Abstract
Helicobacter pylori antibiotic susceptibility in the Dominican Republic has not been monitored. We assessed H. pylori antibiotic susceptibility in the Dominican Republic, and analyzed H. pylori mutations associated with antibiotic resistance. We recruited 158 dyspeptic patients in Santo Domingo and used agar dilution to test susceptibility to five antibiotics. Polymerase chain reaction–based sequencing was used to assess gyrA, gyrB, rdxA, frxA, and 23S rRNA mutations; next-generation sequencing was used to identify other metronidazole resistance–associated genes. Among 64 H. pylori strains isolated, we identified two (3.1%), one (1.6%), and no strains with clarithromycin, amoxicillin, and tetracycline resistance, respectively. Moreover, high frequency of metronidazole resistance (53/64, 82.8%) was observed, whereas levofloxacin resistance is emerging (23/64, 35.9%). We identified many rdxA and frxA mutations in metronidazole-resistant strains, but no synergistic effect was apparent. We revealed novel mutations in dppA, dppB, fdxA, and fdxB, irrespective of rdxA and frxA mutations. Novel mutations at Ser-14 of trx1 and Arg-221 of dapF were associated with different levels of metronidazole resistance. Most levofloxacin-resistant strains had a substitution at Asn-87 of gyrA, including the strain with the highest levofloxacin resistance, whereas only three substitutions were found at Ser-479 of gyrB with no synergistic effect. Besides the 23S rRNA A2142G mutation, we observed another mutation at T1958G in both clarithromycin-resistant strains. We confirmed high metronidazole and levofloxacin resistance associated with genetic mutations in the Dominican Republic. However, prevalence of clarithromycin resistance was low, suggesting that standard clarithromycin-based triple therapy remains useful as initial treatment of H. pylori infection.
Introduction
The emergence of drug resistance to antibiotics used to treat Helicobacter pylori, the Gram-negative bacterium responsible for severe gastroduodenal diseases, is a serious problem. According to current guidelines, triple therapy composed of a proton pump inhibitor and two antibiotics, amoxicillin (AMX) and clarithromycin (CAM) or metronidazole (MNZ), remains the standard first-line regimen for treatment of H. pylori infection,1,2 although caution is advised with its use.2 In recent years, efficacy of this regimen has been seriously challenged, and a cure rate below 70% has been reported in many countries.3 Therefore, optimizing the first-line regimen based on local antibiotic resistance patterns is critical to prevent repeated courses of treatment and the spread of secondary antibiotic resistance.4
Helicobacter pylori genes with mutations implicated in drug resistance have been identified and can be detected by molecular methods. An understanding of H. pylori antibiotic resistance mechanisms is important in considerations of more rational antibiotic combinations. CAM resistance has been shown to be associated with any one of five recognized point mutations in H. pylori 23S rRNA. These mutations comprise an A to G substitution at nucleotide positions 2142 or 2143, A to C at 2142, A to T at 2144, T to C at 2717, and C to A at 2694.5,6 The mechanisms of MNZ resistance are complex but largely associated with inactivation of rdxA (hp0954 in the H. pylori 26695 genome) and frxA (hp0642), which, together, have a synergistic effect that results in a high level of resistance.7,8 Other identified mutations, including rpsU (hp0562),9 dppA (hp0298), dppB (hp0299), rps4 (hp1294), ackA (hp0903), rnc (hp0662), dapF (hp0566),10 recA (hp0153),11 fdxA (hp0277),12 fdxB (hp0284),13 and trx1 (hp0824),14,15 have also been associated with MNZ resistance. On the other hand, mutations in gyrase subunit A (gyrA) (hp0641), and gyrB (hp0501), have been associated with fluoroquinolone resistance.16
The prevalence of antibiotic resistance in Latin America varies by geographic region. The lowest resistance to CAM was reported to be 2% in Paraguay, and the highest was reported to be 50% in Peru.17 A various prevalence of antibiotic resistance has also been reported in Latin American countries, 13–95%, 0–39%, and 0–86% resistance, respectively, to MNZ, AMX, and tetracycline (TCN).17 The Dominican Republic is a nation that occupies the eastern part of the second largest island, Hispaniola, in the Caribbean Sea, with a total population of 10.41 million in 2014. The age-standardized rate of gastric cancer in the Dominican Republic is reported to be 7.3 per 100,000 per year (http://globocan.iarc.fr/). Although a recent study regarding the prevalence of H. pylori infection was reported (58.9%),18 H. pylori antibiotic susceptibility in the Dominican Republic strains has not be monitored. According to the European Maastricht Consensus,2,19 CAM-containing triple therapies administered without prior susceptibility testing should be abandoned in a region when the local CAM resistance frequency is greater than 15–20%. In populations with < 40% MNZ resistance, MNZ-based triple therapy is preferable.19,20 Therefore, it is critical to examine current drug resistance frequencies to select the appropriate first-line regimen for H. pylori treatment in the Dominican Republic. In this study, we aimed to determine the antibiotic susceptibility of H. pylori and to identify mutations associated with antibiotic resistance in this pathogen in the Dominican Republic.
Materials and Methods
Patients and H. pylori.
This study included 158 consecutive patients (55 males and 103 females; age range, 17–91 years; mean age, 47.1 ± 16.2 years) who underwent endoscopy examination at the Digestive Disease Center, Dr. Luis E. Aybar Health and Hygiene City, Santo Domingo, Dominican Republic. Peptic ulcer diseases, including gastric and duodenal ulcers, were diagnosed by endoscopic observation, whereas chronic gastritis and gastric cancer were determined by histologic examination. Written informed consent was obtained from all participants, and the study protocol was approved by the ethics committees of Dr. Luis E. Aybar Health and Hygiene City; Institute of Microbiology and Parasitology, Autonomous University of Santo Domingo, Santo Domingo, Dominican Republic; and the Oita University Faculty of Medicine, Japan.
For H. pylori culture, antral biopsy specimens were homogenized and inoculated onto antibiotic selection plates, and then subcultured on Mueller Hinton II Agar medium (Becton Dickinson, Sparks, MD) supplemented with 7% horse blood without antibiotics. The plates were incubated up to 10 days at 37°C under microaerophilic conditions (10% O2, 5% CO2, and 85% N2). Helicobacter pylori isolates were identified based on colony morphology; Gram staining results; and oxidase, catalase, and urease reactions. Isolated strains were stored at −80°C in Brucella broth (Becton Dickinson, Sparks, MD) containing 10% dimethyl sulfoxide and 10% horse serum.
Antibiotic susceptibility testing.
The serial 2-fold agar dilution method was used to determine the minimum inhibitory concentrations (MICs) of AMX (Sigma Chemical Co., St. Louis, MO), CAM (Abbott Laboratories, Abbott Park, IL), MNZ (Sigma), TCN (Sigma), and levofloxacin (LVX) (Sigma). Briefly, bacteria were subcultured on Mueller Hinton II Agar medium (Becton Dickinson) supplemented with 10% defibrinated horse blood. The bacterial suspension, adjusted to a McFarland opacity standard of 3.0, was inoculated onto the plates. After 72 hours of incubation, the MIC of each antibiotic was determined. Helicobacter pylori ATCC 43504 was used for quality control testing. The resistance breakpoints were determined as described by the European Committee on Antimicrobial Susceptibility Testing (http://www.eucast.org/). Strains were considered resistant with MICs > 0.125 mg/L for AMX, > 0.25 mg/L for CAM, > 8 mg/L MNZ, and > 1 mg/L for TCN and LVX.
Molecular analysis of resistant strains.
DNA was extracted from cultured H. pylori using a commercially available kit (Qiagen, Hilden, Germany) for amplification of rdxA and frxA from MNZ-resistant strains, gyrA and gyrB from LVX-resistant strains, and 23S rRNA peptidyl transferase from CAM-resistant strains using the primers as described previously.21–23 As a control, we sequenced five randomly selected strains that were sensitive to MNZ and LVX and one sensitive to CAM from the Dominican Republic isolates. The sequences were then compared with the published sequence of H. pylori 26695 (GenBank accession number AE000511.1 GI: 6626253) using MAFFT version 7 (http://mafft.cbrc.jp/alignment/server/) and confirmed by visual inspection.
To find genetic mutations other than the typical rdxA and frxA mutations, we also obtained full-length rdxA, frxA, rpsU, dppA, dppB, rps4, ackA, rnc, dapF, recA, fdxA, fdxB, and trx1 sequences by next-generation sequencing (NGS) (MiSeq next-generation sequencer; Illumina, Inc., San Diego, CA). MiSeq output was integrated into contig sequences using CLC Genomics Workbench 7.0.4. (CLC Bio-Qiagen, Aarhus, Denmark) Genomics Workbench was also used for gene predictions and translation to protein sequences.
Statistical analysis.
Discrete variables were tested using the χ2 test, whereas continuous variables were tested using Mann–Whitney U and t tests. P values < 0.05 were considered statistically significant. SPSS statistical software package version 18.0 (SPSS, Inc., Chicago, IL) was used for all statistical analyses.
Ethical standards.
We declare that all procedures performed for this work comply with the ethical standards of the relevant national and institutional committees on human experimentation and with the Helsinki Declaration of 1975, as revised in 2008.
Results
Prevalence of antibiotic resistance.
A total of 64 H. pylori strains were isolated from 19 male (age range, 21 to 69 years; mean age, 43.9 ± 12.4 years) and 45 female patients (age range, 17 to 82 years; mean age 43.8 ± 14.9 years). Of these patients, 47 had chronic gastritis, 16 had peptic ulcer diseases, and one had gastric cancer. The prevalence of CAM resistance was only 3.1% (2/64), in contrast to the higher prevalence reported in many other regions.24 A low prevalence of AMX resistance (1/64, 1.6%) was observed, and no TCN resistance was detected (Table 1). However, similar to data reported from Africa, Asia, and Europe,24 the frequency of MNZ resistance was high (53/64, 82.8%). This frequency was also much higher than the neighboring countries of Costa Rica (42.0%) and Venezuela (59.0%).17 Moreover, the prevalence of LVX resistance was also high (23/64, 35.9%). The distribution of patients' ages and antimicrobial resistance of isolates is shown in Table 2. Antibiotic resistance did not differ by age (P = 0.46, 0.51, 0.07, and 0.20 for AMX, CAM, MNZ, and LVX, respectively), sex (P = 0.51, 0.35, 0.85, and 0.11 for AMX, CAM, MNZ, and LVX, respectively), or disease group (P = 0.22, 0.69, 0.59, and 0.41 for AMX, CAM, MNZ, and LVX, respectively).
Table 1.
Antibiotic susceptibility of 64 Helicobacter pylori strains isolated from the Dominican Republic
| Antibiotic | Number (%) of resistant isolates from patients | |||||
|---|---|---|---|---|---|---|
| All patients (N = 64) | Gastritis (N = 47) | PUD (N = 16) | GC (N = 1) | Male (N = 19) | Female (N = 45) | |
| AMX | 1 (1.6) | 0 (0.0) | 1 (6.3) | 0 (0.0) | 0 (0.0) | 1 (2.2) |
| CAM | 2 (3.1) | 2 (4.3) | 0 (0.0) | 0 (0.0) | 0 (0.0) | 2 (4.4) |
| MNZ | 53 (82.8) | 40 (85.1) | 12 (75.0) | 1 (100.0) | 16 (84.2) | 37 (82.2) |
| TCN | 0 (0.0) | 0 (0.0) | 0 (0) | 0 (0.0) | 0 (0.0) | 0 (0.0) |
| LVX | 23 (35.9) | 19 (40.4) | 4 (25.0) | 0 (0.0) | 4 (21.1) | 19 (42.2) |
AMX = amoxicillin; CAM = clarithromycin; GC = gastric cancer; LVX = levofloxacin; MNZ = metronidazole; PUD = peptic ulcer disease; TCN = tetracycline.
Table 2.
Distribution of antibiotic resistance in patients of the Dominican Republic by age
| Antibiotic | Number (%) per age group (years) | |||||
|---|---|---|---|---|---|---|
| 17–29 | 30–39 | 40–49 | 50–59 | 60–91 | Total | |
| Total | 10 | 14 | 20 | 11 | 9 | 64 |
| AMX | 0 (0.0) | 1 (7.1) | 0 (0.0) | 0 (0.0) | 0 (0.0) | 1 |
| CAM | 0 (0.0) | 1 (7.1) | 0 (0.0) | 1 (9.1) | 0 (0.0) | 2 |
| MNZ | 8 (80.0) | 14 (100.0) | 17 (85.0) | 7 (63.6) | 7 (77.8) | 53 |
| TCN | 0 (0.0) | 0 (0.0) | 0 (0.0) | 0 (0.0) | 0 (0.0) | 0 |
| LVX | 2 (20.0) | 5 (35.7) | 4 (20.0) | 6 (54.5) | 6 (66.7) | 24 |
AMX = amoxicillin; CAM = clarithromycin; LVX = levofloxacin; MNZ = metronidazole; TCN = tetracycline.
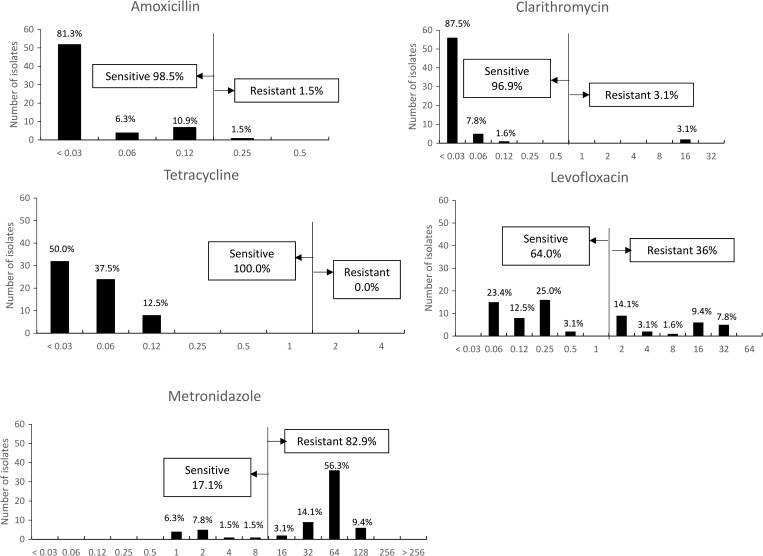
Overall, no strain was resistant to all antibiotics tested, and only two strains were resistant to three antibiotics (AMX, MNZ, and LVX or CAM, MNZ, and LVX) (Table 3). Of the study strains, 31.3% (20/64) showed dual-drug resistance to MNZ and LVX. No differences were observed in clinical outcomes between single-drug- and multidrug-resistant H. pylori infections (P = 0.13). There was no association between antibiotic resistance and clinical outcomes (P > 0.05). The MIC values for each antibiotic are shown in Figure 1 . Helicobacter pylori showed a high level of resistance (with MIC values of 128 mg/L or greater) to MNZ in 23.2% (6/64) of isolates and to LVX (with MIC values of 32 mg/L or greater) in 7.8% (5/64) of isolates.
Table 3.
Multidrug resistance patterns of Helicobacter pylori in the Dominican Republic
| Resistance patterns | N |
|---|---|
| Dual drugs | |
| CAM + MNZ | 1 (1.6) |
| MNZ + LVX | 20 (31.3) |
| Triple drugs | |
| AMX + MNZ + LVX | 1 (1.6) |
| CAM + MNZ + LVX | 1 (1.6) |
AMX = amoxicillin; CAM = clarithromycin; LVX = levofloxacin; MNZ = metronidazole.
Figure 1.
Antibiotic minimum inhibitory concentration values. The frequencies of resistance to metronidazole and levofloxacin were high; in contrast, the frequencies of resistance to clarithromycin, amoxicillin, and tetracycline were very low.
Detection of H. pylori gene mutations associated with antimicrobial resistance.
The five MNZ-resistant strains did not show specific bands for rdxA by polymerase chain reaction, and for four strains, insufficient sequence data resulted. Therefore, a total of 44 MNZ-resistant and five sensitive (control) strains were analyzed in this study. A DNA sequence analysis of rdxA from MNZ-sensitive strains revealed intact reading frames (lacking nonsense mutations). A pairwise sequence alignment indicated that the MNZ-sensitive strains shared 94.6–97.1% identity with the reference strain, H. pylori 26695. On ignoring mutations that were present in both sensitive and resistant strains, all resistant strains but one contained mutations in rdxA. However, the one strain (DM4) that did not have an rdxA mutation had an frxA mutation (Table 4). Among resistant strains, 68.2% (30/44), 38.6% (17/44), and 31.8% (14/44) harbored missense mutations, premature stop codons, and translational frameshifts, respectively. Several rdxA alterations were categorized as class 1 mutations, which are expected to reduce affinity of the apoprotein for the flavin mononucleotide factor25; for example, an amino acid substitution at R16 was found in nine strains, at R200 in seven strains, and at S18 in DM118. Class II mutations, which are expected to destabilize dimer formation, were found at S43 in strains DM10 and DM118; Q50 in strains DM50 and DM143; and G145 in strains DM51 and DM87. Class III mutations were detected at C19 in strain DM103. No class IV mutations were found. Although a previous study21 revealed the importance of amino acid substitutions A80H, A118T, A118S, Q197K, and V204I in resistance, we found these substitutions in both sensitive and resistant strains. In contrast, we confirmed the importance of a threonine replacement at position 51.26 Other substitutions at E35, V85, G122, K190, and K198 might also be important in strains from the Dominican Republic.
Table 4.
Mutations in rdxA and frxA that were associated with metronidazole resistance
| No. | Strains | MIC (mg/L) | Profile | rdxA mutations | frxA mutations |
|---|---|---|---|---|---|
| 1 | DM5 | 2 | Sensitive | None | None |
| 2 | DM20 | 1 | Sensitive | None | None |
| 3 | DM63 | 4 | Sensitive | P44T, K168E, W209C | V34A |
| 4 | DM92 | 2 | Sensitive | E194Q | Poor sequence quality |
| 5 | DM95 | 1 | Sensitive | H25R | None |
| 6 | DM4 | 64 | Resistant | None | Q27E, N111H, N129T, S130D |
| 7 | DM7 | 64 | Resistant | A68T, V85A, 150 frameshift | Poor sequence quality |
| 8 | DM8 | 128 | Resistant | R16C, S108A, D205A | A70V |
| 9 | DM10 | 64 | Resistant | S43W | Q27E, A70G, A85V, N111H, N129T, S130D, E176K, N182D |
| 10 | DM14 | 64 | Resistant | R10K, P44L, K168R, K198I, R200* | Q27E, M126I, N129T, S130D, A153V, E176K, N182D |
| 11 | DM32 | 64 | Resistant | G170S | G45R |
| 12 | DM39 | 64 | Resistant | K198I, K201E | Q27E, N111H, N129T, S130D, E176K |
| 13 | DM43 | 32 | Resistant | 41frameshift | Q27E, N111H, N129T, S130D, N182D |
| 14 | DM44 | 64 | Resistant | E75* | Q27E, N111H, N129T, S130D, E176K |
| 15 | DM50 | 64 | Resistant | Q50* | Q27E, A32E, N111H, N129T, S130D, E176K |
| 16 | DM51 | 32 | Resistant | G145R, E174G, | A70V |
| 17 | DM52 | 64 | Resistant | 120frameshift | Q27E, A70G, N111H, N129H, S130D, E169A, E176K |
| 18 | DM57 | 64 | Resistant | 3frameshift | Q27E, A70G, 86frameshift |
| 19 | DM60 | 64 | Resistant | R16C, P96L | None |
| 20 | DM62 | 64 | Resistant | A68T, G189S | Q27E, P41L, A70G, N111H, N129T, S130D, M146I |
| 21 | DM64 | 64 | Resistant | 9frameshift | Q27E, A32E, N111H, N129T, S130D, E176K, N182D |
| 22 | DM73 | 64 | Resistant | L188F, K190* | None |
| 23 | DM75 | 64 | Resistant | K190Y, A193* | Q27E, A32E, T110A, N111H, N129T, S130D, E176K |
| 24 | DM76 | 128 | Resistant | 122–149del, D205A | Poor sequence quality |
| 25 | DM79 | 64 | Resistant | S30G, E38* | Q27E, N111H, N129T, S130D, E176K |
| 26 | DM81 | 128 | Resistant | 42frameshift | V103I |
| 27 | DM84 | 64 | Resistant | G122S | Q27E, A32E, N111H, N129T, S130D, E176K, N182D |
| 28 | DM86 | 64 | Resistant | T31H, V57A, L62V, K64N, P106S, G155* | None |
| 29 | DM87 | 64 | Resistant | V85G, G145E, V151A, V192Q, A193G, E194R, Q197E, R200* | Q27E, N111H, N129T, S130D, E176K |
| 30 | DM88 | 32 | Resistant | R16H, R200M, S202I, 204frameshift | None |
| 31 | DM89 | 64 | Resistant | 65frameshift | Q27E, N111H, N129T, S130D, E176K, N182D |
| 32 | DM96 | 64 | Resistant | E35* | Q27E, N111H, M126I, N129T, S130D, E176K, N182D |
| 33 | DM100 | 32 | Resistant | S108A, K190E | A153V, E176K |
| 34 | DM103 | 32 | Resistant | C19* | Poor sequence quality |
| 35 | DM104 | 64 | Resistant | R16H, 207del | 22frameshift |
| 36 | DM105 | 16 | Resistant | P51S, K203Q | 18frameshift |
| 37 | DM112 | 64 | Resistant | 27frameshift | Q27E, N111H, N129T, S130D, E176K, N182D |
| 38 | DM115 | 64 | Resistant | R16H, H97Y, V182A, R200* | A85V, N111H |
| 39 | DM117 | 32 | Resistant | H69P, V85G, V86E, Q139H, L153* | Q27E, L72I, N111H, M126I, N129T, S130D, E169A, E176K, N182D |
| 40 | DM118 | 32 | Resistant | S18F, S43L, K63Q, G155R, K198T, R200* | Q27E, N111H, M126I, N129T, S130D, E176K, N182D |
| 41 | DM126 | 64 | Resistant | V85G, F117V, G122A, I147L, K198I, 208frameshift | Q27E, N111H, N129T, S130D, E169A, N182D |
| 42 | DM142 | 64 | Resistant | R16H, G122A, P180T, 197frameshift | Q27E, N111H, N129T, S130D, N182D |
| 43 | DM143 | 64 | Resistant | Q50* | 18frameshift |
| 44 | DM146 | 64 | Resistant | T31H, 192frameshift | Poor sequence quality |
| 45 | DM147 | 64 | Resistant | R16Y, H25Y, E35D, L62F, Q146H, R200* | 18frameshift |
| 46 | DM148 | 128 | Resistant | R16C, R166S, 198frameshift | A153V |
| 47 | DM150 | 16 | Resistant | Q197H, 200frameshift | R38I |
| 48 | DM151 | 128 | Resistant | C140Y, Q197* | N111H, N129T, S130D, E176K |
| 49 | DM152 | 64 | Resistant | E35* | Q27E, N111H, N129T, S130D, N182D |
MIC = minimum inhibitory concentration. None: no specific mutations were identified; A68T means threonine replaced alanine in amino acid position 68; R200* means a stop codon replaced arginine in amino acid position 200; 150frameshift means there was a frameshift mutation in amino acid position 150. We ignored mutations that were present in both sensitive and resistant strains.
We analyzed frxA in 40 resistant strains, because poor sequencing results were obtained for four strains (Table 4). Similar to mutational patterns in rdxA, frxA in MNZ-resistant strains also contained missense mutations and translational frameshifts (36/40, 90.0% and 5/40, 12.5%, respectively). In contrast with results of a previous study,27 Q27E (57.5%, 23/40), N111H (57.5%, 23/40), N129T (55.0%, 22/40), S130D (57.5%, 23/40), E176K (45.0%, 18/40), and N182D (32.5%, 13/40) were predominant among the frxA mutations. Overall, although these two genes showed similarities in their patterns of mutation, no synergistic effect on MIC values was observed; for example, one MNZ-resistant strain with an MIC of 32 mg/L but showed no mutation in frxA, and this could not explain the different levels of MNZ resistance. Therefore, we randomly selected two sensitive strains, with MIC values of 32, 64, and 128 mg/L each and performed NGS (Table 5). Using the H. pylori strain 26695 and the MNZ-sensitive control strain, irrespective of their rdxA and frxA mutations, we revealed novel mutations associated with MNZ resistance in the full-length sequences of dppA (E473G), dppB (I322V), fdxA (N32S and S79I), and fdxB (V64I, M65L, and P245A). We did not obtain full-length rdxA, frxA, or ackA sequences from the NGS data. Moreover, we proposed that novel amino acid substitutions at Ser-14 of trx1 and Arg-221 of dapF were associated with different levels of MNZ resistance.
Table 5.
Mutations in other genes that were associated with metronidazole resistance
| No. | Strains | MIC (mg/L) | Profile | rpsU (hp0562) | rps4 (hp1294) | dppA (hp0298) | dppB (hp0299) | rnc (hp0662) | dapF (hp0566) | recA (hp0153) | fdxA (hp0277) | fdxB (hp1508) | trx1 (hp0824) |
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| 1 | DM30 | 1 | Sensitive | None | None | 35del, 36del, 37del, 179H | None | None | None | None | None | None | E9G |
| 2 | DM116 | 1 | Sensitive | None | None | None | None | None | None | None | None | None | None |
| 3 | DM76 | 128 | Resistant | None | A48T, V170A | E473G | I322V | P15L | V16A, A84N, I109V, K117E, E161G, G162L, R221C, G248E, A257V, R266G, E269K | None | N32S, S79I | V64I, M65L, P245A, I304T, A428I, A439T | S14N |
| 4 | DM151 | 128 | Resistant | A34T | None | T32A, E473G | R124C | 1del, N152K, | K50N, N164F, E177A, R221C, G248E, A257I, R266G, G271H | None | 43del, 48del, S79I | F125L, A227V, N231K, P245A, R307H, M321L, A325T | S14N |
| 5 | DM4 | 64 | Resistant | None | None | None | K199R, I322V | 1del | V16A, K50R, D51N, A84N, K117E, E161G, G162L, G248E, A245V, R266G, G271H | None | N32S, S79I | A38V, V64I, M65L, A227V | None |
| 6 | DM64 | 64 | Resistant | C32Y | None | E473G | I322V | P15S | V16A, K50R, D51N, A84N, I109V, K117E, E161G, G162L, G248E, A257V, R266G, E269K | None | N32S, S79I | V64I, M65L, P245A, H311A, N395S, N424H | None |
| 7 | DM18 | 32 | Resistant | None | None | E473G | I21V, V127M, I322V | I13A, R109H | V16A, A84N, I109V, K117E, E161G, G162L, G248D, A257V, R266G, E269K | None | N32S, S79I | V64I, M65L, M163V, P245T, S417G, A428I | N102S |
| 8 | DM51 | 32 | Resistant | None | None | Q383E | None | None | K129N, A254T, V267I | S40D, I191V | N32S, S79I | S122R, P123S, D170E, I173V, P245A, N424S | I40V |
MIC = minimum inhibitory concentration; None: no specific mutations were identified; 35del means there was a deletion at amino acid position 35; I79H means histidine replaced isoleucine in amino acid position 79. We ignored mutations that were present in both sensitive and resistant strains.
Among the 23 LVX-resistant strains, 18 had amino acid substitutions associated with gyrA mutations (Table 6). Eleven of LVX-resistant strains (47.8%) showed an amino acid substitution at Asp-91, and 13 strains showed an amino acid substitution at Asn-87 (56.5%), including four strains with the highest LVX MIC values (32 mg/L). In addition, three strains exhibited amino acid substitutions associated with mutations in gyrB, including a substitution of interest at Ser-479. Although two strains with high levels of resistance showed mutations in both genes, one strain (DM62) that contained two substitutions of interest (Asp-91 [gyrA] and Ser-479 [gyrB]) had an LVX MIC value of 2 mg/L, suggesting there was no correlation between the degree of LVX resistance and the number of mutations in these two genes. Interestingly, there were no mutations in either gyrA or gyrB in three strains, including in the strain with the highest LVX MIC.
Table 6.
Mutations in gyrA and gyrB that were associated with levofloxacin resistance
| No. | Strain | MIC (mg/L) | Profile | gyrA mutations | gyrB mutations |
|---|---|---|---|---|---|
| 1 | DM8 | 0.25 | Sensitive | None | None |
| 2 | DM32 | 0.25 | Sensitive | None | None |
| 3 | DM57 | 0.25 | Sensitive | None | None |
| 4 | DM86 | 0.25 | Sensitive | None | None |
| 5 | DM95 | 0.25 | Sensitive | None | None |
| 6 | DM9 | 16 | Resistant | None | None |
| 7 | DM10 | 32 | Resistant | N87I | None |
| 8 | DM14 | 2 | Resistant | D91G, E193D, I194F, A197F | None |
| 9 | DM18 | 16 | Resistant | N87T, D91N | None |
| 10 | DM43 | 2 | Resistant | N87T, D91N | None |
| 11 | DM44 | 16 | Resistant | N87T, A97V | None |
| 12 | DM49 | 32 | Resistant | N87A | D435N, S479G |
| 13 | DM50 | 2 | Resistant | E58G, G75V, D91G, Q98L | None |
| 14 | DM51 | 2 | Resistant | D91N | None |
| 15 | DM60 | 2 | Resistant | D91Y, A134V, D145N | None |
| 16 | DM62 | 2 | Resistant | D91N | S479G |
| 17 | DM71 | 32 | Resistant | N87I | S479G |
| 18 | DM73 | 32 | Resistant | None | None |
| 19 | DM75 | 32 | Resistant | N87I | None |
| 20 | DM96 | 4 | Resistant | None | None |
| 21 | DM103 | 16 | Resistant | N87I, D91N, I162T | None |
| 22 | DM115 | 2 | Resistant | N87K | None |
| 23 | DM142 | 4 | Resistant | N87T, D91N | None |
| 24 | DM146 | 16 | Resistant | N87T | None |
| 25 | DM147 | 2 | Resistant | D91Y | None |
| 26 | DM148 | 2 | Resistant | None | None |
| 27 | DM150 | 16 | Resistant | N87T, D91N, F149V | None |
| 28 | DM152 | 8 | Resistant | N87I | Y514F |
MIC = minimum inhibitory concentration. None: no specific mutations were identified; N87I means isoleucine replaced asparagine in amino acid position 87. We ignored mutations that were present in both sensitive and resistant strains.
Based on 23S rRNA sequencing of the two CAM-resistant strains, one strain had a point mutation resulting in the A2142G substitution. In addition, a T to G substitution at site 1958 was observed in both strains (Table 7). In contrast, there were no nucleotide mutations in the CAM-sensitive strains.
Table 7.
Mutations in the 23S rRNA that were associated with clarithromycin resistance
| Strain | MIC (mg/L) | Profile | Mutations |
|---|---|---|---|
| DM44 | ≤ 0.03 | Sensitive | None |
| DM62 | 16 | Resistant | T1958G |
| DM105 | 16 | Resistant | A1957G, T1958G, G1964T, A1968T, A2142G* |
MIC = minimum inhibitory concentration.
A recognized mutation; T1958G means guanine replaced thymine at amino acid position 1958.
Nucleotide sequencing.
Nucleotide sequence data from this study are available under DDBJ accession numbers LC176131–LC176133 (23S rRNA), LC176134–LC176161 (gyrA), LC176162–LC176189 (gyrB), LC176190–LC176238 (rdxA), LC200542-LC200585 (frxA), LC199398-LC199405 (rpsU), LC199406-199413 (rpsD), LC199414-LC199421 (dppA), LC199422-LC199429 (dppB), LC199430-LC199437 (rnc), LC199438-LC199445 (dapF), LC199446-LC199453 (recA), LC199454-LC199461 (fdxA), LC199462-LC199469 (fdxB), LC199470-LC199477 (trxA).
Discussion
In this study, we revealed the low prevalence of CAM and AMX resistance. In addition, there were no TCN-resistant strains, suggesting that the standard CAM-based triple therapy may still be useful as an initial treatment of H. pylori infection in the Dominican Republic. Our results agree with those of a meta-analysis, which reported that the overall prevalence (13%) of CAM resistance in Latin America was below the prevalence reported in some European and Asian countries.17 The authors suggested that empirical use of CAM might not be appropriate in Peru and perhaps in Colombia.17 These differences of resistance frequency may be associated with local utilization of antibiotics.28,29 In fact, the 10-year sales trends for macrolides showed large increases in Peru, Brazil, and Argentina.30 In general, macrolides are frequently used for a number of infectious diseases in the Dominican Republic (M. Cruz and J. A. Jiménez Abreu, personal communication). However, there is no reliable report regarding CAM utilization in the Dominican Republic.
Interestingly, although A2142G substitution associated with rdxA is responsible for 90% of primary CAM-resistant H. pylori cases in western countries,31 we observed a T to G substitution at site 1958 in all CAM-resistant strains obtained (N = 2). We need more evidence to conclude that this mutation was directly responsible for or synergistically associated with other CAM-resistance mutations. Point mutations resulting in substitutions other than A2142G detected in this study have been described previously. The substitutions T2183C and A2223G have been more frequently associated with CAM resistance in Asian countries than in Europe and North America.32 The A2143G substitution has much stronger impact on CAM resistance compared with A2142G or A2142C.33
In agreement with reports from most Latin American countries,17 the frequency of resistance to AMX in isolates from the Dominican Republic is low, despite AMX being one of the most commonly used antibiotics around the world. Previous reports have indicated that a low detected frequency of resistance was associated with the phenotypic loss of pbp1A after storage or freezing of isolates.34,35 However, Hu and others showed that resistance to AMX was stable before and after the storage at −80°C for 3 months or even years.36 Indeed, H. pylori resistance to AMX does not necessarily decrease treatment efficacy.37 These could compromise the utilization of AMX in most regimens, where it is commonly used owing to its adequacy and low cost. AMX-based triple or quadruple therapies that include TCN may be a useful alternative first-line regimen in the Dominican Republic.
The extremely high frequency of resistance to MNZ found in the Dominican Republic in this study could be attributable to widespread over-the-counter use of this drug. MNZ is a drug with a modest cost that is used to treat not only H. pylori infections but also many other diseases; for example, intestinal parasites and periodontal and gynecologic diseases that are common in developing countries such as the Dominican Republic.28,38 Therefore, regimens that include MNZ should not be chosen as a first-line treatment therapy in this population. However, in vitro resistance to MNZ may not accurately reflect in vivo resistance; therefore, MNZ resistance should be confirmed by determining concentrations of the drug in the blood of treated patients.39 In addition, because most MNZ-resistant isolates showed MNZ MICs > 32 mg/L, this might represent an appropriate breakpoint, although clinical trials are required to confirm MNZ resistance in the Dominican Republic. Importantly, MNZ resistance is not more clinically relevant than CAM resistance. Furthermore, resistance to MNZ can be overcome by prolonging treatment and adding bismuth to the treatment regimen.40
By comparing the Dominican Republic MNZ-sensitive and MNZ-resistant strains, we recognized a high number of rdxA mutations in the MNZ-resistant strains. We also confirmed some mutations that could be explained by resistance structure (class mutations).25 In contrast with results of a previous study, we found other mutations (E35, V85, G122, K190, and K198) that might be important to resistance in the Dominican strains. This agrees with a previous report, which showed that rdxA mutations were not uniform across all geographical regions.41 Additionally, several frxA mutations were associated with MNZ resistance. We observed one strain without a mutation in rdxA, but that contained an frxA mutation (MNZ MIC of 64 mg/L), suggesting a role for frxA, irrespective of rdxA, although the combination of mutations in these two genes was not associated with higher-level resistance. Finally, we identified novel mutations in dppA, dppB, fdxA, and fdxB that were not associated with mutations in rdxA and frxA. Importantly, mutations in trx1 and dapF were associated with difference levels of MNZ resistance, indicated by MIC values. Unlike dapF, which is associated with lysine and peptidoglycan biosynthesis,42 trx1 has a role as an electron donor for alkyl-hydroperoxide reductase, which is associated with MNZ resistance in vitro.43
LVX has recently been prescribed as a second-line drug to eradicate H. pylori in patients who experienced failed first-line therapies.44,45 However, the frequency of LVX resistance is expanding worldwide, including Latin America.17 A similar expansion of resistance is observed in urinary tract Escherichia coli isolates from Latin America.46 This may lead to cross-resistance with fluoroquinolone of different generations, including nalidixic acid, ciprofloxacin, and ofloxacin. Furthermore, the frequency of LVX resistance in the Dominican Republic is much higher than the average frequencies of fluoroquinolone resistance reported in Latin America (15.0%),17 Europe (14.1%),28 and Asia (11.6%).24 According to European, Asia-Pacific, and American guidelines, LVX should be used in rescue treatments only after antibiotic susceptibility testing.1,19,47
We observed substitutions at Asn-87 and Asp-91 that were associated with LVX resistance as previously reported.41,48,49 In addition, few mutations and the co-occurrence of the Ser-479 substitution in the gyrB subunit with substitutions in gyrA suggested that gyrB mutations had minimal influence on LVX-resistant strains in this study. Three resistant strains showed no changes in gyrA or gyrB. This suggests that substitutions at other sites might act synergistically in LVX resistance in the Dominican Republic strains.
Although only two strains were resistant to three antibiotics, 31.3% of the strains showed dual antibiotic resistance: this resistance was exclusive to MNZ and LVX. The Dominican Republic has a high prevalence of H. pylori infections,18 and increased resistance to antibiotics used to treat these infections might result in an increased frequency of disease recurrence. With high rates of morbidity and mortality due to H. pylori infection–associated pathologies, prevention is the ultimate solution. Our results are very important as a guide for antibiotic use as follow-up to first-line treatment regimen failures. Mutations associated with antibiotic resistance (e.g., 23S rRNA point mutations at A2143G and A2144G) was determined using a fully automated rapid genetic analyzer within 60–120 minutes, compared with the 7–10 days required for testing by culture.50 These results also suggest that alternative strategies such as bismuth or non-bismuth-based four-drug regimens or sequential therapies may be more effective in cases of CAM-based triple therapy failure in the Dominican Republic.
The number of samples analyzed here was relatively low, which presents a limitation to this study. In addition, we did not consider the socioeconomic status, ethnicity, or residence of patients. The Digestive Disease Center is the only specialized public medical center for digestive diseases in the Dominican Republic, with half of patients coming from the countryside and most patients living at a low-middle socioeconomic level. Further studies are required to determine the association of sociodemographic data with antibiotic resistance patterns in the Dominican Republic.
In conclusion, the frequencies of resistance to MNZ and LVX were high in the Dominican Republic, and resistance to these antibiotics was associated with genetic mutations. It has been suggested that MNZ- and LVX-based triple therapies are not useful as a first-line treatment regimen in the Dominican Republic. Fortunately, the prevalence of CAM resistance was found to be low in this study, suggesting that the CAM-based triple therapy may still be useful as an initial treatment of H. pylori infection in the Dominican Republic. Epidemiological surveillance of antibiotic resistance in H. pylori strains will be required to determine optimal treatment strategies for use in the Dominican Republic.
Footnotes
Financial support: This work was supported in part by grants from the National Institutes of Health (DK62813) and the Grants-in-Aid for Scientific Research from the Ministry of Education, Culture, Sports, Science, and Technology (MEXT) of Japan (25293104, 26640114, 15H02657, and 221S0002) (Yoshio Yamaoka). It was also supported by the Japan Society for the Promotion of Science (JSPS) Institutional Program for Young Researcher Overseas Visits (Yoshio Yamaoka), Strategic Funds for the Promotion of Science and Technology from the Japan Science and Technology Agency (JST) (Yoshio Yamaoka). In addition, it was supported in part by a grant from The National Fund for Innovation and Development of Science and Technology (FONDOCYT) from the Ministry of Higher Education Science and Technology (MESCyT) of the Dominican Republic (2012-2013-2A1-65 and 2015-3A1-182) (Modesto Cruz). Muhammad Miftahussurur was a PhD student and Phawinee Subsomwong is a PhD student supported by The Japanese Government (MEXT) Scholarship Program for 2012 and 2013, respectively.
Authors' addresses: Muhammad Miftahussurur, Phawinee Subsomwong, Hiroyuki Nagashima, Junko Akada, and Yoshio Yamaoka, Department of Environmental and Preventive Medicine, Oita University Faculty of Medicine, Yufu, Japan, E-mails: miphto@oita-u.ac.jp, phawinee@oita-u.ac.jp, hnagashi@oita-u.ac.jp, akadajk@oita-u.ac.jp, and yyamaoka@oita-u.ac.jp. Modesto Cruz and Celso Hosking, Institute of Microbiology and Parasitology, Faculty of Science, Autonomous University of Santo Domingo, Santo Domingo, Dominican Republic, E-mails: modesto_cruz@yahoo.com and impa.uasd@gmail.com. José A. Jiménez Abreu, Dominican–Japanese Digestive Disease Center, Dr. Luis E. Aybar Health and Hygiene City, Santo Domingo, Dominican Republic, E-mail: jojis17@gmail.com.
References
- 1.Fock KM, Katelaris P, Sugano K, Ang TL, Hunt R, Talley NJ, Lam SK, Xiao SD, Tan HJ, Wu CY, Jung HC, Hoang BH, Kachintorn U, Goh KL, Chiba T, Rani AA, Second Asia-Pacific Conference. Second Asia-Pacific Consensus Guidelines for Helicobacter pylori infection. J Gastroenterol Hepatol. 2009;24:1587–1600. doi: 10.1111/j.1440-1746.2009.05982.x. [DOI] [PubMed] [Google Scholar]
- 2.Malfertheiner P, Megraud F, O'Morain CA, Atherton J, Axon AT, Bazzoli F, Gensini GF, Gisbert JP, Graham DY, Rokkas T, El-Omar EM, Kuipers EJ. European Helicobacter Study Group Management of Helicobacter pylori infection: the Maastricht IV/ Florence Consensus Report. Gut. 2012;61:646–664. doi: 10.1136/gutjnl-2012-302084. [DOI] [PubMed] [Google Scholar]
- 3.Papastergiou V, Georgopoulos SD, Karatapanis S. Treatment of Helicobacter pylori infection: meeting the challenge of antimicrobial resistance. World J Gastroenterol. 2014;20:9898–9911. doi: 10.3748/wjg.v20.i29.9898. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Miftahussurur M, Yamaoka Y. Appropriate first-line regimens to combat Helicobacter pylori antibiotic resistance: an Asian perspective. Molecules. 2015;20:6068–6092. doi: 10.3390/molecules20046068. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Suzuki RB, Lopes RA, da Camara Lopes GA, Hung Ho T, Speranca MA. Low Helicobacter pylori primary resistance to clarithromycin in gastric biopsy specimens from dyspeptic patients of a city in the interior of Sao Paulo, Brazil. BMC Gastroenterol. 2013;13:164. doi: 10.1186/1471-230X-13-164. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Stone GG, Shortridge D, Flamm RK, Versalovic J, Beyer J, Idler K, Zulawinski L, Tanaka SK. Identification of a 23S rRNA gene mutation in clarithromycin-resistant Helicobacter pylori. Helicobacter. 1996;1:227–228. doi: 10.1111/j.1523-5378.1996.tb00043.x. [DOI] [PubMed] [Google Scholar]
- 7.Goodwin A, Kersulyte D, Sisson G, Veldhuyzen van Zanten SJ, Berg DE, Hoffman PS. Metronidazole resistance in Helicobacter pylori is due to null mutations in a gene (rdxA) that encodes an oxygen-insensitive NADPH nitroreductase. Mol Microbiol. 1998;28:383–393. doi: 10.1046/j.1365-2958.1998.00806.x. [DOI] [PubMed] [Google Scholar]
- 8.Jeong JY, Mukhopadhyay AK, Dailidiene D, Wang Y, Velapatino B, Gilman RH, Parkinson AJ, Nair GB, Wong BC, Lam SK, Mistry R, Segal I, Yuan Y, Gao H, Alarcon T, Brea ML, Ito Y, Kersulyte D, Lee HK, Gong Y, Goodwin A, Hoffman PS, Berg DE. Sequential inactivation of rdxA (HP0954) and frxA (HP0642) nitroreductase genes causes moderate and high-level metronidazole resistance in Helicobacter pylori. J Bacteriol. 2000;182:5082–5090. doi: 10.1128/jb.182.18.5082-5090.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Binh TT, Suzuki R, Trang TT, Kwon DH, Yamaoka Y. Search for novel candidate mutations for metronidazole resistance in Helicobacter pylori using next-generation sequencing. Antimicrob Agents Chemother. 2015;59:2343–2348. doi: 10.1128/AAC.04852-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Li AQ, Dai N, Yan J, Zhu YL. Screening for metronidazole-resistance associated gene fragments of H. pylori by suppression subtractive hybridization. World J Gastroenterol. 2007;13:1847–1850. doi: 10.3748/wjg.v13.i12.1847. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Chang KC, Ho SW, Yang JC, Wang JT. Isolation of a genetic locus associated with metronidazole resistance in Helicobacter pylori. Biochem Biophys Res Commun. 1997;236:785–788. doi: 10.1006/bbrc.1997.7050. [DOI] [PubMed] [Google Scholar]
- 12.Mukhopadhyay AK, Jeong JY, Dailidiene D, Hoffman PS, Berg DE. The fdxA ferredoxin gene can down-regulate frxA nitroreductase gene expression and is essential in many strains of Helicobacter pylori. J Bacteriol. 2003;185:2927–2935. doi: 10.1128/JB.185.9.2927-2935.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Kwon DH, El-Zaatari FA, Kato M, Osato MS, Reddy R, Yamaoka Y, Graham DY. Analysis of rdxA and involvement of additional genes encoding NAD(P)H flavin oxidoreductase (FrxA) and ferredoxin-like protein (FdxB) in metronidazole resistance of Helicobacter pylori. Antimicrob Agents Chemother. 2000;44:2133–2142. doi: 10.1128/aac.44.8.2133-2142.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Baker LM, Raudonikiene A, Hoffman PS, Poole LB. Essential thioredoxin-dependent peroxiredoxin system from Helicobacter pylori: genetic and kinetic characterization. J Bacteriol. 2001;183:1961–1973. doi: 10.1128/JB.183.6.1961-1973.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Jones KR, Cha JH, Merrell DS. Who's winning the war? Molecular mechanisms of antibiotic resistance in Helicobacter pylori. Curr Drug Ther. 2008;3:190–203. doi: 10.2174/157488508785747899. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Rimbara E, Noguchi N, Kawai T, Sasatsu M. Fluoroquinolone resistance in Helicobacter pylori: role of mutations at position 87 and 91 of GyrA on the level of resistance and identification of a resistance conferring mutation in GyrB. Helicobacter. 2012;17:36–42. doi: 10.1111/j.1523-5378.2011.00912.x. [DOI] [PubMed] [Google Scholar]
- 17.Camargo MC, Garcia A, Riquelme A, Otero W, Camargo CA, Hernandez-Garcia T, Candia R, Bruce MG, Rabkin CS. The problem of Helicobacter pylori resistance to antibiotics: a systematic review in Latin America. Am J Gastroenterol. 2014;109:485–495. doi: 10.1038/ajg.2014.24. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Shiota S, Cruz M, Abreu JA, Mitsui T, Terao H, Disla M, Iwatani S, Nagashima H, Matsuda M, Uchida T, Tronilo L, Rodriguez E, Yamaoka Y. Virulence genes of Helicobacter pylori in the Dominican Republic. J Med Microbiol. 2014;63:1189–1196. doi: 10.1099/jmm.0.075275-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Malfertheiner P, Megraud F, O'Morain C, Bazzoli F, El-Omar E, Graham D, Hunt R, Rokkas T, Vakil N, Kuipers EJ. Current concepts in the management of Helicobacter pylori infection: the Maastricht III Consensus Report. Gut. 2007;56:772–781. doi: 10.1136/gut.2006.101634. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Malfertheiner P, Megraud F, O'Morain CA, Gisbert JP, Kuipers EJ, Axon AT, Bazzoli F, Gasbarrini A, Atherton J, Graham DY, Hunt R, Moayyedi P, Rokkas T, Rugge M, Selgrad M, Suerbaum S, Sugano K, El-Omar EM. Management of Helicobacter pylori infection: the Maastricht V/Florence Consensus Report. Gut. 2017;66:6–30. doi: 10.1136/gutjnl-2016-312288. [DOI] [PubMed] [Google Scholar]
- 21.Kwon DH, Pena JA, Osato MS, Fox JG, Graham DY, Versalovic J. Frameshift mutations in rdxA and metronidazole resistance in North American Helicobacter pylori isolates. J Antimicrob Chemother. 2000;46:793–796. doi: 10.1093/jac/46.5.793. [DOI] [PubMed] [Google Scholar]
- 22.Wang LH, Cheng H, Hu FL, Li J. Distribution of gyrA mutations in fluoroquinolone-resistant Helicobacter pylori strains. World J Gastroenterol. 2010;16:2272–2277. doi: 10.3748/wjg.v16.i18.2272. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Ho SL, Tan EL, Sam CK, Goh KL. Clarithromycin resistance and point mutations in the 23S rRNA gene in Helicobacter pylori isolates from Malaysia. J Dig Dis. 2010;11:101–105. doi: 10.1111/j.1751-2980.2010.00423.x. [DOI] [PubMed] [Google Scholar]
- 24.De Francesco V, Giorgio F, Hassan C, Manes G, Vannella L, Panella C, Ierardi E, Zullo A. Worldwide H. pylori antibiotic resistance: a systematic review. J Gastrointestin Liver Dis. 2010;19:409–414. [PubMed] [Google Scholar]
- 25.Martinez-Julvez M, Rojas AL, Olekhnovich I, Espinosa Angarica V, Hoffman PS, Sancho J. Structure of RdxA: an oxygen-insensitive nitroreductase essential for metronidazole activation in Helicobacter pylori. FEBS J. 2012;279:4306–4317. doi: 10.1111/febs.12020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Jenks PJ, Ferrero RL, Labigne A. The role of the rdxA gene in the evolution of metronidazole resistance in Helicobacter pylori. J Antimicrob Chemother. 1999;43:753–758. doi: 10.1093/jac/43.6.753. [DOI] [PubMed] [Google Scholar]
- 27.Kwon DH, Hulten K, Kato M, Kim JJ, Lee M, El-Zaatari FA, Osato MS, Graham DY. DNA sequence analysis of rdxA and frxA from 12 pairs of metronidazole-sensitive and -resistant clinical Helicobacter pylori isolates. Antimicrob Agents Chemother. 2001;45:2609–2615. doi: 10.1128/AAC.45.9.2609-2615.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Megraud F, Coenen S, Versporten A, Kist M, Lopez-Brea M, Hirschl AM, Andersen LP, Goossens H, Glupczynski Y. Study Group Participants Helicobacter pylori resistance to antibiotics in Europe and its relationship to antibiotic consumption. Gut. 2013;62:34–42. doi: 10.1136/gutjnl-2012-302254. [DOI] [PubMed] [Google Scholar]
- 29.Boyanova L, Mitov I. Geographic map and evolution of primary Helicobacter pylori resistance to antibacterial agents. Expert Rev Anti Infect Ther. 2010;8:59–70. doi: 10.1586/eri.09.113. [DOI] [PubMed] [Google Scholar]
- 30.Wirtz VJ, Dreser A, Gonzales R. Trends in antibiotic utilization in eight Latin American countries, 1997–2007. Rev Panam Salud Publica. 2010;27:219–225. doi: 10.1590/s1020-49892010000300009. [DOI] [PubMed] [Google Scholar]
- 31.Oleastro M, Ménard A, Santos A, Lamouliatte H, Monteiro L, Barthélémy P, Mégraud F. Real-time PCR assay for rapid and accurate detection of point mutations conferring resistance to clarithromycin in Helicobacter pylori. J Clin Microbiol. 2003;41:397–402. doi: 10.1128/JCM.41.1.397-402.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Ierardi E, Giorgio F, Losurdo G, Di Leo A, Principi M. How antibiotic resistances could change Helicobacter pylori treatment: a matter of geography? World J Gastroenterol. 2013;19:8168–8180. doi: 10.3748/wjg.v19.i45.8168. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.De Francesco V, Margiotta M, Zullo A, Hassan C, Troiani L, Burattini O, Stella F, Di Leo A, Russo F, Marangi S, Monno R, Stoppino V, Morini S, Panella C, Ierardi E. Clarithromycin-resistant genotypes and eradication of Helicobacter pylori. Ann Intern Med. 2006;144:94–100. doi: 10.7326/0003-4819-144-2-200601170-00006. [DOI] [PubMed] [Google Scholar]
- 34.Dore MP, Osato MS, Realdi G, Mura I, Graham DY, Sepulveda AR. Amoxycillin tolerance in Helicobacter pylori. J Antimicrob Chemother. 1999;43:47–54. doi: 10.1093/jac/43.1.47. [DOI] [PubMed] [Google Scholar]
- 35.Yakoob J, Fan X, Hu G, Liu L, Zhang Z. Antibiotic susceptibility of Helicobacter pylori in the Chinese population. J Gastroenterol Hepatol. 2001;16:981–985. doi: 10.1046/j.1440-1746.2001.02553.x. [DOI] [PubMed] [Google Scholar]
- 36.Hu CT, Wu CC, Lin CY, Cheng CC, Su SC, Tseng YH, Lin NT. Resistance rate to antibiotics of Helicobacter pylori isolates in eastern Taiwan. J Gastroenterol Hepatol. 2007;22:720–723. doi: 10.1111/j.1440-1746.2006.04743.x. [DOI] [PubMed] [Google Scholar]
- 37.Kim N, Kim JM, Kim CH, Park YS, Lee DH, Kim JS, Jung HC, Song IS. Institutional difference of antibiotic resistance of Helicobacter pylori strains in Korea. J Clin Gastroenterol. 2006;40:683–687. doi: 10.1097/00004836-200609000-00004. [DOI] [PubMed] [Google Scholar]
- 38.Megraud F. H. pylori antibiotic resistance: prevalence, importance, and advances in testing. Gut. 2004;53:1374–1384. doi: 10.1136/gut.2003.022111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Farshad S, Alborzi A, Japoni A, Ranjbar R, Hosseini Asl K, Badiee P, Amin Shahidi M, Hosseini M. Antimicrobial susceptibility of Helicobacter pylori strains isolated from patients in Shiraz, southern Iran. World J Gastroenterol. 2010;16:5746–5751. doi: 10.3748/wjg.v16.i45.5746. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Malfertheiner P, Bazzoli F, Delchier JC, Celinski K, Giguere M, Riviere M, Megraud F. Pylera Study Group Helicobacter pylori eradication with a capsule containing bismuth subcitrate potassium, metronidazole, and tetracycline given with omeprazole versus clarithromycin-based triple therapy: a randomised, open-label, non-inferiority, phase 3 trial. Lancet. 2011;377:905–913. doi: 10.1016/S0140-6736(11)60020-2. [DOI] [PubMed] [Google Scholar]
- 41.Teh X, Khosravi Y, Lee WC, Leow AH, Loke MF, Vadivelu J, Goh KL. Functional and molecular surveillance of Helicobacter pylori antibiotic resistance in Kuala Lumpur. PLoS One. 2014;9:e101481. doi: 10.1371/journal.pone.0101481. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Richaud C, Higgins W, Mengin-Lecreulx D, Stragier P. Molecular cloning, characterization, and chromosomal localization of dapF, the Escherichia coli gene for diaminopimelate epimerase. J Bacteriol. 1987;169:1454–1459. doi: 10.1128/jb.169.4.1454-1459.1987. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Trend MA, Jorgensen MA, Hazell SL, Mendz GL. Oxidases and reductases are involved in metronidazole sensitivity in Helicobacter pylori. Int J Biochem Cell Biol. 2001;33:143–153. doi: 10.1016/s1357-2725(00)00085-6. [DOI] [PubMed] [Google Scholar]
- 44.Gisbert JP, Gonzalez L, Calvet X. Systematic review and meta-analysis: proton pump inhibitor vs. ranitidine bismuth citrate plus two antibiotics in Helicobacter pylori eradication. Helicobacter. 2005;10:157–171. doi: 10.1111/j.1523-5378.2005.00307.x. [DOI] [PubMed] [Google Scholar]
- 45.Gisbert JP. Second-line rescue therapy of Helicobacter pylori infection. Therap Adv Gastroenterol. 2009;2:331–356. doi: 10.1177/1756283X09347109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Andrade SS, Sader HS, Jones RN, Pereira AS, Pignatari AC, Gales AC. Increased resistance to first-line agents among bacterial pathogens isolated from urinary tract infections in Latin America: time for local guidelines? Mem Inst Oswaldo Cruz. 2006;101:741–748. doi: 10.1590/s0074-02762006000700006. [DOI] [PubMed] [Google Scholar]
- 47.Chey WD, Wong BC. Practice Parameters Committee of the American College of Gastroenterology American College of Gastroenterology guideline on the management of Helicobacter pylori infection. Am J Gastroenterol. 2007;102:1808–1825. doi: 10.1111/j.1572-0241.2007.01393.x. [DOI] [PubMed] [Google Scholar]
- 48.Nishizawa T, Suzuki H, Kurabayashi K, Masaoka T, Muraoka H, Mori M, Iwasaki E, Kobayashi I, Hibi T. Gatifloxacin resistance and mutations in gyra after unsuccessful Helicobacter pylori eradication in Japan. Antimicrob Agents Chemother. 2006;50:1538–1540. doi: 10.1128/AAC.50.4.1538-1540.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Garcia M, Raymond J, Garnier M, Cremniter J, Burucoa C. Distribution of spontaneous gyrA mutations in 97 fluoroquinolone-resistant Helicobacter pylori isolates collected in France. Antimicrob Agents Chemother. 2012;56:550–551. doi: 10.1128/AAC.05243-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Sugimoto M, Furuta T. Efficacy of tailored Helicobacter pylori eradication therapy based on antibiotic susceptibility and CYP2C19 genotype. World J Gastroenterol. 2014;20:6400–6411. doi: 10.3748/wjg.v20.i21.6400. [DOI] [PMC free article] [PubMed] [Google Scholar]