Abstract
The MYO7A gene encodes a protein belonging to the unconventional myosin super family. Mutations within MYO7A can lead to either non syndromic hearing loss or to the Usher syndrome type 1B (USH1B). Here, we report the results of genetic analyses performed on Moroccan families with autosomal recessive non syndromic hearing loss that identified two families with compound heterozygous MYO7A mutations. Five mutations (c.6025delG, c.6229T>A, c.3500T>A, c.5617C>T and c.4487C>A) were identified in these families, the latter presenting two differently affected branches. Multiple bioinformatics programs and molecular modelling predicted the pathogenic effect of these mutations. In conclusion, the absence of vestibular and retinal symptom in the affected patients suggests that these families have the isolated non-syndromic hearing loss DFNB2 (nonsyndromic autosomal recessive hearing loss) presentation, instead of USH1B.
Introduction
Sensorineural hearing loss (SNHL) is the most prevalent human genetic sensory defect. It is estimated that globally 2 out of 1000 newborns have profound hearing loss [1]. Hereditary hearing loss is divided into two groups, syndromic and non-syndromic. To date, over 150 genes responsible for hearing loss have been identified, among which 70 are implicated in the non-syndromic hearing loss, whereas the others lead to syndromic presentations [2]. MYO7A is an unconventional myosin with a predicted 2215 amino acid sequence. Myosins are motor molecules that play an important role in intracellular movements; these proteins bind to actin filaments and use ATPase activity to generate the energy required for movements [3]. The human MYO7A gene contains 49 coding exons [4] and is expressed in the retina, lung, testis, kidney, and outer and inner hair cells of the cochlea [5]. In the latter, MYO7A is found in the actin-rich stereocilia bundles, cuticular plate, pericuticular necklace, and cell body [5]. MYO7A mutations are responsible for nonsyndromic autosomal recessive hearing loss (DFNB2) [6], autosomal dominant hearing loss (DFNA11) [7–9], and Usher syndrome [10]. Usher syndrome is an autosomal recessive disorder defined by the association of sensorineural hearing loss, retinitis pigmentosa (RP) and variable vestibular areflexia. Clinically, Usher syndrome can be classified into three types, USH type I (USH1), USH type II (USH2) and USH type III (USH3) [11]. Syndromic MYO7A mutations are inherited in a recessive manner, leading to a diagnosis of Usher type 1B (USH1B) [12,13], which is the most severe, and characterized by congenital profound hearing loss, prepuberal onset of retinitis pigmentosa, and vestibular dysfunction. However MYO7A variants have also been associated with USH2 [8,14], which is characterized by less severe features: moderate deafness without vestibular dysfunction and RP with postpubertal onset. Until now, only seven MYO7A mutations have been characterized in cases of recessive non-syndromic hearing loss: p.R244P in Chinese pedigree [15], a compound heterozygote mutation c.133-2A>G (IVS3nt-2A>G) and p.V1199insT in another Chinese pedigree [15], p.E1716del in Pakistani pedigree [16], p.R395H in Iranian pedigree [17], p.Cys652Glyfs*11 (p.C652fsX11) in Iraqi pedigree [18], p.P1887L in Palestinian pedigree [18].
In this study, we report the clinical, genetic and molecular characterization of two Moroccan families with Autosomal Recessive Non-Syndromic Hearing Loss (ARNSHL), thus disclosing the 7th and 8th DFNB2 families described to date. It is the first time that MYO7A mutations are identified with hearing loss in Morocco.
Patients and methods
Family enrolment and clinical evaluation
Autosomal recessive non-syndromic sensorineural hearing loss has been ascertained in two large Moroccan families: SF01 and SF42 (Fig 1A). Written informed consent was obtained from all patients and controls, and the committee on research ethics of the Pasteur Institute of Morocco approved the genetic study. Audiological testing was assessed by pure tone audiometry, speech audiometry and tympanometry, using a pure tone audiometer and a tympanometer. Hearing impairment was classified as mild (20–40 dB), moderate (41–70 dB), severe (71–95 dB), and profound (>95 dB). Tandem gait and Romberg testing were performed for vestibular function evaluation.
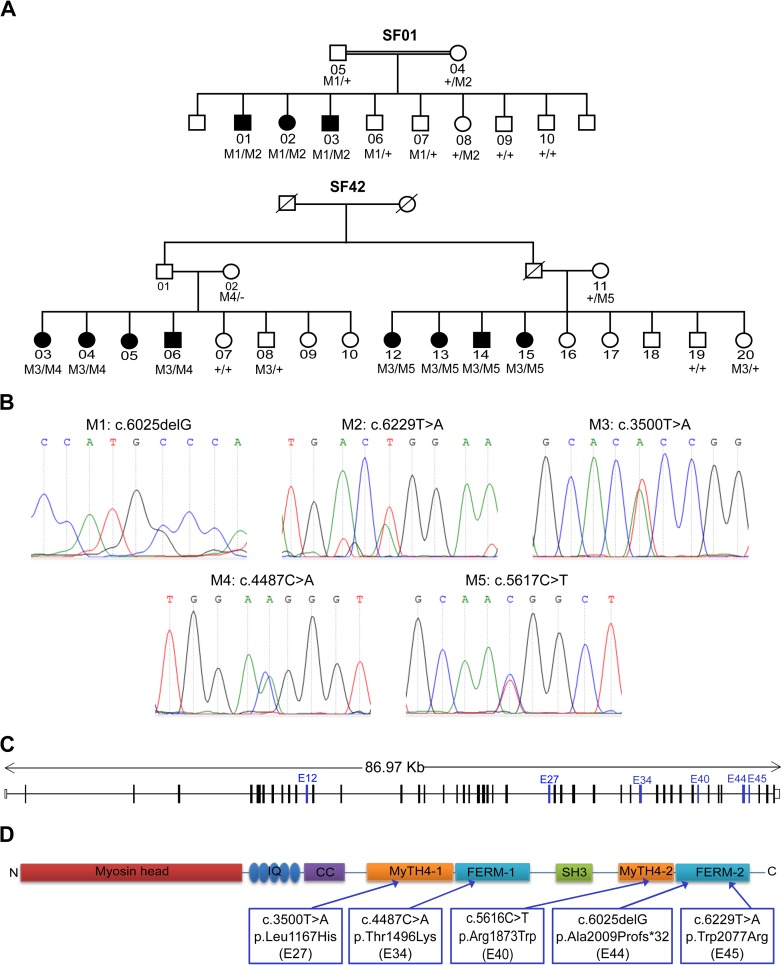
Fig 1. Molecular analysis of families SF01 and SF42 families.
A: Pedigrees and genotype data for each member of the SF01 and SF42 families: M1 to M5: mutation1 to mutation 5. B: Electrophoregram presenting the MYO7A heterozygous mutations identified for each family: Family SF01 (mutations M1 and M2) and Family SF42 (mutations M3, M4 and M5). C: Mutated MYO7A exons. D: Mutated domains in the MYO7A protein.
In addition, 60 families with severe to profound congenital bilateral sensorineural hearing loss and 100 healthy controls from different regions of Morocco with no history of hearing loss, were included in this study to screen the identified MYO7A mutations.
Whole exome sequencing
Genomic DNA was extracted from peripheral blood of the patient SF01.01 by phenol chloroform method following standard protocol [19]. Whole exome sequencing was performed at Otogenetics Corporation (Norcross, GA, USA) to identify the causative mutation in this patient. Genomic DNA was fragmented, end-repaired, and ligated with specific adaptors for library preparation using NEBNext reagents (New England Biolabs, Ipswich, MA, USA). Then, the resulting libraries were captured using Agilent Human exome V5 (51 Mb) capture kit and sequenced on a HiSeq 2000 platform (Illumina, San Diego, USA).
Read mapping and variant analysis
Whole-exome sequencing generated about 25.2 million short reads, comprising 3.2 billion bases. After quality control, short reads were mapped to the human genome reference sequence (University of California Santa Cruz hg19) using the DNAnexus software package (DNAnexus, Inc, Mountain View, CA, USA). Short reads were mapped with an average exome coverage of 30x. Variant calling and annotation were performed using the DNAnexus software package. After quality control of the identified variants, the next step was to exclude all non-exonic and synonymous variants from further analysis. Then, the frequency of each variant was obtained from dbSNP database (version 132) and all variants with a frequency higher than 1% were filtered. Finally, the functional effects of missense variants were predicted using SIFT (Sorting Intolerant From Tolerant) and PolyPhen-2 (Polymorphism Phenotyping).
Mutation confirmation and segregation analysis
To validate candidate mutations, segregation analysis was performed in all family members by Sanger sequencing, using the ABI prism Big Dye Terminator cycle sequencing Ready Reaction kit V 3.1 (ABI Prism/ Apllied Biosystems, Foster City, CA). The sequences were obtained through an ABI Prism 3100 Genetic Analyser (Applied Biosystem).
Additional unrelated control individuals were also sequenced, to determine the frequency of the candidate mutations. The primers used to screen the entire MYO7A coding region were designed using the Primer3 software (S1 Table).
Molecular modelling
The 3D structure of the wild type MYO7A protein was modelled using CPHmodels-3.2, an automated homology modelling program [20]. Then, the FOLD-X program was used to generate mutated structures [21]. Structural analysis and visualization of the predicted structures were performed using YASARA software [22]. To analyse the impact of amino acid substitutions on MYO7A protein structure stability, we used MAESTROweb and SDM (Site Directed Mutator) bioinformatics tools [23,24].
Results
Clinical evaluation
Audiological testing was completed to document the degree of hearing loss, which disclosed severe to profound congenital bilateral hearing loss in all the affected members from the SF01 family. However, in the SF42 family, audiological evaluation showed mild progressive hearing loss in the left branch, while the deafness was congenital and severe in the right one. Normal vestibular function was revealed by caloric tests performed on patients SF01.02, SF42.03 and SF42.14. The visual acuity was normal in both eyes in all affected persons, and fundus examination in patient SF01.02 showed intact fundi, as late as 36 years of age (Table 1).
Table 1. Clinical characteristics of patients SF01.02, SF42.03 and SF42.14.
| Patient | Age | Sex | Onset of hearing loss | Hearing threshold (dB) | Fundus examination | Visual acuity |
|---|---|---|---|---|---|---|
| SF01.02 | 36 | F | Congenital | 70 | Normal retina, blood vessels and optic disc | 20/20 |
| SF42.03 | 33 | F | Congenital | 60 | ND | 20/20 |
| SF42.14 | 30 | M | Congenital | 90 | ND | 20/20 |
Whole exome sequencing
Whole-exome sequencing (WES) of a DNA sample from patient SF1.01 generated a total of 122725 single nucleotide variants and 8622 short insertions and deletions (indels). To select candidate causative mutations, we focused on non-synonymous variants, frameshift in/del and variants in splice sites. Common variants with a frequency greater than 1% were filtered based on dbSNP137, 1000 Genomes Project and HapMap project databases. Considering an autosomal recessive mode of inheritance, homozygous or compound heterozygous variants were selected for further analysis. The remaining variants were filtered using Polyphen-2 and SIFT bioinformatics tools; only variants predicted as damaging were retained (S2 Table). Finally, we performed variant screening in candidate genes by prioritizing previously reported hereditary hearing loss genes.
WES analysis revealed two novel compound heterozygous mutations in MYO7A coding region. The c.6025delG deletion in exon 44 leads to a frameshift and a premature stop codon, truncating the last 206 amino-acids of the protein (p.Ala2009Profs*32). The c.6229T>A missense substitution occurs in exon 45, resulting in an amino acid substitution (p.Trp2077Arg). Sanger sequencing confirmed the co-segregation of these MYO7A mutations with the pathological phenotype, as they were present in all affected individuals at compound heterozygous state. The parents were heterozygous carriers of the c.6025delG (mother) and the c.6229T>A (father) mutations, whereas unaffected family members had only one of these mutations at heterozygous state, or none (Fig 1A and 1B).
In addition, sequencing of all MYO7A exons in 60 families with hereditary hearing loss, led to the identification of further mutations in SF42 family. All affected persons presented the c.3500T>A mutation (p.Leu1167His) in addition to one of the following mutations: c.5617C>T (p.Arg1873Trp) on the right side of the family and c.4487C>A (p.Thr1496Lys) on the left side of the family, at compound heterozygous state (Fig 1A and 1B). These MYO7A mutations were absent in 100 healthy controls.
Among the four missense mutations, three were predicted to be deleterious (p.Leu1167His, p.Arg1873Trp and p.Trp2077Arg) as revealed by multiple bioinformatics software, including SIFT [25], Polyphen [26], CONDEL [27], MutationAssessor [28] and MutationTaster [29]. In contrast, SIFT and Mutation Assessor predicted that the p.Thr1496Lys missense mutation is not damaging, while other prediction tools classified this substitution as disease causing (Table 2).
Table 2. Characteristics of MYO7A mutations.
| cDNA Change | Protein Change | dbSNP rs ID | MAF in Exome Variant Server and Exac | SIFT | Polyphen 2 | Condel | Mutation Assessor |
Mutation Taster | DDIG-in |
|---|---|---|---|---|---|---|---|---|---|
| M3: c.3500T>A | p.Leu1167His | - | - | Deleterious | Probably damaging | Deleterious | High | Disease causing | - |
| M4: c.4487C>A | p.Thr1496Lys | rs373651847 | 0.0079 8.328e-06 |
Tolerated | Possibly damaging | Deleterious | Low | Disease causing | - |
| M5: c.5617C>T | p.Arg1873Trp | rs397516321 | - 8.817e-06 |
Deleterious | Probably damaging | Deleterious | Medium | Disease causing | - |
| M1: c.6025delG | p.Ala2009Profs*32 | - | - | - | - | - | - | Disease causing | Disease causing |
| M2: c.6229T>A | p.Trp2077Arg | - | - | Deleterious | Probably damaging | Deleterious | Medium | Disease causing | - |
All missense mutations as well as the frameshift mutation p.Ala2009Profs*32 are located in the MyTH4 domain and the FERM domain (Fig 1C and 1D). In addition, multiple sequence alignment of MYO7A orthologous proteins from different metazoan species showed that the missense mutations affected highly conserved residues (Fig 2).
Fig 2. Alignment of MYO7A amino acid sequences from different species.
The mutated amino-acids which are highly conserved residues are highlighted in blue.
Structure modelling
A 3D structure of MYO7A head domain was constructed based on the crystal structure of the myosin head domain and of the heavy chain 2 protein (MYH2) (PDB ID: 2XEL). The sequence identity between the target and template proteins is 43.07%, and the constructed model covered 98% of the target sequence. The crystal structure of MYO7A MyTH4-FERM (PDB ID: 3PVL) was used as a template to build a molecular model of the second MyTH4-FERM tandem domain. The identity and convergence between the target and template proteins were 26.54% and 87%, respectively (Fig 3).
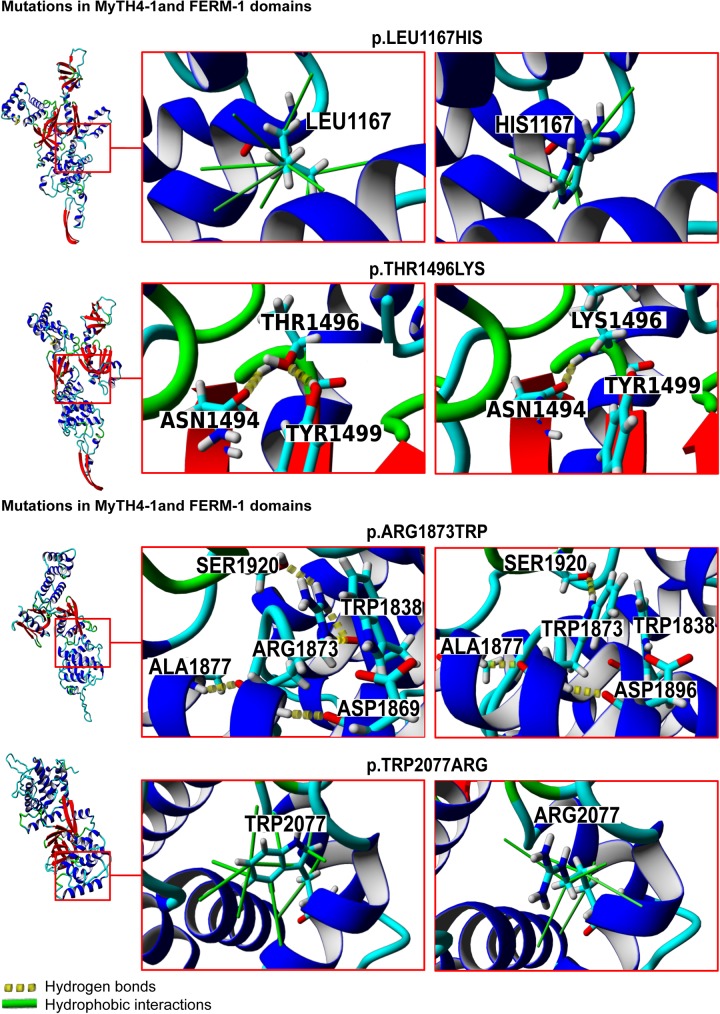
Fig 3. Structural and functional impacts of MYO7A missense mutations, as predicted by molecular modelling and amino acid conservation analysis.
Hydrogen bonds and hydrophobic interactions predicted by the Yasara software. Yellow dotted lines represent hydrogen bonds, and green lines represent hydrophobic interactions.
The substitution of p.Leu1167His is likely to disrupt the hydrophobic interaction between Leu1167 and its neighbouring residues. The p.Thr1496Lys substitution may induce a loss of hydrogen bonds, thus altering the interaction between residues Thr1496 and Tyr1499. The mutation p.Arg1873Trp is predicted to remove the hydrogen bond between Arg1873 and Trp1838 residue. The amino acid change p.Trp2077Arg may disrupt the hydrophobic interactions between Trp2077 and the neighbouring residues.
The bioinformatics tools SDM and MAESTROweb have been used to predict MYO7A missense mutation effects on the protein stability. Two mutations, p.Leu1167His and p.Trp2077Arg, were predicted to be destabilizing by both tools. The p.Arg1873Trp mutation was found to be highly stabilizing causing protein malfunction. The p.Thr1496Lys is predicted to be destabilizing by SDM, in contrast MAESTROweb predicted that this substitution may stabilise the protein (Table 3).
Table 3. Mutation effects on MYO7A 3D structure stability.
| cDNA Change | Protein Change | SDM | MAESTROweb |
|---|---|---|---|
| c.3500T>A | p.Leu1167His | Destabilising | Destabilising |
| c.4487C>A | p.Thr1496Lys | Destabilizing | Stabilising |
| c.5617C>T | p.Arg1873Trp | Highly stabilising | Stabilising |
| c.6229T>A | p.Trp2077Arg | Highly destabilising | Destabilising |
Discussion
In the present study, we performed whole-exome sequencing on a Moroccan family with autosomal recessive, non-syndromic hearing loss (ARNSHL), which did not show mutation in the most common genes involved in the Moroccan deaf population, like GJB2 [30], LRTOMT2 [31], TBC1D24 [32] and TMC1 [33]. We identified two novel MYO7A mutations that prompted us to explore MYO7A sequence in additional Moroccan families. In a second family including two branches, we further revealed three novel MYO7A mutations. All five MYO7A mutations that are predicted to be pathogenic, co-segregated with the phenotype and were absent in 100 healthy individuals. The MYO7A gene encodes a protein belonging to the unconventional myosin superfamily. The MYO7A protein is characterized by the presence of a head domain in the N-terminal region, this domain contains an ATP and an actin binding sites, followed by the neck domain, containing five isoleucine-glutamine (IQ) motifs, which function as binding sites for partners. The tail domain has an important role in the regulation of MYO7A movement, it contains two myosin tail homology 4 (MyTH4) domains, two band 4.1-ezrinradixin-moesin (FERM) domains and an SH3 domain [34]. MYO7A has been reported to be involved in multiple sensory functions, such as vision and hearing. Consequently, mutations in this gene are responsible for the Usher syndrome type 1B, and the homozygous c.1687G>A mutation predicted to result in aberrant splicing was described in a large Moroccan consanguineous USH1 family [35].
In this study, two novel mutations at compound heterozygous state c.6025delG (p.Ala2009Profs*32) and c.6229T>A (p.Trp2077Arg) were identified in patients from family SF01. The c.6025delG deletion leads to a frameshift mutation predicted to introduce a premature stop codon truncating a large part from the second FERM domain. Within the same family, the c.6229T>A, resulting in a substitution of tryptophan, a neutral hydrophobic amino acid residue, by arginine, a positive charged hydrophilic amino acid at 2077 position, may lead to a change in the protein structure by disrupting hydrophobic interactions between the alpha helix containing Trp2077 and the neighbouring residues. Interestingly, in the family SF42, we identified three different compound heterozygous mutations in the two branches of the family. All affected members share the c.3500T>A (p.Leu1167His) mutation at compound heterozygote state, and one of the following mutations: c.5617C>T and c.4487C>A. These missense mutations are located in conserved regions and are putatively damaging, while not found in 100 Moroccan controls. Since the p.Thr1496Lys amino acid change found in the left branch of the family is predicted to be mild, whereas the p.Arg1873Trp amino acid change is predicted to be severe (Table 1), different auditory phenotypes were observed in the two branches: mild and progressive in the left branch, while being congenital severe in the right one. In the first MyTH4 domain, the substitution of Leucine, a hydrophobic uncharged residue, to Histidine a positively charged residue at position 1167 is likely to disrupt the hydrophobic interactions between Leu1167 and its neighbouring residues. The missense mutation p.Thr1496Lys affects the first FERM domain by replacing a neutral residue with a positive charged amino acid. This substitution may eliminate a hydrogen bond between Thr1496 and Tyr1499, thus perturbing the interactions between the adjacent beta strand containing residues Asn1494 and Tyr1499. The substitution p.Arg1873Trp involves the change of a positive charged hydrophilic amino acid to a neutral hydrophobic amino acid. It occurs within the second MyTH4 domain and may change the protein folding by perturbing the hydrogen bond between Arg1873 and Trp1838 residues. The possible pathogenic effect of the mutations identified in this study on the MYO7A protein structure and multiple bioinformatics programs, amino acid conservation analysis and molecular modelling reinforce function. All mutations were located in functionally important protein domains, and are suspected to disrupt the normal MYO7A function, preventing its interaction with other proteins, such as Harmonin (USH1C) and Cadherin 23 (CDH23), SANS (USH1G) [36,37]. The MYO7A and SANS proteins form a complex involved in the formation of the stereo-cilia of hair cells, while the MyTH4-FERM tandem domains mediate the interaction between both proteins [36]. Through its first and second FERM domains; MYO7A binds to the integrin b5 subunit, and regulates cell adhesion and migration in non-stere-ocilia cells [38].
In conclusion, we describe here the first two Moroccan families with MYO7A compound heterozygote mutations causing hearing loss. In addition, our genotype-phenotype correlations suggest that these mutations are leading to the DFNB2 rather than USHIB clinical presentation. Elucidation of the molecular causes of these recessive disorders is crucial to implement prevention programs, especially for highly consanguineous populations.
Supporting information
(DOCX)
(XLSX)
Acknowledgments
The authors would like to thank all patients and their families for their collaboration. Pasteur Institute of Morocco (IPM) and a collaborative project between the French National Institute of Health and Medical Research (INSERM) supported this work and the Moroccan National Centre supported this work for Scientific and Technical Research (CNRST).
Data Availability
All relevant data are within the paper and its Supporting Information files.
Funding Statement
This work was supported by Pasteur Institute of Morocco (IPM) and a collaborative project between the French National Institute of Health and Medical Research (INSERM) and the Moroccan National Centre for Scientific and Technical Research (CNRST).
References
- 1.Smith RJH, Bale JF, White KR. Sensorineural hearing loss in children. Lancet Lond Engl. 2005;365: 879–890. [DOI] [PubMed] [Google Scholar]
- 2.Finsterer J, Fellinger J. Nuclear and mitochondrial genes mutated in nonsyndromic impaired hearing. Int J Pediatr Otorhinolaryngol. 2005;69: 621–647. doi: 10.1016/j.ijporl.2004.12.002 [DOI] [PubMed] [Google Scholar]
- 3.Inoue A, Ikebe M. Characterization of the motor activity of mammalian myosin VIIA. J Biol Chem. 2003;278: 5478–5487. doi: 10.1074/jbc.M210489200 [DOI] [PubMed] [Google Scholar]
- 4.Weil D, Küssel P, Blanchard S, Lévy G, Levi-Acobas F, Drira M, et al. The autosomal recessive isolated deafness, DFNB2, and the Usher 1B syndrome are allelic defects of the myosin-VIIA gene. Nat Genet. 1997;16: 191–193. doi: 10.1038/ng0697-191 [DOI] [PubMed] [Google Scholar]
- 5.Hasson T, Heintzelman MB, Santos-Sacchi J, Corey DP, Mooseker MS. Expression in cochlea and retina of myosin VIIa, the gene product defective in Usher syndrome type 1B. Proc Natl Acad Sci U S A. 1995;92: 9815–9819. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Riazuddin S, Nazli S, Ahmed ZM, Yang Y, Zulfiqar F, Shaikh RS, et al. Mutation spectrum of MYO7A and evaluation of a novel nonsyndromic deafness DFNB2 allele with residual function. Hum Mutat. 2008;29: 502–511. doi: 10.1002/humu.20677 [DOI] [PubMed] [Google Scholar]
- 7.Tamagawa Y, Ishikawa K, Ishikawa K, Ishida T, Kitamura K, Makino S, et al. Phenotype of DFNA11: a nonsyndromic hearing loss caused by a myosin VIIA mutation. The Laryngoscope. 2002;112: 292–297. doi: 10.1097/00005537-200202000-00017 [DOI] [PubMed] [Google Scholar]
- 8.Zhai W, Jin X, Gong Y, Qu L-H, Zhao C, Li Z-H. Phenotype of Usher syndrome type II assosiated with compound missense mutations of c.721 C>T and c.1969 C>T in MYO7A in a Chinese Usher syndrome family. Int J Ophthalmol. 2015;8: 670–674. doi: 10.3980/j.issn.2222-3959.2015.04.05 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Liu XZ, Walsh J, Tamagawa Y, Kitamura K, Nishizawa M, Steel KP, et al. Autosomal dominant non-syndromic deafness caused by a mutation in the myosin VIIA gene. Nat Genet. 1997;17: 268–269. doi: 10.1038/ng1197-268 [DOI] [PubMed] [Google Scholar]
- 10.Weil D, Blanchard S, Kaplan J, Guilford P, Gibson F, Walsh J, et al. Defective myosin VIIA gene responsible for Usher syndrome type 1B. Nature. 1995;374: 60–61. doi: 10.1038/374060a0 [DOI] [PubMed] [Google Scholar]
- 11.Millán JM, Aller E, Jaijo T, Blanco-Kelly F, Gimenez-Pardo A, Ayuso C. An update on the genetics of usher syndrome. J Ophthalmol. 2011;2011: 417217 doi: 10.1155/2011/417217 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Yoshimura H, Iwasaki S, Nishio S, Kumakawa K, Tono T, Kobayashi Y, et al. Massively Parallel DNA Sequencing Facilitates Diagnosis of Patients with Usher Syndrome Type 1. PLOS ONE. 2014;9: e90688 doi: 10.1371/journal.pone.0090688 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Petit C. Usher syndrome: from genetics to pathogenesis. Annu Rev Genomics Hum Genet. 2001;2: 271–297. doi: 10.1146/annurev.genom.2.1.271 [DOI] [PubMed] [Google Scholar]
- 14.Rong W, Chen X, Zhao K, Liu Y, Liu X, Ha S, et al. Novel and recurrent MYO7A mutations in Usher syndrome type 1 and type 2. PloS One. 2014;9: e97808 doi: 10.1371/journal.pone.0097808 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Liu XZ, Walsh J, Mburu P, Kendrick-Jones J, Cope MJ, Steel KP, et al. Mutations in the myosin VIIA gene cause non-syndromic recessive deafness. Nat Genet. 1997;16: 188–190. doi: 10.1038/ng0697-188 [DOI] [PubMed] [Google Scholar]
- 16.Riazuddin S, Nazli S, Ahmed ZM, Yang Y, Zulfiqar F, Shaikh RS, et al. Mutation spectrum of MYO7A and evaluation of a novel nonsyndromic deafness DFNB2 allele with residual function. Hum Mutat. 2008;29: 502–511. doi: 10.1002/humu.20677 [DOI] [PubMed] [Google Scholar]
- 17.Hildebrand MS, Thorne NP, Bromhead CJ, Kahrizi K, Webster JA, Fattahi Z, et al. Variable hearing impairment in a DFNB2 family with a novel MYO7A missense mutation. Clin Genet. 2010;77: 563–571. doi: 10.1111/j.1399-0004.2009.01344.x [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Ben-Salem S, Rehm HL, Willems PJ, Tamimi ZA, Ayadi H, Ali BR, et al. Analysis of two Arab families reveals additional support for a DFNB2 nonsyndromic phenotype of MYO7A. Mol Biol Rep. 2014;41: 193–200. doi: 10.1007/s11033-013-2851-5 [DOI] [PubMed] [Google Scholar]
- 19.Sambrook J, Russell DW. Molecular Cloning: A Laboratory Manual [Internet]. Cold Spring Harbor Laboratory Press; 2001. Available: https://books.google.fr/books?id=Bosc5JVxNpkC [Google Scholar]
- 20.Nielsen M, Lundegaard C, Lund O, Petersen TN. CPHmodels-3.0—remote homology modeling using structure-guided sequence profiles. Nucleic Acids Res. 2010;38: W576–581. doi: 10.1093/nar/gkq535 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Van Durme J, Delgado J, Stricher F, Serrano L, Schymkowitz J, Rousseau F. A graphical interface for the FoldX forcefield. Bioinforma Oxf Engl. 2011;27: 1711–1712. [DOI] [PubMed] [Google Scholar]
- 22.Krieger E, Vriend G. YASARA View—molecular graphics for all devices—from smartphones to workstations. Bioinforma Oxf Engl. 2014;30: 2981–2982. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Laimer J, Hiebl-Flach J, Lengauer D, Lackner P. MAESTROweb: a web server for structure-based protein stability prediction. Bioinforma Oxf Engl. 2016;32: 1414–1416. [DOI] [PubMed] [Google Scholar]
- 24.Worth CL, Preissner R, Blundell TL. SDM—a server for predicting effects of mutations on protein stability and malfunction. Nucleic Acids Res. 2011;39: W215–W222. doi: 10.1093/nar/gkr363 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Kumar P, Henikoff S, Ng PC. Predicting the effects of coding non-synonymous variants on protein function using the SIFT algorithm. Nat Protoc. 2009;4: 1073–1081. doi: 10.1038/nprot.2009.86 [DOI] [PubMed] [Google Scholar]
- 26.Adzhubei IA, Schmidt S, Peshkin L, Ramensky VE, Gerasimova A, Bork P, et al. A method and server for predicting damaging missense mutations. Nat Methods. 2010;7: 248–249. doi: 10.1038/nmeth0410-248 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.González-Pérez A, López-Bigas N. Improving the assessment of the outcome of nonsynonymous SNVs with a consensus deleteriousness score, Condel. Am J Hum Genet. 2011;88: 440–449. doi: 10.1016/j.ajhg.2011.03.004 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Reva B, Antipin Y, Sander C. Predicting the functional impact of protein mutations: application to cancer genomics. Nucleic Acids Res. 2011;39: e118 doi: 10.1093/nar/gkr407 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Schwarz JM, Rödelsperger C, Schuelke M, Seelow D. MutationTaster evaluates disease-causing potential of sequence alterations. Nat Methods. 2010;7: 575–576. doi: 10.1038/nmeth0810-575 [DOI] [PubMed] [Google Scholar]
- 30.Bakhchane A, Bousfiha A, Charoute H, Salime S, Detsouli M, Snoussi K, et al. Update of the spectrum of GJB2 gene mutations in 152 Moroccan families with autosomal recessive nonsyndromic hearing loss. Eur J Med Genet. 2016;59: 325–329. doi: 10.1016/j.ejmg.2016.05.002 [DOI] [PubMed] [Google Scholar]
- 31.Charif M, Bounaceur S, Abidi O, Nahili H, Rouba H, Kandil M, et al. The c.242G>A mutation in LRTOMT gene is responsible for a high prevalence of deafness in the Moroccan population. Mol Biol Rep. 2012;39: 11011–11016. doi: 10.1007/s11033-012-2003-3 [DOI] [PubMed] [Google Scholar]
- 32.Bakhchane A, Charif M, Salime S, Boulouiz R, Nahili H, Roky R, et al. Recessive TBC1D24 Mutations Are Frequent in Moroccan Non-Syndromic Hearing Loss Pedigrees. PloS One. 2015;10: e0138072 doi: 10.1371/journal.pone.0138072 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Bakhchane A, Charoute H, Nahili H, Roky R, Rouba H, Charif M, et al. A novel mutation in the TMC1 gene causes non-syndromic hearing loss in a Moroccan family. Gene. 2015;574: 28–33. doi: 10.1016/j.gene.2015.07.075 [DOI] [PubMed] [Google Scholar]
- 34.Sakai T, Jung HS, Sato O, Yamada MD, You D-J, Ikebe R, et al. Structure and Regulation of the Movement of Human Myosin VIIA. J Biol Chem. 2015;290: 17587–17598. doi: 10.1074/jbc.M114.599365 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Boulouiz R, Li Y, Abidi O, Bolz H, Chafik A, Kubisch C, et al. Analysis of MYO7A in a Moroccan family with Usher syndrome type 1B: novel loss-of-function mutation and non-pathogenicity of p.Y1719C. Mol Vis. 2007;13: 1862–1865. [PubMed] [Google Scholar]
- 36.Wu L, Pan L, Wei Z, Zhang M. Structure of MyTH4-FERM domains in myosin VIIa tail bound to cargo. Science. 2011;331: 757–760. doi: 10.1126/science.1198848 [DOI] [PubMed] [Google Scholar]
- 37.Boëda B, El-Amraoui A, Bahloul A, Goodyear R, Daviet L, Blanchard S, et al. Myosin VIIa, harmonin and cadherin 23, three Usher I gene products that cooperate to shape the sensory hair cell bundle. EMBO J. 2002;21: 6689–6699. doi: 10.1093/emboj/cdf689 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Liu Y, Guan L, Zhan J, Lu D, Wan J, Zhang H. FERM domain-containing unconventional myosin VIIA interacts with integrin β5 subunit and regulates αvβ5-mediated cell adhesion and migration. FEBS Lett. 2014;588: 2859–2866. doi: 10.1016/j.febslet.2014.06.049 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
(DOCX)
(XLSX)
Data Availability Statement
All relevant data are within the paper and its Supporting Information files.