Abstract
Immunoglobulin G (IgG) proteins are known for the huge diversity of the variable domains of their heavy and light chains, aimed at protecting each individual against foreign antigens. The IgG also harbor specific polymorphism concentrated in the CH2 and CH3-CHS constant regions located on the Fc fragment of their heavy chains. But this individual particularity relies only on a few amino acids among which some could make accurate sequence determination a challenge for mass spectrometry-based techniques.
The purpose of the study was to bring a molecular validation of proteomic results by the sequencing of encoding DNA fragments. It was performed using ten individual samples (DNA and sera) selected on the basis of their Gm (gamma marker) allotype polymorphism in order to cover the main immunoglobulin heavy gamma (IGHG) gene diversity. Gm allotypes, reflecting part of this diversity, were determined by a serological method. On its side, the IGH locus comprises four functional IGHG genes totalizing 34 alleles and encoding the four IgG subclasses. The genomic study focused on the nucleotide polymorphism of the CH2 and CH3-CHS exons and of the intron. Despite strong sequence identity, four pairs of specific gene amplification primers could be designed. Additional primers were identified to perform the subsequent sequencing. The nucleotide sequences obtained were first assigned to a specific IGHG gene, and then IGHG alleles were deduced using a home-made decision tree reading of the nucleotide sequences. IGHG amino acid (AA) alleles were determined by mass spectrometry. Identical results were found at 95% between alleles identified by proteomics and those deduced from genomics. These results validate the proteomic approach which could be used for diagnostic purposes, namely for a mother-and-child differential IGHG detection in a context of suspicion of congenital infection.
The human immune response mediated by the antibodies relies essentially on IgG, subdivided into four subclasses IgG1, IgG2, IgG3, and IgG4, ordered by decreasing concentrations in the circulating blood (1). The specificity of this immune response is ensured by the immense diversity of the repertoire of antigenic recognition carried by the paratopes, i.e. the variable domains of the heavy and light chains (2). The immunoglobulin (IG)1 heavy gamma chains exhibit polymorphisms, mainly localized on the CH2 and CH3-CHS regions of the Fc fragment. This diversity relies on the number of heavy gamma (IGHG) nucleotide substitutions and amino acid (AA) changes listed in IMGT®, the international ImMunoGeneTics information system® (http://www.imgt.org) (3). The polymorphism of the IG gamma chains is both isotypic (there are four functional IGHG genes) and allelic. To date 34 IMGT alleles (5 IGHG1, 6 IGHG2, 19 IGHG3, and 4 IGHG4) are identified, which correspond to 25 alleles with amino acid changes in the coding regions or IMGT AA alleles (3 IGHG1, 4 IGHG2, 15 IGHG3, and 3 IGHG4 IMGT AA alleles, respectively) (4, 5). Several of the gamma chain polymorphisms are genetic variants detected serologically (or allotypes) and the combination of these gamma markers (Gm) carried by the gamma1, gamma2, and gamma3 chains constitute the G1m alleles, G2m alleles, and G3m alleles (4). The peptide diversity is subtle and is based on only a few amino acids sequence changes, some of which being very close when using mass measurement.
It may be important to take into account the IGHG polymorphism in several, nonexhaustive, cases. First, the use of monoclonal antibodies and related products is growing rapidly as therapeutic agents in disease areas such as cancer, rheumatoid arthritis, and Alzheimer's disease (6–8). Second, observations have been made on the dependence on some Fc conserved but also polymorphic AA localized at the CH2-CH3 domain interface, of the IgG Fc binding affinity to the FCGRT (neonatal Fc receptor, FcRn) (9). Indeed, apart from its role in transferring maternal IgG from mother to fetus via the placenta, the FCGRT contributes to enhance the IgG half-life in serum (10), and this aspect may be critical in the context of IgG-based therapeutics (11). Last but not least, the knowledge and the use of the in vivo IGHG polymorphism may lead to an important improvement of the biological diagnosis in neonates of certain congenital pathologies. Indeed, difficulties are encountered in the serologic detection of IgG neosynthesized by the fetus in cases where congenital infections are suspected, because of the systemic transfer of maternal IgG that occurs in utero across the placenta, resulting in about 90% of the maternal serum level of IgG in the full-term newborns at delivery (12). It is namely the case of parasitic infections such as congenital toxoplasmosis and congenital Chagas disease (13) for which combinations of parasitological, molecular or immunological tests are required to confirm the infection status (14, 15). As proof of concept, a proteomic approach exploiting the individual IGHG3 AA polymorphism has been patented in our laboratory, that aimed at distinguishing maternal from fetal total IgG in a newborn's blood sample by means of the detection by bottom-up mass spectrometry of proteotypic peptides allowing an assignation to IGHG3 alleles attributable to the IgG3 of either maternal or fetal origin (16, 17).
However, the strong sequence homogeneity between the different IGHG AA alleles imposes a strict verification of the results obtained by mass spectrometry (MS). For this reason, the aim of the present study was to bring a molecular confirmation of IGHG AA alleles determined by bottom-up mass spectrometry. A panel of ten plasma samples was selected from a previous study on immunogenetics of malaria performed in Benin, on the basis of the inter-individual diversity in their Gm alleles (4), deduced by the phenotypic characterization of their Gm allotypes using a serological technique (18). In parallel, the corresponding genomic DNA was extracted and the gene portions comprising the polymorphic CH2 and CH3-CHS exons of the IGHG1, IGHG2, IGHG3, and IGHG4 genes were amplified and then sequenced in order to determine individual IGHG alleles. This study allowed assessing the degree of concordance between serological, proteomic and genomic determinations performed for each sample. Correlations have already been established between Gm alleles and IGHG AA alleles (4) as well as between G3m alleles deduced from serology and IGHG3 nucleotide alleles determined by genomic sequencing (19). The present study is the first to validate IGHG alleles identified by MS using DNA sequencing on samples selected on the basis of their Gm allele diversity, itself deduced from the serological determination of Gm allotypes. Outside the originality of the double confrontation of results presented here, this work aims at reinforcing the necessity to determine unambiguously IGHG AA sequences in view of the various therapeutic and/or diagnostic applications under development cited above.
EXPERIMENTAL PROCEDURES
Samples Collection
Plasma and corresponding DNA samples were obtained from a study on human genetic determinants of malaria that was performed in 2006–2007 in the south of Benin by the UMR 216 (18). This study was conducted among 155 children belonging mainly to the Fon ethnic group and was authorized by the institutional “Ethics Committee of the Faculté des Sciences de la Santé″ (FSS) from the Université d'Abomey-Calavi (UAC) in Benin. For the purpose of the present study, samples from 10 children were selected on the basis of their Gm allotype polymorphism, determined by a serological method.
Children PA01, PA07, PA09, PA16, PA31, PA42, PA45, PA48 were asymptomatic carriers of the parasites responsible for malaria and were recruited in a primary school of Ouidah with the approval of the coordinating doctor of the sanitary zone and the inspector of education. For these schoolchildren, a collective written informed consent was obtained from the responsible person in charge of the Parents Association, after having dispensed oral information on the study to the school director, the teachers and the members of the Parents Association. The remaining two children had different clinical presentations of malaria, i.e. mild malaria attack (AS50) and severe malaria (NP49) and were recruited in the pediatric service of the National University Hospital of Cotonou. For these children, a written informed consent was obtained from their parents or guardians.
Blood was collected into 5 ml EDTA Vacutainer® tubes and after centrifugation, plasma samples and isolated peripheral buffy coat were frozen at −20 °C. Genomic DNA was extracted from peripheral blood buffy-coats using the QIAamp DNA Blood Mini Kit (Qiagen GmbH, Hilden, Germany) (20).
Serological Determination of G1m and G3m Allotypes
Gm allotypes for G1m (1, 2, 3, 17) and G3m (5, 6, 10, 11, 13, 14, 15, 16, 21, 24, 28) determinants (4) were analyzed in plasma samples by a qualitative standard hemagglutination inhibition method (21). Whereas G3m28 is a marker of the gamma 3 chains in European and Asian populations, Gm28 is often present on the gamma 1 chains in African populations (22). In brief, human blood group O Rh (D) erythrocytes were coated with anti-Rh (D) antibodies of known Gm allotypes. Plasma sample and reagent monospecific anti-allotype antibodies were added. Plasma containing IgG with a particular Gm allotype inhibited hemagglutination by the corresponding reagent anti-allotype antibody, whereas plasma sample that was negative for this allotype did not.
Mass Spectrometry (MS) Analysis of IGHG Tryptic Peptides
Sample Preparation
For each sample, total IgG was purified by injecting 200 μl of plasma into a Protein G column (Protein G Sepharose HP SpinTrap, GE Healthcare, France) which presents high affinity for the Fc fragment of all IgG subclasses. Following manufacturer's instructions, IgG binding was performed at neutral pH, elution was performed by lowering the pH, and the eluted material was collected in neutralization buffer to preserve the integrity of acid-labile IgG. Dithiothreitol (DTT, 20 mm final) was added to 47 μl of purified samples for 30 min at 56 °C in order to disrupt disulfide bonds. Proteolysis was performed by incubation with trypsin (Promega, France, 10 ng/μl final) for 3 h at 37 °C, and stopped with trifluoroacetic acid (TFA, Pierce, France, 0.5% final).
Bottom-up LC-MS/MS Analysis
Peptides were concentrated and separated by nano High-Performance Liquid Chromatography (nHPLC) hyphenated with an orbitrap mass spectrometer. For each sample, 4 μl were then analyzed in LC-MS/MS. Analyzes were realized using an RSLC Ultimate 3000 Rapid Separation liquid chromatographic system coupled to a hybrid Q Exactive mass spectrometer (Thermo Fisher Scientific). Briefly, peptides were loaded and washed on a C18 reverse phase precolumn (3 μm particle size, 100 Å pore size, 75 μm i.d., 2 cm length). The loading buffer contained 98% H2O, 2% acetonitrile (ACN) and 0.1% TFA. Peptides were then separated on a C18 reverse phase resin (2 μm particle size, 100 Å pore size, 75 μm i.d.,15 cm length) with a 35 min “effective” gradient from 99% A (0.1% formic acid and 100% H2O) to 40% B (80% ACN, 0.085% formic acid and 20% H2O).
The mass spectrometer acquired data throughout the elution process and operated in a data dependent scheme with full MS scans acquired in the orbitrap analyzer, followed by up to 15 MS/MS higher-energy collisional dissociation (HCD) spectra on the most abundant ions detected in the MS scan. Mass spectrometer settings were: full MS (AGC: 3.10e6, resolution: 70,000, m/z range 400–2000, maximum ion injection time: 100 ms); MS/MS (Normalized Collision Energy: 30, resolution: 17,500, intensity threshold: 2.10e4, isolation window: 4.0 m/z, dynamic exclusion time setting: 12 s, AGC Target: 1.10e5 and maximum injection time: 100 ms). The fragmentation was permitted for precursor with a charge state of 2 to 4 excluding isotopes. The software used to generate Mascot generic format (MGF) files was Proteome Discoverer 1.3.
Database Queries of Extracted Experimental Data
Three FASTA-formatted protein databases were used in combination in order to assign a majority of fragmentation spectra: 1- the Homo sapiens entries from the UniProt/SwissProt database release 2016–02 (20,273 sequences), 2- the 2014 IMGT® IGHG allele database (2, 3, 5) (IMGT Repertoire. Sections: Protein displays, Alignments of alleles, http://www.imgt.org) (60 sequences; 20,789 amino acids) and 3- the individual databases of IGHG CH2 and CH3-CHS sequences obtained following the molecular investigation described above for the 10 patients incorporated in this study and supplied in supplemental Table S1 (60 sequences; 13,004 amino acids). Peaklists extracted from MS/MS spectra were compared with the above databases using Mascot search engine 2.5.1 (Matrix Science). The following settings were applied: mass tolerances were 4 ppm for precursors and 10 mmu for fragments, a significance threshold score corresponding to p < 0.05 was applied to filter identifications but a minimum individual peptide Mascot score value of 25 excluded poorly annotated spectra. Based on these criteria, a nontargeted (bottom up) analysis of the samples was performed. In order to avoid possible ambiguities, the following restrictive conditions were applied for mascot queries: trypsin proteolysis specificity without missed cleavage, methionine oxidation as unique modification allowed (as partial) and mass-to-charge (m/z) states with z>2 or more. Under these conditions false positive rates were usually under 1%. Mascot searches resulted in protein groups sharing several peptides but showing specific (unique) peptides demonstrating their presence. Peptide information regarding AA diversity of IGHG1 to IGHG4 genes and their alleles were carefully collected.
Molecular Investigation of the IGHG Diversity
DNA Amplification
The nucleotide sequences of the 34 IGHG alleles which cover the diversity of the four functional IGHG genes were extracted from IMGT/GENE-DB (5) and Alignment of alleles, IMGT Repertoire® (3) (http://www.imgt.org) and aligned using the Multalin website (http://multalin.toulouse.inra.fr). Two polymorphic areas framing the CH2 and the CH3-CHS exons, respectively, allowed designing 4 pairs of primers (IDT, Leuven Belgium) for the amplification of each IGHG gene separately, as represented in Fig. 1 and supplemental Table S2.
Fig. 1.
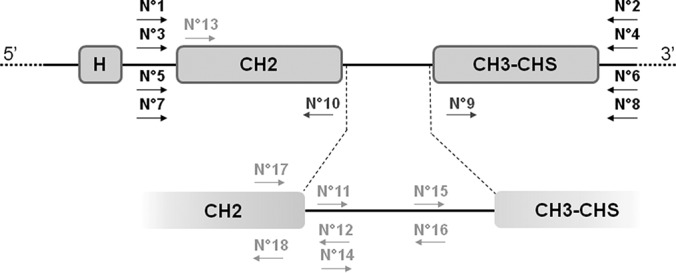
Position of primers on a partial representation of the Homo sapiens IGHG genes. The CH1 exon is not shown. H represents the single hinge exon of IGHG1, IGHG2 and IGHG4, and the most 3′ hinge exon of IGHG3 H4 (Gene tables > IGHC, IMGT Repertoire, http://www.imgt.org). A focus on the intron separating CH2 and CH3-CHS exons is also shown. The primers are represented by arrows which indicate their position and 5′ > 3′ sequence orientation. Sequence numbering positions are for the allele*01 of each IGHG gene (IMGT/LIGM-DB Accession numbers (24): J00228 for IGHG1, J00230 for IGHG2, X03604 for IGHG3, and K01316 for IGHG4) according to IMGT/BlastSearch (http://www.imgt.org). Black arrows represent primers used for PCR and sequencing: N°1/IGHG1_1017–1036 (FWD 5′-gccgggtgctgacacgtcca-3′), N°2/IGHG1_1820–1801 (REV 5′-cttgccggccgtcgcactca-3′), N°3/IGHG2_1018–1037 (FWD 5′-gctgggtgctgacacgtcca-3′), N°4/IGHG2_1819-1800 (REV 5′-cttgcyggccgtggcactca-3′), N°5/IGHG3_1596–1615 (FWD 5′-gtcgggtgctgacacatctg-3′), N°6/IGHG3_2400–2381 (REV 5′-cttgccggcyrtsgcactca-3′), N°7/IGHG4_1029–1048 (FWD 5′-gcatccacctccatctcttc-3′), N°8/IGHG4_1820–1801 (REV 5′-cttgccggccctggcactca-3′). Dark gray arrows represented IGHG consensual sequencing primers, they are common for the 4 IGHG genes: N°9/IGHGseqF (FWD 5′-aggtcagcctgacctgcctg-3′) localized 1544–1563 for IGHG1, 1543–1562 for IGHG2, 2124–2143 for IGHG3, and 1544–1563 for IGHG4, and N°10/IGHGseqR (REV 5′-tggagaccttgcacttgtac-3′) localized 1336–1317 for IGHG1, 1334–1315 for IGHG2, 1915–1896 for IGHG3, and 1335–1316 for IGHG4. Light gray arrows represented IGHG specific sequencing primers: N°11/IGHG1_1390–1405 (FWD 5′-acccgtggggtgcgag-3′), N°12/IGHG1_1405–1390 (REV 5′-ctcgcaccccacgggt-3′), N°13/IGHG2_1058–1073 (FWD 5′-ccacctgtggcaggac-3′), N°14/IGHG2_1388–1403 (FWD 5′-acccgcggggtatgag-3′), N°15/IGHG3_1999–2014 (FWD 5′-aggccagcttgaccca-3′), N°16/IGHG3_2010–1995 (REV 5′-tcaagctggcctctgt-3′), N°17/IGHG4_1348–1366 (FWD 5′-cgtcctccatcgagaaaac-3′), N°18/IGHG4_1356–1340 (REV 5′-tggaggacgggaggcct-3′). FWD = forward; REV = reverse; r = a or g; y = c or t; s = g or c.
Genomic DNA amplification was performed using the AccuPrimeTM TaqDNA Polymerase High Fidelity kit (Invitrogen, France) according to the following conditions: water (10.9 μl) + 10× Buffer II (2 μl) + 100% DMSO (0.4 μl) + 5 U/μl enzyme (0.1 μl) + 5 μm forward primer (0.8 μl) + 5 μm reverse primer (0.8 μl) + 5 ng/μl DNA sample (5 μl). For IGHG1 and IGHG2 the amplification conditions were 94 °C during 30 s (preliminary denaturation); 94 °C during 30 s (denaturation), 69 °C during 30 s (annealing), 72 °C during 1 min (elongation) 35 times; 72 °C during 7 min (final elongation) and then maintaining at 10 °C. Similar conditions were applied for IGHG3 and IGHG4 gene amplification except for annealing and elongation temperatures, which were set at 60 °C and 68 °C, respectively. PCR products of 804 bp (IGHG1), 802 bp (IGHG2), 805 bp (IGHG3), and 762 bp (IGHG4) were obtained. Their quality was checked by gel migration on 1% agarose solubilized in TBE 0.5× at 100 V during 40 min in presence of a 100 bp molecular weight marker (Invitrogen, France). Then, the PCR products were purified using Wizard SV Gel and PCR Clean Up system kit (Promega, France). The DNA concentration was measured on a NanoDrop 2000c spectrophotometer (Thermo Scientific, France).
DNA Sequencing
Sequencing of the PCR products was performed using each of the four pairs of amplification primers with the addition of one consensual pair of primers located inside the CH2 and CH3-CHS region (Fig. 1). A control sequencing of the same PCR products was performed using sequencing primers specific for each of the four IGHG genes and located either inside the CH2 exon, or in the intron between the CH2 and CH3-CHS exons. As recommended by the “Genome and Sequencing Platform” (Institut Cochin-Centre de Recherche, Paris, France), sample templates were prepared as follows: 800 ng of purified PCR product and 30 ng (4 pmoles) primers (forward or reverse) in a 15 μl final volume. Sequencing was performed at the Platform on a 3730XL DNA Analyzer (Applied Biosystems) and raw sequences made available on the Platform website.
Sequence Analysis
Raw sequences were read and corrected with the “4peaks” free software (http://nucleobytes.com/index.php/4peaks). Contigs were then created using the “Sequencher® 5.0” software (Gene Codes Corporation, Ann Arbor, MI), allowing a visualization of sequence chromatograms to be compared with the IGHG1*01, IGHG2*01, IGHG3*01, and IGHG4*01 IMGT® reference alleles (http://www.imgt.org) (5). Although the intron localized between the CH2 and CH3-CHS exons was amplified and sequenced, the sequence analysis was focused on the CH2 and CH3-CHS exons. A first analysis step consisted to check, in addition to the observation of a single band on agarose gel, that the amplification products were IGHG gene specific. For this purpose, verification was made of the presence of several specific codons located along the CH2 and CH3-CHS exons of each IGHG gene, as summarized in supplemental Table S3. Once this done, each IGHG allele could be attributed by moving successively from a polymorphic nucleotide to another along the coding sequence covering the CH2 and CH3-CHS exons. This work was facilitated by the implementation of a decision tree reading of the IGHG nucleotide sequences, represented in supplemental Table S4.
Proteogenomic Analysis
The IGHG nucleotide sequences resulting from the sequencing of amplified DNA fragments of all experimental samples were artificially spliced between the CH2 and CH3-CHS exons and translated using the converter software (http://didac.free.fr/seq/dna2pro.htm). This allowed to create for each sample an experimental “sample database” to be imported in Mascot software for a new peptide query aiming at detecting single amino acid variants (SAAV) unknown to date (26, 27) and therefore not listed in the concatenated IMGT® IGHG allele and SwissProt database used in the first MS analysis. The peptide interrogation was performed according to the same conditions as previously used. For each sample of purified IgG, the analysis proceeded in two steps, a first query of IGHG peptides on each sample-specific database preceding a second query where the sample-specific databases were added to the IMGT® and SwissProt databases.
RESULTS
Gm Alleles Deduced from Serological Determination of Gm Allotypes
Experimental samples originated from 7 boys and 3 girls aged from 4 to 10 years (mean age ± S.D. = 6.9 ± 1.6 years) all belonging to the Fon ethnic group which prevails in South-Benin except one child (PA09) who belonged to the Yoruba ethnic group originating from Nigeria. These samples were selected out of a series of 155 samples for the presence in their plasma of IgG characterized by the four main G3m alleles (G3m5*, G3m6*, G3m15*, and G3m24*) that may be encountered among sub-Saharan African populations (28). Table I presents the results of the serological determination of G3m allotypes, from which G3m alleles were deduced. It appears that IgG from each plasma sample were representative of one particular G3m allele combination, at the homozygous or heterozygous state, Gm allotypes being co-dominantly expressed (4, 19). Otherwise, all samples presented the G1m17,1 allele, known to be encoded by IGHG1*01, IGHG1*02, or IGHG1*05 (Gene table Homo sapiens IGHC, IMGT Repertoire) (http://www.imgt.org) (4). NP49 and PA31 samples were Gm28-positive with a plausible attribution of this allotype to the gamma 1 chain (IGHG1*05p, IGHG1*06p) (4) (Allotypes, IMGT Repertoire, http://www.imgt.org).
Table I. G3m and IGHG3 AA alleles deduced from the G3m allotype determination.
| Plasma samples | G3m allelesa | G3m simplified formb | Attributable IGHG3 AA alleles |
|---|---|---|---|
| AS50 | 5,10,11,13,14/5,10,11,13,14 | G3m5*/G3m5* | IGHG3*01*05*06*07*09*10*11*12/IGHG3*01*05*06*07*09*10*11*12 |
| NP49 | 10,11,13,15/10,11,13,15 | G3m15*/G3m15* | IGHG3*17/IGHG3*17 |
| PA01 | 5,10,11,13,14/10,11,13,15 | G3m5*/G3m15* | IGHG3*01*05*06*07*09*10*11*12/IGHG3*17 |
| PA07 | 5,10,11,13,14/5,6,10,11,14 | G3m5*/G3m6* | IGHG3*01*05*06*07*09*10*11*12/IGHG3*13 |
| PA09 | 5,10,11,13,14/5,6,11,24 | G3m5*/G3m24* | IGHG3*01*05*06*07*09*10*11*12/IGHG3*03 |
| PA16 | 5,6,10,11,14/5,6,10,11,14 | G3m6*/G3m6* | IGHG3*13/IGHG3*13 |
| PA31 | 10,11,13,15/5,6,11,24 | G3m15*/G3m24* | IGHG3*17/IGHG3*03 |
| PA42 | 5,6,10,11,14/10,11,13,15 | G3m6*/G3m15* | IGHG3*13/IGHG3*17 |
| PA45 | 5,10,11,13,14/5,10,11,13,14 | G3m5*/G3m5* | IGHG3*01*05*06*07*09*10*11*12/IGHG3*01*05*06*07*09*10*11*12 |
| PA48 | 5,6,11,24/5,6,11,24 | G3m24*/G3m24* | IGHG3*03/IGHG3*03 |
a G3m alleles are characterized by specific combinations of G3m allotypes and are written by increasing numbers of markers (separated by commas) (2, 4). As Gm allotypes are encoded on chromosome 14 (14q 32.3), two G3m alleles are expressed and mentioned using a slash mark.
b the simplified G3m form contains the number of one representative allotype followed by *.
IGHG AA Alleles Deduced from Mass Spectrometry Analysis Using Usual Peptide Databases
As we routinely use the SwissProt Homo sapiens FASTA protein database we quickly realized it was not pertinent regarding the IGHG diversity. Thus we added the IMGT® database of all IGHG AA alleles. Using two FASTA sequence databases with overlapping sequences can be confusing. Indeed if unique peptides were attributable to specific IGHG genes and alleles to each IG heavy chain, as listed in Table II, many peptides were shared between all alleles of a given IGHG gene. Because Mascot sorts resulting identifications according to the hits with the best matching set of peptides, only peptides comprised in the same sets of the Mascot analysis were considered and by default not those included in the subsets. This allowed exclusion of possible ambiguities corresponding to alleles with lesser occurrence probability. Following these rules, two heterozygous IGHG allele pairings could be easily deduced. It was for example the case of IGHG3*03/IGHG3*13 for AS50 and PA16 where the unambiguous attribution of IGHG3*03 (based on the presence of the discriminatory R.WQEGNVFSCSVMHEALHNR.F peptide) led to the deduction of IGHG3*13 as second IGHG3 allele (based on the presence of both K.GFYPSDIAVEWESSGQPENNYK.T and R.WQEGNIFSCSVMHEALHNR.F issued from IGHG3*06*07*13 and IGHG3*13, respectively). In other cases, allele pairings could be partly deduced, such as IGHG2*06/IGHG2*01*03*04*05*06 for A550, PA07, PA09, PA16, and PA42. By lack of a sufficient coverage of discriminatory peptides, MS analysis did not allow defining the alleles for IGHG1 of all samples and IGHG4 of a majority of them, however for PA01, PA42, and PA45 the K.TTPPVLDSDGSFFLYSR.L peptide allowed to exclude IGHG4*03.
Table II. IGHG AA alleles deduced from mass spectrometry analysis of proteotypic peptides after trypsin digestion of the IG heavy chain. “Observed proteotypic peptides” correspond to a trypsin digestion (cleavage site indicated by a point) of the constant region of the IG gamma chains encoded by a given Homo sapiens IGHG gene; “m/z” is the mass-to-charge ratio corresponding to the peptide, in oxidized form or not, observed with the maximal score; “z” represents the peptide charge state; “Ox” indicates whether the peptide was found at least once in oxidized form (if so, number of occurrences/number of observations) or not (N); “Corresponding IGHG alleles” are mentioned according to amino acid sequences and positions from IMGT repertoire (http://www.imgt.org) and (25); “Attributable IGHG allele combinations” are identified by a slash mark separating the two attributable alleles or groups of alleles.
| Purified IgG samples | IGHG gene | Observed proteotypic peptides | m/z | z | Ox | Scoremax | Position | Corresponding IGHG alleles | Attributable IGHG allele combinations |
|---|---|---|---|---|---|---|---|---|---|
| AS50 | IGHG1 | K.GPSVFPLAPSSK.S | 593.83 | 2 | N | 66 | CH1 1.1–13 | all IGHG1 | all IGHG1 / |
| K.THTCPPCPAPELLGGPSVFLFPPKPK.D | 683.36 | 4 | N | 62 | H 7-CH2 13 | all IGHG1 | all IGHG1 | ||
| R.TPEVTCVVVDVSHEDPEVK.F | 1041.51 | 2 | N | 81 | CH2 17–39 | all IGHG1 | |||
| R.EEQYNSTYR.V | 595.26 | 2 | N | 30 | CH2 83–85 | all IGHG1 | |||
| K.TTPPVLDSDGSFFLYSK.La | 937.46 | 2 | N | 42 | CH3 79–89 | all IGHG1 | |||
| IGHG2 | R.TPEVTCVVVDVSHEDPEVQFNWYVDGVEVHNAK.T | 935.94 | 4 | N | 67 | CH2 17–80 | all IGHG2 except IGHG2*02 | IGHG2*06 / | |
| R.EEQFNSTFR.V b | 579.26 | 2 | N | 31 | CH2 83–85 | all IGHG2 | IGHG2*01*03*04*05*06 | ||
| R.VVSVLTVVHQDWLNGK.E | 897.50 | 2 | N | 79 | CH2 85.1–101 | all IGHG2 | |||
| K.GFYPSDISVEWESNGQPENNYK.T | 1280.57 | 2 | N | 54 | CH3 26–80 | IGHG2*06 | |||
| IGHG3 | R.WQEGNVFSCSVMHEALHNR.F | 1122.51 | 2 | 5/13 | 110 | CH3 95–116 | IGHG3*03 | IGHG3*03 / | |
| R.WQEGNIFSCSVMHEALHNR.F | 1129.51 | 2 | 2/7 | 111 | CH3 95–116 | IGHG3*13 | IGHG3*13 | ||
| IGHG4 | K.YGPPCPSCPAPEFLGGPSVFLFPPKPK.D | 943.81 | 3 | N | 66 | H 3-CH2 13 | all IGHG4 | all IGHG4 / | |
| R.TPEVTCVVVDVSQEDPEVQFNWYVDGVEVHNAK.T | 1244.59 | 3 | N | 54 | CH2 17–80 | all IGHG4 | all IGHG4 | ||
| R.WQEGNVFSCSVMHEALHNHYTQK.S | 915.75 | 3 | N | 55 | CH3 95–120 | all IGHG4 | |||
| NP49 | IGHG1 | K.GPSVFPLAPSSK.S | 593.83 | 2 | N | 67 | CH1 1.1–13 | all IGHG1 | all IGHG1 / |
| K.THTCPPCPAPELLGGPSVFLFPPKPK.D | 910.81 | 3 | N | 68 | H 7-CH2 13 | all IGHG1 | all IGHG1 | ||
| R.TPEVTCVVVDVSHEDPEVK.F | 1041.51 | 2 | N | 81 | CH2 17–39 | all IGHG1 | |||
| IGHG2 | R.TPEVTCVVVDVSHEDPEVQFNWYVDGVEVHNAK.T | 935.94 | 4 | N | 59 | CH2 17–80 | all IGHG2 except IGHG2*02 | IGHG2*01*03*04*05*06 / | |
| R.EEQFNSTFR.V b | 579.27 | 2 | N | 32 | CH2 83–85 | all IGHG2 | all IGHG2 | ||
| R.VVSVLTVVHQDWLNGK.E | 897.50 | 2 | N | 83 | CH2 85.1–101 | all IGHG2 | |||
| K.TTPPMLDSDGSFFLYSK.Lc | 961.45 | 2 | 1/4 | 48 | CH3 79–89 | all IGHG2 | |||
| IGHG3 | K.GFYPSDIAMEWESSGQPENNYK.T | 1275.05 | 2 | 1/2 | 40 | CH3 26–80 | IGHG3*17*18*19 | IGHG3*03 / | |
| R.WQEGNVFSCSVMHEALHNR.F | 748.67 | 3 | N | 37 | CH3 95–116 | IGHG3*03 | IGHG3*17*18*19 | ||
| R.WQQGNIFSCSVMHEALHNHYTQK.S | 920.09 | 3 | 3/8 | 66 | CH3 95–120 | IGHG3*17*18*19 | |||
| IGHG4 | K.YGPPCPSCPAPEFLGGPSVFLFPPKPK.D | 943.81 | 3 | N | 78 | H 3-CH2 13 | all IGHG4 | all IGHG4 / | |
| R.TPEVTCVVVDVSQEDPEVQFNWYVDGVEVHNAK.T | 1244.59 | 3 | N | 71 | CH2 17–80 | all IGHG4 | all IGHG4 | ||
| R.EPQVYTLPPSQEEMTK.N | 938.95 | 2 | N | 36 | CH3 1.1–17 | all IGHG4 | |||
| PA01 | IGHG1 | K.GPSVFPLAPSSK.S | 593.83 | 2 | N | 61 | CH1 1.1–13 | all IGHG1 | all IGHG1 / |
| K.THTCPPCPAPELLGGPSVFLFPPKPK.D | 910.81 | 3 | N | 64 | H 7-CH2 13 | all IGHG1 | all IGHG1 | ||
| R.TPEVTCVVVDVSHEDPEVK.F | 1041.51 | 2 | N | 83 | CH2 17–39 | all IGHG1 | |||
| K.TTPPVLDSDGSFFLYSK.La | 937.46 | 2 | N | 60 | CH3 79–89 | all IGHG1 | |||
| IGHG2 | R.TPEVTCVVVDVSHEDPEVQFNWYVDGVEVHNAK.T | 935.94 | 4 | N | 71 | CH2 17–80 | all IGHG2 except IGHG2*02 | IGHG2*01*03*04*05*06 / | |
| IGHG2*01*03*04*05*06 | |||||||||
| IGHG3 | R.WQQGNIFSCSVMHEALHNR.F | 753.02 | 3 | 1/6 | 76 | CH3 95–116 | IGHG3*01*02*04 to *12 | IGHG3*01*02*04 to *12 / | |
| IGHG3*01*02*04 to *12 | |||||||||
| IGHG4 | K.YGPPCPSCPAPEFLGGPSVFLFPPKPK.D | 943.81 | 3 | N | 68 | H 3-CH2 13 | all IGHG4 | IGHG4*01*02*04 / | |
| R.TPEVTCVVVDVSQEDPEVQFNWYVDGVEVHNAK.T | 1244.59 | 3 | N | 30 | CH2 17–80 | all IGHG4 | all IGHG4 | ||
| R.EPQVYTLPPSQEEMTK.N | 938.95 | 2 | N | 48 | CH3 1.1–17 | all IGHG4 | |||
| K.TTPPVLDSDGSFFLYSR.L | 634.65 | 3 | N | 34 | CH3 79–89 | all IGHG4 except IGHG4*03 | |||
| R.WQEGNVFSCSVMHEALHNHYTQK.S | 915.75 | 3 | 1/6 | 52 | CH3 95–120 | all IGHG4 | |||
| PA07 | IGHG1 | K.GPSVFPLAPSSK.S | 593.82 | 2 | N | 67 | CH1 1.1–13 | all IGHG1 | all IGHG1 / |
| K.THTCPPCPAPELLGGPSVFLFPPKPK.D | 910.81 | 3 | N | 75 | H 7-CH2 13 | all IGHG1 | all IGHG1 | ||
| R.TPEVTCVVVDVSHEDPEVK.F | 1041.50 | 2 | N | 88 | CH2 17–39 | all IGHG1 | |||
| R.EEQYNSTYR.V | 595.26 | 2 | N | 31 | CH2 83–85 | all IGHG1 | |||
| K.TTPPVLDSDGSFFLYSK.La | 937.46 | 2 | N | 60 | CH3 79–89 | all IGHG1 | |||
| IGHG2 | R.TPEVTCVVVDVSHEDPEVQFNWYVDGVEVHNAK.T | 935.95 | 4 | N | 86 | CH2 17–80 | all IGHG2 except IGHG2*02 | IGHG2*06 / | |
| R.EEQFNSTFR.Vb | 579.26 | 2 | N | 31 | CH2 83–85 | all IGHG2 | IGHG2*01*03*04*05*06 | ||
| K.GFYPSDISVEWESNGQPENNYK.T | 1280.57 | 2 | N | 54 | CH3 26–80 | IGHG2*06 | |||
| IGHG3 | R.WQQGNIFSCSVMHEALHNR.F | 753.02 | 3 | N | 105 | CH3 95–116 | IGHG3*01*02*04 to *12 | IGHG3*13 / | |
| R.WQEGNIFSCSVMHEALHNR.F | 1129.51 | 2 | 2/9 | 112 | CH3 95–116 | IGHG3*13 | IGHG3*01*02*04 to *12 | ||
| IGHG4 | K.YGPPCPSCPAPEFLGGPSVFLFPPKPK.D | 943.81 | 3 | N | 87 | H 3-CH2 13 | all IGHG4 | all IGHG4 / | |
| R.TPEVTCVVVDVSQEDPEVQFNWYVDGVEVHNAK.T | 1244.59 | 3 | N | 85 | CH2 17–80 | all IGHG4 | all IGHG4 | ||
| R.EEQFNSTYR.V | 587.26 | 2 | N | 31 | CH2 83–85 | all IGHG4 | |||
| R.EPQVYTLPPSQEEMTK.N | 938.95 | 2 | N | 46 | CH3 1.1–17 | all IGHG4 | |||
| R.WQEGNVFSCSVMHEALHNHYTQK.S | 915.75 | 3 | 2/10 | 72 | CH3 95–120 | all IGHG4 | |||
| PA09 | IGHG1 | K.GPSVFPLAPSSK.S | 593.83 | 2 | N | 67 | CH1 1.1–13 | all IGHG1 | all IGHG1 / |
| K.THTCPPCPAPELLGGPSVFLFPPKPK.D | 910.81 | 3 | N | 63 | H 7-CH2 13 | all IGHG1 | all IGHG1 | ||
| R.TPEVTCVVVDVSHEDPEVK.F | 1041.50 | 2 | N | 82 | CH2 17–39 | all IGHG1 | |||
| R.EEQYNSTYR.V | 595.26 | 2 | N | 25 | CH2 83–85 | all IGHG1 | |||
| K.TTPPVLDSDGSFFLYSK.La | 937.46 | 2 | N | 60 | CH3 79–89 | all IGHG1 | |||
| IGHG2 | R.TPEVTCVVVDVSHEDPEVQFNWYVDGVEVHNAK.T | 935.94 | 4 | N | 61 | CH2 17–80 | all IGHG2 except IGHG2*02 | IGHG2*06 / | |
| K.GFYPSDISVEWESNGQPENNYK.T | 854.05 | 3 | N | 41 | CH3 26–80 | IGHG2*06 | IGHG2*01*03*04*05*06 | ||
| IGHG3 | R.WQEGNVFSCSVMHEALHNR.F | 1122.51 | 2 | 2/8 | 110 | CH3 95–116 | IGHG3*03 | IGHG3*03 / | |
| R.WQQGNIFSCSVMHEALHNR.F | 1129.02 | 2 | 1/8 | 76 | CH3 95–116 | IGHG3*01*02*04 to *12 | IGHG3*01*02*04 to *12 | ||
| IGHG4 | K.YGPPCPSCPAPEFLGGPSVFLFPPKPK.D | 943.81 | 3 | N | 65 | H 3-CH2 13 | all IGHG4 | all IGHG4 / | |
| R.TPEVTCVVVDVSQEDPEVQFNWYVDGVEVHNAK.T | 1244.59 | 3 | N | 40 | CH2 17–80 | all IGHG4 | all IGHG4 | ||
| R.EPQVYTLPPSQEEMTK.N | 938.95 | 2 | N | 48 | CH3 1.1–17 | all IGHG4 | |||
| R.WQEGNVFSCSVMHEALHNHYTQK.S | 915.75 | 3 | 1/4 | 61 | CH3 95–120 | all IGHG4 | |||
| PA16 | IGHG1 | K.GPSVFPLAPSSK.S | 593.83 | 2 | N | 58 | CH1 1.1–13 | all IGHG1 | all IGHG1 / |
| K.THTCPPCPAPELLGGPSVFLFPPKPK.D | 910.81 | 3 | N | 62 | H 7-CH2 13 | all IGHG1 | all IGHG1 | ||
| R.TPEVTCVVVDVSHEDPEVK.F | 1041.51 | 2 | N | 92 | CH2 17–39 | all IGHG1 | |||
| K.TTPPVLDSDGSFFLYSK.La | 937.46 | 2 | N | 51 | CH3 79–89 | all IGHG1 | |||
| IGHG2 | R.TPEVTCVVVDVSHEDPEVQFNWYVDGVEVHNAK.T | 935.94 | 4 | N | 64 | CH2 17–80 | all IGHG2 except IGHG2*02 | IGHG2*06 / | |
| K.GFYPSDISVEWESNGQPENNYK.T | 1280.57 | 2 | N | 53 | CH3 26–80 | IGHG2*06 | IGHG2*01*03*04*05*06 | ||
| IGHG3 | K.GFYPSDIAVEWESSGQPENNYK.T | 1259.07 | 2 | N | 40 | CH3 26–80 | IGHG3*06*07*13 | IGHG3*03 / | |
| R.WQEGNVFSCSVMHEALHNR.F | 1122.51 | 2 | 6/15 | 108 | CH3 95–116 | IGHG3*03 | IGHG3*13 | ||
| R.WQEGNIFSCSVMHEALHNR.F | 753.35 | 3 | N | 88 | CH3 95–116 | IGHG3*13 | |||
| IGHG4 | K.YGPPCPSCPAPEFLGGPSVFLFPPKPK.D | 943.81 | 3 | N | 78 | H 3-CH2 13 | all IGHG4 | all IGHG4 / | |
| R.TPEVTCVVVDVSQEDPEVQFNWYVDGVEVHNAK.T | 1244.59 | 3 | N | 28 | CH2 17–80 | all IGHG4 | all IGHG4 | ||
| R.EPQVYTLPPSQEEMTK.N | 938.95 | 2 | N | 42 | CH3 1.1–17 | all IGHG4 | |||
| R.WQEGNVFSCSVMHEALHNHYTQK.S | 915.75 | 3 | N | 56 | CH3 95–120 | all IGHG4 | |||
| PA31 | IGHG1 | K.GPSVFPLAPSSK.S | 593.83 | 2 | N | 60 | CH1 1.1–13 | all IGHG1 | all IGHG1 / |
| K.THTCPPCPAPELLGGPSVFLFPPKPK.D | 910.81 | 3 | N | 59 | H 7-CH2 13 | all IGHG1 | all IGHG1 | ||
| R.TPEVTCVVVDVSHEDPEVK.F | 1041.51 | 2 | N | 81 | CH2 17–39 | all IGHG1 | |||
| R.EEQYNSTYR.V | 595.26 | 2 | N | 30 | CH2 83–85 | all IGHG1 | |||
| IGHG2 | R.TPEVTCVVVDVSHEDPEVQFNWYVDGVEVHNAK.T | 935.94 | 4 | N | 65 | CH2 17–80 | all IGHG2 except IGHG2*02 | IGHG2*01*03*04*05*06 / | |
| IGHG2*01*03*04*05*06 | |||||||||
| IGHG3 | K.GFYPSDIAMEWESSGQPENNYK.T | 1283.05 | 2 | 1/1 | 39 | CH3 26–80 | IGHG3*17*18*19 | IGHG3*03 / | |
| R.WQEGNVFSCSVMHEALHNR.F | 1122.51 | 2 | 6/12 | 111 | CH3 95–116 | IGHG3*03 | IGHG3*17*18*19 | ||
| R.WQQGNIFSCSVMHEALHNHYTQK.S | 920.09 | 3 | 2/7 | 62 | CH3 95–120 | IGHG3*17*18*19 | |||
| IGHG4 | K.YGPPCPSCPAPEFLGGPSVFLFPPKPK.D | 943.81 | 3 | N | 74 | H 3-CH2 13 | all IGHG4 | all IGHG4 / | |
| R.TPEVTCVVVDVSQEDPEVQFNWYVDGVEVHNAK.T | 1244.59 | 3 | N | 84 | CH2 17–80 | all IGHG4 | all IGHG4 | ||
| R.EEQFNSTYR.V | 587.26 | 2 | N | 30 | CH2 83–85 | all IGHG4 | |||
| R.EPQVYTLPPSQEEMTK.N | 938.95 | 2 | N | 51 | CH3 1.1–17 | all IGHG4 | |||
| R.WQEGNVFSCSVMHEALHNHYTQK.S | 915.75 | 3 | N | 61 | CH3 95–120 | all IGHG4 | |||
| PA42 | IGHG1 | K.GPSVFPLAPSSK.S | 593.83 | 2 | N | 64 | CH1 1.1–13 | all IGHG1 | all IGHG1 / |
| K.THTCPPCPAPELLGGPSVFLFPPKPK.D | 910.81 | 3 | N | 60 | H 7-CH2 13 | all IGHG1 | all IGHG1 | ||
| R.TPEVTCVVVDVSHEDPEVK.F | 1041.50 | 2 | N | 84 | CH2 17–39 | all IGHG1 | |||
| R.EEQYNSTYR.V | 595.26 | 2 | N | 30 | CH2 83–85 | all IGHG1 | |||
| K.TTPPVLDSDGSFFLYSK.L | 937.46 | 2 | N | 64 | CH3 79–89 | all IGHG1 | |||
| IGHG2 | R.TPEVTCVVVDVSHEDPEVQFNWYVDGVEVHNAK.T | 935.94 | 4 | N | 74 | CH2 17–80 | all IGHG2 except IGHG2*02 | IGHG2*06 / | |
| K.GFYPSDISVEWESNGQPENNYK.T | 1280.57 | 2 | N | 53 | CH3 26–80 | IGHG2*06 | IGHG2*01*03*04*05*06 | ||
| IGHG3 | K.TPLGDTTHTCPR.C | 649.81 | 2 | N | 75 | H1 3–16 | all IGHG3 | IGHG3*01*02*04 to *12 / | |
| K.SCDTPPPCPR.C | 536.73 | 2 | N | 45 | H2 H3 H4 3–14 | all IGHG3 | all IGHG3 | ||
| R.TPEVTCVVVDVSHEDPEVQFK.W | 1179.02 | 2 | N | 81 | CH2 17–41 | all IGHG3 | |||
| R.WQQGNIFSCSVMHEALHNR.F | 753.02 | 3 | 3/13 | 124 | CH3 95–116 | IGHG3*01*02*04 to *12 | |||
| IGHG4 | K.YGPPCPSCPAPEFLGGPSVFLFPPKPK.D | 943.81 | 3 | N | 73 | H 3-CH2 13 | all IGHG4 | IGHG4*01*02*04 / | |
| R.TPEVTCVVVDVSQEDPEVQFNWYVDGVEVHNAK.T | 933.69 | 4 | N | 76 | CH2 17–80 | all IGHG4 | all IGHG4 | ||
| R.EPQVYTLPPSQEEMTK.N | 938.95 | 2 | N | 53 | CH3 1.1–17 | all IGHG4 | |||
| K.TTPPVLDSDGSFFLYSR.L | 951.47 | 2 | N | 46 | CH3 79–89 | all IGHG4 except IGHG4*03 | |||
| R.WQEGNVFSCSVMHEALHNHYTQK.S | 915.75 | 3 | 3/9 | 70 | CH3 95–120 | all IGHG4 | |||
| PA45 | IGHG1 | K.GPSVFPLAPSSK.S | 593.83 | 2 | N | 64 | CH1 1.1–13 | all IGHG1 | all IGHG1 / |
| K.THTCPPCPAPELLGGPSVFLFPPKPK.D | 910.81 | 3 | N | 64 | H 7-CH2 13 | all IGHG1 | all IGHG1 | ||
| R.TPEVTCVVVDVSHEDPEVK.F | 1041.51 | 2 | N | 89 | CH2 17–39 | all IGHG1 | |||
| R.EEQYNSTYR.V | 595.26 | 2 | N | 40 | CH2 83–85 | all IGHG1 | |||
| R.VVSVLTVLHQDWLNGK.E d | 904.50 | 2 | N | 68 | CH2 85.1–101 | all IGHG1 | |||
| K.TTPPVLDSDGSFFLYSK.L | 937.46 | 2 | N | 60 | CH3 79–89 | all IGHG1 | |||
| IGHG2 | R.TPEVTCVVVDVSHEDPEVQFNWYVDGVEVHNAK.T | 935.94 | 4 | N | 62 | CH2 17–80 | all IGHG2 except IGHG2*02 | IGHG2*01*03*04*05*06 / | |
| IGHG2*01*03*04*05*06 | |||||||||
| IGHG3 | K.TPLGDTTHTCPR.C | 649.81 | 2 | N | 81 | H1 3–16 | all IGHG3 | IGHG3*01*02*04 to *12 / | |
| K.SCDTPPPCPR.C | 536.73 | 2 | N | 45 | H2 H3 H4 3–14 | all IGHG3 | all IGHG3 | ||
| R.TPEVTCVVVDVSHEDPEVQFK.W | 1179.07 | 2 | N | 75 | CH2 17–41 | all IGHG3 | |||
| R.WQQGNIFSCSVMHEALHNR.F | 1129.03 | 2 | 2/8 | 82 | CH3 95–116 | IGHG3*01*02*04 to *12 | |||
| IGHG4 | K.YGPPCPSCPAPEFLGGPSVFLFPPKPK.D | 943.81 | 3 | N | 51 | H 3-CH2 13 | all IGHG4 | IGHG4*01*02*04 / | |
| R.EPQVYTLPPSQEEMTK.N | 938.95 | 2 | N | 50 | CH3 1.1–17 | all IGHG4 | all IGHG4 | ||
| K.TTPPVLDSDGSFFLYSR.L | 951.47 | 2 | N | 60 | CH3 79–89 | all IGHG4 except IGHG4*03 | |||
| R.WQEGNVFSCSVMHEALHNHYTQK.S | 915.75 | 3 | N | 52 | CH3 95–120 | all IGHG4 | |||
| PA48 | IGHG1 | K.GPSVFPLAPSSK.S | 593.83 | 2 | N | 67 | CH1 1.1–13 | all IGHG1 | all IGHG1 / |
| K.THTCPPCPAPELLGGPSVFLFPPKPK.D | 910.81 | 3 | N | 63 | H 7-CH2 13 | all IGHG1 | all IGHG1 | ||
| R.TPEVTCVVVDVSHEDPEVK.F | 1041.50 | 2 | N | 88 | CH2 17–39 | all IGHG1 | |||
| K.TTPPVLDSDGSFFLYSK.La | 937.46 | 2 | N | 45 | CH3 79–89 | all IGHG1 | |||
| IGHG2 | R.TPEVTCVVVDVSHEDPEVQFNWYVDGVEVHNAK.T | 1247.59 | 3 | N | 72 | CH2 17–80 | all IGHG2 except IGHG2*02 | IGHG2*01*03*04*05*06 / | |
| R.EEQFNSTFR.V b | 579.27 | 2 | N | 25 | CH2 83–85 | all IGHG2 | all IGHG2 | ||
| R.VVSVLTVVHQDWLNGK.E | 897.50 | 2 | N | 75 | CH2 85.1–101 | all IGHG2 | |||
| K.TTPPMLDSDGSFFLYSK.Lc | 953.45 | 2 | 1/4 | 50 | CH3 79–89 | all IGHG2 | |||
| IGHG3 | K.TPLGDTTHTCPR.C | 649.81 | 2 | N | 94 | H1 3–16 | all IGHG3 | IGHG3*03 / | |
| K.SCDTPPPCPR.C | 536.73 | 2 | N | 45 | H2 H3 H4 3–14 | all IGHG3 | all IGHG3 | ||
| R.TPEVTCVVVDVSHEDPEVQFK.W | 1179.07 | 2 | N | 81 | CH2 17–41 | all IGHG3 | |||
| R.WQEGNVFSCSVMHEALHNR.F | 1122.50 | 2 | 4/11 | 110 | CH3 95–116 | IGHG3*03 | |||
| IGHG4 | K.YGPPCPSCPAPEFLGGPSVFLFPPKPK.D | 943.81 | 3 | N | 64 | H 3-CH2 13 | all IGHG4 | all IGHG4 / | |
| R.TPEVTCVVVDVSQEDPEVQFNWYVDGVEVHNAK.T | 933.69 | 4 | N | 66 | CH2 17–80 | all IGHG4 | all IGHG4 | ||
| R.EPQVYTLPPSQEEMTK.N | 938.95 | 2 | N | 53 | CH3 1.1–17 | all IGHG4 | |||
| R.WQEGNVFSCSVMHEALHNHYTQK.S | 1373.12 | 2 | N | 59 | CH3 95–120 | all IGHG4 |
a peptide CH3 79–89 attributed to all IGHG1 alleles and not to IGHG4*03 also proposed in subsets (not considered in the deduction strategy).
b peptide CH2 83–85 attributed to all IGHG2 alleles and not to IGHG3*11 and IGHG3*12 also proposed in subsets (not considered in the deduction strategy).
c peptide CH3 79–89 attributed to IGHG2*01*03*04*05*06 group of alleles and not to IGHG3*06, IGHG3*07, IGHG3*13 or IGHG3*15 alleles proposed in subsets (not considered in the deduction strategy).
d peptide CH2 85.1–101 attributed to all IGHG1 alleles and not to IGHG4 (all alleles except IGHG4*02) or IGHG3 (all alleles except IGHG3*09) proposed in subsets (not considered in the deduction strategy).
IGHG Alleles Deduced from Sequencing of the CH2 and CH3-CHS Region
Critical nucleotide positions listed in the supplemental Table S4 allowed discriminating alleles assigned to each IGHG gene. The 10 samples were homozygous for IGHG1*02 (Alignment of alleles IGHG1, IMGT Repertoire, http://www.imgt.org), as demonstrated by the reading of the IGHG1 CH2 to CH3-CHS nucleotide sequences. Following the same strategy, the IGHG2*06 allele (Alignment of alleles IGHG2, IMGT Repertoire, http://www.imgt.org) could be deduced unambiguously, for 9 out of 20 IGHG2 alleles (Table III). In 10 other cases, the IGHG2*01*03*04*05 could be assigned to four types of alleles departing from the IGHG2*01 IMGT® reference allele by some silent nucleotide substitutions (all comprising at least CH2 t273>c, based on the IMGT® unique numbering (25)), which have not yet been described. In the last case, that concerned the PA01 sample, a nonsynonymous CH2 g274>c substitution (associated with two silent ones, CH2 t273>c and CH3 a243>g) generated a CH2 V92>L AA change defining, for the sequence CH2 and CH3-CHS, a putative new AA allele of IGHG2. Regarding IGHG3 (Alignment of alleles IGHG3, IMGT Repertoire, http://www.imgt.org), AA alleles could be directly deduced from the sequencing, and were in order of decreasing frequency IGHG3*01*04*05*10, IGHG3*03, IGHG3*13, and IGHG3*17 (Table IV). Lastly, for IGHG4 (Alignment of alleles IGHG4, IMGT Repertoire, http://www.imgt.org), the IGHG4*01 or IGHG4*04 AA alleles could be unambiguously assigned from the sequencing in 8 out of 20 cases (Table V). In nine other cases, two types of silent substitutions (including at least CH2 g3.4>a) led to the identification of putative alleles which have not yet been described, without impeding a common deduction of the IGHG4*01*04. For the PA07 sample, two nonsynonymous substitutions (CH2 g271>a and a322>c) generated the AA changes CH2 V91>I and N108>H, respectively, defining for the sequence CH2 and CH3-CHS a putative new AA allele of IGHG4. An additional CH3 a32>g substitution corresponding to a CH3 Q11>R AA change occurred for one allele from both AS50 and PA16 samples, defining for the sequence CH2 and CH3-CHS an additional putative novel IGHG4 AA allele.
Table III. IGHG2 AA alleles deduced from genomic sequencing of the CH2 and CH3-CHS exons. As IGHG2 is encoded on chromosome 14 (14q 32.3), two IGHG2 alleles are expressed per sample. Amino acid changes not referenced in IMGT® are mentioned in bold.
| DNA samples | Nt substitutions and AA changes with reference to IGHG2*01 (J00230) |
Genomic IMGT IGHG2 alleles determined | IGHG2 AA alleles deduced | ||
|---|---|---|---|---|---|
| Nt IMGTa | AA IMGTa | AA EUb | |||
| AS50 | CH2 t273>c; CH3 g112>t, g1173>a | CH2 V91; CH3 A38>S, T117 | CH2 V308 =; CH3 A378S, T437 = | IGHG2*06 | IGHG2*06 |
| CH2 t273>c; CH3 g112>t, g1173>a | CH2 V91; CH3 A38>S, T117 | CH2 V308 = ; CH3 A378S, T437 = | IGHG2*06 | IGHG2*06 | |
| NP49 | CH2 t273>c; CH3 g112>t, g1173>a | CH2 V91; CH3 A38>S, T117 | CH2 V308 =; CH3 A378S, T437 = | IGHG2*06 | IGHG2*06 |
| CH2 t273>c; CH3 g112>t, g1173>a | CH2 V91; CH3 A38>S, T117 | CH2 V308 =; CH3 A378S, T437 = | IGHG2*06 | IGHG2*06 | |
| PA01 | CH2 t273>c; CH3 a243>g | CH2 V91; CH3 T81 | CH2 V308 =; CH3 T394 = | not referenced | IGHG2*01*03*04*05 |
| CH2 t273>c, g274>c; CH3 a243>g | CH2 V91, V92>L; CH3 T81 | CH2 V308 =, V309L; CH3 T394 = | not referenced | IGHG2*[V92>L] | |
| PA07 | CH2 t273>c; CH3 g112>t, g1173>a | CH2 V91; CH3 A38>S, T117 | CH2 V308 =; CH3 A378S, T437 = | IGHG2*06 | IGHG2*06 |
| CH2 t273>c | CH2 V91 | CH2 V308 = | not referenced | IGHG2*01*03*04*05 | |
| PA09 | CH2 t273>c; CH3 g112>t, g1173>a | CH2 V91; CH3 A38>S, T117 | CH2 V308 =; CH3 A378S, T437 = | IGHG2*06 | IGHG2*06 |
| CH2 t273>c | CH2 V91 | CH2 V308 = | not referenced | IGHG2*01*03*04*05 | |
| PA16 | CH2 t273>c; CH3 g112>t, g1173>a | CH2 V91; CH3 A38>S, T117 | CH2 V308 =; CH3 A378S, T437 = | IGHG2*06 | IGHG2*06 |
| CH2 t273>c | CH2 V91 | CH2 V308 = | not referenced | IGHG2*01*03*04*05 | |
| PA31 | CH2 t273>c | CH2 V91 | CH2 V308 = | not referenced | IGHG2*01*03*04*05 |
| CH2 c105>t, t273>c; CH3 a243>g, g1173>a | CH2 P35, V91; CH3 T81, T117 | CH2 P271 =, V308 =; CH3 T394 =, T437 = | not referenced | IGHG2*01*03*04*05 | |
| PA42 | CH2 t273>c; CH3 g112>t, g1173>a | CH2 V91; CH3 A38>S, T117 | CH2 V308 =; CH3 A378S, T437 = | IGHG2*06 | IGHG2*06 |
| CH2 t273>c; CH3 g112>t, g1173>a | CH2 V91; CH3 A38>S, T117 | CH2 V308 =; CH3 A378S, T437 = | IGHG2*06 | IGHG2*06 | |
| PA45 | CH2 t273>c; CH3 a243>g | CH2 V91; CH3 T81 | CH2 V308 =; CH3 T394 = | not referenced | IGHG2*01*03*04*05 |
| CH2 t273>c; CH3 a243>g, g1173>a | CH2 V91; CH3 T81, T117 | CH2 V308 =; CH3 T394 =, T437 = | not referenced | IGHG2*01*03*04*05 | |
| PA48 | CH2 t273>c | CH2 V91 | CH2 V308 = | not referenced | IGHG2*01*03*04*05 |
| CH2 t273>c | CH2 V91 | CH2 V308 = | not referenced | IGHG2*01*03*04*05 | |
a Nomenclature according to the IMGT unique numbering for C-DOMAIN (2, 25) and IMGT Allele alignment Homo sapiens IGHG2 (http://www.imgt.org/IMGTrepertoire/Proteins/alleles/index.php?species=Homo%20sapiens&group=IGHC&gene=IGHG2).
b Nomenclature adapted from Human Genome Variation Society (www.hgvs.org) and (29); amino acid (AA) EU numbering according to IMGT® www.imgt.org (23).
Table IV. IGHG3 AA alleles deduced from genomic sequencing of the CH2 and CH3-CHS exons. As IGHG3 is encoded on chromosome 14 (14q 32.3), two IGHG3 alleles are expressed per sample.
| DNA samples | Nt substitutions and AA changes with reference to IGHG3*01 (X03604) |
Genomic IMGT IGHG3 alleles determined | IGHG3 AA alleles deduced | ||
|---|---|---|---|---|---|
| Nt IMGTa | AA IMGTa | AA EUb | |||
| AS50 | CH3 a250>g, a263>g, c267>a, c292>g, c300>t, a301>g | CH3 M84>V, K88>R, L89, Q98>E, N100, I101>V | CH3 M397V, K409R, L410 =, Q419E, N421 =, I422V | IGHG3*03 | IGHG3*03 |
| CH3 c237>g, g243>a, c292>g | CH3 N79>K, T81, Q98>E | CH3 N392K, T394 =, Q419E | IGHG3*13 | IGHG3*13 | |
| NP49 | CH3 a250>g, a263>g, c267>a, c292>g, c300>t, a301>g | CH3 M84>V, K88>R, L89, Q98>E, N100, I101>V | CH3 M397V, K409R, L410 =, Q419E, N421 =, I422V | IGHG3*03 | IGHG3*03 |
| - | - | - | IGHG3*01*04*05*10 | IGHG3*01*04*05*10 | |
| PA01 | - | - | - | IGHG3*01*04*05*10 | IGHG3*01*04*05*10 |
| - | - | - | IGHG3*01*04*05*10 | IGHG3*01*04*05*10 | |
| PA07 | CH3 c237>g, g243>a, c292>g | CH3 N79>K, T81, Q98>E | CH3 N392K, T394 =, Q419E | IGHG3*13 | IGHG3*13 |
| - | - | - | IGHG3*01*04*05*10 | IGHG3*01*04*05*10 | |
| PA09 | CH3 a250>g, a263>g, c267>a, c292>g, c300>t, a301>g | CH3 M84>V, K88>R, L89, Q98>E, N100, I101>V | CH3 M397V, K409R, L410 =, Q419E, N421 =, I422V | IGHG3*03 | IGHG3*03 |
| - | - | - | IGHG3*01*04*05*10 | IGHG3*01*04*05*10 | |
| PA16 | CH3 a250>g, a263>g, c267>a, c292>g, c300>t, a301>g | CH3 M84>V, K88>R, L89, Q98>E, N100, I101>V | CH3 M397V, K409R, L410 =, Q419E, N421 =, I422V | IGHG3*03 | IGHG3*03 |
| CH3 c237>g, g243>a, c292>g | CH3 N79>K, T81, Q98>E | CH3 N392K, T394 =, Q419E | IGHG3*13 | IGHG3*13 | |
| PA31 | CH3 a250>g, a263>g, c267>a, c292>g, c300>t, a301>g | CH3 M84>V, K88>R, L89, Q98>E, N100, I101>V | CH3 M397V, K409R, L410 =, Q419E, N421 =, I422V | IGHG3*03 | IGHG3*03 |
| CH3 g115>a, c237>g, a250>g, g344>a, t347>a | CH3 V39>M, N79>K, M84>V, R115>H, F116>Y | CH3 V379M, N392K, M397V, R435H, F436Y | IGHG3*17 | IGHG3*17 | |
| PA42 | - | - | - | IGHG3*01*04*05*10 | IGHG3*01*04*05*10 |
| - | - | - | IGHG3*01*04*05*10 | IGHG3*01*04*05*10 | |
| PA45 | - | - | - | IGHG3*01*04*05*10 | IGHG3*01*04*05*10 |
| - | - | - | IGHG3*01*04*05*10 | IGHG3*01*04*05*10 | |
| PA48 | CH3 a250>g, a263>g, c267>a, c292>g, c300>t, a301>g | CH3 M84>V, K88>R, L89, Q98>E, N100, I101>V | CH3 M397V, K409R, L410 =, Q419E, N421 =, I422V | IGHG3*03 | IGHG3*03 |
| CH3 a250>g, a263>g, c267>a, c292>g, c300>t, a301>g | CH3 M84>V, K88>R, L89, Q98>E, N100, I101>V | CH3 M397V, K409R, L410 =, Q419E, N421 =, I422V | IGHG3*03 | IGHG3*03 | |
a Nomenclature according to the IMGT unique numbering for C-DOMAIN (2, 25) and IMGT Allele alignment Homo sapiens IGHG3 (http://www.imgt.org/IMGTrepertoire/Proteins/alleles/index.php?species=Homo%20sapiens&group=IGHC&gene=IGHG3).
b Nomenclature adapted from Human Genome Variation Society (www.hgvs.org) and (29); amino acid (AA) EU numbering according to IMGT® www.imgt.org (23).
Table V. IGHG4 AA alleles deduced from genomic sequencing of the CH2 and CH3-CHS exons. As IGHG4 is encoded on chromosome 14 (14q 32.3), two IGHG4 alleles are expressed per sample. Amino acid changes not referenced in IMGT® are mentioned in bold.
| DNA samples | Nt substitutions and AA changes with reference to IGHG4*01 (K01316) |
Genomic IMGT IGHG4 alleles determined | IGHG4 AA alleles deduced | ||
|---|---|---|---|---|---|
| Nt IMGTa | AA IMGTa | AA EUb | |||
| AS50 | CH3 a267>c | CH3 L89 | CH3 L410 = | IGHG4*04 | IGHG4*04 |
| CH2 g271>a, a322>c; CH3 a32>g, g243>a, a267>c | CH2 V91>I, N108>H; CH3 Q11>R, T81, L89 | CH2 V308I, N325H; CH3 Q355R, T394 =, L410 = | not referenced | IGHG4*[V91>I;N108>H;Q11>R] | |
| NP49 | CH3 a267>c | CH3 L89 | CH3 L410 = | IGHG4*04 | IGHG4*04 |
| CH2 g3.4>a | CH2 E1.4 | CH2 E233 = | not referenced | IGHG4*01*04 | |
| PA01 | CH2 g3.4>a | CH2 E1.4 | CH2 E233 = | not referenced | IGHG4*01*04 |
| CH2 g3.4>a, t42>c | CH2 E1.4, T14 | CH2 E233 =, T250 = | not referenced | IGHG4*01*04 | |
| PA07 | CH2 g271>a, a322>c; CH3 a267>c | CH2 V91>I, N108>H; CH3 L89 | CH2 V308I, N325H; CH3 L410 = | not referenced | IGHG4*[V91>I;N108>H] |
| CH2 g3.4>a, t42>c | CH2 E1.4, T14 | CH2 E233 =, T250 = | not referenced | IGHG4*01*04 | |
| PA09 | CH3 a267>c | CH3 L89 | CH3 L410 = | IGHG4*04 | IGHG4*04 |
| CH2 g3.4>a | CH2 E1.4 | CH2 E233 = | not referenced | IGHG4*01*04 | |
| PA16 | CH2 g271>a, a322>c; CH3 a32>g, g243>a, a267>c | CH2 V91>I, N108>H; CH3 Q11>R, T81, L89 | CH2 V308I, N325H; CH3 Q355R, T394 =, L410 = | not referenced | IGHG4*[V91>I;N108>H;Q11>R] |
| CH2 g3.4>a | CH2 E1.4 | CH2 E233 = | not referenced | IGHG4*01*04 | |
| PA31 | CH3 a267>c | CH3 L89 | CH3 L410 = | IGHG4*04 | IGHG4*04 |
| CH2 g3.4>a | CH2 E1.4 | CH2 E233 = | not referenced | IGHG4*01*04 | |
| PA42 | - | - | - | IGHG4*01 | IGHG4*01 |
| CH3 a267>c | CH3 L89 | CH3 L410 = | IGHG4*04 | IGHG4*04 | |
| PA45 | - | - | - | IGHG4*01 | IGHG4*01 |
| - | - | - | IGHG4*01 | IGHG4*01 | |
| PA48 | CH2 g3.4>a | CH2 E1.4 | CH2 E233 = | not referenced | IGHG4*01*04 |
| CH2 g3.4>a | CH2 E1.4 | CH2 E233 = | not referenced | IGHG4*01*04 | |
a Nomenclature according to the IMGT unique numbering for C-DOMAIN (2, 25) and IMGT Allele alignment Homo sapiens IGHG4 (http://www.imgt.org/IMGTrepertoire/Proteins/alleles/index.php?species=Homo%20sapiens&group=IGHC&gene=IGHG4).
b Nomenclature adapted from Human Genome Variation Society (www.hgvs.org) and (29); amino acid (AA) EU numbering according to IMGT® www.imgt.org (23).
Confrontation of Serological, Proteomic, and Genomic Approaches of IGHG Allele Deduction or Determination
IGHG1
Whatever the method employed, all samples were monomorphic for IGHG1 alleles. The genomic determination was the most accurate, leading to IGHG1*02, whereas serology identified G1m17,1 allotypes (that may correspond to either IGHG1*01 or IGHG1*02) and proteomics could not yield distinguishable peptides from specific IGHG1 alleles because of lack of distinguishing peptide coverage. In contrary to serological results which indicated a Gm28 carriage for NP49 and PA31, the combined CH3 g344, 115R and CH3 a347, 116Y positions corresponding to this marker were not found in any sample either on IGHG1 or IGHG3 alleles. However, it is not excluded that only one of the two alleles present per individual was sequenced and that the allele carrying Gm28 was missed in the amplification.
IGHG2
No serological determination was performed for IGHG2 but prevalent Gm haplotypes are characteristic of given populations, and because of the African origin of the individuals under study, an absence of G2m23 allotype is the most plausible configuration, consistent with the IGHG2*01*03*04*05*06 (4, 28). Proteomics helped to restrict the possibilities with the assignment of IGHG2*06 combined to IGHG2*01*03*04*05*06 for AS50, PA07, PA09, PA16, and PA42. Interestingly, genomics contributed to further specify these results by attributing IGHG2*06 at the homozygous state to AS50, NP49, and PA42 and at the heterozygous one to PA07, PA09, and PA16. Moreover, it is worth to notice that a new IGHG2 AA allele was suggested for PA01 on the basis of a nonsynonymous substitution.
IGHG3
For the much more polymorphic IGHG3 alleles, results from the different methods were challenging. They are summarized in the Table VI. Considering that a whole number of 20 IGHG3 alleles were deduced, the greatest number of discordances (n = 8) was recorded between serological and genomic approaches, and 6 of them appeared also between serology and proteomics. Strikingly, only one discrepancy could be noted between proteomics and genomics regarding the IGHG3*01*04*05*10 nucleotide allele which was not identified neither by serology (deduction of homozygous IGHG3*17) nor proteomics (deduction of heterozygous IGHG3*03/IGHG3*17*18*19, as shown by MS/MS fragmentation spectra of the supplemental Fig. S1).
Table VI. Confrontation of serological, proteomic and genomic approaches for IGHG3 allele deduction or determination. As IGHG3 is encoded on chromosome 14 (14q 32.3), two IGHG3 alleles are expressed for each sample.
| Samplesa | Combination of IGHG3 alleles according to the method |
Discordances between methodsb |
||||
|---|---|---|---|---|---|---|
| Serology (Ser) | Proteomics (Pro) | Genomics (Gen) | Ser vs. Pro | Ser vs. Gen | Pro vs. Gen | |
| AS50 | IGHG3*01*05*06*07*09*10*11*12 | IGHG3*03 | IGHG3*03 | X | X | |
| IGHG3*01*05*06*07*09*10*11*12 | IGHG3*13 | IGHG3*13 | X | X | ||
| NP49 | IGHG3*17 | IGHG3*03 | IGHG3*03 | X | X | |
| IGHG3*17 | IGHG3*17*18*19 | IGHG3*01*04*05*10 | X | X | ||
| PA01 | IGHG3*17 | IGHG3*01*02*04 to *12 | IGHG3*01*04*05*10 | X | X | |
| IGHG3*01*05*06*07*09*10*11*12 | IGHG3*01*02*04 to *12 | IGHG3*01*04*05*10 | ||||
| PA07 | IGHG3*13 | IGHG3*13 | IGHG3*13 | |||
| IGHG3*01*05*06*07*09*10*11*12 | IGHG3*01*02*04 to *12 | IGHG3*01*04*05*10 | ||||
| PA09 | IGHG3*03 | IGHG3*03 | IGHG3*03 | |||
| IGHG3*01*05*06*07*09*10*11*12 | IGHG3*01*02*04 to *12 | IGHG3*01*04*05*10 | ||||
| PA16 | IGHG3*13 | IGHG3*03 | IGHG3*03 | X | X | |
| IGHG3*13 | IGHG3*13 | IGHG3*13 | ||||
| PA31 | IGHG3*03 | IGHG3*03 | IGHG3*03 | |||
| IGHG3*17 | IGHG3*17*18*19 | IGHG3*17 | ||||
| PA42 | IGHG3*13 | IGHG3*01*02*04 to *12 | IGHG3*01*04*05*10 | X | X | |
| IGHG3*17 | all IGHG3 | IGHG3*01*04*05*10 | X | |||
| PA45 | IGHG3*01*05*06*07*09*10*11*12 | IGHG3*01*02*04 to *12 | IGHG3*01*04*05*10 | |||
| IGHG3*01*05*06*07*09*10*11*12 | all IGHG3 | IGHG3*01*04*05*10 | ||||
| PA48 | IGHG3*03 | IGHG3*03 | IGHG3*03 | |||
| IGHG3*03 | all IGHG3 | IGHG3*03 | ||||
a Individual samples consisted in plasma (serology), purified IgG (proteomics) or genomic DNA (genomics).
b Discordances between methods are mentioned by “X”.
IGHG4
Regarding IGHG4 alleles, only proteomics and genomics could be compared: as for IGHG2 above, results were concordant, with a greater accuracy afforded by the genomic sequencing, which allowed identification of IGHG4*01, IGHG4*04, or IGHG4*01*04. Again, here two new IGHG4 AA alleles (one for AS50 and PA16 and another for PA07) resulting from three nonsynonymous substitutions were uncovered by both techniques.
IGHG AA Alleles Deduced from Mass Spectrometry Analysis Using Sample-specific Databases
Molecular analysis allowed the identification of novel IGHG2 (6) (Table III and supplemental Table S5) and IGHG4 (4) (Table V and supplemental Table S6) nucleotide sequences, and among them, 1 IGHG2 and 2 IGHG4 sequences were associated with a total of four AA changes. As the previous Mascot searches could only match known sequences, we compiled a sample-specific database with the aim to perform the Mascot query again for checking the presence of putatively four new trypsin-cut peptides in the purified IgG samples. The same conditions as already described earlier were used, except that query results considered sets, samesets but also subsets of peptides. The use of the SwissProt Homo sapiens FASTA database from Expasy was kept to match all nonrelevant proteins that were co-purified, thus preventing an increase of false positive matching. The transcribed R.VVSVLTVLHQDWLNGK.E peptide (CH2 85.1–101, V92>L) was assigned to an IGHG2 subset by Mascot following analysis of purified IgG from the PA01 sample. This observation was striking as this peptide is usually found on all IGHG1, all IGHG3 (except IGHG3*09) and all IGHG4 (except IGHG4*02) peptide alleles but never on IGHG2. Similarly, in the case of AS50 and PA16 samples, the transcribed R.EPQVYTLPPSR.E peptide (CH3 1.1–11, Q11>R) was assigned to an IGHG4 set of peptides whereas this peptide is usually exclusive of IGHG1, IGHG2, and IGHG3 peptide alleles. A potentially new IGHG4 peptide, K.VSHK.G (CH2 106–108, N108>H), never referenced among IGHG sequences, was too small to be seen by MS because of the tryptic cleavage specificity, from the AS50, PA07 and PA16 samples. It was not the case of the transcribed R.VVSVLTILHQDWLNGK.E peptide (CH2 85.1–101, V91>I), never referenced among IGHG sequences, that Mascot observed and assigned to an IGHG4 set of peptides for purified IgG from both AS50 and PA16 samples, and not in PA07 where it was also expected.
In order to check the accuracy of these last results, a final analysis consisted for each sample in a Mascot query among pooled IMGT®, SwissProt and sample-specific databases. The same relevant unexpected (PA01 sample: R.VVSVLTVLHQDWLNGK.E, IGHG2 CH2 85.1–101, V92>L, subset level; AS50 and PA16 samples: R.EPQVYTLPPSR.E, IGHG4 CH3 1.1–11, Q11>R, set level) or at yet undescribed (AS50 and PA16 samples: R.VVSVLTILHQDWLNGK.E, IGHG4 CH2 85.1–101, V91>I, set level) peptides were identified unambiguously (supplemental Fig. S2).
DISCUSSION
The present study was carried in an attempt to check with another technique the adequacy of bottom-up mass spectrometry for the identification of the IGHG allelic diversity. Therefore, a nucleotide sequencing of the CH2 and CH3-CHS domains, which concentrate the allelic diversity corresponding to the Fc fragment of the IG heavy gamma chain, was undertaken among DNA samples from ten individuals, and results were compared with those obtained in the corresponding sera by a hemagglutination inhibition method as well as in the corresponding purified IgG by bottom-up MS. For IGHG3, the highest concordance was found between proteomics under nonambiguous conditions and genomics (19 identical alleles/groups of alleles out of 20) whereas identical results were found in only 14 and 12 out of 20 alleles when comparing serology versus proteomics and serology versus genomics, respectively (Table VI). Moreover, the sequencing of the IGHG among these samples originating from Beninese individuals led to the identification of 6 IGHG2 and 4 IGHG4 nucleotide sequences not yet described. When translated, two of these sequences led to 2 putative IGHG peptides not yet described and assigned to IGHG4 by Mascot, that were R.VVSVLTILHQDWLNGK.E (CH2 85.1–101, V91>I) and K.VSHK.G (CH2 106–108, N108>H). By means of proteogenomics, R.VVSVLTILHQDWLNGK.E was identified in the genomically modified set peptide group of the Mascot analysis for 2 (AS50 and PA16) out of 3 samples where this peptide was expected. Regarding this last peptide, a sequence alignment performed by a BLAST search on NCBI® database (National Center for Biotechnology Information, http://blast.ncbi.nlm.nih.gov/Blast.cgi) did not identify any referenced peptide allowing 100% sequence cover: whatever the aligned peptide, the amino acid at position 308 of CH2 85.1–101 is always V instead of I. As the monoisotopic mass difference of 14.015650 Da between V and I is exactly the same as a methylation (UniMod, http://www.unimod.org/, protein modifications for mass spectrometry), the possibility of an artifact was considered but immediately ruled out for two reasons: (1) valine is a neutral amino acid not subject to methylation and (2) in both AS50 and PA16 samples, the MS/MS fragmentation reported y and or b fragments circumscribing exactly the residues of interest (supplemental Fig. S2). An experimental proof would consist in the addition to the test sample before injection of an identical synthetic peptide harboring a CH2 85.1–101, V91>I substitution, in order to compare the m/z and retention time values of endogenous versus synthetic peptides. Similarly, deamidation of Asn (N) or Gln (Q) as possible amino acids modifications were not considered in the Mascot queries to avoid erroneous appearance of irrelevant allele sequences in the Mascot results. The sample preparation and handling methods must therefore avoid the conditions of appearance of such particularly misleading artefactual modifications (30, 31).
To summarize, proteomic and genomic results were highly concordant for all IGHG sequences, with the pointing of defined AA changes suggestive of new IGHG2 and/or IGHG4 peptide sequences. For IGHG3, genomic results always consolidated proteomic ones except in one case (Table VI). It concerned the NP49 sample issued from a 6-year old boy hospitalized at Cotonou (Benin) for cerebral malaria combined with clinically diagnosed anemia. As blood transfusion is currently used in African developing countries to prevent worsening of the malaria pathology (32), it is plausible that the IGHG3*17/IGHG3*17*18*19 detected by serology/proteomics may originate from a donor's IgG3 circulating in the blood of the NP49 recipient. The other discordances recorded between IGHG3 alleles deduced by serology versus proteomics or genomics may be attributable to the following reasons. First, IGHG3 alleles are encoded by codominant genes (4), but in case of heterozygous carriage, it is plausible that one allele may be expressed more abundantly than the other, as already shown for IGHG1 alleles (33), leading to its only detection by serology, such as may be the case for the PA16 sample, where the IGHG3*13 production may exceed that of IGHG3*03 (found by MS and not by hemagglutination inhibition). In some other cases (PA01 and PA42) the serological attribution of IGHG3*17 seems unlikely because absent from both MS and molecular deductions. It must be kept in mind that IGHG3*17 results from the G3m10,11,13,15,27 combination of Gm allotypes and differs from IGHG3*01 and related alleles from the G3m5*, by a concomitant presence of G3m15 and absence of G3m5 and G3m14 allotypes. It can be argued that the difficulty of obtaining well characterized reagent monoclonal antibodies may lead to unstable agglutinates (we used polyclonal reagents coming from blood donors) (34). Among other possible explanations for discrepancies between serological and proteomic/genomic results, are the access to plasma (containing fibrinogen) rather than serum, combined to availability of limited plasma volumes (implying dilutions), that hampered an optimal realization of the hemagglutination inhibition method. However, an eventual depletion in IgG bearing particular IGHG3 alleles during IgG purification on Protein-G column was not an option, as in any case among the results presented here (Table VI) was an IGHG3 allele found by both serology and genomics and not by proteomics.
The excellent correlation between proteomic and genomic results was partly inherent to the analysis setting used in Mascot, where removing dynamic parameters such a as missed cleavages, methionine oxidation and especially deamidation of N or Q prevented a misclassification of alleles linked for example to the identification of the proteotypic peptide R.WQEGNIFSCSVMHEALHNR.F (peptide signature of IGHG3*13) instead of R.WQQGNIFSCSVMHEALHNR.F (peptide signature of IGHG3*01*02*04 to *12). Nonetheless, these precautions did not prevent a mismatch of short peptide sequences (averaging 20 AA) during the probabilistic reconstitution by Mascot of the polypeptide (more than 200 AA) that covers the CH2 and CH3-CHS domains of the Fc fragment. For example, a misalignment by Mascot of K.PREEQYNSTYR.V and R.WQQGNIFSCSVMHEALHNHYTQK.S may lead to identification of IGHG1*04 whereas the second peptide may also be relevant to IGHG3*17 or IGHG3*18*19 alleles when associated with other short discriminatory peptides all present in the mixture resulting from the trypsin digestion of purified IgG from the four IgG subclasses.
A methodological effort of simplification would consist in performing a specific enzymatic cleavage of all Fc/2 fragments by a cysteine proteinase from Streptococcus pyogenes (IdeS) (35). This enzyme, combined to PNGase F for the hydrolysis of all glycans attached on IgG heavy chain and subject to inter- and intraindividual variations (36), generates polypeptide fragments of about 24 kDa (211 AA) concentrating all possible polymorphic AA combinations on the IGHG CH2 and CH3-CHS domains. This results in 21 possible Fc/2 peptides differing by at least 1 Da which could be analyzed using a middle-down MS strategy (37, 38). This new technology that combines aspects of top-down (intact protein) and bottom-up (enzymatic proteolysis) strategies aims to achieve both high resolution and high mass accuracy (39). It presents the advantage of minimizing wrong assignment to a particular AA IGHG allele which could result from erroneous combinations of small peptides when using the bottom-up process. Moreover, as the identification of one IGHG allele will resume in the characterization of one polypeptide, it is conceivable that the discriminatory peptides under analysis will be more frequent than when dissected by trypsin into small peptides necessitating a probabilistic reconstitution in a single sequence (40).
In the context of the present study, a middle-down MS approach would have allowed to assign VVSVLTVLHQDWLNGK (CH2 85.1–101, V92>L) to IGHG2 (sample PA01) as well as EPQVYTLPPSR to IGHG4 (samples AS50 and PA16) by identifying these infrequent sequences within a polypeptide harboring other IGHG2 or IGHG4 AA signatures, respectively. It would also have been possible to make analyzable the short VSHK (CH2 106–108, N108>H) new peptide assignable to IGHG4 (samples AS50, PA07 and PA16). Lastly, despite very high sequence percentage of identity, there is no formal proof that the newly identified R.VVSVLTILHQDWLNGK.E (CH2 85.1–101, V91>I) peptide originates from an IGHG4 polypeptide sequence. Indeed, the preparative treatment of plasma samples using Protein G columns led more to IgG enrichment than to an exclusive IgG purification, and middle-down MS could refute the very low probability for this peptide to belong to a residual plasma protein bearing an unknown to date amino acid polymorphism. In fact, the advent of middle-down MS combined to proteogenomics will contribute to move forward an increasingly detailed description of the Fc fragment diversity, in support of demonstrations like the one presented here.
In conclusion, this study confirms the reliability of the MS approach for investigating the IGHG AA diversity under stringent conditions of analysis, and brings new molecular tools adapted to a fast screening of this diversity. Many applications can result from an accurate determination of these polymorphisms, such as the full validation of therapeutic antibody sequences whose technology is booming (41). Another promising application would consist in the diagnosis of congenital infections in neonates by a differential detection of maternal and fetal IgG on the basis of the IGHG individual diversity (16, 17). Work is underway in our laboratory to apply the middle-down MS approach to polymorphic Fc/2 fragments obtained after complete isolation of parasite-specific IgG from neonates suspected of congenital toxoplasmosis or Chagas disease. If successful, this new way to neonatal serological diagnosis using proteomics could also benefit to congenital infections of bacterial or viral origin.
DATA AVAILABILITY
The newly described IGHG2 nucleotide sequences have been deposited in the GenBank database under GEDI (GenBank/ENA/DDBJ/IMGT/LIGM-DB) Accession Numbers KX670549 to KX670554, sequences KX670550 and KX670551 differing in the intron. Similarly, the newly described IGHG4 exon nucleotide sequences have been deposited in the GenBank database under GEDI Accession Numbers KX670555 to KX670558. The mass spectrometry proteomics data have been deposited to the ProteomeXchange Consortium via the PRIDE (42) partner repository with the dataset identifier PXD005021.
Supplementary Material
Acknowledgments
We thank the participating children and their families. We are grateful to Evelyne Guitard (UMR 5288, CNRS, Université Paul Sabatier Toulouse III, France) for technical advice.
Footnotes
Author contributions: MD, CD, JV, FG and FMN designed research; MD, AE, FA, ML, FG and FMN performed research; ML, JMD, MPL and FG contributed reagents or analytical tools; MD, CD, AE, CG, FG and FMN analyzed data; MD, FG and FMN wrote the paper.
* This work was supported by DVS-Maturation-IRD grant DVS-2012. A PhD scholarship was awarded to Alexandra Emmanuel by the Quisqueya University, Port-au-Prince, Haiti. The authors have declared that no competing interests exist.
 This article contains supplemental material.
This article contains supplemental material.
1 The abbreviations used are:
- IG
- immunoglobulin
- AA
- amino acid
- CH
- constant exon or domain of IG heavy chain
- Fc
- fragment crystallizable
- Gm
- gamma marker
- IGHG
- immunoglobulin heavy constant gamma gene or chain
- SAAV
- single amino acid variant.
REFERENCES
- 1. Lefranc M. P., and Lefranc G. (2001) The Immunoglobulin Factsbook, pp. 1–458, Academic Press, London, UK [Google Scholar]
- 2. Lefranc M. P. (2014) Immunoglobulin (IG) and T cell receptor genes (TR): IMGT® and the birth and rise of immunoinformatics. Front. Immunol. 5, 22. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Lefranc M. P., Giudicelli V., Duroux P., Jabado-Michaloud J., Folch G., Aouinti S., Carillon E., Duvergey H., Houles A., Paysan-Lafosse T., Hadi-Saljoqi S., Sasorith S., Lefranc G., and Kossida S. (2015) IMGT®, the international ImMunoGeneTics information system® 25 years on. Nucleic Acids Res. 43, D413–D422 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Lefranc M. P., and Lefranc G. (2012) Human Gm, Km, and Am allotypes and their molecular characterization: a remarkable demonstration of polymorphism. Methods Mol. Biol. 882, 635–680 [DOI] [PubMed] [Google Scholar]
- 5. Giudicelli V., Chaume D., and Lefranc M. P. (2005) IMGT/GENE-DB: a comprehensive database for human and mouse immunoglobulin and T cell receptor genes. Nucleic Acids Res. 33, D256–D261 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Jefferis R., and Lefranc M. P. (2009) Human immunoglobulin allotypes: possible implications for immunogenicity. MAbs 1, 332–338 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Magdelaine-Beuzelin C., Vermeire S., Goodall M., Baert F., Noman M., Assche G. V., Ohresser M., Degenne D., Dugoujon J.-M., Jefferis R., Rutgeerts P., Lefranc M. P., and Watier H. (2009) IgG1 heavy chain-coding gene polymorphism (G1m allotypes) and development of antibodies-to-infliximab. Pharmacogenet. Genomics 19, 383–387 [DOI] [PubMed] [Google Scholar]
- 8. Beck A., Wagner-Rousset E., Ayoub D., Van Dorsselaer A., and Sanglier-Cianférani S. (2013) Characterization of therapeutic antibodies and related products. Anal. Chem. 85, 715–736 [DOI] [PubMed] [Google Scholar]
- 9. Ward E. S., and Ober R. J. (2009) Multitasking by exploitation of intracellular transport functions the many faces of FcRn. Adv. Immunol. 103, 77–115 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Stapleton N. M., Einarsdóttir H. K., Stemerding A. M., and Vidarsson G. (2015) The multiple facets of FcRn in immunity. Immunol. Rev. 268, 253–268 [DOI] [PubMed] [Google Scholar]
- 11. Jefferis R. (2012) Isotype and glycoform selection for antibody therapeutics. Arch. Biochem. Biophys. 526, 159–166 [DOI] [PubMed] [Google Scholar]
- 12. Palmeira P., Quinello C., Silveira-Lessa A. L., Zago C. A., and Carneiro-Sampaio M. (2012) IgG placental transfer in healthy and pathological pregnancies. Clin. Dev. Immunol. :985646, 2012 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Carlier Y., Truyens C., Deloron P., and Peyron F. (2012) Congenital parasitic infections: a review. Acta Trop. 121, 55–70 [DOI] [PubMed] [Google Scholar]
- 14. Carlier Y., Sosa-Estani S., Luquetti A. O., and Buekens P. (2015) Congenital Chagas disease: an update. Mem. Inst. Oswaldo Cruz 110, 363–368 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Villard O., Cimon B., L'Ollivier C., Fricker-Hidalgo H., Godineau N., Houze S., Paris L., Pelloux H., Villena I., and Candolfi E. (2016) Serological diagnosis of Toxoplasma gondii infection: Recommendations from the French National Reference Center for Toxoplasmosis. Diagn. Microbiol. Infect. Dis. 84, 22–33 [DOI] [PubMed] [Google Scholar]
- 16. Migot-Nabias F., Dechavanne C., Guillonneau F., Dugoujon J. M., and Lefranc M. P. (2014) Method of neonatal serological diagnosis. Patents FR 11 57296. (2011), PCT/EP2012065737. (2012), WO2013021057. (2013) and US2014178916A1
- 17. Dechavanne C., Guillonneau F., Chiappetta G., Sago L., Lévy P., Salnot V., Guitard E., Ehrenmann F., Broussard C., Chafey P., Le Port A., Vinh J., Mayeux P., Dugoujon J. M., Lefranc M. P., and Migot-Nabias F. (2012) Mass spectrometry detection of G3m and IGHG3 alleles and follow-up of differential mother and neonate IgG3. PLoS ONE 7, e46097. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Migot-Nabias F., Noukpo J. M., Guitard E., Doritchamou J., Garcia A., and Dugoujon J. M. (2008) Imbalanced distribution of GM immunoglobulin allotypes according to the clinical presentation of Plasmodium falciparum malaria in Beninese children. J. Infect. Dis. 198, 1892–1895 [DOI] [PubMed] [Google Scholar]
- 19. Dard P., Lefranc M.-P., Osipova L., and Sanchez-Mazas A. (2001) DNA sequence variability of IGHG3 alleles associated to the main G3m haplotypes in human populations. Eur. J. Hum. Genet. 9, 765–772 [DOI] [PubMed] [Google Scholar]
- 20. Brucato N., Cassar O., Tonasso L., Tortevoye P., Migot-Nabias F., Plancoulaine S., Guitard E., Larrouy G., Gessain A., and Dugoujon J. M. (2010) The imprint of the Slave Trade in an African American population: mitochondrial DNA, Y chromosome and HTLV-1 analysis in the Noir Marron of French Guiana. BMC Evol. Biol. 10, 314. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Field L. L., and Dugoujon J. M. (1989) Immunoglobulin allotyping (Gm, Km) of GAW5 families. Genet. Epidemiol. 6, 31–33 [DOI] [PubMed] [Google Scholar]
- 22. van Loghem E., de Lange G., van Leeuwen A. M., van Eede P. H., Nijenhuis L. E., Lefranc M. P., and Lefranc G. (1982) Human IgG allotypes co-occurring in more than one IgG subclass. Vox Sang. 43, 301–309 [DOI] [PubMed] [Google Scholar]
- 23. Edelman G. M., Cunningham B. A., Gall W. E., Gottlieb P. D., Rutishauser U., and Waxdal M. J. (1969) The covalent structure of an entire gammaG immunoglobulin molecule. Proc. Natl. Acad. Sci. U.S.A. 63, 78–85 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Giudicelli V., Duroux P., Ginestoux C., Folch G., Jabado-Michaloud J., Chaume D., and Lefranc M.-P. (2006) IMGT/LIGM-DB, the IMGT comprehensive database of immunoglobulin and T cell receptor nucleotide sequences. Nucleic Acids Res. 34, D781–D784 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Lefranc M.-P., Pommié C., Kaas Q., Duprat E., Bosc N., Guiraudou D., Jean C., Ruiz M., Da Piédade I., Rouard M., Foulquier E., Thouvenin V., and Lefranc G. (2005) IMGT unique numbering for immunoglobulin and T cell receptor constant domains and Ig superfamily C-like domains. Dev. Comp. Immunol. 29, 185–203 [DOI] [PubMed] [Google Scholar]
- 26. Hernandez C., Waridel P., and Quadroni M. (2014) Database construction and peptide identification strategies for proteogenomic studies on sequenced genomes. Curr. Top. Med. Chem. 14, 425–434 [DOI] [PubMed] [Google Scholar]
- 27. Nesvizhskii A. I. (2014) Proteogenomics: concepts, applications and computational strategies. Nat. Methods 11, 1114–1125 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Dugoujon J.-M., Hazout S., Loirat F., Mourrieras B., Crouau-Roy B., and Sanchez-Mazas A. (2004) GM haplotype diversity of 82 populations over the world suggests a centrifugal model of human migrations. Am. J. Phys. Anthropol. 125, 175–192 [DOI] [PubMed] [Google Scholar]
- 29. den Dunnen J. T., Dalgleish R., Maglott D. R., Hart R. K., Greenblatt M. S., McGowan-Jordan J., Roux A. F., Smith T., Antonarakis S. E., and Taschner P. E. (2016) HGVS Recommendations for the description of sequence variants: 2016 Update. Hum. Mutat. 37, 564–569 [DOI] [PubMed] [Google Scholar]
- 30. Sinha S., Zhang L., Duan S., Williams T. D., Vlasak J., Ionescu R., and Topp E. M. (2009) Effect of protein structure on deamidation rate in the Fc fragment of an IgG1 monoclonal antibody. Protein Sci. 18, 1573–1584 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Li X., Cournoyer J. J., Lin C., and O'Connor P. B. (2008) Use of 18O labels to monitor deamidation during protein and peptide sample processing. J. Am. Soc. Mass. Spectrom. 19, 855–864 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Orish V. N., Ilechie A., Combey T., Onyeabor O. S., Okorie C., and Sanyaolu A. O. (2016) Evaluation of blood transfusions in anemic children in Effia Nkwanta Regional Hospital, Sekondi-Takoradi, Ghana. Am. J. Trop. Med. Hyg. 94, 691–694 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Goetze A. M., Zhang Z., Liu L., Jacobsen F. W., and Flynn G. C. (2011) Rapid LC-MS screening for IgG Fc modifications and allelic variants in blood. Mol. Immunol. 49, 338–352 [DOI] [PubMed] [Google Scholar]
- 34. Jefferis R., Reimer C. B., Skvaril F., de Lange G. G., Goodall D. M., Bentley T. L., Phillips D. J., Vlug A., Harada S., Radl J., Claassen E., Boersma J. A., and Coolen J. (1992) Evaluation of monoclonal antibodies having specificity for human IgG subclasses: results of the 2nd IUIS/WHO collaborative study. Immunol. Lett. 31, 143–168 [DOI] [PubMed] [Google Scholar]
- 35. Vincents B., von Pawel-Rammingen U., Björck L., and Abrahamson M. (2004) Enzymatic characterization of the streptococcal endopeptidase, IdeS, reveals that it is a cysteine protease with strict specificity for IgG cleavage due to exosite binding. Biochemistry 43, 15540–15549 [DOI] [PubMed] [Google Scholar]
- 36. Pucić M., Knezević A., Vidic J., Adamczyk B., Novokmet M., Polasek O., Gornik O., Supraha-Goreta S., Wormald M. R., Redzić I., Campbell H., Wright A., Hastie N. D., Wilson J. F., Rudan I., Wuhrer M., Rudd P. M., Josić D., and Lauc G. (2011) High throughput isolation and glycosylation analysis of IgG-variability and heritability of the IgG glycome in three isolated human populations. Mol. Cell. Proteomics 10, M111.010090. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37. Leblanc Y., Romanin M., Bihoreau N., and Chevreux G. (2014) LC-MS analysis of polyclonal IgGs using IdeS enzymatic proteolysis for oxidation monitoring. J. Chromatogr. B Analyt. Technol. Biomed. Life Sci. 961, 1–4 [DOI] [PubMed] [Google Scholar]
- 38. He X. A., Washburn N., Arevalo E., and Robblee J. H. (2015) Analytical characterization of IgG Fc subclass variants through high-resolution separation combined with multiple LC-MS identification. Anal. Bioanal. Chem. 407, 7055–7066 [DOI] [PubMed] [Google Scholar]
- 39. Tsiatsiani L., and Heck A. J. (2015) Proteomics beyond trypsin. FEBS J. 282, 2612–2626 [DOI] [PubMed] [Google Scholar]
- 40. Nesvizhskii A. I., and Aebersold R. (2005) Interpretation of shotgun proteomic data: the protein inference problem. Mol. Cell. Proteomics 4, 1419–1440 [DOI] [PubMed] [Google Scholar]
- 41. Resemann A., Jabs W., Wiechmann A., Wagner E., Colas O., Evers W., Belau E., Vorwerg L., Evans C., Beck A., and Suckau D. (2016) Full validation of therapeutic antibody sequences by middle-up mass measurements and middle-down protein sequencing. MAbs. 8, 318–330 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42. Vizcaíno J. A., Csordas A., del-Toro N., Dianes J. A., Griss J., Lavidas I., Mayer G., Perez-Riverol Y., Reisinger F., Ternent T., Xu Q. W., Wang R., and Hermjakob H. (2016) 2016 update of the PRIDE database and related tools. Nucleic Acids Res. 44(D1), D447–D456 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
The newly described IGHG2 nucleotide sequences have been deposited in the GenBank database under GEDI (GenBank/ENA/DDBJ/IMGT/LIGM-DB) Accession Numbers KX670549 to KX670554, sequences KX670550 and KX670551 differing in the intron. Similarly, the newly described IGHG4 exon nucleotide sequences have been deposited in the GenBank database under GEDI Accession Numbers KX670555 to KX670558. The mass spectrometry proteomics data have been deposited to the ProteomeXchange Consortium via the PRIDE (42) partner repository with the dataset identifier PXD005021.


