Abstract
Multiple (selected) reaction monitoring (MRM/SRM) of peptides is a growing technology for target protein quantification because it is more robust, precise, accurate, high-throughput, and multiplex-capable than antibody-based techniques. The technique has been applied clinically to the large-scale quantification of multiple target proteins in different types of fluids. However, previous MRM-based studies have placed less focus on sample-preparation workflow and analytical performance in the precise quantification of proteins in saliva, a noninvasively sampled body fluid. In this study, we evaluated the analytical performance of a simple and robust multiple reaction monitoring (MRM)-based targeted proteomics approach incorporating liquid chromatography with mass spectrometry detection (LC-MRM/MS). This platform was used to quantitatively assess the biomarker potential of a group of 56 salivary proteins that have previously been associated with human cancers. To further enhance the development of this technology for assay of salivary samples, we optimized the workflow for salivary protein digestion and evaluated quantification performance, robustness and technical limitations in analyzing clinical samples. Using a clinically well-characterized cohort of two independent clinical sample sets (total n = 119), we quantitatively characterized these protein biomarker candidates in saliva specimens from controls and oral squamous cell carcinoma (OSCC) patients. The results clearly showed a significant elevation of most targeted proteins in saliva samples from OSCC patients compared with controls. Overall, this platform was capable of assaying the most highly multiplexed panel of salivary protein biomarkers, highlighting the clinical utility of MRM in oral cancer biomarker research.
Multiple reaction monitoring (MRM)1, also known as selected reaction monitoring (SRM), of peptides is a growing technology for target protein quantification because it is more robust, precise, accurate, high-throughput, and multiplex-capable than antibody-based techniques, such as enzyme-linked immunosorbent assays (ELISAs) (1–4). In this MRM-based technique, which incorporates liquid chromatography with mass spectrometry detection (LC-MRM/MS), specific transitions of precursors selected in Q1 to fragment ions selected in Q3 are monitored using a triple-quadrupole MS instrument, generating signals for quantification. The technique has been applied clinically to the large-scale quantification of multiple target proteins in different types of fluids, including plasma/serum (3, 5–7), urine (1, 8, 9), and cerebrospinal fluid (10). However, less focus has been placed on sample-preparation workflow and analytical performance of LC-MRM/MS in the precise quantification of proteins in saliva, particularly in the context of screening for oral cancer biomarkers.
Oral cancer is a type of head and neck cancer in which cancerous tissue growth is located in the oral cavity. A total of 263,900 new cases of oral cancer were reported worldwide in 2008 (11, 12), and this number increased to 300,400 in 2012 (13). Oral cancer is the four-most frequent cancer of incidence rate in Taiwanese males and also had the fourth fastest rate of increase on a case-per-year basis in Taiwan in 2013 (14). The incidence of oral cancer in Taiwan has increased over the past two decades, with the age-standardized incidence in males reaching 42.16/100,000 in 2013, a rate that is higher than that in the United States and countries in Europe (14). Oral squamous cell carcinoma (OSCC) is the most common subtype of oral cavity cancer. The use of betel nut, tobacco, and alcohol products are important risk factors that correlate with the occurrence of oral cancer (15–17). Thus, limiting contact with risk factors for oral cancers and early diagnosis are expected to significantly reduced mortality and morbidity in oral cancer patients (18). Saliva, as the proximal fluid of oral tissue, has been found to contain more than 1000 proteins, mRNAs, microRNAs, and metabolites that span a wide range of biological functions (19, 20). The abnormalities of cancerous oral tissue are associated with dysregulated secretion of molecules into saliva, providing an alternative for non-invasive detection and clinical management of oral diseases. Some proteomic studies have profiled the saliva proteome to discover disease biomarkers, including those for cancer (21, 22), periodontal disease (23), and orthodontic tooth movement (24). Relative quantification of saliva proteins between different groups of samples has been achieved (21, 25, 26), 35 and 158 proteins were quantified by targeted mass spectrometry in normal human saliva recently (27, 28). However, precise quantification of the concentration ranges and clinical utilities of protein biomarkers in disease states were limited.
In this work, we report an optimized workflow for precise quantitation of 56 saliva proteins in a single run using LC-MRM/MS coupled with the use of stable isotope-labeled standard (SIS) peptides. To demonstrate the potential clinical application of this workflow, we evaluated its analytical performance and quantitation ability in two sample sets and compared them between demographically matched control and oral cancer groups for verifying the clinical utility.
Experimental Section
Collection of Clinical Saliva Samples
Ambulatory subjects were recruited from the Oral and Maxillofacial Surgery Clinic, Chi-Mei Medical Center, Liouying, Tainan, Taiwan from 2008 to 2013. The study protocol was approved by the Medical Ethics and Human Clinical Trial Committee of Chi-Mei Medical Center. These patients sought oral cancer screening because of a history of smoking and/or betel quid chewing. Subjects with a history of major salivary gland extirpation or severe mucositis were excluded. Subjects were advised to refrain from eating, drinking, or oral hygiene procedures for at least 1 h before the sample collections. The mucosa of all subjects was categorized as healthy or oral carcinoma based on an oral mucosal examination. After a water mouth rinse, ∼3–4 ml unstimulated whole saliva was collected from all subjects. The collected saliva samples were centrifuged at 910 × g for 15 min at 4 °C to remove the pellet. The resulting cleared supernatants were treated with protease inhibitor mixture (Sigma Chemical Co., St. Louis, MO) and then aliquoted for storage at −80 °C until use. The freeze thaw cycles before sample preparation was not more than two times.
Supplemental Table S1 summarizes the demographic data of the 119 individuals who provided saliva samples used in this study. The diagnoses of OSCC were confirmed by biopsy. The study participants were randomly divided into two independent groups—clinical set 1 contained 32 OSCC samples and set 2 contained 29 OSCC samples; each group contained 29 control samples. The purpose of dividing these samples into two sets was to allow a statistical evaluation of biomarker performance in two independent pools of individual samples. The average concentration values of three technical repeats (independent digestion and MS analysis) for target proteins in individual samples were reported in subsequent analyses. The final performance of the platform, including area under the curve (AUC) and p values of target proteins, were calculated for the total sample (n = 119) by combining clinical sets 1 and 2.
Optimization of Tryptic Digestion of Salivary Proteins and Addition of SIS Peptides
The protein concentration in saliva sample was measured using a BCA Protein Assay Kit (Thermo Scientific Pierce, Rockford, IL). The optimized salivary protein digestion protocol is a modification of a procedure previously reported by Proc et al. (7). In the optimized digestion workflow, the dried sample (15 μg protein) was dissolved in 15 μl of 25 mm ammonium bicarbonate After addition of 15 μl 10% sodium deoxycholate (DOC) to achieve complete dissolution, 81.4 μl of 25 mm ammonium was added to dilute the final DOC concentration. The sample was reduced by incubating with 12.4 μl of 50 mm tris(2-carboxyethyl)phosphine (TCEP) at 60 °C for 30 min, and then alkylated by incubating with 13.8 μl of 100 mm iodoacetamide at 37 °C for 30 min. Modified sequencing-grade trypsin (Promega, Madison, WI) was added to the reduced, alkylated samples at a 20:1 protein/enzyme ratio, and samples were digested at 37 °C for 9 h. The tryptic digestion was stopped by acidifying the sample with 6 μl of 10% formic acid and 1.5 μl of 10% trifluoroacetic acid (TFA) solution that contains a mixture of 56 SIS peptide standards. Each individual saliva protein digest was spiked with a constant amount of an SIS mixture containing 56 [13C6]Lys-, [13C615N2]Lys-, or [13C615N4]Arg-coded SIS peptides, as shown in supplemental Table S2. SIS peptides were synthesized and purified at the UVic-Genome BC Proteomics Centre, and their concentrations were determined as previously described (29) The digested samples were centrifuged for 10 min at 16,000 × g and 4 °C, effectively pelleting DOC and removing it from the digest. The sample was stored at −20 °C before subsequent processing. Samples were desalted and concentrated by solid-phase extraction using a Waters Oasis HLB μElution Plate (Waters, Milford, MA) using the procedure modified according to the manufacturer's protocol. Briefly, the resin in each well was rinsed with acetonitrile and equilibrated with 200 μl equilibration buffer (0.1% TFA and 0.1% formic acid). The salivary protein digest was loaded onto the plate, then washed with water and eluted twice with 25 μl of 70% acetonitrile. Eluted samples were frozen and lyophilized to dryness. The number of the freeze-thaw cycles was consistent across samples. Samples were rehydrated with 0.1% formic acid (v/v) to produce a 0.25 μg/μl concentration for LC-MRM/MS analysis.
LC-MRM/MS Analysis and Data Acquisition
A nanoACQUITY UPLC system was used for the injection and separation of salivary peptides. LC-MRM/MS analysis of each sample took 70 min. Four-microliter samples (representing 1 μg peptides) were injected onto a resolving analytical column (nanoACQUITY UPLC C18, 150 μm × 10 mm, 1.7-μm particle size; Waters) at a flow rate of 1 μl/min in 97% buffer A (0.1% formic acid in water; J.T. Baker, Center Valley, PA) and 3% buffer B (0.1% formic acid in acetonitrile; J.T. Baker) for 10 min. Samples were then separated at a flow rate of 400 nL/min with a 48-min linear gradient from 3% to 28% buffer B, then a 5-min linear gradient from 28% to 38% buffer B, and finally a 1-min linear gradient from 38% to 95% buffer B. The analytical column was then reconditioned by holding buffer B at 95% for 5 min prior to ramping back down to 3% solvent B over 1 min and re-equilibrating for 10 min with 3% buffer B. A blank solvent injection (25-min analysis at 400 nL/min) was run between all samples to prevent sample carryover on the UPLC column.
An AB/MDS Sciex 5500 QTRAP with a nano-electrospray ionization source controlled by Analyst 1.5.1 software (AB Sciex, Framingham, MA) was used for all scheduled LC-MRM/MS analyses. All acquisition methods used the following parameters: ion spray voltage, 1900–2200 V; curtain gas setting, 20 psi (UHP nitrogen); interface heater temperature, 150 °C; and MS operating pressure, 3.5 × 10−5 Torr. Q1 and Q3 were set to unit resolution (0.6–0.8 Da full width at half height). The MRM transition pairs were selected by firstly consideration of criteria mentioned in the Kuzyk's work (30) including the amino acid sequences of the peptides must be unique to the target proteins and contain no missed cleavage sites. The targeted peptide should not contain amino acids Cys and Met. The peptide length was limited between 8–20 amino acids with Q1 ion less than 1000 Da. The pooled saliva samples for calibration curves were then used for manually evaluation of the specificity of the quantifier Q1/Q3 pairs in clinical samples. The Q1/Q3 transition with extensive matrix interference was replaced by another transition for quantification. The MRM acquisition method was constructed using three MRM ion pairs per peptide with fragment-ion-specific tuned declustering potential (DP), entrance potential (EP), collision energy (CE) voltages, collision cell exit potential (CXP), and retention time constraints. For data acquisition, scheduled MRM function was used to reduce cycle times and generate more points per peak for more accurate quantitation. The scheduled MRM option was used for all data acquisition, with a target cycle time of 1 s and a 4-min MRM detection window. Transitions of 56 peptides corresponding to 56 target proteins were quantified in an LC-MRM/MS run.
MRM Data Analysis and Generation of Calibration Curves
All MRM data were processed with MultiQuant software (version 2.1; AB Sciex) using the MQ4 algorithm for peak integration. All the peak areas for concentration calculation were inspected manually to avoid possible interference in the mass spectra. The peaks with intensities lower than 150 in the MRM spectra were not considered for further concentration calculation.
Generation of calibration curves for the 56 peptides was performed essentially as previously described (29, 31). Briefly, a standard curve was generated for each target peptide using different amounts of a tryptic digest from a standard saliva sample, prepared by pooling three individuals (from two OSCC patients and one control) and trypsin-digesting using the same protocol used for all clinical samples. This standard saliva sample, which contained a large number of endogenous proteins, was spiked with a constant amount of SIS peptides, as indicated in supplemental Table S2. The composition of SIS mixture was adjusted according to the concentration levels and signal intensities of endogenous salivary peptides to warrant the quantitation accuracy (31). This standard saliva sample was then used to generate an 11-point dilution curve (including a blank) in which the SIS peptide concentration was held constant and the light peptide concentration was varied by appropriately diluting the tryptic digest. The standard saliva sample (sample I) was kept at the same concentration as the unknowns (0.9 μg endogenous peptides injected). The amounts of the endogenous peptides injected on column were 9.0 pg, 90.0 pg, 4.5 ng, 9.0 ng, 45.0 ng, 90.0 ng, 180.0 ng, 450.0 ng, 0.9 μg, and 1.8 μg for samples A–J, respectively. A fixed amount of the 56-SIS-peptide mixture was spiked into samples A to J. The concentration of each SIS peptide is accurately known, and the concentration of the protein in the unknown samples can be determined from the observed peak area ratios. Three technical repeats were performed independently (from digestion to the final LC-MRM/MS step) for each of the individual clinical samples and for each of the concentration points of calibration curves. The average concentration values of the three technical repeats for each concentration point were reported in subsequent analyses. A linear regression was performed on all calibration curves using a standard 1/x (x = concentration ratio) weighting option to assist in covering a wide dynamic range. Three MRM ion pairs were measured per peptide, with one being used as the quantifier and the other two being used to verify retention times and reveal any signal interference. All MRM peaks were inspected manually to ensure correct peak detection and accurate integration.
A fixed amount of the 56-SIS-peptide mixture was added to each of the individual clinical saliva sample as internal standards for quantification. To perform statistical analyses, we assigned concentrations of proteins without detectable peaks a value of zero (29). The concentration of each target protein was expressed as fmol/μg and ng/ml of salivary protein, as derived from the determined molar concentration of each proteotypic peptide, assuming complete tryptic digestion and 100% peptide recovery.
Assessments of Coefficient of Variation, Limit of Detection, and Limit of Quantitation Values for Detection of 56-plexed Salivary Peptides by MRM/MS
The limit of detection (LOD) was defined as the lowest level at which a signal is observed for the endogenous target peptide in all three replicates with a signal-to-noise ratio (S/N) > 20. The low limit of quantitation (LLOQ) was defined as the lowest concentration of endogenous peptide that could be measured with a coefficient of variation (CV) < 20% (n = 3) and quantitative error less than 20% compared with the theoretical concentration (29). Reproducibility performance (CV) was determined by performing MRM/MS analyses three times using the three processed replicates of the standard saliva sample (sample I of calibration curves).
Immunobead-based Suspension Array System for Detection of Saliva Apolipoproteins
The saliva levels of three apolipoproteins (APOA1, A2, and E) were determined with the MILLIPLEX MAP Human Apolipoprotein Magnetic Bead Panel kit (A customized product of Catalogue Number: APOMAG-62K, Millipore, MA) using the Bio-Plex system (Bio-Rad Laboratories, Taipei, Taiwan). The assay procedure was a modification of the blood sample-suitable protocol provided by Millipore. Briefly, filter-bottom, 96-well microplates were pre-wetted with wash buffer and blocked for 10 min with assay buffer. A standard curve was generated by preparing 5-fold serial dilutions of appropriate standards in assay buffer. Ten microliters of prepared standards and saliva samples (2-fold dilution) were added into wells containing 90 μl of assay buffer with immunobead mixtures. Microplates were incubated for 50 min at room temperature on a microtiter shaker in the dark and then washed three times with wash buffer using a vacuum manifold (Millipore). The mixture of biotin-conjugated detection antibodies was added to the wells and plates were incubated for 30 min. After washing, phycoerythrin-streptavidin was added and plates were incubated for 30 min. Finally, the washed immunobeads were resuspended in sheath buffer and analyzed using the Bio-Plex 200 system. Standard curves and analyte concentrations were determined using Bio-Plex Manager software version 4.2 (Bio-Rad Laboratories).
ELISA for Quantification of Salivary Albumin Protein
The salivary concentration of albumin protein was measured using a commercial ELISA kit from the R&D Systems (Catalog Number: DY1455) for comparison with MRM-MS data. All saliva samples were prepared as 4000-fold dilution. Several dilution ratios were further tested for the samples that were not within the linear range of the calibration curve. The assay was processed according to the instruction provided by the kit manufacturer (R&D). ELISA plates were coated with capture antibody (2 μg/ml in PBS, 100 μl per well) and incubated overnight at room temperature. Each well on the plate was then washed with 400 μl wash buffer (0.05% Tween® 20 in PBS, pH 7.2–7.4) and blocked with 300 μl of Reagent Diluent (1% bovine serum albumin in PBS, pH 7.2–7.4) at room temperature for 1 h. A total of three washes were performed before addition of 100 μl human albumin standard solutions (2.5–160 ng/ml) or diluted saliva samples in each well. The plates were sealed, incubated for 2 h at room temperature, and washed. Subsequently, 100 μl of the detection antibody prepared as 125 ng/ml was added into each well at room temperature for incubation of 2 h. After three washes, 100 μl of the working dilution of streptavidin-HRP (1:200) was added to each well and incubated for 20 min. After three washes, 100 μl of substrate solution was then added to each well for incubation of 20 min. After three washes, 50 μl of stop solution was added to each well. The optical density was determined using an ELISA reader at a wavelength of 450 nm.
Statistical Analysis for Verification of a Single Biomarker
The statistical package SPSS 13.0 (SPSS Inc., Chicago, IL) was used for all analyses. No outlier was rejected when performing statistical analysis of the 56 proteins for differentiation of oral cancer from control samples. Differences in concentration levels of targeted salivary proteins between control and cancer states, measured by MRM/MS assay, were analyzed using the nonparametric Mann-Whitney test. A receiver operator characteristic (ROC) curve analysis and AUC were used to determine the ability to discriminate between the two clinical groups. The optimal cut-off point was determined using Youden's index (J), calculated as J = 1 - (false positive rate + false negative rate) = 1 - [(1 - sensitivity) + (1 - specificity)] = sensitivity + specificity - 1 (32). All tests were two-sided, and p values < 0.05 were considered statistically significant. Because many of the targeted proteins have overlapping distributions of concentration levels in saliva between control and OSCC, another outlier-cut-off value was also defined using control specimens by outlier analysis based on Tn-test, and is determined as the closest concentration in the ROC curve for presentation of sensitivity/specificity values. For those clinical saliva specimens who showed higher concentrations than the outlier-cutoff values of control specimens were classified as OSCC cases. The additional outlier-cutoff value for each targeted protein was also used to determine the alternative sensitivity and specificity for OSCC diagnosis.
Statistical Analysis of Multiple Biomarkers
Multivariate logistic regression was used for evaluating the synergistic effect of multiple salivary proteins on the classification of OSCC patients and control subjects. In multivariate logistic regression, a vector X contains the measured values of a set of salivary proteins (x1, x2, x3 …), whereas a binary variable Y encodes the status of OSCC and non-OSCC with 1 and 0 respectively. The estimated probability of OSCC of the subject i was represented as a logistic function of t:
| (Eq. 1) |
and t is a linear function of X. The forward stepwise method was performed to determine the variables in X. This was done by choosing one single variable xj which manifested the strongest correlation with Y, then adding other variables progressively for the improvement of correlations. SPSS software was used for the stepwise regression analysis.
RESULTS
Selection of protein candidates and salivary protein digestion workflow
To evaluate the sample-preparation workflow, we digested five processed replicates that had been independently digested from a pooled saliva sample (from one control and three OSCC saliva specimens) using four different protocols. We first chose 56 common proteins that were shown to be detectable in body fluids and were capable of providing significant signals in MS analysis. Many of these proteins are present in body fluids at differential concentrations in the context of diseases (supplemental Table S3) (2, 3, 6, 10, 33, 34). Clinical saliva samples were collected for precise quantification of 56 target proteins by LC-MRM/MS for evaluation as OSCC biomarkers.
The best digestion process may vary from protein to protein because of structural factors, including protein hydrophobicity, folding status, disulfide bonds, denaturing conditions, and accessibility of proteolytic enzymes to the cleavage sites. Trypsin is the most common enzyme used for proteomic studies. Broek et al. reported that trypsin digestion is a key point in MRM workflows and showed that the presence of matrix affects digestion efficiency (35). The incorporation of stable-isotope labeled protein is currently costly, although this step can minimize imprecisions caused by the matrix effect and incomplete digestion. Stable-isotope-labeled peptides are still widely used as internal standards for most MRM-based multiplexed protein quantification strategies (1, 3, 5).
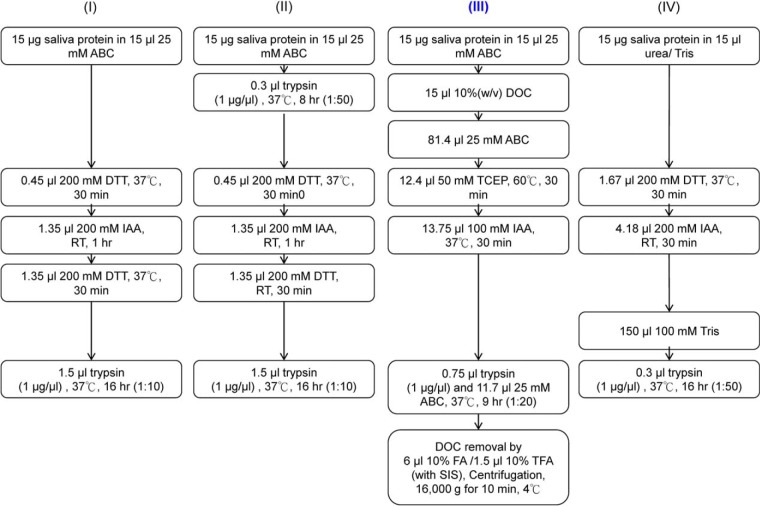
In this study, four common trypsin digestion workflows (protocols I-IV) used for protein quantification by MRM analysis (1, 7, 36) were modified for comparison of digestion yield and reproducibility of salivary protein digestion based on LC-MRM/MS results (Fig. 1). The optimized protocol (protocol III) was chosen for the digestion of all saliva samples used for the calibration curve as well as individual clinical samples. Protocols I and II were used for quantification of urinary proteins by MRM in our previous work (1). Protocol III was modified from a recommended procedure, originally optimized for plasma protein quantification, that used a 9-hour digestion time and DOC as a denaturant (7). The DOC concentration during digestion was reduced to 1% (w/v) as suggested for plasma protein digestion (7). Protocol IV was modified from a digestion procedure used by Whiteaker (36) for peptide immunoaffinity followed by multiplexed MRM/MS, a process termed immuno-MRM/MS or stable isotope standard capture with anti-peptide antibodies (SISCAPA) (37).
Fig. 1.
Four procedures used for optimization of tryptic digestion of salivary proteins. Procedure III was the most reproducible method, generating the highest peaks for the largest number of proteins. Therefore, all individual saliva samples were digested using protocol III prior to fractionation and LC-MS/MS analyses. Multiplexed MRM quantification of 56 proteins (using 56 peptides) was performed in a single multiplexed LC-MRM-MS analysis.
Precision
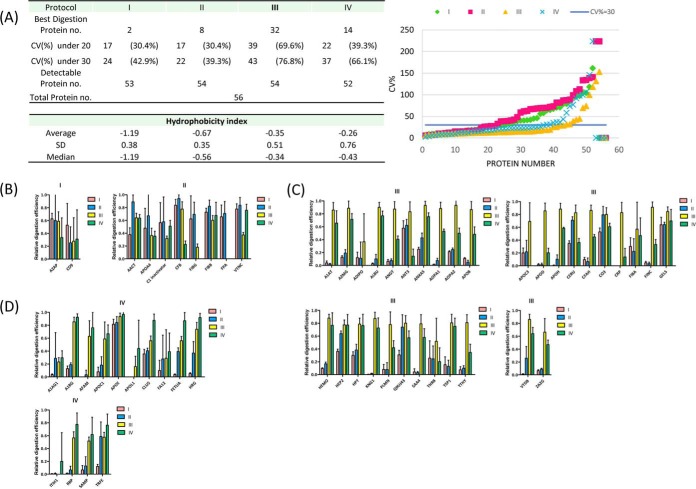
The optimal protocol for salivary protein digestion was determined based on reproducibility as well as signal intensities. Using five independently digested peptides samples from a pooled saliva sample, we found that 39 (69.6%) and 43 (76.8%) of the 56 proteins—the highest percentage of the four protocols—could be quantified using protocol III with CV values less than 20 and 30%, respectively. For protocols I, II, and IV, 24 (42.9%), 22 (39.3%), and 37 (66.1%) proteins, respectively, showed reproducibility within 30% (Fig. 2A). Therefore, protocol III was the sample preparation method with the best precision.
Fig. 2.
A, Reproducibility of the four digestion protocols, represented by the CV of light-to-heavy peak area ratios. The completeness of digestion of the 56 proteins using (B) protocol I and II, (C) protocol III, and (D) protocol IV, determined based on the highest signals obtained.
Sensitivity
A total of 53, 54, 54, and 52 proteins were detectable (S/N > 20) using protocols I, II, III, and IV, respectively. To evaluate platform sensitivity, we compared the signal intensities of the detectable proteins using the four protocols. The largest number of targeted saliva proteins with the highest peak intensities—32 (57.1%)—was obtained using protocol III. For protocols I, II, and IV, MRM peak intensities were highest for 2 (3.6%), 8 (14.3%) and 14 (25.0%) proteins, respectively. The results of precision and signal intensity analyses of the 56 proteins are summarized in Fig. 2B–2D. These results show that protocol III is capable of digesting the most proteins completely; accordingly, it was used for subsequent experimental processing of clinical samples. The average hydrophobicity index values (38) of peptides with highest intensities using protocols I, II, III, and IV were −1.19, −0.67, −0.35, and −0.26, respectively (Fig. 2A), where the larger the value, the more hydrophobic the peptide. For more hydrophilic peptides, digestion efficiencies were generally similar using the four protocols, although protocols I and II tended to be more efficient (Fig. 2B). However, using a surfactant (DOC in protocol III) and a stronger denaturation reagent (urea in protocol IV) more dramatically improved the digestion efficiency of hydrophobic proteins compared with hydrophilic proteins (Fig. 2C and 2D).
Technical Performance of MRM/MS on Salivary Proteins, as Applied to a Pooled Saliva Sample Used for Calibration Curves
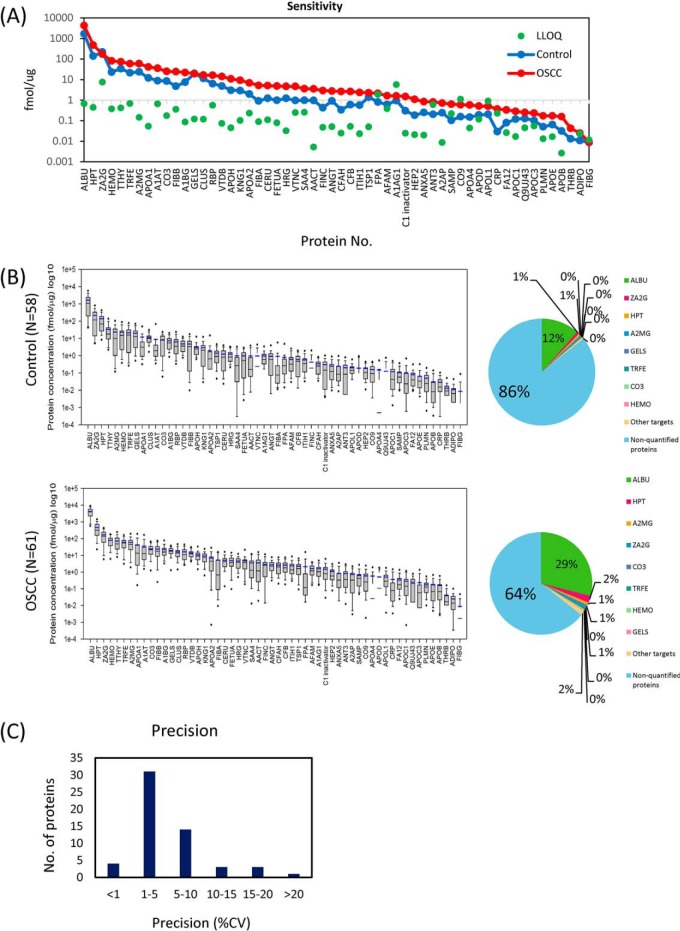
Applying protocol III, we used calibration curves to determine the LOD and LLOQ of 56 salivary proteins, starting from 15 μg of each protein. A total of 336 transitions were monitored with a 53-min liquid chromatography gradient coupled with a scheduled LC-multiplexed MRM/MS run. The LLOQ was defined as the lowest concentration of endogenous peptide that could be measured with a CV < 20% (n = 3 in this study) and accuracy within 80–120%, and the LOD was defined as the lowest concentration at which a signal was observed for the endogenous target peptide with an S/N ratio > 20 in all three replicates. LOD and LLOQ data for the detection of salivary peptides in the pooled saliva sample are summarized in supplemental Table S4. Signals for all 56 proteins in the standard curve were detectable with a S/N ratio > 20. The determined LLOQ values for endogenous salivary proteins ranged from 2.6 amol/μg for APOB (apolipoprotein B) to 7.4 fmol/μg for ZA2G (zinc α2 glycoprotein), whereas LOD values ranged from 0.1 for KNG1 (kininogen 1) to 5628.0 amol/μg for A1AG1 (α1-acid glycoprotein) (Fig. 3A). The calibration curves of the 56 proteins are summarized in supplemental Fig. S1A and supplemental Table S4. The LLOD and quantifiable concentration points in the standard curves with a contribution from the proteolytic peptides of the 56 endogenous salivary proteins are shown as supplemental Fig. S1B.
Fig. 3.
A, The distributions of average concentrations of the 56 proteins in control and OSCC saliva samples, and a comparison with LLOQ values. B, The concentration distributions of the 56 target peptides in the control group (n = 58) and oral cancer (n = 61) determined using the MRM/MS approach. The calculated concentration is minimum estimated concentration in saliva by assumption of 100% yield of trypsin digestion. The horizontal black lines in each box plot denote the 10th, 25th, 50th, 75th and 90th percentiles of the data distribution. The horizontal blue line in each box plot denotes the average concentration value for each protein. The two dots denote the 5th and 95th percentiles of the data distribution. C, Reproducibility of protein quantification is showed as CV values.
The “I” concentration point (i.e. the sample prepared to contain exactly the same light and heavy peptides as individual samples) in the calibration curve was used to evaluate reproducibility (Fig. 3C). Of the 56 proteins, 49 (87.5%) were quantified with a CV less than 10%, and 55 (98.2%) were quantified with a CV less than 20%. Only one protein, THRB (thyroid hormone receptor-β) exhibited a CV > 20%.
The optimized protocol was then used for quantifying the 56 proteins in individual clinical saliva specimens (n = 119), which showed an average protein concentration of ∼2 mg/ml. In each case, 15 μg protein (∼7.5 μl of saliva on average) was used as starting material. The major advantages of the multiplexed LC-MS/MS method over other published methods include (1) the ability to analyze a large number of salivary proteins in a single LC-MRM run, making it particularly useful for marker panel development; (2) separation of all proteins in a single run and unambiguous identification by SIS peptides, which provide more precise quantitation data than immunoassays; and (3) the requirement for only small sample volumes—an advantage for clinical samples, where amounts are limited. We have developed and validated a scalable, sensitive assay for multiplexed quantitation of 56 proteins in human saliva samples in a single assay using an optimized sample preparation method and multiplexed MRM/MS coupling incorporating SIS for accurate quantification. These target proteins might represent novel biomarkers for a variety of oral diseases for future translational research efforts.
Quantitation of Protein Biomarker Candidates in Individual Salivary Samples from Two Independent Clinical Sample Sets
Among the 56 target proteins, 55 were quantified using the corresponding calibration curves. A qualified calibration curve (i.e. S/N ratio > 20 and CV < 20% in three processed replicates) with the required multiple concentration points and acceptable LLOQ value could not be established for THRB. Therefore, THRB was quantified in individual clinical samples based on known SIS concentrations using light-to-heavy peak area ratios. The quantification of 56 proteins in the two clinical sample sets was completed by LC-MRM/MS. The raw files of MRM data for individual clinical saliva samples have been deposited in a storage server ftp.peptideatlas.org (Full URL: ftp://PASS00938:MR7594j@ftp.peptideatlas.org/) provided by PeptideAtlas with Username (Dataset Identifier): PASS00938 and Password: MR7594j. Official URL for this dataset is http://www.peptideatlas.org/PASS/PASS00938. If we assume that the yield of trypsin digestion is 100%, the minimum estimated concentrations of the 56 salivary proteins by MRM-MS in individual specimens can provide a valuable starting point for future development of salivary protein biomarkers. The protein concentration unit (mol/μg total protein) quantified by MRM/MS was further transformed to ng/ml, as commonly used for immunoassays. Average and standard deviation values (n = 29–32) for concentrations of the 56 proteins in these 119 clinical specimens from the two separate clinical groups are shown in supplemental Tables S5A–S5D. Detectable and quantifiable case numbers of each protein are summarized in detail in Supplemental Tables S5A, S5C, and S5E. A comparison of the average concentration of each protein in control and OSCC patients with associated LOD and LLOQ values is shown in Fig. 3A. THRB was not quantifiable (i.e. with a measured concentration lower than the LLOQ) in any clinical sample. Of the 56 proteins, 45 (80%) were quantifiable at concentrations higher than the corresponding LLOQ values in more than 50% of OSCC patients, indicating the potential of this assay for routine clinical analysis. The LLOQ values of three proteins, A1AG1, CO9 (Complement C9) and APOL1 (apolipoprotein L1), were significantly higher than the average concentration in OSCC patient samples, which may result in reduced accuracy of the determined concentrations of these three proteins in clinical specimens. This phenomenon may be addressed by changing to another signature peptide with ionization properties better suited to quantification. The average concentrations of the remaining 53 proteins were all higher than or close to their LLOQ values. The results in Fig. 3A demonstrate that the average concentration for most proteins is higher than the corresponding LLOQ value. Therefore, this highly multiplexed LC-MRM/MS assay is suitable for development for routine use in quantification of salivary proteins.
A total of 14 and 36% of salivary proteins were contributed from the proteins measured by this multiplexed LC-MRM-MS analysis in control and OSCC patients, respectively. Among the 56 target proteins, albumin was the most abundant protein detected in all individuals in both normal (n = 58) and oral cancer (n = 61) groups, as shown in supplemental Tables S5E and S5F. The average concentration of albumin was 1.66 and 4.18 pmol/μg in control and OSCC, respectively. These values translate to 114.9 and 289.8 ng/μg, which account for 11.5% and 29.0% of total salivary protein in control and OSCC, respectively (Fig. 3B). Thus, the concentration of albumin in normal saliva as a percentage is lower than that in the plasma proteome (55%) (24, 39), but similar to that in the urine proteome of control patients (10.3%) (40). The albumin concentration in saliva increased dramatically among oral cancer patients, a finding similar to that observed for the urine proteome of bladder cancer patients, where the percentage of albumin was increased to 39.6% (40). The total concentration of the 56 targeted proteins accounted for 14.3% and 35.6% of total salivary proteins in control and oral cancer samples, respectively. The average concentration of targeted salivary proteins was found to cover six orders of magnitudes, ranging from 441.6 μg/ml (LVNEVTEFAK from albumin) for cancer patients to 0.300 ng/ml (IFYNQQNHYDGSTGK from HDIPO) for control samples. A large number of other salivary proteins that have not been studied systematically and were not targeted for accurate concentration determination in this study are worthy of further investigation. To understand the reliability of the reported concentrations in this study, quantified data by MRM-MS were also compared with concentrations measured by commercially available Bio-Plex and ELISA assays. APOA1, APOA2, APOE, and albumin were selected as the model proteins to determine the quantified correlation between MRM-MS (peptide-level quantitation) and two immune-assays (protein-level quantitation). The correlation results are summarized as supplemental Fig. S2. All the comparisons between MRM-MS and immune assays show high correlations (γ = 0.779–0.871) with statistical significance (p < 0.001, n = 116 or 117). The determined concentration levels by two types of assays for these target proteins were similar in the order of magnitude that indicate the reliability of MRM-MS data. The precise quantification of salivary proteins shown here indicates that LC-MRM/MS is an efficient tool for exploring the concentration distribution of a large number of disease-associated proteins in saliva, and as such will be useful for monitoring disease status using saliva as a clinical sample source.
Assessment of the Diagnostic Efficacy of Salivary Proteins in Detecting Oral Cancer Using Two Independent Sample Sets
Discovery proteomics is a field that is commonly dominated by data-dependent and -independent workflows. MRM was recognized in 2012 as the Nature Method of the Year (41) with its instrumental robustness, multiplex capacity and simple data format. To evaluate the biomarker performance of these target proteins for OSCC detection and measurement of individual effects, we analyzed 58 randomly selected individual control samples and 61 OSCC samples by LC-MRM/MS, and separated results into two pools of clinical sample sets. A statistical analysis (Table I) revealed that, of the 56 salivary proteins, 44 were significantly increased in the OSCC group compared with normal controls (p < 0.05, n = 61 and 58) in clinical sample set 1, and 53 were increased in clinical sample set 2. As a final measure of performance, data for the two clinical sample sets were combined and analyzed as a group. This analysis showed that the levels of 53 salivary proteins were significantly increased in the OSCC group (p < 0.05, n = 119).
Table I. Statistical results of the 56 proteins for differentiation of oral cancer from control samples.
| Protein | Clinical set 1 |
Clinical set 2 |
All |
Clinical significancea |
||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Control vs. OSCC (n = 29 vs. 32, ng/ml) |
Control vs. OSCC (n = 29 vs. 29, ng/ml) |
Control vs. OSCC (n = 58 vs. 61, ng/ml) |
Cut-off value (ng /ml) | Sensitivity | Specificity | |||||||
| fold-change | p value | AUC value | fold-change | p value | AUC value | fold-change | p value | AUC value | ||||
| A1AG1 | 2.44 | 0.01 | 0.69 | 2.18 | 0.01 | 0.71 | 2.42 | 0.00 | 0.67 | |||
| A1AT | 3.24 | 0.00 | 0.81 | 14.28 | 0.00 | 0.92 | 6.34 | 0.00 | 0.87 | * (213.04) | 0.869 | 0.793 |
| A1BG | 3.56 | 0.00 | 0.81 | 5.88 | 0.00 | 0.84 | 4.48 | 0.00 | 0.82 | |||
| A2AP | 3.74 | 0.03 | 0.66 | 4.46 | 0.01 | 0.70 | 4.11 | 0.00 | 0.68 | |||
| A2MG | 2.56 | 0.00 | 0.72 | 7.33 | 0.00 | 0.84 | 3.83 | 0.00 | 0.77 | |||
| AACT | 2.90 | 0.02 | 0.68 | 10.24 | 0.00 | 0.80 | 5.40 | 0.00 | 0.75 | * (29.44) | 0.639 | 0.759 |
| ADIPO | 4.99 | 0.00 | 0.73 | 2.58 | 0.04 | 0.66 | 3.79 | 0.00 | 0.70 | |||
| AFAM | 4.37 | 0.00 | 0.82 | 3.49 | 0.00 | 0.79 | 3.94 | 0.00 | 0.81 | |||
| ALBU | 3.37 | 0.00 | 0.81 | 4.85 | 0.00 | 0.85 | 3.92 | 0.00 | 0.81 | |||
| ANGT | 4.16 | 0.00 | 0.82 | 6.01 | 0.00 | 0.83 | 5.03 | 0.00 | 0.82 | *(82.41) | 0.672 | 0.828 |
| ANT3 | 6.05 | 0.00 | 0.75 | 6.47 | 0.00 | 0.82 | 6.28 | 0.00 | 0.78 | *(12.64) | 0.672 | 0.776 |
| ANXA5 | 4.52 | 0.00 | 0.77 | 3.10 | 0.05 | 0.65 | 3.68 | 0.00 | 0.70 | |||
| APOA1 | 5.09 | 0.01 | 0.68 | 5.11 | 0.00 | 0.77 | 5.01 | 0.00 | 0.73 | |||
| APOA2 | 5.03 | 0.00 | 0.75 | 4.66 | 0.00 | 0.79 | 4.79 | 0.00 | 0.77 | |||
| APOA4 | 2.91 | 0.07 | 0.63 | 10.17 | 0.01 | 0.71 | 6.07 | 0.00 | 0.67 | |||
| APOB | 6.62 | 0.00 | 0.77 | 8.35 | 0.00 | 0.81 | 7.50 | 0.00 | 0.79 | *(14.04) | 0.738 | 0.776 |
| APOC1 | 2.89 | 0.06 | 0.64 | 4.28 | 0.00 | 0.74 | 3.52 | 0.00 | 0.69 | |||
| APOC3 | 2.61 | 0.66 | 0.53 | 4.23 | 0.04 | 0.65 | 3.19 | 0.08 | 0.59 | |||
| APOD | 4.05 | 0.01 | 0.69 | 47.89 | 0.01 | 0.64 | 4.82 | 0.00 | 0.65 | |||
| APOE | 2.49 | 0.16 | 0.61 | 5.18 | 0.00 | 0.73 | 3.83 | 0.00 | 0.67 | |||
| APOH | 5.33 | 0.00 | 0.89 | 5.23 | 0.00 | 0.83 | 5.26 | 0.00 | 0.86 | *(178.93) | 0.803 | 0.845 |
| APOL1 | 3.00 | 0.09 | 0.63 | 4.39 | 0.00 | 0.79 | 3.62 | 0.00 | 0.71 | |||
| C1 inactivator | 3.81 | 0.00 | 0.77 | 12.69 | 0.00 | 0.88 | 7.27 | 0.00 | 0.83 | *(36.73) | 0.639 | 0.914 |
| CERU | 4.81 | 0.00 | 0.84 | 9.91 | 0.00 | 0.87 | 6.64 | 0.00 | 0.85 | *(310.05) | 0.705 | 0.897 |
| CFAH | 10.09 | 0.00 | 0.91 | 12.07 | 0.00 | 0.91 | 11.01 | 0.00 | 0.91 | *(69.70) | 0.869 | 0.845 |
| CFB | 4.51 | 0.00 | 0.80 | 10.58 | 0.00 | 0.90 | 6.73 | 0.00 | 0.85 | *(93.75) | 0.754 | 0.828 |
| CLUS | 1.72 | 0.06 | 0.64 | 2.47 | 0.01 | 0.70 | 2.04 | 0.00 | 0.67 | |||
| CO3 | 3.33 | 0.00 | 0.77 | 6.68 | 0.00 | 0.85 | 4.69 | 0.00 | 0.81 | |||
| CO9 | 3.50 | 0.06 | 0.64 | 8.79 | 0.00 | 0.76 | 5.71 | 0.00 | 0.70 | |||
| CRP | 8.65 | 0.00 | 0.76 | 35.68 | 0.00 | 0.77 | 22.19 | 0.00 | 0.77 | *(2.21) | 0.541 | 0.931 |
| FA12 | 7.54 | 0.00 | 0.85 | 5.91 | 0.00 | 0.82 | 6.49 | 0.00 | 0.83 | *(6.24) | 0.787 | 0.741 |
| FETUA | 6.08 | 0.00 | 0.84 | 9.30 | 0.00 | 0.86 | 7.45 | 0.00 | 0.85 | *(86.51) | 0.705 | 0.897 |
| FIBA | 10.50 | 0.01 | 0.71 | 6.54 | 0.00 | 0.77 | 7.06 | 0.00 | 0.74 | |||
| FIBB | 6.20 | 0.00 | 0.81 | 10.40 | 0.00 | 0.84 | 7.92 | 0.00 | 0.83 | *(522.82) | 0.721 | 0.810 |
| FIBG | 6.99 | 0.00 | 0.78 | 0.19 | 0.33 | 0.43 | 1.88 | 0.03 | 0.61 | |||
| FINC | 8.24 | 0.00 | 0.82 | 16.06 | 0.00 | 0.86 | 11.50 | 0.00 | 0.84 | *(137.81) | 0.787 | 0.810 |
| FPA | 1.52 | 0.12 | 0.38 | 3.40 | 0.19 | 0.60 | 2.86 | 0.80 | 0.49 | |||
| GELS | 1.16 | 0.36 | 0.57 | 2.34 | 0.01 | 0.70 | 1.57 | 0.01 | 0.63 | |||
| HEMO | 4.46 | 0.00 | 0.84 | 7.04 | 0.00 | 0.87 | 5.36 | 0.00 | 0.85 | *(1607.47) | 0.754 | 0.810 |
| HEP2 | 8.13 | 0.00 | 0.88 | 9.15 | 0.00 | 0.92 | 8.63 | 0.00 | 0.90 | *(17.46) | 0.803 | 0.914 |
| HPT | 5.00 | 0.00 | 0.79 | 7.03 | 0.00 | 0.87 | 5.60 | 0.00 | 0.79 | *(6798.48) | 0.721 | 0.741 |
| HRG | 4.78 | 0.00 | 0.82 | 6.34 | 0.00 | 0.89 | 5.38 | 0.00 | 0.85 | *(139.51) | 0.689 | 0.897 |
| ITIH1 | 5.89 | 0.00 | 0.84 | 7.06 | 0.00 | 0.87 | 6.49 | 0.00 | 0.85 | *(87.01) | 0.754 | 0.828 |
| KNG1 | 4.64 | 0.00 | 0.83 | 5.40 | 0.00 | 0.83 | 5.00 | 0.00 | 0.83 | *(289.63) | 0.738 | 0.793 |
| PLMN | 4.37 | 0.00 | 0.80 | 5.79 | 0.00 | 0.84 | 5.04 | 0.00 | 0.82 | *(7.22) | 0.672 | 0.828 |
| Q9UJ43 | 2.64 | 0.01 | 0.70 | 3.59 | 0.00 | 0.72 | 3.11 | 0.00 | 0.71 | |||
| RBP | 3.16 | 0.00 | 0.77 | 7.22 | 0.00 | 0.86 | 4.08 | 0.00 | 0.79 | |||
| SAA4 | 5.81 | 0.00 | 0.73 | 5.66 | 0.00 | 0.81 | 5.92 | 0.00 | 0.76 | *(13.03) | 0.639 | 0.793 |
| SAMP | 6.99 | 0.00 | 0.88 | 10.54 | 0.00 | 0.90 | 8.16 | 0.00 | 0.88 | *(5.21) | 0.770 | 0.897 |
| THRB | 3.35 | 0.07 | 0.63 | 5.00 | 0.00 | 0.79 | 4.24 | 0.00 | 0.71 | |||
| TRFE | 3.25 | 0.00 | 0.79 | 6.37 | 0.00 | 0.82 | 4.50 | 0.00 | 0.80 | |||
| TSP1 | 1.43 | 0.22 | 0.59 | 2.86 | 0.01 | 0.71 | 1.90 | 0.00 | 0.65 | |||
| TTHY | 2.59 | 0.01 | 0.70 | 5.75 | 0.00 | 0.85 | 3.21 | 0.00 | 0.74 | |||
| VTDB | 4.12 | 0.00 | 0.87 | 4.96 | 0.00 | 0.87 | 4.51 | 0.00 | 0.87 | |||
| VTNC | 4.58 | 0.00 | 0.87 | 12.00 | 0.00 | 0.85 | 7.79 | 0.00 | 0.87 | *(108.38) | 0.721 | 0.914 |
| ZA2G | 0.78 | 0.89 | 0.49 | 2.33 | 0.01 | 0.69 | 0.96 | 0.10 | 0.59 | |||
* defined as fold-change > 5, p value < 0.05, and AUC > 0.75.
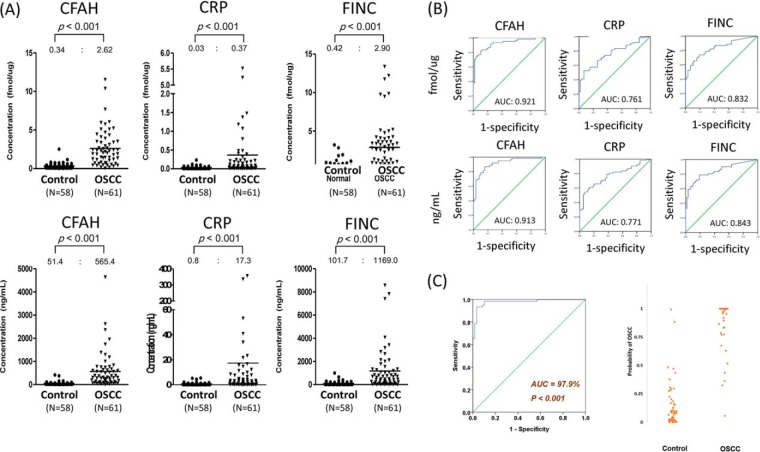
Twenty-five proteins were significantly different between OSCC samples and controls (fold change > 5, p < 0.05, and AUC > 0.75, n = 119), and thus are potential biomarkers of OSCC. These included A1AT (alpha-1-antitrypsin), AACT (alpha-1-antichymotrypsin), ANGT (angiotensinogen), ANT3 (antithrombin-III), APOB (apolipoprotein B-100), APOH (apolipoprotein H), C1 inactivator (Complement C1 inactivator), CERU (ceruloplasmin), CFAH (complement factor H), CFB (complement factor B), CRP (C-Reactive protein), FA12 (coagulation factor XIIa LC), FETUA (alpha-2-HS-glycoprotein), FIBB (fibrinogen beta chain), FINC (fibronectin), HEMO (hemopexin), HEP2 (heparin cofactor II), HPT (haptoglobin beta chain), HRG (histidine-rich glycoprotein), ITIH1 (inter-α-trypsin inhibitor heavy chain 1), KNG1 (kininogen 1), PLMN (plasminogen), SAA4 (serum amyloid A-4 protein), SAMP (serum amyloid P-component), and VTNC (vitronectin). Fold changes, p values, AUC values, cut-off concentrations determined by Youden's index and outlier analyses, corresponding sensitivity, specificity, and ROC curves for discrimination of OSCC from control specimens are summarized in Table I, supplemental Table S6, and supplemental Fig. S1E. The concentration distributions and ROC curves of three biomarker candidates (CFAH, CRP, and FINC) with large changes in concentration (>10-fold, p < 0.05, and AUC > 0.75, n = 119) are shown in Fig. 4. The complete concentration distributions of the 56 proteins in the two clinical states (control and oral cancer), expressed as fmol/μg and ng/ml, are shown in supplemental Figs. S1C and S1D, respectively. To the best of our knowledge, this is the first demonstration of multiplex quantification of these target proteins in individual saliva samples in oral cancer. Additionally, our quantitation results suggest the potential of these proteins for non-invasive screening of OSCC using saliva specimens, a possibility worthy of further validation in a larger sample set.
Fig. 4.
A, Concentrations and (B) ROC curves of CFAH, CRP, and FINC protein biomarkers in 119 individuals from control and oral cancer groups. The measured average concentration values are labeled above each plot. These three proteins show the best ability to differentiate controls from oral cancer patient (>10-fold change, p < 0.05, and AUC > 0.8). C, The performance of the five-biomarker panel (CFAH, AFAM, GELS, SAMP, VTDB) for the classification of OSCC patients (n = 61) and controls (n = 58). The AUC value is 97.9% of the classification using the 5-peptide panel. The right figure shows the estimated probability of OSCC for each patient and control subject. By use of the optimum cutoff of 0.5, the sensitivity of the classification was 93.44%, and the specificity was 96.55%.
A Panel of Multiple Biomarkers for OSCC Diagnosis
For the classification of OSCC (n = 61) and control (n = 58) subjects, the maximum AUC achieved by one single salivary protein was 91% (CFAH, in Table I). Here we evaluated the utilization of multiple salivary proteins for an improved diagnostic performance. The forward stepwise multivariate logistic regression method was applied to the entire dataset of 56 salivary proteins, and a panel of 5 biomarkers (CFAH, AFAM, GELS, SAMP, VTDB) was identified. All the 5 biomarkers remained statistically significant when they were incorporated together in the multivariate analysis (supplemental Table S7). A logistic regression model was thus constructed for estimating Pr(OSCC) using the 5 biomarkers
| (Eq. 2) |
and
| (Eq. 3) |
The range of Pr was between 0 and 1, showing low and high risk of OSCC respectively. The AUC of the ROC curve was 97.9%, showing a good classification performance using the 5-peptide panel (Fig. 4C). The optimum cut-off value of Pr was calibrated to be 0.5 using the Youden's J index. With this cut-off, the sensitivity of the classification was 93.44%, and the specificity was 96.55%.
DISCUSSION
Saliva, an oral fluid derived from the salivary glands that is 99% water, also contains electrolytes, metabolites and proteins, including enzymes, immunoglobulins and antimicrobial factors, mucosal glycoproteins, albumin, and polypeptides (42). It has been used in recent decades as a diagnostic fluid (23). Amylase is the most abundant protein in human saliva and accounts for 40–50% of the total salivary protein concentration (43). In this work, we concluded that the weight percentage of salivary albumin ranged from 11.5% (114.9 ± 123.0 ng/μg total protein) of total proteins for control individuals to 29.0% (289.8 ± 186.7 ng/μg total proteins) for oral cancer patients (Fig. 3B). This range can also be expressed as 112.6 ± 139.5 μg/ml for control individuals to 441.6 ± 457.7 μg/ml for oral cancer patients. The concentrations of salivary albumin in oral cancer patients measured using MRM are within the concentration range (200–501 μg/ml) reported previously in an elderly population using a spectrophotometric method (44). Although saliva can be collected non-invasively, obtaining a sufficient volume for multiplexed measurement of multiple proteins by ELISAs remains difficult in the clinic. The concentration data for saliva proteins in healthy controls and cancer patients reported here, precisely and accurately quantified using SIS, can serve as reference data for molecular diagnostics and biomarker marker development in future studies.
The relative saliva proteome has been widely explored for the discovery of biomarkers of multiple diseases (25, 45–49). However, accurately assessing the concentration of saliva proteins with an immune-based approach is limited to a few target proteins (50, 51). In a recent study, Hirtz et al. quantified 35 proteins by multiplexed LC-MRM/MS, 32 of which were quantified in human saliva with calibration curves that achieved good linearity (27). Percy et al. using a 158-plex MRM assay to quantify the saliva proteins in the disease-free pooled sample (28). In the current work, we applied targeted MS through optimization of sample preparation workflow, particularly the digestion procedure for routine measurements in daily practice, to further explore the clinical utility of a larger number of protein targets in healthy individuals and patients with oral cancer. The comparison of targeted proteins between this study and those of Hirtz (27) and Percy (28) are showed as supplemental Fig. S3. Thirteen salivary proteins of this study were not quantified in the two previous works. The optimized workflow, together with triplicate LC-MRM/MS, was used to analyze clinical salivary samples. Additionally, many emerging methods are using targeted quantification strategies with SIS peptides nested inside discovery LC-MS/MS experiments with no significant loss of performance. Compared with previous immune-based approaches, the new method increased sample analysis throughput and showed flexible multiplex capability. Moreover, this study provides the largest saliva concentration dataset available to date.
CONCLUSIONS
A sensitive and precise platform based on highly multiplexed MRM/MS analysis was developed by optimizing analytical workflow and evaluating technical and clinical performances, and was tested by quantification of 56 proteins. We found that many of these targeted proteins were elevated in OSCC saliva specimens, indicating that these proteins are potential biomarkers that could assist in oral cancer screening. The results of this study provide information regarding the concentrations of many salivary proteins in the context of healthy and cancer states, and as such could serve as a basis for translational research for clinical applications.
DATA AVAILABILITY
The raw files of MRM data for individual clinical saliva samples have been deposited in a storage server ftp.peptideatlas.org (ftp://PASS00938:MR7594j@ftp.peptideatlas.org/) provided by PeptideAtlas with Username (Dataset Identifier): PASS00938 and Password: MR7594j. Official URL for this dataset is http://www.peptideatlas.org/PASS/PASS00938.
Supplementary Material
Footnotes
Author contributions: Y.T. Chen and J.S. Yu designed research; Y.T. Chen, H.W. Chen, C.F. Wu, C.C. Wu, C.H. Tsai, M. Wu, and W.T. Ou Yang performed research; W.F. Chiang, J.S. Yu, and Y.S. Chang contributed new reagents or analytic tools; Y.T. Chen, H.W. Chen, C.F. Wu, L.J. Chu, and K.H. Liang analyzed data; Y.T. Chen, C.C. Wu, and K.H. Liang wrote the paper.
* This research was supported by grants from the Ministry of Science and Technology of Taiwan, Republic of China (102-2113-M-182-001-MY2, and 104-2113-M-182-001-MY2), Chang Gung Memorial Hospital (CMRPD3E0251, CMRPD3E0252, CMRPD3E0031, CMRPD3E0032, CMRPD3E0033, CIRPD3B0012, CMRPD1G0131, and BMRPD78). The study was also supported by grant from Ministry of Health and Welfare, Taiwan (PMRPD1B0102, 0103, and 0104). The instrumental and data analysis resources were supported by the proteomic core lab (CLRPD190016 and CLRPD190015) at Chang Gung University, Taoyuan, Taiwan.
 This article contains supplemental material.
This article contains supplemental material.
1 The abbreviations used are:
- MRM
- multiple reaction monitoring
- A1AG1
- alpha-1-acid glycoprotein
- A1AT
- alpha-1-anti-trypsin
- AACT
- alpha-1-antichymotrypsin
- ANGT
- angiotensinogen
- ANT3
- antithrombin-III
- APOB
- apolipoprotein B
- APOH
- apolipoprotein H
- AUC
- area under the curve
- C1 inactivator
- complement C1 inactivator
- CE
- collision energy
- CERU
- ceruloplasmin
- CFAH
- complement factor H
- CFB
- complement factor B
- CRP
- C-Reactive protein
- CV
- coefficient of variation
- CXP
- collision cell exit potential
- DOC
- sodium deoxycholate
- DP
- declustering potential
- ELISA
- enzyme-linked immunosorbent assay
- EP
- entrance potential
- FA12
- coagulation factor XIIa LC
- FETUA
- alpha-2-HS-glycoprotein
- FIBB
- fibrinogen beta chain
- FINC
- fibronectin
- HEMO
- hemopexin
- HEP2
- heparin cofactor II
- HPT
- haptoglobin beta chain
- HRG
- histidine-rich glycoprotein
- ITIH1
- inter-alpha-trypsin inhibitor heavy chain 1
- LC
- liquid chromatography
- LOD
- limit of detection
- LLOQ
- low limit of quantitation
- KNG1
- kininogen 1
- OSCC
- oral squamous cell carcinoma
- PLMN
- plasminogen
- ROC
- receiver operator characteristic
- SAA4
- serum amyloid A-4 protein
- SAMP
- serum amyloid P-component
- SRM
- selected reaction monitoring
- SIS
- stable isotope-labeled standard
- SISCAPA
- stable isotope standard capture with anti-peptide antibodies
- TCEP
- tris(2-carboxyethyl)phosphine
- TFA
- trifluoroacetic acid
- VTNC
- vitronectin
- ZA2G
- zinc alpha-2 glycoprotein.
REFERENCES
- 1. Chen Y. T., Chen H. W., Domanski D., Smith D. S., Liang K. H., Wu C. C., Chen C. L., Chung T., Chen M. C., Chang Y. S., Parker C. E., Borchers C. H., and Yu J. S. (2012) Multiplexed quantification of 63 proteins in human urine by multiple reaction monitoring-based mass spectrometry for discovery of potential bladder cancer biomarkers. J. Proteomics 75, 3529–3545 [DOI] [PubMed] [Google Scholar]
- 2. Darde V. M., Barderas M. G., and Vivanco F. (2013) Multiple reaction monitoring (MRM) of plasma proteins in cardiovascular proteomics. Methods Mol. Biol. 1000, 191–199 [DOI] [PubMed] [Google Scholar]
- 3. Domanski D., Percy A. J., Yang J., Chambers A. G., Hill J. S., Freue G. V., and Borchers C. H. (2012) MRM-based multiplexed quantitation of 67 putative cardiovascular disease biomarkers in human plasma. Proteomics 12, 1222–1243 [DOI] [PubMed] [Google Scholar]
- 4. Pan S., Chen R., Brand R. E., Hawley S., Tamura Y., Gafken P. R., Milless B. P., Goodlett D. R., Rush J., and Brentnall T. A. (2012) Multiplex targeted proteomic assay for biomarker detection in plasma: a pancreatic cancer biomarker case study. J. Proteome Res. 11, 1937–1948 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Abbatiello S. E., Schilling B., Mani D. R., Zimmerman L. J., Hall S. C., MacLean B., Albertolle M., Allen S., Burgess M., Cusack M. P., Ghosh M., Hedrick V., Held J. M., Inerowicz H. D., Jackson A., Keshishian H., Kinsinger C. R., Lyssand J., Makowski L., Mesri M., Rodriguez H., Rudnick P., Sadowski P., Sedransk N., Shaddox K., Skates S. J., Kuhn E., Smith D., Whiteaker J. R., Whitwell C., Zhang S., Borchers C. H., Fisher S. J., Gibson B. W., Liebler D. C., MacCoss M. J., Neubert T. A., Paulovich A. G., Regnier F. E., Tempst P., and Carr S. A. (2015) Large-scale inter-laboratory study to develop, analytically validate and apply highly multiplexed, quantitative peptide assays to measure cancer-relevant proteins in plasma. Mol. Cell. Proteomics 14, 2357–2374 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Chambers A. G., Percy A. J., Simon R., and Borchers C. H. (2013) MRM for the verification of cancer biomarker proteins: recent applications to human plasma and serum. Expert Rev. Proteomics 11, 137–148 [DOI] [PubMed] [Google Scholar]
- 7. Proc J. L., Kuzyk M. A., Hardie D. B., Yang J., Smith D. S., Jackson A. M., Parker C. E., and Borchers C. H. (2010) A quantitative study of the effects of chaotropic agents, surfactants, and solvents on the digestion efficiency of human plasma proteins by trypsin. J. Proteome Res. 9, 5422–5437 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Chen C. L., Lai Y. F., Tang P., Chien K. Y., Yu J. S., Tsai C. H., Chen H. W., Wu C. C., Chung T., Hsu C. W., Chen C. D., Chang Y. S., Chang P. L., and Chen Y. T. (2012) Comparative and targeted proteomic analyses of urinary microparticles from bladder cancer and hernia patients. J. Proteome Res. 11, 5611–5629 [DOI] [PubMed] [Google Scholar]
- 9. Selevsek N., Matondo M., Sanchez Carbayo M., Aebersold R., and Domon B. (2011) Systematic quantification of peptides/proteins in urine using selected reaction monitoring. Proteomics 11, 1135–1147 [DOI] [PubMed] [Google Scholar]
- 10. Percy A. J., Yang J., Chambers A. G., Simon R., Hardie D. B., and Borchers C. H. (2014) Multiplexed MRM with Internal Standards for Cerebrospinal Fluid Candidate Protein Biomarker Quantitation. J. Proteome Res. 13, 3733–3747 [DOI] [PubMed] [Google Scholar]
- 11. Ferlay J., Shin H. R., Bray F., Forman D., Mathers C., and Parkin D. M. (2010) Estimates of worldwide burden of cancer in 2008: GLOBOCAN 2008. Int. J. Cancer 127, 2893–2917 [DOI] [PubMed] [Google Scholar]
- 12. Jemal A., Bray F., Center M. M., Ferlay J., Ward E., and Forman D. (2011) Global cancer statistics. CA 61, 69–90 [DOI] [PubMed] [Google Scholar]
- 13. Torre L. A., Bray F., Siegel R. L., Ferlay J., Lortet-Tieulent J., and Jemal A. (2015) Global cancer statistics, 2012. CA 65, 87–108 [DOI] [PubMed] [Google Scholar]
- 14. Health Promotion Administration, MOHAW, TAIWAN JANUARY 2016. (2016) Cancer Registry Annual ReporT, 2013, Taiwan. [Google Scholar]
- 15. Guo S. E., Huang T. J., Huang J. C., Lin M. S., Hong R. M., Chang C. H., and Chen M. Y. (2013) Alcohol, betel-nut and cigarette consumption are negatively associated with health promoting behaviors in Taiwan: a cross-sectional study. BMC Public Health 13, 257. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Gupta S., Singh R., Gupta O. P., and Tripathi A. (2014) Prevalence of oral cancer and pre-cancerous lesions and the association with numerous risk factors in North India: A hospital based study. Natl. J. Maxillofacial Surg. 5, 142–148 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Mahapatra S., Kamath R., Shetty B. K., and Binu V. S. (2015) Risk of oral cancer associated with gutka and other tobacco products: A hospital-based case-control study. J. Cancer Res. Therap. 11, 199–203 [DOI] [PubMed] [Google Scholar]
- 18. Mangalath U., Aslam S. A., Abdul Khadar A. H., Francis P. G., Mikacha M. S., and Kalathingal J. H. (2014) Recent trends in prevention of oral cancer. J. Int. Soc. Prev. Community Dent. 4, S131–S138 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Bonne N. J., and Wong D. T. (2012) Salivary biomarker development using genomic, proteomic and metabolomic approaches. Genome Med. 4, 82. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Lau C., Kim Y., Chia D., Spielmann N., Eibl G., Elashoff D., Wei F., Lin Y. L., Moro A., Grogan T., Chiang S., Feinstein E., Schafer C., Farrell J., and Wong D. T. (2013) Role of pancreatic cancer-derived exosomes in salivary biomarker development. J. Biol. Chem. 288, 26888–26897 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Hu S., Arellano M., Boontheung P., Wang J., Zhou H., Jiang J., Elashoff D., Wei R., Loo J. A., and Wong D. T. (2008) Salivary proteomics for oral cancer biomarker discovery. Clin. Cancer Res. 14, 6246–6252 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Jou Y. J., Hua C. H., Lin C. D., Lai C. H., Huang S. H., Tsai M. H., Kao J. Y., and Lin C. W. (2014) S100A8 as potential salivary biomarker of oral squamous cell carcinoma using nanoLC-MS/MS. Clin. Chim. Acta 436, 121–129 [DOI] [PubMed] [Google Scholar]
- 23. AlMoharib H. S., AlMubarak A., AlRowis R., Geevarghese A., Preethanath R. S., and Anil S. (2014) Oral fluid based biomarkers in periodontal disease: part 1. Saliva. J. Int. Oral Health 6, 95–103 [PMC free article] [PubMed] [Google Scholar]
- 24. Ellias M. F., Zainal Ariffin S. H., Karsani S. A., Abdul Rahman M., Senafi S., and Megat Abdul Wahab R. (2012) Proteomic analysis of saliva identifies potential biomarkers for orthodontic tooth movement. Sci. World J. 647240. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Messana I., Cabras T., Iavarone F., Manconi B., Huang L., Martelli C., Olianas A., Sanna M. T., Pisano E., Sanna M., Arba M., D'Alessandro A., Desiderio C., Vitali A., Pirolli D., Tirone C., Lio A., Vento G., Romagnoli C., Cordaro M., Manni A., Gallenzi P., Fiorita A., Scarano E., Calo L., Passali G. C., Picciotti P. M., Paludetti G., Fanos V., Faa G., and Castagnola M. (2015) Chrono-proteomics of human saliva: variations of the salivary proteome during human development. J. Proteome Res. 14, 1666–1677 [DOI] [PubMed] [Google Scholar]
- 26. Sun S., Zhao F., Wang Q., Zhong Y., Cai T., Wu P., Yang F., and Li Z. (2014) Analysis of age and gender associated N-glycoproteome in human whole saliva. Clin. Proteomics 11, 25. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Hirtz C., Vialaret J., Nowak N., Gabelle A., Deville de Periere D., and Lehmann S. (2016) Absolute quantification of 35 plasma biomarkers in human saliva using targeted MS. Bioanalysis 8, 43–53 [DOI] [PubMed] [Google Scholar]
- 28. Percy A. J., Hardie D. B., Jardim A., Yang J., Elliott M. H., Zhang S., and Borchers C. H. (2016) Multiplexed panel of precisely quantified salivary proteins for biomarker assessment. Proteomics doi: 10.1002/pmic.201600230 [Epub ahead of print] [DOI] [PubMed] [Google Scholar]
- 29. Chen Y. T., Chen H. W., Domanski D., Smith D. S., Liang K. H., Wu C. C., Chen C. L., Chung T., Chen M. C., Chang Y. S., Parker C. E., Borchers C. H., and Yu J. S. (2012) Multiplexed quantification of 63 proteins in human urine by multiple reaction monitoring-based mass spectrometry for discovery of potential bladder cancer biomarkers. J. Proteomics 75, 3529–3545 [DOI] [PubMed] [Google Scholar]
- 30. Kuzyk M. A., Smith D., Yang J., Cross T. J., Jackson A. M., Hardie D. B., Anderson N. L., and Borchers C. H. (2009) Multiple reaction monitoring-based, multiplexed, absolute quantitation of 45 proteins in human plasma. Mol. Cell. Proteomics 8, 1860–1877 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Kuzyk M. A., Smith D., Yang J., Cross T. J., Jackson A. M., Hardie D. B., Anderson N. L., and Borchers C. H. (2009) Multiple reaction monitoring-based, multiplexed, absolute quantitation of 45 proteins in human plasma. Mol. Cell. Proteomics 8, 1860–1877 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Youden W. J. (1950) Index for rating diagnostic tests. Cancer 3, 32–35 [DOI] [PubMed] [Google Scholar]
- 33. Chen Y. T., Chen C. L., Chen H. W., Chung T., Wu C. C., Chen C. D., Hsu C. W., Chen M. C., Tsui K. H., Chang P. L., Chang Y. S., and Yu J. S. (2010) Discovery of novel bladder cancer biomarkers by comparative urine proteomics using iTRAQ technology. J. Proteome Res. 9, 5803–5815 [DOI] [PubMed] [Google Scholar]
- 34. Percy A. J., Chambers A. G., Yang J., and Borchers C. H. (2013) Multiplexed MRM-based quantitation of candidate cancer biomarker proteins in undepleted and non-enriched human plasma. Proteomics 13, 2202–2215 [DOI] [PubMed] [Google Scholar]
- 35. van den Broek I., Romijn F. P., Smit N. P., van der Laarse A., Drijfhout J. W., van der Burgt Y. E., and Cobbaert C. M. (2015) Quantifying protein measurands by Peptide measurements: where do errors arise? J. Proteome Res. 14, 928–942 [DOI] [PubMed] [Google Scholar]
- 36. Whiteaker J. R., Zhao L., Anderson L., and Paulovich A. G. (2010) An automated and multiplexed method for high throughput peptide immunoaffinity enrichment and multiple reaction monitoring mass spectrometry-based quantification of protein biomarkers. Mol. Cell. Proteomics 9, 184–196 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37. Anderson N. L., Anderson N. G., Haines L. R., Hardie D. B., Olafson R. W., and Pearson T. W. (2004) Mass spectrometric quantitation of peptides and proteins using Stable Isotope Standards and Capture by Anti-Peptide Antibodies (SISCAPA). J. Proteome Res. 3, 235–244 [DOI] [PubMed] [Google Scholar]
- 38. http://www.alphalyse.com/customer-support/gpmaw-lite/start-gpmaw-lite/http://www.alphalyse.com/gpmaw_lite.html.
- 39. Anderson N. L., and Anderson N. G. (2002) The human plasma proteome: history, character, and diagnostic prospects. Mol. Cell. Proteomics 1, 845–867 [DOI] [PubMed] [Google Scholar]
- 40. Chen Yi-Ting, P. C. E., Chen Hsiao-Wei, Chen Chien-Lun, Domanski Dominik, Smith Derek S., Wu Chih-Ching, Chung Ting, Liang Kung-Hao, Chen Min-Chi, Chang Yu-Sun, Borchers Christoph H. and Yu Jau-Song (2013) Discovery and Validation Case Studies, Recommendations: A Pipeline that Integrates the Discovery and Verification Studies of Urinary Protein Biomarkers Reveals Candidate Markers for Bladder Cancer, RSC [Google Scholar]
- 41. (2012) Method of the Year 2012. Nat. Methods 10, 1. [DOI] [PubMed] [Google Scholar]
- 42. de Almeida Pdel V., Gregio A. M., Machado M. A., de Lima A. A., and Azevedo L. R. (2008) Saliva composition and functions: a comprehensive review. J. Contemporary Dental Practice 9, 72–80 [PubMed] [Google Scholar]
- 43. Mandel A. L., Peyrot des Gachons C., Plank K. L., Alarcon S., and Breslin P. A. (2010) Individual differences in AMY1 gene copy number, salivary alpha-amylase levels, and the perception of oral starch. PloS one 5, e13352. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44. Meurman J. H., Rantonen P., Pajukoski H., and Sulkava R. (2002) Salivary albumin and other constituents and their relation to oral and general health in the elderly. Oral Surgery, Oral Med. Oral Pathol. Oral Radiol. Endodontics 94, 432–438 [DOI] [PubMed] [Google Scholar]
- 45. Schulz B. L., Cooper-White J., and Punyadeera C. K. (2013) Saliva proteome research: current status and future outlook. Crit. Rev. Biotechnol. 33, 246–259 [DOI] [PubMed] [Google Scholar]
- 46. Krapfenbauer K., Drucker E., and Thurnher D. (2014) Identification of tumour-related proteins as potential screening markers by proteome analysis-protein profiles of human saliva as a predictive and prognostic tool. EPMA J. 5, 20. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47. Winck F. V., Prado Ribeiro A. C., Ramos Domingues R., Ling L. Y., Riano-Pachon D. M., Rivera C., Brandao T. B., Gouvea A. F., Santos-Silva A. R., Coletta R. D., and Paes Leme A. F. (2015) Insights into immune responses in oral cancer through proteomic analysis of saliva and salivary extracellular vesicles. Sci. Reports 5, 16305. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48. Wu C. C., Chu H. W., Hsu C. W., Chang K. P., and Liu H. P. (2015) Saliva proteome profiling reveals potential salivary biomarkers for detection of oral cavity squamous cell carcinoma. Proteomics 15, 3394–3404 [DOI] [PubMed] [Google Scholar]
- 49. Grassl N., Kulak N. A., Pichler G., Geyer P. E., Jung J., Schubert S., Sinitcyn P., Cox J., and Mann M. (2016) Ultra-deep and quantitative saliva proteome reveals dynamics of the oral microbiome. Genome Med. 8, 44. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50. Aboodi G. M., Sima C., Moffa E. B., Crosara K. T., Xiao Y., Siqueira W. L., and Glogauer M. (2015) Salivary cytoprotective proteins in inflammation and resolution during experimental gingivitis–A pilot study. Front. Cell. Infect. Microbiol. 5, 92. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51. Schicht M., Stengl C., Sel S., Heinemann F., Gotz W., Petschelt A., Pelka M., Scholz M., Rausch F., Paulsen F., and Brauer L. (2015) The distribution of human surfactant proteins within the oral cavity and their role during infectious diseases of the gingiva. Ann. Anat. 199, 92–97 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
The raw files of MRM data for individual clinical saliva samples have been deposited in a storage server ftp.peptideatlas.org (ftp://PASS00938:MR7594j@ftp.peptideatlas.org/) provided by PeptideAtlas with Username (Dataset Identifier): PASS00938 and Password: MR7594j. Official URL for this dataset is http://www.peptideatlas.org/PASS/PASS00938.