Abstract
Steroidogenic acute regulatory protein-related lipid transfer domain containing 7 (StarD7) is a poorly characterized member of the steroidogenic acute regulatory protein-related lipid transfer proteins, up-regulated in JEG-3 cells, involved in intracellular transport and metabolism of lipids. Previous studies dealing with the mechanisms underlying the human StarD7 gene expression led us to define the cis-acting regulatory sequences in the StarD7 promoter using as a model JEG-3 cells. These include a functional T cell-specific transcription factor 4 (TCF4) site involved in Wnt-β-catenin signaling. To understand these mechanisms in more depth, we examined the steroidogenic factor 1 (SF-1) contribution to StarD7 expression. Cotransfection experiments in JEG-3 cells point out that the StarD7 promoter is activated by SF-1, and this effect is increased by forskolin. EMSA using JEG-3 nuclear proteins demonstrated that SF-1 binds to the StarD7 promoter. Additionally, chromatin immunoprecipitation analysis indicated that SF-1 and β-catenin are bound in vivo to the StarD7 promoter. Reporter gene assays in combination with mutations in the SF-1 and TCF4 binding sites revealed that the StarD7 promoter is synergistically activated by SF-1 and β-catenin and that the TCF4 binding site (−614/−608) plays an important role in this activation. SF-1 amino acid mutations involved in the physical interaction with β-catenin abolished this activation; thus demonstrating that the contact between the two proteins is necessary for an efficient StarD7 transcriptional induction. Finally, these data suggest that β-catenin could function as a bridge between SF-1 and TCF4 forming a ternary complex, which would stimulate StarD7 expression. The SF-1 and β-catenin pathway convergence on StarD7 expression may have important implications in the phospholipid uptake and transport, contributing to the normal trophoblast development.
Trophoblast performs the majority of the placental absorptive, immunoprotective and endocrinological functions, regulating the exchange of nutrients, gases, and other factors between the maternal and fetal circulations. The trophoblast differentiates in two ways: the villous and the extravillous trophoblast. Placental villous cytotrophoblasts proliferate and differentiate, by fusion, to form a syncytiotrophoblast layer. This event starts with modifications of the plasma membranes of both cell partners such as expression of syncytin, connexin 43, and enrichment of phosphatidylserine on the cell surface (1).
Lipid movement across and between the two layers of cell membrane is a biochemical event, in which the transport process can be attributed to the functions of specific proteins (2). Among these proteins is the steroidogenic acute regulatory protein (StAR)-related lipid transfer domain (StarD) superfamily that encompasses a 200-amino acid globular domain implicated in lipid/sterol binding (3, 4).
StarD7 mRNA was first identified as a JEG-3 overexpressed gene compared with normal and benign trophoblastic samples (5). In a previous study, we demonstrated a predominant cytoplasmic localization of StarD7 in human cytotrophoblast cells with a clear and partial relocalization toward the plasma membrane after the syncytialization process (6). Additionally, StarD7 recombinant protein forms stable Gibbs and Langmuir monolayers at the air-buffer interface, showing marked surface activity and interaction with phospholipid monolayers, mainly with phosphatidylserine, cholesterol, and phosphatidylglycerol (7). A recent report demonstrated StarD7 protein expression in a mouse hepatoma cell line (HEPA-1) and in rat liver, suggesting that it facilitates the delivery of phosphatidylcholine to mitochondria (8).
Previous studies indicate that the regulation of StarD7 expression in JEG-3 cells occurs through a β-catenin-mediated activation mechanism that involves transcriptional induction (9). Sequence analysis revealed that, within the 5′ upstream region of StarD7 gene, there are, in addition to the previously characterized T cell-specific transcription factor (TCF)4 binding site, consensus motives for the binding of steroidogenic factor 1 (SF-1) and cAMP response elements.
SF-1 is a member of the nuclear receptor family that plays multiple roles in development and metabolism. This transcription factor, identified in all steroidogenic tissues, including placenta, is required for the differentiation of mammalian endocrine glands and sexual development (10, 11). SF-1 plays a role in the expression control of a number of cAMP responsive genes, such as the human StAR (12–14). Even though SF-1 was described as an orphan receptor, strong evidence indicates that SF-1 is regulated by endogenous ligands and suggests an unexpected relationship between phospholipids and endocrine development and function (15–18).
The Wnt/β-catenin signaling pathway controls gene expression to coordinate many cellular processes, such as proliferation, differentiation, and cell motility of normal development and cancer cell progression by TCF/lymphoid enhancer-binding factor 1 family (19). Wnt molecules comprise a large family of secreted glycoproteins, which interact with specific surface receptors, the different members of the frizzled family and low-density lipoprotein receptor-related protein-5/6 (20). A major target of the canonical Wnt signaling pathway is the coactivator β-catenin. Without Wnt signaling, β-catenin is phosphorylated by a complex containing glycogen synthase kinase 3β. This marks β-catenin for proteosomal degradation by the so-called β-catenin destruction complex. Active Wnt signaling disrupts this complex, which results in β-catenin stabilization and nuclear localization. In the nucleus, it regulates target gene expression through partnerships with the TCF/lymphoid enhancer-binding factor 1 transcription factors (19). Several reports have documented a cross-regulation between the Wnt-signaling pathways and the nuclear receptor family, including SF-1 (21–29).
Based on these findings, we have investigated the role of SF-1 and β-catenin/TCF proteins in the transcriptional regulation of StarD7 gene expression. This study demonstrates that SF-1 induces StarD7 expression by interaction with the StarD7 promoter region, and this effect is increased by the addition of forskolin (FSK). Moreover, these findings indicate that β-catenin synergizes with SF-1 to activate the StarD7 promoter.
Results
The 5′-flanking region of the StarD7 gene drives cell-selective expression
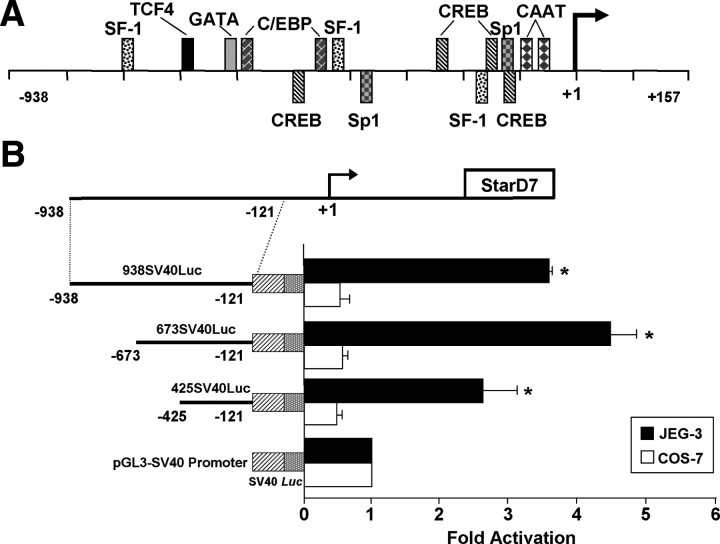
We previously reported the isolation and structural characterization of the human StarD7 gene promoter (9). To verify whether the sequence between nucleotides −938 and −121 has cell-specific regulatory elements, three different constructs containing the −938/−121, −673/−121, and −425/−121 sequences were subcloned 5′ upstream of the heterologous simian virus 40 (SV40) promoter in the pGL3-SV40 promoter vector and transfected into JEG-3 (derived from trophoblast tissue) and COS-7 (derived from African green monkey kidney) cells. Each of the StarD7 construct showed an increase in luciferase activity up to 4-fold over the activity of the pGL3-SV40 promoter vector in JEG-3 cells. On the contrary, no enhance in transcriptional activity over the SV40 promoter vector was observed when these constructs were transfected into COS-7 cells (Fig. 1B). These findings indicate that enhancer activities functionally dependent on transcription factors present in JEG-3 cells are located between positions −938 and −121. These results are in agreement with the very low StarD7 expression found in COS-7 compared with JEG-3 cells (6).
Fig. 1.
A, Schematic diagram of the 5′-flanking region of the StarD7 gene. The putative transcription start site is designated +1 and is shown by the arrow. Sequence analysis of the −938/+157 region using the MatInspector software (56) revealed many putative cis-elements, including two CAAT-boxes (−96/−92 and −66/−62), two Sp1 binding sites (−376/−371 and −121/−116), three SF-1 elements (−792/−785, −493/−486, and −169/−162), four CRE elements (−510/−489, −235/−214, −160/−139, and −116/−96), one TCF4 binding site (−614/−608), and various other response elements. Consensus binding sites are represented. B, The 5′ flanking region of StarD7 promoter gene drives cell-specific expression. The nucleotide −938/−121, −673/−121, and −425/−121 fragments of the StarD7 promoter were ligated upstream of the minimal SV40 promoter, each coupled to a Luc reporter gene. The chimeric constructs were transiently transfected into JEG-3 trophoblast cell line or nontrophoblast COS-7 cell line along with a phRL-TK-Renilla. Luciferase activity of each construct was expressed relative to Renilla values and the x-axis shows fold-induction relative to the luciferase activity of the pGL3-SV40 promoter vector. Each value represents the mean ± sem of three separate transfections, each performed in triplicate. *, P < 0.05 compared with cells transfected with the pGL3-SV40 promoter vector.
Expression levels of SF-1 affect StarD7 promoter activity
A TCF4 binding site located at −614/−608 bp relative to the transcription start site, required to activate StarD7 gene promoter by the β-catenin/TCF4 transcription factor, was previously identified (9). Further examination of the promoter region revealed three potential binding motives for the orphan nuclear receptor SF-1: a sequence at −792/−785 (CAAGGTCA, upper strand) and two other potential binding sites, the sequence (CAAGGACA, upper strand) located at −493/−486 and a region at −169/−162 (CTACCTTG, lower strand) (Fig. 1A). In addition, four putative cAMP response element (CRE) elements (−510/−489, −235/−214, −160/−139, and −116/−96) were identified.
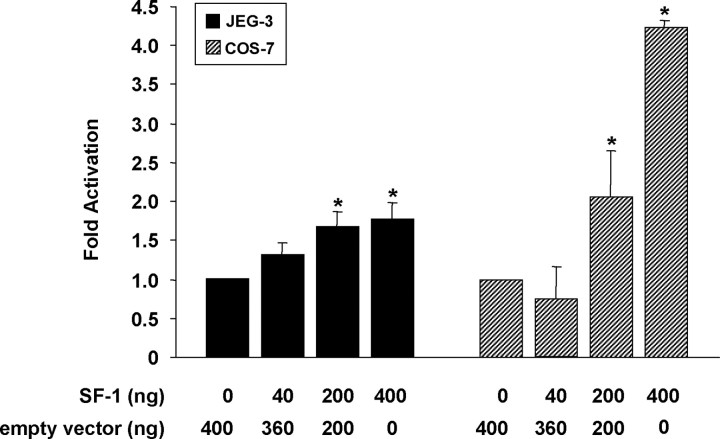
Because we could not detect SF-1 transcript in COS-7 cells by RT-PCR of total RNA, but found SF-1 mRNA in JEG-3 cell cultures (data not shown), we postulated that the relative absence of SF-1 expression in COS-7 compared with JEG-3 cells (30, 31) could explain, in part, the lack of enhancer activity of the StarD7 constructs in these cells. To gain insight into the functional responsiveness of the StarD7 gene promoter with respect to the SF-1 transcription factor, cotransfection experiments were performed in COS-7 and JEG-3 cells using the 938StarD7Luc construct (−938/+157, StarD7 promoter) together with an expression plasmid for SF-1. The results were clear and consistent, demonstrating a modest but significant increase in the reporter activity above baseline dependent upon the cotransfected SF-1 levels in both cell types (Fig. 2).
Fig. 2.
JEG-3 and COS-7 cells were transiently cotransfected with 938StarD7luc and phRL-TK Renilla constructs and the empty vector or SF-1 expression plasmid as indicated. Relative luciferase reporter gene activity was normalized against the Renilla activity, and results were expressed as the fold increase related to the activity in cells cotransfected with the empty expression vector defined as 1-fold. The values represent the mean ± sem of triplicate experiments. *, P < 0.05 compared with cells cotransfected with the empty expression vector.
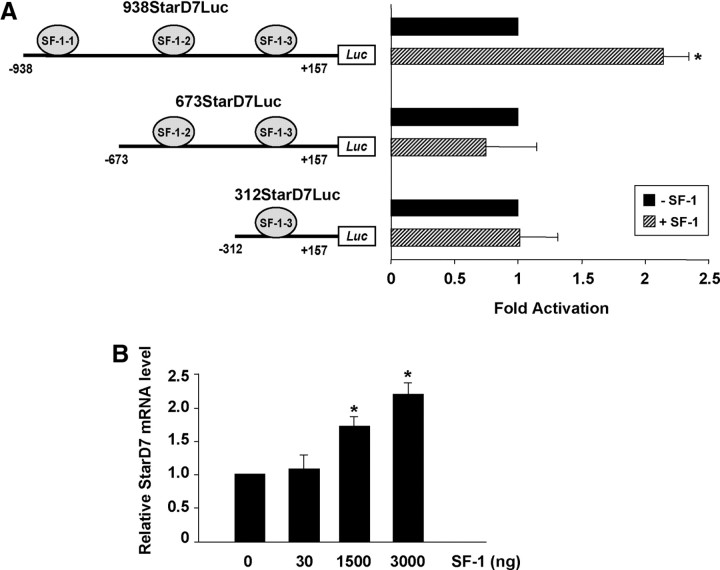
To examine the relevance of the SF-1 consensus binding sites on the transcriptional activation of StarD7 gene by SF-1, JEG-3 cells were cotransfected with the SF-1 expression plasmid and one of following StarD7 promoter deletion constructs: 673StarD7Luc, 312StarD7Luc, where one or two of SF-1 binding sites were removed, in comparison with 973StarD7Luc. Overexpression of SF-1 transcription factor enhanced reporter activity only in those cells that were transfected with the promoter fragment containing the three SF-1 consensus binding sites (Fig. 3A). Furthermore, the influence of SF-1 overexpression on endogenous StarD7 mRNA expression in JEG-3 cells was evaluated. Cell transfection with the control SF-1 empty vector did not induce StarD7 mRNA, whereas induction of endogenous mRNA was observed with SF-1 overexpression (Fig. 3B). Altogether, these results suggest that StarD7 expression is transcriptionally activated by SF-1 in JEG-3 cells, through functional responsive elements located at −792/−785.
Fig. 3.
Expression levels of SF-1 affect StarD7 promoter activity. A, JEG-3 cells were transiently cotransfected with 938StarD7Luc, 673StarD7Luc, or 312StarD7Luc and phRL-TK Renilla constructs and SF-1 expression plasmid as indicated. Relative luciferase reporter gene activity was normalized against the Renilla activity, and results were expressed as the fold increase related to the activity in cells cotransfected with the empty expression vector defined as 1-fold. The values represent the mean ± sem of triplicate experiments. B, Quantitative real-time PCR of StarD7. Total RNA was extracted from JEG-3 cells 24 h after transfection with SF-1 expression plasmid as indicated. StarD7 mRNA level was determined by real-time RT-PCR, normalized against cyclophilin A using the comparative 2−ΔΔCt method (57). Values for StarD7 mRNA represent the mean ± sem of triplicate experiments. *, P < 0.05 compared with cells cotransfected with the empty expression vector.
cAMP and SF-1 mediate StarD7 promoter response
We next examined the ability of cAMP signaling pathway to modulate StarD7 transcriptional response to the SF-1 transcription factor. JEG-3 cells transfected with the 938StarD7Luc construct were incubated in the presence or absence of the adenylyl cyclase activator FSK for 24 h. Treatment with FSK 10 μm increased basal promoter activity near 2-fold. However, in the presence of this agent, SF-1 overexpression (200 ng) further induced StarD7 promoter activity, suggesting a cooperative effect (Fig. 4).
Fig. 4.
cAMP and SF-1 mediate StarD7 promoter response. JEG-3 cells were transiently cotransfected with 938StarD7luc, and pRL-TK Renilla constructs and SF-1 expression plasmid (200 ng). Twenty-four hours after transfection, the medium was replaced, and where indicated, the cells were treated or not with 10 μm of FSK. Relative luciferase reporter gene activity normalized against the Renilla activity was calculated, and results were expressed as the fold increase related to the activity in cells cotransfected with the empty expression vector and without FSK treatment defined as 1-fold. *, P < 0.05 compared with cells transfected with the empty expression vector and without FSK treatment; #, P < 0.05 compared with cells transfected with the SF-1 expression vector and without FSK treatment.
SF-1 binds to the human StarD7 gene
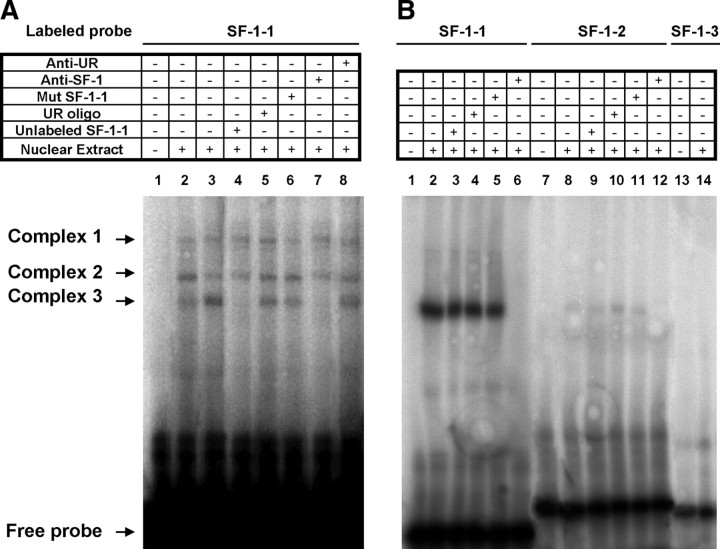
Because we demonstrated that the 5′ upstream region deletion of the 938StarD7Luc construct containing the −792/−785 SF-1 binding site (called SF-1-1) abolished the effect of SF-1 on StarD7 promoter activity (Fig. 3A), we decide to examine whether SF-1 present in JEG-3 cells is able to bind to this cis-element. To do this, a synthetic oligonucleotide probe encompassing the sequence containing this SF-1 consensus binding site was prepared and used in EMSA. In the presence of JEG-3 nuclear extracts made from cells transfected (Fig. 5A, lanes 3–8) or not (Fig. 5A, lane 2) with the SF-1 expression plasmid, three protein-DNA complexes were formed (Fig. 5A, lanes 2 and 3). Incubation of JEG-3 nuclear extracts made from cells transfected with the SF-1 expression plasmid resulted in an increased intensity of the specific complex 3. Formation of this complex was abolished by the addition of 500-fold molar excess of the unlabeled wild-type probe (Fig. 5A, lane 4), but not by 500-fold molar excess of an unrelated oligonucleotide sequence, confirming sequence-specific DNA binding (Fig. 5A, lane 5). Competition with a mutated SF-1-1 oligonucleotide did not modify significantly the intensity of this band (Fig. 5A, lane 6). Addition of anti-SF-1 antibody clearly abolished the complex (Fig. 5A, lane 7), effect that was not observed with an unrelated antibody (Fig. 5A, lane 8), confirming the presence of SF-1 in this complex. Similar results were observed when COS-7 nuclear extracts made from cells transfected with the SF-1 expression plasmid were used (Fig. 5B, lanes 1–6).
Fig. 5.
Identification of SF-1 binding motives in StarD7 promoter. EMSA of nuclear protein interactions with DNA fragments derived from the StarD7 promoter region. A, Ten micrograms of nuclear extract from JEG-3 cells transfected (lanes 3–8) or not (lane 2) with SF-1 transcription factor were incubated with the end-labeled double-stranded oligonucleotide containing the wild-type SF-1-1 binding site located at −792/−785. B, Ten micrograms of nuclear extract from COS-7 cells transfected with SF-1 transcription factor were incubated with the end-labeled double-stranded oligonucleotide containing the following wild-type SF-1-1 (lanes 1–6), SF-1-2 (lanes 7–12), or SF-1-3 (lanes 13 and 14) binding sites located at −792/−785, −493/−486, and −169/−162, respectively. Competition assays with unlabeled wild-type SF-1-1, unlabeled mutated SF-1-1, or unrelated double-stranded oligonucleotides were performed at 500-fold excess to determine binding specificity. Monoclonal antibody against SF-1 was used to create a supershift in EMSA. The presence (+) or absence (−) of unlabeled SF-1-1 oligonucleotide, unrelated oligonucleotide (UR oligo), SF-1 antibody (anti-SF-1), unlabeled mutated SF-1-1 (MutSF-1-1), unrelated antibody (anti-UR), or cell nuclear extract was as indicated. Free probe and protein-DNA complex are marked with arrows.
Furthermore, labeled oligonucleotide probes encompassing the sequence containing the SF-1-2 (−493/−486) or SF-1-3 (−169/−162) consensus binding sites were prepared and used in EMSA. In the presence of COS-7 nuclear extracts made from cells transfected with the SF-1 expression plasmid, a faint protein-SF-1-2-DNA complex was formed, indicating a very low ability of in vitro interaction of SF-1 transcription factor with the SF-1-2 consensus site (Fig. 5B, lanes 7–12). Additionally, no complex was detected with SF-1-3 oligonucleotide and COS-7 cell extracts (Fig. 5B, lanes 13 and 14). Similar results were observed when JEG-3 nuclear extracts made from cells transfected with the SF-1 expression plasmid were used (data not shown).
Site-directed mutagenesis defines the role of the −792/−785 SF-1-binding site in the StarD7 promoter activity
Although the 5′-deletion and gel shift assays (Figs. 3A and 5) suggested that SF-1 regulates StarD7 promoter activity interacting principally with SF-1-1 site, they did not directly address the in vivo role of this site in StarD7 expression. Therefore, JEG-3 cells were transfected with the SF-1 expression plasmid plus either the wild-type 938StarD7Luc reporter construct or the 938MutSF-1Luc construct harboring a mutation in this SF-1 motif. Even though the deletion of nucleotides located between −938 and −673 (Fig. 3A) abolished the SF-1 effect on StarD7 promoter activity, mutation of the −792/−785 SF-1-binding site (SF-1-1) exhibited only a slight diminution on the response to SF-1 (Fig. 6, A and B, black bars). These results suggest that, in addition to the −792 SF-1 binding motif (SF-1-1), the SF-1 transcription factor may be recruited to the StarD7 promoter region through different promoter elements or by the interaction with other proteins.
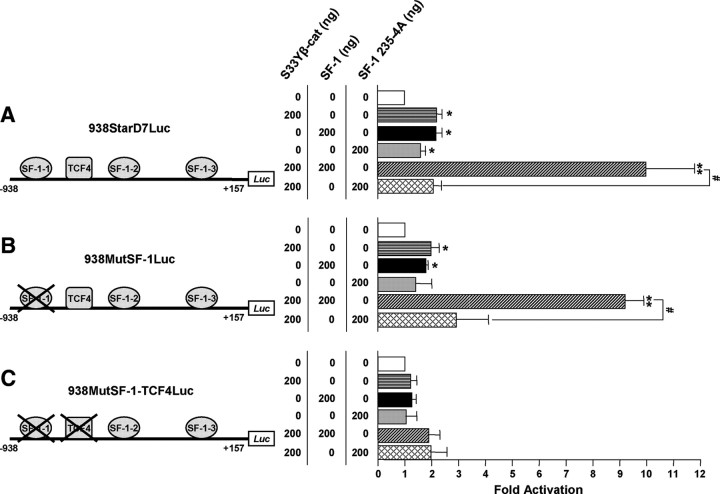
Fig. 6.
The StarD7 promoter is synergistically activated by SF-1 and β-catenin. The 938StarD7Luc (A), 938MutSF-1Luc (B), and 938MutSF-1-TCF4Luc (C) promoter constructs were transfected alone or in combination with S33Y β-catenin, SF-1 or SF-1 235-4A, or a combination of S33Y β-catenin with SF-1 or with SF-1 235-4A expression constructs (see Materials and Methods). Cells were lysed, and luciferase activity was determined 48 h after cotransfection. Relative luciferase reporter gene activity normalized against total proteins was calculated, and results were expressed as the fold increase related to the activity in cells cotransfected with the empty expression vector defined as 1-fold. The values represent the mean ± sem of triplicate experiments. *, P < 0.05; **, P < 0.01 compared with cells cotransfected with the empty expression vector; #, P < 0.05 as indicated.
Activation of StarD7 promoter by SF-1 and β-catenin overexpression
It has previously been shown that Wnt/β-catenin pathway activates the human StarD7 promoter (9). Based on these data and those from the literature that underline the ability of the Wnt glycoprotein family to affect SF-1-dependent transcription via its association with β-catenin, we focused on examining whether β-catenin modulates activity of SF-1-mediated transcription of the StarD7 gene.
Transactivation properties of β-catenin and SF-1 were assessed by transient cotransfection experiments in JEG-3 cells. The promoter activity of both the 938StarD7Luc and 938MutSF-1Luc constructs was activated more than 4-fold in the presence of overexpressed SF-1 and S33Y β-catenin (a constitutively activated form), compared with the activation observed with either SF-1 or S33Y β-catenin alone (Fig. 6, A and B, diagonally hatched bars vs. horizontally hatched and black bars). These results show that SF-1 and β-catenin act in synergy, rather than additively, to activate StarD7 gene transcription.
To investigate the importance of the SF-1-1 and TCF4/β-catenin binding sites, we generated the 938MutSF-1-TCF4Luc construct, in which both consensus binding motives were mutated. When JEG-3 cells were cotransfected with this mutant construct and the SF-1 or the S33Y β-catenin expression plasmids alone or a combination of both plasmids, StarD7 promoter activation was abolished, suggesting that the TCF4 consensus binding site (−614/−608) plays an important role in this activation (Fig. 6C, black, horizontal hatched, and diagonally hatched bars).
SF-1 ligand-binding domain mediates the functional interaction with β-catenin
It was reported that the SF-1 is able to interact with β-catenin via an acidic amino acid cluster located at 235–238 residues (25). We explored whether the synergist transactivation of StarD7 promoter depends on the interaction between SF-1 and β-catenin. To this end, JEG-3 cells were transfected with the SF-1 235-4A expression plasmid, where the 235–238-amino acid residues are substituted by four alanines, alone or together with the S33Y β-catenin plasmid and the following StarD7 luciferase report plasmids: 938StarD7Luc, 938MutSF-1Luc, and 938MutSF-1-TCF4Luc (Fig. 6). The data indicate that any significant difference in promoter induction of 938StarD7Luc-transfected JEG-3 cells was found either the wild-type SF-1 transcription factor or the SF-1 mutated version (which retains the DNA-binding motif) was used (Fig. 6A, gray and black bars). Conversely, the SF-1 235-4A expression plasmid was unable to induce the promoter activity of the 938MutSF-1Luc and the 938MutSF-1-TCF4Luc constructs, (Fig. 6, B and C, gray bars). These results suggest that in the absence of a functional SF-1-1 binding site, transactivation by SF-1 requires the interaction with the β-catenin coactivator. Further support to the existence of a physical interaction between both transcriptional factors, such as it was demonstrated for other promoter genes, was obtained in the assays performed to address the synergistic activation observed between SF-1 and β-catenin. Compared with the results found with the overexpression of the wild-type SF-1 transcription factor, the mutated SF-1 235-4A version was unable to increase the S33Y β-catenin-stimulated StarD7 promoter activity of all the StarD7 promoter constructs (Fig. 6, A–C, strippled bars). Collectively, these observations strongly indicate the involvement of TCF4/β-catenin and SF-1 proteins in the regulation of StarD7 promoter activity in JEG-3 cells.
SF-1 and β-catenin localize to the endogenous StarD7 promoter
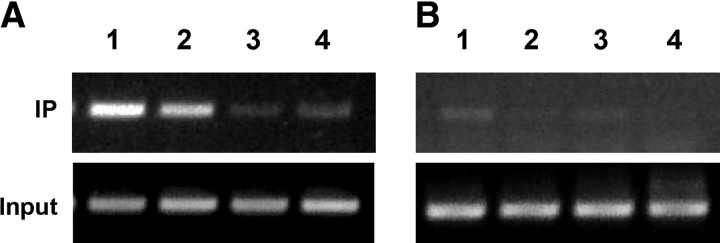
Finally, chromatin immunoprecipitation (ChIP) analysis of JEG-3 cells with β-catenin or SF-1 antibodies revealed consistent association of the SF-1 and β-catenin cofactor with StarD7 chromatin, suggesting that both proteins are present in the transcription complexes assembled on the endogenous StarD7 promoter (Fig. 7A, lanes 1 and 2). A weak background was observed when an unrelated antibody was used to immunoprecipitate StarD7 DNA (Fig. 7A, lane 3).
Fig. 7.
SF-1 transcription factor and β-catenin specifically binds to the StarD7 promoter. S33Y β-catenin and SF-1 transfected-JEG-3 cells cultured during 24 h were fixed with formaldehyde, and chromatin solutions were subjected to ChIP analyses using the following antibodies: lane 1, anti-SF-1; lane 2, anti-β-catenin; lane 3, unrelated anti-PSG antibody; and lane 4, control without antibody. The obtained DNA was analyzed by PCR using primers to amplify the StarD7 promoter region (A) or the control genomic region of iNOS (B). The results are representative of three independent experiments. IP, Immunoprecipitated samples.
To further demonstrate the specificity of the ChIP assay, we performed a PCR assay on the immunoprecipitated DNA using primers spanning 238 bp of the inducible nitric oxide synthase (iNOS) gene located 19 kb far away from its promoter sequence. As expected, this region was absent in the anti-SF-1 (Fig. 7B, lane 1) and anti-β-catenin immunoprecipitated DNA (Fig. 7B, lane 2). These findings indicate that SF-1/β-catenin/TCF4 specifically bind to the StarD7 promoter in vivo.
Discussion
Numerous reports have recently highlighted the role of the Wnt signaling pathway in implantation, placentation, and trophoblast differentiation (32–39). In this regard, we previously described that the activation of β-catenin, a transcriptional coactivator and member of the canonical Wnt signaling pathway, up-regulates StarD7 gene expression in JEG-3 cells (9). Here, we further characterized the SF-1 transcription factor effect on the human StarD7 promoter activity and analyzed the convergence of this factor and β-catenin in StarD7 gene regulation. Initially, it was demonstrated that this promoter activity is functionally dependent on transcription factors present in JEG-3 cells but not COS-7 cells. An increase in StarD7 promoter activity was observed in both JEG-3 and COS-7 cell lines when SF-1 was overexpressed, underscoring the importance that this transcription factor has in StarD7 gene expression. These results are consistent with the notion that the activation of StarD7 promoter is most likely dependent on the SF-1 amount. Moreover, a significant increase in endogenous StarD7 mRNA was observed in JEG-3 cell line after SF-1 overexpression.
Deletion analysis of the StarD7 promoter region revealed that the SF-1 consensus binding site located at −792/−785 is required for the transcriptional induction mediated by SF-1. Besides, it was demonstrated that this effect is increased by the addition of FSK, suggesting the involvement of protein kinase activity (PKA) activation. Although the exact mechanism of action of FSK reinforcing the effect of SF-1 on StarD7 gene expression was not explored, this could be exerted by an increase in the stability of SF-1 protein as it was demonstrated by Aesøy et al. (40). An interaction with CRE binding protein (CREB) (41) is another possibility, because several putative CRE binding sites are present in the StarD7 promoter region (Fig. 1). CREB is phosphorylated by PKA, and the interaction between SF-1 and phosphorylated CREB has been suggested as a mechanism for the cooperation between the SF-1 and cAMP (42). Similar SF-1 cooperation with other factors was reported for many cAMP-regulated genes, including StAR, another member of START domain family (43–46). Chen et al. (47) described that in response to the cAMP, p300 regulates SF-1 transcriptional activity by means of increased acetylation, DNA binding, and recruitment to nuclear foci. Furthermore, cAMP/PKA can regulate the stability and/or activity of glycogen synthase kinase 3β/β-catenin by phosphorylation events (48–50).
EMSA demonstrate that an oligonucleotide probe containing the SF-1-1 binding motif of StarD7 promoter region (−792/−785) can bind to JEG-3 cell protein extracts and also to proteins from JEG-3 and COS-7 SF-1-transfected cells. This in vitro protein-DNA complex was affected by an excess of the wild-type unlabeled SF-1-1 consensus sequence and by anti-SF-1 antibody addition. Moreover, only a very faint complex was found with SF-1-2 binding site. These data suggest that SF-1 transcription factor is capable of associating with the StarD7 promoter region mainly through SF-1-1 motif. However, disruption of the SF-1-1 binding site lead to an approximately 30% decrease in StarD7 reporter activity, demonstrating a modest contribution of this SF-1 cis-element in StarD7 gene expression, suggesting that the regulation by SF-1 transcription factor might occur, in addition to this cis-element, through different promoter interactions. Previously, we demonstrated the importance of a TCF4 binding site, located at −614/−608 of StarD7 promoter, in StarD7 gene expression regulated by Wnt signaling (9). The identification of SF-1-1 binding motif involved in StarD7 gene regulation and the several reports that documented cross-regulation of nuclear receptors and Wnt signaling pathways (21–29, 51) provided us the opportunity to characterize the roles of SF-1 and β-catenin coactivator in StarD7 gene transcription in more depth.
Evidence is accumulating that nuclear hormone receptor and canonical Wnt pathway interact at different levels regulating cell growth and proliferation, differentiation, apoptosis, and metastatic potential in numerous tissues (26, 51). Particularly, several reports indicate that β-catenin acts as a coactivator of SF-1 when it transduces Wnt signals to Dax1 (25), StAR (23, 49, 52), aromatase (27), GnRH receptor (53), Müllerian inhibiting substance type II receptor (22), LHB/Lhb (28, 29), and inhibin (21) target genes.
Our current data demonstrate the functional involvement of SF-1 and β-catenin in controlling transcription of the StarD7 gene. In particular, we demonstrate that β-catenin is able to synergize with SF-1 the StarD7 gene transcription activation.
In addition, the in vivo interaction between SF-1, β-catenin, and TCF4 to the StarD7 promoter was further supported by ChIP assay in conditions of SF-1- and β-catenin-induced StarD7 transcription, addressing a positive transcriptional regulation mediated by these proteins. Finally, site-directed mutagenesis revealed that SF-1-1 binding motif mutation had scarce effects on increasing StarD7 promoter activity exerted by a combination of SF-1 and β-catenin overexpression. On the contrary, mutation of both SF-1-1 and TCF4 binding elements abolished the transcriptional activation induced by individually SF-1 and β-catenin or both proteins together. In a similar way, cotransfection experiments performed with a mutated version of SF-1 transcription factor, unable to interact with β-catenin (25), lead to a full loss of the synergistically effect of both proteins on StarD7 gene expression. These findings disclose that the activation of StarD7 gene expression requires the binding of the β-catenin to TCF4 transcription factor, suggesting that β-catenin could function as a bridge between SF-1 and TCF4 forming a ternary complex, which would activate StarD7 expression.
Finally, accumulating evidence reveals that SF-1 has a lipid-binding pocket able to bind different phospholipids (15, 16, 44). A recent study suggests that the activity of SF-1 may be regulated through different potential ligands, influencing its transcriptional capacity (54). In this sense, it is interesting to point out the possibility of an interregulation pathway between the phospholipid-bound SF-1 and the capacity of StarD7 to interact with lipids.
In summary, we demonstrated that SF-1 induces StarD7 expression by interacting with the StarD7 promoter, and this effect is increased by the addition of FSK. In addition, these findings indicate that β-catenin synergizes with SF-1 to activate the StarD7 promoter. The auto/paracrine actions of Wnt signaling in combinations with SF-1 transcription factor on StarD7 gene expression may have important implications in the phospholipid uptake and transport contributing to the normal development of trophoblast cells.
Materials and Methods
Reagents
All reagents were purchased from Sigma, Co. (Buenos Aires, Argentina) unless otherwise indicated.
Antibodies
Rabbit anti-SF-1 antibody was kindly provided by Morohashi (National Institute for Basic Biology, Aichi, Japan). Mouse monoclonal anti-β-catenin (E-5) sc-7963 and mouse monoclonal anti pregnancy-specific glycoprotein (PSG)1 (BAP3) sc-59348 were purchased from Santa Cruz Biotechnology, Inc. (Santa Cruz, CA).
Plasmids and oligonucleotides
Bovine SF-1 transcription factor sequence cloned into the expression vector pSG5 was kindly provided by Keith L. Parker (University of Texas Southwestern Medical Center, Dallas, TX). The expression vectors pCINeoS33Yβ-catenin (encoding a constitutively activated β-catenin) and pCMX SF-1 (235-4A) (encoding a SF-1 form with the 235–238 amino acid residues substituted by four alanines) were kindly provided by Bert Vogelstein (The Johns Hopkins University Medical Institutions, Baltimore, MD) and Morohashi, (National Institute for Basic Biology), respectively. The oligonucleotides used in the PCR, EMSA, and ChIP assays are listed in Table 1. They were produced in an automated DNA synthesizer by Sigma, Co.
Table 1.
Oligonucleotide sequences
| Name | Sequence (5′ to 3′) |
|---|---|
| For cloning | |
| 938Fw | GCATTCCGGGATCAGAGTG |
| 673Fw | CCTCCAGCTGCATGACTCCT |
| 425Fw | TGGCGGTTATCACTGGGAGC |
| 312Fw | CACCTGGCAGGATTCTAACAGG |
| -121Rv | CCTCCTGCCCCTTCATGCAT |
| +157Rv | ACTGCTACACCAGGCACTCC |
| Mut SF-1Fw | CCTGGGTGGGGGCCATGGTTTCAGATGAGCCAGTTTATC |
| MutSF-1Rv | GATAAACTGGCTCATCTGAAACCATGGCCCCCACCCAGG |
| TCF4MutFv | GGCTAACTTATTAGTCCCCTGGGTGCAGTCTAGTTCCCTAA |
| TCF4MutRv | GGGAACTAGACTGCACCCAGGGGACTAATAAGTTAGCC |
| For EMSA | |
| SF-1–1 (S) | CTGGGTGGGGGCCACAAGGTCAGATG |
| SF-1–1 (AS) | CATCTGACCTTGTGGCCCCCACCCAG |
| SF-1–2 (S) | TCAGGAATG AACAAGGACAGAAGCAAGATGG |
| SF-1–2 (AS) | TTCCATCTTGCTTCTGTCCTTGTTCATTCCTG |
| SF-1–3 (S) | TCTCGCCGCCGAGCCACCTACCTTGCTAGC |
| SF-1–3 (AS) | TGCTAGCAAGGTAGGTGGCTCGGCGGC |
| NR (S) | AGGAAGGGTCCCTCTCACCAGGC |
| NR (AS) | GCCTGGTGAGAGGGACCCTTCCT |
| MutSF-1–1 (S) | CTGGGTGGGGGCCATGGTTTCAGATG |
| MutSF-1–1 (AS) | TCATCTGAAACCATGGCCCCCACCC |
| For RT-PCR | |
| SF-1Fw | GCATCTTGGGCTGCCTGCAG |
| SF-1Rv | CCTTGCCGTGCTGGACCTGG |
| GAPDHFw | GGTGAAGGTCGGAGTCAACG |
| GAPDHRv | GATCTCGCTCCTGGAAGATGG |
| For ChIP | |
| StarD7pFw | AGCCCACTATGAGACTGGAGTTTG |
| StarD7pRv | GGAACTAGACTGCCTTTGTAGGAC |
| iNOSFw | TGGCAGCATCAGAGGGGACC |
| iNOSRv | GCAGGACAGGGGACCAGATCGAA |
The consensus sequences are shown in boldface. The mutated nucleotides are shown underlined. S, Sense; AS, antisense.
Promoter constructs
The sequence of the 5′-flanking region of the human StarD7 gene (Gene ID, 56910) was used to obtain different StarD7 constructs by a PCR-based approach. To generate the −938/−121, −673/−121, and −425/−121 DNA fragments, the 5′-forward (Fw) 938Fw, 673Fw, 425Fw, and the −121 reverse (Rv) primer were included in a PCR using the human genomic RZPD 737H022155D clone (RZPD, Berlin, Germany) as template. The amplified products were cloned into the pCRII-TOPO vector followed by digestion with SacI and XhoI and subcloned into the SacI/XhoI sites of the pGL3-SV40 Promoter vector (Promega, Madison, WI). The resulting constructs were designated 938SV40Luc, 673SV40Luc and 425SV40Luc. Similarly, the −938/+157, −673/+157, and −312/+157 DNA fragments were amplified with the appropriate 5′-forward primers: 938Fw, 673Fw, 312Fw, and the +157Rv primer. In this case, the amplified products were cloned into the pCRII-TOPO vector (Promega) followed by digestion with KpnI and XhoI and subcloned into the KpnI/XhoI sites of the pGL3-Basic vector (Promega). The resulting constructs were designated 938StarD7Luc, 673StarD7Luc, and 312StarD7Luc.
A mutant construct in the SF-1-1 binding site located at the position −792/−785 was generated by a PCR-based approach with two amplification rounds using the 938StarD7Luc construct as template. Two primer sets were used for the first amplification round: MutSF-1Fw and +157Rv; and 938Fw and MutSF-1Rv. The products of the first amplification round were then subjected to a second round of amplification using the 938Fw and +157Rv primers. The resulting product was cloned into the pCRII-TOPO vector followed by digestion with KpnI and XhoI and subcloned into the KpnI /XhoI sites of the pGL3-Basic vector. The resulting construct was designated 938MutSF-1Luc. Likewise, to generate the construct 938MutSF-1-TCF4Luc bearing point mutations in the SF-1 and the TCF4 binding sites, the TCF4MutFv and TCF4MutRv oligonucleotides were included in the PCR using the 938MutSF-1Luc clone as template.
Cell lines and transient transfections
JEG-3 and COS-7 cell lines were cultured in DMEM, 10% (vol/vol) fetal calf serum, 100 μg/ml penicillin, and 100 μg/ml streptomycin (Invitrogen, Carlsbad, CA). The cells were seeded in 24 multiwell plates at 1 × 105 cells/ well, cultured for 24 h, and afterwards transfected with 2 μl of Lipofectamine (2000) and 400 ng of the different firefly luciferase reporter constructs. After 4 h, medium was replaced with DMEM-10% fetal calf serum and antibiotics and cultured for a further 24 or 48 h (as indicated) and treated or not with FSK 10 μm. Alternatively, the cells were cotransfected with different amounts of the expression vectors (as indicated) and 400 ng of the specified StarD7 promoter firefly luciferase reporter construct and incubated for a further 48 h. After that, the cells were harvested in 100 μl of Reporter Lysis Buffer (Promega), and firefly luciferase and control Renilla luciferase were simultaneously assayed, as described in the dual luciferase reporter assay system (Promega). Firefly luciferase activity was normalized to Renilla luciferase or total proteins as indicated.
Nuclear protein preparations for EMSA assays
JEG-3 and COS-7 cells (∼4 × 106) were washed twice with 4 ml of TNE buffer [50 mm Tris-HCl (pH 7.5), 150 mm NaCl, and 1 mm EDTA], scraped, and resuspended in 1 ml of TNE. Cells were centrifuged for 5 min at 1500 rpm at 4 C. The pellet was resuspended in 200 μl of lysis buffer [10 mm Tris-HCl (pH 7.5), 10 mm NaCl, 2 mm MgCl2, containing 1 mm phenylmethylsulfonylfluoride, and 2 μl of inhibitor protease cocktail (Sigma, Co.)] and 200 μl of 1% vol/vol Nonidet P40. The suspension was incubated for 10 min on ice and centrifuged for 3 min at 3000 rpm at 4 C. After centrifugation, the supernatants were taken as cytosolic extracts and frozen. The nuclear pellet from lysis was washed briefly in 200 μl of TNE buffer and then subjected to high salt extraction with 45 μl of 0.2 m NaCl and 45 μl of 0.6 m NaCl. Subsequently, 180 μl of H2O (supplemented with 1 mm dithiothreitol, 1 mm phenylmethylsulfonylfluoride, and 2 μl protease inhibitors) were added. Nuclear extracts were cleared for 10 min at 10,000 rpm at 4 C and supernatants frozen at −80 C. The protein concentration was determined using the Bradford method.
Electrophoretic mobility shift assay
EMSA were performed following the previously described protocol (55). Complementary SF-1-1, SF-1-2, and SF-1-3 oligonucleotides were annealed and radiolabeled by the fill-in reaction with Klenow in the presence of [α32P]ATP. The specific activity of radiolabeled probes was determined. Nuclear extracts obtained from JEG-3 or COS-7 cells, previously transfected or not with SF-1 expression vector, were incubated with radiolabeled probes at room temperature for 20 min in 10 μl of binding buffer containing 10 mm HEPES (pH 7.9), 120 mm KCl, 0.1 mm dithiothreitol, 0.125 mm EDTA, 10% glycerol, 1 μg of salmon sperm DNA, and 1 μg of poly(dI-dC). The samples were electrophoresed in 5% polyacrylamide gel, followed by drying of the gel and autoradiography. Competition for binding was performed by adding a 100- to 500-fold excess of unlabeled DNA, 100- to 500-fold excess of unrelated DNA, and 100- to 500-fold excess of MutSF-1-1 DNA, and supershift was detected by 1 μg anti-SF-1 antibody.
Semiquantitative RT-PCR
Single-stranded cDNA were synthesized in a single tube with random primers (Invitrogen) in 20 μl of reaction. Briefly, 1 μg of total RNA was incubated with these primers (1.25 ng/μl), and the RT reaction was performed as previously described (5). PCR amplification was carried out with two sets of PCR primers, including the SF-1Fw and SF-1Rv primers for SF-1 gene, and the glyceraldehyde-3-phosphate dehydrogenase (GAPDH)Fw and GAPDHRv primers for the control GAPDH gene. The amplification protocol for all the cDNA species included: 1 cycle at 95 C for 3 min; 20–35 cycles at 95 C for 30 sec, 60 C for 30 sec, and 72 C for 30 sec; 1 cycle at 72 C for 10 min; and 1 cycle at 4 C for 10 min. Experiments were conducted to confirm that all PCR amplifications were in the linear range. The amplification products were visualized by ethidium bromide staining.
Quantitative RT-PCR
StarD7 expression was quantified by real-time PCR (ABI 7500; Applied Biosystems, Carlsbad, CA) with Sequence Detection Software version 1.4. Experiments were performed as previously described (9).
Chromatin immunoprecipitation
ChIP assays were performed following the protocol outlined by the manufacturer (Upstate, Waltham, MA) with minor modifications (9). Briefly, approximately 1 × 106 JEG-3 cells previously transfected with SF-1 and constitutively active S33Y β-catenin expression plasmids were fixed in culture medium with formaldehyde (final concentration of 1%) for 20 min at 37 C to cross-link chromatin and nuclear proteins. After sonication, the nuclear lysates were immunoprecipitated with a rabbit anti-SF-1 antibody or a mouse monoclonal anti-β-catenin antibody. A mouse monoclonal anti-PSG antibody was used as a negative control. After washing, elution, reverse cross-linking, and purification, approximately one-twentieth of the purified DNA (2 μl) was used in each PCR. The StarD7pFw and StarD7pRv primers amplify a 396-bp DNA fragment that contains the SF-1-1 (−792/−785) and TCF4 (−614/−608) consensus binding elements of the human StarD7 gene promoter, based on the human StarD7 gene sequence (GenBank access no.: NT_026970.9). The control iNOSFw and iNOSRv primers amplify a 238-bp fragment that is part of exons 8 and 9 of the human iNOS gene, located approximately 19 kb from the transcription start point, which does not contain SF-1 and TCF binding sites.
Data analysis
Pair-wise comparison between groups was evaluated with a two-tailed Student's t test. Data are expressed as mean ± sem from at least three independent experiments. Significance was taken as P < 0.05.
Acknowledgments
We thank Dr. Bert Volgestein (The Johns Hopkins University Medical Institutions, Baltimore, MD) for providing the pCINeoS33Yβ-catenin expression plasmid, Dr. Keith L. Parker (University of Texas Southwestern Medical Center, Dallas, TX) for kindly providing the SF-1 expression plasmid, and Dr. Morohashi (National Institute for Basic Biology, Aichi, Japan) for providing pCMX SF-1 (235-4A) and anti-SF-1 antibody.
This work was supported by fellowships and grants from the Consejo Nacional de Investigaciones Científicas y Técnicas de Argentina (V.R., J.F.-M., S.A.), the Agencia Nacional de Promoción Ciencia y Técnica (V.R.), the Ministerio de Ciencia y Tecnología de la Provincia de Córdoba and the Secretaría de Ciencia y Tecnología de la Universidad Nacional de Córdoba (S.A.). S.G-R. and G.M.P-D. are Career Investigators of CONICET.
Disclosure Summary: The authors have nothing to disclose.
Footnotes
- ChIP
- Chromatin immunoprecipitation
- CRE
- cAMP response element
- CREB
- cAMP response element binding protein
- FSK
- forskolin
- Fw
- forward
- GAPDH
- glyceraldehyde-3-phosphate dehydrogenase
- iNOS
- inducible nitric oxide synthase
- PKA
- protein kinase activity
- PSG
- pregnancy-specific glycoprotein
- Rv
- reverse
- SF-1
- steroidogenic factor 1
- StAR
- steroidogenic acute regulatory protein
- StarD
- StAR-related lipid transfer domain
- SV40
- simian virus 40
- TCF
- T cell-specific transcription factor.
References
- 1. Huppertz B , Frank HG , Kingdom JC , Reister F , Kaufmann P. 1998. Villous cytotrophoblast regulation of the syncytial apoptotic cascade in the human placenta. Histochem Cell Biol 110:495–508 [DOI] [PubMed] [Google Scholar]
- 2. Holthuis JC , Levine TP. 2005. Lipid traffic: floppy drives and a superhighway. Nat Rev Mol Cell Biol 6:209–220 [DOI] [PubMed] [Google Scholar]
- 3. Ponting CP , Aravind L. 1999. START: a lipid-binding domain in StAR, HD-ZIP and signalling proteins. Trends Biochem Sci 24:130–132 [DOI] [PubMed] [Google Scholar]
- 4. Tsujishita Y , Hurley JH. 2000. Structure and lipid transport mechanism of a StAR-related domain. Nat Struct Biol 7:408–414 [DOI] [PubMed] [Google Scholar]
- 5. Durand S , Angeletti S , Genti-Raimondi S. 2004. GTT1/StarD7, a novel phosphatidylcholine transfer protein-like highly expressed in gestational trophoblastic tumour: cloning and characterization. Placenta 25:37–44 [DOI] [PubMed] [Google Scholar]
- 6. Angeletti S , Rena V , Nores R , Fretes R , Panzetta-Dutari GM , Genti-Raimondi S. 2008. Expression and localization of StarD7 in trophoblast cells. Placenta 29:396–404 [DOI] [PubMed] [Google Scholar]
- 7. Angeletti S , Maggio B , Genti-Raimondi S. 2004. Surface activity and interaction of StarD7 with phospholipid monolayers. Biochem Biophys Res Commun 314:181–185 [DOI] [PubMed] [Google Scholar]
- 8. Horibata Y , Sugimoto H. 2010. StarD7 mediates the intracellular trafficking of phosphatidylcholine to mitochondria. J Biol Chem 285:7358–7365 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Rena V , Angeletti S , Panzetta-Dutari G , Genti-Raimondi S. 2009. Activation of β-catenin signalling increases StarD7 gene expression in JEG-3 cells. Placenta 30:876–883 [DOI] [PubMed] [Google Scholar]
- 10. Parker KL , Rice DA , Lala DS , Ikeda Y , Luo X , Wong M , Bakke M , Zhao L , Frigeri C , Hanley NA , Stallings N , Schimmer BP. 2002. Steroidogenic factor 1: an essential mediator of endocrine development. Recent Prog Horm Res 57:19–36 [DOI] [PubMed] [Google Scholar]
- 11. Sadovsky Y , Dorn C. 2000. Function of steroidogenic factor 1 during development and differentiation of the reproductive system. Rev Reprod 5:136–142 [DOI] [PubMed] [Google Scholar]
- 12. Lavoie HA , King SR. 2009. Transcriptional regulation of steroidogenic genes: STARD1, CYP11A1 and HSD3B. Exp Biol Med 234:880–907 [DOI] [PubMed] [Google Scholar]
- 13. Parker KL , Schimmer BP. 1997. Steroidogenic factor 1: a key determinant of endocrine development and function. Endocr Rev 18:361–377 [DOI] [PubMed] [Google Scholar]
- 14. Val P , Aigueperse C , Ragazzon B , Veyssière G , Lefrançois-Martinez AM , Martinez A. 2004. Adrenocorticotropin/3′,5′-cyclic AMP-mediated transcription of the scavenger akr1–b7 gene in adrenocortical cells is dependent on three functionally distinct steroidogenic factor-1-responsive elements. Endocrinology 145:508–518 [DOI] [PubMed] [Google Scholar]
- 15. Forman BM. 2005. Are those phospholipids in your pocket? Cell Metab 1:153–155 [DOI] [PubMed] [Google Scholar]
- 16. Li Y , Choi M , Cavey G , Daugherty J , Suino K , Kovach A , Bingham NC , Kliewer SA , Xu HE. 2005. Crystallographic identification and functional characterization of phospholipids as ligands for the orphan nuclear receptor steroidogenic factor-1. Mol Cell 17:491–502 [DOI] [PubMed] [Google Scholar]
- 17. Urs AN , Dammer E , Kelly S , Wang E , Merrill AH , Sewer MB. 2007. Steroidogenic factor-1 is a sphingolipid binding protein. Mol Cell Endocrinol 265- 266:174–178 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Wang W , Zhang C , Marimuthu A , Krupka HI , Tabrizizad M , Shelloe R , Mehra U , Eng K , Nguyen H , Settachatgul C , Powell B , Milburn MV , West BL. 2005. The crystal structures of human steroidogenic factor-1 and liver receptor homologue-1. Proc Natl Acad Sci USA 102:7505–7510 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Fuerer C , Nusse R , Ten Berge D. 2008. Wnt signalling in development and disease. Max Delbruck center for molecular medicine meeting on Wnt signalling in development and disease. EMBO Rep 9:134–138 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Mikels AJ , Nusse R. 2006. Wnts as ligands: processing, secretion and reception. Oncogene 25:7461–7468 [DOI] [PubMed] [Google Scholar]
- 21. Gummow BM , Winnay JN , Hammer GD. 2003. Convergence of Wnt signalling and steroidogenic factor-1 (SF-1) on transcription of the rat inhibin α gene. J Biol Chem 278:26572–26579 [DOI] [PubMed] [Google Scholar]
- 22. Hossain A , Saunders GF. 2003. Synergistic cooperation between the β-catenin signalling pathway and steroidogenic factor 1 in the activation of the Mullerian inhibiting substance type II receptor. J Biol Chem 278:26511–26516 [DOI] [PubMed] [Google Scholar]
- 23. Jordan BK , Shen JH , Olaso R , Ingraham HA , Vilain E. 2003. Wnt4 overexpression disrupts normal testicular vasculature and inhibits testosterone synthesis by repressing steroidogenic factor 1/β-catenin synergy. Proc Natl Acad Sci USA 100:10866–10871 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Kennell JA , O'Leary EE , Gummow BM , Hammer GD , MacDougald OA. 2003. T-cell factor 4N (TCF-4N), a novel isoform of mouse TCF-4, synergizes with β-catenin to coactivate C/EBPα and steroidogenic factor 1 transcription factors. Mol Cell Biol 23:5366–5375 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Mizusaki H , Kawabe K , Mukai T , Ariyoshi E , Kasahara M , Yoshioka H , Swain A , Morohashi K. 2003. Dax-1 (dosage-sensitive sex reversal-adrenal hypoplasia congenita critical region on the X chromosome, gene 1) gene transcription is regulated by wnt4 in the female developing gonad. Mol Endocrinol 17:507–519 [DOI] [PubMed] [Google Scholar]
- 26. Mulholland DJ , Dedhar S , Coetzee GA , Nelson CC. 2005. Interaction of nuclear receptors with the Wnt/β-catenin/Tcf signalling axis: Wnt you like to know? Endocr Rev 26:898–915 [DOI] [PubMed] [Google Scholar]
- 27. Parakh TN , Hernandez JA , Grammer JC , Weck J , Hunzicker-Dunn M , Zeleznik AJ , Nilson JH. 2006. Follicle-stimulating hormone/cAMP regulation of aromatase gene expression requires β-catenin. Proc Natl Acad Sci USA 103:12435–12440 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Salisbury TB , Binder AK , Grammer JC , Nilson JH. 2007. Maximal activity of the luteinizing hormone β-subunit gene requires β-catenin. Mol Endocrinol 21:963–971 [DOI] [PubMed] [Google Scholar]
- 29. Salisbury TB , Binder AK , Nilson JH. 2008. Welcoming β-catenin to the gonadotropin-releasing hormone transcriptional network in gonadotropes. Mol Endocrinol 22:1295–1303 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Bamberger AM , Ezzat S , Cao B , Wong M , Parker KL , Schulte HM , Asa SL. 1996. Expression of steroidogenic factor-1 (SF-1) mRNA and protein in the human placenta. Mol Hum Reprod 2:457–461 [DOI] [PubMed] [Google Scholar]
- 31. Ngan ES , Cheng PK , Leung PC , Chow BK. 1999. Steroidogenic factor-1 interacts with a gonadotrope-specific element within the first exon of the human gonadotropin-releasing hormone receptor gene to mediate gonadotrope-specific expression. Endocrinology 140:2452–2462 [DOI] [PubMed] [Google Scholar]
- 32. Fitzgerald JS , Germeyer A , Huppertz B , Jeschke U , Knöfler M , Moser G , Scholz C , Sonderegger S , Toth B , Markert UR. 2010. Governing the invasive trophoblast: current aspects on intra- and extracellular regulation. Am J Reprod Immunol 63:492–505 [DOI] [PubMed] [Google Scholar]
- 33. Getsios S , Chen GT , MacCalman CD. 2000. Regulation of β-catenin mRNA and protein levels in human villous cytotrophoblasts undergoing aggregation and fusion in vitro: correlation with E-cadherin expression. J Reprod Fertil 119:59–68 [DOI] [PubMed] [Google Scholar]
- 34. Hewitt DP , Mark PJ , Dharmarajan AM , Waddell BJ. 2006. Placental expression of secreted frizzled related protein-4 in the rat and the impact of glucocorticoid-induced fetal and placental growth restriction. Biol Reprod 75:75–81 [DOI] [PubMed] [Google Scholar]
- 35. Pollheimer J , Loregger T , Sonderegger S , Saleh L , Bauer S , Bilban M , Czerwenka K , Husslein P , Knöfler M. 2006. Activation of the canonical wingless/T-cell factor signalling pathway promotes invasive differentiation of human trophoblast. Am J Pathol 168:1134–1147 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36. Sonderegger S , Haslinger P , Sabri A , Leisser C , Otten JV , Fiala C , Knöfler M. 2010. Wingless (Wnt)-3A induces trophoblast migration and matrix metalloproteinase-2 secretion through canonical Wnt signalling and protein kinase B/AKT activation. Endocrinology 151:211–220 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37. Sonderegger S , Husslein H , Leisser C , Knöfler M. 2007. Complex expression pattern of Wnt ligands and frizzled receptors in human placenta and its trophoblast subtypes. Placenta 28(Suppl A):S97–S102 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38. Sonderegger S , Pollheimer J , Knöfler M. 2010. Wnt signalling in implantation, decidualisation and placental differentiation—review. Placenta 31:839–847 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39. Wong NC , Novakovic B , Weinrich B , Dewi C , Andronikos R , Sibson M , Macrae F , Morley R , Pertile MD , Craig JM , Saffery R. 2008. Methylation of the adenomatous polyposis coli (APC) gene in human placenta and hypermethylation in choriocarcinoma cells. Cancer Lett 268:56–62 [DOI] [PubMed] [Google Scholar]
- 40. Aesøy R , Mellgren G , Morohashi K , Lund J. 2002. Activation of cAMP-dependent protein kinase increases the protein level of steroidogenic factor-1. Endocrinology 143:295–303 [DOI] [PubMed] [Google Scholar]
- 41. Carlone DL , Richards JS. 1997. Functional interactions, phosphorylation, and levels of 3′,5′-cyclic adenosine monophosphate-regulatory element binding protein and steroidogenic factor-1 mediate hormone-regulated and constitutive expression of aromatase in gonadal cells. Mol Endocrinol 11:292–304 [DOI] [PubMed] [Google Scholar]
- 42. Ito M , Park Y , Weck J , Mayo KE , Jameson JL. 2000. Synergistic activation of the inhibin α-promoter by steroidogenic factor-1 and cyclic adenosine 3′,5′-monophosphate. Mol Endocrinol 14:66–81 [DOI] [PubMed] [Google Scholar]
- 43. Clemens JW , Lala DS , Parker KL , Richards JS. 1994. Steroidogenic factor-1 binding and transcriptional activity of the cholesterol side-chain cleavage promoter in rat granulosa cells. Endocrinology 134:1499–1508 [DOI] [PubMed] [Google Scholar]
- 44. Hoivik EA , Lewis AE , Aumo L , Bakke M. 2010. Molecular aspects of steroidogenic factor 1 (SF-1). Mol Cell Endocrinol 315:27–39 [DOI] [PubMed] [Google Scholar]
- 45. Liu Z , Simpson ER. 1997. Steroidogenic factor 1 (SF-1) and SP1 are required for regulation of bovine CYP11A gene expression in bovine luteal cells and adrenal Y1 cells. Mol Endocrinol 11:127–137 [DOI] [PubMed] [Google Scholar]
- 46. Manna PR , Eubank DW , Stocco DM. 2004. Assessment of the role of activator protein-1 on transcription of the mouse steroidogenic acute regulatory protein gene. Mol Endocrinol 18:558–573 [DOI] [PubMed] [Google Scholar]
- 47. Chen WY , Juan LJ , Chung BC. 2005. SF-1 (nuclear receptor 5A1) activity is activated by cyclic AMP via p300-mediated recruitment to active foci, acetylation, and increased DNA binding. Mol Cell Biol 25:10442–10453 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48. Hino S , Tanji C , Nakayama KI , Kikuchi A. 2005. Phosphorylation of β-catenin by cyclic AMP-dependent protein kinase stabilizes β-catenin through inhibition of its ubiquitination. Mol Cell Biol 25:9063–9072 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49. Roy L , McDonald CA , Jiang C , Maroni D , Zeleznik AJ , Wyatt TA , Hou X , Davis JS. 2009. Convergence of 3′,5′-cyclic adenosine 5′-monophosphate/protein kinase A and glycogen synthase kinase-3β/β-catenin signalling in corpus luteum progesterone synthesis. Endocrinology 150:5036–5045 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50. Taurin S , Sandbo N , Qin Y , Browning D , Dulin NO. 2006. Phosphorylation of β-catenin by cyclic AMP-dependent protein kinase. J Biol Chem 281:9971–9976 [DOI] [PubMed] [Google Scholar]
- 51. Beildeck ME , Gelmann EP , Byers SW. 2010. Cross-regulation of signalling pathways: an example of nuclear hormone receptors and the canonical Wnt pathway. Exp Cell Res 316:1763–1772 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52. Schinner S , Willenberg HS , Krause D , Schott M , Lamounier-Zepter V , Krug AW , Ehrhart-Bornstein M , Bornstein SR , Scherbaum WA. 2007. Adipocyte-derived products induce the transcription of the StAR promoter and stimulate aldosterone and cortisol secretion from adrenocortical cells through the Wnt-signalling pathway. Int J Obes 31:864–870 [DOI] [PubMed] [Google Scholar]
- 53. Gardner S , Stavrou E , Rischitor PE , Faccenda E , Pawson AJ. 2010. Targeting mediators of Wnt signalling pathways by GnRH in gonadotropes. J Mol Endocrinol 44:195–201 [DOI] [PubMed] [Google Scholar]
- 54. Sablin EP , Blind RD , Krylova IN , Ingraham JG , Cai F , Williams JD , Fletterick RJ , Ingraham HA. 2009. Structure of SF-1 bound by different phospholipids: evidence for regulatory ligands. Mol Endocrinol 23:25–34 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55. Nores R , Blanchon L , López-Díaz F , Bocco JL , Patrito LC , Sapin V , Panzetta-Dutari GM. 2004. Transcriptional control of the human pregnancy-specific glycoprotein 5 gene is dependent on two GT-boxes recognized by the ubiquitous specificity protein 1 (Sp1) transcription factor. Placenta 25:9–19 [DOI] [PubMed] [Google Scholar]
- 56. Quandt K , Frech K , Karas H , Wingender E , Werner T. 1995. MatInd and MatInspector: new fast and versatile tools for detection of consensus matches in nucleotide sequence data. Nucleic Acids Res 23:4878–4684 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57. Livak KJ , Schmittgen TD. 2001. Analysis of relative gene expression data using real-time quantitative PCR and the 2(−ΔΔC(T)) Method. Methods 25:402–408 [DOI] [PubMed] [Google Scholar]