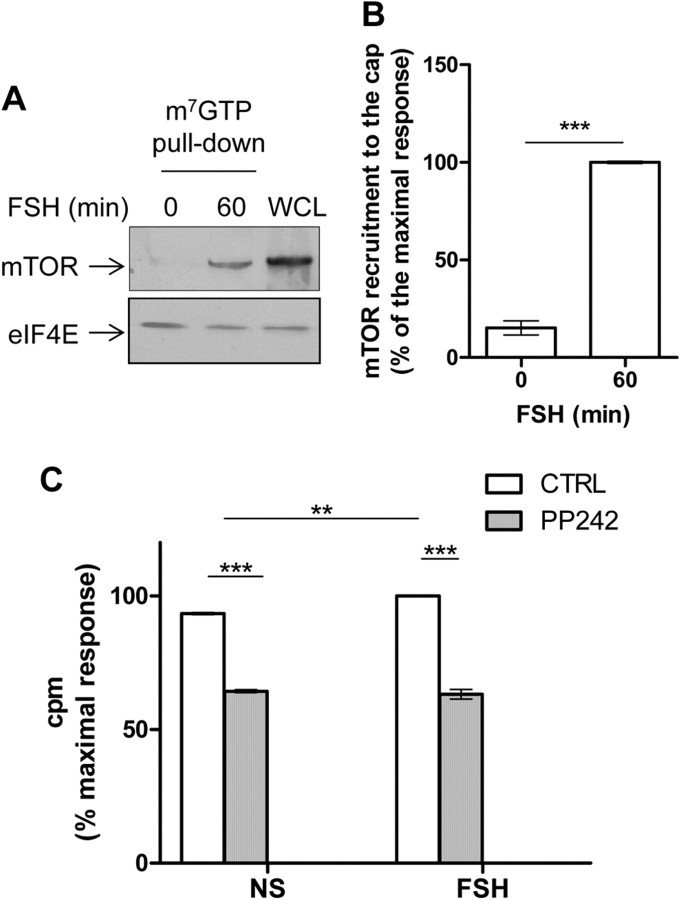
Because FSH stimulation enhanced polysome formation, hence mRNA translation, we assumed that the hormone might induce signaling events triggering the translational machinery. FSH stimulation led to a robust and significant recruitment of mTOR to the cap (Fig. 3, A and B). In agreement with mTOR-mediated translational activity, PP242, a potent mTOR pharmacological inhibitor, totally impaired FSH-induced protein neosynthesis (Fig. 3C). In this set of experiments, the stimulatory effect of FSH was weak yet significant. Cell viability appeared unaltered upon PP242 treatment when compared with control conditions, as monitored by the Bradford quantification of total proteins (data not shown).
Fig. 3.
FSH increases the recruitment of mTOR to the mRNA 5′cap. A, Sertoli cells were treated with FSH for 60 min before lysis. Cap-binding proteins were precipitated using m7GTP-sepharose-coated beads and analyzed by Western blot to detect the recruitment of mTOR. WCL, Whole cell lysate. B, Densitometry analysis of Western blots probed with antibodies raised against mTOR normalized by the eIF4E amount. Values are expressed as means ± sem of three independent experiments. Statistical difference was calculated using a two-tailed unpaired t test in control vs. FSH-stimulated cells. ***, P < 0.005. C, 35S-labeled methionine and cysteine incorporation. Cells were pretreated for 10 min with PP242 (100 μm) before FSH stimulation for 90 min. The radioactivity was counted as in Fig. 1. Results were expressed as means ± sem of triplicates. Statistical difference was calculated using one-way ANOVA with a Bonferroni posttest to compare unstimulated vs. FSH-stimulated cells and control vs. PP242-treated cells. **, P < 0.01; ***, P < 0.005.
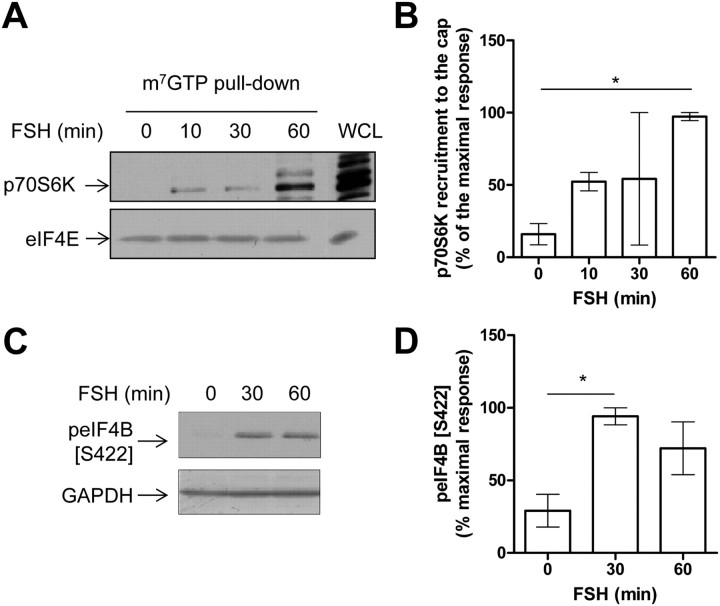
A prominent target of mTOR within the initiation complex is p70S6K. Our group has previously shown that the kinase was activated in Sertoli cells stimulated by FSH (15, 16). Here we observed that p70S6K was also recruited to the 5′cap, upon FSH stimulation (Fig. 4, A and B), as soon as 10 min of stimulation and even more at 60 min. This recruitment appeared to be functional because one of the p70S6K targets within the preinitiation complex, eIF4B, was inducibly phosphorylated (Fig. 4, C and D). Likewise, we previously reported that another p70S6K ribosomal target, ribosomal protein S6, was inducibly phosphorylated on one of its regulatory phosphorylation sites, i.e. Ser235/236 (15, 16) upon FSH stimulation.
Fig. 4.
FSH increases the functional recruitment of p70S6K to the mRNA 5′cap. A, Cap-binding proteins were precipitated from lysates of Sertoli cells treated with FSH for the indicated period of time using m7GTP-sepharose-coated beads and analyzed by Western blot to detect the recruitment of p70S6K. C, Western blot analysis of eIF4B phosphorylation on Ser 422 in FSH-stimulated Sertoli cells. Membranes were reprobed with an anti-GAPDH antibody to monitor gel loading. B and D, Densitometry analysis of Western blots probed with antibodies raised against p70S6K normalized by the eIF4E amount or raised against phosphorylated Ser422 of eIF4B, respectively. Values are expressed as means ± sem of three independent experiments. Statistical difference was calculated using one-way ANOVA with a Bonferroni posttest in control vs. FSH-stimulated cells. *, P < 0.05 (B and D).
FSH induces the recruitment of eIF4G to the preinitiation complex but bypasses 4E-BP1 release from the 5′cap
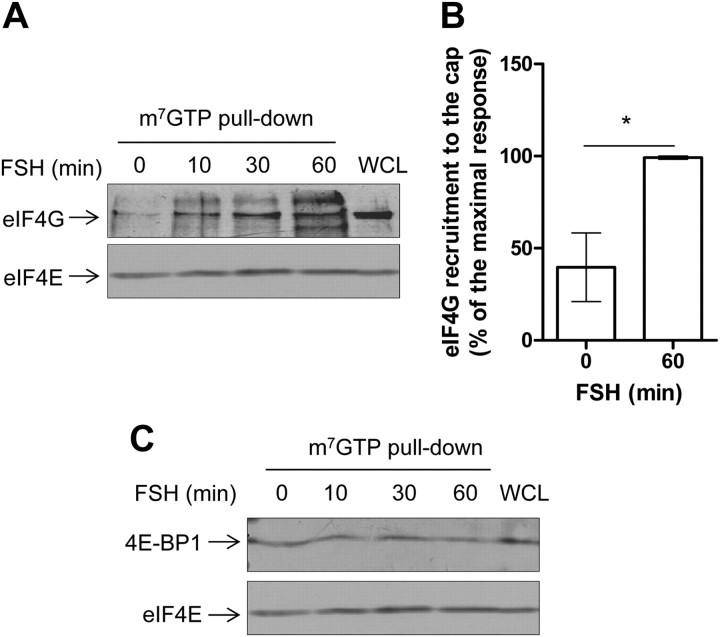
Next, we investigated whether FSH-induced signaling could directly impact on proteins of the translation machinery. m7GTP pull-down experiments were performed with extracts from Sertoli cells stimulated with FSH at various time points. eIF4G was clearly recruited by eIF4E (Fig. 5, A and B), as early as 10 min of FSH stimulation. When the association of 4E-BP1 to eIF4E was examined by m7GTP column affinity, it appeared that 4E-BP1 was retained on eIF4E (Fig. 5C). As expected, eIF4E was constitutively resident on the 5′cap. We verified that 4E-BP1 could not specifically bind to Sepharose beads (data not shown). This result was surprising because 4E-BP1 binding to the cap-binding protein eIF4E precludes further recruitment of other components of the translation initiation complex, such as eIF4G, which competes with 4E-BP1 for a common docking site on eIF4E (25) (see Discussion).
Fig. 5.
FSH stimulation leads to the recruitment of eIF4G to the mRNA 5′cap, without apparent displacement of 4E-BP1. Sertoli cells were stimulated with FSH for 10 to 60 min before lysis. Cap-binding proteins were precipitated using m7GTP-sepharose-coated beads and analyzed by Western blot to detect the recruitment of eIF4G and eIF4E (A) and 4E-BP1 and eIF-4E (C), as indicated. B, Quantification of the eIF4G signal normalized by the eIF4E amount in three independent experiments. Values are expressed as means ± sem. Statistical difference was calculated using a two-tailed unpaired t test in control vs. FSH-stimulated cells. *, P < 0.05. C. Here is shown one representative experiment of five.
4E-BP1 phosphorylation profile in FSH-stimulated Sertoli cells
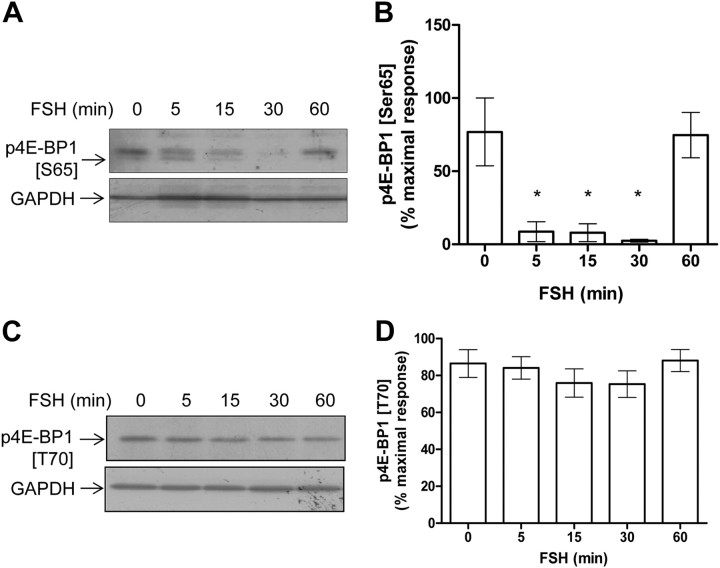
4E-BP1 is a prominent target of signaling pathways, which ultimately regulates mRNA translation (26). It has been shown to be phosphorylated in a sequential manner, with phosphorylation of Ser65 being the ultimate event before 4E-BP1 dissociates from eIF4E (27). Surprisingly, however, in Sertoli cells, we observed that Ser65 was dephosphorylated (Fig. 6, A and B), whereas the Thr70 site remained unaffected (Fig. 6, C and D) upon FSH stimulation. Altogether these results suggest that, be 4E-BP1 phosphorylated or not upon FSH stimulation, it remains associated to the mRNA 5′cap in Sertoli cells.
Fig. 6.
Analysis of 4E-BP1 phosphorylation profile in Sertoli cells. Lysates were prepared from cells stimulated in time course of FSH stimulation. A, The phosphorylation of 4E-BP1 was analyzed by Western blot using a specific antibody raised against phospho-Ser65. B, The densitometry analysis of three independent kinetics, and the results are expressed as mean values ± sem. C, Representative Western blot using a specific antibody raised against phosphorylated Thr70. D, Densitometry analysis of six independent kinetics. Membranes were reprobed with an anti-GAPDH antibody to monitor gel loading. Values are expressed as means ± sem. Statistical difference was calculated using one-way ANOVA with a Bonferroni posttest in control vs. FSH-stimulated cells. *, P < 0.05.
FSH stimulation enhances the translatability of selective mRNA
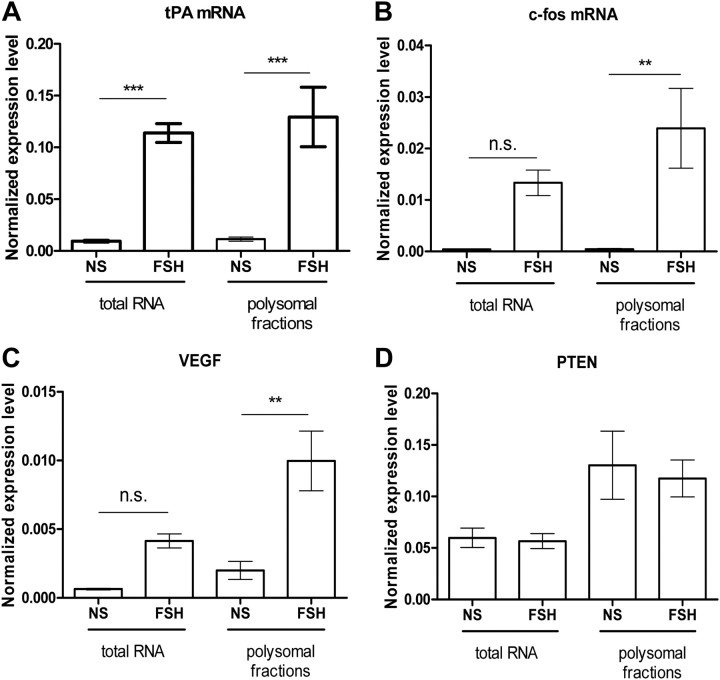
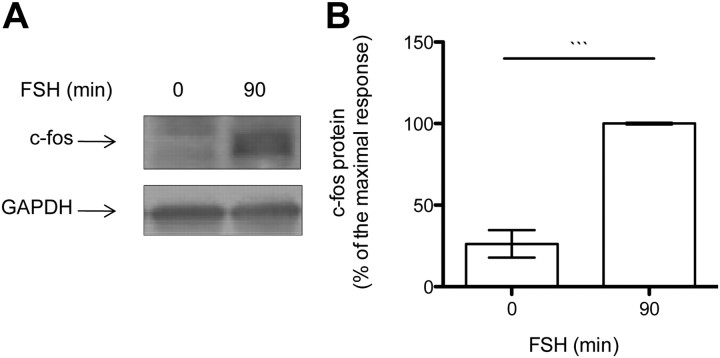
Our data on 4E-BP1 phosphorylation and binding to the 5′ cap, even when eIF4G is recruited, are not consistent with many other reports, indicating that their interaction with eIF4E is mutually exclusive (25). On this basis we hypothesized that only a small fraction of mRNA is being translated upon FSH stimulation. Therefore, quantitative RT-PCR (qRT-PCR) experiments were carried out with the actively translated mRNA isolated in the polysomal fractionation (Fig. 7 and Supplemental Table 2 for the raw data). A set of genes known to have their expression enhanced at the mRNA and/ or protein level after FSH stimulation has been chosen for this analysis. In these conditions, three subclasses of mRNA could be distinguished. First, mRNA such as tPA (Fig. 7A) or LRPR1 (Supplemental Fig. 4A), for which transcriptional regulation and polysome recruitment are stimulated by FSH to a similar extent; second, mRNA such as c-fos (Fig. 7B) and VEGF (Fig. 7C) whose polysome recruitment predominates over transcriptional regulation. In agreement, the level of total c-fos protein was enhanced after 90 min of FSH stimulation (Fig. 8, A and B). The third category comprises mRNA known to be regulated by FSH, such as transferrin, AMH, cyclin D2 (Supplemental Fig. 4, B, C, and D, respectively) and PTEN (Fig. 7D) but whose regulation cannot be visible after 90 min of stimulation. For those, no further effect on polysomal recruitment could be observed. Altogether these results indicate that FSH stimulation enhances the translation rate of some selective mRNA.
Fig. 7.
Selective recruitment of mRNA to the polysomes in FSH-stimulated Sertoli cells. The presence of the tPA (A), c-fos (B), VEGF (C), and PTEN (D) mRNA was assayed by qRT-PCR in the polysomal fractions. Results are expressed as the mean relative expression level relative to GAPDH expression ± sem in three to five independent experiments. Each qRT-PCR reaction was done twice. Statistical difference was calculated using one-way ANOVA with a Bonferroni posttest in control vs. FSH-stimulated cells. ** P < 0.01; ***, P < 0.005.
Fig. 8.
The level of c-fos protein is enhanced after as little as 90 min of FSH stimulation. A, The expression of c-fos was analyzed by Western blot. Membranes were reprobed with an anti-GAPDH antibody to monitor gel loading. B, The densitometry analysis of three independent kinetics, and the results are expressed as mean values ± sem. Statistical difference was calculated using two-tailed unpaired t test in control vs. FSH-stimulated cells. *, P < 0.05.
Discussion
Here we provide the first demonstration that FSH enhances protein translation of a subset of mRNA. Furthermore, we show that FSH stimulation enhances the functional recruitment of signaling kinases involved in the control of translation, such as mTOR and p70S6K, to the mRNA 5′ cap. We propose that the main regulatory step is the recruitment of the eIF4G scaffolding protein and that the 4E-BP1 inhibitor does not seem to be a major player.
Our initial choice for mRNA candidates was based on previous observations that the corresponding genes were responsive to FSH, as seen by detection of the mRNA or of the protein, regardless the level of regulation. We confirmed that the effect of FSH is in the same range on total and in polysome-associated tPA and LRPR1 mRNA. This result suggests that in both cases, the effect of FSH is primarily at the level of transcriptional and that all the mRNA that are transcribed are also translated, without any further active role of FSH signaling. Besides, the c-fos and VEGF mRNA are up-regulated after 90 min of FSH stimulation but to a much lesser extent than their expression level in the polysomes. Hence, this result underscores an active role of FSH signaling in the ability of the c-fos and VEGF mRNA to be selectively translated. Accordingly, an increase in c-fos protein level is visible as soon as 90 min of FSH stimulation. A third class of mRNA, i.e. the transferrin, AMH, cyclin D2 mRNA, is not up-regulated at all and are not actively recruited to the polysomes. For some, if not all, of them, it can then be speculated that over longer FSH exposure, an effect on translatability might have been observed without any detectable increase in total mRNA level. The PTEN mRNA also falls into this class. It was assayed here because in a previous report, we had demonstrated that the PTEN protein level was increased very rapidly, within a few minutes of FSH stimulation, presumably from a preexisting pool of mRNA (17). Here we confirm that the PTEN gene is not regulated by FSH signaling at the transcriptional level, and we report here that the regulatory mechanism does not seem to involve an increased recruitment of the PTEN mRNA to the polysomes, suggesting a posttranslational regulation of PTEN by FSH. Actually, it has been recently confirmed that hormone-triggered PTEN expression was not regulated at the transcriptional level, but rather by posttranscriptional recognition of the PTEN mRNA by microRNA, in seminiferous tubules from adult rats and in primary Sertoli cells from prepubertal animals stimulated with FSH and testosterone (28).
What is the point of stimulating the translation of some mRNA and not of all of them? In Sertoli cells, it is an appealing hypothesis that selective mRNA translation could be related to site-specific translation. Local translation of spatially restricted mRNA has emerged as a powerful means to functionally compartmentalize neosynthesized proteins within polarized cells. This mechanism implicates that mRNA translation is silenced during their intracellular transport and switched on when they reach their ultimate destination (29). Epithelial-like Sertoli cells are polarized feeding cells that constitute a niche for spermatogonial maintenance and differentiation throughout life. They provide appropriate nutrients to the maturating germ cells that subsequently take place all along Sertoli cell body, from the lamina to the lumen. It is tempting to speculate that FSH could locally and tightly regulate translation of preexisting regionalized pools of mRNA in Sertoli cells. This assumption is supported by the observation that tPA is locally produced in Sertoli cells to remodel the blood-testis barrier during spermatogenesis. Likewise, in the seminiferous tubules, the VEGF mRNA is expressed only by Sertoli cells, whereas its receptors are found at the surface of germ cells. The role of this protein might be restricted to proliferation of spermatogonia, which are in intimate contact with the basal part of Sertoli cells (30).
Although prepubertal rat Sertoli cells rarely divide in vivo, for an unclear reason they resume proliferation in vitro (31, 32). Cells proliferate more actively at low density. However, cap-dependent translation is inhibited when cell undergo mitosis at the benefit of internal ribosome entry site-dependent translation, which accounts for about 10% of the translation of cellular mRNA (33) and hence may fail to be detected in our incorporation assay. Therefore, we hypothesize that the FSH stimulatory effect on protein neosynthesis that we observed in cells seeded at high density occurs mainly by regulation of cap-dependent translation. Accordingly, cap-dependent protein neosynthesis is totally abolished by cell preincubation when mTOR activity is inhibited.
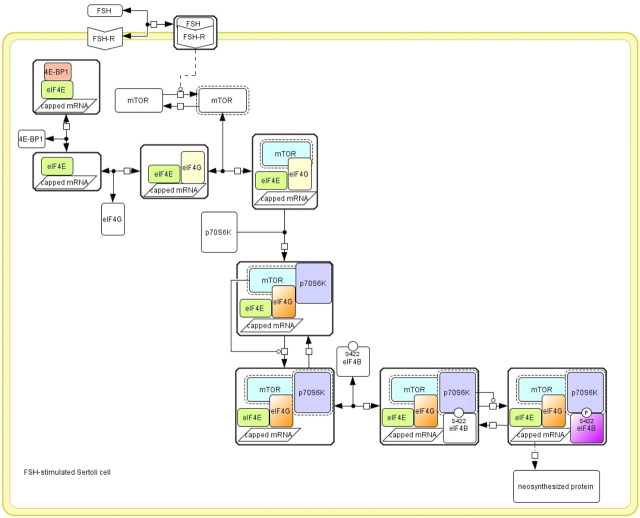
We observed that eIF4E associates with both 4E-BP1 and eIF4G in m7GTP pull-down experiments. Simultaneous interaction is practically not feasible for the main reason that eIF4G and 4E-BP1 both recognize a similar docking site on eIF4E (25). These observations led us to speculate that there could be different populations of eIF4E, one bound to 4E-BP1 and the other one not bound to 4E-BP1 and then free to interact with eIF4G (Fig. 9). Supplemental Fig. 5 provides the mathematical demonstration that several populations of eIF4E are presumably involved, one being constant and the other one evolving through the different complexes induced by FSH. Consistently, in Mammals, eIF4E-2 associates with 4E-BP1 and not with eIF4G, and conversely, eIF4E-3 associates with eIF4G and not with 4E-BP1 (34). In line with our above-mentioned hypothesis that FSH stimulation could direct site-specific mRNA translation, the eIF4E pool bound to 4E-BP1 could be linked to silenced, localizing mRNA, whereas the eIF4E/eIF4G interaction would permit the formation of functional translation preinitiation complexes on mRNA being translated at the site at which the encoded protein needs to be produced.
Fig. 9.
Hypothetical scheme of the regulation of the mRNA translational machinery by FSH-induced signaling. The Cell Designer software (http://celldesigner.org) (46) has been used to formalize the topology of the signaling network. The legend is as follows: stimulatory catalyzes are indicated by a line ended in a circle. Dashed lines mean indirect reactions. The active molecular species are surrounded by black dots. P, Phosphorylation, with the position of the phosphorylated residue being indicated when known. Complexes are surrounded by a box. Double arrows indicate reversible reactions. FSH triggers cap-dependent translation by enhancing p70S6K, mTOR, and eIF4G recruitment to the cap. p70S6K activation by FSH has been reported elsewhere (15).
Release of 4E-BP1 from the mRNA 5′cap is primed by growth factor-induced sequential phosphorylation on Thr37/46, before phosphorylation of Thr70 and of Ser65 (35). In Sertoli cells, 4E-BP1 gets dephosphorylated and rephosphorylated on its Ser65 and remains constitutively phosphorylated on its Thr70. Recently it has been proposed that 4E-BP1 sequence of phosphorylation was different in physiological situations (36). In line with our results, a mutant mimicking 4E-BP1 hyperphosphorylation can still interact with eIF4E, in in vitro translation assays, suggesting that other modifications are required for full eIF4E release (37). Therefore, our results suggest that, in some conditions, phosphorylated 4E-BP1 can still bind eIF4E.
In Sertoli cells, mTOR was inducibly recruited to the cap, upon FSH stimulation. In previous works, mTOR has been shown to be recruited to the translation preinitiation complex to phosphorylate its targets, i.e. p70S6K and 4E-BP1, in situ (38). Given that 4E-BP1 phosphorylation is not enhanced upon FSH stimulation, mTOR recruitment may serve here only to phosphorylate p70S6K. Actually we have previously shown that p70S6K is phosphorylated on T389 by a phosphoinositide-3 kinase- and mTOR-dependent manner (15). Furthermore, we report here that p70S6K was functionally recruited to the 5′cap (Fig. 4). This observation contrasts with previous data showing that active p70S6K is released from the cap upon insulin stimulation of human embryonic kidney 293 cells (38). Altogether these observations support the notion that, in cell models close to physiology, translation may be regulated by mechanisms that do not entirely match those observed in paradigmatic starved cell lines stimulated with bona fide growth factors. In addition, FSH acts via a GPCR, and we previously reported that, like LH (39), FSH signaling mediates an unexpected activation mechanism of p70S6K in Sertoli cells (15, 16), which may be related to its association pattern to the 5′cap. So far, few studies on how GPCR control translation have been extensive enough to provide a clear picture of the whole translational network (40). This is quite puzzling because GPCR are involved in most physiological processes by binding a vast array of structurally different agonists, in fully differentiated cells, whose physiology demands regulation of protein neosynthesis. GPCR couple to adaptor proteins of their own, such as β-arrestins, whereby they launch intracellular signaling cascades. Once bound to its cognate receptor, each agonist promotes a conformational change causing the physical association of the GPCR with G proteins. In addition, ligand binding also induces a range of conformational changes that overcomes G coupling and leads to β-arrestin-mediated signaling. Recently, in response to activation of the angiotensin II receptor type 1A, β-arrestins have been shown to intervene in the translational control by phosphorylating Mnk-1 (MAPK-interacting protein 1) in an ERK-dependent manner (41). Initially known to convey GPCR toward the endocytic machinery, β-arrestins support a temporal hierarchy of signaling events by scaffolding various signaling modules (42). Because these observations also come true for the FSH-R (13), whether β-arrestins play a role in FSH-dependent regulation of translation remains to be addressed.
In conclusion, our results provide new avenues in understanding cell responses triggered by FSH. In vivo, regulation of gene expression occurring at the translational level could serve to fine-tune Sertoli cell protein content, in response to subtle variations of the FSH input, regarding its concentration, glycosylated isoform (43), or receptor density at the cell surface (14) and could serve to compartmentalize the mRNA to be translated at each spermatogenic stage.
Acknowledgments
We thank the technical assistance of the rat breeders Deborah Crespin, Marine Cirot, and Claude Cahier.
This work was funded by the Institut National de la Recherche Agronomique (INRA) the Action Incitative Prioritaire AgroBI, the Action d'Envergure Institut National de Recherche en Informatique et en Automatique/Institut National de la Recherche Agronomique REGATE, the Institut Fédératif de Recherche 135, and the Centre National de la Recherche Scientifique. A.M. and K.L. were funded by fellowships from the Région Centre and from the INRA, N.O. was funded by a fellowship from the French Ministère de la Recherche et de la Technologie, and V.C. was funded by a fellowship from the Région Bretagne.
Disclosure Summary: The authors have nothing to disclose.
Footnotes
- AMH
- Anti-Mullerian hormone
- CHX
- cycloheximide
- Cp
- crossing point
- DTT
- dithiothreitol
- 4E-BP1
- eukaryotic initiation factor 4E-binding protein
- eIF4B
- eukaryotic initiation factor 4B
- eIF4E
- eukaryotic initiation factor 4E
- eIF4G
- eukaryotic initiation factor 4G
- FSH-R
- FSH receptor
- GAPDH
- glyceraldehyde-3-phosphate dehydrogenase
- GPCR
- G protein-coupled receptor
- m7-GTP
- methylguanosine triphosphate
- mTOR
- mammalian target of rapamycin
- p70S6K
- p70 S6 kinase
- PTEN
- phosphatase and tensin homolog deleted on chromosome 10
- qRT-PCR
- quantitative RT-PCR
- tPA
- tissue-type plasminogen activator
- VEGF
- vascular endothelial growth factor.
References
- 1. Means AR , Hall PF. 1967. Effect of FSH on protein biosynthesis in testes of the immature rat. Endocrinology 81:1151–1160 [DOI] [PubMed] [Google Scholar]
- 2. DePhilip RM , Kierszenbaum AL. 1982. Hormonal regulation of protein synthesis, secretion, and phosphorylation in cultured rat Sertoli cells. Proc Natl Acad Sci USA 79:6551–6555 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Sanborn BM , Caston LA , Chang C , Liao S , Speller R , Porter LD , Ku CY. 1991. Regulation of androgen receptor mRNA in rat Sertoli and peritubular cells. Biol Reprod 45:634–641 [DOI] [PubMed] [Google Scholar]
- 4. McLean DJ , Friel PJ , Pouchnik D , Griswold MD. 2002. Oligonucleotide microarray analysis of gene expression in follicle-stimulating hormone-treated rat Sertoli cells. Mol Endocrinol 16:2780–2792 [DOI] [PubMed] [Google Scholar]
- 5. Sadate-Ngatchou PI , Pouchnik DJ , Griswold MD. 2004. Follicle-stimulating hormone induced changes in gene expression of murine testis. Mol Endocrinol 18:2805–2816 [DOI] [PubMed] [Google Scholar]
- 6. Schwänhausser B , Busse D , Li N , Dittmar G , Schuchhardt J , Wolf J , Chen W , Selbach M. 2011. Global quantification of mammalian gene expression control. Nature 473:337–342 [DOI] [PubMed] [Google Scholar]
- 7. Cobb MH. 1986. An insulin-stimulated ribosomal protein S6 kinase in 3T3-L1 cells. J Biol Chem 261:12994–12999 [PubMed] [Google Scholar]
- 8. Tabarini D , Garcia de Herreros A , Heinrich J , Rosen OM. 1987. Purification of a bovine liver S6 kinase. Biochem Biophys Res Commun 144:891–899 [DOI] [PubMed] [Google Scholar]
- 9. Jenö P , Ballou LM , Novak-Hofer I , Thomas G. 1988. Identification and characterization of a mitogen-activated S6 kinase. Proc Natl Acad Sci USA 85:406–410 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Raught B , Peiretti F , Gingras AC , Livingstone M , Shahbazian D , Mayeur GL , Polakiewicz RD , Sonenberg N , Hershey JW. 2004. Phosphorylation of eucaryotic translation initiation factor 4B Ser422 is modulated by S6 kinases. EMBO J 23:1761–1769 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Brunn GJ , Hudson CC , Sekulić A , Williams JM , Hosoi H , Houghton PJ , Lawrence JC , Abraham RT. 1997. Phosphorylation of the translational repressor PHAS-I by the mammalian target of rapamycin. Science 277:99–101 [DOI] [PubMed] [Google Scholar]
- 12. Prévôt D , Darlix JL , Ohlmann T. 2003. Conducting the initiation of protein synthesis: the role of eIF4G. Biol Cell 95:141–156 [DOI] [PubMed] [Google Scholar]
- 13. Kara E , Crépieux P , Gauthier C , Martinat N , Piketty V , Guillou F , Reiter E. 2006. A phosphorylation cluster of five serine and threonine residues in the C-terminus of the follicle-stimulating hormone receptor is important for desensitization but not for β-arrestin-mediated ERK activation. Mol Endocrinol 20:3014–3026 [DOI] [PubMed] [Google Scholar]
- 14. Tranchant T , Durand G , Gauthier C , Crépieux P , Ulloa-Aguirre A , Royère D , Reiter E. 2011. Preferential β-arrestin signalling at low receptor density revealed by functional characterization of the human FSH receptor A189 V mutation. Mol Cell Endocrinol 331:109–118 [DOI] [PubMed] [Google Scholar]
- 15. Lécureuil C , Tesseraud S , Kara E , Martinat N , Sow A , Fontaine I , Gauthier C , Reiter E , Guillou F , Crépieux P. 2005. Follicle-stimulating hormone activates p70 ribosomal protein S6 kinase by protein kinase A-mediated dephosphorylation of Thr 421/Ser 424 in primary Sertoli cells. Mol Endocrinol 19:1812–1820 [DOI] [PubMed] [Google Scholar]
- 16. Musnier A , Heitzler D , Boulo T , Tesseraud S , Durand G , Lécureuil C , Guillou H , Poupon A , Reiter E , Crépieux P. 2009. Developmental regulation of p70 S6 kinase by a G protein-coupled receptor dynamically modelized in primary cells. Cell Mol Life Sci 66:3487–3503 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Dupont J , Musnier A , Decourtye J , Boulo T , Lécureuil C , Guillou H , Valet S , Fouchécourt S , Pitetti JL , Nef S , Reiter E , Crépieux P. 2010. FSH-stimulated PTEN activity accounts for the lack of FSH mitogenic effect in prepubertal rat Sertoli cells. Mol Cell Endocrinol 315:271–276 [DOI] [PubMed] [Google Scholar]
- 18. Kayampilly PP , Menon KM. 2007. Follicle-stimulating hormone increases tuberin phosphorylation and mammalian target of rapamycin signaling through an extracellular signal-regulated kinase-dependent pathway in rat granulosa cells. Endocrinology 148:3950–3957 [DOI] [PubMed] [Google Scholar]
- 19. Alam H , Maizels ET , Park Y , Ghaey S , Feiger ZJ , Chandel NS , Hunzicker-Dunn M. 2004. FSH activation of HIF-1 by the PI3-kinase/AKT/Rheb/mTOR pathway is necessary for induction of select protein markers of follicular differentiation. J Biol Chem 279:19431–19440 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Alam H , Weck J , Maizels E , Park Y , Lee EJ , Ashcroft M , Hunzicker-Dunn M. 2009. Role of the phosphatidylinositol-3-kinase and extracellular regulated kinase pathways in the induction of hypoxia-inducible factor (HIF)-1 activity and the HIF-1 target vascular endothelial growth factor in ovarian granulosa cells in response to follicle-stimulating hormone. Endocrinology 150:915–928 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Kayampilly PP , Menon KM. 2009. Follicle-stimulating hormone inhibits adenosine 5′-monophosphate-activated protein kinase activation and promotes cell proliferation of primary granulosa cells in culture through an Akt-dependent pathway. Endocrinology 150:929–935 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Rattan R , Giri S , Singh AK , Singh I. 2005. 5-Aminoimidazole-4-carboxamide-1-β-D-ribofuranoside inhibits cancer cell proliferation in vitro and in vivo via AMP-activated protein kinase. J Biol Chem 280:39582–39593 [DOI] [PubMed] [Google Scholar]
- 23. Inoki K , Zhu T , Guan KL. 2003. TSC2 mediates cellular energy response to control cell growth and survival. Cell 115:577–590 [DOI] [PubMed] [Google Scholar]
- 24. Raught B , Gingras AC. 1999. eIF4E activity is regulated at multiple levels. Int J Biochem Cell Biol 31:43–57 [DOI] [PubMed] [Google Scholar]
- 25. Haghighat A , Mader S , Pause A , Sonenberg N. 1995. Repression of cap-dependent translation by 4E-binding protein 1: competition with p220 for binding to eukaryotic initiation factor-4E. EMBO J 14:5701–5709 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Pause A , Belsham GJ , Gingras AC , Donzé O , Lin TA , Lawrence JC , Sonenberg N. 1994. Insulin-dependent stimulation of protein synthesis by phosphorylation of a regulator of 5′-cap function. Nature 371:762–767 [DOI] [PubMed] [Google Scholar]
- 27. Gingras AC , Raught B , Sonenberg N. 2001. Regulation of translation initiation by FRAP/mTOR. Genes Dev 15:807–826 [DOI] [PubMed] [Google Scholar]
- 28. Nicholls PK , Harrison CA , Walton KL , McLachlan RI , O'Donnell L , Stanton PG. 2011. Hormonal regulation of Sertoli cell micro-RNAs at spermiation. Endocrinology 152:1670–1683 [DOI] [PubMed] [Google Scholar]
- 29. Besse F , Ephrussi A. 2008. Translational control of localized mRNAs: restricting protein synthesis in space and time. Nat Rev Mol Cell Biol 9:971–980 [DOI] [PubMed] [Google Scholar]
- 30. Nalbandian A , Dettin L , Dym M , Ravindranath N. 2003. Expression of vascular endothelial growth factor receptors during male germ cell differentiation in the mouse. Biol Reprod 69:985–994 [DOI] [PubMed] [Google Scholar]
- 31. Crépieux P , Marion S , Martinat N , Fafeur V , Vern YL , Kerboeuf D , Guillou F , Reiter E. 2001. The ERK-dependent signalling is stage-specifically modulated by FSH, during primary Sertoli cell maturation. Oncogene 20:4696–4709 [DOI] [PubMed] [Google Scholar]
- 32. Ahmed EA , Barten-van Rijbroek AD , Kal HB , Sadri-Ardekani H , Mizrak SC , van Pelt AM , de Rooij DG. 2009. Proliferative activity in vitro and DNA repair indicate that adult mouse and human Sertoli cells are not terminally differentiated, quiescent cells. Biol Reprod 80:1084–1091 [DOI] [PubMed] [Google Scholar]
- 33. Pyronnet S , Dostie J , Sonenberg N. 2001. Suppression of cap-dependent translation in mitosis. Genes Dev 15:2083–2093 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Joshi B , Cameron A , Jagus R. 2004. Characterization of mammalian eIF4E-family members. Eur J Biochem 271:2189–2203 [DOI] [PubMed] [Google Scholar]
- 35. Gingras AC , Raught B , Gygi SP , Niedzwiecka A , Miron M , Burley SK , Polakiewicz RD , Wyslouch-Cieszynska A , Aebersold R , Sonenberg N. 2001. Hierarchical phosphorylation of the translation inhibitor 4E-BP1. Genes Dev 15:2852–2864 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36. Ayuso MI , Hernández-Jiménez M , Martín ME , Salinas M , Alcázar A. 2010. New hierarchical phosphorylation pathway of the translational repressor eIF4E-binding protein 1 (4E-BP1) in ischemia-reperfusion stress. J Biol Chem 285:34355–34363 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37. Oulhen N , Boulben S , Bidinosti M , Morales J , Cormier P , Cosson B. 2009. A variant mimicking hyperphosphorylated 4E-BP inhibits protein synthesis in a sea urchin cell-free, cap-dependent translation system. PLoS One 4:e5070. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38. Holz MK , Ballif BA , Gygi SP , Blenis J. 2005. mTOR and S6K1 mediate assembly of the translation preinitiation complex through dynamic protein interchange and ordered phosphorylation events. Cell 123:569–580 [DOI] [PubMed] [Google Scholar]
- 39. Hou X , Arvisais EW , Davis JS. 2010. Luteinizing hormone stimulates mammalian target of rapamycin signaling in bovine luteal cells via pathways independent of AKT and mitogen-activated protein kinase: modulation of glycogen synthase kinase 3 and AMP-activated protein kinase. Endocrinology 151:2846–2857 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40. Musnier A , Blanchot B , Reiter E , Crépieux P. 2010. GPCR signalling to the translation machinery. Cell Signal 22:707–716 [DOI] [PubMed] [Google Scholar]
- 41. DeWire SM , Kim J , Whalen EJ , Ahn S , Chen M , Lefkowitz RJ. 2008. β-Arrestin-mediated signaling regulates protein synthesis. J Biol Chem 283:10611–10620 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42. Reiter E , Lefkowitz RJ. 2006. GRKs and β-arrestins: roles in receptor silencing, trafficking and signaling. Trends Endocrinol Metab 17:159–165 [DOI] [PubMed] [Google Scholar]
- 43. Ulloa-Aguirre A , Crépieux P , Poupon A , Maurel MC , Reiter E. 2011. Novel pathways in gonadotropin receptor signaling and biased agonism. Rev Endocr Metab Disord 12:259–274 [DOI] [PubMed] [Google Scholar]
- 44. Guillou F , Martinat N , Combarnous Y. 1986. Study of the superactivity of equine follicle-stimulating hormone in in vitro stimulation of rat Sertoli cells. Biochim Biophys Acta 887:196–203 [DOI] [PubMed] [Google Scholar]
- 45. Schmittgen TD , Livak KJ. 2008. Analyzing real-time PCR data by the comparative C(T) method. Nat Protoc 3:1101–1108 [DOI] [PubMed] [Google Scholar]
- 46. Kitano H , Funahashi A , Matsuoka Y , Oda K. 2005. Using process diagrams for the graphical representation of biological networks. Nat Biotechnol 23:961–966 [DOI] [PubMed] [Google Scholar]