Abstract
Human blood plasma is a complex biological fluid containing soluble proteins, sugars, hormones, electrolytes, and dissolved gasses. As plasma interacts with a wide array of bodily systems, changes in protein expression, or the presence or absence of specific proteins are regularly used in the clinic as a molecular biomarker tool. A large body of literature exists detailing proteomic changes in pathologic contexts, however little research has been conducted on the quantitation of the plasma proteome in age-specific, healthy subjects, especially in pediatrics. In this study, we utilized SWATH-MS to identify and quantify proteins in the blood plasma of healthy neonates, infants under 1 year of age, children between 1–5 years, and adults. We identified more than 100 proteins that showed significant differential expression levels across these age groups, and we analyzed variation in protein expression across the age spectrum. The plasma proteomic profiles of neonates were strikingly dissimilar to the older children and adults. By extracting the SWATH data against a large human spectral library we increased protein identification more than 6-fold (940 proteins) and confirmed the concentrations of several of these using ELISA. The results of this study map the variation in expression of proteins and pathways often implicated in disease, and so have significant clinical implication.
Human blood plasma is a complex but ideal biological system containing proteins from a variety of cellular localizations. As such plasma is one of the best reporter systems in the clinical setting, as it communicates with most parts of the body and reflects changes in the state of the organism as a whole. Unlike serum, which is depleted of coagulation proteins that play a role not only in coagulation but also in processes such as inflammation, human plasma contains all the soluble proteins found in blood. Hence, human plasma is a commonly used sample for biomarker discovery.
Proteomics studies utilizing human plasma have, to date, focused largely on specific disease settings, on detecting the highest number of proteins or on the effect of certain drugs on changes in plasma protein expression. In addition, most such studies have focused on adults, with limited number of studies and hence limited knowledge of the plasma proteome in infants and children. The plasma proteomic studies in children have focused on examining changes in protein expression that occur during Cardiopulmonary Bypass Surgery (1), as well as in several disease states including: blood disorders (2), sickle cell disease (3), leukemia (4), and cystic fibrosis (5). There has, to date, only been one study that has focused on the physiological development of the plasma proteome by examining the changes in plasma protein expression that occur with age, from neonates through to adults (6). That study utilized two-dimensional difference in gel electrophoresis (2D-DIGE) methodology and described the concept of aging of the human plasma proteome by identifying significant age-specific changes in expression of up to 100 proteins spots.
The Human Proteome Organization (HUPO)1 Plasma Proteome Project (PPP) recognizes the importance of analyzing and understanding age related differences in the plasma proteome by identifying this as one of their scientific aims and research priorities (7). In addition, the main aim of the recently established Pediatric Proteomics (PediOme) initiative, a part of HUPO, is to advance the use of proteomics techniques to solve major issues in pediatric medicine through characterization of the pediatric proteome across a wide-variety of tissues and biological samples.
In 2012, a new proteomics approach known as SWATH-MS became available (8). This high-throughput, data independent acquisition (DIA) method allows confident identification and quantification of query peptides across large sample sets with a dynamic range of 4 orders of magnitude, and is as such well suited to studies of human plasma. A particular advantage of SWATH-MS is the ability to re-mine previously acquired data sets. As an emerging technology, to date only a handful of studies have utilized SWATH-MS for investigations of human plasma (9, 10). The first study investigated the performance and suitability of SWATH-MS for quantification of N-linked glycoproteins in human plasma and suggested that this combined approach is suitable for biomarker discovery and verification (9). The second study utilized SWATH-MS to quantify the variability in the plasma proteome in monozygotic and dizygotic twins, demonstrating large variability in protein abundance between twins (10).
The use of SWATH-MS technology in neonatal and pediatric plasma samples to determine variability and subsequent comparison to adult plasma remains an exciting research opportunity, and one that has, to date, not been explored. Therefore, this study aimed to utilize SWATH-MS to quantitate age-specific variability of plasma proteins in healthy neonates, children, and adults.
EXPERIMENTAL PROCEDURES
Participant Recruitment, Sample Collection and Storage
Blood samples used in this study were collected and processed according to established protocols (6, 11–15). Neonatal samples (within 72 h postbirth) were collected from healthy term neonates from the Family Birthing Unit or post-natal wards at the Royal Women's Hospital, Melbourne. Eligibility criteria included: gestation >37 weeks, vaginal delivery, birth-weight >2500gm, APGAR at 5min ≥7 and absence of systemic abnormalities. Pediatric samples were obtained from healthy children attending the hospital for minor elective day-surgery (e.g. tongue-tie release and circumcision) who were not receiving any medications and had no significant family history of major diseases, particularly diseases known to be associated with the process of aging (e.g. diabetes and cardiovascular disease). The pediatric cohort excluded adolescents taking oral contraceptive and smokers. Adult samples were obtained from healthy volunteers aged between 20 and 45 years of age who were not taking any medications (including oral contraceptive), were not smokers, and similarly, with no past medical history. Family history was assessed via a brief interview with the parents of the children and the adult volunteers themselves. Blood samples were collected in S-Monovette® tubes (Sarstedt, Australia), containing 1 volume of citrate per 9 volumes of blood and were centrifuged at 3000 rpm, 10 min, 10°C, with plasma stored at −80 °C, until testing.
This study was approved by the Royal Children's Hospital Ethics in Human Research Committee, reference number 20031; and the Royal Women's Hospital Human Research Ethics Committee (#02/08). Written informed consent was obtained from parents of children and from adult participants themselves.
Plasma Sample Preparation
25 μl of human plasma sample was diluted in 475 μl of 50 mm ammonium bicarbonate (Sigma, St. Louis, MO) solution. Samples were reduced with 5 mm dithiothreitol (Bio-Rad, Hercules, CA) at 65 °C for 30 min, followed by alkylation with 10 mm iodoacetamide (Sigma) at room temperature for 30 min in the dark. One fifth of the reduced and alkylated sample (121 μl) was digested with sequencing-grade porcine trypsin (20 μg, Promega) overnight at 37 °C. The digested sample was diluted (×10.5) in 0.1% formic acid prior to all mass spectrometry analyses.
Information Dependent Acquisition Mass Spectrometry
A 5600 TripleTOF mass spectrometer (Sciex, Framingham, MA) coupled to an Eksigent Ultra-nanoLC-1D system (Eksigent Technologies, Dublin, CA) was employed for both IDA and SWATH-MS analysis. For IDA data, 10 μl of digested sample from each group was pooled, making four group pools (Neonate, < 1 year, 1–5 year, adult) and 5 μl from each group was injected.
Peptides were loaded onto a reverse phase peptide C18 Captrap (Bruker, Billerica, MA) for pre-concentration and desalted for 5 min with the loading buffer (0.1% (v/v) formic acid, 2% (v/v) acetonitrile) at a flow rate of 10 μl per minute. After desalting, the peptide trap was switched in-line with an in-house packed analytical column (150 μm × 10 cm, solid core Halo C18, 160 Å, 2.7 μm (Bruker)). Peptides were eluted and separated from the column using the buffer B (99.9% (v/v) acetonitrile, 0.1% (v/v) formic acid) gradient starting from 2% and increasing to 10% in 10 min then to 35% over 78 min at a flow rate of 500 nL per minute. After peptide elution, the column was flushed with 95% buffer B for 15 min and re-equilibrated with 98% buffer A (0.1% (v/v) formic acid) for 15 min before next injection. Blanks were run between samples. In IDA mode, a TOFMS survey scan was acquired at m/z 350–1500 with 0.25 s accumulation time, with the ten most intense precursor ions (2+–5+; counts > 150) in the survey scan consecutively isolated for subsequent product ion scans. Dynamic exclusion was used with a window of 20 s. Product ion spectra were accumulated for 50 milliseconds in the mass range m/z 100—1500 with rolling collision energy.
IDA Data Analysis
Protein identification from IDA data was performed with ProteinPilot (v4.2, Sciex) using the Paragon algorithm. The search parameters were as follows: sample type: identification; cys alkylation: iodoacetamide; digestion: trypsin; instrument: TripleTOF 5600; special factors: none; ID focus: biological modifications; miss-cleavages: two. The Homo sapiens search database was obtained from UniProt (20,265 entries, Jan 2015). A reversed-decoy database search strategy was used with ProteinPilot, with the calculated protein FDR equaling 0.68%.
SWATH Library Construction
The ProteinPilot group file from the IDA search result of the 4 sample groups was imported into PeakView (v2.1) (Sciex) and used as a local peptide assay library. This library contained 151 proteins identified from un-depleted plasma samples. An extended peptide assay library was constructed using the SWATHXtend script (16), which merged the local peptide assay library with a 10,000 protein human (17) spectral library downloaded from SWATHAtlas.
SWATH-MS
For SWATH-MS experiments 5 μl of each digested sample was injected in sample order, with two technical replicates for each injection; one sample was run in triplicates once at the beginning and twice at the end of the series to check for potential instrument drift as detailed in Supplemental Information. Identical LC conditions were used as described above, with m/z window sizes determined based on precursor m/z frequencies in previous IDA data. SWATH variable window acquisition with a set of 60 overlapping windows (1 amu for the window overlap) was constructed covering the mass range of m/z 399.5 - 1249.5. In SWATH mode, first a TOFMS survey scan was acquired (m/z 350–1500, 0.05 s) then the 60 predefined m/z ranges were sequentially subjected to MS/MS analysis. Product ion spectra were accumulated for 50 milliseconds in the mass range m/z 350–1500 with rolling collision energy optimized for lowed m/z in m/z window +10%, with the total duty cycle of 3.7 s.
SWATH Data Analysis
SWATH peaks were extracted using PeakView (v.2.1). Shared and modified peptides were excluded. Peak extraction parameters were set as the following: 100 peptides per protein, 6 transition ions per peptide, peptide confidence threshold 99%, FDR extraction threshold 1%, XIC (Extract Ion Chromatogram) retention time window 10 min and mass tolerance 75 ppm. The extracted transition ion peak areas, peptide peak areas and protein peak areas were exported in Excel for further statistical analysis.
Experimental Design and Statistical Rationale
Quantitative MS data was obtained from ten independent biological samples for each of the four age cohort groups. Two technical replicate were obtained for each biological sample in SWATH acquisition. The peptide ion peak areas were averaged for the replicate technical injections, then further scaled so that the sum of the ion intensities of each sample equal the maximum total ion intensity. The normalized ion intensities were summed for each peptide and protein. The entire protein level data was clustered using hierarchical clustering (Euclidean distance and complete linkage), and visualized using a principal component analysis to examine global trends. The data distribution was checked visually using boxplots and density plots (Supplemental information: data quality).
Two approaches were undertaken for determining differential expression: an analysis of variance on the log-transformed normalized protein peak areas, and pairwise comparisons of pairs of samples. For the analysis of variance, proteins were deemed to be differentially expressed if the ANOVA p value was less than 0.05, and the maximum protein fold change exceeded 1.5. We evaluated adjusting the ANOVA p values for multiple testing using the fdr criterion of Benjamini and Hochberg, however the combination of unadjusted p value with fold change thresholds was utilized as it represented a stricter requirement in this case.
For the pairwise comparisons, samples from all selected pairs of conditions were compared first by a two-sample Student's t test of the log-transformed protein areas, and secondly by combining individual peptide-level ratios for each protein. The reporting threshold required a Student's t test p value less than 0.05 and fold change exceeding 1.5.
For ELISA testing we used the same 40 independent biological samples utilized for the SWATH analysis, as well as 10 additional individuals for each age group. A two-sample Student's t test was applied to detect significant differences between groups.
Pathway Analysis
Differentially expressed proteins were examined in the context of biological data using the Ingenuity Pathway Analysis tool with the abundance ratio of neonates to adults as the featured observation, and using default settings for determining enrichment. The top five most enriched pathways, functions and diseases and the top three networks were tabulated and described.
Enzyme-Linked Immunosorbent Assay (ELISA) validation
We utilized the following ELISA kits according to the manufacturer's specifications: GPI-specific phospholipase D (GPLD1); (CSB-EL009721HU, Cusabio, Wuhan, China), Periostin (POSTN) (CSB-E16444H, Cusabio, Wuhan, China), Alpha-2-macroglobulin (A2M) (ab108888, Abcam, Cambridge, United Kingdom), Histidine rich glycoprotein (HRG) (CSB-E13159H, Cusabio, Wuhan, China).
RESULTS
Sample Characteristics
SWATH-MS analysis was performed on 10 individual samples from each of the following age groups: Neonates (5F/5M, within 72 h post-birth), < 1 year olds (3F/7M, median age 7.7 months, range 6 months to 11 months), 1–5 year olds (5F/5M, median age 3.12 years, range 1.59 to 4.39 years) and adults (5F/5M, median age 31.40 years, range 22.02 to 44.97 years).
SWATH-MS
We used nondepleted plasma samples from the sample cohort and IDA to establish a local peptide assay library containing 146 proteins. SWATH acquisition of the 40 individual samples were obtained, and then matched to the local assay library for quantitation of peptide areas (supplemental Table S1).
Overall Differences Across the Age-spectrum From Neonates to Adults
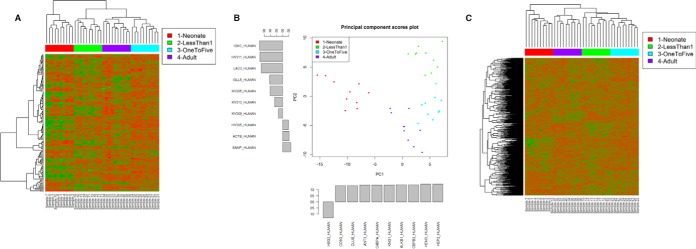
Hierarchical clustering of the 40 plasma samples using SWATH quantitation of 146 proteins showed well separated clusters corresponding to the experimental age groups. Interestingly, this analysis clearly showed that plasma protein levels from the neonates were quite distinct from the three remaining age groups, with adults and one to five year olds clustering next to each other (Fig. 1). These trends were confirmed by principal component analysis, showing clear segregation based on age, with neonates clustering separately compared with all other age groups.
Fig. 1.
A, Unsupervised hierarchical clustering analysis for the 146 protein identifications using the local assay library. Relative expression patterns obtained using Euclidean distance. Green - proteins with decreased expression; Red - proteins with increased expression. B, Principal Component Analysis of data from A. The top 10 highest loadings for PC1 and PC2 are shown. C, Unsupervised hierarchical clustering for 940 proteins using an extended assay library.
Given the strong separation of the samples with unsupervised analysis (clustering and PCA), not surprisingly a large percentage of the proteins were found to be differentially expressed based on the analysis of variance (ANOVA). A total of 107 proteins were found to have ANOVA p value < 0.05 and maximum fold change (ratio of highest average to the lowest average) at least 1.5 (Table I). This was a stricter approach compared with the 121 proteins identified using a Benjamini and Hochberg adjusted p value less than 0.05 (data not shown). The minimum fold change, of 1.51 was observed for Apolipoprotein-M, ranging up to greater than 300-fold change for Hemoglobin subunit gamma-2 which is significantly elevated in neonates. Specifically, there were 86 (80%) proteins with a maximum fold change ranging between 1.5 and 5; 11 (10%) proteins with a maximum fold change between 5- and 10-fold; 6 (6%) proteins with a maximum fold change between 10- and 50-fold and 4 (4%) proteins with a maximum fold change >50-fold.
Table I. ANOVA analysis showing the variation in protein expression across the age spectrum (neonates to adults). Results are expressed as mean variation in protein expression.
| Uniprot accession | Protein | Mean–neonate | Mean–<1 y.o. | Mean–1–5 y.o. | Mean–adult | Fold change | ANOVA adjusted p value | Protein name |
|---|---|---|---|---|---|---|---|---|
| O95445 | APOM_HUMAN | 1.53E+06 | 2.20E+06 | 1.89E+06 | 1.45E+06 | 1.51 | 3.21E-03 | Apolipoprotein M |
| P01008 | ANT3_HUMAN | 6.58E+06 | 9.94E+06 | 9.63E+06 | 7.91E+06 | 1.51 | 1.73E-08 | Antithrombin-III |
| P08603 | CFAH_HUMAN | 1.25E+07 | 1.46E+07 | 1.88E+07 | 1.48E+07 | 1.51 | 1.26E-05 | Complement factor H |
| P02675 | FIBB_HUMAN | 1.61E+08 | 1.06E+08 | 1.39E+08 | 1.24E+08 | 1.51 | 2.86E-05 | Fibrinopeptide B |
| P05156 | CFAI_HUMAN | 3.05E+05 | 3.33E+05 | 4.66E+05 | 3.54E+05 | 1.53 | 1.09E-05 | Complement factor I |
| P02671 | FIBA_HUMAN | 1.43E+08 | 9.35E+07 | 1.30E+08 | 1.22E+08 | 1.54 | 8.05E-07 | Fibrinogen alpha chain |
| P05543 | THBG_HUMAN | 9.95E+05 | 9.78E+05 | 1.05E+06 | 6.80E+05 | 1.55 | 1.39E-02 | Thyroxine-binding globulin |
| P0C0L5 | CO4B_HUMAN | 8.41E+05 | 6.76E+05 | 1.05E+06 | 8.66E+05 | 1.55 | 4.51E-02 | Complement C4-B alpha chain |
| P02679 | FIBG_HUMAN | 1.23E+08 | 7.93E+07 | 1.10E+08 | 1.00E+08 | 1.56 | 7.00E-07 | Fibrinogen gamma chain |
| P04004 | VTNC_HUMAN | 8.32E+06 | 1.07E+07 | 1.30E+07 | 1.09E+07 | 1.57 | 5.54E-04 | Vitronectin V10 subunit |
| P06331 | HV209_HUMAN | 9.09E+05 | 1.44E+06 | 1.32E+06 | 9.62E+05 | 1.59 | 1.19E-03 | Ig heavy chain V-II region ARH-77 |
| P18136 | KV313_HUMAN | 1.21E+07 | 8.66E+06 | 1.20E+07 | 1.37E+07 | 1.59 | 8.99E-04 | Ig kappa chain V-III region HIC |
| P08697 | A2AP_HUMAN | 4.26E+06 | 5.03E+06 | 4.64E+06 | 3.13E+06 | 1.60 | 8.34E-04 | Alpha-2-antiplasmin |
| O75636 | FCN3_HUMAN | 3.01E+05 | 4.17E+05 | 4.11E+05 | 2.57E+05 | 1.62 | 5.03E-04 | Ficolin-3 |
| P05090 | APOD_HUMAN | 2.93E+06 | 2.67E+06 | 2.84E+06 | 4.35E+06 | 1.63 | 1.37E-03 | Apolipoprotein D |
| P05452 | TETN_HUMAN | 2.67E+05 | 3.39E+05 | 4.35E+05 | 2.89E+05 | 1.63 | 8.46E-06 | Tetranectin |
| P01031 | CO5_HUMAN | 3.57E+06 | 2.17E+06 | 2.88E+06 | 2.78E+06 | 1.64 | 1.70E-06 | Complement C5 beta chain |
| P36955 | PEDF_HUMAN | 1.08E+06 | 8.38E+05 | 1.31E+06 | 1.38E+06 | 1.66 | 5.48E-03 | Pigment epithelium-derived factor |
| P00734 | THRB_HUMAN | 8.60E+06 | 1.32E+07 | 1.43E+07 | 1.15E+07 | 1.66 | 6.11E-05 | Activation peptide fragment 1 |
| P04433 | KV309_HUMAN | 3.27E+06 | 2.18E+06 | 3.51E+06 | 3.68E+06 | 1.69 | 1.97E-03 | Ig kappa chain V-III region VG |
| P02652 | APOA2_HUMAN | 1.13E+07 | 1.92E+07 | 1.55E+07 | 1.81E+07 | 1.69 | 3.72E-09 | Proapolipoprotein A-II |
| P01042 | KNG1_HUMAN | 4.99E+06 | 8.05E+06 | 8.44E+06 | 7.33E+06 | 1.69 | 1.96E-06 | T-kinin |
| P22792 | CPN2_HUMAN | 7.20E+05 | 8.92E+05 | 1.24E+06 | 7.56E+05 | 1.73 | 2.11E-06 | Carboxypeptidase N subunit 2 |
| P06727 | APOA4_HUMAN | 1.05E+07 | 1.83E+07 | 1.44E+07 | 1.09E+07 | 1.74 | 3.53E-03 | Apolipoprotein A-IV |
| Q14624 | ITIH4_HUMAN | 1.26E+07 | 1.64E+07 | 2.20E+07 | 1.77E+07 | 1.74 | 1.99E-07 | 35 kDa inter-alpha-trypsin inhibitor heavy chain H4 |
| P02751 | FINC_HUMAN | 1.63E+07 | 2.27E+07 | 2.84E+07 | 2.32E+07 | 1.74 | 1.32E-05 | Anastellin |
| P04208 | LV106_HUMAN | 1.82E+06 | 1.04E+06 | 1.67E+06 | 1.70E+06 | 1.74 | 3.82E-04 | Ig lambda chain V-I region WAH |
| P00747 | PLMN_HUMAN | 7.07E+06 | 1.04E+07 | 1.24E+07 | 1.16E+07 | 1.75 | 1.20E-07 | Plasmin heavy chain A, short form |
| P10643 | CO7_HUMAN | 1.02E+06 | 8.59E+05 | 9.18E+05 | 5.74E+05 | 1.78 | 1.61E-05 | Complement component C7 |
| P13671 | CO6_HUMAN | 8.29E+05 | 1.11E+06 | 1.49E+06 | 8.71E+05 | 1.80 | 4.73E-07 | Complement component C6 |
| P00751 | CFAB_HUMAN | 1.10E+07 | 1.37E+07 | 2.00E+07 | 1.46E+07 | 1.81 | 1.38E-06 | Complement factor B Bb fragment |
| P10909 | CLUS_HUMAN | 7.90E+06 | 1.15E+07 | 1.45E+07 | 1.29E+07 | 1.83 | 4.09E-11 | Clusterin alpha chain |
| P01611 | KV119_HUMAN | 5.58E+05 | 3.00E+05 | 3.13E+05 | 4.90E+05 | 1.85 | 2.49E-05 | Ig kappa chain V-I region Wes |
| P05155 | IC1_HUMAN | 1.12E+07 | 1.84E+07 | 2.13E+07 | 1.33E+07 | 1.88 | 6.91E-08 | Plasma protease C1 inhibitor |
| P04207 | KV308_HUMAN | 1.20E+06 | 8.12E+05 | 1.54E+06 | 1.10E+06 | 1.90 | 5.77E-02 | Ig kappa chain V-III region CLL |
| P01602 | KV110_HUMAN | 2.52E+06 | 1.32E+06 | 1.62E+06 | 2.14E+06 | 1.90 | 1.66E-03 | Ig kappa chain V-I region HK102 |
| P06319 | LV605_HUMAN | 1.25E+05 | 1.83E+05 | 1.49E+05 | 8.97E+04 | 2.04 | 5.61E-02 | Ig lambda chain V-VI region EB4 |
| P0C0L4 | CO4A_HUMAN | 1.63E+06 | 1.18E+06 | 2.45E+06 | 2.12E+06 | 2.07 | 2.67E-04 | C4a anaphylatoxin |
| P01609 | KV117_HUMAN | 4.10E+06 | 1.94E+06 | 2.40E+06 | 3.31E+06 | 2.10 | 2.11E-06 | Ig kappa chain V-I region Scw |
| P04217 | A1BG_HUMAN | 8.36E+06 | 1.52E+07 | 1.77E+07 | 1.38E+07 | 2.12 | 7.59E-08 | Alpha-1B-glycoprotein |
| P02753 | RET4_HUMAN | 1.69E+06 | 1.87E+06 | 1.30E+06 | 2.79E+06 | 2.15 | 7.16E-03 | Plasma retinol-binding protein(1–176) |
| Q96PD5 | PGRP2_HUMAN | 1.33E+06 | 2.75E+06 | 2.88E+06 | 1.60E+06 | 2.16 | 1.89E-09 | N-acetylmuramoyl-l-alanine amidase |
| P03952 | KLKB1_HUMAN | 8.74E+05 | 1.68E+06 | 1.90E+06 | 1.55E+06 | 2.17 | 3.99E-08 | Plasma kallikrein light chain |
| P01880 | IGHD_HUMAN | 2.08E+05 | 2.02E+05 | 2.58E+05 | 4.43E+05 | 2.18 | 4.77E-02 | Ig delta chain C region |
| P01019 | ANGT_HUMAN | 1.79E+07 | 9.18E+06 | 8.12E+06 | 9.37E+06 | 2.20 | 1.36E-04 | Angiotensin-2 |
| P07358 | CO8B_HUMAN | 4.22E+05 | 7.55E+05 | 9.47E+05 | 8.63E+05 | 2.24 | 1.68E-10 | Complement component C8 beta chain |
| P51884 | LUM_HUMAN | 1.20E+06 | 1.54E+06 | 1.44E+06 | 6.87E+05 | 2.25 | 1.04E-06 | Lumican |
| P02763 | A1AG1_HUMAN | 3.04E+07 | 3.54E+07 | 6.88E+07 | 3.79E+07 | 2.25 | 2.42E-05 | Alpha-1-acid glycoprotein 1 |
| P06314 | KV404_HUMAN | 1.97E+06 | 1.26E+06 | 1.56E+06 | 2.84E+06 | 2.26 | 2.65E-07 | Ig kappa chain V-IV region B17 |
| P00450 | CERU_HUMAN | 1.99E+07 | 3.79E+07 | 4.62E+07 | 3.25E+07 | 2.31 | 4.66E-06 | Ceruloplasmin |
| P33151 | CADH5_HUMAN | 7.02E+04 | 4.50E+04 | 5.37E+04 | 2.95E+04 | 2.37 | 9.62E-07 | Cadherin-5 |
| P02750 | A2GL_HUMAN | 9.15E+05 | 1.18E+06 | 2.19E+06 | 1.19E+06 | 2.39 | 1.15E-05 | Leucine-rich alpha-2-glycoprotein |
| P01857 | IGHG1_HUMAN | 1.99E+08 | 8.27E+07 | 1.38E+08 | 1.43E+08 | 2.40 | 2.71E-08 | Ig gamma-1 chain C region |
| P06309 | KV205_HUMAN | 6.33E+06 | 3.19E+06 | 5.93E+06 | 7.74E+06 | 2.42 | 7.59E-08 | Ig kappa chain V-II region GM607 |
| P02748 | CO9_HUMAN | 1.19E+06 | 1.73E+06 | 2.96E+06 | 2.67E+06 | 2.48 | 6.00E-07 | Complement component C9a |
| P19652 | A1AG2_HUMAN | 3.82E+06 | 9.53E+06 | 9.21E+06 | 8.34E+06 | 2.49 | 6.60E-09 | Alpha-1-acid glycoprotein 2 |
| P07225 | PROS_HUMAN | 2.42E+05 | 5.61E+05 | 6.05E+05 | 5.72E+05 | 2.50 | 3.85E-14 | Vitamin K-dependent protein S |
| P01023 | A2MG_HUMAN | 3.89E+08 | 4.58E+08 | 4.29E+08 | 1.76E+08 | 2.59 | 1.53E-14 | Alpha-2-macroglobulin |
| P01860 | IGHG3_HUMAN | 5.40E+07 | 2.08E+07 | 4.30E+07 | 4.30E+07 | 2.59 | 3.33E-10 | Ig gamma-3 chain C region |
| Q96IY4 | CBPB2_HUMAN | 9.14E+04 | 1.77E+05 | 2.37E+05 | 1.72E+05 | 2.59 | 3.98E-10 | Carboxypeptidase B2 |
| P07357 | CO8A_HUMAN | 3.90E+04 | 1.02E+05 | 9.24E+04 | 8.66E+04 | 2.62 | 4.38E-05 | Complement component C8 alpha chain |
| P05546 | HEP2_HUMAN | 1.95E+06 | 4.89E+06 | 5.14E+06 | 3.92E+06 | 2.63 | 3.45E-12 | Heparin cofactor 2 |
| P18428 | LBP_HUMAN | 8.15E+05 | 3.08E+05 | 4.34E+05 | 4.81E+05 | 2.64 | 1.25E-05 | Lipopolysaccharide-binding protein |
| Q06033 | ITIH3_HUMAN | 1.41E+06 | 8.36E+05 | 1.06E+06 | 5.33E+05 | 2.64 | 1.79E-06 | Inter-alpha-trypsin inhibitor heavy chain H3 |
| P02649 | APOE_HUMAN | 1.43E+07 | 6.68E+06 | 8.18E+06 | 5.32E+06 | 2.69 | 1.24E-08 | Apolipoprotein E |
| P27169 | PON1_HUMAN | 1.91E+06 | 4.67E+06 | 5.15E+06 | 4.73E+06 | 2.69 | 4.77E-10 | Serum paraoxonase/arylesterase 1 |
| P07360 | CO8G_HUMAN | 2.91E+05 | 5.49E+05 | 7.87E+05 | 6.43E+05 | 2.70 | 2.87E-09 | Complement component C8 gamma chain |
| P35858 | ALS_HUMAN | 3.42E+05 | 6.82E+05 | 7.48E+05 | 9.25E+05 | 2.70 | 2.45E-07 | Insulin-like growth factor-binding protein complex acid labile subunit |
| P02743 | SAMP_HUMAN | 4.59E+05 | 3.68E+05 | 6.63E+05 | 1.03E+06 | 2.81 | 1.73E-06 | Serum amyloid P-component |
| P02790 | HEMO_HUMAN | 2.99E+07 | 7.40E+07 | 8.43E+07 | 6.99E+07 | 2.82 | 1.19E-15 | Hemopexin |
| P02775 | CXCL7_HUMAN | 5.48E+05 | 1.88E+05 | 2.10E+05 | 4.15E+05 | 2.91 | 2.85E-05 | Platelet basic protein |
| P00739 | HPTR_HUMAN | 1.64E+05 | 3.87E+05 | 4.34E+05 | 4.95E+05 | 3.01 | 5.68E-05 | Haptoglobin-related protein |
| P07996 | TSP1_HUMAN | 3.69E+04 | 1.21E+04 | 1.58E+04 | 1.86E+04 | 3.03 | 3.02E-03 | Thrombospondin-1 |
| P0CG06 | LAC3_HUMAN | 8.26E+04 | 2.68E+04 | 6.46E+04 | 7.97E+04 | 3.08 | 1.88E-10 | Ig lambda-3 chain C regions |
| B9A064 | IGLL5_HUMAN | 9.10E+06 | 3.56E+06 | 9.82E+06 | 1.11E+07 | 3.12 | 4.57E-10 | Immunoglobulin lambda-like polypeptide 5 |
| P01834 | IGKC_HUMAN | 2.04E+08 | 6.88E+07 | 1.43E+08 | 2.41E+08 | 3.49 | 3.13E-13 | Ig kappa chain C region |
| P0DJI8 | SAA1_HUMAN | 3.50E+05 | 9.70E+04 | 1.36E+05 | 2.87E+05 | 3.60 | 1.90E-02 | Serum amyloid protein A(2–104) |
| P80748 | LV302_HUMAN | 3.91E+06 | 1.04E+06 | 2.22E+06 | 4.05E+06 | 3.90 | 7.50E-12 | Ig lambda chain V-III region LOI |
| P60709 | ACTB_HUMAN | 7.96E+05 | 5.29E+05 | 1.37E+06 | 2.29E+06 | 4.34 | 1.19E-07 | Actin, cytoplasmic 1, N-terminally processed |
| P06889 | LV405_HUMAN | 1.63E+05 | 5.00E+05 | 3.63E+05 | 1.15E+05 | 4.34 | 7.32E-08 | Ig lambda chain V-IV region MOL |
| P04430 | KV122_HUMAN | 3.85E+05 | 8.68E+04 | 1.99E+05 | 3.58E+05 | 4.44 | 6.73E-04 | Ig kappa chain V-I region BAN |
| P04003 | C4BPA_HUMAN | 2.60E+06 | 8.24E+06 | 1.17E+07 | 8.11E+06 | 4.48 | 1.32E-11 | C4b-binding protein alpha chain |
| P04278 | SHBG_HUMAN | 5.77E+04 | 2.62E+05 | 2.39E+05 | 8.93E+04 | 4.55 | 7.64E-07 | Sex hormone-binding globulin |
| Q13790 | APOF_HUMAN | 6.85E+04 | 2.39E+05 | 2.73E+05 | 3.20E+05 | 4.67 | 3.98E-10 | Apolipoprotein F |
| P68871 | HBB_HUMAN | 4.94E+06 | 3.57E+06 | 4.91E+06 | 1.02E+06 | 4.83 | 5.25E-04 | Hemoglobin subunit beta |
| P01859 | IGHG2_HUMAN | 1.01E+07 | 3.12E+06 | 4.78E+06 | 1.52E+07 | 4.86 | 5.25E-15 | Ig gamma-2 chain C region |
| P04196 | HRG_HUMAN | 1.93E+06 | 6.49E+06 | 9.87E+06 | 6.88E+06 | 5.09 | 2.89E-11 | Histidine-rich glycoprotein |
| Q9Y490 | TLN1_HUMAN | 2.81E+03 | 2.46E+03 | 5.14E+03 | 1.31E+04 | 5.34 | 5.50E-07 | Talin-1 |
| P06311 | KV311_HUMAN | 3.83E+04 | 1.12E+04 | 2.80E+04 | 7.75E+04 | 6.90 | 4.60E-05 | Ig kappa chain V-III region IARC/BL41 |
| P01772 | HV311_HUMAN | 3.50E+05 | 5.31E+04 | 2.05E+05 | 3.74E+05 | 7.04 | 4.57E-10 | Ig heavy chain V-III region KOL |
| P01861 | IGHG4_HUMAN | 4.65E+06 | 8.49E+05 | 2.26E+06 | 6.18E+06 | 7.27 | 1.91E-06 | Ig gamma-4 chain C region |
| P02771 | FETA_HUMAN | 3.35E+06 | 4.84E+05 | 4.28E+05 | 4.90E+05 | 7.83 | 1.44E-10 | Alpha-fetoprotein |
| O43866 | CD5L_HUMAN | 1.60E+05 | 1.04E+06 | 1.23E+06 | 1.30E+06 | 8.10 | 3.51E-11 | CD5 antigen-like |
| P02452 | CO1A1_HUMAN | 1.40E+05 | 1.06E+05 | 6.81E+04 | 1.66E+04 | 8.42 | 5.90E-16 | Collagen alpha-1(I) chain |
| P01780 | HV319_HUMAN | 1.17E+05 | 1.72E+04 | 1.07E+05 | 1.49E+05 | 8.68 | 7.83E-09 | Ig heavy chain V-III region JON |
| P01871 | IGHM_HUMAN | 1.49E+06 | 9.65E+06 | 1.19E+07 | 1.36E+07 | 9.14 | 2.09E-12 | Ig mu chain C region |
| P04438 | HV208_HUMAN | 7.01E+05 | 7.02E+04 | 3.18E+05 | 2.36E+05 | 9.98 | 2.14E-02 | Ig heavy chain V-II region SESS |
| P00738 | HPT_HUMAN | 1.00E+07 | 6.96E+07 | 1.09E+08 | 5.99E+07 | 10.83 | 4.72E-06 | Haptoglobin alpha chain |
| P04220 | MUCB_HUMAN | 1.55E+05 | 1.24E+06 | 1.20E+06 | 1.81E+06 | 11.67 | 3.13E-13 | Ig mu heavy chain disease protein |
| P01591 | IGJ_HUMAN | 1.34E+05 | 7.78E+05 | 1.11E+06 | 1.76E+06 | 13.11 | 5.39E-15 | Immunoglobulin J chain |
| P69905 | HBA_HUMAN | 1.66E+07 | 2.67E+06 | 3.33E+06 | 8.51E+05 | 19.55 | 3.99E-08 | Hemoglobin subunit alpha |
| P07951 | TPM2_HUMAN | 3.35E+04 | 1.47E+04 | 1.08E+05 | 3.22E+05 | 21.83 | 1.20E-07 | Tropomyosin beta chain |
| P01877 | IGHA2_HUMAN | 7.27E+04 | 1.64E+05 | 4.55E+05 | 2.03E+06 | 27.98 | 2.69E-15 | Ig alpha-2 chain C region |
| A0M8Q6 | LAC7_HUMAN | 5.29E+04 | 9.09E+04 | 2.07E+06 | 2.93E+06 | 55.33 | 8.49E-06 | Ig lambda-7 chain C region |
| P01876 | IGHA1_HUMAN | 6.11E+05 | 5.10E+06 | 1.24E+07 | 3.58E+07 | 58.59 | 1.39E-20 | Ig alpha-1 chain C region |
| P69891 | HBG1_HUMAN | 1.90E+06 | 1.88E+04 | 2.85E+04 | 2.03E+04 | 101.25 | 1.96E-13 | Hemoglobin subunit gamma-1 |
| P69892 | HBG2_HUMAN | 4.87E+06 | 2.49E+04 | 1.87E+04 | 1.48E+04 | 328.58 | 9.58E-22 | Hemoglobin subunit gamma-2 |
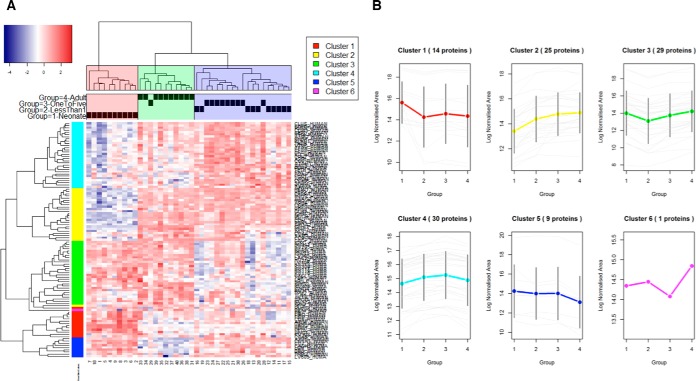
The resulting hierarchical clustering of these 107 proteins confirmed a similar pattern to that observed for the unsupervised clustering of the overall data (Fig. 2A). These proteins could be grouped into 6 specific clusters of expression (Fig. 2B). The list of proteins included in each cluster is presented in supplemental Table S2. The most interesting of these were clusters 2 and 4, with 25 and 30 proteins, respectively, showing increased expression with increasing age; and clusters 1 and 5, with 14 and 9 proteins, respectively, showing decreased expression with increasing age. Proteins included in these clusters represent the KEGG pathways of complement and coagulation cascade, with the most significant gene ontology functions being: acute inflammatory response, protein activation cascade, complement activation and regulation of humoral immune response.
Fig. 2.
A, Heatmap of the 107 differentially expressed proteins from the local assay library identified using ANOVA. The clustering patterns were obtained using a correlation-based distance and complete linkage. Blue - proteins with decreased expression; Red - proteins with increased expression. B, Clusters and protein abundance trends obtained by hierarchical clustering of the differentially expressed proteins identified using an ANOVA. x-axis represents the four age groups (1, Neonates; 2, <1 year olds; 3, 1–5 year olds; 4, Adults). y-axis represents log Normalized Area of the expression of each relevant protein (gray), with the overall mean presented as a colored line incorporating standard deviation (2SD) bars. The separation in three clusters was based on cutting the dendrogram at a distance of 0.1 and coloring the resulting sample clustering.
Ingenuity Pathway Analysis
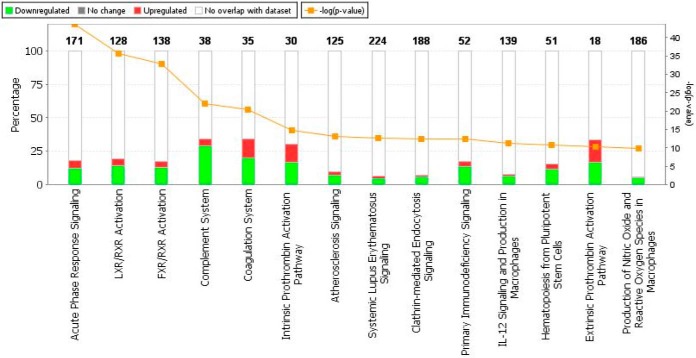
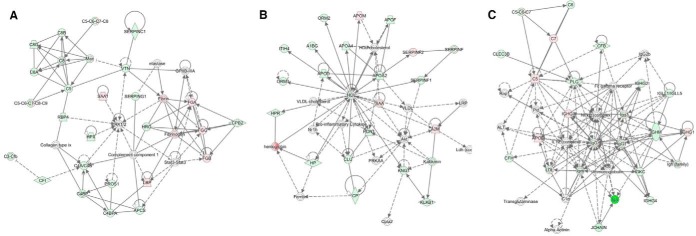
The top enriched pathways for the 107 differentially expressed proteins (based on the ANOVA) are shown in Fig. 3. Although 14 pathways were shown to be represented by the differentially expressed proteins, the top 5 pathways in order of significance were: Acute phase response signaling (18.1% overlap); LXR/RXR Activation (19.5% overlap); FXR/RXR Activation (17.4% overlap); Complement system (34.2% overlap) and Coagulation system (34.3% overlap). Fig. 4 shows the top 3 identified networks as Hematological system development and function, Organismal functions, Developmental disorder (Fig. 4A); Neurological disease, Lipid metabolism, Molecular transport (Fig. 4B) and Humoral Immune response, Inflammatory disease, Immunological disease (Fig. 4C). Diseases and disorders, as well as physiological system development and functions identified to be of significance are listed in Table II.
Fig. 3.
Top enriched pathways for the 107 differentially expressed proteins based on the ANOVA analysis, ranked in increasing order of their Fisher exact test p value (Ingenuity pathway analysis, default background comparison). Green - proteins with decreased expression in neonates; Red - proteins with increased expression in neonates; White - no overlap with the data set; Orange - log (p value), Fisher exact test, right tailed.
Fig. 4.
Most significant networks identified using Ingenuity Pathway Analysis. A, Hematological System Development and Function, Organismal Functions, Developmental Disorder; B, Neurological Disease, Lipid Metabolism, Molecular Transport; C, Humoral Immune Response, Inflammatory Response, Immunological Disease. Red denotes up-regulation in neonates compared with adults, green denotes down-regulation in neonates. White denotes a molecule not in the data set. The legend for the shapes is as follows: Double circle, Complex/Group; Trapezium, transporter; Diamond, enzyme; Horizontal diamond, Peptidase; Circle, other.
Table II. Ingenuity Pathway Analysis of diseases and disorders and development and function based on the comparison of the 108 differentially expressed proteins to the default background.
| p value | # Molecules | |
|---|---|---|
| Diseases and Disorders | ||
| Inflammatory response | 1.01E-04–5.02E-23 | 58 |
| Metabolic disease | 1.20E-04–4.73E-22 | 46 |
| Immunological disease | 9.85E-05–6.15E-21 | 39 |
| Neurological disease | 1.23E-04–3.86E-19 | 46 |
| Psychological disorders | 2.73E-08–3.86E-19 | 3 |
| Cell-to-cell signaling and interaction | 1.49E-04–1.38E-20 | 52 |
| Cellular movement | 1.39E-04–3.29E-18 | 41 |
| Cell death and survival | 1.15E-04–5.86E-12 | 49 |
| Cellular development | 6.94E-05–2.94E-11 | 27 |
| Lipid metabolism | 1.49E-04–1.55E-10 | 36 |
| Physiological system development and function | ||
| Humoral immune response | 8.95E-05–5.02E-23 | 20 |
| Hematological system development and function | 1.04E-04–8.00E-20 | 51 |
| Immune cell trafficking | 1.04E-04–3.28E-19 | 43 |
| Organismal functions | 7.21E-05–4.35E-17 | 22 |
| Tissue morphology | 9.53E-05–6.84E-13 | 4 |
Extending the PediOme With a Large Repository Based Spectral Library
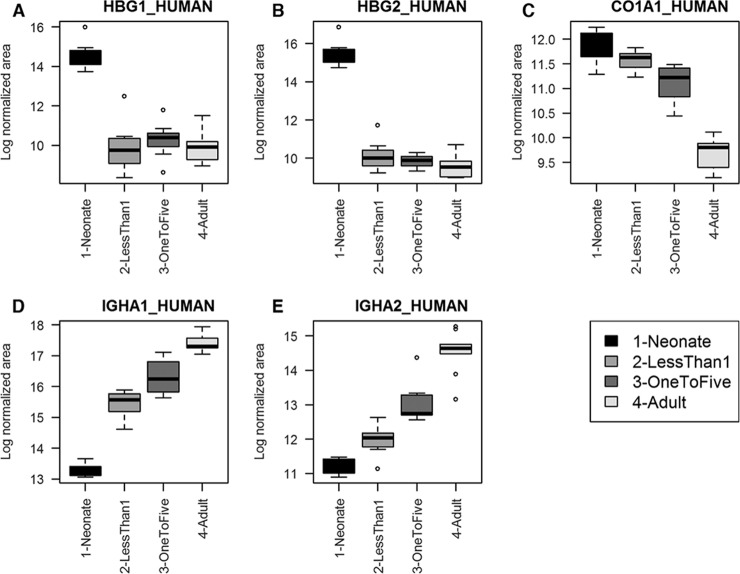
As it is notoriously difficult to generate an extensive protein profile of nondepleted plasma by MS because of the presence of many highly abundant proteins we evaluated whether more proteins could be detected by searching the SWATH data against a large human repository based spectral library containing spectra for more than 10,000 human proteins (17). We used SWATHXtend software to merge this library with our local library, ensuring reliable retention time alignments for extraction of SWATH acquired data. With this approach using nondepleted, nonfractionated plasma we extracted quantitative information from 940 proteins (extraction FDR 1%, two or more peptides required) (supplemental Table S3, Fig. 1C), a greater than 6-fold increase compared with the locally generated IDA based spectral library. We selected two proteins periostin (POSTN) and GPI-specific phospholipase 1 (GPLD1) which had high quality peptide spectral matches across multiple peptides in the extended library. These proteins are reported to be secreted but are commonly under-reported in proteomic analyses of plasma, so we used ELISA to confirm their presence and concentrations. The expression patterns for five proteins with the largest age-specific changes in expression are shown in Fig. 5. These are: Hemoglobin subunit gamma-1 (HBG1); Hemoglobin subunit gamma-2 (HBG2); collagen alpha-1I chain (CO1A1); Ig alpha-1 chain C region (IGHA1), and Ig alpha-2 chain C region (IGHA2).
Fig. 5.
Age-specific expression patterns of five proteins of interest selected for discussion. Expression patterns for five proteins with the largest age-specific changes in expression. A, Hemoglobin subunit gamma-1 (HBG1); B, Hemoglobin subunit gamma-2 (HBG2); C, collagen alpha-1I chain (CO1A1); D, Ig alpha-1 chain C region (IGHA1) and E, Ig alpha-2 chain C region (IGHA2).
ELISA Validation
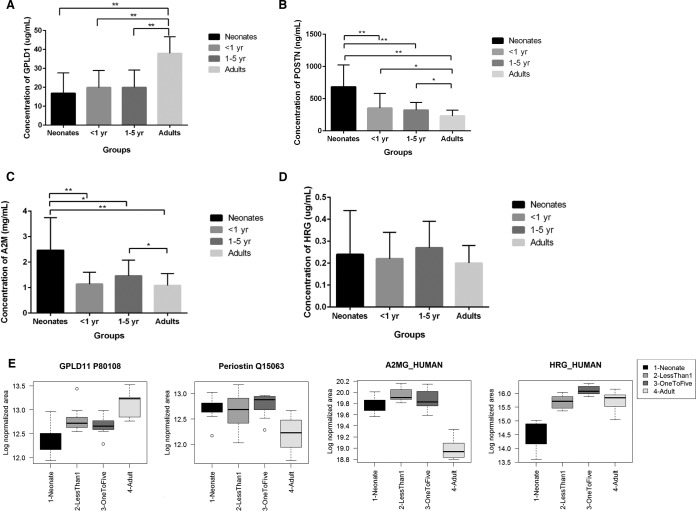
ELISA confirmed the expression of periostin and GPLD1 in plasma samples from each age group. GPLD1 expression (Fig. 6A) was greatest in adults with declining abundance with age (40 μg/ml in adults, 18 μg/ml neonates). Periostin showed the inverse trend, but with much lower expression (0.6 μg/ml in neonates, 0.01 μg/ml in adults). It was significantly downregulated (Fig. 6B) in all age groups in comparison to neonates, and significantly downregulated in Adults in comparison to <1 and 1–5-year age groups. This agrees with the SWATH data and demonstrates that an extended human library is a useful strategy for deeper plasma profiling.
Fig. 6.
ELISA validation of differentially expressed proteins. A, GPI-specific phospholipase 1 (GPLD1), B, Periostin (POSTN), C, Alpha-2-macroglobulin (A2M), D, Histidine rich glycoprotein (HRG). E, For comparison, boxplots showing the respective abundance patterns detected from the SWATH data using the extended library (GPLD1 and POSTN) and the local library (A2M and HRG). All error bars are standard deviation (SD). * Denotes significance ≤ 0.05; ** denotes significance ≤ 0.01.
We also validated expression of two other proteins, with our findings in agreement with the SWATH data Alpha-2-macroglobulin (A2M) was found to be significantly downregulated (Fig. 6C) in all age groups (<1 year, 1–5 years and Adult) in comparison to neonates. The 1–5-year group was found to be significantly up-regulated in comparison to adults. Histidine rich glycoprotein (HRG) (Fig. 6D) was shown to be up-regulated in the 1–5-year group when compared with neonates, but downregulated in <1 year and Adults.
Pairwise Comparisons of Protein Expression Using an Extended Plasma Library
Pairwise comparisons between specific age groups were also performed. Proteins that were differentially expressed in the local as well as the extended library are listed as: neonates compared with < 1 year olds (supplemental Table S4), with 26 and 25 proteins downregulated and 17 and 23 proteins up-regulated in local and extended libraries; neonates compared with 1–5 year olds (supplemental Table S5), with 35 and 38 proteins downregulated and 8 and 11 proteins up-regulated in local and extended libraries; neonates compared with adults (supplemental Table S6), with 25 and 24 proteins downregulated and 9 and 18 proteins up-regulated in local and extended libraries; < 1 year olds compared with 1 - 5 year olds (supplemental Table S7), with 15 and 16 proteins downregulated and 0 and 2 proteins up-regulated in local and extended libraries; < 1 year olds compared with adults (supplemental Table S8), with 15 and 20 proteins downregulated and 9 and 12 proteins up-regulated in local and extended libraries; 1 - 5 year olds compared with adults (supplemental Table S9), with 8 and 7 proteins downregulated and 11 and 15 proteins up-regulated in local and extended libraries.
DISCUSSION
Although previous studies have assessed the aging of the plasma proteome in pediatric and adult populations (6), this is the first study to have utilized SWATH-MS technology to quantify and analyze variability in differential protein expression in both adult and pediatric age-specific contexts. An added advantage of this methodology was the ability to utilize ingenuity pathway analysis to elucidate enriched biochemical pathways and pathways involved in disease processes. Indeed, ANOVA analysis of the variation in protein expression across multiple age groups shows that proteins involved in hematopoietic development and function, the immune system, and physical growth undergo significant change as the plasma proteome ages.
We identified 107 proteins that varied in abundance over the four age groups examined. The most significant as identified by ANOVA analysis play a role in hematopoietic development, and immune function. Hemoglobin subunit gamma-2 (HBG2) is a protein encoded by the HBG2 gene, and has been shown to play an important role in the development of the hematopoietic system (18). In the developing fetus, HBG1 and HBG2 code for two gamma chain proteins, which along with corresponding alpha chains, make up fetal hemoglobin (HbF). At birth, HbF undergoes replacement by mature adult hemoglobin (HbA), in a process that continues over the first 6 months of age (19). As shown in Table I, the mean expression levels of HBG2 decreased significantly with age as HbA replaced HbF. In a similar manner, collagen alpha-1I chain (CO1A1), a protein that forms part of the alpha-1I chain in Type I collagen also undergoes decreased expression from neonates to adults. As Type I collagen is found in abundance in bone, tendons and the dermis (20), a likely explanation for this is that CO1A1 expression spikes early in life as a result of growth, and is then downregulated in adulthood to a maintenance function. In contrast, Ig alpha-1 chain C region (IGHA1), a protein that plays a role in the formation of IgA antibodies (21), was shown in our analysis to exhibit higher expression as the proteome aged, with largest expressional changes observed in the neonatal to 1–5 age groups. This suggests that rapid development of the immune system occurs in the neonatal to early childhood period, and continues to mature in adolescent years and adulthood.
An advantage to utilizing SWATH-MS was the ability to re-mine the data by using a significantly larger assay library for matching. Here we took advantage of an extensive human assay library containing spectra from over 10,000 proteins and re-extracted quality matching peptides from SWATH data of undepleted and nonfractionated plasma. This increased the detected peptides by greater than 6-fold, enabling more thorough peptidome quantification, representing the largest set to date of SWATH quantified proteins in nondepleted human plasma. ELISA validation of two putatively secreted proteins, periostin and GPLD-1 confirmed their SWATH measured levels in plasma. Liu et al. (10) used SWATH analysis of human plasma and reported the quantitation of 342 proteins in 232 plasma samples collected longitudinally from pairs of twins. They detected GPLD-1, but not periostin and the annotated subcellular localization of these 342 protein was varied. This included nuclear proteins (e.g. Transcription elongation regulator 1(TCERG1), Heterogeneous nuclear ribonucleoproteins), mitochondrial proteins (e.g. GRP75, Trifunctional enzyme subunit alpha (ECHA), Adenylate kinase 2) and Golgi proteins (e.g. Golgi membrane protein GP73), among others with nonobvious functional roles in circulation. With these observations there is good reason to utilize the large-scale, proteome wide library approach as we have shown here as this readily increases the depth of plasma proteome coverage using LC-MS without sample immunodepletion. Interestingly, a related approach enabled quantitation of over 1500 proteins in mouse plasma, many of which were of cellular origin (22). Of course, secondary approaches should attempt to validate the detection of cellular proteins observed in plasma. Type 1 error cannot be excluded when utilizing large-scale libraries of this heterogeneous nature, and one strategy is to add stringency by requiring evidence for multiple peptides, in this case at least 2 per protein.
We used pathway knowledge to identify the most enriched pathways, as well as, functions and diseases. Acute phase response signaling, involved in the acute nonspecific immune response, constituted the most enriched canonical pathway. Given the previous evidence that IGHA1 expression increased as the subjects aged, this is further indication that immune development is a dynamic and continuously evolving system. Moreover, identification of IL-6 as the top upstream regulator (a cytokine involved in the inflammatory response, and a mediator of the acute phase response (23)), the inflammatory response and the humoral immune response as the most enriched disease context and physiological function respectively, further supports this contention.
Indeed, quantitation of age-specific variability of plasma proteins has clear and relevant application to the clinic. Although changes in proteomic expression in disease-specific contexts have been well characterized in the literature and mentioned previously in this paper, identification of expression changes in age-specific contexts is a potentially powerful tool for clinical use. For example, blood borne pathologies such as beta-thalassemias and other related conditions often feature prolonged gamma chain HBG2 production, long into early childhood (24). Thus, proteomic analysis of patient plasma, in reference to age-specific protein reference ranges, may assist clinicians in the diagnostic phase, or help to elucidate the severity of the relevant pathology.
In this study we have shown that the plasma proteome undergoes significant change as the system ages. Apart from identifying proteins with various functional roles in systems such as hematopoietic development, the immune response and growth, we also identified the most highly enriched pathways, upstream regulators, disease contexts and functions in comparison to the default background. Future studies should be directed at further elucidating age-specific variability in protein expression and, importantly, its links to disease processes.
DATA AVAILABLILITY
The mass spectrometry proteomics data have been deposited to the ProteomeXchange Consortium via the PRIDE [1] partner repository with the data set identifier PXD005661.
Reviewer account details: Username: reviewer18033@ebi.ac.uk, Password: auk6Amra.
Supplementary Material
Acknowledgments
We thank members of the anesthetic and surgical departments of the Royal Children's Hospital for facilitation of sample collection.
Footnotes
Author contributions: M.P.M., P.M., and V.I. designed research; S.B., D.P., H.P., V.K., T.Z., and X.S. performed research; S.B., D.P., V.K., X.S., M.P.M., P.M., and V.I. analyzed data; S.B., D.P., M.P.M., P.M., and V.I. wrote the paper.
* This study was supported by the Victorian Government's Operational Infrastructure Support Program. This research has been supported in part by Bioplatforms Australia Ltd and facilitated by access to Australian Proteome Analysis Facility supported by the Australian Government's National Collaborative Research Infrastructure Scheme.
 This article contains supplemental material.
This article contains supplemental material.
1 The abbreviations used are:
- ELISA
- enzyme linked immunosorbent assay
- 2D-DIGE
- two-dimensional difference in-gel electrophoresis
- HUPO
- Human proteome Organization
- PediOme
- Pediatric Proteomics
- DIA
- data independent acquisition
- TOFMS
- time of flight mass spectrometry
- ANOVA
- analysis of variance
- GPLD1
- GPI-specific phospholipase D
- POSTN
- periostin
- A2M
- alpha-2-macroglobulin
- HRG
- histidine rich glycoprotein
- IDA
- information dependent acquisition
- PCA
- principal component analysis
- HBG1
- hemoglobin subunit gamma-1
- HBG2
- hemoglobin subunit gamma-2
- CO1A1
- collagen alpha-1(I) chain
- IGHA1
- Ig alpha-1 chain C region
- IGHA2
- Ig alpha-2 chain C region
- HbF
- fetal hemoglobin
- HgA
- adult hemoglobin
- TCERG1
- transcription elongation regulator 1
- ECHA
- trifunctional enzyme subunit alpha
- LC-MS
- liquid chromatography mass spectrometry.
REFERENCES
- 1. Lull M. E., Freeman W. M., Myers J. L., Midgley F., Kimatian S. J., Undar A., and Vrana K. E. (2006) Plasma proteomics: a noninvasive window on pathology and pediatric cardiac surgery. ASAIO J. 52, 562–566 [DOI] [PubMed] [Google Scholar]
- 2. Michiels J. J., Brenneman Z., Gadisseur A., van der Planken M., Schroyens W., van de Velde A., and van Vliet H. (2006) Classification and characterization of herediary types 2A, 2B, 2C, 2D, 2E, 2M, 2N and 2U (unclassifiable) von Wilebrand disease. Clin. App. Thromb. Hemost. 12, 397–420 [DOI] [PubMed] [Google Scholar]
- 3. Kakhniashvili D. G., Griko N. B., Bulla L. A. Jr., and Goodman S. R. (2005) The proteomics of sickle cell disease: profiling of erythrocyte membrane proteins by 2D-DIGE and tandem mass spectrometry. Exp. Biol. Med. 230, 787–792 [DOI] [PubMed] [Google Scholar]
- 4. Yocum A. K., Busch C. M., and Felix C. A. (2006) Proteomics-based strategy to identify biomarkers and pharmacological targets in leukemias with t(4;11) translocations. J. Proteome Res. 5, 2743–2753 [DOI] [PubMed] [Google Scholar]
- 5. Roxo-Rosa M., da Costa G., and Luider T. M. (2006) Proteomic analysis of nasal cells from cystic fibrosis patients and noncystic fibrosis control individuals: search for novel biomarkers of cystic fibrosis lung disease. Proteomics 6, 2314–2325 [DOI] [PubMed] [Google Scholar]
- 6. Ignjatovic V., Lai C., Summerhayes R., Mathesius U., Tawfilis S., Perugini M. A., and Monagle P. (2011) Age-related differences in plasma proteins: how plasma proteins change from neonates to adults. PLoS ONE 6, e17213. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Omenn G. S., ed HUPO. (2006) Exploring the Human Plasma Proteome, pp. 1–33, Wiley-VCH Verlag GmbH&Co., Weinheim. [Google Scholar]
- 8. Gillet L. C., Navarro P., Tate S., Rost H., Selevsek N., Reiter L., Bonner R., and Aebersold R. (2012) Targeted data extraction of the MS/MS spectra generated by data-independent acquisition: a new concept for consistent and accurate proteome analysis. Mol. Cell. Proteomics 11, 1535–9484 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Liu Y., Huttenhain R., Surinova S., Gillet L. C., Mouritsen J., Brunner R., Navarro P., and Aebersold R. (2013) Quantitative measurements of N-linked glycoproteins in human plasma by SWATH-MS. Proteomicsc 13, 1247–1256 [DOI] [PubMed] [Google Scholar]
- 10. Liu Y., Buil A., Collins B. C., Gillet L. C., Blum L. C., Cheng L. Y., Vitek O., Mouritsen J., Lachance G., Spector T. D., Dermitzakis E. T., and Aebersold R. (2015) Quantitative variability of 342 plasma proteins in a human twin population. Mol. Syst. Biol. 11, 786. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Monagle P., Barnes C., Ignjatovic V., Furmedge J., Newall F., Chan A., De Rosa L., Hamilton S., Ragg P., Robinson S., Auldist A., Crock C., Roy N., and Rowlands S. (2006) Developmental haemostasis: Impact for clinical haemostasis laboratories. Thromb. Haemost. 95, 362–372 [DOI] [PubMed] [Google Scholar]
- 12. Ignjatovic V., Straka E., Summerhayes R., and Monagle P. (2010) Age-specific differences in binding of heparin to plasma proteins. J. Thromb. Haemost. 6, 1290–1294 [DOI] [PubMed] [Google Scholar]
- 13. Ignjatovic V., Summerhayes R., Newall F., and Monagle P. (2009) The in vitro response to low-molecular-weight heparin is not age-dependent in children. Thromb. Haemost. 103, 855–856 [DOI] [PubMed] [Google Scholar]
- 14. Attard C., van der Straaten T., Karlaftis V., Monagle P., and Ignjatovic V. (2013) Developmental haemostasis: age-specific differences in the quantity of haemostatic proteins. J. Thromb. Haemost. 11, 1850–1854 [DOI] [PubMed] [Google Scholar]
- 15. Attard C., Kubitza D., Monagle P., and Ignjatovic V. (2014) The in vitro anticoagulant effect of Rivaroxaban in Neonates. Blood Coagulation Fibrinolysis 25, 237–240 [DOI] [PubMed] [Google Scholar]
- 16. Wu J. X., Song X., Pascovici D., Zaw T., Care N., Krisp C., and Molloy M. P. (2016) SWATH Mass Spectrometry Performance Using Extended Peptide MS/MS Assay Libraries. Mol. Cell. Proteomics 15, 2501–2514 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Rosenberger G., Koh C. C., Guo T., Rost H. L., Kouvonen P., Collins B. C., Heusel M., Liu Y., Caron E., Vichalkovski A., Faini M., Schubert O. T., Faridi P., Ebhardt H. A., Matondo M., Lam H., Bader S. L., Campbell D. S., Deutsch E. W., Moritz R. L., Tate S., and Aebersold R. (2014) A repository of assays to quantify 10,000 human proteins by SWATH-MS. Sci. Data 1, 140031. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Higgs D. R., Vickers M. A., Wilkie A. O., Pretorius I. M., Jarman A. P., and Weatherall D. J. (1989) A review of the molecular genetics of the human alpha-globin gene cluster. Blood 73, 1081–104 [PubMed] [Google Scholar]
- 19. Pereira C., Relvas L., Bento C., Abade A., Ribeiro M. L., and Manco .L. (2015) Polymorphic variations influencing fetal hemoglobin levels: association study in beta-thalassemia carriers and in normal individuals of Portuguese origin. Blood Cells Mol. Dis. 54, 315–320 [DOI] [PubMed] [Google Scholar]
- 20. Mann V., and Ralston S. H. (2003) Meta-analysis of COL1A1 Sp1 polymorphism in relation to bone mineral density and osteoporotic fracture. Bone 32, 711–717 [DOI] [PubMed] [Google Scholar]
- 21. Takahashi K., Raska M., Stuchlova Horynova M., Hall S. D., Poulsen K., Kilian M., Hiki Y., Yuzawa Y., Moldoveanu Z., Julian B. A., Renfrow M. B., and Novak J. (2014) Enzymatic sialylation of IgA1 O-glycans: implications for studies of IgA nephropathy. PloS One 9, e99026. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Malmström E., Kilsgård O., Hauri S., Smeds E., Herwald H., Malmström L., Malmström J. (2016) Large-scale inference of protein tissue origin in gram-positive sepsis plasma using quantitative targeted proteomics. Nat. Commun. 6, 10261. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Heinrich P. C., Castell J. V., and Andus T. (1990) Interleukin-6 and the acute phase response. Biochem. J. 265, 621–636 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Neishabury M, Zamani S, Azarkeivan A, Abedini SS, Darvish H, Zamani F, and Najmabadi H. (2012) The modifying effect of Xmn1-HBG2 on thalassemic phenotype is associated with its linked elements in the beta globin locus control region, including the palindromic site at 5′HS4. Blood cells Mol Dis. 48, 1–5. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.