Abstract
Conventional plasmid vectors are incapable of achieving sustained levels of transgene expression in vivo even in quiescent mammalian tissues because the transgene expression cassette is silenced. Transcriptional silencing results from the presence of the bacterial plasmid backbone or virtually any DNA sequence of >1 kb in length placed outside of the expression cassette. Here, we show that transcriptional silencing can be substantially forestalled by increasing the An/Tn sequence composition in the plasmid bacterial backbone. Increasing numbers of An/Tn sequences increased sustained transcription of both backbone sequences and adjacent expression cassettes. In order to recapitulate these expression profiles in compact and portable plasmid DNA backbones, we engineered the standard kanamycin or ampicillin antibiotic resistance genes, optimizing the number of An/Tn sequence without altering the encoded amino acids. The resulting vector backbones yield sustained transgene expression from mouse liver, providing generic DNA vectors capable of sustained transgene expression without additional genes or mammalian regulatory elements.
Keywords: plasmid vector, nucleosomes, non-coding transcription, plasmid silencing, An/Tn tracts, antibiotic resistance gene
The plasmid backbone contained in DNA vectors silences expression of the transgene. By increasing the An/Tn content (nucleosome-relaxation sequences) in the ampicillin/kanamycin resistance genes, Kay and colleagues have achieved persistent plasmid-mediated transgene expression from a mouse liver. These plasmids are propagated by routine methods and show similar expression profiles compared to minicircle DNA vectors.
Introduction
Generic plasmid DNA vectors containing a classical promoter, coding sequence, and polyadenylation signal (polyA) are often used for expressing therapeutic, reporter, or proteins of interest in a myriad of cells and tissues. Even in quiescent tissues in which plasmid DNA levels remain static, transgene expression is transcriptionally extinguished over a period of several weeks. We have previously referred and defined this as transcription silencing.1 Having explored the mechanism of transcriptional silencing in the mouse liver following hydrodynamic injection, we have found that CpG content and DNA methylation have little or no influence in transcription silencing.2 Instead, the length of the DNA outside of the eukaryotic expression cassette (between the 5′ end of the promoter and the 3′ end of the poly A site) was determined to be a critical factor that dictates transcriptional silencing in this system.1, 3 Silencing begins to be observed with ∼1 kb or more of backbone DNA, even if the bacterial plasmid DNA sequences are replaced with random DNA sequences.1 These observations led to the production of alternative plasmid vectors commonly referred to as minicircle DNAs (MCs)4, 5, 6, 7, 8, 9 (devoid of a bacterial backbone) or mini-intronic plasmid DNAs (MIPs),3 in which the bacterial components required for plasmid propagation in bacteria are engineered into the mammalian expression cassette. However, each of these products requires additional steps and/or specific reagents for their production.
Previous work has shown nucleosomal patterns to be a key element in the establishment and maintenance of silenced chromatin.10 A set of An/Tn sequence motifs is known to substantially modify nucleosomal positioning and mobility.11 The An/Tn tracts are thought to decrease the structural stability to the basic 147-bp nucleosome core wrapped around a histone octamer, which allows for greater accessibility of RNA polymerase II (RNAPII) complexes.11, 12, 13, 14, 15, 16, 17 Such sequences, sometimes referred to as nucleosome exclusion but perhaps more appropriately as nucleosome relaxation or loosing, remain a mystery in terms of their mode of action.
Certain sequences with a high content of An/Tn segments have been shown to counteract silencing in an invertebrate system,18 although we note that the particular sequences used in the nematode system involved shorter An/Tn segments and a phased occurrence rather than the longer An/Tn runs described as exclusionary in vertebrate and yeast systems.
As part of our efforts to further explore plasmid-mediated transgene silencing, we asked if the addition of short An/Tn tracts would influence plasmid-mediated transgene expression. Ultimately, we used the results of these experiments to design sequence-modified bacterial antibiotic resistance genes to abrogate transcriptional silencing, providing prolonged transgene expression from canonical plasmid vectors in vivo.
Results
Increasing An/Tn Sequence Composition in Plasmid Bacterial Backbone Abrogated Plasmid-Mediated Transcriptional Silencing In Vivo
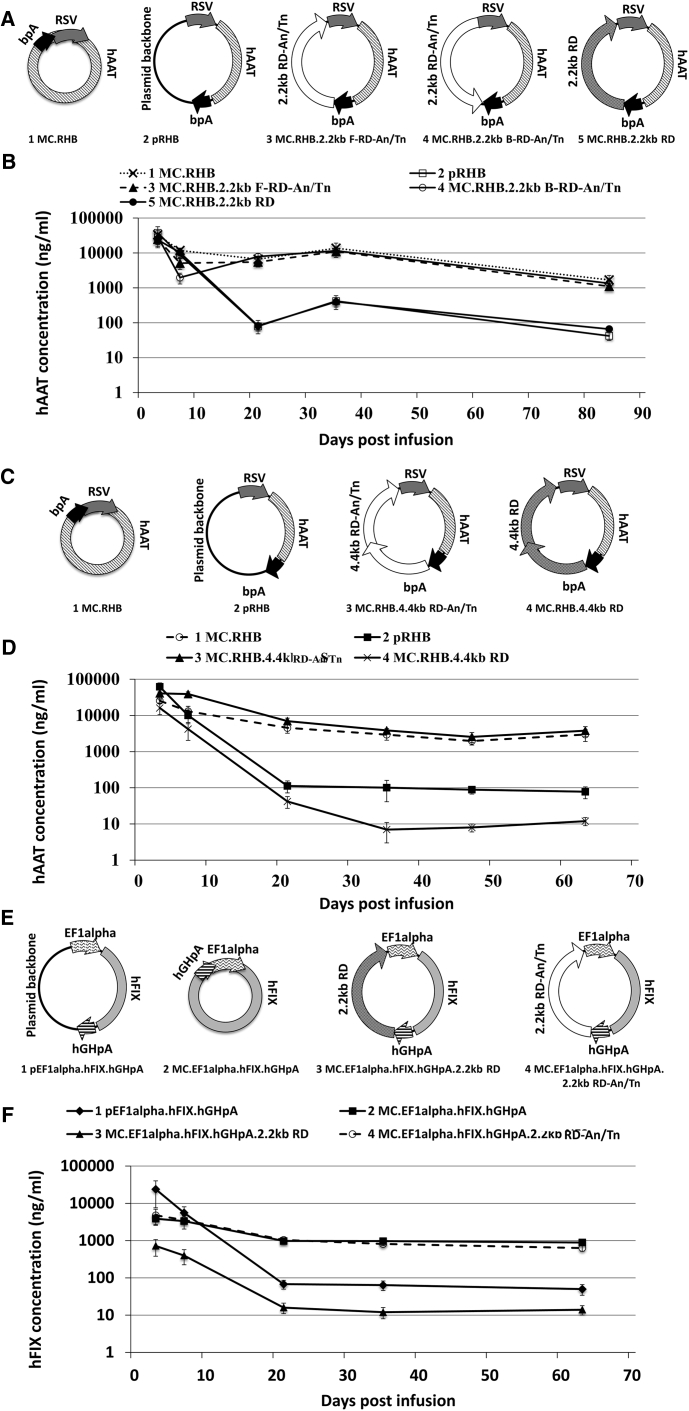
In order to establish how the sequences outside of the canonical eukaryotic expression cassette affect plasmid-mediated transgene expression, we replaced the bacterial plasmid backbone with a 2.2 kb random DNA (RD) fragment using minicircle DNA technology (Figure 1). To study how the An/Tn motifs affected transgene expression, we cloned a stretch of 20 “T” nucleotides into every 60 bp of the RD sequence (RD-An/Tn) (Figure S1) and then compared these vectors for transgene expression in a mouse liver.
Figure 1.
Effects of RD-An/Tn Sequence in Maintaining Transgene Expression In Vivo
(A) Schematic of the hAAT DNA plasmid constructs. The RD-An/Tn segment was inserted as the backbone in either forward (MC.RHB.2.2kb F-RD-An/Tn) or backward orientation (MC.RHB.2.2kb B-RD-An/Tn). (B) Serum hAAT levels measured by ELISA at various time points after equimolar infusion of the DNA vectors into the mouse liver shown in (A) (n = 5/group). Error bars represent the SD in all figures. Serum hAAT levels on day 84 post infusion were used to calculate p values. The p value of each group was calculated by comparing with the non-silencing control group (MC.RHB). If p < 0.005, the tested group is considered as silenced. Otherwise the tested group is considered as non-silenced. For the pRHB group, p = 0.002. For the MC.RHB.2.2kb F-RD-An/Tn group, p = 0.06. For the MC.RHB.2.2kb B-RD-An/Tn group, p = 0.2. For the MC.RHB.2.2kb RD group, p = 0.0008. (C) Schematic of vectors used for the experiments shown in (D). Two copies of 2.2-kb RD-An/Tn or RD sequences were placed after the bpA sequence as the backbone. (D) Serum hAAT levels after equimolar transfection of vectors presented in (C). Serum hAAT levels on day 63 post infusion were used to calculate p values. The MC.RHB group was used as the non-silencing control. For the pRHB group, p = 0.002. For MC.RHB.4.4kb RD-An/Tn, p = 0.3. For MC.RHB.4.4kb RD, p = 0.002. (E) Schematic representation of the DNA constructs expressing hFIX. 2.2-kb RD-An/Tn or 2.2-kb RD sequence was used as backbone in these constructs. (F) The same molar amounts of DNA constructs pictured in (E) were infused into mice, and the plasma hFIX levels were measured by ELISA at various time points. Serum hFIX levels on day 63 post infusion were used to calculate p values. The MC.EF1alpha.hFIX.hGpA group was used as non-silencing control. For the pEF1alpha.hFIX.hGHpA group, p = 0.0001. For MC.EF1alpha.hFIX.hGHpA.2.2kb RD, p = 0.0001. For the MC.EF1alpha.hFIX.hGHpA.2.2kb RD-An/Tn group, p = 0.007 (hAAT and hFIX have been defined in the text; hGHpA, human growth hormone polyA; bpA, bovine growth hormone polyA).
Plasmids consisting of the human alpha 1-antitrypsin (hAAT) cDNA driven by the Rous sarcoma virus (RSV) promoter containing the RD or RD-An/Tn fragment cloned in either direction were delivered into the livers of 6- to 8-week-old C57BL/6J mice by hydrodynamic transfection (Figures 1A and 1B). A conventional plasmid vector (pRHB) and minicircle vector (MC.RHB) were used as the silencing and non-silencing controls, respectively. Transgene expression monitored by serial serum hAAT measurements is shown in Figure 1B. We followed transgene expression levels over time. Generally, by 3 to 5 weeks after infusion, the steady-state expression level is observed. To ensure steady-state conditions, we used the 2-month time point (day 63 or later if the results for day 63 were not available) for statistical comparisons (Figure 1B). The RD backbone containing and canonical plasmids were quickly silenced, whereas the RD-An/Tn backbone containing and canonical minicircles resulted in sustained expression levels. We found that the effect on transgene expression was independent of the orientation of the backbone DNA (Figures 1A and 1B) and that even when two copies of the RD-An/Tn segment (4.4 kb) were used to replace the plasmid backbone, the transgene expression levels were maintained (Figures 1C and 1D). To establish these results were not dependent on the expression cassette, the RD-An/Tn backbone was also tested in a human Factor IX (hFIX) reporter system driven by the EF1-alpha promoter. Consistent with the previous results, the hFIX expression was sustained at high levels in minicircles containing the RD-An/Tn backbone (Figures 1E and 1F).
Plasmid Vectors Carrying An/Tn-Sequence-Modified Antibiotic Resistance Genes Provide Sustained Plasmid-Mediated Transgene Expression In Vivo
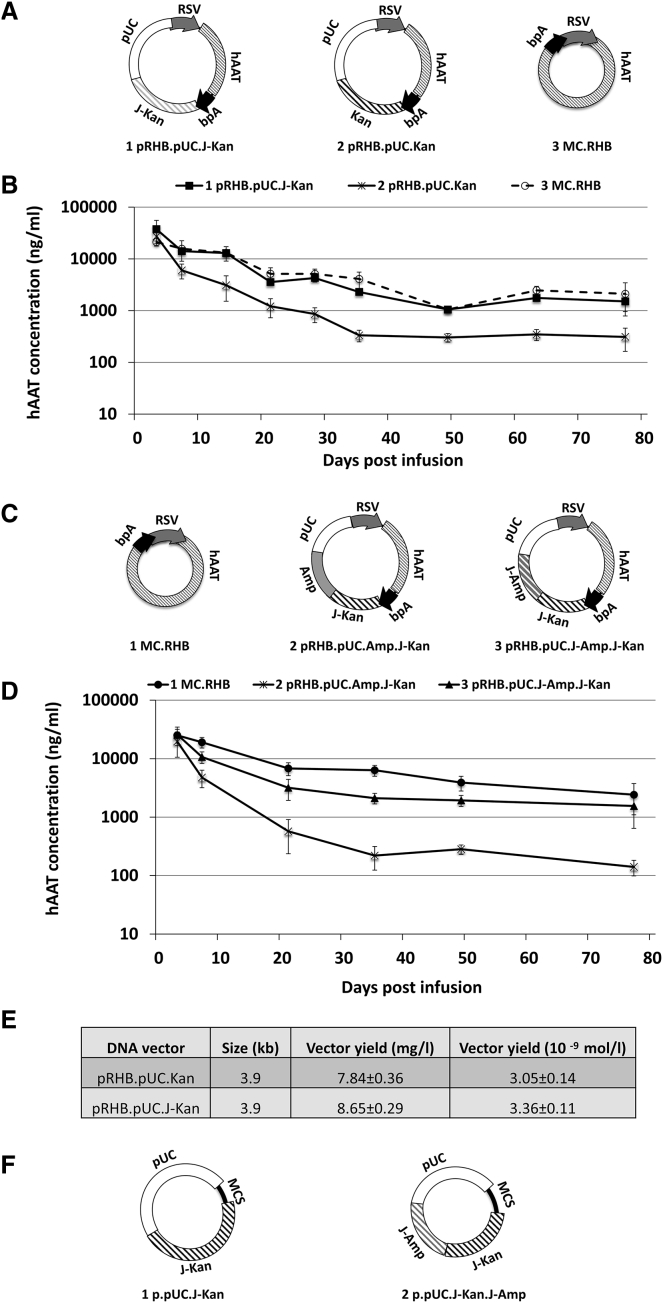
We hypothesized that perhaps optimizing the An/Tn content of canonical plasmids might result in vectors providing the same benefit of sustained transgene expression. Therefore, we engineered two antibiotic resistance genes commonly used in plasmid propagation vectors for maximal An/Tn content. The following rules were followed while modifying the sequences: (1) select the codons that incorporate more “An/Tn” but maintain the same amino acid sequence; (2) create as many “An/Tn” tracts as possible; (3) avoid RNAPII pause or cleavage/polyA-addition sequences, such as “AATAAA,” “TTTATT,” “AATAA,” or “TTATT”; and (4) avoid the use of rare codons. About 30% of the kanamycin resistance gene bases were modified (J-Kan). After modification, the GC content dropped from 60.6% in wild type to 39.0% in J-Kan (Figure S2A). The codon adaptation index (CAI), a measure of codon usage bias, remained the same (Figure S2B). Similarly, for the ampicillin resistance gene, the GC content dropped from 49.8% in wild type to 34.8% in J-Amp, with the CAI remaining nearly constant (0.67 from 0.63) after modification (Figures S3A and S3B).
The plasmid vector with J-Kan sequences was compared in parallel with the plasmid vector containing the wild-type kanamycin resistance gene (Kan) sequences in animals for their abilities to sustain transgene expression. The incorporation of J-Kan sequences into the plasmid backbone resulted in enhanced duration of hAAT transgene expression in vivo to a level similar to that expressed from the corresponding minicircle vector and about six times higher than that expressed from a plasmid with wild-type Kan (Figures 2A and 2B). Likewise, a plasmid (pRHB.pUC.J-Amp.J-Kan) with a modified “An/Tn” rich ampicillin resistance gene (J-Amp) was also able to maintain transgene expression to the same level as the minicircle vector and enhanced transgene expression to about ten times higher compared to the corresponding plasmid containing the wild-type ampicillin resistance gene pRHB.pUC.Amp.J-Kan (Figures 2C and 2D).
Figure 2.
Effect of Sequence-Modified Antibiotic Resistance Genes on Transgene Expression
(A) Schematic of plasmid vectors used for infusion pictured in (B). (B) Serum hAAT levels at various time points after equimolar infusion into the mouse liver. Serum hAAT levels on day 63 post infusion were used to calculate p values. The MC.RHB group was used as the non-silencing control. For the pRHB.pUC.J-Kan group, p = 0.06. For the pRHB.pUC.Kan group, p = 0.0001. (C) Schematic of plasmid vectors with either wild-type Ampicillin resistance gene (Amp) or modified Amplicillin (J-Amp) as part of the plasmid backbone sequence. (D) Serum levels of transgene hAAT at various time points after the infusion of plasmids pictured in (C). Serum hAAT levels on day 77 post infusion were used to calculate p values. The MC.RHB group was used as the non-silencing control. For the pRHB.pUC.Amp.J-Kan group, p = 0.0049. For pRHB.pUC.J-Amp.J-Kan, p = 0.3. (E) The yield of plasmid vectors with modified antibiotic resistance genes as part of the plasmid backbone sequence and conventional plasmid vectors (n = 4/vector). The yield was derived from quadruplicate 100-mL overnight cultures. The following formula was used to convert the yield of mg/L to mol/L: mol/L = [yield (mg/L) × 10E−3 g/L]/[size (kb) × 1,000 × 330 × 2 g/mol], where 330 is the average molecular weight of dNTP. (F) Structures of the ready-to-use cloning vector with sequence-modified antibiotic resistance genes.
The modified antibiotic resistance genes did not alter the ability of the corresponding plasmids to be propagated in standard bacterial culture (Figure 2E). To make the modified plasmid suited for routine cloning of any expression cassette, we constructed universal cloning vectors (p.pUC.J-Kan and p.pUC.J-Kan.J-Amp) containing the multiple cloning sites (MCSs) (Figure 2F).
Abrogation of Transcriptional Silencing by An/Tn Sequences Is Associated with Enhanced Plasmid Backbone Transcriptional Activity
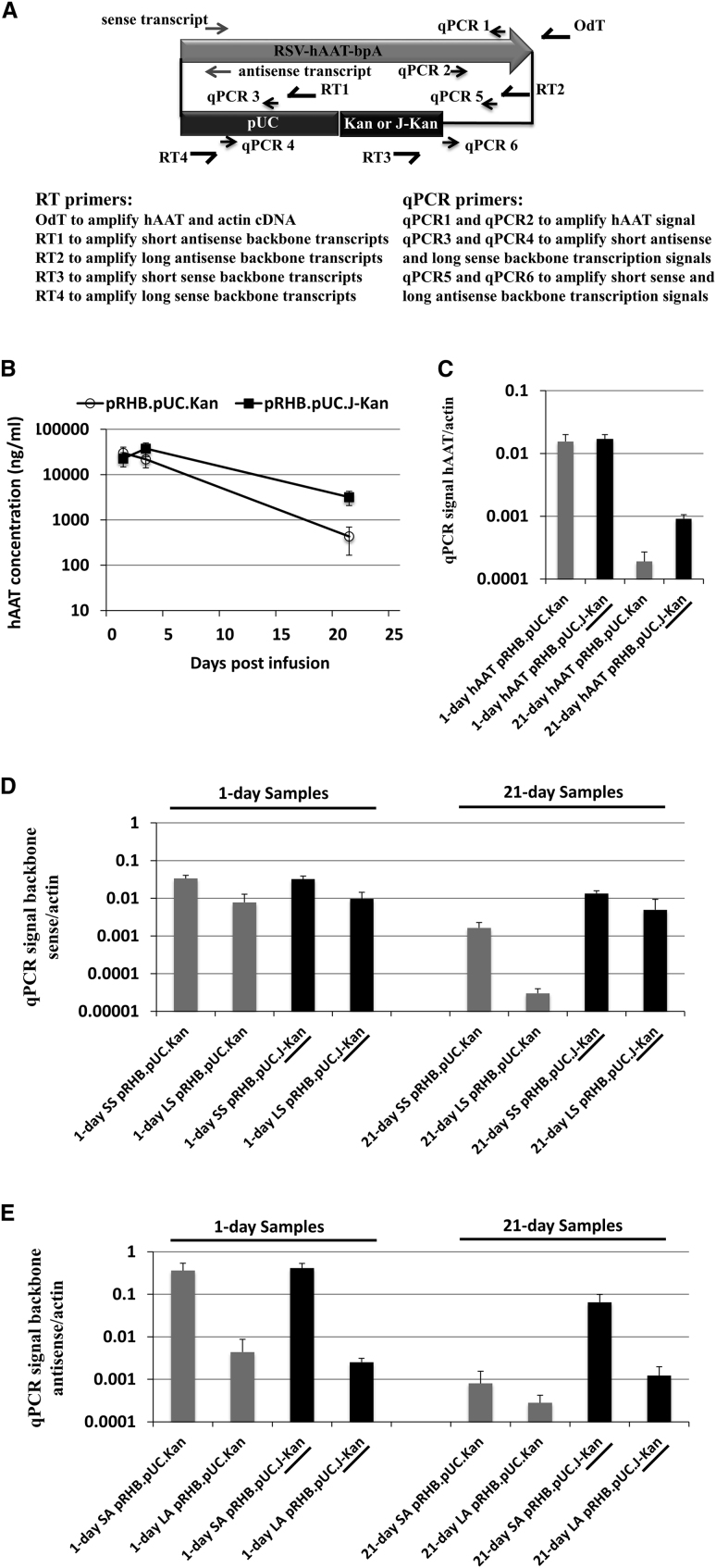
The fact that nucleosome relaxation sequences allow for persistent expression led us to hypothesize that maintenance of transcription might require RNAPII activity to be maintained on the DNA plasmid templates. Recent studies have provided evidence of widespread divergent transcription at protein-encoding gene promoters.19 It has been proposed that transcription factors must first nucleate a sense-oriented preinitiation complex at the transcription start site (TSS) but can initiate both sense and antisense-oriented transcription.19 The RNAPII pauses at the AAUAAA hexamer in the polyA signal.20, 21 RNAPII does not fall off the DNA template immediately upon reaching the hexamer. In many situations, RNAPII molecules present well over 1 kb downstream of the hexamer and continue to exhibit delayed progress down the template in vivo.20 One possible source for a sustainable transcriptional state on the plasmid DNA would be a condition in which transcription would continue past the canonical expression cassette into the backbone, with the continuing transcription serving to keep the plasmid in an open (and accessible) chromatin configuration. To explore the distribution of plasmid-derived RNA transcripts within the plasmid sequence, we used reverse transcription and real-time PCR (RT-qPCR) to compare the entire transcription activities of silencing and non-silencing plasmid vectors.
In Figure 3A, separate primer pairs were designed to detect transcripts from the expression cassette and backbone regions of the pRHB.pUC.Kan and pRHB.pUC.J-Kan vectors. These two vectors were infused into the livers of 6- to 8-week-old C57BL/6J, and total liver RNA samples were extracted on day 1 and day 21 after infusion (Figure 3B). As shown in Figure 3C, and consistent with protein expression (Figure 3B), substantially higher hAAT mRNA concentrations from pRHB.pUC.J-Kan (non-silenced) versus pRHB.pUC.Kan (silenced) infused animals was observed on day 21 after infusion. Interestingly, both sense and antisense transcripts originating from the plasmid backbone regions were present. The non-silencing vector backbone (pRHB.pUC.J-Kan) generated significantly higher transcription activities at both sense and antisense orientations when compared with transcription from the silencing vector backbone (pRHB.pUC.Kan) on day 21 (Figures 3D and 3E).
Figure 3.
Detection of Plasmid Backbone Transcription from Both Sense and Antisense Orientation
(A) Schematic of primer designs used for RT and qPCR experiments to detect transcripts from the expression cassette and backbone regions. The actin transcript was used as a standardization control. (B) Serum levels of the hAAT product on days 1, 3, and 21 after infusion of pRHB.pUC.Kan and pRHB.pUC.J-Kan plasmids (n = 6/group). (C) The hAAT transcripts detected from RNA samples extracted from animals shown in (B) on day 1 and day 21 after infusion. Primers used for the RT and qPCR experiment were shown in (A). (D) Short and long backbone transcripts in the sense orientation detected by using primers listed in (A) from RNA samples extracted on day 1 and day 21 after infusion (SS, short sense; LS, long sense). (E) Short and long backbone transcripts in the antisense orientation are detected by using primers shown in (A) from RNA samples extracted on day 1 and day 21 after infusion (SA, short antisense; LA, long antisense). Three liver RNA samples from each group were extracted on day 1 and day 21. Each sample was tested with qPCR in duplicate. Error bars represent standard deviation of six qPCR test tubes of each sample.
The silencing MC.RHB.2.2kb RD and non-silencing MC.RHB.2.2kb RD-An/Tn vectors gave the same pattern of sense and antisense transcripts from the expression cassette and plasmid backbone (Figure S4), as did the pRHB.pUC.Kan and pRHB.pUC.J-Kan vectors (Figure 3), respectively. This further supports the idea that the substitution of J-Kan for Kan overcomes silencing by the same mechanism as including nucleosome-relaxation sequences in the plasmid DNA backbone.
Incorporation of RNAPII Arrest Sites in Plasmid Backbone Silenced the Transgene Expression
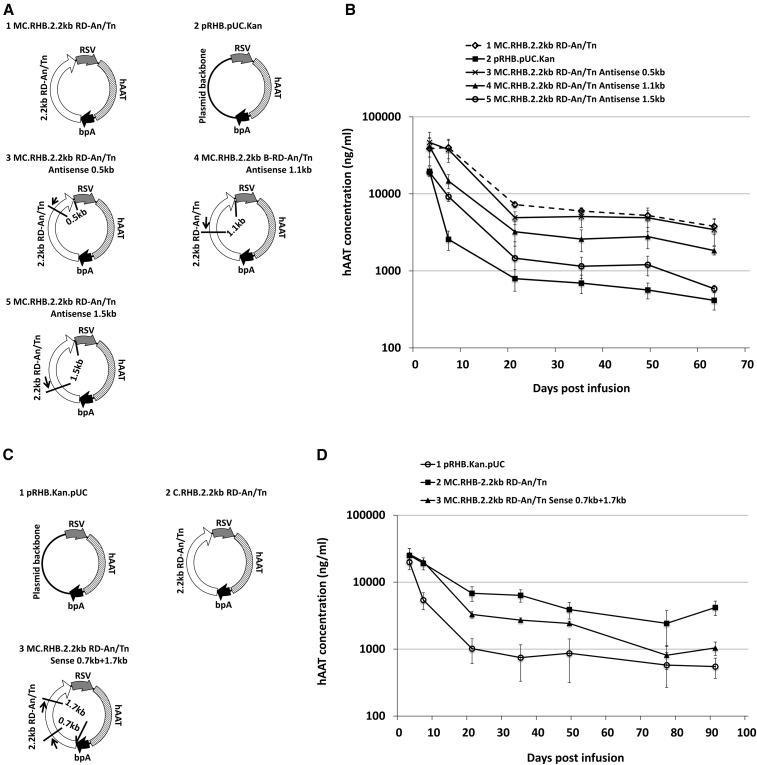
To further provide support for the transcriptional dependence of the plasmid backbone sequences on persistence of transgene expression, we asked whether transcriptional pausing could alter expression profiles from the non-silenced plasmids. Transcription elongation by RNAPII does not proceed uniformly, and polymerase pausing is thought to be a mechanism to regulate transcription. RNAPII arrest sites identified by studies performed in cell-free systems are primarily localized across the coding region. Pause sites block a fraction of transcribing polymerases, making them unable to continue RNA synthesis.22 In addition, RNAPII arrest sites function in an orientation-dependent manner.23, 24 Because pause sites are frequently located in the coding sequence, it has been difficult to manipulate and study them in living cells. In this system, we have the ability to place these sequences into plasmid backbones for in vivo study. We hypothesized that if the plasmid backbone transcription activities influence the expression of transgene, the incorporation of an RNAPII arrest site in the plasmid backbone of non-silencing vectors should block backbone transcription, causing silencing of the transgene expression cassette. To test this hypothesis, we incorporated the mammalian histone H3.3 RNAPII arrest site “TTTTTTTCCCTTTTTT”25 into the non-silencing backbone sequence RD-An/Tn. This RNAPII arrest site was chosen because it contains a Tn sequence that is consistent with the RD-An/Tn design used in our experiments described above.
Because RNAPII arrest sites function in an orientation-dependent manner, one copy of the H3.3 RNAPII arrest site was first incorporated at different locations in the backbone at the antisense orientation (Figure 4A) to block backbone transcription in the antisense orientation and tested the expression profiles of these vectors in mice. We constructed vectors with this site at various distances from the RSV promoter. “MC.RHB.2.2kb RD-An/Tn Antisense 0.5kb,” “MC.RHB.2.2kb RD-An/Tn Antisense 1.1kb,” and “MC.RHB.2.2kb RD-An/Tn Antisense 1.5kb” had one copy of the H3.3 RNAPII arrest site incorporated into the backbone at the antisense orientation at 0.5 kb, 1.1 kb, and 1.5 kb away from the RSV promoter, respectively. As shown by protein expression in Figure 4B, transgene silencing was observed once the arrest site was placed further away from the promoter (1.1 kb and 1.5 kb) in the antisense orientation. Thus, the H3.3 RNAPII arrest site at certain positions in the backbone is capable of blocking backbone transcription and silencing transgene expression.
Figure 4.
Influence of Incorporating the H3.3 RNAPII Arrest Site in the Backbone on Transgene Expression
(A) Schematic of plasmid vectors used for the infusion pictured in (B). The arrow indicated the orientation of transcription that the RNAPII arrest site was designed to block (antisense). The short line indicated the location of the H3.3 RNAPII arrest site. The number indicated the distance of the RNAPII arrest site from the RSV promoter. (B) Serum hAAT levels at various time points after equimolar transfection into the mouse liver (n = 5/group). Serum hAAT levels on day 63 post infusion were used to calculate p values. The MC.RHB.2.2kb RD-An/Tn group was used as the non-silencing control. For the pRHB.pUC.Kan group, p = 0.0002. For the MC.RHB.2.2kb RD-An/Tn Antisense 0.5kb group, p = 0.66. For the MC.RHB.2.2kb RD-An/Tn Antisense 1.1kb group, p = 0.004. For the MC.RHB.2.2kb RD-An/Tn Antisense 1.5kb group, p = 0.0003. (C) Schematic of plasmid vectors used for injection pictured in (D). Each copy of the H3.3 RNAPII arrest site was incorporated into two locations on the backbone (0.7 kb and 1.7 kb away from bpA) at the sense orientation. (D) Serum hAAT levels at various time points after equimolar transfection into the mouse liver (n = 5/group). Serum hAAT levels on day 91 post infusion were used to calculate p values. The MC.RHB.2.2kb RD-An/Tn group was used as the non-silencing control. For the pRHB.pUC.Kan group, p = 0.0002. For the MC.RHB.2.2kb RD-An/Tn Sense 0.7kb+1.7kb group, p = 0.0002.
When one copy of the same RNAPII arrest site was incorporated into various locations of the backbone at the sense orientation, the transgene expression profile was not affected (Figures S5A and S5B). However, when two copies of the H3.3 RNAPII arrest site were incorporated into the MC.RHB.2.2kb RD-An/Tn backbone in the sense orientation (0.7 kb and 1.7 kb away from polyA) (Figure 4C), transgene expression was silenced (Figure 4D). Nevertheless, pausing of antisense backbone transcription had a more substantial effect on transgene silencing, indicating weaker backbone transcription at the antisense orientation than at the sense orientation. Establishing this result was not just due to the specific sequences; a second An/Tn-rich c-Myc RNAPII arrest site, “TTTTAATTTATTTTTTTAT,” was also tested and found to give similar results (Figures S5C and S5D). These results support the idea that transcription in both directions may be important for long-term maintenance of expression.26, 27, 28
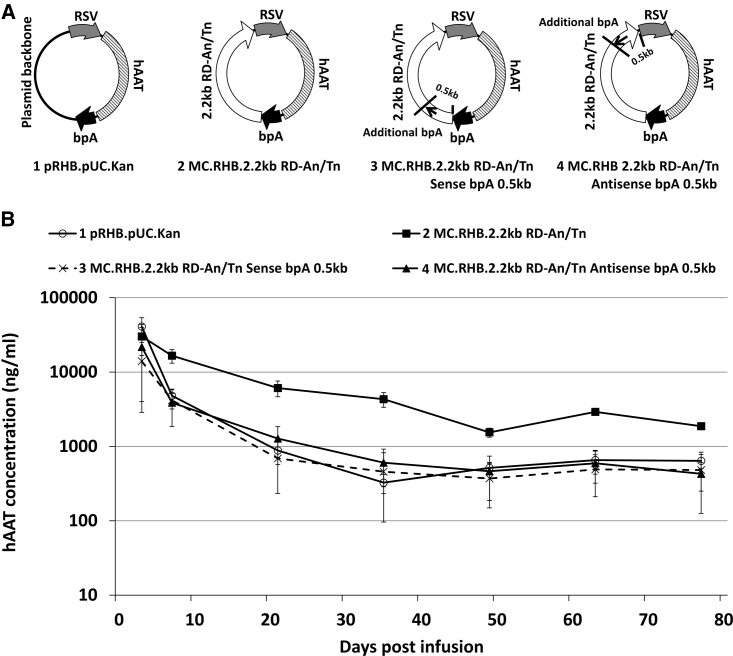
Finally, because polyA signals are efficient in inducing an RNAPII pause, one copy of 230-bp-long bpA was inserted into the RD-An/Tn backbone at each orientation (Figure 5A). As expected, that additional copy of bpA signal at either orientation in the RD-An/Tn backbone was sufficient to silence transgene expression (Figure 5B). Taken together, our data are consistent with the concept that maintaining non-canonical transcription from regions outside of the transgene expression cassette may indeed be required to maintain persistent expression.
Figure 5.
Effect of Additional bpA Signal Inserted into the Plasmid Backbone on Transgene Expression
(A) Schematic of plasmid constructs infused into animals for experiment pictured in (B). The arrow indicated the transcription orientation that the additional bpA signal was designed to block. The short line indicated the location of the additional bpA signal. The number indicated the distance of additional bpA signal from the expression cassette end at the same orientation. (B) Serum hAAT levels at various time points after equimolar transfection into the mouse liver (n = 5/group). Serum hAAT levels on day 63 post infusion were used to calculate p values. The MC.RHB.2.2kb RD-An/Tn group was used as the non-silencing control. For the pRHB.pUC.Kan group, p = 0.0001. For the MC.RHB.2.2kb RD-An/Tn Sense bpA 0.5kb group, p = 0.0001. For the MC.RHB.2.2kb RD-An/Tn Antisense bpA 0.5kb group, p = 0.0001.
Discussion
The persistence of episomal vector-mediated transgene expression is dictated by a number of parameters. In the case of plasmid-mediated gene transfer, transgene expression is dependent on the length of the plasmid backbone makeup and, in some cases, the CpG content. Canonical plasmids have a high CpG content, which can trigger innate immune responses that promote the loss of the vector DNA.2, 29, 30, 31, 32 However, the CpG-mediated loss of transgene expression is only observed when lipid-based vehicles are used for DNA delivery.2, 33 The hydrodynamic transfection without lipids used in this work avoids the innate response.2, 33 Consistent with the expectation of a CpG-independent foreign DNA response and of identifying a means toward its mitigation, in the present study, we compared plasmids with or without AT-rich RNAPII arrest sites in their backbone regions (Figures 4, S5C, and S5D). These plasmids have identical CpG content. However, the plasmids with RNAPII arrest sites in their backbone silenced transgene, whereas the plasmids without RNAPII arrest sites were able to sustain transgene expression. These results are consistent with our previous findings that CpG content has little or no influence in transcription silencing.2
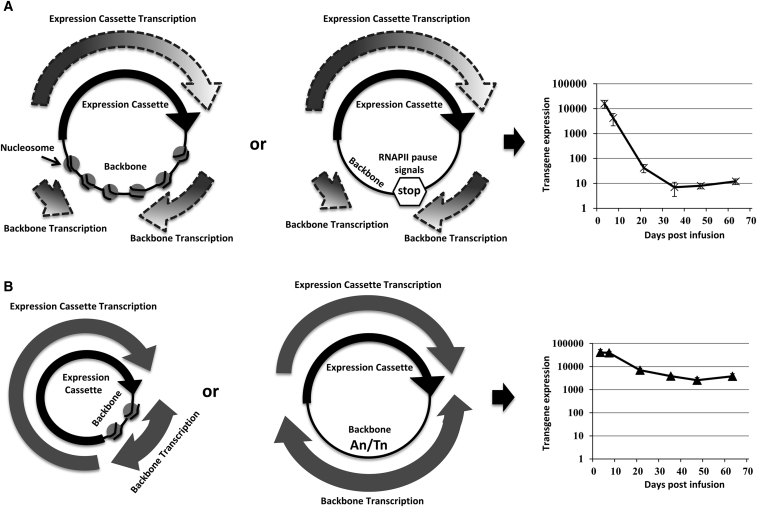
From our previous study, we demonstrated that the length and not the sequence of the intervening DNA contained outside of the expression cassette was responsible for plasmid-mediated transgene silencing.1 Our experiments here show that including a high proportion of An/Tn in the plasmid bacterial backbone allows for the maintenance of transgene transcription, and this is associated with enhanced backbone transcripts. Because A-T base pairs have fewer hydrogen bonds than G-C base pairs, the An/Tn rich region requires fewer forces for unzipping the double-stranded DNA.34 Such features can enhance the AT-rich region in bacterial replication origin to form replication complexes.35 In terms of plasmid backbone transcription, a possible source of the observations would also come from an ability of An/Tn sequences to make the nucleosome unwinding easier for RNAPII through the backbone, permitting the RNAPII complex to remain on plasmid and continue to re-initiate transcription (Figure 6). In the absence of sufficient An/Tn sequences, RNAPII could stochastically stall or pause, eventually falling off the plasmid DNA and leading to an unprotected DNA that could be silenced. We propose a model whereby the ability to maintain RNAPII on the DNA sequences outside of the expression cassette is the primary factor that determines transgene persistence (Figure 6). If the DNA is unfavorable and RNAPII engagement is lost, then the plasmid vectors take on a “heterochromatic” chromatin state.36 Expression of basally transcribed plasmid vectors would then persist under conditions in which euchromatic chromatin is the default state for transcribed DNA. However, further studies are required to establish the stepwise mechanism and whether the degree of RNAPII occupancy based on the backbone An/Tn sequence composition is the direct cause of plasmid-mediated transcriptional silencing.
Figure 6.
Proposed Model That the Ability to Maintain RNAPII on the DNA Sequences Outside of the Expression Cassette Is the Primary Factor That Determines Transgene Persistence
(A) Schematic of transgene silencing model for the silenced plasmids. When the plasmid backbone is long enough to sufficiently recruit enough numbers of nucleosomes (left panel) or contains RNAPII pause signals (middle panel), the backbone transcription is blocked. This consequently prevents an efficient reinitiation of transcription in the plasmid expression cassette region and causes plasmid silencing (right panel). (B) Schematic of transgene persisting model for the non-silenced plasmids. When the plasmid backbone is short enough to avoid recruiting a sufficient amount of nucleosomes (left panel) or contains An/Tn rich sequences (middle panel), the backbone transcription is sustained. Therefore, RNAPII is able to be reinitiated efficiently in the plasmid expression cassette region to maintain persistent transcription (right panel).
The pEPI-1 vector that contains a chromosomal scaffold/matrix-attached region (S/MAR) has been shown to have a major influence on transcriptional rate and might influence plasmid DNA replication when tightly associated with the nuclear matrix.37, 38, 39 The matrix-associated regions (MARs) are defined as AT-rich DNA sequences that are preferentially retained by the nuclear matrix.40, 41 This raises a question of whether the An/Tn-rich backbone sequences would serve as MAR elements. Arguing against this as the primary mechanism are observations in which the insertion of an RNAPII arrest site resulted in plasmids that are identical, except for the insertion of this site (RD-An/Tn backbone with and without the RNAPII arrest site; Figures 4, S5C, and S5D). The plasmid with an RNAPII arrest site has an even higher content of An/Tn sequences, and both of these backbones should have the potential to serve as MARs. However, our results demonstrate that the RD-An/Tn backbone with RNAPII arrest site silences transgene expression (Figures 4, S5C, and S5D). These data point away from a single protective element (such as a MAR) in sustaining plasmid function in this system, leading us to the working model of continuous baseline transcription as a major determinant for sustained activity.
To be suitable in molecular engineering applications, plasmid vectors would need to be stable for amplification in bacterial hosts. Although certain unusual sequences can affect growth or integrity in E. coli (such as GT-rich “recombinant islands” that locally increase the RecA-mediated DNA recombination42), we found no evidence that the modified antibiotic resistance genes (J-Kan and J-Amp) described in this study had any such effects. Indeed, experiences in construction, growth, yield, and homogeneity of the expression clones based on the J-Kan and J-Amp vectors were comparable to our extensive experiences with the parental pUC vectors.
In our study, all vectors were infused into C57BL/6J mice through hydrodynamic tail vein injection. The data collected through our experiments thus only represent plasmid expression levels from the mouse livers. Although additional testing in other tissues will be required for evaluation of suitability for diverse tissue types, there is a precedent for generalization of anti-silencing effects beyond liver and hydrodynamic injection.43, 44, 45 Importantly, we applied our mechanistic findings to make a canonical plasmid that is resilient to transcriptional silencing. Our previous approaches to enhance transgene expression used minicircle4 or MIP3 plasmid vector technology. Although each of these have their advantages, the fact that special DNAs, bacterial cell lines, and/or purification schemes were required limited their use by some laboratories as well as limited their production in large-scale manufacturing protocols. The reagents described in this study will be beneficial to many in order to optimize transgene expression studies.
Materials and Methods
Materials
The RD-An/Tn and modified J-Kan and J-Amp sequences were synthesized by Thermo Fisher Scientific. A SacI site and an XbaI site were incorporated to the 5′ and 3′ sequences of J-Kan, respectively. An XbaI site was incorporated to both the 5′ and 3′ sequences of J-Amp.
Vector Construction
pRHB and MC.RHB were previously described.3, 4 To generate vectors using synthesized RD-An/Tn sequence and Random DNA sequence as backbone, minicircle vectors MC.RHB.2.2kb RD-An/Tn and MC.RHB.2.2kb RD were created. Because neither RD-An/Tn nor RD is able to support DNA replication and selection in bacterial host strains, conventional plasmid vectors are not suitable for our purpose. The RD-An/Tn sequence was inserted into the SpeI site of minicircle MC.RHB, producing parental plasmid on both orientations. The resulting minicircle parental plasmids were further prepared into minicircle vectors MC.RHB.2.2kb F-RD-An/Tn (forward) and MC.RHB.2.2kb B-RD-An/Tn (backward). The RD sequence was also inserted into the SpeI site of minicircle MC.RHB, producing parental plasmid to produce MC.RHB.2.2kb RD. In order to build the minicircle vector carrying 4.4 kb of RD-An/Tn as the backbone, an additional copy of 2.2 kb of RD-An/Tn sequence was inserted into the XbaI site of the MC.RHB.2.2kb RD-An/Tn parental plasmid. The same restriction enzyme was used to insert a second copy of 2.2 kb of RD to generate the MC.RHB.4.4kb RD vector. To generate the pRHB.pUC.J-Kan vector, the pRHB was digested by SacI and XbaI to remove the wild-type kanamycin resistance gene sequence, followed by ligating to the SacI-XbaI double-digested synthesized J-Kan fragment. To build the pRHB.pUC.Amp.J-Kan vector, the XbaI site containing forward primer 5′ ATATATTCTAGAATGAGTATTCAACATTTCCGTG 3′ and the XbaI site containing backward primer 5′ ATATATTCTAGATTACCAATGCTTAATCAGTGAG 3′ were used to amplify the wild-type ampicillin resistance gene sequence through PCR. This PCR product was further treated with XbaI, and then ligated with the XbaI-digested pRHB.pUC.J-Kan vector. The pRHB.pUC.J-Amp.J-Kan vector was constructed through ligating the XbaI-digested pRHB.pUC.J-Kan vector and XbaI-digested synthesized J-Amp fragment. The orientation of insertion was confirmed through DNA sequencing. The RNAPII arrest site was inserted into RD-An/Tn backbone through site-directed mutagenesis by using QuikChange II XL Site-Directed Mutagenesis Kit (Agilent Technologies). The following primers were used. The forward primer 5′ GATTCCACCGGGCGCAGCAAAAAAGGGAAAAAAAGTTCCTCTCGAGCGGCGG 3′ and the backward primer 5′ CCGCCGCTCGAGAGGAACTTTTTTTCCCTTTTTTGCTGCGCCCGGTGGAATC 3′ were used to generate MC.RHB.2.2kb RD-An/Tn Antisense 0.5kb. The forward primer 5′ GCCAGGCAGGAGTCTCAAAAAAAAGGGAAAAAAAGTTCTCGCGAGTATCTGC 3′ and the backward primer 5′ GCAGATACTCGCGAGAACTTTTTTTCCCTTTTTTTTGAGACTCCTGCCTGGC 3′ were used to construct MC.RHB.2.2kb RD-An/Tn Antisense 1.1kb. The forward primer 5′ CTTGAATTCTTCTATTCCAAAAAAGGGAAAAAAACGCCGCGGGATGAGGCCG 3′ and the backward primer 5′ CGGCCTCATCCCGCGGCGTTTTTTTCCCTTTTTTGGAATAGAAGAATTCAAG 3′ were used to build MC.RHB.2.2kb RD-An/Tn Antisense 1.5kb.
Animal Studies
6- to 8-week-old female C57BL/6J mice purchased from Jackson Laboratory were used for DNA or rAAV vector infusion. To ensure the same molar amount of DNA was injected into each animal, 3.63 μg/kb DNA was used for various sized constructs. Each DNA construct was diluted into 1.8 mL of 0.9% NaCl for each animal, and was delivered through hydrodynamic tail vein injection. After infusion, blood samples were collected periodically by a retro-orbital technique. The serum hAAT and the plasma hFIX levels were quantified by ELISA measurements. All animal studies performed within this paper were oversighted by the Stanford review board.
ELISA
ELISA for hAAT and hFIX was performed as previously described46 with the following antibodies: goat anti-human alpha-1 antitrypsin primary antibody at 1:1,000 (Strategic Biosolutions S0311G000-S4), goat pAB to alpha-1 antitrypsin (HRP) (Abcam ab7635), mouse anti-human F9 IgG primary antibody at 1:1,000 (Sigma F2645), and polyclonal goat anti-human F9 peroxidase-conjugated IgG secondary antibody at 1:4,200 (Enzyme Research GAFIX-APHRP). The detection limit of ELISA for hAAT and hFIX under our experimental condition is 1 ng/mL, and the background for non-infused negative control mouse serum is zero.
RT-qPCR
Liver samples of animals tested in Figure 3B were used for RT-qPCR experiments in Figures 3C–3E. Liver total RNA samples were extracted by using the mirVana miRNA Isolation Kit (Ambion). 5 μg of total RNA of each sample was used to generate cDNA through the SuperScriptIII First-Strand Synthesis System for RT-PCR (Invitrogen). The RT1 primer 5′ TTACCAGTGGCTGCTGCCA 3′ was used to amplify short antisense backbone transcripts. The RT2 primer 5′ CAGGCATGCTGGGGATGCGGT 3′ was used to amplify long antisense backbone transcripts. The RT3 primer 5′ CGCCGCATTGCATCAGCCAT 3′ was used to amplify short sense backbone transcripts. The RT4 primer 5′ ATACCTGTCCGCCTTTCTCC 3′ was used to amplify long sense backbone transcripts. The standard OdT was used to amplify hAAT and actin cDNA. 5 μL of each cDNA product was further used for each 20 μL qPCR reaction. qPCR was performed by using the QuantiFast SYBR Green PCR Kit (QIAGEN) with the Corbett Research RG6000 PCR instrument (Corbett Research). Forward primer (qPCR2) 5′ AAGGCAAATGGGAGAGACCT 3′ and reverse primer (qPCR1) 5′ TACCCAGCTGGACAGCTTCT 3′ oligos were used to amplify about a 150 bp fragment from the hAAT cDNA region. Forward primer 5′ TTGCTGACAGGATGCAGAAG 3′ and reverse primer 5′ TGATCCAC ATCTGCTGGAAG 3′ oligos were used to amplify a 150 bp fragment from β-actin as the loading control. Forward primer (qPCR3) 5′ AGTCGTGTCTTACCGGGTTG 3′ and reverse primer (qPCR4) 5′ GGCGCTTTCTCATAGCTCAC 3′ were used to amplify about 150 bp of short antisense and long sense backbone transcription signals. Forward primer (qPCR5) 5′ ATGGACAGCAAGCGAACCG 3′ and the reverse primer (qPCR6) 5′ GCGAAACGATCCTCATCCTG 3′ were used to amplify about 150 bp of short sense and long antisense backbone transcription signals. The tested transgene signal was then normalized to the β-actin signal. Liver samples of animals tested in Figure S4A were used for RT-qPCR experiments in Figures S4B–S4F. The hAAT and actin signals were amplified as described in Figure 3. The RT primers 5′ CCCTCTCCTATTCTGGCTC3′, 5′TGACTCCCGAAGGCCGGT3′, and 5′CGACTATATTATCAGCTTACGA3′ were used to amplify short, medium, and long sense transcripts from the RD-An/Tn backbone, respectively. The RT primers 5′ACGAACTCCGACGTCTGTAT3′, 5′AAAGCCGCATGGAAAGTTGAG3′, and 5′TGTTTAATGGCTGGAGTACAAA3′ were used to amplify short, medium, and long antisense transcripts from the RD-An/Tn backbone, respectively. For the RD backbone, RT primers 5′CCAGTCAATTATGAGTTACTCC3′ and 5′TGCGCGTACACCACCAGC3′ were used to amplify short and long sense transcripts, whereas RT primers 5′TGGAGCCGCGCCCGGC3′ and 5′TTAGCCGCGAGGGAGATACA3′ were used to amplify short and long antisense transcripts from the RD backbone region. Forward primer 5′TTGGTGGCTCCCTAAAGTTG3′ and reverse primer 5′TCACGCGTGGAAAACATAAA3′ were used to amplify about 150 bp of short sense and long antisense transcription signals from the RD-An/Tn backbone. Forward primer 5′GCGAGCGAATATGCTTTGTT3′ and reverse primer 5′GTATAGGGATGGTAGGCGCA3′ were used to amplify about 150 bp of short antisense and long sense transcription signals from the RD-An/Tn backbone. Forward primer 5′GGAGCCAGAATAGGAGAGGG3′ and reverse primer 5′TCCGGAATCCGTAGTACGTC3′ were used to amplify about 150 bp of medium sense and medium antisense transcription signals from the RD-An/Tn backbone. Forward primer 5′TAAAGACTGCGCCGGTAACT3′ and reverse primer 5′GCGTGGTGAATATCTCGGTT3′ were used to amplify about 150 bp of short sense transcription signals from the RD backbone. Forward primer 5′GAAGCCGGAAGAACTGTCTG3′ and reverse primer 5′GCGCCTACCATCCCTATACA3′ were used to amplify about 150 bp of long sense and short antisense transcription signals from the RD backbone. Forward primer 5′AACCGAGATATTCACCACGC3′ and reverse primer 5′TCCCTCGTAGCTCTCCTCAA3′ were used to amplify about 150 bp of long antisense transcription signals from the RD backbone.
Author Contributions
J.L. came up with the initial idea, designed, performed and analyzed experiments. F.Z. performed experiments. A.Z.F. designed experiments and contributed to manuscript preparation in conjunction with J.L. and M.A.K. M.A.K. designed experiments, supervised the work and contributed to manuscript preparation in conjunction with J.L. and A.Z.F.
Acknowledgments
We wish to express our sincere thanks to Professor Roger Kornberg, Dr. Christian Frøkjaer Jensen, Dr. Lia Gracey Maniar, and Dr. Loren Hansen for their suggestions.
Footnotes
Supplemental Information includes five figures and can be found with this article online at http://dx.doi.org/10.1016/j.ymthe.2017.03.003.
Supplemental Information
References
- 1.Lu J., Zhang F., Xu S., Fire A.Z., Kay M.A. The extragenic spacer length between the 5′ and 3′ ends of the transgene expression cassette affects transgene silencing from plasmid-based vectors. Mol. Ther. 2012;20:2111–2119. doi: 10.1038/mt.2012.65. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Chen Z.Y., Riu E., He C.Y., Xu H., Kay M.A. Silencing of episomal transgene expression in liver by plasmid bacterial backbone DNA is independent of CpG methylation. Mol. Ther. 2008;16:548–556. doi: 10.1038/sj.mt.6300399. [DOI] [PubMed] [Google Scholar]
- 3.Lu J., Zhang F., Kay M.A. A mini-intronic plasmid (MIP): a novel robust transgene expression vector in vivo and in vitro. Mol. Ther. 2013;21:954–963. doi: 10.1038/mt.2013.33. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Chen Z.Y., He C.Y., Ehrhardt A., Kay M.A. Minicircle DNA vectors devoid of bacterial DNA result in persistent and high-level transgene expression in vivo. Mol. Ther. 2003;8:495–500. doi: 10.1016/s1525-0016(03)00168-0. [DOI] [PubMed] [Google Scholar]
- 5.Chen Z.Y., He C.Y., Kay M.A. Improved production and purification of minicircle DNA vector free of plasmid bacterial sequences and capable of persistent transgene expression in vivo. Hum. Gene Ther. 2005;16:126–131. doi: 10.1089/hum.2005.16.126. [DOI] [PubMed] [Google Scholar]
- 6.Bigger B.W., Tolmachov O., Collombet J.M., Fragkos M., Palaszewski I., Coutelle C. An araC-controlled bacterial cre expression system to produce DNA minicircle vectors for nuclear and mitochondrial gene therapy. J. Biol. Chem. 2001;276:23018–23027. doi: 10.1074/jbc.M010873200. [DOI] [PubMed] [Google Scholar]
- 7.Darquet A.M., Cameron B., Wils P., Scherman D., Crouzet J. A new DNA vehicle for nonviral gene delivery: supercoiled minicircle. Gene Ther. 1997;4:1341–1349. doi: 10.1038/sj.gt.3300540. [DOI] [PubMed] [Google Scholar]
- 8.Mayrhofer P., Blaesen M., Schleef M., Jechlinger W. Minicircle-DNA production by site specific recombination and protein-DNA interaction chromatography. J. Gene Med. 2008;10:1253–1269. doi: 10.1002/jgm.1243. [DOI] [PubMed] [Google Scholar]
- 9.Schakowski F., Gorschlüter M., Buttgereit P., Märten A., Lilienfeld-Toal M.V., Junghans C., Schroff M., König-Merediz S.A., Ziske C., Strehl J. Minimal size MIDGE vectors improve transgene expression in vivo. In Vivo. 2007;21:17–23. [PubMed] [Google Scholar]
- 10.Beisel C., Paro R. Silencing chromatin: comparing modes and mechanisms. Nat. Rev. Genet. 2011;12:123–135. doi: 10.1038/nrg2932. [DOI] [PubMed] [Google Scholar]
- 11.Iyer V., Struhl K. Poly(dA:dT), a ubiquitous promoter element that stimulates transcription via its intrinsic DNA structure. EMBO J. 1995;14:2570–2579. doi: 10.1002/j.1460-2075.1995.tb07255.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Segal E., Widom J. Poly(dA:dT) tracts: major determinants of nucleosome organization. Curr. Opin. Struct. Biol. 2009;19:65–71. doi: 10.1016/j.sbi.2009.01.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Sekinger E.A., Moqtaderi Z., Struhl K. Intrinsic histone-DNA interactions and low nucleosome density are important for preferential accessibility of promoter regions in yeast. Mol. Cell. 2005;18:735–748. doi: 10.1016/j.molcel.2005.05.003. [DOI] [PubMed] [Google Scholar]
- 14.Field Y., Kaplan N., Fondufe-Mittendorf Y., Moore I.K., Sharon E., Lubling Y., Widom J., Segal E. Distinct modes of regulation by chromatin encoded through nucleosome positioning signals. PLoS Comput Biol. 2008;4:e1000216. doi: 10.1371/journal.pcbi.1000216. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Yuan G.C., Liu Y.J., Dion M.F., Slack M.D., Wu L.F., Altschuler S.J., Rando O.J. Genome-scale identification of nucleosome positions in S. cerevisiae. Science. 2005;309:626–630. doi: 10.1126/science.1112178. [DOI] [PubMed] [Google Scholar]
- 16.Mavrich T.N., Jiang C., Ioshikhes I.P., Li X., Venters B.J., Zanton S.J., Tomsho L.P., Qi J., Glaser R.L., Schuster S.C. Nucleosome organization in the Drosophila genome. Nature. 2008;453:358–362. doi: 10.1038/nature06929. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Schones D.E., Cui K., Cuddapah S., Roh T.Y., Barski A., Wang Z., Wei G., Zhao K. Dynamic regulation of nucleosome positioning in the human genome. Cell. 2008;132:887–898. doi: 10.1016/j.cell.2008.02.022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Frøkjær-Jensen C., Jain N., Hansen L., Davis M.W., Li Y., Zhao D., Rebora K., Millet J.R., Liu X., Kim S.K. An abundant class of non-coding DNA can prevent stochastic gene silencing in the C. elegans germline. Cell. 2016;166:343–357. doi: 10.1016/j.cell.2016.05.072. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Seila A.C., Calabrese J.M., Levine S.S., Yeo G.W., Rahl P.B., Flynn R.A., Young R.A., Sharp P.A. Divergent transcription from active promoters. Science. 2008;322:1849–1851. doi: 10.1126/science.1162253. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Orozco I.J., Kim S.J., Martinson H.G. The poly(A) signal, without the assistance of any downstream element, directs RNA polymerase II to pause in vivo and then to release stochastically from the template. J. Biol. Chem. 2002;277:42899–42911. doi: 10.1074/jbc.M207415200. [DOI] [PubMed] [Google Scholar]
- 21.Park N.J., Tsao D.C., Martinson H.G. The two steps of poly(A)-dependent termination, pausing and release, can be uncoupled by truncation of the RNA polymerase II carboxyl-terminal repeat domain. Mol. Cell. Biol. 2004;24:4092–4103. doi: 10.1128/MCB.24.10.4092-4103.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Hawryluk P.J., Ujvári A., Luse D.S. Characterization of a novel RNA polymerase II arrest site which lacks a weak 3′ RNA-DNA hybrid. Nucleic Acids Res. 2004;32:1904–1916. doi: 10.1093/nar/gkh505. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Wiest D.K., Hawley D.K. In vitro analysis of a transcription termination site for RNA polymerase II. Mol. Cell. Biol. 1990;10:5782–5795. doi: 10.1128/mcb.10.11.5782. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Kash S.F., Innis J.W., Jackson A.U., Kellems R.E. Functional analysis of a stable transcription arrest site in the first intron of the murine adenosine deaminase gene. Mol. Cell. Biol. 1993;13:2718–2729. doi: 10.1128/mcb.13.5.2718. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Reines D., Wells D., Chamberlin M.J., Kane C.M. Identification of intrinsic termination sites in vitro for RNA polymerase II within eukaryotic gene sequences. J. Mol. Biol. 1987;196:299–312. doi: 10.1016/0022-2836(87)90691-7. [DOI] [PubMed] [Google Scholar]
- 26.Xu Z., Wei W., Gagneur J., Perocchi F., Clauder-Münster S., Camblong J., Guffanti E., Stutz F., Huber W., Steinmetz L.M. Bidirectional promoters generate pervasive transcription in yeast. Nature. 2009;457:1033–1037. doi: 10.1038/nature07728. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Kowalczyk M.S., Higgs D.R., Gingeras T.R. Molecular biology: RNA discrimination. Nature. 2012;482:310–311. doi: 10.1038/482310a. [DOI] [PubMed] [Google Scholar]
- 28.Wang G.Z., Lercher M.J., Hurst L.D. Transcriptional coupling of neighboring genes and gene expression noise: evidence that gene orientation and noncoding transcripts are modulators of noise. Genome Biol. Evol. 2011;3:320–331. doi: 10.1093/gbe/evr025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Tan Y., Li S., Pitt B.R., Huang L. The inhibitory role of CpG immunostimulatory motifs in cationic lipid vector-mediated transgene expression in vivo. Hum. Gene Ther. 1999;10:2153–2161. doi: 10.1089/10430349950017149. [DOI] [PubMed] [Google Scholar]
- 30.Yew N.S., Wang K.X., Przybylska M., Bagley R.G., Stedman M., Marshall J., Scheule R.K., Cheng S.H. Contribution of plasmid DNA to inflammation in the lung after administration of cationic lipid:pDNA complexes. Hum. Gene Ther. 1999;10:223–234. doi: 10.1089/10430349950019011. [DOI] [PubMed] [Google Scholar]
- 31.Yew N.S., Cheng S.H. Reducing the immunostimulatory activity of CpG-containing plasmid DNA vectors for non-viral gene therapy. Expert Opin. Drug Deliv. 2004;1:115–125. doi: 10.1517/17425247.1.1.115. [DOI] [PubMed] [Google Scholar]
- 32.Chen Z.Y., He C.Y., Meuse L., Kay M.A. Silencing of episomal transgene expression by plasmid bacterial DNA elements in vivo. Gene Ther. 2004;11:856–864. doi: 10.1038/sj.gt.3302231. [DOI] [PubMed] [Google Scholar]
- 33.Yew N.S., Zhao H., Przybylska M., Wu I.H., Tousignant J.D., Scheule R.K., Cheng S.H. CpG-depleted plasmid DNA vectors with enhanced safety and long-term gene expression in vivo. Mol. Ther. 2002;5:731–738. doi: 10.1006/mthe.2002.0598. [DOI] [PubMed] [Google Scholar]
- 34.Essevaz-Roulet B., Bockelmann U., Heslot F. Mechanical separation of the complementary strands of DNA. Proc. Natl. Acad. Sci. USA. 1997;94:11935–11940. doi: 10.1073/pnas.94.22.11935. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Rajewska M., Wegrzyn K., Konieczny I. AT-rich region and repeated sequences - the essential elements of replication origins of bacterial replicons. FEMS Microbiol. Rev. 2012;36:408–434. doi: 10.1111/j.1574-6976.2011.00300.x. [DOI] [PubMed] [Google Scholar]
- 36.Gracey Maniar L.E., Maniar J.M., Chen Z.Y., Lu J., Fire A.Z., Kay M.A. Minicircle DNA vectors achieve sustained expression reflected by active chromatin and transcriptional level. Mol. Ther. 2013;21:131–138. doi: 10.1038/mt.2012.244. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Piechaczek C., Fetzer C., Baiker A., Bode J., Lipps H.J. A vector based on the SV40 origin of replication and chromosomal S/MARs replicates episomally in CHO cells. Nucleic Acids Res. 1999;27:426–428. doi: 10.1093/nar/27.2.426. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Cook G.A., Wilkinson D.A., Crossno J.T., Jr., Raghow R., Jennings L.K. The tetraspanin CD9 influences the adhesion, spreading, and pericellular fibronectin matrix assembly of Chinese hamster ovary cells on human plasma fibronectin. Exp. Cell Res. 1999;251:356–371. doi: 10.1006/excr.1999.4596. [DOI] [PubMed] [Google Scholar]
- 39.Baiker A., Maercker C., Piechaczek C., Schmidt S.B., Bode J., Benham C., Lipps H.J. Mitotic stability of an episomal vector containing a human scaffold/matrix-attached region is provided by association with nuclear matrix. Nat. Cell Biol. 2000;2:182–184. doi: 10.1038/35004061. [DOI] [PubMed] [Google Scholar]
- 40.Cockerill P.N., Garrard W.T. Chromosomal loop anchorage of the kappa immunoglobulin gene occurs next to the enhancer in a region containing topoisomerase II sites. Cell. 1986;44:273–282. doi: 10.1016/0092-8674(86)90761-0. [DOI] [PubMed] [Google Scholar]
- 41.Gasser S.M., Laemmli U.K. Cohabitation of scaffold binding regions with upstream/enhancer elements of three developmentally regulated genes of D. melanogaster. Cell. 1986;46:521–530. doi: 10.1016/0092-8674(86)90877-9. [DOI] [PubMed] [Google Scholar]
- 42.Tracy R.B., Chédin F., Kowalczykowski S.C. The recombination hot spot chi is embedded within islands of preferred DNA pairing sequences in the E. coli genome. Cell. 1997;90:205–206. doi: 10.1016/s0092-8674(00)80328-1. [DOI] [PubMed] [Google Scholar]
- 43.Dietz W.M., Skinner N.E., Hamilton S.E., Jund M.D., Heitfeld S.M., Litterman A.J., Hwu P., Chen Z.Y., Salazar A.M., Ohlfest J.R. Minicircle DNA is superior to plasmid DNA in eliciting antigen-specific CD8+ T-cell responses. Mol. Ther. 2013;21:1526–1535. doi: 10.1038/mt.2013.85. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Chang T.Y., Chung C.Y., Chuang W.M., Li L.Y., Jeng L.B., Ma W.L. Durable expression of minicircle DNA-liposome-delivered androgen receptor cDNA in mice with hepatocellular carcinoma. BioMed Res. Int. 2014;2014:156356. doi: 10.1155/2014/156356. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Huang M., Chen Z., Hu S., Jia F., Li Z., Hoyt G., Robbins R.C., Kay M.A., Wu J.C. Novel minicircle vector for gene therapy in murine myocardial infarction. Circulation. 2009;120:S230–S237. doi: 10.1161/CIRCULATIONAHA.108.841155. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Kay M.A., Graham F., Leland F., Woo S.L. Therapeutic serum concentrations of human alpha-1-antitrypsin after adenoviral-mediated gene transfer into mouse hepatocytes. Hepatology. 1995;21:815–819. [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.