Abstract
Single cells respond heterogeneously to biochemical treatments, which can complicate the analysis of in vitro and in vivo experiments. In particular, stressful perturbations may induce the epithelial-mesenchymal transition (EMT), a transformation through which compact, sensitive cells adopt an elongated, resistant phenotype. However, classical biochemical measurements based on population averages over large numbers cannot resolve single cell heterogeneity and plasticity. Here, we use high content imaging of single cell morphology to classify distinct phenotypic subpopulations after EMT. We first characterize a well-defined EMT induction through the master regulator Snail in mammary epithelial cells over 72 h. We find that EMT is associated with increased vimentin area as well as elongation of the nucleus and cytoplasm. These morphological features were integrated into a Gaussian mixture model that classified epithelial and mesenchymal phenotypes with >92% accuracy. We then applied this analysis to heterogeneous populations generated from less controlled EMT-inducing stimuli, including growth factors (TGF-β1), cell density, and chemotherapeutics (Taxol). Our quantitative, single cell approach has the potential to screen large heterogeneous cell populations for many types of phenotypic variability, and may thus provide a predictive assay for the preclinical assessment of targeted therapeutics.
Introduction
The epithelial-mesenchymal transition (EMT) transforms compact, adherent cells into an elongated, motile phenotype and has been associated with tumor dissemination and drug resistance.1, 2 During EMT, cell-cell contacts are downregulated (e.g. E-cadherin) and cytoskeletal proteins are upregulated (e.g. vimentin), which promote scattering and local dissemination from the tumor surface.3 EMT has also been associated with resistance to apoptosis, particularly in the context of conventional chemotherapies.4 One “master regulator” of EMT is the Snail family of zinc-finger transcription factors, which directly repress E-cadherin transcription5 and are associated with poor clinical outcome.6 More generally, aberrant extracellular stimuli from the microenvironment such as growth factors (e.g. TGF-β1)7 or sublethal stresses (e.g. cytotoxic treatments)8 can trigger various EMT pathways and associated programs of phenotypic changes.9 Such phenotypic plasticity is challenging to measure using classical biochemical assays based on population averages at endpoints. Instead, single cell measurements are essential to reveal distinct subpopulations or rare and exceptional phenotypes.10, 11 These deeper insights into heterogeneity may improve the predictive capability of preclinical drug testing and guide the design of targeted therapies.12
High-content screening (HCS) is a promising approach to quantify heterogeneous single cell morphology through optical fluorescence microscopy.13 HCS incorporates automated segmentation of cellular and subcellular features from digital images, enabling a complete readout of all single cell behaviors within a population, including outliers and infrequent events.14 This approach can elucidate the intrinsic heterogeneity in a given cell population and reveal the effects of controlled perturbations, based on immunostaining of fixed cells or fluorescent live cell reporters.15–25 Moreover, HCS has been applied to screen candidate anticancer compounds against cell lines in a multiwell plate format.18, 23, 24, 26–28 Several studies have recently characterized the effect of inhibitors on epithelial or mesenchymal cell lines,23 and EMT has been used to calibrate HCS analyses.25, 27 However, HCS has not been applied for longitudinal studies of EMT in response to defined stimuli such as growth factor or chemical stimulation. Such quantitative measurements of single cell heterogeneity are crucial given previous observations of phenotypic plasticity that occur spontaneously29 and may be amplified in response to sublethal drug treatment.30 Indeed, drug response can vary with cell plating density.31 which may be a consequence of EMT. For instance, basal epithelial cells such as MCF-10A have been observed to display mesenchymal-like phenotypes in low-density cultures, measured through transcriptional profiling or fluorescent reporters.32–34 At increasing densities, the cell population tends to be more epithelial, suggesting some cells may revert back from a mesenchymal phenotype. Single cell profiling could elucidate these density dependent phenotypic changes, which may occur in physiological or pathological contexts.
In this article, we measure single cell morphology and biomarker expression during EMT induction. First, we apply a highly controlled EMT stimulus through an inducible Snail-1 construct in human mammary epithelial cells, which results in a dramatic elongation of subcellular nuclear, vimentin, and cytoplasmic features. These quantitative changes in shape features define a reference model to classify the combined population of control and induced EMT cells as a mixture of two phenotypic subpopulations. Next, this reference model is applied to classify heterogeneous populations undergoing EMT at varying time points after Snail-1 induction, treatment with TGF-β1, and at varying densities. Finally, we show that treatment with the microtubule inhibitor Taxol can exacerbate EMT. This overall approach establishes a general framework for reconstructing EMT and single cell heterogeneity in the context of biochemical perturbations.
Methods
Cell Culture
Human mammary epithelial cells stably transfected with an ER-Snail-16SA retroviral construct (MCF-10A Snail) as well as the parental cells (MCF-10A) were a gift from D.A. Haber (Massachusetts General Hospital).35 The Snail-16SA variant is refractory to phosphorylation and is thus stably expressed and localized in the nucleus, where it initiates EMT induction.36 Both MCF-10A variants were cultured following Brugge and coworkers.37 Cells were plated at low density (500 cells/well, unless specified) in 96-half-well high content imaging microplates (Fisher Scientific) coated with 5 μg/cm2 fibronectin (Sigma-Aldrich). After settling and adhering overnight, cells were subjected to exogenous stimuli or drugs as indicated. Special care was taken to ensure that cells remained subconfluent at the conclusion of the experiments, unless noted. T-47D breast carcinoma cells were acquired from the Developmental Therapeutics Program (DTP) of the National Cancer Institute, cultured in RPMI 1640 (ATCC) supplemented with 10% Fetal Bovine Serum (Fisher Scientific), 0.2U/mL bovine insulin (Sigma-Aldrich), 100 units/mL penicillin, and 100 μg/mL streptomycin, and were passaged following standard ATCC protocol. MDA-MB-231 cells were purchased from ATCC and were cultured in DMEM containing L-Glutamine, L-glucose, and sodium pyruvate (Fisher Scientific), and supplemented with 10% FBS (Fisher Scientific), 100 units/mL penicillin, and 100 μg/mL streptomycin. T-47D and MDA-MB-231 cells were cultured for 96 h in high content imaging plates as described and plated with 3,000 and 1,500 cells/well, respectively. These higher seeding densities with respect to MCF-10A experiments were experimentally determined, and compensate for differences in the characteristic doubling times of each cell line.
EMT Induction with Snail-1 or TGF-β1
Snail-1 expression was induced in MCF-10A Snail cells through the addition of 4-hydroxytamoxifen (OHT; Sigma-Aldrich) resuspended in DMSO and used at a final concentration of 500 nM in media. To induce EMT to varying extents at the population level, a time course of Snail-1 induction was prepared with four conditions, all over a course of 72 h. The durations of OHT exposure included 0, 24, 48, and 72 h, where DMSO (0.05% final) was applied during the remainder of the 72 h time course. Separately, EMT was induced in MCF-10A cells through treatment with 5 ng/mL recombinant human TGF-β1 (R&D Systems) in growth media. A comparable time course was conducted with four conditions over 72 h. All conditions were initially treated with growth media and TGF-β1 was added to different wells for durations of 0, 24, 48 and 72 h. For both EMT induction experiments, media was replenished at the 48 h time point, to prevent nutrient deprivation and growth factor depletion.
Density Dependent Induction of EMT
MCF-10A cells were plated in high content imaging plates, as described, where 375 cells/well was chosen as the lowest seeding density. This density was first validated during routine culture (equivalent to ~1:20 split) as it maintained the ability to produce characteristic epithelial monolayers several days after passaging cells. Thus, to generate a range of terminal densities, cells were plated at either 375 or 500 cells/well (the typical density for all other experiments) and subsequently cultured for either 72 or 96 h before fixation and immunostaining.
Drug Treatment with Taxol for EMT Induction
MCF-10A Snail cells were cultured in media with OHT for 72 hours to induce a mesenchymal phenotype (preinduced) or with DMSO to maintain an epithelial phenotype (uninduced). Both preinduced and uninduced cells were seeded at a density of 750 cells/well, allowed to adhere to fibronectin-coated multiwell plates overnight as described, and the growth media containing OHT or DMSO was removed. Both cell types were then treated with either DMSO (0.05%, control) or 4 nM paclitaxel (Taxol) for 48 hours, and subsequently fixed and immunostained.
Immunostaining and Fluorescent Imaging
At the conclusion of time course experiments, cells were fixed for 20 minutes at 4°C with 4% paraformaldehyde in 1X PBS (all solutions are in 1X PBS unless specified). Cells were permeabilized with 0.1% Triton X-100, washed, and blocked with 10% goat serum. Cells were washed with sodium acetate buffer for 15 minutes and then rinsed several times with 1% nonfat dry milk. Cells were incubated overnight at 4°C with primary antibodies: 250 μg/mL E-cadherin (Fisher Scientific) diluted at 1:500 and vimentin (Cell Signaling Technology) diluted at 1:200 in 1% milk. Cells were then washed with 1% milk and incubated in the dark at room temperature with suitably matched secondary antibodies: 2 mg/mL Alexa Fluor 488 and Alexa Fluor 555 (Invitrogen) diluted at 1:500 in 1% milk. Lastly, cells were washed with 1X PBS, incubated with 2 μg/mL Hoechst Pentahydrate (Invitrogen) and 2 μg/mL HCS CellMask Deep Red (ThermoFisher Scientific) for 30 minutes at room temperature, and washed again.
Immunostained cells were imaged using an inverted epifluorescence microscope (Nikon TiE). Images were acquired with a 20X Super Plan Fluor objective (NA = 0.45, extra long working distance) and with a 14-bit resolution sCMOS camera (Andor Neo). Fluorescence illumination was provided by a light-guide coupled Lumencore Sola white light excitation system. Care was taken to ensure all images were acquired with identical acquisition parameters (exposure time, camera gain/gamma control, and microscope aperature settings). Post-acquisition, fluorescence thresholds for each experiment were set for qualitative comparison across conditions using builtin NIS Elements AR settings to eliminate background noise and reduce pixel saturation. Detailed settings are described in Supporting Information.
Image Analysis
Cell detection and shape feature analysis were performed using CellProfiler 2.1 (Broad Institute).38 First, the uneven illumination was corrected across channels and image sets. Next, fluorescently labeled nuclei (DAPI channel) were segmented as primary objects. Based on the nuclei positions, fluorescently labeled vimentin and cytoplasm were segmented as separate secondary objects. These fluorescent objects were manually verified and corrected as needed. Detailed segmentation parameters are described in the Supporting Information (Fig. S2). Finally, shape measurements were extracted from the segmented objects, particularly nuclear max radius, vimentin area, cytoplasm form factor, and cytoplasm max feret diameter.
Phenotype Classification using a Gaussian Mixture Model (GMM)
An initial training set was developed by segmenting cells treated either with DMSO or OHT for 72 h, corresponding to putative epithelial and mesenchymal phenotypes. For each condition, CellProfiler was used to segment >100 cells for 15 nuclear, vimentin, and cytoplasmic shape metrics, which were rescaled between 0 and 1 for consistent comparison (see Supporting Information for additional details, Fig. S1–3). Next, the single cell features from both conditions were combined into a single dataset and an expectation maximization (EM) algorithm was used to determine maximum likelihood estimates of the parameters for a GMM39 assuming two subpopulations (gmdistribution.fit, MATLAB R2013b). Based on the 15 metrics, all possible combinations of metrics (up to 5) were used to train the GMM. This classifier was then tested against a second dataset of control and induced cells prepared under consistent experimental conditions. A third dataset based on epithelial (T-47D) and mesenchymal (MDA-MB-231) cell lines were also tested. An optimized 4 metric GMM distribution was used to partition all subsequent datasets into epithelial and mesenchymal subpopulations, respectively. For visualization purposes, the phenotypic distributions are displayed by normalized cytoplasmic maximum feret diameter, which is highly predictive.
Results
Profiling Epithelial and Mesenchymal Phenotypes after Snail Induction
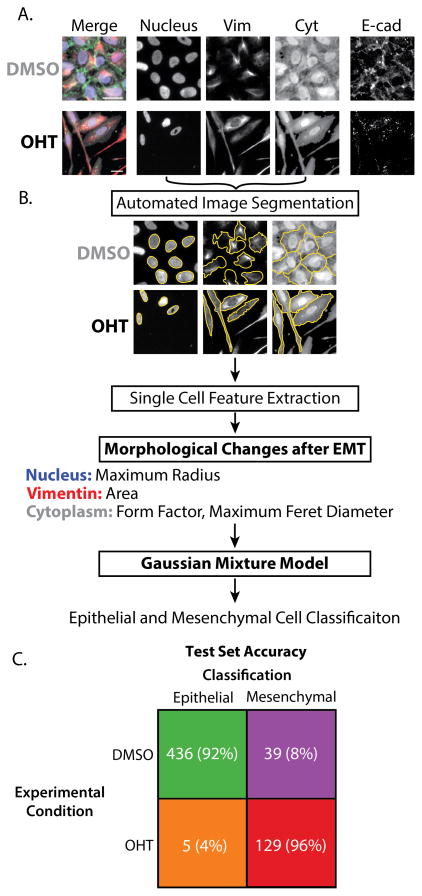
To establish a training set of epithelial and mesenchymal phenotypes, a well-defined EMT stimulus was applied through the master regulator Snail. Human mammary epithelial cells (MCF-10A Snail) transfected with a constitutively active variant of Snail-136 fused to an estrogen receptor response element were exposed to 4-hydroxytamoxifen (OHT) for 72 h. Previous work using this inducible Snail construct has demonstrated that 72 h treatment with OHT results in nearly complete EMT in the population based on cell morphology and biomarker expression levels.35 Based on these results, cells were treated with either DMSO or OHT for 72 h, followed by an additional 72 h of DMSO treatment to generate pronounced differences in control and induced populations, respectively (Fig. S1). Cells from both conditions were then fixed for immunofluorescent staining of the nucleus (Hoechst), epithelial (E-cadherin) and mesenchymal (vimentin) biomarkers, and cytoplasmic morphology (CellMask). Subsequently, widefield fluorescence microscopy revealed that cells in the control condition (DMSO) displayed a cobblestone morphology with relatively compact nuclei, vimentin, and cytoplasm, as well as enhanced E-cadherin expression at the edges (Fig. 1A). In contrast, cells in the induced condition (OHT) displayed a spindle-like morphology with more elongated nuclei, vimentin, and cytoplasmic features, as well as nearly nonexistent E-cadherin expression (Fig. 1A). These dramatic differences in cell morphology and biomarker expression are consistent with an epithelial phenotype for the control condition (DMSO) and a mesenchymal phenotype for the induced condition (OHT).
Figure 1. Classification of epithelial and mesenchymal phenotypes after 72 h DMSO (control) and OHT (Induced Snail-1), followed by 72 h DMSO.
(A) Immunofluorescent staining of nuclei, vimentin, cytoplasm and E-cadherin (blue, red, gray, and green, respectively). Scale = 25 μm. (B) Process flow of single cell segmentation and feature extraction. (C) Performance classifier of actual and predicted subpopulations with the number of cells and percent ( ) of cells in each group.
These qualitative changes in morphology and biomarker expression were then quantified at the single cell level through automated object segmentation and measurement (Fig. 1B, S2 and Supporting Information). CellProfiler was used to identify single cells based on their nuclei, which were then used to locate the corresponding vimentin and cytoplasmic features. Based on these segmented nuclear, vimentin, and cytoplasmic features, a total of 44 metrics were extracted, including 15 notable shape features (Fig. S3A, S3B). These metrics included fluorescence intensities, which were somewhat variable across repeated experiments, perhaps due to antibodies40 and slight variations in imaging conditions.41, 42 For instance, cells displayed substantial differences in integrated vimentin intensity for control (DMSO) and induced EMT (OHT) conditions, but this was largely due to changes in area, since the mean and median vimentin intensities were relatively similar (Fig. S3C). Similarly, E-cadherin quantification proved to be challenging due to its inconsistent or minimal expression after EMT, so it was only used qualitatively for confirmation of cell phenotype. Thus, we focused primarily on shape metrics, which are standardized and should be broadly applicable across experimental conditions. Well over 100 cells were analyzed per condition, allowing good sampling of the statistical distribution associated with each phenotype.
Gaussian mixture models (GMM) were fit to different training datasets based on all possible combinations of single cell metrics (up to five) combined from the control (DMSO) and induced (OHT) conditions. An optimal combination of 4 metrics was selected: nuclear maximum radius, vimentin area, cytoplasmic form factor, and cytoplasmic maximum feret diameter (Fig. S4). This classifier was then tested on a separate dataset with comparable experimental conditions. For the control (DMSO) condition, 92% of cells were classified as epithelial. Instead, for the induced (OHT) condition, 96% of cells were classified as mesenchymal (Fig. 1C, S5A, S5B). This discrepancy between the experimental condition and phenotype classification may accurately represent the heterogeneity in each experimental condition. For instance, MCF-10A cells can undergo a spontaneous but transient EMT,32, 33 which could explain why the control (DMSO) condition had a small percentage of cells which were classified as mesenchymal (Fig. S5C). In addition, we observed that some cells in the induced (OHT) condition displayed smaller nuclei and vimentin features due to recent division, which could explain why these cells were classified as epithelial (Fig. S5D).
The classifier was then tested against cell lines with established epithelial or mesenchymal phenotypes. In particular, T-47D is a human breast carcinoma line with a strong luminal epithelial phenotype, and a biomarker profile distinct from basal and fibroblast phenotypes.20 T-47D cells displayed compact, rounded morphologies, and formed epithelial clusters with prominent E-cadherin expression and lack of vimentin expression (Fig. S6A). Instead, MDA-MB-231 is a highly metastatic human breast adenocarcinoma line with a highly mesenchymal phenotype. MDA-MB-231 cells were dispersed in culture with elongated, spindle-like morphologies, as well as strong vimentin expression, and absence of E-cadherin expression (Fig. S6A). For the same combination of four metrics, the GMM classifier displayed 100% agreement for actual and predicted T-47D, as well as 95% for actual and predicted MDA-MB-231 (Fig. S6B, C). This improved accuracy is consistent with the phenotypic homogeneity that would be expected from these two cell lines. Since T-47D did not exhibit noticeable vimentin expression, the accuracy of a classifier based on only nuclear maximum radius, cytoplasmic form factor, and cytoplasmic maximum feret diameter was also evaluated. This reduced set of 3 metrics was highly accurate for T-47D cells, with 99% agreement (Fig. S6D). However, the absence of vimentin area in this reduced set was significantly worse for classifying MDA-MB-231 cells, with only 63% agreement. Further reduction to two metrics for classification revealed notable combinations including vimentin area and cytoplasmic max feret diameter, which resulted in slightly increased accuracy for epithelial phenotypes (MCF-10A DMSO, T-47D), but slightly decreased accuracy for mesenchymal phenotypes (MCF-10A OHT, MDA-MB-231) (Fig. S7). In general, combinations of vimentin and cytoplasmic metrics were good predictors of EMT phenotype, with 93–100% accuracy for each epithelial and mesenchymal phenotype. However combinations of nuclear and vimentin or nuclear and cytoplasmic were slightly worse predictors, with 90–100% and 87–94% accuracy per condition, respectively. Further, nuclear, vimentin, or cytoplasmic metrics alone were even worse predictors with accuracy ranges including 61–88%, 87–97%, and 86–95%, respectively. Thus, through a process of combinatorial optimization, a set of 4 metrics (nuclear max radius, vimentin area, cytoplasm form factor, cytoplasm max feret diameter) was chosen for evaluating subsequent experiments and demonstrates the applicability of GMM for cell classification across cell lines. The high accuracies achieved for distinguishing between epithelial and mesenchymal subpopulations reflect the dramatic differences in the chosen metrics across phenotypes. For instance, between the control and induced MCF-10A cells, there was a four-fold increase in mean vimentin area and a two-fold increase in mean cytoplasmic max feret diameter. Similarly, between the epithelial T-47D and mesenchymal MDA-MB-231 cells, there was a five-fold increase in mean cytoplasmic max feret diameter, as well as a 33% decrease in cytoplasmic form factor. Thus, this single cell profiling can detect subtle variations within a population, but these do not affect the accuracy of probabilistic assignment to epithelial and mesenchymal classes. Overall, this analysis establishes a robust classification scheme for epithelial and mesenchymal phenotypes based on biomarker and morphology.
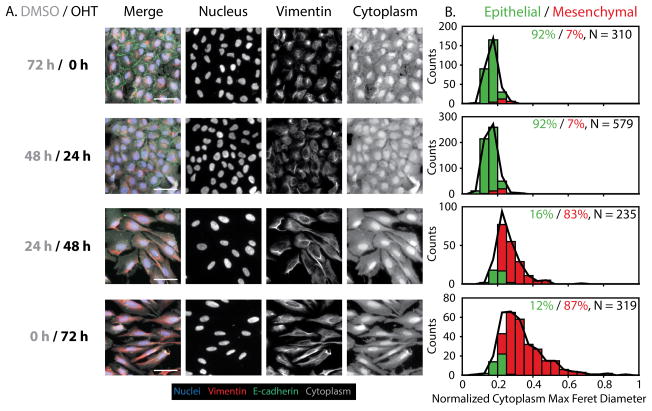
Snail Induction Drives Rapid EMT over 72 h
Next, this classification scheme was applied to reconstruct single cell phenotypes during a time course of Snail-mediated EMT induction over 72 h. MCF-10A Snail cells were cultured in four different conditions with varying OHT treatment times over 72 h. For clarity, each condition is denoted by the duration of OHT exposure, e.g. 0 h (control), 24 h, 48 h and 72 h, where the remainder of the 72 h experiment corresponds to treatment with DMSO (Fig. S8A).
Between 0 h and 24 h, a noticeable loss of E-cadherin was visible, although the nuclei and vimentin remained relatively similar in size and shape (Fig. 2A). From 24 h to 48 h, the cytoplasmic and vimentin features became dramatically elongated with brighter vimentin intensities and there was some subtle elongation of the nuclei. This trend continued between 48 h and 72 h, although the differences in morphology were less dramatic. Overall, these trends are consistent with previous bulk measurements of Snail-mediated EMT induction, which showed an initial loss of E-cadherin and other epithelial markers, followed by a rapid gain of vimentin and mesenchymal markers.35 At the single cell level, the quantitative changes in segmented nuclear, vimentin, and cytoplasmic shape features were consistent with the qualitative trends. In particular, vimentin and cytoplasmic associated features (vimentin area and cytoplasm max feret diameter) remained relatively consistent between 0 h and 24 h, increased dramatically between 24 h and 48 h, then remained relatively consistent between 48 h and 72 h. Instead, nuclear maximum radius displayed an increase as early as 24 h, but did not continue to change appreciably at later time points. In addition, cytoplasmic form factor (object “roundness”) remained unchanged between 0 h and 24 h, but gradually decreased from 24 to 72 h (Fig. S9).
Figure 2. Time course measurements of cell morphology and biomarker expression with Snail-1 induction (OHT).
(A) Control cells (DMSO) show characteristic epithelial features, including compact, cobblestone like morphologies with high E-cadherin at the cell edges and low vimentin expression. 24 h OHT treatment results in decreased E-cadherin at the edges. 48 h and 72 h OHT treatment result in vimentin and cytoplasmic features with increased area and elongation. Scale = 50 μm. (B) GMM phenotypic classification shows that 0 h and 24 h are primarily epithelial (green bars and %), with rapid transition to mesenchymal phenotype at 48 h and 72 h (red bars and %), relative to the complete population (black line). N indicates the number of individual cells pooled per condition.
Next, these morphological features were used to classify cells from each condition into epithelial (E) and mesenchymal phenotypes (M). Initially at 0 h and 24 h, the population was almost entirely epithelial (92%) (Fig. 2B, S8B). At 48 h, there was a dramatic shift in the distribution of phenotypes, such that the population was now primarily mesenchymal (83%). At 72 h, this distribution remained comparable and was dominated by the mesenchymal phenotype (87%) (Fig. 2B, S8B). Altogether, these results suggest that Snail-mediated EMT induction is rapid with relatively consistent kinetics across single cells. Further, 72 h OHT treatment followed by 72 h withdrawal of the stimulus (training and test conditions) yielded a 96% mesenchymal population. This result suggests that 72 h of OHT treatment is sufficient to induce a stable mesenchymal phenotype, likely sustained through epigenetic modifications (Fig. S8C, S8D).35 Further examination of the posterior probability distribution revealed that 93–96% of the population classified as either epithelial or mesenchymal phenotype with high probability (Fig. S10). This serves as an effective validation for this GMM classification scheme, which will subsequently be applied to quantify EMT induction in response to less defined biochemical and environmental stimuli.
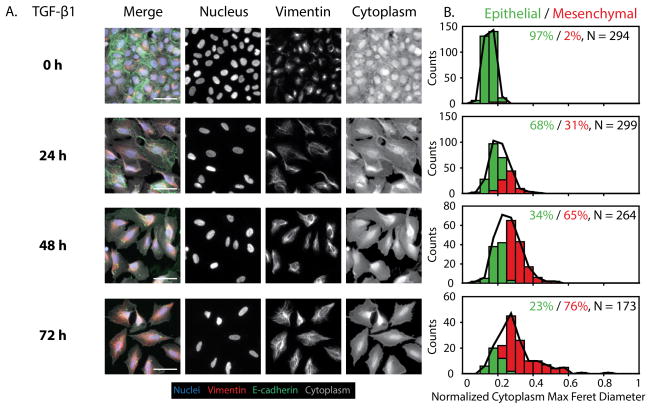
TGF-β1 Induction Drives Gradual EMT over 72 h
The cytokine transforming growth factor TGF-β1 is well-known to induce EMT, although the cellular response can vary in context and remains poorly understood.7 Since the kinetics of EMT induction through TGF-β1 are likely to be less consistent from cell to cell, the use of single cell profiling may yield new insights that cannot be ascertained from population-averaged measurements. To characterize TGF-β1 induced EMT, wild type MCF-10A cells were cultured in four different conditions with varying treatment times (5 ng/mL TGF-β1), again ranging from 0 h to 72 h (Fig. S11A).
Between 0 h and 24 h, there was a noticeable, but incomplete loss of E-cadherin at the cell edges with some elongation and overexpression of vimentin (Fig. 3A). From 24 h through 72 h, the E-cadherin at the cell-cell junctions was largely internalized, while vimentin was brighter and more elongated. In comparison to the previous Snail induction, this TGF-β1 induction resulted in a widened rectangular shape, with less elongated vimentin and cytoplasmic features (Fig. 2A, 3A). Overall, these qualitative trends indicate that TGF-β1 induction results in more variable kinetics at the single cell level relative to Snail induction through OHT. The quantitative single cell metrics also showed more gradual changes from 0 h to 72 h with TGF-β1, instead of the rapid EMT observed with Snail-induction from 24 h to 48 h. In particular, the vimentin area and cytoplasm max feret diameter displayed a gradual increase as early as 24 h, which continued to rise over 48 h and 72 h (Fig. S12). Additionally, there was an obvious decrease in cytoplasmic form factor from 0 to 24 h, which remained low across 48 and 72 h, relative to the 0 h condition. On the other hand, nuclear max radius did not change appreciably across TGF-β1 induction conditions, thus representing a subtle distinction between the morphologic changes that take place for Snail vs. TGF-β1 mediated EMT induction (Fig. S12). At the single cell level, it should be noted that the morphological changes are subtler with TGF-β1 induction relative to Snail induction, so that the overall distributions of vimentin, and cytoplasmic associated features display less of a shift through 72 h. The smaller shift of the statistical distribution may arise from the less elongated and more teardrop shaped vimentin features at the single cell level.
Figure 3. Time course measurements of cell morphology and biomarker expression with TGF-β1 induction.
(A) Control cells (0 h) also show characteristic epithelial features with high E-cadherin at the cell edges and low vimentin expression. 24 h TGF-β1 treatment results in loss of E-cadherin for some cells, compared with increased vimentin expression. 48 h and 72 h OHT treatment result in vimentin and cytoplasmic features with increased area and elongation. Interestingly, TGF-β1 results in a more boxy shape than with Snail-1 induction. Scale = 50 μm. (B) GMM phenotypic classification shows that 0 h is primarily epithelial (green bars and %), with a gradual increase in mesenchymal phenotypes (red bars and %) over 72 h, relative to the complete population (black line). N indicates the number of individual cells pooled per condition.
The classification of these single cell morphological features into epithelial and mesenchymal phenotypes further corroborates with these trends. At 0 h, the population was primarily epithelial (97%) (Fig. 3B, S11B). However, at 24 h, some of the population underwent EMT, resulting in a diminished epithelial subpopulation (68%) and increased mesenchymal subpopulation (31%). This trend continues at 48 h, with a further decrease in epithelial subpopulation (34%) and increase in mesenchymal subpopulation (65%). By 72 h, there is an additional shift in the population, with the fewest epithelial cells (23%) and a mostly mesenchymal subpopulation (76%) (Fig. 3B, S11B). This incomplete EMT at the population level after 72 h is consistent with flow cytometry measurements reported elsewhere, which show that complete loss of epithelial biomarkers and gain of mesenchymal markers requires ~ 7–15 days, the kinetics of which are more rapid with increasing dosage.43 Thus, to examine whether or not an initial TGF-β1 exposure stimulates long term EMT induction, TGF-β1 was pulsed for 72 h, then withdrawn for an additional 72 h (Fig. S11C). Interestingly, 72 h withdrawal of TGF-β1 after 72 h treatment yielded a primarily mesenchymal population (97%) (Fig. S11D). These results are in agreement with OHT pulse/withdrawal experiments, suggesting that 72 h exposure to either stimulus (Snail induction via OHT or TGF-β1) is sufficient to initiate an EMT program that is self-sustained over a 72 h period of withdrawal from the stimulus (Fig. S8C, S8D, S11C, S11D).35 Further examination of the posterior probability distribution revealed that ~90% of the treated populations classified as either epithelial or mesenchymal with high probability, but there was an appreciable ~10% where classification was less definitive (Fig. S13). Thus, the GMM used here was able to reveal distinct differences in EMT induction between the potent Snail stimulus, displaying rapid EMT kinetics, and the exogenous application of the growth factor TGF-β1, displaying a more gradual EMT induction.
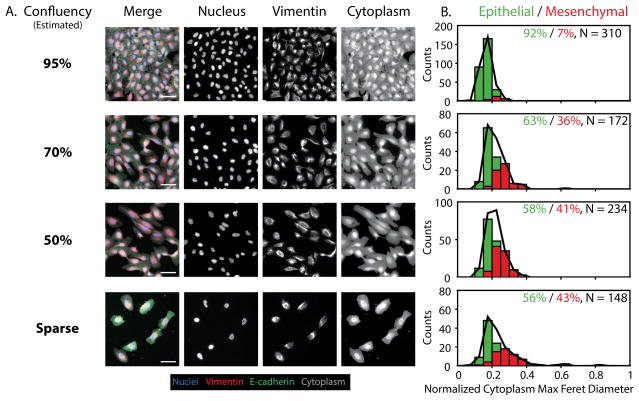
MCF-10A Cells Exhibit Plasticity and Undergo EMT in Subconfluent Cultures
Cell plating density has been observed to affect epithelial phenotype, as well as proliferation rate and drug sensitivity. Density-dependent effects are challenging to resolve using conventional bulk assays, but may be evaluated using single cell profiling. To characterize the role of cell density on EMT, wild type MCF-10A cells were cultured under four different conditions to yield varying levels of confluency ranging from ~15% to ~95% (Fig. S14A, S14B). Cells at high confluency (95%) formed a monolayer with the characteristic epithelial phenotype including strong E-cadherin expression at cell-cell junctions, low levels of vimentin, and cuboidal shaped cells (Fig. 4A). As cell density decreased, cells became elongated with a gradual loss of E-cadherin at cell junctions, an increase in internalized E-cadherin, and a noticeable upregulation of vimentin expression (Fig. 4A). Furthermore, single cell segmentation revealed that vimentin area, cytoplasm maximum feret diameter, and nuclear maximum radius all increased considerably from 95% to 70% confluence (Fig. S15). At the population level, these vimentin and cytoplasm morphologic features were relatively consistent across the subconfluent densities (70% and 50%), while few cells at 50% confluence had even larger vimentin areas, suggesting a slight shift towards more EMT at the lower density. Finally, the sparse confluency condition represented the most extreme increase in vimentin area, cytoplasm maximum feret diameter, and nuclear maximum radius relative to higher density conditions (Fig. S15). Surprisingly, there was no clear trend for cytoplasmic form factor, which may result from the more boxy morphologies.
Figure 4. Changes in cell morphology and biomarker expression with cell density.
(A) Cells at high density (95%) form a confluent monolayer with cobblestone morphology, tight E-cadherin junctions and low vimentin. Cells at lower, subconfluent densities (70%, 50%) display some loss of E-cadherin, with some elongation. Sparse cells (15%) display no E-cadherin junctions, with vimentin and cytoplasmic features that display increased area and elongation. Scale = 50 μm. (B) GMM phenotypic classification shows that 95% density is primarily epithelial (green bars and %), with a gradual increase in mesenchymal (red bars and %) phenotypes up to 51% at 15% density, relative to the complete population (black line). N indicates the number of individual cells pooled per condition.
Classification of epithelial or mesenchymal phenotypes by GMM revealed that the high confluency condition (~95%) was classified as primarily epithelial (92%) (Fig. 4B). However, at less confluent densities, an appreciable number of cells were classified as mesenchymal. In particular, cells at 70% confluency were classified as 36% mesenchymal. Similarly, cells at 50% confluency were classified as 41% mesenchymal. Finally, at sparse coverage (~15%), approximately 43% of the population was mesenchymal. This comparable percentage of mesenchymal cells may result from local aggregation, so that the effective local density is similar despite differences in overall density. It is also noteworthy that the statistical distribution of these two subpopulations remained relatively narrow across these conditions, particularly in terms of their cytoplasmic maximum feret diameter. Nevertheless, at sparse coverage (15%), the mesenchymal subpopulation displayed significant kurtosis with a heavy right tail, indicative of more extreme mesenchymal phenotypes. This result was corroborated by the distribution of posterior probabilities, since 11–14% of the population was less definitively classified, relative to the 86–89% of the population classified as epithelial or mesenchymal with high probability (Fig. S16). Overall, these results indicate that MCF-10A cells can display less epithelial phenotypes at subconfluent densities. Thus, during the course of cell culture experiments, as MCF-10A cells progress from sparse to subconfluent to confluent densities, the statistical distribution of phenotypes will initially be more mesenchymal before reverting to more epithelial. Thus, EMT can occur transiently in culture even in the absence of other exogenous stimuli. Such a transient EMT is likely to be advantageous for epithelial wound healing, permitting a rapid migratory response from cells at high density to damaged areas with diminished cell density.34 These results are thus likely to be relevant for investigations of directed epithelial cell migration both in vitro and in vivo.
Sublethal Taxol Enhances EMT in Uninduced and Induced Snail Populations
More generally, EMT activation is associated with stressful microenvironmental conditions, including exposure to sublethal doses of cytotoxic drugs.8 In particular, it has been demonstrated that the conventional chemotherapeutic paclitaxel (Taxol) is capable of inducing EMT and enhancing metastatic potential.44 Further, even short term, 24 h exposures to Taxol have been demonstrated to induce EMT and stem-like properties via Snail-1 in various breast cancer cell lines.45 Such a perturbation is likely to result in a highly heterogeneous response at the single cell level, which can be sensitively profiled with this technique. To measure the effect of sublethal chemotherapeutics on morphological phenotype, MCF-10A Snail cells were treated for 72 h with DMSO or OHT, followed by treatment for 48 h with DMSO or 4 nM Taxol (Fig. S17A).
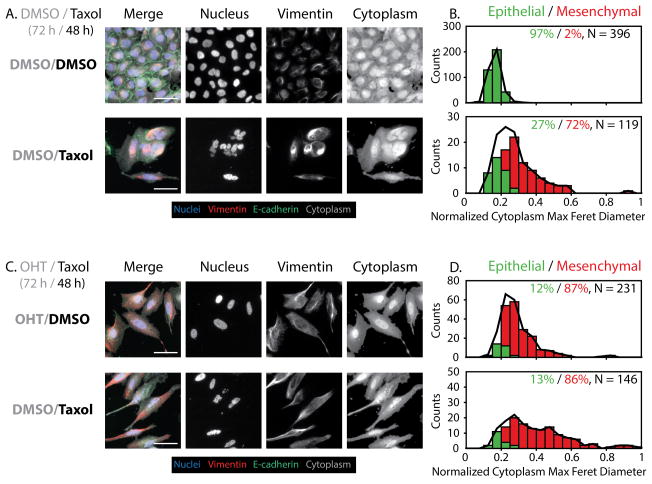
For control epithelial cells (72 h DMSO), subsequent treatment with sublethal Taxol resulted in a visible loss of E-cadherin and a dramatic increase in vimentin area with brighter intensity for a subset of cells in the population (Fig. 5A, Fig. S19). The nuclei now displayed a wide range of aberrant and distorted morphologies, while the vimentin organization displayed characteristic bundling and tangling morphologies (Fig. 5A). The corresponding single cell metrics showed a pronounced shift in cytoplasm-associated features, especially a significant increase in cytoplasm max feret diameter and decrease in cytoplasm form factor for a subset of cells in the population. The average nuclear max radius decreased slightly with Taxol treatment, but the variability increased noticeably. In addition, the vimentin area increased markedly with Taxol treatment, and displayed a much wider distribution than the control (Fig. S18). These trends are in agreement with the morphological classification by GMM. In particular, the uninduced control condition (DMSO/DMSO) as primarily epithelial (97%), while the uninduced Taxol condition was largely mesenchymal (72%) with a smaller epithelial (27%) subset (Fig. 5B, S17B).
Figure 5. Changes in cell morphology and biomarker expression with sublethal Taxol treatment.
(A) Uninduced control cells (DMSO/DMSO) show epithelial features, but uninduced treated cells (DMSO/Taxol) show increased vimentin expression and elongation with aberrant nuclear morphologies. Scale = 50 μm. (B) GMM phenotypic classification shows an increase in mesenchymal (red bars and %) phenotype after Taxol treatment compared to the untreated control, which is primarily epithelial (green bars and %), relative to the complete population (black line). (C) Induced control cells (OHT/DMSO) show more mesenchymal features. Induced treated cells (OHT/Taxol) show strong mesenchymal features, with aberrant nuclei and extremely elongated morphologies. Scale = 50 μm. (D) GMM phenotypic classification shows a strong shift towards mesenchymal phenotype after Taxol treatment. N indicates the number of individual cells pooled per condition.
For preinduced EMT cells (72 h OHT), subsequent treatment with sublethal Taxol further exacerbated the mesenchymal phenotype, with highly elongated vimentin and cytoplasmic features (Fig. 5C). The corresponding single cell metrics showed further substantial increases in vimentin area and cytoplasm max feret diameter, as well as a significant decrease in cytoplasm form factor. The average nuclear max radius did not change appreciably with the addition of taxol, however, the distribution widened, indicating the nuclear variability that results from taxol treatment (Fig. S18). These induced Taxol conditions correspond to single cell morphologies that are comparable to, or even more pronounced than the 72 h OHT induction of Snail (Fig. S9). Classification of epithelial or mesenchymal phenotype by GMM showed that the preinduced control condition (OHT/DMSO) was distributed as 12% epithelial and 87% mesenchymal (Fig. 5D). Similarly, the preinduced Taxol condition (OHT/Taxol) was primarily mesenchymal (86%) with some residual epithelial phenotype (13%). Indeed, the statistical distribution of the mesenchymal subpopulation in the induced Taxol condition displays a relatively wide variance (Fig. 5D, S18), likely resulting from an overall increase in the vimentin and cytoplasm-associated features. Biologically, sublethal Taxol treatment is likely to stress cells to activate EMT, conferring drug resistance, but also to enhance the stability of polymerized vimentin.44 It should be noted that the overall cell numbers are noticeably diminished relative to untreated controls (Fig. 5A, 5B). Although 4 nM Taxol is sublethal, there may be some anti-proliferative effects on a fraction of the subpopulation (Fig. S19). Moreover, this treatment may be driving some phenotypic selection. As a consequence, the variability of the nuclear, vimentin, and cytoplasmic morphologies results in a very wide distribution of mesenchymal phenotypes relative to previous induction techniques. This result was corroborated by the distribution of posterior probabilities, where 94–98% of the population classified as epithelial or mesenchymal with high probability (Fig. S20). Altogether, these results indicate that Taxol treatment is sufficient to induce EMT and further enhance EMT when applied in combination with endogenous stimuli.
Discussion and Conclusions
EMT has been hypothesized to drive phenotypic heterogeneity and plasticity in cancer cell populations, which can be challenging to measure using population-averaged assays at endpoints. Nevertheless, cancer cells often display characteristic patterns of functional phenotypes, suggesting that a reductionist approach could be sufficient to capture tumor complexity. For instance, the overall population could be approximated as a weighted mixture of subpopulations with distinct phenotypes.10 In principle, the statistical distributions of these subpopulations could be inferred by measuring large numbers of single cell phenotypes at discrete snapshots in time.12
Our probabilistic approach classifies a heterogeneous population with a Gaussian mixture model based on two phenotypes: epithelial and mesenchymal. These two extreme states were based on well-defined training phenotypes from controlled Snail induction. In contrast, EMT induction through TGF-β1, varying plating density and Taxol resulted in greater variability in activation kinetics and cell morphology. Indeed, EMT induction with TGF-β1 and plating density resulted in less elongated morphologies, while Taxol treatment resulted in highly aberrant morphology. Nevertheless, further examination of the posterior probability distribution revealed that at least 90% of cells could be classified as epithelial or mesenchymal with very high probability, even with sample sizes of hundreds of cells. Interestingly, a small subpopulation was classified less definitively, reaching ~10% for TGF-β1 treatment and subconfluent density. It has been recently been demonstrated that “partial” EMT phenotypes can occur with partial loss of both E-cadherin and vimentin after treatment with moderate concentrations of TGF-β1.43 In addition, circulating tumor cells in breast cancer can display a spectrum of epithelial to mesenchymal phenotypes, based on the coexpression of epithelial and mesenchymal transcripts (from RNA-in situ hybridization).46 We interpret our results conservatively to avoid overfitting with too many phenotypes, but it is conceivable that this subpopulation represents a partial EMT phenotype. Further work is needed to define a partial EMT phenotype as well as experimental conditions where it may occur. Overall, we envision our approach based on morphology can be widely utilized by researchers with different antibodies and instrumentation, as well as complementing absolute measurements of molecular biomarkers.
More generally, improved analyses for single cell heterogeneity will help to interpret preclinical drug testing and predict patient therapies. Currently, the use of large panels of cell lines to systematically identify biomarkers of drug sensitivity is performed at the population level.47 Nevertheless, sublethal dosing can actually exacerbate drug resistance, perhaps through EMT.8 The use of HCS to measure single cell and subpopulation behaviors may yield new insights into the emergence of heterogeneity and the efficacy of combinatorial or sequential therapeutic regimens. A complementary approach is to reconstruct phenotypic heterogeneity and plasticity by lineage tracing of single cells over extended periods.48 Live cell imaging using fluorescent proteins and reporters could more sensitively resolve the kinetics of EMT, as well as the associated consequences for migration and proliferation.49 Finally, qualitative analysis of cell morphology is already a standard practice to interpret tumor histology. Cells with elongated morphology are frequently observed in malignant tumor sections from human patients, although they cannot be unambiguously distinguished from stromal cells with mesenchymal biomarkers (e.g. fibroblasts).50 Understanding the extraordinary intratumoral diversity that occurs in vivo due to genomic instability and the aberrant tumor microenvironment represents a fundamentally important challenge.51 Future work will examine the applicability of our classifiers to analyze live cell imaging, 3D culture as well as tissue histology.
In conclusion, we have demonstrated a probabilistic classification scheme for EMT based on single cell morphology and biomarkers. This approach was validated with controlled Snail induction and identified distinct epithelial and mesenchymal phenotypes associated with dramatic elongation and overexpression of vimentin, as well as some elongation of the nuclei. This analysis was applied to longitudinal measurements of Snail induction, showing that it was relatively rapid and switch-like. In contrast, TGF-β1 induction kinetics were more variable at the single cell level and occurred more gradually. Even without exogenous stimuli, EMT was observed at low cell densities. Finally, sublethal treatment with the chemotherapeutic Taxol exacerbated EMT for both uninduced and induced Snail populations, resulting in highly elongated vimentin and aberrant nuclear morphologies. Overall, these single cell approaches offer unique biological insights into phenotypic heterogeneity and plasticity that would be overlooked in higher-throughput assays based on population averages and endpoints.
Supplementary Material
Insight, innovation, integration.
The epithelial-mesenchymal transition (EMT) drives phenotypic changes in cancer cell morphology, drug resistance, and invasiveness. However, this heterogeneity and plasticity is challenging to quantify using conventional bulk assays. Here, we use high content imaging to profile single cell morphologies as they undergo EMT. We develop a probabilistic morphological signature that accurately classifies heterogeneous populations into distinct epithelial or mesenchymal phenotypes. We apply this approach to quantitatively reconstruct EMT kinetics after treatment with growth factors, sublethal chemotherapeutics, and varying cell density. We anticipate this single cell approach will reveal new insights into tumor heterogeneity and drug resistance in preclinical assays for drug screening and precision medicine.
Acknowledgments
We thank D.A. Haber (Massachusetts General Hospital) for the gift of cell lines, M. Gamboa Castro and A.B. Kane (Brown University) for careful reading of the manuscript. This research was supported by the National Institutes of Health through the T32 Training Grant in Environmental Pathology (5T32ES007272-24), the COBRE Center for Cancer Research Development at Rhode Island Hospital (1P30GM110759-01A1), a Rhode Island Foundation Medical Research Grant, Jason and Donna McGraw Weiss ‘89 as well as Brown University (Karen T. Romer Undergraduate Teaching and Research Awards, BioMed DEANS Award, OVPR Research Seed Award and Faculty Start-Up Funds).
References
- 1.Thiery JP, Acloque H, Huang RYJ, Nieto MA. Cell. 2009;139:871–890. doi: 10.1016/j.cell.2009.11.007. [DOI] [PubMed] [Google Scholar]
- 2.Kalluri R, Weinberg RA. J Clin Invest. 2009;119:1420–1428. doi: 10.1172/JCI39104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Christofori G. Nature. 2006;441:444–450. doi: 10.1038/nature04872. [DOI] [PubMed] [Google Scholar]
- 4.Singh A, Settleman J. Oncogene. 2010;29:4741–4751. doi: 10.1038/onc.2010.215. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Peinado H, Olmeda D, Cano A. Nat Rev Cancer. 2007;7:415–428. doi: 10.1038/nrc2131. [DOI] [PubMed] [Google Scholar]
- 6.Blanco MJ, Moreno-Bueno G, Sarrio D, Locascio A, Cano A, Palacios J, Nieto MA. Oncogene. 2002;21:3241–3246. doi: 10.1038/sj.onc.1205416. [DOI] [PubMed] [Google Scholar]
- 7.Massagué J. Nat Rev Mol Cell Biol. 2012;13:616–630. doi: 10.1038/nrm3434. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Holohan C, Van Schaeybroeck S, Longley DB, Johnston PG. Nat Rev Cancer. 2013;13:714–726. doi: 10.1038/nrc3599. [DOI] [PubMed] [Google Scholar]
- 9.Gonzalez DM, Medici D. Sci Signal. 2014;7:re8–re8. doi: 10.1126/scisignal.2005189. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Altschuler SJ, Wu LF. Cell. 2010;141:559–563. doi: 10.1016/j.cell.2010.04.033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Kolitz SE, Lauffenburger DA. Biochemistry. 2012;51:7433–7443. doi: 10.1021/bi300846p. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Yang R, Niepel M, Mitchison TK, Sorger PK. Clin Pharmacol Ther. 2010;88:34–38. doi: 10.1038/clpt.2010.96. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Zanella F, Lorens JB, Link W. Trends in Biotechnology. 2010;28:237–245. doi: 10.1016/j.tibtech.2010.02.005. [DOI] [PubMed] [Google Scholar]
- 14.Danuser G. Cell. 2011;147:973–978. doi: 10.1016/j.cell.2011.11.001. [DOI] [PubMed] [Google Scholar]
- 15.Perlman ZE, Slack MD, Feng Y, Mitchison TJ, Wu LF, Altschuler SJ. Science. 2004;306:1194–1198. doi: 10.1126/science.1100709. [DOI] [PubMed] [Google Scholar]
- 16.Slack MD, Martinez ED, Wu LF, Altschuler SJ. Proc Natl Acad Sci USA. 2008;105:19306–19311. doi: 10.1073/pnas.0807038105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Cohen AA, Geva-Zatorsky N, Eden E, Frenkel-Morgenstern M, Issaeva I, Sigal A, Milo R, Cohen-Saidon C, Liron Y, Kam Z, Cohen L, Danon T, Perzov N, Alon U. Science. 2008;322:1511–1516. doi: 10.1126/science.1160165. [DOI] [PubMed] [Google Scholar]
- 18.Gascoigne KE, Taylor SS. Cancer Cell. 2008;14:111–122. doi: 10.1016/j.ccr.2008.07.002. [DOI] [PubMed] [Google Scholar]
- 19.Spencer SL, Gaudet S, Albeck JG, Burke JM, Sorger PK. Nature. 2009;459:428–432. doi: 10.1038/nature08012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Jones TR, Carpenter AE, Lamprecht MR, Moffat J, Silver SJ, Grenier JK, Castoreno AB, Eggert US, Root DE, Golland P, Sabatini DM. Proc Natl Acad Sci USA. 2009;106:1826–1831. doi: 10.1073/pnas.0808843106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Paszek P, Ryan S, Ashall L, Sillitoe K, Harper CV, Spiller DG, Rand DA, White MRH. Proc Natl Acad Sci USA. 2010;107:11644–11649. doi: 10.1073/pnas.0913798107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Yin Z, Sadok A, Sailem H, McCarthy A, Xia X, Li F, Garcia MA, vans LE, Barr AR, Perrimon N, Marshall CJ, Wong STC, Bakal C. Nat Cell Biol. 2013 doi: 10.1038/ncb2764. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Sero JE, Sailem HZ, Ardy RC, Almuttaqi H, Zhang T, Bakal C. Molecular Systems Biology. 2015;11:790–790. doi: 10.15252/msb.20145644. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Quintavalle M, Elia L, Price JH, Heynen-Genel S, Courtneidge SA. Sci Signal. 2011;4:ra49–ra49. doi: 10.1126/scisignal.2002032. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Steininger RJ, Rajaram S, Girard L, Minna JD, Wu LF, Altschuler SJ. Cytometry A. 2015;87:558–567. doi: 10.1002/cyto.a.22599. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Low J, Huang S, Blosser W, Dowless M, Burch J, Neubauer B, Stancato L. Molecular Cancer Therapeutics. 2008;7:2455–2463. doi: 10.1158/1535-7163.MCT-08-0328. [DOI] [PubMed] [Google Scholar]
- 27.Caie PD, Walls RE, Ingleston-Orme A, Daya S, Houslay T, Eagle R, Roberts ME, Carragher NO. Molecular Cancer Therapeutics. 2010;9:1913–1926. doi: 10.1158/1535-7163.MCT-09-1148. [DOI] [PubMed] [Google Scholar]
- 28.Gough AH, Chen N, Shun TY, Lezon TR, Boltz RC, Reese CE, Wagner J, Vernetti LA, Grandis JR, Lee AV, Stern AM, Schurdak ME, Taylor DL. PLOS ONE. 2014;9:e102678–102616. doi: 10.1371/journal.pone.0102678. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Gupta PB, Fillmore CM, Jiang G, Shapira SD, Tao K, Kuperwasser C, Lander ES. Cell. 2011;146:633–644. doi: 10.1016/j.cell.2011.07.026. [DOI] [PubMed] [Google Scholar]
- 30.Sharma SV, Lee DY, Li B, Quinlan MP, Takahashi F, Maheswaran S, McDermott U, Azizian N, Zou L, Fischbach MA, Wong KK, Brandstetter K, Wittner B, Ramaswamy S, Classon M, Settleman J. Cell. 2010;141:69–80. doi: 10.1016/j.cell.2010.02.027. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Hafner M, Niepel M, Chung M, Sorger PK. Nat Meth. 2016;13:521–527. doi: 10.1038/nmeth.3853. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Sarrió D, Rodriguez-Pinilla SM, Hardisson D, Cano A, Moreno-Bueno G, Palacios J. Cancer Res. 2008;68:989–997. doi: 10.1158/0008-5472.CAN-07-2017. [DOI] [PubMed] [Google Scholar]
- 33.Cichon MA, Nelson CM, Radisky DC, Radisky D. Cancer Inform. 2015;14:1–13. doi: 10.4137/CIN.S18965. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Gilles C, Polette M, Zahm JM, Tournier JM, Volders L, Foidart JM, Birembaut P. J Cell Sci. 1999;112(Pt 24):4615–4625. doi: 10.1242/jcs.112.24.4615. [DOI] [PubMed] [Google Scholar]
- 35.Javaid S, Zhang J, Anderssen E, Black JC, Wittner BS, Tajima K, Ting DT, Smolen GA, Zubrowski M, Desai R, Maheswaran S, Ramaswamy S, Whetstine JR, Haber DA. Cell Rep. 2013;5:1679–1689. doi: 10.1016/j.celrep.2013.11.034. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Zhou BP, Deng J, Xia W, Xu J, Li YM, Gunduz M, Hung MC. Nat Cell Biol. 2004;6:931–940. doi: 10.1038/ncb1173. [DOI] [PubMed] [Google Scholar]
- 37.Debnath J, Muthuswamy SK, Brugge JS. Methods. 2003;30:256–268. doi: 10.1016/s1046-2023(03)00032-x. [DOI] [PubMed] [Google Scholar]
- 38.Kamentsky L, Jones TR, Fraser A, Bray MA, Logan DJ, Madden KL, Ljosa V, Rueden C, Eliceiri KW, Carpenter AE. Bioinformatics. 2011;27:1179–1180. doi: 10.1093/bioinformatics/btr095. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Dempster AP, Laird NM, Rubin DB. JR Stat Soc. 1977;39:1–38. [Google Scholar]
- 40.Bradbury A, Plückthun A. Nature. 2015;518:27–29. doi: 10.1038/518027a. [DOI] [PubMed] [Google Scholar]
- 41.Coffman VC, Wu JQ. Trends Biochem Sci. 2012;37:499–506. doi: 10.1016/j.tibs.2012.08.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Waters JC, Wittmann T. In: Methods in Cell Biology. Jennifer CW, Torsten W, editors. Vol. 123. Academic Press; 2014. pp. 1–18. [DOI] [PubMed] [Google Scholar]
- 43.Zhang J, Tian XJ, Zhang H, Teng Y, Li R, Bai F, Elankumaran S, Xing J. Sci Signal. 2014;7:ra91–ra91. doi: 10.1126/scisignal.2005304. [DOI] [PubMed] [Google Scholar]
- 44.Kajiyama H, Shibata K, Terauchi M, Yamashita M, Ino K, Nawa A, Kikkawa F. Int J Oncol. 2007;31:277–283. [PubMed] [Google Scholar]
- 45.Park SY, Kim MJ, Park SA, Kim JS, Min KN, Kim DK, Lim W, Nam JS, Sheen YY. Oncotarget. 2015;6(35) doi: 10.18632/oncotarget.6063. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Yu M, Bardia A, Wittner BS, Stott SL, Smas ME, Ting DT, Isakoff SJ, Ciciliano JC, Wells MN, Shah AM, Concannon KF, Donaldson MC, Sequist LV, Brachtel E, Sgroi D, Baselga J, Ramaswamy S, Toner M, Haber DA, Maheswaran S. Science. 2013;339:580–584. doi: 10.1126/science.1228522. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Sharma SV, Haber DA, Settleman J. Nat Rev Cancer. 2010;10:241–253. doi: 10.1038/nrc2820. [DOI] [PubMed] [Google Scholar]
- 48.Alcolea MP, Jones PH. Nat Rev Cancer. 2013;13:161–171. doi: 10.1038/nrc3460. [DOI] [PubMed] [Google Scholar]
- 49.Wong IY, Javaid S, Wong EA, Perk S, Haber DA, Toner M, Irimia D. Nature Mat. 2014;13:1063–1071. doi: 10.1038/nmat4062. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Bill R, Christofori G. FEBS Lett. 2015;589:1577–1587. doi: 10.1016/j.febslet.2015.05.002. [DOI] [PubMed] [Google Scholar]
- 51.Almendro V, Marusyk A, Polyak K. Annu Rev Pathol Mech Dis. 2013;8:277–302. doi: 10.1146/annurev-pathol-020712-163923. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.