Abstract
We have previously shown that the glucocorticoid receptor (GR) is required for skin homeostasis and epidermal barrier competence. To understand the transcriptional program by which GR regulates skin development, we performed a microarray analysis using the skin of GR−/− and GR+/+ mice of embryonic d 18.5 and identified 442 differentially expressed genes. Functional clustering demonstrated overrepresentation of genes involved in ectoderm/epidermis development. We found strong repression of genes encoding proteins associated with the later stages of epidermal differentiation, such as several small proline-rich proteins (Sprrs) and corneodesmosin (Cdsn). This, together with the up-regulation of genes induced earlier during epidermal development, including the epithelial-specific gene transcripts E74-like factor 5 (Elf5) and keratin 77 (Krt77), fits with the phenotype of defective epidermal differentiation observed in the GR−/− mice. We also found down-regulation of the antimicrobial peptide defensin β 1 (Defb1) and FK506-binding protein 51 (Fkbp51). Skin developmental expression profiling of these genes and studies in cultured keratinocytes from GR−/− and wild type embryos demonstrated that gene regulation occurred in a cell-autonomous manner. To investigate the consequences of GR loss in adult epidermis, we generated mice with inducible inactivation of GR restricted to keratinocytes (K14-cre-ERT2//GRloxP/loxP mice). K14-cre-ERT2//GRloxP/loxP mice featured thickened skin with increased keratinocyte proliferation and impaired differentiation. Whereas Krt77 and Elf5 expression remained unaffected by loss of GR in adult epidermis, Fkbp51, Sprr2d, and Defb1 were strongly repressed. Importantly, we have identified both Fkbp51 and Defb1 as direct transcriptional targets of GR, and we have shown that GR-mediated regulation of these genes occurs in both developing and adult epidermis. We conclude that both overlapping and differential GR targets are regulated in developing vs. adult skin.
Glucocorticoid receptor regulates overlapping and differential gene subsets in developing and adult skin, as demonstrated by loss-of-function mouse models.
The glucocorticoid receptor (GR) is a ubiquitous intracellular protein that belongs to the superfamily of steroid nuclear receptors and functions as a ligand-dependent transcription factor. Upon binding to glucocorticoid (GC) ligand, GR dissociates from cytoplasmic complexes, dimerizes, and translocates to the nucleus, where it can modulate gene transcription in a cell type-specific manner (reviewed in Ref. 1). The determinants that achieve the gene- and cell type specificity of GR-transcriptional regulation are not well established. In fact, transcriptomic approaches have found little overlap in GC-regulated genes between different cell types (2, 3). GR can also regulate gene expression through DNA-binding-independent mechanisms that involve its interference with other transcription factors, such as nuclear factor-κB or activator protein 1 (AP-1) (4). In addition, GR has nongenomic actions that involve physical interaction of the receptor at the plasma membrane with p85α/phosphatidylinositol 3-kinase, thus modulating AKT activity (5).
Given its pleiotropic actions in many vital organs, GR is required for survival and also plays a crucial role in skin pathophysiology (6, 7). GC derivatives are the most widely used therapeutic agents for treating numerous cutaneous diseases (8). However, GR is also required for proper skin development, as shown by complementary approaches of genetically modified mice with gain- and loss of function of GR (9, 10, 11, 12). In mammals, the skin acts as a barrier between the individual and the environment. Proper acquisition of a functional skin barrier during embryonic development requires a correct balance between proliferation, differentiation, and controlled apoptosis of epidermal keratinocytes (reviewed in Ref. 13). Impairment of these processes can cause skin disorders of keratinization and cornification, the consequences of which range from postnatal lethality to increased susceptibility to cutaneous infections and development of inflammatory skin diseases (14, 15, 16).
The epidermis is a stratified epithelium consisting of a basal layer formed by proliferative keratinocytes and several upper layers in which keratinocytes become progressively more differentiated as they migrate outward (13). During mouse epidermal development upon commitment to terminal differentiation, genes expressed by basal keratinocytes, such as keratin K5, are repressed and differentiation-specific proteins, including keratins K1 and K10, are up-regulated. Epidermal terminal differentiation represents a specialized form of programmed cell death, in which viable keratinocytes convert into dead, flattened squames of the stratum corneum. These processes need to be tightly coordinated to form a competent epidermal barrier in vivo; however, the underlying transcriptional program is not completely understood (15). It is known, however, that the calcium gradient of the mammalian epidermis, with low levels in the basal/intermediate layers and high levels in the upper layers, modulates the expression of differentiation-specific proteins (17, 18). We recently demonstrated that GR−/− late embryos [embryonic d 18.5 (E18.5)] feature incomplete epidermal stratification and impaired keratinocyte differentiation, resulting in a defective skin barrier (12). The use of cultured mouse primary keratinocytes (MPKs) from GR−/− mice demonstrated that GR regulates keratinocyte growth and apoptosis in a cell-autonomous manner. However, although epidermal differentiation was severely impaired in vivo, cultured MPKs from GR-deficient mice were able to differentiate in the presence of high calcium, a well-established model of in vitro keratinocyte differentiation (19). These results suggest that GR−/− skin might display a defective calcium gradient that can be overcome in cell culture by adding calcium. Alternatively, cultured GR−/− keratinocytes could be able to differentiate due to growth factors and hormones present in the culture medium that are lacking in the GR−/− embryonic skin in vivo.
Previous reports identified putative GR transcriptional targets in keratinocytes by indirect approaches that evaluated the effects of the synthetic ligand dexamethasone (Dex) in vitro or in wild type (wt) embryos that were exposed to pharmacological doses of the corticosteroid during development (20, 21). In this work, we have analyzed the transcriptional profile of GR−/− embryonic skin using a microarray approach to identify GR transcriptional targets in vivo that are relevant for epidermal morphogenesis.
Given the perinatal lethality of the GR−/− mice and to further investigate the role of GR specifically in adult keratinocytes in vivo, we have generated an inducible conditional knock-out mouse model, the K14-cre-ERT2//GRloxP/loxP mouse. This model allows us to distinguish the consequences of the loss of GR in epidermal keratinocytes from those of the complete knockout and demonstrate that GR is not only required for normal epidermal development but also for adult skin homeostasis. Additionally, our analysis of gene expression suggests that GR regulates common and differential gene subsets in embryonic and adult skin.
Results
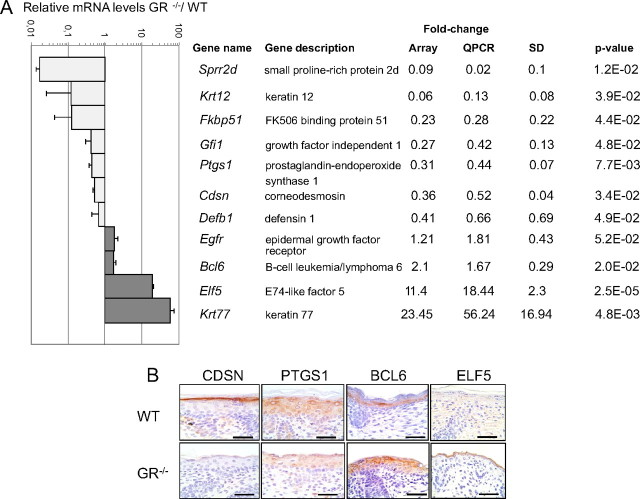
To understand the mechanisms by which GR regulates skin development, we performed a transcriptomic analysis using the skin of GR−/− mouse embryos (E18.5) as compared with wt littermates. We found 442 differentially expressed genes (DEGs) in GR−/− vs. GR+/+ skin [false discovery rate (FDR) < 0.05]; of these, 206 were repressed and 236 were induced (for the complete list see Supplemental Table 1 published on The Endocrine Society’s Journals Online web site at http://mend.endojournals.org). DEGs were grouped by functional clustering according to the Gene Ontology category of biological process (Supplemental Fig. 1). Although genes were categorized based on protein function, many genes likely fit into more than one category. Consistent with the previously described phenotype of GR−/− mice, with impaired keratinocyte terminal differentiation and defective epidermal development (12), the functional category of ectoderm/epidermis development was overrepresented in the microarray analysis (4.6%) (Supplemental Fig. 1 and Supplemental Table 2). We performed quantitative RT-PCR (QPCR) to validate the microarray results and found a very good correlation for the genes tested (Fig. 1). Among these genes, we found several components of the cornified envelope that were strongly repressed, including the small proline-rich protein (Sprr) gene family members Sprr1b, Sprr2d, and Sprr2h as well as corneodesmosin (Cdsn) (Fig. 1 and Supplemental Table 2; fold change: 0.06, 0.09, 0.13, 0.36, respectively). It is worth noting, that expression of the components of the epidermal cornified envelope has been previously correlated with barrier initiation (reviewed in Ref. 22), which is defective in GR−/− skin (12). We detected the expression of keratin 12 (Krt12) mRNA in developing epidermis in contrast to previous reports of expression restricted to cornea epithelium (23). We also found down-regulation of FK506-binding protein 51 (Fkbp51), growth factor independent 1 (Gfi1), prostaglandin-endoperoxide synthase 1 (Ptgs1), and defensin β 1 (Defb1). In addition, we found a modest but consistent up-regulation of epidermal growth factor receptor (Egfr), a key player in epithelial development (24), as well as B-cell leukemia/lymphoma 6 (Bcl6), an immediate early gene that is activated in lymphocytes but the expression of which has also been described in keratinocytes (25). Notably, the highest up-regulated genes were two epithelial-specific gene transcripts: E74-like factor 5 (Elf5) and the keratin 77 (Krt77) (26, 27).
Fig. 1.
Differential transcriptomic profile of E18.5 GR−/− embryonic skin compared with wt (WT) littermates. A, Relative mRNA levels of a subset of DEGs in GR−/− vs. wt skin as determined by microarray analysis (quadruplicate skin samples of each genotype) and QPCR validation in an independent experiment with at least three individuals of each genotype. Fold changes (relative to wt) in the array vs. QPCR are shown. sds for QPCR data are shown as well as P values calculated using Student’s t test. B, Protein expression of CDSN, PTGS1 BCL6, and ELF5 was demonstrated in paraffin-embedded dorsal skin of GR−/− and wt E18.5 littermates by immunostaining using specific antibodies. Bars, 50 μm.
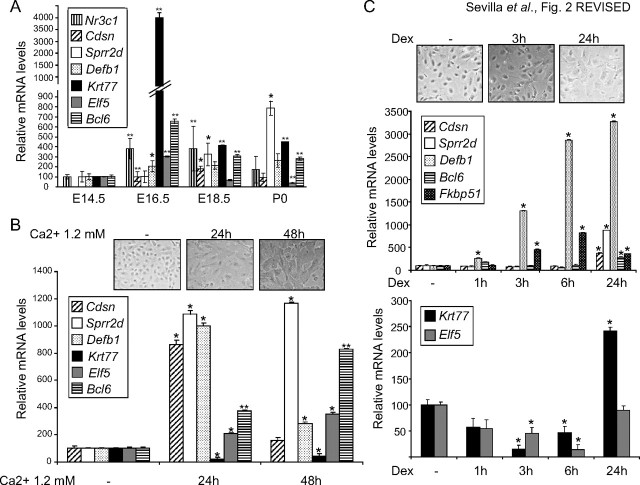
Our analysis of the transcriptional profile of GR−/− developing skin has identified genes previously unknown as GR-regulated in keratinocytes such as Sprr2d, Krt12, Defb1, Egfr, Krt77, and Elf5. We have also detected other genes, including Cdsn, Ptgs1, Bcl6, and Fkbp51, which were previously reported to be affected by Dex treatment in keratinocytes (20, 21, 28). We confirmed transcript changes at the protein level for CDSN, PTGS1, BCL6, and ELF5, by immunohistochemistry in GR−/− embryonic skin using specific antibodies (Fig. 1B). These proteins are normally detected in the cornified envelope (CDSN) and upper layers of the developing skin (PTGS1, BCL6) and have been previously reported to be involved in keratinocyte differentiation (29, 30, 25). ELF5 expression in skin is controversial because some reports describe Elf5 expression in mouse epidermis and oral keratinocyte cell lines (26, 31) whereas others describe Elf5 exclusively in the inner root sheath of the hair follicle (32). To better understand the contribution of the identified genes in epidermal development, we focused on a subset of the novel GR targets, using the previously described Dex-regulated genes as a control of transcriptional regulation mediated by GR in vivo. We analyzed the expression of Cdsn, Sprr2d, Defb1, Krt77, Elf5, and Bcl6 in the skin of wt developing embryos and newborn mice by QPCR using at least four individuals of each age (E14.5–P0 (newborn), Fig. 2A). Cdsn mRNA levels were undetectable at E14.5 but induced at E16.5 and E18.5. Sprr2d transcripts were up-regulated at E18.5, and further up-regulated at P0. These data are in agreement with the reported role of these genes in late epidermal differentiation (22). In contrast, Krt77, Elf5, and Bcl6 mRNA levels peaked at E16.5 and were subsequently repressed at E18.5 and P0 (Fig. 2A). Defb1 transcript levels were induced at E16.5 and remained at similar levels at P0. The fact that Krt77, Elf5, and Bcl6 are expressed with relatively high levels in normal developing epidermis at E16.5, when the epidermis is still immature (Fig. 2A), suggests that these genes are part of the transcriptional program of epidermal maturation at early stages. Their subsequent repression suggests that this expression has to be reduced for the proper completion of differentiation. Importantly, the observed changes in these GR-regulated genes paralleled the mRNA expression of Nr3c1 (the GR-encoding gene) during skin development (Fig. 2A).
Fig. 2.
Expression of DEGs during in vivo and in vitro keratinocyte differentiation. A, Gene expression of Nr3c1 and the indicated DEGs during skin development in E14.5, E16.5, E18.5, and newborn (P0) wt mice. B, Kinetics of gene expression during keratinocyte differentiation induced by treatment of wt MPKs with high calcium (1.2 mm). Cell differentiation was assessed by phase contrast (upper panels), and expression of the indicated genes was determined by QPCR. C, Kinetics of gene expression during keratinocyte differentiation induced by treating wt MPKs with either vehicle (−) or Dex (100 nm). Cell differentiation was assessed by phase contrast (upper panels), and gene expression was determined by QPCR. Experiments were performed using at least three culture replicates for each treatment and timepoint. Statistically significant differences were assessed by Student′s t test (*, P < 0.005; **, P < 0.001).
To evaluate gene expression changes during differentiation, we isolated and cultured MPKs from wt embryos (E18.5) and treated them with media containing high concentrations of calcium (1.2 mm). After calcium exposure, we detected characteristic changes in cell morphology including cell flattening and increased cell-cell contacts (Fig. 2B, upper panel). We analyzed the changes in gene expression at 24 h and 48 h after calcium treatment and found, as expected, that the differentiation-associated markers Cdsn and Sprr2d were markedly up-regulated at 24 h (Fig. 2B, 8- to 11-fold increase, respectively). After 48 h of high calcium exposure, and in agreement with their observed mRNA expression pattern in wt developing skin (Fig. 2A), Cdsn returned to basal levels whereas Sprr2d transcription was further increased (Fig. 2B, 1.4- to 12-fold increase, respectively). We also found strongly increased Defb1 transcript levels that peaked at 24 h and calcium induction of Elf5 and Bcl6, although to a lesser extent (2- to 4-fold increase) and with delayed kinetics. In contrast, Krt77 mRNA was repressed by high calcium in wt cultured keratinocytes (Fig. 2B), which is in agreement with the observed repression of this gene in late wt embryos (Fig. 2A).
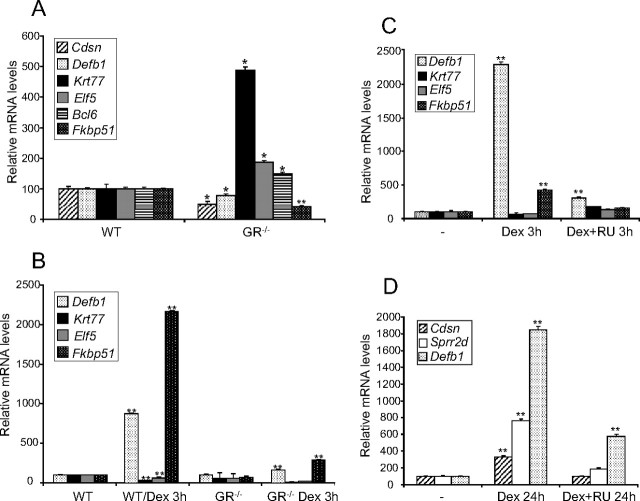
Because the lack of GR has an impact on the expression of the identified genes (Fig. 1), we took the complementary approach to study the consequences of ligand activation of the GR in a time-course experiment using wt MPKs (Fig. 2C). Dex (100 nm) elicited morphological features of differentiation of cultured keratinocytes at 24 h, as shown in Fig. 2C, upper panel, and previous reports (12). Dex treatment up-regulated Cdsn, Sprr2d, Defb1, and Fkbp51 in cultured MPKs, in agreement with their repression in GR null skin (Fig. 2C, middle panel). However, Bcl6 induction after Dex treatment was not consistent with its up-regulation in GR−/− skin. The observed kinetics of Dex induction for Cdsn, Sprr2d, and Bcl6 were late (24 h) and for Defb1 and Fkbp51, early and sustained (3–24 h). In contrast, Krt77 and Elf5 were repressed by Dex at 3–6 h (2- to 4-fold) in a transient manner, consistent with the up-regulation observed in GR−/− skin (Fig. 2C, lower panel). Although the expression of many of the identified genes is restricted to keratinocytes, such as Cdsn, Sprr1b, Sprr2d, Sprr2 h, ectodysplasin-A receptor (Edar), Egfr, Krt77, and Elf5, other genes were expressed in cell types in addition to epithelia, as with Defb1, Bcl6, and Fkbp51. The changes we observed in vivo might be keratinocyte autonomous or due to dermal contribution. To ascertain this, we cultured GR−/− and wt MPKs in media with normal serum and evaluated gene expression (Fig. 3A). The differences in the transcript levels of Cdsn, Defb1, Krt77, Elf5, Bcl6, and Fkbp51 between GR−/− and wt MPKs grown in normal serum-containing media were consistent with those found in the microarray and validation experiments (Fig. 3A). It is also possible that other hormones and growth factors, including corticosterone, contribute in some way to the transcriptional profile of the developing skin. To assess this, GR−/− and wt MPKs were grown in media with steroid-depleted serum, and gene expression was evaluated in the presence and absence of treatment with Dex (Fig. 3B). Notably, when MPKs isolated from GR knockout mice were cultured in steroid-depleted serum, Krt77 and Elf5 were not up-regulated, thus suggesting that additional hormones and growth factors in the medium are relevant for the basal expression levels of these genes (Fig. 3B). However, most importantly, all of the genes we analyzed in GR−/− MPKs treated with Dex showed either no response or marginal changes as compared with wt (Fig. 3B and data not shown). These data demonstrate that GR is required for transcriptional regulation of the identified genes in a keratinocyte-specific manner.
Fig. 3.
Gene regulation of relevant DEGs in cultured MPKs isolated from GR−/− skin and responses to GR agonist and antagonists. A and B, GR−/− and wt MPKs isolated from E18.5 embryonic skin were cultured in either serum-containing media (A) or steroid-depleted serum (B), and gene expression was determined by QPCR. In addition, Dex response at 3 h was evaluated in GR−/− and wt MPKs (B). Note the drastically reduced response to Dex of GR−/− MPKs relative to wt controls. C and D, Effects of the GR antagonist RU486 in combination with Dex in wt cultured keratinocytes at 3 h (C) and 24 h (D). Note that RU486 reverses the effects of Dex on induced and repressed genes. Experiments were performed using at least three culture replicates for each genotype and treatment, and statistical significance was assessed by Student′s t test (*, P < 0.005; **, P < 0.001). RU, RU486.
We further investigated whether the identified genes are regulated through GR activity by evaluating the effects of the GR antagonist RU486 in combination with Dex in wt cultured keratinocytes (Fig. 3, C and D). Although the genes regulated by Dex with early kinetics are more likely to be primary GR-transcriptional targets, we analyzed the expression of both early (Krt77, Elf5, Fkbp51) and late (Cdsn, Sprr2d) genes. In the case of Defb1, Dex induced mRNA expression at 3 h and 24 h (Fig. 2C). For all of the tested genes, RU486 was able to reverse the effects of Dex, regardless of whether the ligand-activated GR induced or repressed gene expression (Fig. 3, C and D). These data suggest that gene regulation is primarily due to GR activity. To differentiate direct GR-transcriptional targets from those regulated via other GR-controlled proteins, we used the inhibitor of protein synthesis cycloheximide (CHX) to check the mRNA levels of Defb1, Fkbp51, Krt77, and Elf5, which are regulated by Dex with early kinetics (Fig. 4A). We observed that Defb1 and Fkbp51 show a robust induction in the presence of Dex plus CHX, indicating that their regulation is due to GR-mediated transcriptional activation (Fig. 4A). In the case of Krt77 and Elf5, both genes were still repressed in the presence of Dex plus CHX (Fig. 4A). To discriminate whether these genes were regulated via direct binding of GR to their regulatory elements, we performed chromatin immunoprecipitation (ChIP) assays using wt MPKs treated with vehicle or Dex (Fig. 4B). We first examined Fkbp51 because the Dex-dependent binding of the GR at an Fkbp51 enhancer has been previously demonstrated in other cell types (3). For these experiments, given that Fkbp51 mRNA induction was detected at 3 h upon Dex treatment (Fig. 2C and Fig. 3, B and C), we evaluated GR binding after 2 h of incubation with Dex (Fig. 4). Our data show that GR was recruited to the Fkbp51 enhancer exclusively after ligand activation of GR in cultured keratinocytes.
Fig. 4.
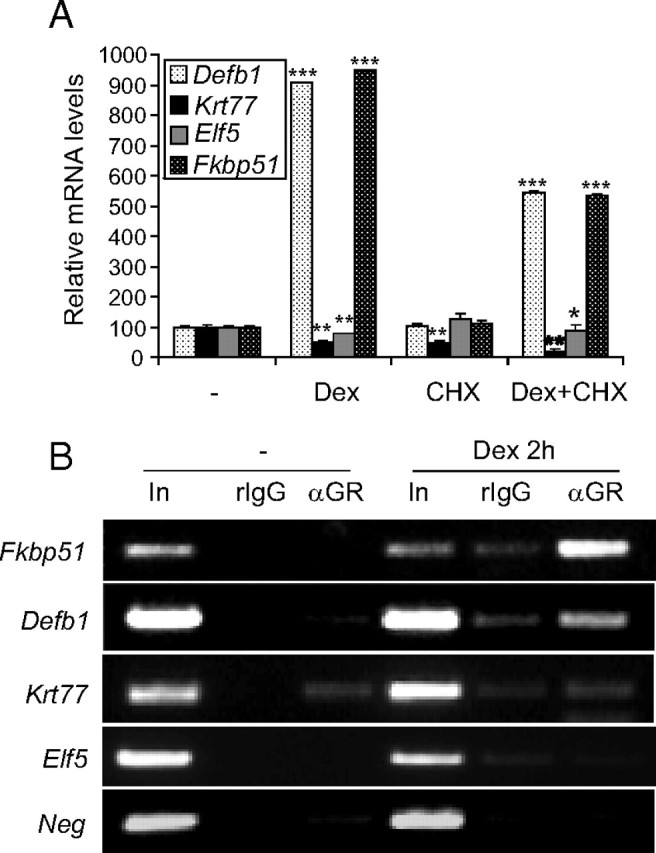
Identification of Fkbp51 and Defb1 as primary GR target genes in keratinocytes. A, wt MPKs were cultured in the presence of vehicle (−), Dex (100 nm), CHX (2.5 μg/ml), or Dex plus CHX for 3 h to assess gene expression of the indicated genes by QPCR (*, P < 0.05; **, P < 0.005; ***, P < 0.0001). B, GR binding to the indicated genes was assessed by ChIP assays using wt MPKs treated with vehicle (−) or Dex (100 nm) for 2 h. Chromatin was immunoprecipitated with either rabbit IgG (rIgG, 2 μg) or rabbit anti-GR antibody (2 μg) followed by PCR using primers specific for sequences that contained putative GBS, as described in the text. GR bound to Fkbp51 and Defb1 but not Krt77- or Elf5-regulatory sequences.
It has been demonstrated that most of the GR-binding sites (GBS) in a natural gene context are represented by nonconventional GC response elements (GREs) and/or composite elements (3, 33, 34). We first searched for predicted GBS in conserved regions proximal to genes of interest using Alibaba 2.1. We then compared the conservation of putative GBS between human and mouse, because it has been reported that, for GC-induced genes, conservancy is a good predictor of GR binding (33). Using this strategy, we found putative GBS with high degrees of conservancy between mouse and human in the proximal promoter of Krt77 and the first intron of Elf5. However we were unable to detect binding of the GR to these regions in ChIP assays (Fig. 4). Neither the upstream region nor the introns of the Defb1 gene are conserved between human and mouse; nevertheless, we searched for putative GBS in its proximal promoter and found two putative GR half-sites, with the sequence AGAACA. We detected GR bound to the Defb1 proximal promoter in Dex-treated vs. untreated wt MPKs, as shown by ChIP assays (Fig. 4). These data suggest that in the presence of ligand, GR may transactivate Defb1 by its recruitment to a GBS half-site.
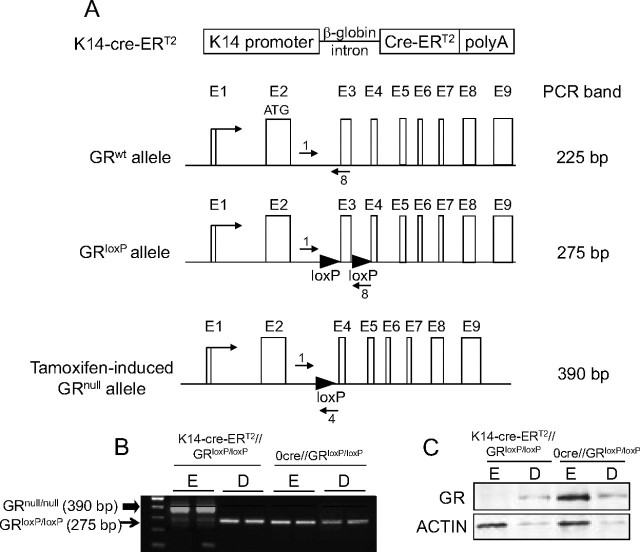
Studies of the role of GR in gene regulation and homeostasis in adult skin were not feasible due to the perinatal death of GR−/− mice. Therefore, we generated mice with an inducible conditional inactivation of GR restricted to keratinocytes. This model also allowed us to compare the consequences of GR inactivation restricted to the epidermis with that of the complete knockout. We crossed mice with both gr alleles floxed [GRloxP/loxP (35)] with mice expressing a Cre recombinase-ER fusion protein under the control of the keratinocyte-specific keratin 14 promoter [K14-cre-ERT2 (36)] (Fig. 5A). To induce activation of Cre, the resulting progeny, K14-cre-ERT2//GRloxP/loxP and 0-cre-ERT2//GRloxP/loxP littermates, were treated with tamoxifen (TAM) at 8 wk of age (37). Recombination was tested in the epidermis and dermis from tail biopsies by PCR (Fig. 5B). Our data demonstrate that Cre-mediated recombination was specifically detected in the epidermis of TAM-treated K14-cre-ERT2//GRloxP/loxP and not 0-cre-ERT2//GRloxP/loxP mice, as shown by the appearance of the null band (390 bp), whereas recombination did not occur in the dermis (Fig. 5B). Accordingly, we detected loss of the GR protein only in the epidermis of TAM- treated K14-cre-ERT2//GRloxP/loxP mice, as shown by immunoblotting (Fig. 5C).
Fig. 5.
Generation of mice with inducible GR loss-of-function restricted to adult keratinocytes (K14-cre-ERT2//GRloxP/loxP mice). A, Scheme depicting the structure of K14-Cre-ERT2 and GRloxP/loxP transgenic lines. The K14-Cre-ERT2 transgene contains the human K14 promoter, the rabbit β-globin intron, the Cre-ERT2 coding sequence, and the SV40 polyadenylation (polyA) signal. The GRloxP allele was generated by introducing two loxP sequences in the second and third introns. In the double transgenic K14-cre-ERT2//GRloxP/loxP mice treated with TAM, the recombinase under the control of the K14 promoter deletes the DNA fragment flanked by the loxP sites, rendering a GRnull allele exclusively in keratinocytes. The size of the PCR products corresponding to the GRwt (225 bp), GRloxP (275 bp), and GRnull (390 bp) alleles is indicated at the right. B, K14-cre-ERT2//GRloxP/loxP and 0-cre-ERT2//GRloxP/loxP adult mouse littermates were treated with TAM, and keratinocyte-specific recombination was assessed in epidermis (E) and dermis (D) from tail biopsies. Recombination was exclusively detected in the epidermis of TAM-treated K14-cre-ERT2//GRloxP/loxP and not 0-cre-ERT2//GRloxP/loxP mice. C, Immunoblotting using specific anti-GR antibody demonstrates the absence of GR protein specifically in the epidermis of K14-cre-ERT2//GRloxP/loxP mice. Actin was used as a loading control.
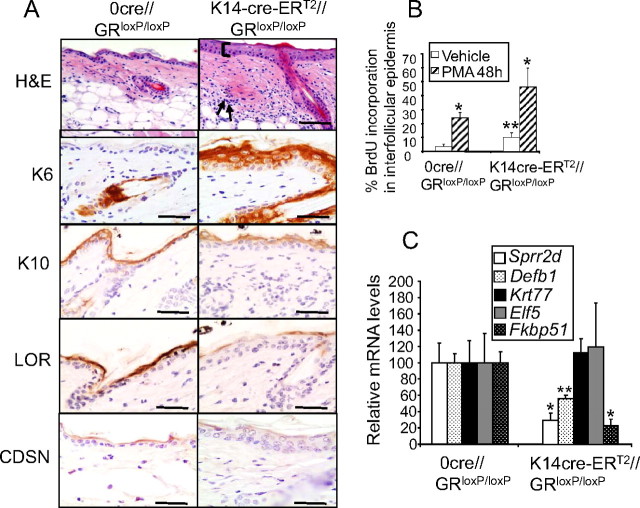
We analyzed the skin phenotype of TAM-treated K14-cre-ERT2//GRloxP/loxP mice in comparison with 0-cre-ERT2//GRloxP/loxP littermates (Fig. 6A) by hematoxylin/eosin (H&E) staining. Adult skin with keratinocyte-restricted GR loss of function was characterized by thickened epidermis and a marked dermal infiltrate (Fig. 6A, H&E). These findings demonstrate that GR is not only required for normal epidermal development but also for adult skin homeostasis, even in nonpathological conditions. A more detailed immunohistochemical analysis demonstrated that several markers of epidermal proliferation and differentiation had alterations in their expression and/or localization (Fig. 6A). The expression of keratin K6, normally restricted to the hair follicle, was up-regulated in the interfollicular epidermis of the conditional GR knockout, a feature suggestive of hyperproliferation (11). In addition, the expression of the early marker of epidermal differentiation K10 was confined to the most suprabasal layer of the epidermis of K14- cre-ERT2//GRloxP/loxP mice instead of being localized throughout all suprabasal layers. The expression of loricrin and CDSN, both markers of late epidermal differentiation, was reduced in K14-cre-ERT2//GRloxP/loxP, with a patchy appearance. To quantitate proliferation in K14-cre-ERT2//GRloxP/loxP mice, we analyzed bromodeoxyuridine (BrdU) incorporation in adult epidermis that was either untreated or treated with the phorbol ester phorbol 12-myristate 13-acetate (PMA) for 48 h (Fig. 6B). This acute treatment is known to induce hyperproliferation and inflammation when applied topically (11). The average basal rate of epidermal proliferation in adult skin is 2%, and we found a statistically significant increase of around 3-fold in mice with GR inactivation in keratinocytes (Fig. 6B). In addition, the proliferative response to PMA was more pronounced in K14-cre-ERT2//GRloxP/loxP as compared with control littermates (∼2-fold). Overall, our findings underline the key role of GR in the adult to inhibit keratinocyte proliferation both in basal and stress conditions.
Fig. 6.
Alterations in proliferation, differentiation, and gene expression of K14-cre-ERT2//GRloxP/loxP mice. A, Impaired proliferation and differentiation of adult skin with keratinocyte-restricted GR inactivation. H&E staining of adult K14-cre-ERT2//GRloxP/loxP mice showed thickened epidermis (bracket) and a marked dermal infiltrate (arrows). Immunostaining using specific antibodies against keratin 6 (K6), keratin 10 (K10), loricrin (LOR), and CDSN shows abnormal epidermal proliferation and differentiation in the conditional knockout mice. Bars, 50 μm. B, Percentage of BrdU incorporation in adult interfollicular epidermis treated with vehicle or the phorbol ester PMA for 48 h. C, Gene regulation in the absence of GR in adult keratinocytes was determined by QPCR using the epidermis of adult conditional knockouts and control tail biopsies. Coincidences and divergences in gene regulation due to the lack of GR were found in adult vs. embryonic skin. Fkbp51, Sprr2d, and Defb1 mRNA levels were strongly repressed in K14-cre-ERT2//GRloxP/loxP mice whereas no changes in Krt77 or Elf5 transcripts were detected. Statistically significant differences were assessed by Student′s t test (*, P < 0.01; **, P < 0.005).
Because the histopathology of K14-cre-ERT2//GRloxP/loxP skin demonstrated impaired proliferation and differentiation, we checked whether this phenotype correlated with the dysregulation of genes identified in our microarray study. QPCR assays using the epidermis of adult transgenic and nontransgenic tail biopsies showed that Sprr2d, Defb1, and Fkbp51 mRNA levels were strongly repressed in mice with GR inactivation restricted to keratinocytes, indicating that GR regulates several genes in embryonic and adult skin in a similar manner (Fig. 6C). In addition, our data demonstrate that the expression of these genes is specifically due to GR function in keratinocytes. However, neither Krt77 nor Elf5 transcripts exhibited statistically significant changes in adult epidermis with specific ablation of GR (Fig. 6C), suggesting that GR modulates these genes exclusively during epidermal development.
Discussion
Skin barrier function must be complete at the time of birth to preserve mammals from changes in body temperature, dehydration, and entrance of microorganisms (14, 15). Recently, we demonstrated that GR is required during embryonic development for correct epidermal stratification and formation of a competent skin barrier (12). In the present work, we aimed to investigate whether an altered transcriptional program may underlie the defects in GR−/− skin. Our microarray analysis has identified, among others, the dysregulation of genes that belong to the functional category of ectoderm/epidermis development. In the developing GR−/− skin, we found strong down-regulation of some of the major constituents of the cornified envelope (Sprr1b, Sprr2d, Sprr2h, and Cdsn), as well as repression of genes involved in epidermal terminal differentiation (Ptgs1 and Bcl6). We also detected a modest but consistent up-regulation of several genes that are key players in epithelial development, such as Egfr, Edar, and forkhead box N1 (Foxn1) (Supplemental Table 2 and Fig. 1; 1.2-, 1.2- and 1.5-fold, respectively). The highest up-regulated genes in the GR-deficient skin were Krt77 and the epithelial-specific transcription factor Elf5 (23- and 11-fold, respectively). Both genes are expressed at relatively high levels during earlier stages (E16.5) and down-regulated in later stages of epidermal development (Fig. 2A). Our findings are in agreement with previous reports indicating that Krt77 mRNA is abruptly induced at E15.5 and exclusively localized in the developing suprabasal layers of the epidermis (27). Elf5 and Bcl6 were also transiently up-regulated at E16.5, and their mRNA levels declined thereafter (Fig. 2A). The fact that Elf5 and Bcl6 were inhibited at later stages in differentiating skin yet up-regulated by high calcium in cultured keratinocytes (Fig. 2, A and B) is most likely due to the limitations of this in vitro model for keratinocyte differentiation, which is unable to fully recapitulate the physiological circumstances. Moreover, this model only accounts for the differentiation of the interfollicular epidermis, but not for other epidermal compartments such as sebaceous glands and hair follicles. This may explain the apparent discrepancies of Elf5 and possibly Bcl6 gene expression in vivo vs. in vitro.
It is known that Elf5 is also expressed in other epithelial cells and has a prominent role in mammary gland and lung morphogenesis (38, 39). Given that GR also plays key roles in the development of both tissues (6), it cannot be ruled out that ELF5 acts as a mediator of GR signaling in other epithelia in addition to epidermis. In addition, both Krt77 and Elf5 have glandular expression (40, 41). The fact that some of the identified genes are expressed in other stratified epithelia deriving from the embryonic ectoderm is in agreement with the phenotype reported in ectodermal derivatives of two GR gain-of-function transgenic mouse models (10, 11). In this regard, our data showing Krt12 mRNA in the developing epidermis instead of exclusively restricted to cornea epithelium (23) indicates a wider and developmentally regulated expression pattern of this keratin.
In a previous report that analyzed the transcriptomic profile of cultured human keratinocytes in response to Dex, the authors proposed that GCs may have a dual effect on epidermal differentiation by promoting the late stages of terminal differentiation while simultaneously inhibiting the early stages (20). The authors reported that Dex up-regulates genes involved in stratum corneum formation, such as transglutaminase 1, filaggrin, and Cdsn. Their results fit with the opposite regulation of these genes that we detect in vivo in GR−/− skin. The augmented expression of Krt77, Elf5, and Bcl6 in GR null skin (Fig. 1) can be explained either by the lack of inhibition of early markers of keratinocyte differentiation and/or reflect a delay in epidermal development of these mice, because these transcripts peak at early embryonic stages (E15.5–E16.5). However, given our observations of regulation of Krt77 and Elf5 by Dex in vitro, a lack of GR-mediated inhibition of these markers likely contributes to their overexpression in the GR−/− keratinocytes.
A study by Patel and colleagues (21) reported that exposure of wt early embryos (E15.5) to Dex in utero accelerated epidermal barrier formation by approximately 12 h relative to untreated embryos. It was described that the transcriptional profile of Dex-treated skin was significantly changed only at this time point, when the skin is still immature, and not in subsequent developmental stages. Based on these data, the authors postulated that GC actions seem to be crucial for epidermal barrier acquisition in a critical developmental window (E15– E15.5), before the observed accelerated barrier formation (21). In contrast, we have detected transcriptional changes in GR−/− embryonic skin at E18.5, suggesting a broader field of competence of GR action. Several of the genes described as corticosteroid-regulated in E15.5 Dex-treated embryos are coincident with genes identified in our microarray and are regulated in an opposite manner relative to GR-deficient skin. These genes include the keratinocyte differentiation markers Cdsn and Ptgs1 and the inflammatory cytokine Il-18 (Fig. 1 and Supplemental Table 1). However, many other genes identified by Patel et al. do not overlap with our data set. The different transcriptomic signatures may be explained by the diverse experimental settings, including the embryo developmental stage (E18.5 vs. E15.5) and/or the complete ablation of GR vs. the use of pharmacological doses of GCs. The advantage of our analysis of the transcriptional profile of GR−/− developing skin is that it provides a relevant biological setting to identify GR transcriptional targets in vivo. In fact, this analysis has allowed us to unveil genes previously unknown as GR-regulated in keratinocytes.
There are several mouse models with impaired barrier formation showing an aberrant expression of epidermal differentiation markers, including members of the SPRR family, K1, loricrin, filaggrin, involucrin, repetin, and the transcriptional regulator Krüppel-like factor 4 (KLF4) (22). Although we found that the mRNA levels of loricrin, filaggrin, involucrin, and repetin, components of the so-called epidermal differentiation complex, were reduced in GR−/− skin with statistical significance (fold change: 0.7, 0.22, 0.49, 0.048, respectively; P < 0.01); these genes were excluded from our list with a more restrictive cutoff (FDR < 0.05).
The histological evaluation of GR null skin did not show a phenotype compatible with a chronic inflammatory state and, accordingly, the genes of inflammatory and immune responses were not overrepresented in our data set (Supplemental Table 1). The cytokine IL18 has been shown to play a role in human keratinocyte survival (42), which is in agreement with its strong repression in GR null skin, which has increased keratinocyte apoptosis (12). Defb1, a gene related to the innate immune response and epithelial defense, was also repressed in GR−/− skin. This contrasts with other mouse models with defective epidermal barriers, including knockout mice for the gene-encoding transcription factors Gata-3 and Klf4, in which the innate immune response was activated by up-regulation of different antimicrobial peptides (Defb1 in the Gata-3 null and Defb-3 and -6 in the Klf4 null), most likely as a compensatory mechanism (43). In addition to its role in the innate immune skin response, it was shown that overexpression of Defb1 in vitro promotes terminal differentiation of a keratinocyte cell line (44). The decreased Defb1 mRNA expression and reduced terminal differentiation observed in the embryonic GR−/− and the adult K14-cre-ERT2//GRloxP/loxP skin suggest that this gene may also play a role in keratinocyte differentiation in vivo. Recently, it has been postulated that the loss of human Defb1 function contributes to the malignant progression of oral squamous cell carcinoma (45). Although human Defb1 was shown to be induced by Dex in airway epithelial cultures (46), to our knowledge this gene has not been shown to be regulated by GR in keratinocytes. Similarly, Fkbp51 is known as a classical GR-target in other cell types (3) and described as the highest up-regulated gene in follicular stem cells of adult transgenic K5-GR mice (28). However, it was not previously reported as a direct GR transcriptional target in keratinocytes (Fig. 4). Our findings that Defb1 and Fkbp51 are regulated by GR in GR−/− embryonic skin and adult epidermis of K14-cre-ERT2//GRloxP/loxP (Figs. 1 and 5) and that they are primary GR transcriptional targets (Fig. 4) are suggestive of an important role in the skin.
It was recently shown that most GBS in the natural gene context are represented by nonconventional GRE sites and/or composite elements that are distributed evenly through the genomic DNA (3, 31, 32). In A549 cells, the majority of GBS (63%) were found to be more than 10 kb distal from transcriptional start sites (31). This explains the difficulty of identifying GR transcriptional targets by conventional approaches searching for consensus GBS sequences in proximal promoters. In addition, the low overlap of GC-regulated genes among different cell types found by transcriptomic approaches further supports that the contribution of cell type-specific transcription factors and coregulators is crucial to modulate GR-mediated transcription. It has been reported that for GC-induced genes, conservation of a GRE sequence among species is a good predictor of GR binding (3). We first searched for putative GBS in gene sequences that were highly conserved between human and mouse. Our search retrieved no putative conserved GBS in the first intron of Defb1. However, we observed the Dex-dependent binding of GR to a sequence in the Defb1 proximal promoter (−1 kb) that contained two GRE half-sites (Fig. 4). These results suggest that ligand-bound GR can transactivate Defb1 by its recruitment to a GRE half-site, without requiring dimerization-dependent DNA binding of the receptor. The presence of AP-1 and Sp-1 sites near the GRE hexameric sequences suggests that the binding of GR is mediated through a composite element (data not shown). In this context, the fact that GR−/− skin, but not GRdim/dim, featured abnormal skin architecture (12) must be reinterpreted considering that, as previously demonstrated, several genes can be transcriptionally regulated in the GRdim/dim mice through direct binding of GR monomers to DNA (47, 48, 49).
We also assessed whether GR bound to Krt77 and Elf5 gene-regulatory sequences by ChIP assays (Fig. 4). For these experiments, we used sequences highly conserved between human and mouse (Krt77, 8/10 nucleotide conservancy; Elf5, 10/10 nucleotide conservancy). We could not detect GR binding to either of these sites, likely because these genes are GR repressed and thus, the rule of nucleotide conservation does not apply. However, it cannot be ruled out that GR binds to other genomic sequences and/or that it transcriptionally regulates these genes through transrepression mechanisms that do not require the binding of the receptor to DNA. In fact, Krt77 and Elf5 have predicted AP-1 sites located within the 1-kb region upstream of their transcriptional start sites (data not shown).
A major point of this study is the demonstration that GR is not only required for embryonic epidermal development but also for adult skin homeostasis, as shown by the increased keratinocyte proliferation and impaired differentiation of TAM-treated K14-cre-ERT2//GRloxP/loxP mice in comparison with 0-cre-ERT2//GRloxP/loxP littermates (Fig. 6, A and B). When we checked whether this adult skin phenotype correlated with the altered transcriptional profile identified in our microarray analysis using GR-deficient embryos, we found similar repression of Sprr2d, Defb1, and Fkbp51 transcripts; however, Krt77 and Elf5 transcripts were GR regulated exclusively in developing skin (Fig. 6C). The adult skin of K14-cre-ERT2//GRloxP/loxP mice showed increased dermal inflammation with numerous polymorphonuclear cells, which stained positive for an anti-Ly6G antibody (data not shown). Our data indicate that GR plays different roles in developing vs. adult skin.
Impaired epidermal barrier function can underlie inflammatory skin diseases such as psoriasis and atopic dermatitis, and topical GCs are one of the main treatments for these disorders (15, 16). Given the crucial role of GR in epidermal barrier function and skin homeostasis, we set out to determine its transcriptional targets in the skin during development. Our results show that many of the genes regulated by GR during skin development are similarly controlled in the adult tissue. Therefore, the data from this study will be useful for future research on the effects of GC treatment in normal and diseased adult skin.
Materials and Methods
Animal experimentation and treatments
GR−/−, GRloxP/loxP, and K14-Cre-ERT2 mice have been previously reported (6, 35, 36). Genotyping of the transgenic colonies was performed using DNA isolated from mouse tail biopsies and analyzed by PCR, as described elsewhere (12, 35). GR−/− and GR+/+ embryo littermates (n = 36) were obtained by cesarean derivation at 18.5 d after conception (the morning of the day that the vaginal plug was seen was considered as d 0.5). For the microarray analysis, we analyzed four embryo dorsal skin samples of each genotype and six additional skin samples were independently collected for the validation by quantitative QPCR. We also analyzed 22 skin samples from wt E16.5, E18.5, and P0 mice. K14-cre-ERT2//GRloxP/loxP male mice were crossed with 0-cre-ERT2//GRloxP/loxP females, and the resulting adult progeny were used for the experiments (n = 32). Dorsal skin was excised and either rapidly frozen in liquid nitrogen to isolate RNA and protein, fixed for immunohistochemistry, or processed for preparation of mouse primary keratinocytes (MPKs) as described below.
Mice were housed in standard temperature and humidity conditions, and animal experimentation was conducted with accepted standards of humane animal care in our animal facility (Instituto de Biomedicina de Valencia IBV-CSIC). Experiments were performed in accordance with the current Spanish and European normative that governs research with animals (Real Decreto 1201/2005, B.O.E. no. 252, October 10, 2005 and Convenio Europeo 1–2-3 del 18/3/1986).
PMA (Sigma, St. Louis, MO) was applied topically as a single dose of 8 μg/mouse for 48 h. For tamoxifen (TAM)-induced recombination, K14-cre-ERT2//GRloxP/loxP and 0-cre-ERT2//GRloxP/loxP adult mice (8-wk) were ip injected with 0.2 mg TAM diluted in corn oil (C8267, Sigma; 100 μl/mouse) daily for five consecutive days, recombination was tested in tail epidermis biopsies by PCR 10 d after the last injection using specific primers (35).
RNA isolation and microarray analysis
Total RNA was isolated from dorsal skin of mice of the indicated genotypes and corresponding control littermates (at least four animals of each genotype) or from cultured keratinocytes (at least three replicates were used per experimental group) using Trizol reagent (Invitrogen, Carlsbad, CA), following manufacturer′s recommendations. The differential transcriptomic profile of GR−/− vs. GR+/+ E18.5 skin was analyzed using four microarrays (Affymetrix GeneChip Mouse Genome 430 2.0) per genotype. RNA was purified by standard procedures (QIAGEN, Valencia, CA) and its integrity tested by the Bioanalyzer 2100 (Agilent Technologies, Palo Alto, CA). Microarray hybridization, data normalization, and analysis were carried on the genomic platform at the Centro de Investigación del Cáncer de Salamanca, as described on their webpage (http://ubioinfo. cicancer.org). The quality control for microarray hybridization includes an estimation of the signal detected for each probeset using the algorithm RMA (Robust Microarray Analysis), performed in three steps: 1) background correction, 2) quantile normalization and 3) summarized estimation by probeset. The values resulting from this algorithm are read as absolute expression values in a logarithmic scale (log 2). To estimate the DEGs with statistically significant value between control and transgenic samples, the algoritm SAM (Significance Analysis of Microarrays) was used. SAM is a modified test of differential contrast that includes the discrimination of the P values by permutations and has an adjusted P value by multiple tests using the False Discovery Rate or FDR (50). The FDR cutoff uses the δ value (d) of the algoritm. Each δ is associated to a given FDR and a number of significant genes (http://ubioinfo.cicancer. org). The gene list with the 442 DEGs with statistical significance (FDR < 0.5) in GR−/− vs. GR+/+ is in Supplemental Table 1. Functional clustering was performed according to the Gene Ontology (GO) category: biological process using the tool set Babelomics v.3.1 (http://www.babelomics.org/) (Supplemental Fig. 1). Genes that belong to the functional category ectoderm/epidermis development appear in Supplemental Table 2.
QPCR
Total RNA (1 μg) was reverse transcribed by using oligo-dT (Fermentas, Inc., Burlington, Ontario, Canada) followed by QPCR using specific oligonucleotides for each of the genes tested and FastStart Universal SYBR Green Master ROX (Roche, Indianapolis, IN) in an Applied Biosystems 7500 Fast real time PCR system (Applied Biosystems, Foster City, CA). Oligonucleotide sequences are in Supplemental Table 3. At least three biological replicates were used for each experimental group (mouse skin or cultured keratinocytes), and technical triplicates were assessed to calculate the mean value ± sd. Statistical significance was calculated using Student’s t test, P < 0.05.
Antibodies
Polyclonal antibodies to GR (sc-1004; Santa Cruz Biotechnology, Inc., Santa Cruz, CA) and an antibody specific for actin (A-2066, Sigma Chemical Co.) were used for immunoblotting. Secondary peroxidase-conjugated antirabbit antibody was from Amersham (Aylesbury, UK), and secondary peroxidase-conjugated antimouse antibody was from Jackson ImmunoResearch Laboratories, Inc. (West Grove, PA).
For imunohistochemistry, we used anti-BCL6 (sc-858) and anti-ELF5 (sc-9645) from Santa Cruz, anti-PTGS1 (160109) from Cayman Chemical (Ann Arbor, MI), and the monoclonal antibody F28-27 specific for CDSN [a gift from Serre′s laboratory, Centre National de la Recherche Scientifique (51)]. The antikeratin K5 (PRB-160P), K6 (PRB-169P), K10 (PRB-159P), and loricrin (PRB-145P) antibodies were from Covance (Babco, Berkeley, CA). Secondary biotin-conjugated antirabbit or antimouse antibodies were from Jackson ImmunoResearch Laboratories, Inc. (West Grove, PA).
Histological and immunohistochemical analysis
Skin samples were fixed in 4% paraformaldehyde or 70% ethanol and embedded in paraffin. Consecutive 4 μm-thick sections were obtained. For histopathology, sections were stained with H&E. Before immunostaining, paraffin sections were dewaxed and microwaved in 10 mm citrate solution. For immunohistochemistry, paraffin sections were blocked with 5% fetal bovine serum and then incubated with the primary antibody for at least 1 h. Slides were washed three times with PBS and then incubated with conjugated secondary antibodies for 1 h. The reaction was visualized with the Avidin-Biotin-Complex (ABC) kit from DAKO (Vectastain Elite; Vector Laboratories, Inc, Burlingame, CA) using diamino-benzidine as chromogenic substrate for peroxidase. Slides were mounted and analyzed by light microscopy (Leica DM RXA2; Leica Corp., Deerfield, IL), and images were taken at 40× magnification.
In vivo epidermal BrdU labeling
Epithelial cell proliferation was measured by ip injection of BrdU (130 μg/g of body weight, Roche Diagnostics GmbH, Mannheim, Germany) 1 h before animals were killed. BrdU incorporation was detected by immunohistochemistry of paraffin-embedded sections using a mouse anti-BrdU monoclonal antibody (Roche). The number of BrdU-positive cells and the number of total cells were determined per 200 μm of interfollicular epithelium in each section. Experiments were performed at least in five individuals of each genotype, and statistical significance was assessed by using Student’s t test, (P < 0.05).
MPK isolation, culture, and treatments
Skin of E18.5 embryos was incubated in 0.25% trypsin at 4 C overnight. The epidermis was separated from the dermis, minced, and homogenized in complete low calcium medium. The filtered solution contained MPKs, which were collected by centrifugation. MPKs were pooled (two mice of each genotype), and 106 cells were plated into one 35-mm diameter collagen I-coated tissue culture dish (BD Biosciences, Palo Alto, CA) and cultured at 37 C in standard medium. After 24 h, the medium was replaced with low calcium medium, and cells were grown until subconfluency. The composition of standard medium was: Essential MEM (EMEM; (BioWhitakker, Inc., Walkersville, MD), supplemented with 4% fetal calf serum (BioWhitakker, Inc.) plus 0.6 mm CaCl2 and antibiotics. To prepare low-calcium medium, fetal calf serum was depleted of divalent cations by treatment with Chelex deionizing resin (Bio-Rad, Hempstead, UK) and supplemented with CaCl2 to a final concentration of 0.05 mm. EGF (Sigma) (10 ng/ml) and antibiotics were added to growth medium.
To assess in vitro differentiation, equal numbers of MPKs were plated and grown to confluency under low calcium (0.05 mm) conditions and then shifted to high calcium (1.2 mm) for 24 h or 48 h.
Dex (100 nm; Sigma;), RU486 (1 μm; BIOMOL Research Laboratories, Inc., Plymouth Meeting, PA), CHX (2.5 μg/ml; Sigma), or vehicle was added for the indicated times to confluent wt MPKs that had been incubated in charcoal-stripped serum overnight to deplete steroid hormones.
Immunoblotting
Whole-cell extracts (20 μg) were prepared as previously described (12), boiled in Laemmli buffer, and separated on 10% sodium dodecyl sulfate-polyacrylamide gels, and then transferred to nitrocellulose filters (Hybond ECL, Amersham, Aylesbury, UK). The membranes were stained with Ponceau S (Sigma) to verify equal protein loading and transfer. Filters were blocked with 5% nonfat dry milk in PBS-0.1% Tween 20 at 4 C overnight, washed three times in PBS-0.1% Tween 20, and incubated with the indicated antibodies. After washing, membranes were incubated with a peroxidase-conjugated secondary antibody (Amersham), washed again, and analyzed using the enhanced chemiluminescence method (Amersham), according to manufacturer’s recommendations. Experiments were performed in at least three individuals of each genotype.
Chromatin immunoprecipitations
ChIPs were performed using the ChIP assay kit from Millipore Corp. (Billerica, MA) following manufacturer’s instructions, with slight modifications. MPKs were seeded at a density of 750,000 cells per 35-mm dish and grown to approximately 85% confluency in low calcium medium, as indicated above. After overnight incubation in low-calcium medium containing charcoal-stripped serum, cells were treated in the presence or absence of Dex (100 nm) for 2 h. Keratinocytes were cross linked by incubation with 1% formaldehyde for 10 min at room temperature. Chromatin shearing was performed in Eppendorf tubes using a Bioruptor sonicator (Diagenode, Liege, Belgium) set to HI for 12 min with 30 sec on/off cycles. Sonicated chromatin was immunoprecipitated with 2 μg of either rabbit IgG (Sigma) or rabbit anti-GR (sc-1004; Santa Cruz Biotechnology). After ChIP, reversion of cross-linking, and treatment with proteinase K and RNase, DNA was purified using a DNA clean-up spin kit (Genomed, Löhne, Germany). PCR was performed with primers specific for regions containing putative GBS (Supplemental Table 4).
Alibaba 2.1 was used to search for putative GBS (http://www.gene-regulation.com/pub/programs.html#alibaba2). Conservation between species was assessed using the Vista Genome Browser (http://pipeline.lbl.gov/cgi-bin/gateway2). The primers used for ChIP assays were designed using Primer BLAST (http://www.ncbi.nlm.nih.gov/primer-blast).
Acknowledgments
We thank Dr. Serre, Centre National de la Recherche Scientifique, for the gift of anti-CDSN antibody. We acknowledge Fátima Riveiro and Silvia Fuentes for histological work and also thank S. García-Palomares for collection of skin samples from early embryos and newborn mice. We are grateful to Professor Günther Schütz and Jan Tuckermann for providing us with GR−/−, K14Cre-ERT2, and GRloxP/loxP mice.
NURSA Molecule Pages:
Ligands: Dexamethasone | RU486;
Nuclear Receptors: GR.
Footnotes
This work was supported by Grant SAF2008-00540 of the Ministerio de Ciencia e Innovación from the Spanish government and Fundación Ramón Areces (050507070007). L.M.S. is recipient of a contract, JAE-Doc, cofunded by European Funds, and V.L. holds a fellowship from the Ministerio de Ciencia e Innovación (BES-2009-021944).
Disclosure Summary: The authors have nothing to disclose
First Published Online September 29, 2010
L.M.S. and P.B. contributed equally to this work.
Abbreviations: AP-1, Activator protein 1; BrdU, bromodeoxyuridine; CDSN, corneodesmosin; ChIP, chromatin immunoprecipitation; CHX, cycloheximide; DEGs, differentially expressed genes; Dex, dexamethasone; E18.5, embryonic d 18.5; ELF5, E74-like factor 5; FDR, false discovery rate; GBS, GR-binding sites; GC, glucocorticoid; GR, glucocorticoid receptor; GRE, GC response element; H&E, hematoxylin/eosin; KLF4, Krüppel-like factor 4; MPK, mouse primary keratinocyte; PMA, phorbol 12-myristate 13-acetate; P0, newborn; QPCR, quantitative RT-PCR; TAM, tamoxifen; wt, wild type.
References
- 1.Nicolaides NC, Galata Z, Kino T, Chrousos GP Charmandari E2010. The human glucocorticoid receptor: molecular basis of biologic function. Steroids 75:1–12 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Revollo JR, Cidlowski JA2009. Mechanisms generating diversity in glucocorticoid receptor signaling. Ann NY Acad Sci 1179:167–178 [DOI] [PubMed] [Google Scholar]
- 3.So A, Chaivorapol C, Bolton EC, Li H, Yamamoto KR2007. Determinants of cell- and gene-specific transcriptional regulation by the glucocorticoid receptor. PLoS Genet 23:927–938 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.De Bosscher K, Vanden berghe W, Haegeman G2003. The interplay between the glucocorticoid receptor and nuclear factor-kB or activator protein-1: molecular mechanisms for gene repression. Endocr Rev 24:488–522 [DOI] [PubMed] [Google Scholar]
- 5.Davies L, Karthikeyan N, Lynch JT, Sial EA, Gkourtsa A, Demonacos C, Krstic-Demonacos M2008. Cross talk of signaling pathways in the regulation of the glucocorticoid receptor function. Mol Endocrinol 22:1331–1344 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Cole TJ, Blendy JA, Monaghan AP, Krieglstein K, Schmid W, Aguzzi A, Fantuzzi G, Hummler E, Unsicker K, Schütz G1995. Targeted disruption of the glucocorticoid receptor gene blocks adrenergic chromaffin cell development and severely retards lung maturation. Genes Dev 9:1608–1621 [DOI] [PubMed] [Google Scholar]
- 7.Wintermantel TM, Berger S, Greiner EF, Schütz G2004. Genetic dissection of corticosteroid receptor function in mice. Horm Metab Res 36:387–391 [DOI] [PubMed] [Google Scholar]
- 8.De Bosscher K, Haegeman G2009. Minireview: Latest perspectives on antiinflammatory actions of glucocorticoids. Mol Endocrinol 23:281–291 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Pérez P, Page A, Bravo A, Del Río M, Giménez-Conti I, Budunova L, Slaga TJ, Jorcano JL2001. Altered skin development and impaired proliferative and inflammatory responses in transgenic mice overexpressing the glucocorticoid receptor. FASEB J 15:2030– 2032 [DOI] [PubMed] [Google Scholar]
- 10.Cascallana JL, Bravo A, Donet E, Leis H, Lara MF, Paramio JM, Jorcano JL, Pérez P2005. Ectoderm-targeted overexpression of the glucocorticoid receptor induces hypohidrotic ectodermal dysplasia. Endocrinology 146:2629–2638 [DOI] [PubMed] [Google Scholar]
- 11.Donet E, Bosch P, Sanchis A, Bayo P, Ramírez A, Cascallana JL, Bravo A, Pérez P2008. Transrepression function of the glucocorticoid receptor regulates eyelid development and keratinocyte proliferation but is not sufficient to prevent skin chronic inflammation. Mol Endocrinol 22:799–812 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Bayo P, Sanchis A, Bravo A, Cascallana JL, Buder K, Tuckermann J, Schütz G, Pérez P2008. Glucocorticoid receptor is required for skin barrier competence. Endocrinology 149:1377–1388 [DOI] [PubMed] [Google Scholar]
- 13.Fuchs E2007. Scratching the surface of skin development. Nature 445:834–842 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.O′Shaughnessy RF, Christiano AM2004. Inherited disorders of the skin in human and mouse: from development to differentiation. Int J Dev Biol 48:171–179 [DOI] [PubMed] [Google Scholar]
- 15.Segre JA2006. Epidermal barrier formation and recovery in skin disorders. J Clin Invest 116:1150–1158 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Palmer CN, Irvine AD, Terron-Kwiatkowski A, Zhao H, Liao H, Lee SP, Goudie DR, Sandilands A, Campbell LE, Smith FJ, O'Regan GM, Watson RM, Cecil JE, Bale SJ, Compton JG, DiGiovanna JJ, Fleckman P, Lewis-Jones S, Arseculeratne G, Sergeant A, Munro CS, El Houate B, McElreavey K, Halkjaer LB, Bisgaard H, Mukhopadhyay S, McLean WH2006. Common loss-of-function variants of the epidermal barrier protein filaggrin are a major predisposing factor for atopic dermatitis. Nat Genet 38:441–446 [DOI] [PubMed] [Google Scholar]
- 17.Elias P, Ahn S, Brown B, Crumrine D, Feingold KR2002. Origin of the epidermal calcium gradient: regulation by barrier status and role of active vs passive mechanisms. J Invest Dermatol 119:1269–1274 [DOI] [PubMed] [Google Scholar]
- 18.Elias PM, Ahn SK, Denda M, Brown BE, Crumrine D, Kimutai LK, Kömüves L, Lee SH, Feingold KR2002. Modulations in epidermal calcium regulate the expression of differentiation-specific markers. J Invest Dermatol 119:1128–1136 [DOI] [PubMed] [Google Scholar]
- 19.Hennings H, Michael D, Cheng C, Steinert P, Holbrook K, Yuspa SH1980. Calcium regulation of growth and differentiation of mouse epidermal cells in culture. Cell 19:245–254 [DOI] [PubMed] [Google Scholar]
- 20.Stojadinovic O, Lee B, Vouthounis C, Vukelic S, Pastar I, Blumenberg M, Brem H, Tomic-Canic M2007. Novel genomic effects of glucocorticoids in epidermal keratinocytes. J Biol Chem 282:4021–4034 [DOI] [PubMed] [Google Scholar]
- 21.Patel S, Xi ZF, Seo EY, McGaughey D, Segre JA2006. Klf4 and corticosteroids activate an overlapping set of transcriptional targets to accelerate in utero epidermal barrier acquisition. Proc Natl Acad Sci USA 103:18668–18673 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Candi E, Schmidt R, Melino G2005. The cornified envelope: a model of cell death in the skin. Nat Rev 6:328–340 [DOI] [PubMed] [Google Scholar]
- 23.Tanifuji-Terai N, Terai K, Hayashi Y, Chikama T, Kao W2006. Expression of keratin 12 and maturation of corneal epithelium during development and postnatal growth. Investigative Ophthalmol Visual Sci 47:545–551 [DOI] [PubMed] [Google Scholar]
- 24.Schneider MR, Werner S, Paus R, Wolf E2008. Beyond wavy hairs: the epidermal growth factor receptor and its ligands in skin biology and pathology. Am J Pathol 173:14–24 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Yoshida T, Fukuda T, Okabe S, Hatano M, Miki T, Hirosawa S, Miyasaka N, Isono K, Tokuhisa T1996. The BCL6 gene is predominantly expressed in keratinocytes at their terminal differentiation stage. Biochem Biophys Res Commun 228:216–220 [DOI] [PubMed] [Google Scholar]
- 26.Tummala R, Sinha S2006. Differentiation-specific transcriptional regulation of the ESE-2 gene by a novel keratinocyte-restricted factor. J Cell Biochem 97:766–781 [DOI] [PubMed] [Google Scholar]
- 27.Bazzi H, Fantauzzo KA, Richardson GD, Jahoda CAB, Christiano AM2007. Transcriptional profiling of developing mouse epidermis reveals novel patterns of coordinated gene expression. Dev Dynam 236:961–970 [DOI] [PubMed] [Google Scholar]
- 28.Chebotaev D, Yemelyanov A, Zhu L, Lavker RM, Buduova I2007. The tumor suppressor effect of the glucocorticoid receptor in skin is mediated via its effect on follicular stem cells. Oncogene 26:3060–3068 [DOI] [PubMed] [Google Scholar]
- 29.Leclerc EA, Huchenq A, Mattiuzzo NR, Metzger D, Chambon P, Ghyselinck NB, Serre G, Jonca N, Guerrin M2009. Corneodesmosin gene ablation induces lethal skin barrier disruption and hair-follicle degeneration related to desmosome dysfunction. J Cell Sci 122:2699–2709 [DOI] [PubMed] [Google Scholar]
- 30.Tiano HF, Loftin CD, Akunda J, Lee CA, Spalding J, Sessoms A, Dunson DB, Rogan EG, Morham SG, Smart RC, Langenbach R2002. Deficiency of either cyclooxygenase (COX)-1 or COX-2 alters epidermal differentiation and reduces mouse skin tumorigenesis. Cancer Res 62:3395–3401 [PubMed] [Google Scholar]
- 31.Parikh N, Nagarajan P, Sei-Ichi M, Sinha S, Garrett-Sinha LA2008. Isolation and characterization of an immortalized oral keratinocyte cell line of mouse origin. Arch Oral Biol 53:1091–1100 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Choi YS, Cheng J, Segre J, Sinha S2008. Generation and analysis of Elf5-LacZ mouse: unique and dynamic expression of Elf5 (ESE-2) in the inner root sheath of cycling hair follicles. Histochem Cell Biol 129:85–94 [DOI] [PubMed] [Google Scholar]
- 33.So AY, Cooper SB, Feldman BJ, Manuchehri M, Yamamoto KR2008. Conservation analysis predicts in vivo occupancy of glucocorticoid receptor-binding sequences at glucocorticoid-induced genes. Proc Natl Acad Sci USA 105:5745–5749 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Meijsing SH, Pufall MA, So A, Bates DL, Chen L, Yamamoto KR2009. DNA binding site sequence directs glucocorticoid receptor structure and activity. Science 24:407–410 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Tronche F, Kellendonk C, Kretz O, Gass P, Anlag K, Orban PC, Bock R, Klein R, Schütz G1999. Disruption of the glucocorticoid receptor gene in the nervous system results in reduced anxiety. Nat Genet 23:99–103 [DOI] [PubMed] [Google Scholar]
- 36.Metzger D, Indra AK, Li M, Chapellier B, Calleja C, Ghyselinck NB, Chambon P2003. Targeted conditional somatic mutagenesis in the mouse: temporally-controlled knock out of retinoid receptors in epidermal keratinocytes. Methods Enzymol 364:379–408 [DOI] [PubMed] [Google Scholar]
- 37.Tuckermann JP, Kleiman A, Moriggl R, Spanbroek R, Neumann A, Illing A, Clausen BE, Stride B, Förster I, Habenicht AJ, Reichardt HM, Tronche F, Schmid W, Schütz G2007. Macrophages and neutrophils are the targets for immune suppression by glucocorticoids in contact allergy. J Clin Invest 117:1381–1390 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Choi YS, Chakrabarti R, Escamilla-Hernandez R, Sinha S2009. Elf5 conditional knockout mice reveal its role as a master regulator in mammary alveolar development: failure of Stat5 activation and functional differentiation in the absence of Elf5. Dev Biol 329:227–241 [DOI] [PubMed] [Google Scholar]
- 39.Metzger DE, Stahlman MT, Shannon JM2008. Misexpression of ELF5 disrupts lung branching and inhibits epithelial differentiation. Dev Biol 320:149–160 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Langbein L, Cribier B, Schirmacher P, Praetzel-Wunder S, Peltre B, Schweizer J2008. New concepts on the histogenesis of eccrine neoplasia from keratin expression in the normal eccrine gland, syringoma and poroma. Br J Dermatol 159:633–645 [DOI] [PubMed] [Google Scholar]
- 41.Lapinskas EJ, Palmer J, Ricardo S, Hertzog PJ, Hammacher A, Pritchard MA2004. A major site of expression of the ets transcription factor Elf5 is epithelia of exocrine glands. Histochem Cell Biol 122:521–526 [DOI] [PubMed] [Google Scholar]
- 42.Hosotani Y, Kashiwamura S, Kimura-Shimmyo A, Sekiyama A, Ueda H, Ikeda T, Mimura O, Okamura H2008. Interleukin-18 prevents apoptosis via PI3K/Akt pathway in normal human keratinocytes. J Dermatol 35:514–524 [DOI] [PubMed] [Google Scholar]
- 43.de Guzman-Strong C, Wertz PW, Wang C, Yang F, Meltzer PS, Andl T, Millar SE, Ho IC, Pai SY, Segre JA2006. Lipid defect underlies selective skin barrier impairment of an epidermal-specific deletion of Gata-3. J Cell Biol 175:661–670 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Frye M, Bargon J, Gropp R2001. Expression of human βdefensin-1 promotes differentiation of keratinocytes. J Mol Med 79:275–282 [DOI] [PubMed] [Google Scholar]
- 45.Wenghoefer M, Pantelis A, Dommisch H, Reich R, Martini M, Allam JP, Novak N, Bergé S, Jepsen S, Winter J2008. Decreased gene expression of human β-defensin-1 in the development of squamous cell carcinoma of the oral cavity. Int J Oral Maxillofac Surg 37:660–663 [DOI] [PubMed] [Google Scholar]
- 46.Starner TD, Agerberth B, Gudmundsson GH, McCray Jr PB2005. Expression and activity of β-defensins and LL-37 in the developing human lung. J Immunol 174:1608–1615 [DOI] [PubMed] [Google Scholar]
- 47.Reichardt HM, Kaestner KH, Tuckermann J, Kretz O, Wessely O, Bock R, Gass P, Schmid W, Herrlich P, Angel P, Schütz, G1998. DNA binding of the glucocorticoid receptor is not essential for survival. Cell 93:531–541 [DOI] [PubMed] [Google Scholar]
- 48.Adams M, Meijer OC, Wang J, Bhargava A, Pearce D2003. Homodimerization of the glucocorticoid receptor is not essential for response element binding: activation of the phenylethanolamine N-methyltransferase gene by dimerization-defective mutants. Mol Endocrinol 17:2583–2592 [DOI] [PubMed] [Google Scholar]
- 49.Rogatsky I, Wang JC, Derynck MK, Nonaka DF, Khodabakhsh DB, Ha CM, Darimont BD, Garabedian MJ, Yamamoto KR2003. Target-specific utilization of transcriptional regulatory surfaces by the glucocorticoid receptor. Proc Natl Acad Sci USA 100:13845–138450 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Benjamini Y, Hochberg Y 1995 Controlling the false discovery rate: a practical and powerful approach to multiple testing. JR Stat Soc (Ser B) 57:289–300 [Google Scholar]
- 51.Serre G, Mils V, Haftek M, Vincent C, Croute F, Réano A, Ouhayoun JP, Bettinger S., Soleilhavoup JP1991. Identification of late differentiation antigens of human cornified epithelia, expressed in re-organized desmosomes and bound to cross-linked envelope. J Invest Dermatol 97:1061–1072 [DOI] [PubMed] [Google Scholar]