Agonist-bound progesterone receptor activates the cSrc/Kras/Raf-1/Erk2/NF-kB signaling cascade leading to up-regulated expression of p53 in vascular endothelial cells.
Abstract
We previously showed that progesterone (P4) inhibited the proliferation of human umbilical vein endothelial cells (HUVECs) through a p53-dependent pathway. Now we investigated further the molecular mechanism underlying the hormone activity. In cultured HUVECs, P4 increased the protein levels of phosphorylated Src (p-Src), Raf-1, and ERK. The levels of p-Src and p-Src-progesterone receptor complex in HUVECs were increased by P4 treatment. These effects were blocked by pretreatment with a progesterone receptor antagonist, RU486. The P4-induced increase in p53 transactivity was abolished by pretreatment with Src kinase inhibitors. Moreover, administration with cSrc antisense oligonucleotide prevented the P4-induced increases of the levels of p53 mRNA and protein. These data suggest that P4-induced up-regulation of p53 might be mediated through activation of cSrc. Pretreatment with Src kinase inhibitors also prevented P4-induced membrane translocation of Kras and increases of the protein levels of phosphorylated Raf and phosphorylated ERK. Transfection with dominant-negative ERK2 prevented the P4-induced increases of protein level and promoter activity of p53 and a decrease of thymidine incorporation. P4 also increased nuclear factor-κB (NF-κB) nuclear translocation and NF-κB binding onto the p53 promoter. These effects were abolished by pretreatment with ERK inhibitors. The P4-induced up-regulation of the p53 promoter activity was prevented by preadministration with dominant-negative ERK2 or NF-κB inhibitors. Taken together, our data suggest that the cSrc/Kras/Raf-1/ERK2/NF-κB signaling pathway contributes to the P4-induced up-regulation of p53 in HUVECs. These findings highlight progesterone receptor activation of extranuclear signaling pathways in regulating p53 and cell cycle progression in HUVECs.
Angiogenesis, a process of forming new blood vessels, produced by endothelial cells of preexisting blood vessels (1, 2), occurs in many physiological and pathological conditions (3–5). In adults, physiological angiogenesis in all tissues is decreased with few exceptions during the life span. The ovary and endometrium are two of the few tissues in the adult organism in which angiogenesis is a prominent occurrence under normal conditions. A preponderance body of experimental results indicates that female sex hormones play an important role in regulating angiogenesis in the female reproductive organs (6, 7).
Progesterone (P4), which has been called a paradoxical hormone, exhibits stimulatory or inhibitory effects on growth, depending on the tissue and the treatment regimen (8). The effects of P4 on endothelial cells have been documented. Vázquez et al. (9) used the progesterone receptor (PR) knockout mice animal model to demonstrate that P4 could inhibit the proliferation of vascular endothelial cells and the rate of reendothelialization and thus impair vascular repair processes through PR-dependent pathways. Our previous in vitro and in vivo studies also demonstrated that P4 alone could inhibit the proliferation of cultured human umbilical vein endothelial cells (HUVECs) through a p53-mediated pathway (10). However, the molecular mechanism underlying this P4-induced up-regulation of p53 is still not clear.
In the present study, we described the molecular mechanism underlying P4-induced up-regulation of p53 in HUVECs. Our results showed that P4 bound to the PR in the cytoplasm, subsequently induced cSrc activation, which in turn activated the Kras/Raf-1/ERK2/nuclear factor-κB (NF-κB) signaling pathway and eventually increased p53 expression in HUVECs. The findings of this study will provide some novel insights into the molecular mechanism underlying P4-induced inhibition of angiogenesis. Perhaps such better understanding could lead to useful new therapies for neoplastic-related disorders that might be attached by suppression of vascular overgrowth.
Results
Effect of PR activation on the P4-induced increase of the cSrc activity
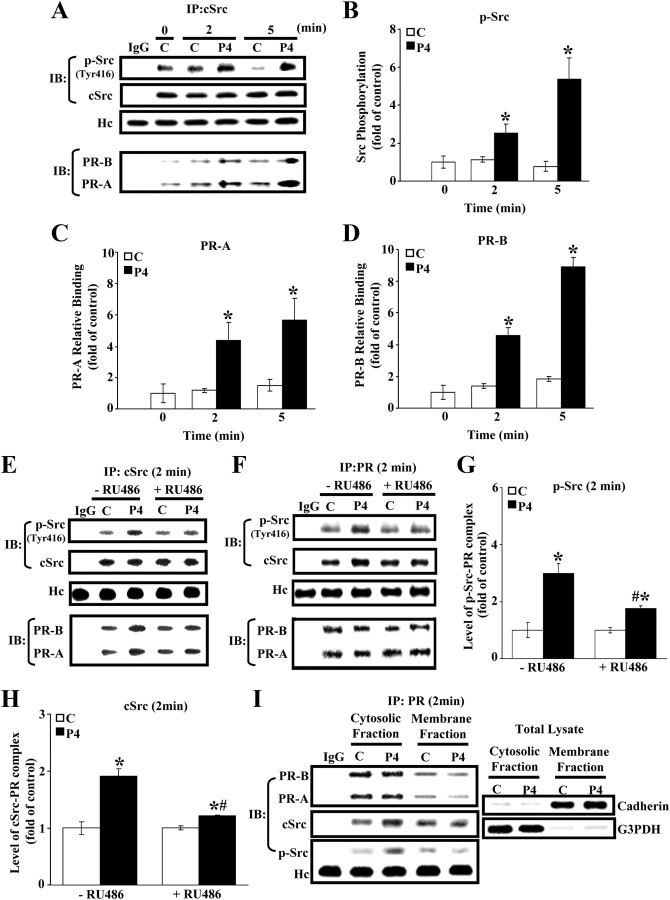
To observe the effect of P4 treatment on the cSrc activation and the interaction between PR and cSrc, the immunoprecipitation assay was conducted to show that the levels of p-Src and the formation of the PR-cSrc and PR-p-Src complexes in HUVECs were increased at 2 min after P4 (500 nm) treatment (Fig. 1, A–D). Pretreatment of HUVECs with a PR antagonist, RU486, prevented the P4-induced increases of the levels of p-Src (Fig. 1E) and the formation of PR-cSrc and PR-p-Src complex (Fig. 1, F–H). The increased PR-p-Src and PR-cSrc complexes were recovered mainly in the cytosolic fraction (Fig. 1I); therefore, this rapid action of the P4 was localized to the cell cytoplasm but not at intranuclear gene loci.
Fig. 1.
Role of PR in the P4-induced increase of cSrc activity. Treatment of HUVECs with P4 (500 nm) for 2 or 5 min increased the level of p-Src and the formation of PR-A-cSrc and PR-B-cSrc complexes as shown by electrophoretograms (A). Quantitative results (B) for p-Src levels, the formation of PR-A-cSrc complex (C), and the formation of PR-B-cSrc complex (D) were adjusted with corresponding Hc level and expressed as induction ratio over control. Values represent the means ± sem (n = 3). *, P < 0.05, different from corresponding control. E, P4 increased the level of p-Src, and this effect was blocked by PR antagonist, RU486 (50 nm). F, P4 increased the formation of PR-p-Src and PR-cSrc, and these effects were blocked by PR antagonist, RU486 (50 nm). The quantitative data from F were adjusted with corresponding Hc level and expressed as induction over controls to show the suppression activity of RU486 on PR-p-Src (G) and PR-cSrc (H). Values represent the means ± sem (n = 3). *, P < 0.05, different from corresponding control; #, P < 0.05, different from P4-treated group without RU486. I, Subcellular fractionation of HUVECs showed that the stimulatory effect of P4 on the formation of PR-p-Src and PR-cSrc complexes was localized to the cytoplasmic fraction and not in the membrane fraction. The levels of Hc were used to verify equivalent protein loading. The levels of cadherin and G3PDH in total lysate were used as a membrane and cytosolic protein marker, respectively, to confirm the purities of isolation. C, Control; Hc, heavy chain of IgG; IP, immunoprecipitation; IB, immuoblotting.
Effect of cSrc activation in the P4-induced increase of p53
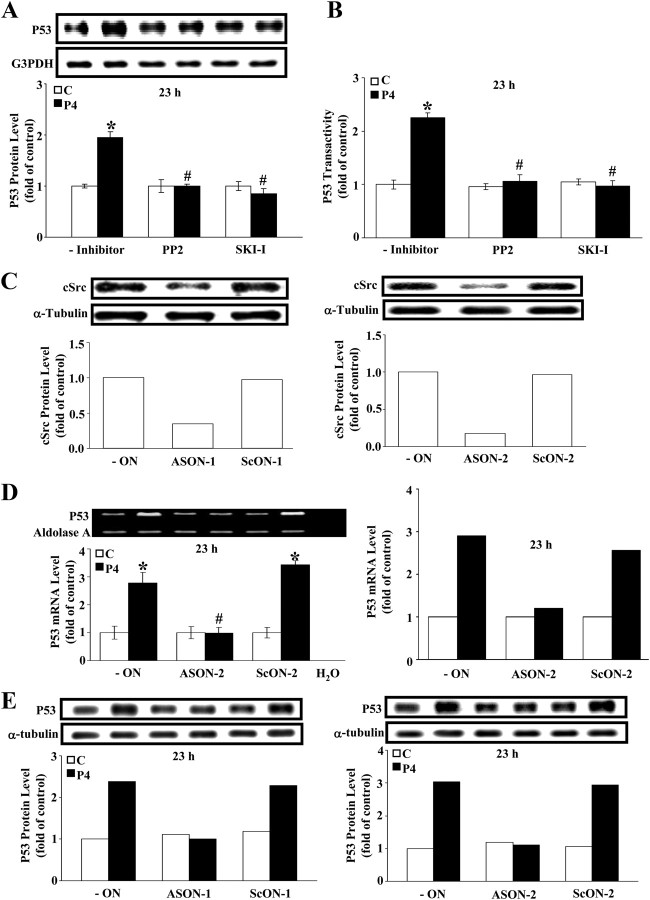
Because cSrc has been implicated in the regulation of cell proliferation (11), we studied whether activation of cSrc is involved in the P4-induced up-regulation of p53 in these HUVECs. As illustrated in Fig. 2A, pretreatment of HUVECs with a Src kinase inhibitor (400 nm), 4-amino-5-(4-chlorophenyl)-7-(t-butyl)pyrazolo[3,4-d]pyramidine (PP2), or Src kinase inhibitor I (SKI-I), prevented the P4-induced increases of p53 protein level. Because P4 treatment increased the promoter activities of p21 and p27 and the binding of p53 protein to the p21 and the p27 promoter (10), HUVECs were transfected with the p53-luciferase reporter DNA vector, which contains p53 response enhancer element, for assaying alterations in p53 transcriptional activity. As shown in Fig. 2B, the P4-induced increase of transactivation of p53 was prevented by pretreatment of the cell with PP2 or SKI-I. Furthermore, pretreatment of HUVECs with two different antisense oligonucleotides (ASs) (1 μm), which targeted different regions on the cSrc mRNA, resulted in a 70 and 90% decrease of the basal cSrc protein level, respectively (Fig. 2C) and completely prevented the P4-induced up-regulation of the levels of p53 mRNA demonstrated by semiquantitative PCR (Fig. 2D, left panel) as well as quantitative real-time PCR (Fig. 2D, right panel) and protein (Fig. 2E). Evidently activation of cSrc might be a preceding step in the P4-induced up-regulation of p53.
Fig. 2.
cSrc activation in the P4-induced increases of p53 protein, mRNA, and transactivity. A, P4 increased the level of p53 protein, but this P4 effect is blocked by a Src inhibitor, PP2 (400 nm), or SKI-I (400 nm). Top panel shows Western blot analysis; lower panel shows a graph of quantitation of these data adjusted with G3PDH protein level and expressed as ratio over control (without inhibitor). Values represent the means ± sem (n = 3). *, P < 0.05, different from corresponding control; #, P < 0.05, different from P4-treated group without inhibitor. B, P4 increased p53 transactivity, but this P4 effect was blocked by a Src inhibitor, PP2 (400 nm) or SKI-I (400 nm). HUVECs were pretransfected with p53-luciferase vector that contains encoded p53 response element. Renilla luciferase expression vector was used as the control and cotransfected with p53-luciferase vector in the luciferase activity assay. Values represent the means ± sem (n = 8). *, P < 0.05, different from corresponding control; #, P < 0.05, different from P4-treated group without inhibitor. Two different targeting AS oligonucleotides of cSrc were used to study the involvement of cSrc in the P4-induced increases of p53. Pretreatment of HUVECs with each AS oligonucleotide of cSrc abolished the P4-induced increases of the levels of cSrc protein (C), p53 mRNA (D, left panel was examined by semiquantitative PCR, whereas right panel was examined by quantitative real time PCR), and p53 protein (E). Western blot analysis and semiquantitative RT-PCR were shown on top, and data, which were adjusted with aldolase A mRNA level (for RT-PCR) or α-tubulin protein level (for Western blot) and expressed as ratio over corresponding control, were plotted in figure below. Values represent the means ± sem (n =3). *, P < 0.05, different from corresponding control; #, P < 0.05, different from P4-treated group without ON. C, Control; ON, oligonucleotide; AsON, cSrc AS; ScON, cSrc scramble oligonucleotide.
Effect of Kras activation on the P4-induced up-regulation of p53 in HUVECs
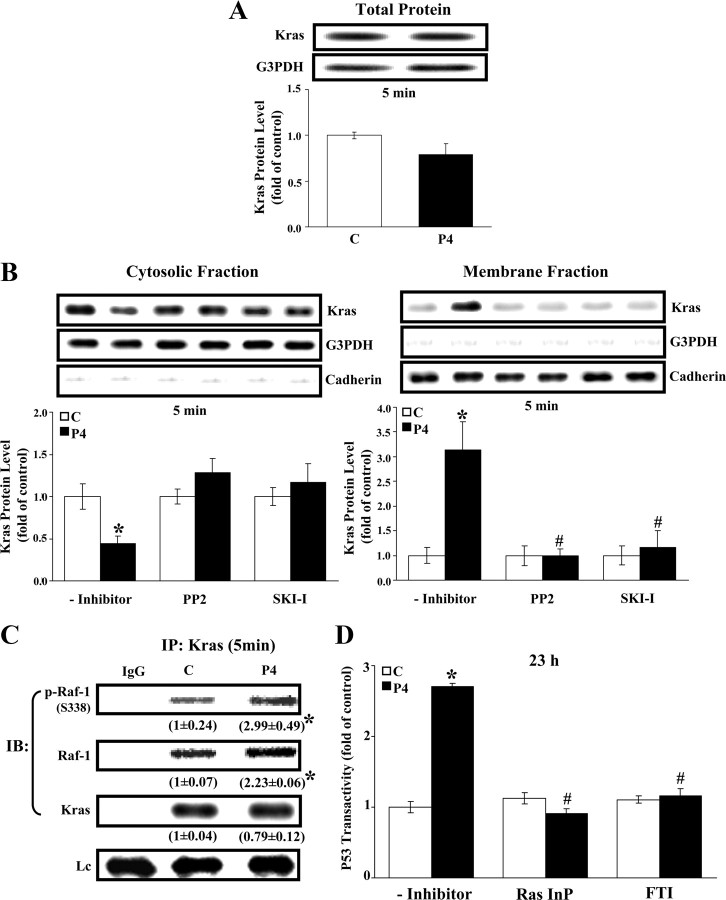
We further investigated the involvement of Kras in the P4-induced increase of transactivation activity of p53 in HUVECs. As illustrated in Fig. 3B, P4 (500 nm) treatment increased the membrane translocation of Kras at 5 min but did not significantly affect the total protein level of Kras (Fig. 3A). Pretreatment of HUVECs with 400 nm of PP2 or SKI-I prevented the P4-induced membrane translocation of Kras. P4 treatment for 5 min induced the recruitment and phosphorylation (site Ser 338) of Raf-1, the direct effector protein of Kras (Fig. 3C). Pretreatment of HUVECs with a Kras inhibitor, Ras inhibitory peptide (Ras InP; 5 μm), or farnesyltransferase inhibitor (FTI; 10 μm) abolished the P4-induced increases of p53 transactivity (Fig. 3D). These results suggest that P4 induced cSrc activation, which in turn activated Kras, and finally caused p53 up-regulation.
Fig. 3.
Activation of Kras in the P4-induced increase of p53 transactivity. A, Treatment of HUVECs with P4 (500 nm) for 5 min did not significantly alter the level of Kras. Top panel, A representative result of Kras and G3PDH protein levels determined by Western blot analysis. Bottom panel, Quantitative result of Kras protein level, which was adjusted with G3PDH protein level and expressed as ratio over its own control. Values represent the means ± sem (n = 3). B, Kras protein was translocated from cytosolic sites onto membrane components, and this Kras translocation in HUVECs was increased by 5 min treatment with P4 (500 nm) and completely blocked by pretreatment with a Src inhibitor (PP2 or SKI-I). Left panel, Cytosolic fraction. Right panel, Membrane fraction. Western blots were shown on top and data, which were adjusted with G3PDH (cytosolic fraction) or cadherin (membrane fraction) protein level and expressed as ratio over corresponding control, were plotted in graph below. Values represent the means ± sem (n = 3). *, P < 0.05, different from corresponding control; #, P < 0.05, different from P4-treated group without inhibitor. C, P4 treatment for 5 min increased the Kras-Raf-1 and Kras-p-Raf-1 complexes in HUVECs. Western blots show a representative result of p-Raf-1, Raf-1, and Kras protein levels. Values shown in parentheses represent the quantified results (means ± sem, n = 3) after adjusted with their own Lc level and expressed as ratio over control. *, P < 0.05, different from corresponding control. D, P4-increased p53 transactivity was abolished by pretreatment with Ras InP or FTI. HUVECs were pretransfected with p53-luc vector that contains encoded p53 response element. Renilla luciferase expression vector, used as the control, was cotransfected with p53-luc vector in the luciferase activity assay. Values represent the means ± sem (n = 4). *, P < 0.05, different from corresponding control; #, P < 0.05, different from P4-treated group without inhibitor. C, Control; IP, immunoprecipitation; IB, immunoblotting; Lc, light chain of IgG.
Effect of ERK1/2 activation in the P4-induced increases of the protein levels of p53, p21, and p27 and a decrease of DNA synthesis in HUVECs
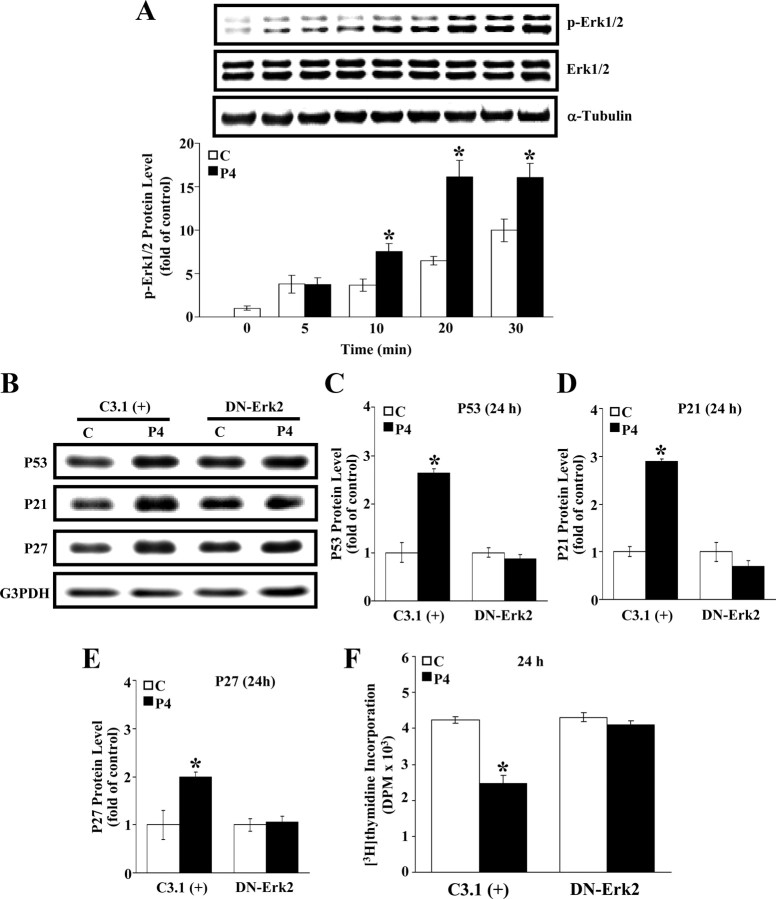
Because the Raf/ERK-regulated signaling has been indicated to be involved in the hormone-regulated cell growth and proliferation and regulated by the Ras-mediated signaling, we further examined the involvement of ERK in the P4-induced cell cycle arrest in HUVECs. As shown in Fig. 4A, the level of p-ERK 1/2 in HUVECs was increased at 10 min after P4 (500 nm) treatment. To confirm the involvement of ERK2 activation in the P4-induced p53 up-regulation, HUVECs were transfected with dominant-negative (DN) ERK2 cDNA to block the ERK2 function. Western blot analysis demonstrated that pretransfection of HUVECs with DN-ERK2 cDNA prevented the P4-induced up-regulation of the protein levels of p53, p21, and p27 (Fig. 4, B–E). DN-ERK2 transfection also prevented the P4-induced inhibition of [3H]thymidine incorporation in HUVECs (Fig. 4F). These results implicate that ERK2 activation might be involved in the regulation of P4-induced proliferation inhibition in HUVECs. Pretreatment of HUVECs with 5 μm of Ras InP (a Kras inhibitor) or 10 μm of sulindac sulfide [a Raf-1 inhibitor (Raf-1 In)] prevented the P4-induced increases of the protein levels of p-Raf-1 and p-ERK1/2 (Fig. 5). Hence, the Raf-1/ERK1/2-mediated signaling pathway might be the downstream signaling molecules initiated by cSrc/Kras activation.
Fig. 4.
Phosphorylation of ERK in the P4-induced increases of p53, p21, and p27 protein level and a decrease of DNA synthesis in HUVECs. A, P4 (500 nm) time dependently increased ERK1/2 phosphorylation. Top panel, A representative result of p-ERK1/2, ERK1/2, and α-tubulin protein levels. Bottom panel, Quantitative results of p-ERK1/2 and ERK1/2 protein level, which was adjusted with corresponding α-tubulin level and expressed as ratio over control (0 min). Values represent the means ± sem (n = 3). *, P < 0.05, different from corresponding control. B, Pretransfection with DN-ERK2 prevented P4-increased p53, p21, and p27 protein levels. A representative result of p53, p21, p27, and G3PDH protein levels are shown. Quantitative results of p53 (C), p21 (D), and p27 (E) protein levels were adjusted with G3PDH protein level and expressed as ratio over corresponding control. Values represent the means ± sem (n = 3). *, P < 0.05, different from corresponding control. F, Pretransfection of HUVECs with DN-ERK2 prevented the P4-induced inhibition in [3H]thymidine incorporation. Values represent the means ± sem (n = 4). *, P < 0.05 different from corresponding control. C, Control; C3.1 (+), pcDNA 3.1 (+).
Fig. 5.

Raf-1 and ERK1/2 are the downstream molecules of the Ras-mediated signaling. Pretreatment of HUVECs with Ras InP (5 μm) or Raf-1 In (sulindac sulfide; 10 μm) prevented the P4-induced increases of the levels of p-Raf-1 and p-ERK1/2. A, A representative result of ERK, p-ERK, Raf-1, and p-Raf-1 protein levels determined by Western blot analysis. The electrophoresis membrane was probed with α-tubulin antibody to verify equivalent sample loading. Quantitative results of p-ERK1/2 (B) and p-Raf-1 (C) were shown after adjusted with α-tubulin protein level and expressed as fold induction of corresponding control. Values represent the means ± sem (n = 3). *, P < 0.05, different from corresponding control; #, P < 0.05, different from P4-treated without inhibitor. C, Control.
Effect of P4 on nuclear translocation of NF-κB (p65) and the p53 promoter activity in HUVECs
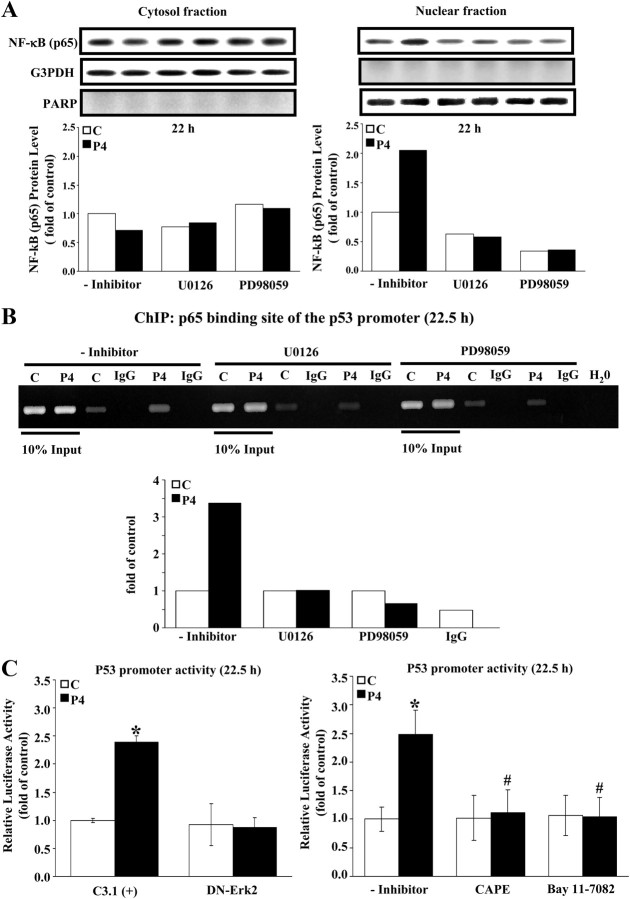
To examine whether NF-κB translocation is involved in the P4-induced increase of the p53 promoter activity, we analyzed the potential binding motif of NF-κB (p65) on the p53 promoter region which spans from −361 to +1 (the most 3′ end of this fragment, which is defined as +1, in this study is a XbaI restriction site at the exon 1 of the p53 gene) and found a NF-κB (p65) binding motif at −64 to approximately −55 site of this p53 promoter. As shown in Fig. 6A, the translocation of p65 protein from cytosol to nuclear loci was increased at 22 h after P4 (500 nm) treatment, and this effect was abolished by pretreatment of HUEVCs with an ERK inhibitor, U0126 (5 μm) or PD98059 (10 μm), suggesting that nuclear translocation of the p65 protein might be a downstream event of ERK1/2 activation. To examine the regulatory capability of p65 protein on the p53 promoter activity, the chromatin immunoprecipitation (ChIP) assay was performed. As shown in Fig. 6B, P4 increased the binding of the p65 protein onto the p53 promoter at 22.5 h after treatment, and this increased binding activity was blocked by the pretreatment of HUVECs with an ERK inhibitor, U0126 (5 μm) or PD98059 (10 μm). To confirm the regulatory effect of ERK2/p65 on p53 expression in the P4-treated HUVECs, the cells were transfected with DN-ERK2 cDNA to block the ERK2 function or pretreated with a NF-κB (p65) inhibitor, caffeic acid phenethyl ester (CAPE; 10 μm) or Bay 11-7081 (50 nm), to impede the p65 activation. Luciferase activity assay showed that the P4 (500 nm)-induced increase of p53 promoter activity was prevented by pretransfection of HUVECs with DN-ERK2 cDNA (Fig. 6C, left panel) or by pretreatment with CAPE (10 μm) or Bay 11-7081 (50 nm) (Fig. 6C, right panel).
Fig. 6.
Involvement of NF-κB (p65) in regulation of the p53 promoter activity. A, P4 (500 nm) induced NF-κB translocation from cytosolic components into nuclear components in HUVECs, and this increased translocation was completely blocked by pretreatment with an ERK2 inhibitor, U0126 (5 μm), or PD98059 (10 μm). Left panel, Cytosolic fraction. Right panel, Nuclear fraction. Western blots were shown on top and data, which were adjusted with G3PDH (cytosolic fraction) or poly(ADP-ribose) polymerase (PARP) (nuclear fraction) protein level and expressed as ratio over control (without inhibitor treatment), were plotted in graph below. B, The P4 (500 nm)-induced increase of the NF-κB DNA binding activity on the p53 promoter was completely blocked by pretreatment with an ERK2 inhibitor, U0126 (5 μm) or PD98059 (10 μm). The p53 DNA binding activity was assessed by using ChIP assay and detected by semiquantitative PCR (top panel) and quantitative real-time PCR (bottom panel). C, The P4 (500 nm)-induced increases of the p53 promoter activity was prevented by pretransfection of HUVECs with DN-ERK2 (left panel) or by pretreating the cells with a NF-κB inhibitor, CAPE (10 μm) or Bay 11-7082 (50 nm) (right panel). Renilla luciferase expression vector was used as a control and cotransfected with promoter in the luciferase activity assay. Twenty-four hours after transfection, the cells were rendered quiescent and then treated with 10% FBS and vehicle/inhibitor without or with P4 (500 nm) for 22.5 h. Subsequently the cells were processed for the luciferase activity assay. Quantitative results of the p53 promoter activity were shown and expressed as fold induction of corresponding control. Values represent the means ± sem (n = 4). *, P < 0.05, different from corresponding control; #, P < 0.05, different from P4-treated without inhibitor. C, Control; C3.1 (+), pcDNA 3.1 (+).
The relationship between p53 and human homolog of mouse double-minute 2 (MDM2) in the P4-treated HUVECs
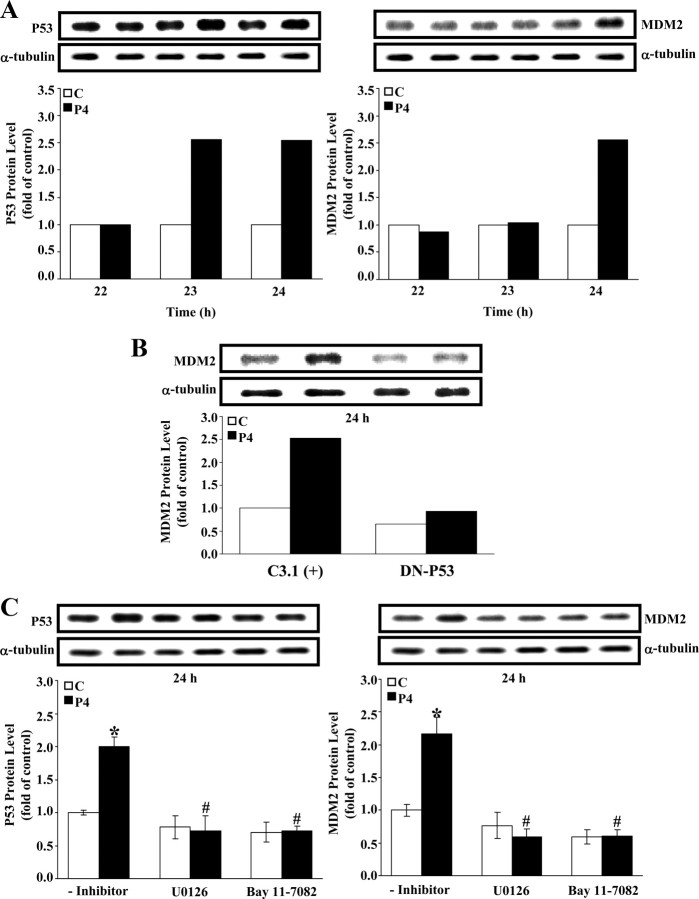
Because MDM2 protein is a primary negative regulator of the p53 protein and the activated p53 protein has been indicated to serve as a transcriptional factor to increase the MDM2 protein level, we examined the interaction between p53 and MDM2 protein in the P4-treated HUVECs. As shown in Fig. 7A, the P4 (500 nm)-induced increases of the p53 protein level were observed at 23 and 24 h after P4 treatment (Fig. 7A, left panel), whereas increases of the MDM2 protein level were observed at 24 h (Fig. 7A, right panel), suggesting that the P4-induced increase of MDM2 protein level might be mediated by an increased p53 protein level. To examine this possibility, HUVECs were pretransfected with DN-p53 cDNA (V143A mutant) followed by P4 (500 nm) treatment. As shown in Fig. 7B, pretransfection of HUVEC with DN-P53 cDNA prevented the P4-induced increase of the MDM2 protein level. In addition, the P4 (500 nm)-induced increases of the protein level of p53 (Fig. 7C, left panel) and MDM2 (Fig. 7C, right panel) were abolished by pretreatment of the cells with 5 μm U0126 (an ERK inhibitor) or 50 nm Bay 11-7082 (a p65 inhibitor).
Fig. 7.
p53 increases the level of MDM2 protein in HUVECs. A, P4 (500 nm) time dependently increased p53 and MDM2 protein levels. The P4-induced increases of the p53 protein level were observed at 23 and 24 h after P4 treatment, whereas increases of MDM2 protein level were observed at 24 h. Western blots of p53 and MDM2 protein were shown on top and data, which were adjusted with α-tubulin protein level and expressed as ratio over corresponding control, were plotted in graph below. B, Pretransfection of HUVECs with DN-p53 prevented the P4-induced increases of MDM2 protein. Top panel shows Western blot analysis; graph below shows data from Western blot, which were adjusted with α-tubulin and expressed as a ratio over control [C3.1(+)-transfected]. C, The P4 (500 nm)-induced increases of p53 and MDM2 protein level in HUVECs were abolished by pretreatment with U0126 (5 μm) or Bay 11-7082 (50 nm). Western blots of p53 and MDM2 proteins were shown on top and data, which were adjusted with α-tubulin protein level and expressed as ratio over control (without treatment of inhibitor), were plotted in graph below. Values represent the means ± sem (n = 3). *, P < 0.05, different from corresponding control; #, P < 0.05, different from P4-treated group without inhibitor. C, Control; DN-P53, DN-p53; C3.1 (+), pcDNA 3.1 (+).
Discussion
Previously we demonstrated that P4 at physiological levels (5–500 nm) caused cell cycle arrest in HUVECs through induction of the p53 protein (10). In the present study, we further delineated the molecular mechanisms underlying P4-induced up-regulation of p53. The cSrc/Kras/Raf-1/ERK2/NF-κB signaling pathway in HUVECs is activated by P4. The blockade of this pathway abolished the P4-induced up-regulation of p53. To our knowledge, this is the first demonstration that P4 up-regulated the p53 expression in HUVECs through activation of the cSrc/Kras/Raf-1/ERK2/NF-κB signaling pathway mediated by PR located in the cytoplasm.
Customarily, P4 is thought to mediate its biological effects through binding to its nuclear receptors, PR-A or PR-B, to alter gene transcription. Lately a growing body of data suggests that P4 could also bind to the PR presented in cytoplasm or membrane to initiate a quick acting nongenomic signaling cascade (12). Such a nonclassic, nongenomic, or extranuclear putative membrane-associated PR has been identified in several species including porcine (13, 14), human (15), and rat (16). The classic steroid receptors are members of the steroid/thyroid hormone receptor superfamily (17). After binding to the ligand, PR in the cytosol forms dimers and then translocates to the nucleus, in which the steroid/steroid receptor complex binds to DNA and acts as a transcription factor. In contrast, the putative membrane-associated PR initiates rapid effects on diverse signaling pathways, independently of transcriptional or genomic action (18). The results from the present study showed that the formation of PR-cSrc and PR-p-Src complexes in HUVECs was increased at 2 and 5 min after P4 treatment, respectively (Fig. 1, A–D). These findings suggest that P4-induced activation of cSrc might be mediated through a nongenomic action of P4 to increase the formation of PR-cSrc complex. These effects were blocked by pretreatment of HUVECs with RU486 (Fig. 1, E–H). Our data also show that the formation of both PR-A-cSrc and PR-B-cSrc complexes was increased by P4 treatment (Fig. 1, A–D). This finding is consistent with a previous report showing that the activated PR directly binds to the SH3 domain of cSrc through physical interaction with its proline-rich (PXXP) motif (19). Interestingly, our data show that, in the absence of P4, the formation of PR-cSrc and PR-p-Src complexes was observed in both cytosolic and membrane fractions (Fig. 1I). In the presence of P4, the levels of PR-cSrc and PR-p-Src complexes were increased. However, the P4-induced increases of PR-cSrc and PR-p-Src complexes were observed only in the cytosolic fraction. Evidently, P4 acts by binding to PR located in the cytoplasm. This rapid action of P4 in the cytosol and consequent activation of cSrc has also been implicated in the regulation of cell cycle function (12, 20). Further indication that cSrc activation is involved in the P4-induced up-regulation of p53 was evidenced by failure of these effects of P4 to occur in HUVECs pretreated with a Src inhibitor, PP2 or SKI-I (Fig. 2, A and B), or with cSrc ASs (Fig. 2, D and E). Taken together, these results suggest that activation of the cSrc-mediated signaling pathway might be involved in the P4-induced up-regulation of p53.
The present study shows that P4 treatment increased the membrane translocation of Kras (Fig. 3B), the formation of Kras-Raf and Kras-p-Raf complexes (Fig. 3C), and the levels of p-ERK1/2 (Fig. 4A). Pretreatment of HUVECs with a Src kinase inhibitor, PP2 or SKI-I, blocked the P4-induced increase of membrane translocation of Kras (Fig. 3B). Pretreatment of the cells with Ras InP or FTI blocked the P4-induced increase of the p53 transactivity (Fig. 3D). These findings suggest that Kras is the downstream molecule of cSrc involved in the P4-induced up-regulation of p53. Moreover, the P4-induced increases of the phosphorylated (p)-ERK1/2 and p-Raf-1 protein were blocked by pretreatment with Ras InP or Raf-1 In (Fig. 5, B and C), suggesting that ERK1/2 is the downstream molecule of Raf-1. The involvement of ERK1/2 activation in the P4-induced p53 up-regulation and DNA synthesis inhibition in HUVECs was further supported by the evidence that DN-ERK2 transfection prevented the P4-induced increases of the protein levels of p53 (Fig. 4C), p21 (Fig. 4D), and p27 (Fig. 4E) and a decrease of DNA synthesis (Fig. 4F). These findings further suggest that P4-induced inhibition of DNA synthesis in HUVECs was mediated through a nongenomic action of PR.
The results from our previous and present study suggest that a transcriptional regulation is involved in the P4-induced increase of the protein level of p53. It is generally accepted that the half-life of p53 is primarily regulated by MDM2, an important negative regulator of p53. MDM2 targets to ubiquitin residues of p53 and elicits p53 ubiquitination and proteasomal degradation. Mdm2 is one of the p53's target genes, and any increase of p53 usually leads to an increase in Mdm2 levels (21, 22). In the present study, the increased p53 protein level was observed at 23 h after P4 treatment, whereas the MDM2 protein level was not increased until 24 h after P4 treatment (Fig. 7A). Failure in the DNA binding activity of p53 by pretransfection of HUVECs with DN-P53 cDNA abolished the P4-increased protein level of MDM2 (Fig. 7B). Taken together, these data suggest that increases of the p53 protein level induced by P4 treatment might not be due to a dysfunction of MDM2. Several transcriptional factors have been suggested to be involved in the regulation of p53 gene transcription. For example, CAAT/enhancer-binding protein-β could efficiently bind to the p53 promoter and increase p53 expression (23). HoxA5 (a homeodomain containing gene) could bind to the p53 promoter, up-regulate the p53 gene transcription, and lead to apoptosis in the human breast tumor (24). Pituitary homeobox 1 transcriptionally activates the p53 expression and results in apoptosis in MCF-7 cells (25). Human p53 promoter activity can be autoactivated by p53 protein itself, and this activity can be enhanced in the presence of nuclear NF-κB (26). In a high-throughput screening, four genes including CRMP-2, GAS41, RPS6K4, and RUNDC1, have been identified to negatively regulate the p53 transcriptional activity (27). In addition to several transcriptional factors regulating p53 transcription via direct binding to the promoter region of the p53 gene, some studies have demonstrated the correlation between certain cytosolic molecules and p53 protein levels. In colon cancer SW480 cells, transfection with Kras AS increases the protein level of p53, suggesting a Ras-dependent regulation in p53 transcription (28). In human fibrosarcoma AP14 cells, a defect of p53 expression was restored after overexpression of ERK2. Oncogenic Ras increases the levels of p53 mRNA and protein, whereas treatment with a MAPK inhibitor U0126 time dependently decreased the protein level of p53 (29). The present results show that P4 treatment increased NF-κB nuclear translocation (Fig. 6A) and binding onto the p53 promoter (Fig. 6B). These effects caused by P4 treatment were abolished by pretreating the cells with ERK inhibitors. The P4-induced increase on the p53 promoter activity was blocked by pretreatment of the HUVECs with a NF-κB inhibitor (Bay11-7082 or CAPE) or pretransfection with DN-ERK2 (Fig. 6C), suggesting the involvement of NF-κB in the P4-induced increase of the p53 promoter activity.
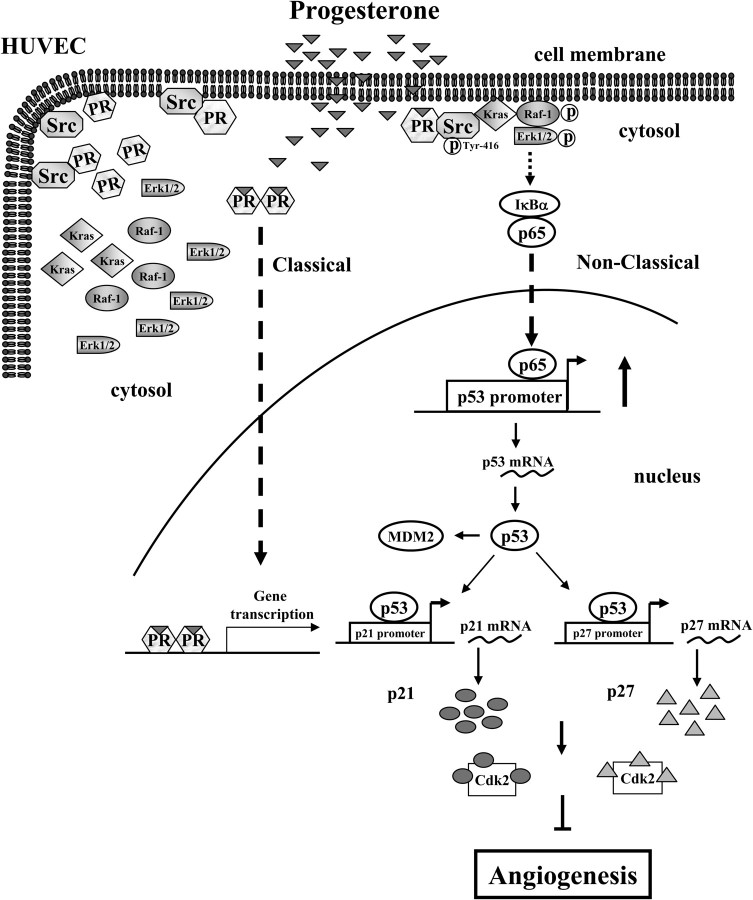
In conclusion, this study provides evidence that P4 bound to PR, subsequently activated the cSrc/Kras/Raf-1/ERK2/NF-κB signaling cascade, which in turn up-regulated the expression of p53, and eventually resulted in the inhibition of endothelial cell proliferation. This signaling cascade was also found to be involved in the folic acid-induced up-regulation of p53 and inhibition of endothelial cell proliferation (Lee, W.-S., et al unpublished data), suggesting that the cSrc/Kras/Raf-1/ERK2/NF-κB signaling cascade is a new pathway in regulating angiogenesis but not the unique progesterone action. Although the results of the present study strongly suggest that P4-induced increase of p53 and inhibition of cell cycle progression in HUVECs were mainly mediated through PR activation of extranuclear signaling pathways, the possible involvement of nuclear PR in this event still can not be completely ruled out. Based on the results from the present study and our previous study, we propose a model of the molecular mechanism underlying P4-induced cell cycle arrest in HUVECs as shown in Fig. 8.
Fig. 8.
Model for P4-induced up-regulation of p53 in HUVECs. P4 activated cSrc, subsequently caused membrane translocation of Kras, which in turn activated Raf-1 and ERK1/2 and increased nuclear translocation of NF-κB (p65), and finally up-regulated the levels of p53 mRNA and protein and inhibited DNA synthesis in HUVECs. Cdk2, Cyclin-dependent kinase 2.
Materials and Methods
Cell and cell culture
HUVECs were grown in M199 (GIBCO, Grand Island, NY) containing 10% fetal bovine serum (FBS; GIBCO), endothelial cell growth supplement (30 mg/liter) (Biomedical Technologies, Stoughton, MA), sodium heparin (5 U/ml; Sigma-Aldrich, St. Louis, MO), 10 mm HEPES (Sigma-Aldrich), and kanamycin (10 mg/liter; GIBCO) in a humidified 37 C incubator. Cells from passages 3–6 were used.
Inhibitors
PP2 was purchased from A. G. Scientific, Inc. (San Diego, CA). Src kinase inhibitor I was purchased from EMD Biosciences, Inc. (La Jolla, CA). Ras InP (H-val-pro-pro-pro-val-pro-pro-arg-arg-arg-OH) was purchased from Alexis (Lausen, Switzerland). FTI trifluoroacetate salt was purchased from Sigma-Aldrich. Sulindac sulfide was purchased from Enzo Life Sciences (San Diego, CA). PD98059, U0126, CAPE, and Bay 11-7082 were purchased from Cayman Chemical (Ann Arbor, MI).
Subcellular fractionation
To observe subcellular localization of P4-induced changes in PR and cSrc intermediates, the cells were washed with ice-cold PBS and resuspended in hypotonic buffer (20 mm Tris-HCl, pH 7.5; 5 mm EGTA; 2 mm EDTA; 1 mm NaVO3; 1 mm dithiothreitol; 10 mm NaF; 10 mm Na2H2P2O7) plus protease inhibitor cocktail (Sigma-Aldrich). After freezing at −80 C for 2 h, the cell lysates were disrupted by passing through a 3/10 ml BD insulin syringe and 30-gauge needle 120 times and then incubated on ice with repeated vortex mixing six times (5 min each time). The supernatant was collected as the cytosolic fraction after being centrifuged at 14,000 × g for 10 min at 4 C. Pellets were washed with ice-cold PBS two times and then lysed in the extract buffer (50 mm HEPES, pH 7.0; 250 mm NaCl; 2.5 mm EDTA; 1% Nonidet P-40; 5% glycerol; 1 mm NaVO3; 10 mm NaF; 10 mm Na2H2P2O7) plus protease inhibitor cocktail (Sigma-Aldrich) on ice with repeated vortex mixing nine times (5 min for each) and then centrifuged at 14,000 × g for 30 min at 4 C. The supernatant was collected as the particulate (membrane) fraction.
Nuclear extraction
To examine the effect of P4 on nuclear translocation of NF-κB (p65), the NE-PER® nuclear and cytoplasmic extraction reagents (Thermo Fisher Scientific, Rockford, IL) was used and the extraction was performed according to the manufacturer's protocol. Briefly, the cell pellet was disrupted in cytoplasmic extraction reagent I buffer plus protease inhibitor cocktail (Sigma-Aldrich) and then lysed in cytoplasmic extraction reagent II buffer to release cytoplasmic proteins. After being centrifuged at 16,000 × g for 5 min at 4 C, the insoluble pellet, which contained nuclear proteins, was washed with ice-cold PBS two times and then incubated in nuclear extraction reagent buffer plus protease inhibitor cocktail (Sigma-Aldrich).
Cell transfection
For transient transfection of the indicated constructs into HUVECs, jetPEI-HUVEC transfection reagent (Polyplus Transfection, Bioparc, France) was used, and the transfection was performed according to the manufacturer's protocol. Briefly, jetPEI-HUVEC/DNA mixture was added dropwise into the DMEM+Glutamax TM I medium (GIBCO) containing 2% FBS, mixed gently, and incubated in a humidified 37 C incubator for 4 h. The growth medium was then replaced and the cells were incubated for the next 24 h. The transfection efficiency was approximately 55% as determined by percentage of the cells transfected with enhanced green fluorescent protein.
Luciferase assay
After treatment with 0.1% dimethylsulfoxide (DMSO; Sigma-Aldrich) (control group) or 500 nm P4 (Sigma-Aldrich) in 0.1% DMSO, the cells were washed twice with ice-cold PBS and then lysed in the passive lysis buffer (Promega, Madison, WI). Luciferase activities were recorded in a 20/20n luminometer (Turner Biosystems, Sunnyvale, CA) using the dual-luciferase reporter assay kit (Promega) according to the manufacturer's instructions. Briefly, individual measurement of firefly luciferase activity and Renilla luciferase activity was assayed sequentially in the same reaction tube. The cell lysate was gently mixed with luciferase assay reagent and then the luciferase activity was recorded. The Stop & Glo reagent (Promega) was then added into the mixture of luciferase assay reagent and cell lysate to measure the Renilla luciferase activity.
[3H]thymidine incorporation
The [3H]thymidine incorporation was performed as previously described (10). Briefly, HUVECs were applied to 24-well plates in growth medium. After the cells had grown to 60–70% confluence, they were rendered quiescent by incubation for 24 h in M199 containing 2% FBS. M199 (phenol red free) supplemented with 10% charcoal-stripped FBS and DMSO (0.1%) with or without P4 (500 nm) was then added to the cells, and the cultures were allowed to incubate for an additional 24 h. During the last 3 h of the incubation, [3H]thymidine was added at 1 μCi ml−1 (1 μCi = 37 kBq). Incorporated [3H]thymidine was extracted in 0.2 n NaOH and measured in a liquid scintillation counter.
Protein extraction and Western blot analysis
Western blot analyses were applied to determine the protein levels in HUVECs and were performed as described previously (10). Electrophoresis was performed using 7.5% or 12% sodium dodecyl sulfate (SDS)-polyacrylamide gel (3 h, 70 V). Separated proteins were transferred onto polyvinyl difluoride membranes (4 h, 400 mA), treated with 5% fat-free milk powder (Anchor, Auckland, NZ) to block the nonspecific IgGs and incubated overnight at 4 C with specific antibody against p53, p21, p27, Kras, p-Raf-1, p-ERK1/2, p-Src, cadherin, NF-κB (Santa Cruz Biotechnology, Santa Cruz, CA); Raf-1 (Epitomics, Burlingame, CA); cSrc (Abcam, Cambridge, MA); PR (Thermo Fisher Scientific, Fremont, CA); Kras (Novus Biologicals, Littleton, CO); MDM2 (Millipore, Temecula, CA); ERK1/2, α-tubulin (Sigma-Aldrich), or glycerol-3-phosphate dehydrogenase (G3PDH; Jackson ImmunoResearch Laboratories, West Grove, PA). The blot was then incubated with antimouse or antirabbit IgG (Jackson ImmunoResearch Laboratories) conjugated to horseradish peroxidase for 1 h. Subsequently the polyvinyl difluoride membrane was developed with chemiluminescence reagent plus (PerkinElmer Life Sciences, Boston, MA). To quantify the intensity of the protein expression levels, the signal was recorded by Berthold WinLight 32 system (Berthold Technologies GmbH & Co KG, Bad Wildbad, Germany), and the band densities were determined as arbitrary absorption units using the Image-Pro Plus 4.5 software program (Media Cybernetics, Silver Spring, MD).
Immunoprecipitation
As previously described (10), the P4-treated HUVECs were lysed in gold lysis buffer [137 mm NaCl; 20 mm Tris, pH 7.9; 10 mm NaF; 5 mm EDTA; 1 mm EGTA; 10% (vol/vol) glycerol; 1% Triton X-100; 1 mm sodium orthovanadate; 1 mm sodium pyrophosphate; 100 μm β-glycerophosphate; 1 mm phenylmethylsulfonyl fluoride (PMSF); 10 μg/ml aprotinin; 10 μg/ml leupeptin] and then immunoprecipitated with anti-cSrc (Abcam), anti-PR (Thermo Fisher Scientific), or anti-Kras (Novus Biologicals) antibody. Cell lysate was precleared by being incubated with protein A agarose beads (Millipore) containing 1 μg of normal mouse IgG (Santa Cruz Biotechnology) at 4 C for 30 min. The beads were discarded by pulsing vortex in the centrifuge at 12,000 × g for 10 sec. The specific antibody (2 μg) was added to 350 μg of precleared cell lysate, and the mixture was then gently shaken overnight at 4 C. The antibody-protein complex was captured by being incubated with PBS prewashed beads at 4 C with gentle shaking. After 4 h incubation, the precipitates were collected by pulsing vortex, washed with ice-cold PBS three times, resuspended in 2× sample buffer (125 mm Tris-HCl, pH 6.8; 4% SDS; 20% glycerol; 10% β-mercaptoethanol; and 0.02% bromophenol blue), and then incubated at 98 C for 10 min to release the proteins from the beads before electrophoresis.
RNA isolation, semiquantitative RT-PCR, and quantitative real-time PCR
As previously described (10), 2 μg of total RNA isolated from HUVECs with Trizol reagent (Invitrogen, Carlsbad, CA) was reverse transcribed to synthesize cDNA using SuperScript III first-strand synthesis system for RT-PCR (Invitrogen). The following primers were used for semiquantitative RT-PCR: p53, 5′-CCTCACCATCATCACACTGG-3′ and 5′-CCTCATTCAGCTCTCGGAAC-3′; and aldolase A, 5′-GGCACCGAGAACACCGAGGAGA-3′ and 5′-TTGATGCCCACAACACCGCCC-3′. PCR amplification with p53 primers, together with the presence of the aldolase A primers, was carried out in a linearly increasing product amp/cycle curve. The PCR products were analyzed using 2% agarose gels containing ethidium bromide. A LightCycler thermocycler (Roche, Mannheim, Germany) was used for the quantitative real-time PCR. Primers specific for p53 gene (forward: 5′-TCAACAAGATGTTTTGCCAACTG-3′ and reverse: 5′-ATGTGCTGTGACTGCTTGTAGATG-3′) and NF-κB binding site within the p53 promoter (forward: 5′-CTTACTTGTCATGGCGACTGTCCA-3′ and reverse: 5′-GACGGTGGCTCTAGACTTTTGAGA-3′) were used. The aldolase A was used as a control to normalize the expression of p53 gene. The fluorescence intensity was measured using the built-in LightCycler software (version 4; Roche).
p53 promoter construct
The 361-bp fragment of human p53 promoter was amplified by genomic PCR using genomic DNA from HUVECs as the template. The 5′ end of this fragment is a BamHI site, which is 231 bp upstream of the exon 1 of the p53 gene, whereas the 3′ end of this fragment is an XbaI site, which locates in exon 1 of the p53 gene (30). The following primers were used: 5′-AGAAGAAAGGATCCAGCTGAGAGC-3′ and 5′-GACGGTGGCTCTAGACTTTTGAGA-3′. This PCR fragment was ligated into pcDNA3.1+ vector (Invitrogen) and then digested with KpnI and XbaI. After blunting the XbaI site of the fragment and the HindIII site of pGL3-basic vector (Promega) with Klenow fragment (Promega), the p53 promoter fragment was then subcloned into pGL3-basic luciferase reporter vector at the KpnI/XbaI sites. The sequence fidelity was confirmed by ABI PRISM 377 DNA analysis system (Applied Biosystems Inc., Foster City, CA).
ChIP assay
The ChIP assay was performed using a standard protocol with minor modifications. Briefly, HUVECs were fixed with 1% (vol/vol) formaldehyde in room temperature for 10 min, stopped by adding glycine to a final concentration of 0.125 m, washed twice with ice-cold PBS, resuspended, and then homogenized in 0.5 ml swelling buffer (5 mm 1,4-piperazine diethane sulfonic acid, pH 8.0; 0.5% Triton X-100; 0.5 mm PMSF) plus Complete protease inhibitor (Roche, Mannheim, Germany). The nuclei pellet was sonicated in 0.5 ml sonication buffer (10 mm EDTA; 50 mm Tris-HCl, pH 8.0; 0.5 mm PMSF) plus Complete protease inhibitor (Roche). A solution containing 0.1% SDS and 85 mm KCl was added to reduce the nonspecific binding. Samples were divided into two equal amounts and individually incubated for 1 h at 4 C with 1 μg of anti-NF-κB antibody (Santa Cruz Biotechnology) or 1 μg of normal mouse IgG (Santa Cruz Biotechnology) as a control, added 30 μl of protein A agarose/salmon sperm DNA slurry (Upstate, Temecula, CA), and then incubated at 4 C overnight with constant rotation. The protein A agarose/antibody/chromatin complex was washed and then dissociated using the Upstate ChIP protocol. DNA was purified using the QIAquick PCR purification kit (QIAGEN, Valencia, CA) and eluted in a 50 μl elution buffer [10 mm Tris-HCl (pH 8.5)]. One microliter of the DNA was assayed in a PCR cycle. The NF-κB binding site within the p53 promoter was analyzed in MOTIF Search (http://motif.genome.jp/) and the primers designed using Primer-BLAST of National Center for Biotechnology Information (Bethesda, MD) were: forward, CTTACTTGTCATGGCGACTGTCCA; reverse, GACGGTGGCTCTAGACTTTTGAGA. All the PCR products were analyzed using 2% agarose gels containing ethidium bromide.
Antisense oligonucleotide
Two individual phosphorothiolated AS oligonucleotides were used to target the cSrc mRNA and a phosphorothiolated scramble (Sc) oligonucleotide was used for the control. One is complementary to the region of active motif around amino acid 416, 5′-ACGTAGTTGCTGGGGATGTA-3′ is for AS cSrc (ASON-1) and 5′-GCCCACACGATCACTTCTAA-3′ is for Sc cSrc (ScON-1). The other one is complementary to nucleotides 349–372 (31), 5′-GGGCTTGCTCTTGTTGCTACCCAT-3′ is for AS cSrc (ASON-2) and 5′-GTTTCCTGCCATGGTGCCTGATCT-3′ is for Sc cSrc (ScON-2). The cSrc oligonucleotides purchased from MDBio, Inc. (Taipei, Taiwan). were administrated to the cell culture for 48 h at a final concentration up to 1 μm before the cells were treated with 500 nm P4.
Statistical procedures
All data were expressed as the mean value ± sem. Comparisons were subjected to one-way ANOVA followed by Fisher's least significant difference test. Significance was accepted at P < 0.05.
Acknowledgments
This work was supported by the National Science Council Grant NSC 96-2320-B-038-023 and NSC 97-2320-B-038-033-MY3(to W.S.-L.).
Disclosure Summary: The authors have nothing to declare.
NURSA Molecule Pages:
Nuclear Receptors: PR;
Ligands: Progesterone | RU486.
Footnotes
- AS
- Antisense oligonucleotide
- CAPE
- caffeic acid phenethyl ester
- ChIP
- chromatin immunoprecipitation
- DMSO
- dimethylsulfoxide
- DN
- dominant negative
- FBS
- fetal bovine serum
- FTI
- farnesyltransferase inhibitor
- G3PDH
- glycerol-3-phosphate dehydrogenase
- HUVEC
- human umbilical vein endothelial cell
- MDM2
- human homolog of mouse double-minute 2
- NF-κB
- nuclear factor-κB
- P4
- progesterone
- PMSF
- phenylmethylsulfonyl fluoride
- PP2
- 4-amino-5-(4-chlorophenyl)-7-(t-butyl)pyrazolo[3,4-d]pyramidine
- PR
- progesterone receptor
- p
- phosphorylated
- Raf-1 In
- Raf-1 inhibitor
- Ras InP
- Ras inhibitory peptide
- Sc
- scramble
- SDS
- sodium dodecyl sulfate
- SKI-I
- Src kinase inhibitor I.
References
- 1. Folkman J , Shing Y. 1992. Angiogenesis. J Biol Chem 267:10931–10934 [PubMed] [Google Scholar]
- 2. Risau W. 1997. Mechanisms of angiogenesis. Nature 386:671–674 [DOI] [PubMed] [Google Scholar]
- 3. Polverini PJ. 1995. The pathophysiology of angiogenesis. Crit Rev Oral Biol Med 6:230–247 [DOI] [PubMed] [Google Scholar]
- 4. Papetti M , Herman IM. 2002. Mechanisms of normal and tumor-derived angiogenesis. Am J Physiol Cell Physiol 282:C947–C970 [DOI] [PubMed] [Google Scholar]
- 5. Zygmunt M , Herr F , Münstedt K , Lang U , Liang OD. 2003. Angiogenesis and vasculogenesis in pregnancy. Eur J Obstet Gynecol Reprod Biol 110:S10–S18 [DOI] [PubMed] [Google Scholar]
- 6. Plendl J. 2000. Angiogenesis and vascular regression in the ovary. Anat Histol Embryol 29:257–266 [DOI] [PubMed] [Google Scholar]
- 7. Girling JE , Rogers PA. 2005. Recent advances in endometrial angiogenesis research. Angiogenesis 8:89–99 [DOI] [PubMed] [Google Scholar]
- 8. Graham JD , Clarke CL. 1997. Physiological action of progesterone in target tissues. Endocr Rev 18:502–519 [DOI] [PubMed] [Google Scholar]
- 9. Vázquez F , Rodríguez-Manzaneque JC , Lydon JP , Edwards DP , O'Malley BW , Iruela-Arispe ML. 1999. Progesterone regulates proliferation of endothelial cells. J Biol Chem 274:2185–2192 [DOI] [PubMed] [Google Scholar]
- 10. Hsu SP , Ho PY , Juan SH , Liang YC , Lee WS. 2008. Progesterone inhibits human endothelial cell proliferation through a p53-dependent pathway. Cell Mol Life Sci 65:3839–3850 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Shupnik MA. 2004. Crosstalk between steroid receptors and the c-Src-receptor tyrosine kinase pathways: implications for cell proliferation. Oncogene 23:7979–7989 [DOI] [PubMed] [Google Scholar]
- 12. Boonyaratanakornkit V , McGowan E , Sherman L , Mancini MA , Cheskis BJ , Edwards DP. 2007. The role of extranuclear signaling actions of progesterone receptor in mediating progesterone regulation of gene expression and the cell cycle. Mol Endocrinol 21:359–375 [DOI] [PubMed] [Google Scholar]
- 13. Meyer C , Schmid R , Scriba PC , Wehling M. 1996. Purification and partial sequencing of high-affinity progesterone-binding site(s) from porcine liver membranes. Eur J Biochem 239:726–731 [DOI] [PubMed] [Google Scholar]
- 14. Falkenstein E , Meyer C , Eisen C , Scriba PC , Wehling M. 1996. Full-length cDNA sequence of a progesterone membrane-binding protein from porcine vascular smooth muscle cells. Biochem Biophys Res Commun 229:86–89 [DOI] [PubMed] [Google Scholar]
- 15. Gerdes D , Wehling M , Leube B , Falkenstein E. 1998. Cloning and tissue expression of two putative steroid membrane receptors. Biol Chem 379:907–911 [DOI] [PubMed] [Google Scholar]
- 16. Nölte I , Jeckel D , Wieland FT , Sohn K. 2000. Localization and topology of ratp28, a member of a novel family of putative steroid-binding proteins. Biochim Biophys Acta 1543:123–130 [DOI] [PubMed] [Google Scholar]
- 17. Evans RM. 1988. The steroid and thyroid hormone receptor superfamily. Science 240:889–895 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Gellersen B , Fernandes MS , Brosens JJ. 2009. Non-genomic progesterone actions in female reproduction. Hum Reprod Update 15:119–138 [DOI] [PubMed] [Google Scholar]
- 19. Boonyaratanakornkit V , Scott MP , Ribon V , Sherman L , Anderson SM , Maller JL , Miller WT , Edwards DP. 2001. Progesterone receptor contains a proline-rich motif that directly interacts with SH3 domains and activates c-Src family tyrosine kinases. Mol Cell 8:269–280 [DOI] [PubMed] [Google Scholar]
- 20. Boonyaratanakornkit V , Bi Y , Rudd M , Edwards DP. 2008. The role and mechanism of progesterone receptor activation of extra-nuclear signaling pathways in regulating gene transcription and cell cycle progression. Steroids 73:922–928 [DOI] [PubMed] [Google Scholar]
- 21. Honda R , Yasuda H. 1999. Association of p19(ARF) with Mdm2 inhibits ubiquitin ligase activity of Mdm2 for tumor suppressor p53. EMBO J 18:22–27 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Fang S , Jensen JP , Ludwig RL , Vousden KH , Weissman AM. 2000. Mdm2 is a RING finger-dependent ubiquitin protein ligase for itself and p53. J Biol Chem 275:8945–8951 [DOI] [PubMed] [Google Scholar]
- 23. Boggs K , Reisman D. 2007. C/EBPβ participates in regulating transcription of the p53 gene in response to mitogen stimulation. J Biol Chem 282:7982–7990 [DOI] [PubMed] [Google Scholar]
- 24. Raman V , Martensen SA , Reisman D , Evron E , Odenwald WF , Jaffee E , Marks J , Sukumar S. 2000. Compromised HOXA5 function can limit p53 expression in human breast tumours. Nature 405:974–978 [DOI] [PubMed] [Google Scholar]
- 25. Liu DX , Lobie PE. 2007. Transcriptional activation of p53 by Pitx1. Cell Death Differ 14:1893–1907 [DOI] [PubMed] [Google Scholar]
- 26. Benoit V , Hellin AC , Huygen S , Gielen J , Bours V , Merville MP. 2000. Additive effect between NF-κB subunits and p53 protein for transcriptional activation of human p53 promoter. Oncogene 19:4787–4794 [DOI] [PubMed] [Google Scholar]
- 27. Llanos S , Efeyan A , Monsech J , Dominguez O , Serrano M. 2006. A high-throughput loss-of-function screening identifies novel p53 regulators. Cell Cycle 5:1880–1885 [DOI] [PubMed] [Google Scholar]
- 28. Halaschek-Wiener J , Wacheck V , Kloog Y , Jansen B. 2004. Ras inhibition leads to transcriptional activation of p53 and down-regulation of Mdm2: two mechanisms that cooperatively increase p53 function in colon cancer cells. Cell Signal 16:1319–1327 [DOI] [PubMed] [Google Scholar]
- 29. Agarwal ML , Ramana CV , Hamilton M , Taylor WR , DePrimo SE , Bean LJ , Agarwal A , Agarwal MK , Wolfman A , Stark GR. 2001. Regulation of p53 expression by the RAS-MAP kinase pathway. Oncogene 20:2527–2536 [DOI] [PubMed] [Google Scholar]
- 30. Reisman D , Greenberg M , Rotter V. 1988. Human p53 oncogene contains one promoter upstream of exon 1 and a second, stronger promoter within intron 1. Proc Natl Acad Sci USA 85:5146–5150 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Marzia M , Sims NA , Voit S , Migliaccio S , Taranta A , Bernardini S , Faraggiana T , Yoneda T , Mundy GR , Boyce BF , Baron R , Teti A. 2000. Decreased cSrc expression enhances osteoblast differentiation and bone formation. J Cell Biol 151:311–320 [DOI] [PMC free article] [PubMed] [Google Scholar]