Abstract
Metabolism coordinates the conversion of available nutrients toward energy, biosynthetic intermediates, and signaling molecules to mediate virtually all biological functions. Dysregulation of metabolic pathways contributes to many diseases, so a detailed understanding of human metabolism has significant therapeutic implications. Over the last decade major technological advances in the areas of analytical chemistry, computational estimation of intracellular fluxes, and biological engineering have improved our ability to observe and engineer metabolic pathways. These approaches are reminiscent of the design, operation, and control of industrial chemical plants. Immune cells have emerged as an intriguing system in which metabolism influences diverse biological functions. Application of metabolic flux analysis and related approaches to macrophages and T cells offers great therapeutic opportunities to biochemical engineers.
Graphical abstract
Introduction
Metabolism comprises the set of coordinated biochemical reactions that are executed by cells. Thus, metabolic processes represent a critical link between a cell’s genetic program (which encodes mRNA and proteins/enzymes) and the surrounding chemical microenvironment, where substrates are converted to energy and the biosynthetic intermediates required for cell division. The metabolic state of a cell or organism is therefore tightly linked to its health, and such information is of particular use in the field of biomedicine. Many, but not all, of the most active metabolic pathways in mammalian cells have been documented and characterized over the last century. For example, new pathways associated with the tricarboxylic acid (TCA) cycle [1,2] and pentose phosphate pathway [3,4] have recently been characterized in mammals, providing new potential targets for controlling inflammation (Immune-responsive gene 1/cis-aconitate decarboxylase; IRG1/CAD) and cancer cell growth (transketolase-like 1; TKTL1), respectively. However, beyond such basic pathway discoveries we also have much to learn about the functional regulation of many biochemical pathways in human cells. Engineers have solved analogous problems in designing mechanical, electrical, and chemical systems, offering lessons for biomedicine which can enhance our understanding of disease pathogenesis. Neither engineering or biology alone can succeed in this task. Critical insights into the phenotype of metabolic disorders and diseases in general will come from the clinic [5]. Detailed information on the behavior of such interconnected metabolic networks will come from systems-based analytics [6]. In the coming years these approaches will become increasingly integrated to advance our understanding of human physiology in the coming years [7].
In this review we highlight the utility of engineering concepts in studying cellular metabolism. We relate cellular functions and human metabolic physiology to that of an industrial chemical plant, highlighting the utility of real-time process parameters in operation of the latter while pointing out the need for analogous data in human metabolism. We highlight recent advances in the areas of analytical chemistry, computational analysis of metabolomics datasets, and biological engineering that are now facilitating the acquisition of human biochemical process conditions. Finally, we discuss recent studies that have explored the role of metabolism in regulating the immune system, an area of intense interest within the biomedical community that holds great therapeutic potential. The convergence of engineering and biomedical science on these problems is likely to catalyze many discoveries in the coming years.
Chemical process plant as an engineered model system
The regulation of cellular biochemical processes has grown more complex throughout evolution, with functional specialization increasing from prokaryotes to eukaryotes and, in turn, to multicellular organisms. In a simplified form, human cell and tissue metabolism can be viewed as a set of interacting chemical reaction sequences. Conceptually, a chemical plant functions similarly to the human body in that both use interconnected chemical processes to execute specialized functions. Chemical engineers design, troubleshoot, and optimize such systems by breaking them down into smaller unit operations, as such plants consist of various units that execute specific steps of the overall chemical process. Piping of transfer fluids from one unit to the next connects each unit operation, and products from one reactor are substrates in downstream chemical reactions and/or separations processes. Since malfunction in a single unit operation can affect the entire process, chemical plants are highly controlled and regulated. Detailed study of individual unit operations as well as systems-level process control analysis are therefore critical for designing robust and productive chemical processes.
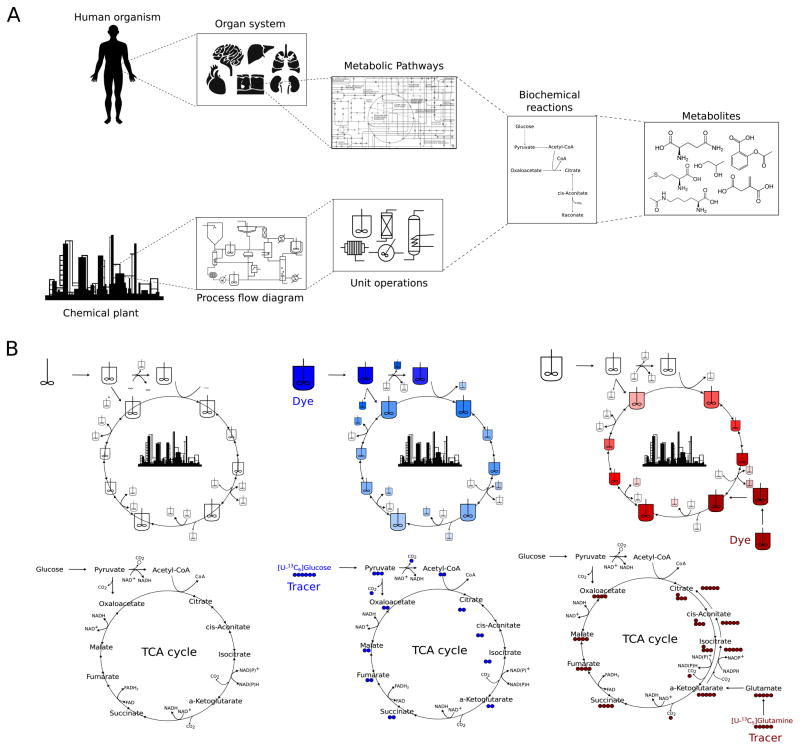
A number of parallels between chemical plants and the human body emerge with respect to their function and analysis (Fig. 1A). Organs such as the pancreas, liver, and kidney regulate nutrients and metabolic waste products to ensure that adequate energy and chemical building blocks are supplied to other tissues, such as the heart and brain. The vascular system serves to physiologically link each of these operation centers. At the subcellular level, distinct metabolic pathways are catalyzed by enzymes often localized in specific organelles. For example, mitochondria are the site of numerous biosynthetic and bioenergetic reactions, the lysosome and peroxisome are sites of recycling and detoxification, and the nucleus is the center of genetic control. Process control analysis is a fundamental tool of chemical engineers used to design and optimize industrial plants. Metabolic control analysis (MCA) has been analogously applied to characterize the regulation of individual enzymes and metabolic pathways [8]. However, key differences between chemical processes and cells or tissues highlight the challenges facing the biomedical community but also provide insights into approaches that can improve our understanding of human metabolism and disease pathology.
Figure 1.
Human physiology viewed as an industrial chemical process
A The human body consists of numerous functional systems with interacting components that execute metabolic reactions. Conceptually, chemical plants function similarly, and both can be characterized in terms of distinct unit operations. Quantitative information on physiological states within the body are therefore required to optimize and improve human health.
B Stable isotope-assisted metabolomics facilitates visualization of metabolic fluxes. Injection of dyes generates a color distribution that can be used to calculate flows across continuous stirred tank reactors (CSTRs). Isotope tracers and metabolomics allow visualization of metabolic dynamics from one metabolic pool to the next. Labeling of TCA intermediates is shown when metabolizing [U-13C6]glucose and [U-13C5]glutamine, with open circles depicting 12C atoms and closed circles depicting 13C isotopes.
Since the design of a chemical plant is based on a priori knowledge, the flow of substrates, products, waste, and energy is well known. Furthermore, gauges present throughout the system provide engineers with real-time data on current process conditions (e.g. temperature, pressure) and deviations from targeted values. In contrast, human metabolic processes and their regulation are not fully characterized, and many unknowns remain to be discovered. Furthermore, given the challenges of clinical work it is difficult if not impossible to know the time-dependent concentrations and sources of metabolites within human tissues, cells, and subcellular compartments. As such, a major limiting factor in advancing our understanding of how metabolism contributes to human disease is the acquisition and analysis of biochemical information in cells and tissues. In particular the most valuable information lies in metabolic fluxes, which are the ultimate metric describing an enzyme’s function. Dysregulation of fluxes (e.g. limited oxygen transport into tissues during ischemia, phosphorylation of glucose in cancer cells) is a key factor in virtually all diseases that in some cases can be used as a diagnostic biomarker (i.e. FDG-PET in cancer) [9]. Therefore, the acquisition of quantitative data on metabolic fluxes is needed to understand the mechanisms through which metabolism impacts or drives disease. Indeed, Lazebnik first related the function of apoptotic signaling pathways to electrical engineering concepts applicable to the circuitry of a transistor radio [10]. Rather than approach the integrated circuitry of such pathways by knocking out components one-by-one, a more systematic method was proposed using quantitative information on pathway function. In terms of metabolism the situation is similar, as fluxes cannot be effectively characterized as “ON” or “OFF.” Instead, the molar rates of reactions (in some cases relative to other pathways) are most informative. Fortunately, technological advances have now greatly improved our ability to estimate metabolic fluxes in complex biological systems.
Technological advances in studying metabolism
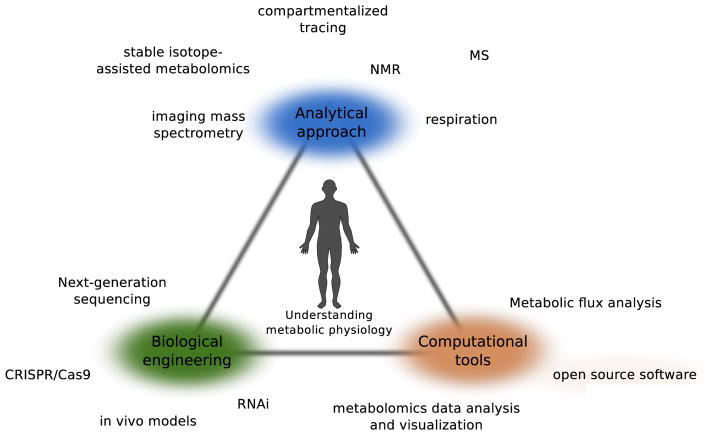
In the last few decades, major advances in the areas of analytical chemistry, biological engineering, and computational interpretation of fluxes has greatly improved our ability to quantify metabolism in cells and organisms (Fig. 2). Striking improvements in mass spectrometry and other analytical platforms is increasing the chemical information available to biomedical researchers. New software tools are allowing metabolic researchers to interpret and catalog these data and resolve pathway fluxes in unprecedented detail. Next-generation tools for genome engineering have enabled researchers to screen for critical metabolic pathways in certain cell populations and interrogate the function of enzymes and pathways [11,12]. These advances are beginning to impact our understanding of human metabolic physiology and are reviewed in detail below. We subsequently highlight some examples where metabolic flux analysis (MFA) and related approaches have contributed to our understanding of immune cell regulation by metabolic pathways. Importantly, our knowledge of metabolic pathways and their regulation are by far not complete, and continued innovation in our ability to probe these phenomena are required to elucidate the physiological mechanisms of disease.
Figure 2.
Key technological advances that facilitate quantitation of metabolic processes.
A number of innovative technologies allow for detailed metabolic analysis, including analytical chemistry approaches (blue), new methods for engineering biological systems (green), and computational tools for interpreting metabolomics data or estimating flux (red).
Analytical measurements of metabolic pathways
A wide variety of analytical platforms, especially nuclear magnetic resonance spectrometry (NMR) and mass spectrometry (MS)-based techniques, have improved the sensitivity and resolution of metabolite quantitation in biological systems. 1H, 13C, and 31P NMR-based techniques offer critical information to biomedical scientists given their non-invasiveness [13]. Magnetic resonance imaging (MRI) techniques have successfully been applied to measure metabolites in vivo, such as 2-hydroxyglutarate in the brains of patients with gliomas carrying isocitrate dehydrogenase (IDH) mutations [14]. Although the sensitivity of NMR is relatively low compared to MS-analytics, when used with stable isotope tracers NMR provides information on the position of labeling (isotopomers) which is particularly informative in pathways such as the TCA cycle [15]. To further enhance the sensitivity of such measurements 13C substrates can be hyperpolarized prior to administration. In this manner, dynamic nuclear polarization (DNP) can greatly improve both in vivo and ex vivo quantitation of isotopomers and thus metabolic activity associated with pyruvate and TCA metabolism [16–18]. More detailed and comprehensive descriptions of this technology with applications in perfused tissues and cell cultures are available [19–21], as NMR continues to be an indispensable tool for biochemists.
Mass spectrometry (MS) has emerged as a versatile tool for quantifying small molecules in biological systems at varying levels of mass resolution. For greater separation, mass spectrometry is typically combined with other separation methods, including gas chromatography (GC-MS), liquid-chromatography (LC-MS) or capillary electrophoresis (CE-MS) [22,23]. Whereas GC provides high chromatographic resolution that is particularly suited for volatile analytes and molecules such as fatty acids [24], chemical derivatization is often required. LC or direct infusion into high resolution MS instruments is increasingly used in metabolomics and flux-based applications as well. Some approaches can provide spatial information on metabolites via imaging mass spectrometry, including matrix-assisted laser desorption ionization (MALDI), desorption electrospray ionization MS (DESI-MS), and secondary ion mass spectrometry (SIMS), as reviewed in detail elsewhere [25]. Given the diversity of chemistries within the metabolome, no single approach can provide a complete metabolic profile to the user. As such, the choice of technology should be considered carefully and tailored to the system and hypothesis.
Importantly, conventional metabolomics studies provide only a snapshot of metabolite levels rather than valuable, quantitative information on fluxes noted above. Therefore, stable-isotope tracing is often combined with MS or NMR analysis of isotopologue or isotopomer quantitation, respectively [23]. In principal, the interconversion of metabolites from one pool to the next is similar to the flow through the unit operations or tanks of a chemical plant. In the same way that an inputted dye can be used to determine residence times or generate color distributions that are a function of the flow from one unit to the next, isotopic labeling allows biomedical researchers to visualize the dynamics and interconnectivity of metabolic pathways (Fig. 1B). The most useful stable isotopes used for observing intermediary metabolism in mammalian systems are 13C, 15N, and 2H (deuterium), though any “label” carried through a reaction can be employed [26]. Administration of tracer in mammalian cell culture is fairly straightforward, though the choice of labeled substrate and undefined nature of some medium components must be considered. In vivo applications are increasingly common and can be enhanced by infusion of tracer to achieve steady-state enrichment in plasma [27]. Combinations of different tracers may also be employed to increase the information available for calculating fluxes [28]. In addition to measuring free metabolites, isotopic enrichment is readily quantified into components of cellular biomass, which can be separated and analyzed to increase flux observability. Recent studies have isolated RNA, DNA, glycogen, and glycans to quantify isotope enrichment in metabolic precursors to enhance signal and provide additional information for flux calculations [29–32], drawing on related approaches that quantify acetyl-coenzyme A labeling from fatty acid measurements [33,34]. Finally, improved quantitation of metabolite labeling via tandem mass spectrometry has the potential to enhance flux resolution in MFA applications [35–37]. Broadly, these analytical approaches are increasing the data available for modeling intracellular metabolism, with the hope that more information increases the fidelity of results.
In addition to the metabolomics approaches noted above, direct measurements of fluxes also provide useful information when characterizing the metabolic state of higher cells. Measurement of uptake and secretion fluxes from cultured cells can provide valuable information on amino acid and glycolytic metabolism [38]. On the other hand, microplate-based assays have been increasingly used to monitor respiration rates to assess mitochondrial dysfunction [39]. Notably, in combination with plasma membrane permeabilization, measurements of mitochondrial respiration in permeabilized cells can allow for control of mitochondrial substrate provision, resulting in more in-depth mitochondrial studies using respirometry [40] or metabolomics [41] approaches. Increasingly, combinations of one or more analytical approaches are used to decipher the metabolism of complex systems, as each measurement may provide specific information for improved flux resolution [32,42]. Such orthogonal measurements facilitate a more integrative view on metabolism but also require increasingly complicated software for analysis.
Computational advances
The analytical approaches described above generate highly complex data sets that can include absolute or relative metabolite levels, direct flux measurements (e.g. glucose uptake, respiration), and tracer-specific isotopologue or isotopomer abundances. As a result, researchers have developed numerous algorithms for interpreting these data, resolving fluxes, and designing experiments [43,44]. Some of the most complex software is designed to estimate fluxes and associated sensitivities or confidence intervals for a given system. The user must input a metabolic network (often pared down to focus on intermediary metabolism) and measurements of fluxes, isotopologues, and when required metabolite abundances. The latter data must be considered when conducting kinetic or non-stationary metabolic flux analysis (NSMFA) [45]. Versatile software packages are becoming available to resolve fluxes using various tracers (e.g. 13C, 15N, 2H), and these have been reviewed in detail elsewhere [46]. Often, the most informative data lies in flux confidence intervals obtained via parameter continuation [47], as not all fluxes will be resolvable for a given network and experimental dataset.
Tracer choice directly impacts the isotopologues and fluxes that can be determined from a particular experiment, and computational approaches have been developed to evaluate, optimize, and design tracer combinations with enhanced resolution [48–50]. For example, combinations of glucose tracers are useful for studying the pentose phosphate pathway [51], and glutamine tracers are particularly informative when studying tricarboxylic acid (TCA) metabolism in proliferating cancer cells [48]. However, post-mitotic cells such as differentiated myoblasts (i.e. myotubes) exhibit very low glutamine anaplerosis [52], and alternate tracers should be considered depending on the metabolic state of the system to be studied.
Various software are available for extracting information from metabolomics datasets generated on different platforms. Here we focus on software used to determine isotope enrichment in analytes. Mathematical correction of natural isotope abundance using sum formula and theoretical abundance within targeted metabolomics datasets is fairly straightforward [53], and opensource software is available for this purpose [54,55]. Algorithms have recently been developed to identify labeled compounds in an untargeted manner. Non-targeted tracer fate detection (NTFD) facilitates the identification and quantitation of isotopic labeling of all detectable metabolites downstream of a given tracer in GC/MS datasets [56]. Similar software has been developed for LC/MS platforms [57]. Increasingly, researchers are incorporating tools for extracting and interpreting isotopologue data within their metabolomics software platforms, which will facilitate the use of isotope tracing and MFA in more complex biological systems [58].
These data-driven approaches are critical tools for understanding the metabolic state of a cell, tissue, animal, or patient. Various software-based approaches that balance fluxes based on network stoichiometry and gene or protein expression are also available, allowing researchers to explore the importance of pathways in silico. While challenges in the choice of objective function (i.e. how to optimize metabolism) and modeling of compartmentalized systems remain, these tools provide a unique means of generating metabolic hypotheses to be functionally tested [59,60]. Importantly, knowledge of metabolism is a requirement for interpreting results from such models, as software algorithms alone are unlikely to provide useful data for researchers.
Engineering biological systems
Metabolism represents the biochemical phenotype of a biological system, though genetic mutations, transcription, translation, and post-translational modifications all exert significant control over these pathways. As such, it is increasingly important to study metabolism in the context of engineered biological systems to shed new light on metabolic regulation. While the technologies described above have significantly improved our ability to study metabolism, perhaps even greater advances have been made in technologies used to engineer genes, proteins, and the microenvironment in mammalian systems. For example, numerous techniques are now available to control gene expression, including conventional RNA interference [61] or recently developed clustered regularly interspaced short palindromic repeats (CRISPRs)/Cas-system gene editing tools. The latter provides a powerful tool for modifying DNA sequences in a site-specific manner for numerous applications [62,63]. Indeed, researchers have applied CRISPR/Cas9 in vivo to correct mutations associated with the human metabolic disease hereditary tyrosinemia [64] and muscular dystrophy [65–67]. Furthermore, CRISPR-Cas9-based genetic screens have been applied to identify synthetic lethalities in metabolism [12]. On the other hand, engineering cells with targeted knockouts in folate-mediated one carbon metabolism has improved our ability to characterize this pathway [11]. While further research is needed to increase efficiency and decrease off-target effects [68], this technology has already been applied to improve our understanding of metabolic pathway function in mammalian cells.
Another major challenge in deciphering the metabolism of higher cells is their compartmentation [69], as many reactions and enzymes are localized to one or more subcellular organelles. Analysis of isolated mitochondria or selective cell permeabilization applied in conjunction with metabolomics, isotopic tracing, and/or respirometry can provide some information on the function of these organelles [41,52,70]. However, mitochondria in “isolation” likely exhibit different phenotypes compared to those within active cells. Also, given the fast turnover rates of many metabolites, separation of organelles prior to MS or NMR analysis is not ideal. Subcellular compartmentalization is increasingly incorporated into MFA models [52], often to account for labeling discrepancies in related metabolites like pyruvate, lactate, and alanine. Microbial co-cultures present analogous problems to MFA studies, as similar reactions may operate differently in adjacent cells (or organelles). Incorporation of biomass labeling into more complex models has been effective for resolving fluxes in microbial co-cultures [71], and such approaches may be effective in studying compartmentalized and/or multi-cellular mammalian tissue systems. Recently we developed a genetically encoded reporter system that works in conjunction with 2H tracers (and in theory other isotopes) to provide information on compartment-specific NAD(P)H metabolism [72]. By inducibly expressing mutant IDH1 or IDH2 in the cytosol or mitochondria, respectively, one can quantify 2H-labeling on (D)2HG produced in each compartment to determine how folate-mediated one carbon metabolism contributes to compartment-specific NADPH pools. This method also provided insights into the function of reductive carboxylation in anchorage-independent cancer cells [73].
Cell metabolism is commonly studied in 2D-cell culture models, but this microenvironment does not necessarily reflect the actual in vivo environment, e.g. of cancer cells [27]. Therefore, in vivo model organisms, in particular rodents, are valuable tools that help to advance our understanding of metabolism. The ability to engineer model organisms is increasing further as the CRISPR-Cas9 genetic toolbox enables rapid generation of new genetically engineered in vivo model systems [74]. In addition to in vivo model organisms, substantial progress has been made in engineering cellular microenvironments which more accurately reflect in vivo situations, such as human organs-on-chips [75] or HuMiX, a model of gastrointestinal human-microbe interface to study complex interactions between human cells and bacteria [76]. Given the complexities of metabolic systems and their regulatory requirements, it is unlikely that such engineered models will replace in vivo testing completely. While, the relative simplicity of in vitro systems can provide a better means of elucidating molecular mechanisms, any conclusions should be viewed in the context of the system used.
Case study: immunometabolism
A large number of detailed studies employing metabolomics and/or flux analysis approaches to study disease pathogenesis have been published recently. Notably, the large number of metabolic investigations and discoveries makes it impossible to comprehensively review studies in all human tissues. In particular, analyses of the heart, liver, brain, and tumors have been described in detail. Here we focus on applications in the immune system, an area of emerging interest and tremendous therapeutic potential.
The immune system is comprised of diverse cell types present in various tissue microenvironments around the body. As such, immune cells must sense and respond to a highly complex set of physiological settings. We are now beginning to appreciate that many of these signals converge on metabolic enzymes and pathways to exert control over immune cell function. During an immune response cells of the innate and adaptive immune system become activated and reprogram metabolism to execute their diverse functions, which may involve rapid proliferation, regulatory cross-talk amongst different cell types, or clearance of dead tissue and pathogens. Upon differentiation to their downstream lineages immune cells therefore exhibit strikingly distinct metabolic phenotypes, so there is much to learn in each situation. To date, most studies have focused on metabolic changes occurring within T cells and macrophages [77], but the many cell types present in the immune system present immunologists and biochemical engineers with a deep set of questions to be addressed in the coming years.
While naïve T cells rely on oxidative phosphorylation and fatty acid oxidation for energy production, activated T cells must reprogram metabolism to fulfill the metabolic requirements of proliferation and cytokine production [78]. Therefore, activated T cells increase glucose uptake, glycolytic rates and glutamine catabolism. The mammalian target of rapamycin complex 1 (mTORC1), a central regulator of metabolism and cell growth, has emerged as a critical regulator of T cell function [79]. Modulation of metabolic pathways, including glycolysis and components of the electron transport chain strongly influence T cell expansion and function [80,81]. More unique mechanisms associated with these pathways are now coming to light. For example, aerobic glycolysis facilitates binding of the glycolytic enzyme glyceraldehyde 3-phosphate dehydrogenase (GAPDH) to effector cytokine mRNA to influence T cell function [82]. Furthermore, the glycolytic metabolite phosphoenolpyruvate (PEP) acts as a signal under nutrient replete conditions to mediate T cell function, and in low-glucose such as the tumor microenvironment low PEP levels can compromise the anti-tumor effects of the immune system [83]. Metabolic studies have also recently demonstrated a role for serine and folate-mediated one carbon metabolism in T cell proliferation [84]. On the other hand, a functional electron transport chain is also required for expansion of activated T cell populations [85]. Notably, changes in lipid metabolism elicit strong effects on immune cell function. In response to pathogens, induction of lipogenic pathways is an integral part of antigen-driven blastogenesis and clonal expansion in CD8+ T cells [86]. In memory T cells, regulation of mitochondrial fatty acid oxidation by cytokines is critical for generating energy and cell survival [87]. In addition, isotopic tracer analysis has revealed co-regulation of cholesterol metabolism and the type 1 interferon (IFN) pathway which allows macrophages to coordinate antiviral responses [88].
Macrophages are present in almost all tissues and play critical roles in the immune response, as they facilitate clearance of invading pathogens and mediate tissue homeostasis associated with inflammation. Recently, an integrated analysis of transcriptomic and metabolomic data revealed new insights into the distinct metabolic signatures of classical (M1) and alternative (M2) polarized macrophages. Unlike alternatively polarized macrophages, M1-like macrophages have a distinct metabolic pattern characterized by high glycolytic flux and an impaired TCA cycle reminiscent of decreased IDH and succinate dehydrogenase (SDH) activities [89]. This regulation allows for accumulation of cis-aconitate and succinate. The former is a precursor of the antimicrobial metabolite itaconate, which can accumulate to mM levels in stimulated macrophages and microglia [90–92]. Succinate can mediate various biological functions and is thought to enhance interleukin (IL)-1β expression via stabilization of hypoxia-inducible factor (HIF)-1α [93]. Recently, we and others applied metabolomics, stable isotope tracing, and respiratory to uncover a link between these two phenomena, where itaconate acts as an endogenous SDH inhibitor to reprogram immune metabolism and modulate succinate levels [1,2,94]. Notably, though HIF is stabilized under pro-inflammatory conditions, metabolic tracing has demonstrated that stimulated macrophages maintain pyruvate flux into the TCA cycle via pyruvate dehydrogenase (PDH) to sustain itaconate production [95]. It is therefore quite valuable to understand how these molecules are produced during immune cell stimulation, as itaconate and other molecules with immunomodulatory function may emerge as promising therapies.
Conclusions
The dynamics of immune cell populations in the body presents both challenges and opportunities to the biomedical research community. Ultimately, MFA studies using models of increasing complexity will become important for elucidating the regulation and function of metabolic pathways within the immune system. Application to cell models and in vivo systems will be required, with an eye to deciphering mechanisms and interesting phenotypes. Given the importance of immune cell function in combatting infections, clearing tumor cells, and autoimmune diseases, immunometabolism will remain an active area of study in the foreseeable future. Engineers and immunologists will need to work together to understand and control these systems effectively.
Highlights.
Chemical engineering approaches offer lessons in understanding metabolic physiology
Technological advances have greatly improved our ability to quantify metabolism
Isotope tracing and metabolic flux analysis are advancing biomedical discoveries
Metabolic regulation in immune cells is an emerging area therapeutic potential
Metabolic regulation in immune cells is an emerging area therapeutic potential
Acknowledgments
We apologize to those authors who we were unable to cite in the fields of biochemical engineering and immunometabolism. We thank members of the Metallo lab for insightful discussions. This work was supported by NIH grant CA188652 to C.M.M., California Institute of Regenerative Medicine (CIRM) Award RB5-07356 to C.M.M., NSF CAREER Award 1454425 to C.M.M., and Deutsche Forschungsgesellschaft (German Research Foundation) grant CO1488/1-1 to T.C.
Footnotes
Conflict of interests
The authors have declared that no competing interests exist.
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References and recommended reading
Papers of particular interest, published within the period of review, have been highlighted as:
• of special interest
•• of outstanding interest
- 1.Cordes T, Wallace M, Michelucci A, Divakaruni AS, Sapcariu SC, Sousa C, Koseki H, Cabrales P, Murphy AN, Hiller K, et al. Immunoresponsive gene 1 and itaconate inhibit succinate dehydrogenase to modulate intracellular succinate levels. J Biol Chem. 2016;291:14274–14284. doi: 10.1074/jbc.M115.685792. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Lampropoulou V, Sergushichev A, Bambouskova M, Nair S, Vincent EE, Loginicheva E, Cervantes-Barragan L, Ma X, Huang SC-C, Griss T, et al. Itaconate Links Inhibition of Succinate Dehydrogenase with Macrophage Metabolic Remodeling and Regulation of Inflammation. Cell Metab. 2016;24:1–9. doi: 10.1016/j.cmet.2016.06.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Ahn WS, Crown SB, Antoniewicz MR. Evidence for transketolase-like TKTL1 flux in CHO cells based on parallel labeling experiments and (13)C-metabolic flux analysis. Metab Eng. 2016;37:72–78. doi: 10.1016/j.ymben.2016.05.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Diaz-Moralli S, Aguilar E, Marin S, Coy JF, Dewerchin M, Antoniewicz MR, Meca-Cortés O, Notebaert L, Ghesquière B, Eelen G, et al. A key role for transketolase-like 1 in tumor metabolic reprogramming. Oncotarget. 2016;7:51875–97. doi: 10.18632/oncotarget.10429. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.DeBerardinis RJ, Thompson CB. Cellular metabolism and disease: what do metabolic outliers teach us? Cell. 2012;148:1132–44. doi: 10.1016/j.cell.2012.02.032. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Orth JD, Thiele I, Palsson BØ. What is flux balance analysis? Nat Biotechnol. 2010;28:245–248. doi: 10.1038/nbt.1614. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Metallo CM, Vander Heiden MG. Understanding metabolic regulation and its influence on cell physiology. Mol Cell. 2013;49:388–98. doi: 10.1016/j.molcel.2013.01.018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Moreno-Sánchez R, Saavedra E, Rodríguez-Enríquez S, Olín-Sandoval V, Moreno-Sánchez R, Saavedra E, Rodríguez-Enríquez S, Olín-Sandoval V. Metabolic Control Analysis: A Tool for Designing Strategies to Manipulate Metabolic Pathways. J Biomed Biotechnol. 2008;2008:1–30. doi: 10.1155/2008/597913. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Farwell MD, Pryma DA, Mankoff DA. PET/CT imaging in cancer: current applications and future directions. Cancer. 2014;120:3433–45. doi: 10.1002/cncr.28860. [DOI] [PubMed] [Google Scholar]
- 10.Lazebnik Y. Can a biologist fix a radio? - or, What I learned while studying apoptosis. Biochem. 2004;69:1403–1406. doi: 10.1007/s10541-005-0088-1. [DOI] [PubMed] [Google Scholar]
- 11.Ducker GS, Chen L, Morscher RJ, Ghergurovich JM, Esposito M, Teng X, Kang Y, Rabinowitz JD. Reversal of Cytosolic One-Carbon Flux Compensates for Loss of the Mitochondrial Folate Pathway. Cell Metab. 2016;23:1140–1153. doi: 10.1016/j.cmet.2016.04.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Birsoy K, Wang T, Chen WW, Freinkman E, Abu-Remaileh M, Sabatini DM. An Essential Role of the Mitochondrial Electron Transport Chain in Cell Proliferation Is to Enable Aspartate Synthesis. Cell. 2015;162:540–551. doi: 10.1016/j.cell.2015.07.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Hwang J-H, Choi CS. Use of in vivo magnetic resonance spectroscopy for studying metabolic diseases. Exp Mol Med. 2015;47:e139. doi: 10.1038/emm.2014.101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Choi C, Ganji SK, DeBerardinis RJ, Hatanpaa KJ, Rakheja D, Kovacs Z, Yang X-L, Mashimo T, Raisanen JM, Marin-Valencia I, et al. 2-hydroxyglutarate detection by magnetic resonance spectroscopy in IDH-mutated patients with gliomas. Nat Med. 2012;18:624–9. doi: 10.1038/nm.2682. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Malloy CR, Sherry AD, Jeffrey FMH. Evaluation of carbon flux and substrate selection through alternate pathways involving the citric acid cycle of the heart by 13C NMR spectroscopy. J Biol Chem. 1988;263:6964–6971. [PubMed] [Google Scholar]
- 16.Mishkovsky M, Comment A, Gruetter R. In vivo detection of brain Krebs cycle intermediate by hyperpolarized magnetic resonance. J Cereb Blood Flow Metab. 2012;32:2108–13. doi: 10.1038/jcbfm.2012.136. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Hensley CT, Faubert B, Yuan Q, Lev-Cohain N, Jin E, Kim J, Jiang L, Ko B, Skelton R, Loudat L, et al. Metabolic Heterogeneity in Human Lung Tumors. Cell. 2016;164:681–694. doi: 10.1016/j.cell.2015.12.034. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Chaumeil MM, Larson PEZ, Yoshihara HAI, Danforth OM, Vigneron DB, Nelson SJ, Pieper RO, Phillips JJ, Ronen SM. Non-invasive in vivo assessment of IDH1 mutational status in glioma. Nat Commun. 2013;4:2429. doi: 10.1038/ncomms3429. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Chaumeil MM, Najac C, Ronen SM. Chapter One – Studies of Metabolism Using 13C MRS of Hyperpolarized Probes. Methods in Enzymology. 2015:1–71. doi: 10.1016/bs.mie.2015.04.001. [DOI] [PubMed] [Google Scholar]
- 20.Rodrigues TB, Serrao EM, Kennedy BWC, Hu D-E, Kettunen MI, Brindle KM. Magnetic resonance imaging of tumor glycolysis using hyperpolarized 13C-labeled glucose. Nat Med. 2013;20:93–97. doi: 10.1038/nm.3416. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Lumata L, Yang C, Ragavan M, Carpenter N, DeBerardinis RJ, Merritt ME. Chapter Two –Hyperpolarized 13C Magnetic Resonance and Its Use in Metabolic Assessment of Cultured Cells and Perfused Organs. Methods in Enzymology. 2015:73–106. doi: 10.1016/bs.mie.2015.04.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Wishart DS. Emerging applications of metabolomics in drug discovery and precision medicine. Nat Rev Drug Discov. 2016;15:473–84. doi: 10.1038/nrd.2016.32. [DOI] [PubMed] [Google Scholar]
- 23.Zamboni N, Saghatelian A, Patti GJ. Defining the metabolome: size, flux, and regulation. Mol Cell. 2015;58:699–706. doi: 10.1016/j.molcel.2015.04.021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Brenna JT. Fatty acid analysis by high resolution gas chromatography and mass spectrometry for clinical and experimental applications. Curr Opin Clin Nutr Metab Care. 2013;16:548–54. doi: 10.1097/MCO.0b013e328363bc0a. [DOI] [PubMed] [Google Scholar]
- 25.Vickerman JC. Molecular imaging and depth profiling by mass spectrometry--SIMS, MALDI or DESI? Analyst. 2011;136:2199–217. doi: 10.1039/c1an00008j. [DOI] [PubMed] [Google Scholar]
- 26.Buescher JM, Antoniewicz MR, Boros LG, Burgess SC, Brunengraber H, Clish CB, DeBerardinis RJ, Feron O, Frezza C, Ghesquiere B, et al. A roadmap for interpreting 13C metabolite labeling patterns from cells. Curr Opin Biotechnol. 2015;34:189–201. doi: 10.1016/j.copbio.2015.02.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27••.Davidson SM, Papagiannakopoulos T, Olenchock BA, Heyman JE, Keibler MA, Luengo A, Bauer MR, Jha AK, O’Brien JP, Pierce KA, et al. Environment Impacts the Metabolic Dependencies of Ras-Driven Non-Small Cell Lung Cancer. Cell Metab. 2016;23:517–528. doi: 10.1016/j.cmet.2016.01.007. Tracer-based study using genetically engineered mouse models that highlighted key differences in the metabolism of tumor cells in vivo versus those cultured in vitro. Differences in glutamine anaplerosis were quite significant, as plasma glutamine did not contribute significantly to TCA intermediates in vivo. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Hasenour CM, Wall ML, Ridley DE, Hughey CC, James FD, Wasserman DH, Young JD. Mass Spectrometry-Based Microassay of 2H and 13C Plasma Glucose Labeling to Quantify Liver Metabolic Fluxes In vivo. Am J Physiol Endocrinol Metab. 2015;309:E191–203. doi: 10.1152/ajpendo.00003.2015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Badur MG, Zhang H, Metallo CM. Enzymatic passaging of human embryonic stem cells alters central carbon metabolism and glycan abundance. Biotechnol J. 2015;10:1600–1611. doi: 10.1002/biot.201400749. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Miranda-Santos I, Gramacho S, Pineiro M, Martinez-Gomez K, Fritz M, Hollemeyer K, Salvador A, Heinzle E. Mass Isotopomer Analysis of Nucleosides Isolated from RNA and DNA Using GC/MS. Anal Chem. 2015;87:617–623. doi: 10.1021/ac503305w. [DOI] [PubMed] [Google Scholar]
- 31.Long CP, Au J, Gonzalez JE, Antoniewicz MR. 13C metabolic flux analysis of microbial and mammalian systems is enhanced with GC-MS measurements of glycogen and RNA labeling. Metab Eng. 2016;38:65–72. doi: 10.1016/j.ymben.2016.06.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Murphy TA, Dang CV, Young JD. Isotopically nonstationary 13C flux analysis of Myc-induced metabolic reprogramming in B-cells. Metab Eng. 2013;15:206–217. doi: 10.1016/j.ymben.2012.07.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Crown SB, Marze N, Antoniewicz MR. Catabolism of Branched Chain Amino Acids Contributes Significantly to Synthesis of Odd-Chain and Even-Chain Fatty Acids in 3T3-L1 Adipocytes. PLoS One. 2015;10:e0145850. doi: 10.1371/journal.pone.0145850. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Green CR, Wallace M, Divakaruni AS, Phillips SA, Murphy AN, Ciaraldi TP, Metallo CM. Branched-chain amino acid catabolism fuels adipocyte differentiation and lipogenesis. Nat Chem Biol. 2016;12:15–21. doi: 10.1038/nchembio.1961. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Alves TC, Pongratz RL, Zhao X, Yarborough O, Sereda S, Shirihai O, Cline GW, Mason G, Kibbey RG. Integrated, Step-Wise, Mass-Isotopomeric Flux Analysis of the TCA Cycle. Cell Metab. 2015;22:936–47. doi: 10.1016/j.cmet.2015.08.021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Choi J, Grossbach MT, Antoniewicz MR. Measuring complete isotopomer distribution of aspartate using gas chromatography/tandem mass spectrometry. Anal Chem. 2012;84:4628–32. doi: 10.1021/ac300611n. [DOI] [PubMed] [Google Scholar]
- 37.Rühl M, Rupp B, Nöh K, Wiechert W, Sauer U, Zamboni N. Collisional fragmentation of central carbon metabolites in LC-MS/MS increases precision of 13C metabolic flux analysis. Biotechnol Bioeng. 2012;109:763–71. doi: 10.1002/bit.24344. [DOI] [PubMed] [Google Scholar]
- 38.Jain M, Nilsson R, Sharma S, Madhusudhan N, Kitami T, Souza AL, Kafri R, Kirschner MW, Clish CB, Mootha VK. Metabolite profiling identifies a key role for glycine in rapid cancer cell proliferation. Science. 2012;336:1040–4. doi: 10.1126/science.1218595. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Ribeiro SM, Giménez-Cassina A, Danial NN. Measurement of mitochondrial oxygen consumption rates in mouse primary neurons and astrocytes. Methods Mol Biol. 2015;1241:59–69. doi: 10.1007/978-1-4939-1875-1_6. [DOI] [PubMed] [Google Scholar]
- 40.Divakaruni AS, Rogers GW, Murphy AN. Measuring Mitochondrial Function in Permeabilized Cells Using the Seahorse XF Analyzer or a Clark-Type Oxygen Electrode. Curr Protoc Toxicol. 2014;60:25.2.1–25.2.16. doi: 10.1002/0471140856.tx2502s60. [DOI] [PubMed] [Google Scholar]
- 41.Nicolae A, Wahrheit J, Nonnenmacher Y, Weyler C, Heinzle E. Identification of active elementary flux modes in mitochondria using selectively permeabilized CHO cells. Metab Eng. 2015;32:95–105. doi: 10.1016/j.ymben.2015.09.014. [DOI] [PubMed] [Google Scholar]
- 42••.Fan J, Kamphorst JJ, Mathew R, Chung MK, White E, Shlomi T, Rabinowitz JD. Glutamine-driven oxidative phosphorylation is a major ATP source in transformed mammalian cells in both normoxia and hypoxia. Mol Syst Biol. 2013;9:1–11. doi: 10.1038/msb.2013.65. The authors integrated stable isotope tracing with respiratory measurements and a mammalian metabolic flux model to determine the role of oncogene activation and low oxygen levels on energy metabolism. This work demonstrated that oxidative phosphorylation produces the majority of ATP, even in hypoxic or Ras activated conditions highlighting the important role of oxidative glutamine metabolism in most cancer. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43••.Young JD. INCA: a computational platform for isotopically non-stationary metabolic flux analysis. Bioinformatics. 2014;30:1333–5. doi: 10.1093/bioinformatics/btu015. This manuscript introduces a software platform for stationary or non-stationary MFA applications which has been applied to mammalian and other systems. After estimation of fluxes, parameter continuation to determine flux confidence intervals is readily executed. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Wiechert W. 13C metabolic flux analysis. Metab Eng. 2001;3:195–206. doi: 10.1006/mben.2001.0187. [DOI] [PubMed] [Google Scholar]
- 45.Munger J, Bennett BD, Parikh A, Feng X-J, McArdle J, Rabitz HA, Shenk T, Rabinowitz JD. Systems-level metabolic flux profiling identifies fatty acid synthesis as a target for antiviral therapy. Nat Biotechnol. 2008;26:1179–1186. doi: 10.1038/nbt.1500. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Dandekar T, Fieselmann A, Majeed S, Ahmed Z. Software applications toward quantitative metabolic flux analysis and modeling. Brief Bioinform. 2014;15:91–107. doi: 10.1093/bib/bbs065. [DOI] [PubMed] [Google Scholar]
- 47.Antoniewicz MR, Kelleher JK, Stephanopoulos G. Determination of confidence intervals of metabolic fluxes estimated from stable isotope measurements. Metab Eng. 2006;8:324–337. doi: 10.1016/j.ymben.2006.01.004. [DOI] [PubMed] [Google Scholar]
- 48.Metallo CM, Walther JL, Stephanopoulos G. Evaluation of 13C isotopic tracers for metabolic flux analysis in mammalian cells. J Biotechnol. 2009;144:167–74. doi: 10.1016/j.jbiotec.2009.07.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Walther JL, Metallo CM, Zhang J, Stephanopoulos G. Optimization of 13C isotopic tracers for metabolic flux analysis in mammalian cells. Metab Eng. 2012;14:162–171. doi: 10.1016/j.ymben.2011.12.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Crown SB, Long CP, Antoniewicz MR. Optimal tracers for parallel labeling experiments and 13C metabolic flux analysis: A new precision and synergy scoring system. Metab Eng. 2016;38:10–18. doi: 10.1016/j.ymben.2016.06.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Crown SB, Ahn WS, Antoniewicz MR. Rational design of 13C-labeling experiments for metabolic flux analysis in mammalian cells. BMC Syst Biol. 2012;6:43. doi: 10.1186/1752-0509-6-43. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52•.Vacanti NM, Divakaruni AS, Green CR, Parker SJ, Henry RR, Ciaraldi TP, Murphy AN, Metallo CM. Regulation of substrate utilization by the mitochondrial pyruvate carrier. Mol Cell. 2014;56:425–35. doi: 10.1016/j.molcel.2014.09.024. A combination of isotope tracer study, metabolic flux analysis (MFA), and respiratory assays that studied the role of mitochondrial pyruvate carrier for mitochondrial substrate utilization. Suppression of mitochondrial pyruvate transport activity resulted in a metabolic reprogramming to meet the bioenergetic demands of cancer cells suggesting pyruvate transport as potential therapeutic target in cancer cells. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Fernandez CA, Des Rosiers C, Previs SF, David F, Brunengraber H. Correction of 13C mass isotopomer distributions for natural stable isotope abundance. J Mass Spectrom. 1996;31:255–62. doi: 10.1002/(SICI)1096-9888(199603)31:3<255::AID-JMS290>3.0.CO;2-3. [DOI] [PubMed] [Google Scholar]
- 54.Hiller K, Hangebrauk J, Jäger C, Spura J, Schreiber K, Schomburg D. Metabolite Detector: comprehensive analysis tool for targeted and nontargeted GC/MS based metabolome analysis. Anal Chem. 2009;81:3429–39. doi: 10.1021/ac802689c. [DOI] [PubMed] [Google Scholar]
- 55.Millard P, Letisse F, Sokol S, Portais J-C. IsoCor: correcting MS data in isotope labeling experiments. Bioinformatics. 2012;28:1294–6. doi: 10.1093/bioinformatics/bts127. [DOI] [PubMed] [Google Scholar]
- 56.Hiller K, Metallo CM, Kelleher JK, Stephanopoulous G. Nontargeted elucidation of metabolic pathways using stable-isotope tracers and mass spectrometry. Anal Chem. 2010;82:6621–8. doi: 10.1021/ac1011574. [DOI] [PubMed] [Google Scholar]
- 57.Bueschl C, Krska R, Kluger B, Schuhmacher R. Isotopic labeling-assisted metabolomics using LC-MS. Anal Bioanal Chem. 2013;405:27–33. doi: 10.1007/s00216-012-6375-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Cho K, Mahieu N, Ivanisevic J, Uritboonthai W, Chen Y-J, Siuzdak G, Patti GJ. isoMETLIN: A Database for Isotope-Based Metabolomics. Anal Chem. 2014;86:9358–9361. doi: 10.1021/ac5029177. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59•.Hyötyläinen T, Jerby L, Petäjä EM, Mattila I, Jäntti S, Auvinen P, Gastaldelli A, Yki-Järvinen H, Ruppin E, Orešič M. Genome-scale study reveals reduced metabolic adaptability in patients with non-alcoholic fatty liver disease. Nat Commun. 2016;7:8994. doi: 10.1038/ncomms9994. Genome-scale human metabolic model that revealed that accumulation of liver fat modulates mitochondrial metabolism, lipolysis, and gluconeogenesis. This approach uncovered reduced metabolic adaptability in conditions with increased amounts of liver fat, e.g. non-alcoholic fatty liver disease (NAFLD) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Mardinoglu A, Agren R, Kampf C, Asplund A, Uhlen M, Nielsen J. Genome-scale metabolic modelling of hepatocytes reveals serine deficiency in patients with non-alcoholic fatty liver disease. Nat Commun. 2014;5:3083. doi: 10.1038/ncomms4083. [DOI] [PubMed] [Google Scholar]
- 61.Ipsaro JJ, Joshua-Tor L. From guide to target: molecular insights into eukaryotic RNA-interference machinery. Nat Struct Mol Biol. 2015;22:20–28. doi: 10.1038/nsmb.2931. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Mali P, Yang L, Esvelt KM, Aach J, Guell M, DiCarlo JE, Norville JE, Church GM. RNA-guided human genome engineering via Cas9. Science. 2013;339:823–6. doi: 10.1126/science.1232033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Zhou Y, Zhu S, Cai C, Yuan P, Li C, Huang Y, Wei W. High-throughput screening of a CRISPR/Cas9 library for functional genomics in human cells. Nature. 2014;509:487–91. doi: 10.1038/nature13166. [DOI] [PubMed] [Google Scholar]
- 64.Yin H, Xue W, Chen S, Bogorad RL, Benedetti E, Grompe M, Koteliansky V, Sharp PA, Jacks T, Anderson DG. Genome editing with Cas9 in adult mice corrects a disease mutation and phenotype. Nat Biotechnol. 2014;32:551–3. doi: 10.1038/nbt.2884. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Long C, Amoasii L, Mireault AA, McAnally JR, Li H, Sanchez-Ortiz E, Bhattacharyya S, Shelton JM, Bassel-Duby R, Olson EN. Postnatal genome editing partially restores dystrophin expression in a mouse model of muscular dystrophy. Science. 2016;351:400–3. doi: 10.1126/science.aad5725. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Nelson CE, Hakim CH, Ousterout DG, Thakore PI, Moreb EA, Castellanos Rivera RM, Madhavan S, Pan X, Ran FA, Yan WX, et al. In vivo genome editing improves muscle function in a mouse model of Duchenne muscular dystrophy. Science. 2016;351:403–7. doi: 10.1126/science.aad5143. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Tabebordbar M, Zhu K, Cheng JKW, Chew WL, Widrick JJ, Yan WX, Maesner C, Wu EY, Xiao R, Ran FA, et al. In vivo gene editing in dystrophic mouse muscle and muscle stem cells. Science. 2016;351:407–11. doi: 10.1126/science.aad5177. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Zhang X-H, Tee LY, Wang X-G, Huang Q-S, Yang S-H. Off-target Effects in CRISPR/Cas9-mediated Genome Engineering. Mol Ther Acids. 2015;4:e264. doi: 10.1038/mtna.2015.37. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Zamboni N. 13C metabolic flux analysis in complex systems. Curr Opin Biotechnol. 2011;22:103–108. doi: 10.1016/j.copbio.2010.08.009. [DOI] [PubMed] [Google Scholar]
- 70.Yang C, Ko B, Hensley CT, Jiang L, Wasti AT, Kim J, Sudderth J, Calvaruso MA, Lumata L, Mitsche M, et al. Glutamine oxidation maintains the TCA cycle and cell survival during impaired mitochondrial pyruvate transport. Mol Cell. 2014;56:414–424. doi: 10.1016/j.molcel.2014.09.025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Gebreselassie NA, Antoniewicz MR. 13C-metabolic flux analysis of co-cultures: A novel approach. Metab Eng. 2015;31:132–139. doi: 10.1016/j.ymben.2015.07.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72••.Lewis CA, Parker SJ, Fiske BP, McCloskey D, Gui DY, Green CR, Vokes NI, Feist AM, Vander Heiden MG, Metallo CM. Tracing compartmentalized NADPH metabolism in the cytosol and mitochondria of mammalian cells. Mol Cell. 2014;55:253–63. doi: 10.1016/j.molcel.2014.05.008. Compartment-specific metabolic tracing methodology that integrated 2H isotope tracer study in intact cells with a reporter system to visualize compartmentalized reaction activities. By applying this approach the authors demonstrated that serine conversion to glycine occurs primarily in the mitochondria. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73•.Jiang L, Shestov AA, Swain P, Yang C, Parker SJ, Wang QA, Terada LS, Adams ND, McCabe MT, Pietrak B, et al. Reductive carboxylation supports redox homeostasis during anchorage-independent growth. Nature. 2016;532:255–258. doi: 10.1038/nature17393. This paper applied MFA to study anchorage-independent cancer cell cultures as a model of tumor metastasis. The authors demonstrate that reductive carboxylation is upregulated under such conditions to transfer reducing equivalents into mitochondria, making use of compartment-specific metabolic tracing. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Papagiannakopoulos T, Bauer MR, Davidson SM, Heimann M, Subbaraj L, Bhutkar A, Bartlebaugh J, Vander Heiden MG, Jacks T. Circadian Rhythm Disruption Promotes Lung Tumorigenesis. Cell Metab. 2016;24:324–331. doi: 10.1016/j.cmet.2016.07.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Bhatia SN, Ingber DE. Microfluidic organs-on-chips. Nat Biotechnol. 2014;32:760–772. doi: 10.1038/nbt.2989. [DOI] [PubMed] [Google Scholar]
- 76.Shah P, Fritz JV, Glaab E, Desai MS, Greenhalgh K, Frachet A, Niegowska M, Estes M, Jäger C, Seguin-Devaux C, et al. A microfluidics-based in vitro model of the gastrointestinal human–microbe interface. Nat Commun. 2016;7:11535. doi: 10.1038/ncomms11535. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Ghesquière B, Wong BW, Kuchnio A, Carmeliet P. Metabolism of stromal and immune cells in health and disease. Nature. 2014;511:167–176. doi: 10.1038/nature13312. [DOI] [PubMed] [Google Scholar]
- 78.O’Neill LAJ, Kishton RJ, Rathmell JC. A guide to immunometabolism for immunologists. Nat Rev Immunol. 2016 doi: 10.1038/nrmicro.2016.15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Chi H. Regulation and function of mTOR signalling in T cell fate decisions. Nat Rev Immunol. 2012;12:325. doi: 10.1038/nri3198. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80•.Macintyre AN, Gerriets VA, Nichols AG, Michalek RD, Rudolph MC, Deoliveira D, Anderson SM, Abel ED, Chen BJ, Hale LP, et al. The Glucose Transporter Glut1 Is Selectively Essential for CD4 T Cell Activation and Effector Function. Cell Metab. 2014;20:61–72. doi: 10.1016/j.cmet.2014.05.004. Metabolic study on T cell metabolism that analyzed the impact of nutrient transport on immune functions. The authors modulated glucose transporter activity and demonstrated that glucose transport is required for T cell activation and regulates immunologic diseases in vivo. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Blagih J, Coulombe F, Vincent EE, Dupuy F, Galicia-Vázquez G, Yurchenko E, Raissi TC, van der Windt GJW, Viollet B, Pearce EL, et al. The Energy Sensor AMPK Regulates T Cell Metabolic Adaptation and Effector Responses In Vivo. Immunity. 2015;42:41–54. doi: 10.1016/j.immuni.2014.12.030. [DOI] [PubMed] [Google Scholar]
- 82.Chang CH, Curtis JD, Maggi LB, Faubert B, Villarino AV, O’Sullivan D, Huang SCC, Van Der Windt GJW, Blagih J, Qiu J, et al. XPosttranscriptional control of T cell effector function by aerobic glycolysis. Cell. 2013;153:1239–1251. doi: 10.1016/j.cell.2013.05.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.Ho PC, Bihuniak JD, MacIntyre AN, Staron M, Liu X, Amezquita R, Tsui YC, Cui G, Micevic G, Perales JC, et al. Phosphoenolpyruvate Is a Metabolic Checkpoint of Anti-tumor T Cell Responses. Cell. 2015;162:1217–1228. doi: 10.1016/j.cell.2015.08.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84.Ron-Harel N, Santos D, Ghergurovich JM, Sage PT, Reddy A, Lovitch SB, Dephoure N, Satterstrom FK, Sheffer M, Spinelli JB, et al. Mitochondrial Biogenesis and Proteome Remodeling Promote One-Carbon Metabolism for T Cell Activation. Cell Metab. 2016;24:104–117. doi: 10.1016/j.cmet.2016.06.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85.Sena LA, Li S, Jairaman A, Prakriya M, Ezponda T, Hildeman DA, Wang C-R, Schumacker PT, Licht JD, Perlman H, et al. Mitochondria Are Required for Antigen-Specific T Cell Activation through Reactive Oxygen Species Signaling. Immunity. 2013;38:225–236. doi: 10.1016/j.immuni.2012.10.020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86.Kidani Y, Elsaesser H, Hock MB, Vergnes L, Williams KJ, Argus JP, Marbois BN, Komisopoulou E, Wilson EB, Osborne TF, et al. Sterol regulatory element–binding proteins are essential for the metabolic programming of effector T cells and adaptive immunity. Nat Immunol. 2013;14:489–499. doi: 10.1038/ni.2570. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87.van der Windt GJW, Everts B, Chang C-H, Curtis JD, Freitas TC, Amiel E, Pearce EJ, Pearce EL. Mitochondrial Respiratory Capacity Is a Critical Regulator of CD8+ T Cell Memory Development. Immunity. 2012;36:68–78. doi: 10.1016/j.immuni.2011.12.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88•.York AG, Williams KJ, Argus JP, Zhou QD, Brar G, Vergnes L, Gray EE, Zhen A, Wu NC, Yamada DH, et al. Limiting Cholesterol Biosynthetic Flux Spontaneously Engages Type i IFN Signaling. Cell. 2015;163:1716–1729. doi: 10.1016/j.cell.2015.11.045. In this study, the authors integrated RNA sequencing data with stable isotope-assisted metabolomics to analyze the role of cellular lipid metabolism in macrophage metabolism and uncovered an inflammatory circuit important for immune function. This work demonstrated that metabolic reprogramming during the immune response to pathogens, in particular low cholesterol levels, triggers type I interferon signaling to driving antiviral activity. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 89••.Jha AK, Huang SC-C, Sergushichev A, Lampropoulou V, Ivanova Y, Loginicheva E, Chmielewski K, Stewart KM, Ashall J, Everts B, et al. Network Integration of Parallel Metabolic and Transcriptional Data Reveals Metabolic Modules that Regulate Macrophage Polarization. Immunity. 2015;42:419–430. doi: 10.1016/j.immuni.2015.02.005. Integrated high-throughput analysis with metabolic and transcriptional data that identified metabolic alterations in macrophage phenotypes. This systems approach revealed distinct metabolic patterns between classically (M1) and alternative (M2) polarized macrophages, in particular an impaired TCA cycle in M1 polarized macrophages. [DOI] [PubMed] [Google Scholar]
- 90.Michelucci A, Cordes T, Ghelfi J, Pailot A, Reiling N, Goldmann O, Binz T, Wegner A, Tallam A, Rausell A, et al. Immune-responsive gene 1 protein links metabolism to immunity by catalyzing itaconic acid production. Proc Natl Acad Sci U S A. 2013;110:7820–5. doi: 10.1073/pnas.1218599110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 91.Strelko CL, Lu W, Dufort FJ, Seyfried TN, Chiles TC, Rabinowitz JD, Roberts MF. Itaconic acid is a mammalian metabolite induced during macrophage activation. J Am Chem Soc. 2011;133:16386–9. doi: 10.1021/ja2070889. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 92.Cordes T, Michelucci A, Hiller K. Itaconic Acid: The Surprising Role of an Industrial Compound as a Mammalian Antimicrobial Metabolite. Annu Rev Nutr. 2015;35:451–473. doi: 10.1146/annurev-nutr-071714-034243. [DOI] [PubMed] [Google Scholar]
- 93.Tannahill GM, Curtis aM, Adamik J, Palsson-McDermott EM, McGettrick aF, Goel G, Frezza C, Bernard NJ, Kelly B, Foley NH, et al. Succinate is an inflammatory signal that induces IL-1β through HIF-1α. Nature. 2013;496:238–42. doi: 10.1038/nature11986. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 94.Németh B, Doczi J, Csete D, Kacso G, Ravasz D, Adams D, Kiss G, Nagy AM, Horvath G, Tretter L, et al. Abolition of mitochondrial substrate-level phosphorylation by itaconic acid produced by LPS-induced Irg1 expression in cells of murine macrophage lineage. FASEB J. 2016;30:286–300. doi: 10.1096/fj.15-279398. [DOI] [PubMed] [Google Scholar]
- 95.Meiser J, Krämer L, Sapcariu SC, Battello N, Ghelfi J, D’Herouel AF, Skupin A, Hiller K. Pro-inflammatory Macrophages Sustain Pyruvate Oxidation through Pyruvate Dehydrogenase for the Synthesis of Itaconate and to Enable Cytokine Expression. J Biol Chem. 2016;291:3932–3946. doi: 10.1074/jbc.M115.676817. [DOI] [PMC free article] [PubMed] [Google Scholar]