Abstract
The transcriptional transactivator Pax6 binds the pancreatic islet cell-specific enhancer sequence (PISCES) of the rat insulin I gene. However the human, mouse, and rat insulin gene II promoters do not contain a PISCES element. To analyze the role of Pax6 in those PISCES-less promoters, we investigated its influence on rat insulin gene II expression and included in our studies the main activators: pancreatic and duodenal homeobox protein-1 (PDX-1) and BETA2/E47. Luciferase assays, Northern blots, and RIA were used to study effects of Pax6 overexpression, gel shift and chromatin precipitation assays to study its binding to the DNA, and yeast two-hybrid assays and glutathione S transferase capture assays to investigate its interactions with PDX-1 and BETA2. Finally, glucose-dependent intracellular transport of Pax6 was demonstrated by fluorescence microscopy. Overexpression of Pax6 prevents activation of the rat insulin II gene by BETA2 and PDX-1 and hence suppresses insulin synthesis and secretion. In vitro, Pax6 binds to the A-boxes, thereby blocking binding of PDX-1, and at the same time, its paired domain interacts with BETA2. Fluorescence microscopy demonstrated that the nuclear-cytoplasmic localization of Pax6 and PDX-1 are oppositely regulated by glucose. From the results, it is suggested that at low concentrations of glucose, Pax6 is localized in the nucleus and prevents the activation of the insulin gene by occupying the PDX-1 binding site and by interacting with BETA2.
The insulin gene promoter is repressed by the transcription factor Pax6, which binds to A-box DNA elements competing with Pdx1 and interacting with BETA2.
A unique combination of nuclear regulatory proteins acts in concert to control β-cell-specific expression of the insulin gene. The most crucial sequence elements within the insulin promoter are the A- and E-boxes, which are highly conserved in the human, mouse, and rat insulin genes (1) (see Fig. 1A). The E-boxes, containing the consensus motif CANNTG, are binding sites for the insulin enhancer factor 1, a protein complex composed of the ubiquitous basic helix-loop-helix protein E12/E47 and an islet cell-specific factor BETA2 (2, 3, 4, 5, 6, 7). The A-elements, harboring a TAAT motif, are able to bind members of the homeodomain class of transcription factors like pancreatic and duodenal homeobox protein-1 (PDX-1). PDX-1 is expressed preferentially but not exclusively in β-cells (8). It plays a major role in the maintenance of the β-cell phenotype and in modulating insulin gene transcription in response to glucose. In conjunction with several other transcription factors, like the rat insulin promoter element 3b (RIPE3b) binding factor musculoaponeurotic fibrosarcoma A (MafA) (9), binding of PDX-1 to the key A-boxes leads to insulin gene activation (5, 10, 11, 12, 13, 14). Synergistic activation of insulin promoter activity is induced by simultaneous binding of PDX-1, insulin enhancer factor 1, and MafA (3, 15, 16).
Fig. 1.
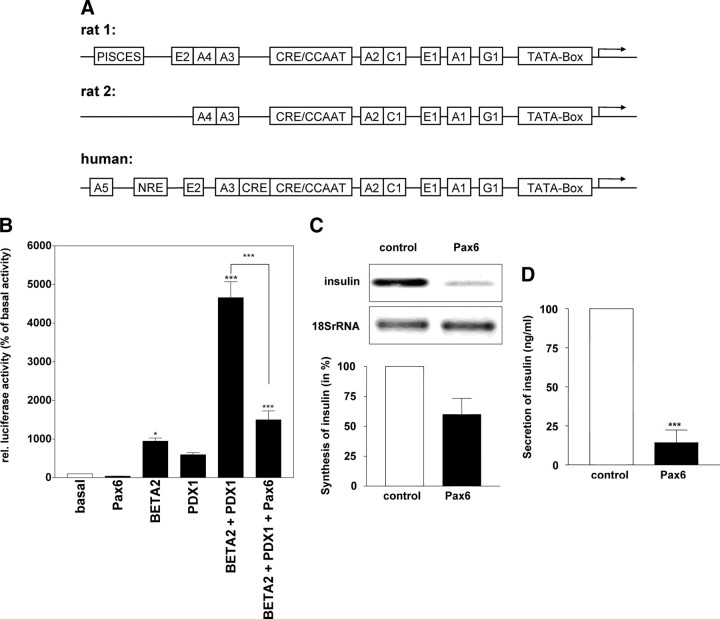
Pax6 acts as a repressor of insulin synthesis and secretion. A, Schematic overview of the promoters from the rat insulin genes 1 and 2 and the human insulin gene. B, HepG2 cells were transiently transfected with 23 ng pBAT-Pax6, 2 μg pCDM8-BETA2, and 0.6 μg pCDM8-PDX-1 expression vectors in the indicated combinations using calcium phosphate precipitation to investigate their effect on the RINSII promoter −451 to +127 (pGL3-RINSII) using the Dual-Luciferase Reporter Assay System. Results are presented as means ± sem of three independent experiments performed in triplicate. The values are expressed as percentage of the basal activity obtained from cells with pGL3-RINSII alone. C, Using Lipofectamine 2000, MIN6 cells were transiently transfected with 4 μg pCDM8-Pax6 or 4 μg Tet repressor (control). After 48 h, the RNA was isolated and analyzed using Northern blots with DIG-labeled insulin RNA probes. D,The medium (500 μl) of transfected MIN6 cells was used to analyze the basal insulin secretion by RIA. Statistical analyses were carried out using one-way ANOVA (B) or Student’s unpaired t test (C and D). Differences are considered statistically significant at P < 0.05: *, P < 0.05; ***, P < 0.0001.
Pax6 is a critical transcription factor in the development of the eye, the central nervous system, and the pancreas. In the islets of Langerhans, Pax6 was found to be expressed in all of the four distinct cell phenotypes (17). This transcription factor contains two DNA-binding domains (BD), the paired box and the homeodomain. It has been demonstrated that each motif can function independently or together to regulate different genes (18, 19). Regulation of downstream targets by Pax6 likely involves different interactions with multiple proteins, resulting in the generation of different effects in different cells. In mature mouse or rat islet cells, Pax6 activates gene expression by binding with its paired box at the pancreatic islet cell-specific enhancer sequence (PISCES) element, present within the glucagon, somatostatin, and rat insulin I promoters (20, 21, 22). In the rat insulin gene II promoter, as well as in mouse and human insulin gene promoters, the sequence of the homologous region differs in one or two bases. This prevents the binding of Pax6, as we observed in gel-shift analyses (data not shown). Thus, there must be another mechanism by which Pax6 may be involved in the regulation of the insulin gene expression. It has been reported that the homeobox of Pax6 is able to bind to AT-rich motifs (23, 24), which are to be found within the A-boxes of the insulin gene promoter and within the G1-element of the glucagon promoter. This suggests a competition between PDX-1 and Pax6, which has been demonstrated for the glucagon gene but neglected so far with respect to the insulin gene (24).
In the present studies, we investigated the involvement of Pax6 in the regulation of the expression of the rat insulin gene II. In addition, we analyzed interactions of Pax6 with the most relevant promoter elements and transcription factors using gel-shift analyses, chromatin immunoprecipitations, the yeast two-hybrid system, and glutathione S transferase (GST) capture assays. Finally, we studied the intracellular localization of Pax6 using fluorescence microscopy.
Results
Pax6 acts as a repressor of the insulin gene II expression
To study the effects of Pax6 on the expression of the rat insulin gene II, we used the Dual-Luciferase Reporter Assay System. Rat hepatoma HepG2 cells were transfected with the firefly-luciferase gene under control of the RINSII promoter (−451 to +127), and the transcription factors PDX-1, BETA2, and Pax6 were overexpressed in several combinations using optimal amounts of expression plasmids determined for each separately (Fig. 1A). Overexpression of Pax6 alone did not activate the RINSII promoter, but an approximately 10-fold activation by BETA2 alone and an approximately 5- to 8-fold activation by PDX-1 alone have been observed. Coexpression of PDX-1 and BETA2 resulted in an approximately 50-fold activation of the rat insulin II promoter. This induction of promoter activity was suppressed highly significantly by overexpression of Pax6. To confirm the results of the luciferase assays, we transfected MIN6 cells for overexpression of Pax6 or of reverse Tet-responsive transactivator as a negative control. Northern blots (Fig. 1B) and RIAs (Fig. 1C) demonstrated repressed synthesis and secretion of insulin by overexpressed Pax6.
Pax6 binds to A-boxes within the rat insulin gene II promoter
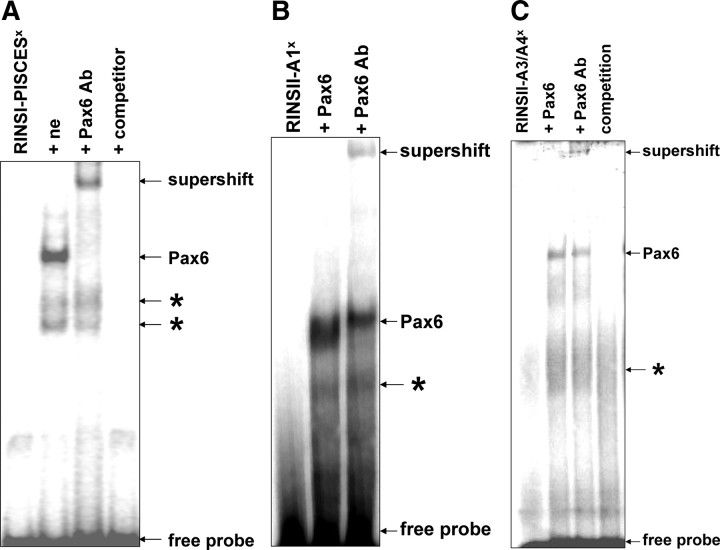
The effects of Pax6 on rat insulin gene II promoter indicate that there might be a Pax6 binding site different from PISCES in the promoter sequence. First we examined in an EMSA with a 32P-labeled oligonucleotide containing the PISCES element of the rat insulin gene I promoter whether Pax6 contained in nuclear extracts from INS-1 cells specifically binds to this oligonucleotide. We observed a complex that could be supershifted by the use of an antibody directed against Pax6 (Fig. 2A). Additionally, EMSA using radiolabeled RINSII-A1 (Fig. 2B) or RINSII-A3/A4 (Fig. 2C) oligonucleotides, and in vitro-translated Pax6 confirmed the binding of Pax6 to these A-boxes.
Fig. 2.
Pax6 binds to the A1-box of the rat insulin II promoter. A, For EMSA, nuclear extracts (ne) obtained from INS-1 cells were incubated with the double-stranded radiolabeled (superscript x) oligonucleotide RINSI-PISCES. The DNA-Pax6 complex (lane 2) was supershifted by a monoclonal antibody directed against Pax6 (lane 3). As a competitor, a 100-fold excess of unlabeled RINSI-PISCES was used (lane 4). B, Double-stranded radiolabeled oligonucleotide RINSII-A1 or C, RINSII-A3/A4 was incubated with 4 μl in vitro translation product Pax6. The DNA-Pax6 complex (lane 2) was supershifted by a monoclonal antibody directed against Pax6 (lane 3). Asterisks indicate unspecific signals.
Pax6 is able to replace PDX-1 from the RINSII-A3-box
Because the A-boxes are also binding sites for PDX-1, we investigated the competitive binding behavior of PDX-1 and Pax6. In the EMSA with a RINSII-A3/A4 oligonucleotide and INS1 nuclear extracts from cells grown at 11 mmol/liter glucose, the DNA-protein complex corresponding to PDX-1 was competitively replaced by increasing amounts of in vitro-translated Pax6 (Fig. 3A). The in vitro translation mixture itself was without effect on this EMSA (data not shown). Chromatin immunoprecipitations confirmed the interaction of both transcription factors with the RINSII-A3/A4-box in living cells. When cells were grown in the presence of glucose, PDX-1 has been found to bind strongly to the A3/A4 element, whereas in contrast, Pax6 seems to interact only weakly. However, using nuclear extracts from cells grown without glucose, binding of PDX-1 was not detectable, whereas Pax6 has been found to bind much more strongly under such conditions (Fig. 3B). Statistical analyses show that the differential binding was maximal when cells grown in 17 mmol/liter glucose were compared with those grown without glucose.
Fig. 3.
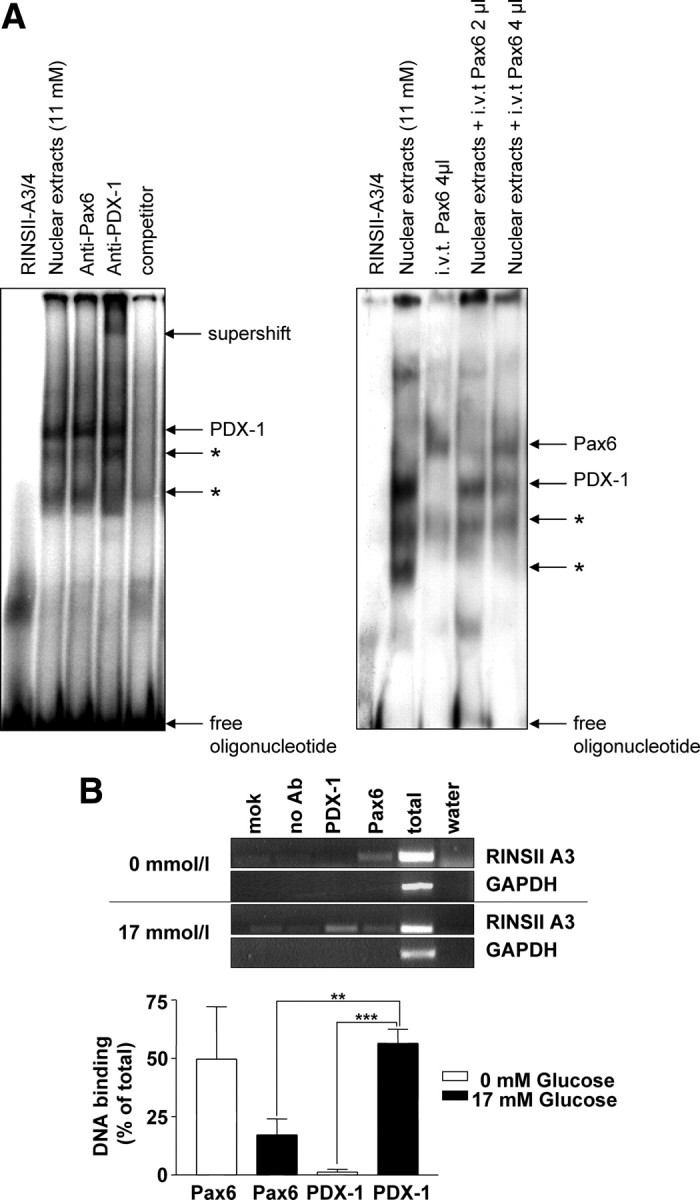
PDX-1 is displaced from the A3/A4-box by increasing amounts of Pax6. A, For EMSA, nuclear extracts from INS-1 cells containing endogenous PDX-1 and the indicated amounts of in vitro-translated (i.v.t) Pax6 were incubated with the double-stranded radiolabeled oligonucleotide RINSII-A3/A4. B, Chromatin from INS-1 cells cultivated for 15 h at 0 or 17 mmol/liter glucose was used for immunoprecipitation with polyclonal antibodies directed against Pax6 or PDX-1. Precipitated and purified DNA was subjected to PCR using the primer A3for together with A3rev and GAPDHfor together with GAPDHrev. As a positive control, chromatin before immunoprecipitation (total) was used. Negative controls were samples without chromatin [mock (mok)], without antibody (no Ab), and for PCR with water instead of samples (water). A quantitative analysis from four independent chromatin immunoprecipitations is shown in the diagram. Results are presented as means ± sem. Statistical analysis was carried out using Student’s unpaired t test. Differences are considered statistically significant at P < 0.05: *, P < 0.05; **, P < 0.005; ***, P < 0.0001.
Pax6 interacts with BETA2
By recognizing A-boxes, Pax6 would bind close to the adjacent E-box targets of BETA2, which might permit a direct interaction of both transcription factors. We demonstrated the occurrence of a Pax6-BETA2 interaction using the yeast two-hybrid system (Fig. 4A). This interaction was quantified in a liquid assay using o-nitrophenyl-β-D-galactopyranoside (ONPG) as substrate. Figure 4B shows for BETA2 and Pax6 a result of nearly 3% of Gal4 activity in comparison with about 5% measured for the well-proven interaction of p53 and the large T antigen of simian virus 40 (SV40). In controls, BETA2 and Pax6 alone did significantly lower activate reporter gene expression.
Fig. 4.
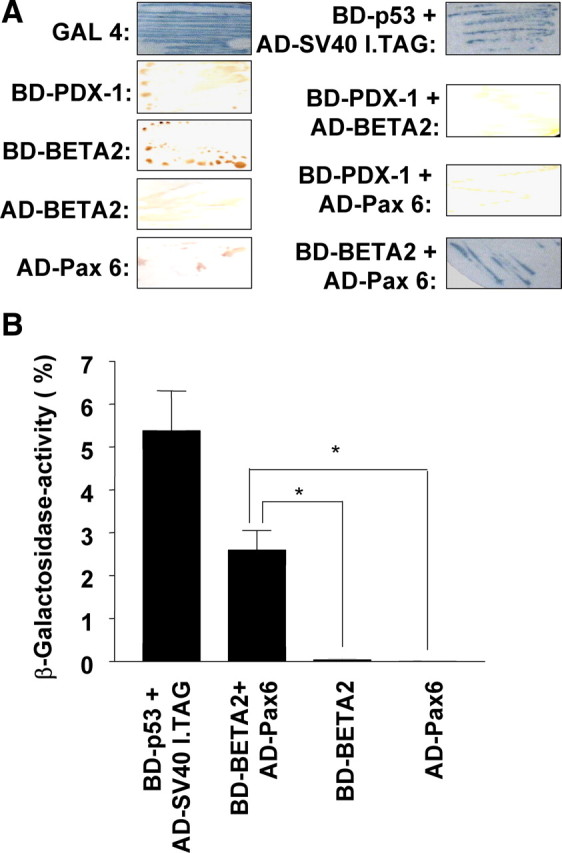
Pax6 interacts in living cells with BETA2. A, In the two-hybrid system, yeast cells (Y187), transformed with 0.1 μg plasmid, expressing the indicated proteins, fused with Gal4 BD or Gal4 AD, were treated with X-Gal. Development of blue color indicates an interaction. The complete GAL4 transcription factor as well as BD-p53 combined with AD-SV40 large T antigen protein was used as a positive control. Autonomous activation was excluded by using yeast cells expressing the hybrid proteins separately. B, The detected interaction was quantified by a liquid culture assay using ONPG as substrate. The galactosidase units are presented as percentage of β-galactosidase activity in yeast cells expressing the complete Gal4 transcription factor. Statistical analyses of three independent experiments performed in triplicate were done using Student’s unpaired t test. Differences are considered statistically significant at P < 0.05. *, P < 0.05.
To determine the domain of Pax6 that interacts with BETA2, we studied wild-type and mutant proteins in GST capture assays. Figure 5A shows that in vitro-translated 35S-labeled BETA2 interacts with GST-Pax6 but not with GST alone. The strongest signal was detected using a truncated GST-Pax6 fragment containing only the paired box (Fig. 5B). The homeobox-containing C-terminal GST-Pax6 fragment does not bind to BETA2. These results were confirmed using 35S-labeled in vitro-translated Pax6 or both truncated domains and GST-BETA2 (Fig. 5, C–E). The GST pull-down experiment in Fig. 5F demonstrates the interaction of GST-BETA2 with endogenous Pax6 from RINm5F cells.
Fig. 5.
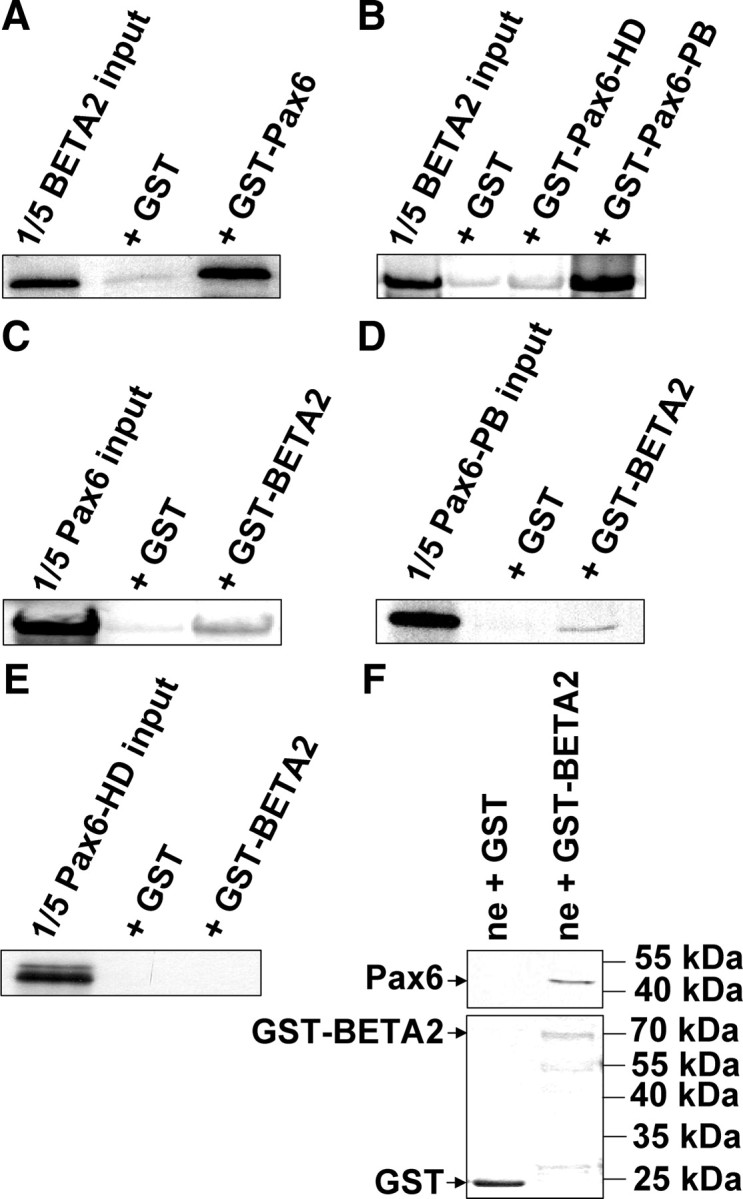
BETA2 interacts in vitro with the paired box of Pax6. A–E, The radiolabeled in vitro translation products BETA2 (A and B), Pax6 (C), Pax6 paired box (D), and Pax6 homeodomain protein (E) were incubated with purified GST or the indicated GST fusion proteins bound to glutathione-Sepharose beads followed by four washing steps. Recovered bound radiolabeled protein was separated by SDS-PAGE and detected by autoradiography. Each first lane represents 20% of the amount of the radiolabeled protein added to each sample. F, GST or GST-BETA2 fusion protein bound to glutathione-Sepharose beads was incubated with nuclear extracts (ne) from RINm5F cells followed by four washing steps. Recovered bound protein was separated by SDS-PAGE and detected by Western blot analyses using rabbit anti-Pax6 IgG and horseradish peroxidase-linked antirabbit IgG. For control, the used amounts of GST and GST-BETA2 were presented in a Coomassie-stained SDS gel.
Pax6 is colocalized with BETA2 in the nucleus mainly at low concentrations of glucose
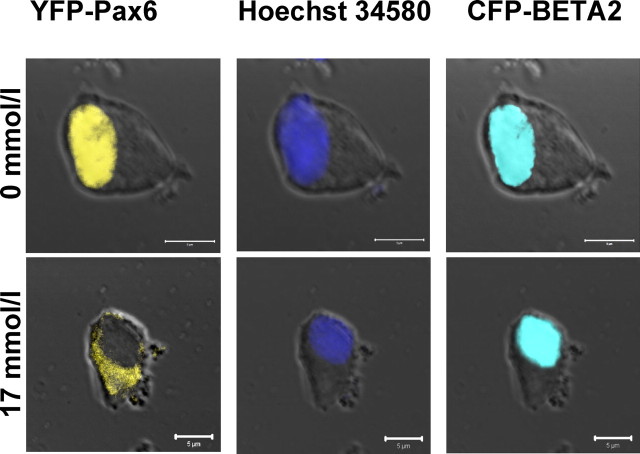
Interaction of Pax6 and BETA2 presupposes that both proteins are present in the same cellular compartment. By confocal microscopy, we found the overexpressed yellow fluorescent fusion protein (YFP)-Pax6 predominantly in the cytoplasm of transfected RINm5F cells at 17 mmol/liter glucose. However, if the cells were starved, Pax6 was detectable in the nucleus. pECFP-BETA2, showing cyan fluorescence, was cotransfected to demonstrate that the nuclear localization of this transcription factor is not influenced by different concentrations of glucose (Fig. 6).
Fig. 6.
Intracellular localization of Pax6 depends on glucose. RINm5F cells coexpressing yellow fluorescent YFP-Pax6 and cyan fluorescent CFP-BETA2 were cultured 15 h with the indicated concentrations of glucose (scale bars, 5 μm). Nuclear staining was done with Hoechst 34580. The images represent approximately 1-μm-thick confocal slices of whole cells.
The nucleocytoplasmic transport of Pax6 and PDX-1 is oppositely regulated by glucose
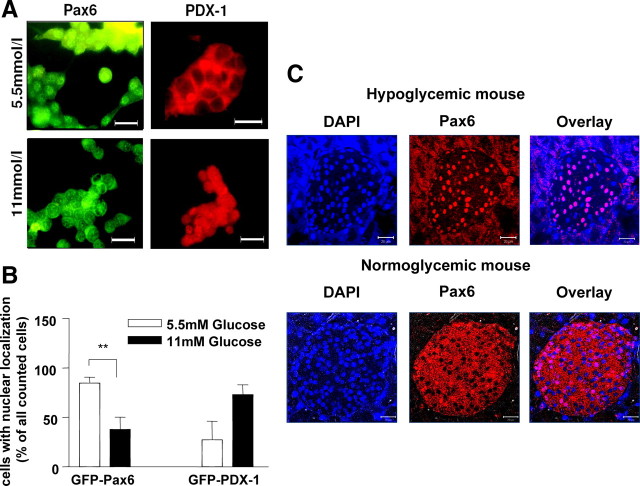
To confirm this differential behavior of Pax6 and PDX-1 in response to glucose, we examined cells grown at different physiological concentrations of glucose by fluorescence microscopy. Detection of Pax6 (green) in INS-1 cells showed a cytoplasmic localization under normal culture conditions (11 mmol/liter glucose). However, at low concentrations of glucose (5.5 mmol/liter), Pax6 was predominantly detectable in the nucleus (Fig. 7A). In contrast to Pax6, the PDX-1 was preferentially localized within the nucleus at high concentrations of glucose. For statistical analyses, we transfected INS-1 cells with pEGFP-Pax6 or pEGFP-PDX-1 and assessed the localization of the expression product in 200 cells for each experiment. Under low glucose conditions, we found significantly more cells with GFP-Pax6 localized in the nucleus in comparison with 11 mmol/liter glucose. GFP-PDX-1 was translocated oppositely (Fig. 7B). A kinetic experiment revealed that GFP-Pax6 is translocated into the cytoplasm within 2 h after addition of glucose to starved INS-1 cells (data not shown). To demonstrate the physiological relevance of glucose-dependent nucleocytoplasmic distribution of Pax6 in vivo, immunohistochemistry on pancreas sections of normo- or hypoglycemic C57BL/6 mice was carried out (Fig. 7C) using the purified polyclonal rabbit anti-Pax6 IgG and an Alexa 555-linked secondary antibody. In the normoglycemic mice, the cytoplasm of islet cells was clearly stained with anti-Pax6, and most of the nuclei were not. Hypoglycemia resulted in nuclear localization of Pax6.
Fig. 7.
The intracellular localization of Pax6 and PDX-1 is regulated by glucose in an opposite manner. A, INS-1 cells cultured with the indicated concentrations of glucose were stained with an antibody directed against Pax6 and antimouse IgG coupled to green fluorescent Cy2 or an antibody directed against PDX-1 and antirabbit IgG coupled to the red fluorescent Cy3 (scale bars, 20 μm). B, INS-1 cells overexpressing green fluorescent Pax6 or PDX-1 were cultured 15 h with the indicated concentrations of glucose, and 200 cells of each experiment were analyzed. Cells with stained nuclei are expressed as percentage of the counted cells. Data are presented as mean ± sem of three independent experiments. Statistical analysis was carried out using Student’s unpaired t test. Differences are considered statistically significant at P < 0.05. **, P < 0.005. C, Paraffin slices of pancreas from normoglycemic or hypoglycemic C57BL/6 mice were used for immunofluorescence studies with polyclonal rabbit anti-Pax6. Nuclei were stained with 4′,6-diamidino-2-phenylindole (DAPI). The images represent approximately 2.4-μm-thick confocal slices of the fluorescent samples (scale bars, 20 μm).
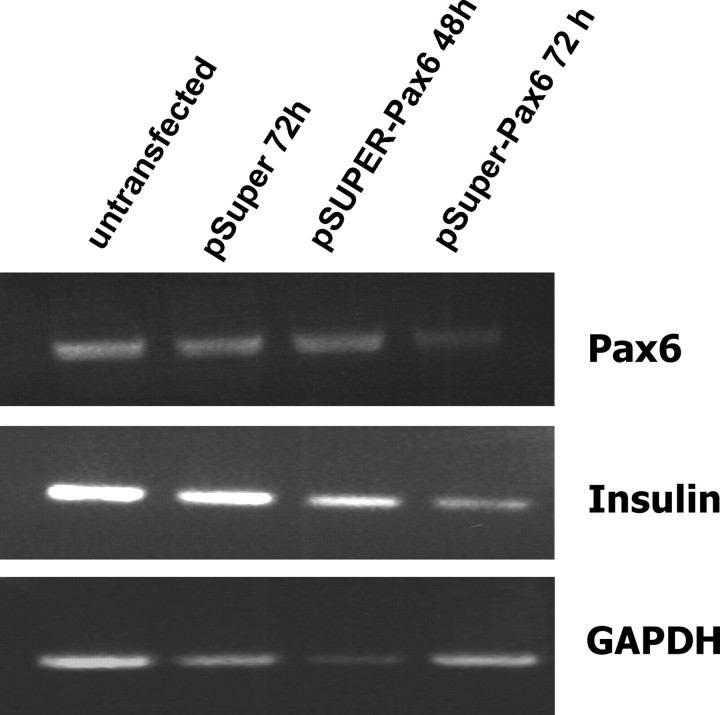
Down-regulation of Pax6 reduces the expression of rat insulin gene I and II
The transient transfection of INS-1 cells with pSUPER-Pax6 resulted in an expression of an interfering RNA (RNAi) against Pax6, and 72 h after transfection, the amount of Pax6 mRNA was reduced. RT-PCR further showed that the expression of rat insulin genes I and II were also reduced after 72 h. The control with the empty pSUPER vector showed no effects.
Discussion
The transcription factors PDX-1, BETA2, and Pax6 have been implicated in the regulation of expression of genes coding for pancreatic peptide hormones. However, all three factors are simultaneously present only in β-cells. In adult animals, PDX-1 expression is largely limited to the insulin- and somatostatin-producing cells of pancreatic islets and cells of the pancreatic ducts and the duodenal epithelium (17, 26, 27, 28). In mature β-cells, PDX-1 binds to A-boxes of the insulin gene promoter and plays a major role in modulating insulin gene transcription in response to glucose (2, 11, 12). BETA2/NeuroD expression is restricted to neuronal and pancreatic α- and β-cells (4, 29). In β-cells, BETA2 binds to E-boxes in the insulin promoter together with E12/47 (2, 4, 6, 30).
It has been shown that simultaneous binding of E12/47 and BETA2 to E-boxes and PDX-1 to the adjacent A-boxes leads to synergistic activation of the insulin promoter (2, 8, 15, 16). A direct interaction between PDX-1 and BETA2 has been observed only in vitro using GST capture assays (8, 14). Although the molecular mechanism underlying that synergistic activation is not well understood, transcriptional coactivators may be involved in the formation of a larger transcriptional activation complex. One candidate may be p300/CBP (cAMP response element binding protein), which is proposed to stimulate insulin gene transcription via direct interaction with E12/47 and BETA2 (7). A second candidate may be Bridge1. This protein coactivates the E12/47-mediated transactivation of the insulin promoter activity (30).
Pax6 is expressed in embryonic and differentiated cells (20) in all four cell types of the islet (18). A mutated Pax6 correlates with glucose intolerance, hyperglycemia, and hypoinsulinemia. In the line of our argumentation, mutations, for example affecting export of Pax6 from the nucleus, could be responsible for these effects. Nevertheless, Pax6 is important for the maintenance of β-cell function and is involved in the regulation of the expression of glucose transporter 2, prohormone convertase 1/3, PDX-1, MafA, and BETA2 (31, 32, 33, 34, 35, 36).
Concerning the rat insulin I gene transcription, it has been reported that Pax6 acts as activator (21), but our data are providing evidence that Pax6 is an inhibitor of insulin gene expression and secretion. The first evidence is the observed highly significant repression of the PDX-1/BETA2-mediated promoter activity in our reporter gene assays, confirmed by Northern blots with cells overexpressing Pax6 (Fig. 1).
As has been reported, Pax6 activates the rat insulin gene I promoter by binding to the PISCES element (21). But neither the rat insulin II nor the human or mouse insulin gene promoter contain such a sequence element (Fig. 1A). However, a binding of Pax6 to the promoters of mouse insulin I and II genes in βTC-3 cells has been reported (37). Beside the PISCES element, Pax6 additionally binds at TAAT motifs (23, 38) as we confirmed for the A-boxes of the rat insulin gene II promoter (Fig. 2). This implies that under starved conditions, increasing amounts of nuclear localized Pax6 could displace PDX-1 from that site, which we have shown as a second indication for the inhibitory action of Pax6 (Fig. 3). Under those conditions, binding of Pax6 may be stabilized by interactions with adjacent factors. Using the yeast two-hybrid system to analyze protein-protein interactions together with the GST capture assays, we demonstrate that BETA2 interacts directly with the paired box of Pax6 (Figs. 4 and 5). Because of different expression levels in bacteria, the amount of GST fusion proteins varied among the GST capture assays (lower intensities seen in Fig. 5, C and D compared with A and B). But within one assay, the amount of GST fusion protein and GST was comparable as has been shown in advance. Moreover, it cannot be excluded that the GST tag on BETA2 could influence the interaction. Beside the demonstrated associations, we could not confirm a direct interaction between PDX-1 and BETA2 or between PDX-1 and Pax6 (Fig. 4). The binding of Pax6 to the A-box and its association with BETA2 may prevent the formation of a transcriptional activation complex at the RINSII promoter. The subcellular localization of Pax6 and PDX-1, which is regulated by glucose concentration in an opposite manner, suggests that Pax6 acts as an inhibitor of insulin gene expression. Pax6 is predominantly localized in the cytoplasm under stimulating glucose conditions and is found in the nucleus, colocalized with BETA2, mainly in starved cells (Figs. 6–8), whereas the transactivator PDX-1 showed the opposite behavior (39, 40, 41) (Fig. 8). The detected localization of Pax6 in the cytoplasm of islet cells in the pancreas of normoglycemic mice is in contrast to the published data of Zhou et al. (42), although they used the same polyclonal rabbit anti-Pax6 IgG from Chemicon (Fig. 7C) and C57BL/6 mice. Perhaps the differences are age related because we used adult wild-type animals, whereas they used transgenic embryos. Pax6, as a regulator of development, may be in the nucleus during the embryonic stage for reasons unrelated to insulin regulation. For the chromatin immunoprecipitation assay experiments, we used rigorous conditions to get an unambiguous effect but, as can be seen from the immunofluorescence experiment shown in Fig. 7, the movement of Pax6 from cytoplasm to nucleus and the transfer of PDX-1 in the opposite direction also takes place under physiologically relevant glucose concentrations.
Fig. 8.
Down-regulation of Pax6 reduces the expression of rat insulin gene I and II. RNA from INS-1 cells transfected with pSUPER or pSUPER-Pax6 was prepared 48 or 72 h after transfection and was used for RT-PCR amplifying cDNA of Pax6, insulin, or GAPDH.
Hence we conclude that at low concentrations of glucose, Pax6 is localized within the nucleus and prevents the activation of insulin gene expression by interacting with BETA2 and occupying the PDX-1 binding site. During phases with low levels of glucose, Pax6 inhibits insulin synthesis but may additionally maintain the expression of important factors like glucose transporter 2, glucokinase, PDX-1, BETA2, and MafA necessary to maintain β-cell function. Consistent with this view, diminishing of Pax6 will result in lowered insulin gene expression as observed in response to RNAi-mediated down-regulation of Pax6 (Fig. 8). It is possible that longitudinal hyperglycemia prevents this recovery of regulators and results in glycotoxicity, the same effects that are described for down-regulated Pax6. Our hypothesis is not in conflict with the results of German et al. (15), Sander et al. (5), Yasuda et al. (31), or Ashery-Padan et al. (32) and supports the observation of Olson and colleagues (43, 44, 45, 46) that chronic exposure to high glucose reduces insulin gene transcription as well as synthesis and DNA binding of important activators of insulin gene expression (e.g. PDX-1) in HIT-T15β, β-TC6, and INS-1 cells.
Interestingly, whereas Pax6 acts as a repressor of insulin gene expression in β-cells, it enhances the islet-specific glucose-6-phosphatase catalytic subunit-related protein gene transcription together with PDX-1 in these cells (47). Pax6 is involved in the control of several factors crucial for maintaining the islet phenotype of β- and α-cells by interacting as repressor or activator. Probably, the two protein domains (paired box and homeodomain) of Pax6 that recognize and bind different DNA elements may contribute to the different effects of Pax6. In α-cells, Pax6 enhances the glucagon gene expression. In these cells, Pax6 binds as a monomer to the G3 element (PISCES motif) and as both monomer and heterotrimer with Cdx-2/3 and p300/CBP (cAMP response element binding protein) to the G1 element (TAAT motif) of the glucagon promoter (24, 48, 49). At high concentrations of glucose, the Pax6-dependent activation of the glucagon gene expression is down-regulated by insulin, whereas glucose plays only a minor, if any, role (50, 51). Moreover, it has been demonstrated that ectopic expression of PDX-1 in glucagonoma InR1G9 cells inhibits glucagon gene transcription via Pax6 (24). PDX-1 and Pax6 thus seem to act antagonistically. Insulin and glucagon synthesis are inversely regulated, and our results strongly suggest that the opposite effects of Pax6 on glucagon and insulin gene transcription may contribute to this regulation. Further work and in vivo experiments are needed to confirm our results and to understand the mechanisms by which Pax6 acts as inducer or repressor of transcription.
Materials and Methods
Materials
Purified polyclonal rabbit anti-PDX-1 and anti-Pax6 antibodies were obtained from Chemicon International Inc. (Temecula, CA). Monoclonal mouse anti-Pax6 antibody developed by Atsushi Kawakami was obtained from the Developmental Studies Hybridoma Bank developed under the auspices of the Eunice Kennedy Shriver National Institute of Child Health and Human Development and maintained by the University of Iowa, Department of Biological Sciences (Iowa City, IA). Antiserum from goat directed against PDX-1 was supplied by Christopher V. E. Wright, Vanderbilt University Medical Center (Nashville, TN). Polyclonal goat IgG directed against BETA2 was obtained from Santa Cruz Biotechnology Inc. (Santa Cruz, CA). The secondary antibodies antimouse Cy2, antigoat Cy3, and antirabbit Cy3 were obtained from Dianova (Hamburg, Germany) and Alexa Fluor 555 rabbit antigoat IgG and Alexa Fluor 647 goat antirabbit IgG were from Invitrogen (Eugene, OR). All other chemicals were from commercially available sources. C57BL/6 mice were purchased from the Department of Medical Microbiology of the Greifswald University (Greifswald, Germany).
Cell lines and transfections
Rat hepatoma HepG2 cells and rat insulinoma INS-1 and RINm5F cells were propagated in RPMI-1640 medium containing 11 mmol/liter glucose, supplemented with 10% heat-inactivated fetal bovine serum, 100 U/ml penicillin, and 100 μg/ml streptomycin at 37 C in a humidified (5% CO2, 95% air) atmosphere, and 10 mmol/liter HEPES buffer, 2 mmol/liter l-glutamine, 1 mmol/liter sodium pyruvate, and 50 μmol/liter β-mercaptoethanol were added to the medium for INS-1 cells. Mouse insulinoma MIN6 cells were cultured in DMEM with 25 mmol/liter glucose, 2 mmol/liter l-glutamine, 10% heat-inactivated fetal bovine serum, 5% heat-inactivated horse serum, 100 U/ml penicillin, 100 μg/ml streptomycin, and 0.25 μmol/liter β-mercaptoethanol.
For reporter gene analyses, optimal amounts of expression plasmids generating maximal effects without squelching have been estimated and used. HepG2 cells were transfected by the calcium phosphate-DNA coprecipitation method using 6 μg pGL3-RINSII, 0.2 μg pRL-TK, 23 ng pBAT-Pax6, 0.6 μg pCDM8-PDX-1, and 2 μg pCDM8-BETA2 constructs in combinations as indicated. DNA (375 μl) in 0.25 mol/liter CaCl2 was carefully added to 375 μl 2× HBS [0.28 mol/liter NaCl, 25 mmol/liter HEPES, 1.4 mmol/liter Na2HPO4 (pH 7.16)] and incubated for 30 min at room temperature, and 250 μl was added to 106 cells with 1 ml medium. Eighteen hours after transfection, cells were glycerol shocked for 2 min with 15% glycerol. Forty-eight hours after transfection, the cells were harvested, and luciferase assays were done using the Dual-Luciferase Reporter Assay System (Promega GmbH, Mannheim, Germany) according to the manufacturer’s instructions.
For all other transfections, Lipofectamine 2000 (Invitrogen GmbH, Karlsruhe, Germany) was used according to the manufacturer’s protocol. For fluorescence microscopy, RINm5F cells or INS-1 cells were transfected with 4 μg pEGFP, pECFP, and pEYFP constructs. MIN6 cells (3 × 106) were transfected with 4 μg pCDM8-Pax6 or 4 μg pUHD172-1neo (Clontech, Mountain View, CA) for expression of a reverse Tet-responsive transactivator as a negative control. Forty-eight hours after transfection, 500 μl of the supernatant was used for RIA. INS-1 cells were transfected with 3 μg pSUPER (Oligo Engine, Seattle, WA) or pSUPER-Pax6 to express RNAi against Pax6. For Northern blot analyses or RT-PCR, the RNA was isolated from these cells using TRIzol reagent (Invitrogen, Germany) according to the manufacturer’s protocol.
Constructs
Murine pdx-1 cDNA was obtained from Dr. H. Edlund (Umea, Sweden). Hamster beta2 cDNA was a gift from Dr. M.-J. Tsai (Houston, TX). pBAT-Pax6 was supplied by Dr. W. Knepel (Göttingen, Germany). This construct, containing the complete mouse pax6 cDNA, was used in expression studies and as a template for PCR amplification. The following primers for PCR were synthesized with an adjacent restriction site indicated as (X): Pax6for, 5′-CTAGCTAGAATT(X)ATCCGGAGGCTGCCAACCAGC-3′; Pax6revStop, 5′-CCGATCACT(X)GCATGCTCTCTCCTTCTCTCTTTACTG-3′; Pax6rev, 5′-CCGATCACT(X)CTGTAATCGAGGCCA-3′; Pax6homeodomainfor, 5′-GCCCCGG(X)GAATACCCAGTGTGTCATCAATAA-3′; Pax6pairedboxrev, 5′-GATCTCTC(X)TTATTGATGACACACTGGGTAT-3′; BETA2for, 5′-CCGCTAGCTA(X)ACATGACCAAATCATACAGCGAG-3′; BETA2revStop, 5′-CCGATCACTT(X)CTAATCGTGAAAGATGGCATT-3′; BETA2rev, 5′-CCGATCACTT(X)CGATCGTGAAAGATGGCATTC-3′; PDX-1for, 5′-CCCCTAGCTA(X)CCATGAACAGTGAGGAGCAGTAC-3′; and PDX-1rev, 5′-CCGATCACTT(X)CTCAGACTGCTGTCCTCACCG-3′.
pGL3-RINSII (KpnI, HindIII) contains the firefly luciferase gene under control of the rat insulin II gene promoter (−451/+127) in pGL3 basic (Promega) and was used for the Dual-Luciferase Reporter Assay System (Promega). pdx-1, beta2, and pax6 cDNAs were inserted into pCDM8 (Invitrogen, Germany) leading to the following constructs: pCDM8-PDX-1 (HindIII, XbaI), pCDM8-BETA2 (blunt end on EcoRI site), pCDM8-Pax6-paired-box amino acids 1-123 (HindIII, NotI), and pCDM8-Pax6 homeodomain amino acids 117-422 (HindIII, NotI).
We performed yeast two-hybrid screens using the following constructs of the plasmids pAS2-1, which contains the GAL4 BD and pACT2 with the GAL4 activation domain (AD): BD-PDX-1 (NcoI, BamHI), BD-BETA2 (SfiI, BamHI), AD-BETA2 (NcoI, BamHI), and AD-Pax6 (BamHI). pCL1 (containing the complete Gal4 cDNA), pVA3-1 (BD-murine P53) and pTD1-1 (AD-SV40 large T antigen protein) were used as positive controls. All plasmids are components of the MATCHMAKER two-hybrid system 2 (Clontech Laboratories, Inc., BD Biosciences, Heidelberg, Germany). The two-hybrid screens were done following the manufacturer‘s protocol.
For in vitro translation, pRc/CMV-Pax6 (HindIII, NotI) was constructed by inserting the pax6 cDNA into pRC/CMV (Invitrogen, Germany).
To create GST fusion proteins, pGex-Pax6 (BamHI), pGex-Pax6 paired box amino acids 1-123 (BamHI), pGex-Pax6 homeodomain amino acids 117-422 (BamHI), and pGex-BETA2 (BamHI) were constructed using the pGEX-3X vector (APB Europe GmbH, Freiburg, Germany).
Using the vectors pEYFP-N1, pECFP-N1, pEGFP-N2, and pEGFP-C2 (BD Biosciences), we created the constructs pEYFP-Pax6 (HindIII, BamHI), pEGFP-Pax6 (HindIII, BamHI in pEGFP-N2), pEGFP-PDX-1 (BamHI in pEGFP-C2), and pECFP-BETA2 (SalI, BamHI).
pSUPER-Pax6 contains a RNAi oligonucleotide against the transactivation domain of Pax6 (TADfor, 5′-GAT CCC CAA TAA CCT GCC TAT GCA ACC CTT CAA GAG AGG GTT GCA TAG GCA GGT TAT TTT TTT-3′; TADrev, 5′-AGC TTA AAA AAA TAA CCT GCC TAT GCA ACC CTC TCT TGA AGG GTT GCA TAG GCA GGT TAT TGG G-3′) inserted at HindIII/BglII restriction sites.
Immunohistochemistry
INS-1 cells grown on coverslips were fixed and incubated with mouse anti-Pax6 or rabbit anti-PDX-1 antibody (dilution 1:200). The primary antibodies were visualized with green fluorescent Cy2-conjugated goat antimouse IgG (1:400) or red fluorescent Cy3-conjugated goat antirabbit IgG (1:400). Images were acquired with an Olympus IX-70 microscope (Olympus Deutschland GmbH, Hamburg, Germany).
For confocal microscopy, RINm5F cells grown on glass coverslips were transfected with pEYFP-Pax6 and pECFP-BETA2. The cells were stained with Hoechst 34580, fixed using paraformaldehyde, and covered with Mowiol.
Paraffin slices of pancreas from C57/BL6 mice were successively incubated for 5 min in xylol, 95% (vol/vol) and 75% (vol/vol) ethanol, aqua bidest and 30 min in immune fluorescence (IF) buffer [0.2% (wt/vol) BSA, 0.05% (wt/vol) saponin, 0.1% (wt/vol) sodium azide in PBS (pH 7.4)] followed by an overnight incubation at 4 C with polyclonal rabbit anti-Pax6, diluted 1000-fold in IF buffer. The immunoreacted primary antibody was visualized with Alexa Fluor 555 rabbit antigoat IgG diluted 200-fold in IF buffer, by incubation for 2 h at room temperature in the dark. Nuclei were stained with 42 μg/ml 4′,6-diamidino-2-phenylindole (Invitrogen) in IF buffer for 1 min. Slices were covered with DakoCytomation fluorescent mounting medium (DakoCytomation, Hamburg, Germany).
Laser scanning confocal microscopy was carried out using a Zeiss LSM 510 confocal microscope (Zeiss, Jena, Germany) fitted with a ×40 1.2 NA Zeiss oil immersion objective or a ×63 1.2 NA Zeiss water immersion objective. Images were evaluated using Zeiss Image Browser software (Zeiss, Jena, Germany).
Northern blots
The RNA was isolated from transfected MIN6 cells using TRIzol reagent (Invitrogen) according to the manufacturer’s protocol. Two micrograms of RNA were used for Northern blot analyses. To produce a digoxigenin (DIG)-labeled RNA probe directed against insulin mRNA, a 333-bp PCR product created with the primers Insfor 5′-CAACATGGCCCTGTGGATGC-3′ and Insrev 5′-AGTTGCAGTAGTTCTCCAGC-3′ was inserted into pGEM-T-Easy (Promega). The linearized construct was used for in vitro transcription with a SP6/T7 DIG RNA labeling kit (Roche, Mannheim, Germany). The RNA were separated on a formaldehyde gel and blotted onto a nylon membrane. After hybridization with the DIG-labeled probes overnight at 68 C, the detection was carried out using the DIG wash and block buffer set and CDP-Star (Roche, Mannheim, Germany) following the manufacturer’s protocol.
Radioimmunoassay
Supernatant (500 μl) of transfected MIN6 cells were used to analyze the insulin secretion using a rat insulin RIA kit (Linco Research, St. Charles, MO) according to the manufacturer’s protocol. The samples were counted using a multi-crystal γ-counter LB2104 (Berthold, Bad Wildbad, Germany) for 1 min.
In vitro translation
In vitro translation was performed using pRC/CMV-Pax6, pCDM8-Pax6 paired box, pCDM8-Pax6 homeodomain, or pCDM8-BETA2 in the TNT Quick coupled transcription/translation system (Promega) following the manufacturer‘s protocol. Autoradiography of the products after SDS-PAGE shows a 48-kDa protein for Pax6, 16-kDa protein for Pax6 paired box, 33-kDa protein for Pax6 homeodomain, and 40-kDa protein for BETA2.
Preparation of GST fusion proteins
An overnight culture of Escherichia coli strain Bl21 codon plus transformed with a pGex-3X construct was induced with 0.2 mmol/liter isopropyl-β-D-1-thiogalactopyranoside and lysed in PBS with 1% Nonidet P-40 by sonication. After centrifugation, the lysate was incubated with PBS-equilibrated glutathione Sepharose 4B beads (APB Europe) for 30 min at room temperature. The beads were washed and resuspended in PBS. Yield and purity were checked on a Coomassie-stained SDS gel.
GST capture assay
GST fusion protein coupled to glutathione Sepharose beads was incubated with 5 μl 35S-labeled in vitro-translated protein or 50 μl nuclear extracts for 1 h at room temperature in 1 ml PBS. The beads were washed four times with PBS, and the bound radioactive proteins were subjected to SDS-PAGE and visualized by a phosphor imager. Interacting Pax6 from nuclear extracts was detected by Western blot analyses.
Preparation of nuclear extracts and EMSA
The preparation of nuclear extracts was done as described previously (25). For EMSA, we used oligonucleotides corresponding to the PISCES element of the rat insulin I promoter (RINSI-PISCES, 5′-TTCTGGGAAATGAGGTGGAAAATGCTCAGCC-3′), to the A1 element of the rat insulin II promoter (RINSII-A1, 5′-CCCCTCTGGCCATCTGCTGATCCACCCTTAATGGGACAAA-3′), and to the A3/A4 element of the rat insulin II promoter (RINSII-A3/A4, 5′-GCCCCTATTAAGACTCTAATTACCCTAAGGCTAAG-3′). Double-stranded oligonucleotides were end labeled using [γ-32P]dATP and T4 polynucleotide kinase. Up to 4 μl in vitro-translated Pax6 or 5 μl nuclear extracts from INS-1 cells, 4 pmol double-stranded unlabeled oligonucleotide for competition, and 0.2 μg anti-Pax6-antibody for supershifts were incubated with 200 fmol labeled oligonucleotide (1300 Bq) and 0.05 μg/μl polydeoxyinosine-polydeoxycytidine in binding buffer containing 100 mmol/liter KCl, 5 mmol/liter MgCl2, 1 mmol/liter dithiothreitol (DTT), 10 mmol/liter Tris-HCl, and 10% glycerol for 30 min at room temperature before separating on a 6% polyacrylamide gel in 0.5× Tris/Borate/EDTA buffer. The gel was dried and analyzed using a phosphor imager.
Chromatin immunoprecipitation
INS-1 cells, incubated overnight with 11 mmol/liter or 5.5 mmol/liter glucose, were fixed in 1% formaldehyde for 2 min, and fixation was stopped by adding glycine to a final concentration of 0.1 m. After washing with PBS, the cells were harvested, snap frozen in liquid nitrogen, and after addition of 100 μl lysis buffer [20 mmol/liter HEPES (pH 7.9), 25% glycerol, 420 mmol/liter NaCl, 1.5 mmol/liter MgCl2, 0.2 mmol/liter EDTA, protease inhibitors], thawed by incubation on ice for 20 min. After centrifugation, the precipitated nuclei were lysed by 100 μl nucleus lysis buffer [50 mmol/liter Tris-HCl (pH 8), 1 mmol/liter EDTA, 150 mmol/liter NaCl, 1% SDS, 2% Triton X-100]. Isolated and sonicated chromatin was precleared with 20 μl protein A Sepharose in 1 ml immunoprecipitation buffer [50 mmol/liter Tris-HCl (pH 8), 1 mmol/liter EDTA, 150 mmol/liter NaCl, 0.1% Triton X-100] overnight. After centrifugation, 200 μl of the supernatant was subjected to immunoprecipitation with polyclonal antibodies directed against Pax6 or PDX-1 and 40 μl protein A Sepharose overnight. The Sepharose beads were washed four times in 1.5 ml immunoprecipitation buffer and incubated overnight with 200 μl SDS-NaCl-DTT buffer [62.5 mmol/liter Tris-HCl (pH 6.8), 200 mmol/liter NaCl, 2% SDS, 10 mmol/liter DTT] at 65 C. After phenol chloroform extraction, DNA was dissolved in 20 μl bidest, and 5 μl was used for PCR analyses with the primers A3for 5′-GCTGTGAACTGGTTCATCAG-3′ and A3rev 5′-CTGCAGAAAGTGCTCATTGG-3′ amplifying the rat insulin promoter −148 to −248 from the transcription start site. As a control, we used the primers GAPDHfor 5′-GTCGTGGAGTCTACTGGCGTCTTC-3′ and GAPDHrev 5′-GTTGTCATTGAGAGCAATGCCAGC-3′ amplifying the coding region +276 to +911 from the translation start site of GAPDH. PCR products were separated by agarose gel electrophoresis.
Semiquantitative RT-PCR
cDNA was prepared by reverse transcription of 2 μg total RNA using 0.5 μg BamTT primer (3′-CGC GGA TCC TTT TTT TTT TTT TTT TTT-5′) and Superscript II reverse transcriptase. Pax6, insulin, or GAPDH was amplified with Taq DNA polymerase in a DNA thermal cycler (Eppendorf, Hamburg, Germany) using the following gene-specific primers: Pax6for, 5′-GCC CCG GGG ATC CGA ATA CCC AGT GTG TCA TCA ATA A-3′, and Pax6rev, 5′-CCG ATC ACT GGA TCC CTG TAA TCG AGG CCA-3′, with an expected product of 948 bp; insulinfor, 5′-TTC CAG GTC ATT GTT CCA ACA TG-3′, and insulinrev, 5′-GTA GTT CTC CAG TTG GTA GAG G-3′ with an expected product of 355 bp; and GAPDHfor, 5′-GTC GTG GAG TCT ACT GGC GTC TTC-3′, and GAPDHrev, 5′-GTT GTC ATT GAG AGC AAT GCC AGC-3′ with an expected product of 650 bp. The conditions for each cycle were 30 sec at 94 C, 40 sec at 57 C, and 60 sec at 72 C (25 cycles).
Statistical analysis
Means ± sem were calculated from three or more different experiments using Student’s unpaired t test or one-way ANOVA. Differences are considered statistically significant at P < 0.05.
Acknowledgments
We thank Ines Schmidt for skillful technical assistance and Christopher V. E. Wright for supplying antiserum from goat directed against PDX-1.
Footnotes
This work was supported by the foundation Deutsche Forschungsgemeinschaft and by the German Federal Ministry of Education and Research (BMBF) within the program “Entrepreneurial Regions: Centre for Innovation Competence” (Project No. ZIK 011).
Disclosure Summary: The authors have nothing to disclose.
First Published Online October 13, 2010
Abbreviations: AD, Activation domain; BD, DNA-binding domain; DIG, digoxigenin; DTT, dithiothreitol; GST, glutathione S transferase; IF, immune fluorescence; MafA, musculoaponeurotic fibrosarcoma A; PDX-1, pancreatic and duodenal homeobox protein-1; PISCES, pancreatic islet cell-specific enhancer sequence; RIPE3b, rat insulin promoter element 3b; RNAi, interfering RNA; SV40, simian virus 40; YFP, yellow fluorescent protein.
References
- 1.German M, Ashcroft S, Docherty K, Edlund H, Edlund T, Goodison S, Imura H, Kennedy G, Madsen O, Melloul D, Moss L, Olson K, Permutt MA, Philippe J, Robertson P, Rutter WJ, Serup P, Stein R, Steiner D, Tsai MJ, Walker MD1995. The insulin gene promoter. Diabetes 44:1002–1004 [DOI] [PubMed] [Google Scholar]
- 2.Dumonteil E, Philippe J1996. Insulin gene: organisation, expression and regulation. Diabetes Metab 22:164–173 [PubMed] [Google Scholar]
- 3.Dumonteil E, Laser B, Constant I, Philippe J1998. Differential regulation of the glucagon and Insulin I gene promoters by the basic helix-loop-helix transcription factors E47 and BETA2. J Biol Chem 273:19945–19954 [DOI] [PubMed] [Google Scholar]
- 4.Naya F J, Stellrecht CM, Tsai MJ1995. Tissue specific regulation of the insulin gene by a novel basic helix-loop-helix transcription factor. Genes Dev 9:1009–1019 [DOI] [PubMed] [Google Scholar]
- 5.Sander M, German MS1997. The β-cell transcription factors and development of the pancreas. J Mol Med 75:327–340 [DOI] [PubMed] [Google Scholar]
- 6.Sharma A, Moore M, Marcora E, Lee JE, Qiu Y, Samaras S, Stein R1999. The NeuroD/BETA2 sequences essential for insulin gene transcription colocalize with those necessary for neurogenesis and p300/CREB binding protein binding. Mol Cell Biol 19:704–713 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Qiu Y, Sharma A, Stein R1998. p300 mediates transcriptional stimulation by the basic helix-loop-helix activators of the insulin gene. Mol Cell Biol 18:2957–2964 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Peers B, Leonard J, Sharma S, Teitelmann G, Montminy MR1994. Insulin expression in pancreatic islet cells relies on cooperative interactions between the helix loop helix factor E47 and the homeobox factor STF-1. Mol Endocrinol 8:1798–1806 [DOI] [PubMed] [Google Scholar]
- 9.Zhao L, Guo M, Matsuoka TA, Hagman DK, Parazzoli SD, Poitout V, Stein R2005. The islet β-cell-enriched MafA activator is a key regulator of insulin gene transcription. J Biol Chem 280:11887–11894 [DOI] [PubMed] [Google Scholar]
- 10.Ohlsson H, Karlsson K, Edlund T1993. IPF1, a homeodomain-containing transactivator of the insulin gene. EMBO J 12:4251–4259 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Leibiger IB, Schwarz T, Leibiger B, Walther R1995. Functional analysis of a newly identified TAAT-box of rat insulin-II gene promoter. FEBS Lett 362:210–214 [DOI] [PubMed] [Google Scholar]
- 12.Petersen HV, Serup P, Leonard J, Michelsen BK, Madsen OD1994. Transcriptional regulation of the human insulin gene is dependent on the homeodomain protein STF1/IPF1 acting through the CT boxes. Proc Natl Acad Sci USA 91:10465–10469 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Qiu Y, Guo M, Huang S, Stein R2002. Insulin gene transcription is mediated by interaction between the p300 coactivator and PDX-1, BETA2, and E47. Mol Cell Biol 22:412–420 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Ohneda K, Mirmira RG, Wang J, Johnson JD, German MS2000. The homeodomain of PDX-1 mediates multiple protein-protein interactions in the formation of a transcriptional activation complex on the insulin promoter. Mol Cell Biol 20:900–911 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.German MS, Wang J, Chadwick RB, Rutter WJ1992. Synergistic activation of the insulin gene by a IM-homeodomain protein and a basic helix-loop-helix protein building a functional insulin minienhancer complex. Genes Dev 6:2165–2176 [DOI] [PubMed] [Google Scholar]
- 16.Glick E, Leshkowitz D, Walker MD2000. Transcription factor BETA2 acts cooperatively with E2A and PDX1 to activate the insulin gene promoter. J Biol Chem 275:2199–2204 [DOI] [PubMed] [Google Scholar]
- 17.Madsen OD, Jensen J, Petersen HV, Pedersen EE, Oster A, Andersen FG, Jørgensen MC, Jensen PB, Larsson LI, Serup P1997. Transcription factors contributing to the pancreatic β-cell phenotype. Horm Metab Res 29:265–270 [DOI] [PubMed] [Google Scholar]
- 18.St-Onge L, Sosa-Pineda B, Chowdhury K, Mansouri A, Gruss P1997. Pax6 is required for differentiation of glucagon-producing α-cells in mouse pancreas. Nature 387:406–409 [DOI] [PubMed] [Google Scholar]
- 19.Sheng G, Harris E, Bertuccioli C, Desplan C1997. Modular organization of Pax/homeodomain proteins in transcriptional regulation. Biol Chem 378:863–872 [DOI] [PubMed] [Google Scholar]
- 20.Beimesche S, Neubauer A, Herzig S, Grzeskowiak R, Diedrich T, Cierny I, Scholz D, Alejel T, Knepel W1999. Tissue-specific transcriptional activity of a pancreatic islet cell-specific enhancer sequence/Pax6-binding site determined in normal adult tissues in vivo using transgenic mice. Mol Endocrinol 13:718–728 [DOI] [PubMed] [Google Scholar]
- 21.Sander M, Neubüser A, Kalamaras J, Ee HC, Martin GR, German MS1997. Genetic analysis reveals that Pax6 is required for normal transcription of pancreatic hormone genes and islet development. Genes Dev 11:1662–1673 [DOI] [PubMed] [Google Scholar]
- 22.Wrege A, Diedrich T, Hochhoth C, Knepel W1995. Transcriptional activity domain A of the rat glucagon G3 element conferred by an islet-specific nuclear protein that also binds to similar pancreatic islet cell-specific enhancer sequences (PISCES). Gene Expr 4: 205–216. [PMC free article] [PubMed]
- 23.Kalousová A, Benes V, Paces J, Paces V, Kozmik Z1999. DNA binding and transactivating properties of the paired and homeobox protein Pax4. Biochem Biophys Res Commun 259:510–518 [DOI] [PubMed] [Google Scholar]
- 24.Ritz-Laser B, Gauthier BR, Estreicher A, Mamin A, Brun T, Ris F, Salmon P, Halban PA, Trono D, Philippe J2003. Ectopic expression of the β-cell specific transcription factor Pdx1 inhibits glucagon gene transcription. Diabetologia 46:810–821 [DOI] [PubMed] [Google Scholar]
- 25.Hessabi B, Ziegler P, Schmidt I, Hessabi C, Walther R1999. The nuclear localization signal (NLS) of PDX-1 is part of the homeodomain and represents a novel type of NLS. Eur J Biochem 263:170–177 [DOI] [PubMed] [Google Scholar]
- 26.Marshak S, Totary H, Cerasi E, Melloul D1996. Purification of the β-cell glucose-sensitive factor that transactivates the insulin gene differentially in normal and transformed islet cells. Proc Natl Acad Sci USA 93:15057–15062 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Miller CP, McGehee Jr RE, Habener JF1994. IDX-1: a new homeodomain transcription factor expressed in rat pancreatic islets and duodenum that transactivates the somatostatin gene. EMBO J 13:1145–1156 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Serup P, Petersen HV, Pedersen EE, Edlund H, Leonard J, Petersen JS, Larsson LI, Madsen OD1995. The homeodomain protein IPF-1/STF-1 is expressed in a subset of islet cells and promotes rat insulin 1 gene expression dependent on an intact E1 helix-loop-helix factor binding site. Biochem J 310(Pt 3):997–1003 [DOI] [PMC free article] [PubMed]
- 29.Naya FJ, Huang HP, Qiu Y, Mutoh H, DeMayo FJ, Leiter AB, Tsai MJ1997. Diabetes, defective pancreatic morphogenesis, and abnormal enteroendocrine differentiation in BETA2/NeuroD-deficient mice. Genes Dev 11:2323–2334 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Thomas MK, Yao KM, Tenser MS, Wong GG, Habener JF1999. Bridge-1, a novel PDZ-domain coactivator of E2A-mediated regulation of insulin gene transcription. Mol Cell Biol 19:8492–8504 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Yasuda T, Kajimoto Y, Fujitani Y, Watada H, Yamamoto S, Watarai T, Umayahara Y, Matsuhisa M, Gorogawa S, Kuwayama Y, Tano Y, Yamasaki Y, Hori M2002. Pax6 mutation as a genetic factor common to aniridia and glucose intolerance. Diabetes 51:224–230 [DOI] [PubMed] [Google Scholar]
- 32.Ashery-Padan R, Zhou X, Marquardt T, Herrera P, Toube L, Berry A, Gruss P2004. Conditional inactivation of Pax6 in the pancreas causes early onset of diabetes. Dev Biol 269:479–488 [DOI] [PubMed] [Google Scholar]
- 33.Wen JH, Chen YY, Song SJ, Ding J, Gao Y, Hu QK, Feng RP, Liu YZ, Ren GC, Zhang CY, Hong TP, Gao X, Li LS2009. Paired box 6 (PAX6) regulates glucose metabolism via proinsulin processing mediated by prohormone convertase 1/3 (PC1/3). Diabetologia 52:504–513 [DOI] [PubMed] [Google Scholar]
- 34.Samaras SE, Cissell MA, Gerrish K, Wright CV, Gannon M, Stein R2002. Conserved sequences in a tissue-specific regulatory region of the pdx-1 gene mediate transcription in pancreatic β-cells: role for hepatocyte nuclear factor 3β and Pax6. Mol Cell Biol 22:4702–4713 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Raum JC, Hunter CS, Artner I, Henderson E, Guo M, Elghazi L, Sosa-Pineda B, Ogihara T, Mirmira RG, Sussel L, Stein R2010. Islet β-cell-specific MafA transcription requires the 5`-flanking conserved Region 3 control domain. Mol Cell Biol 30:4234–4244 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Gosmain Y, Martinet E, Cheyssac C, Guerardel A, Mamin A, Katz LS, Karim B, Philippe J 30 June 2010. Pax6 controls the expression of critical genes involved in pancreatic α-cell differentiation and function. J Biol Chem 10.1074/jbc.M110.147215 [DOI] [PMC free article] [PubMed]
- 37.Cissell MA, Zhao L, Sussel L, Henderson E, Stein R2003. Transcription factor occupancy of the insulin gene in vivo. J Biol Chem 278:751–756 [DOI] [PubMed] [Google Scholar]
- 38.Smith SB, Ee HC, Conners JR, German MS1999. Paired-hoeodomain transcription factor PAX4 acts as a transcriptional repressor in early pancreatic development. Mol Cell Biol 19:8272–8280 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Rafiq I, Kennedy HJ, Rutter GA1998. Glucose dependent translocation of insulin promoter factor-1 (IPF-1) between the nuclear periphery and the nucleoplasm of single MIN6 β-cells. J Biol Chem 273:23241–23247 [DOI] [PubMed] [Google Scholar]
- 40.Macfarlane WM, McKinnon CM, Felton-Edkins ZA, Cragg H, James RF, Docherty K1999. Glucose stimulates translocation of the homeodomain transcription factor PDX1 from the cytoplasm to the nucleus in pancreatic β-cells. J Biol Chem 274:1011–1016 [DOI] [PubMed] [Google Scholar]
- 41.Guillemain G, Da Silva Xavier G, Rafiq I, Leturque A, Rutter GA2004. Importin β1 mediates the glucose-stimulated nuclear import of pancreatic and duodenal homeobox-1 in pancreatic islet β-cells (MIN6). Biochem J 378:219–227 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Zhou Q, Law AC, Rajagopal J, Anderson WJ, Gray PA, Melton DA2007. A multipotent progenitor domain guides pancreatic organogenesis. Dev Cell 13:103–114 [DOI] [PubMed] [Google Scholar]
- 43.Olson LK, Redmon JB, Towle HC, Robertson RP1993. Chronic exposure of HIT cells to high glucose concentrations paradoxically decreases insulin gene transcription and alters binding of insulin gene regulatory protein. J Clin Invest 92:514–519 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Sharma A, Olson LK, Robertson RP, Stein R1995. The reduction of insulin gene transcription in HIT-T15 β cells chronically exposed to high glucose concentration is associated with the loss of RIPE3b1 and STF-1 transcription factor expression. Mol Endocrinol 9:1127–1134 [DOI] [PubMed] [Google Scholar]
- 45.Poitout V, Olson LK, Robertson RP1996. Chronic exposure of βTC-6 cells to supraphysiologic concentrations of glucose decreases binding of the RIPE3b1 insulin gene transcription activator. J Clin Invest 97:1041–1046 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Olson LK, Qian J, Poitout V1998. Glucose rapidly and reversibly decreases INS-1 cell insulin gene transcription via decrements in STF-1 and C1 activator transcription factor activity. Mol Endocrinol 12:207–219 [DOI] [PubMed] [Google Scholar]
- 47.Martin CC, Oeser JK, O'Brien RM2004. Differential regulation of islet-specific glucose-6-phosphatase catalytic subunit-related protein gene transcription by Pax-6 and Pdx-1. J Biol Chem 279: 34277–34289 [DOI] [PubMed]
- 48.Hussain MA, Habener JF1999. Glucagon gene transcription activation mediated by synergistic interactions of pax-6 and cdx-2 with the p300 co-activator. J Biol Chem 274:28950–28957 [DOI] [PubMed] [Google Scholar]
- 49.Ritz-Laser B, Estreicher A, Klages N, Saule S, Philippe J1999. Pax6 and Cdx2/3 interact to activate glucagon gene expression on the G1 control element. J Biol Chem 274:4124–4132 [DOI] [PubMed] [Google Scholar]
- 50.Dumonteil E, Magnan C, Ritz-Laser B, Ktorza A, Meda P, Philippe J2000. Glucose regulates proinsulin and prosomatostatin but not proglucagon messenger ribonucleic acid levels in rat pancreatic islets. Endocrinology 141:174–180 [DOI] [PubMed] [Google Scholar]
- 51.Grzeskowiak R, Amin J, Oetjen E, Knepel W2000. Insulin responsiveness of the glucagon gene conferred by interactions between proximal promoter and more distal enhancer-like elements involving the paired domain transcription factor Pax6. J Biol Chem 275:30037–30045 [DOI] [PubMed] [Google Scholar]