Abstract
Lysine acetylation is a protein posttranslational modification (PTM) that occurs on thousands of lysine residues in diverse organisms from bacteria to humans. Accurate measurement of acetylation stoichiometry on a proteome-wide scale remains challenging. Most methods employ a comparison of chemically acetylated peptides to native acetylated peptides, however, the potentially large differences in abundance between these peptides presents a challenge for accurate quantification. Stable isotope labeling by amino acids in cell culture (SILAC)-based mass spectrometry (MS) is one of the most widely used quantitative proteomic methods. Here we show that serial dilution of SILAC-labeled peptides (SD-SILAC) can be used to identify accurately quantified peptides and to estimate the quantification error rate. We applied SD-SILAC to determine absolute acetylation stoichiometry in exponentially-growing and stationary-phase wild-type and Sirtuin deacetylase CobB-deficient cells. To further analyze CobB-regulated sites under conditions of globally increased or decreased acetylation, we measured stoichiometry in phophotransacetylase (ptaΔ) and acetate kinase (ackAΔ) mutant strains in the presence and absence of the Sirtuin inhibitor nicotinamide. We measured acetylation stoichiometry at 3,669 unique sites and found that the vast majority of acetylation occurred at a low stoichiometry. Manipulations that cause increased nonenzymatic acetylation by acetyl-phosphate (AcP), such as stationary-phase arrest and deletion of ackA, resulted in globally increased acetylation stoichiometry. Comparison to relative quantification under the same conditions validated our stoichiometry estimates at hundreds of sites, demonstrating the accuracy of our method. Similar to Sirtuin deacetylase 3 (SIRT3) in mitochondria, CobB suppressed acetylation to lower than median stoichiometry in WT, ptaΔ, and ackAΔ cells. Together, our results provide a detailed view of acetylation stoichiometry in E. coli and suggest an evolutionarily conserved function of Sirtuin deacetylases in suppressing low stoichiometry acetylation.
Quantitative MS is a powerful technique used to measure the abundance of thousands of peptides in a single experiment. One of the most commonly used methods to quantify relative differences in peptide abundance is stable isotope labeling by amino acids in cell culture (SILAC)1 (1). Previous studies showed that large differences in peptide abundance are less accurately quantified (2), and that signal-to-noise limits quantification accuracy (3). However, a method to identify accurately quantified peptides and to estimate quantification error has not been developed. Here we present a method to estimate quantification error in a SILAC-based quantitative MS experiment and to identify accurately quantified peptides.
Lysine acetylation is a reversible protein posttranslational modification (PTM) that occurs on thousands of lysine residues in diverse organisms from bacteria to humans (4, 5). Acetylation regulates various cellular functions ranging from transcription, DNA damage repair, splicing, autophagy, and microtubule organization (6), and is prominently implicated in the regulation of metabolism (7). Acetylation can be catalyzed by acetyltransferases and removed by deacetylases (8). However, acetylation also occurs nonenzymatically by reaction with acetyl-CoA (9–11), and with acetyl-phosphate (AcP) in bacteria (12).
We previously developed a method to estimate the stoichiometry of lysine acetylation by comparing the abundance of native acetylated peptides to partially chemically acetylated peptides (11, 13). This method has several technical challenges; it is difficult to determine the degree and variability of partial chemical acetylation, and the abundance of chemically acetylated peptides is often substantially different from native acetylated peptides, complicating accurate quantification of these differences. Other groups have compared native acetylated peptides to complete (100%) chemically acetylated peptides to determine acetylation stoichiometry (14–16). However, the difference in abundance between 100% acetylated peptides and native acetylated peptides may be too large to quantify accurately. In one study, the authors found that >75% of their stoichiometry estimates were affected by false quantification (16), highlighting the challenges in obtaining accurate quantification under these conditions. A recent study employed MS2-based quantification of 100% acetylated protein to measure stoichiometry (15), and argued that MS1-based quantification (as used by (14, 16)) was less accurate under these conditions.
Here we improved our previous method to estimate acetylation stoichiometry in two ways. We applied serial dilution-SILAC (SD-SILAC) to ensure that the differences between chemically acetylated and native acetylated peptides were accurately quantified, and we used a combination of complete and partial chemical acetylation to directly determine the degree and variability of partial chemical acetylation.
EXPERIMENTAL PROCEDURES
Cell growth, protein extraction, and peptide preparation
E. coli MG1655 (lysAΔ, argAΔ) and CobB-deficient MG1655 (cobBΔ, lysAΔ, argAΔ) were constructed previously (12). ackAΔ (JW2293–1), ptaΔ (JW2294–1) were obtained from the E. coli Genetic Resource Center (http://cgsc.biology.yale.edu). All strains were cultured in M9 minimal media supplemented with heavy isotopes of arginine and lysine (13C615N4-arginine and 13C615N2-lysine, Cambridge Isotope Laboratories, Tewksbury, MA). Unless otherwise stated, cells were harvested during exponential growth (OD 600 nm ∼0.5), washed once with PBS, and frozen in liquid nitrogen. Stationary phase cells were cultured for ∼48h following growth arrest as monitored by OD 600 nm. Nicotinamide (Sigma) treatment was performed as follows; overnight cultures were diluted to an OD 600 nm of 0.1 and allowed to double to an OD 600 nm of 0.2, 25 mm nicotinamide (or H2O for control treatment) was added and the cells were allowed to double twice more to a final OD 600 nm of 0.8 before collecting. Frozen cell pellets were resuspended in 8 m guanidine HCl (GuHCl), 50 mm Hepes pH 8.5, followed by sonication for 30 s. Chemical acetylation was performed by adding acetyl-phosphate (Sigma) to 150 mm (partial acetylation), or NHS-acetate (Thermo-Fisher Scientific) to 40 mm (complete acetylation). Reactions were incubated at 37 °C for ∼1 h and were quenched by the addition of Tris buffer (pH 7.5) to 250 mm. Samples were mixed based on empirically determined SILAC ratios for lysine-independent tryptic peptides (which are unaffected by chemical acetylation). For experiments using partial chemical acetylation (supplementary Data Set S1, experiments 4–11) the samples were diluted to 2 m GuHCl with 50 mm Hepes, pH 8.5 and pre-digested with 1/200 (weight/weight) Lys-C protease for 4 h. Otherwise, samples were diluted to 1 m GuHCl with 50 mm Hepes, pH 8.5 and the protein digested to peptides by addition of 1/100 (weight/weight) trypsin protease for 16 h. Digested protein samples were cleared of precipitates by centrifugation (2500 × g, 5 min) and loaded onto reversed-phase C18 Sep-Pak columns (Waters, Milford, MA), pre-equilibrated with 5 ml acetonitrile and 2 × 5 ml 0.1% TFA. Peptides were washed with 1 × 5 ml 0.1% TFA and 1 × 5 ml H2O, and then eluted with 2.5 ml 50% acetonitrile (ACN). For acetyllysine peptide enrichment the eluate was mixed with 100 μl 10x IAP buffer (500 mm MOPS; pH 7.2, 100 mm Na-phosphate, 500 mm NaCl, 2.5% Nonidet P-40) for final concentration of 2× IAP in 500 μl. Subsequently, ACN was removed by vacuum centrifugation and the volume of samples was adjusted to 1 ml by H2O. Acetylated peptide enrichment was performed using the PTMscan acetyllysine enrichment kit (Cell Signaling Technologies, Danvers, MA). Acetylated peptides were fractionated by micro-SCX columns in a Stage-Tip format (17, 18). Peptide fractions were purified and concentrated with reversed-phase Stage-Tips as described (17).
Mass Spectrometry
Peptide fractions were analyzed by online nanoflow LC-MS/MS using a Proxeon easy nLC system connected to a Q-Exactive plus HF mass spectrometer (Thermo Scientific), as described (19). Raw data was computationally processed using MaxQuant (version 1.5.5.4) and searched against all 4305 entries in the UniProt E. coli K12 database (downloaded June 30, 2014) using the integrated Andromeda search engine (http://www.maxquant.org/) (20). The Andromeda default settings were used; an initial search was performed using a mass tolerance of 20 ppm, followed by mass recalibration and a main search with a mass tolerance of 6 ppm for parent ions and 20 ppm (HCD) for fragment ions. Peptide sequences were searched using trypsin specificity and allowing a maximum of two missed cleavages. Cysteine carbamidomethylation was added as a fixed modification, and N-acetylation of proteins, oxidized methionine, and acetylated lysine were added as variable modifications. Acetylated peptides were filtered for a minimum Andromeda score of 40, as per the default settings for modified peptides. Known contaminants were removed based on classification by MaxQuant. The false discovery rate (FDR) was estimated using a target-decoy approach (21) allowing a maximum of 1% false identifications from a reversed sequence database. The re-quantify feature was used where indicated. The raw data, MaxQuant output files, and annotated MS2 spectra for all acetylated peptides found in supplementary Data Set S1 have been deposited to the ProteomeXchange Consortium (http://proteomecentral.proteomexchange.org) via the PRIDE partner repository (22) with the data set identifier PXD005757.
Data Analysis
Acetylation stoichiometry was analyzed only for singly acetylated peptides using the modification specific peptide tables from the MaxQuant output. Peptides with more than one acetylation site were omitted because the degree of initial stoichiometry at each position could not be accounted for. When analyzing experiments using 100% acetylation, we omitted peptides generated by cleaving C-terminal to lysine residues because acetylation blocks these cleavage events. To calculate stoichiometry (S) using SILAC light-labeled, 100% acetylated peptides the following formula was used; S = (100%*dilution factor)*(SILAC Ratio H/L). For example, if the 100% acetylated protein was diluted 10x, S = (100%*0.1)*(SILAC Ratio H/L). To calculate the degree of partial chemical acetylation the following formula was used; Degree (%) of chemical acetylation = (SPartial Acetylation - SNative)/(1 - SNative). To calculate stoichiometry using SILAC heavy-labeled 7.9% acetylated peptides, the following formula was used; Stoichiometry = (0.079)/((SILAC Ratio H/L)-(1–0.079)). In the preceding formula, the SILAC Ratios H/L were corrected according to the serial dilution series. A detailed description of how to process the MaxQuant output tables for stoichiometry calculation can be found in the supplementary Materials. For comparison to previously published SILAC quantification, we reanalyzed our previously published data (12) using the same E. coli sequence database and version of MaxQuant that was used in this study. Statistical analysis was performed using R (version 3.1.1).
Experimental Design and Statistical Rationale
To determine quantification error rates, we used peptides that were isolated from a single biological sample. Because these experiments were designed to examine quantification error and not biological differences, the biological source and associated variability has no bearing on the analysis. Each sample was measured three times at different dilutions to uncover peptide SILAC ratios that were inconsistent with the dilution series; hence each measurement can be considered a technical replicate. For the analysis of acetylation stoichiometry in E. coli, the proteins were isolated from a single culture (a single biological replicate for each cell type). For each quantitative MS experiment the proteins were mixed at three different ratios (three technical replicates). The S.D.-SILAC method described in this study was used to identify accurately quantified peptides and only those peptides were used to calculate stoichiometry. Statistical tests used to analyze data are indicated in the respective figure legends and/or manuscript sections.
RESULTS
SD-SILAC To Determine Accurate Quantification
Serial dilution is widely used in biochemical studies to ensure accurate measurements. Here we applied this concept to SILAC-based quantitative MS to identify accurately quantified peptides and to estimate quantification error. We serially diluted SILAC-labeled proteins so that the difference between the measured SILAC ratios should equal the dilution ratios (DRs) (Fig. 1A). We defined inaccurate quantification as peptides whose DRs deviated more than 2-fold from the expected value, a difference that was greater than three standard deviations from the median of the DR distribution in a test experiment (supplemental Figs. S1A and S1B). We applied this method to assay the degree of quantification error when comparing unperturbed (control versus control) samples from E. coli, HeLa cells, and liver tissue (supplementary Text S1, supplemental Figs. S1 and S2, and supplemental Table S1). Together, these data show that we can estimate the amount of quantification error, the observed error rates show sample-specific differences, different computational approaches affect the error rate, and we can filter our results to include only accurately quantified peptides.
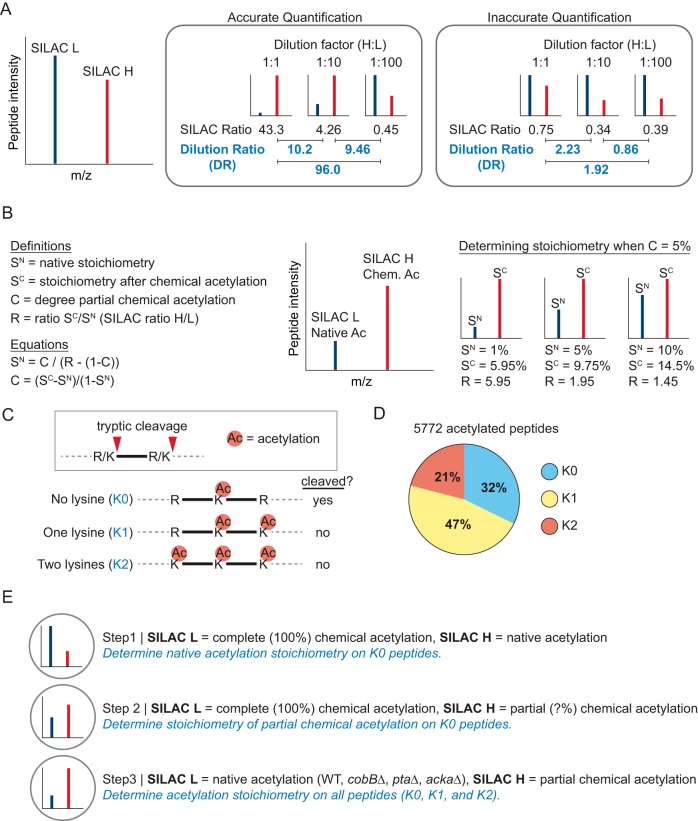
Fig. 1.
Serial-dilution SILAC (SD-SILAC) and its application to measure acetylation stoichiometry. A, Model showing the relationship between SILAC ratios (H/L) and dilution ratios (DRs) in a dilution series of SILAC peptides. The examples of accurate and inaccurate quantification are based on data used in this study. B, Diagram illustrating the use of 5% partial chemical acetylation to determine acetylation stoichiometry. Native acetylation stoichiometry (SN) can be calculated based on the SILAC ratio (R) when the degree of partial chemical acetylation (C) is known. C, Diagram showing the effect of lysine acetylation on tryptic cleavage. D, The pie chart shows the fraction of acetylated peptides generated by tryptic cleavage independent of lysine (K0) at a single lysine (K1) or at two lysines (K2). E, Outline of the quantitative mass spectrometry experiments used to determine acetylation stoichiometry.
Measuring Acetylation Stoichiometry
We previously measured acetylation stoichiometry by comparing the abundance of partially chemically acetylated peptides to native acetylated peptides (11, 13) (Fig. 1B). Other groups have measured stoichiometry by comparing 100% chemically acetylated peptides to native acetylated peptides (14–16). However, because acetylation blocks trypsin cleavage at lysine residues (Fig. 1C), 100% chemical acetylation limits the analysis to peptides generated by cleaving at arginine residues. Most (68%) of the native acetylation sites we detected in E. coli occurred on tryptic peptides generated by cleaving at one or more lysine residues (Fig. 1D), indicating that these sites may not be detected when using 100% chemical acetylation. In this study, we used a combination of 100% and partial chemical acetylation to determine acetylation stoichiometry (Fig. 1E). By combining this approach with serial dilution of SILAC labeled proteins (SD-SILAC) we provide further confidence in the accuracy of our stoichiometry measurements.
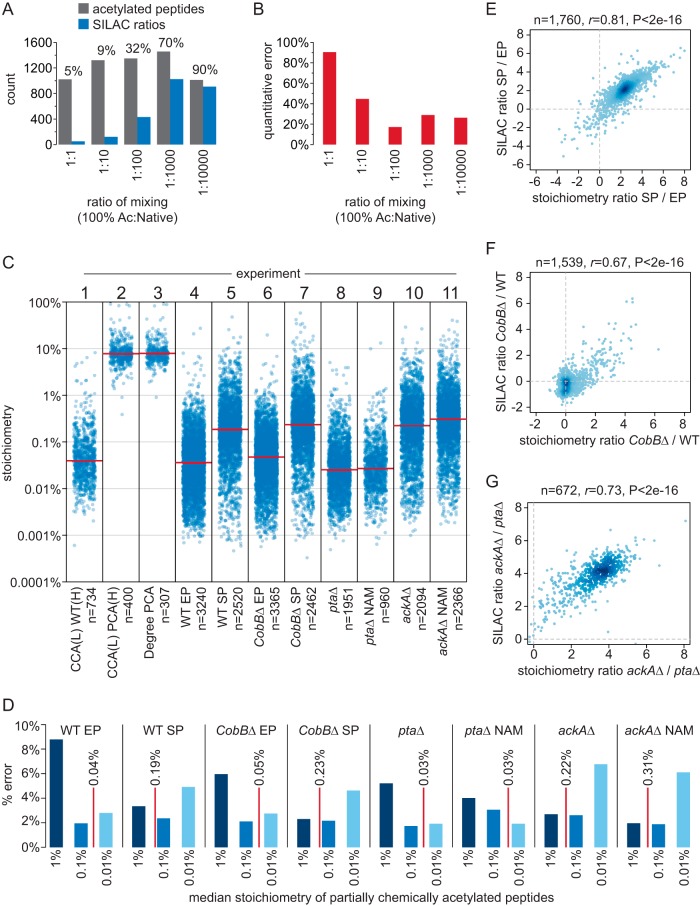
In the first step, we used 100% chemical acetylation to determine native acetylation stoichiometry. SILAC-light-labeled 100% chemically acetylated protein was mixed with SILAC-heavy-labeled native acetylated protein in a 10-fold serial dilution at a ratio of 1:1, 1:10, 1:100, 1:1000, and 1:10000; which corresponds to the presence of chemically acetylated peptides at a stoichiometry of 100%, 10%, 1%, 0.1%, and 0.01%. Acetylated peptides were enriched from these samples and quantified by MS (supplementary Data Set S1, experiment 1). At a 1:1 mixing ratio only 5% of the identified peptides were quantified (had SILAC ratios), and the quantification error rate was 90% (Fig. 2A). As the concentration of chemically acetylated peptides decreased, the fraction of peptides with SILAC ratios increased, and the quantification error decreased (Figs. 2A and 2B). Using only accurately quantified peptides, we calculated a median stoichiometry of 0.04% (Fig. 2C, experiment 1). These data show that when the concentration of chemically acetylated peptides is similar to native acetylated peptides, we identify a greater fraction of peptides with SILAC ratios, and the quantification error is reduced. In addition, we found that 100% chemical acetylation interfered with tryptic digestion, which is likely to result in inaccurate calculation of stoichiometry at some sites (supplemental Fig. S3A).
Fig. 2.
Measuring acetylation stoichiometry. A, The column graph shows the number of acetylated peptides, and acetylated peptides with SILAC ratios, that were identified at the indicated mixing ratios of 100% acetylated (Ac) peptides and native peptides. The fraction (%) of peptides with SILAC ratios is indicated. B, The column graph shows the quantification error for acetylated peptides that were identified at the indicated mixing ratios of 100% acetylated (Ac) peptides and native peptides. C, The category scatterplot shows the distributions of acetylated peptide stoichiometry or the degree of partial chemical acetylation (Degree PCA, experiment 3) for the indicated experiments. CCA = complete chemical acetylation, PCA = partial chemical acetylation, (L) = SILAC light labeled, (H) = SILAC heavy labeled, WT = wild type, EP = exponential phase, SP = stationary phase, NAM = nicotinamide, knock out (Δ) strains are as indicated. The median of the distributions are indicated by the red lines. D, The column graph shows the percent quantification error (% error) for the indicated experiments. The median stoichiometry of native acetylation for each experiment is indicated by the red lines. E, The scatterplot shows the correlation between differences in acetylation stoichiometry between SP and EP cells (stoichiometry ratio SP/EP) and independent SILAC quantification of those same differences (SILAC ratio SP/EP). The number of acetylated peptides analyzed (n), Pearson's correlation (r), and p value (P) of correlation is shown. F, The scatterplot shows the correlation between differences in acetylation stoichiometry between cobBΔ and WT cells (stoichiometry ratio cobBΔ/WT) and previous SILAC quantification (12) of the same acetylated peptides (SILAC ratio cobBΔ/WT). G, The scatterplot shows the correlation between differences in acetylation stoichiometry between ackAΔ and ptaΔ cells (stoichiometry ratio ackAΔ/ptaΔ) and previous SILAC quantification (12) of the same acetylated peptides (SILAC ratio ackAΔ/ptaΔ).
In the second step, we compared 100% acetylated protein to partially chemically acetylated protein to directly measure the degree and variability of partial chemical acetylation (supplementary Data Set S1, experiment 2). Using SD-SILAC to ensure quantitative accuracy, we determined a median stoichiometry of partial chemical acetylation that was 7.9% (Fig. 2C, experiment 2). For a subset of these peptides, we also determined native stoichiometry (in experiment 1), and comparison of these data allowed us to calculate the degree of partial chemical acetylation (Fig. 2C, experiment 3). The median degree of partial chemical acetylation was 7.9%, which is identical to the median stoichiometry after partial chemical acetylation because the initial stoichiometry (median 0.04%) is negligible. We further find that partial chemical acetylation has a relatively low variability, as most (84%) of the acetylated peptides differed by less than a factor of two from the median (were 3.9% to 15.7% acetylated). Some of this variability can be attributed to the reduced tryptic digestion of 100% acetylated proteins (supplemental Fig. S3A), and the actual degree of partial chemical acetylation is likely to be less variable than these results suggest. Supporting this idea, we calculated stoichiometry that exceeded 100% on nine acetylated peptides (2% of the peptides analyzed), a result that is consistent with overestimation of stoichiometry because of incomplete tryptic digestion. To independently verify these findings, we examined the median decrease in lysine-dependent tryptic peptides (supplemental Fig. S3B). This indirect method provided an estimate of partial chemical acetylation that was similar (7.1–7.6%) to that determined above.
In the third step, we used partial chemical acetylation and SD-SILAC to determine stoichiometry under conditions that cause robust changes in acetylation at hundreds of sites. We compared wild-type (WT) and Sirtuin deacetylase CobB-deficient (cobBΔ) cells during exponential phase (EP) growth and after 48 h stationary phase (SP) arrest, which we previously showed causes globally increased acetylation (12). We examined phosphotransacetylase-deficient (ptaΔ) cells which block AcP formation and suppress acetylation, and acetate kinase-deficient (ackAΔ) cells which accumulate AcP and have globally elevated acetylation levels (12). ptaΔ and ackAΔ cells were additionally treated with the Sirtuin deacetylase inhibitor nicotinamide (NAM) to inhibit CobB activity. Partially chemically acetylated proteins were mixed with control-treated proteins in a 10-fold dilution series and only accurately quantified peptides were used to calculate acetylation stoichiometry (supplementary Data Set S1). We determined stoichiometry on thousands of acetylated peptides in each experiment (Fig. 2C, experiments 4–11). Quantification error was highest for the samples whose concentration of partially chemically acetylated peptides differed the most from the median stoichiometry of native acetylation (Fig. 2D), highlighting the importance of controlling for accurate quantification. To confirm our findings, we compared absolute changes based on our acetylation stoichiometry measurements to relative changes that were independently measured using SILAC quantification (Fig. 2E–2G). We found a high correlation between stoichiometry measurements and SILAC quantification of these same differences, validating our results at hundreds of sites. In addition, differences in acetylation stoichiometry in NAM-treated ptaΔ and ackAΔ cells were correlated with loss of CobB (supplemental Fig. S3C), indicating that NAM targeted CobB-regulated sites and further supporting our stoichiometry measurements. To provide additional validation, we compared acetylation site intensity (the summed intensity of all peptides harboring the acetylation site) to acetylation stoichiometry on six individual proteins (supplemental Fig. S3D). The analysis was performed on individual proteins to account for differences in protein abundance that affect peptide intensity. Site intensity and site stoichiometry were significantly correlated (median Pearson's correlation of 0.69), even though peptide intensity is known to be inherently variable. Together, these data independently support the accuracy of our stoichiometry measurements.
Acetylation Site Stoichiometry in E. coli
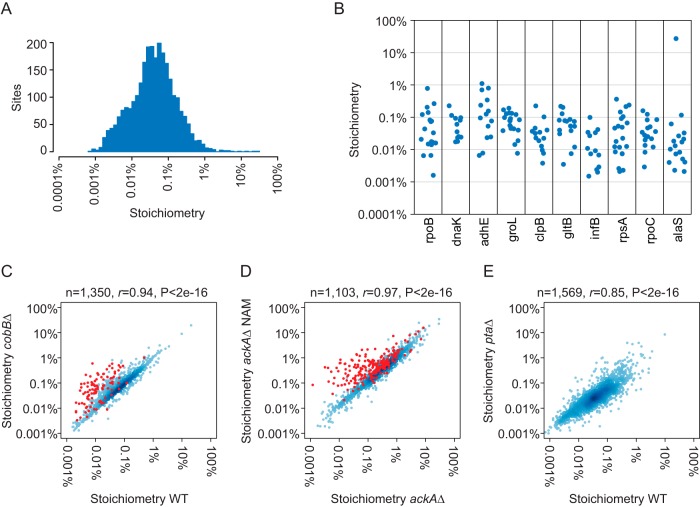
We determined acetylation stoichiometry at the level of acetylated peptides; however, some sites occur on several different peptide species because of missed tryptic cleavages and oxidized methionine. We therefore generated a list of acetylation site stoichiometry based on the summed stoichiometry of acetylated peptides that harbor the same acetylation site (supplementary Data Set S2). We determined stoichiometry at 3,669 unique sites in all conditions combined; 2,700 of which were measured in exponentially growing WT cells (Fig. 3A, Table I). Median stoichiometry was only 0.04% in WT cells and median stoichiometry in cobBΔ and ptaΔ cells was similar to WT (0.06% and 0.03%, respectively). Stoichiometry varied by two to three orders of magnitude within individual proteins (Fig. 3B), indicating that stoichiometry is variable at the site-level. Our measurements were highly correlated between different experiments and E. coli strains, supporting the reproducibility of our method. Excluding CobB-regulated sites (shown in red), stoichiometry in WT and cobBΔ cells had a Pearsons's correlation (r) of 0.94 (Fig. 3C). Similarly excluding CobB-regulated sites, stoichiometry in ackAΔ and NAM-treated ackAΔ cells was highly correlated (r = 0.97) (Fig. 3D). Stoichiometry in WT and ptaΔ was also well-correlated (r = 0.85) (Fig. 3E), even though these cells are derived from different strain backgrounds.
Fig. 3.
Acetylation site stoichiometry in E. coli. A, The histogram shows the distribution of acetylation site stoichiometry in WT EP cells (n = 2700). B, The category scatterplot shows the distributions of acetylation site stoichiometry for the indicated proteins in WT EP cells. C, The scatterplot shows the correlation between acetylation stoichiometry in WT and cobBΔ cells. CobB-regulated sites (as defined by SILAC quantification (12)) are shown in red and were excluded from the Pearson's correlation. The number of sites analyzed (n), Pearson's correlation (r), and p value (P) of correlation is shown. D, The scatterplot shows the correlation between acetylation stoichiometry in ackAΔ and nicotinamide (NAM)-treated ackAΔ cells. CobB-regulated sites (as defined by a stoichiometry ratio cobBΔ/WT >2) are shown in red and were excluded from the Pearson's correlation. E, The scatterplot shows the correlation between acetylation stoichiometry in WT and ptaΔ cells.
Table I. Acetylation site stoichiometry in E. coli. The table summarizes stoichiometry measurements in the indicated experiments. WT = wild type, EP = exponential phase, SP = stationary phase, NAM = nicotinamide, knock out (Δ) strains are as indicated. Median stoichiometry, and the number and fraction of sites with greater than 1% and 10% stoichiometry are shown.
| Experiment | Number of sites | Median stoichiometry | Number > 1% | Fraction >1% | Number > 10% | Fraction >10% |
|---|---|---|---|---|---|---|
| WT EP | 2700 | 0.04% | 29 | 1.1% | 3 | 0.11% |
| WT SP | 2155 | 0.22% | 235 | 10.9% | 10 | 0.46% |
| cobBΔ EP | 2789 | 0.05% | 31 | 1.1% | 1 | 0.04% |
| cobBΔ SP | 2121 | 0.26% | 297 | 14.0% | 25 | 1.18% |
| ptaΔ | 1772 | 0.03% | 14 | 0.8% | 0 | 0.00% |
| ptaΔ NAM | 910 | 0.03% | 10 | 1.1% | 1 | 0.11% |
| ackAΔ | 1802 | 0.25% | 263 | 14.6% | 5 | 0.28% |
| ackAΔ NAM | 2045 | 0.35% | 388 | 19.0% | 21 | 1.03% |
To better understand differences in the acetylation stoichiometry at individual sites, we performed bioinformatic analyses to study the impact of amino acid sequence and protein structure on stoichiometry. High stoichiometry acetylation was not biased to occur within a particular linear amino acid sequence (supplemental Fig. S4A). A small number of sites occurring within predicted disordered regions had significantly lower stoichiometry, as did predicted buried sites (supplemental Fig. S4B). However, sites occurring within various predicted secondary structures did not show significantly different stoichiometry (supplemental Fig. S4B). We used protein crystal structures to directly examine the impact of structure on stoichiometry at a smaller number of sites. We found no significant correlation to lysine pKa (supplemental Fig. S4C) and a weak, but significant, correlation to the solvent-accessible surface area of acetylated lysine residues (supplemental Fig. S4D). These data suggest that lysine accessibility may influence the degree of acetylation at any given site; however, we were unable to identify a strong correlation between stoichiometry and amino acid sequence or protein structure.
High Stoichiometry Acetylation
Very few sites had high stoichiometry in WT EP, cobBΔ, and ptaΔ cells, whereas conditions that promote increased nonenzymatic acetylation, such as SP and ackAΔ, resulted in substantially higher median stoichiometry and a greater fraction of high stoichiometry sites (Table I). These results suggest that high stoichiometry acetylation occurs infrequently, at least under the laboratory conditions used in this study. Although we observe high stoichiometry acetylation in SP cells, we previously showed that increased acetylation depends on growth arrest because of nitrogen depletion in the presence of glucose, conditions that may not occur outside of the laboratory. Whether acetylation accumulates to high levels in nature remains an important open question.
Accurate measurement of high-stoichiometry acetylation presents a technical challenge. For high stoichiometry acetylation, the difference between partially chemically acetylated and native acetylated peptides will be small and difficult to quantify accurately. In our experiments, sites with >7.3% stoichiometry will be less than 2-fold increased by partial chemical acetylation. SILAC may not accurately resolve these small differences, as shown by the distributions of peptide SILAC ratios comparing unperturbed (control versus control) samples (supplemental Fig. S5A). Furthermore, small errors in quantification within this range will yield large differences in the measured stoichiometry, particularly as the SILAC ratio approaches a value of 1 (supplemental Fig. S5B). However, we find little evidence for such peptides in our data set. We quantified 5487 acetylated peptides in WT EP cells, of which 11 (0.2%) had SILAC ratios < 2 (after adjusting for serial dilution). Of those 11 peptides, six were accurately quantified and stoichiometry was calculated. Of the remaining five peptides, four were only quantified at a single dilution so that quantification accuracy could not be assessed, and one was determined to be inaccurately quantified. A small number (3 in WT EP cells, 0.1% of the total) of chemically acetylated peptides were quantified as less abundant than native acetylation (SILAC ratio < 1), which resulted in negative stoichiometry. Sites with negative stoichiometry are reported as “NaN” (supplementary Data Sets S1 and S2), and may be considered as putative high stoichiometry sites.
Analysis of stoichiometry at CobB-regulated acetylation sites
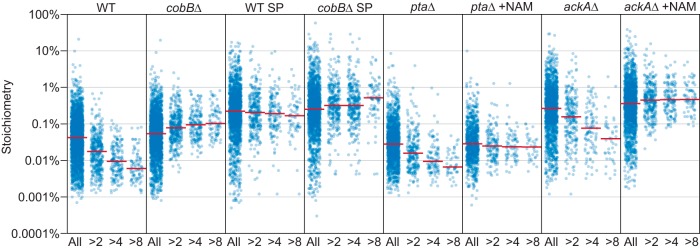
CobB is the only known Sirtuin deacetylase in E. coli. The deacetylase activity of CobB was first shown to regulate the activity of acetyl-CoA synthetase (23), and subsequent work linked CobB with regulating central metabolism in Salmonella enterica (24). We previously mapped CobB-regulated acetylation sites in E. coli and showed that loss of CobB increased acetylation at ∼10% of sites (12). Our analysis of acetylation stoichiometry shows that sites affected by loss of CobB have a significantly lower stoichiometry than sites not affected by loss of CobB, in a manner that is proportional to the degree of increased acetylation in the absence of CobB (Fig. 4A and Table II). In WT SP cells CobB did not suppress acetylation at regulated sites to the same degree as in WT, ptaΔ, and ackAΔ cells, suggesting reduced CobB activity in these cells. This does not appear to be because of the higher stoichiometry of acetylation in SP cells, as ackAΔ cells had a similar median stoichiometry and a greater number of high stoichiometry sites (Table I). In cobBΔ cells, acetylation stoichiometry was slightly elevated at CobB-regulated sites compared with unregulated sites, indicating that the sites that show the greatest degree of increased acetylation in the absence of CobB do so because CobB suppresses acetylation to a lower stoichiometry at these sites (Fig. 4A and Table II, compare WT to cobBΔ, ptaΔ to ptaΔ NAM, and ackAΔ to ackAΔ NAM). The median stoichiometry of the sites most affected by loss of CobB (>8-fold increased) was just 0.006% in WT cells, seven times lower than the median stoichiometry of unregulated sites. This result could be explained by a greater degree of chemical acetylation at CobB-regulated sites, which would cause us to systematically underestimate native stoichiometry at these sites. To test this idea, we compared the reduced cleavage of lysine residues following partial chemical acetylation at unregulated and CobB-regulated sites (supplemental Fig. S5C). These data show that CobB-regulated sites were not chemically acetylated in vitro to a greater degree than unregulated sites. In addition, CobB-regulated sites did not display reduced stoichiometry in WT SP cells, suggesting that the reduced stoichiometry in WT, ptaΔ, and ackAΔ cells is because of the deacetylase activity of CobB in vivo rather than the reactivity of these sites to chemical acetylation in vitro (the same in vitro chemically acetylated sample was used to determine stoichiometry in all experiments). Together these data support our finding that CobB suppresses acetylation to lower than median stoichiometry, and indicates that the amount of increased acetylation in the absence of CobB is related to the degree that CobB suppresses acetylation at that site.
Fig. 4.
Acetylation stoichiometry at CobB-regulated sites. The category scatterplot shows the distributions of acetylation site stoichiometry for the indicated experiments. WT = wild type, EP = exponential phase, SP = stationary phase, NAM = nicotinamide, and knock out (Δ) strains are as indicated. Sites that are more than 2-fold (>2), 4-fold (>4), or 8-fold (>8) increased in the absence of CobB (during EP growth) are indicated. The median of the distributions are indicated by the red lines.
Table II. Acetylation site stoichiometry at CobB-regulated sites. The table shows the number of acetylation sites and their median stoichiometry in the indicated experiments, classified according to increased acetylation in cobBΔ EP cells compared to WT EP cells. Sites with greater than 2-fold (CobB >2), 4-fold (CobB >4), and 8-fold (CobB >8) increased acetylation are indicated. WT = wild type, EP = exponential phase, SP = stationary phase, NAM = nicotinamide, and knock out (Δ) strains are as indicated.
| All sites | CobB >2 | CobB >4 | CobB >8 | |
|---|---|---|---|---|
| WT EP | ||||
| Number | 2243 | 357 | 167 | 87 |
| Median stoichiometry | 0.042% | 0.018% | 0.009% | 0.006% |
| cobBΔ EP | ||||
| Number | 1663 | 285 | 138 | 68 |
| Median stoichiometry | 0.223% | 0.206% | 0.192% | 0.168% |
| WT SP | ||||
| Number | 2243 | 357 | 167 | 87 |
| Median stoichiometry | 0.054% | 0.078% | 0.094% | 0.103% |
| cobBΔ SP | ||||
| Number | 1657 | 293 | 293 | 80 |
| Median stoichiometry | 0.252% | 0.319% | 0.319% | 0.517% |
| ptaΔ | ||||
| Number | 1462 | 228 | 101 | 49 |
| Median stoichiometry | 0.028% | 0.016% | 0.009% | 0.007% |
| ptaΔ NAM | ||||
| Number | 832 | 141 | 74 | 36 |
| Median stoichiometry | 0.029% | 0.025% | 0.024% | 0.023% |
| ackAΔ | ||||
| Number | 1474 | 225 | 99 | 44 |
| Median stoichiometry | 0.266% | 0.157% | 0.076% | 0.039% |
| ackAΔ NAM | ||||
| Number | 1594 | 282 | 139 | 73 |
| Median stoichiometry | 0.360% | 0.447% | 0.459% | 0.465% |
DISCUSSION
Here we describe a method to identify accurately quantified peptides and to estimate quantification error in a SILAC MS experiment. Our method relies on identifying disagreement between SILAC ratios measured at two or more different dilutions; if these ratios disagree it indicates that at least one of the measurements is inaccurate. However, one of the measurements may be accurate. Therefore, the strength in our method is its ability to identify accurately quantified peptides, but not to provide a precise measurement of error. SD-SILAC may not be generally applicable in a typical SILAC experiment, as the amount of additional MS measurement time, and the loss of data points not measured in replicate experiments, may not be justified—particularly if quantification accuracy is not critical to interpreting the data. However, SD-SILAC can enable more accurate quantification if peptide ratios are highly divergent, as shown here for native acetylated and chemically acetylated peptides. SD-SILAC could also enable more accurate quantification in other settings that require quantification over a large dynamic range, such as pulse-chase SILAC methods that are used to determine rates of protein synthesis or degradation.
The method used here to determine stoichiometry is an improvement of our previous methods (11, 13). We directly measured the degree and variability of partial chemical acetylation and we used SD-SILAC to control for quantification error. We found that partial chemical acetylation did not vary substantially, suggesting a small effect on the accuracy of our calculations. Our analysis of quantification error underscores the importance of controlling for quantification accuracy under conditions where the abundances of quantified peptides are substantially different. Previous studies that employed 100% chemical acetylation to estimate stoichiometry used heavy-isotope-labeled acetylating agents to specifically label the chemically acetylated peptides (14–16). However, the difference in abundance between the 100% acetylated and native acetylated peptides leads to errors in quantification, as shown here, and in previous studies (15, 16). Furthermore, because heavy-isotope-labeling is not complete (100%), stoichiometry estimates in these studies were limited by the unlabeled fraction, which was reported to be as high as 2% (16) and 4% (14), making it impossible to quantify sites with less than 1% stoichiometry. The serial dilution approach described here can overcome these limitations and provides a method to ensure quantification accuracy. Independent validation of stoichiometry measurements using orthogonal approaches, such as heavy-labeled peptide standards, is critical given the novelty of the methods used to measure stoichiometry. We previously validated our method using heavy-labeled peptide standards (11, 13), and in this study, we supported our findings by comparing measurements of absolute stoichiometry under different conditions to relative quantification of the same differences, and by comparing acetylation stoichiometry to acetylation site intensity within individual proteins.
We found that CobB suppresses acetylation to lower than median levels at its target sites. These results mirror our findings for the mitochondrial Sirtuin SIRT3 (13), and suggest that CobB may have a general function in suppressing acetylation. SIRT3 also suppresses acetylation to very low stoichiometry, and SIRT3 may be important for protecting mitochondria from nonenzymatic acetylation stress. Nonenzymatic acetylation can be thought of as a type of protein lesion and Sirtuin deacetylases as protein repair factors that remove acetylation lesions (27). SIRT3 has been shown to have greater impact on metabolic function under conditions that promote nonenzymatic acetylation in mitochondria, such as calorie restriction, fasting, and high fat diet (28). Similarly, loss of CobB had a greater impact on cellular growth and acetylation levels during growth on acetate media (25, 29). Given the endosymbiotic theory that mitochondria evolved from a bacterium, it is interesting to note the similarities between CobB and SIRT3, and our data suggest an evolutionarily origin of Sirtuin deacetylases as protein repair factors targeting acetylation. In addition to suppressing low stoichiometry acetylation, CobB may specifically regulate protein activity through deacetylation. CobB has been shown to regulate acetyl-CoA synthetase (23), RNase II (32), and evidence suggests it may regulate additional enzymes (25, 33), as well as restricting phage replication (34). However, it is unlikely that CobB regulates every protein that it targets for deacetylation. In this study, we found that 31% (112) of the CobB-regulated sites occurred on 69 essential proteins (as defined by (35)). Even though CobB regulates acetylation of these essential proteins, cobBΔ cells have a comparable growth rate to WT cells in glucose-containing minimal media (29). These observations suggest that deacetylation of these essential proteins by CobB is largely dispensable for their function under laboratory conditions. However, the promiscuous activity of CobB and other Sirtuin deacetylases may be important under yet unidentified conditions that result in high levels of acetylation stress.
DATA AVAILABILITY
The raw data, MaxQuant output files, and annotated MS2 spectra for all acetylated peptides found in supplementary Data set S1 have been deposited to the ProteomeXchange Consortium (http://proteomecentral.proteomexchange.org) via the PRIDE partner repository (22) with the data set identifier PXD005757.
Supplementary Material
Acknowledgments
We thank the members of the department of proteomics for discussion of these data. We thank the laboratory of Matthias Mann for providing SILAC labeled mouse liver tissue and the laboratory of Jeremy Daniel for providing unlabeled mouse liver tissue. We thank Juergen Cox for providing feedback on the manuscript.
Footnotes
Author Contributions: BTW and SS designed and performed experiments, analyzed data, and prepared the manuscript. BKH performed experiments. DL and LJJ provided bioinformatics support for secondary structure prediction and for data analysis. BTW and CC directed and supervised all aspects of the study. All authors contributed to editing the manuscript.
* BTW is supported by a grant from the Novo Nordisk Foundation (NNF15OC0017774). CC is supported by the Hallas Møller Investigator Fellowship from the Novo Nordisk Foundation (NNF14OC0008541). The Novo Nordisk Foundation Center for Protein Research is supported financially by the Novo Nordisk Foundation (Grant agreement NNF14CC0001).
 This article contains supplemental material.
This article contains supplemental material.
Competing Financial Interests: The authors declare no competing financial interests.
1 The abbreviations used are:
- SILAC
- stable isotope labeling by amino acids in cell culture
- SD-SILAC
- serial dilution-SILAC
- PTM
- post-translational modification
- DR
- dilution ratio
- SCX
- strong cation exchange.
REFERENCES
- 1. Ong S. E., Blagoev B., Kratchmarova I., Kristensen D. B., Steen H., Pandey A., and Mann M. (2002) Stable isotope labeling by amino acids in cell culture, SILAC, as a simple and accurate approach to expression proteomics. Mol. Cell Proteomics 1, 376–386 [DOI] [PubMed] [Google Scholar]
- 2. Lau H. T., Suh H. W., Golkowski M., and Ong S. E. (2014) Comparing SILAC- and stable isotope dimethyl-labeling approaches for quantitative proteomics. J. Proteome Res. 13, 4164–4174 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Bakalarski C. E., Elias J. E., Villen J., Haas W., Gerber S. A., Everley P. A., and Gygi S. P. (2008) The impact of peptide abundance and dynamic range on stable-isotope-based quantitative proteomic analyses. J. Proteome Res. 7, 4756–4765 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Choudhary C., Kumar C., Gnad F., Nielsen M. L., Rehman M., Walther T. C., Olsen J. V., and Mann M. (2009) Lysine acetylation targets protein complexes and co-regulates major cellular functions. Science 325, 834–840 [DOI] [PubMed] [Google Scholar]
- 5. Zhang J., Sprung R., Pei J., Tan X., Kim S., Zhu H., Liu C. F., Grishin N. V., and Zhao Y. (2009) Lysine acetylation is a highly abundant and evolutionarily conserved modification in Escherichia coli. Mol. Cell Proteomics 8, 215–225 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Verdin E., and Ott M. (2015) 50 years of protein acetylation: from gene regulation to epigenetics, metabolism and beyond. Nat. Rev. 16, 258–264 [DOI] [PubMed] [Google Scholar]
- 7. Choudhary C., Weinert B. T., Nishida Y., Verdin E., and Mann M. (2014) The growing landscape of lysine acetylation links metabolism and cell signalling. Nat. Rev. 15, 536–550 [DOI] [PubMed] [Google Scholar]
- 8. Shahbazian M. D., and Grunstein M. (2007) Functions of site-specific histone acetylation and deacetylation. Annu. Rev. Biochem. 76, 75–100 [DOI] [PubMed] [Google Scholar]
- 9. Paik W. K., Pearson D., Lee H. W., and Kim S. (1970) Nonenzymatic acetylation of histones with acetyl-CoA. Biochim. Biophys. Acta 213, 513–522 [DOI] [PubMed] [Google Scholar]
- 10. Wagner G. R., and Payne R. M. (2013) Widespread and Enzyme-independent N{epsilon}-Acetylation and N{epsilon}-Succinylation of Proteins in the Chemical Conditions of the Mitochondrial Matrix. J. Biol. Chem. 288, 29036–29045 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Weinert B. T., Iesmantavicius V., Moustafa T., Scholz C., Wagner S. A., Magnes C., Zechner R., and Choudhary C. (2014) Acetylation dynamics and stoichiometry in Saccharomyces cerevisiae. Mol. Syst. Biol. 10, 716. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Weinert B. T., Iesmantavicius V., Wagner S. A., Scholz C., Gummesson B., Beli P., Nystrom T., and Choudhary C. (2013) Acetyl-phosphate is a critical determinant of lysine acetylation in E. coli. Mol. Cell 51, 265–272 [DOI] [PubMed] [Google Scholar]
- 13. Weinert B. T., Moustafa T., Iesmantavicius V., Zechner R., and Choudhary C. (2015) Analysis of acetylation stoichiometry suggests that SIRT3 repairs nonenzymatic acetylation lesions. EMBO J. 34, 2620–2632 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Baeza J., Dowell J. A., Smallegan M. J., Fan J., Amador-Noguez D., Khan Z., and Denu J. M. (2014) Stoichiometry of site-specific lysine acetylation in an entire proteome. J. Biol. Chem. 289, 21326–21338 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Meyer J. G., D'Souza A. K., Sorensen D. J., Rardin M. J., Wolfe A. J., Gibson B. W., and Schilling B. (2016) Quantification of lysine acetylation and succinylation stoichiometry in proteins using mass spectrometric data-independent acquisitions (SWATH). J. Am. Soc. Mass Spectrom. 27, 1758–1771 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Zhou T., Chung Y. H., Chen J., and Chen Y. (2016) Site-Specific Identification of Lysine Acetylation Stoichiometries in Mammalian Cells. J. Proteome Res. 15, 1103–1113 [DOI] [PubMed] [Google Scholar]
- 17. Rappsilber J., Mann M., and Ishihama Y. (2007) Protocol for micro-purification, enrichment, pre-fractionation and storage of peptides for proteomics using StageTips. Nat. Protoc. 2, 1896–1906 [DOI] [PubMed] [Google Scholar]
- 18. Weinert B. T., Scholz C., Wagner S. A., Iesmantavicius V., Su D., Daniel J. A., and Choudhary C. (2013) Lysine succinylation is a frequently occurring modification in prokaryotes and eukaryotes and extensively overlaps with acetylation. Cell Reports 4, 842–851 [DOI] [PubMed] [Google Scholar]
- 19. Kelstrup C. D., Jersie-Christensen R. R., Batth T. S., Arrey T. N., Kuehn A., Kellmann M., and Olsen J. V. (2014) Rapid and deep proteomes by faster sequencing on a benchtop quadrupole ultra-high-field Orbitrap mass spectrometer. J. Proteome Res. 13, 6187–6195 [DOI] [PubMed] [Google Scholar]
- 20. Cox J., Neuhauser N., Michalski A., Scheltema R. A., Olsen J. V., and Mann M. (2011) Andromeda: a peptide search engine integrated into the MaxQuant environment. J. Proteome Res. 10, 1794–1805 [DOI] [PubMed] [Google Scholar]
- 21. Elias J. E., and Gygi S. P. (2007) Target-decoy search strategy for increased confidence in large-scale protein identifications by mass spectrometry. Nat. Methods 4, 207–214 [DOI] [PubMed] [Google Scholar]
- 22. Vizcaino J. A., Cote R. G., Csordas A., Dianes J. A., Fabregat A., Foster J. M., Griss J., Alpi E., Birim M., Contell J., O'Kelly G., Schoenegger A., Ovelleiro D., Perez-Riverol Y., Reisinger F., Rios D., Wang R., and Hermjakob H. (2013) The PRoteomics IDEntifications (PRIDE) database and associated tools: status in 2013. Nucleic Acids Res. 41, D1063–D1069 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Starai V. J., Celic I., Cole R. N., Boeke J. D., and Escalante-Semerena J. C. (2002) Sir2-dependent activation of acetyl-CoA synthetase by deacetylation of active lysine. Science 298, 2390–2392 [DOI] [PubMed] [Google Scholar]
- 24. Wang Q., Zhang Y., Yang C., Xiong H., Lin Y., Yao J., Li H., Xie L., Zhao W., Yao Y., Ning Z. B., Zeng R., Xiong Y., Guan K. L., Zhao S., and Zhao G. P. (2010) Acetylation of metabolic enzymes coordinates carbon source utilization and metabolic flux. Science 327, 1004–1007 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Castano-Cerezo S., Bernal V., Post H., Fuhrer T., Cappadona S., Sanchez-Diaz N. C., Sauer U., Heck A. J., Altelaar A. F., and Canovas M. (2014) Protein acetylation affects acetate metabolism, motility and acid stress response in Escherichia coli. Mol. Syst. Biol. 10, 762. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Scholz C., Lyon D., Refsgaard J. C., Jensen L. J., Choudhary C., and Weinert B. T. (2015) Avoiding abundance bias in the functional annotation of posttranslationally modified proteins. Nat. Methods 12, 1003–1004 [DOI] [PubMed] [Google Scholar]
- 27. Wagner G. R., and Hirschey M. D. (2014) Nonenzymatic protein acylation as a carbon stress regulated by sirtuin deacylases. Mol. Cell 54, 5–16 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Newman J. C., He W., and Verdin E. (2012) Mitochondrial protein acylation and intermediary metabolism: regulation by sirtuins and implications for metabolic disease. J. Biol. Chem. 287, 42436–42443 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Castano-Cerezo S., Bernal V., Blanco-Catala J., Iborra J. L., and Canovas M. (2011) cAMP-CRP co-ordinates the expression of the protein acetylation pathway with central metabolism in Escherichia coli. Mol. Microbiol. 82, 1110–1128 [DOI] [PubMed] [Google Scholar]
- 30. Madsen C. T., Sylvestersen K. B., Young C., Larsen S. C., Poulsen J. W., Andersen M. A., Palmqvist E. A., Hey-Mogensen M., Jensen P. B., Treebak J. T., Lisby M., and Nielsen M. L. (2015) Biotin starvation causes mitochondrial protein hyperacetylation and partial rescue by the SIRT3-like deacetylase Hst4p. Nat. Commun. 6, 7726. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Bedalov A., Chowdhury S., and Simon J. A. (2016) Biology, chemistry, and pharmacology of sirtuins. Methods Enzymol. 574, 183–211 [DOI] [PubMed] [Google Scholar]
- 32. Song L., Wang G., Malhotra A., Deutscher M. P., and Liang W. (2016) Reversible acetylation on Lys501 regulates the activity of RNase II. Nucleic Acids Res. 44, 1979–1988 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Li R., Gu J., Chen Y. Y., Xiao C. L., Wang L. W., Zhang Z. P., Bi L. J., Wei H. P., Wang X. D., Deng J. Y., and Zhang X. E. (2010) CobB regulates Escherichia coli chemotaxis by deacetylating the response regulator CheY. Mol. Microbiol. 76, 1162–1174 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Koyuncu E., Budayeva H. G., Miteva Y. V., Ricci D. P., Silhavy T. J., Shenk T., and Cristea I. M. (2014) Sirtuins are evolutionarily conserved viral restriction factors. mBio 5 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. Gerdes S. Y., Scholle M. D., Campbell J. W., Balazsi G., Ravasz E., Daugherty M. D., Somera A. L., Kyrpides N. C., Anderson I., Gelfand M. S., Bhattacharya A., Kapatral V., D'Souza M., Baev M. V., Grechkin Y., Mseeh F., Fonstein M. Y., Overbeek R., Barabasi A. L., Oltvai Z. N., and Osterman A. L. (2003) Experimental determination and system level analysis of essential genes in Escherichia coli MG1655. J. Bacteriol. 185, 5673–5684 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
The raw data, MaxQuant output files, and annotated MS2 spectra for all acetylated peptides found in supplementary Data set S1 have been deposited to the ProteomeXchange Consortium (http://proteomecentral.proteomexchange.org) via the PRIDE partner repository (22) with the data set identifier PXD005757.