Abstract
CRISPR-associated protein 9 (Cas9)-mediated genome editing provides a promising cure for HIV-1/AIDS; however, gene delivery efficiency in vivo remains an obstacle to overcome. Here, we demonstrate the feasibility and efficiency of excising the HIV-1 provirus in three different animal models using an all-in-one adeno-associated virus (AAV) vector to deliver multiplex single-guide RNAs (sgRNAs) plus Staphylococcus aureus Cas9 (saCas9). The quadruplex sgRNAs/saCas9 vector outperformed the duplex vector in excising the integrated HIV-1 genome in cultured neural stem/progenitor cells from HIV-1 Tg26 transgenic mice. Intravenously injected quadruplex sgRNAs/saCas9 AAV-DJ/8 excised HIV-1 proviral DNA and significantly reduced viral RNA expression in several organs/tissues of Tg26 mice. In EcoHIV acutely infected mice, intravenously injected quadruplex sgRNAs/saCas9 AAV-DJ/8 reduced systemic EcoHIV infection, as determined by live bioluminescence imaging. Additionally, this quadruplex vector induced efficient proviral excision, as determined by PCR genotyping in the liver, lungs, brain, and spleen. Finally, in humanized bone marrow/liver/thymus (BLT) mice with chronic HIV-1 infection, successful proviral excision was detected by PCR genotyping in the spleen, lungs, heart, colon, and brain after a single intravenous injection of quadruplex sgRNAs/saCas9 AAV-DJ/8. In conclusion, in vivo excision of HIV-1 proviral DNA by sgRNAs/saCas9 in solid tissues/organs can be achieved via AAV delivery, a significant step toward human clinical trials.
Keywords: Cas9, gRNA, HIV, viral latency, gene therapy, AAV, humanized BLT mice, Tg26 mice, genome editing, animal models
Graphical Abstract
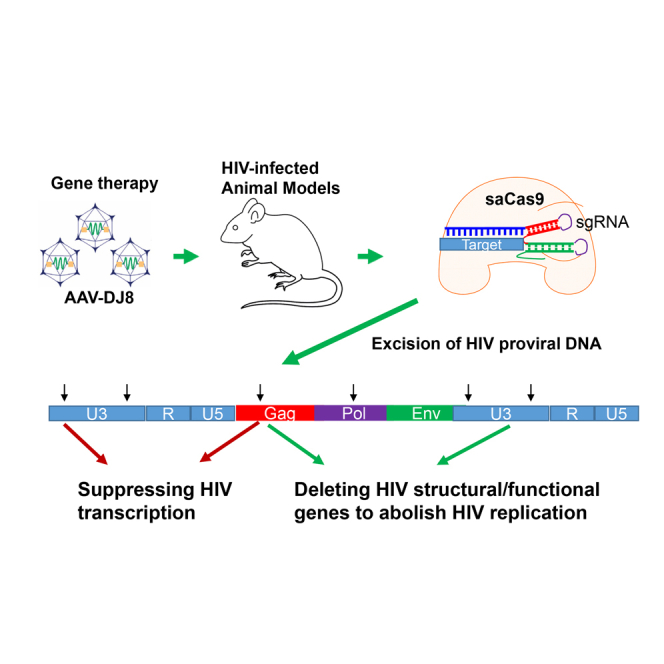
Yin et al. use multiplex CRISPR/Cas9 genome editing technology to excise the HIV-1 provirus in a precise manner in three different HIV-1 animal models via in vivo AAV gene delivery. The feasibility of HIV excision in infected cells in vivo paves the way toward human clinical trials to cure HIV-1 infection.
Introduction
The CRISPR-associated protein 9 (Cas9) is an RNA-guided endonuclease originally discovered as an integral mediator of bacterial adaptive immunity. The Cas9-mediated genome editing system generates site-specific double-strand DNA breaks (DSBs) that remarkably increase homologous recombination efficacy and thus render easy, fast, and economical manipulation of the genome in various cells and organs. In the past 4 years, Cas9 technology has been extensively utilized in basic science and pre-clinical settings for the potential treatment of genetic diseases, cancer, and infectious diseases. The majority of these applications are attributed to the successful development of the Streptococcus pyogenes Cas9 (spCas9), a 1,368-amino acid (aa) protein that recognizes a protospacer adjacent motif (PAM) of NRG nucleotides to the 3′ target site.1 However, several smaller Cas9 proteins from other bacterial species have been garnering more attention for their greater feasibility in viral gene therapy, specifically from Streptococcus thermophiles, Neisseria meningitides, and Staphylococcus aureus (saCas9). The efficiency and specificity of saCas9 (1,053 aa) in mammalian cells was characterized recently.2 The smaller size renders adeno-associated virus (AAV)-mediated gene delivery systems more feasible in clinical trials.3 The AAV-saCas9 system has been successfully tested in gene knockout2, 4, 5 and knock-in6, 7 studies in experimental animals.
Currently, combined antiretroviral therapy (cART) has been widely used to suppress active HIV-1 replication; however, a permanent cure is still lacking due to the existence of latent reservoirs, in which HIV proviral DNA integrates in the host genome. Upon cART cessation, the HIV viral load rebounds to cause a catastrophic resurgence of HIV/AIDS. Thus, several promising and novel strategies are being developed to achieve a permanent or “sterile” cure for HIV/AIDS. The versatile Cas9/single-guide RNA (sgRNA) technology is one strategy employed to eradicate the integrated proviral DNA in HIV-1 latent reservoirs and prevents HIV-1 (re)infection. For example, the spCas9 system has been successfully used to disrupt HIV-1 entry co-receptors CCR5 and CXCR4, along with several proviral structural proteins.8, 9, 10 We reported that stable transfection of cultured human cells with plasmids encoding spCas9 and HIV-1 sgRNAs targeting four different sites of the HIV-1 long terminal repeat (LTR) could successfully excise HIV-1 proviral DNA.11 Pre-treatment of cells with spCas9/LTR sgRNAs renders them resistant to new HIV-1 infection in vitro,11 which was also observed by others.12, 13, 14 We further demonstrated the feasibility and efficiency of HIV-1 excision by applying the spCas9/sgRNA system via lentivirus-mediated delivery to HIV-1-infected primary CD4+ T cells from human healthy subjects and patients with HIV-1.15 Finally, we demonstrated that a combination of sgRNAs targeting LTRs and the viral structural genes in the spCas9 system provides a more efficient means of HIV-1 excision in cultured cells.16 Using high-coverage whole-genome sequencing and cell-based functional assays, we demonstrated the absence of off-target effects when the multiplex sgRNAs/spCas9 system was used.11, 15, 16 Due to the limitations of spCas9 for AAV gene delivery in the clinical setting, we adapted the saCas9/sgRNA system in an all-in-one AAV vector for HIV excision in a recent report. We demonstrated the proof of concept that AAV9-mediated saCas9/duplex sgRNAs can be used to excise the integrated HIV-1 genome in HIV-1 transgenic mice and rats.17 In this study, we further tested the feasibility and efficiency of HIV-1 excision by saCas9/sgRNA-mediated genome editing in humanized bone marrow/liver/thymus (BLT) mice inoculated with HIV-1 via the vaginal mucosa or intraperitoneal (i.p.) transmission. The same genome editing system was also introduced in conventional mice during an acute infection of EcoHIV18, 19 to demonstrate its ability to block active HIV-1 replication.
Results
The saCas9/sgRNA System Efficiently Excises HIV-1 Proviral DNA
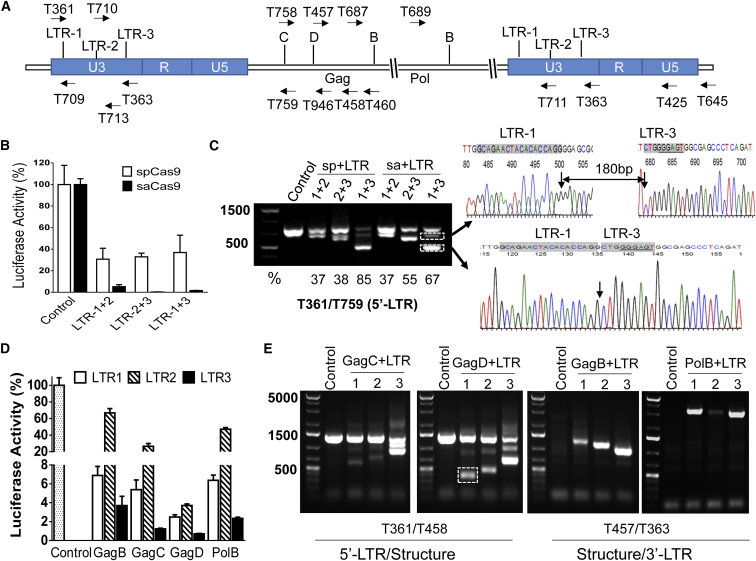
To determine the excision efficiency of the saCas9 system, we selected three sgRNAs targeting the HIV LTR (Figure 1A) using the optimal PAM NNGRRN2 and we performed EcoHIV-enhanced firefly luciferase (eLuc) reporter assays and direct PCR genotyping as we described previously.16 We employed an sgRNA pairing approach because it is more reliable for identifying successful excision of the HIV-1 proviral genome by functional reporter assays and PCR genotyping.11, 15, 16, 20 As shown in Figure 1B, all combinations between the selected three LTR sgRNAs in the presence of saCas9 almost completely eliminated EcoHIV-eLuc activity (by 95%–99%). Since these identified seed (target) sequences were designed as NGGRRN that exactly match the spCas9 PAM (NGG), we cloned them into a lentiviral WG vector,16 in which spCas9-specific sgRNA sequence differs from the sgRNA sequence for saCas9 (except the seed sequence),2, 21 and compared the HIV-1 excision efficiency between saCas9/sgRNA and spCas9/sgRNA. Side-by-side transfection and EcoHIV-eLuc reporter studies showed that the spCas9/sgRNA with these three pairing methods was less effective at reducing luciferase activity (Figure 1B) and excising EcoHIV DNA (Figure 1C). The higher excision efficiency of the saCas9/sgRNA system may be attributed to the higher co-expression level of sgRNA and saCas9 in the same cells because they are in a single vector. Additionally, the smaller size of saCas9 allows higher gene delivery efficiency, and the intrinsic nuclease activity of saCas9 may be more efficient at making DSBs than spCas9.2, 21 The different sgRNA sequences used between the two different bacterial Cas9s may also contribute to saCas9’s better cleaving efficiency.2, 21
Figure 1.
saCas9/sgRNA Efficiently Excises the HIV-1 Genome and Suppresses HIV-1 Luciferase Reporter Expression
(A) Location of sgRNA target sites and PCR primers (arrows). (B and C) Efficiency comparison between saCas9/sgRNA and the spCas9/sgRNA system using the EcoHIV-eLuc reporter assay (B) and direct PCR genotyping (C). The excision efficiency (%) indicates the density of the fragmental deletion band(s) over the total bands. Arrows point to the representative fragmental 296-bp deletion between LTR-1 and LTR-3 target sites at the third nucleotide from PAM and to an additional 180-bp insertion, which were validated by TA cloning and Sanger sequencing (C). (D and E) Three LTR sgRNAs paired with viral structural sgRNAs induced robust inhibition of EcoHIV-eLuc activity by the ONE-Glo Luciferase Assay (D) and fragmental deletion/insertion by direct PCR genotyping with the indicated primers (E). The representative fragmental deletion in the dotted box in (E) was corroborated with Sanger sequencing (see Figure S1). HEK293T cells in six wells of the 96-well plate were co-transfected with the pNL4-3-EcoHIV-eLuc reporter (10 ng/well), the pCMV-renilla-luciferase reporter (2 ng/well), and the indicated Cas9/sgRNA-expressing vectors as follows: for the spCas9 system (B), pLV-EF1a-spCas9-T2A-RFP (80 ng/well) and the indicated sgRNA expression vectors (60 ng/well each for paired gRNAs); for the saCas9 system (B and D), pX601-saCas9/LTR sgRNA expression vectors (100 ng/well each for indicted pairs). After 48 hr, eLuc activity in the cell lysates was measured with the ONE-Glo luciferase assay system and renilla-luciferase activity was measured with the Renilla-Luciferase Assay System. Data represent the mean ± SD of four independent transfections with percentage changes in eLuc after renilla-luciferase normalization as compared with the corresponding empty pX601 vector. Two additional wells of the transfected cells were used for direct PCR genotyping (C and E). Similar experiments were repeated two to three times.
Combination of LTR sgRNAs with Gag or Pol sgRNA Induces Potent Excision
Using the spCas9/sgRNA system, we demonstrated easier PCR genotyping and more efficient HIV-1 excision through the combination of sgRNAs targeting the LTRs and the viral structural genes (e.g., Gag or Pol).16 To test whether this is also applicable to the saCas9/sgRNA system, we paired sgRNAs targeting Gag or Pol with one of the three LTR sgRNAs. All combinations of LTR-1 or LTR-3 with Gag or Pol sgRNAs induced a robust reduction of EcoHIV-eLuc activity in HEK293T cells (Figure 1D). The pairing of LTR-2 and GagD also induced a robust reduction, but pairing with GagB, GagC, and PolB exhibited lower excision efficiency in reducing the reporter activity (Figure 1D). Direct PCR genotyping with 5′-LTR or 3′-LTR primers paired with the structural primers validated the cleaving efficiency of all of the sgRNA combinations (Figure 1E). To further validate the fragmental deletion and provide a rationale for quadruplex sgRNAs (see below), we selected a combination of sgRNAs of LTR-1 and GagD for TA cloning and Sanger sequencing. The results showed the expected sequence of the fragment after deletion between the two target sites (Figure S1 for LTR-1 plus GagD). These data suggest that all of the sgRNAs selected for the saCas9 system are highly efficient in making DSBs at their predicted target sites and that pairing the LTR sgRNAs with sgRNAs targeting viral structural regions induced varying degrees of functional excision.
Multiplex sgRNAs in an All-in-One AAV Vector Result in More Potent Excision
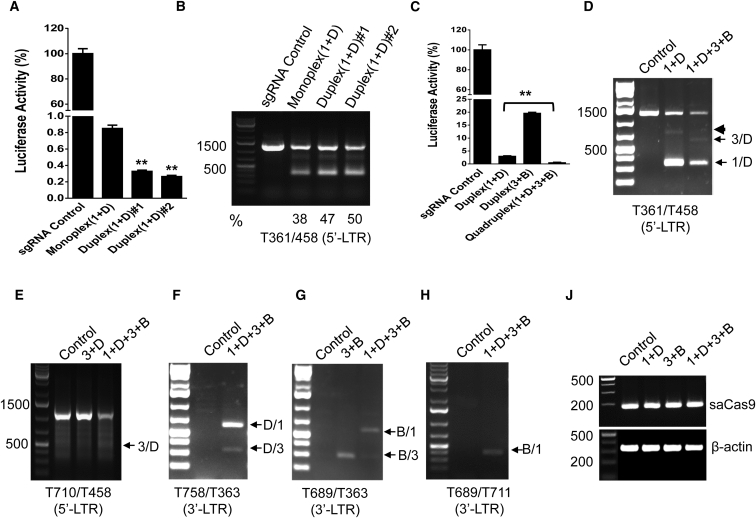
To ensure maximum efficiency of HIV-1 excision and suppression, we selected the pair of LTR-1 and GagD to compare the excision efficiency from co-transfection of two single sgRNA-expressing vectors versus transfection of one duplex sgRNA-expressing vector. The smaller size of saCas9 (3.159 kb) allows a single AAV vector to harbor two sgRNA-expressing cassettes and an saCas9-expressing cassette for efficient AAV packaging and gene delivery.22 Transfection of the duplex sgRNAs/saCas9 vector induced a further reduction of luciferase reporter activity as compared with the co-transfection of two separate single sgRNA/saCas9 vectors (Figure 2A). Cleavage of the reporter gene was confirmed by PCR genotyping, and the ratio of the cleaved fragmental band over the wild-type band exhibited the stronger cleaving capability of the duplex sgRNAs/saCas9 vector than the two separate sgRNA/saCas9 vectors (Figure 2B). The outperforming effect of the duplex sgRNAs/saCas9 may result from the co-expression of these three components in the same cells.
Figure 2.
Efficacy of Multiplex sgRNAs in an All-in-One AAV Vector
(A and B) Duplex sgRNAs/saCas9 exhibited stronger inhibition of EcoHIV-eLuc activity (A) and excision of the HIV-1 genome (B). The excision efficiency (%) indicates the density of the fragmental deletion band over the total bands (B). (C–J) Quadruplex sgRNAs/saCas9 exhibited stronger inhibition of EcoHIV-eLuc activity (C) and higher excision efficiency at both 5′-LTR-1/LTR-3 with GagD (D and E) or GagD/PolB with 3′-LTR (F–J). The primer T361/T458 (D) detected fragmental deletions between 5′-LTR-1/LTR-3 and GagD (arrows) and an additional insertion was observed in both the duplex and quadruplex groups (arrowhead). T710/T458 detected the deletion between 5′-LTR-3 and GagD (E). Deletions between GagD or PolB and 3′-LTR-1/LTR-3 were detected by the primer T758/T363 (F) and T689/T363 (G), respectively. The primer pair T689/T711 detected the deletion between PolB and 3′-LTR-1 (H). The genomic DNA was normalized with saCas9 and β-actin (J). HEK293T cells in a 96-well plate were co-transfected with the EcoHIV-eLuc reporter plus two individual monoplex sgRNA-expressing vectors (100 ng each), one duplex sgRNA-expressing vector (100 ng) with an empty pX601 vector (100 ng) to achieve an equal amount of DNA (200 ng, A and B), or one duplex/quadruplex sgRNA-expressing vector and the sgRNA control (100 ng each, C–J). After 48 hr, the ONE-Glo Luciferase Assay (A and C) was performed as described in Figure 1, and direct PCR genotyping (B and D–J) was performed using the indicated primer pairs.
As demonstrated above and previously,11, 15, 16 two LTR sgRNAs are capable of excising the entire HIV-1 genome in addition to the fragmental deletion of both LTRs, and a combination of sgRNAs targeting to LTRs and structural protein resulted in higher proviral cleavage efficiency and easier PCR genotyping. To combine these two features for optimized efficiency, we selected a cocktail of LTR-1, LTR-3, GagD, and PolB sgRNAs and cloned their expression cassettes into an all-in-one saCas9 AAV vector using a novel interchangeable infusion cloning strategy. By transient transfection with equal amounts of the duplex or quadruplex sgRNAs/saCas9-expressing single plasmid, we found that the quadruplex sgRNAs/saCas9 plasmid was more effective at reducing EcoHIV-eLuc reporter activity (Figure 2C). This was validated by the PCR genotyping, which showed a stronger reduction of the wild-type band generated by PCR primers across the 5′-LTR and Gag (Figures 2D and 2E). The primers T361/T458 (Figure 1A) detected a fragmental deletion between 5′-LTR-1 and GagD and an additional insertion in the duplex group, while another fragmental deletion expected between 5′-LTR-3 and GagD was observed in the quadruplex group (Figure 2D). The primer pair T710/T458 (Figure 1A) detected the predicted fragment after deletion between the 5′-LTR-3 and GagD sites (Figure 2E). Further PCR genotyping analysis of the Gag/Pol and 3′-LTR regions showed that the quadruplex sgRNA system had greater potential for cleaving the entire HIV-1 genome. The primer T758/T363 detected the two deletions between GagD and 3′-LTR-1 or LTR-3 (Figure 2F), while T689/T363 detected the two deletions between PolB and LTR-1 or LTR-3 (Figure 2G). The primer T689/T711 detected one deletion as predicted (Figure 2H). Comparative analysis of the band intensity for the fragment deletions showed that LTR-1 and GagD were most effective, while LTR-3 was less efficient in the quadruplex sgRNAs/saCas9 system (Figures 2D, 2F, and 2G). These cleaving patterns suggest that the quadruplex cocktail strategy is the most effective at excising HIV-1 entire genome due to multiplex fragmental deletions and multiple indel mutations around the target sequence.
Quadruplex sgRNAs/saCas9 Can Be Packaged in a Single AAV-DJ Vector for More Effective Genome Editing
The successful packaging of monoplex and duplex sgRNAs/saCas9 into a single AAV vector was reported recently.22 The duplex sgRNAs/saCas9-expressing cassette (4.969 kb) almost reaches the packaging limit of AAV viruses (∼5.0–5.2 kb).23 However, genome sizes ranging from 5.2 to 8.9 kb have been reported for efficient AAV packaging and gene delivery both in vitro and in vivo.24, 25, 26 Therefore, we tested whether the quadruplex sgRNAs/saCas9 expression cassettes (5.716 kb) can be packaged efficiently into an AAV virus. We selected the AAV-DJ serotype because it combines the features of eight native AAV serotypes (AAV-2, AAV-4, AAV-5, AAV-8, AAV-9, avian AAV, bovine AAV, and caprine AAV) for a higher transduction efficiency in a broader range of cells/tissues,27 escapes from antibody neutralization,28 and provides the highest liver transduction efficiency in vivo. Liver enrichment is integral for demonstrating the proof of concept that the HIV-1 provirus can be excised in vivo. For the duplex sgRNAs/saCas9, a viral titer of ∼4.15–4.3 × 1013 genomic copies (GC)/mL (unpurified crude lysate) was obtained as determined by PCR, with a functional titer of 4.4–9.7 × 108 transduction units (TU)/mL, as determined by immunocytochemistry with an anti-hemagglutinin (HA) antibody that detects the HA-tagged saCas9 protein. For the quadruplex sgRNAs/saCas9, a similar genomic titer of 4.2 × 1013 GC/mL was obtained with a functional titer of 2.5 × 108 TU/mL, which is 1.8- to 3.9-fold less than that from the duplex sgRNAs/saCas9 (Figure S2). These data suggest that the quadruplex sgRNAs/saCas9 AAV can be successfully packaged into an AAV-DJ virus that can efficiently infect mammalian cells with functional expression of the saCas9 protein.
AAV-Mediated Gene Delivery of Multiplex sgRNAs/saCas9 Effectively Excises HIV-1 Proviral DNA in Neural Stem Cells from HIV-1 Tg26 Transgenic Mice
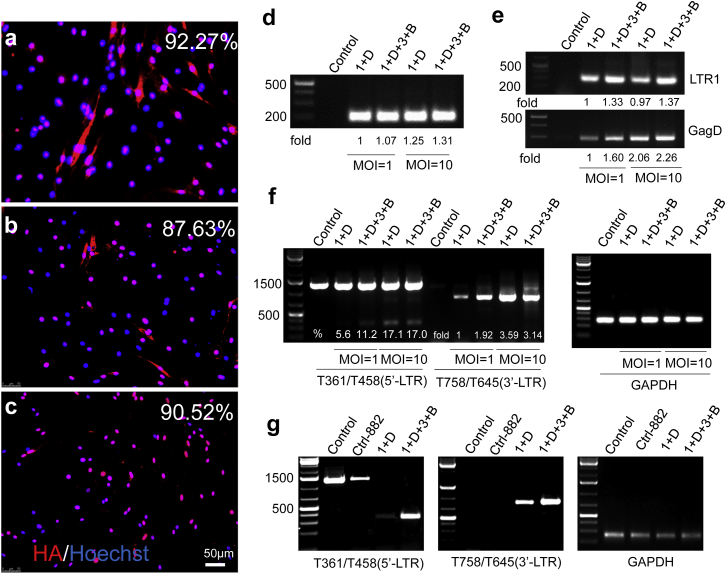
The HIV-1 Tg26 transgenic mouse was previously generated to contain a loci integrated with more than 10 copies of the pNL4-3 proviral genome with a 3.1-kb deletion spanning the partial Gag and Pol genes to render virus replication incompetent.29 These Tg26 mice develop a well-characterized kidney disease associated with HIV-1 viral gene expression30, 31 and potential uncharacterized neurological and cardiovascular deficits.32 Before in vivo studies, we evaluated the HIV-1 excision efficiency of multiplex sgRNAs/saCas9 using neural stem cells (NSCs)/neural progenitor cells (NPCs) isolated from Tg26 transgenic mice. The rationale for using NSCs/NPCs is the increasing evidence showing the potential HIV infection/latency in NSCs/NPCs,33, 34, 35, 36, 37, 38, 39, 40 which may relate to HIV-associated neurocognitive disorders (HANDs). Immunocytochemistry with an anti-HA antibody in mouse NSCs/NPCs after sgRNAs/saCas9 AAV-DJ infection showed that the monoplex sgRNA/saCas9 AAV-DJ infected 92.3% of cells determined at 20 days post-inoculation with a functional MOI (fMOI) of 10, which equals 1.8 × 105 genomic MOI (gMOI), suggesting that the AAV-DJ virus is highly effective at infecting NSCs/NPCs. Equal fMOIs of duplex (6.5 × 105 gMOI) or quadruplex (2.1 × 106 gMOI) sgRNAs/saCas9 AAV-DJ (Figure S2) produced similar infection efficiencies of around 87.63% and 90.52%, respectively (Figures 3A–3C). Afterward, we determined the efficiency of both duplex and quadruplex sgRNAs/saCas9 in cleaving the integrated HIV-1 proviral DNA in cultured NSCs/NPCs. We first validated the dose-dependent delivery of saCas9 and sgRNAs by PCR analysis at 2 days after AAV-DJ infection (Figures 3D and 3E). Then, we evaluated the HIV-1 excision efficacy by PCR genotyping. Both duplex and quadruplex sgRNAs/saCas9 induced a dose-dependent deletion of the predicted DNA fragments at 2 days after AAV infection, while quadruplex sgRNAs/saCas9 induced a higher cleaving efficiency when an equal fMOI was used (Figure 3F). The cleavage efficiency was dramatically increased at 20 days post-infection (Figure 3G) because of the long-term AAV transduction leading to sustained expression of saCas9/sgRNAs and cumulative genome editing. Again, the quadruplex induced a higher efficiency of HIV-1 excision (Figure 3G) despite the similar infection efficiency (Figures 3B and 3C). These data suggest that AAV-mediated multiplex sgRNAs/saCas9 can effectively excise the integrated HIV-1 proviral genome in cultured NSCs/NPCs from HIV-1 Tg26 transgenic mice and the oversized quadruplex sgRNAs/saCas9 AAV-DJ retains high efficiency of gene transduction and functional genome editing.
Figure 3.
AAV-DJ-Mediated Delivery of the Multiplex sgRNAs/saCas9-Expressing Vector Effectively Excises the HIV-1 Integrated Genome in NSCs from Tg26 Transgenic Mice
(A–C) Similar infection efficiency of monoplex (A), duplex (B), and quadruplex (C) in NSCs at 20 days post-infection with a fMOI of 10. The infected cells were determined by immunofluorescence cytochemistry with anti-HA tag antibody. (D–F) Direct PCR analysis validated the efficient delivery of transgene saCas9 (D) and LTR-1/GagD (E) and excision of 5′-LTR/GagD and GagD/3′-LTR (F) at 2 days post-infection. Relative fold changes or excision efficiency (%) were calculated via densitometry. (G) Quadruplex sgRNAs/saCas9 showed stronger cleavage efficacy than the duplex sgRNAs/saCas9 at 20 days post-infection with an fMOI of 10. The primers T361/T458 detected the deletions between 5′-LTR-1/LTR-3 and GagD, and the primers T758/T645 detected the deletions between GagD and 3′-LTR-1/LTR-3 (F and G). GAPDH was used for normalization of genomic DNA. Control-882 AAV-DJ virus was used as a nonfunctional control. GAPDH, glyceraldehyde 3-phosphate dehydrogenase; NSC, neural stem cell.
Multiplex sgRNAs/saCas9 Effectively Excises HIV-1 Proviral DNA in Various Tissues/Organs of HIV-1 Tg26 Transgenic Mice
To test whether the Cas9/sgRNA system is capable of excising integrated HIV-1 proviral DNA in vivo, we selected the AAV-DJ serotype for in vivo gene delivery.27, 41 As shown in Figure S3A, one single dose of AAV-DJ virus (4.15–4.20 × 1012 GC) via tail-vein injection could efficiently deliver sgRNAs- and saCas9-expressing cDNA into the liver and spleen in the first week alone. Histopathological and immunohistochemical examination of the liver did not identify any AAV-related tissue toxicity. The observed high transduction efficiency and minimal tissue toxicity is consistent with a previous report using similar dosages in adult mice.42 One additional injection of the same dose at 1 week after the first injection did show a consistent pattern of transgene distribution (Figure S3B). The AAV-DJ carrying duplex or quadruplex sgRNAs along with saCas9 results in similar gene delivery efficiencies in several organs (Figures S3A and S3B), even though the insertion of quadruplex sgRNAs/saCas9 appears to be beyond the limits of AAV packaging capacity. To determine the efficacy of sgRNAs/saCas9 in cleaving the integrated HIV-1 genome in animal tissues/organs, PCR genotyping was performed with a pair of primers that can amplify the fragmental deletion and ascertain the excision event.11, 15, 16 For the 5′-LTR/Gag deletion, we did not observe any fragmental deletions 1 week after infection by using nested PCR amplification, which is likely due to a low expression level of saCas9 in the first week after in vivo AAV delivery (data not shown). However, after conducting the optimization of the nested PCR analysis using the indicated primer pairs (Figure S3C), we observed the fragmental deletion to various extents in most organs/tissues, particularly in the liver, bone marrow, and spleen (Figure S3D). The additional AAV injection at 7 days after the first infection did increase the cleaving efficiency in most organs/tissues (Figure S3D). Because there are more than 10 copies of the HIV-1 transgene in a single tandem repeat in Tg26 mice,29 the size of the fragmental deletion, which is based on where the excision occurs in the tandem repeat, varied; some were larger than predicted, implying potential differences in DNA repairing events between tissues/organs. These fragmental deletions were verified by TA cloning of representative bands and Sanger sequencing (Figure S3E). Again, both duplex and quadruplex sgRNAs/saCas9 AAV-DJ induced a similar pattern of fragmental deletion mainly in the liver, while other organs showed a variable pattern of fragmental deletions (Figure S3D).
To expand the tissue ranges and potential cell types of HIV-1 excision in Tg26 transgenic mice, we packaged the quadruplex sgRNAs/saCas9 all-in-one vector into AAV-DJ/8, which increases gene delivery efficiency in other tissues/organs rather than the liver.27, 41 To maximize the in vivo transduction efficiency, we purified and concentrated the AAV-DJ/8 virus using large-scale preparation.43 Packaging efficiency for the oversized quadruplex sgRNAs/saCas9 reached 3.07 × 1014 GC/mL, which was close to the titer of a control AAV-Cre vector (4.21 × 1014 GC/mL) that was prepared at the same time. One intravenous (i.v.) injection of the purified quadruplex sgRNAs/saCas9 AAV-DJ/8 (1.5 × 1012 GC/mouse) resulted in efficient transduction of the saCas9 transgene in most organs/tissues collected (Figure S4A). The expression of saCas9 mRNA and protein in transduced organs/tissues was validated by RT-qPCR with primers targeting saCas9 and immunohistochemistry with an anti-HA antibody (Figure S5A). For the 5′-LTR/Gag primer pair, we observed visible fragmental deletions of the designed size using conventional PCR conditions (Figure 4A). Using the Gag/3′-LTR primer pair, we observed the fragmental deletion in liver, lung, and brain tissues under conventional PCR conditions (Figure 4B). TA cloning and Sanger sequencing validated the fragmental deletion of the designed size in both the brain and liver (Figure S6). One additional infection (1.5 × 1012 GC/mouse) 2 weeks after the first injection expanded the range of transduced tissues/organs and increased the gene transduction efficiency of saCas9 and sgRNAs 4 weeks after the first injection (Figures S4B and S5B). The excision efficiency as shown by the fragmental deletion with the Gag/3′-LTR primer pair for PCR genotyping was also increased in the liver and lungs and expanded to other organs such as the kidney and spleen (Figure 4C). RT-qPCR analysis revealed a significant reduction in mRNA expression of structural viral proteins Gag and Env in the brain, kidneys, liver, and lungs (Figures 4D–4F) as well as the regulatory protein Tat in the brain and kidneys (Figures 4G and S4C). This is supported by our other studies in HIV-1 transgenic mice and rats.17 Taken together, both AAV-DJ and AAV-DJ/8 viruses harboring either duplex or quadruplex sgRNAs/saCas9 achieved sufficient gene delivery in vivo and subsequent excision of the integrated HIV-1 genome in various organs/tissues of Tg26 transgenic mice.
Figure 4.
Quadruplex sgRNAs/saCas9 AAV-DJ/8 Induces HIV-1 Proviral DNA Excision and a Robust Reduction in HIV-1 RNA Transcription in Most Organs/Tissues of Tg26 Transgenic Mice
(A and B) Tg26 mice were injected via the tail vein with purified AAV-DJ/8 virus (1.535 × 1012 GC/mouse). Two weeks after injection, mice were euthanized and their tissues were collected for genomic DNA extraction and PCR genotyping. The positive control represented excision of the EcoHIV-eLuc reporter in HEK293T cells transfected with AAV-saCas9/sgRNA (LTR-1 plus GagD) vector. The negative control represented the mice injected with mock AAV virus. (C) An additional injection of AAV-DJ/8 was given at 2 weeks after the first injection and tissue samples were harvested 2 weeks later for PCR genotyping with the indicated primers. The representative deletion fragments, indicated by the dotted boxes in (B), were verified with TA cloning and Sanger sequencing (see Figure S6). (D) Diagram showing the location of RT-qPCR primer pairs for HIV transcripts. (E–G) RT-qPCR analysis showing a robust decrease (p < 0.001) in the levels of HIV-1 RNA transcripts in sgRNAs/saCas9-treated mice as compared with the control mice injected with mock AAV virus. Data represent the mean ± SD of triplicate experiments after normalization with housekeeping gene Ppia.
Whole-genome sequencing and off-target analysis in our previous studies11, 15, 16 and other reports2, 44, 45, 46, 47 did not detect any apparent off-target effects of spCas9 with multiplex sgRNAs in cultured primary cells or cell lines. To examine any potential in vivo off-target effects in this study, we performed a T7E1 assay using genomic DNA from the tissues/organs of Tg26 mice treated with quadruplex sgRNAs/saCas9. Based on potential off-target sites predicted by bioinformatics analysis as we previously described,11, 48 we selected two to three predicted sites for each sgRNA target (LTR-1, LTR-3, and GagD) with the highest score of specificity and we performed high-fidelity PCR generating a 500- to 800-bp product for T7E1 evaluation. As shown in Figure S7, no mutations were found in seven predicted off-target sites within the mouse genome but a clear mutation was detected in the on-target PCR product.
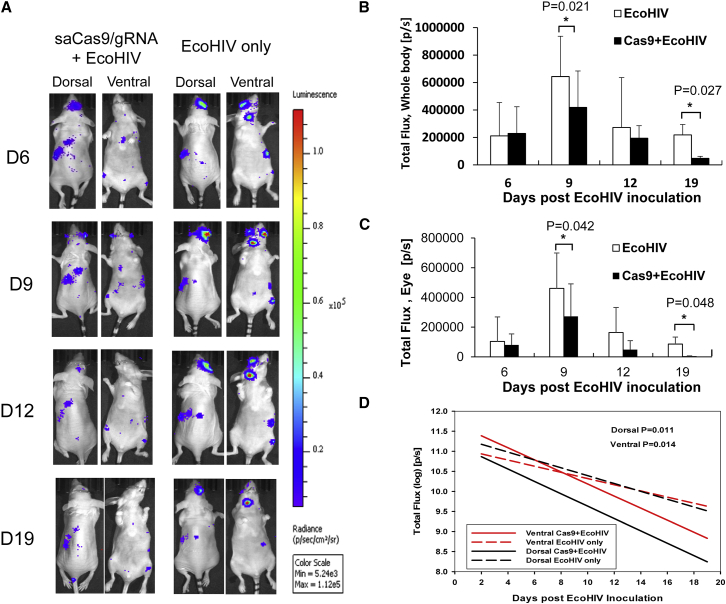
In Vivo HIV-1 Excision Is Visualized in an Experimental Mouse Model Inoculated with Cell-Free EcoHIV-eLuc Reporter Virus
A transgenic mouse model, such as the aforementioned Tg26 mouse, can be used to test our saCas9 system on HIV proviral excision but does not recapitulate clinical HIV infection/latency in terms of HIV-1 proviral copies/loci, random integration, various infected cell types, and the high rate of HIV mutation. To further test the feasibility and efficiency of HIV-1 excision by our approach using AAV-sgRNA/saCas9 after systemic HIV-1 infection, we utilized an EcoHIV-eLuc reporter virus to infect a conventional NCr strain of nude mice.19, 49, 50 Right after EcoHIV-eLuc inoculation (total 250 ng HIV p24/mouse, n = 3) via retro-orbital injection in the right eye, a single dose of AAV-DJ/8 (3.07 × 1012 GC/mouse) was injected into the same mice via the same injection route immediately after EcoHIV inoculation. Another three NCr mice were injected with only EcoHIV-eLuc of the same dosage as a negative control group. We performed longitudinal bioluminescence imaging (BLI) of the live animals over 19 days after EcoHIV-eLuc inoculation (Figure 5A). We observed a significant reduction in EcoHIV-eLuc reporter activity on both days 9 and 19 from the entire mouse or the right eye area (Figures 5B and 5C). Compared with the negative control group, the reduction in EcoHIV-eLuc infection by AAV-sgRNAs/saCas9 treatment was statistically significant, as measured by the bioluminescence output from the neck lymph node at the dorsal (p < 0.011) and ventral (p < 0.014) sides during the entire course of the experiment (19 days) using a linear mixed-effects model for statistical significance (Figure 5D).
Figure 5.
Bioluminescence Imaging Analysis Shows that Quadruplex sgRNAs/saCas9 AAV-DJ/8 Induces Excision of EcoHIV-eLuc In Vivo
(A) AAV-DJ/8 was administered via a retro-orbital injection into the blood sinus of the right eye of each mouse (n = 3) right after EcoHIV-eLuc inoculation via the same injection route. Representative bioluminescence imaging of one mouse on days 6, 9, 12, and 19 post-EcoHIV inoculation is shown. All of the images were measured for radiance (photon/second/Sr2) and pseudocolored with the same rainbow scale for fair comparison. (B) Total flux of bioluminescence (total photons/second, P/S) measured from the entire body of each group (n = 3/group until day 19 [n = 2]) on each indicated day post-EcoHIV-eLuc inoculation. (C) Bioluminescence output from EcoHIV-eLuc-infected cells in each mouse was measured from the ROI defined on the right eye for comparison. Data represent the mean ± SD, with statistical significance indicated by an asterisk in each comparison (p < 0.05, Student’s one-sided t test). (D) The bioluminescence output from the neck lymph node of each mouse (n = 3 until day 19 [n = 2]) was used for comparison of the efficacy of saCas9/gRNA treatment longitudinally on HIV excision (p values are two-sided using a linear mixed-effects model).
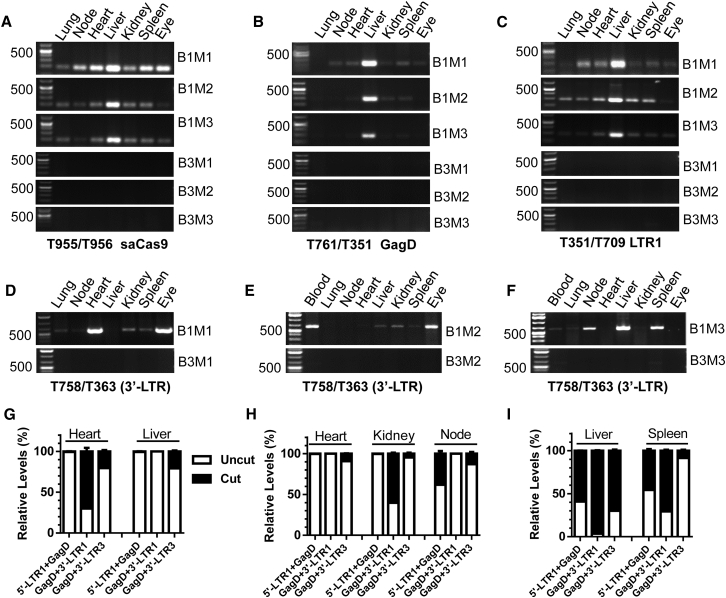
To validate the in vivo excision efficiency, we performed PCR genotyping using the established above-mentioned PCR conditions at the endpoint of experiments. The efficient transduction of saCas9, GagD, and LTR-1 transgenes by AAV-DJ/8 was detected mainly in the liver, while it varied slightly in other organs and tissues (Figures 6A–6C). Using conventional PCR, we observed an efficient and robust excision of the predicted fragment in most tissues/organs, with the highest efficacy observed in the injected right eye area, heart, blood, liver, spleen, and lymph nodes (Figures 6D–6F). The variability in edited tissues from different animals may result from the random cellular/tissue distribution of EcoHIV-eLuc infection, AAV-mediated gene delivery, and Cas9-mediated excision. Again, some of the fragmental deletions were validated by TA cloning and Sanger sequencing (Figure S8). To quantify the excision efficiency of quadruplex sgRNAs/saCas9 in vivo, we performed qPCR analysis of the excised fragments using three different primer combinations (Figure S9). The melting peak curve analysis clearly identified three types of proviral DNA excisions as designed: 5′-LTR-1 to GagD, GagD to 3′-LTR-1, or GagD to 3′-LTR-3. Using the uncut (non-targeting) region (the sequence flanking both target sites) as internal normalization, we assessed the excision efficiency in selected tissues/organs (Figures 6G–6I). Although the individual number may underestimate the efficiency due to various combinations of excision, the results clearly showed that the excision efficiency (Figures 6G–6I) was consistent with PCR genotyping (Figures 6D–6F). The excision types and efficiency varied with different organs/tissues (Figures 6G–6I). Interestingly, all three types of excision occurred with 96% efficiency for the GagD/3′-LTR in the B1M3 mouse liver (Figure 6I). These data suggest that the quadruplex sgRNAs/saCas9 AAV-DJ/8 is highly effective at excising the HIV proviral genome even in newly infected cells during in vivo systematic HIV infection.
Figure 6.
PCR Genotyping and qPCR Analysis Validated the Gene Transduction and Excision Efficiency of EcoHIV-eLuc by Quadruplex sgRNAs/saCas9 AAV-DJ/8 in Various Organs/Tissues of Mice
(A–C) Various extents of gene transduction in indicated tissues/organs for saCas9 (A), GagD (B), and LTR-1 (C) at 19 days after infection with quadruplex sgRNAs/saCas9 AAV-DJ/8 (upper panel) or empty control AAV-Cre-DJ/8 (lower panel). (D–F) Conventional PCR genotyping showing various extents of EcoHIV DNA excision between GagD and the 3′-LTR in each indicated tissue/organ (upper panel) compared with the negative control (lower panel). (G–I) qPCR analysis of three indicated fragmental excision types in different tissues/organs selected from B1M1 (G), B1M2 (H) and B1M3 (I). Data represent the mean ± SD of triplicate reactions and are expressed as relative levels over the internal uncut (non-targeting) region of EcoHIV DNA in the same tissues/organs. BxMx, box and mouse number.
In Vivo Excision of HIV-1 Provirus from Latently Infected Human Cells Is Detected in Humanized BLT Mice
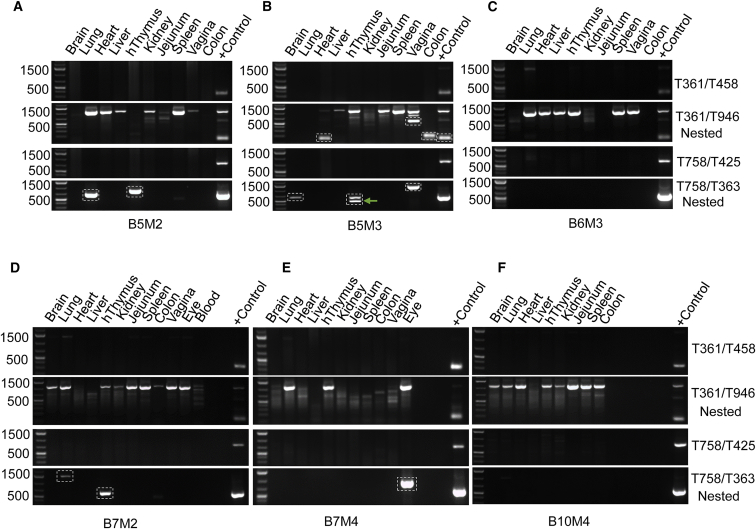
To demonstrate the feasibility of saCas9/sgRNA genome editing on excising the HIV-1 provirus in a more clinically relevant animal model, we administered AAV-DJ/8 carrying sgRNAs/saCas9 into humanized BLT mice inoculated with a R5-, M-tropic HIVNL-BaL-eLuc reporter virus via vaginal mucosal transmission (n = 3; mouse identification [ID] nos. B5M2, B5M3, and B6M3) or intraperitoneal injection (n = 3; mouse ID nos. B7M2, B7M4, and B10M4). Whole-body BLI (unpublished data) with a sensitivity of visualizing five single cells in a niche was used to longitudinally visualize and analyze the spatiotemporal dynamics of HIV-1 dissemination in these BLT mice.50 These BLT mice showed a certain degree of either local or systemic HIV dissemination, but the infection diminished and completely ceased after the first 60 days without any visible infection determined by whole-body BLI examination in the following 70 days or longer. This phenomenon could possibly be attributed to T-cell depletion, HIV-1 latency, or the possible silencing of eLuc reporter by mutations during HIV-1 replication. Since no HIV-1 genome was detected by nested PCR in some of the tissues (Figure 7), the contribution of mutation to the silenced reporter expression is less likely, which is further confirmed by our observation in another ongoing study that BLT mice showed successful systemic infection and latency under cART (unpublished data). The quadruplex sgRNAs/saCas9 AAV-DJ/8 was administered via both intravenous and intravaginal routes in HIV-infected BLT mice B5M2 and B5M3, but not B6M3, which served as the negative control. A single intravenous injection of AAV-DJ/8 was administered to HIV-infected BLT mice B7M2 and B7M4, but not B10M4, which served as the negative control. At 2–4 weeks after AAV delivery, BLT mice were euthanized and genomic DNA samples were extracted from major organs/tissues for PCR genotyping as described above. The presence of HIV-1 proviral DNA in the harvested organs/tissues was validated with nested PCR using the 5′-LTR/Gag primer pairs (T361/T458 then T361/T946) (Figure 7). Importantly, we detected the fragmental deletion with both the 5′-LTR/Gag primer pair T361/T946 and the Gag/3′-LTR primer pair T758/T363 (Figures 7A, 7B, 7D, and 7E) in the vagina, heart, lungs, right eye (AAV injection site), colon, and human thymic organoid under the left kidney capsule. In particular, we observed a much higher intensity of the band representing a fragmental deletion, while the band representing the uncut proviral DNA was barely visible in the same tissue (e.g., the heart and colon of the B5M3 mouse) (Figure 7B). This result clearly suggests an efficient excision of HIV-1 proviral DNA in latently infected human cells within a solid tissue via a single i.v. injection of quadruplex sgRNAs/saCas9 AAV-DJ/8. TA cloning of these PCR products and Sanger sequencing validated the fragmental deletions (Figure S10 for PCR products from 5′-nested PCR using primer pair T361/T946; Figure S11 for PCR products from 3′-nested PCR using primer pair T758/T363). Within the cleaved residual sequence at both the 5′-LTR/Gag region (Figure S10) and the Gag/3′-LTR region (Figure S11), various degrees of insertion or deletion were observed between the predicted cleaving sites at the third nucleotide from PAM of the targeting sgRNAs (Figures S10A and S11A). These sets of pre-clinical data demonstrate the proof of principle that quadruplex sgRNAs/saCas9 can be delivered by AAV-DJ/8 into resident HIV-1 latently infected human cells in major tissues and organs in a humanized mouse model to excise integrated HIV-1 proviral DNA.
Figure 7.
Excision of HIV Proviral DNA by Quadruplex sgRNAs/saCas9 AAV-DJ/8 in Humanized BLT Mice Inoculated with HIVNL-BaL-eLuc at 2, 3, and 4 Weeks after AAV Treatment
Conventional PCR (35 cycles) using primer pair T361/T458 (for amplifying 5′-LTR/Gag) was used to detect the presence of HIV proviral DNA, which showed a weak wild-type band (1.4 kb) in the lung tissue of the B6M3 negative control without saCas9/sgRNA treatment (C). Nested PCR (35 cycles) using primer pair T361/T946 (for 5′-LTR/Gag) with the template from the first-round PCR (22 cycles with the primer pair T361/T458) identified the wild-type DNA fragment (1.13 kb) of HIV-1 proviral DNA in most, if not all, organs/tissues of each BLT mouse (A–F) and the PCR product of fragmental deletions resulted from HIV-1 excision in the heart, colon, and vagina of HIV-infected BLT mice treated with saCas9/sgRNA (A). Nested PCR with the primer pair T758/T363 (for Gag/3′-LTR) using the template from the first-round PCR (25 cycles, T758/T425) identified the fragmental deletion bands in organs/tissues of saCas9/sgRNA-treated BLT mice (A, B, D, and E) but not the untreated BLT mice (C and F). The fragmental deletion bands resulted from saCas9 editing were framed with dotted boxes followed by TA cloning and Sanger sequencing for validation (see Figures S10 and S11). The positive control represents the excision of the EcoHIV-eLuc reporter in HEK293T cells transfected with AAV-saCas9/sgRNA (LTR-1 plus GagD) vector. BxMx, box and mouse number.
Discussion
To our knowledge, this study is the first to demonstrate the effective excision of HIV-1 proviral DNA from the host genome in pre-clinical animal models using saCas9 and multiplex sgRNAs delivered by an all-in-one AAV-DJ/8 vector. These AAV viruses induce effective gene delivery and HIV-1 excision in vitro as well as in vivo in HIV-1 Tg26 transgenic mice, EcoHIV-eLuc acutely infected mice, and HIV-1-infected humanized BLT mice. Based on the extensive and highly promising clinical application of AAVs in gene therapy (for review, see Fuchs and Desrosiers51 and Brady et al.52), our study demonstrates the proof of principle for treating HIV-1/AIDS with the CRISPR/Cas9 genome editing approach. Furthermore, the combination of easy/fast multiplex sgRNA cloning, rapid reporter screening, reliable direct PCR genotyping, and highly efficient AAV virus production provides potential for using saCas9/sgRNA genome editing in personalized and precision medicine. Using multiplex sgRNAs to target multiple HIV proviral DNA simultaneously is beneficial to prevent the potential escape of HIV due to the high mutation rate during HIV-1 replication because the probability of double mutations in two different target sequences in the same HIV proviral genome is much lower than that of single mutation in a target site. Finally, we demonstrate that the quadruplex sgRNAs/saCas9 strategy combining sgRNAs targeting two LTR sites (at both the 5′ and 3′ ends) and two structural proteins (Gag and Pol) is a promising therapeutic candidate for HIV-1 excision, due to maximization of the possibility of multiple indel mutations on six on-target sites and fragmental deletions among these sites. This quadruplex strategy also provides additional advantages, including (1) its ability to greatly reduce the potential of HIV-1 escape,53, 54 (2) the high possibility of HIV-1 excision despite the continuous proviral mutation in the clinical HIV-1 patient population, (3) reliable loss-of-function achievement due to removal of a substantial portion of the target gene or genome,55 and (4) optimal efficiency of excision.16
In vivo genome editing efficiency relies mainly on effective gene delivery. Lentivirus-mediated Cas9/gRNA delivery has been preferentially and extensively employed in most laboratories;56, 57 however, safety and immunogenicity concerns limit its clinical applications.58 AAV-mediated gene therapy is becoming a promising approach in clinical trials because it reduces the risks of mutagenesis, toxicity, and severe host immune response.3, 59 The feasibility and efficiency of AAV-mediated Cas9 delivery has been tested extensively both in vitro and in vivo (for review, see Moser and Hirsch60). Several laboratories using minimal promoters and poly(A) sequences have reported success in applying AAV-spCas9 in cultured cells and animal models.4, 61, 62, 63, 64 Splitting spCas9 into functional N-terminal and C-terminal parts65, 66 provides a new way to deliver spCas9 more efficiently in pre-clinical animal models.67, 68 The AAV-split-spCas9 nickase vector using the GyrA intein system dramatically increases the specificity of genome editing.69 Among several smaller Cas9 orthologs, saCas9 has been well characterized and is highly promising for animal studies and clinical applications. The high efficiency of AAV packaging and gene transduction in animal models using saCas9 plus one or two sgRNA-expressing cassettes has been tested in an all-in-one AAV vector with various AAV serotypes.2, 4, 5, 6, 7 Here, we demonstrate that duplex HIV-sgRNAs with saCas9 in an all-in-one AAV-DJ vector are capable of delivering transgenes and excising the HIV-1 genome in cultured cells and particularly in various organs/tissues of Tg26 transgenic mice. This is supported by our recent study using duplex sgRNAs/saCas9 AAV9 in HIV-1 transgenic mice and rats.17 Furthermore, we demonstrate that the oversized all-in-one AAV vector encoding quadruplex sgRNAs and saCas9 is similarly capable of achieving high-titer packaging into the AAV-DJ/8 virus and inducing effective excision of the HIV-1 genome in vitro and in vivo.
Several approaches to place multiplex sgRNA-expressing cassettes in an all-in-one vector have been reported. Using the RNA processing property of the Csy4 protein or ribozymes, a single transcript under a Pol II promoter can be processed into multiple sgRNAs.70, 71 Multiplex vectors with up to seven sgRNAs under the same U6 promoter have been successfully constructed using the Golden Gate cloning method, and their multiplex targeting efficiency has been tested for multiplex genome/epigenome editing and simultaneous activation/repression of multiple genes.72 Multiplex sgRNAs driven by four independent Pol III promoters (human U6 promoter, mouse U6 promoter, 7SK, and H1) have been also tested.57 Here we developed a novel In-Fusion (Clontech) cloning strategy with interrogated exchange of two restriction enzymes, rendering easy and fast cloning of any number of multiplex sgRNAs into a single vector. This approach is applicable to any multiplex sgRNA under any Pol III promoter and to various viral gene delivery systems. Although we showed that four sgRNAs under four identical U6 promoters exhibited sufficient activity to induce target DNA cleavage, we found that the sgRNA LTR-1, GagD, and PolB in the quadruplex sgRNAs/saCas9 system are best, while LTR-3 is weaker in cleaving HIV-1 provirus although it does show a high potency of cleavage in the duplex sgRNA system. Whether the location of each sgRNA among the multiplex sgRNA cascade affects sgRNA expression efficiency warrants further investigation. In addition, it would be interesting to optimize the combination of various sgRNA-driven promoters57 for maximal AAV packaging and gene transduction.
This study established the feasibility of using quadruplex sgRNAs/saCas9 AAV gene therapy in excising the integrated HIV-1 provirus in both Tg26 transgenic mice and humanized BLT mice. sgRNAs/saCas9 can be considered as a pre-exposure prophylaxis (PrEP) during EcoHIV inoculation in conventional mice. The intravenous administration of AAV-DJ or AAV-DJ/8 achieved extensive gene transduction and HIV-1 excision in most tissues/organs, with the highest in liver followed by the heart, lungs, spleen, vagina, colon, and lymph nodes. Using outgrowth assays, it was estimated that only ∼30 HIV-1 latently infected resting T cells (total 3 × 106 resting T cells/BLT mouse × 9.9 infectious units per million cells) can be detected in each HIV-1-infected BLT mouse.73 Because of the difficulty in isolating HIV-infected resting T cells from the high density of solid lymph tissue or somatic organs of humanized BLT mice or EcoHIV-infected mice, we employed a novel in vivo imaging system as we previously described50 to monitor suppressed HIV-1 expression longitudinally to establish a latency model, followed by PCR genotyping to detect the existence or absence of proviral DNA in tissues/organs. One of the priorities in the HIV/AIDS field is to eliminate proviruses from latently infected resting CD4+ T cells and other latent cellular reservoirs in vivo. This study demonstrates the clear excision of Gag/3′-LTR and 5′-LTR/Gag fragments in latently infected human cells within mouse multiple organs/tissues by a single AAV-DJ/8 administration in HIV-infected humanized BLT mice. This finding also validates the vast extent of tissue reservoirs harboring HIV-1 viruses. However, further studies are needed to target latently infected T cells in order to eliminate viremia rebound after cART.
The findings reported here provide justification for further clinical development of the Cas9/sgRNA approach for HIV-1 excision in preventing and improving HIV-1 diseases. However, its pharmacological index and window need to be evaluated further in a more clinically relevant animal model, such as simian immunodeficiency virus (SIV)-infected non-human primates. In addition, the future of the CRISPR/Cas9 genome editing system in vivo would rely on a very effective gene/protein delivery vehicle such as nanoparticles, which can be manufactured in different sizes with different targeting properties.74, 75 Gene delivery efficiency will be improved over time for HIV-1 latent cells, such as resting CD4+ T cells, monocytes/macrophages, or hematopoietic stem cells, via the continuous and extensive exploration of new AAV serotypes,62 integrase-deficient lentiviruses,76 and nanoparticles.7, 74, 77
Potential off-target effects of the Cas9/sgRNA system in mammalian cells have been a great concern in this field. Many technologies and strategies have been developed in recent years and Cas9 specificity has been improved significantly; however, continued vigilance is still required (for review, see Tycko et al.78). Although plenty of studies, including ours, have used various strategies, such as whole-genome sequencing,11, 15, 16 genome-wide, unbiased identification of DSBs enabled by sequencing (GUIDE-seq),44, 79 digenome sequencing (digenome-seq),80, 81, and direct in situ breaks labeling, enrichment on streptavidin, and next-generation sequencing (BLESS)2 to measure off-target effects, none or very few off-target effects have been detected thus far.2, 11, 44, 45, 46, 47, 82, 83 However, further comprehensive and deep analysis on the potential off-target effects for Cas9/sgRNA-mediated HIV excision is still needed before clinical trials.
In conclusion, the saCas9 protein’s smaller size allows duplex or even quadruplex sgRNA-expressing cassettes to be delivered in an all-in-one AAV vector for high-titer packaging and robust gene transduction in cell cultures and animal models. The saCas9 with quadruplex sgRNAs targeting both 5′- and 3′-end LTRs and structural Gag and Pol regions induces versatile patterns of small indel mutations and large fragmental deletions and thus provides optimal efficiency of HIV-1 genome excision. The feasibility and efficacy of HIV-1 excision in animals via this highly promising AAV gene therapy will pave the way toward clinical trials in the near future.
Materials and Methods
Bioinformatics Design of gRNAs
We utilized the Broad Institute gRNA designer tool for highly effective gRNA design (https://www.broadinstitute.org/rnai/public/analysis-tools/sgrna-design) because this tool also provides the extended spacer sequence (NNNN[20nt]NGGNNN) for the spCas9 system. The saCas9 PAM sequence for optimal on-target cleaving is NNGRR(N). We selected only NGGRR(N) because it contains NGG PAM and thus the designed seed sequence for saCas9 system can also be used to the well-established spCas9 system. To test this, we picked three sgRNAs targeting the HIV-1 LTR promoter region, three sgRNAs targeting Gag, and one sgRNA targeting Pol. The oligonucleotides for these sgRNA targets are listed in Table 1.
Table 1.
Oligonucleotides for sgRNAs Targeting HIV-1 LTR, Gag, and Pol and PCR Primers
| Target Name | Direction | Sequence |
|---|---|---|
| sgRNA-Targeting Oligonucleotides | ||
| LTR-1 | T708: sense | caccGCAGAACTACACACCAGGGCC |
| T709: antisense | aaacGGCCCTGGTGTGTAGTTCTGC | |
| LTR-2 | T710: sense | caccGTTACACCCTATGAGCCAGCA |
| T711: antisense | aaacTGCTGGCTCATAGGGTGTAAC | |
| LTR-3 | T712: sense | caccGTGTGGCCTGGGCGGGACTG |
| T713: antisense | aaacCAGTCCCGCCCAGGCCACAC | |
| GagB | T714: sense | caccGCCTTCCCACAAGGGAAGGCCA |
| T715: antisense | aaacTGGCCTTCCCTTGTGGGAAGGC | |
| GagC | T758: sense | caccGCGAGAGCGTCGGTATTAAGCG |
| T759: antisense | aaacCGCTTAATACCGACGCTCTCGC | |
| GagD | T760: sense | caccGGATAGATGTAAAAGACACCA |
| T761: antisense | aaacTGGTGTCTTTTACATCTATCC | |
| PolB | T716: sense | caccGCATGGGTACCAGCACACAA |
| T717: antisense | aaacTTGTGTGCTGGTACCCATGC | |
| Primers for Conventional PCR | ||
| Gag | T458: antisense | CCCACTGTGTTTAGCATGGTATT |
| T457: sense | AATGGTACATCAGGCCATATCAC | |
| GagC | T758: sense | CACCGCGAGAGCGTCGGTATTAAGCG |
| GagD | T761: antisense | CACCTGGTGTCTTTTACATCTATCC |
| PolA | T689: sense | CACCGCAGGATATGTAACTGACAG |
| LTR-1 | T709: antisense | AAACGGCCCTGGTGTGTAGTTCTGC |
| LTR-2 | T710: sense | CACCGTTACACCCTATGAGCCAGCA |
| T711: antisense | AAACTGCTGGCTCATAGGGTGTAAC | |
| LTR-E | T361: sense | CACCGATCTGTGGATCTACCACACACA |
| LTR-F | T363: antisense | CACCGCTGCTTATATGCAGCATCTGAG |
| Cas-hU6 | T351: sense | CGCCTCGAGGATCCGAGGGCCTATTTCCCATGATTCC |
| Tg26-3 vector | T645: antisense | TGGAATGCAGTGGCGCGATCTTGGC |
| SaCas9 | T955: sense | AACAGATTCAAGACCAGCGACTAC |
| T956: antisense | TACCATTCTTTGATGTCCTTCCAG | |
| GAPDH | T623: antisense | GCTAAGCAGTTGGTGGTGCAGGA |
| #40: sense | TCACCATCTTCCAGGAGCGA | |
| β-actin | sense | GGACTTCGAGCAAGAGATGG |
| antisense | ACACTGTGTTGGCGTACAG | |
| Gag-Nest | T946: antisense | ACCTGGCTGTTGTTTCCTGTGTC |
| HIV-U5a | T425: antisense | AAACGAGTCACACAACAGACGGGC |
| LTR-1-Off-1 | T991: sense | TCAGCCATGAGGAAGAACTTGGA |
| T992: antisense | TCTCCAGAGTGCTGGCAAGGTCC | |
| LTR-1-Off-2 | T993: sense | TCACCTGGTGCCAGTGTCTGCGG |
| T994: antisense | TATGAATGAGTTTGGCGTGTATG | |
| LTR-1-Off-3 | T995: sense | ATCGATGAGGCTCTCAGCATCACC |
| T996: antisense | TGGTGAGGCCTCTGGGCCACTTGAG | |
| LTR-3-Off-1 | T1013: sense | AGCCACACTCTGGCACTGAGACAAG |
| T1014: antisense | AGTAAGCATAGGTATGGAGAGGC | |
| LTR-3-Off-3 | T1017: sense | ACAGCCACATGCAGGAGGTGACCAC |
| T1018: antisense | ACATGTGCCTTGGCTTGTATGTGG | |
| GagD-Off-1 | T1019: sense | TTGAAGCAGAGTTAAGGAATCTTGG |
| T1020: antisense | TGCCATGTTCCTTCTGTAATCATAG | |
| GagD-Off-2 | T1021: sense | TCTCTATGTAGTCTTGGCTGTCCTG |
| T1022: antisense | ACCATGGCATCTAGCTGTGCTGAC | |
| Primers for Real-Time PCR | ||
| Gag | T760: sense | CACCGGATAGATGTAAAAGACACCA |
| T946: antisense | ACCTGGCTGTTGTTTCCTGTGTC | |
| Env | T876: sense | CCGAAGGAATAGAAGAAGAAG |
| T691: antisense | CACCGAGAGTAAGTCTCTCAAGCGG | |
| Tat | T1002: sense | TGGAAGCATCCAGGAAGTCAGCC |
| T1003: antisense | TTCTTCTTCTATTCCTTCGGGCC | |
| Mouse Ppia | T979: sense | GCCCAGTATGCTTGGGTATC |
| T980: antisense | TGCTGACTCCCAGAACAGA | |
| Human β2M | sense | TGCTGTCTCCATGTTTGATGTATCT |
| antisense | TCTCTGCTCCCCACCTCTAAGT | |
| 5′-LTR-1 plus GagD uncut | T872 | GGACTCGGCTTGCTGAAG |
| T759 | aaacCGCTTAATACCGACGCTCTCGC | |
| 5′-LTR-1 plus GagD cut | T361 | caccGATCTGTGGATCTACCACACACA |
| T946 | ACCTGGCTGTTGTTTCCTGTGTC | |
| 5′-LTR-1 plus GagD internal | T457 | AATGGTACATCAGGCCATATCAC |
| T458 | CCCACTGTGTTTAGCATGGTATT | |
| GagD plus 3′-LTR-1 uncut | T873 | ATCTCTGCTGTCCCTGTAA |
| T874 | AATCCCCAAAGTCAAGGAGTAA | |
| GagD plus 3′-LTR-1 cut | T758 | caccGCGAGAGCGTCGGTATTAAGCG |
| T535 | AAACAAGGTCAGTGGATATCTGATC | |
| GagD plus 3′-LTR-1 internal | T872 | GGACTCGGCTTGCTGAAG |
| T759 | aaacCGCTTAATACCGACGCTCTCGC | |
| GagD plus 3′-LTR-3 uncut | T873 | ATCTCTGCTGTCCCTGTAA |
| T874 | AATCCCCAAAGTCAAGGAGTAA | |
| GagD plus 3′-LTR-3 cut | T758 | caccGCGAGAGCGTCGGTATTAAGCG |
| T363 | caccGCTGCTTATATGCAGCATCTGAG | |
| GagD plus 3′-LTR-3 internal | T872 | GGACTCGGCTTGCTGAAG |
| T759 | aaacCGCTTAATACCGACGCTCTCGC | |
Plasmids and Cloning of Multiplex gRNA Expression AAV Vectors
The pNL4-3-EcoHIV-eLuc and pHIVNL-BaL-eLuc reporter vectors were constructed as previously described.16, 48, 50 A pair of oligonucleotides for each targeting site with 5′-CACC and 3′-AAAC overhang (Table 1) was obtained from AlphaDNA. The target seed sequence was cloned via BsaI sites into pX601-AAV-CMV::NLS-saCas9-NLS-3xHA-bGHpA;U6::BsaI-sgRNA (a gift from Feng Zhang via Addgene) (61591; Addgene).2 The pX601 AAV was digested with BsaI, treated with antarctic phosphatase, and purified with a Quick nucleotide removal kit (QIAGEN). An equal amount of complementary oligonucleotide was mixed in T4 polynucleotide kinase (PNK) buffer for annealing. These annealed seed pairs were phosphorylated with T4 PNK and ligated into the BsaI-digested AAV using T7 DNA ligase. The ligation mixture was transformed into Stabl3 competent cells. Positive clones were identified by PCR screening and verified by Sanger sequencing. For multiplex sgRNA cloning, we established two approaches: double digestion traditional cloning and In-Fusion seamless PCR cloning. In the double digestion strategy, the target AAV vector was first digested with KpnI and blunted, and then digested with EcoRI, while the transfer insert harboring the expected sgRNA expression cassette was first digested with NotI and blunted and then digested with EcoRI. The insert and the vector were purified with the QIAquick Gel Extraction Kit (QIAGEN) followed by standard overhang/blunt end cloning. The positive duplex sgRNA-expressing AAV clones were identified by double digestion with NotI and BamHI or EcoRI. In the In-Fusion PCR cloning strategy, the target AAV vector was linearized with EcoRI and KpnI. The insertion fragment was produced via PCR using the primer pair T795/T796 (Table 1) with a mutation of the 3′-end KpnI site (for further addition of new sgRNAs) and the transfer sgRNA AAV vector as the template. After purification, the linearized vector and the insertion PCR product were ligated using an In-Fusion HD Cloning Kit (Clontech). The positive duplex sgRNA-expressing AAV clones were identified by double digestion with NotI and BamHI or EcoRI. Using a similar proof of principle for both cloning strategies, the duplex sgRNAs/saCas9 vector can serve as a target vector for inserting additional sgRNA expression cassettes. One necessary precaution is to ensure the absence of these digestion sites (KpnI, NotI, EcoRI) in the seed sequence of the selected sgRNAs.
Cell Culture, Transfection, and Firefly-Luciferase Reporter Assay
HEK293T cells were cultured in high-glucose DMEM containing 10% FBS and antibiotics (100 U/mL penicillin and 100 μg/mL streptomycin) in a humidified atmosphere with 5% CO2 at 37°C. For the luciferase reporter assay, cells were cultured in a 96-well plate (3 × 104 cells/well) and transfected with the indicated plasmids using the standard calcium phosphate precipitation protocol. After 48 hr, the cell were lysed and assayed with the ONE-Glo luciferase assay system (Promega) in a 2104 EnVision Multilabel Reader (PerkinElmer).
Terra PCR Direct Genotyping and Nested PCR
To perform high-throughput genotyping using PCR, cells were seeded in a 96-well plate and transfected with indicated vectors. After 48 hr, the media were removed and the cells were treated with 45 μL 50 mM NaOH per well and incubated for 10 min at 95°C. After neutralization with 5 μL 1 M Tris-HCl (pH 8.0), 0.5 μL DNA extract was used in a 10-μL PCR reaction using Terra PCR Direct Polymerase Mix (Clontech) and the indicated primers. To genotype the excised HIV proviral DNA from the isolated tissues and organs from EcoHIV-eLuc inoculated mice, each tissue/organ was triturated by each individual pair of scissors to avoid cross-contamination and was then digested with proteinase K in a lysis buffer consisting of 1% SDS, 10 mM Tris (pH 8.0), 5 mM EDTA, and 100 mM NaCl in a 55°C water bath. Genomic DNA was then extracted from the tissue lysates using conventional phenol/chloroform extraction methods. Tissue samples were broken up mechanically with a pestle and mortar and were then processed with the Nucleospin tissue kit and Nucleospin RNA/Protein kit (Clontech) according to the manufacturer’s protocols to extract genomic DNA, RNA, and protein from specific Tg26 mouse tissues. To perform PCR genotyping, 100-ng DNA samples were denatured at 98°C for 3 min, followed by two steps of conventional PCR for 35 cycles with an annealing/extension step at 68°C for 1 min/kb. For nested PCR, the first round of reaction was run for 22–25 cycles with the indicated primers and the second round of PCR was run for 35 cycles with the nested PCR primers.
RT-qPCR
Total RNAs from the indicated organs/tissues were extracted with an RNeasy Mini Kit (QIAGEN) per the manufacturer’s instructions. The potentially residual genomic DNA was removed through on-column DNase digestion with an RNase-Free DNase Set (QIAGEN). One μg of RNA for each sample was reverse transcribed into cDNAs using random hexanucleotide primers with a High-Capacity cDNA Reverse Transcription Kit (Invitrogen). qPCR analyses of cDNA or HIV genomic DNA were carried out in a LightCycler480 (Roche) using a SYBR Green PCR Master Mix Kit (Applied Biosystems). The primer pairs for saCas9, Gag, Env, and Tat were designed as shown in Table 1. The primers for human β2 microglobulin and mouse Ppia housekeeping genes were obtained from RealTimePrimers. The primers for HIV excision quantification were designed as shown in Figure S8. Each sample was tested in triplicate. Cycle threshold (Ct) values were obtained graphically for the target genes and housekeeping genes. Differences in Ct values between the housekeeping gene and target genes were represented as ΔCt values. ΔΔCt values were obtained by subtracting the ΔCt values of control samples from those of experimental samples. Relative fold or percentage change in gene expression or HIV DNA excision was calculated as 2−ΔΔCt. In some cases, the amplification curve and the melting peak curve were used for the comparative analysis.
Viral Packaging and Purification
Small- and large-scale preparation of AAV-DJ and AAV-DJ/8 was performed via an AAV production service (https://vigenebio.com/) following a previously published protocol.84 Briefly, HEK293T cells were co-transfected with three vectors and the viral particles were harvested and purified by iodixanol gradient ultracentrifugation. Viruses were concentrated and formulated in PBS. Virus titers were determined by the viral genome copy number in a 1-mL sample (GC/mL) using real-time PCR with a linearized genome plasmid as a standard.
For packaging replication competent EcoHIV-eLuc and HIVNL-BaL-eLuc, plasmid DNA encoding a full-length molecular clone of either EcoHIV or HIV-1 carrying the BLI reporter (Figure S12) was transfected into HEK293T cells using Lipofectamine 3000 (Invitrogen). At 24 hr post-transfection, media containing transfection reagent and DNA were aspired and cells were washed twice with PBS. Fresh media were added. At 48 and 60 hr post-transfection, supernatants were collected and filtered through a 0.45-μm filter. The lentiviral supernatant was concentrated using the Lenti-X Concentrator (Clontech) following the manufacturer’s instructions or using 20% sucrose density centrifugation at 20,000g, 4°C, for 4 hr.85 For EcoHIV-eLuc, the quantity of p24 was determined by ELISA (XpressBio) according to the manufacturer’s protocol. The titer of HIVNL-BaL-eLuc was determined on GHOST(3) X4/R5 (requested from Drs. Vineet N. Kewal Ramani and Dan R. Littman via the NIH AIDS Reagent Program) for GFP expression analyzed by fluorescence-activated cell sorting (FACS) analysis.
Viral Inoculation of Humanized BLT Mice
The University of Pittsburgh or Temple University Institutional Animal Care and Use Committee (IACUC) approved all animal care and procedures. Humanized BLT mice based on an NOD scid gamma strain transplanted with human fetal liver, thymus, and CD34+ cells isolated from the syngeneic liver were purchased from the Jackson Laboratory. For infection, mice were anesthetized via isoflurane inhalation (2%–2.5% with a 2-L/min oxygen flow rate). Intravaginal inoculations (total 1 × 106 tissue culture infectious units [TCIU] of HIVNL-BaL-eLuc per mouse) were performed by slowly pipetting 20 μL viral supernatant into the vaginal canal with a mid-size pipette tip. Intraperitoneal inoculations (total 1 × 105 TCIU of HIVNL-BaL-eLuc per mouse) were performed by injecting 100 μL indicated viral titer into the intraperitoneal cavity with a 29-gauge needle and insulin syringe. HIV infection in BLT mice was visualized using in vivo BLI to reveal longitudinal HIV dissemination. The chosen HIV-infected BLT mice were previously determined to show possible latency without detectable HIV-reporter activity in vivo for at least 70 days or longer prior to AAV-DJ/8/Cas9-sgRNA administration.
In Vivo AAV Injection
For the sgRNAs/saCas9 gene delivery efficiency and functional assay in Tg26 transgenic mice, 100 μL unpurified AAV- DJ serotype or 5 μL purified AAV- DJ/8 serotype diluted in 1× DMEM without serum in a total volume of 100 μL was injected into animals via the tail vein. For mice injected with AAV-DJ, group 1 was euthanized and group 2 received an additional injection 1 week after the first injection. Group 2 was euthanized 1 week after the second injection. Control mice were injected with AAV virus carrying an empty vector. For mice injected with AAV-DJ/8, the interval time between group 1 and group 2 was 2 weeks.
For in vivo injection of AAV-DJ/8 into NCr nude mice inoculated with EcoHIV-eLuc, a total of 10 μL purified AAV-DJ/8 containing saCas9/sgRNA (3.07 × 1014 GC/mL) was first diluted in 90 μL PBS for retro-orbital injection into the blood sinus of the right eye of each mouse right after a retro-orbital injection of a total of 100 μL EcoHIV-eLuc supernatant containing a total of 250 ng p24 determined by ELISA (ZeptoMetrix) at the same injection site.
The quadruplex sgRNAs/saCas9 AAV-DJ/8 delivery in BLT mice was performed via the intravaginal or intravenous route or via both routes, which is described in the text. Intravaginal inoculation was performed similarly to the aforementioned HIV inoculation, with a total of 20 μL PBS containing a total of 6.14 × 1012 GC of AAV-DJ/8/saCas9-sgRNA per BLT mouse. Intravenous inoculation was administered using an insulin syringe with a 29-gauge needle via the blood sinus of the right eye (retro-orbital injection) with 100 μL PBS containing total 6.14 × 1012 GC of AAV-DJ/8/saCas9-sgRNA per BLT mouse.
Bioluminescence Imaging
Imaging commenced at the indicated time points after viral inoculation. During all imaging, mice were anesthetized via isoflurane inhalation (2%–2.5% with a 2-L/min oxygen flow rate). D-luciferin potassium salt (Gold BioTechnology) was dissolved in sterile PBS and injected intraperitoneally at a dose of 150 mg/kg body weight. Bioluminescence images were acquired for 2 min (open light filter, binning 8 × 8, f/stop 1, 100-mm field of view) using an IVIS Lumina XR small animal optical imaging system (PerkinElmer).
Image Analysis
All image analysis was performed using Living Image 4.3.1 software. Regions of interest (ROIs) were drawn around the measured area or around the entire animal for comparison and average radiance values (in p/s/cm2/str) were used for all image evaluation. For display, images were windowed so that all light output above background levels adjacent to the mouse was seen. A maximum radiance value of 110,000 p/s/cm2/str was selected for the high end of the imaging scale and the background cutoff was set at 6,000 p/s/cm2/str, which was the average light emission of the mice imaged at two different time points prior to viral inoculation.
Genomic DNA, RNA, and Protein Extraction
Tissue samples of selected organs were broken up mechanically with a pestle and mortar, then processed with the Nucleospin tissue kit and the Nucleospin RNA/protein kit (Clontech) following the user manuals to extract genomic DNA, RNA, and protein. Extracted tissues were minced with sterilized and contaminant-free scissors and resulting tissue was added to a lysis buffer (2.5 mM Tris base, 5 mM EDTA, and 5 mM NaCl, pH 8.0) containing proteinase K and 1% SDS and incubated at 55°C overnight. Following incubation, the resulting mixture was extracted with one volume of phenol/CHCl3/isoamyl alcohol (25:24:1) via inversion for 10 min. The mixture was centrifuged at 12,000 rpm for 10 min, and the aqueous layer was removed and subjected to extraction once more with one volume of CHCl3/isoamyl alcohol (24:1) and separated via centrifugation as previously described. To precipitate the extracted DNA, one volume of isopropanol was added to the removed aqueous layer and the mixture was inverted for 15 min. The DNA was pelleted via centrifugation at 12,000 rpm for 15 min. The DNA pellet was then washed with 75% ethanol and air dried. Once nearly dry, TRIS-EDTA (TE) buffer (pH 8.0) was added to the pellet and was resolved at 55°C overnight.
TA Cloning and Sanger Sequencing
The bands of interest in agarose gels were excised and gel purified for cloning into the pCRII T-A vector (Invitrogen), and the nucleotide sequence of individual clones was determined by sequencing at GENEWIZ using universal T7 and/or SP6 primers.
Immunocytochemistry and Immunohistochemistry
Cells were fixed with 4% paraformaldehyde followed by standard immunocytochemistry with a monoclonal anti-HA antibody (1:200, catalog no. 66006-1-Ig; Proteintech). The ratio of saCas9-HA-positive cells to Hoechst-positive nuclei was quantified in six random fields per well for three wells. For immunohistochemistry, snap-frozen tissues/organs underwent cryostatic sectioning at 10 μm and were fixed with 4% paraformaldehyde for 10 min. After washing, permeation with 0.5% Triton X-100, and blocking with 10% normal donkey serum, the sections were incubated with rabbit anti-HA polyclonal antibody (1:100, catalog no. 51064-2-AP; Proteintech) in PBS with 0.1% Triton X-100 overnight at 4°C. After washing three times each for 10 min, the sections were incubated with corresponding Alexa Fluor-conjugated donkey secondary antibody (1:500; Invitrogen) and phalloidin (100 nM, catalog no. PHDR1; Cytoskeleton, Inc.) for 1 hr at room temperature. Hoechst 33258 was used for nuclear counterstaining.
Statistical Analysis
The quantitative data represent the mean ± SD from two to four independent experiments and were evaluated by the Student’s t test. p values that were < 0.05 or 0.01 were considered statistically significant. For BLI data in the EcoHIV infection mouse model, the Student’s t test and linear mixed-effects models were used to compare total photon flux change over time between saCas9 plus EcoHIV versus EcoHIV only, which was measured by BLI from both the dorsal and ventral sides, respectively.
Author Contributions
C.Y., T.Z., Y.Z., X.X., and F.L. performed the vector cloning, sequencing, cell cultures, PCR, and immunostaining. F.L., R.P., S.D., and Xu.Q. did animal breeding, genotyping, and tail-vein injection. W.H., K.K., W.-B.Y., W.X., and X.M. conceived the experimental design and helped with data analysis/interpretation and preparation of the manuscript. W.-B.Y. and Xi.Q. performed in vivo studies and longitudinal live imaging of EcoHIVNL4.3-eLuc and HIV-NL-BaL-eLuc infection in NCr mice and humanized BLT mice for AAV/saCas9 treatment. C.Y., T.Z., W.-B.Y., and W.H. prepared the figures. H.Z. performed the statistical analysis. W.H., W.-B.Y., C.Y., and T.Z. prepared the manuscript. The final manuscript was reviewed and approved by all authors.
Conflicts of Interest
W.H., K.K., and Y.Z. are named inventors on patents that cover the viral gene editing technology that is the subject of this manuscript. In addition to the foregoing interests, K.K. is a co-founder and scientific advisor and holds equity in Excision Biotherapeutics, a biotech start-up that has licensed the viral gene editing technology from Temple University for commercial development and clinical trials. W.H. is a scientific advisor for Excision Biotherapeutics. The authors declare that this work was produced solely by the authors and that no other individuals or entities influenced any aspects of the work including, but not limited to, the study conception and design; data acquisition, analysis and interpretation; and writing of the manuscript.
Acknowledgments
We thank Dr. Jennifer Gordon for expertise, advice, and support of the studies with Tg26 transgenic mice, as well as Jessica Otte for assistance with Tg26 mice. This work was supported by NIH grants R01NS087971 (to W.H. and K.K.), P30MH092177 (to K.K.), and R21MH100949 and R21NS094084 (to W.-B.Y.). R.P. was supported by NIH grant 5T32MH079785. This study utilized services offered by core facilities of the Comprehensive NeuroAIDS Center (CNAC) (NIH grant P30MH092177) at Temple University Lewis Katz School of Medicine.
Footnotes
Supplemental Information includes twelve figures and can be found with this article online at http://dx.doi.org/10.1016/j.ymthe.2017.03.012.
Contributor Information
Won-Bin Young, Email: bioimaging@mail.com.
Kamel Khalili, Email: kamel.khalili@temple.edu.
Wenhui Hu, Email: whu@temple.edu.
Supplemental Information
References
- 1.Kleinstiver B.P., Prew M.S., Tsai S.Q., Topkar V.V., Nguyen N.T., Zheng Z., Gonzales A.P., Li Z., Peterson R.T., Yeh J.R. Engineered CRISPR-Cas9 nucleases with altered PAM specificities. Nature. 2015;523:481–485. doi: 10.1038/nature14592. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Ran F.A., Cong L., Yan W.X., Scott D.A., Gootenberg J.S., Kriz A.J., Zetsche B., Shalem O., Wu X., Makarova K.S. In vivo genome editing using Staphylococcus aureus Cas9. Nature. 2015;520:186–191. doi: 10.1038/nature14299. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Hastie E., Samulski R.J. Adeno-associated virus at 50: a golden anniversary of discovery, research, and gene therapy success--a personal perspective. Hum. Gene Ther. 2015;26:257–265. doi: 10.1089/hum.2015.025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Swiech L., Heidenreich M., Banerjee A., Habib N., Li Y., Trombetta J., Sur M., Zhang F. In vivo interrogation of gene function in the mammalian brain using CRISPR-Cas9. Nat. Biotechnol. 2015;33:102–106. doi: 10.1038/nbt.3055. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Zuris J.A., Thompson D.B., Shu Y., Guilinger J.P., Bessen J.L., Hu J.H., Maeder M.L., Joung J.K., Chen Z.Y., Liu D.R. Cationic lipid-mediated delivery of proteins enables efficient protein-based genome editing in vitro and in vivo. Nat. Biotechnol. 2015;33:73–80. doi: 10.1038/nbt.3081. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Yin H., Xue W., Chen S., Bogorad R.L., Benedetti E., Grompe M., Koteliansky V., Sharp P.A., Jacks T., Anderson D.G. Genome editing with Cas9 in adult mice corrects a disease mutation and phenotype. Nat. Biotechnol. 2014;32:551–553. doi: 10.1038/nbt.2884. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Yin H., Song C.Q., Dorkin J.R., Zhu L.J., Li Y., Wu Q., Park A., Yang J., Suresh S., Bizhanova A. Therapeutic genome editing by combined viral and non-viral delivery of CRISPR system components in vivo. Nat. Biotechnol. 2016;34:328–333. doi: 10.1038/nbt.3471. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Manjunath N., Yi G., Dang Y., Shankar P. Newer gene editing technologies toward HIV gene therapy. Viruses. 2013;5:2748–2766. doi: 10.3390/v5112748. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Stone D., Kiem H.P., Jerome K.R. Targeted gene disruption to cure HIV. Curr. Opin. HIV AIDS. 2013;8:217–223. doi: 10.1097/COH.0b013e32835f736c. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Wang W., Ye C., Liu J., Zhang D., Kimata J.T., Zhou P. CCR5 gene disruption via lentiviral vectors expressing Cas9 and single guided RNA renders cells resistant to HIV-1 infection. PLoS ONE. 2014;9:e115987. doi: 10.1371/journal.pone.0115987. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Hu W., Kaminski R., Yang F., Zhang Y., Cosentino L., Li F., Luo B., Alvarez-Carbonell D., Garcia-Mesa Y., Karn J. RNA-directed gene editing specifically eradicates latent and prevents new HIV-1 infection. Proc. Natl. Acad. Sci. USA. 2014;111:11461–11466. doi: 10.1073/pnas.1405186111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Ebina H., Misawa N., Kanemura Y., Koyanagi Y. Harnessing the CRISPR/Cas9 system to disrupt latent HIV-1 provirus. Sci. Rep. 2013;3:2510. doi: 10.1038/srep02510. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Liao H.K., Gu Y., Diaz A., Marlett J., Takahashi Y., Li M., Suzuki K., Xu R., Hishida T., Chang C.J. Use of the CRISPR/Cas9 system as an intracellular defense against HIV-1 infection in human cells. Nat. Commun. 2015;6:6413. doi: 10.1038/ncomms7413. [DOI] [PubMed] [Google Scholar]
- 14.Zhu W., Lei R., Le Duff Y., Li J., Guo F., Wainberg M.A., Liang C. The CRISPR/Cas9 system inactivates latent HIV-1 proviral DNA. Retrovirology. 2015;12:22. doi: 10.1186/s12977-015-0150-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Kaminski R., Chen Y., Fischer T., Tedaldi E., Napoli A., Zhang Y., Karn J., Hu W., Khalili K. Elimination of HIV-1 genomes from human T-lymphoid cells by CRISPR/Cas9 gene editing. Sci. Rep. 2016;6:22555. doi: 10.1038/srep22555. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Yin C., Zhang T., Li F., Yang F., Putatunda R., Young W.B., Khalili K., Hu W., Zhang Y. Functional screening of guide RNAs targeting the regulatory and structural HIV-1 viral genome for a cure of AIDS. AIDS. 2016;30:1163–1174. doi: 10.1097/QAD.0000000000001079. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Kaminski R., Bella R., Yin C., Otte J., Ferrante P., Gendelman H.E., Li H., Booze R., Gordon J., Hu W., Khalili K. Excision of HIV-1 DNA by gene editing: a proof-of-concept in vivo study. Gene Ther. 2016;23:690–695. doi: 10.1038/gt.2016.41. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Rabinovich B.A., Ye Y., Etto T., Chen J.Q., Levitsky H.I., Overwijk W.W., Cooper L.J., Gelovani J., Hwu P. Visualizing fewer than 10 mouse T cells with an enhanced firefly luciferase in immunocompetent mouse models of cancer. Proc. Natl. Acad. Sci. USA. 2008;105:14342–14346. doi: 10.1073/pnas.0804105105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Potash M.J., Chao W., Bentsman G., Paris N., Saini M., Nitkiewicz J., Belem P., Sharer L., Brooks A.I., Volsky D.J. A mouse model for study of systemic HIV-1 infection, antiviral immune responses, and neuroinvasiveness. Proc. Natl. Acad. Sci. USA. 2005;102:3760–3765. doi: 10.1073/pnas.0500649102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Khalili K., Kaminski R., Gordon J., Cosentino L., Hu W. Genome editing strategies: potential tools for eradicating HIV-1/AIDS. J. Neurovirol. 2015;21:310–321. doi: 10.1007/s13365-014-0308-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Nishimasu H., Cong L., Yan W.X., Ran F.A., Zetsche B., Li Y., Kurabayashi A., Ishitani R., Zhang F., Nureki O. Crystal structure of Staphylococcus aureus Cas9. Cell. 2015;162:1113–1126. doi: 10.1016/j.cell.2015.08.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Friedland A.E., Baral R., Singhal P., Loveluck K., Shen S., Sanchez M., Marco E., Gotta G.M., Maeder M.L., Kennedy E.M. Characterization of Staphylococcus aureus Cas9: a smaller Cas9 for all-in-one adeno-associated virus delivery and paired nickase applications. Genome Biol. 2015;16:257. doi: 10.1186/s13059-015-0817-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Wu Z., Yang H., Colosi P. Effect of genome size on AAV vector packaging. Mol. Ther. 2010;18:80–86. doi: 10.1038/mt.2009.255. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Allocca M., Doria M., Petrillo M., Colella P., Garcia-Hoyos M., Gibbs D., Kim S.R., Maguire A., Rex T.S., Di Vicino U. Serotype-dependent packaging of large genes in adeno-associated viral vectors results in effective gene delivery in mice. J. Clin. Invest. 2008;118:1955–1964. doi: 10.1172/JCI34316. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Grieger J.C., Samulski R.J. Packaging capacity of adeno-associated virus serotypes: impact of larger genomes on infectivity and postentry steps. J. Virol. 2005;79:9933–9944. doi: 10.1128/JVI.79.15.9933-9944.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Wu J., Zhao W., Zhong L., Han Z., Li B., Ma W., Weigel-Kelley K.A., Warrington K.H., Srivastava A. Self-complementary recombinant adeno-associated viral vectors: packaging capacity and the role of rep proteins in vector purity. Hum. Gene Ther. 2007;18:171–182. doi: 10.1089/hum.2006.088. [DOI] [PubMed] [Google Scholar]
- 27.Grimm D., Lee J.S., Wang L., Desai T., Akache B., Storm T.A., Kay M.A. In vitro and in vivo gene therapy vector evolution via multispecies interbreeding and retargeting of adeno-associated viruses. J. Virol. 2008;82:5887–5911. doi: 10.1128/JVI.00254-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Bartel M., Schaffer D., Büning H. Enhancing the clinical potential of AAV vectors by capsid engineering to evade pre-existing immunity. Front. Microbiol. 2011;2:204. doi: 10.3389/fmicb.2011.00204. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Dickie P., Felser J., Eckhaus M., Bryant J., Silver J., Marinos N., Notkins A.L. HIV-associated nephropathy in transgenic mice expressing HIV-1 genes. Virology. 1991;185:109–119. doi: 10.1016/0042-6822(91)90759-5. [DOI] [PubMed] [Google Scholar]
- 30.Kopp J.B., Klotman M.E., Adler S.H., Bruggeman L.A., Dickie P., Marinos N.J., Eckhaus M., Bryant J.L., Notkins A.L., Klotman P.E. Progressive glomerulosclerosis and enhanced renal accumulation of basement membrane components in mice transgenic for human immunodeficiency virus type 1 genes. Proc. Natl. Acad. Sci. USA. 1992;89:1577–1581. doi: 10.1073/pnas.89.5.1577. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Haque S., Lan X., Wen H., Lederman R., Chawla A., Attia M., Bongu R.P., Husain M., Mikulak J., Saleem M.A. HIV promotes NLRP3 inflammasome complex activation in murine HIV-associated nephropathy. Am. J. Pathol. 2016;186:347–358. doi: 10.1016/j.ajpath.2015.10.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Cheung J.Y., Gordon J., Wang J., Song J., Zhang X.Q., Tilley D.G., Gao E., Koch W.J., Rabinowitz J., Klotman P.E. Cardiac dysfunction in HIV-1 transgenic mouse: role of stress and BAG3. Clin. Transl. Sci. 2015;8:305–310. doi: 10.1111/cts.12331. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Hahn Y.K., Podhaizer E.M., Hauser K.F., Knapp P.E. HIV-1 alters neural and glial progenitor cell dynamics in the central nervous system: coordinated response to opiates during maturation. Glia. 2012;60:1871–1887. doi: 10.1002/glia.22403. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Das S., Basu A. Viral infection and neural stem/progenitor cell’s fate: implications in brain development and neurological disorders. Neurochem. Int. 2011;59:357–366. doi: 10.1016/j.neuint.2011.02.020. [DOI] [PubMed] [Google Scholar]
- 35.Mishra M., Taneja M., Malik S., Khalique H., Seth P. Human immunodeficiency virus type 1 Tat modulates proliferation and differentiation of human neural precursor cells: implication in NeuroAIDS. J. Neurovirol. 2010;16:355–367. doi: 10.3109/13550284.2010.513028. [DOI] [PubMed] [Google Scholar]
- 36.Rothenaigner I., Kramer S., Ziegler M., Wolff H., Kleinschmidt A., Brack-Werner R. Long-term HIV-1 infection of neural progenitor populations. AIDS. 2007;21:2271–2281. doi: 10.1097/QAD.0b013e3282f12f27. [DOI] [PubMed] [Google Scholar]
- 37.Schwartz L., Civitello L., Dunn-Pirio A., Ryschkewitsch S., Berry E., Cavert W., Kinzel N., Lawrence D.M., Hazra R., Major E.O. Evidence of human immunodeficiency virus type 1 infection of nestin-positive neural progenitors in archival pediatric brain tissue. J. Neurovirol. 2007;13:274–283. doi: 10.1080/13550280701344975. [DOI] [PubMed] [Google Scholar]
- 38.Fatima M., Kumari R., Schwamborn J.C., Mahadevan A., Shankar S.K., Raja R., Seth P. Tripartite containing motif 32 modulates proliferation of human neural precursor cells in HIV-1 neurodegeneration. Cell Death Differ. 2016;23:776–786. doi: 10.1038/cdd.2015.138. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Schneider M., Tigges B., Meggendorfer M., Helfer M., Ziegenhain C., Brack-Werner R. A new model for post-integration latency in macroglial cells to study HIV-1 reservoirs of the brain. AIDS. 2015;29:1147–1159. doi: 10.1097/QAD.0000000000000691. [DOI] [PubMed] [Google Scholar]
- 40.Balinang J.M., Masvekar R.R., Hauser K.F., Knapp P.E. Productive infection of human neural progenitor cells by R5 tropic HIV-1: opiate co-exposure heightens infectivity and functional vulnerability. AIDS. 2017;31:753–764. doi: 10.1097/QAD.0000000000001398. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Mao Y., Wang X., Yan R., Hu W., Li A., Wang S., Li H. Single point mutation in adeno-associated viral vectors -DJ capsid leads to improvement for gene delivery in vivo. BMC Biotechnol. 2016;16:1. doi: 10.1186/s12896-015-0230-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Yang Y., Wang L., Bell P., McMenamin D., He Z., White J., Yu H., Xu C., Morizono H., Musunuru K. A dual AAV system enables the Cas9-mediated correction of a metabolic liver disease in newborn mice. Nat. Biotechnol. 2016;34:334–338. doi: 10.1038/nbt.3469. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Holehonnur R., Luong J.A., Chaturvedi D., Ho A., Lella S.K., Hosek M.P., Ploski J.E. Adeno-associated viral serotypes produce differing titers and differentially transduce neurons within the rat basal and lateral amygdala. BMC Neurosci. 2014;15:28. doi: 10.1186/1471-2202-15-28. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Tsai S.Q., Zheng Z., Nguyen N.T., Liebers M., Topkar V.V., Thapar V., Wyvekens N., Khayter C., Iafrate A.J., Le L.P. GUIDE-seq enables genome-wide profiling of off-target cleavage by CRISPR-Cas nucleases. Nat. Biotechnol. 2015;33:187–197. doi: 10.1038/nbt.3117. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Frock R.L., Hu J., Meyers R.M., Ho Y.J., Kii E., Alt F.W. Genome-wide detection of DNA double-stranded breaks induced by engineered nucleases. Nat. Biotechnol. 2015;33:179–186. doi: 10.1038/nbt.3101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Smith C., Gore A., Yan W., Abalde-Atristain L., Li Z., He C., Wang Y., Brodsky R.A., Zhang K., Cheng L., Ye Z. Whole-genome sequencing analysis reveals high specificity of CRISPR/Cas9 and TALEN-based genome editing in human iPSCs. Cell Stem Cell. 2014;15:12–13. doi: 10.1016/j.stem.2014.06.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Veres A., Gosis B.S., Ding Q., Collins R., Ragavendran A., Brand H., Erdin S., Cowan C.A., Talkowski M.E., Musunuru K. Low incidence of off-target mutations in individual CRISPR-Cas9 and TALEN targeted human stem cell clones detected by whole-genome sequencing. Cell Stem Cell. 2014;15:27–30. doi: 10.1016/j.stem.2014.04.020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Zhang Y., Yin C., Zhang T., Li F., Yang W., Kaminski R., Fagan P.R., Putatunda R., Young W.B., Khalili K., Hu W. CRISPR/gRNA-directed synergistic activation mediator (SAM) induces specific, persistent and robust reactivation of the HIV-1 latent reservoirs. Sci. Rep. 2015;5:16277. doi: 10.1038/srep16277. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Kelschenbach J.L., Saini M., Hadas E., Gu C.J., Chao W., Bentsman G., Hong J.P., Hanke T., Sharer L.R., Potash M.J. Mice chronically infected with chimeric HIV resist peripheral and brain superinfection: a model of protective immunity to HIV. J. Neuroimmune Pharmacol. 2012;7:380–387. doi: 10.1007/s11481-011-9316-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Song J., Cai Z., White A.G., Jin T., Wang X., Kadayakkara D., Anderson C.J., Ambrose Z., Young W.B. Visualization and quantification of simian immunodeficiency virus-infected cells using non-invasive molecular imaging. J. Gen. Virol. 2015;96:3131–3142. doi: 10.1099/jgv.0.000245. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Fuchs S.P., Desrosiers R.C. Promise and problems associated with the use of recombinant AAV for the delivery of anti-HIV antibodies. Mol. Ther. Methods Clin. Dev. 2016;3:16068. doi: 10.1038/mtm.2016.68. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Brady J.M., Baltimore D., Balazs A.B. Antibody gene transfer with adeno-associated viral vectors as a method for HIV prevention. Immunol. Rev. 2017;275:324–333. doi: 10.1111/imr.12478. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Wang Z., Pan Q., Gendron P., Zhu W., Guo F., Cen S., Wainberg M.A., Liang C. CRISPR/Cas9-derived mutations both inhibit HIV-1 replication and accelerate viral escape. Cell Rep. 2016;15:481–489. doi: 10.1016/j.celrep.2016.03.042. [DOI] [PubMed] [Google Scholar]
- 54.Wang G., Zhao N., Berkhout B., Das A.T. CRISPR-Cas9 can inhibit HIV-1 replication but NHEJ repair facilitates virus escape. Mol. Ther. 2016;24:522–526. doi: 10.1038/mt.2016.24. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Bauer D.E., Canver M.C., Orkin S.H. Generation of genomic deletions in mammalian cell lines via CRISPR/Cas9. J. Vis. Exp. 2015;95:e52118. doi: 10.3791/52118. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Peng J., Zhou Y., Zhu S., Wei W. High-throughput screens in mammalian cells using the CRISPR-Cas9 system. FEBS J. 2015;282:2089–2096. doi: 10.1111/febs.13251. [DOI] [PubMed] [Google Scholar]
- 57.Kabadi A.M., Ousterout D.G., Hilton I.B., Gersbach C.A. Multiplex CRISPR/Cas9-based genome engineering from a single lentiviral vector. Nucleic Acids Res. 2014;42:e147. doi: 10.1093/nar/gku749. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Rothe M., Schambach A., Biasco L. Safety of gene therapy: new insights to a puzzling case. Curr. Gene Ther. 2014;14:429–436. doi: 10.2174/1566523214666140918110905. [DOI] [PubMed] [Google Scholar]
- 59.Mingozzi F., High K.A. Immune responses to AAV vectors: overcoming barriers to successful gene therapy. Blood. 2013;122:23–36. doi: 10.1182/blood-2013-01-306647. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Moser R.J., Hirsch M.L. AAV vectorization of DSB-mediated gene editing technologies. Curr. Gene Ther. 2016;16:207–219. doi: 10.2174/1566523216666160602213738. [DOI] [PubMed] [Google Scholar]
- 61.Platt R.J., Chen S., Zhou Y., Yim M.J., Swiech L., Kempton H.R., Dahlman J.E., Parnas O., Eisenhaure T.M., Jovanovic M. CRISPR-Cas9 knockin mice for genome editing and cancer modeling. Cell. 2014;159:440–455. doi: 10.1016/j.cell.2014.09.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Senís E., Fatouros C., Große S., Wiedtke E., Niopek D., Mueller A.K., Börner K., Grimm D. CRISPR/Cas9-mediated genome engineering: an adeno-associated viral (AAV) vector toolbox. Biotechnol. J. 2014;9:1402–1412. doi: 10.1002/biot.201400046. [DOI] [PubMed] [Google Scholar]
- 63.Howes R., Schofield C. Genome engineering using adeno-associated virus (AAV) Methods Mol. Biol. 2015;1239:75–103. doi: 10.1007/978-1-4939-1862-1_5. [DOI] [PubMed] [Google Scholar]
- 64.Hung S.S., Chrysostomou V., Li F., Lim J.K., Wang J.H., Powell J.E., Tu L., Daniszewski M., Lo C., Wong R.C. AAV-mediated CRISPR/Cas gene editing of retinal cells in vivo. Invest. Ophthalmol. Vis. Sci. 2016;57:3470–3476. doi: 10.1167/iovs.16-19316. [DOI] [PubMed] [Google Scholar]
- 65.Wright A.V., Sternberg S.H., Taylor D.W., Staahl B.T., Bardales J.A., Kornfeld J.E., Doudna J.A. Rational design of a split-Cas9 enzyme complex. Proc. Natl. Acad. Sci. USA. 2015;112:2984–2989. doi: 10.1073/pnas.1501698112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Zetsche B., Volz S.E., Zhang F. A split-Cas9 architecture for inducible genome editing and transcription modulation. Nat. Biotechnol. 2015;33:139–142. doi: 10.1038/nbt.3149. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Nguyen D.P., Miyaoka Y., Gilbert L.A., Mayerl S.J., Lee B.H., Weissman J.S., Conklin B.R., Wells J.A. Ligand-binding domains of nuclear receptors facilitate tight control of split CRISPR activity. Nat. Commun. 2016;7:12009. doi: 10.1038/ncomms12009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Nuñez J.K., Harrington L.B., Doudna J.A. Chemical and biophysical modulation of Cas9 for tunable genome engineering. ACS Chem. Biol. 2016;11:681–688. doi: 10.1021/acschembio.5b01019. [DOI] [PubMed] [Google Scholar]
- 69.Fine E.J., Appleton C.M., White D.E., Brown M.T., Deshmukh H., Kemp M.L., Bao G. Trans-spliced Cas9 allows cleavage of HBB and CCR5 genes in human cells using compact expression cassettes. Sci. Rep. 2015;5:10777. doi: 10.1038/srep10777. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Nissim L., Perli S.D., Fridkin A., Perez-Pinera P., Lu T.K. Multiplexed and programmable regulation of gene networks with an integrated RNA and CRISPR/Cas toolkit in human cells. Mol. Cell. 2014;54:698–710. doi: 10.1016/j.molcel.2014.04.022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Gao Y., Zhao Y. Self-processing of ribozyme-flanked RNAs into guide RNAs in vitro and in vivo for CRISPR-mediated genome editing. J. Integr. Plant Biol. 2014;56:343–349. doi: 10.1111/jipb.12152. [DOI] [PubMed] [Google Scholar]
- 72.Sakuma T., Nishikawa A., Kume S., Chayama K., Yamamoto T. Multiplex genome engineering in human cells using all-in-one CRISPR/Cas9 vector system. Sci. Rep. 2014;4:5400. doi: 10.1038/srep05400. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Denton P.W., Olesen R., Choudhary S.K., Archin N.M., Wahl A., Swanson M.D., Chateau M., Nochi T., Krisko J.F., Spagnuolo R.A. Generation of HIV latency in humanized BLT mice. J. Virol. 2012;86:630–634. doi: 10.1128/JVI.06120-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Wang M., Zuris J.A., Meng F., Rees H., Sun S., Deng P., Han Y., Gao X., Pouli D., Wu Q. Efficient delivery of genome-editing proteins using bioreducible lipid nanoparticles. Proc. Natl. Acad. Sci. USA. 2016;113:2868–2873. doi: 10.1073/pnas.1520244113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Sun W., Ji W., Hall J.M., Hu Q., Wang C., Beisel C.L., Gu Z. Self-assembled DNA nanoclews for the efficient delivery of CRISPR-Cas9 for genome editing. Angew. Chem. Int. Ed. Engl. 2015;54:12029–12033. doi: 10.1002/anie.201506030. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Hu P., Li Y., Sands M.S., McCown T., Kafri T. Generation of a stable packaging cell line producing high-titer PPT-deleted integration-deficient lentiviral vectors. Mol. Ther. Methods Clin. Dev. 2015;2:15025. doi: 10.1038/mtm.2015.25. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Yu X., Liang X., Xie H., Kumar S., Ravinder N., Potter J., de Mollerat du Jeu X., Chesnut J.D. Improved delivery of Cas9 protein/gRNA complexes using lipofectamine CRISPRMAX. Biotechnol. Lett. 2016;38:919–929. doi: 10.1007/s10529-016-2064-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Tycko J., Myer V.E., Hsu P.D. Methods for optimizing CRISPR-Cas9 genome editing specificity. Mol. Cell. 2016;63:355–370. doi: 10.1016/j.molcel.2016.07.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Kleinstiver B.P., Prew M.S., Tsai S.Q., Nguyen N.T., Topkar V.V., Zheng Z., Joung J.K. Broadening the targeting range of Staphylococcus aureus CRISPR-Cas9 by modifying PAM recognition. Nat. Biotechnol. 2015;33:1293–1298. doi: 10.1038/nbt.3404. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Kim D., Kim S., Kim S., Park J., Kim J.S. Genome-wide target specificities of CRISPR-Cas9 nucleases revealed by multiplex Digenome-seq. Genome Res. 2016;26:406–415. doi: 10.1101/gr.199588.115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Kim D., Bae S., Park J., Kim E., Kim S., Yu H.R., Hwang J., Kim J.I., Kim J.S. Digenome-seq: genome-wide profiling of CRISPR-Cas9 off-target effects in human cells. Nat. Methods. 2015;12:237–243. doi: 10.1038/nmeth.3284. [DOI] [PubMed] [Google Scholar]
- 82.Zuckermann M., Hovestadt V., Knobbe-Thomsen C.B., Zapatka M., Northcott P.A., Schramm K., Belic J., Jones D.T., Tschida B., Moriarity B. Somatic CRISPR/Cas9-mediated tumour suppressor disruption enables versatile brain tumour modelling. Nat. Commun. 2015;6:7391. doi: 10.1038/ncomms8391. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.Yang L., Grishin D., Wang G., Aach J., Zhang C.Z., Chari R., Homsy J., Cai X., Zhao Y., Fan J.B. Targeted and genome-wide sequencing reveal single nucleotide variations impacting specificity of Cas9 in human stem cells. Nat. Commun. 2014;5:5507. doi: 10.1038/ncomms6507. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84.Zolotukhin S., Byrne B.J., Mason E., Zolotukhin I., Potter M., Chesnut K., Summerford C., Samulski R.J., Muzyczka N. Recombinant adeno-associated virus purification using novel methods improves infectious titer and yield. Gene Ther. 1999;6:973–985. doi: 10.1038/sj.gt.3300938. [DOI] [PubMed] [Google Scholar]
- 85.al Yacoub N., Romanowska M., Haritonova N., Foerster J. Optimized production and concentration of lentiviral vectors containing large inserts. J. Gene Med. 2007;9:579–584. doi: 10.1002/jgm.1052. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.