The NS3s of Dengue virus serotypes 2 and 4 are elongated and dynamic in solution. The protease domain of NS3 undergoes substantial conformational alterations owing to RNA binding. The interactions and effects of quercetin binding in NS3 are described.
Keywords: flavivirus, dengue, Dengue virus, nonstructural proteins, NS3, protease, helicase, small-angle X-ray scattering, RNA binding
Abstract
Dengue virus (DENV), which has four serotypes (DENV-1 to DENV-4), is the causative agent of the viral infection dengue. DENV nonstructural protein 3 (NS3) comprises a serine protease domain and an RNA helicase domain which has nucleotide triphosphatase activities that are essential for RNA replication and viral assembly. Here, solution X-ray scattering was used to provide insight into the overall structure and flexibility of the entire NS3 and its recombinant helicase and protease domains for Dengue virus serotypes 2 and 4 in solution. The DENV-2 and DENV-4 NS3 forms are elongated and flexible in solution. The importance of the linker residues in flexibility and domain–domain arrangement was shown by the compactness of the individual protease and helicase domains. Swapping of the 174PPAVP179 linker stretch of the related Hepatitis C virus (HCV) NS3 into DENV-2 NS3 did not alter the elongated shape of the engineered mutant. Conformational alterations owing to RNA binding are described in the protease domain, which undergoes substantial conformational alterations that are required for the optimal catalysis of bound RNA. Finally, the effects of ATPase inhibitors on the enzymatically active DENV-2 and DENV-4 NS3 and the individual helicases are presented, and insight into the allosteric effect of the inhibitor quercetin is provided.
1. Introduction
Dengue, which is caused by Dengue virus (DENV), is an infection threat to nearly 40% of the world’s population (World Health Organization, 2009 ▸). The virus belongs to the genus Flavivirus, and can be further subcategorized into four serotypes (DENV-1 to DENV-4) based on their antigenic properties. The transmission of DENV involves humans and Aedes mosquitoes, where A. aegypti is the main vector of DENV. The virus is equipped with an ∼11 kb single-stranded positive-sense genomic RNA, which becomes released into the host-cell cytoplasm after viral infection. This RNA carries a single open reading frame (ORF) encoding the information for three structural proteins (envelope, membrane and capsid) and seven nonstructural proteins (NS1, NS2A, NS2B, NS3, NS4A, NS4B and NS5) (Lindenbach et al., 2007 ▸). The structural proteins are responsible for virion formation, while the nonstructural proteins play roles in the synthesis of new viral RNA.
Replication of viral RNA takes place in a replication complex (RC), which is located in a vesicle-like structure that originates from the endoplasmic reticulum (ER) membrane (Junjhon et al., 2014 ▸). Two nonstructural proteins, NS3 and NS5, form the catalytic core of the RC and are enzymes consisting of two domains connected by a linker. While NS5 consists of a methyltransferase (MTase) domain responsible for RNA-cap synthesis and an RNA-dependent RNA polymerase (RdRp) domain, which is the core enzyme in the synthesis of a new RNA strand, its counterpart NS3 is composed of a protease domain and a helicase domain. NS3 and NS5 interact via the bNLS region of NS5 (residues 320–368; Johansson et al., 2001 ▸) and amino acids 566–585 of NS3 (Tay et al., 2015 ▸).
The N-terminal residues 1–168 of the 70 kDa NS3 (618 residues) form the protease domain, which is connected to the helicase domain by an 11/12-residue linker (Fig. 1 ▸). The protease domain is a chymotrypsin-like serine protease that participates in proteolysis of the polyprotein to release the individual viral proteins from a long polyprotein. The protease domain requires the central domain of NS2B as a cofactor for proper folding. The presence of residues 49–67 of NS2B is sufficient to stabilize the protease domain (Erbel et al., 2006 ▸; Luo, Xu, Hunke et al., 2008 ▸), but residues 49–96 are essential for enzymatic activity and stability (Clum et al., 1997 ▸). The helicase domain (residues 180–618) possesses an NTPase-dependent RNA helicase activity, which may unwind the double-stranded RNA during RNA replication using energy supplied by NTP hydrolysis (Lindenbach et al., 2007 ▸; Luo et al., 2015 ▸; Warrener et al., 1993 ▸). The exact mechanism of coupling of the unwinding events remains unclear. Besides the unwinding activity, the helicase has also been suggested to play a role in RNA capping. It catalyzes the first RNA-capping reaction, termed the RNA triphosphatase activity, in which the γ-phosphate, which is located at the 5′-end of the newly synthesized RNA, is hydrolyzed to create a diphosphate RNA, which is then directed to the methyltransferase to complete RNA capping (Ferron et al., 2012 ▸; Wengler & Wengler, 1993 ▸).
Figure 1.
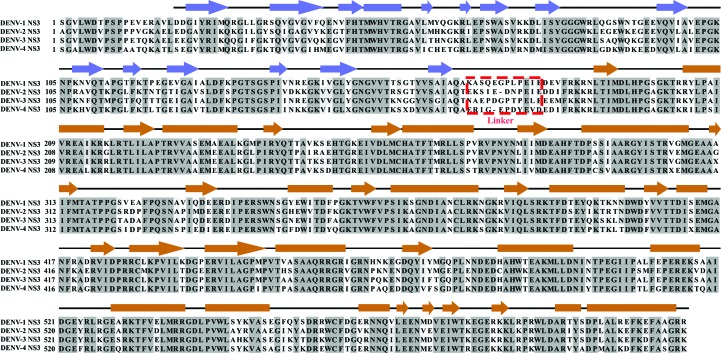
Multiple sequence alignment of full-length NS3 proteins from DENV-1 (GenBank EU081230), DENV-2 (GenBank GQ398264), DENV-3 (GenBank AY662691) and DENV-4 (GenBank AY776330), with secondary structures derived from the crystallographic structure of DENV-4 NS2B18NS3 (PDB entry 2vbc; Luo, Xu, Hunke et al., 2008 ▸). The secondary structures are indicated by boxes (α-helices), lines (loops) and arrows (β-sheets), respectively, and are coloured blue for the protease domain and brown for the helicase domain. These two domains are connected via a 11-residue or 12-residue inter-domain linker with low sequence conservation (Luo, Xu, Hunke et al., 2008 ▸), which is shown in a red dashed box.
The crystallographic structures of DENV-4 NS2B18NS3, consisting of full-length NS3 and a short stretch of NS2B (residues 49–66), revealed two different protease–helicase arrangements, termed conformation I [PDB entries 2vbc (Luo, Xu, Hunke et al., 2008 ▸) and 2wzq (Luo et al., 2010 ▸)] and conformation II (PDB entry 2whx; Luo et al., 2010 ▸). Compared with conformation I, the protease domain in conformation II is rotated by 161° relative to the helicase domain, allowing the binding of ADP–Mn2+ to the helicase domain. When the product ADP was computationally docked into the helicase domain in conformation I, steric clashes were observed (Luo et al., 2010 ▸). Furthermore, the related NS3s of Murray Valley encephalitis virus (MVES; PDB entry 2wv9; Assenberg et al., 2009 ▸) and Hepatitis C virus (HCV; PDB entry 1cu1; Yao et al., 1999 ▸) revealed two additional protease–helicase orientations, in which the protease domain is spatially separated from the helicase domain as in the MVES NS3, or the protease and helicase domains come into closer proximity, resulting in a more compact HCV NS3.
To carry out both protease and helicase activities optimally while the domains are physically linked by an 11/12-residue linker, NS3 requires intermolecular and intramolecular changes in addition to the concerted interactions with viral and host proteins and the viral RNA in the replication complex. Recently, small-angle X-ray scattering (SAXS) studies of the NS3 counterpart, NS5, of all four DENV serotypes revealed that NS5 adopts multiple conformers in solution via its flexible ten-residue linker, with DENV-4 NS5 being more compact compared with the NS5s from DENV-1, DENV-2 and DENV-3; the Lys271, Ser266 and Thr267 residues of DENV-4 NS5 cause this difference in compactness of the protein (Saw et al., 2015 ▸; Subramanian Manimekalai et al., 2016 ▸). Here, a systematic SAXS approach was used to study NS2B18NS3, as well as the individual protease and helicase domains, from DENV-2 and DENV-4 in order to gain insight into the structural traits, compactness and flexibility of NS3 of both serotypes in solution. Binding of the substrate ATP or its analogue did not alter the overall dimensions of NS3, while a slight and significant effect of ADP or RNA (5′-AGA CUA ACA ACU-3′) binding was observed, with the latter causing movement of the protease domain with respect to the helicase domain. Finally, enzymatic studies unravel the interaction with quercetin and the effects of its binding, thereby providing insight into the action of a potent ATPase inhibitor.
2. Materials and methods
2.1. Purification of NS2B18NS3 and single domains of NS3 from DENV-2 and DENV-4
The construction of the gene encoding NS2B18NS3 from DENV-4 (GenBank AY776330) into the pET-32b plasmid has been described previously (Luo, Xu, Hunke et al., 2008 ▸). The clone was kindly provided by Professor Luo Dahai (Lee Kong Chian School of Medicine, Nanyang Technological University, Singapore). The gene encoding NS2B18NS3 from DENV-2 (GenBank GQ398264) was generated using the overlap-extension PCR cloning method. The respective DNA was kindly provided by Professor Liu Ding Xiang, School of Biological Sciences, Nanyang Technological University, Singapore.
The plasmids carrying the respective DENV-2 NS3 genes were transformed into Escherichia coli BL21-CodonPlus (DE3)-RIL cells (Stratagene, USA) for protein production. To produce 15N,13C-labelled DENV-2 protease, a 15 ml LB seeder culture of the transformed cells was grown overnight and the culture was centrifuged at 3000g for 5 min at 4°C to pellet the cells. The cells were washed and subsequently suspended in 1 l M9 minimal medium supplemented with MgSO4, CaCl2, thiamine, FeCl3, trace elements and kanamycin. Starting at an OD600 value of 0.1, the cells were allowed to grow to an OD600 of 0.6 at 37°C, at which point the culture was induced with isopropyl β-d-1-thiogalactopyranoside (IPTG) at a final concentration of 1 mM. The culture was grown overnight at 20°C and the cells were harvested by centrifugation at 8500g for 15 min.
The cells were lysed on ice by sonication for 3 × 1 min in buffer A [20 mM sodium phosphate pH 7.5, 500 mM NaCl, 0.8 mM DTT, 2 mM Pefabloc SC (Biomol)]. The cell lysate was centrifuged at 12 500g for 25 min and the supernatant was filtered (0.45 µm; Millipore). The filtered supernatant was incubated with Ni–NTA agarose (Qiagen) for 1 h at 4°C and the Trx-His6-tagged protein was eluted with an imidazole gradient from 20 to 500 mM in buffer A. Fractions containing the recombinant protein were pooled and dialyzed overnight with thrombin in buffer B (20 mM sodium phosphate pH 7.5, 200 mM NaCl, 1 mM DTT). The sample was then incubated with Ni–NTA agarose for 1 h at 4°C to remove cleaved Trx-His6 tag. Recombinant proteins were applied onto a Superdex 200 HR 10/300 column (GE Healthcare) or a Superdex 75 HR 10/300 column (GE Healthcare) in buffer C (20 mM Tris–HCl pH 7.5, 200 mM NaCl, 1 mM DTT). The fractions containing the respective protein were pooled and concentrated using Amicon Ultra-4 centrifugal units (50, 30 and 10 kDa molecular-mass cutoff; Millipore). In the case of the 15N,13C-labelled DENV-2 NS3 protease domain, the sample was applied onto a Superdex 75 HR 10/300 column (GE Healthcare), which was equilibrated in buffer D (20 mM sodium phosphate pH 6.8, 150 mM NaCl, 1 mM DTT). Fractions containing the labelled protein were pooled, buffer-exchanged to buffer E (20 mM sodium phosphate pH 6.8, 50 mM NaCl, 1 mM DTT) and concentrated using an Amicon Ultra-4 Centrifugal Unit (10 kDa molecular-weight cutoff; Millipore).
The protein concentrations for all samples were measured from the absorption of the protein solutions at 280 nm using a Shimadzu BioSpec-nano spectrophotometer. The extinction coefficient and molecular weight of the protein were calculated using the online ProtParam tool (Gasteiger et al., 2005 ▸) and are shown in Supplementary Table S1.
2.2. ATP-hydrolysis assay
A continuous ATP-hydrolysis assay (Ho et al., 2015 ▸) was used to measure the specific ATP-hydrolysis activities of NS2B18NS3 and the NS3 helicase domains from DENV-2 and DENV-4. In this assay, ATP was constantly regenerated by an enzymatic reaction, while the consumption of NADH was measured spectroscopically at 340 nm. The change in absorbance was measured for 250 s in 2 s intervals at 37°C after adding 5 µg NS2B18NS3 or helicase domain to 1 ml reaction solution (25 mM HEPES pH 7.5, 25 mM KCl, 5 mM MgCl2, 5 mM KCN, 2 mM phosphoenolpyruvate, 2 mM ATP, 0.35 mM NADH, 30 units of l-lactic acid dehydrogenase, 30 units of pyruvate kinase). All of the measurements were performed in triplicate and the activity was derived by fitting the linear section of the slope from the averaged data.
The inhibitory effects of the known ATPase inhibitors resveratrol, quercetin and NBD-Cl (Gledhill & Walker, 2005 ▸) on DENV-2 NS2B18NS3 and its helicase domain were studied by mixing 150 µM inhibitor with the reaction solution stated above. After adding 5 µg DENV-2 NS2B18NS3 or helicase domain to the reaction mixture, the change in absorbance was recorded for 250 s in 2 s intervals at 37°C.
2.3. Nuclear magnetic resonance (NMR) analysis of DENV-2 NS3 protease domain
NMR experiments were performed at 298 K on a 700 MHz NMR spectrometer (Bruker Avance) equipped with a 5 mm cryoprobe. Uniformly 13C/15N-labelled DENV-2 NS3 protease domain was prepared at 0.3 mM in 20 mM phosphate buffer pH 6.8, 50 mM NaCl, 1 mM DTT, 10% D2O. To assign the backbone resonances of the DENV-2 NS3 protease domain, heteronuclear NMR experiments were performed, including 1H–15N HSQC, HNCACB and CBCACONH. In order to reduce the experimental time, three-dimensional HNCACB and CBCACONH data were recorded in non-uniform sampling (NUS) of the indirect dimension at 25% sampling rates and were reconstructed using the compressed sensing (CS) scheme by MddNMR (Kazimierczuk & Orekhov, 2011 ▸; Orekhov & Jaravine, 2011 ▸). All NMR spectra were processed with NMRPipe and NMRDraw (Delaglio et al., 1995 ▸) and were analyzed using SPARKY (Goddard & Kneller, 2002 ▸).
2.4. Small-angle X-ray scattering data collection
SAXS data for NS2B18NS3 from DENV-2 and DENV-4 and for the linker mutant of DENV-2 NS2B18NS3, as well as for the DENV-2 NS3 protease and helicase domains, were measured using a Bruker NANOSTAR SAXS instrument equipped with a MetalJet X-ray source (Excillum, Germany) and a VÅNTEC 2000 detector system as described in Balakrishna et al. (2015 ▸), Dip et al. (2014 ▸) and Tay et al. (2015 ▸). SAXS experiments were carried out at 15°C in a concentration series ranging from 1.2 to 5.4 mg ml−1 in buffer C (20 mM Tris–HCl pH 7.5, 200 mM NaCl, 1 mM DTT) with a sample volume of 40 µl in a vacuum-tight quartz capillary. The scattering of the buffer, obtained from the column flowthrough, was subtracted from the scattering of the sample, and all scattering data were normalized by the concentration.
SAXS data for DENV-2 NS2B18NS3 in the absence or presence of MgATP, MgADP, MgAMPPNP [adenosine 5′-(β,γ-imido)triphosphate] and RNA (5′-AGA CUA ACA ACU-3′) were collected on the Bio-SAXS beamline BL4-2 at Stanford Synchrotron Radiation Lightsource (SSRL; Smolsky et al., 2007 ▸). Data were collected using a Rayonix MX225-HE CCD detector (Rayonix, Evanston, Illinois, USA) with a 1.7 m sample-to-detector distance and a beam energy of 11 keV (wavelength λ = 0.1127 nm), covering a range of momentum transfer of 0.068 < q < 4.7 nm−1 [q = 4πsin(θ)/λ, where 2θ is the scattering angle]. SAXS experiments were performed using the BL4-2 autosampler (Martel et al., 2012 ▸). An aliquot of 30 µl of buffer and sample was subsequently exposed to the X-ray beam via a 1.5 mm quartz capillary cell and oscillated during exposure to minimize radiation damage. For each sample two protein concentrations, 1.13 and 4.5 mg ml−1, were measured in buffer C (20 mM Tris–HCl pH 7.5, 200 mM NaCl, 1 mM DTT). 5 mM DTT, which acts as radical scavenger, was added to all samples freshly prior to data collection. The data for each sample were recorded as a total of 15 frames at 1 s intervals. The data were tested for possible radiation damage by comparing all 15 data frames, and the data without radiation damage were subsequently averaged and the buffer scattering intensity was subtracted using the SasTool software (http://ssrl.slac.stanford.edu/~saxs/analysis/sastool.htm).
2.5. SAXS data analysis
All data-processing steps were performed using the PRIMUS program package (Konarev et al., 2003 ▸, 2006 ▸) from the ATSAS package version 2.7.1. The experimental data obtained for all protein samples were analyzed for aggregation using the Guinier region (Guinier, 1939 ▸). The forward scattering I(0) and the radius of gyration R g were computed using the Guinier approximation, assuming that at very small angles (q < 1.3/R g) the intensity is represented as I(q) = I(0)exp[(qR g)2/3] (Guinier, 1939 ▸). These parameters were also computed from the extended scattering patterns using the indirect transform package GNOM (Svergun, 1992 ▸), which provides the distance distribution function P(r) and hence the maximum particle dimension D max, as well as the radius of gyration R g. The D max was determined by visually inspecting the P(r) curves at both limits, r = 0 and r = D max, for a smooth and concave distribution that is positive everywhere and provides a good fit to the data. Qualitative particle motion was inferred by plotting the scattering patterns in the normalized Kratky plot {(qR g)2[I(q)/I(0)] versus qR g; Durand et al., 2010 ▸}. The Porod–Debye plot was generated by transforming scattering data as q 4 I(q) versus q 4 (Rambo & Tainer, 2011 ▸). Ab initio low-resolution models of the proteins were built by DAMMIF (Franke & Svergun, 2009 ▸) considering low-angle data (q < 2 nm−1). 20 independent ab initio reconstructions were performed for each protein and were then averaged using DAMAVER (Volkov & Svergun, 2003 ▸). The averaged and filtered ab initio model was superimposed with the atomic model using SUPCOMB (Kozin & Svergun, 2001 ▸). The average excluded volume V Ex computed using DAMAVER was further used to estimate the molecular mass of the protein (Petoukhov et al., 2012 ▸). Superimposition between the ab initio reconstruction and the atomic model was performed using SUPCOMB (Kozin & Svergun, 2001 ▸). The ab initio reconstructions were clustered using DAMCLUST (Petoukhov et al., 2012 ▸).
In order to determine the oligomeric state of the protein, the molecular mass (MM) of the protein can be estimated in several ways from the SAXS data. The MM was determined from the forward scattering of the sample, I(0), by comparison with lysozyme (Mylonas & Svergun, 2007 ▸). Alternatively, the MM was estimated from the scattering data based on the excluded (i.e. hydrated) particle volume V p (Mertens & Svergun, 2010 ▸; Petoukhov et al., 2012 ▸) and using the volume of correlation V c [defined as the ratio of I(0) to its total scattered intensity; Rambo & Tainer, 2013 ▸]. The MM estimated for NS2B18NS3 from DENV-2 and DENV-4 as well as the DENV-2 NS3 protease and helicase domains (Tables 1 ▸ and 2 ▸) confirmed a monomeric state of the protein.
Table 1. Data-collection and scattering-derived parameters for NS2B18NS3 from DENV-2 and DENV-4 as well as the DENV-2 NS3 mutant 174PPAVP179 .
| Protein sample | DENV-2 NS2B18NS3 | DENV-4 NS2B18NS3 | DENV-2 NS2B18NS3 mutant 174PPAVP179 |
|---|---|---|---|
| Data-collection parameters | |||
| Instrument (source and detector) | Bruker NANOSTAR equipped with MetalJet eXcillum X-ray source and VÅNTEC-2000 detector | ||
| Beam geometry | 100 µm slit | ||
| Wavelength (nm) | 0.134 | ||
| q range (nm−1) | 0.16–4.0 | ||
| Exposure time (min) | 30 (6 frames × 5 min) | ||
| Concentration range (mg ml−1) | 1.20–5.40 | 1.50–5.40 | 1.1–5.0 |
| Temperature (K) | 288.15 | 288.15 | 288.15 |
| Structural parameters† | |||
| I(0) [from P(r)] (arbitrary units) | 103.1 ± 0.76 | 79.31 ± 0.68 | 96.73 ± 1.2 |
| R g [from P(r)] (nm) | 3.25 ± 0.02 | 3.23 ± 0.03 | 3.28 ± 0.03 |
| I(0) (from Guinier) (arbitrary units) | 102.2 ± 1.20 | 79.39 ± 1.12 | 97.58 ± 2.36 |
| R g (from Guinier) (nm) | 3.18 ± 0.05 | 3.19 ± 0.07 | 3.25 ± 0.08 |
| D max (nm) | 10.3 ± 1 | 10.3 ± 1 | 10.8 ± 1 |
| Porod volume estimate (V p) (nm3) | ∼98 | ∼98 | ∼92 |
| DAMMIF excluded volume (V ex) (nm3) | ∼134 | ∼134 | ∼127 |
| Dry volume from sequence‡ (nm3) | ∼87.2 | ∼86.4 | ∼86.9 |
| Molecular-mass (MM) determination† | |||
| Calculated monomeric MM (from sequence§) (kDa) | ∼72.0 | ∼71.4 | ∼71.8 |
| MM from Porod invariant (V p/1.6) (kDa) | 61.3 ± 6.1 | 61.3 ± 6.1 | 57.5 ± 5.8 |
| MM from excluded volume (V ex/2) (kDa) | 66.5 ± 6.7 | 67.0 ± 6.7 | 63.5 ± 6.4 |
| MM from volume of correlation (V c) (kDa) | 70.3 ± 7.0 | 69.9 ± 7.0 | 78.6 ± 7.8 |
| Software employed | |||
| Primary data reduction | BRUKER SAS | ||
| Data processing | PRIMUS | ||
| Ab initio analysis | DAMMIF | ||
| Validation and averaging | DAMAVER | ||
| Computation of model intensities | CRYSOL | ||
| Flexibility | EOM 2.0 | ||
| Three-dimensional graphics representations | PyMOL | ||
Reported for merged data; the uncertainties for the R g and I(0) values were calculated from the variance of the least-squares fit, and that for the D max value was estimated by testing P(r) near the best-fitting value. For the MM, the uncertainty was taken from known sources (Mylonas & Svergun, 2007 ▸).
Table 2. Data-collection and scattering-derived parameters for the protease (NS2B18NS31–179) and helicase (NS3169–618) domains of DENV-2 NS3.
| Protein sample | NS2B18NS31–179 | NS3169–618 |
|---|---|---|
| Data-collection parameters | ||
| Instrument (source and detector) | Bruker NANOSTAR equipped with MetalJet eXcillum X-ray source and VÅNTEC-2000 detector | |
| Beam geometry | 100 µm slit | |
| Wavelength (nm) | 0.134 | |
| q range (nm−1) | 0.16–4.0 | |
| Exposure time (min) | 30 (6 frames × 5 min) | |
| Concentration range (mg ml−1) | 1.60–4.70 | 2.50–5.00 |
| Temperature (K) | 288.15 | 288.15 |
| Structural parameters† | ||
| I(0) [from P(r)] (arbitrary units) | 41.49 ± 0.3 | 97.08 ± 0.56 |
| R g [from P(r)] (nm) | 1.95 ± 0.02 | 2.55 ± 0.02 |
| I(0) (from Guinier) (arbitrary units) | 41.28 ± 0.3 | 98.13 ± 0.78 |
| R g (from Guinier) (nm) | 1.92 ± 0.02 | 2.59 ± 0.03 |
| D max (nm) | 6.3 ± 1 | 8.4 ± 1 |
| Porod volume estimate (V p) (nm3) | ∼31.5 | ∼73 |
| DAMMIF excluded volume (V ex) (nm3) | ∼45.4 | ∼94 |
| Dry volume from sequence‡ (nm3) | ∼26.4 | ∼62.3 |
| Molecular-mass (MM) determination† | ||
| Calculated monomeric MM (from sequence§) (kDa) | ∼21.8 | ∼51.4 |
| MM from Porod invariant (V p/1.6) (kDa) | 18.9 ± 2 | 43.8 ± 4.4 |
| MM from excluded volume (V ex/2) (kDa) | 22.6 ± 2 | 46.7 ± 4.7 |
| MM from volume of correlation (V c) (kDa) | 24.3 ± 2 | 50.1 ± 5.0 |
| Software employed | ||
| Primary data reduction | BRUKER SAS | |
| Data processing | PRIMUS | |
| Ab initio analysis | DAMMIF | |
| Validation and averaging | DAMAVER | |
| Rigid-body modelling | CORAL | |
| Computation of model intensities | CRYSOL | |
| Three-dimensional graphics representations | PyMOL | |
Reported for 3.0 and 2.5 mg ml−1 measurements for the protease and helicase domains, respectively, of DENV-2 NS3; the uncertainties for the R g and I(0) values were calculated from the variance of the least-squares fit, and that for the D max value was estimated by testing P(r) near the best-fitting value. For the MM, the uncertainty was taken from known sources (Mylonas & Svergun, 2007 ▸).
The theoretical scattering curves from atomic structures were generated and evaluated against experimental scattering curves using CRYSOL (Svergun et al., 1995 ▸). Quantitative assessment of the flexibility was performed using the ensemble-optimization method (EOM 2.0; Tria et al., 2015 ▸), which assumes the coexistence of a range of conformations in solution, for which an average scattering intensity fits the experimental data (Bernadó et al., 2007 ▸; Svergun et al., 2013 ▸; Tria et al., 2015 ▸). The single domains (protease and helicase) were extracted from the crystallographic structure of DENV-4 NS2B18NS3 (PDB entry 2vbc) and used as rigid bodies in EOM model generation, with the 11 residues of the interdomain linker being flexible in EOM. 10 000 models were generated by fixing the helicase domain and allowing free movement of the protease domain to approximate the conformational space for a protein exhibiting considerable motion. For each model in the pool the theoretical scattering curve was automatically computed using CRYSOL (Svergun et al., 1995 ▸). A genetic algorithm (GA) was then used to select ensembles with varying numbers of conformers by calculating the average theoretical profile and fitting this to the experimental SAXS data. The GA was repeated 100 independent times and the ensemble with the lowest discrepancy value (χ2) was reported to represent the coexistence of possible conformers in solution. Independent repetitions of the GA allow the computation of R g and D max distributions, from which structural information about the motion of the particle can be extracted. Distributions with R g average values above the R g average values from the random pool are considered to be extended, whereas models with values below the average are considered to be compact. Quantification of the flexibility is computed as the Shannon information entropy of the R g distributions and is reported as R flex. Ensemble R flex values close to that extracted from the random pool (pool R flex) are considered to be an indication of random motion. Potential artifactual solutions were identified by the complementary metric R σ, which represents the ratio of the standard deviation of the solution distribution to that from the pool distribution. Values of R σ of lower than 1.0 are expected when the ensemble R flex is smaller than the pool R flex, and values greater than 1.0 otherwise.
The SREFLEX program (Panjkovich & Svergun, 2016 ▸) was used to improve the agreement between the experimental scattering data and the high-resolution model for the flexible protein using normal-mode analysis (NMA). The program splits the structure into pseudo-domains and proceeds to probe large rearrangements and smaller localized movements in order to improve the agreement with the experimental data (Panjkovich & Svergun, 2016 ▸).
3. Results
3.1. Purification of NS2B18NS3 from DENV-2 and DENV-4
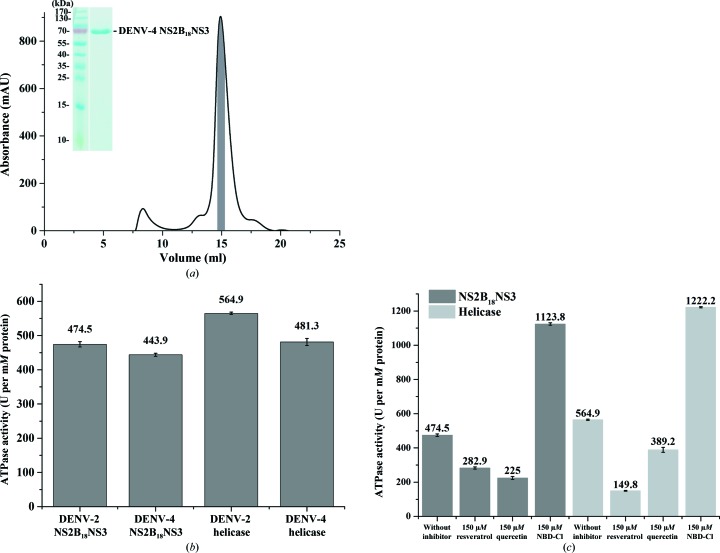
NS2B18NS3 from DENV-2 and DENV-4 was purified as described in §2.1 (Supplementary Fig. S1a and Fig. 2a ▸). The peaks containing NS2B18NS3 were identified by SDS–PAGE and the fractions from the top 15% of the peak (highlighted as a shaded area in Supplementary Fig. S1a and Fig. 2 ▸ a) were pooled and concentrated to three protein concentrations. Inspection of the SDS gels revealed high purity of DENV-4 NS2B18NS3 (inset in Fig. 2 ▸ a) and DENV-2 NS2B18NS3 (inset in Supplementary Fig. S1a). The DENV-2 protease and helicase domains were purified to high purity, as shown in the SDS gels (insets in Supplementary Figs. S1b and S1c).
Figure 2.
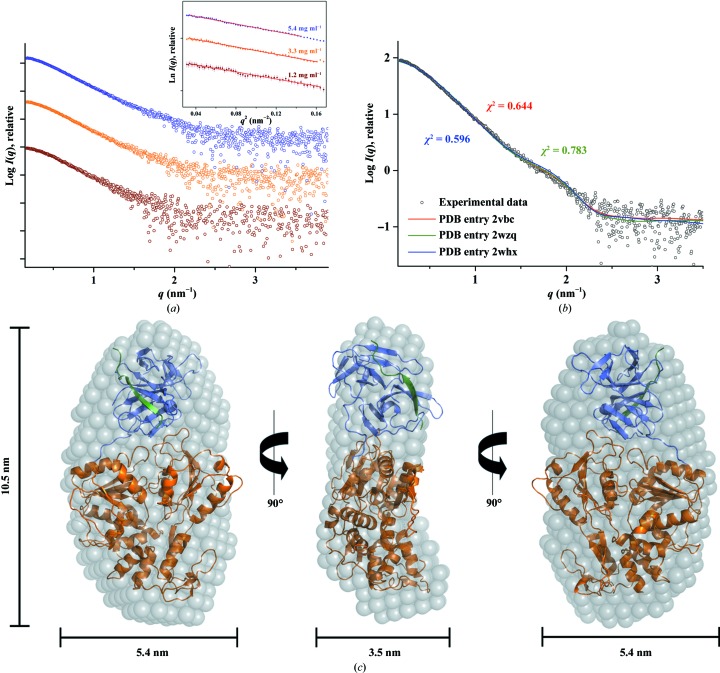
ATP-hydrolysis activity assay of DENV-2 and DENV-4 NS3 proteins. (a) An elution diagram of purified DENV-4 NS2B18NS3 using a Superdex 200 HR 10/300 column. The fractions from the shaded region were pooled and concentrated. Inset: the SDS gel shows a highly pure protein, which is suitable for SAXS studies. (b) ATP-hydrolysis activity data reveal that the recombinant NS2B18NS3 and NS3 helicase domains from DENV-2 and DENV-4 are active. (c) ATP-hydrolysis activity bar diagram for DENV-2 NS2B18NS3 and its helicase domain in the absence or presence of the effectors resveratrol, quercetin and NBD-Cl. In the presence of resveratrol and quercetin ATP hydrolysis was inhibited in both DENV-2 NS2B18NS3 and the helicase domain, whereas in the presence of NBD-Cl ATP hydrolysis was enhanced in both NS2B18NS3 and the helicase domain.
3.2. ATP-hydrolysis activity of NS2B18NS3 and NS3 helicase domains from DENV-2 and DENV-4
Since NTP hydrolysis provides the energy to power the translocation and the unwinding process of NS3, ATP hydrolysis of recombinant NS2B18NS3 as well as the NS3 helicase domains from DENV-2 and DENV-4 were measured to confirm the activity of the recombinant proteins. ATPase activities of 474.5 and 443.9 U per mM of protein were determined for DENV-2 and DENV-4 NS2B18NS3, respectively (Fig. 2 ▸ b). The DENV-2 NS3 helicase domain (NS3168–618) and the DENV-4 NS3 helicase domain (NS3172–618) showed ATPase activities of 564.9 and 481.3 U per mM of protein, respectively. The obtained values indicate that the NS2B18NS3 proteins as well as the NS3 helicase domains from DENV-2 and DENV-4 are active and have comparable activity. Interestingly, the ATP-hydrolysis activity of the DENV-2 helicase domain is 17% higher compared with that of the entire DENV-2 NS2B18NS3 (Fig. 2 ▸ b).
To further analyze the ATP-hydrolysis activity of DENV-2 NS2B18NS3 and its helicase domain, the three ATPase inhibitors resveratrol, quercetin and NBD-Cl (Gledhill & Walker, 2005 ▸) were used. As shown in Fig. 2 ▸(c), resveratrol (at 150 µM) and quercetin (150 µM) reduced the ATPase activity by ∼40 and ∼52%, respectively, while NBD-Cl (at 150 µM) enhanced the ATP hydrolysis of both DENV-2 NS2B18NS3 and the helicase domain (Fig. 2 ▸ c). Whereas the inhibitory effect of resveratrol is higher for the DENV-2 NS3 helicase domain compared with DENV-2 NS2B18NS3, quercetin reduced the ATPase activity more strongly in DENV-2 NS2B18NS3 than in the DENV-2 NS3 helicase domain. This is line with recent findings derived from computational studies, which predict that quercetin binds to the protease domain (de Sousa et al., 2015 ▸).
3.3. Solution X-ray scattering experiments of DENV-4 NS2B18NS3
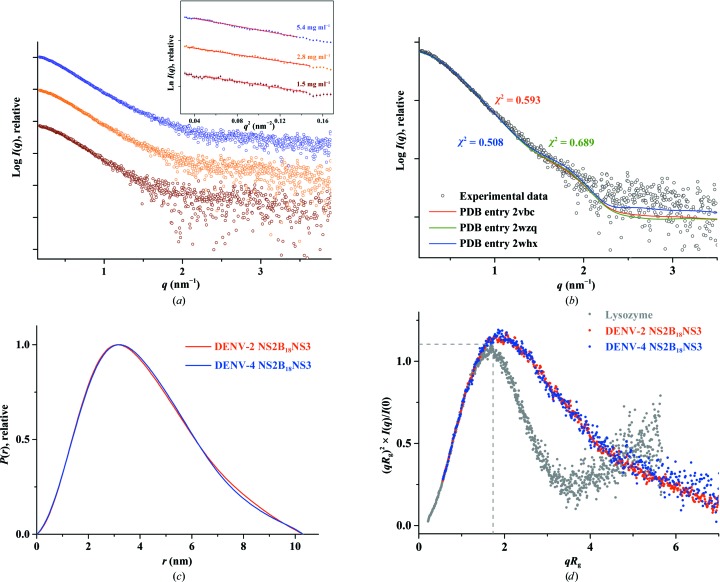
In order to characterize DENV-4 NS2B18NS3 in solution and to understand the orientation as well as the flexibility of the two enzyme domains, protease and helicase, of NS2B18NS3 in solution, SAXS experiments were performed with freshly purified proteins. SAXS patterns of DENV-4 NS2B18NS3 were recorded at 1.5, 2.8 and 5.4 mg ml−1. The derived R g for DENV-4 NS2B18NS3 was determined to be 3.19 ± 0.07 nm (Table 1 ▸). The P(r) function showed a single peak with a small tail (Fig. 3 ▸ c), with a maximum particle dimension (D max) of 10.3 ± 1 nm, indicating that DENV-4 NS2B18NS3 has an elongated conformation in solution (Supplementary Fig. S4a). The R g value extracted from the P(r) function was 3.23 ± 0.03 nm, which is in agreement with the R g values extracted from the Guinier region.
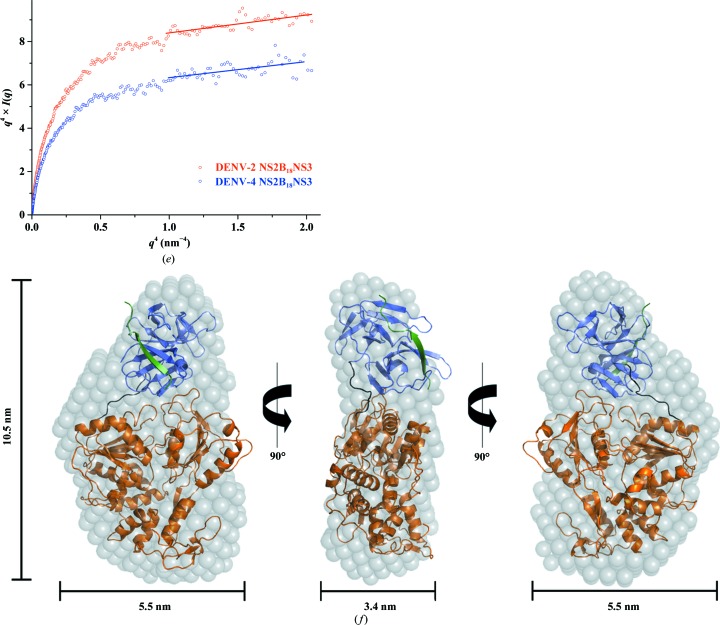
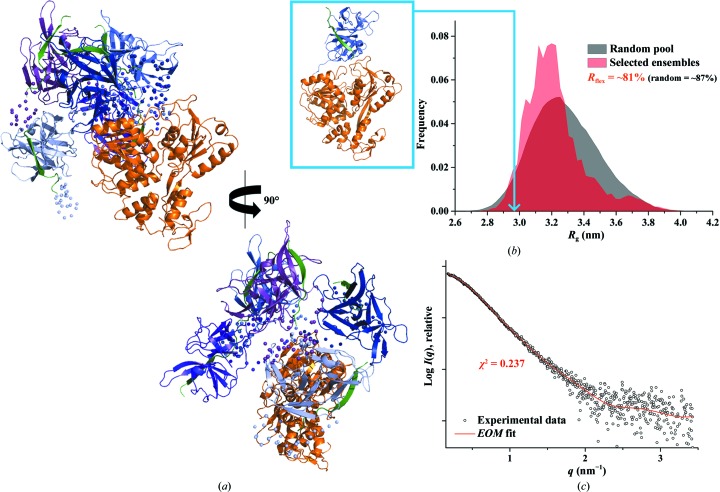
Figure 3.
Solution X-ray scattering studies of DENV-4 NS2B18NS3. (a) SAXS pattern (circles) of DENV-4 NS2B18NS3 at concentrations of 1.5 mg ml−1 (brown), 2.8 mg ml−1 (orange) and 5.4 mg ml−1 (blue). Inset: Guinier plots show linearity at all concentrations used, indicating no aggregation (Supplementary Fig. S3a). A slight concentration dependency was observed at the highest concentration (5.4 mg ml−1; Supplementary Fig. S2a). Therefore, the SAXS patterns of DENV-4 NS2B18NS3 at 1.5 and 2.8 mg ml−1 were merged to improve the data quality. The scattering profiles are offset for clarity by applying arbitrary scale factors. (b) Small-angle X-ray scattering pattern (black circles) and calculated scattering profiles from crystal structures (red line, PDB entry 2vbc; green line, PDB entry 2wzq; blue line, PDB entry 2whx) of DENV-4 NS2B18NS3. (c) Overlapping of the pair-distance distribution functions P(r) of NS2B18NS3 of DENV-2 (red line) and DENV-4 (blue line) shows no difference between the two serotypes. (d) Normalized Kratky plot of DENV-2 NS2B18NS3 (red dots) compared with the DENV-4 NS2B18NS3 protein (blue dots) and the compact globular lysozyme (grey dots) with a peak (grey dotted line) representing the theoretical peak and assuming an ideal Guinier region of a globular particle. The scattering pattern of DENV-4 and DENV-2 NS3 exhibits a broad bell-shaped profile shifted towards the right with respect to standard globular proteins, indicating the presence of motion in the protein. (e) Porod–Debye plots (blue circles, DENV-4 NS2B18NS3; red circles, DENV-2 NS2B18NS3) and their fit lines. The absence of a Porod–Debye plateau in the plot suggests that DENV-4 NS2B18NS3 (Porod exponent = 3.5) and DENV-2 NS2B18NS3 (Porod exponent = 3.5) are flexible in solution. (f) Ab initio DAMMIF envelopes superimposed onto a cartoon representation of the crystallographic structure of DENV-4 NS2B18NS3 (PDB entry 2vbc), with the protease coloured blue and the helicase coloured orange. Front (left), side (middle) and bottom (right) views are displayed.
The low-resolution shapes of DENV-4 NS2B18NS3 were reconstructed ab initio using DAMMIF, and the final averaged and filtered model with a normalized spatial discrepancy (NSD) of 0.717 ± 0.04 is shown in Fig. 3 ▸(f). The DENV-4 NS2B18NS3 solution model showed an elongated conformation (Fig. 3 ▸ f), and superimposed well with the crystallographic structure of DENV-4 NS2B18NS3 (PDB entry 2vbc; Luo, Xu, Hunke et al., 2008 ▸). To date, three DENV-4 NS2B18NS3 crystallographic structures with two conformations have been reported, in which the crystallographic structures of the wild-type protein (PDB entry 2vbc) and the E173GP174 mutant protein (PDB entry 2wzq; Luo et al., 2010 ▸) adopted the so-called conformation I. In comparison, wild-type NS2B18NS3 in complex with ADP–Mn2+ (PDB entry 2whx; Luo et al., 2010 ▸) showed a second conformation in which the protease domain was rotated by approximately 161° with respect to the helicase domain (Luo et al., 2010 ▸). In order to understand which conformation is more predominant in solution, all three structures were used as input to CRYSOL to evaluate the fit between these crystallographic structures and the experimental solution data. As revealed in Fig. 3 ▸(b), the generated theoretical scattering patterns of PDB entries 2vbc, 2wzq and 2whx fit well to the experimental curve of DENV-4 NS2B18NS3, with χ2 discrepancy values of 0.593, 0.689 and 0.508, respectively, reflecting that the ADP–Mn2+-bound NS2BNS3 structure is slightly more favoured in solution. The 20 individual ab initio reconstructed SAXS shapes were clustered using DAMCLUST (Petoukhov et al., 2012 ▸), which produced five clusters with an NSD of 0.618 ± 0.029 within the cluster members. Three clusters contained two reconstructions, and the remaining two contained six and seven reconstructions.
The SAXS data were further analyzed to assess the potential flexibility of the protein. The normalized Kratky plot provides information about the conformational behaviour of the examined particle. For a well folded globular protein such as lysozyme, the plot will show a well defined bell-curve profile as seen in Fig. 3 ▸(d). In comparison, the normalized Kratky plot of DENV-4 NS2B18NS3 exhibited a broad bell-shaped profile shifted towards the right with respect to standard globular proteins, indicating the presence of motion as well as DENV-4 NS2B18NS3 being extended (Durand et al., 2010 ▸; Receveur-Brechot & Durand, 2012 ▸).
To assess the potential flexibility of NS2B18NS3, a Porod–Debye plot was generated, with Porod exponents of 3.5 for serotype DENV-4 and 3.5 for serotype DENV-2 (Fig. 3 ▸ e). The shift in the normalized Kratky plot and the absence of a Porod plateau in the Porod–Debye plot suggest that NS2B18NS3 from DENV-2 and DENV-4 is flexible in solution.
3.4. Ensemble formation of DENV-4 NS2B18NS3
In order to characterize the dynamic behaviour of DENV-4 NS2B18NS3 with respect to the potential flexibility of the protease domain relative to the helicase domain, the ensemble-optimization method (Tria et al., 2015 ▸) was used (Fig. 4 ▸). The ensemble solution selected by EOM 2.0 provided a χ2 discrepancy value of 0.237 (Fig. 4 ▸ c). Five representative models with different orientations of the protease domain with respect to the helicase domain are shown in Fig. 4 ▸(a), demonstrating the possible coexistence of different arrangements of these two domains in solution. A quantification of the flexibility (ensemble R flex = ∼81% versus pool R flex = ∼87%) suggested random motion of the protease domain with respect to the helicase domain (Fig. 4 ▸ b). The quality of the ensemble solution was further confirmed by the control value R σ = 0.90 (expected to be lower than 1.0 when ensemble R flex < pool R flex). Of the five EOM models that were used to fit the experimental data, three representative models with R g values of 3.65, 3.11 and 3.01 nm and D max values of 12.5, 10.7 and 10.1 nm, respectively, made major contributions (25% each), with the remaining two models contributing 13% each to the fitting.
Figure 4.
Flexibility characterization of DENV-4 NS2B18NS3, showing an ensemble of five possible positions for the protease domain based on EOM 2.0 analysis. (a) Different views of the ensemble models in cartoon representation with the helicase domain of DENV-4 NS2B18NS3 in orange and the flexible protease domain in blue, together with their conformational contributions (purple, 25%; light blue, 25%; purple blue, 25%; blue, 13%; slate blue, 13%; the connecting linkers are rendered semi-transparent and NS2B18 is in green). (b) Comparison between the R g distributions (pool, black; ensemble, red) shows a random flexibility as confirmed by R flex (∼81% versus ∼87%, with 87% as the threshold value for randomness) and R σ = 0.90. The R g corresponding to the DENV-4 NS2B18NS3 crystal structure (PDB entry 2vbc) is labelled on the curve (light blue arrow). (c) Small-angle X-ray scattering pattern (black circles) and calculated ensemble scattering profiles (red line) with a discrepancy χ2 of 0.237.
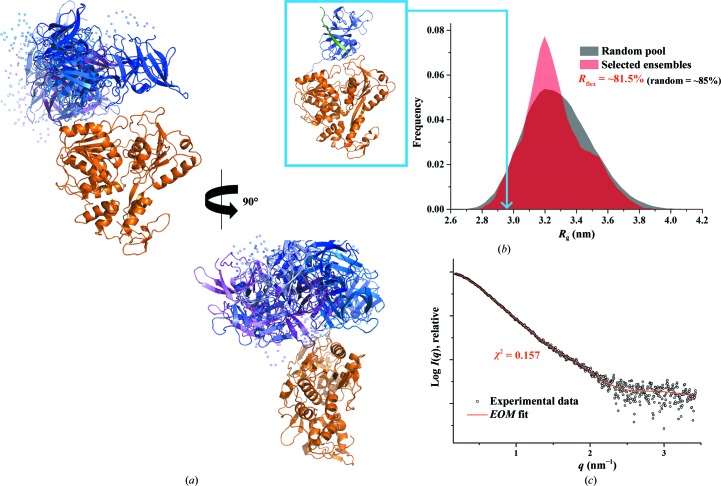
3.5. Solution X-ray studies of DENV-2 NS2B18NS3
Recent solution studies of the NS3 counterpart NS5 showed that all four serotypes of DENV NS5 adopt multiple conformations owing to the flexible linker. However, among the different serotypes, the DENV-4 NS5 is more compact and less flexible compared with NS5s of DENV-1, DENV-2 and DENV-3 (Saw et al., 2015 ▸). A multiple sequence alignment of DENV NS3 proteins from all four serotypes showed low sequence similarity in the 11-residue linker region (red dashed box in Fig. 1 ▸). In analogy to the recent DENV NS5 studies (Saw et al., 2015 ▸), the NS3 serotype DENV-2, which is one of the most predominant serotypes in Southeast Asia, was selected and compared with the DENV-4 NS3 using SAXS. The merged data reveal similar structural parameters for DENV-2 NS2B18NS3 and DENV-4 NS2B18NS3, as summarized in Table 1 ▸ (Figs. 5 ▸ a and 3 ▸ c and Supplementary Figs. S2b and 3b). The low-resolution shapes of DENV-2 NS2B18NS3 (NSD = 0.709 ± 0.05; Fig. 5 ▸ c) are also similar to those of DENV-4 NS2B18NS3; however, clustering analysis of all 20 ab initio models identified only four clusters (NSD = 0.587 ± 0.02 within the cluster members), with a major cluster containing about half of the reconstructions (ten) while the three minor clusters have four, three or two reconstructions. The normalized Kratky plot of DENV-2 NS2B18NS3 (Fig. 3 ▸ d) had a similar profile to DENV-4 NS2B18NS3 (Fig. 3 ▸ d), and EOM analysis revealed that the protease domain adopts a different orientation with respect to the helicase domain (Fig. 6 ▸), with one major representative model (R g = 3.15 nm and D max = 11.1 nm) contributing 50% to the experimental data fitting and six other minor models contributing equally (∼8%).
Figure 5.
Solution X-ray scattering studies of DENV-2 NS2B18NS3. (a) SAXS patterns (circles) of DENV-2 NS2B18NS3 at 1.2 mg ml−1 (brown), 3.3 mg ml−1 (orange) and 5.4 mg ml−1 (blue) concentration. Inset: Guinier plots show linearity at all concentrations used, indicating no aggregation. The pure recombinant DENV-2 NS2B18NS3 (Supplementary Fig. S1a) showed no concentration dependence (1.2, 3.3 and 5.4 mg ml−1; Supplementary Fig. S2a); however, since the Porod volume and molecular-weight estimate are slightly larger at higher concentration, the scattering curves were merged. The increase in Porod volume and estimated molecular weight may be owing to a concentration effect or to improved data quality at the higher concentration. The scattering profiles are offset for clarity by applying arbitrary scale factors. (b) SAXS pattern (black circles) and calculated scattering profiles from crystal structures (red lines, PDB entry 2vbc; green, PDB entry 2wzq; blue, PDB entry 2whx) of DENV-2 NS2B18NS3. (c) Ab initio DAMMIF envelopes superimposed onto the cartoon representation of the crystallographic structure of DENV-2 NS2B18NS3 (PDB entry 2vbc), with the protease coloured blue and the helicase coloured orange. Front (left), side (middle) and bottom (right) views are displayed.
Figure 6.
Flexibility characterization of DENV-2 NS2B18NS3, showing an ensemble of seven possible positions for the protease domain based on EOM 2.0 analysis. (a) Different views of the ensemble models in cartoon representation with the helicase domain of DENV-2 NS2B18NS3 in orange and the flexible protease domain in blue, together with their conformational contributions (purple blue, 50%; blue, 8%; light blue, 8%; slate blue, 8%; marine blue, 8%; violet purple, 8%; light purple, 8%; the connecting linkers are rendered semi-transparent and NS2B18 is in green). (b) Comparison between the R g distributions (pool, black; ensemble, red) shows a random flexibility as confirmed by R flex (∼81.5% versus ∼85%, with 85% as the threshold value for randomness) and R σ = 0.89. The R g which corresponds to the DENV-4 NS2B18NS3 crystal structure (PDB entry 2vbc) is labelled with a light blue arrow. (c) SAXS pattern (black circles) and calculated ensemble scattering profiles (red line) with a discrepancy χ2 of 0.157.
3.6. Solution X-ray studies of the DENV-2 NS2B18NS3 linker mutant
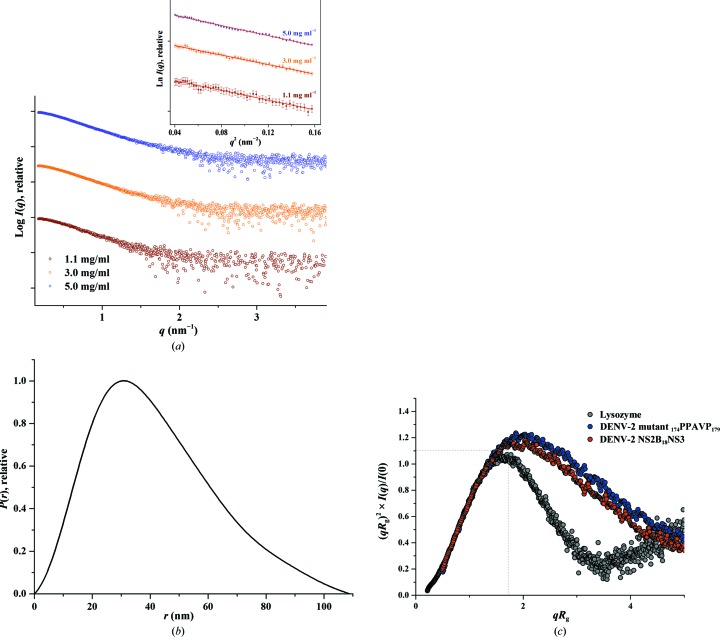
A linker mutant was generated by replacing the amino-acid residues in the linker region (residues 174–179) of DENV-2 NS2B18NS3 with the invariant PPXXP (PPAVP in this case; DENV-2 NS3 mutant 174PPAVP179) motif from the Hepatitis C virus (HCV) NS3 linker. The linker from HCV NS3, which is rich in proline residues, is believed to be responsible for the compact globular structure of HCV NS3 (Li et al., 2014 ▸). To investigate whether this invariant motif from the HCV NS3 linker has any effect on the compactness and flexibility of DENV-2 NS2B18NS3, the DENV-2 NS3 mutant 174PPAVP179 was generated and SAXS experiments were performed with the freshly purified mutant at concentrations of 1.1, 3.0 and 5.0 mg ml−1 (Fig. 7 ▸ a). The derived structural parameters of DENV-2 NS3 mutant 174PPAVP179 were comparable with those of DENV-2 NS2B18NS3 (Table 1 ▸; Fig. 7 ▸; Supplementary Figs. S3c and S4c) and the normalized Kratky plot showed a slightly higher flexibility and/or extended nature of the DENV-2 NS3 mutant 174PPAVP179 (Fig. 7 ▸ c).
Figure 7.
SAXS studies of the DENV-2 NS3 mutant 174PPAVP179. (a) SAXS scattering patterns (circles) of the linker mutant at 1.1 mg ml−1 (brown), 3.0 mg ml−1 (orange) and 5.0 mg ml−1 (blue) concentration, which showed no concentration dependency (Supplementary Fig. S2c); all three concentrations were merged to avoid any unwanted effects arising from the slightly larger Porod volume and molecular-weight estimate at higher concentration. Inset, Guinier plots show linearity at all concentrations used, indicating no aggregation. The scattering profiles are offset for clarity by applying arbitrary scale factors. (b) P(r) function (line) of the linker mutant. (c) Comparison of normalized Kratky plots (dots) between the linker mutant (blue) and DENV-2 NS2B18NS3 (red).
3.7. Solution X-ray studies of single domains of DENV-2 NS2B18NS3
In order to understand whether the helicase and/or the protease domain contribute to the flexibility of NS2B18NS3, the protease domain (NS2B18NS31–179) and helicase domain (NS3169–618) of DENV-2 NS3 were generated and purified (Supplementary Figs. S1b and S1c). In the absence of the cofactor NS2B, the protease is known to be unfolded in solution (Gupta et al., 2015 ▸). Therefore, NMR spectroscopy was performed on the purified DENV-2 protease to assess its folding state. The well dispersed 1H–15N HSQC spectrum of the protease indicated proper folding and monodispersity of the protein (Supplementary Fig. S5). SAXS patterns of DENV-2 NS3 protease (1.6, 3.0 and 4.7 mg ml−1) and helicase domains (2.5, 3.0 and 5.0 mg ml−1) were recorded at various protein concentrations, and the overall structural parameters of the protease and helicase domains of DENV-2 NS3 are shown in Table 2 ▸ (Supplementary Figs. S3d, S3e, S2d, S2e, S6a, S6b and S6c). Normalized Kratky plots of the DENV-2 protease and helicase domains reflect that both domains are more compact than the entire DENV-2 NS2B18NS3 (Supplementary Fig. S6d) and the slight shift indicates some flexibility inside the domains and/or an extended form. The missing 17 residues at the N-terminus and 13 residues at the C-terminus (including the linker) in the crystal structure of the DENV-2 protease domain (PDB entry 2fom; Erbel et al., 2006 ▸) were modelled using CORAL (Fig. 8 ▸ a). The scattering data of the helicase fit well to the crystal structure (PDB entry 2bmf; Xu et al., 2005 ▸) (Supplementary Fig. S6b).
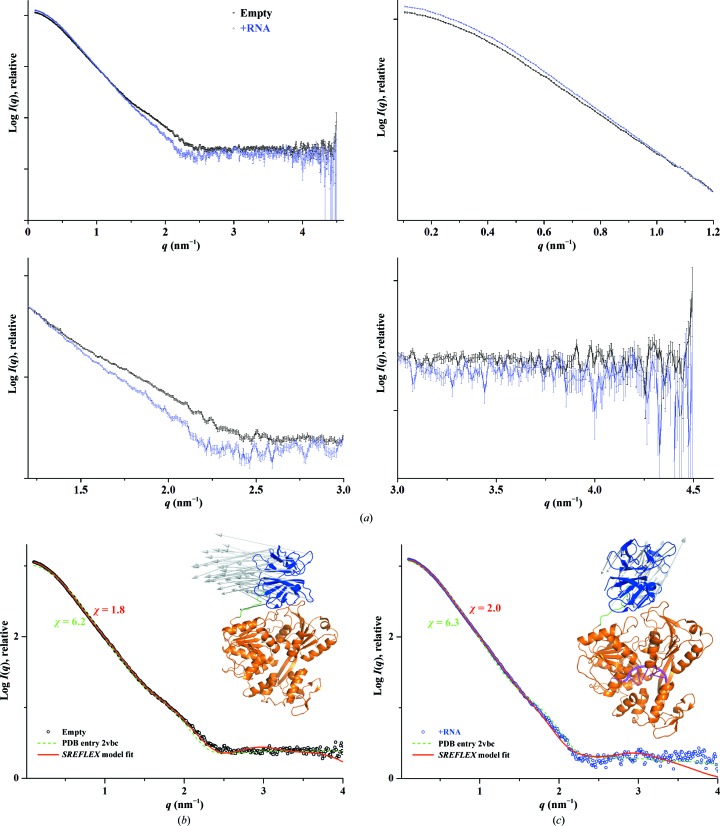
Figure 8.
(a) Comparison of SAXS scattering patterns (circles) between the empty form (black) and that in the presence of 12-mer RNA (blue), with an enlarged view at lower, medium and higher angles clearly indicating the difference at lower and medium angles. (b) SAXS profile of empty DENV-2 NS2B18NS3 (black circles), showing better consistency of the SREFLEX model (red line) than the crystal structure (PDB entry 2vbc; green dashed line). The inset shows the complete SREFLEX simulated movement after the refinement stage. Vectors are drawn connecting the equivalent residues from the crystal structure to the SREFLEX model, which is not shown for clarity. (c) SAXS profile in the presence of the 12-mer RNA (blue circles), showing better fitting of the SREFLEX model (red line) than the crystal structure (PDB entry 2vbc; green dashed line). Inset: the complete SREFLEX simulated movement of the protease domain in the RNA-bound form (magenta).
3.8. Solution X-ray studies of DENV-2 NS2B18NS3 with substrates
In order to understand the effect of substrate binding to NS3, DENV-2 NS2B18NS3 was studied in the presence of MgATP, MgADP, MgAMPPNP (a nonhydrolyzable analogue of ATP) and RNA (5′-AGA CUA ACA ACU-3′). The 12-mer RNA was chosen based on previous structural studies of this RNA with the DENV-4 NS3 helicase domain (Luo, Xu, Watson et al., 2008 ▸), which demonstrated that the single-stranded bound RNA induced structural alterations in the helicase domain. Here, we are aiming to understand the structural changes upon RNA binding to the entire DENV NS3. To observe any changes in DENV-2 NS2B18NS3 in the absence (empty form) or the presence of substrates, high-resolution SAXS data were collected using a synchrotron source (§2.4). Since DENV-2 NS2B18NS3 did not have any concentration-dependent interparticle interaction (Supplementary Fig. S2b), the data for each sample measurement were collected at only two protein concentrations (1.13 and 4.5 mg ml−1) and were merged. The resulted scattering profile superimposed well with the scattering profile collected using an in-house X-ray instrument (Supplementary Fig. S7). The molar ratio of protein to substrate for ATP, ADP and AMPPNP was 1:3, whereas an equimolar ratio was used in the presence of RNA. The merged scattering profiles of the DENV-2 NS2B18NS3 empty form and in the presence of different substrates are shown in Supplementary Fig. 8 ▸(a). No protein aggregation was observed in any of the data collected. As shown in Table 3 ▸, no significant change in the R g or D max value (Supplementary Fig. S7b) could be observed between the nucleotide-free DENV-2 NS2B18NS3 (R g = 3.22 ± 0.01 nm) and the protein in the presence of MgATP (R g = 3.23 ± 0.01 nm) or MgAMPPNP (R g = 3.22 ± 0.01 nm) (Supplementary Fig. S3f). However, a slight decrease in the R g value was determined for the ADP-bound form (R g = 3.20 ± 0.01 nm) as well as a slight increase in the R g value (R g = 3.25 ± 0.01 nm) of the RNA-bound complex (5′-AGA CUA ACA ACU-3′) of DENV-2 NS3 (Supplementary Fig. S3f). The normalized Kratky plots of DENV-2 NS2B18NS3 in the absence or presence of nucleotides exhibit similar profiles, indicating no flexibility change upon nucleotide binding (Supplementary Fig. S8c).
Table 3. Data-collection and scattering-derived parameters for DENV-2 NS2B18NS3 in the absence or presence of substrates.
| Protein sample | Empty form | + MgATP | + MgADP | + MgAMPPNP | + RNA (AGA CUA ACA ACU) |
|---|---|---|---|---|---|
| Data-collection parameters | |||||
| Instrument (source and detector) | Bio-SAXS beamline BL4-2 at SSRL with Rayonix MX225-HE CCD detector | ||||
| Beam geometry | 200 µm slit | ||||
| Wavelength (nm) | 0.1127 | ||||
| q range (nm−1) | 0.068–4.7 | ||||
| Exposure time (s) | 15 (15 frames × 1 s) | ||||
| Concentration range (mg ml−1) | 1.13 and 4.5 | ||||
| Temperature (K) | 288.15 | ||||
| Structural parameters† | |||||
| I(0) (from Guinier) (arbitrary units) | 1173.7 ± 2.2 | 1163.5 ± 1.8 | 1153.3 ± 1.2 | 1195.5 ± 2.1 | 1301.3 ± 1.7 |
| R g (from Guinier) (nm) | 3.22 ± 0.01 | 3.23 ± 0.01 | 3.20 ± 0.01 | 3.22 ± 0.01 | 3.25 ± 0.01 |
| D max (nm) | 10.3 ± 1 | 10.3 ± 1 | 10.2 ± 1 | 10.5 ± 1 | 10.5 ± 1 |
| Porod volume estimate (V p) (nm3) | ∼95.1 | ∼94.6 | ∼95.1 | ∼95.5 | ∼108.3 |
| Dry volume from sequence‡ (nm3) | ∼87.2 | ∼87.2 | ∼87.2 | ∼87.2 | ∼87.2 |
| Software employed | |||||
| Primary data reduction | SasTool | ||||
| Data processing | PRIMUS | ||||
Reported for merged data; the uncertainties for the R g and I(0) values were calculated from the variance of the least-squares fit, and that for the D max value was estimated by testing P(r) near the best-fitting value. For the MM, the uncertainty was taken from known sources (Mylonas & Svergun, 2007 ▸).
A comparison of the scattering profiles of the empty and RNA-bound forms at higher q values revealed a small difference at lower q values (reflected in an increase in the R g value) and significant deviations in the q range 1.5–2.5 nm−1 (Fig. 8 ▸ c). The divergence noted in the q range 1.5–2.5 nm−1 is typical of large conformational changes occurring in the protein (Putnam et al., 2007 ▸), leading us to infer that structural alterations have indeed occurred owing to RNA binding. In order to rule out the contribution of free species of RNA and DENV-2 NS2B18NS3 to the scattering profile of the RNA-bound form, the sum of the scattering from free RNA and the empty form are compared with the RNA-bound complex data (Supplementary Fig. S9), similar to the recently described scattering studies for the RNA-binding domain of a protein kinase (Patel et al., 2012 ▸). The scattering profiles clearly demonstrate that the RNA-bound complex and the sum of the free species of the RNA and empty form are not equivalent, suggesting that a bound complex has formed and the data for the RNA-bound form are not merely from the free species in solution (Supplementary Fig. S9). The scattering data did not agree well with the crystal structures, with the empty DENV-2 NS2B18NS3 having similar χ values of 6.2 and 6.3 for conformation I (PDB entry 2vbc) and conformation II (PDB entry 2whx), respectively. The RNA-bound DENV-2 NS2B18NS3 showed a slight preference for conformation I, with a χ value of 5.8, over conformation II (χ = 6.9). This suggested that the position of the protease domain with respect to the helicase domain might be different in the RNA-bound DENV-2 NS2B18NS3, as observed in the crystal structures of conformations I and II (Luo et al., 2010 ▸). Although the EOM analysis of DENV-2 NS2B18NS3 described above revealed that the protein is flexible and that the protease domain adopts many different orientations with respect to the helicase domain in solution, it showed that one major conformational model with an R g value of 3.15 nm and a D max of 11.1 nm contributes 50%. In addition, six other minor conformational models have a contribution of about 8% each. Furthermore, a clustering analysis of the shape reconstruction identified one major cluster. The DENV-2 NS2B18NS3 scattering profile had a reasonable agreement (χ2 = 0.7–0.8) for the single conformations I or II (Fig. 5 ▸ b), suggesting the presence of one major conformation in addition to a few minor conformations in solution. Therefore, we proceeded to determine one of the probable conformations for both the empty and the RNA-bound forms that has a reasonable fit to the high-resolution SAXS data using SREFLEX (Panjkovich & Svergun, 2016 ▸). The program uses normal-mode analysis (NMA) to estimate the flexibility of the crystal structure and then tries to probe large conformational rearrangements and smaller localized movements representing possible conformational changes, and thereby improves the fitting to the experimental data (Panjkovich & Svergun, 2016 ▸). For the RNA-bound data, the single-stranded RNA molecule (seven nucleotides) was modelled into the helicase domain of NS3 based on the crystal structure of the RNA-bound helicase (Luo, Xu, Watson et al., 2008 ▸), in which only seven out of 12 nucleotides were ordered. One of the probable models identified by SREFLEX suggested that in the empty form the protease domain is positioned such as to have a larger translation and a smaller twist movement from the crystal structure (vectors representing the movements are shown in the inset in Fig. 8 ▸ b). This model agrees with the experimental data with a χ value of 1.8 (Fig. 8 ▸ b). However, in one of the possible RNA-bound SREFLEX models the protease domain adopts a larger twist and marginal translation movements in the opposite direction when compared with the empty form in order to fit the experimental data (χ = 2.0; Fig. 8 ▸ c). A small discrepancy is noted in the fitting of the RNA-bound form in the q range 1.8–2.5 nm−1. This could indicate that some structural alteration took place in the helicase domain owing to RNA binding, similar to that observed in the crystal structure of the RNA-bound helicase (Luo, Xu, Watson et al., 2008 ▸), or it may possibly arise from the scattering of additional unaccounted nucleotides present in the RNA that were not modelled. Moreover, splitting the helicase domain into its respective three subdomains and rerunning the SREFLEX program did not yield a better fit, which could be attributed to the inherent limitation of SAXS-based refinement approaches for smaller conformational variations (Panjkovich & Svergun, 2016 ▸). Nevertheless, with the reasonable agreement between the SREFLEX model and the experimental data, it can be suggested that the binding of RNA has induced substantial conformational alterations in the protease domain of DENV-2 NS2B18NS3, without excluding the possibility of some significant modifications in the helicase domain.
4. Discussion
DENV NS3 is a two-domain enzyme complex that interacts with several viral proteins and host proteins to enable viral replication. Its protease carries out viral polyprotein cleavage, which is a key step in viral protein maturation (Falgout et al., 1991 ▸). The NS3 helicase domain, as well as the MTase and RdRp of NS5, form the catalytic core in the DENV genome-replication machinery, and perform the essential sequential steps for viral RNA replication and capping, including dsRNA unwinding, genomic RNA replication, NTPase/RTPase activity, guanylyltransferase activity and methyltransferase activity (Egloff et al., 2002 ▸; Tay et al., 2015 ▸). To accomplish these tasks, the individual domains inside NS3 and NS5 as well as the NS3–NS5 ensemble have to communicate during genome-replication events.
Previous enzymatic studies of NS3 and its helicase domain reported that DENV-2 NS3 without the NS2B cofactor exhibited a higher unwinding activity but a lower ATP-hydrolysis activity compared with the helicase domain (residues 171–618; Xu et al., 2005 ▸; Yon et al., 2005 ▸), while DENV-4 NS2B18NS3 revealed a tenfold higher affinity for ATP analogues than the helicase domain (residues 177–618; Luo, Xu, Hunke et al., 2008 ▸). The ATP-hydrolysis data in this study showed a comparable hydrolytic activity between NS2B18NS3 and the NS3 helicase domains from DENV-2 and DENV-4, with a slightly higher ATPase activity of the DENV-2 NS3 helicase domain (Fig. 2 ▸ b). Comparable ATPase activities have also been reported for an MVEV NS2B47NS3 construct (Assenberg et al., 2009 ▸), as well as for DENV-2 NS2B47NS3 and its helicase domain (Gebhard et al., 2012 ▸). These observed differences in helicase activity were proposed to be caused by the different protein constructs and/or assays utilized in these experiments (Luo et al., 2015 ▸).
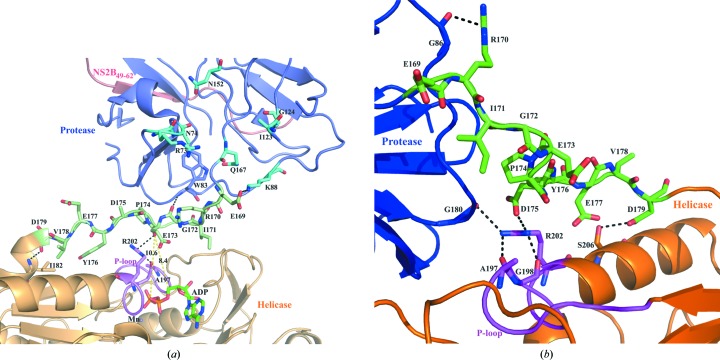
Enzymatic studies of recombinant NS2B18NS3 and the NS3 helicase domains of DENV-2 and DENV-4 provided insights into the effects of the ATPase inhibitors resveratrol and quercetin, which significantly reduce the ATP-hydrolysis activity of the helicase domain regardless of the absence or presence of the protease domain. Of particular interest is that resveratrol has a stronger inhibitory effect on the helicase domain alone compared with full-length NS3 (NS2B18NS3), whereas quercetin inhibits the entire NS2B18NS3 more strongly than the helicase domain only. The inhibitory role of quercetin in the RNA-replication machinery has been shown previously, in which a reduction of DENV-2 RNA levels of 67% was observed (Zandi et al., 2011 ▸). In a computational docking study, quercetin was shown to bind to an allosteric site of the DENV-3 protease domain (de Sousa et al., 2015 ▸) via the predicted residues Asn74, Arg73, Lys88, Ile123, Gly124, Asn152 and Gln167 (Fig. 9 ▸ a). A closer look at the linker region inside the DENV-4 NS3 structure (PDB entry 2whx; Luo et al., 2010 ▸) highlights that these residues are in close proximity to the linker residues, which interact with the phosphate-loop (P-loop) of the helicase domain (Fig. 9 ▸ a). This interaction occurs via a salt bridge formed by the linker residue Glu169 and Lys88 of the protease domain as well as the linker residue Gly172, which makes a hydrogen-bonding interaction with Trp83 of the protease domain. The linker residues Glu173 and Asp179 form hydrogen bonds to the helicase-domain residues Arg202 and Ile182, respectively. Although the side chain of Asp175 was not identified in the crystal structure, this residue might also interact with Arg202 as observed in conformer I (Fig. 9 ▸ b). Arg202 belongs to the P-loop region that stabilizes the transition state during ATP hydrolysis. Therefore, any changes in the linker residues will directly affect ATP hydrolysis. Surprisingly, Lys88, which interacts with the linker, is also predicted to interact with quercetin. It can be speculated that the binding of quercetin might affect the linker residues, leading to altered interaction(s) with the P-loop residues described above, followed by inhibition of ATP cleavage. Therefore, the allosteric effect of quercetin binding to the protease domain may strengthen the inhibition of ATP hydrolysis in NS3 by inducing a conformational change in the P-loop of the helicase domain, which may affect the transition state of ATP hydrolysis. These results provide new insight into how the well known ATPase inhibitor quercetin acts in NS3 and may shed light for lead-optimization experiments in the future.
Figure 9.
Interaction profile of linker residues (169ERIGEPDYEVD179) in DENV4 NS3 crystal structures with conformation I and II. The protease and helicase domains are identified by blue and orange colours, respectively. The linker region is shown in green and the P-loop of the helicase domain is in magenta. The NS2B49–62 peptide is coloured red. (a) Enlarged view of the linker region in the crystal structure of conformation II (PDB entry 2whx) containing ADP (green sticks) and manganese (violet spheres). The residues that are predicted to interact with quercetin are shown in cyan (de Sousa et al., 2015 ▸). The ADP molecule is at a distance of 8.4 Å and the β-phosphate of ADP is at a distance of 10.6 Å from the linker region. The linker residue Glu169 interacts with Lys88 of the protease domain though a salt bridge and Gly172 interacts with Trp83 through backbone hydrogen bonding. Glu173 and Asp179 belonging to the linker region form hydrogen bonds to Arg202 and Ile182 of the helicase domain, respectively. (b) The linker residue Arg170 makes hydrogen bonds to Gly86 of the protease domain in the crystal structure of conformation I (PDB entry 2vbc). Moreover, Asp175 of the linker region makes a salt-bridge interaction with Arg202 of the P-loop in the helicase. Asp179 makes a hydrogen-bond interaction with Ser206 of the helicase. In addition to the salt bridge above, Arg202 also makes a hydrogen-bonding interaction with Gly80 of the protease and Ala197 and Gly198 of the P-loop in the helicase.
To date, two conformations have been identified from crystallographic structures of DENV-4 NS2B18NS3, termed conformation I (PDB entry 2vbc; Luo, Xu, Hunke et al., 2008 ▸) and conformation II (PDB entry 2whx; Luo et al., 2010 ▸). Both structures have an extended conformation, which is in agreement with the determined solution shape. By aligning both conformations centred on the helicase domain, it is revealed that the protease domain in conformation II rotates 161° compared with conformation I (Luo et al., 2010 ▸). This rotation does not alter the overall extended conformation observed in both structures, but the ATP-binding site in conformation II is now accessible for ADP to bind. In conformation I, the protease domain blocks the NTP-binding site of the helicase domain. Interestingly, by analyzing the two conformations together with the SAXS data for DENV-2 and DENV-4 NS2B18NS3, both conformations are favourable in solution.
The advantage of studying NS2B18NS3 from both serotypes in solution is the information that is provided about the flexibility of both enzymes, which demonstrates that the protease domain of NS2B18NS3 adopts random motion with respect to the helicase domain. This also underlines the higher B factor of the protease determined in the crystallographic structure, as well as the accessibility of this domain for proteolytic cleavage (Luo et al., 2010 ▸). The calculated R g value (2.98 nm) of the NS2B18NS3 structure (PDB entry 2vbc) is lower than the R g distribution values (3.1–3.3 nm) derived from EOM analysis using solution SAXS data of DENV-4 NS2B18NS3 (Fig. 4 ▸ b). Together with the normalized Kratky (Fig. 3 ▸ b) and Porod–Debye plots (Fig. 3 ▸ e), the R g distribution observed indicates that the DENV-2 and DENV-4 NS2B18NS3s are flexible, with the inter-domain distance between the protease and helicase domains being predominantly extended in solution. The linker region of NS2B18NS3 connecting the two domains will mainly contribute to the flexibility, as shown for its enzymatic counterpart NS5 (Saw et al., 2015 ▸; Subramanian Manimekalai et al., 2016 ▸; Zhao et al., 2015 ▸). Considering that the protease domain of NS3 is able to cleave in cis at the NS2A–NS2B junction and in trans at the NS3–NS4A junction (Falgout et al., 1991 ▸; Preugschat et al., 1990 ▸), the two-domain NS3 should adopt a flexible motion so that the protease domain can reorientate and extend itself to carry out its proteolytic function.
In contrast to the extended conformation of DENV NS2B18NS3, the crystallographic structure of the related NS33–631NS4A21–32 from HCV adopts a more globular conformation, with the NS3 protease-domain active site positioned towards the molecular interior. The compactness of this protein has been proposed to be caused in part by the 174PPAVP179 linker stretch (Li et al., 2014 ▸). As presented, substitution of the DENV-2 NS2B18NS3 linker by 174PPAVP179 did not significantly alter the extended feature of the DENV-2 NS3 mutant 174PPAVP179, but slightly increased the flexibility of the mutant protein (Fig. 7 ▸). Although a solution study of the entire HCV NS33–631NS4A21–32 is lacking, we speculate that the more compact form observed in the crystal structure of NS33–631NS4A21–32 may be caused by crystal packing, in which the C-terminus of NS3 comes into proximity to the protease-domain active site (Yao et al., 1999 ▸).
Conformational changes in macromolecules upon substrate binding and catalysis is a common feature of enzymatic mechanisms, and is required for optimal enzyme turnover (Hammes, 2002 ▸; Eisenmesser et al., 2005 ▸). SAXS data for DENV-2 NS2B18NS3 in the presence of MgATP and its analogue MgAMPPNP did not show a change in the overall dimensions of the enzyme, while the binding of MgADP led to a slight decrease in the R g value. Interestingly, RNA caused a significant change in NS3, as indicated in the lower and medium q ranges of the scattering profile (Fig. 8 ▸ a). The SREFLEX models calculated based on these data adopted different movements of the protease domain with respect to the helicase domain in the absence or presence of RNA (Figs. 8 ▸ b and 8 ▸ c). One of the plausible models of the empty form reveals large translational movements of the protease domain when compared with the crystal structure (PDB entry 2vbc; Luo, Xu, Hunke et al., 2008 ▸), while in the case of the RNA-bound form the protease domain had largely rotational movement. Recently, binding of ssRNA to the helicase triggered a slight conformation change within the helicase domain and changed the P-loop into the NTP-bound conformation (Luo, Xu, Watson et al., 2008 ▸). Investigating the DENV-4 NS2B18NS3 crystallographic structure that adopted conformation II (Luo et al., 2010 ▸), a few interactions were identified among the protease domain, linker region and the P-loop of the helicase domain (Fig. 9 ▸ a). Therefore, the interaction of RNA and NS3 may induce a conformational change in the P-loop of the helicase domain and thereby effect ATP hydrolysis, followed by a cascade of conformational changes to the linker and the protease domain, which undergoes the substantial conformational alterations required for optimal catalysis of bound RNA.
In conclusion, the solution studies have revealed that DENV NS3 is extended in solution and suggest the functional conformation to be the nucleotide-bound conformation irrespective of the serotype. The importance of the linker residues in protein flexibility and domain–domain arrangement was shown by the compactness of the individual protease and helicase domains as well as the DENV-2 NS3 mutant 174PPAVP179. The study also confirms that binding of RNA caused conformational alteration(s) inside NS3 in solution, signifying the necessity for high-level flexibility for this multifunctional protein. Finally, insight into the allosteric effect of the ATPase inhibitor quercetin is provided, which may pave the way for new derivatives in the future.
Supplementary Material
Supplementary Table and Figures file. DOI: 10.1107/S2059798317003849/wa5111sup1.pdf
Acknowledgments
WGS thanks the authority of Nanyang Technological University for awarding a research scholarship. This work, as well as the research scholarship of AP, was supported by the Ministry of Education MOE Tier 3 (MOE2012-T3-1-008), Singapore to GG. Use of the Stanford Synchrotron Radiation Lightsource, SLAC National Accelerator Laboratory is supported by the US Department of Energy, Office of Science, Office of Basic Energy Sciences under Contract No. DE-AC02-76SF00515. The SSRL Structural Molecular Biology Program is supported by the DOE Office of Biological and Environmental Research and by the National Institutes of Health, National Institute of General Medical Sciences (including P41GM103393). The contents of this publication are solely the responsibility of the authors and do not necessarily represent the official views of NIGMS or NIH.
References
- Assenberg, R., Mastrangelo, E., Walter, T. S., Verma, A., Milani, M., Owens, R. J., Stuart, D. I., Grimes, J. M. & Mancini, E. J. (2009). J. Virol. 83, 12895–12906. [DOI] [PMC free article] [PubMed]
- Balakrishna, A. M., Basak, S., Manimekalai, M. S. S. & Grüber, G. (2015). J. Biol. Chem. 290, 3183–3196. [DOI] [PMC free article] [PubMed]
- Bernadó, P., Mylonas, E., Petoukhov, M. V., Blackledge, M. & Svergun, D. I. (2007). J. Am. Chem. Soc. 129, 5656–5664. [DOI] [PubMed]
- Clum, S., Ebner, K. E. & Padmanabhan, R. (1997). J. Biol. Chem. 272, 30715–30723. [DOI] [PubMed]
- Delaglio, F., Grzesiek, S., Vuister, G. W., Zhu, G., Pfeifer, J. & Bax, A. (1995). J. Biomol. NMR, 6, 277–293. [DOI] [PubMed]
- Dip, P. V., Kamariah, N., Subramanian Manimekalai, M. S., Nartey, W., Balakrishna, A. M., Eisenhaber, F., Eisenhaber, B. & Grüber, G. (2014). Acta Cryst. D70, 2848–2862. [DOI] [PubMed]
- Durand, D., Vivès, C., Cannella, D., Pérez, J., Pebay-Peyroula, E., Vachette, P. & Fieschi, F. (2010). J. Struct. Biol. 169, 45–53. [DOI] [PubMed]
- Egloff, M.-P., Benarroch, D., Selisko, B., Romette, J.-L. & Canard, B. (2002). EMBO J. 21, 2757–2768. [DOI] [PMC free article] [PubMed]
- Eisenmesser, E. Z., Millet, O., Labeikovsky, W., Korzhnev, D. M., Wolf-Watz, M., Bosco, D. A., Skalicky, J. J., Kay, L. E. & Kern, D. (2005). Nature (London), 438, 117–121. [DOI] [PubMed]
- Erbel, P., Schiering, N., D’Arcy, A., Renatus, M., Kroemer, M., Lim, S. P., Yin, Z., Keller, T. H., Vasudevan, S. G. & Hommel, U. (2006). Nature Struct. Mol. Biol. 13, 372–373. [DOI] [PubMed]
- Falgout, B., Pethel, M., Zhang, Y. M. & Lai, C. J. (1991). J. Virol. 65, 2467–2475. [DOI] [PMC free article] [PubMed]
- Ferron, F., Decroly, E., Selisko, B. & Canard, B. (2012). Antiviral Res. 96, 21–31. [DOI] [PMC free article] [PubMed]
- Franke, D. & Svergun, D. I. (2009). J. Appl. Cryst. 42, 342–346. [DOI] [PMC free article] [PubMed]
- Gasteiger, E., Hoogland, C., Gattiker, A., Duvaud, S., Wilkins, M. R., Appel, R. D. & Bairoch, A. (2005). The Proteomics Protocols Handbook, edited by J. M. Walker, pp. 571–607. Totowa: Humana Press.
- Gebhard, L. G., Kaufman, S. B. & Gamarnik, A. V. (2012). PLoS One, 7, e36244. [DOI] [PMC free article] [PubMed]
- Gledhill, J. R. & Walker, J. E. (2005). Biochem. J. 386, 591–598. [DOI] [PMC free article] [PubMed]
- Goddard, T. D. & Kneller, D. G. (2002). SPARKY 3. University of California, San Francisco, USA.
- Guinier, A. (1939). Ann. Phys. (Paris), 12, 161–237.
- Gupta, G., Lim, L. & Song, J. (2015). PLoS One, 10, e0134823. [DOI] [PMC free article] [PubMed]
- Hammes, G. G. (2002). Biochemistry, 41, 8221–8228. [DOI] [PubMed]
- Ho, J., Sielaff, H., Nadeem, A., Svanborg, C. & Grüber, G. (2015). J. Mol. Biol. 427, 1866–1874. [DOI] [PubMed]
- Johansson, M., Brooks, A. J., Jans, D. A. & Vasudevan, S. G. (2001). J. Gen. Virol. 82, 735–745. [DOI] [PubMed]
- Junjhon, J., Pennington, J. G., Edwards, T. J., Perera, R., Lanman, J. & Kuhn, R. J. (2014). J. Virol. 88, 4687–4697. [DOI] [PMC free article] [PubMed]
- Kazimierczuk, K. & Orekhov, V. Y. (2011). Angew. Chem. Int. Ed. 50, 5556–5559. [DOI] [PubMed]
- Konarev, P. V., Petoukhov, M. V., Volkov, V. V. & Svergun, D. I. (2006). J. Appl. Cryst. 39, 277–286.
- Konarev, P. V., Volkov, V. V., Sokolova, A. V., Koch, M. H. J. & Svergun, D. I. (2003). J. Appl. Cryst. 36, 1277–1282.
- Kozin, M. B. & Svergun, D. I. (2001). J. Appl. Cryst. 34, 33–41.
- Li, K., Phoo, W. W. & Luo, D. (2014). Virol. Sin. 29, 74–85. [DOI] [PMC free article] [PubMed]
- Lindenbach, B. D., Thiel, H.-J. & Rice, C. M. (2007). Fields Virology, 5th ed., edited by D. M. Knipe & P. M. Howley, pp. 1101–1152. Philadelphia: Lippincott-Raven.
- Luo, D., Vasudevan, S. G. & Lescar, J. (2015). Antiviral Res. 118, 148–158. [DOI] [PubMed]
- Luo, D., Wei, N., Doan, D. N., Paradkar, P. N., Chong, Y., Davidson, A. D., Kotaka, M., Lescar, J. & Vasudevan, S. G. (2010). J. Biol. Chem. 285, 18817–18827. [DOI] [PMC free article] [PubMed]
- Luo, D., Xu, T., Hunke, C., Grüber, G., Vasudevan, S. G. & Lescar, J. (2008). J. Virol. 82, 173–183. [DOI] [PMC free article] [PubMed]
- Luo, D., Xu, T., Watson, R. P., Scherer-Becker, D., Sampath, A., Jahnke, W., Yeong, S. S., Wang, C. H., Lim, S. P., Strongin, A., Vasudevan, S. G. & Lescar, J. (2008). EMBO J. 27, 3209–3219. [DOI] [PMC free article] [PubMed]
- Martel, A., Liu, P., Weiss, T. M., Niebuhr, M. & Tsuruta, H. (2012). J. Synchrotron Rad. 19, 431–434. [DOI] [PMC free article] [PubMed]
- Mertens, H. D. T. & Svergun, D. I. (2010). J. Struct. Biol. 172, 128–141. [DOI] [PubMed]
- Mylonas, E. & Svergun, D. I. (2007). J. Appl. Cryst. 40, s245–s249.
- Orekhov, V. Y. & Jaravine, V. A. (2011). Prog. Nucl. Magn. Reson. Spectrosc. 59, 271–292. [DOI] [PubMed]
- Panjkovich, A. & Svergun, D. I. (2016). Phys. Chem. Chem. Phys. 18, 5707–5719. [DOI] [PubMed]
- Patel, S., Blose, J. M., Sokoloski, J. E., Pollack, L. & Bevilacqua, P. C. (2012). Biochemistry, 51, 9312–9322. [DOI] [PMC free article] [PubMed]
- Petoukhov, M. V., Franke, D., Shkumatov, A. V., Tria, G., Kikhney, A. G., Gajda, M., Gorba, C., Mertens, H. D. T., Konarev, P. V. & Svergun, D. I. (2012). J. Appl. Cryst. 45, 342–350. [DOI] [PMC free article] [PubMed]
- Preugschat, F., Yao, C. W. & Strauss, J. H. (1990). J. Virol. 64, 4364–4374. [DOI] [PMC free article] [PubMed]
- Putnam, C. D., Hammel, M., Hura, G. L. & Tainer, J. A. (2007). Q. Rev. Biophys. 40, 191–285. [DOI] [PubMed]
- Rambo, R. P. & Tainer, J. A. (2011). Biopolymers, 95, 559–571. [DOI] [PMC free article] [PubMed]
- Rambo, R. P. & Tainer, J. A. (2013). Nature (London), 496, 477–481. [DOI] [PMC free article] [PubMed]
- Receveur-Brechot, V. & Durand, D. (2012). Curr. Protein Pept. Sci. 13, 55–75. [DOI] [PMC free article] [PubMed]
- Saw, W. G., Tria, G., Grüber, A., Subramanian Manimekalai, M. S., Zhao, Y., Chandramohan, A., Srinivasan Anand, G., Matsui, T., Weiss, T. M., Vasudevan, S. G. & Grüber, G. (2015). Acta Cryst. D71, 2309–2327. [DOI] [PMC free article] [PubMed]
- Smolsky, I. L., Liu, P., Niebuhr, M., Ito, K., Weiss, T. M. & Tsuruta, H. (2007). J. Appl. Cryst. 40, s453–s458.
- Sousa, L. R. F. de, Wu, H., Nebo, L., Fernandes, J. B., da Silva, M. F. G. F., Kiefer, W., Kanitz, M., Bodem, J., Diederich, W. E., Schirmeister, T. & Vieira, P. C. (2015). Bioorg. Med. Chem. 23, 466–470. [DOI] [PubMed]
- Subramanian Manimekalai, M. S., Saw, W. G., Pan, A., Grüber, A. & Grüber, G. (2016). Acta Cryst. D72, 795–807. [DOI] [PubMed]
- Svergun, D. I. (1992). J. Appl. Cryst. 25, 495–503.
- Svergun, D., Barberato, C. & Koch, M. H. J. (1995). J. Appl. Cryst. 28, 768–773.
- Svergun, D. I., Koch, M. H. J., Timmins, P. A. & May, R. P. (2013). Small Angle X-ray and Neutron Scattering from Biomacromolecular Solutions. Oxford University Press.
- Tay, M. Y. F., Saw, W. G., Zhao, Y., Chan, K. W. K., Singh, D., Chong, Y., Forwood, J. K., Ooi, E. E., Grüber, G., Lescar, J., Luo, D. & Vasudevan, S. G. (2015). J. Biol. Chem. 290, 2379–2394. [DOI] [PMC free article] [PubMed]
- Tria, G., Mertens, H. D. T., Kachala, M. & Svergun, D. I. (2015). IUCrJ, 2, 207–217. [DOI] [PMC free article] [PubMed]
- Volkov, V. V. & Svergun, D. I. (2003). J. Appl. Cryst. 36, 860–864.
- Warrener, P., Tamura, J. K. & Collett, M. S. (1993). J. Virol. 67, 989–996. [DOI] [PMC free article] [PubMed]
- Wengler, G. & Wengler, G. (1993). Virology, 197, 265–273. [DOI] [PubMed]
- World Health Organization (2009). Dengue: Guidelines for Diagnosis, Treatment, Prevention and Control. Geneva: World Health Organization. http://www.who.int/tdr/publications/documents/dengue-diagnosis.pdf. [PubMed]
- Xu, T., Sampath, A., Chao, A., Wen, D., Nanao, M., Chene, P., Vasudevan, S. G. & Lescar, J. (2005). J. Virol. 79, 10278–10288. [DOI] [PMC free article] [PubMed]
- Yao, N., Reichert, P., Taremi, S. S., Prosise, W. W. & Weber, P. C. (1999). Structure, 7, 1353–1363. [DOI] [PubMed]
- Yon, C., Teramoto, T., Mueller, N., Phelan, J., Ganesh, V. K., Murthy, K. H. M. & Padmanabhan, R. (2005). J. Biol. Chem. 280, 27412–27419. [DOI] [PubMed]
- Zandi, K., Teoh, B.-T., Sam, S.-S., Wong, P.-F., Mustafa, M. R. & AbuBakar, S. (2011). Virol. J. 8, 560. [DOI] [PMC free article] [PubMed]
- Zhao, Y., Soh, T. S., Chan, K. W. K., Fung, S. S. Y., Swaminathan, K., Lim, S. P., Shi, P.-Y., Huber, T., Lescar, J., Luo, D. & Vasudevan, S. G. (2015). J. Virol. 89, 10717–10721. [DOI] [PMC free article] [PubMed]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Supplementary Table and Figures file. DOI: 10.1107/S2059798317003849/wa5111sup1.pdf