Abstract
The plasma membrane proteome plays a crucial role in inter- and intracellular signaling, cell survival, and cell identity. As such, it is a prominent target for pharmacological intervention. The relatively low abundance of this subproteome in conjunction with challenging extractability and solubility still hampers its comprehensive analysis. Here, we combined a chemical glycoprotein-tagging strategy with mass spectrometry to enable comprehensive analysis of the cell-surface glycoproteome. To benchmark this workflow and to provide guidance for cell line selection for functional experiments, we generated an inventory of the N-linked cell-surface glycoproteomes of 15 standard laboratory human cell lines and three primary lymphocytic cell types. On average, about 900 plasma membrane and secreted proteins were identified per experiment, including more than 300 transporters and ion channels. Primary cells displayed distinct expression of surface markers and transporters underpinning the importance of carefully validating model cell lines selected for the study of cell surface-mediated processes. To monitor dynamic changes of the cell-surface proteome in a highly multiplexed experiment, we employed an isobaric mass tag-based chemical labeling strategy. This enabled the time-resolved analysis of plasma membrane protein presentation during differentiation of the monocytic suspension cell line THP-1 into macrophage-like adherent cells. Time-dependent changes observed in membrane protein presentation reflect functional remodeling during the phenotypic transition in three distinct phases: rapid surface presentation and secretion of proteins from intracellular pools concurrent with rapid internalization of no longer needed proteins and finally delayed presentation of newly synthesized macrophage markers. Perturbation of this process using marketed receptor tyrosine kinase inhibitors revealed dasatinib to severely compromise macrophage differentiation due to an off-target activity. This finding suggests that dynamic processes can be highly vulnerable to drug treatment and should be monitored more rigorously to identify adverse drug effects.
The plasma membrane constitutes the physical interface between a cell and its environment and plays a key role in intercellular interactions, intercellular communications, and cell adhesion, as well as in outside-in and inside-out signal transmission. These crucial functions are performed by integral plasma membrane proteins as well as membrane-associated proteins with extracellular domains. For instance, adhesion proteins define cell shape and motility (1); transporters are involved in the transport of molecules, including nutrients, salts, and bioactive compounds (2, 3); and growth factor receptors as well as G-protein-coupled receptors (GPCRs)1 transfer external stimuli into the cell (4). Importantly, the composition of the plasma membrane proteome is not static, but cells adjust according to their developmental status, nutrient availability, or external signals (5). During malignant cell transformation, major changes in the plasma membrane proteome enable cancerous cells to invade tissue, trigger vascularization, evade the immune system, and become independent of external survival signals (6). Approximately 60% of all Food and Drug Administration-approved drugs target transmembrane proteins reflecting the pharmacological relevance of cell-surface proteins. GPCRs (>25%) and ion channels (>10%) are the most frequently targeted protein classes (7, 8). Among the top 10 globally selling drugs in 2014, eight drugs were directly or indirectly targeting membrane proteins (9). For drugs with intracellular targets, the plasma membrane poses a barrier for influx and efflux. Drugs can be actively transported out of cells by ATP-binding cassette (ABC) transporters that couple ATP hydrolysis to the active efflux of small molecules. Amplified ABC transporter activity is a common mechanism towards resistance of cancer cells against chemotherapeutics (10). Drug uptake is controversially discussed in the literature. In addition to the broadly accepted uptake by passive diffusion (11), there is growing evidence for active transport mechanisms (12, 13) via solute carriers (SLC) (12, 14, 15). Recently, Winter et al. (16) demonstrated that the cell toxicity of the chemotherapeutic YM155 is dependent on protein-mediated transport via SLC35F2 thus linking expression of this transporter to efficacy of the drug.
Systematic investigation of cell-surface proteins is commonly performed by mass spectrometry-based proteomics. Whereas early work on the plasma membrane proteome provided first insights into the protein diversity in this cellular compartment (17), the relatively low abundance of many plasma membrane proteins and the limited compatibility with generic proteomics protocols still hamper the comprehensive analysis of this subproteome. Sensitivity can be increased by affinity-tagging of N-linked glycoproteins on the cell surface, which enables selective enrichment of a major fraction of the plasma membrane proteome. This approach has been shown to primarily label sialylated proteins and has been successfully applied to identify cell-surface markers or to map the cell-surface proteome of cell lines (5, 18). Yet, a comparison with the coverage achieved by RNA microarrays suggests that these initial studies may have offered only an initial glimpse on the true complexity of the cell-surface proteome (19). Furthermore, dynamic changes in the cell-surface proteome during cellular differentiation or upon perturbation by bioactive compounds in a multiplexed, time-resolved manner have not been addressed so far.
Here, we present a simple and robust N-linked glycoproteome enrichment protocol optimized for deep coverage of the plasma membrane proteome. The procedure was applied to characterize the cell-surface proteome of 15 standard laboratory human cell lines and three primary cell types. We observed significant differences in receptor diversity and transporter abundances between primary cells and corresponding cell lines. Relative quantification using isobaric mass tagging enabled quantitative monitoring of dynamic processes on the cell surface in multiplexed experiments. We, for the first time, report a comprehensive analysis of the profound remodeling of the plasma membrane proteome during monocyte to macrophage differentiation of THP-1 cells. Treatment of these cells with the kinase inhibitor dasatinib (Sprycel) during differentiation severely compromised macrophage differentiation due to an off-target activity resulting in substantial morphological and functional changes and the loss of many macrophage-associated proteins.
EXPERIMENTAL PROCEDURES
Reagents and Cell Culture
Reagents and media were purchased from Life Technologies, Inc., or Thermo Fisher Scientific (Waltham, MA) unless otherwise noted. All cell lines were purchased from ATCC. Ramos, PC-3, YT, THP-1, and K562 cells were cultured in RPMI 1640 medium containing 10% fetal calf serum (FCS). Jurkat E6.1 cells were cultured in RPMI 1640 medium supplemented with 4.5 g/liter glucose, 10 mm HEPES, 1 mm sodium pyruvate, and 10% FCS. Panc 08.13 cells were cultured in RPMI 1640 medium containing 15% FCS and 10 μg/liter insulin. A549 and HEK293 cells were cultured in DMEM containing 10% FCS. CaCo-2 cells were cultured in MEM supplemented with 10% FCS. HeLa and HepG2 cells were cultured in DMEM supplemented with 20% FCS, 1 mm sodium pyruvate, and 1% non-essential amino acids. MCF-7 cells were cultured in MEM containing 10% FCS, 1 mm sodium pyruvate, 1% non-essential amino acids, and 10 μg/liter insulin. Saos2 were cultured in McCoy's 5a media containing 15% FCS. U87MG were cultured in RPMI 1640 medium containing 10% FCS, 1% Glutamax and 1% Pyruvate. All cell lines were cultured at 37 °C and 5% CO2 except for A549 cells that were cultured at 37 °C and 10% CO2. Phorbol 12-myristate 13-acetate (PMA) and imatinib (Gleevec, STI-571) were purchased from Sigma-Aldrich (St. Louis, MO); dasatinib (Sprycel, BMS-354825) and sunitinib (Sutent, SU11248) were from Santa Cruz Biotechnology (Santa Cruz, CA), and vitamin D3 was from Cayman Chemicals (Biomol, Hamburg, Germany). Human primary B-cells, T-cells, and NK cells were extracted from buffy coats (DRK Mannheim, Germany) by Histopaque density gradient centrifugation and the EasySep enrichment kits (StemCell Technologies Inc., Vancouver, BC) according to the manufacturer's instructions.
Kinobeads and Competition Assay
Kinobeads profiling was performed essentially as described (64). Briefly, cells were homogenized in lysis buffer (50 mm Tris-HCl, pH 7.5, 5% glycerol, 1.5 mm MgCl2, 150 mm NaCl, 20 mm NaF, 1 mm Na3VO4, 1 mm DTT, 5 mm calyculin A, 0.8% Igepal-CA630 and protease inhibitor mixture (Roche Diagnostics, Basel Switzerland)) using a Dounce homogenizer on ice. Lysates were cleared by ultracentrifugation and adjusted to 5 mg/ml protein concentration using the Bradford assay. Compounds were dissolved in dimethyl sulfoxide, and various concentrations were added to 5-ml lysate samples followed by 50 μl of kinobeads suspension. Samples were agitated for 30 min at 4 °C. Subsequently, the beads were washed and collected by centrifugation, and bound material was eluted with SDS sample buffer, fractionated by SDS-gel electrophoresis on 4–12% NuPAGE gels (Invitrogen, Carlsbad, USA), and stained with colloidal Coomassie. Samples were in-gel digested, TMT-labeled and analyzed by mass spectrometry, as described in detail below.
THP-1 Differentiation and Drug Treatment
THP-1 cells were diluted to 0.5 × 106 cells/ml into fresh culture media 1 day prior to differentiation experiments. For each experimental condition, 1 × 107 cells were seeded in RPMI 1640 medium supplemented with 10% FCS and 100 nm PMA or 100 nm vitamin D3 (calcitriol) on a 15-cm Petri dish and incubated at 37 °C and 5% CO2. PMA-differentiated cells were further treated with 1 μm kinase inhibitor (dasatinib, sunitinib, and imatinib) over 1 h at 37 °C in RPMI 1640 medium supplemented with 10% FCS prior to the addition of 100 nm PMA. PMA differentiation was verified by Western blotting for up-regulation of CD11b (ab52478, Abcam, Cambridge, UK) or down-regulation of MRC2 (ab70132, Abcam).
Cell-surface Labeling and Enrichment of Labeled Proteins
For each experiment, 1 × 107 suspension cells or a 15-cm Petri dish of adherent cells at 70–80% confluency were washed twice with PBS followed by oxidation of carbohydrates with 1 mm sodium metaperiodate in pH 6.5 adjusted PBS at 4 °C for 10 min in the dark. Cells were washed twice with PBS followed by biotinylation with 1 mm alkoxyamine-PEG4-biotin in presence of 10 mm aniline for 10 min at 4 °C in the dark. Cells were washed three times with PBS. Suspension cells were pelleted at 340 × g; adherent cells were detached by scraping and pelleted at 340 × g. Cell pellets were frozen in liquid nitrogen and stored at −80 °C. Cell pellets were lysed by boiling for 10 min at 95 °C in 200 μl of SDS lysis buffer (4% SDS, 60 mm Tris, pH 7.6, 50 mm DTT). Samples were cooled and sonicated on ice once (Bandelin Sonopuls HD 2200) at 50% power output with one burst of 10 s. Lysates were diluted 1:10 in PBS and enrichment of biotinylated proteins was performed in deep 96-well filter plates (MicruLute Combinatorial, Porvair Sciences, Leatherhead, UK). For each enrichment, 15 μl of high capacity streptavidin-agarose resin was prepared by washing twice with wash buffer 1 (WB1: 0.4% SDS, 20 mm Tris, pH 7.5, 400 mm NaCl). Resin was incubated with lysate for 60 min at 25 °C while shaking, followed by 3× washing with WB1, 8× washing with wash buffer 2 (WB2: 20 mm Tris, pH 7.5, and 400 mm NaCl); and 8× washing with wash buffer 3 (WB3: 50 mm triethylammonium bicarbonate (TEAB), 2 m urea). Subsequently, resin was incubated for 45 min with 45 mm DTT in 50 mm TEAB at 25 °C followed by 30 min of incubation at 25 °C with 100 mm iodoacetamide in 50 mm TEAB. Resin was washed five times with WB3 and digested overnight at 25 °C in 60 μl of 50 mm TEAB, 1.33 m urea, and 0.4 μg of trypsin. The following day, additional 0.4 μg of trypsin in 50 mm TEAB were added, and the resin was incubated for an additional 4 h at 25 °C. Peptides were eluted by centrifugation. For additional PNGase F elution, the resin was washed five times with wash buffer 4 (WB4: 50 mm HEPES, pH 7.5) followed by incubation with 0.5 units of PNGase F in WB4 for 3 h at 37 °C. Peptides were eluted by centrifugation.
Sample Preparation for MS
For TMT quantification experiments, peptide samples were dried in vacuo, resuspended in 100 μl of 200 mm TEAB 10% acetonitrile, and labeled with 10-plex TMT (TMT10, Thermo Fisher Scientific) reagents. The labeling reaction was performed by adding 50 μl of 25 mm TMT-label reagent dissolved in acetonitrile, incubating for 90 min at 25 °C, and quenching with 2.5% hydroxylamine in 100 mm TEAB for 15 min at 25 °C. Labeled peptides were pooled and desalted using C18 stage tips (21).
LC-MS/MS Analysis
Samples from the THP-1 differentiation time course experiment were separated into nine fractions, by using reversed-phase chromatography at pH 12 (1-mm Xbridge column (Waters, Milford, MA)), as described previously (22). All other samples were measured without fractionation. Samples were dried in vacuo and resuspended in 0.05% trifluoroacetic acid in water. 30% of the sample was injected into an Ultimate3000 nanoRLSC (Dionex, Sunnyvale, CA) coupled to a Q Exactive or Q Exactive Plus (Thermo Fisher Scientific). Peptides were separated on custom-made 50 cm × 100 μm (inner diameter) C18 reversed-phase columns (Reprosil, Dr. Maisch GmbH, Ammerbuch-Entringen, Germany) at 40 °C. Gradient elution was performed from 2–40% acetonitrile in 0.1% formic acid over 2 or 4 h. Samples were on-line injected into Q-Exactive mass spectrometers operating with a data-dependent top10 method. MS spectra were acquired by using 70,000 resolution and an ion target of 3E6 for MS1 scans. Higher energy collisional dissociation (HCD) scans were performed with 25% (label-free) or 35% (TMT quantification) normalized collision energy at 35,000 resolution (at m/z 200), and the ion target setting was set to 2E5.
Peptide and Protein Identification
Raw data were processed using an in-house pipeline (23). Mascot 2.4 (Matrix Science, Boston, MA) was used for protein identification by using a 10 ppm mass tolerance for peptide precursors and 20 mDa (HCD) mass tolerance for fragment ions. Enzyme specificity was set to trypsin with up to three missed cleavages. For label-free experiments, carbamidomethylation of cysteine residues was set as fixed modification and methionine oxidation, and N-terminal acetylation of proteins was set as variable modifications. For TMT quantification experiments, carbamidomethylation of cysteine residues and TMT modification of lysine residues were set as fixed modifications, and methionine oxidation, N-terminal acetylation of proteins, and TMT modification of peptide N-termini were set as variable modifications. Deamidation of asparagines was set as additional variable modification for PNGase F samples. The search database consisted of a customized version of the International Protein Index protein sequence database (numbers of entries, 108,790; year, 2009) combined with a decoy version of this database created by using a script supplied by Matrix Science. IPI accession numbers of all identified proteins were mapped to the UniProtKB database (December 14, 2016) by matching the corresponding protein sequences. Unless stated otherwise, we accepted protein identifications as follows. (i) For single-spectrum to sequence assignments, we required this assignment to be the best match and a minimum Mascot score of 31 and a 10× difference of this assignment over the next best assignment. Based on these criteria, the decoy search results indicated <1% false discovery rate (FDR). (ii) For multiple spectrum to sequence assignments and using the same parameters, the decoy search results indicate <0.1% FDR. All identified proteins were quantified; FDR for quantified proteins was below 1%.
Peptide and Protein Quantification
In the case of label-free quantification, the Top3 method (24) was applied in a slightly modified form (25). In brief, for each protein the maximum intensity of the extracted ion current of the three most abundant peptide sequences was log10 transformed and averaged. The derived value is denoted as MS1 abundance and is a good proxy for protein abundance in the sample.
For TMT quantification experiments, reporter ion intensities were read from raw data and multiplied with ion accumulation times (the unit is milliseconds) to yield a measure proportional to the number of ions; this measure is referred to as “ion area” (26). Spectra matching the peptides were filtered according to the following criteria: mascot ion score >15, signal-to-background of the precursor ion >4, and signal-to-interference >0.5 (27). Fold-changes were corrected for isotope purity as described and adjusted for interference caused by co-eluting nearly isobaric peaks as estimated by the signal-to-interference measure (28). Protein quantification was derived from individual spectra matching distinct peptides by using a sum-based bootstrap algorithm.
Data Analysis
Annotations of proteins were based on the UniProtKB database (December 14, 2016). Identified proteins in trypsin digest samples were filtered for MS1 abundance of >0 and total number of peptide to spectrum matches of ≥2. Identified proteins in PNGase F digest samples were filtered for MS1 abundance of >0 and a mascot score of ≥20. Relative abundance estimations for trypsin and PNGase F samples were normalized to the average density maximum of all samples (supplemental Fig. S1, G and H).
For identification of proteins differentially expressed between cell lines and cognate primary cells, p values were calculated by t test based on imputed data (estimated protein abundance values of 0 replaced by normal distributed data simulating signals of low abundant proteins, normal distribution standard deviation 0.3, normal distribution mean downshifted by 1.8 to lower end of the distribution of measured values, separate for each column) (29). Significant proteins were filtered for Benjamini-Hochberg corrected p values of <0.05. For GO-Term enrichment analysis, the enriched GO-Terms were filtered for Benjamini-Hochberg corrected p values of <0.01. In TMT quantification samples, proteins were filtered for three or more quantified spectra and two or more unique peptides. The 0-h time point was set as reference. For identification of significant proteins regulated during differentiation of THP-1 cells, a modified t test (30) was applied, and data were filtered for log2-transformed relative abundances of ≥2 or ≤−2 at least once between 24 and 72 h of the time course with Benjamini-Hochberg corrected p values <0.01 at the respective time point. For identification of proteins significantly altered in abundance compared with cells differentiated in the presence or absence of kinase inhibitors, the data were filtered for log2-transformed relative abundances of ≥1 or ≤−1 between these differentiation outcomes for both replicates and p values <0.05. Data analysis and visualization were performed in TIBCO Spotfire 6.0.2 or R. Gene ontology enrichment was obtained from DAVID Bioinformatics Resources 6.7 (31) or in R (topGO, Bioconductor) (32). Principal component analyses were performed in Perseus 1.5.1.6. Common bead background proteins were annotated based on CRAPome data (filters used for CRAPome: streptavidin-agarose beads, proteins with spectral counts of ≥5) (33). All proteins annotated with intracellular localization or without any UniProt annotated localizations were considered as bead background if they were annotated as common bead background proteins in the CRAPome database.
Phagocytosis Assay
The phagocytosis assay was conducted based on the CytoSelect 96-well phagocytosis assay from Cell Biolabs (San Diego, CA). After 48 h of differentiation in the presence or absence of kinase inhibitor, adherent cells were detached with trypsin, and 5 × 104 cells in 100 μl of fresh RPMI 1640 medium supplemented with 10% FCS, 100 nm PMA, and 1 μm kinase inhibitor or DMSO were seeded per well in a 96-well microtiter plate. After 24 h of incubation at 37 °C and 5% CO2, 2 μm cytochalasin D was added to respective phagocytosis inhibition wells, and the plate was incubated for an additional 1 h. Non-opsonized zymosan particles (10 μl/sample) were added, and cells were incubated for 1 h. The amount of engulfed zymosan particles was determined by measuring the absorbance at 405 nm with a 96-well microtiter plate reader (EnVision Plate Reader, PerkinElmer Life Sciences), and the relative phagocytotic activity was referenced to normally differentiated THP-1 cells.
Experimental Design and Statistical Rationale
For the cell-surface proteome mappings of cell lines and primary cells, we performed label-free experiments and analyzed each sample in two independent biological replicates by mass spectrometry. For principal component analysis and hierarchical clustering, the protein abundance (based on a modified Top3 method) was used. In addition, biological quadruplicates of four cell lines were analyzed to assess saturation of proteome coverage and robustness of segregation.
All differentiation and drug treatment experiments were performed using a TMT labeling strategy. For the differentiation time course experiment, we combined two experimental designs to achieve high precision for short as well as late time points. In the first experiment block (denoted as experimental group 1), the differentiation process was split into two separate experiments, one covering the short incubation times (0, 4, and 8 h, each in biological triplicates) and the second experiment covering the long incubation times (0, 24, 48, and 72 h, each in biological duplicates). For further confirmation of results, the differentiation was repeated in two additional experiments (denoted as experimental group 2 and experimental group 3) covering all six time points in single TMT experiments. Proteins identified with more than two quantified spectra and more than one unique peptide were considered quantified. Statistical analysis was performed on experimental group 1. p values were calculated using a modified t test (30) and Benjamini-Hochberg corrected. A combined fold-change and p value cutoff was applied to determine significant outliers: log2 relative abundances of ≥2 or ≤−2 at least once between 24 and 72 h of the time course and p values of <0.01. Experimental groups 2 and 3 were used for confirming regulated protein identifications in further biological replicates.
For identification of proteins significantly regulated due to the presence of kinase inhibitor during differentiation, we compared the 48-h time point of normally differentiated cells and cells differentiated in the presence of kinase inhibitors from two biological replicates. A combined fold-change and p value cutoff was applied to determine significant outliers: log2 relative abundances of ≥1 or ≤−1 in both replicates and t test p values <0.05.
RESULTS
Optimization and Validation of Plasma Membrane Proteome Enrichment
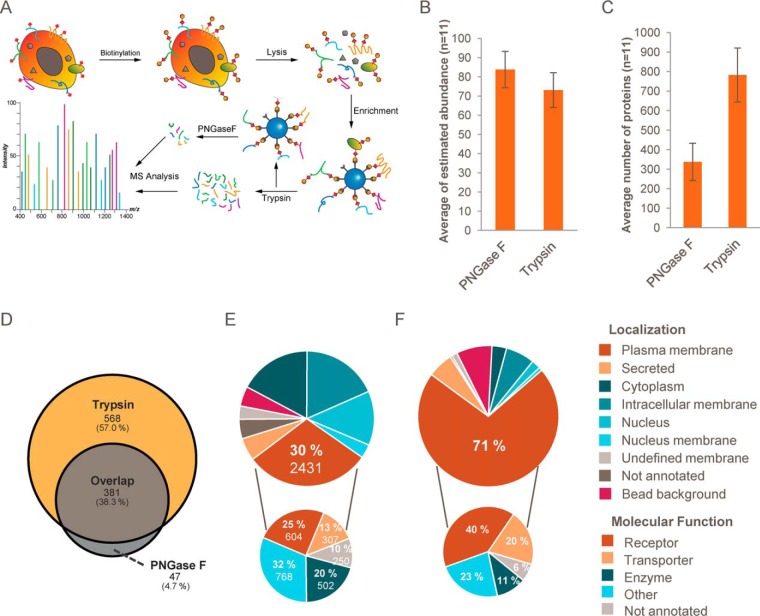
To enable comprehensive profiling of N-glycan-modified cell-surface proteins, we implemented an alkoxyamine-based surface biotinylation strategy based on published procedures (Fig. 1A) (34). First, carbohydrates are oxidized with sodium metaperiodate and subsequently biotinylated with alkoxyamine-PEG4-biotin in the presence of aniline. After cell lysis in 4% SDS, biotinylated proteins are enriched on streptavidin-coated beads and trypsinized followed by LC-MS/MS analysis. Alternatively, glycosylated peptides can be eluted upon deglycosylation with PNGase F (5). We compared the selectivity, sensitivity, and coverage achieved with both approaches by label-free LC-MS/MS analysis of duplicate experiments with 11 different cell lines/primary cells using 4-h gradient separation on an Orbitrap QExactive platform. Proteins associated with the plasma membrane (according to UniProtKB database, December 2016) were the most abundant proteins for all tested cell lines, indicating an efficient enrichment strategy. When analyzing the peptides generated by on-bead trypsinization, on average 73% of the total protein abundance could be attributed to plasma membrane-associated proteins. The specificity of the approach could be further improved to 84% protein abundance of plasma membrane proteins when peptides were specifically eluted with PNGase F after trypsinization (Fig. 1B and supplemental Tables S1 and S2). In both cases, an additional 6% of the total protein abundance could be attributed to secreted proteins. In UniProt, the remaining fraction of identified proteins was annotated as intracellular. PNGase F elution of bound peptides could enable the identification of glycosylation sites, as PNGase F cleavage induces deamidation of Asn to Asp with a mass shift of +0.98 Da. However, even without deglycosylation, we observed a high rate of spontaneous deamidation of asparagine residues after trypsin elution. About 5% of all identified peptides in trypsin elution samples were identified with deamidated asparagine. As this high background could compromise the ability to correctly identify glycosylation sites, we decided not to further investigate these post-translational modifications. We note that the specific release of intact proteins from streptavidin beads by PNGase F was not successful in our hands. When comparing both elution methods, analyzing all tryptic peptides yielded more than twice as many plasma membrane proteins as the PNGase F strategy (on average 781 versus 337, Fig. 1, C and D), and protein identifications were based on nearly twice as many unique peptides, on average (10.2 versus 5.7, supplemental Fig. S1C). Because of the substantially higher coverage achieved and the modest loss in specificity, the PNGase F digestion step was omitted in the following experiments, and analyses were focused on the complete sets of tryptic peptides generated by on-bead proteolysis from individual samples.
Fig. 1.
Selective biotinylation of plasma membrane glycoproteins enables robust identification and quantification of cell-surface proteins by LC-MS/MS. A, schematic illustration of the workflow. Plasma membrane proteins with N-glycosylations (red squares) are oxidized with sodium metaperiodate and subsequently biotinylated by alkoxyamine-PEG4-biotin. Cells are lysed in presence of SDS and DTT followed by the enrichment of biotinylated proteins with streptavidin-coated beads. Beads are stringently washed, and bound proteins are trypsinized. The resulting peptide mixture is analyzed by LC-MS/MS. Additionally, glycosylated peptides can be specifically eluted by the N-glycosidase PNGase F. B, comparison of the on-bead trypsinization procedure (trypsin) and the PNGase F elution protocol based on the average abundance of captured plasma membrane proteins (Top3 method) of 11 cell lines (n = 2). Error bars indicate standard deviations. C, as in B, but the comparison is based on the average number of identified plasma membrane proteins. D, representative Venn diagram illustrating the overlap of identified plasma membrane proteins using trypsin and PNGase F protocols for cell-surface biotinylated CaCo2 samples (n = 2). E, total number of identified proteins (large cake diagram) grouped by their annotated subcellular localization (UniProtKB) and annotated common bead background proteins (CRAPome). Plasma membrane proteins grouped by their annotated molecular function are shown in the small cake diagram. F, as in E but based on estimated fractional abundance of identified proteins. See also supplemental Fig. S1.
Among the particular strengths of this cell surface labeling method for analyzing the plasma membrane proteome are its sensitivity and low resource requirements. We compared the plasma membrane proteome coverage of our N-glycoproteome enrichment strategy to published full expression proteomics analyses for three cell lines (Jurkat, MCF7, and HeLa) (35). Whereas cell surface-enriched samples were analyzed in a single 4-h LC-MS/MS run, the published whole proteome data was based on three replicates each fractionated into six fractions corresponding to 60 h of total analysis time per cell line. On average, whole proteome analyses yielded 7520 proteins of which 1402 were annotated to be plasma membrane-associated compared with on average 835 plasma membrane-associated proteins identified with the surface protein enrichment strategy (supplemental Fig. S1D). However, the majority of the plasma membrane-associated proteins found in the expression proteomics analyses are not integral membrane proteins but include enzymes and other proteins that are localized at the cytosolic side of the plasma membrane. In contrast, for proteins with annotated transmembrane domains, the cell-surface labeling strategy achieved a 20% higher coverage (445 in expression proteomics versus 535 cell surface enrichment), including 40% more plasma membrane receptors and transporters (230 expression proteomics versus 322 cell surface enrichment). In summary, due to the significantly lower complexity of the cell surface enriched samples, a higher cell surface-presented plasma membrane proteome coverage (with comparable sequence coverage) was achieved with less than 10% of the MS analysis time compared with whole proteome analysis (supplemental Fig. S1, E and F). To determine the number of replicates necessary for comprehensive coverage of cell-surface proteomes and to investigate the reproducibility of the assay, four replicate cell-surface mapping experiments from four cell lines were performed. Although the second replicate on average added 9% coverage, the third and fourth replicate only added 3 and 2%, respectively (supplemental Fig. S1J and supplemental Table S1). This indicates that saturation is almost reached after two replicates. Tight clustering of all four replicates in a principal component analysis demonstrated high reproducibility (supplemental Fig. S1I).
Cell-surface Proteome of Cell Lines and Primary Cells
To demonstrate the applicability of the cell-surface labeling strategy for large scale studies and to generate a cell line-specific inventory of the plasma membrane proteome, we analyzed 15 commonly used human cell lines and three primary cell types isolated from human blood (supplemental Table S1). This selection included 10 adherent epithelial cell lines from various tissue origins, four lymphoblastic suspension cell lines, and one monocytic cell line (THP-1) that can be differentiated into macrophage- and osteoclast-like adherent cells. For comparison with primary cells, B-cells, T-cells, and NK cells were isolated from human blood and compared with the lymphoblastic cell lines Ramos, Jurkat, and YT, respectively.
Cell-surface proteome profiling experiments were performed in duplicates for each cell type and yielded on average 861 plasma membrane and secreted proteins with high reproducibility (supplemental Fig. S1B). Relative protein abundances were estimated from MS1 signal abundances based on a modified Top3 method (24, 25). Coefficients of correlation of estimated protein abundances between replicates were typically above 0.9 (supplemental Fig. S1A). In the combined dataset, we identified a total of 2431 plasma membrane proteins, including 604 receptors, 307 transporters, 502 enzymes, 768 proteins with other molecular functions such as GTPase activation, developmental protein, cytokine, or chaperone, and 250 proteins with no annotated molecular function (Fig. 1E). In total, 192 out of 397 known SLC transporters and 29 out of 47 known ABC transporters were identified. Semi-quantitative assessment of protein abundances based on the Top3 method (24) revealed that on average 77% of the total protein abundance per sample could be attributed to plasma membrane-associated and secreted proteins, and less than 10% was derived from common bead background-binding proteins, as annotated by the contaminant repository of affinity purifications (CRAPome) (Fig. 1F) (33). The majority of the remaining 15% are derived from proteins with intracellular membrane or unknown membrane localization. Potentially these proteins have a missing cell surface annotation. Finally other non-membrane proteins could be co-enriched by association to cytosolic parts of cell surface transmembrane proteins. As the enrichment is based on protein glycosylation, we analyzed the presence of annotated glycosylation sites on identified proteins. On average, 72% of the total protein abundance in the samples could be attributed to proteins with annotated glycosylation sites (supplemental Fig. 1K). The remaining 28% originated mostly from proteins with intracellular localization according to UniProt. Focusing on cell surface-localized proteins, for 50% of identified plasma membrane and 80% of secreted proteins, glycosylation sites are annotated in UniProt. These glycosylated cell-surface proteins account for 89% of the total protein abundance of cell-surface proteins (supplemental Fig. 1L). Among the remaining 11%, mostly proteins without annotated transmembrane domains (7%) were identified. The other 4% accounted for cell-surface proteins without an annotated glycosylation but annotated transmembrane domains. These proteins are potentially directly enriched proteins but with missing glycosylation site annotations. In summary, both analyses suggest that ∼25% of the total protein abundance measured with our glycoprotein enrichment strategy is due to background binders.
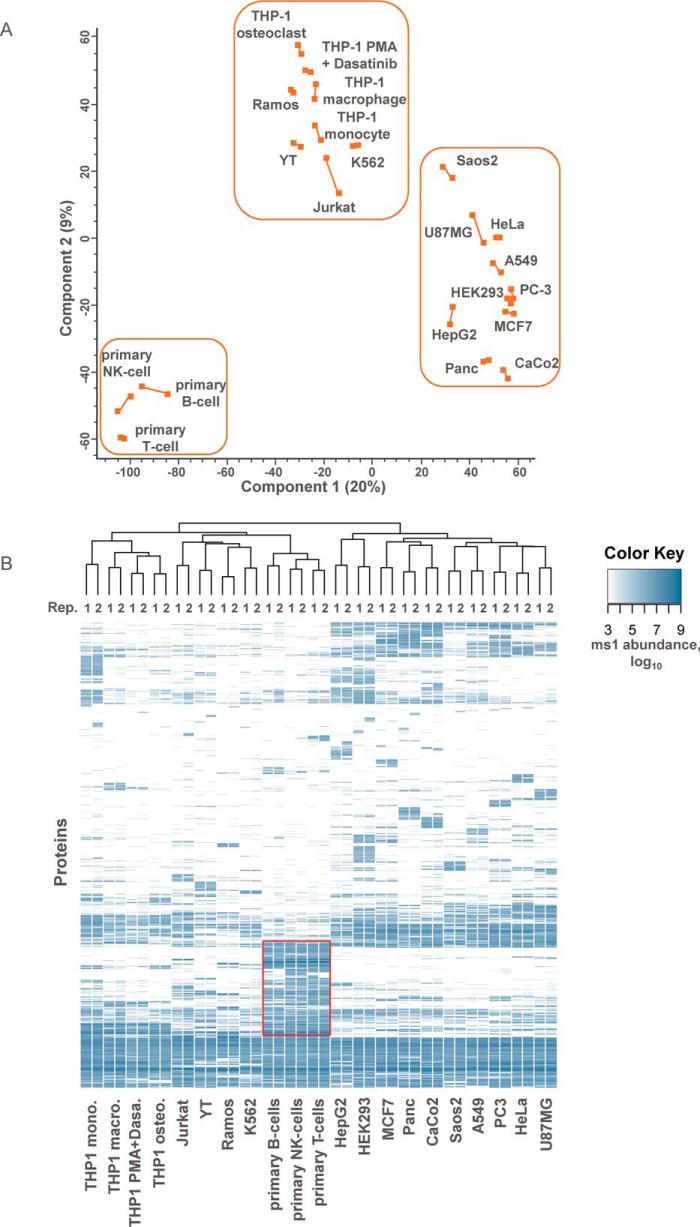
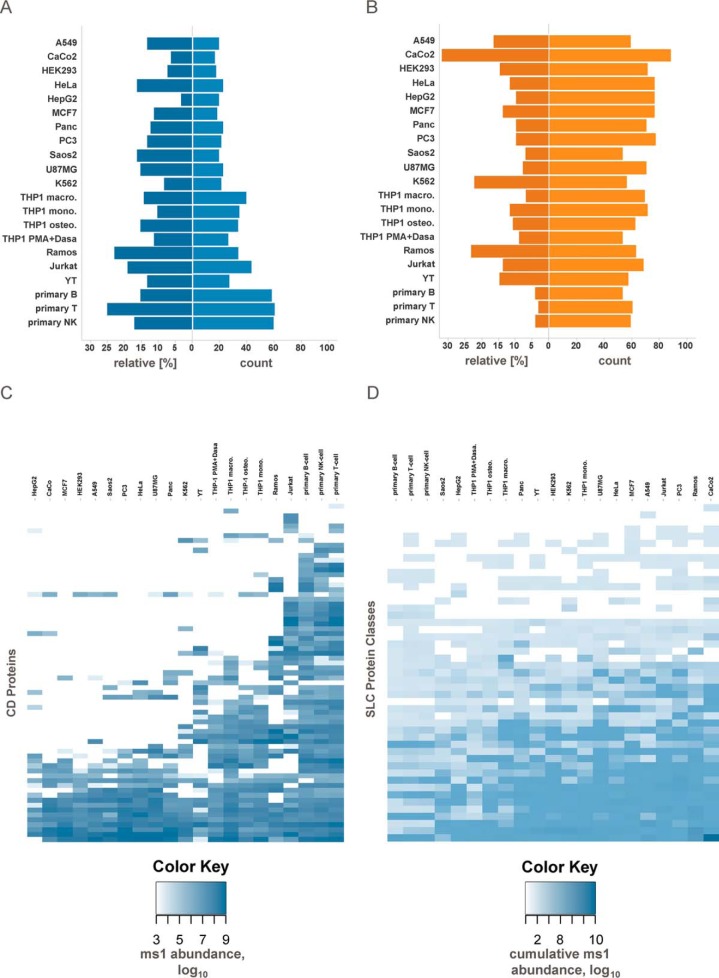
Principal component analysis based on the measured plasma membrane protein abundances clustered replicates in close proximity along the first and the second principal component axes (Fig. 2A). On the first principal component, cell systems were grouped in three distinct clusters separating epithelial from lymphoid cell lines, and interestingly, primary cells from cell lines typically used as a model for these primary cells (e.g. Jurkat cells are a model for T-cells, Ramos cells are a model for B-cells, etc.). The second principal component further separates the cells within each cluster, e.g. the osteoclast and macrophage-like cell lines derived from the monocytic cell line THP-1. Hierarchical clustering yielded similar groupings of cell lines (Fig. 2B). Clustering on the protein level identified differential presentation of surface proteins in line with expected differences between epithelial and lymphoid cell lines. A distinct protein cluster was common to the tested primary cells and included chemokine receptors, clusters of differentiation, interleukin receptors, integrins, and also a number of solute carrier proteins, e.g. SLC18A2 (amine transporter), SLC2A5 (fructose transporter), and SLC2A9 (fructose and glucose transporter). GO terms for immune response, inflammatory response, or platelet activation were strongly enriched among the proteins more abundantly expressed (p value < 0.05) in primary cells as compared with cell lines. Differences between cell lines and primary cells were also observed when comparing protein abundances of selected protein families like ABC transporters, clusters of differentiation (CD), G-protein-coupled receptors (GPCR), integrins, and solute carriers (SLC) (Fig. 3 and supplemental Fig. S2). For primary cell types, on average 59 CD proteins were detected compared with 34 in the corresponding cell lines, but the cumulative relative abundance was similar among the different cells (Fig. 3, A and C). In contrast, primary and immortalized cells on average showed similar numbers of expressed SLC proteins (58 versus 64), but their cumulative relative abundance was much lower in primary cells compared with their cognate cell lines (4% versus 18%) (Fig. 3, B and D). Exceptions are facilitative GLUT transporters (SLC2 (36)), choline-like transporters (SLC44 (37)), and the SLC25 mitochondrial carrier family (38), which were present with similar abundances on all selected cell lines and primary cells. Other SLC transporter groups like the amino acid and nucleoside transporters SLC1, SLC7, SLC38, and SLC29 (39–42) were substantially more abundant in cell lines. Indeed, cancer cells have been described to show different and enhanced expression profiles of SLC transporters compared with normal cells (43); however, so far a detailed analysis about which SLCs are presented on the surface has been missing. Furthermore, primary cells showed more GPCRs (56 versus 23) (supplemental Fig. S2), which is in line with previously published transcriptomic profiles (44) and a higher relative cumulative abundance of integrins (17% versus 9%).
Fig. 2.
Comparison of plasma membrane proteomes of cell lines and primary cells. A, principal component analysis of cell line and primary cell-surface proteomes in two biological replicates. The first two principal components account for 29% of the total variation. Axes are labeled with the percent total variance of the corresponding principal component. Biological replicates are connected by lines. Epithelial, lymphoblastic, and primary cells are grouped in boxes. B, heat map representation of the cell-surface proteomes based on protein class abundances (Top3 method) and average linkage clustering using a Euclidian distance matrix.
Fig. 3.
Differences of cell-surface proteomes of primary cells and cell lines reflect lower metabolism and higher functional specialization. A, relative abundance and absolute number of identified CD proteins. Relative abundance is defined as the sum of MS1 signal abundance of CD proteins divided by the MS1 signal abundance summed over all plasma membrane proteins. B, relative abundance and absolute number of identified SLC transporters. C, heat map representation based on CD protein abundances. D, heat map representation based on SLC protein class abundances. See also supplemental Fig. S2.
Monitoring the Cell-surface Proteome of Differentiating Monocytes
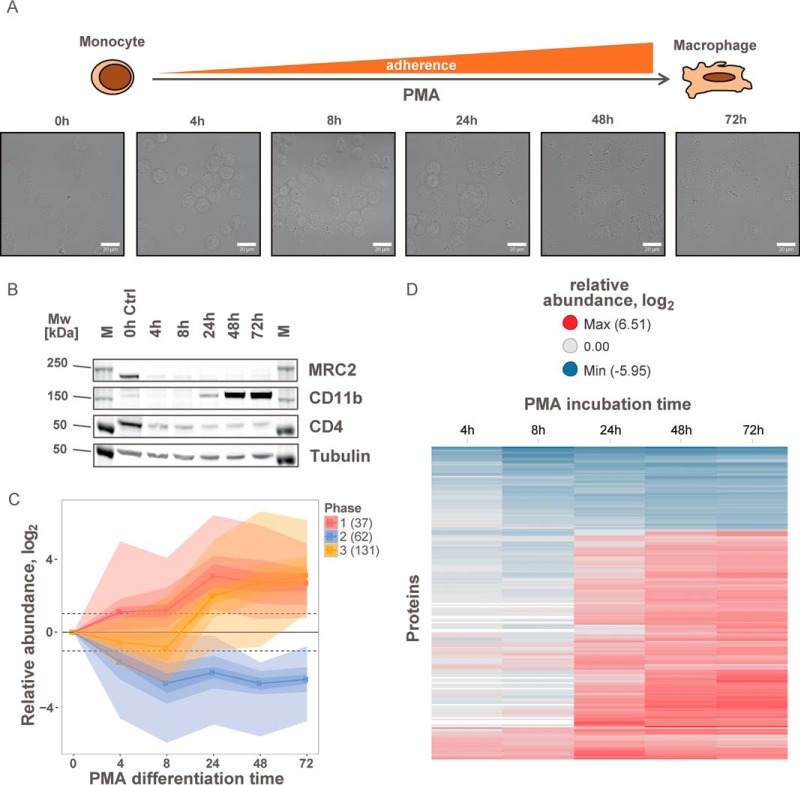
Cell-surface proteome maps are typically static representations of cellular states. However, human biology is determined by dynamic processes, in which environmental changes or external stimuli induce cellular adaptation or differentiation. Monocytes, for example, circulate in the bloodstream and upon recruitment to damaged or infected tissues differentiate into macrophages (45). To monitor dynamic processes occurring at the cell surface during such transitions, we devised a tandem mass tag (TMT)-based cell-surface proteomics strategy that enables multiplexed measurements of up to 10 samples without missing values (46). This strategy was applied for the time-resolved analysis of dynamic processes of the cell-surface proteome of THP-1 cells during phorbol-12-myristate-13-acetate (PMA)-induced differentiation from monocytic into macrophage-like cells (Fig. 4A). In total, we performed at least three biological replicates for each time point (0, 4, 8, 24, 48, and 72 h), which were analyzed in three experimental groups after labeling with TMT reagents (supplemental Table S3).
Fig. 4.
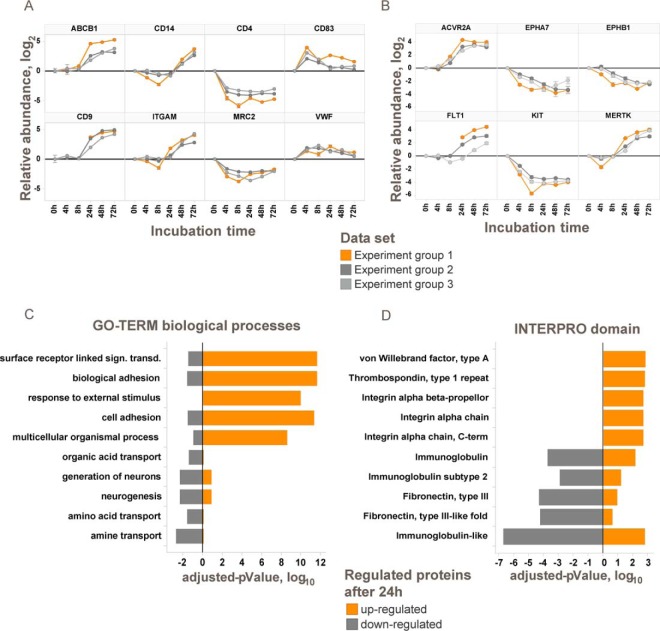
Time-dependent monitoring of the plasma membrane proteome during differentiation of THP-1 cells. A, morphological changes of suspension monocytic THP-1 cells to adherent macrophage-like cells during differentiation in the presence of 100 nm PMA monitored by light microscopy. B, immunoblot of regulated cell-surface markers during differentiation of monocytic THP-1 cells to macrophage-like cells by PMA (M: molecular weight marker). C, abundance changes of significantly regulated plasma membrane proteins during differentiation grouped into three phases. Color shadings indicate data density. The numbers of proteins in each cluster are indicated. D, heat map of relative abundances for significantly regulated plasma membrane proteins during differentiation. See also supplemental Fig. S3.
To identify differences between the two differentiation states, we tested for proteins that were significantly altered between the start and end point with a p value below 0.01 (modified t test (30)) and a change of ≥4-fold in at least one of the three late time points (24, 48, or 72 h). Significant changes between undifferentiated and differentiated THP-1 cells were detected for 20% of the cell-surface proteome (230 out of 1106 quantified plasma membrane and secreted proteins; Fig. 4, supplemental Fig. S3, and supplemental Table S3). Regulated proteins include known markers for monocyte to macrophage differentiation such as CD11b (47, 48) and CD4 (49) (supplemental Table S3, Fig. 5A, and supplemental Fig. S3A). Changes on the cell-surface proteome can be correlated to functional differences between monocytes and macrophages. Macrophages invade into tissue, and in line with this function we observed, for example, down-regulation of PODXL2 (−8-fold), which is necessary for leukocyte rolling over vascular surfaces (50) and up-regulation of cell adhesion proteins like CD9 (+24-fold), CD22 (+32-fold), and ITGAV (+12-fold) (51–53). After differentiation, the macrophage-like THP-1 cells are non-dividing. Consequently, proteins involved in the control of cell growth and cell differentiation like PTPRF, SEMA4A, and EPHA7 are significantly down-regulated. Altered display of cell-surface transporters included down-regulated sugar (SLC2A5 and SLC2A9) and amino acid transporters (SLC38A5, SLC43A1, and SLC7A2) probably reflecting the lower nutrient demand of differentiated and resting cells. The multidrug resistance transporter ABCB1 was also strongly up-regulated (+37-fold up) enabling ATP hydrolysis-driven efflux of small molecules.
Fig. 5.
Changes on the THP-1 cell-surface proteome during differentiation with PMA reflect differences between monocyte and macrophage functions. A, examples for significantly regulated proteins. Log2 transformed abundance changes relative to 0 h are shown for three independent experiments. B, examples for significantly regulated plasma membrane-associated kinases. C, GO-Term enrichment analysis of up- and down-regulated proteins after 72 h. Bars indicate logarithmic Benjamini-Hochberg corrected p values. D, INTERPRO domain enrichment analysis of up- and down-regulated proteins after 72 h. Bars indicate logarithmic Benjamini-Hochberg corrected p values. See also supplemental Fig. S3.
Interestingly, functional dependences between proteins can also be observed. For example, the urokinase plasminogen activator surface receptor PLAUR was found 28-fold up-regulated, whereas its negative regulator, the c-type mannose receptor MRC2 that controls the extracellular level of PLAUR (54), was 5-fold reduced during differentiation. GO-term enrichment analysis was used to identify biological processes associated with this cellular transition. For up-regulated proteins, cell-surface receptor-linked signal transduction, biological adhesion and response to external stimuli were identified as the most significant processes, whereas down-regulated proteins are involved in amine transport, amino acid transport, or neurogenesis (Fig. 5C). Protein domain enrichment analysis suggests up-regulation of mainly integrin domain-containing proteins, whereas proteins with immunoglobulin-like or fibronectin domains were down-regulated (Fig. 5D).
The multiplexed experimental design allows for a thorough analysis of transient and permanent abundance changes of the cell-surface proteome. The entire differentiation process can be clustered into three time-dependent major phases (Fig. 4C): rapid presentation/secretion, concurrent internalization, and delayed presentation of new cell-surface proteins. Immediately after stimulation with PMA, cells start to secrete ECM proteins, including collagens COL1A1 and COL1A2, von Willebrand factor, and fibrillin-1 thus priming the cells for settling and cell adhesion (supplemental Fig. S3). A rapid and substantial increase was also observed for the transmembrane proteins CD83, PLVAP, and tumor necrosis factor receptor superfamily member 21, which is involved in B-cell activation and vascular permeability (Fig. 5A). Because of this rapid response to the stimulus, this process must be to a large extent independent of de novo protein synthesis. For example, CD83 is stored inside monocytes and macrophages and rapidly relocates to the cell surface upon LPS-induced activation (55). In our data, CD83 abundance peaked at the 4-h time point. Upon completion of the differentiation process after 72 h, the abundance was 2–3-fold lower, suggesting that the adhesion receptor function of this protein is particularly critical for the early stages of differentiation. The second step comprises internalization of a larger set of proteins, including nutrient transporters and proteins involved in regulation of cell differentiation like PTPRF, IL6R, and KIT reflecting the transition from dividing to non-dividing cells. Finally, the de novo synthesis of cell-surface proteins leads to the presentation of a new subset of the plasma membrane proteome beginning 24 h after the onset of differentiation (Fig. 4D). These proteins fulfill macrophage-specific functions such as cell adhesion and phagocytosis.
Influence of Kinase Inhibitors on the Cell-surface Proteome of Differentiating Monocytes
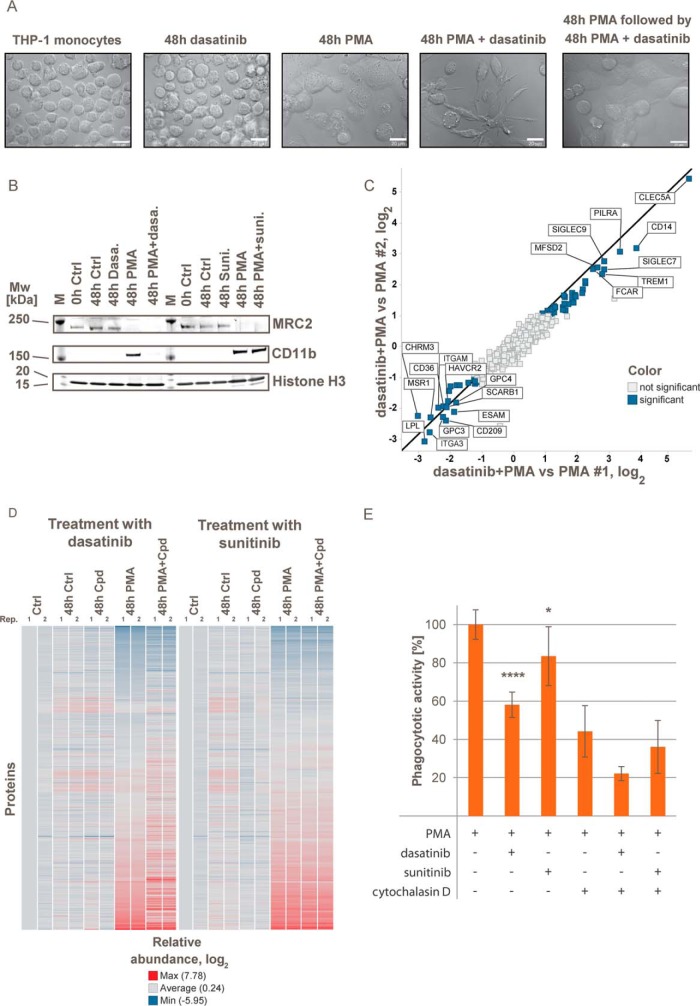
In the combined dataset, abundance profiles were generated for 68 plasma membrane-associated kinases. Among these, 14 kinases were significantly regulated after 24 h during monocyte to macrophage differentiation, including KIT and ephrin receptors EPHA3, EPHA7, and EPHB1 (Fig. 5B and supplemental Fig. S3B and supplemental Table S3) suggesting substantial rewiring of signaling networks. We next tested how cells react to perturbation of signaling processes specific to the monocyte to macrophage differentiation with small molecule kinase inhibitors. Treatment of THP-1 cells with the BCR-Abl inhibitor imatinib (56), the broad specificity inhibitor sunitinib (57), and the ABL and Src family kinase inhibitor dasatinib (58) did not affect the morphology, viability, and proliferation of undifferentiated THP-1 cells within 48 h (Fig. 6A and supplemental Fig. S4A). However, dasatinib but none of the other two inhibitors induced severe morphological changes when present for 48 h during PMA-induced differentiation leading to cells being more dendritic, less flat, and less adherent. In contrast, when dasatinib was applied to already differentiated cells, it had no influence on the morphology.
Fig. 6.
THP-1 differentiation in presence of dasatinib causes changes in cellular morphology, cell-surface marker presentation and phagocytotic activity. A, morphological differences between monocytic THP-1 cells after cultivation or PMA differentiation in the presence or absence of 1 μm dasatinib for 48 h monitored by light microscopy. Differentiation in the presence of dasatinib results in a less adherent and more dendritic cell shape. Addition of dasatinib to already differentiated cells has no influence on the morphology. B, immunoblots for cell-surface markers regulated during differentiation of THP-1 cells by PMA (M: molecular weight marker). C, proteins with significantly altered abundance in THP-1 cells differentiated in presence or absence of dasatinib. D, heat map representation of quantified plasma membrane-associated proteins during differentiation of monocytic THP-1 to macrophage-like cells in presence or absence of dasatinib and sunitinib. Colors represent log2 relative abundance to untreated controls. E, phagocytosis assay in differentiated THP-1 cells in absence or presence of 2 μm cytochalasin D. Mean of four replicates; error bars represent standard deviations. See also supplemental Fig. S4.
We next compared how dasatinib and sunitinib affected the surface proteomes of monocytic or macrophage-like THP-1 cells as well as during PMA-induced differentiation using a TMT-based quantitative mass pectrometry strategy and immunoblotting. Sunitinib did not induce any significant changes when applied during cell differentiation (Fig. 6D and supplemental Fig. S4B). In contrast, if dasatinib was present during differentiation up-regulation of CD11b was blocked, although MRC2 down-regulation was not affected (Fig. 6B). In total, abundances of 56 cell-surface proteins were significantly altered after differentiation in the presence of dasatinib compared with control differentiation experiments (Fig. 6C and supplemental Table S4). Pronounced up-regulation was observed for 35 proteins, including CLEC5a (47-fold up), a positive regulator of osteoclastogenesis (59), as well as TREM1, PILRA, and CD14, which together with CLEC5a are markers for mature myeloid cells (60–62). Substantial down-regulation of 21 proteins was observed, including macrophage markers like CD209, CD22, HAVCR2, and TLR4 suggesting compromised macrophage-specific functions. Up-regulation of cell adhesion proteins like ESAM, ITGA3, ITGAE, and ITGAM during differentiation from monocytes to macrophages was reduced or completely blocked in the presence of dasatinib in line with a less adherent cellular morphology (supplemental Table S4). In addition, we observed reduced expression of SCARB1, MSR1, CD36, and LPL, which are involved in phagocytosis as well as fatty acid, cholesterol, and lipid endocytosis. In agreement with these findings, the phagocytotic activity of THP-1 cells differentiated in the presence of dasatinib was reduced by >40% but was not significantly changed for cells differentiated in the presence of sunitinib (Fig. 6E). Addition of the phagocytosis inhibitor cytochalasin D (2 μm) resulted in partial but significant reduction of the phagocytotic activity in all samples. Because CLEC5a has a role in osteoclastogenesis, we went back to the above described cell-surface proteome inventory in which we compared the cell-surface proteome of THP-1 cells differentiated in the presence of dasatinib with osteoclast-like THP-1 cells obtained by differentiation using vitamin D3 (63) and with monocytic and macrophage-like THP-1 cells. Principal component analysis of plasma membrane protein abundances showed cells differentiated with PMA in the presence of dasatinib to be related to both osteoclast-like and macrophage-like THP-1 cells. This indicates dysregulation of PMA-induced differentiation by dasatinib rather than mere inhibition (Fig. 2A).
The identification of the targets responsible for dysregulation of THP-1 cell differentiation is challenging due to the limited selectivity of the marketed BCR-Abl inhibitor dasatinib. Differential analysis of the selectivity profiles obtained for the tested inhibitors using kinobeads (supplemental Fig. S5 and supplemental Table S5) (64) suggested that neither Abl kinase inhibition nor any of the other kinases affected by imatinib or sunitinib are responsible for the observed dysregulation of the differentiation program. Ephrin receptor tyrosine kinases are potently inhibited by dasatinib but not by imatinib or sunitinib. Because of the observed regulation of ephrin receptors during macrophage differentiation, and the established role of ephrins for regulation of cell differentiation (65) and regulation of bone homeostasis (66), it seems very likely that off-target inhibition of ephrin kinases by dasatinib caused dysregulated macrophage differentiation.
DISCUSSION
We mapped the cell-surface proteomes of commonly used established cell lines and primary cells at a previously unprecedented depth detecting on average >800 plasma membrane proteins per cell type and >2400 distinct cell-surface proteins in 21 different cell systems. These numbers compared very favorably to the 300 proteins per cell type and in total 1492 cell-surface proteins from 41 human cell types detected in a recent publication (18). The increased coverage achieved in this study can be attributed to more efficient labeling of surface proteins by including aniline as catalyst, enrichment on protein level, and improved MS instrumentation. Our cell-surface compendium facilitates selecting the appropriate cell system in which proteins of interest are expressed in sufficient amounts to monitor responsiveness to external stimuli, cell signaling, or drug transport. The marked differences observed in surface proteome composition between primary cell systems and corresponding cell lines of similar origin are in line with altered responsiveness to many stimuli and differences in drug sensitivity (67–69). Growth rate is an intrinsic selection criterion for cell lines in culture favoring high nutrient uptake. This can be achieved by increased transporter expression (70), e.g. of solute carrier proteins that might in turn be hijacked for transporter-mediated drug uptake (12, 13, 16). Lymphoid primary cells display much lower expression of transporters than corresponding cell lines suggesting that data on drug uptake and drug potency for such cell lines might not be predictive for the in vivo situation and that candidate drug molecules should be tested in primary systems resembling the in vivo state as closely as possible (71).
Previous studies (5), as well as the data presented here, demonstrated major changes in the cell-surface proteome between the end points of cellular differentiation. However, relatively little is known about the mechanisms underlying such differentiation processes, and published studies are mainly based on transcript profiling (72). With our time-resolved cell-surface proteomics approach, we analyzed the changes of monocyte to macrophage differentiation by making use of the high multiplexing capabilities of tandem mass tag-based quantitative mass spectrometry. Our approach for the first time enabled the comprehensive analysis of the dynamic remodeling processes of the cell-surface proteome during differentiation of THP-1 cells into macrophages and provided unique insights into the underlying mechanisms. Remodeling on the cell surface occurs in three major steps. In a first step, collagens and other extracellular matrix proteins are secreted, and proteins such as CD83 that are stored intracellularly are relocalized to the plasma membrane (55). Morphologically, the initial step manifests as the cells start to become adherent, suggesting that remodeling of the ECM and relocalization of PLAVB, CD83, and other proteins induce initial adherence of THP-1 cells. The second step that partially overlaps with step 1 is dominated by internalization or degradation of no longer required surface markers, transporters, and proteins regulating cell proliferation. Although internalization of some monocyte markers such as CD4 is very fast, abundances of some receptor tyrosine kinases, such as the ephrin receptors, steadily decrease over the course of 2 days. The third step is dominated by delayed presentation of newly synthesized proteins at the plasma membrane that reflect altered nutritional requirements and function of non-dividing macrophages. It is surprising to see that with a few exceptions, such as CD83, hardly any proteins are transiently regulated. The delayed de novo synthesis following degradation of no longer needed proteins suggests a highly energy efficient process that enables differentiation without temporarily increasing biomass.
We observed a major change in morphology and function of macrophages, when differentiation of monocytes by PMA was performed in the presence of the marketed kinase inhibitor dasatinib. In recent reports, dasatinib has been suggested to influence differentiation processes in vivo and in vitro, e.g. enhancing megakaryocyte differentiation and inhibition of platelet formation in mice (73) and promotion of retinoic acid-induced differentiation of AML cells (74). Here, we provide a detailed description of alterations in the cell-surface proteome when monocytes are differentiated in the presence of dasatinib. We found up-regulation of CLEC5A described to be involved in osteoclastogenesis. Interestingly, dasatinib has already been described to affect osteoclasts by acting as a bone-modifying agent through enhancing the differentiation and function of osteoblasts while inhibiting osteoclast differentiation (75). These observations suggest that patients treated with dasatinib might suffer from a weakened immune system due to impaired replenishment with newly differentiated functional macrophages. As only some of the classic macrophage cell-surface markers are affected, targeted methods using immunostaining or FACS might not spot this drug effect. Sunitinib and imatinib, two other marketed kinase inhibitors, do not elicit this response. Because imatinib and dasatinib were both designed as Abl kinase inhibitors, an off-target activity of dasatinib must be responsible for the impaired differentiation. Ephrin receptors are potently hit by dasatinib, they are expressed in these cells, and their presentation is regulated during differentiation. These data and the known role of ephrin receptor tyrosine kinases in differentiation processes (65) are suggestive for these proteins being the dasatinib off-targets responsible for the observed dysregulation of the differentiation into macrophages. However, yet no selective ephrin receptor inhibitors are available hampering further evaluation of this hypothesis.
The methods presented here allow the detailed analysis of time- and stimulus-dependent effects on the cell-surface proteome to a depth of more than 1000 cell-surface proteins in a single experiment. In contrast to methods monitoring transcript levels of known surface proteins, our approach enables detection of altered subcellular localization and proteins secreted from intracellular vesicles. The coverage of the cell-surface proteome that can be achieved with this mass spectrometry-based strategy greatly exceeds multiplexed immunohistochemistry-based approaches (76) and is unbiased in nature. Using conventional fluorescence-based detection methods such as FACS, depletion of a specific cell population upon drug treatment can hardly be distinguished from perturbations in the cell-surface proteome masking detection. It is important to note, however, that the presented enrichment technique is primarily (5) based on selective enrichment of sialylated cell-surface proteins, and non-sialylated glycoproteins may be overlooked. Also potential stimulus-dependent effects on protein sialylation or modifications might impact results. Quantitative mass spectrometry allows for the comprehensive analysis of perturbations yielding a defined cell surface proteotype that is characteristic for off-target-mediated dysregulation of cell differentiation by a bioactive compound, for example. These findings suggest to include cell differentiation assays more rigorously in preclinical testing of candidate drugs to detect such adverse drug effects at an early stage in drug discovery.
DATA AVAILABILITY
The mass spectrometry proteomics data have been deposited to the ProteomeXchange Consortium via the PRIDE partner repository with the dataset identifier PXD005846 (https://www.ebi.ac.uk/pride).
Supplementary Material
Acknowledgments
We are grateful to A. Sinz for discussions and support. We thank J. Stuhlfauth for support in cell culture; M. Bösche, K. Kammerer, M. Klös-Hudak, T. Rudi, and M. Steidel for expert technical assistance and T. Mathieson and C. Fufezan for providing scripts for data upload and spectra annotation.
Footnotes
Author contributions: M.B. and H.C.E. conceived and supervised the project. M.K. performed experiments. M.K., S.G., H.C.E., and M.B. analyzed data and M.K., H.C.E., and M.B. wrote the manuscript.
* This work was funded by Cellzome GmbH / GlaxoSmithKline. All authors are employees of this company.
 This article contains supplemental material.
This article contains supplemental material.
1 The abbreviations used are:
- GPCR
- G-protein coupled receptor
- ABC transporter
- ATP-binding cassette transporter
- CD
- clusters of differentiation
- FCS
- fetal calf serum
- FDA
- food and drug administration
- FDR
- false discovery rate
- GO
- gene onthology
- HCD
- higher energy collisional dissociation
- IAA
- iodoacetamide
- NCE
- normalized collision energy
- PMA
- phorbol-12-myristate-13-acetate
- SDS
- sodium dodecyl sulfate
- SLC
- solute carrier
- TEAB
- triethylammonium bicarbonate
- TMT
- tandem mass tag
- MEM
- minimum essential medium
- DMEM
- Dulbecco's modified eagle medium
- PNGase
- peptide:N-glycosidase
- ECM
- extracellular matrix.
REFERENCES
- 1. Neugebauer K. M., and Reichardt L. F. (1991) Cell-surface regulation of β1-integrin activity on developing retinal neurons. Nature 350, 68–71 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2. Anderle P., Huang Y., and Sadée W. (2004) Intestinal membrane transport of drugs and nutrients: genomics of membrane transporters using expression microarrays. Eur. J. Pharm. Sci. 21, 17–24 [DOI] [PubMed] [Google Scholar]
- 3. Li Y., Massey K., Witkiewicz H., and Schnitzer J. E. (2011) Systems analysis of endothelial cell plasma membrane proteome of rat lung microvasculature. Proteome Sci. 9, 15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Navarro G., Cordomí A., Zelman-Femiak M., Brugarolas M., Moreno E., Aguinaga D., Perez-Benito L., Cortés A., Casadó V., Mallol J., Canela E. I., Lluís C., Pardo L., García-Sáez A. J., McCormick P. J., and Franco R. (2016) Quaternary structure of a G-protein-coupled receptor heterotetramer in complex with Gi and Gs. BMC Biol. 14, 26. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Wollscheid B., Bausch-Fluck D., Henderson C., O'Brien R., Bibel M., Schiess R., Aebersold R., and Watts J. D. (2009) Mass-spectrometric identification and relative quantification of N-linked cell surface glycoproteins. Nat. Biotechnol. 27, 378–386 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Hanahan D., and Weinberg R. A. (2000) The hallmarks of cancer. Cell 100, 57–70 [DOI] [PubMed] [Google Scholar]
- 7. Klabunde T., and Hessler G. (2002) Drug design strategies for targeting G-protein-coupled receptors. Chembiochem 3, 928–944 [DOI] [PubMed] [Google Scholar]
- 8. Overington J. P., Al-Lazikani B., and Hopkins A. L. (2006) How many drug targets are there? Nat. Rev. Drug Discov. 5, 993–996 [DOI] [PubMed] [Google Scholar]
- 9. Lindsley C. W. (2015) 2014 global prescription medication statistics: strong growth and CNS well represented. ACS Chem. Neurosci. 6, 505–506 [DOI] [PubMed] [Google Scholar]
- 10. Gottesman M. M., Fojo T., and Bates S. E. (2002) Multidrug resistance in cancer: role of ATP-dependent transporters. Nat. Rev. Cancer 2, 48–58 [DOI] [PubMed] [Google Scholar]
- 11. Di L., Artursson P., Avdeef A., Ecker G. F., Faller B., Fischer H., Houston J. B., Kansy M., Kerns E. H., Krämer S. D., Lennernäs H., and Sugano K. (2012) Evidence-based approach to assess passive diffusion and carrier-mediated drug transport. Drug Discov. Today 17, 905–912 [DOI] [PubMed] [Google Scholar]
- 12. Dobson P. D., and Kell D. B. (2008) Carrier-mediated cellular uptake of pharmaceutical drugs: an exception or the rule? Nat. Rev. Drug Discov. 7, 205–220 [DOI] [PubMed] [Google Scholar]
- 13. Kell D. B., Dobson P. D., Bilsland E., and Oliver S. G. (2013) The promiscuous binding of pharmaceutical drugs and their transporter-mediated uptake into cells: what we (need to) know and how we can do so. Drug Discov. Today 18, 218–239 [DOI] [PubMed] [Google Scholar]
- 14. Cha S. H., Sekine T., Fukushima J. I., Kanai Y., Kobayashi Y., Goya T., and Endou H. (2001) Identification and characterization of human organic anion transporter 3 expressing predominantly in the kidney. Mol. Pharmacol. 59, 1277–1286 [DOI] [PubMed] [Google Scholar]
- 15. Kimura N., Masuda S., Tanihara Y., Ueo H., Okuda M., Katsura T., and Inui K. (2005) Metformin is a superior substrate for renal organic cation transporter OCT2 rather than hepatic OCT1. Drug Metab. Pharmacokinet. 20, 379–386 [DOI] [PubMed] [Google Scholar]
- 16. Winter G. E., Radic B., Mayor-Ruiz C., Blomen V. A., Trefzer C., Kandasamy R. K., Huber K. V., Gridling M., Chen D., Klampfl T., Kralovics R., Kubicek S., Fernandez-Capetillo O., Brummelkamp T. R., and Superti-Furga G. (2014) The solute carrier SLC35F2 enables YM155-mediated DNA damage toxicity. Nat. Chem. Biol. 10, 768–773 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Pshezhetsky A. V., Fedjaev M., Ashmarina L., Mazur A., Budman L., Sinnett D., Labuda D., Beaulieu J. F., Ménard D., Nifant'ev I., and Levy E. (2007) Subcellular proteomics of cell differentiation: quantitative analysis of the plasma membrane proteome of Caco-2 cells. Proteomics 7, 2201–2215 [DOI] [PubMed] [Google Scholar]
- 18. Bausch-Fluck D., Hofmann A., Bock T., Frei A. P., Cerciello F., Jacobs A., Moest H., Omasits U., Gundry R. L., Yoon C., Schiess R., Schmidt A., Mirkowska P., Härtlová A., Van Eyk J. E., et al. (2015) A mass spectrometric-derived cell-surface protein atlas. PLoS One 10, e0121314. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Graessel A., Hauck S. M., von Toerne C., Kloppmann E., Goldberg T., Koppensteiner H., Schindler M., Knapp B., Krause L., Dietz K., Schmidt-Weber C. B., and Suttner K. (2015) A combined omics approach to generate the surface atlas of human naive CD4+ T cells during early T-cell receptor activation. Mol. Cell. Proteomics 14, 2085–2102 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Deleted in proof.
- 21. Rappsilber J., Mann M., and Ishihama Y. (2007) Protocol for micro-purification, enrichment, pre-fractionation and storage of peptides for proteomics using StageTips. Nat. Protoc. 2, 1896–1906 [DOI] [PubMed] [Google Scholar]
- 22. Kruse U., Pallasch C. P., Bantscheff M., Eberhard D., Frenzel L., Ghidelli S., Maier S. K., Werner T., Wendtner C. M., and Drewes G. (2011) Chemoproteomics-based kinome profiling and target deconvolution of clinical multi-kinase inhibitors in primary chronic lymphocytic leukemia cells. Leukemia 25, 89–100 [DOI] [PubMed] [Google Scholar]
- 23. Franken H., Mathieson T., Childs D., Sweetman G. M., Werner T., Tögel I., Doce C., Gade S., Bantscheff M., Drewes G., Reinhard F. B., Huber W., and Savitski M. M. (2015) Thermal proteome profiling for unbiased identification of direct and indirect drug targets using multiplexed quantitative mass spectrometry. Nat. Protoc. 10, 1567–1593 [DOI] [PubMed] [Google Scholar]
- 24. Silva J. C., Gorenstein M. V., Li G. Z., Vissers J. P., and Geromanos S. J. (2006) Absolute quantification of proteins by LCMSE: a virtue of parallel MS acquisition. Mol. Cell. Proteomics 5, 144–156 [DOI] [PubMed] [Google Scholar]
- 25. Becher I., Savitski M. M., Savitski M. F., Hopf C., Bantscheff M., and Drewes G. (2013) Affinity profiling of the cellular kinome for the nucleotide cofactors ATP, ADP, and GTP. ACS Chem. Biol. 8, 599–607 [DOI] [PubMed] [Google Scholar]
- 26. Savitski M. M., Sweetman G., Askenazi M., Marto J. A., Lang M., Zinn N., and Bantscheff M. (2011) Delayed fragmentation and optimized isolation width settings for improvement of protein identification and accuracy of isobaric mass tag quantification on orbitrap-type mass spectrometers. Anal. Chem. 83, 8959–8967 [DOI] [PubMed] [Google Scholar]
- 27. Savitski M. M., Fischer F., Mathieson T., Sweetman G., Lang M., and Bantscheff M. (2010) Targeted data acquisition for improved reproducibility and robustness of proteomic mass spectrometry assays. J. Am. Soc. Mass Spectrom. 21, 1668–1679 [DOI] [PubMed] [Google Scholar]
- 28. Savitski M. M., Mathieson T., Zinn N., Sweetman G., Doce C., Becher I., Pachl F., Kuster B., and Bantscheff M. (2013) Measuring and managing ratio compression for accurate iTRAQ/TMT quantification. J. Proteome Res. 12, 3586–3598 [DOI] [PubMed] [Google Scholar]
- 29. Deeb S. J., D'Souza R. C., Cox J., Schmidt-Supprian M., and Mann M. (2012) Super-SILAC allows classification of diffuse large B-cell lymphoma subtypes by their protein expression profiles. Mol. Cell. Proteomics 11, 77–89 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Smyth G. K. (2004) Linear models and empirical Bayes methods for assessing differential expression in microarray experiments. Stat. Appl. Genet. Mol. Biol. 3, Article3. [DOI] [PubMed] [Google Scholar]
- 31. Huang da W., Sherman B. T., and Lempicki R. A. (2009) Systematic and integrative analysis of large gene lists using DAVID bioinformatics resources. Nat. Protoc. 4, 44–57 [DOI] [PubMed] [Google Scholar]
- 32. Alexa A., and Rahnenfuhrer J. (2015) topGO: topGO: enrichment analysis for gene ontology, version 2.18.0 [Google Scholar]
- 33. Mellacheruvu D., Wright Z., Couzens A. L., Lambert J. P., St-Denis N. A., Li T., Miteva Y. V., Hauri S., Sardiu M. E., Low T. Y., Halim V. A., Bagshaw R. D., Hubner N. C., Al-Hakim A., Bouchard A., et al. (2013) The CRAPome: a contaminant repository for affinity purification-mass spectrometry data. Nat. Methods 10, 730–736 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Zeng Y., Ramya T. N., Dirksen A., Dawson P. E., and Paulson J. C. (2009) High-efficiency labeling of sialylated glycoproteins on living cells. Nat. Methods 6, 207–209 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. Geiger T., Wehner A., Schaab C., Cox J., and Mann M. (2012) Comparative proteomic analysis of 11 common cell lines reveals ubiquitous but varying expression of most proteins. Mol. Cell. Proteomics 11, M111.014050. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36. Mueckler M., and Thorens B. (2013) The SLC2 (GLUT) family of membrane transporters. Mol. Aspects Med. 34, 121–138 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37. Traiffort E., O'Regan S., and Ruat M. (2013) The choline transporter-like family SLC44: properties and roles in human diseases. Mol. Aspects Med. 34, 646–654 [DOI] [PubMed] [Google Scholar]
- 38. Palmieri F. (2013) The mitochondrial transporter family SLC25: identification, properties and physiopathology. Mol. Aspects Med. 34, 465–484 [DOI] [PubMed] [Google Scholar]
- 39. Kanai Y., Clémençon B., Simonin A., Leuenberger M., Lochner M., Weisstanner M., and Hediger M. A. (2013) The SLC1 high-affinity glutamate and neutral amino acid transporter family. Mol. Aspects Med. 34, 108–120 [DOI] [PubMed] [Google Scholar]
- 40. Fotiadis D., Kanai Y., and Palacín M. (2013) The SLC3 and SLC7 families of amino acid transporters. Mol. Aspects Med. 34, 139–158 [DOI] [PubMed] [Google Scholar]
- 41. Schiöth H. B., Roshanbin S., Hägglund M. G., and Fredriksson R. (2013) Evolutionary origin of amino acid transporter families SLC32, SLC36, and SLC38 and physiological, pathological, and therapeutic aspects. Mol. Aspects Med. 34, 571–585 [DOI] [PubMed] [Google Scholar]
- 42. Young J. D., Yao S. Y., Baldwin J. M., Cass C. E., and Baldwin S. A. (2013) The human concentrative and equilibrative nucleoside transporter families, SLC28 and SLC29. Mol. Aspects Med. 34, 529–547 [DOI] [PubMed] [Google Scholar]
- 43. Li Q., and Shu Y. (2014) Role of solute carriers in response to anticancer drugs. Mol. Cell Ther. 2, 15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44. Groot-Kormelink P. J., Fawcett L., Wright P. D., Gosling M., and Kent T. C. (2012) Quantitative GPCR and ion channel transcriptomics in primary alveolar macrophages and macrophage surrogates. BMC. Immunol. 13, 57. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45. Daigneault M., Preston J. A., Marriott H. M., Whyte M. K., and Dockrell D. H. (2010) The identification of markers of macrophage differentiation in PMA-stimulated THP-1 cells and monocyte-derived macrophages. PLoS One 5, e8668. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46. Werner T., Sweetman G., Savitski M. F., Mathieson T., Bantscheff M., and Savitski M. M. (2014) Ion coalescence of neutron encoded TMT 10-plex reporter ions. Anal. Chem. 86, 3594–3601 [DOI] [PubMed] [Google Scholar]
- 47. Schwende H., Fitzke E., Ambs P., and Dieter P. (1996) Differences in the state of differentiation of THP-1 cells induced by phorbol ester and 1,25-dihydroxyvitamin D3. J. Leukocyte Biol. 59, 555–561 [PubMed] [Google Scholar]
- 48. Sintiprungrat K., Singhto N., Sinchaikul S., Chen S. T., and Thongboonkerd V. (2010) Alterations in cellular proteome and secretome upon differentiation from monocyte to macrophage by treatment with phorbol myristate acetate: insights into biological processes. J. Proteomics 73, 602–618 [DOI] [PubMed] [Google Scholar]
- 49. Auwerx J., Staels B., Van Vaeck F., and Ceuppens J. L. (1992) Changes in IgG Fc receptor expression induced by phorbol 12-myristate 13-acetate treatment of THP-1 monocytic leukemia cells. Leuk. Res. 16, 317–327 [DOI] [PubMed] [Google Scholar]
- 50. Kerr S. C., Fieger C. B., Snapp K. R., and Rosen S. D. (2008) Endoglycan, a member of the CD34 family of sialomucins, is a ligand for the vascular selectins. J. Immunol. 181, 1480–1490 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51. Cook G. A., Longhurst C. M., Grgurevich S., Cholera S., Crossno J. T. Jr., and Jennings L. K. (2002) Identification of CD9 extracellular domains important in regulation of CHO cell adhesion to fibronectin and fibronectin pericellular matrix assembly. Blood 100, 4502–4511 [DOI] [PubMed] [Google Scholar]
- 52. Sgroi D., and Stamenkovic I. (1994) The B-cell adhesion molecule CD22 is cross-species reactive and recognizes distinct sialoglycoproteins on different functional T-cell sub-populations. Scand. J. Immunol. 39, 433–438 [DOI] [PubMed] [Google Scholar]
- 53. Sims M. A., Field S. D., Barnes M. R., Shaikh N., Ellington K., Murphy K. E., Spurr N., and Campbell D. A. (2000) Cloning and characterisation of ITGAV, the genomic sequence for human cell adhesion protein (vitronectin) receptor α subunit, CD51. Cytogenet. Cell Genet. 89, 268–271 [DOI] [PubMed] [Google Scholar]
- 54. Messaritou G., East L., Roghi C., Isacke C. M., and Yarwood H. (2009) Membrane type-1 matrix metalloproteinase activity is regulated by the endocytic collagen receptor Endo180. J. Cell Sci. 122, 4042–4048 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55. Cao W., Lee S. H., and Lu J. (2005) CD83 is preformed inside monocytes, macrophages and dendritic cells, but it is only stably expressed on activated dendritic cells. Biochem. J. 385, 85–93 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56. Capdeville R., Buchdunger E., Zimmermann J., and Matter A. (2002) Glivec (STI571, imatinib), a rationally developed, targeted anticancer drug. Nat. Rev. Drug Discov. 1, 493–502 [DOI] [PubMed] [Google Scholar]
- 57. Sun L., Liang C., Shirazian S., Zhou Y., Miller T., Cui J., Fukuda J. Y., Chu J. Y., Nematalla A., Wang X., Chen H., Sistla A., Luu T. C., Tang F., Wei J., and Tang C. (2003) Discovery of 5-[5-fluoro-2-oxo-1,2-dihydroindol-(3Z)-ylidenemethyl]-2,4-dimethyl-1H-pyrrole-3-carboxylic acid (2-diethylaminoethyl)amide, a novel tyrosine kinase inhibitor targeting vascular endothelial and platelet-derived growth factor receptor tyrosine kinase. J. Med. Chem. 46, 1116–1119 [DOI] [PubMed] [Google Scholar]
- 58. Kantarjian H., Jabbour E., Grimley J., and Kirkpatrick P. (2006) Dasatinib. Nat. Rev. Drug Discov. 5, 717–718 [DOI] [PubMed] [Google Scholar]
- 59. Inui M., Kikuchi Y., Aoki N., Endo S., Maeda T., Sugahara-Tobinai A., Fujimura S., Nakamura A., Kumanogoh A., Colonna M., and Takai T. (2009) Signal adaptor DAP10 associates with MDL-1 and triggers osteoclastogenesis in cooperation with DAP12. Proc. Natl. Acad. Sci. U.S.A. 106, 4816–4821 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60. Fournier N., Chalus L., Durand I., Garcia E., Pin J. J., Churakova T., Patel S., Zlot C., Gorman D., Zurawski S., Abrams J., Bates E. E., and Garrone P. (2000) FDF03, a novel inhibitory receptor of the immunoglobulin superfamily, is expressed by human dendritic and myeloid cells. J. Immunol. 165, 1197–1209 [DOI] [PubMed] [Google Scholar]
- 61. Gingras M. C., Lapillonne H., and Margolin J. F. (2002) TREM-1, MDL-1, and DAP12 expression is associated with a mature stage of myeloid development. Mol. Immunol. 38, 817–824 [DOI] [PubMed] [Google Scholar]
- 62. Ritter M. R., Reinisch J., Friedlander S. F., and Friedlander M. (2006) Myeloid cells in infantile hemangioma. Am. J. Pathol. 168, 621–628 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63. Park K. Y., Li W. A., and Platt M. O. (2012) Patient specific proteolytic activity of monocyte-derived macrophages and osteoclasts predicted with temporal kinase activation states during differentiation. Integr. Biol. 4, 1459–1469 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64. Bantscheff M., Eberhard D., Abraham Y., Bastuck S., Boesche M., Hobson S., Mathieson T., Perrin J., Raida M., Rau C., Reader V., Sweetman G., Bauer A., Bouwmeester T., Hopf C., et al. (2007) Quantitative chemical proteomics reveals mechanisms of action of clinical ABL kinase inhibitors. Nat. Biotechnol. 25, 1035–1044 [DOI] [PubMed] [Google Scholar]
- 65. Wilkinson D. G. (2014) Regulation of cell differentiation by Eph receptor and ephrin signaling. Cell Adh. Migr. 8, 339–348 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66. Edwards C. M., and Mundy G. R. (2008) Eph receptors and ephrin signaling pathways: a role in bone homeostasis. Int. J. Med. Sci. 5, 263–272 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67. Dobson-Belaire W. N., Cochrane A., Ostrowski M. A., and Gray-Owen S. D. (2011) Differential response of primary and immortalized CD4+ T cells to Neisseria gonorrhoeae-induced cytokines determines the effect on HIV-1 replication. PLoS One 6, e18133. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68. Lidington E. A., Moyes D. L., McCormack A. M., and Rose M. L. (1999) A comparison of primary endothelial cells and endothelial cell lines for studies of immune interactions. Transpl. Immunol. 7, 239–246 [DOI] [PubMed] [Google Scholar]
- 69. Thonemann B., Schmalz G., Hiller K. A., and Schweikl H. (2002) Responses of L929 mouse fibroblasts, primary and immortalized bovine dental papilla-derived cell lines to dental resin components. Dent. Mater. 18, 318–323 [DOI] [PubMed] [Google Scholar]
- 70. McCracken A. N., and Edinger A. L. (2013) Nutrient transporters: the Achilles' heel of anabolism. Trends Endocrinol. Metab. 24, 200–208 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71. Cree I. A., Glaysher S., and Harvey A. L. (2010) Efficacy of anti-cancer agents in cell lines versus human primary tumour tissue. Curr. Opin. Pharmacol. 10, 375–379 [DOI] [PubMed] [Google Scholar]
- 72. Martinez F. O., Gordon S., Locati M., and Mantovani A. (2006) Transcriptional profiling of the human monocyte-to-macrophage differentiation and polarization: new molecules and patterns of gene expression. J. Immunol. 177, 7303–7311 [DOI] [PubMed] [Google Scholar]
- 73. Mazharian A., Ghevaert C., Zhang L., Massberg S., and Watson S. P. (2011) Dasatinib enhances megakaryocyte differentiation but inhibits platelet formation. Blood 117, 5198–5206 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74. Kropf P. L., Wang L., Zang Y., Redner R. L., and Johnson D. E. (2010) Dasatinib promotes ATRA-induced differentiation of AML cells. Leukemia 24, 663–665 [DOI] [PubMed] [Google Scholar]
- 75. Garcia-Gomez A., Ocio E. M., Crusoe E., Santamaria C., Hernández-Campo P., Blanco J. F., Sanchez-Guijo F. M., Hernández-Iglesias T., Briñón J. G., Fisac-Herrero R. M., Lee F. Y., Pandiella A., San Miguel J. F., and Garayoa M. (2012) Dasatinib as a bone-modifying agent: anabolic and anti-resorptive effects. PLoS One 7, e34914. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76. Levenson R. M., Borowsky A. D., and Angelo M. (2015) Immunohistochemistry and mass spectrometry for highly multiplexed cellular molecular imaging. Lab. Invest. 95, 397–405 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
The mass spectrometry proteomics data have been deposited to the ProteomeXchange Consortium via the PRIDE partner repository with the dataset identifier PXD005846 (https://www.ebi.ac.uk/pride).