Abstract
SUMOylation is a critical regulator of a broad range of cellular processes, and is thought to do so in part by modulation of protein interaction. To comprehensively identify human proteins whose interaction is modulated by SUMOylation, we developed an in vitro binding assay using human proteome microarrays to identify targets of SUMO1 and SUMO2. We then integrated these results with protein SUMOylation and protein-protein interaction data to perform network motif analysis. We focused on a single network motif we termed a SUMOmodPPI (SUMO-modulated Protein-Protein Interaction) that included the INO80 chromatin remodeling complex subunits TFPT and INO80E. We validated the SUMO-binding activity of INO80E, and showed that TFPT is a SUMO substrate both in vitro and in vivo. We then demonstrated a key role for SUMOylation in mediating the interaction between these two proteins, both in vitro and in vivo. By demonstrating a key role for SUMOylation in regulating the INO80 chromatin remodeling complex, this work illustrates the power of bioinformatics analysis of large data sets in predicting novel biological phenomena.
The functional protein microarray is a powerful and versatile systems biology and proteomics tool that allows the profiling of the activity of thousands of proteins in parallel. Applications of functional protein microarrays range from the profiling of protein interactions, construction of enzyme-substrate networks, and identification of novel enzymatic activities. Since the development of the yeast proteome microarray over 15 years ago (1), more recent work has seen the development of complete and near-complete proteome arrays representing viruses, bacteria and plants (2–4). However, most existing human protein microarrays are comprised of only a minority of the human proteome (5–9). We have recently developed a human proteome microarray, the HuProt array, which includes >17,000 full-length human proteins (10). The proteins used to generate this microarray were expressed in yeast and purified under native conditions. Expressing recombinant eukaryotic proteins in yeast improves the likelihood that proteins will retain their biological activity relative to prokaryotic and in vitro expression systems.
Numerous studies have harnessed the power of the HuProt array to profile a wide range of protein activities, including protein-protein interactions (11, 12), protein-RNA interactions (13, 14), analysis of monoclonal antibody specificity (10), biomarker discovery (15), identification of substrates of protein kinases (16), and S-nitrosylation (17).
Recently, we have conducted SUMOylation assays using the HuProt microarray to identify numerous previously uncharacterized SUMO E3 ligase-dependent substrates using a subset of human SUMO E3 ligases.1 Protein SUMOylation is an essential post-translational modification in most organisms, including yeast (19), C. elegans (20), Arabidopsis (21), and mice (22, 23). The reversible SUMO-modification of target proteins involves an enzymatic cascade chemically similar to ubiquitylation, involving E1 activating, E2 conjugating, and E3 ligating enzymes, and SUMO proteases. As the only classes of SUMOylation enzymes for which multiple members have been identified, the SUMO E3 ligases and the SUMO proteases have been proposed to be the major factors determining substrate specificity.
Although SUMOylation can regulate a diverse array of protein activities, at the molecular level SUMOylation alters protein surfaces and their ability to interact with other proteins (23). It has been proposed that SUMO acts like a “molecular glue” that mediates noncovalent interactions between modified substrates and SUMO-binding proteins (24). This is illustrated by the interaction between the promyelocytic leukemia (PML) protein and thymine DNA glycosylase (TDG), in which SUMO-modification as well as SUMO-interaction motifs (SIMs)2 on both proteins contribute to binding (24). Similarly, SUMO1 modification of RanGAP1 promotes its interaction with RanBP2 (25). The canonical SIM contains a hydrophobic core consisting of four hydrophobic amino acids that can be either preceded or followed by multiple negatively charged amino acids. Recently, other protein motifs have been shown to mediate SUMO-binding, including MYM-type and ZZ-type zinc finger domains (26, 27).
Previous efforts to identify SUMO-binding proteins were limited either by their focus on a small number of cell or tissue types, or else were restricted to identifying nuclear proteins that bound to only a single SUMO isoform (24, 28, 29). One such study relied on the use of an MS-coupled pulldown (30), whereas others used yeast two-hybrid analysis, which typically correlates only moderately well with biochemical protein-protein interaction assays (24, 28). These methods, although powerful, often do not distinguish between direct and indirect interactions.
For our studies, we utilized the HuProt microarray containing >17,000 individually purified human proteins, thus avoiding bias in favor of a particular cell or tissue type or subcellular compartment, and allowing identification of direct and/or low-abundance target proteins. We also looked at binding to both SUMO1 and SUMO2 monomers, as well as SUMO1 and SUMO2 trimers, which were used as model SUMO chains. Our data set represents a nearly 2-fold increase in the number of known SUMO-interacting proteins. The vast majority of these proteins had not been previously reported as having SUMO binding activity.
To use our data set to make novel predictions about SUMOylation function, we integrated our SUMO-binding and SUMOylation data with protein-protein interaction data from publicly available databases to perform network motif analysis. We identified 21 three-component network motifs. Nine of these network motifs were significantly enriched in this data set, whereas four network motifs were significantly depleted. We focused on a single network motif containing a SUMO-binding protein and a SUMO-modified protein that were previously reported to interact, along with SUMO itself (31, 32). This specific network motif suggests a potential role for SUMO in modulating the protein-protein interaction between the other two nodes; and we thus termed this motif a SUMOmodPPI (SUMO-modulated Protein-Protein Interaction). We then validated an example of this network motif that comprised the INO80 chromatin remodeling complex subunits INO80E and TFPT, along with SUMO2. We found that TFPT could be SUMOylated in transfected cells and identified the relevant SUMOylation site on the protein. We also validated the SUMO-binding activity of INO80E and identified the region of INO80E important for this interaction. Finally, we demonstrated that SUMO helps facilitate the interaction between INO80E and TFPT. This work demonstrates the power of integrative analysis of large data sets in predicting novel biological phenomena.
MATERIALS AND METHODS
SUMO Binding Assay on Human Proteome Microarrays
SUMO binding experiments were conducted with four probes representing the SUMO1, SUMO2, and the SUMO1 and SUMO2 trimers, all in triplicate. Protein chips were incubated overnight in blocking buffer (20 mm HEPES-KOH pH 7.3, 100 mm potassium acetate, 2 mm magnesium acetate, 1 mm EGTA, 0.05% Tween-20, 2% BSA) at 4 °C. Chips were then incubated with SUMO1 or SUMO2 monomer or trimer in assay buffer (20 mm HEPES-KOH pH 7.3, 100 mm potassium acetate, 2 mm magnesium acetate, 1 mm EGTA, 0.05% Tween-20) for 1 h at room temperature. Following incubation with SUMO, chips were washed 3× with assay buffer and 3× with PBS, followed by incubation with DyLight 549-anti-SUMO1 (21C7) or DyLight 649-anti-SUMO2 (8A2). Negative control experiments were done in parallel using antibodies only without SUMO. Chips were then washed 3× with 1× TBS with 0.05% Tween-20, 1× with water, then spun to dry and scanned.
Immunoprecipitation and Immunoblotting
HEK 293T cells were transfected with Fugene 6 (Promega Madison, WI) and HCT 116 cells were transfected with Fugene HD (Promega). Both cell types were plated in 6-well plates, and harvested 24–48 h following transfection. In order to detect protein SUMOylation, cells were washed with 1X PBS, then lysed in SUMO IP lysis buffer (50 mm Tris-HCl pH 7.4, 150 mm NaCl, 2 mm EDTA, 1% Triton X-100, 0.5% sodium deoxycholate, 20 mm N-ethyl-maleimide, 1% SDS, and Roche protease inhibitor mixture) for 10 min at 4 °C. Then SUMO IP binding buffer (50 mm Tris-HCl pH 7.4, 150 mm NaCl, 2 mm EDTA, 1% Triton X-100, 0.5% sodium deoxycholate) was then added to dilute the lysate 10-fold, and lysates were transferred to cold microcentrifuge tubes. Cells were then sonicated on ice and centrifuged at 15,000 × g at 4 °C for 10 min. Soluble cell lysates were incubated with Dynabeads (Life Technologies Carlsbad, CA) pre-bound to either anti-Flag (F1804, Sigma St. Louis, MO), anti-myc (R95025, Life Technologies), or anti-V5 (R9605, Life Technologies) antibodies for 2 h at 4 °C and washed 3 times with SUMO IP wash buffer (50 mm Tris-HCl pH 7.4, 150 mm NaCl, 2 mm EDTA, 1% Triton X-100, 0.5% sodium deoxycholate, and 0.1% SDS). Bound proteins were eluted with 1× LDS-sample buffer (NP0008, Life Technologies) with 5% beta-mercaptoethanol. In order to look at protein interactions between INO80E and TFPT by coimmunoprecipitation, cells were transfected and harvested as above, but lysed in INO80 lysis buffer (40 mm HEPES-NaOH pH 7.9, 0.3 m NaCl, 0.2% Triton X-100, and 10% glycerol), followed by centrifugation. Lysate supernatants were then bound to Dynabeads linked to anti-FLAG or anti-V5 for 2 h at 4 °C and then washed 3× with INO80 wash buffer (40 mm HEPES-NaOH 7.9, 0.25 m NaCl, 0.2% Triton X-100, and 10% glycerol). In both immunoprecipitation to detect SUMOylated proteins and coimmunoprecipitation experiments to detect interacting proteins, samples were resolved on 4–12% NuPage Bis-Tris gels (Life Technologies) in MES buffer and subject to immunoblotting using HRP-anti-V5 and HRP-anti-myc antibodies (Life Technologies).
Recombinant Proteins
GST, GST-tagged SUMO1, SUMO2, and SUMO1 and SUMO2 trimers were expressed in bacteria and GST-INO80E was expressed in yeast. All GST-tagged proteins were purified by affinity chromatography on glutathione-Sepharose 4B beads (GE Healthcare Chicago, IL) according to manufacturer's instructions.
In Vitro Binding Assays
Recombinant GST, GST-tagged SUMO1, SUMO2, or SUMO2 trimer were diluted into 100 μl of 1× PBS, 0.05% Tween 20 and incubated with either glutathione-Sepharose or alternatively glutathione-coated 96-well plates (Thermo Fisher Scientific, Waltham, MA). Following overnight incubation at 4 °C, wells were blocked for 1 h at room temperature with 2% bovine serum albumin in assay buffer (20 mm HEPES-KOH pH 7.3, 100 mm potassium acetate, 2 mm magnesium acetate, 1 mm EGTA, 0.05% Tween-20). INO80E and TFPT were produced by in vitro transcription and translation in rabbit reticulocyte lysate in the presence of [35S] methionine according to manufacturer's instructions (Promega). In vitro translated proteins (10 μl) were diluted into 100 μl of assay buffer, when appropriate, incubated with SUMO1 or SUMO2 and the SUMO E1 and E2 enzymes, then incubated with the immobilized GST-tagged proteins for 1 h at room temperature. Unbound proteins were removed by washing, and bound proteins were eluted with SDS sample buffer and resolved by SDS-PAGE and autoradiography. Equal loading of immobilized proteins was verified by staining with SimplyBlue SafeStain (Life Technologies). Quantitative binding was determined by scanning densitometry measurements of radiolabeled protein relative to the amount of binding partner protein as indicated by Coomassie stain.
In Vitro SUMOylation Assays
TFPT was produced by in vitro transcription and translation in rabbit reticulocute lysate in the presence of [35S] methionine according to manufacturer's instructions (Promega). Radiolabeled protein was then incubated either with or without SUMO1 or SUMO2 and the SUMO E1 and E2 enzymes for 1 h at 37 °C to yield a mixture of SUMOylated and unmodified TFPT for subsequent binding experiments.
Protein Microarray Data Analysis
To identify positive hits for each chip, uninformative spots, such as those in damaged or high-background regions of the chip, were first identified as having a coefficient of variation (CV) value >1.5, and excluded from analysis. A background correction was then applied to the remaining spots, defined as the signal intensity (SI) value of each spot as the odds ratio of the foreground median divided by the background median. If a spot has a weak signal, foreground median and background median signal intensity will be similar, and its SI will thus be ∼1. To normalize the signal, we assumed that true hits are relatively rare and almost evenly dispersed in each block, thus we force each block on a chip to have a median SI of 1. To be considered a positive hit, the duplicate spots of each protein need to both have signal intensities (foreground/background ratio) 5 S.D. above the mean value. Additionally, positive hits need to be detected in 2 out of 3 triplicate experiments to be considered positive. Positive hits that were detected in negative control chips were also excluded from analysis.
Network Motif Analysis
Protein microarray data from both SUMO-binding experiments and SUMOylation experiments (18) was integrated with protein-protein interaction data from publicly available databases, including BioGRID, DIP, MIPS, IntAct, and HPRD and network motifs were identified and analyzed as previously described (38). The Z-value of a motif is calculated as the difference of its observed occurrence in a real network and its averaged occurrence in several hundred random networks, normalized with the standard deviation.
RESULTS
Identification of SUMO-binding Partners Using HuProt Arrays
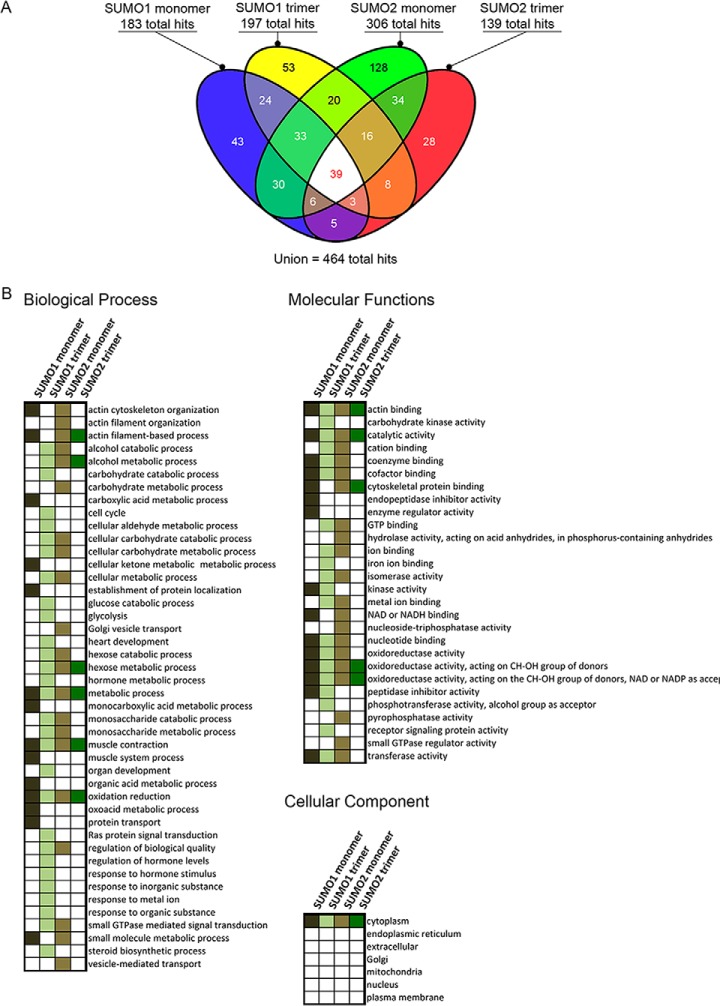
Previous work reported that the first alpha helix and second beta strand of both SUMO1 and SUMO2 can act as a SUMO paralog recognition region that interacts with the SIMs of SUMO paralog-specific binding proteins (26, 33, 34). Although only a handful of SUMO paralog-specific binding proteins have been identified to date, this suggests that more paralog-specific binding proteins remain to be discovered. In order to identify novel SUMO paralog-specific binding proteins, we purified SUMO1 and SUMO2 monomers, as well as SUMO1 and SUMO2 trimers, to be used as polySUMO chain mimics (Fig. 1A). We then used these protein probes in a novel human proteome microarray-based SUMO-binding assay (Fig. 1B). For detection of SUMO binding proteins on the HuProt microarrays, we prepared fluorescently labeled SUMO isoform-specific monoclonal antibodies. We then performed SUMO-binding assays in triplicate for each of the four probes. Among 457 total SUMO binding proteins that were identified by our assay, only 13 had been previously identified, among 270 previously known SUMO-binding proteins (supplemental Data Set S1 and supplemental Fig. S1). Eleven of these 13 previously identified SUMO-binding proteins were found by two SUMO2-focused affinity purification and mass spectrometry based screens (29, 35). The four SUMO protein probes that we tested showed wide variation in their binding specificity (Fig. 2A). The SUMO2 monomer probe bound to the largest number of proteins (306), as well as the largest number of unique proteins (128). The SUMO1 trimer bound 197, the SUMO1 monomer bound 183, and the SUMO2 trimer bound 139 proteins in total. The pairwise probe combinations of SUMO1 trimer and SUMO2 monomer, and SUMO1 monomer and SUMO2 monomer, each bound the largest number of proteins (108). On the other hand, the SUMO1 trimer and SUMO2 trimer bound to the smallest number of shared proteins (66). Thirty-nine proteins bound to all four SUMO probes.
Fig. 1.
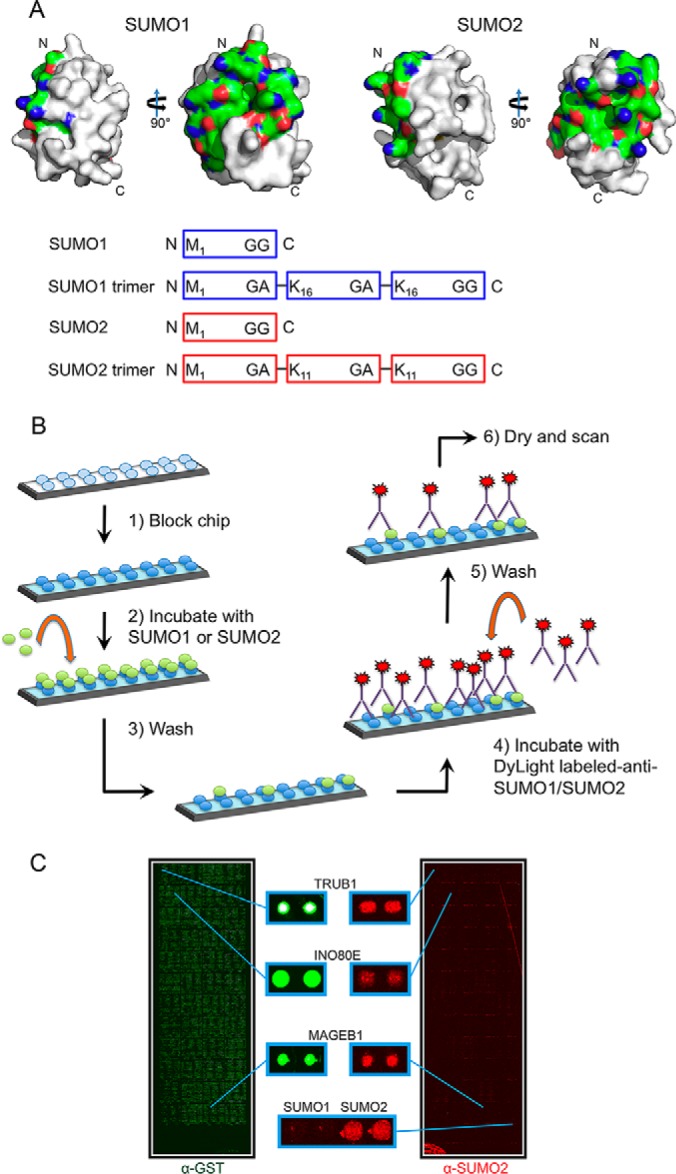
Design of protein microarray-based assay for identification of human SUMO binding proteins. A, (Top) SUMO1 and SUMO2 protein surfaces generated using MacPyMOL with previously reported crystal structures (SUMO1 PDB ID 2PE6 and SUMO2 PDB ID 4BKG). The surfaces corresponding to the first alpha-helix and second beta-strand for both SUMO1 and SUMO2 are shown in color (nitrogens are shown in red, oxygens in blue and carbons in green). This surface has been reported to be important for interactions with other proteins. (Bottom) Cartoon showing 4 probes that were used in our experiments. SUMO1 and SUMO2 monomer probes were based on the proteolytically processed (mature) forms of SUMO1 and SUMO2. The SUMO1 and SUMO2 trimer probes, described previously (Zhu 2008), were expressed as trimeric fusion proteins. B, Schematic of the SUMO-binding protocol used with the human proteome microarray. C, Representative microarray images showing human proteome microarray visualized with an antibody to GST, showing all proteins (left, in green), and an antibody to SUMO2 following a SUMO-binding experiment with the SUMO2 trimer (right, in red).
Fig. 2.
Identification of SUMO binding proteins using human proteome microarrays. A, Venn diagram showing the number of SUMO-binding experiment hits for each probe tested. B, Gene ontology analysis on proteins identified in our screen for SUMO-binding proteins. Gene ontology categories that are significantly enriched among proteins that bound to one of our four probes are shown as a colored box, color-coded by probe.
GO Analysis Reveals Potential New Function of SUMOylation
We next conducted gene ontology (GO) analysis on the novel SUMO binding proteins (Fig. 2B). This analysis revealed several notable properties of these proteins. With regard to cellular component, we observed significant enrichment for cytoplasmic localization among proteins that were bound by all four probes. Considering that many previously identified SUMO-modified proteins are localized to the nucleus, including many transcription factors, this enrichment of cytoplasmic proteins suggests that the importance of SUMOylation in the cytoplasm may be underappreciated.
We also found that some GO categories that were selectively enriched among proteins that bound to all 4 probes, different combinations of 2 or 3 probes, or else a single probe. For example, proteins associated with the GO categories of “actin binding,” “catalytic activity” and “oxidoreductase activity” were selectively enriched among targets bound by all 4 probes. A particularly surprising result of this analysis was the finding that of all pairwise combinations of probes tested, proteins that bound the SUMO1 trimer and SUMO2 monomer, which are two of the most dissimilar probes, shared the largest number of molecular function GO terms.
With regard to biological process, enriched GO terms common to proteins that bound all four probes included three terms: “metabolic process,” “muscle contraction,” and “oxidation reduction.” Among the proteins that we identified that are associated with muscle contraction, including myoglobin, myosin light chain 6B, sarcospan and alpha-1-syntrophin, each of these contained at least one predicted canonical SIM. Sarcospan and alpha-1-syntrophin are members of the dystrophin-glycoprotein complex (DGC), which plays a role in membrane integrity and signal transduction in heart and skeletal muscle and links the actin cytoskeleton to various receptors. Although a role for SUMO in muscle function has been largely unexplored, skeletal muscle actin is SUMOylated and altered cellular localization of SUMO1 has been observed in skeletal muscle in response to physical exercise (36, 37). As with molecular function, the SUMO1 trimer and SUMO2 monomer binding proteins likewise shared the largest number of GO categories compared with any other pairwise probe combination.
Analysis of Integrated SUMO Networks
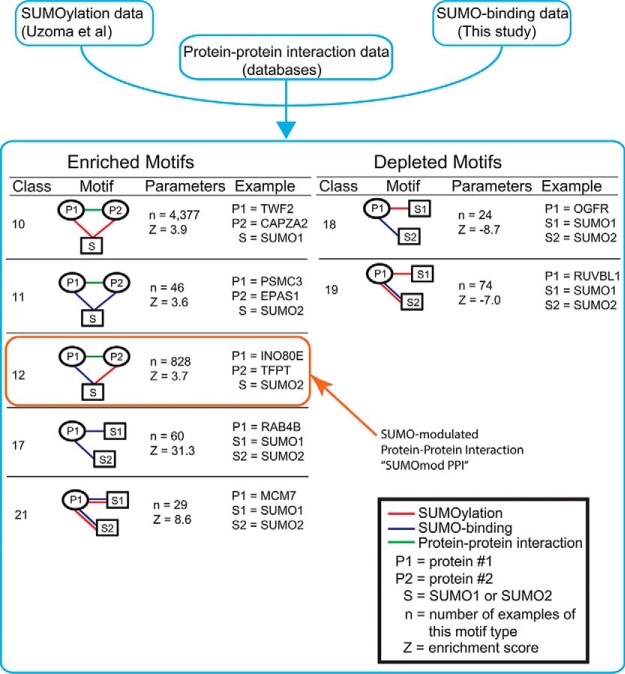
We recently conducted SUMOylation assays using the HuProt microarray to identify numerous previously uncharacterized SUMO E3 ligase-dependent substrates using a subset of human SUMO E3 ligases (supplemental Data Set S2). We hypothesized that by integrating our SUMO-binding data with our in vitroSUMOylation data (18), as well as protein-protein interaction data from publicly available databases, we could make novel predictions about SUMOylation function. This combined data set includes a total of 2910 high-confidence SUMOylation substrates, 489 high-confidence SUMO binding proteins, and 6121 reported protein interactions for proteins that are either SUMOylation substrates or SUMO binding proteins, or both. The integration of these three types of data sets resulted in a network of 2503 nodes, connected by 9520 edges. To gain biological insights from this complex network, we searched for tripartite network motifs that were statistically overrepresented. These network motifs can be considered to be the basic building blocks for the network and provide the information underlying the design principle of SUMO-dependent networks. We can describe each network motif using two parameters, n, the number of occurrences of the motif, and Z, an enrichment score (Fig. 3, supplemental Fig. S4 and supplemental Data Set S3). This enrichment score reflects the difference between the observed occurrence of this network motif in the integrated SUMOylation network and its expected occurrence in a set of random networks (38). In these network motifs, at least one node represents SUMO and at least one node represents a protein from either the SUMO-binding or SUMOylation data sets. These three proteins are then connected by edges that represent one of three types of protein-protein interaction, including (1) noncovalent SUMO-binding identified in our assay or (2) covalent SUMOylation, or (3) a protein-protein interaction from one of several publicly available databases. Among the 21 classes of tripartite network motifs, we found nine enriched (Z>0) and four depleted (Z<0) motifs (Fig. 3). One representative example is shown for each class of the tripartite motif. For example, network motif class 10 represents two interacting proteins that are both SUMOylated. One such instance of this network motif includes the proteins TWF2 and CAPZA2, which interact with each other and are both SUMOylated by SUMO1. In total, we found 4,377 instances of this motif (n = 4377), and this number of occurrences is significantly more than what we would expect from a random network, as indicated by a positive enrichment score (Z = 3.9).
Fig. 3.
Network analysis of SUMO binding and SUMO-conjugated human proteins. Schematic of network motif analysis approach showing the three data sets that were used and all the enriched and depleted network motifs. The “SUMOmod PPI” network motif that was the focus of subsequent experiments is boxed in orange.
INO80E, TFPT, and SUMO2 Form the SUMOmodPPI Network Motif
We then selected one network motif that included two proteins previously reported to interact and included a novel SUMO-binding protein (this data set) and a SUMOylated protein (38). This network motif immediately suggests a potential role for SUMO in modulating this protein-protein interaction and thus, we termed this network motif a “SUMO-modulated protein-protein interaction” (SUMOmodPPI). One such SUMOmodPPI consists of the INO80 chromatin remodeling complex subunits INO80E and TFPT, and SUMO2 (Fig. 3). Our proteome microarray assay revealed that INO80E binds specifically to the SUMO2 monomer and the SUMO2 trimer. The other protein in this triad, TFPT, was previously found to be specifically modified by SUMO2 in the presence of the SUMO E3 ligases PIAS1 and PIAS3 (38). As a candidate SUMOmodPPI, we hypothesized that SUMO may play a role in modulating the interaction between INO80E and TFPT. Before we could proceed with testing our hypothesis, we first set out to validate their SUMO-binding and SUMOylation properties, using conventional assays.
INO80E Preferentially Interacts with SUMO2 Via Its SIM Domain
First, we evaluated the binding of INO80E, translated in vitro in rabbit reticulocyte lysate in the presence of [35S]-methionine, to immobilized GST-tagged SUMO1 or SUMO2 polymers. We observed a strong interaction between INO80E and the SUMO2 polymer in contrast to a very weak interaction between INO80E and the SUMO1 polymer (Fig. 4A), in close agreement with our human proteome microarray data.
Fig. 4.
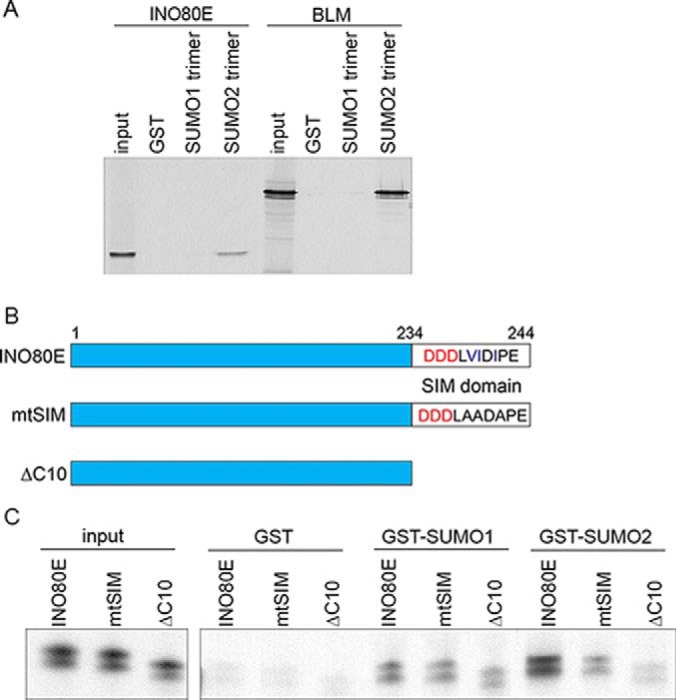
INO80E binds to SUMO1 and SUMO2 through a C-terminal SIM domain. A, Cartoon showing INO80E, the location of the SUMO interaction motif (SIM) at the C-terminus of the protein, and the two SIM-deficient INO80E mutants generated for this study. B, Results from in vitro binding assay. INO80E, the INO80E SIM mutant and the INO80EΔC10 were generated by in vitro translation in the presence of [35S]-methionine and incubated with glutathione sepharose-bound GST-SUMO1 and GST-SUMO2. C, INO80E mtSIM shows reduced binding to GST-SUMO2, but not GST-SUMO1.
Visual inspection of the amino acid sequence of INO80E revealed a canonical SIM at the protein C terminus containing a hydrophobic core “VIDI” preceded by three negatively charged aspartate residues (Fig. 4B). In order to test whether this predicted SIM was important for SUMO binding, we made two mutant forms of the INO80E protein. In the first mutant, we mutated the hydrophobic residues in the hydrophobic core of the SIM to alanines (mtSIM). In the second mutant, we made a truncation mutant that lacked the ten C-terminal residues, including the hydrophobic core sequence and the preceding negatively charged residues (ΔC10). The mtSIM mutant showed a 70% reduction in binding of INO80E to GST-SUMO2, whereas binding to GST-SUMO1 was unchanged (Fig. 4C). However, although the INO80E truncation mutant (ΔC10) lacking the ten C-terminal amino acids showed a 20% reduction in binding to GST-SUMO1, it showed an 80% reduction in binding to GST-SUMO2, approaching the lower limit of detection in our assay. This stands in contrast to previous work reporting the importance of acidic residues specifically in SUMO1 binding (28). This result demonstrates the importance of both the charged and hydrophobic residues in mediating interaction between INO80E and SUMO, both for SUMO1 and, particularly, for SUMO2.
TFPT is SUMOylated at Lys216
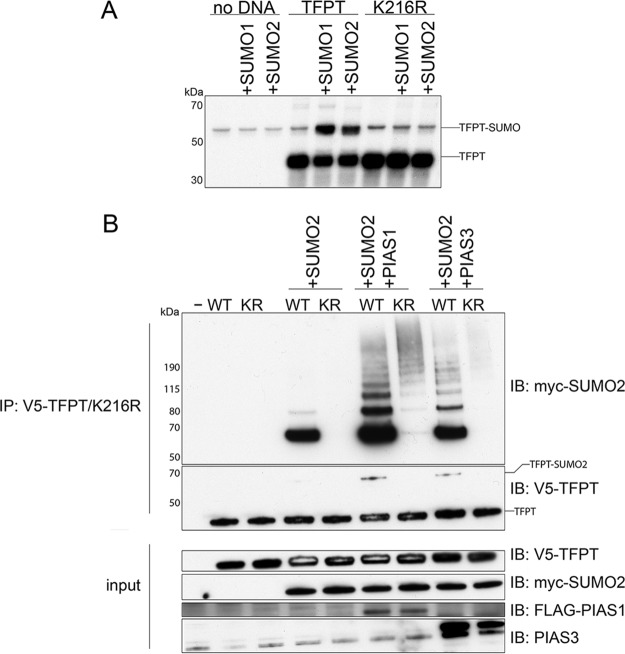
Next, we turned to the other protein in this network, TFPT, and tested whether it could be SUMO-modified. Amino acid sequence analysis using GPS-SUMO software (39) revealed the presence of a consensus SUMOylation motif at K216 near the C terminus of the protein. To test whether this lysine residue was important for SUMOylation, we created a lysine-to-arginine TFPT mutant, TFPTK216R. Then we conducted in vitro SUMOylation assays by expressing TFPT in rabbit reticulocyte lysate in the presence of [35S]-methionine, followed by incubation with or without the E1 and E2 SUMOylation enzymes with either SUMO1 or SUMO2 (Fig. 5A). When wildtype TFPT was incubated with the SUMOylation enzymes and either SUMO1 or SUMO2, we observed a strong band representing a 20-kDa molecular weight shift consistent with a SUMO-modified form of TFPT. However, when we incubated the in vitro translated TFPTK216R with the SUMOylation enzyme cocktails in the presence of either SUMO1 or SUMO2, no such shifted band was observed. This observation indicated that Lys216 is the major lysine residue for TFPT SUMO-modification by both SUMO isoforms SUMO1 and SUMO2.
Fig. 5.
PIAS1 and PIAS3 induce modification of TFPT by SUMO2. A, Results from in vitro SUMOylation assay for TFPT. Wildtype TFPT and the K216R mutant were translated in vitro in the presence of [35S]-methionine and incubated either with or without the E1 and E2 SUMOylation enzymes, and with either SUMO1 or SUMO2. B, Results from the in vivo SUMOylation assay for TFPT. HEK 293T cells were cotransfected with TFPT and the K216R mutants both in the presence or absence of SUMO2, and in the presence or absence of either PIAS1 or PIAS3. We immunoprecipitated V5-tagged TFPT and immunoblotted against myc-tagged SUMO2 to detect SUMOylated TFPT.
To validate this result in vivo, we tested the ability of TFPT to be SUMOylated in transfected mammalian cells. We cotransfected V5-tagged TFPT with or without myc-tagged SUMO2 into HEK 293T cells, then immunoprecipitated V5-TFPT using an anti-V5 antibody, followed by immunoblotting using an anti-myc antibody to detect SUMOylated TFPT. In the absence of myc-SUMO2, no bands were observed in the anti-myc Western blot (Fig. 5B). However, when we cotransfected SUMO2, we saw a strong band representing SUMO2-modified TFPT. Next, we tested the ability of TFPTK216R to be SUMOylated in mammalian cells. Although TFPTK216R was expressed (see input anti-V5 blot) at equal levels and immunoprecipitated (see IP:V5/IB:V5 blot) with equal efficiency, we failed to see any signals in the anti-myc blot, indicating that TFPTK216R was not SUMOylated. This shows that Lys216 is essential for SUMOylation of TFPT in mammalian cells.
TFPT SUMOylation at Lys216 is Enhanced by PIAS1 and PIAS3 In Vivo
Our group had shown significant enhancement of TFPT SUMOylation in the presence of the SUMO E3 ligases PIAS1 and PIAS3 using HuProt microarray based SUMOylation assays (18). In order to test whether PIAS1 and PIAS3 enhanced SUMOylation of TFPT in mammalian cells, we cotransfected TFPT constructs with either FLAG-tagged PIAS1 or untagged PIAS3 constructs (Fig. 5B). In cells cotransfected with TFPT, SUMO2 and PIAS1 constructs, we observed a strong signal corresponding to mono-SUMOylated TFPT with several additional bands representing several higher molecular weight forms of SUMOylated TFPT, presumably because of either poly SUMOylation at multiple lysine residues or modification by polymeric SUMO2. However, in cells cotransfected with TFPTK216R, SUMO2 and PIAS1 constructs, the anti-myc immunoblot showed only weak signal corresponding to the highest range of molecular weights. Consistent with our above observation, Lys216 is the major site in TFPT to be SUMOylated by PIAS1. Finally, we tested the ability of TFPT SUMOylation to be enhanced by the SUMO E3 ligase PIAS3. As with PIAS1, we saw a strong band representing mono-SUMOylated TFPT as well as a laddering band pattern representing several higher molecular weight forms of SUMOylated TFPT. This pattern was not observed when transfecting TFPTK216R. Taken together, these results indicate that both SUMO E3 ligases PIAS1 and PIAS3 could effectively enhance TFPT's SUMOylated on Lys216 in mammalian cells, and that PIAS1 and PIAS3 could further stimulate formation of higher molecular weight forms of SUMOylated TFPT.
SUMOylation Enhances Interaction Between TFPT and ION80E
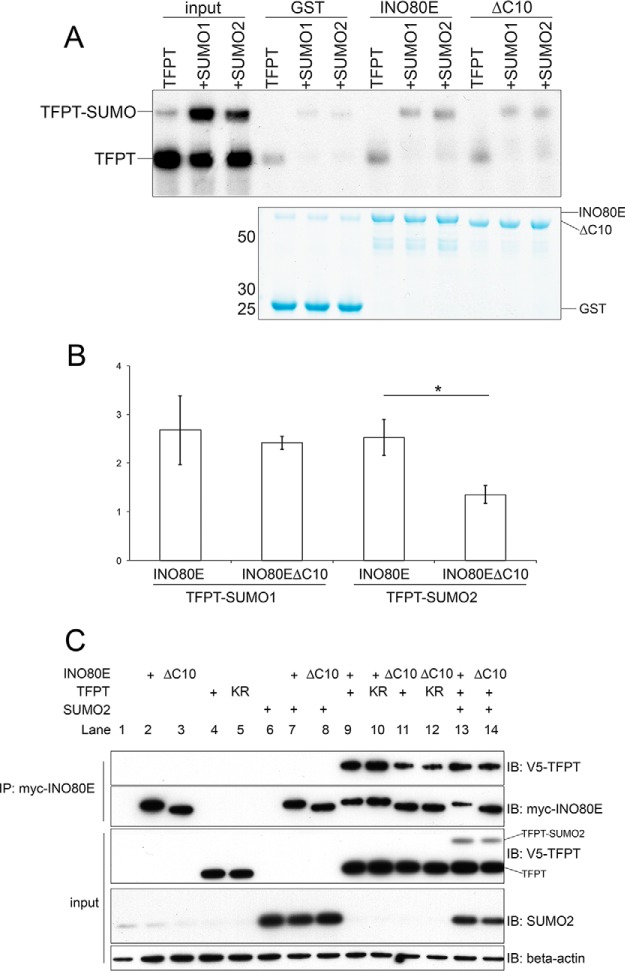
INO80E and TFPT are known subunits of the human INO80 chromatin-remodeling complex and interact with the N-terminal domain of the INO80 ATPase (31). The SUMOmodPPI network motif containing INO80E, TFPT and SUMO2 predicts that SUMOylation may enhance interaction between INO80E and TFPT. Considering that the INO80E C-terminal SIM is important for the ability of INO80E to interact with SUMO2, and that TFPT is modified by SUMO2, we hypothesized that the INO80E SIM may mediate the interaction between INO80E and SUMOylated TFPT. In order to test this hypothesis, we conducted an in vitro binding assay to allow us to measure binding between INO80E and SUMO-modified TFPT. We first generated recombinant radiolabeled TFPT using in vitro translation in the presence of [35S]-methionine, then incubated the reaction with or without SUMO1 or SUMO2 and the E1 and E2, to give us a mixture of unmodified and SUMO-modified TFPT (Fig. 6A). We then used the resulting protein mixture to conduct binding assays with purified recombinant GST-INO80E bound to glutathione Sepharose. Although the mixture of TFPT and SUMO2-modified TFPT forms contained a larger fraction of the unmodified form than the SUMOylated form, following binding to GST-INO80E, we only recovered the SUMO2-modified form of TFPT, suggesting that INO80E preferentially interacts with the SUMOylated form of TFPT.
Fig. 6.
INO80E selectively binds to SUMO-conjugated TFPT. A, Results from in vitro binding assay of [35S]-methionine-labeled TFPT to wildtype and SIM-deficient INO80E in the presence and absence of E1/E2 enzymes and SUMO. B, Quantitative analysis of these results by scanning densitometry. C, Results from in vivo binding assay of wildtype and mutant TFPT and INO80E the presence and absence of SUMO2. Myc-tagged INO80E was immunoprecipitated, then immunoblotted with anti-V5 to detect coimmunoprecipitated TFPT.
When we conducted a binding reaction using the INO80EΔC10 mutant lacking the C-terminal SIM, we saw a reduction in the recovery of SUMOylated TFPT. We then repeated this experiment in triplicate. We found a significant reduction in binding of the INO80EΔC10 mutant to the SUMO2-modified form of TFPT, whereas binding to the SUMO1-modified form of TFPT was not significantly reduced (Fig. 6B). Additionally, the INO80EΔC10 mutant did not show any difference in its interaction with unmodified TFPT, relative to wildtype INO80E (data not shown). This suggests that the C-terminal SIM of INO80E is specifically required for interacting with the SUMO2-modified form of TFPT.
Next, we wanted to determine whether the INO80E C-terminal SIM was important for the interaction between INO80E and TFPT in mammalian cells. We conducted coimmunoprecipitation experiments in HEK293T cells to investigate the interaction between INO80E and TFPT. We transfected cells with myc-INO80E and V5-TFPT, then conducted immunoprecipitation using anti-myc, and Western blotting using anti-V5 (Fig. 6C). This showed that wildtype INO80E could coimmunoprecipitate both wildtype TFPT and TFPTK216R (lanes 9 and 10). However, when we cotransfected the INO80EΔC10 mutant with TFPT, we observed a greatly reduced ability of mutant INO80E to coimmunoprecipitate both TFPT and TFPTK216R (lanes 11 and 12). We were unable to detect the SUMO-modified form of TFPT in the coimmunoprecipitated fraction, likely because of the low stoichiometry of this form in transfected cells.
Finally we cotransfected INO80E wildtype and INO80EΔC10 mutant with TFPT and SUMO2. We observed enhanced interaction between INO80E and TFPT under conditions of SUMO2 overexpression, and this was observed for both the wildtype INO80E and the INO80EΔC10 mutant (lanes 13 and 14). However, by calculating the ratio of coimmunoprecipitated TFPT relative to the amount of INO80E, we observed a greater than 3-fold increase in the amount of TFPT bound to wildtype INO80E compared with the amount of TFPT bound to INO80EΔC10. These results suggest that SUMO2 promotes the INO80E-TFPT interaction, and that the INO80E C-terminal SIM is important for this interaction.
DISCUSSION
Harnessing the power of the human proteome microarray, we performed the most comprehensive and unbiased search to date for novel SUMO-binding partners. Furthermore, by integrating this SUMO-binding data with SUMO substrate data, as well as protein-protein interactions, we have generated an extensive list of SUMO-centered networks that suggest new functions of SUMO, particularly in mediating protein-protein interactions. This fits with previously proposed models of SUMO functioning as molecular glue that stabilizes protein-protein interaction (24, 40). We have found that a combination of SUMOylation and SUMO-binding mediate the interaction between INO80 complex subunits. SUMOylation of the INO80 complex subunit TFPT primes the protein for interaction with INO80E, via its SUMO interaction motif.
However, there are some important caveats to this model. For example, we observed that INO80E has the ability to interact with unmodified TFPT and that this interaction is enhanced when SUMOylated TFPT is absent from the reaction mixture. Under in vitro conditions in which SUMOylated TFPT is present, there is only limited binding of INO80E to unmodified TFPT. This suggests to us that both SUMO-modified and unmodified forms of TFPT may compete for binding to a single site on INO80E, and that TFPT interaction with this site may be enhanced by TFPT SUMOylation. In mammalian cells, we found that the interaction between INO80E and TFPT may be dependent on the INO80E SIM, regardless of whether the SUMOylation site on TFPT that we identified has been mutated. This could indicate that the INO80E SIM may be required for interaction with unmodified TFPT. Another possibility is that TFPT could be SUMOylated on multiple additional sites that these minor SUMOylation sites may also contribute to the interaction with INO80E. Alternatively, the INO80E SIM may interact with other endogenous SUMO-modified proteins, which may in turn mediate the interaction between INO80E and TFPT.
Although the roles of the INO80E and TFPT subunits in INO80 complex function are unknown, and thus the significance of their interaction is unclear, these proteins have been proposed to serve a regulatory function that is likely specific to metazoans, as there are no known orthologs of these subunits in yeast (32). We propose that the SUMO-SIM interaction may be an important regulator of the INO80 complex.
Furthermore, we confirmed the results of a human proteome microarray-based screen to identify SUMO E3 ligase-dependent SUMOylation substrates that showed that SUMOylation of TFPT is enhanced by the SUMO E3 ligases PIAS1 and PIAS3 (18). Previously, PIAS1 and PIAS3 were identified as upstream regulators of chromatin remodeling through their role in stimulating SUMOylation of the protein MBD1, thus affecting its ability to interact with the histone methyltransferase SETDB1 (41). Thus, regulation of chromatin-associated protein-protein interactions may be a general role of the PIAS1 and PIAS3 SUMO E3 ligases.
There is some evidence to suggest TFPT may function as a transcription factor. TFPT was reported to contain a b-ZIP domain (31), and it was found to possess sequence-specific DNA binding activity in a screen to identify DNA binding proteins using a transcription factor protein microarray (8). The transcription factor YY1, a known SUMO target, was shown to recruit the INO80 complex to specific gene promoters (42, 43). SUMOylated TFPT may play a similar role, by binding to both the genome and to the INO80 complex via the SUMO-binding subunit INO80E, in order to recruit the complex to specific genomic locations. This in turn should regulate chromatin conformation and gene transcription. This is a particularly attractive hypothesis considering the well-established role of SUMOylation in regulating transcription factors by directing the assembly of specific multiprotein complexes.
A recent study found that knockdown of the INO80 ATPase, as well as the other INO80 complex subunits INO80E and TFPT, resulted in embryonic stem cell differentiation (44). Additionally, knockdown of INO80E in mouse embryonic stem cells resulted in significant gene expression changes in >1200 genes as measured by microarray analysis (44). Interestingly, the INO80 complex was found to regulate the gene Sox2 and occupy multiple locations in the Sox2 gene in mouse embryonic stem cells. Interestingly, a YY1 binding site identified by the ENCODE project also overlaps with a TFPT DNA sequence motif identified previously (8) in the orthologous region of the human genome. This points to a potential role of both TFPT and YY1 working redundantly to recruit the INO80 complex to regulatory elements of Sox2 to sustain embryonic stem cell pluripotency.
This work extends the list of known SUMO-binding proteins 10-fold. Considering that many previously identified SUMO-modified proteins are localized to the nucleus, including many transcription factors, this enrichment of cytoplasmic proteins suggests that the importance of SUMOylation in the cytoplasm may be underappreciated. There are alternative explanations for the observed enrichment of cytoplasmic proteins among the SUMO-interacting proteins identified by our approach. It is possible that SUMO's interaction with nuclear proteins could be dependent on the presence of other nuclear proteins or DNA that is not present in the reaction mixture incubated on the protein microarray, thus hindering our ability to identify nuclear SUMO-interacting proteins. Additionally, it is possible that in some cases, other proteins or biomolecules were copurified with proteins that were spotted in the array. Although our analysis of randomly chosen purified proteins indicates that these proteins are generally of high quality and high purity, and we estimate that 98% of proteins have the correct band size, it is impractical to analyze every protein that we have purified and spotted on our protein arrays. Unidentified contaminants, including other interacting proteins, could interact strongly with our SUMO probes, thus giving us a false positive result. We believe that every experimental approach to identify SUMO-interacting proteins has their own inherent biases. Previous analysis of protein quantities spotted on our arrays versus protein subcellular localization and function showed that there are higher amounts of cytoplasmic proteins on our protein arrays compared with proteins localized to other parts of the cell. Additionally, transcription factors, which have frequently been associated with SUMOylation in the literature, are expressed at lower levels in our yeast expression system, which may contribute to the low number of transcription factors identified in our assay.
The in vivo role of the SUMO-binding activity of these proteins must be determined through systematic identification of the relevant SUMO interaction motifs, mutagenesis, and further study of the downstream consequences. We believe that our network motif analysis approach further contributes an important context in which to look at the role of the SUMO-binding activity of these proteins.
Supplementary Material
Acknowledgments
We thank Lizhi Jiang for excellent technical assistance and managing clone collections, and Hee-Sool Rho, Crystal Woodard, Shaohui Hu, and John Neiswinger for help with protein purification.
Footnotes
Author contributions: E.C., I.U., M.M., J.Q., H.Z., and S.B. designed research; E.C., I.U., and J.J. performed research; W.H., J.H., C.G., J.J., and M.M. contributed new reagents or analytic tools; E.C., W.H., I.U., and J.H. analyzed data; E.C., M.M., J.Q., H.Z., and S.B. wrote the paper.
* The work was supported by NIH grant EY017015 (to S.B.), National Institutes of Health Common Fund Grant U54RR020839 to H.Z., and a predoctoral award from the American Heart Association 10PRE3040000 to E.C. The content is solely the responsibility of the authors and does not necessarily represent the official views of the National Institutes of Health.
 This article contains supplemental material.
This article contains supplemental material.
1 I. Uzoma, J. Hu, E. Cox, S. Xia, J. Zhou, H.-S. Rho, C. Guzzo, C. Paul, O. Ajala, C. R. Goodwin, J. Jeong, H. Zhang, P. Meluh, S. Blackshaw, M. J. Matunis, J. Qian, H. Zhu, Global identification of SUMO substrates reveals that crosstalk between SUMOylation and phosphorylation promotes cell migration, manuscript under review.
2 The abbreviations used are:
- SIM
- SUMO-interaction motif.
REFERENCES
- 1. Zhu H., Bilgin M., Bangham R. et al. (2001) Global analysis of protein activities using proteome chips. Science 293, 2101–2105 [DOI] [PubMed] [Google Scholar]
- 2. Zhu H., Hu S., Jona G., Zhu X., Kreiswirth N., Willey B. M., Mazzulli T., Liu G., Song Q., Chen P., Cameron M., Tyler A., Wang J., Wen J., Chen W., Compton S., and Snyder M. (2006) Severe acute respiratory syndrome diagnostics using a coronavirus protein microarray. Proc. Natl. Acad. Sci. USA 103, 4011–4016 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Chen C. S., Korobkova E., Chen H., Zhu J., Jian X., Tao S. C., He C., and Zhu H. (2008) A proteome chip approach reveals new DNA damage recognition activities in Escherichia coli. Nat. Methods 5, 69–74 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Popescu S. C., Popescu G. V., Bachan S., Zhang Z., Seay M., Gerstein M., Snyder M., and Dinesh-Kumar S. P. (2007) Differential binding of calmodulin-related proteins to their targets revealed through high-density Arabidopsis protein microarrays. Proc. Natl. Acad. Sci. USA 104, 4730–4735 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Lueking A., Possling A., Huber O., Beveridge A., Horn M., Eickhoff H., Schuchardt J., Lehrach H., and Cahill D. J. (2003) A nonredundant human protein chip for antibody screening and serum profiling. Mol. Cell. Proteomics 2, 1342–1349 [DOI] [PubMed] [Google Scholar]
- 6. Hu S., Li Y., Liu G., Song Q., Wang L., Han Y., Zhang Y., Song Y., Yao X., Tao Y., Zeng H., Yang H., Wang J., Zhu H., Chen Z. N., and Wu L. (2007) A protein chip approach for high-throughput antigen identification and characterization. Proteomics 7, 2151–2161 [DOI] [PubMed] [Google Scholar]
- 7. Song Q., Liu G., Hu S., Zhang Y., Tao Y., Han Y., Zeng H., Huang W., Li F., Chen P., Zhu J., Hu C., Zhang S., Li Y., Zhu H., and Wu L. (2010) Novel autoimmune hepatitis-specific autoantigens identified using protein microarray technology. J. Proteome Res. 9, 30–39 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Hu S., Xie Z., Onishi A., Yu X., Jiang L., Lin J., Rho H. S., Woodard C., Wang H., Jeong J. S., Long S., He X., Wade H., Blackshaw S., Qian J., and Zhu H. (2009) Profiling the human protein-DNA interactome reveals ERK2 as a transcriptional repressor of interferon signaling. Cell 139, 610–622 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Hu S., Wan J., Su Y., Song Q., Zeng Y., Nguyen H. N., Shin J., Cox E., Rho H. S., Woodard C., Xia S., Liu S., Lyu H., Ming G. L., Wade H., Song H., Qian J., and Zhu H. (2013) DNA methylation presents distinct binding sites for human transcription factors. eLife 2, e00726. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Jeong J. S., Jiang L., Albino E., Marrero J., Rho H. S., Hu J., Hu S., Vera C., Bayron-Poueymiroy D., Rivera-Pacheco Z. A., Ramos L., Torres-Castro C., Qian J., Bonaventura J., Boeke J. D., Yap W. Y., Pino I., Eichinger D. J., Zhu H., and Blackshaw S. (2012) Rapid identification of monospecific monoclonal antibodies using a human proteome microarray. Mol. Cell. Proteomics 11, O111.016253. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Huang Y., Jeong J. S., Okamura J., Sook-Kim M., Zhu H., Guerrero-Preston R., and Ratovitski E. A. (2012) Global tumor protein p53/p63 interactome: making a case for cisplatin chemoresistance. Cell Cycle 11, 2367–2379 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Chen Y., Yang L.-N., Cheng L., Tu S., Guo S.-J., Le H.-Y., Xiong Q., Mo R., Li C.-Y., Jeong J.-S., Jiang L., Blackshaw S., Bi L.-J., Zhu H., Tao S.-C., and Ge F. (2013) Bcl2-associated athanogene 3 interactome analysis reveals a new role in modulating proteasome activity. Mol. Cell. Proteomics 12, 2804–2819 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Barry G., Briggs J. A., Vanichkina D. P., Poth E. M., Beveridge N. J., Ratnu V. S., Nayler S. P., Nones K., Hu J., Bredy T. W., Nakagawa S., Rigo F., Taft R. J., Cairns M. J., Blackshaw S., Wolvetang E. J., and Mattick J. S. (2014) The long non-coding RNA Gomafu is acutely regulated in response to neuronal activation and involved in schizophrenia-associated alternative splicing. Mol. Psychiatry 19, 486–494 [DOI] [PubMed] [Google Scholar]
- 14. Donnelly C. J., Zhang P. W., Pham J. T., Heusler A. R., Mistry N. A., Vidensky S., Daley E. L., Poth E. M., Hoover B., Fines D. M., Maragakis N., Tienari P. J., Petrucelli L., Traynor B. J., Wang J., Rigo F., Bennett C. F., Blackshaw S., Sattler R., and Rothstein J. D. (2013) RNA toxicity from the ALS/FTD C9ORF72 expansion is mitigated by antisense intervention. Neuron 80, 415–428 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Hu C. J., Song G., Huang W., Liu G. Z., Deng C. W., Zeng H. P., Wang L., Zhang F. C., Zhang X., Jeong J. S., Blackshaw S., Jiang L. Z., Zhu H., Wu L., and Li Y. Z. (2012) Identification of new autoantigens for primary biliary cirrhosis using human proteome microarrays. Mol. Cell. Proteomics 11, 669–680 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Tarrant M. K., Rho H. S., Xie Z., Jiang Y. L., Gross C., Culhane J. C., Yan G., Qian J., Ichikawa Y., Matsuoka T., Zachara N., Etzkorn F. A., Hart G. W., Jeong J. S., Blackshaw S., Zhu H., and Cole P. A. (2012) Regulation of CK2 by phosphorylation and O-GlcNAcylation revealed by semisynthesis. Nat. Chem. Biol. 8, 262–269 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Lee Y. I., Giovinazzo D., Kang H. C., Lee Y., Jeong J. S., Doulias P. T., Xie Z., Hu J., Ghasemi M., Ischiropoulos H., Qian J., Zhu H., Blackshaw S., Dawson V. L., and Dawson T. M. (2014) Protein microarray characterization of the S-nitrosoproteome. Mol. Cell. Proteomics 13, 63–72 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Uzoma I., Hu J., Cox E. et al. (2017) Global Identification of SUMO Substrates Reveals that Crosstalk between SUMOylation and Tyrosine Phosphorylation Promotes Cell Migration. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Johnson E. S., Schwienhorst I., Dohmen R. J., and Blobel G. (1997) The ubiquitin-like protein Smt3p is activated for conjugation to other proteins by an Aos1p/Uba2p heterodimer. EMBO J. 16, 5509–5519 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Fraser A. G., Kamath R. S., Zipperlen P., Martinez-Campos M., Sohrmann M., and Ahringer J. (2000) Functional genomic analysis of C. elegans chromosome I by systematic RNA interference. Nature 408, 325–330 [DOI] [PubMed] [Google Scholar]
- 21. Saracco S. A., Miller M. J., Kurepa J., and Vierstra R. D. (2007) Genetic analysis of SUMOylation in Arabidopsis: conjugation of SUMO1 and SUMO2 to nuclear proteins is essential. Plant Physiol. 145, 119–134 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Nacerddine K., Lehembre F., Bhaumik M., Artus J., Cohen-Tannoudji M., Babinet C., Pandolfi P. P., and Dejean A. (2005) The SUMO pathway is essential for nuclear integrity and chromosome segregation in mice. Dev. Cell 9, 769–779 [DOI] [PubMed] [Google Scholar]
- 23. Geiss-Friedlander R., and Melchior F. (2007) Concepts in sumoylation: a decade on. Nat. Rev. Mol. Cell Biol. 8, 947–956 [DOI] [PubMed] [Google Scholar]
- 24. Takahashi H., Hatakeyama S., Saitoh H., and Nakayama K. I. (2005) Noncovalent SUMO-1 binding activity of thymine DNA glycosylase (TDG) is required for its SUMO-1 modification and colocalization with the promyelocytic leukemia protein. J. Biol. Chem. 280, 5611–5621 [DOI] [PubMed] [Google Scholar]
- 25. Matunis M., Wu J., and Blobel G. (1998) SUMO-1 Modification and Its Role in Targeting the Ran GTPase-activating Protein, RanGAP1, to the Nuclear Pore Complex. J. Cell Biol. 140, 499. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Guzzo C. M., Ringel A., Cox E., Uzoma I., Zhu H., Blackshaw S., Wolberger C., and Matunis M. J. (2014) Characterization of the SUMO-Binding Activity of the Myeloproliferative and Mental Retardation (MYM)-Type Zinc Fingers in ZNF261 and ZNF198. PloS One 9, e105271. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Danielsen J. R., Povlsen L. K., Villumsen B. H., Streicher W., Nilsson J., Wikstrom M., Bekker-Jensen S., and Mailand N. (2012) DNA damage-inducible SUMOylation of HERC2 promotes RNF8 binding via a novel SUMO-binding Zinc finger. J. Cell Biol. 197, 179–187 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Hecker C. M., Rabiller M., Haglund K., Bayer P., and Dikic I. (2006) Specification of SUMO1- and SUMO2-interacting motifs. J. Biol. Chem. 281, 16117–16127 [DOI] [PubMed] [Google Scholar]
- 29. Ouyang J., Shi Y., Valin A., Xuan Y., and Gill G. (2009) Direct binding of CoREST1 to SUMO-2/3 contributes to gene-specific repression by the LSD1/CoREST1/HDAC complex. Mol. Cell 34, 145–154 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Prakash A., Piening B., Whiteaker J., Zhang H., Shaffer S. A., Martin D., Hohmann L., Cooke K., Olson J. M., Hansen S., Flory M. R., Lee H., Watts J., Goodlett D. R., Aebersold R., Paulovich A., and Schwikowski B. (2007) Assessing bias in experiment design for large scale mass spectrometry-based quantitative proteomics. Mol. Cell. Proteomics 6, 1741–1748 [DOI] [PubMed] [Google Scholar]
- 31. Jin J., Cai Y., Yao T., Gottschalk A. J., Florens L., Swanson S. K., Gutierrez J. L., Coleman M. K., Workman J. L., Mushegian A., Washburn M. P., Conaway R. C., and Conaway J. W. (2005) A mammalian chromatin remodeling complex with similarities to the yeast INO80 complex. J. Biol. Chem. 280, 41207–41212 [DOI] [PubMed] [Google Scholar]
- 32. Chen L., Cai Y., Jin J., Florens L., Swanson S. K., Washburn M. P., Conaway J. W., and Conaway R. C. (2011) Subunit organization of the human INO80 chromatin remodeling complex: an evolutionarily conserved core complex catalyzes ATP-dependent nucleosome remodeling. J. Biol. Chem. 286, 11283–11289 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Reverter D., and Lima C. D. (2005) Insights into E3 ligase activity revealed by a SUMO-RanGAP1-Ubc9-Nup358 complex. Nature 435, 687–692 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Zhu J., Zhu S., Guzzo C. M., Ellis N. A., Sung K. S., Choi C. Y., and Matunis M. J. (2008) Small ubiquitin-related modifier (SUMO) binding determines substrate recognition and paralog-selective SUMO modification. J. Biol. Chem. 283, 29405–29415 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. Aguilar-Martinez E., Chen X., Webber A., Mould A. P., Seifert A., Hay R. T., and Sharrocks A. D. (2015) Screen for multi-SUMO-binding proteins reveals a multi-SIM-binding mechanism for recruitment of the transcriptional regulator ZMYM2 to chromatin. Proc. Natl. Acad. Sci. USA 112, E4854–E4863 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36. Uda M., Kawasaki H., Iizumi K., Shigenaga A., Baba T., Naito H., Yoshioka T., and Yamakura F. (2015) Sumoylated alpha-skeletal muscle actin in the skeletal muscle of adult rats. Mol. Cell. Biochem. 409, 59–66 [DOI] [PubMed] [Google Scholar]
- 37. Gehlert S., Klinz F. J., Willkomm L., Schiffer T., Suhr F., and Bloch W. (2016) Intense resistance exercise promotes the acute and transient nuclear translocation of small ubiquitin-related modifier (SUMO)-1 in human myofibres. Int. J. Mol. Sci. 17, E646. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38. Hwang W., Hackler L. Jr., Wu G., Ji H., Zack D. J., and Qian J. (2012) Dynamics of regulatory networks in the developing mouse retina. PloS One 7, e46521. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39. Zhao Q., Xie Y., Zheng Y., Jiang S., Liu W., Mu W., Liu Z., Zhao Y., Xue Y., and Ren J. (2014) GPS-SUMO: a tool for the prediction of sumoylation sites and SUMO-interaction motifs. Nucleic Acids Res. 42, W325–W330 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40. Matunis M. J., Zhang X. D., and Ellis N. A. (2006) SUMO: the glue that binds. Dev. Cell 11, 596–597 [DOI] [PubMed] [Google Scholar]
- 41. Lyst M. J., Nan X., and Stancheva I. (2006) Regulation of MBD1-mediated transcriptional repression by SUMO and PIAS proteins. EMBO J. 25, 5317–5328 [DOI] [PMC free article] [PubMed] [Google Scholar] [Retracted]
- 42. Deng Z., Wan M., and Sui G. (2007) PIASy-mediated sumoylation of Yin Yang 1 depends on their interaction but not the RING finger. Mol. Cell. Biol. 27, 3780–3792 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43. Cai Y., Jin J., Yao T., Gottschalk A. J., Swanson S. K., Wu S., Shi Y., Washburn M. P., Florens L., Conaway R. C., and Conaway J. W. (2007) YY1 functions with INO80 to activate transcription. Nat. Struct. Mol. Biol. 14, 872–874 [DOI] [PubMed] [Google Scholar]
- 44. Wang L., Du Y., Ward J. M., Shimbo T., Lackford B., Zheng X., Miao Y. L., Zhou B., Han L., Fargo D. C., Jothi R., Williams C. J., Wade P. A., and Hu G. (2014) INO80 facilitates pluripotency gene activation in embryonic stem cell self-renewal, reprogramming, and blastocyst development. Cell Stem Cell 14, 575–591 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.