Abstract
Changing the position of the poly(A) tail in an mRNA—alternative polyadenylation—is an important mechanism to increase the diversity of gene expression, especially in metazoans. Alternative polyadenylation often occurs in a tissue- or developmental stage-specific manner and can significantly affect gene activity by changing the protein product generated, the stability of the transcript, its localization, or its translatability. Despite the important regulatory effects that alternative polyadenylation have on gene expression, only a sparse few examples have been mechanistically characterized. Here, we review the known mechanisms for the control of alternative polyadenylation, catalog the tissues that demonstrate a propensity for alternative polyadenylation, and focus on the proteins that are known to regulate alternative polyadenylation in specific tissues. We conclude that the field of alternative polyadenylation remains in its infancy, with possibilities for future investigation on the horizon. Given the profound effect alternative polyadenylation can have on gene expression and human health, improved understanding of alternative polyadenylation could lead to numerous advances in control of gene activity.
INTRODUCTION
The goal of this article is to describe many of the known mechanisms of regulated nuclear mRNA polyadenylation in specific metazoan tissues: tissue-specific alternative polyadenylation. With that goal, we will not address (except in passing) cytoplasmic polyadenylation, which has been reviewed elsewhere, including in this WIREs RNA series. Nor will we address polyadenylation in yeast and other developmentally simple organisms, since they are not generally regarded as having tissues. What we will cover is alternative polyadenylation—the meaning of which has evolved since the advent of bioinformatic analyses of genomes and their transcribed products. Where possible in this article, we will identify tissue-specific proteins that are known to be involved with the mechanisms of alternative polyadenylation. By identifying these components in tissue-specific alternative polyadenylation, we will have a better opportunity of learning its principles.
POLYADENYLATION AND ALTERNATIVE POLYADENYLATION
mRNA polyadenylation—the cleavage of a pre-mRNA at its 3′ end, followed by the addition of 200–250 adenosine residues—is essential for the transcription, nuclear export, stability, translation, and quality control of the mRNA. To learn more about polyadenylation, read any of the excellent reviews on its mechanisms (here are two to get you started: Refs 1 and 2). The choice of a transcript’s cleavage/polyadenylation site occurs in the nucleus and is an important regulated process in gene expression. Changes in this site choice—alternative polyadenylation—can lead to changes in transcript stability, localization, splicing pattern, translation, and the protein product generated from the gene. More than 30% of mouse and 50% of human transcripts are alternatively polyadenylated.3 Surprisingly, relatively few mechanisms controlling alternative polyadenylation have been elucidated, despite its prominent role in gene expression and human health.4
Alternative polyadenylation
The definition of alternative polyadenylation has evolved since its discovery. In the early days, it was assumed that the first polyadenylation site identified for a given transcript was the Platonic default site and that any subsequently identified sites were alternatives. Presumably, the early thinking continued, there was also a default polyadenylation machinery (usually conceived of as the machinery in HeLa cells), and tissue-specific auxiliary proteins resulted in the choice of alternative sites. As we will show here, this view is probably too simple. For example, mRNAs in individual tissues will display use of more than one polyadenylation site, although the ratios of the sites may vary from tissue to tissue. Therefore, for the purposes of this article, we will define tissue-specific alternative polyadenylation as predominant use of one or more sites in a tissue that requires augmentation or modification of the core polyadenylation machinery.
Core polyadenylation proteins recognize signals in the pre-mRNA
Several signals in the pre-mRNA direct the polyadenylation machinery to a site. The sequence AAUAAA (or something similar) is called the polyadenylation signal (PAS) and is generally found 15–30 bases upstream of the site of cleavage/polyadenylation. A second signal, downstream of the cleavage site, is the downstream sequence element (DSE), which is U or G/U rich. Other sequences upstream or downstream of the cleavage site also appear to modulate polyadenylation (Figure 1); often these other sequences may be bound by tissue-specific factors.
FIGURE 1.
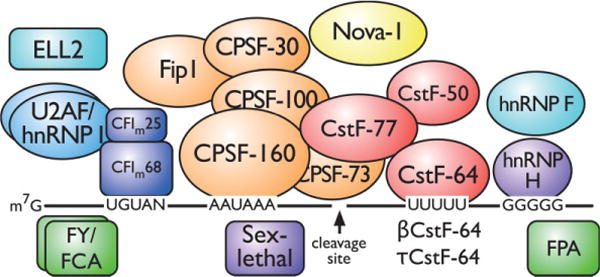
Core and auxiliary proteins involved in tissue-specific alternative polyadenylation. The pre-mRNA (black line) consists of upstream sequence elements (UGUAN), the polyadenylation signal (AAUAAA), a cleavage site (arrow), the downstream sequence element (UUUUU), and the downstream G-rich element (GGGGG). The core polyadenylation proteins consist of the cleavage and polyadenylation specificity factor proteins, the cleavage stimulation factor proteins, and mammalian cleavage factor I. Auxiliary (U2AF, hnRNP F, hnRNP H, and hnRNP I) and tissue-specific (Nova-1, βCstF-64, τCstF-64, ELL2, sex-lethal, FPA, FY, and FCA) proteins are indicated.
Each pre-mRNA signal is bound by a specific component of the polyadenylation machinery. The cleavage and polyadenylation specificity factor (CPSF, consisting of five different polypeptides of 160, 100, 73, and 30 kDa and Fip1) and the cleavage stimulation factor (CstF, consisting of three proteins of 77, 64, and 50 kDa) bind to the AAUAAA and U- or G/U-rich elements, respectively. CPSF-160 and CstF-64 contain RNA-binding domains and bind directly to their respective elements. Additional cleavage factors (CFIm and CFIIm) interact with UGUAN elements upstream of the PAS. The affinity of CPSF and CstF for the pre-mRNA determines whether a polyadenylation site is weak or strong. This affinity depends on the exact sequences of the PAS, DSE, and auxiliary elements and responds to changes in the amounts of the core polyadenylation proteins. Stability of these complexes can also be modified by interactions with other factors, and probably by protein modification, as well. Following cleavage (by CPSF-73), the poly(A) polymerase adds up to 250 adenosine residues.
Mechanisms of alternative polyadenylation
Surprisingly few tissue-specific auxiliary factors have been discovered. To our knowledge, only a small number of mechanisms of alternative polyadenylation have been fully characterized with direct evidence leading to a change in polyadenylation site for that tissue/developmental stage [see the discussions below on CstF-64 in B cells, FY in Arabidopsis, and neuro-oncological ventral antigen (Nova) in brain]. The remaining systems remain mysterious, although there are strong candidate proteins in each. One can imagine additional mechanisms for alternative polyadenylation, including modification of basal polyadenylation machinery (by phosphorylation, for example), blockade of more optimal binding sites by RNA-binding proteins, and others. However, the number of validated alternative polyadenylation mechanisms is currently very small.a
ABUNDANT ALTERNATIVE POLYADENYLATION IN THE BRAIN SUPPORTS NEURAL FUNCTION
Splicing and polyadenylation control calcitonin gene-related peptide expression
The brain and peripheral neurons express a larger proportion of the genome than any other tissue, resulting in a high level of mRNA diversity. Most of this diversity is attributed to alternative splicing and polyadenylation. In fact, the earliest described case of alternative processing of a mammalian gene was the neuronally expressed calcitonin gene-related peptide (CGRP) alpha protein. CGRP is expressed from the Calca gene, which produces calcitonin in nonneurological tissues. The earliest reports proposed that the CGRP pattern of processing was caused by alternative polyadenylation,5 but subsequent studies determined that neuronal processing of CGRP from calcitonin was a complex gallimaufry of alternative splicing and polyadenylation that still holds secrets.6
Three forms of CstF-64 are expressed in brain
Studies from our laboratory have shown that CstF-64 (which is expressed in all tissues) and two variants, τCstF-64 and βCstF-64 (Figure 2), are expressed in brain. τCstF-64 (gene name Cstf2t) is a retrotransposed paralog of CstF-64 that is expressed at highest levels in testis, but is also detected in brain and immune cells.7,8 τCstF-64 will be discussed in more detail below because of its role in spermatogenesis. However, targeted deletion of Cstf2t did not result in obvious neurological or immunological problems, so its roles in the brain and immune systems are either subtle or unnecessary.9
FIGURE 2.
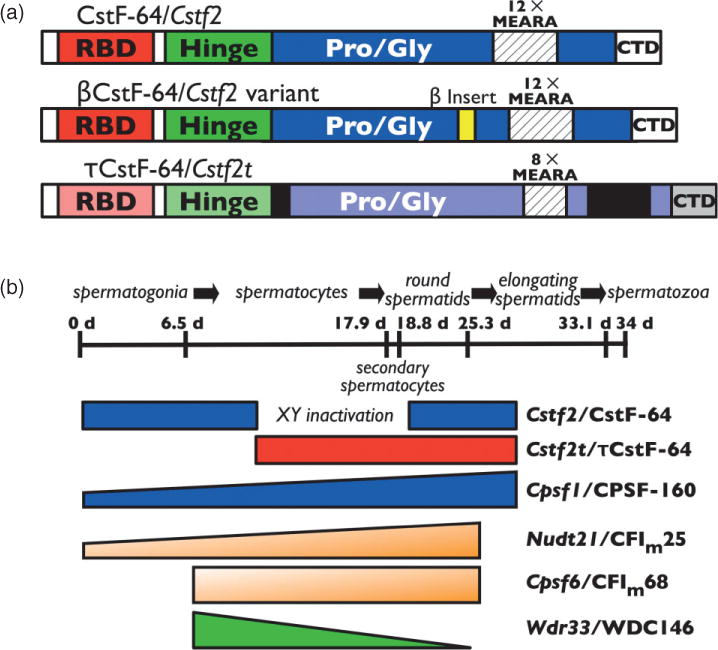
Proteins involved in testis and brain alternative polyadenylation. (a) Three forms of CstF-64 are involved in polyadenylation. βCstF-64 (middle) is an alternatively spliced variant of CstF-64 (top) that is expressed in vertebrate neurons. τCstF-64 (bottom) is an autosomal paralog of CstF-64 that is expressed primarily in male germ cells in mammals. The RNA-binding domain, CstF-77 interaction domain (Hinge), proline- and glycine-rich domain (Pro/Gly), MEARA amino acid repeats, and C-terminal domain are indicated. (b) Expression patterns of alternative polyadenylation proteins during spermatogenesis. A timeline of spermatogenesis in mice has been shown (~34 days). Cells that undergo mitotic division (spermatogonia), meiosis (spermatocytes), and postmeiotic development (round or elongating spermatids, and spermatozoa) are indicated. Expression periods for CstF-64, τCstF-64, CPSF-160, CFIm25, CFIm68, and WDC146 are at bottom. XY inactivation indicates the period of male sex chromosome inactivation.
βCstF-64, however, seems more likely to play an essential role in brain polyadenylation. βCstF-64 (the Greek letter β was chosen to designate its expression in brain) is a neuron-specific splice variant of CstF-64 that results in addition of 49 amino acids within the proline/glycine-rich domain of CstF-64.10 βCstF-64 is expressed in all regions of the brain and in peripheral nerves, suggesting it has a ubiquitous function in the nervous system. Furthermore, it is present in all vertebrates, suggesting that its function is ancient and important. Finally, ectopic expression of βCstF-64 in somatic cells stimulates expression of luciferase reporter mRNAs with neuronal polyadenylation sites (G. S. Shankarling and C. C. MacDonald, unpublished data), demonstrating its probable involvement in mRNA processing. However, a direct role for βCstF-64 in control of alternative polyadenylation has not yet been demonstrated.
Nova controls alternative polyadenylation as well as alternative splicing
The cutely named Nova proteins are a family of RNA-binding proteins that were found associated with paraneoplastic opsoclonus myoclonus ataxia, a neurological disorder that is also associated with neuroblastoma. Analyses of sites of Nova-1 (gene name: Nova1) binding in brain RNA demonstrated a strong association of Nova-1 with intronic regions near alternatively spliced exons as well as changes in splice patterns for key genes.11 Newer analyses discovered an additional association of Nova-1 with 3′ untranslated regions (UTRs) of pre-mRNAs near polyadenylation sites.12 Inclusion of those sites in the 3′ ends of reporter genes decreased polyadenylation in the presence of Nova-1. Licatalosi et al.12 speculate that Nova-1 could suppress polyadenylation at some sites by binding adjacent to CPSF or CstF elements in the pre-mRNA or could enhance polyadenylation at other sites by antagonizing binding of unknown ‘auxiliary factors’. A third possibility the authors did not enumerate was that Nova might attract auxiliary splicing factors to polyadenylation sites and influence site selection in that way. By whatever mechanism, Nova-1 is associated with neuronal control of alternative polyadenylation.
CstF-64 CONTROLS ALTERNATIVE POLYADENYLATION IN THE IMMUNE SYSTEM
The immune system uses myriad mechanisms to diversify gene expression, and alternative polyadenylation is not to be left out. Immunoglobulin (Ig) is expressed in two forms: early B cells make a membrane-bound Ig, while later plasma cells make a secreted form. The difference between these two protein isoforms is a short membrane-spanning domain in B cells but not in plasma cells. This protein domain switch is decided by which of two polyadenylation sites is selected: if the upstream site is selected, then the secreted form is produced; if the downstream site is selected, then the membrane-bound form is produced. Increasing the levels of CstF-64 in a model B cell line is sufficient to switch Ig from the membrane-bound to the secreted form.13 These and similar experiments have led to models in which amounts and activities of proteins of the core polyadenylation machinery (not just CstF-64, although CstF-64 is a common culprit) regulate poly(A) site selection. Subsequent experiments showed that there is even more complexity in Ig switching (involving splicing, hnRNP binding, RNA polymerase loading, and transcriptional elongation) than was first thought.14 Alternative polyadenylation in other tissues likely shares this complexity, as well.
MULTIPLE TESTIS-SPECIFIC POLYADENYLATION PROTEINS ARE EXPRESSED DURING SPERMATOGENESIS
Patterns of polyadenylation are different in male germ cells
There are many reports on mRNAs that use unique polyadenylation sites in mammalian testis.15,16 In addition to alternative polyadenylation leading to 3′ UTRs of different lengths, alternative polyadenylation sites can lead to germ cell-specific protein isoforms and even control long interspersed element (LINE-1) activity.17 Testis-expressed polyadenylation sites display a lower incidence of AAUAAA, contain unique upstream and DSEs, and have shorter 3′ UTRs.15,18 Additionally, polyadenylation regions from testis-enriched polyadenylation sites are used inefficiently in somatic cells.19 These observations suggest that there are testis-specific mechanisms supporting nuclear polyadenylation in male germ cells.
CstF-64 and τCstF-64 are expressed during spermatogenesis
CstF-64 (gene name: Cstf2) and CPSF-160 (gene name: Cpsf1) are overexpressed in postmeiotic male germ cells.20 Overexpression of CstF-64 could influence polyadenylation site choice in some germ-cell genes21 as it does in Ig in immune cells. However, CstF-64 is not expressed during meiosis because of male sex chromosome inactivation (Figure 2). During the critical period of meiosis in males (which lasts almost for 9 days in mice and considerably longer in humans), the X and Y chromosomes are inactivated, so the X-linked CstF-64 is not available to participate in polyadenylation. Thus, researchers in our laboratory discovered a variant of CstF-64 in male germ cells that we named τCstF-6422 (the ‘τ’ was chosen to remind us of its expression in testis, although it is also expressed in brain and immune cells). τCstF-64 (gene name: Cstf2t) is an autosomal paralog of Cstf2t that likely resulted from retrotransposition of the CstF-64 mRNA23 around the time mammals diverged from archosaurs (B. Dass and C. C. MacDonald, unpublished data). Like CstF-64, τCstF-64 coimmunoprecipitates with other members of the CstF polyadenylation complex (K. W. M. and C. C. MacDonald, unpublished data) and therefore likely participates in 3′ end formation of germ-cell mRNAs.
Because of its pattern of expression in testis (Figure 2), it seemed likely that τCstF-64 would be necessary for germ-cell polyadenylation and thus for gene expression during spermatogenesis. Targeted deletion of Cstf2t confirmed that hypothesis: male (but not female) Cstf2t−/− mice were infertile due to aberrant meiotic and postmeiotic development but were otherwise physiologically normal.9 Unexpectedly, spermatogenesis, while defective, did not halt, as might have been expected if polyadenylation (and thus gene expression) was blocked during meiosis. Instead, a few motile spermatozoa were seen, comparable to the human infertility known as oligoasthenoteratozoospermia. Since expression of many thousands of mRNAs was altered in the Cstf2t−/− mouse testes, this suggested that cascades of genes were affected in these mice. The most likely—but by no means the only—hypothesis is that τCstF-64 is critical for the proper polyadenylation of a few key genes and that those key genes then affect a larger number of downstream genes. Determining how τCstF-64 controls expression of those key targets will be important for determining mechanisms that control spermatogenesis as well as polyadenylation.
Core and peripheral polyadenylation proteins in male germ cells
Mammalian cleavage factor I (CFIm) is a heterodimer consisting of a small subunit of 25 kDa (CFIm25, gene name: Nudt21) and a large subunit that is either 59, 68, or 72 kDa (the 68 and 72 kDa forms are formed by alternative splicing of the Cpsf6 gene, while the 59 kDa polypeptide is from a paralogous gene). Together, the large and small subunits of CFIm enhance the recruitment of CPSF to poly(A) sites that lack AAUAAA by binding to upstream UGUAN motifs.24,25 In male germ cells, CFIm25 and CFIm68 are expressed more highly than in other tissues and seem to contribute to their own alternative polyadenylation.26 CFIm25 is also associated with DNA near the sterol regulatory element-binding transcription factor 2 (Srebf2) polyadenylation sites26 and probably contributes to the meiotic and postmeiotic alternative polyadenylation of Srebf2 in germ cells.27
Other proteins are expressed in the testis which could potentially be involved in the testis-specific patterns of polyadenylation. For example, WDC146 (gene name: Wdr33) is a mammalian homolog of the Pfs2 (yeast) and FY (Arabidopsis) proteins.28 In yeast, Pfs2p bridges several core polyadenylation proteins, while in Arabidopsis, FY changes polyadenylation site choice of its own mRNA (see subsequent sections). Since WDC146 is expressed in nuclei of spermatogenic cells,29 it too might be involved in testis-specific polyadenylation.
OTHER ORGANISMS, OTHER TISSUES
Mammals do not have a monopoly on tissue-specific alternative polyadenylation. For example, suppressor-of-forked is the Drosophila homolog of CstF-77. Mutations in su(f) suppress the forked bristle phenotype of f1 in Drosophila epidermal cells.30 CstF-77 also controls its own alternative polyadenylation in Drosophila and possibly mammalian tissues.31,32 Not to be left out, the Drosophila sex-lethal protein, in addition to controlling gender, regulates alternative polyadenylation of enhancer of rudimentary in female germline cells.33
Alternative polyadenylation regulates seasonal plant flowering at several levels
Plants have a complex system controlling seasonal flowering in the floral meristem. The mRNA encoding FCA (a central regulator of flowering) has several alternatively polyadenylated forms, the longer of which encodes an RNA-binding protein.28 Full-length FCA binds FY, and together, this complex redirects polyadenylation from the full-length site back upstream to the alternative site leading to the production of a truncated FCA and the loss of RNA binding. Neither FCA nor FY is a direct homolog of any member of the core mammalian polyadenylation machinery (Arabidopsis has been shown to possess the full arsenal of polyadenylation proteins observed in mammals), although the FY homolog PFS2 is a core member in Saccharomyces cerevisiae. This autoregulatory loop was, therefore, the first example of an auxiliary factor that redirected polyadenylation site choice from one position to another.
In addition, there is so much more going on in flowering control. Functioning similarly to FY, PCFS4 (a homolog of the Pcf11 polyadenylation protein) is an auxiliary protein involved in the alternative polyadenylation of the FCA mRNA.34 Similarly, FPA (an RNA-binding protein of the spen gene family) controls alternative polyadenylation of antisense FCA transcripts35; CstF-64 and CstF-77 are probably also involved in this mechanism.36 Finally, we would be remiss not to mention the considerable research into polyadenylation mechanisms in Arabidopsis and other plants, which affect gene expression in embryo and gametophyte development, flowers, leaves, and other plant tissues.37–39
CONCLUSION—WHERE ARE ALL THE TISSUE-SPECIFIC POLYADENYLATION FACTORS?
Tissue-specific alternative polyadenylation is a widespread phenomenon. Yet, in preparing this review, we were struck by how much remains to be understood about its mechanisms. For example, surprisingly few tissue-specific auxiliary proteins have been discovered that control alternative polyadenylation (FY and FPA are the most prominent). More often, changes in levels of the core polyadenylation proteins seem to be involved (CstF-64 and its variants or CFIm) or splicing vies with polyadenylation for control (the Ig switch or Nova in brain). Authenticated mechanisms for other examples remain tantalizingly incomplete.
Furthermore, none of these mechanisms turn out to be as simple as they might first appear. The calcitonin/CGRP system is still being studied and has uncovered new mechanisms for splicing as well as for polyadenylation. Similarly, CstF-64 is only a small part of the Ig switch in B cells. However, hope remains for understanding these mechanisms. A recent proteomic study of the core polyadenylation machinery uncovered a list of potential central and auxiliary factors.40 Some of these were previously identified as potential factors in alternative polyadenylation. Exciting results from histone mRNA 3′ end formation may reveal new candidates.41 Chromatin modifications likely play a role, as well.42 We look forward to further progress in this arena.
Footnotes
We have used the mouse gene naming conventions throughout this article because the majority of the genetic work, including gene knockouts, has been done in this species.
References
- 1.Mandel CR, Bai Y, Tong L. Protein factors in pre-mRNA 3′-end processing. Cell Mol Life Sci. 2008;65:1099–1122. doi: 10.1007/s00018-007-7474-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Millevoi S, Vagner S. Molecular mechanisms of eukaryotic pre-mRNA 3′ end processing regulation. Nucleic Acids Res. 2010;38:2757–2774. doi: 10.1093/nar/gkp1176. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Tian B, Hu J, Zhang H, Lutz CS. A large-scale analysis of mRNA polyadenylation of human and mouse genes. Nucleic Acids Res. 2005;33:201–212. doi: 10.1093/nar/gki158. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Danckwardt S, Hentze MW, Kulozik AE. 3′ end mRNA processing: molecular mechanisms and implications for health and disease. EMBO J. 2008;27:482–498. doi: 10.1038/sj.emboj.7601932. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Amara SG, Evans RM, Rosenfeld MG. Calci-tonin/calcitonin gene-related peptide transcription unit: tissue-specific expression involves selective use of alternative polyadenylation sites. Mol Cell Biol. 1984;4:2151–2160. doi: 10.1128/mcb.4.10.2151. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Zhou HL, Baraniak AP, Lou H. Role for Fox-1/Fox-2 in mediating the neuronal pathway of calcitonin/calcitonin gene-related peptide alternative RNA processing. Mol Cell Biol. 2007;27:830–841. doi: 10.1128/MCB.01015-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Wallace AM, Dass B, Ravnik SE, Tonk V, Jenkins NA, Gilbert DJ, Copeland NG, MacDonald CC. Two distinct forms of the 64,000 Mr protein of the cleavage stimulation factor are expressed in mouse male germ cells. Proc Natl Acad Sci U S A. 1999;96:6763–6768. doi: 10.1073/pnas.96.12.6763. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Wallace AM, Denison T, Attaya EN, MacDonald CC. Developmental differences in expression of two forms of the CstF-64 polyadenylation protein in rat and mouse. Biol Reprod. 2004;70:1080–1087. doi: 10.1095/biolreprod.103.022947. [DOI] [PubMed] [Google Scholar]
- 9.Dass B, Tardif S, Park JY, Tian B, Weitlauf HM, Hess RA, Carnes K, Griswold MD, Small CL, MacDonald CC. Loss of polyadenylation protein τCstF-64 causes spermatogenic defects and male infertility. Proc Natl Acad Sci U S A. 2007;104:20374–20379. doi: 10.1073/pnas.0707589104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Shankarling GS, Coates PW, Dass B, MacDonald CC. A family of splice variants of CstF-64 expressed in vertebrate nervous systems. BMC Mol Biol. 2009;10:22. doi: 10.1186/1471-2199-10-22. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Ule J, Jensen KB, Ruggiu M, Mele A, Ule A, Darnell RB. CLIP identifies Nova-regulated RNA networks in the brain. Science. 2003;302:1212–1215. doi: 10.1126/science.1090095. [DOI] [PubMed] [Google Scholar]
- 12.Licatalosi DD, Mele A, Fak JJ, Ule J, Kayikci M, Chi SW, Clark TA, Schweitzer AC, Blume JE, Wang X, et al. HITS-CLIP yields genome-wide insights into brain alternative RNA processing. Nature. 2008;456:464–469. doi: 10.1038/nature07488. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Takagaki Y, Seipelt RL, Peterson ML, Manley JL. The polyadenylation factor CstF-64 regulates alternative processing of IgM heavy chain pre-mRNA during B cell differentiation. Cell. 1996;87:941–952. doi: 10.1016/s0092-8674(00)82000-0. [DOI] [PubMed] [Google Scholar]
- 14.Martincic K, Alkan SA, Cheatle A, Borghesi L, Milcarek C. Transcription elongation factor ELL2 directs immunoglobulin secretion in plasma cells by stimulating altered RNA processing. Nat Immunol. 2009;10:1102–1109. doi: 10.1038/ni.1786. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Liu D, Brockman JM, Dass B, Hutchins LN, Singh P, McCarrey JR, MacDonald CC, Graber JH. Systematic variation in mRNA 3′-processing signals during mouse spermatogenesis. Nucleic Acids Res. 2007;35:234–246. doi: 10.1093/nar/gkl919. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Zhang H, Lee JY, Tian B. Biased alternative polyadenylation in human tissues. Genome Biol. 2005;6:R100. doi: 10.1186/gb-2005-6-12-r100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Perepelitsa-Belancio V, Deininger P. RNA truncation by premature polyadenylation attenuates human mobile element activity. Nat Genet. 2003;35:363–366. doi: 10.1038/ng1269. [DOI] [PubMed] [Google Scholar]
- 18.Ji Z, Lee JY, Pan Z, Jiang B, Tian B. Progressive lengthening of 3′ untranslated regions of mRNAs by alternative polyadenylation during mouse embryonic development. Proc Natl Acad Sci U S A. 2009;106:7028–7033. doi: 10.1073/pnas.0900028106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.McMahon KW, Hirsch BA, MacDonald CC. Differences in polyadenylation site choice between somatic and male germ cells. BMC Mol Biol. 2006;7:35. doi: 10.1186/1471-2199-7-35. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Dass B, Attaya EN, Wallace AM, MacDonald CC. Overexpression of the CstF-64 and CPSF-160 polyadenylation protein messenger RNAs in mouse male germ cells. Biol Reprod. 2001;64:1722–1729. doi: 10.1095/biolreprod64.6.1722. [DOI] [PubMed] [Google Scholar]
- 21.Chennathukuzhi VM, Lefrancois S, Morales CR, Syed V, Hecht NB. Elevated levels of the polyadenylation factor CstF 64 enhance formation of the 1 kB testis brain RNA-binding protein (TB-RBP) mRNA in male germ cells. Mol Reprod Dev. 2001;58:460–469. doi: 10.1002/1098-2795(20010401)58:4<460::AID-MRD15>3.0.CO;2-F. [DOI] [PubMed] [Google Scholar]
- 22.Dass B, McMahon KW, Jenkins NA, Gilbert DJ, Copeland NG, MacDonald CC. The gene for a variant form of the polyadenylation protein CstF-64 is on chromosome 19 and is expressed in pachytene spermatocytes in mice. J Biol Chem. 2001;276:8044–8050. doi: 10.1074/jbc.M009091200. [DOI] [PubMed] [Google Scholar]
- 23.Wang PJ. X chromosomes, retrogenes and their role in male reproduction. Trends Endocrinol Metab. 2004;15:79–83. doi: 10.1016/j.tem.2004.01.007. [DOI] [PubMed] [Google Scholar]
- 24.Venkataraman K, Brown KM, Gilmartin GM. Analysis of a noncanonical poly(A) site reveals a tripartite mechanism for vertebrate poly(A) site recognition. Genes Dev. 2005;19:1315–1327. doi: 10.1101/gad.1298605. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Kubo T, Wada T, Yamaguchi Y, Shimizu A, Handa H. Knock-down of 25 kDa subunit of cleavage factor Im in Hela cells alters alternative polyadenylation within 3′-UTRs. Nucleic Acids Res. 2006;34:6264–6271. doi: 10.1093/nar/gkl794. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Sartini BL, Wang H, Wang W, Millette CF, Kilpatrick DL. Pre-messenger RNA cleavage factor I (CFIm): potential role in alternative polyadenylation during spermatogenesis. Biol Reprod. 2008;78:472–482. doi: 10.1095/biolreprod.107.064774. [DOI] [PubMed] [Google Scholar]
- 27.Wang H, Sartini BL, Millette CF, Kilpatrick DL. A developmental switch in transcription factor isoforms during spermatogenesis controlled by alternative messenger RNA 3′-end formation. Biol Reprod. 2006;75:318–323. doi: 10.1095/biolreprod.106.052209. [DOI] [PubMed] [Google Scholar]
- 28.Simpson GG, Dijkwel PP, Quesada V, Henderson I, Dean C. FY is an RNA 3′ end-processing factor that interacts with FCA to control the Arabidopsis floral transition. Cell. 2003;113:777–787. doi: 10.1016/s0092-8674(03)00425-2. [DOI] [PubMed] [Google Scholar]
- 29.Ito S, Sakai A, Nomura T, Miki Y, Ouchida M, Sasaki J, Shimizu K. A novel WD40 repeat protein, WDC146, highly expressed during spermatogenesis in a stage-specific manner. Biochem Biophys Res Commun. 2001;280:656–663. doi: 10.1006/bbrc.2000.4163. [DOI] [PubMed] [Google Scholar]
- 30.Simonelig M, Elliott K, Mitchelson A, O’Hare K. Inter-allelic complementation at the suppressor of forked locus of Drosophila reveals complementation between suppressor of forked proteins mutated in different regions. Genetics. 1996;142:1225–1235. doi: 10.1093/genetics/142.4.1225. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Pan Z, Zhang H, Hague LK, Lee JY, Lutz CS, Tian B. An intronic polyadenylation site in human and mouse CstF-77 genes suggests an evolutionarily conserved regulatory mechanism. Gene. 2006;366:325–334. doi: 10.1016/j.gene.2005.09.024. [DOI] [PubMed] [Google Scholar]
- 32.Hatton LS, Eloranta JJ, Figueiredo LM, Takagaki Y, Manley JL, O’Hare K. The Drosophila homologue of the 64 kDa subunit of cleavage stimulation factor interacts with the 77 kDa subunit encoded by the suppressor of forked gene. Nucleic Acids Res. 2000;28:520–526. doi: 10.1093/nar/28.2.520. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Gawande B, Robida MD, Rahn A, Singh R. Drosophila sex-lethal protein mediates polyadenylation switching in the female germline. EMBO J. 2006;25:1263–1272. doi: 10.1038/sj.emboj.7601022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Xing D, Zhao H, Xu R, Li QQ. Arabidopsis PCFS4, a homologue of yeast polyadenylation factor Pcf11p, regulates FCA alternative processing and promotes flowering time. Plant J. 2008;54:899–910. doi: 10.1111/j.1365-313X.2008.03455.x. [DOI] [PubMed] [Google Scholar]
- 35.Hornyik C, Terzi LC, Simpson GG. The Spen family protein FPA controls alternative cleavage and polyadenylation of RNA. Dev Cell. 2010;18:203–213. doi: 10.1016/j.devcel.2009.12.009. [DOI] [PubMed] [Google Scholar]
- 36.Liu F, Marquardt S, Lister C, Swiezewski S, Dean C. Targeted 3′ processing of antisense transcripts triggers Arabidopsis FLC chromatin silencing. Science. 2010;327:94–97. doi: 10.1126/science.1180278. [DOI] [PubMed] [Google Scholar]
- 37.Lorkovic ZJ. Role of plant RNA-binding proteins in development, stress response and genome organization. Trends Plant Sci. 2009;14:229–236. doi: 10.1016/j.tplants.2009.01.007. [DOI] [PubMed] [Google Scholar]
- 38.Xing D, Zhao H, Li QQ. Arabidopsis CLP1-SIMILAR PROTEIN3, an ortholog of human polyadenylation factor CLP1, functions in gametophyte, embryo, and postembryonic development. Plant Physiol. 2008;148:2059–2069. doi: 10.1104/pp.108.129817. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Hunt AG, Xu R, Addepalli B, Rao S, Forbes KP, Meeks LR, Xing D, Mo M, Zhao H, Bandyopadhyay A, et al. Arabidopsis mRNA polyadenylation machinery: comprehensive analysis of protein–protein interactions and gene expression profiling. BMC Genomics. 2008;9:220. doi: 10.1186/1471-2164-9-220. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Shi Y, Di Giammartino DC, Taylor D, Sarkeshik A, Rice WJ, Yates JR, 3rd, Frank J, Manley JL. Molecular architecture of the human pre-mRNA 3′ processing complex. Mol Cell. 2009;33:365–376. doi: 10.1016/j.molcel.2008.12.028. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Sullivan KD, Steiniger M, Marzluff WF. A core complex of CPSF73, CPSF100, and Symplekin may form two different cleavage factors for processing of poly(A) and histone mRNAs. Mol Cell. 2009;34:322–332. doi: 10.1016/j.molcel.2009.04.024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Spies N, Nielsen CB, Padgett RA, Burge CB. Biased chromatin signatures around polyadenylation sites and exons. Mol Cell. 2009;36:245–254. doi: 10.1016/j.molcel.2009.10.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
FURTHER READING
- Paronetto MP, Sette C. Role of RNA-binding proteins in mammalian spermatogenesis. Int J Androl. 2010;33:2–12. doi: 10.1111/j.1365-2605.2009.00959.x. A recent review of the role of RNA-binding proteins, nuclear and cytoplasmic, in post-transcriptional control of spermatogenesis. [DOI] [PubMed] [Google Scholar]
- Ehrmann I, Elliott DJ. Post-transcriptional control in the male germ line. Reprod Biomed Online. 2005;10:55–63. doi: 10.1016/s1472-6483(10)60804-8. Summary of post-transcriptional mechanisms in males from primordial germ cells to spermatogenesis. [DOI] [PubMed] [Google Scholar]
- Wang ET, Sandberg R, Luo S, Khrebtukova I, Zhang L, Mayr C, Kingsmore SF, Schroth GP, Burge CB. Alternative isoform regulation in human tissue transcriptomes. Nature. 2008;456:470–476. doi: 10.1038/nature07509. An informative microarray-based survey of alternative splicing and polyadenylation in human tissues. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mayr C, Bartel DP. Widespread shortening of 3′ UTRs by alternative cleavage and polyadenylation activates oncogenes in cancer cells. Cell. 2009;138:673–684. doi: 10.1016/j.cell.2009.06.016. Alternative polyadenylation appears to play a role in cancers from a variety of sources. [DOI] [PMC free article] [PubMed] [Google Scholar]


