Abstract
Steroid hormones regulate gene expression by interaction of their receptors with hormone-responsive elements on DNA or with other transcription factors, but they can also activate cytoplasmic signaling cascades. Rapid activation of Erk by progestins via an interaction of the progesterone receptor (PR) with the estrogen receptor is critical for transcriptional activation of the mouse mammary tumor virus (MMTV) promoter and other progesterone target genes. Erk activation leads to the phosphorylation of PR, activation of mitogen- and stress-activated protein kinase 1, and the recruitment of a complex of the three activated proteins and of P300/CBP-associated factor (PCAF) to a single nucleosome, resulting in the phosphoacetylation of histone H3 and the displacement of heterochromatin protein 1γ. Hormone-dependent gene expression requires ATP-dependent chromatin remodeling complexes. Two switch/sucrose nonfermentable-like complexes, Brahma-related gene 1-associated factor (BAF) and polybromo-BAF are present in breast cancer cells, but only BAF is recruited to the MMTV promoter and cooperates with PCAF during activation of hormone-responsive promoters. PCAF acetylates histone H3 at K14, an epigenetic mark recognized by BAF subunits, thus anchoring the complex to chromatin. BAF catalyzes localized displacement of histones H2A and H2B, facilitating access of nuclear factor 1 and additional PR complexes to the hidden hormone-responsive elements on the MMTV promoter. The linker histone H1 is a structural component of chromatin generally regarded as a general repressor of transcription. However, it contributes to a better regulation of the MMTV promoter by favoring a more homogeneous nucleosome positioning, thus reducing basal transcription and actually enhancing hormone induced transcription. During transcriptional activation, H1 is phosphorylated and displaced from the promoter. The kinase cyclin-dependent kinase 2 is activated after progesterone treatment and could catalyze progesterone-induced phosphorylation of histone H1 by chromatin remodeling complexes. The initial steps of gene induction by progestins involve changes in the chromatin organization of target promoters that require the activation of several kinase signaling pathways initiated by membrane anchored PR. Because these pathways also respond to other external signals, they serve to integrate the hormonal response in the global context of the cellular environment.
Gene induction by progestins involve changes in the chromatin organization of target promoters that require the activation of several kinase signaling pathways.
The classical picture of steroid hormone action via their nuclear receptors considered as ligand-regulated transcription factors has undergone dramatic changes in the past few years. It is becoming increasingly evident that “nuclear” hormone receptors participate in multiple interactions within different cellular compartments that are essential to fully understand the response of the cell to variations in hormone levels. In addition, other membrane-bound hormone receptors that have been identified and contribute to hormone action are still not well understood (1). These multiple interactions serve to integrate the hormonal signal into the network of default programs and external signals impinging on the cell at a given time. It is the outcome of this integration that specifies the nature, intensity, and duration of the cellular response.
Estrogen and progesterone influence a variety of functions in different target cells. Many years of attention have been focused on the transcriptional effects of these hormones, because the steroid hormone receptors (SHRs) were seen as ligand-dependent transcription factors. Upon activation by the specific hormone, SHRs were shown to interact with hormone-responsive elements (HREs) in the promoter/enhancer region of target genes. SHRs can also activate genes lacking HREs by virtue of interactions with other sequence-specific transcription factors bound to their target sequences (2).
Important insights into the mechanisms of transcriptional regulation were recently obtained with studies on estrogen receptor (ER)-driven gene expression (3, 4). By using chromatin immunoprecipitation (ChIP), it was demonstrated that, once recruited to target promoters, estradiol-bound ER induces an ordered and cyclical recruitment of coactivator complexes, some of which contain histone acetyl transferases (HATs), histone methyltransferases, or ATP-dependent remodeling activities (3, 5). After these activating complexes, one observed recruitment of components of the ubiquitin-proteasome system, displacement of the receptors, and recruitment of corepressor complexes, containing histone deacetylases (HDACs). Even CpG methylation, generally associated with stable epigenetic silencing of transcription (6), showed cyclical changes critical for transcriptional regulation (7, 8). In the mouse mammary tumor virus (MMTV) promoter, we also detect waves of receptor and factors recruitment associated with histone modification, but they are less pronounced and not repetitious (9). This is likely due to the fact that hormonal activation of the viral promoter leads to a single round of transcription (10).
Although in all hormonally regulated promoters chromatin is remodeled to facilitate transient interactions with transcription factors, much still remains to be understood regarding the role of nuclear organization and the involvement of transcription factories in the coordination of hormonal regulation of gene networks. Both intrachromosomal and interchromosomal interactions between regulatory sequences are likely complex key processes that involved enzymatic protein modifications, as recently shown for the pS2 locus region (11). Global ChIp-Seq, as recently reported for the ER (12), will be useful to start analyzing the function of these processes in the context of hormonal gene regulation.
In addition to these direct genomic effects, steroid hormones induce rapid nongenomic responses similar to those initiated by peptide growth factors (13). For example, estrogens activate the Src/p21ras/Erk and the PI3K/Akt pathways via direct interaction of the ERα with the sarcoma (Src) homology (SH)2 domain of c-Src and the regulatory subunit of PI3K, respectively (14, 15). Activation of these kinase pathways is essential for estrogen-induced cell proliferation in breast cancer cells. Progestins can also activate these signaling cascades, either via an interaction of the progesterone receptor (PR) with ERα, which itself activates c-Src and PI3K, or by direct interaction of PR with the SH3 domain of c-Src (16, 17, 18). The mechanism of this cross talk between SHRs and kinase signaling pathways has been a matter of study in the past few years.
Progestin induces phosphorylation of PR-B at several sites, including a MAPK consensus site, Ser345. Ser345-phosphorylated PR-B receptors strongly associated with the transcription factor specificity protein 1 to regulate cell cycle relevant genes (such as p21) and growth-promoting target genes, such as epidermal growth factor receptor (EGFR), whose promoters lack canonical progesterone response element (19). These events are critical for progestin-stimulated regulation of specificity protein 1 target genes and breast cancer cell proliferation (19). The importance of the Src/MAPK signaling pathway for progesterone-induced transcription has been demonstrated with a PR mutant unable to activate Src signaling. In breast cancer cells expressing this mutant PR, progesterone cannot induce cyclin D1 gene expression and does not stimulate cell cycle progression (20). Complementary studies have been performed with ER using estrogen-dendrimer conjugates, which because of their charge and size remain outside the nucleus and can only initiate extranuclear signaling (21). Genome-wide cDNA microarray analysis showed that around 25% of E2-regulated genes were estrogen-dendrimer conjugates responsive, highlighting the importance of extranuclear ER signaling pathways in regulating patterns of gene expression in breast cancer cells (22).
SHRs are localized predominantly in the nucleus, although they actively shuffle between nucleus and cytoplasm (23). However, distinct pools of functional membrane-localized SHR have been described for estrogens (24), progestins (25, 26), or androgens (27). Overall, little is understood regarding the mechanisms of translocation of SHR to the plasma membrane. A highly conserved nine amino acid motif in the ligand-binding domains of SHRs has been found to mediate their palmitoylation, which facilitates caveolin-1 association, subsequent membrane localization, and steroid signaling (28).
SHRs have been shown to interact with many additional kinase signaling pathways, including CyclinA/Cdk2, JAK/STAT, and EGF receptor. In addition to its role in cell cycle regulation, CyclinA/Cdk2 also participates in the control of the transcriptional activity of steroid receptors (29). In particular, both ERα and PR are activated by CyclinA/Cdk2 complex. The complex directly phosphorylates ERα, potentiating its transcriptional activity (30). In the case of PR, CyclinA/Cdk2 has been reported to increase expression of progesterone target genes (31, 32, 33) by a mechanism involving phosphorylation of the coactivator SRC-1 (32). It seems that in the promoters regulated by these receptors, CyclinA/Cdk2 participates in multiprotein complexes that contain transcription factors, corepressors, and coactivators, including acetyl transferases (34). Interestingly, the ability of CyclinA/Cdk2 to increase PR activity is independent of its ability to phosphorylate PR (32, 34). Because linker histones are main targets of cyclin-dependent kinase 2 (Cdk2) (35, 36), one could postulate that activation of Cdk2 by steroid hormones could influence chromatin structure and gene expression (37, 38, 39, 40).
There is also evidence that SHRs interact with the JAK/STAT pathway and can form complexes with various STAT family members (41, 42, 43). Like PR, signal transducers and activator of transcription (Stat)5a and Stat5b are required for normal mammary gland growth and differentiation. In breast cancer cells, progestin treatment induces translocation of Stat5 to the nucleus, mediated by association with PR (42). Moreover, the inhibition of lactogenic hormone induction of the β-casein gene in normal mammary epithelial cells during pregnancy involves a negative cross talk between PR and STAT5a (44). There are other examples of PR action via STAT5 binding sites, such as in the mouse Bcl-X gene (45) and in the human 11β-HSD2 gene (46).
Several studies have documented a transcriptional and/or proliferative synergy between EGF and progesterone or estrogen (47, 48). Notably, progesterone up-regulates the expression of EGFR family members on the cell surface (48, 49, 50, 51). In addition to increasing the number of high affinity EGFR per cell, progesterone affects the phosphorylation state of both EGF and c-erbB2 receptors (48). EGF signaling can mimic the actions of progestins by inducing changes in PR phosphorylation, nuclear association, and DNA binding (52). Although EGF alone failed to induce appreciable PR transcriptional activity, the actions of ligand-bound PR are dramatically enhanced in the presence of EGF (52).
In this review, we will use progestin regulation of the MMTV promoter to exemplify these issues, and we will focus on two interconnected fields within the wide area of hormonal signaling networks. First, cross talk of nuclear receptors with kinase signaling pathways; and second, chromatin remodeling. We will concentrate on the effect of progesterone on the Src/Erk/mitogen- and stress-activated protein kinase 1 (Msk1) signaling pathway and how this pathway influences chromatin remodeling and the transcriptional activity of PR. We will finish with a brief summary of possible roles of linker histone subtypes on gene expression and how they could be influenced by a cross talk of PR with kinase signaling pathways.
Cross Talk of PR with the Src/Erk/Msk Pathway Participates in Transcriptional Regulation
Traditionally, the genomic and nongenomic actions of steroid hormones have been considered as two independent pathways, but we have found that the two pathways converge in the modification of structural components of chromatin. Five minutes after progestin administration to T47D breast cancer cells, there is an increase in the activity of the components of the Src/Ras/Erk cascade, which is essential for progestin-induced cell proliferation (18). This effect is mediated by a specific interaction between two domains of the N-terminal half of PR and the ligand-binding domain of ERα, which in this situation is activated in the absence of estrogens (16). Activated ERα interacts directly with the SH2 domain c-Src (15), activating its tyrosine kinase activity and consequently initiating the entire MAP kinase cascade. Because some of the kinases that phosphorylate core histones (Msk1 and Msk2) and histone H1 (Cdk2) are downstream substrates of Erk, we hypothesized that the rapid cytoplasmic effects of steroid hormones could influence their downstream chromatin targets.
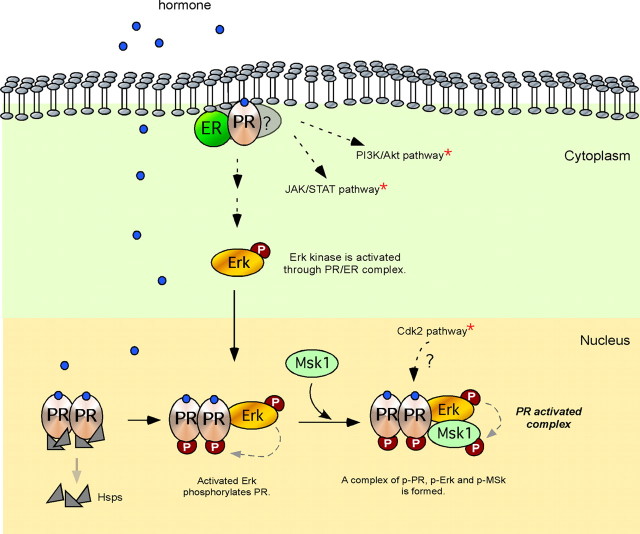
Hormone-dependent activation of the MMTV promoter is blocked by inhibition of the Erk signaling pathway in breast cancer cells (53). Because Erk phosphorylates PR at Ser294 in response to progestins (54), it is possible that the transcriptional inhibition of MMTV induction is due to a lack of PR phosphorylation. However, a similar inhibition of MMTV induction was observed when interfering with the activation of Msk1, which does not compromise PR phosphorylation (53). After 5 min of hormone addition, activated PR, Erk, and Msk1 form a ternary complex, which is selectively recruited to the MMTV promoter nucleosome containing the HREs (Fig. 1) (53). We know that the nonphosphorylated PR can also bind to the exposed HREs on the MMTV promoter nucleosome, but the binding is nonproductive and does not lead to derepression (53).
Fig. 1.
Initial steps of PR activation. Progestins bind to cytoplasmic PR/ER complexes, anchored in the cell membrane by palmitoyl residues, and activate the Src/Ras/Erk pathway, leading to nuclear accumulation of activated pErk. The majority of PR is nuclear and associated with chaperones (Hsps). Upon binding of progestins, PR homodimers dissociate from chaperones, and a fraction of PR is phosphorylated by pErk, which also phosphorylates Msk1. A “PR-activated complex” composed of pPR/pErk/pMsk1 is formed. Progesterone induction also activates other kinase signaling pathways as Janus kinase (JAK)/Stat, phosphatidylinositol kinase (PI3K)/serine-threonine kinase (Akt), and Cdk2 (red asterisk).
Eukaryotic DNA is packaged into chromatin through its association with histone proteins. The nucleosome core particle consists of 146 bp wrapped around a histone octamer consisting of two copies each of the core histone proteins H2A, H2B, H3, and H4. Concomitant with the recruitment of the ternary complex of phospho (p) PR/pErk/pMsk1 to the MMTV promoter, histone H3 becomes phosphorylated at serine 10 and acetylated at lysine 14, only on the nucleosome containing the HREs and not on adjacent nucleosomes (Fig. 2, middle panel) (53). Phosphoacetylation of histone H3 can be blocked by inhibiting Erk or Msk1 activation resulting in a marked reduction of MMTV promoter activation by hormone. Blocking H3 phosphoacetylation precludes displacement of a repressive complex containing HP1γ, as well as the recruitment of the Brg1-containing chromatin remodeling complex, thus preventing displacement of histone H2A/H2B dimers and subsequent promoter activation.
Fig. 2.
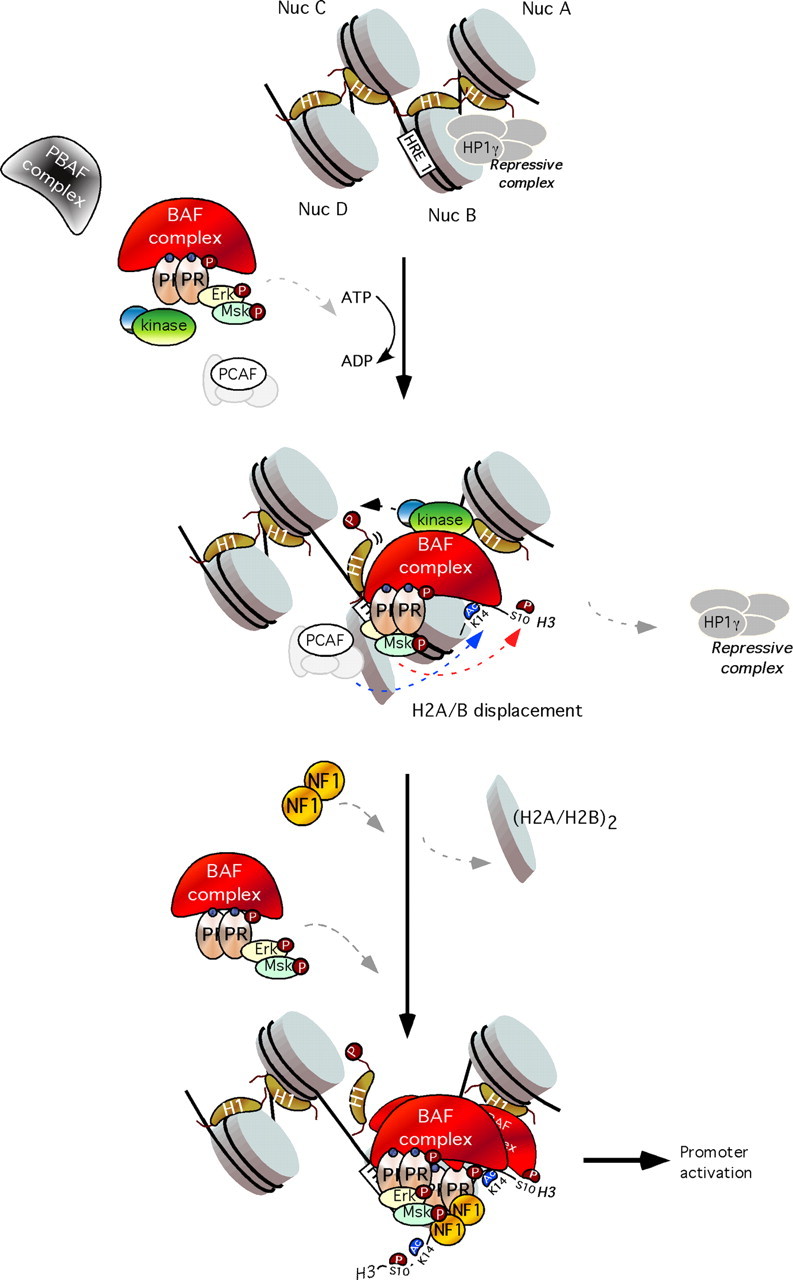
Model for the role of “PR activated complex” in chromatin. Top, In the uninduced state, an HP1γ-containing repressive complex is bound to the promoter, keeping it silent. Middle, Upon hormone induction, activated PR complexes bind BAF and P300/CRE-binding protein-binding protein-associated factor (PCAF) and recruit them to the exposed HRE1 on nucleosome B. This is followed by H3 phosphoacetylation and displacement of the repressive complex and histone H1. Bottom, The BAF complex, stabilized by PCAF-dependent H3K14 acetylation, catalyzes ATP-dependent H2A/H2B displacement and NF1 binding. This facilitates binding to the HREs 2 and 3 of further PR molecules with associated BAF, followed by other coactivators and the basal transcriptional machinery, including RNA polymerase II, leading to promoter activation. PBAF, Polybromo-associated BAF; Nuc, nucleosome.
Most reports on the rapid action of PR have focused in the cell signaling pathways activated by progestins (17, 18, 55), but how these pathways are integrated with the transcriptional function of PR has remained elusive. We have shown that some of the kinases activated by progestins in the cytoplasm phosphorylate PR and form a complex with the activated PR. The complex of activated PR and accompanying kinases is recruited to the target sites in chromatin where the kinases modify chromatin proteins locally as a prerequisite for chromatin remodeling and gene regulation. Thus, we propose that the “nongenomic” and “genomic” pathways of progestin action converge on chromatin to enable gene regulation.
Hormone-Induced ATP-Dependent Chromatin Remodeling Needs Cooperation of Various Enzymatic Activities
Modulation of the structure and dynamics of nucleosomes is an important regulatory mechanism in all DNA-based processes and is primarily catalyzed by chromatin remodeling complexes. Such complexes can either modify histone residues or use the energy of ATP hydrolysis to alter the relationship between histones and DNA (reviewed in Refs. 56, 57, 58, 59, 60). The discovery of ATP-dependent chromatin remodeling complexes has been a major breakthrough in the understanding of chromatin dynamics. There are two human switch/sucrose nonfermentable (SWI/SNF)-like complexes both containing hBRM (human Brahma) or BRG1 (Brahma-related gene 1) as ATPase, as well as a series of other subunits which differ depending on the cell type (61). The stoichiometry of the SWI/SNF subunits are tightly regulated by protein-protein interaction between BRG1-associated factor (BAF)155/BAF170 and BAF57 and by proteasome-mediated protein degradation (62).
There is evidence for a role of the SWI/SNF complex in glucocorticoid gene regulation in yeast (63) and in animal cells (64). Indeed, hSWI/SNF seems to be required for chromatin remodeling initiated by glucocorticoids (65); however, the situation appears to be different for progestins, although they act through the same HREs as glucocorticoids (65). The genome-wide binding of glucocorticoid receptor (GR) occurs mainly at constitutive or hormone-induced deoxyribonuclease (DNase) I hypersensitive sites, some of which require Brg1-containing SWI/SNF complex, whereas others are Brg1 independent (66). The H2A.Z histone variant is highly enriched at both constitutive and inducible DNase I hypersensitive sites.
The MMTV long terminal repeat region encompasses a promoter that in addition to five degenerated HREs also contains a binding site for nuclear factor 1 (NF1), located immediately downstream of the HREs. In chromatin, the MMTV-LTR is organized into positioned nucleosomes (67), with a nucleosome located over the five HREs and the NF1 binding site (68). On this promoter nucleosome, the binding site for NF1 is not accessible (69). Only two of the five HREs, the strong palindromic HRE1 and the weak half-palindrome HRE4, can be bound by hormone receptors. The central HREs, in particular the palindromic HRE2 and the half-palindrome HRE3, are not accessible for receptor binding (70). After hormone induction in vivo, all HREs and the binding site for NF1 are occupied simultaneously on the surface of a nucleosome-like structure, and a functional synergism is observed between PR and NF1 (68).
Progesterone treatment of T47D-MTVL cells carrying a single integrated copy of the MMTV-Luc transgene leads to recruitment of PR, SWI/SNF, and SNF2 h-related complexes to the MMTV promoter, accompanied by selective displacement of histones H2A and H2B from nucleosome B (71, 72). Thus, these two remodeling ATPases could be part of the complexes responsible for the changes in chromatin sensitivity to nucleases detected 30 min after hormone exposure (68). Recently, the acidic N terminus of the Swi3p subunit of yeast SWI/SNF was identified as a novel H2A/H2B-binding domain required for ATP-dependent H2A/H2B dimer displacement (73).
The ySWI/SNF complex could displace H2A and H2B from well-positioned MMTV promoter nucleosomes but not from a mouse ribosomal promoter nucleosome well positioned on DNA fragments of the same length (71). Thus, the nucleotide sequence seems to contain topological information that determines not only nucleosome positioning but also the outcome of the remodeling process.
After hormone activation in breast cancer cells, the SWI/SNF complex named BAF is recruited to the MMTV promoter, whereas the closely related PBAF complex is not (Fig. 2) (74). The BAF250a/ARID1A subunit present in BAF is essential for SWI/SNF-dependent transcriptional activation of the MMTV promoter and is a necessary facilitator of BRG1-mediated chromatin remodeling (75). Interestingly, BAF250 functions as an E3 ubiquitin ligase adapter for histone H2BK120 (76). This new finding could have important implications for gene activation, because H2BK120 ubiquitination could affect SWI/SNF chromatin remodeling (76).
Histone acetylation is a highly dynamic posttranslational modification that plays an important role in gene expression. High doses of histone deacetylase inhibitors, butyrate or TSA, lead to intense hyperacetylation of core histones and inhibit hormone induction of the MMTV promoter (77, 78) without altering nucleosome positioning (78). However, low doses of the inhibitors activate the MMTV promoter in the absence of hormone and generate a DNAse I-hypersensitive site similar to that observed after hormone treatment (77). This suggests that partial acetylation of histones, other chromatin proteins, or other factors can generate a chromatin structure similar to that induced by hormone.
Histone acetylation at the MMTV promoter in response to hormone shows an initial increase followed by an eventual net deacetylation of histone H4 (79). The histone deacetylases HDAC1 and HDAC3 are bound to the MMTV promoter before transcription activation, and their levels fluctuates after hormone treatment (79). The histone acetyltransferase PCAF is required for progestin induction of target genes and catalyzes the acetylation of histone H3 at K14. This epigenetic mark interacts with the BAF subunits anchoring the complex to chromatin (Fig. 2) (74). Thus, for full activation of the MMTV promoter, cooperation between the two chromatin remodelers BAF and PCAF is needed.
Once the BAF complex is recruited to the MMTV promoter, via interaction with PR and with acetylated H3K14, it displaces histones H2A and H2B, thus facilitating NF1 binding (Fig. 2) (74). The presence of NF1 at the promoter favors binding of PR and associated BAF molecules to the previously hidden HREs 2 and 3, not via protein-protein interaction but by exposing these HREs on the surface of an H3/H4 tetramer (80). These sites are not essential for ATP-dependent H2A/H2B displacement or NF1 binding but are critical for complete PR loading and MMTV promoter activation (80).
Fluorescence photobleaching experiments in living cells containing a tandem of 200 MMTV promoters driving reporter genes have provided direct evidence that the hormone-bound GR undergoes rapid exchange between chromatin and the nucleoplasmic compartment with a half-life of less than a minute (81). This rapid exchange has also been reported for the ER and PR (82, 83, 84), further supporting the model of a highly dynamic turnover of transcription complexes at promoters. In agreement with the difficulties in visualizing a footprint of GR on the MMTV promoter in vivo (85) is the formulation of a “hit-and-run” model of receptor action (81). This rapid turnover measured by fluorescence recovery after photobleaching (FRAP) contrasts with the results obtained by ChIP, which reveal cyclical waves of receptor association to promoters with periods of 10–90 min (3, 4, 5). It is likely that the 20-min cycles of receptors detected by ChIP do not reflect the behavior of individual receptor molecules but instead indicate changes in the configuration of the promoter that favor factor binding. It is also possible that many of the receptor interactions detected by FRAP are transient and nonproductive and that more stable binding requires interaction with other transcription factors or other chromatin components. An example of this behavior has been recently demonstrated by FRAP for GR and HMGB1 (86). Conversely, GR appears to bind to preexisting regions of open chromatin, as detected by DNase I hypersensitivity (66). Because the pattern of higher order chromatin architecture that generates DNase I sensitivity is likely established during cell differentiation, cell identity could play a major role in the determination of tissue-selective receptor function (66).
PR and NF1 Synergize on the MMTV Promoter Wrapped Around and H3/H4 Tetramer
A tetramer of histones H3 and H4 would be a feasible structure of the MMTV “remodeled nucleosome” after hormone induction. H3/H4 tetramers position MMTV promoter sequences in a similar way to histone octamers, but NF1 can bind to a H3/H4 tetramer particle with relatively high affinity (87). MMTV sequences positioned around an H3/H4 tetramer can bind PR and NF1 simultaneously reminiscent to what is observed in cells carrying a single copy of the MMTV promoter integrated in chromatin (68). Binding of PR to the MMTV tetramer particle is enhanced if NF1 is prebound, indicating that binding is cooperative. The nature of this synergism is unknown, but one possibility is that the deformation of the DNA imposed by NF1 binding debilitates the interaction of DNA with the H3/H4 tetramer particle, consequently facilitating access of PR to the essential HREs 2 and 3, which in the octamer particle are oriented with their major grooves pointed toward the histones (80). This model provides a direct molecular mechanism for the observed functional synergism between NF1 and PR during induction of the MMTV promoter (88).
Role of Histone H1
Histone H1 is the prototype of the “linker histones,” which are in contact with the linker DNA that joins consecutive nucleosomes (89). Histone H1 participates in nucleosome positioning, nucleosome spacing, and in the higher-order structure of chromatin. H1-containing chromatin is more resistant to nuclease digestion and shows strong inhibition of nucleosome sliding (90). Consequently, H1 is seen as a structural component related to chromatin compaction and inaccessibility to transcription factors or RNA polymerase. In addition, H1 seems to be actively involved in the regulation of gene expression, as it inhibits chromatin remodeling by the ySWI/SNF complex (91). However, the repressive role of histone H1 on gene expression is controversial. Although it has been reported that MMTV promoter chromatin is depleted of histone H1 after hormonal induction (92), overexpression of histone H1 in cultured cells enhances hormonal trans-activation of the promoter (93).
Histone H1 in mammals consists of a family of closely related, single-gene encoded proteins, including five somatic subtypes (from H1.1 to H1.5) and a terminally differentiated expressed isoform (H1.0). Because knockdown of individual somatic H1 subtypes in mouse has no marked phenotype (94), they have been assumed to be highly redundant. However, inducible knockdown of individual somatic H1 subtypes in breast cancer cells altered a different subset of genes, and the majority of them are down-regulated upon H1 depletion (95). This argues against a general repressive role of linker histones and suggests some nonredundant effects on gene expression. Indeed, depletion of individual subtypes had different effects on cell survival. H1.2 depletion specifically causes cell cycle arrest due to repression of key cell cycle genes, whereas H1.4 depleted cells eventually die of necrosis. Moreover, although H1.2 accounts for approximately only 20% of the total H1 content in T47D breast cancer cells, its depletion causes a general decrease in nucleosome spacing (95) that is not compensated by the overexpression of other subtypes. This suggests that individual subtypes have a selective effect on chromatin structure. Thus, specific phenotypes are observed in breast cancer cells depleted of individual histone H1 subtypes, supporting the idea that distinct roles do exist for the linker histone variants in these cells and proposes that theoretically the same situation may also occur in other cell types.
A recent study using Atomic Force Microscope showed that different H1 subtypes exhibit a different affinity for chromatin and differ in their capacity to condense chromatin (96). H1 subtypes can be classified as weak condensers (H1.1 and H1.2), intermediate condensers (H1.3), and strong condensers (H1.0, H1.4, H1.5, and H1x). The variable C-terminal domain is required for nucleosome spacing by H1.4 and is likely responsible for the chromatin condensation properties of the various subtypes, as shown using chimeras between H1.4 and H1.2 (96). Moreover, linker histones do not preclude ATP-dependent remodeling of minichromosomes by yeast SWI/SNF or Drosophila NURF when tested in minichromosomes, a dynamic system that mimics the situation in vivo. Thus, linker histone subtypes can be considered as differential organizers of chromatin, rather than general repressors (96).
Asymmetric binding of histone H1 to chromatin-organized MMTV promoter sequences compacts the nucleosomal structure (97) and leads to repression of basal transcription and reduced binding of NF1 (10). In contrast, H1 containing MMTV chromatin binds PR with higher affinity and is transcribed more efficiently in the presence of PR and NF1 than chromatin free of linker histone (10, 97). Thus, histone H1 represses hormone independent transcription and enhances the synergism between PR and NF1, resulting in tighter hormonal regulation (10).
This positive effect of H1 is likely due to a more homogeneous nucleosome positioning over the MMTV promoter (10). Therefore, H1 plays a key role during the initial hormonal activation of the MMTV promoter in native chromatin by favoring a better nucleosome positioning and a higher binding of PR. However, for transcription to take place, H1 has to be phosphorylated and displaced from the promoter (10). Phosphorylation is likely mediated by a protein kinase recruited to the promoter by activated PR. In view of the reported activation of Cdk2 after progesterone treatment (33), and due to its ability to phosphorylate histone H1 (37), this kinase is a good potential candidate for catalyzing progesterone-induced phosphorylation of histone H1. One could envisage a more general role for the phosphorylation of histone H1 by Cdk2, because this modification disrupts the interaction between H1 and HP1α promoting decondensation of chromatin independent of H3K9 methylation (36). In combination with the in vitro results, these considerations justify the inclusion of histone H1 and its phosphorylation in a hypothetical model of MMTV activation (Fig. 2). Additional experiments will reveal the nature of the PR-containing complexes responsible for phosphorylation and displacement of histone H1 subtypes from progestin-regulated promoters.
PR vs. GR
There are similarities between PR and GR during hormone-regulated events. Briefly, both receptors recognize the same DNA sequence (98), are phosphorylated by Erk kinases after hormone induction (99, 100), show interaction with SWI/SNF (65), recruit the SWI/SNF complex to the target chromatin, are dependent on HATs for its activity, and associate with several common coactivators and corepressors (101, 102). However, although GR exhibits rapid nongenomic effects, it activates signaling pathways different from those activated by PR. GR activates p38 and JNK MAPKs through a PKC-dependent pathway and is also implicated in the inhibition of EGFR signaling in lung epithelial cells (103, 104), whereas this pathways have not been reported to be activated by PR. Despite the fact that both PR and GR share the same DNA binding sequence, clear differences in their interactions with the HRE region of the MMTV promoter have been observed (105). Thus, the binding of the receptors to genome loci depends not only on the DNA sequence. Rather, other factors, such as chromatin state, cell type-specific transcription factors, and signaling pathways, may determine the specific genomic interactions of both receptors.
Both GR and PR depend on HATs for their activity, but they utilize overlapping but not identical sets of coactivators. Progesterone promotes the recruitment by PR of SRC-1/SRC-3, whereas dexamethasone enhances GR interaction mainly with SRC-2 and SRC-3 (106). Thus, the SRC family members could play an important but receptor-specific role in the orchestration of downstream events at target promoters (106).
Conclusions
The initial steps of gene induction by progestins involve mainly changes in chromatin organization of target promoters that require the activation of kinase signaling pathways initiated by membrane-bound PR. These kinases eventually phosphorylate the receptor to which they bind. The receptor complexes containing the kinases and histone modifying enzymes are recruited to the target promoters where they modify the protruding core histone tails and the linker histones. These modifications lead to the displacement of linker histones and a repressive complex containing HP1γ, likely by specialized ATP-dependent remodeling complexes. In a second step, other specialized ATP-dependent remodelers displace histone H2A/H2B dimers from the promoter nucleosome, enabling synergistic access of other transcription factors and additional receptor complexes to previously hidden binding sites on the surface of a histone H3/H4 tetramer particle. It is only after completion of these initial chromatin remodeling steps that complexes containing mediator and RNA-polymerase with associated basal transcription factors are recruited and further steps in transcription initiation, elongation, RNA splicing, etc. can take place. It remains to be seen how these findings can be reconciled with the rapid exchange of GR-green fluorescent protein observed in living cells carrying a cluster of some 200 MMTV reporters (81). The activation of the MMTV promoter seems to be a very dynamic process, where PR, as a master key regulator, directs the rapid exchange of chromatin remodeling complexes on chromatin. One possibility is that the majority of the GR binding events observed in the MMTV cluster in vivo reflect nonproductive or abortive binding of the various PR-containing complexes. Thus, a fully productive interaction would only occur when the different complexes are recruited in appropriate combinations and/or in the correct sequence.
NURSA Molecule Pages:
Coregulators: AIB1 | BAF57 | BRG1 | BRM | GRIP1 | P/CAF | SRC-1;
Ligands: Dexamethasone | Progesterone;
Nuclear Receptors: ER-α | GR | PR.
Footnotes
This work was supported by grants from the European Union (High-throughput Epigenetic Regulatory Organisation In Chromatin integrated project), the Departament d′Innovacio Universitat I Empresas, the Ministerio de Educación y Ciencia Grants BMC 2003-02902 CSD2006-00049, and the Fondo de Investigación Sanitaria Grants PI0411605 and CP04/00087. G.P.V. was a recipient of a fellowship of the Ramón y Cajal Program.
Disclosure Summary: The authors have nothing to disclose.
First Published Online May 19, 2010
Abbreviations: BAF, BRG1-associated factor; BRG1, Brahma-related gene 1; Cdk2, cyclin-dependent kinase 2; ChIP, chromatin immunoprecipitation; DNase, deoxyribonuclease; EGFR, epidermal growth factor receptor; ER, estrogen receptor; FRAP, fluorescence recovery after photobleaching; GR, glucocorticoid receptor; HAT, histone acetyl transferase; HDAC, histone deacetylase; HRE, hormone-responsive element; MMTV, mouse mammary tumor virus; Msk1, mitogen- and stress-activated protein kinase 1; NF1, nuclear factor 1; p, phospho; PR, progesterone receptor; SH, Src homology; SHR, steroid hormone receptor; Src, sarcoma; Stat, signal transducers and activator of transcription; SWI/SNF, switch/sucrose nonfermentable.
References
- 1.Norman AW, Mizwicki MT, Norman DP2004. Steroid-hormone rapid actions, membrane receptors and a conformational ensemble model. Nat Rev Drug Discov 3:27–41 [DOI] [PubMed] [Google Scholar]
- 2.Beato M, Herrlich P, Schütz G1995. Steroid hormone receptors: many actors in search of a plot. Cell 83:851–857 [DOI] [PubMed] [Google Scholar]
- 3.Métivier R, Penot G, Hübner MR, Reid G, Brand H, Kos M, Gannon F2003. Estrogen receptor-α directs ordered, cyclical, and combinatorial recruitment of cofactors on a natural target promoter. Cell 115:751–763 [DOI] [PubMed] [Google Scholar]
- 4.Shang Y, Hu X, DiRenzo J, Lazar MA, Brown M2000. Cofactor dynamics and sufficiency in estrogen receptor-regulated transcription. Cell 103:843–852 [DOI] [PubMed] [Google Scholar]
- 5.Reid G, Hübner MR, Métivier R, Brand H, Denger S, Manu D, Beaudouin J, Ellenberg J, Gannon F2003. Cyclic, proteasome-mediated turnover of unliganded and liganded ERα on responsive promoters is an integral feature of estrogen signaling. Mol Cell 11:695–707 [DOI] [PubMed] [Google Scholar]
- 6.Jones PA, Takai D2001. The role of DNA methylation in mammalian epigenetics. Science 293:1068–1070 [DOI] [PubMed] [Google Scholar]
- 7.Kangaspeska S, Stride B, Métivier R, Polycarpou-Schwarz M, Ibberson D, Carmouche RP, Benes V, Gannon F, Reid G2008. Transient cyclical methylation of promoter DNA. Nature 452:112–115 [DOI] [PubMed] [Google Scholar]
- 8.Métivier R, Gallais R, Tiffoche C, Le Péron C, Jurkowska RZ, Carmouche RP, Ibberson D, Barath P, Demay F, Reid G, Benes V, Jeltsch A, Gannon F, Salbert G2008. Cyclical DNA methylation of a transcriptionally active promoter. Nature 452:45–50 [DOI] [PubMed] [Google Scholar]
- 9.Vicent GP, Ballaré C, Nacht AS, Clausell J, Subtil-Rodríguez A, Quiles I, Jordan A, Beato M2008. Convergence on chromatin of non-genomic and genomic pathways of hormone signaling. J Steroid Biochem Mol Biol 109:344–349 [DOI] [PubMed] [Google Scholar]
- 10.Koop R, Di Croce L, Beato M2003. Histone H1 enhances synergistic activation of the MMTV promoter in chromatin. EMBO J 22:588–599 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Hu Q, Kwon YS, Nunez E, Cardamone MD, Hutt KR, Ohgi KA, Garcia-Bassets I, Rose DW, Glass CK, Rosenfeld MG, Fu XD2008. Enhancing nuclear receptor-induced transcription requires nuclear motor and LSD1-dependent gene networking in interchromatin granules. Proc Natl Acad Sci USA 105:19199–19204 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Welboren WJ, van Driel MA, Janssen-Megens EM, van Heeringen SJ, Sweep FC, Span PN, Stunnenberg HG2009. ChIP-Seq of ERα and RNA polymerase II defines genes differentially responding to ligands. EMBO J 28:1418–1428 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Lösel R, Wehling M2003. Nongenomic actions of steroid hormones. Nat Rev Mol Cell Biol 4:46–56 [DOI] [PubMed] [Google Scholar]
- 14.Castoria G, Migliaccio A, Bilancio A, Di Domenico M, de Falco A, Lombardi M, Fiorentino R, Varricchio L, Barone MV, Auricchio F2001. PI3-kinase in concert with Src promotes the S-phase entry of oestradiol-stimulated MCF-7 cells. EMBO J 20:6050–6059 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Migliaccio A, Di Domenico M, Castoria G, de Falco A, Bontempo P, Nola E, Auricchio F1996. Tyrosine kinase/p21ras/MAP-kinase pathway activation by estradiol-receptor complex in MCF-7 cells. EMBO J 15:1292–1300 [PMC free article] [PubMed] [Google Scholar]
- 16.Ballaré C, Uhrig M, Bechtold T, Sancho E, Di Domenico M, Migliaccio A, Auricchio F, Beato M2003. Two domains of the progesterone receptor interact with the estrogen receptor and are required for progesterone activation of the c-Src/Erk pathway in mammalian cells. Mol Cell Biol 23:1994–2008 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Boonyaratanakornkit V, Scott MP, Ribon V, Sherman L, Anderson SM, Maller JL, Miller WT, Edwards DP2001. Progesterone receptor contains a proline-rich motif that directly interacts with SH3 domains and activates c-Src family tyrosine kinases. Mol Cell 8:269–280 [DOI] [PubMed] [Google Scholar]
- 18.Migliaccio A, Piccolo D, Castoria G, Di Domenico M, Bilancio A, Lombardi M, Gong W, Beato M, Auricchio F1998. Activation of the Src/p21ras/Erk pathway by progesterone receptor via cross-talk with estrogen receptor. EMBO J 17:2008–2018 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Faivre EJ, Daniel AR, Hillard CJ, Lange CA2008. Progesterone receptor rapid signaling mediates serine 345 phosphorylation and tethering to specificity protein 1 transcription factors. Mol Endocrinol 22:823–837 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Boonyaratanakornkit V, McGowan E, Sherman L, Mancini MA, Cheskis BJ, Edwards DP2007. The role of extranuclear signaling actions of progesterone receptor in mediating progesterone regulation of gene expression and the cell cycle. Mol Endocrinol 21:359–375 [DOI] [PubMed] [Google Scholar]
- 21.Harrington WR, Kim SH, Funk CC, Madak-Erdogan Z, Schiff R, Katzenellenbogen JA, Katzenellenbogen BS2006. Estrogen dendrimer conjugates that preferentially activate extranuclear, nongenomic versus genomic pathways of estrogen action. Mol Endocrinol 20:491–502 [DOI] [PubMed] [Google Scholar]
- 22.Madak-Erdogan Z, Kieser KJ, Kim SH, Komm B, Katzenellenbogen JA, Katzenellenbogen BS2008. Nuclear and extranuclear pathway inputs in the regulation of global gene expression by estrogen receptors. Mol Endocrinol 22:2116–2127 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Baumann CT, Lim CS, Hager GL1999. Intracellular localization and trafficking of steroid receptors. Cell Biochem Biophys 31:119–127 [DOI] [PubMed] [Google Scholar]
- 24.Pietras RJ, Szego CM1977. Specific binding sites for oestrogen at the outer surfaces of isolated endometrial cells. Nature 265:69–72 [DOI] [PubMed] [Google Scholar]
- 25.Fernandes MS, Brosens JJ, Gellersen B2008. Honey, we need to talk about the membrane progestin receptors. Steroids 73:942–952 [DOI] [PubMed] [Google Scholar]
- 26.Gellersen B, Fernandes MS, Brosens JJ2009. Non-genomic progesterone actions in female reproduction. Hum Reprod Update 15:119–138 [DOI] [PubMed] [Google Scholar]
- 27.Losel RM, Falkenstein E, Feuring M, Schultz A, Tillmann HC, Rossol-Haseroth K, Wehling M2003. Nongenomic steroid action: controversies, questions, and answers. Physiol Rev 83:965–1016 [DOI] [PubMed] [Google Scholar]
- 28.Pedram A, Razandi M, Sainson RC, Kim JK, Hughes CC, Levin ER2007. A conserved mechanism for steroid receptor translocation to the plasma membrane. J Biol Chem 282:22278–22288 [DOI] [PubMed] [Google Scholar]
- 29.Weigel NL, Moore NL2007. Cyclins, cyclin dependent kinases, and regulation of steroid receptor action. Mol Cell Endocrinol 265- 266:157–161 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Rogatsky I, Trowbridge JM, Garabedian MJ1999. Potentiation of human estrogen receptor α transcriptional activation through phosphorylation of serines 104 and 106 by the cyclin A-CDK2 complex. J Biol Chem 274:22296–22302 [DOI] [PubMed] [Google Scholar]
- 31.Faivre E, Skildum A, Pierson-Mullany L, Lange CA2005. Integration of progesterone receptor mediated rapid signaling and nuclear actions in breast cancer cell models: role of mitogen-activated protein kinases and cell cycle regulators. Steroids 70:418–426 [DOI] [PubMed] [Google Scholar]
- 32.Narayanan R, Adigun AA, Edwards DP, Weigel NL2005. Cyclin-dependent kinase activity is required for progesterone receptor function: novel role for cyclin A/Cdk2 as a progesterone receptor coactivator. Mol Cell Biol 25:264–277 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Pierson-Mullany LK, Lange CA2004. Phosphorylation of progesterone receptor serine 400 mediates ligand-independent transcriptional activity in response to activation of cyclin-dependent protein kinase 2. Mol Cell Biol 24:10542–10557 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Moore NL, Narayanan R, Weigel NL2007. Cyclin dependent kinase 2 and the regulation of human progesterone receptor activity. Steroids 72:202–209 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Contreras A, Hale TK, Stenoien DL, Rosen JM, Mancini MA, Herrera RE2003. The dynamic mobility of histone H1 is regulated by cyclin/CDK phosphorylation. Mol Cell Biol 23:8626–8636 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Hale TK, Contreras A, Morrison AJ, Herrera RE2006. Phosphorylation of the linker histone H1 by CDK regulates its binding to HP1α. Mol Cell 22:693–699 [DOI] [PubMed] [Google Scholar]
- 37.Bhattacharjee RN, Banks GC, Trotter KW, Lee HL, Archer TK2001. Histone H1 phosphorylation by Cdk2 selectively modulates mouse mammary tumor virus transcription through chromatin remodeling. Mol Cell Biol 21:5417–5425 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Dou Y, Gorovsky MA2000. Phosphorylation of linker histone H1 regulates gene expression in vivo by creating a charge patch. Mol Cell 6:225–231 [DOI] [PubMed] [Google Scholar]
- 39.Dou Y, Mizzen CA, Abrams M, Allis CD, Gorovsky MA1999. Phosphorylation of linker histone H1 regulates gene expression in vivo by mimicking H1 removal. Mol Cell 4:641–647 [DOI] [PubMed] [Google Scholar]
- 40.Herrera RE, Chen F, Weinberg RA1996. Increased histone H1 phosphorylation and relaxed chromatin structure in Rb-deficient fibroblasts. Proc Natl Acad Sci USA 93:11510–11515 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Björnström L, Kilic E, Norman M, Parker MG, Sjöberg M2001. Cross-talk between Stat5b and estrogen receptor-α and -β in mammary epithelial cells. J Mol Endocrinol 27:93–106 [DOI] [PubMed] [Google Scholar]
- 42.Richer JK, Lange CA, Manning NG, Owen G, Powell R, Horwitz KB1998. Convergence of progesterone with growth factor and cytokine signaling in breast cancer. Progesterone receptors regulate signal transducers and activators of transcription expression and activity. J Biol Chem 273:31317–31326 [DOI] [PubMed] [Google Scholar]
- 43.Stöcklin E, Wissler M, Gouilleux F, Groner B1996. Functional interactions between Stat5 and the glucocorticoid receptor. Nature 383:726–728 [DOI] [PubMed] [Google Scholar]
- 44.Buser AC, Gass-Handel EK, Wyszomierski SL, Doppler W, Leonhardt SA, Schaack J, Rosen JM, Watkin H, Anderson SM, Edwards DP2007. Progesterone receptor repression of prolactin/signal transducer and activator of transcription 5-mediated transcription of the β-casein gene in mammary epithelial cells. Mol Endocrinol 21:106–125 [DOI] [PubMed] [Google Scholar]
- 45.Rocha-Viegas L, Vicent GP, Barañao JL, Beato M, Pecci A2006. Glucocorticoids repress bcl-X expression in lymphoid cells by recruiting STAT5B to the P4 promoter. J Biol Chem 281:33959–33970 [DOI] [PubMed] [Google Scholar]
- 46.Subtil-Rodríguez A, Millán-Ariño L, Quiles I, Ballaré C, Beato M, Jordan A2008. Progesterone induction of the 11β-hydroxysteroid dehydrogenase type 2 promoter in breast cancer cells involves coordinated recruitment of STAT5A and progesterone receptor to a distal enhancer and polymerase tracking. Mol Cell Biol 28:3830–3849 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Krusekopf S, Chauchereau A, Milgrom E, Henderson D, Cato AC1991. Co-operation of progestational steroids with epidermal growth factor in activation of gene expression in mammary tumor cells. J Steroid Biochem Mol Biol 40:239–245 [DOI] [PubMed] [Google Scholar]
- 48.Modiano JF, Kokai Y, Weiner DB, Pykett MJ, Nowell PC, Lyttle CR1991. Progesterone augments proliferation induced by epidermal growth factor in a feline mammary adenocarcinoma cell line. J Cell Biochem 45:196–206 [DOI] [PubMed] [Google Scholar]
- 49.Chrysogelos SA, Dickson RB1994. EGF receptor expression, regulation, and function in breast cancer. Breast Cancer Res Treat 29:29–40 [DOI] [PubMed] [Google Scholar]
- 50.Murphy LJ, Sutherland RL, Stead B, Murphy LC, Lazarus L1986. Progestin regulation of epidermal growth factor receptor in human mammary carcinoma cells. Cancer Res 46:728–734 [PubMed] [Google Scholar]
- 51.Sarup JC, Rao KV, Fox CF1988. Decreased progesterone binding and attenuated progesterone action in cultured human breast carcinoma cells treated with epidermal growth factor. Cancer Res 48:5071–5078 [PubMed] [Google Scholar]
- 52.Daniel AR, Qiu M, Faivre EJ, Ostrander JH, Skildum A, Lange CA2007. Linkage of progestin and epidermal growth factor signaling: phosphorylation of progesterone receptors mediates transcriptional hypersensitivity and increased ligand-independent breast cancer cell growth. Steroids 72:188–201 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Vicent GP, Ballaré C, Nacht AS, Clausell J, Subtil-Rodríguez A, Quiles I, Jordan A, Beato M2006. Induction of progesterone target genes requires activation of Erk and Msk kinases and phosphorylation of histone H3. Mol Cell 24:367–381 [DOI] [PubMed] [Google Scholar]
- 54.Zhang Y, Beck CA, Poletti A, Edwards DP, Weigel NL1995. Identification of a group of Ser-Pro motif hormone-inducible phosphorylation sites in the human progesterone receptor. Mol Endocrinol 9:1029–1040 [DOI] [PubMed] [Google Scholar]
- 55.Bagowski CP, Myers JW, Ferrell Jr JE2001. The classical progesterone receptor associates with p42 MAPK and is involved in phosphatidylinositol 3-kinase signaling in Xenopus oocytes. J Biol Chem 276:37708–37714 [DOI] [PubMed] [Google Scholar]
- 56.Cairns BR2005. Chromatin remodeling complexes: strength in diversity, precision through specialization. Curr Opin Genet Dev 15:185–190 [DOI] [PubMed] [Google Scholar]
- 57.Fan HY, Narlikar GJ, Kingston RE2004. Noncovalent modification of chromatin: different remodeled products with different ATPase domains. Cold Spring Harb Symp Quant Biol 69:183–192 [DOI] [PubMed] [Google Scholar]
- 58.Mohrmann L, Verrijzer CP2005. Composition and functional specificity of SWI2/SNF2 class chromatin remodeling complexes. Biochim Biophys Acta 1681:59–73 [DOI] [PubMed] [Google Scholar]
- 59.Racki LR, Narlikar GJ2008. ATP-dependent chromatin remodeling enzymes: two heads are not better, just different. Curr Opin Genet Dev 18:137–144 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Whitehouse I, Rando OJ, Delrow J, Tsukiyama T2007. Chromatin remodelling at promoters suppresses antisense transcription. Nature 450:1031–1035 [DOI] [PubMed] [Google Scholar]
- 61.Wang W2003. The SWI/SNF family of ATP-dependent chromatin remodelers: similar mechanisms for diverse functions. Curr Top Microbiol Immunol 274:143–169 [DOI] [PubMed] [Google Scholar]
- 62.Chen J, Archer TK2005. Regulating SWI/SNF subunit levels via protein-protein interactions and proteasomal degradation: BAF155 and BAF170 limit expression of BAF57. Mol Cell Biol 25:9016–9027 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Yoshinaga SK, Peterson CL, Herskowitz I, Yamamoto KR1992. Roles of SWI1, SWI2, and SWI3 proteins for transcriptional enhancement by steroid receptors. Science 258:1598–1604 [DOI] [PubMed] [Google Scholar]
- 64.Muchardt C, Yaniv M1993. A human homologue of Saccharomyces cerevisiae SNF2/SWI2 and Drosophila brm genes potentiates transcriptional activation by the glucocorticoid receptor. EMBO J 12:4279–4290 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Fryer CJ, Archer TK1998. Chromatin remodelling by the glucocorticoid receptor requires the BRG1 complex. Nature 393:88–91 [DOI] [PubMed] [Google Scholar]
- 66.John S, Sabo PJ, Johnson TA, Sung MH, Biddie SC, Lightman SL, Voss TC, Davis SR, Meltzer PS, Stamatoyannopoulos JA, Hager GL2008. Interaction of the glucocorticoid receptor with the chromatin landscape. Mol Cell 29:611–624 [DOI] [PubMed] [Google Scholar]
- 67.Richard-Foy H, Hager GL1987. Sequence-specific positioning of nucleosomes over the steroid-inducible MMTV promoter. EMBO J 6:2321–2328 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Truss M, Bartsch J, Schelbert A, Haché RJ, Beato M1995. Hormone induces binding of receptors and transcription factors to a rearranged nucleosome on the MMTV promoter in vivo. EMBO J 14:1737–1751 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Eisfeld K, Candau R, Truss M, Beato M1997. Binding of NF1 to the MMTV promoter in nucleosomes: influence of rotational phasing, translational positioning and histone H1. Nucleic Acids Res 25:3733–3742 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Piña B, Brüggemeier U, Beato M1990. Nucleosome positioning modulates accessibility of regulatory proteins to the mouse mammary tumor virus promoter. Cell 60:719–731 [DOI] [PubMed] [Google Scholar]
- 71.Vicent GP, Nacht AS, Smith CL, Peterson CL, Dimitrov S, Beato M2004. DNA instructed displacement of histones H2A and H2B at an inducible promoter. Mol Cell 16:439–452 [DOI] [PubMed] [Google Scholar]
- 72.Vicent GP, Ballaré C, Zaurin R, Saragüeta P, Beato M2006. Chromatin remodeling and control of cell proliferation by progestins via cross talk of progesterone receptor with the estrogen receptors and kinase signaling pathways. Ann NY Acad Sci 1089:59–72 [DOI] [PubMed] [Google Scholar]
- 73.Yang X, Zaurin R, Beato M, Peterson CL2007. Swi3p controls SWI/SNF assembly and ATP-dependent H2A-H2B displacement. Nat Struct Mol Biol 14:540–547 [DOI] [PubMed] [Google Scholar]
- 74.Vicent GP, Zaurin R, Nacht AS, Li A, Font-Mateu J, Le Dily F, Vermeulen M, Mann M, Beato M2009. Two chromatin remodeling activities cooperate during activation of hormone responsive promoters. PLoS Genet 5:e1000567 [DOI] [PMC free article] [PubMed]
- 75.Trotter KW, Fan HY, Ivey ML, Kingston RE, Archer TK2008. The HSA domain of BRG1 mediates critical interactions required for glucocorticoid receptor-dependent transcriptional activation in vivo. Mol Cell Biol 28:1413–1426 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Li XS, Trojer P, Matsumura T, Treisman JE, Tanese N2010. Mammalian SWI/SNF-A subunit BAF250/ARID1 is an E3 ubiquitin ligase that targets histone H2B. Mol Cell Biol 30:1673–1688 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Bartsch J, Truss M, Bode J, Beato M1996. Moderate increase in histone acetylation activates the mouse mammary tumor virus promoter and remodels its nucleosome structure. Proc Natl Acad Sci USA 93:10741–10746 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Bresnick EH, John S, Hager GL1991. Histone hyperacetylation does not alter the positioning or stability of phased nucleosomes on the mouse mammary tumor virus long terminal repeat. Biochemistry 30:3490–3497 [DOI] [PubMed] [Google Scholar]
- 79.Aoyagi S, Archer TK2007. Dynamic histone acetylation/deacetylation with progesterone receptor-mediated transcription. Mol Endocrinol 21:843–856 [DOI] [PubMed] [Google Scholar]
- 80.Vicent GP, Zaurin R, Nacht AS, Font-Mateu J, Le Dily F, Beato M2010. NF1 synergizes with progesterone receptor on the MMTV promoter wrapped around a histone H3/H4 tetramer by facilitating access to the central hormone responsive elements. J Biol Chem 285:2622–2631 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.McNally JG, Müller WG, Walker D, Wolford R, Hager GL2000. The glucocorticoid receptor: rapid exchange with regulatory sites in living cells. Science 287:1262–1265 [DOI] [PubMed] [Google Scholar]
- 82.Stenoien DL, Patel K, Mancini MG, Dutertre M, Smith CL, O'Malley BW, Mancini MA2001. FRAP reveals that mobility of oestrogen receptor-α is ligand- and proteasome-dependent. Nat Cell Biol 3:15–23 [DOI] [PubMed] [Google Scholar]
- 83.Stenoien DL, Nye AC, Mancini MG, Patel K, Dutertre M, O'Malley BW, Smith CL, Belmont AS, Mancini MA2001. Ligand-mediated assembly and real-time cellular dynamics of estrogen receptor α-coactivator complexes in living cells. Mol Cell Biol 21:4404–4412 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84.Rayasam GV, Elbi C, Walker DA, Wolford R, Fletcher TM, Edwards DP, Hager GL2005. Ligand-specific dynamics of the progesterone receptor in living cells and during chromatin remodeling in vitro. Mol Cell Biol 25:2406–2418 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85.Becker PB, Gloss B, Schmid W, Strähle U, Schütz G1986. In vivo protein-DNA interactions in a glucocorticoid response element require the presence of the hormone. Nature 324:686–688 [DOI] [PubMed] [Google Scholar]
- 86.Agresti A, Scaffidi P, Riva A, Caiolfa VR, Bianchi ME2005. GR and HMGB1 interact only within chromatin and influence each other’s residence time. Mol Cell 18:109–121 [DOI] [PubMed] [Google Scholar]
- 87.Spangenberg C, Eisfeld K, Stünkel W, Luger K, Flaus A, Richmond TJ, Truss M, Beato M1998. The mouse mammary tumour virus promoter positioned on a tetramer of histones H3 and H4 binds nuclear factor 1 and OTF1. J Mol Biol 278:725–739 [DOI] [PubMed] [Google Scholar]
- 88.Di Croce L, Koop R, Venditti P, Westphal HM, Nightingale KP, Corona DF, Becker P, Beato M1999. Two-steps synegism between progesterone receptor and the DNA binding domain of NF1 on MMTV minichromosomes. Mol Cell 4:45–54 [DOI] [PubMed] [Google Scholar]
- 89.Widom J1998. Chromatin structure: linking structure to function with histone H1. Curr Biol 8:R788–R791 [DOI] [PubMed]
- 90.Hill DA2001. Influence of linker histone H1 on chromatin remodeling. Biochem Cell Biol 79:317–324 [PubMed] [Google Scholar]
- 91.Horn PJ, Carruthers LM, Logie C, Hill DA, Solomon MJ, Wade PA, Imbalzano AN, Hansen JC, Peterson CL2002. Phosphorylation of linker histones regulates ATP-dependent chromatin remodeling enzymes. Nat Struct Biol 9:263–267 [DOI] [PubMed] [Google Scholar]
- 92.Bresnick EH, Bustin M, Marsaud V, Richard-Foy H, Hager GL1992. The transcriptionally-active MMTV promoter is depleted of histone H1. Nucleic Acids Res 20:273–278 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 93.Gunjan A, Brown DT1999. Overproduction of histone H1 variants in vivo increases basal and induced activity of the mouse mammary tumor virus promoter. Nucleic Acids Res 27:3355–3363 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 94.Fan Y, Sirotkin A, Russell RG, Ayala J, Skoultchi AI2001. Individual somatic H1 subtypes are dispensable for mouse development even in mice lacking the H1(0) replacement subtype. Mol Cell Biol 21:7933–7943 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 95.Sancho M, Diani E, Beato M, Jordan A2008. Depletion of human histone H1 variants uncovers specific roles in gene expression and cell growth. PLoS Genet 4:e1000227 [DOI] [PMC free article] [PubMed]
- 96.Clausell J, Happel N, Hale TK, Doenecke D, Beato M2009. Histone H1 subtypes differentially modulate chromatin condensation without preventing ATP-dependent remodeling by SWI/SNF or NURF. PLoS One 4:e0007243 [DOI] [PMC free article] [PubMed]
- 97.Vicent GP, Meliá MJ, Beato M2002. Asymmetric binding of histone H1 stabilizes MMTV nucleosomes and the interaction of progesterone receptor with the exposed HRE. J Mol Biol 324: 501–517 [DOI] [PubMed]
- 98.von der Ahe D, Janich S, Scheidereit C, Renkawitz R, Schütz G, Beato M1985. Glucocorticoid and progesterone receptors bind to the same sites in two hormonally regulated promoters. Nature 313:706–709 [DOI] [PubMed] [Google Scholar]
- 99.Rogatsky I, Logan SK, Garabedian MJ1998. Antagonism of glucocorticoid receptor transcriptional activation by the c-Jun N-terminal kinase. Proc Natl Acad Sci USA 95:2050–2055 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 100.Rogatsky I, Waase CL, Garabedian MJ1998. Phosphorylation and inhibition of rat glucocorticoid receptor transcriptional activation by glycogen synthase kinase-3 (GSK-3). Species-specific differences between human and rat glucocorticoid receptor signaling as revealed through GSK-3 phosphorylation. J Biol Chem 273:14315–14321 [DOI] [PubMed] [Google Scholar]
- 101.Dilworth FJ, Chambon P2001. Nuclear receptors coordinate the activities of chromatin remodeling complexes and coactivators to facilitate initiation of transcription. Oncogene 20:3047–3054 [DOI] [PubMed] [Google Scholar]
- 102.Torchia J, Glass C, Rosenfeld MG1998. Co-activators and co-repressors in the integration of transcriptional responses. Curr Opin Cell Biol 10:373–383 [DOI] [PubMed] [Google Scholar]
- 103.Croxtall JD, Choudhury Q, Flower RJ2000. Glucocorticoids act within minutes to inhibit recruitment of signalling factors to activated EGF receptors through a receptor-dependent, transcription-independent mechanism. Br J Pharmacol 130:289–298 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 104.Croxtall JD, Choudhury Q, Tokumoto H, Flower RJ1995. Lipocortin-1 and the control of arachidonic acid release in cell signalling. Glucocorticoids (changed from glucorticoids) inhibit G protein-dependent activation of cPLA2 activity. Biochem Pharmacol 50:465–474 [DOI] [PubMed] [Google Scholar]
- 105.Chalepakis G, Arnemann J, Slater E, Brüller HJ, Gross B, Beato M1988. Differential gene activation by glucocorticoids and progestins through the hormone regulatory element of mouse mammary tumor virus. Cell 53:371–382 [DOI] [PubMed] [Google Scholar]
- 106.Li X, Wong J, Tsai SY, Tsai MJ, O'Malley BW2003. Progesterone and glucocorticoid receptors recruit distinct coactivator complexes and promote distinct patterns of local chromatin modification. Mol Cell Biol 23:3763–3773 [DOI] [PMC free article] [PubMed] [Google Scholar]