The www.ssfa-gphr.de platform compares Glycoprotein Hormone Receptors and their available mutations. Novel tools enable improved sequence-structure-function analysis and give new clues for intramolecular activation mechanisms.
Abstract
The SSFA-GPHR (Sequence-Structure-Function-Analysis of Glycoprotein Hormone Receptors) database provides a comprehensive set of mutation data for the glycoprotein hormone receptors (covering the lutropin, the FSH, and the TSH receptors). Moreover, it provides a platform for comparison and investigation of these homologous receptors and helps in understanding protein malfunctions associated with several diseases. Besides extending the data set (> 1100 mutations), the database has been completely redesigned and several novel features and analysis tools have been added to the web site. These tools allow the focused extraction of semiquantitative mutant data from the GPHR subtypes and different experimental approaches. Functional and structural data of the GPHRs are now linked interactively at the web interface, and new tools for data visualization (on three-dimensional protein structures) are provided. The interpretation of functional findings is supported by receptor morphings simulating intramolecular changes during the activation process, which thus help to trace the potential function of each amino acid and provide clues to the local structural environment, including potentially relocated spatial counterpart residues. Furthermore, double and triple mutations are newly included to allow the analysis of their functional effects related to their spatial interrelationship in structures or homology models. A new important feature is the search option and data visualization by interactive and user-defined snake-plots. These new tools allow fast and easy searches for specific functional data and thereby give deeper insights in the mechanisms of hormone binding, signal transduction, and signaling regulation. The web application “Sequence-Structure-Function-Analysis of GPHRs” is accessible on the internet at http://www.ssfa-gphr.de/.
The family of glycoprotein hormone receptors (GPHRs), a subgroup of the G protein-coupled receptors (GPCRs), consists of the TSH receptor (TSHR), FSH receptor (FSHR), and LH and choriogonadotrophin receptor (LHCGR). Many experimental and structural studies have been published in recent decades investigating signaling mechanisms, functions, and also malfunctions of the GPHRs (1–14). A huge number of gain- and loss-of-function mutations in GPHRs are known to cause several diseases (15). In fact, the GPHRs are one of the most studied rhodopsin-like GPCR subgroups. The TSH regulates growth and function of thyroid follicular cells. The gonadotropins, LH and choriogonadotrophin and FSH, play an important role in human reproduction. The collection, molecular analysis, and the comparison of different GPHR-related information are therefore important and enable a comprehensive understanding of GPHR functionalities.
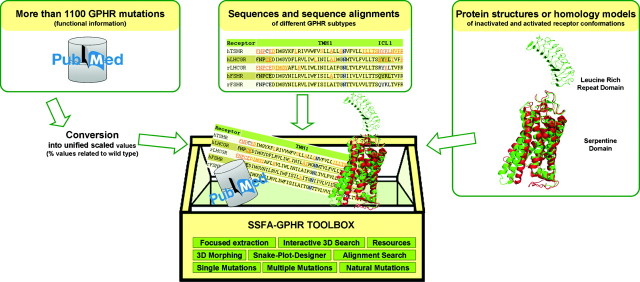
The basic methodology and idea of the SSFA-GPHR (Sequence-Structure-Function-Analysis of Glycoprotein Hormone Receptors) database has been described previously (16). Our freely accessible web application includes data from pathogenic mutations as well as from mutagenesis experiments of GPHRs (>1100 mutations). Strikingly, the functional data are converted into relational percentage values and therefore, in contrast to other information resources, our system allows the comparison, classification, and focused extraction of semiquantitative mutant data from different GPHR subtypes and different experimental approaches, which facilitates further analysis (Fig. 1). This provides a platform for investigation of molecular mechanisms and a better understanding of protein malfunctions. Recently, we described one of the extensions in detail, namely applications for the analysis of pathogenic mutations in three-dimensional (3D) structures (17). Here, we will focus on the complete renewal of the previous database version which includes the addition of several new features as extensions. Our goal is to facilitate the linkage between such unified functional and structural data of the GPHRs interactively at the web interface. We provide new tools for data visualization and interpretation like receptor morphings, which simulate the intramolecular activation process of the 3D protein structure (Fig. 2).
Fig. 1.
Basic architecture of the web application. More than 1100 mutations (functional information) of GPHRs are converted into unified scaled values (% values compared with wild type). This permits a semiquantitative comparison using computational approaches (SSFA-GPHR toolbox), where the mutation data set of different GPHR subtypes together with sequences and sequence alignments are linked to protein structures or homology models of inactivated and activated receptor conformations, resulting in the identification of sequence-structure-function relationships of the GPHRs.
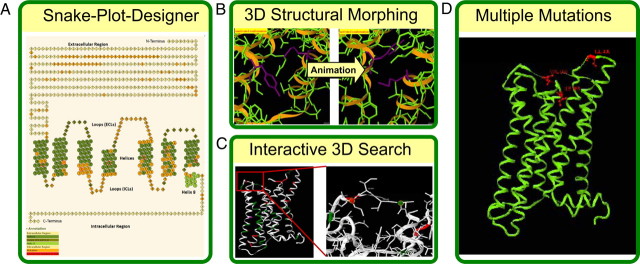
Fig. 2.
Overview of novel features and tools of the web application update. A, 2D – Snake-Plot – Search representing the requested mutation data in a 2D plot of each GPHR sequence and hyperlinking those to the detailed mutation information. The appearance can be modified interactively by the user. B, The 3D Structural Morphing between basal and activated receptor conformations permits an interactive trace of any amino acid changes, including those of the surrounding residues during a potential molecular activation event. C, The Interactive 3D Search allows picking of any particular amino acid positions of interest on a 3D structure or homology model by providing the user with information about available side-chain variations and their particular functional effects that can be mapped onto the selected 3D structure or model. D, Analysis of Multiple Mutations mapped onto 3D structures provides insight into functional effects, such as whether they result from the cooperative effect of double or triple mutations.
Results and Discussion
The database-linked web site is subdivided into five main areas: 1) Description, 2) Search Single Mutations, 3) Search Multiple Mutations, 4) Natural Mutations, and 5) Resources. In brief, the Description Section provides an introduction to the functional data and strategy of the web application, the different search modes and analysis features available, as well as technical hints. The second section, Search Single Mutations, allows querying all data on single point mutations via different tools:
An Alignment Search displays the GPHR sequences aligned to each other, providing a quick means of searching and an overview of available mutations.
The detailed search mode Search by Parameter offers more sophisticated search options: 1) mutations at a specific position, 2) mutations within a restricted region, 3) naturally occurring mutations identified and described to be potentially related to pathogenic activation or inactivation, 4) mutations involving substitution of amino acids of given properties (e.g., a hydrophilic residue is substituted for a hydrophobic), and 5) mutations characterized by a certain functional property.
An alignment which highlights the positions of known constitutively activating mutations on the GPHR sequences is provided, involving amino acid substitutions that are characterized by increased constitutive Gαs activity [cAMP signaling activity ≥180% compared with wild type (100%)].
The 2D – Snake-Plot – Search offers a two-dimensional (2D) “Snake-Plot” representation of the GPHR sequence combined with the functional mutagenesis data.
The 3D – Structural Search links mutagenesis data with structural information.
The newly introduced section, Search Multiple Mutations, has three different entry points to query the database:
1) Double Mutation Overview, 2) Triple Mutation Overview, and 3) Search Multiple Mutations in a distinct position or region.
The Natural Mutations section offers an Alignment-based Search mode, a Tabulated Overview, and a 3D Structural Search for extracting naturally occurring mutations.
All search results are displayed with facilities for further analysis, for example highlighting features, projection onto 3D receptor models or crystal structures, and 3D morphings of receptor models. Morphing simulations permit tracing of potential structural changes from the basal to an activated receptor conformation for any individual wild-type residue. In addition, the results are linked to external web resources [PubMed, UniProtKB/Swiss-Prot (18)] offering online information about the original publication, protein sequence, and annotation data. Important additional information are the template X-ray structures, structural models and morphings that are used and provided in this application, the Snake-Plot-Designer for creating an individual 2D representation of the GPHR sequence of interest, an overview of the GPHR numbering schemes and statistics summarizing the data set for each receptor, and the publications used for analysis.
An innovative new tool: the interactive structure-based search
We developed a search option for mutagenesis data linked directly on the 3D protein structure (or structural model) that allows interactive searching and visualization of functional data and mutant information. This search request is enabled as interactive picking of any particular amino acid position on a 3D structure or homology model followed by providing the user with information about available side-chain variations and their particular functional effects which can be mapped on the 3D structure. This novel feature can be used to identify interacting residues as counterparts and to evaluate potential mechanisms of molecular malfunction caused by specific mutations. We recently described the interactive 3D-Search applet in detail, demonstrating its usefulness in the analysis of naturally occurring mutations (17).
3D structural morphings
To study the conformational transition between a basal and activated state of the serpentine domain of the humTSHR, humFSHR, ratFSHR, humLHCGR, and ratLHCGR (http://www.ssfa-gphr.de/ → Resources → Structural Models), we provide a new applet of so-called structural morphings. The motions in the morphing provide a useful tool for visualizing and analyzing the structural changes that might occur during activation of a GPHR. The morphing is displayed using the Jmol viewer (http://jmol.sourceforge.net/). The appearance of the applet can be changed using the Jmol menu or by using the predefined options for zooming in or coloring of selected side-chains to analyze specific residues.
This tool aims to clarify wild-type amino acid functionalities for each individual residue during the activation process and to give reasonable explanations for significant mutation data like inactivation of the receptor. The utility of the analysis of conformational changes during a possible activation process of the receptor is described for example in Kleinau et al. (20). In this study, so-called “silencing” or “constitutively inactivating mutations” in the TSHR were extracted from the SSFA-GPHR data set and mapped to either the inactive or the active receptor conformation. The aim was to explore molecular reasons for impaired basal activity attributable to specific mutations as observed also for other basally active GPCRs and homologous GPHRs (21, 22). It was shown that the wild-type amino acids are involved in stabilization of the active state, which has indicated important points for receptor activation. Furthermore, such inactivating mutations also highlighted potential direct interfaces of the receptor and G protein (23–25). In conclusion, the 3D morph as well as the distinct inactivated and activated receptor conformations can be used to study processes during signal transduction, as well as postactivation events, like details of G protein interactions. Such insights (receptor/G protein complex model) might be helpful to guide and to explain further experimental data (25, 26), including phosphorylation or interaction with secondary signal transducers.
Analysis of mutational effects by multiple side-chain substitutions
Double and triple mutations are now implemented as a separate data set. Mapping those onto 3D structures enables the analysis of their potential cooperative functional effects with respect to their spatial interrelationship in 3D structures or homology models. Such mutations are designed to reveal deeper insights in the mechanisms of dominant effects of either activating or inactivating effects, respectively.
Analysis of mutational effects via the 2D – Snake-Plot – Search
Most of the analysis tools inspecting the functional mutation data are also applicable to the 2D – Snake-Plot – Search, which provides 2D representations of the GPHR sequence of interest and highlights the requested mutated residues and hyperlinks those to the mutation information (Fig. 2). In contrast to other databases providing snake-plots of GPCRs (27, 28), customization of the snake-plot is possible by choosing user-defined colors for the different receptor regions and mutated residues. In addition, in our interactively designed 2D – Snake-Plot – Search, the absolute position of each amino acid is displayed when dragging the mouse over the amino acid. Clicking on the mutated amino acids hyperlinks to the mutation information. Supporting the facility of inspection, the highly conserved positions of each transmembrane helix are highlighted with a circle around the ball. They represent the reference positions (×.50) of the Ballesteros & Weinstein numbering nomenclature. Our Snake-Plot Tool is unique for GPCRs and other proteins and might be extended for a more general usage. Thus, even without background knowledge of 3D structures or homology models, users can visualize and analyze extracted information from the mutation data set. Moreover, as a more general application we provide the Snake-Plot-Designer in the Resource section where a customized 2D representation of a GPHR sequence can be created. Additionally, any amino acid position or region of interest, for example visualizing specific residues with new experimental data, can be highlighted.
Strikingly, the strength of this database and web tools are the comprehensive GPHR data set of unified scaled mutation data and the link between structural information and functional data that permits a semiquantitative structure function analysis (Figs. 1 and 2). This not only allows fast and easy searches of functional residues but also allows deeper insights into the mechanisms of hormone binding, signal transduction, and signaling regulation to be gained.
Materials and Methods
Technical Aspects
The application SSFA-GPHR is built by combining an Apache web server (http://www.apache.org) hosting PHP Hypertext Preprocessor scripts (PHP 5) and a relational database management system (MySQL 5). 3D structures are displayed using the Jmol structure viewer (http://jmol.sourceforge.net/). To illustrate the glycoprotein hormone receptors in 3D, the viewer is embedded in the application as a Java applet. The Jmol scripting language in combination with JavaScript is used to change the rendering of the molecules to illustrate important structural features.
Nontechnical Aspects
Structures and homology models in different conformations
For the glycoprotein hormone receptors no full receptor structure is available yet. The published LRR domain of FSHR [Protein Data Bank (29) (PDB) code: 1XWD] and TSHR (PDB code: 3G04) were used as template structures for constructing the LRR motifs of the ratFSHR, humLHCGR, and ratLHCGR, each containing N- and C-terminal flanking residues. For model building of the serpentine domain structures of humTSHR, humFSHR, ratFSHR, humLHCGR, and ratLHCGR the 3D structure of bovine rhodopsin (PDB code: 2I35) was used as a basis for the inactivated conformation. For the activated conformation only the active opsin conformation (PDB code: 3DQB) is available as a template. The homology models were produced with the biopolymer module of the SYBYL program package, and for energy minimization and molecular dynamics calculations the AMBER 7.0 force-field was used (31).
Structural Morphing
Activation of receptors induces a reorientation of amino acid side-chains and a relative movement of the transmembrane helices to each other. Different static structural conformations provide the opportunity to simulate structural changes as a dynamic process between both inactivated and activated conformations. For visualization of the activation process in glycoprotein hormone receptors the Morph server (http://molmovdb.org) (32) was used to calculate intermediate frames between the inactive conformation (based on the X-ray structure of bovine rhodopsin; PDB code: 2135) and an activated conformation (based on the X-ray structure of opsin; PDB code: 3DQB). This tool allows the visualization of the intermediate steps of protein transformation between a start conformation and a second different conformation. The interpolation and energy minimization were calculated for each intermediate frame of the animation.
Mutations
Because of the rising amount of mutation data on glycoprotein hormone receptors there is a need for summarizing tools to analyze functional data from different experiments. To allow structure-function analysis such functional data are converted into classifiable unified values. Each original assay resulting from a publication is scaled to percentage values, which are adjusted to the wild-type value of 100% and rounded up or down to the closest decimal place. The data set includes also basic information, like experimental conditions, but also very specific information about the functional assays of humTSHR, humLHCGR, ratLHCGR, humFSHR, and ratFSHR.
Naturally occurring mutations (in vivo) summarize identified gain- and loss-of-function mutations. Many of them are linked to pathological phenomena. The present data set is focused on single side-chain substitutions and the first reported publication for each available mutation is linked to the PubMed/Medline citation, although there are sometimes more publications available for particular mutations. The set describes naturally occurring activating and inactivating mutations. For instance, for the thyrotropin receptor more than 25 naturally occurring mutations are known that increase the constitutive activity dramatically. Such mutations lead to diseases such as toxic adenomas, thyroid cancer, and autosomal dominant forms of hereditary hyperthyroidism (reviewed in Ref. 33).
Constitutively activating mutations (CAMs) of the TSHR are characterized by increased basal Gs (in a few cases also Gq)-mediated signaling activity, which is hormone independent. A new data set comprises double and triple mutations. In the past few years, double and triple mutants, either in spatially close or distant positions of the receptor, have become increasingly interesting because they can provide detailed information about multiple effects or multiple molecular determinants underlying functional effects (19, 20, 30). The database is capable of storing functional data for multiple mutation experiments. The impact of each single mutation, as well as their effect as double or triple mutants and experimental data, is incorporated in the data set.
Acknowledgments
This work was supported by Deutscher Akademischer Austausch Dienst – Kurt Hahn Trust scholarship 2008–2009 (to C.L.W.).
Disclosure Summary: The authors have nothing to declare.
Footnotes
- 2D
- Two-dimensional
- 3D
- three-dimensional
- FSHR
- FSH receptor
- GPCR
- G protein-coupled receptor
- GPHR
- glycoprotein hormone receptor
- LH/CG
- LH and choriogonadotrophin
- LHCGR
- lutropin/choriogonadotropin receptor
- PDB
- Protein Data Bank
- SSFA-GPHR
- Sequence-Structure-Function-Analysis of Glycoprotein Hormone Receptors
- TSHR
- TSH receptor.
References
- 1. Ascoli M , Fanelli F , Segaloff DL. 2002. The lutropin/choriogonadotropin receptor, a 2002 perspective. Endocr Rev 23:141–174 [DOI] [PubMed] [Google Scholar]
- 2. Caltabiano G , Campillo M , De LA , Smits G , Vassart G , Costagliola S , Pardo L. 2008. The specificity of binding of glycoprotein hormones to their receptors. Cell Mol Life Sci 65:2484–2492 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Corvilain B , Van SJ , Dumont JE , Vassart G. 2001. Somatic and germline mutations of the TSH receptor and thyroid diseases. Clin Endocrinol (Oxf) 55:143–158 [PubMed] [Google Scholar]
- 4. Dias JA , Van RP. 2001. Structural biology of human follitropin and its receptor. Arch Med Res 32:510–519 [DOI] [PubMed] [Google Scholar]
- 5. Dias JA. 2005. Endocrinology: fertility hormone in repose. Nature 433:203–204 [DOI] [PubMed] [Google Scholar]
- 6. Garcia-Jimenez C , Santisteban P. 2007. TSH signalling and cancer. Arq Bras Endocrinol Metabol 51:654–671 [DOI] [PubMed] [Google Scholar]
- 7. Gromoll J , Simoni M. 2005. Genetic complexity of FSH receptor function. Trends Endocrinol Metab 16:368–373 [DOI] [PubMed] [Google Scholar]
- 8. Gruters A , Krude H , Biebermann H. 2004. Molecular genetic defects in congenital hypothyroidism. Eur J Endocrinol 151(Suppl 3):U39–U44 [DOI] [PubMed] [Google Scholar]
- 9. Kossack N , Simoni M , Richter-Unruh A , Themmen AP , Gromoll J. 2008. Mutations in a novel, cryptic exon of the luteinizing hormone/chorionic gonadotropin receptor gene cause male pseudohermaphroditism. PLoS Med 5:e88. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Puett D , Li Y , DeMars G , Angelova K , Fanelli F. 2007. A functional transmembrane complex: the luteinizing hormone receptor with bound ligand and G protein. Mol Cell Endocrinol 260–262:126–136 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Simoni M , Gromoll J , Nieschlag E. 1997. The follicle-stimulating hormone receptor: biochemistry, molecular biology, physiology, and pathophysiology. Endocr Rev 18:739–773 [DOI] [PubMed] [Google Scholar]
- 12. Smit MJ , Vischer HF , Bakker RA , Jongejan A , Timmerman H , Pardo L , Leurs R. 2007. Pharmacogenomic and structural analysis of constitutive g protein-coupled receptor activity. Annu Rev Pharmacol Toxicol 47:53–87 [DOI] [PubMed] [Google Scholar]
- 13. Szkudlinski MW , Fremont V , Ronin C , Weintraub BD. 2002. Thyroid-stimulating hormone and thyroid-stimulating hormone receptor structure-function relationships. Physiol Rev 82:473–502 [DOI] [PubMed] [Google Scholar]
- 14. Themmen APN , Huhtaniemi IT. 2000. Mutations of gonadotropins and gonadotropin receptors: elucidating the physiology and pathophysiology of pituitary-gonadal function. Endocr Rev 21:551–583 [DOI] [PubMed] [Google Scholar]
- 15. Schoneberg T , Schulz A , Biebermann H , Hermsdorf T , Rompler H , Sangkuhl K. 2004. Mutant G-protein-coupled receptors as a cause of human diseases. Pharmacol Ther 104:173–206 [DOI] [PubMed] [Google Scholar]
- 16. Kleinau G , Brehm M , Wiedemann U , Labudde D , Leser U , Krause G. 2007. Implications for molecular mechanisms of glycoprotein hormone receptors using a new sequence-structure-function analysis resource. Mol Endocrinol 21:574–580 [DOI] [PubMed] [Google Scholar]
- 17. Kleinau G , Kreuchwig A , Worth CL , Krause G. 2010. An interactive web-tool for molecular analyses links naturally occurring mutation data with three-dimensional structures of the rhodopsin-like glycoprotein hormone receptors. Hum Mutat 31:E1519–E1525 [DOI] [PubMed] [Google Scholar]
- 18. Apweiler R , Martin MJ , O'Donovan C , Magrane M , Alam-Faruque Y , Antunes R , Barrell D , Bely B , Bingley M , Binns D , Bower L , Browne P , Chan WM , Dimmer E , Eberhardt R , Fedotov A , Foulger R , Garavelli J , Huntley R , Jacobsen J , Kleen M , Laiho K , Leinonen R , Legge D , Lin Q, et al. . 2010. The Universal Protein Resource (UniProt) in 2010. Nucleic Acids Res 38:D142–D148 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Jaeschke H , Kleinau G , Sontheimer J , Mueller S , Krause G , Paschke R. 2008. Preferences of transmembrane helices for cooperative amplification of Gαs and Gαq signaling of the thyrotropin receptor. Cell Mol Life Sci 65:4028–4038 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Kleinau G , Jaeschke H , Mueller S , Worth CL , Paschke R , Krause G. 2008. Molecular and structural effects of inverse agonistic mutations on signaling of the thyrotropin receptor–a basally active GPCR. Cell Mol Life Sci 65:3664–3676 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Feng X , Muller T , Mizrachi D , Fanelli F , Segaloff DL. 2008. An intracellular loop (IL2) residue confers different basal constitutive activities to the human lutropin receptor and human thyrotropin receptor through structural communication between IL2 and helix 6, via helix 3. Endocrinology 149:1705–1717 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Tao YX. 2010. The melanocortin-4 receptor: physiology, pharmacology, and pathophysiology. Endocr Rev 31:506–543 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Angelova K , Fanelli F , Puett D. 2008. Contributions of intracellular loops 2 and 3 of the lutropin receptor in Gs coupling. Mol Endocrinol 22:126–138 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Kleinau G , Jaeschke H , Worth CL , Mueller S , Gonzalez J , Paschke R , Krause G. 2010. Principles and determinants of G-protein coupling by the rhodopsin-like thyrotropin receptor. PLoS One 5:e9745. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Timossi C , Maldonado D , Vizcaino A , Lindau-Shepard B , Conn PM , Ulloa-Aguirre A. 2002. Structural determinants in the second intracellular loop of the human follicle-stimulating hormone receptor are involved in G(s) protein activation. Mol Cell Endocrinol 189:157–168 [DOI] [PubMed] [Google Scholar]
- 26. Ulloa-Aguirre A , Uribe A , Zarinan T , Bustos-Jaimes I , Perez-Solis MA , Dias JA. 2007. Role of the intracellular domains of the human FSH receptor in G(alphaS) protein coupling and receptor expression. Mol Cell Endocrinol 260–262:153–162 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Horn F , Weare J , Beukers MW , Horsch S , Bairoch A , Chen W , Edvardsen O , Campagne F , Vriend G. 1998. GPCRDB: an information system for G protein-coupled receptors. Nucleic Acids Res 26:275–279 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Van DJ , Horn F , Costagliola S , Vriend G , Vassart G. 2006. GRIS: glycoprotein-hormone receptor information system. Mol Endocrinol 20:2247–2255 [DOI] [PubMed] [Google Scholar]
- 29. Berman HM , Westbrook J , Feng Z , Gilliland G , Bhat TN , Weissig H , Shindyalov IN , Bourne PE. 2000. The Protein Data Bank. Nucleic Acids Res 28:235–242 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Claus M , Jaeschke H , Kleinau G , Neumann S , Krause G , Paschke R. 2005. A hydrophobic cluster in the center of the third extracellular loop is important for thyrotropin receptor signaling. Endocrinology 146:5197–5203 [DOI] [PubMed] [Google Scholar]
- 31. Case DA. 2002. Molecular dynamics and NMR spin relaxation in proteins. Acc Chem Res 35:325–331 [DOI] [PubMed] [Google Scholar]
- 32. Krebs WG , Gerstein M. 2000. The morph server: a standardized system for analyzing and visualizing macromolecular motions in a database framework. Nucleic Acids Res 28:1665–1675 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Tao YX. 2006. Inactivating mutations of G protein-coupled receptors and diseases: structure-function insights and therapeutic implications. Pharmacol Ther 111:949–973 [DOI] [PubMed] [Google Scholar]