Abstract
To identify biologically relevant groupings or clusters of nuclear receptors (NR) that are associated with breast neoplasia, with potentially diagnostic, discriminant or prognostic value, we quantitated mRNA expression levels of all 48 members of the human NR superfamily by TaqMan low-density array analysis in 116 curated breast tissue samples, including pre- and postmenopausal normal breast and both ERα+ and ERα− tumor tissue. In addition, we have determined NR levels in independent cohorts of tamoxifen-treated ERα+ and ERα− tissue samples. There were differences in relative NR mRNA expression between neoplastic and normal breast, and between ER+ and ER− tumors. First, there is overexpression of the NUR77 subgroup and EAR2 in neoplastic breast. Second, we identify a signature of five NR (ERα, EAR2, NUR77, TRα, and RARγ) that classifies breast samples with more than 97% cross-validated accuracy into normal or cancer classes. Third, we find a novel negative association between five NR (TRβ, NUR77, RORγ, COUP-TFII, and LRH1) and histological grade. Finally, four NR (COUP-TFII, TRβ, PPARγ, and MR) are significant predictors of metastasis-free survival in tamoxifen-treated breast cancers, independent of ER expression. The present study highlights the discriminant and prognostic value of NR in breast cancer; identifies novel, clinically relevant, NR signatures; and highlights NR signaling pathways with potential roles in breast cancer pathophysiology and as new therapeutic targets.
For over a century it has been recognized that many breast cancers are hormone dependent, at least at the initial stages of the disease. The ongoing management of estrogen receptor (ER)α+ tumors after surgery and/or radiotherapy in postmenopausal women involves the administration of the tissue-selective ER antagonist tamoxifen or aromatase inhibitors, which block conversion of adrenal androgens to active estrogens (1). In breast cancer, determining the levels of ERα [and of progesterone receptor (PR)] has been central to management for almost 40 yr, acting as a guide to treatment, and at least in the short term, to prognosis.
Typically, however, tumors that are ER− and/or PR− are unresponsive to hormonal treatments, and most ER+ PR+ tumors contain clones of nonresponsive cells, which may contribute to the development of estrogen resistance and underlie the incidence of ER− metastases from ER+ primaries. There is thus a compelling reason to identify new markers of sensitivity/resistance to hormonal therapy and ultimately leading to new therapeutic targets. Identification of the epidermal growth factor receptor in a proportion to of breast cancers, and antibody therapy directed thereto, is one example of this need being addressed (2). Other possible interventions may thus involve members of the nuclear receptor (NR) superfamily other than those (ER and PR) most commonly studied. For example, up to 50% of ERα− breast cancers are positive for androgen receptor (AR), and currently, there are two clinical trials underway to determine the benefit of targeting this pathway in ERα− tumors (identifiers include NCT00755885, CDR0000614059, CRUK-CR9304-21, and EUDRACT-2007-003240-30).
Members of the NR superfamily play major roles across human biology: in reproduction, development, growth, metabolism, and homeostasis. In terms of pathophysiology, they play key roles in the cardiovascular and immune systems, the central nervous system, the musculoskeletal system and, most relevant to the present study, in the genesis and progression of cancer (3). The 48 members of the human NR superfamily comprise transcription factors responsive to endogenous or exogenous ligands, including steroids, retinoids, dipeptides, and lipids as well as xenobiotics. The NR superfamily also includes the orphan receptors, which lack characterized endogenous and/or synthetic ligands. Rapidly emerging evidence suggests that although many of these orphan NR operate in a ligand-independent manner, endogenous compounds are being increasingly identified that modulate the activity of many orphan NR.
To date, in addition to ER and PR, a number of superfamily members have been reported to be associated with breast cancer, including AR, GR, PPAR, RAR, and RXR (3); we use IUPHAR trivial names for the NR, with alternative names/nomenclature shown in Supplemental Table 1 (published on The Endocrine Society's Journals Online web site at http://mend.endojournals.org). All of these have defined ligands and have been shown either to be aberrantly expressed or to have prognostic value. There have also been studies on orphan NR in breast cancer, including the ERR, DAX-1, NUR77, LRH1, and RORα (4). All of these studies have focused on one NR or a subset of the NR superfamily, with the attendant strengths (and limitations) of such a focus: they have not set out to take a systems approach to the disease, nor addressed the importance of each of the 48 NR superfamily members in that context.
Previous systems approaches to the functional biology of NR have provided novel insights into tissue-specific members of the superfamily, enabling the identification of new subgroupings based on physiological function rather than ligand (5). Furthermore, although Holbeck et al. (6) profiled NR expression in 51 human cancer cell lines including breast cancer, a systematic analysis of NR expression in normal and neoplastic breast has not been undertaken to date. Given the established roles of some receptors (ER and PR) in growth and development in the breast, and the extraordinary metabolic demands on the lactating breast, we postulated that other NR may have as yet unexplored involvement in breast physiology and pathophysiology, most notably in breast cancer.
As a first step to test this hypothesis, we have determined expression levels of the human NR superfamily across a very carefully curated group of samples, including normal pre- and postmenopausal breast tissue and both ER+ and ER− breast tumors. In this way, we hope to identify biologically relevant groupings or clusters of NR that are associated with breast neoplasia, with potentially diagnostic, discriminant, or prognostic value. In addition, this analysis will serve as a scaffold for additional studies on pathways and mechanisms in breast tissues, deepening our understanding of the pathophysiology and allowing the possibility of novel therapeutic interventions.
Materials and Methods
Human breast tissue cohorts
Data and deidentified breast tissue samples (either fresh-frozen tissue or purified total RNA) were obtained from the tissue banks listed in Acknowledgments. The study was approved by the human research ethics committees of the participating institutions.
Of the breast tissue samples used in this study, 66 individual cases of primary invasive ductal carcinoma with associated histopathological grades and clinical information (age at diagnosis; ER, PR, and HER2 status; and hormone replacement therapy history), were the ER+ and ER− cohorts. In terms of ER status as classified immunohistochemically by tissue banks, samples with very high (scores of 2+ or higher) ER expression (ER+ group) or no (negative) ER expression (ER− group) were requested. Fifty normal breast samples, with no known history of breast disease, were collected after breast reduction mammoplasty or from women who had volunteered normal breast biopsy tissue.
In addition to the immunohistochemical ER+/ER− classification provided by the tissue banks, the range of ER mRNA expression values (as generated by the TaqMan PCR assays) in the individual samples was examined. An unambiguous ER mRNA expression value cutoff point between ER+ and ER− samples was chosen, and any samples in the overlap expression zone were not included in the final cohorts.
To be included in the cohort, samples were required to display a defined level of cellularity, low inflammatory cell content, and low fat content. The majority (80%) of cancer tissues contained more than 75% tumor, and most (68%) of the normal tissues contained 50% normal epithelium. For cases that did not meet these cellularity criterion, only those cases with low inflammatory cell and adipocyte content were deemed suitable for inclusion.
The four breast tissue groups in this study were ER+ (n = 33; mean age = 58.8 yr, age range = 36–90 yr), ER− (n = 33; mean age = 53.2 yr, age range = 27–85 yr), premenopausal normal (n = 30; mean age = 37.6 yr, age range = 20–46 yr), and postmenopausal normal (n = 20; mean age = 60.4 yr, age range = 28–78 yr). All analyses in this study used the combined normal sample groups (n = 50) as the normal breast cohort (mean age = 49 yr, age range = 20–78 yr).
Tissue RNA extraction
Fresh-frozen breast tissue was homogenized in QIAzol lysis reagent (QIAGEN, Valencia, CA). Total RNA extraction and purification was conducted using RNeasy lipid tissue mini kit (QIAGEN) according to manufacturer's instructions. Total RNA quantification and quality was assessed using a NanoDrop spectrophotometer (NanoDrop Technologies) and an Agilent 2100 bioanalyzer (Agilent Technologies, Santa Clara, CA), respectively. Thirty breast tissue samples obtained and not included in this study were discarded based on either poor RNA quality and/or cellularity criteria as previously defined.
TaqMan low-density array (TLDA)
Commercial micro fluidic cards, the TaqMan low-density NR gene signature arrays (Applied Biosystems, Foster City, CA; catalog item 4379961), containing an exclusive set of TaqMan gene expression assays for the 48 NRs and 16 internal controls [18S eukaryotic rRNA; ACTB, β-actin; β2M, β2-microglobulin; GAPDH, glyceraldehyde-3-phosphate dehydrogenase; GUSB, β-glucuronidase; HMBS, hydroxymethylbilane synthase; HPRT1, hypoxanthine phosphoribosyltransferase 1 (Lesch-Nyhan syndrome); IPO8, importin 8; PGK1, phosphoglycerate kinase 1; POLR2A, polymerase (RNA) II (DNA directed) polypeptide A, 220 kDa; PPIA, peptidylprolyl isomerase A (cyclophilin A); RPLP0, ribosomal protein, large, P0 (same as 36B4 in mouse); TBP, TATA box-binding protein; TFRC, transferrin receptor (p90, CD71); UBC, ubiquitin C; YWHAZ, tyrosine 3-monooxygenase/tryptophan 5-monooxygenase activation protein, ζ-polypeptide] were used to profile gene expression. These controls include 18S rRNA, GAPDH, and 36B4, which are validated real-time PCR controls from NURSA-supported NR profiling studies (7). The geNorm software embedded within the ABI/Intergromics StatMiner version 4.1 software package was used to compute least expression variation and select the most appropriate, stable, and robust combination of internal control genes with which to normalize the expression data (against the mean of the most stable controls).
For each sample, 1.5 μg total RNA was reverse transcribed with random hexamers with SuperScript III reverse transcriptase (Invitrogen, Carlsbad, CA) in a total volume of 45 μl. A total of 100 μl reaction mixture containing 50 μl cDNA template (333 ng) in ribonuclease-free water and an equal volume of TaqMan universal master mix (Applied Biosystems) was added to each TLDA fill reservoir. Four reservoirs per sample were filled. The TLDA includes all NRs and endogenous controls in triplicate. After sealing the plate, it was run on an ABI 7900HT real-time instrument (Applied Biosystems).
Data analyses
TLDAs were analyzed by the relative quantification method of change in cycle threshold (ΔCt), and comparing the normalized Ct values (ΔCt) of the replicates between the control/calibrator (normal breast) and the target sample (ER+ and ER− breast cancer). Differentially expressed genes were identified by the comparative Ct method and significance assigned by the nonparametric Wilcoxon (Mann-Whitney U) test (8) embedded in the Integromics StatMiner software suite. The geNorm software (within the StatMiner version 4.2 suite) was used to select the normalization controls with the least expression variation. Benjamini-Hochberg false discovery rate (FDR) correction was used to conservatively filter data, adjust P values, and control for false positives (7, 9).
For analysis of relative expression, a workflow was developed to automate the processing of raw TLDA results files and generate N × X relative expression values, where N is the number of genes in the TLDA experiment, and X is the number of samples. We designed a Pipeline Pilot workflow to read the results files from the TLDA analysis (see Supplemental Fig. 1).
Relative quantification, i.e. the calculated fold differences (between the target and the calibrator/reference sample/tissue) are displayed as valid when the Ct values of the gene in the target and calibrator/reference samples were below 35 cycles. Moreover, fold differences are flagged as target not detected, calibrator not detected, and no detection (i.e. expression in either tissue) when the Ct value of the gene(s) in 50% or more of the target, calibrator, and/or both tissues/samples are over 35 cycles (i.e. the arbitrarily selected threshold limit), respectively. When the Ct value of the target or calibrator/reference is greater than 40 (weakly expressed), the algorithm cannot accurately/precisely calculate expression of the transcript in these samples. The fold difference assigned may not therefore, be absolutely correct, but the assignment of an increase or decrease relative to the control is correct (10). The data generated by the ABI SDS software from the ABI7900 instrument do not normally contain missing Ct values, and Ct values are assigned beyond the arbitrarily set threshold (Ct 35) up to a maximum Ct 40. In situations where the sample is undetected, with Ct values beyond the maximum Ct 40, the StatMiner software imputes a value, set to the maximum Ct.
Gene expression analysis
The gene expression dataset has been previously described (11, 12). Normalized data were used. Genes were matched to Entrez-Gene IDs. If multiple probe sets existed, the one with the highest variance was used. A Cox proportional regression model was used to estimate hazard ratios and 95% confidence intervals for distant metastases-free survival in relation to the expression of the various genes as a continuous variable with adjustment for classic prognostic factors: tumor size (T1 vs. T2), nodal status (positive vs. negative), ESR1 gene expression (continuous), and histological grade (grade 1 vs. 2 vs. 3), using the change in the likelihood ratio χ2 value. No adjustment for multiple testing was performed because these analyses were considered exploratory.
Other statistical analyses, including Spearman rank correlation analyses, Mann-Whitney U tests, Jonckheere-Terpstra tests, and discriminant function analysis and univariable ANOVA were conducted using the PASW Statistics Package, versions 17 or 18 (IBM SPSS, Chicago, IL). Taking account of the 820 (=41C2) multiple comparisons, which represents all possible pairwise comparisons of 41 NR, rank associations were deemed to be of interest if the absolute value of the Spearman rank correlation coefficient was greater than or equal to 0.6 (normal cohort) or 0.65 (cancer cohorts), and the P value was <0.00006 (=0.05/41C2).
Hierarchical cluster analysis was carried out using the Broad Institute genomic analysis platform GenePattern (http://www.broadinstitute.org/cancer/software/genepattern).
Results
The majority of the NR superfamily members are expressed in the human breast
To determine the expression of all 48 NR in 116 breast samples [normal breast (n = 50, 30 pre- and 20 postmenopausal); ERα+ (n = 33) and ERα− breast cancer (n = 33)], we used quantitative RT-PCR with TLDA. A flow chart of the analyses that each tissue sample was subjected to is shown in Fig. 1, and the clinical characteristics of the subjects from whom tissue was obtained as Table 1. Figure 2, A–C, shows the relative expression of NR in three breast cohorts (normal, ER+, and ER−) normalized against the median of four controls (IP08, PGK1, PPIA, and UBC). Forty-one of the 48 members of the human NR family were detected in this way; those not detected were DAX-1, SHP, HNF4α, TLX, FXRα, CAR, and SF1. Detectable NR are variably expressed within each group, with levels ranging over three to four orders of magnitude (Fig. 2, A–C). The patterns of gene expression in normal breast are very similar for pre- and postmenopausal tissue (Supplemental Fig. 1, A and B), as illustrated by the small variances in Fig. 2A where both tissues are combined. We analyzed the differential expression of the NR in the premenopausal human breast cohort, relative to the postmenopausal cohort. This analysis revealed that RORβ, ERα, and NUR77 were the only NRs differentially expressed in a significant manner (Supplemental Fig. 1, C and D). RORβ and ERα are significantly increased in postmenopausal breast; in contrast, NUR77 expression is significantly higher in premenopausal breast (Supplemental Fig. 1, C and D). In ER+ tumors, ERα (Fig. 2B), and in ER− tumors, EAR2 (Fig. 2C) are more highly expressed than any other NR (or than ERα and EAR2 in normal breast tissue).
Fig. 1.
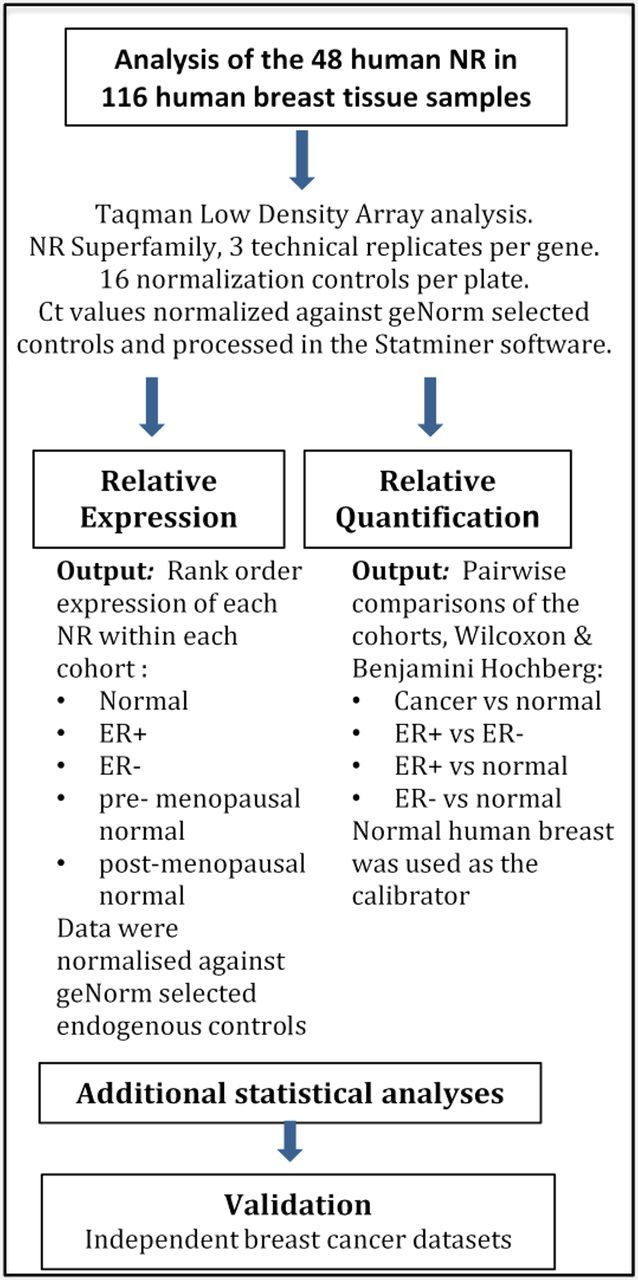
Schematic depiction of the research plan used to analyze 1) the NR expression profiles in normal breast and ER+ and ER− cohorts and 2) the fold differences in NR expression in the ER+ and ER− cohorts relative to the (combined) normal cohort.
Table 1.
Clinical characteristics and patient demographics of the normal and breast cancer cohorts
| Clinical feature | Cohort details |
|---|---|
| Tumor cohorts | |
| ER+ (n) | 33 |
| Grade I (n) | 8 |
| Grade II (n) | 18 |
| Grade III (n) | 7 |
| Age, mean (range) (yr) | 59 (36–90) |
| ER− (n) | 33 |
| Grade II (n) | 6 |
| Grade III (n) | 27 |
| Age, mean (range) (yr) | 53 (27–85) |
| Normal breast cohorts | |
| Premenopausal (n) | 30 |
| Reduction mammoplasty (n) | 9 |
| Breast biopsy (n) | 21 |
| Age, mean (range) (yr) | 38 (20–46) |
| Postmenopausal (n) | 20 |
| Breast biopsy (n) | 20 |
| Age, mean (range) (yr) | 60 (28–78) |
Fig. 2.
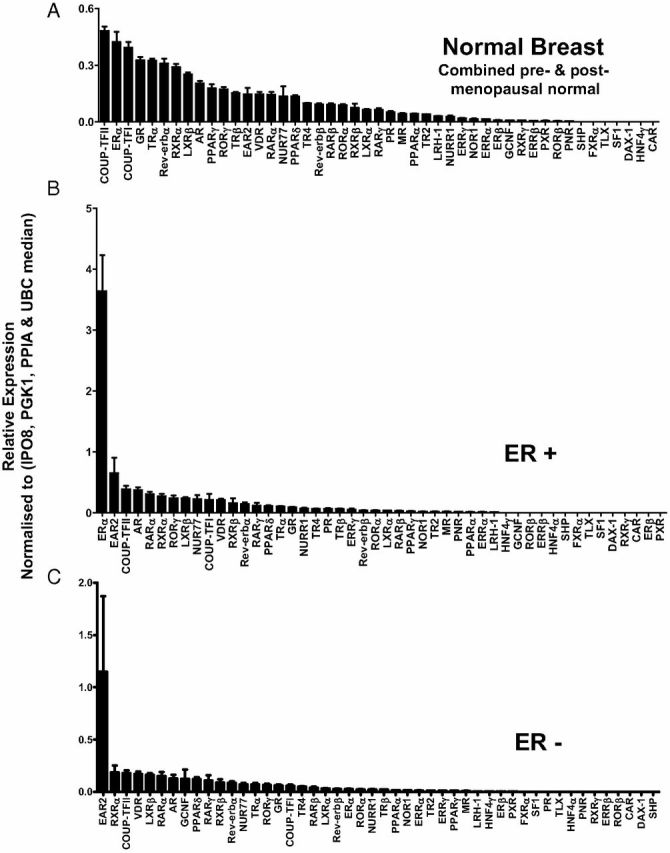
Rank order relative expression (relative expression of NR in each cohort). Data are presented as the relative expression of NR mRNA normalized to the median of the geNorm controls (IPO8, PGK1, PPIA, and UBC). A, Normal breast (combined pre- and postmenopausal normal breast); B, ER+ breast cancers; C, ER− breast cancers.
NR expression differs between normal breast and breast cancer
In addition to ERα and EAR2, there are major differences in NR expression between ER+ and ER− tumors and between tumor and normal tissue. Figure 3 shows supervised cluster analysis illustrating three patterns of NR expression in normal and neoplastic breast tissue. The first cluster (A in Fig. 3) contains 19 NR that are more highly expressed in normal breast than in cancer. The second cluster is of 13 NR (B in Fig. 3) that are more highly expressed in breast cancer. This cluster includes ERRα, and members of the NR1, NR2, and NR4 subgroups; all three NRs in the NR4A subgroup (NUR77, NURR1, and NOR1) are in this cluster. The third cluster (n = 9) (C in Fig. 3) contains NR that discriminate between the two cancer cohorts according to ER status, including RORγ, AR, PNR, and ERα.
Fig. 3.
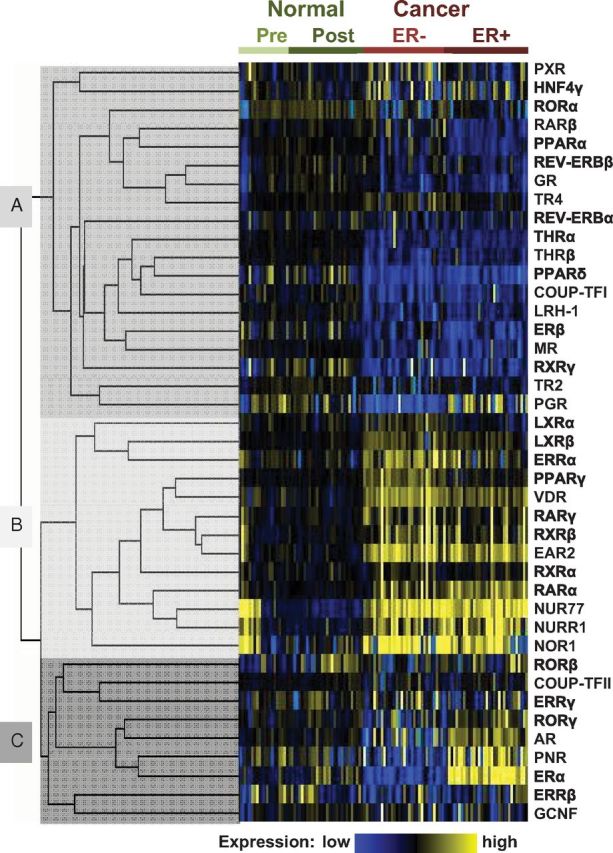
Supervised cluster analysis of NR expression in breast cohorts. The relative expression of NR in normal breast and breast cancer cases, determined by qRT-PCR, was compared by supervised hierarchical clustering analysis using GenePattern (40). The analysis was supervised on cases, and relative expression levels of NR were clustered using a Pearson correlation. Data were median centered both on cases and NR. The resulting clusters were visualized as a heat map, using MapleTree (http://rana.lbl.gov/EisenSoftware.htm), where yellow shading represents NR expression greater than the median and blue represents expression below the median. NR grouped into three major clusters: cluster A contained NR expressed at a lower level in cancer than in normal tissue, cluster B was characterized by NR more highly expressed in malignant than normal tissue, and cluster C identified NR that discriminated between ER+ and ER− breast cancer.
Breast cancer is associated with overexpression of the NUR77/NR4A subgroup and EAR2 and pan-repression of many NR relative to normal breast
We subsequently compared qPCR profiling of the entire NR superfamily in normal breast (n = 50), and in the combined neoplastic (ER+ and ER−) breast cohorts (n = 66) (Fig. 4A and Supplemental Table 2). This shows at least a 3-fold overexpression of both NUR77 and EAR2 (both P < 10−8) and at least 2-fold of NOR1 (P < 10−3) in breast cancer relative to normal breast; the other member of the NR4A subgroup, NURR1, is also significantly induced (P < 10−2 after FDR correction) in breast cancer relative to normal breast (Supplemental Table 2A). In this context, it is interesting to note that NUR77 was expressed approximately 4-fold higher in premenopausal breast relative to postmenopausal breast. Although, NURR1 and NOR-1 were not differentially expressed in these cohorts (Supplemental Fig. 1, C and D). No other members of the NR superfamily are overexpressed in neoplastic compared with normal breast, and over half (23 of 38) are repressed in the breast cancer cohorts (Fig. 4A and Supplemental Table 2A).
Fig. 4.
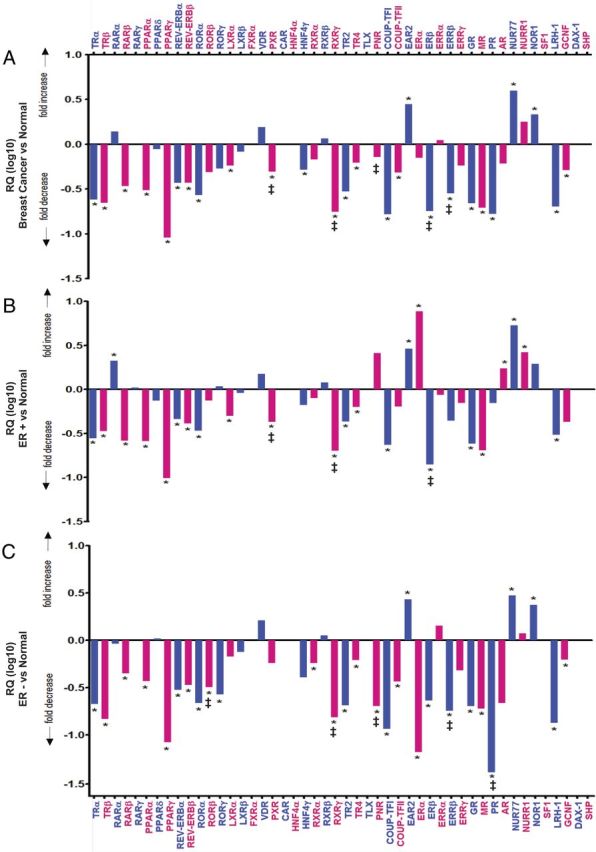
A–C, StatMiner analysis of breast cancer (ER+ and ER−, n = 66) (A), ER+ breast cancer (n =33) (B), and ER− breast cancer (n =33) (C) relative to the calibrator/reference normal (pre- and postmenopausal) breast (n =50). Data are presented as relative quantification (log10), i.e. the calculated fold differences with normal breast as the reference/calibrator normalized to the median of the most stable geNorm (embedded within the Statminer version 4.2 suite) selected controls (IPO8, PGK1, PPIA, and UBC) (selected from the 16 controls assayed on the TLDA) with the least expression variation. Differentially expressed genes were identified by the comparative Ct method, and significance was assigned by the nonparametric Wilcoxon (Mann-Whitney U) test (8) embedded in the Integromics StatMiner software suite. Benjamini-Hochberg FDR correction was used to conservatively filter data, adjust P values, and control for false positives (7, 9). Significance was assigned as P < 0.001 after Benjamini-Hochberg FDR. Relative quantification, i.e. the calculated fold differences [between the target (cancer) and the calibrator/reference sample (normal)] are displayed as valid, when the Ct values of the gene in the target and calibrator/reference samples are below 35 cycles. Fold differences are flagged as target not detected (‡), calibrator not detected (§), and no detection (i.e. expression in either tissue), when the Ct value of the genes in 50% or more of the target, calibrator, and/or both tissues/samples, respectively, are over 35 cycles (the arbitrarily selected threshold limit). NRs (FXRα, CAR, HNF4α, RXRβ, TLX, SF1, DAX-1, and SHP) that were analyzed and denoted as not detected (i.e. Ct value > 35 in cancer and normal breast) have been removed. Alternating blue and pink columns and text are used for clarity and alignment purposes.
Analysis of ER+ breast cancers against normal tissue showed approximately 10-fold overexpression of ERα (P < 10−9), and considerable over expression of NUR77 (P < 10−8), NURR1 (P < 10−4), NOR1 (P < 10−2), AR (P < 10−4), RARα (P < 10−6), and EAR2 (P < 10−8); no increase was seen for any other member of the NR superfamily (Fig. 4B and Supplemental Table 2B). As with the combined cancer cohorts, there is pan-repression of almost half (15 of 36) of the remaining members of the NR superfamily in ER+ breast cancer samples, with a striking decrease in expression of TRα, TRβ, PPARα, PPARγ, GR, MR, TR2, RARβ, and LRH1 in ER+ tumors (adjusted P values < 10−9; Supplemental Table 2B). All other NR with decreased expression show adjusted P values < 10−5 (Supplemental Table 2B).
Analysis of ER− breast cancer against normal breast shows approximately 3-fold overexpression of EAR2 (P < 10−5), NUR77 (P < 10−5), and NOR1 (P < 10−3), with no increase in the expression of any other member of the NR superfamily (Fig. 4C and Supplemental Table 2C). This finding is consistent with our findings for the combined cancer cohorts and ER+ breast cancers; again, there is pan-repression of over half (20 of 38) of the remaining NR. Compared with normal breast, expression of TRα, TRβ, PPARγ, REV-ERBβ, COUP-TFI, ERα, GR, PR, TR2, and LRH1 (P < 10−9) and of RXRα, RXRγ, RORα, RARβ, PPARα, MR, COUP-TFII, REV-ERBα, and ERβ (P < 10−5) is decreased in ER− tumors.
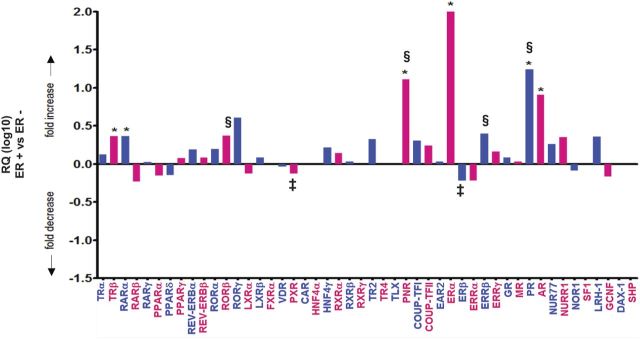
Analysis of ER+ tumors against ER− tumors shows significant overexpression of six NR (AR, RARα, ERα, TRβ, PR, and PNR) compared with ER− (Fig. 5 and Supplemental Table 2D). For a number of these, overexpression is substantial; expression of ERα in ER+ is >100-fold greater than that in ER− (P < 10−9 after FDR), with approximately 10-fold increases in expression of AR, PR, and PNR (P < 10−3 after FDR) (Fig. 5 and Supplemental Table 2D). Of the other NR, expression of RARα and TRβ was approximately 5-fold higher in the ER+ than the ER− group (P < 10−3) (Fig. 5 and Supplemental Table 2D).
Fig. 5.
StatMiner analysis of ER-positive breast cancer (n = 33) relative to the calibrator/reference ER− breast cancer (n = 33). Data are presented as relative quantification (log10), i.e. the calculated fold differences with ER− breast cancer as the reference/calibrator normalized to the median of the most stable geNorm (embedded within the StatMiner version 4.2 suite) selected controls (IPO8, PGK1, PPIA, and UBC), fold differences are flagged as target not detected (‡), calibrator not detected (§), and no detection (i.e. expression in either tissue) as described for Fig. 4. NR (FXRα, CAR, HNF4α, RXRγ, TLX, SF1, DAX-1, and SHP) that were analyzed and denoted as not detected (i.e. Ct value > 35 in ER+ and ER− breast cancer) have been removed.
The overall analysis of the NR differences between ER+ and ER− and between the combined breast cancer and normal cohorts is summarized in tabular form in Fig. 6A. The majority of the 48 human NR (41 of 48) were detected in normal breast. The majority of these (33 of 41) were significantly differentially expressed in neoplastic breast relative to normal breast (Fig. 6A). Of these, seven NR were higher in neoplastic breast, with the rest (26) showing decreased expression (Fig. 6A). Comparison of NR differentially expressed between normal and neoplastic breast revealed that the majority (19 of 33) of NR that differ between neoplastic and normal breast (Fig. 6A) are common to both ER+ and ER− breast cancers, highlighting the major differences in expression of the NR family being between normal and neoplastic tissues. Heterogeneity in NR expression across the three cohorts was confirmed by Kruskal-Wallis nonparametric ANOVA. The majority (31 of 41 = 76%) were differently expressed across the three cohorts (Supplemental Table 3), and those with family-wise error rates less than 0.0005 after Holm's correction for multiple comparisons are shown in Supplemental Table 3.
Fig. 6.
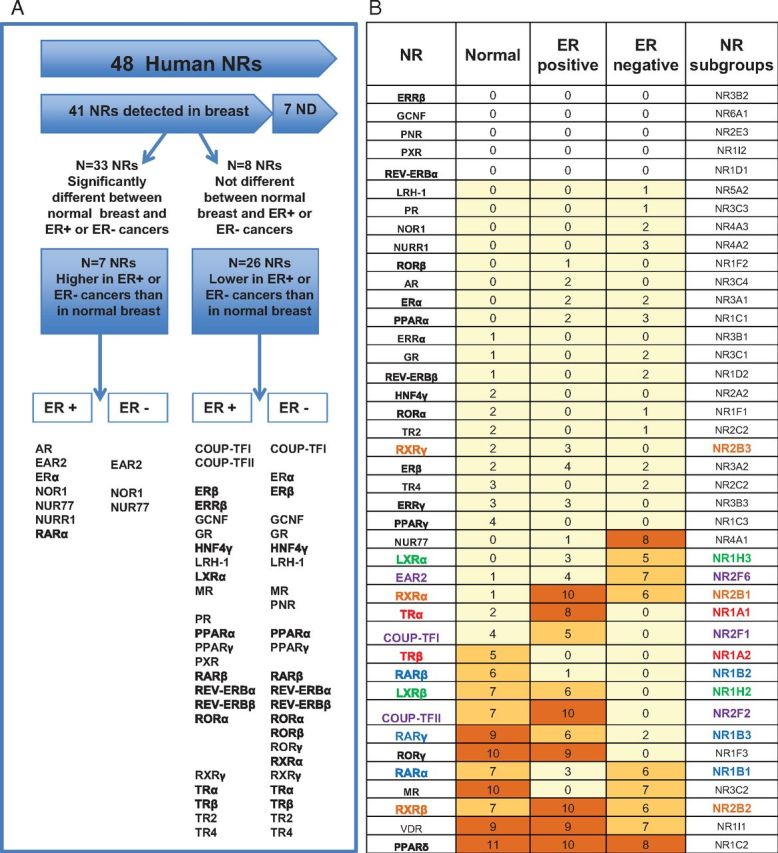
A, Summary of the distribution of the expression of NR in normal breast and the breast cancer. B, Association between the expression of NR in the breast. The table lists all detected NR and their number of significant associations to all other NR in the normal breast and breast cancer cohorts. Colored shading highlights the number of significant associations: yellow, one to five associations, medium orange, five to seven, and dark orange, eight or more associations. NR from subfamilies overrepresented in the group of NR with the highest associations are colored according to subfamily: red, NR1A, blue, NR1B, green, NR1H, orange, NR2B, purple, NR2F. Significant associations had Spearman rank correlation coefficients greater than or equal to 0.6 (normal cohort) or 0.65 (cancer cohorts), and P values < 0.00006. These associations are summarized in Supplemental Figs. 2 and 3.
Specific NR subgroups are coexpressed in the breast
We proceeded to test whether the expression of NR family members was associated by testing the rank correlation of each NR with every other NR in normal breast tissues and in ER+ and ER− breast cancers. Spearman rank correlation coefficients are shown in Supplemental Fig. 2. The different correlation coefficient cutoffs for the normal and cancer cohorts reflected the differences in precision due to different numbers in these cohorts (normal, n = 50; cancers, n = 33 per cohort). Most NR (36 of 41) exhibit associations of interest with at least one other NR in at least one of the breast cohorts, although fewer than half (17 of 41) are characterized by a moderate number (at least five) of associations (Fig. 6B and Supplemental Fig. 2).
Different patterns of association of NR are observed (Fig. 6B and Supplemental Fig. 3, A–C). NUR77, for example, appears highly ER− specific, with eight associations in ER−, one in ER+, and none in normal tissue. Three NR, RXRβ, VDR, and PPARγ, show high levels of association in all three groups. Another three, LXRβ, COUP-TFII, and RORγ are highly associated in normal breast and ER+ breast cancer, with no associations in ER− cancer. TRβ is moderately (n = 5) associated in normal tissues, with no associations in either ER+ or ER− breast cancers (Fig. 6B and Supplemental Fig. 3, A–C).
As shown in Fig. 6B (and in Supplemental Table 4A), sex steroid receptors do not frequently associate with other members of the NR family, either in the normal breast or in breast cancer. For example, PR associates only with ER, AR associates only with RORγ and ERRγ, and only three NR are associated with ERα expression (RARα, EAR2, and PR). ERβ is associated with seven NRs: TRα, PPARβ/δ, PPARγ, LXRα, VDR, RXRα, and MR (Supplemental Table 4A). NR for other steroids vary in their levels of association; although GR is infrequently associated with other NR in any group, and VDR with many other NR in all three groups, MR expression is highly associated with 10 other NR in the normal breast, with Spearman rank correlation coefficients close to or exceeding 0.7 in most instances (Supplemental Table 4B). MR is not correlated with any NR in ER+ breast cancer but with seven NR in ER− breast cancer (Fig. 6B and Supplemental Table 4B). Curiously, only three of the NR associated with MR in ER− breast cancer (RARα, PPARγ, and VDR) also show the association in the normal breast; the remaining four (RXRα, EAR2, ERβ, and NUR77) are associated with MR only in the cancer cohort. Reciprocally, seven of the 10 associations for MR are specific for normal breast (TRβ, RARγ, RORγ, LXRβ, RXRβ, and COUP-TFII).
NRs in addition to the sex steroid receptors ERα, PR, and AR are associated with histological grade
The sex steroid receptors ERα, PR, and AR are classically associated with histological grade, with higher levels indicative of a well-differentiated phenotype. To test whether other NR are associated with clinical characteristics of cancer tissue such as histological grade, the variation in NR expression by grade was determined by the Jonckheere-Terpstra nonparametric test. The known association of ERα, PR, and AR with grade is recapitulated in the cohorts used in this study (Table 2). Five other NR vary in their expression across different grades: LRH1, TRβ, NUR77, RORγ, and COUP-TFII (Table 2). The relationship is negative in that these NR are more highly expressed in better-differentiated, lower-grade lesions. These eight NR identified in our dataset as associated with histological grade (Table 2) were examined in an independent publicly available dataset of 354 tamoxifen-treated ER+ breast cancers (13). Six of the eight NR are significantly associated with histological grade (Table 3). Based on the strong contribution of proliferation to histological grade, we examined the association with the proliferation marker Aurora K, a component of the replication licensing machinery measured at the transcript level and therefore comparable with NR expression in these analyses. Five of the same six NR confirmed as associated with histological grade in the independent dataset were significantly associated with Aurora K expression (Table 3), further implicating this signature in the pathogenesis of these tumors.
Table 2.
NR associated with histological grade in breast cancers
| NR | P value |
|---|---|
| ERα | 0.000003 |
| PR | 0.00007 |
| LRH-1 | 0.00008 |
| TRβ | 0.00009 |
| AR | 0.00009 |
| NUR77 | 0.0002 |
| RORγ | 0.0006 |
| COUP-TFII | 0.0009 |
There was a significant relationship between expression of the indicated NR and increasing histological grade, as identified by the Jonckheere-Terpstra test.
Table 3.
The association of NR with histological grade is confirmed in an independent clinical cohort
| NR | Aurora K |
Histological grade P value | |
|---|---|---|---|
| Pearson's correlation coefficient | P value | ||
| ER | −0.17 | −0.29 | 0.017 |
| PR | −0.29 | 0.00 | 0.000 |
| TRβ | −0.09 | 0.08 | 0.001 |
| NUR77 | −0.19 | 0.00 | 0.100 |
| RORγ | −0.18 | 0.00 | 0.000 |
| COUP-TFII | −0.19 | 0.00 | 0.000 |
| LRH-1 | −0.08 | 0.14 | 0.137 |
| AR | −0.17 | 0.00 | 0.047 |
The eight NR identified in our dataset to be associated with histological grade were tested in an independent publicly available dataset of tamoxifen-treated ER+ breast cancers. All but LRH-1 and TRβ were significantly associated with Aurora K, and all except NUR77 and LRH-1 were significantly associated with histological grade (above).
A signature of five NRs is sufficient to classify breast tissues
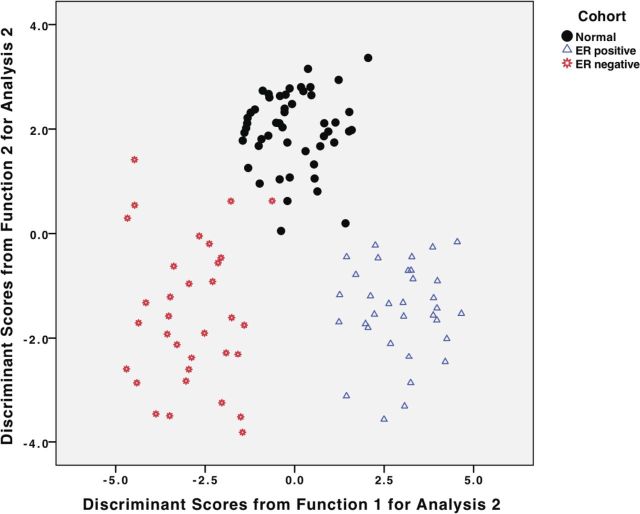
After investigating the pairwise associations between NR summarized in Fig. 6, we explored the possibility that an NR signature could discriminate normal breast from ER+ and ER− tumors. Five-fold cross-validated discriminant function analysis was used to determine whether the combined expression of NRs within an individual tissue sample can effectively predict whether it is normal, ER+, or ER− (Supplemental Table 5, A–C). Five NR (TRα, ERα, NUR77, EAR2, and RARγ) were sufficient for discrimination. ERα was by far the most highly associated with discriminant function 1, which divides ER+ from ER− breast cancers. TRα was by far the most highly associated with discriminant function 2, which discriminates between normal and cancer. The discriminant functions accurately predict the cohort to which a specific case belonged, with cross-validated accuracy of 97.4% (Supplemental Table 5, A–C). Specifically, although ER+ cancers are perfectly classified, one normal case was misclassified as an ER+ breast cancer, and two ER− breast cancers were misclassified as normal. This is also illustrated in a scatter plot of discriminant function scores (Fig. 7), which reinforces the remarkable separation of cases into the three cohorts, with the slight overlap at the boundaries of the normal and ER− scatter plots reflecting the very small percentage of misclassified cases.
Fig. 7.
Discriminant function analysis. The plot demonstrates the classification of all cases into the three clinical cohorts, using the discriminant functions. See Supplemental Table 5, A–C, for complete details.
TRβ, COUP-TFII, MR, and PPARγ are independent predictors of metastasis-free survival in tamoxifen-treated patients
Finally, as an exploratory analysis, we evaluated clinical outcome associations in an independent dataset of ER+ breast cancers from patients with available clinical data, all of whom had received adjuvant tamoxifen monotherapy (13, 14). When TRβ, COUP-TFII, MR, and PPARγ are evaluated individually in a Cox regression hazards model, all are independent predictors of better distant metastasis-free survival after adjustment for expression of ERα (ESR1 mRNA expression) and for other clinical prognostic factors known to be of value in hormone therapy-treated ER+ breast cancers (11). Thus, this novel NR signature provides an additional contribution to long-term prognosis independent of ER+ status and therefore could be interesting for further therapeutic development (Table 4).
Table 4.
NR expression associated with prognosis in tamoxifen-treated ER+ patients
| NR | Univariable (mean, confidence levels) |
Multivariable (each NR adjusted for all others) |
||
|---|---|---|---|---|
| HR (95% CI) | P value | HR | P value | |
| TRβ | 0.48 (0.40–0.79) | 0.001 | 0.51 | 0.010 |
| PPARγ | 0.42 (0.34–0.86) | 0.009 | 0.46 | 0.010 |
| COUP-TFII | 0.50 (0.33–0.77) | 0.002 | 0.59 | 0.034 |
| MR | 0.61 (0.38–0.74) | 0.001 | 0.53 | 0.023 |
Multiple Cox regression analysis unadjusted and adjusted for nodes (negative vs. positive), grade (1 vs. 2 or 3), tumor size (T1 vs. T2), and ERα (continuous). Only those NR significant in the multiple Cox model and whose expression was not correlated with ERα expression are shown. CI, Confidence interval; HR, hazard ratio.
Discussion
Profiling of the human NR superfamily has identified differential and selective expression of NR between normal and neoplastic breast tissues. It has established patterns of association between members of the NR superfamily across normal/ER+/ER− samples and, in addition, shown that particular associations of receptors have both diagnostic and prognostic significance in breast cancer.
In terms of overall expression in normal breast, NR levels are similar in pre- and postmenopausal tissues. Many of the highly expressed NRs in either normal group are common to both. The exceptions are HNF4α, expressed at relatively low levels postmenopausally and at vanishingly low levels in the premenopausal group; NUR77, at relatively high levels premenopausally and at lower levels in postmenopausal patient samples; and ERβ, which shows increased expression in postmenopausal breast. A coherent physiology across the NR superfamily can be attempted only once such questions are answered and also (crucially but with obvious difficulty) when similar biopsy samples are obtained during pregnancy and lactation.
In the context of potential therapeutic exploitation in breast cancer, one notable feature of the data is the aberrant expression of the entire NR4A subgroup in breast cancer. We observed striking overexpression of the NR4A subgroup in ER+ and ER− breast relative to normal breast; high expression of NUR77 was associated with lower histological grade, and NUR77 was one of only five NR with a discriminant role in the classification of breast tissue. The negative correlation of NUR77 expression with histological grade is interesting in the context of significant down-regulation of this NR in the pre- to postmenopausal normal breast (Supplemental Fig. 1, C and D).
NR in the NR4A subgroup are crucial regulators of metabolism, muscle, and energy homeostasis (15). In the brain, they play a role in neuroprotection after stress (16) and have been implicated in vascular remodeling (15). Until recently, NR4A NR were not known to play roles in cells of the epithelial lineage, nor have they been implicated in cancers. Recently, however, NUR77 has been identified in pancreatic cancer, with expression higher than in nontumor tissues (17), and even more recently, modulation of expression of the NUR77 subgroup has been shown in many types of cancers (18). Furthermore, NUR77 transcripts have been identified in a microarray study as up-regulated in primary breast cancer compared with normal breast (19), and in cell lines, NUR77 reduces cell migration (19), suggesting a potential influence on metastasis and consistent with its role as one of five NRs with a discriminant function.
The emerging data on the NUR77 subgroup suggest that they may provide new targets for therapeutic intervention in breast cancer. Further supporting this is the observation that this subgroup of NR can be modulated by the antineoplastic, antiinflammatory, and immunosuppressive purine antimetabolites (e.g. 6-mercapotopurine) (20), 5-fluorouracil, cytosporine B, etc. that have demonstrated therapeutic utility and have been exploited against several cancers (18). Furthermore, in the context of purine antimetabolites, the incidence of breast cancer is decreased in chronically immunosuppressed women (21). Currently, several clinical trials are underway examining the utility and efficacy of 6-meracaptopurine and methotrexate in breast cancer (http://www.controlled-trials.com/ISRCTN63150635).
Another salient finding of this study is the considerable overexpression of EAR2 (an orphan member of the COUP-TF-like family) in ER+ and ER− breast tissue relative to normal and its abundant expression relative to other NR in both the ER+ and ER− cohorts. Our studies show that EAR2 is part of a five NR signature sufficient to accurately classify breast tissue. Although there have been no systematic studies of EAR2 in breast cancer, it has been shown to negatively regulate aromatase expression in breast cancer cell lines by binding directly to the aromatase promoter (22). The other members of the COUP-TF subgroup, COUP-TFI and -II, were significantly decreased in (ER+ and ER−) breast cancer; COUP-TFII expression was negatively correlated with histological grade and was an independent predictor of improved distant metastasis-free survival after adjustment for expression of ERα. In this context, Nagasaki et al. (23) showed that positive COUP-TFII immunohistochemical staining was associated with poor clinical outcome and survival.
The orphan NR LRH-1 was weakly expressed in normal (pre- and postmenopausal) breast tissue and down-regulated in ER+ and ER− breast cancer. Moreover, its expression was negatively correlated with histological grade. LRH-1 is present in breast cancer epithelium and stroma (24) and stimulates breast cancer cell proliferation, invasion, and migration in vitro (25). Known target genes in breast include aromatase (26) and ERα (27). LRH-1 expression is itself regulated by ERα (28) and thus appears to be intimately involved in estrogen signaling. The reasons for this difference between tissue- and cell-based studies remain to be explored, as do the precise roles of LRH-1 in breast cancer in vivo.
Relatively little is known about TR expression in breast cancer. Several studies reported decreased TRα/β expression in breast cancers (29–32) and postulated that TRβ functioned as a tumor suppressor in this cancer. The mechanism of lowered TRβ expression appears to be in part related to aberrant promoter hypermethylation (30). In addition, Guigon et al. (33) generated a knock-in TRβ mutation model and showed that the mice develop accelerated mammary tumors. Lowered expression of TRβ is consistent with the observations that EAR2 operates as a negative coregulator (34). Our data suggest novel previously unrecognized roles for TRα and TRβ in breast cancer, with both TR genes clearly down-regulated in ER+ and ER− cancer compared with normal tissue. TRβ was one of only two NR with expression that associated with that of other NR in normal breast, whereas TRα was associated with other NR only in cancer (ER+/ER−), suggesting divergent and selective expression patterns for each isoform. TRα was one of five NR providing powerful discriminant information to classify breast tissue; TRβ was negatively correlated with histological grade and was an independent predictor of improved distant metastasis-free survival in tamoxifen-treated patients. Taken together, these data provide strong evidence for specific roles for TR in breast cancer; TRβ is seen as a key tumor suppressor molecule, of which restoration and/or preservation of expression may be important clinically. TRα, on the other hand, appears to provide diagnostic information at presentation. TR could potentially be thus useful as biomarkers, as diagnostic tools (TRα) and as prognostic markers of outcome to various therapies (TRβ).
Our data identified PPARγ as a predictor of metastasis-free survival in tamoxifen-treated patients, independent of ER expression. PPARγ has been previously identified as an independent prognostic factor in ductal carcinoma (35), although several clinical studies did not display the expected therapeutic outcomes with glitazones (36–38). This notwithstanding, there is indirect evidence that modulation of PPAR signaling, as occurs in type 2 diabetics being treated with glitazone and/or metformin, can lead to improved clinical outcomes in those diabetics that develop breast cancer (39). Taken together, the results of this study strengthen the inference that there may be potential therapeutic utility of targeting PPARγ in breast cancer in addition to other NRs in combination therapy.
The identified differences, outlined above, in the expression of various NRs between normal and neoplastic breast, or between ER+ and ER− breast cancers, should be considered as markers, and further accrual of data, from genetic and pharmacological studies, will provide insights into the putative mechanistic roles of NR in breast cancer. For example, a recent report into the clinical significance of aberrant NR expression in breast cancer revealed increased PNR in ER+ breast cancers (12). PNR was shown to directly regulate ERα expression and was significantly associated with recurrence-free survival in tamoxifen-treated ER+ breast cancer patients.
To further identify potential interactions between NRs in the breast, we documented the associations between members of the NR superfamily. The results were surprising in three ways. The first is the relative paucity of close associations between the classic NR considered in breast cancer, ER, PR, and AR, with other NR across the three groups, normal, ER+, and ER− tissues. In contrast with the receptors commonly associated with breast cancer, the second finding of interest is the high level of associations shown by three NR (RXRβ, PPARγ, and VDR) across all three groups, normal, ER+, and ER−. One possible (cautious) interpretation of this finding might be that their intracellular roles do not differ substantially between normal and neoplastic tissues. The third finding is perhaps the most striking and potentially in terms of discriminant and prognostic markers the most promising. TRβ is associated with a modest number of other NRs in normal tissue but none in ER+ or ER− breast cancers. COUP-TFI, TRα, LXRβ, COUP-TFII, and RORγ are associated with other NR in both normal and ER+ tissue but none in ER− tissue. This changed association between NR in normal and neoplastic tissues suggests that there are aberrations in the mechanisms controlling NR homeostasis in the breast that may be important for the pathophysiology of breast cancer.
In addition, and very surprisingly, MR (expressed at modest levels in all tissues) shows associations with other NR in normal and ER− tissue but none in ER+ breast cancer. The finding that MR both powerfully discriminates the three cohorts and predicts survival in a cohort of ER+ tumors was unexpected. The physiology of MR in mammary epithelium is obscure; presumably, MR is occupied by cortisol, rather than being mineralocorticoid selective, in these epithelial cells. One possibility is that cortisol-occupied MR may play a crucial role in cellular apoptosis and milk formation under the stimulus of prolactin. A corollary of such a hypothesis is that this action in the late pregnant mammary gland is blocked by the high levels of progesterone. This would suggest that diminished MR expression during the neoplastic process may be in effect antiapoptotic; alternatively, the decreased MR levels in neoplasia may be simply a bystander effect.
In terms of histological grade, the between-NR associations vary across the spectrum. Although degree of significance does not necessarily translate into functional importance, the five NR most significantly associated with histological grade (PR, LRH-1, AR, ERα, and TRβ) show very low levels of association, although the TRβ associations are uniquely in normal tissue. At the other end of the spectrum are NUR77, COUP-TFII, and RORγ. In the Jonckheere-Terpstra test, these three latter NR are significantly associated with histological grade and with the proliferation marker Aurora K as well as the classic ER/PR/AR. The different distribution of association with histological grade, NUR77 vs. COUP-TFII and RORγ in ER− tissue, may point to different roles for NUR77 in terms of histological grade from those of COUP-TFII or RORγ, either driving (NUR77) or protecting against (COUP-TFII and<med> RORγ) dedifferentiation.
The patterns of NR association similarly do not tightly align with the outcomes of the discriminant function analyses, which show five NR (ERα, TRα, NUR77, EAR2, and RARγ) to be sufficient for high-level discrimination between normal breast and ER+ and ER− breast cancer. NUR77 might be held on the basis of association to discriminate normal from ER−, and perhaps EAR2 from cancer, and TRα to discriminate between ER+ and ER− tissue. In the tamoxifen-treated cohort of ER+ tumors, four NR are predictive of metastasis-free survival. Of these four, three (TRβ, MR, and PPARγ) have no associations with other NR in ER+ tumors; a cautious inference might thus be that absence of such associations is protective in terms of disease-free survival. In contrast, COUP-TFII has associations with other NR in ER+ tumors and none in ER− tissue; again, a cautious extrapolation might be that the presence of such associations with COUP-TFII is protective in terms of disease-free survival.
In summary, this systematic study of NR expression and between-NR associations, across the gamut of 48 human NR, has provided very significant evidence for the substantial differential expression of NR in breast cancer. Moreover, they also provide markers for finer histological grading, for high-level distinction between normal, ER+, and ER− tissue, and perhaps most importantly as a prognostic indicator of disease-free survival in ER+ patients.
Current studies on NR-null mice and on tumor cell lines are in progress to examine a range of the questions raised and, furthermore, to define mechanisms and pathways. In the meantime, the value of these novel biomarker signatures for histological stage, discrimination, and prognosis should not be underestimated. The rigorous NR expression analysis expands the horizon of therapeutic intervention and also highlights unanticipated questions; for example, the clinical significance of increased expression of the NUR77 subgroup and the remarkable suppression of MR associations in ER+ tumors compared with its promiscuous association with NR in ER− and normal breast.
Acknowledgments
This work was supported by a Collaborative Program Grant from the National Breast Cancer Foundation Australia. G.E.O.M. was a University of Queensland Vice Chancellors Research Fellow, E.R.S., P.J.F., and C.L.C. were research fellows of the National Health and Medical Research Council of Australia. Breast cancer and normal tissues were provided by Australian Breast Cancer Tissue Bank (ABCTB), which is supported by the National Health and Medical Research Council of Australia, the Cancer Institute NSW, and the National Breast Cancer Foundation; or by the Victorian Cancer BioBank Australia, which is supported by the Victorian Government. ABCTB tissues and samples were made available to researchers on a nonexclusive basis. Normal breast biopsies were obtained from the Susan G. Komen for the Cure Tissue Bank at the IU Simon Cancer Center. We thank contributors to the Susan G. Komen for the Cure Tissue Bank, including Indiana University, which collected the samples used in this study, as well as parents and families whose participation and help made this work possible.
G.E.O.M., N.A.E., K.B., S.L., C.C., J.W.F., E.R.S., M.A.R., E.K., P.J.F., W.D.T., P.J.L., and C.L.C. participated in the design and/or interpretation of the reported experiments or results; G.E.O.M., N.A.E., K.B., S.L., D.G., SJ, M.J.D., E.K., W.D.T., and C.L.C. participated in the acquisition and/or analysis of data; G.E.O.M., N.A.E., K.B., S.L., D.G., C.C., J.W.F., E.R.S., M.A.R., P.J.F., W.D.T., P.J.L., and C.L.C. participated in drafting and/or revising the manuscript; G.E.O.M., N.A.E., K.B., S.L., D.G., S.J., M.J.D., E.K., W.D.T., and C.L.C. were primarily responsible for a particular, specialized role in the research; and G.E.O.M., E.K., and C.L.C. provided administrative, technical, or supervisory support.
Disclosure Summary: The authors have nothing to disclose.
Footnotes
- AR
- androgen receptor
- ΔCt
- change in cycle threshold
- ER
- estrogen receptor
- FDR
- false discovery rate
- NR
- nuclear receptor
- PR
- progesterone receptor
- TLDA
- TaqMan low-density array.
References
- 1. Cuzick J , Sestak I , Baum M , Buzdar A , Howell A , Dowsett M , Forbes JF. 2010. Effect of anastrozole and tamoxifen as adjuvant treatment for early-stage breast cancer: 10-year analysis of the ATAC trial. Lancet Oncol 11:1135–1141 [DOI] [PubMed] [Google Scholar]
- 2. Arteaga CL , Sliwkowski MX , Osborne CK , Perez EA , Puglisi F , Gianni L. 2012. Treatment of HER2-positive breast cancer: current status and future perspectives. Nat Rev Clin Oncol 9:16–32 [DOI] [PubMed] [Google Scholar]
- 3. Conzen SD. 2008. Nuclear receptors and breast cancer. Mol Endocrinol 22:2215–2228 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Riggins RB , Mazzotta MM , Maniya OZ , Clarke R. 2010. Orphan nuclear receptors in breast cancer pathogenesis and therapeutic response. Endocr Relat Cancer 17:R213–R231 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Bookout AL , Jeong Y , Downes M , Yu RT , Evans RM , Mangelsdorf DJ. 2006. Anatomical profiling of nuclear receptor expression reveals a hierarchical transcriptional network. Cell 126:789–799 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Holbeck S , Chang J , Best AM , Bookout AL , Mangelsdorf DJ , Martinez ED. 2010. Expression profiling of nuclear receptors in the NCI60 cancer cell panel reveals receptor-drug and receptor-gene interactions. Mol Endocrinol 24:1287–1296 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Wang SC , Myers SA , Eriksson NA , Fitzsimmons RL , Muscat GE. 2011. Nr4a1 siRNA expression attenuates α-MSH regulated gene expression in 3T3–L1 adipocytes. Mol Endocrinol 25:291–306 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Glover T , Mitchell K. 2008. An introduction to biostatistics. 2nd ed Long Grove, IL: Waveland Press [Google Scholar]
- 9. Crowther LM , Wang SC , Eriksson NA , Myers SA , Murray LA , Muscat GE. 2011. Chicken ovalbumin upstream promoter-transcription factor II regulates nuclear receptor, myogenic, and metabolic gene expression in skeletal muscle cells. Physiol Genomics 43:213–227 [DOI] [PubMed] [Google Scholar]
- 10. Goni R , Garcia P , Foissac S. 2009. The qPCR data statistical analysis. Integromics White Paper [Google Scholar]
- 11. Kim C , Tang G , Pogue-Geile KL , Costantino JP , Baehner FL , Baker J , Cronin MT , Watson D , Shak S , Bohn OL , Fumagalli D , Taniyama Y , Lee A , Reilly ML , Vogel VG , McCaskill-Stevens W , Ford LG , Geyer CE , Wickerham DL , Wolmark N , Paik S. 2011. Estrogen receptor (ESR1) mRNA expression and benefit from tamoxifen in the treatment and prevention of estrogen receptor positive breast cancer. J Clin Oncol 29:4160–4167 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Park YY , Kim K , Kim SB , Hennessy BT , Kim SM , Park ES , Lim JY , Li J , Lu Y , Gonzalez-Angulo AM , Jeong W , Mills GB , Safe S , Lee JS. 2012. Reconstruction of nuclear receptor network reveals that NR2E3 is a novel upstream regulator of ESR1 in breast cancer. EMBO Mol Med 4:52–67 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Loi S , Haibe-Kains B , Desmedt C , Lallemand F , Tutt AM , Gillet C , Ellis P , Harris A , Bergh J , Foekens JA , Klijn JG , Larsimont D , Buyse M , Bontempi G , Delorenzi M , Piccart MJ , Sotiriou C. 2007. Definition of clinically distinct molecular subtypes in estrogen receptor-positive breast carcinomas through genomic grade. J Clin Oncol 25:1239–1246 [DOI] [PubMed] [Google Scholar]
- 14. Loi S , Haibe-Kains B , Desmedt C , Wirapati P , Lallemand F , Tutt AM , Gillet C , Ellis P , Ryder K , Reid JF , Daidone MG , Pierotti MA , Berns EM , Jansen MP , Foekens JA , Delorenzi M , Bontempi G , Piccart MJ , Sotiriou C. 2008. Predicting prognosis using molecular profiling in estrogen receptor-positive breast cancer treated with tamoxifen. BMC Genomics 9:239. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Zhao Y , Bruemmer D. 2010. NR4A orphan nuclear receptors: transcriptional regulators of gene expression in metabolism and vascular biology. Arterioscler Thromb Vasc Biol 30:1535–1541 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Volakakis N , Kadkhodaei B , Joodmardi E , Wallis K , Panman L , Silvaggi J , Spiegelman BM , Perlmann T. 2010. NR4A orphan nuclear receptors as mediators of CREB-dependent neuroprotection. Proc Natl Acad Sci USA 107:12317–12322 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Lee SO , Abdelrahim M , Yoon K , Chintharlapalli S , Papineni S , Kim K , Wang H , Safe S. 2010. Inactivation of the orphan nuclear receptor TR3/Nur77 inhibits pancreatic cancer cell and tumor growth. Cancer Res 70:6824–6836 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Mohan HM , Aherne CM , Rogers AC , Baird AW , Winter DC , Murphy EP. 2012. Molecular pathways: the role of NR4A orphan nuclear receptors in cancer. Clin Cancer Res 18:3223–3228 [DOI] [PubMed] [Google Scholar]
- 19. Alexopoulou AN , Leao M , Caballero OL , Da Silva L , Reid L , Lakhani SR , Simpson AJ , Marshall JF , Neville AM , Jat PS. 2010. Dissecting the transcriptional networks underlying breast cancer: NR4A1 reduces the migration of normal and breast cancer cell lines. Breast Cancer Res 12:R51. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Lennard L. 1992. The clinical pharmacology of 6-mercaptopurine. Eur J Clin Pharmacol 43:329–339 [DOI] [PubMed] [Google Scholar]
- 21. Stewart T , Tsai SC , Grayson H , Henderson R , Opelz G. 1995. Incidence of de-novo breast cancer in women chronically immunosuppressed after organ transplantation. Lancet 346:796–798 [DOI] [PubMed] [Google Scholar]
- 22. Yang C , Yu B , Zhou D , Chen S. 2002. Regulation of aromatase promoter activity in human breast tissue by nuclear receptors. Oncogene 21:2854–2863 [DOI] [PubMed] [Google Scholar]
- 23. Nagasaki S , Suzuki T , Miki Y , Akahira J , Shibata H , Ishida T , Ohuchi N , Sasano H. 2009. Chicken ovalbumin upstream promoter transcription factor II in human breast carcinoma: possible regulator of lymphangiogenesis via vascular endothelial growth factor-C expression. Cancer Sci 100:639–645 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Miki Y , Clyne CD , Suzuki T , Moriya T , Shibuya R , Nakamura Y , Ishida T , Yabuki N , Kitada K , Hayashi S , Sasano H. 2006. Immunolocalization of liver receptor homologue-1 (LRH-1) in human breast carcinoma: possible regulator of insitu steroidogenesis. Cancer Lett 244:24–33 [DOI] [PubMed] [Google Scholar]
- 25. Chand AL , Herridge KA , Thompson EW , Clyne CD. 2010. The orphan nuclear receptor LRH-1 promotes breast cancer motility and invasion. Endocr Relat Cancer 17:965–975 [DOI] [PubMed] [Google Scholar]
- 26. Clyne CD , Speed CJ , Zhou J , Simpson ER. 2002. Liver receptor homologue-1 (LRH-1) regulates expression of aromatase in preadipocytes. J Biol Chem 277:20591–20597 [DOI] [PubMed] [Google Scholar]
- 27. Thiruchelvam PT , Lai CF , Hua H , Thomas RS , Hurtado A , Hudson W , Bayly AR , Kyle FJ , Periyasamy M , Photiou A , Spivey AC , Ortlund EA , Whitby RJ , Carroll JS , Coombes RC , Buluwela L , Ali S. 2011. The liver receptor homolog-1 regulates estrogen receptor expression in breast cancer cells. Breast Cancer Res Treat 127:385–396 [DOI] [PubMed] [Google Scholar]
- 28. Annicotte JS , Chavey C , Servant N , Teyssier J , Bardin A , Licznar A , Badia E , Pujol P , Vignon F , Maudelonde T , Lazennec G , Cavailles V , Fajas L. 2005. The nuclear receptor liver receptor homolog-1 is an estrogen receptor target gene. Oncogene 24:8167–8175 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Li Z , Meng ZH , Chandrasekaran R , Kuo WL , Collins CC , Gray JW , Dairkee SH. 2002. Biallelic inactivation of the thyroid hormone receptor beta1 gene in early stage breast cancer. Cancer Res 62:1939–1943 [PubMed] [Google Scholar]
- 30. Ling Y , Xu X , Hao J , Ling X , Du X , Liu X , Zhao X. 2010. Aberrant methylation of the THRB gene in tissue and plasma of breast cancer patients. Cancer Genet Cytogenet 196:140–145 [DOI] [PubMed] [Google Scholar]
- 31. Ryan J , Curran CE , Hennessy E , Newell J , Morris JC , Kerin MJ , Dwyer RM. 2011. The sodium iodide symporter (NIS) and potential regulators in normal, benign and malignant human breast tissue. PLoS One 6:e16023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Silva JM , Domínguez G , González-Sancho JM , García JM , Silva J , García-Andrade C , Navarro A , Muñoz A , Bonilla F. 2002. Expression of thyroid hormone receptor/erbA genes is altered in human breast cancer. Oncogene 21:4307–4316 [DOI] [PubMed] [Google Scholar]
- 33. Guigon CJ , Kim DW , Willingham MC , Cheng SY. 2011. Mutation of thyroid hormone receptor-beta in mice predisposes to the development of mammary tumors. Oncogene 30:3381–3390 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Zhu XG , Park KS , Kaneshige M , Bhat MK , Zhu Q , Mariash CN , McPhie P , Cheng SY. 2000. The orphan nuclear receptor Ear-2 is a negative coregulator for thyroid hormone nuclear receptor function. Mol Cell Biol 20:2604–2618 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. Papadaki I , Mylona E , Giannopoulou I , Markaki S , Keramopoulos A , Nakopoulou L. 2005. PPARγ expression in breast cancer: clinical value and correlation with ERβ. Histopathology 46:37–42 [DOI] [PubMed] [Google Scholar]
- 36. Burstein HJ , Demetri GD , Mueller E , Sarraf P , Spiegelman BM , Winer EP. 2003. Use of the peroxisome proliferator-activated receptor (PPAR) γ ligand troglitazone as treatment for refractory breast cancer: a phase II study. Breast Cancer Res Treat 79:391–397 [DOI] [PubMed] [Google Scholar]
- 37. Fenner MH , Elstner E. 2005. Peroxisome proliferator-activated receptor-γ ligands for the treatment of breast cancer. Expert Opin Investig Drugs 14:557–568 [DOI] [PubMed] [Google Scholar]
- 38. Rumi MA , Ishihara S , Kazumori H , Kadowaki Y , Kinoshita Y. 2004. Can PPARγ ligands be used in cancer therapy? Curr Med Chem Anticancer Agents 4:465–477 [DOI] [PubMed] [Google Scholar]
- 39. He X , Esteva FJ , Ensor J , Hortobagyi GN , Lee MH , Yeung SC. 2012. Metformin and thiazolidinediones are associated with improved breast cancer-specific survival of diabetic women with HER2+ breast cancer. Ann Oncol 23:1771–1780 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40. Reich M , Liefeld T , Gould J , Lerner J , Tamayo P , Mesirov JP. 2006. GenePattern 2.0. Nat Genet 38:500–501 [DOI] [PubMed] [Google Scholar]