Summary
We examined the effect of diet on pre-hibernation fattening and the gut microbiota of captive arctic ground squirrels (Urocitellus parryii). We measured body composition across time and gut microbiota density, diversity, and function prior to and after five-weeks on control, high-fat, low-fat (18%, 40%, and 10% energy from fat, respectively), or restricted calorie (50% of control) diets. Squirrels fattened at the same rate and to the same degree on all diets. Additionally, we found no differences in gut microbiota diversity or short chain fatty acid production across time or with diet. Analysis of the gut microbial transcriptome indicated differences in community function among diet groups, but not across time, and revealed shifts in the relative contribution of function at a taxonomic level. Our results demonstrate that pre-hibernation fattening of arctic ground squirrels is robust to changes in diet and is accomplished by more than increased food intake. Although our analyses did not uncover a definitive link between host fattening and the gut microbiota, and suggest the squirrels may possess a gut microbial community structure that is unresponsive to dietary changes, studies manipulating diet earlier in the active season may yet uncover a relationship between host diet, fattening and gut microbiota.
Introduction
Seasonal environments that encompass annual cycles of temperature, precipitation, and resource availability have served as selective forces shaping the evolution of molecular, physiological, and behavioral adaptations in indigenous species. Arguably, one of the most extreme examples of this is mammalian hibernation, an adaptation to survive unfavorable conditions characterized by a state of low activity, metabolic rate and body temperature (Tb; Lyman 1982). Obligate seasonal hibernators, such as ground squirrels, annually undergo profound endogenously programmed physiological and behavioral changes to conserve energy during hibernation and to increase endogenous energy reserves in preparation for hibernation (Williams et al., 2014; Dark, 2005). Although a great deal is known about the physiological, metabolic and molecular changes that occur during hibernation (reviewed in Carey et al., 2003; Geiser, 2004; Dark, 2005; van Breukelen and Martin 2015; Williams et al., 2011), few studies have investigated the physiological and behavioral changes of free-living animals that occur across the active season in preparation for hibernation (Buck and Barnes, 1999a; Sheriff et al., 2013; Williams et al., 2014).
For many species, hibernation is fueled principally by endogenous energy reserves, primarily in the form of fat. In these animals, the majority of fat mass is gained late in the active season (Dark, 2005), accumulating over a discrete period following reproduction known as “pre-hibernation fattening” (Körtner and Heidmaier, 1994; Kunz et al., 1998; Sheriff et al., 2013). During this period, animals often double their body mass, and can increase their lipid stores as much as seven-fold (Mrosovsky, 1976; Kunz et al., 1998; Buck and Barnes, 1999a). While the mechanisms involved in such robust fattening are not well understood, seasonal fat-storing hibernators facilitate lipid accumulation through shifts in energy utilization (Sheriff et al., 2012; 2013).
In non-hibernating mammals, fat accumulation and storage has been linked to the gut microbiota. For example, germ-free mice maintain a lower percentage of body fat than conventional mice (Smith et al., 2007) despite consuming equal or greater amounts of food. Additionally, the obese phenotype is transmitted to germ-free (gf) mice through introduction of “obese” gut microbes; a result that is not achieved by the transfer of “lean” gut microbes to gf mice (Ridaura et al., 2013). In some studies, obesity in humans, mice, and rats correlates with reduced gut microbial diversity and a characteristic shift in relative abundance of core gut microbial community members (increased relative abundance of the phyla Firmicutesto Bacteroidetes) compared to lean individuals (Ley et al., 2005; Turnbaugh and Gordon, 2009; Ley et al., 2010); although in others, a different relationship or no relationship between the Firmicutesto Bacteroidetesratio and obesity has been reported (Duncan et al., 2007; Duncan et al., 2008; Murphy et al., 2010; Schwiertz et al., 2010). The “lean” gut microbial metatranscriptome is enriched in transcripts encoding enzymes that participate in carbohydrate metabolism and protein degradation (Riduara et al., 2013), and co-housing obese mice with lean mice prevents additional fattening and transforms their gut microbial metatranscriptome to a lean-like state as a consequence of invasion of lean microbiota (Riduara et al., 2013). Germ-free mice are also able to resist diet-induced obesity when fed a high-fat, sucrose rich “Westernized” diet (Bäckhead et al., 2007; Ridaura et al., 2013). Interestingly, microbial-linked obesity in mice can be rescued with the introduction of “lean” gut microbiota; however, rescue is diet-dependent and occurs in mice maintained on a diet low in fat and rich in dietary fiber, but not on the “Westernized” diet (Ridaura et al., 2013).
Arctic ground squirrels (Urocitellus parryii) serve as an excellent model for studying the effects of diet and gut microbiota in hibernators. As the northernmost hibernating mammal, they have evolved the most extreme annual cycle of physiology and behavior in response to the severity of the arctic environment (Mayer and Roche, 1954; Hock, 1960). The active season lasts a brief three to five months, depending on sex and age (Buck et al., 2008), during which squirrels must compete for mates (males only; Buck and Barnes, 2003; Richter et al. 2017), complete gestation and lactation, and fatten to survive the upcoming hibernation season (both sexes; Buck and Barnes, 1999a). During hibernation, arctic ground squirrels regulate Tbat -2.9°C, reduce metabolic rate to as low as 1% of basal, and are capable of defending a thermal gradient between core Tband ambient temperature of at least 23°C during deep torpor (Barnes, 1989; Buck and Barnes, 2000; Richter et al., 2015). Torpor bouts last for ∼20 days on average while sequestered in their subfreezing hibernacula (Buck and Barnes, 1999b; Buck et al., 2008).
Impressively, arctic ground squirrels can acquire sufficient fat stores to enter hibernation in as little as three-weeks (Buck & Barnes, 1999a). To facilitate fattening, arctic ground squirrels are known to minimize energy expenditure through reduced daily activity, decreased metabolic rate, and lowered Tb(Sheriff et al., 2012; Sheriff et al., 2013). It has also been suggested that shifts in food preference, or availability, toward more energy dense forage such as seeds, berries and mushrooms, may result in an overall increase in energy intake (Batzli and Sobaski, 1980; Mclean, 1984; Gillis et al., 2005; Zazula et al., 2005). While some observational data exists to describe the diet of arctic ground squirrels, the effect of diet on fattening in hibernators remains speculative, and even less is known about the effect of diet on their gut microbial communities. However, given the link between the gut microbial community and obesity, it stands to reason that hibernating mammals may possess a gut microbiota that promotes fattening, and evidence exists to suggest this may be the case. For example, the active season gut microbiota of the arctic ground squirrel is characterized by a high ratio of Firmicutesto Bacteroidetes(Stevenson et al., 2014a) as compared to hibernating squirrels (Stevenson et al., 2014b); a profile that favors fattening in non-hibernating mammals (Ley et al., 2006; Turnbaugh and Gordon, 2009). Additionally, the colonization of germ-free mice with gut microbiota from free-living brown bears effectively transfers metabolic features to the mice, including an increased propensity to fatten (Sommer et al., 2016). To date, no study has explored the influence of diet and the gut microbiota on fattening of a hibernating species.
In this study, we set out to determine if changes in dietary fat content and caloric intake affect the rate and degree of pre-hibernation fattening in wild-caught arctic ground squirrels. Additionally, we sought to uncover the relationship between fattening, diet and the gut microbial community, by characterizing the gut microbial diversity, density, metabolic activity, and metatranscriptome in response to dietary manipulation. We hypothesized that squirrels fed diets higher in fat content or calories would have higher rates and degrees of fattening. Also, given the effect of diet on the gut microbial community of non-hibernators, we postulated that diet composition would impact the gut microbiota of arctic ground squirrels, with high fat diet consumption leading to lower total bacterial diversity and an increase in the relative abundance of Firmicutescompared to Bacteroidetes. Furthermore, we predicted that diet manipulation would alter gut microbial gene expression in a way that reflects the predicted changes in gut microbial community structure (i.e., increased gene transcripts attributable to Firmicutescompared to Bacteroidetesin squirrels consuming either high fat or high calorie diets).
Results
Body Composition
The mean body mass within groups (HF, LF, RD, SD) at T0(5-weeks post-hibernation) did not differ from the mean body mass at 10-weeks post-hibernation. Body mass did not differ among diet treatments (SD, HF, LF, RD; Figure 1A); average mass of animals across diet treatments was 700.8 ± 126.0 g at 10 weeks post-hibernation. The mean percent fat mass within groups (HF, LF, RD, SD) was higher at 10-weeks post-hibernation as compared to T0(p-value < 0.05; Figure 1B). Percent fat mass increased from 23.1 ± 8.9 to 33.0 ± 10.0 in HF (p = 0.037), 23.2 ± 3.6 to 27.8 ± 5.8 in LF (p = 0.005), 25.8 ± 7.0 to 29.3 ± 7.4 in RD (p = 0.037), 23.3 ± 13.4 to 30.1 ± 7.7 in the control (SD; p = 0.037) from T0to 10-weeks post-hibernation (p<0.001; Figure 1B). No significant differences in percent body fat existed among diet treatments.
Figure 1.
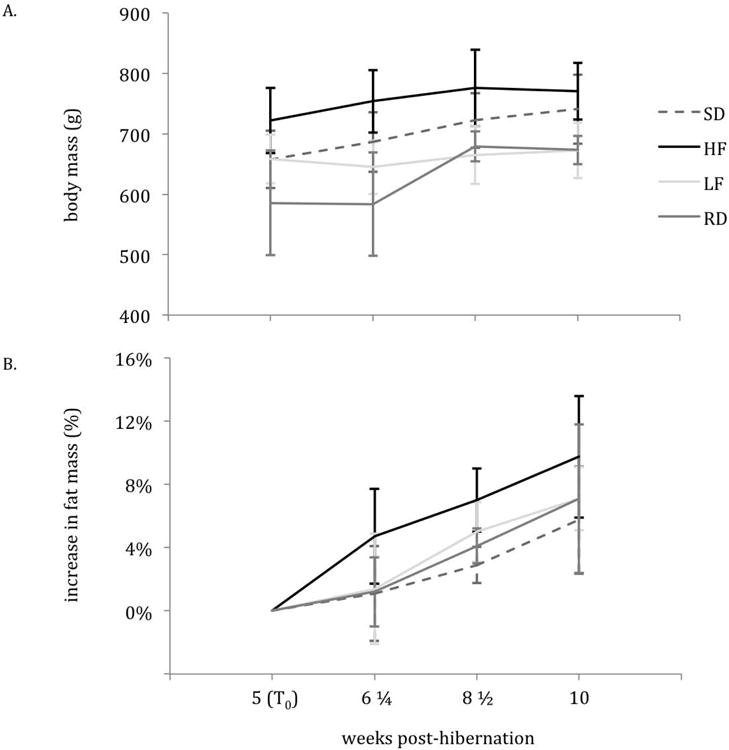
Body mass (in grams) (A) and percent change in fat mass (B) from 5- to 10-weeks post hibernation. Vertical bars at each sampling point (5-, 6¼-, 8½- and 10-weeks post hibernation) represent the standard error from the mean.
The mixed effects model that best described the rate of fattening (change in percent body fat from T0) attributed differences in rate of fattening to random variability between individual squirrels rather than an effect of diet treatment. This model included both a random effect of intercept (ai,d) and slope (bi,d) and had no significant effect of autocorrelation.
Bacterial Density and Viability
Bacterial densities in arctic ground squirrel ceca did not differ across time or among diet treatments with a combined average of 1.93 × 1011± 2.38 × 1011cells per gram of cecal content (Table 1). We also observed no difference in the percentage of live, dead, and injured bacteria among diet treatments. Combining all treatments, cecal bacterial cell viability averaged 80.3% live, 15.8% dead, and 3.9% injured (Table 1).
Table 1. Cecal content pH, bacterial density, and bacterial viability.
| Parameter | T0 | HF | LF | RD | SD |
|---|---|---|---|---|---|
| Bacterial density (cells/gram of cecal content) | 1.8511± 1.8311 | 1.9811± 1.4111 | 1.6611± 1.4411 | 7.0710± 4.8110 | 3.5911± 4.3511 |
| Bacterial viability (%) | |||||
| live | 83.1 ± 4.7 | 80.6 ± 8.3 | 81.5 ± 4.4 | 76.3 ± 10.8 | 80.0 ± 4.8 |
| dead | 13.9 ± 3.8 | 16.0 ± 6 | 14.5 ± 3.9 | 18.1 ± 9.6 | 16.7 ± 4.9 |
| injured | 3.1 ± 1.8 | 3.4 ± 2.4 | 4.0 ± 1.9 | 5.6 ± 2.8 | 3.3 ± 1.4 |
| Cecal pH | 8.04 ± 0.28 | 7.68 ± 0.44 | 7.94 ± 0.46 | 8.25 ± 0.21 | 8.03 ± 0.24 |
Values are the mean ± the standard error.
Bacterial Taxonomic Composition
A total of 672,810 16S rRNA gene reads were sequenced from 40 squirrel ceca. No individual samples were eliminated from diversity analysis. After quality processing, 545,548 sequences remained with an average of 14,091 sequences per sample. Additionally, 8302 OTUs were identified from the processed sequence reads, and rarefaction curve analysis indicated that pyrosequencing captured the majority of microbial diversity in our samples (Figure S1). Additionally, Good's coverage estimates for all samples were greater than 0.95; thus, most of the OTUs present in the samples were detected. We identified 11 phyla among all sample periods (Figure 3; Table S1). The phyla Firmicutesand Bacteroidetescomprised greater than 80% of the gut microbiota identified. The majority of the remaining bacterial community members were assigned to the phyla Verrucomicrobiaand Proteobacteria.
Figure 3.
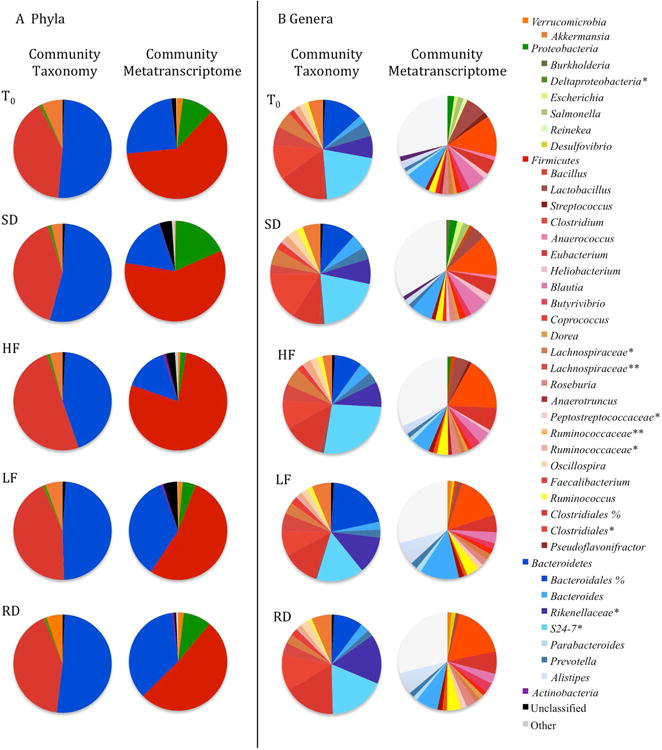
The relative abundance of dominant taxa compared to taxonomic composition of mRNA reads from metatranscriptome at the phylum (A) and genus (B) level where *, **, % represents an unclassified genus, family or order, respectively. Taxa with less than 1% relative abundance were not included.
The majority of Bacteroideteswere assigned to the class Bacteroidiaand order Bacteroidales. The families identified included Bacteroidaceae, Prevotellaceae, Rikenellaceae, S24-7, and an unclassified family belonging to the order of Bacteroidales. Dominant genera identified were Bacteroides, Prevotella, and unclassified genera from the families Rikenellaceaeand S24-7.
The majority of Firmicuteswere assigned to the class Clostridiaand order Clostridiales. The families Lachnospiraceaeand Ruminococcaceaedominated the order Clostridiales. Dominant genera we found included Coprococcus, Oscillospira, and Ruminococcus. The only genus identified that did not belong to Clostridiawas Lactobacillus(belonging to class Bacilli, order Lactrobacillales, and family Lactobacillaceae).
All members that matched to the phylum Verrucomicrobiaidentified to the genus Akkermansia. While the Proteobacteriawas the fourth most prevalent phylum, not one class contributed more than one percent to the overall community.
No differences in the relative abundance of taxa at any level existed between groups sampled at T0and those sampled 10-weeks post-hibernation. We also found no differences in the relative abundance of taxa at any level among diet treatments and the control diet (Figure 3; Table S1).
Microbial Diversity
We found no differences in gut microbial community alpha diversity in animals sampled at T0and those sampled at 10-weeks post-hibernation (SD, HF, LF, RD; Figure S1). Additionally, the alpha diversity of the gut microbial community did not differ among diet treatments (HF, LF, RD) and the control diet (SD).
We found no differences in beta diversity across time (T0and at 10-weeks post-hibernation) or among diet groups. The three distance metrics used to determine beta diversity (Bray-Curtis, weighted and unweighted UniFrac) showed similar trends, and only unweighted UniFrac analyses are presented here. Principal coordinates axes (PC1 vs. PC2, PC2 vs. PC3, PC1 vs. PC3) revealed no distinct trends or separation among gut microbial communities of squirrels sampled at T0and those sampled at 10-weeks post-hibernation, or among diet treatments and the control diet (Figure 2). Furthermore, the gut microbiota did not differ in diet groups sampled at T0and at 10-weeks post-hibernation or among diet treatments and the control diet. Analysis of similarity (ANOSIM) showed that diet groups were not significantly different from each other and dispersion of gut microbiota did not differ among diet groups.
Figure 2.
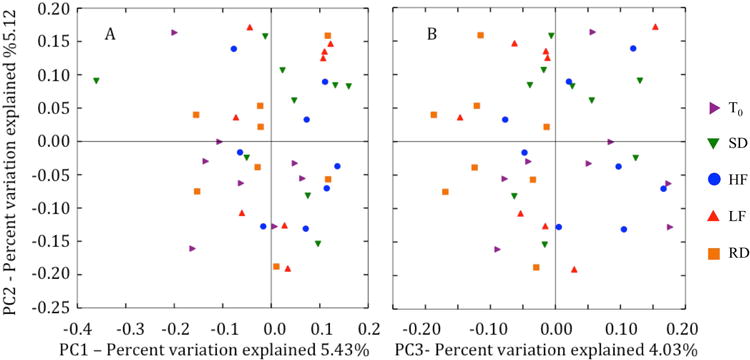
Principle coordinate analysis plots PC1 vs. PC2 (A) and PC2 vs. PC3 (B), of un-weighted UniFrac distance metric of gut microbiotas. Each point (symbol) represents the gut microbial community of an individual arctic ground squirrel at a given sample point or diet group. The distance between two points represents the similarity between the gut microbial communities.
Gut Microbial Transcriptome
RNA libraries were uploaded to MG-RAST (Meyer et al., 2008) and T0, SD, HF, LF and RD contained 1,320,915; 757,580; 1,641,637; 2,148,601 and 1,287,162 sequences with average lengths of 373; 374; 375; 374 and 361 base pairs, respectively. After quality control processing and removal of rRNA gene sequences, 3,208; 2,898; 8,748; 23, 603 and 23,733 sequences contained predicted proteins with known functions for T0, SD, HF, LF and RD, respectively (0.0% contain predicted sequences with unknown function for all diets). RNA sequences were assigned to functional categories using KEGG Orthogonal (KO) annotation. KEGG pathways were assigned for 381, 321, 1901, 5465, 6494 annotated proteins for T0, SD, HF, LF and RD treatments, respectively. The highest abundance of mRNA transcripts matched to the metabolism functional category (KO level 1 subsystem) across all pooled samples.
Taxonomic classification of the transcriptome indicated that the majority of mRNA functional transcripts in all pooled samples were derived from the phyla Firmicutes, Bacteroidetes and Proteobacteria(Figure 3). The Firmicutes-associated transcripts were further assigned to the classes Bacilliand Clostridiawith the majority of Bacilli-associated transcripts identified to the genus Lactobacillus(order Lactobacillalesand family Lactobacillaceae) and the majority of Clostridia-associated transcripts identified to the genus Clostridium(order Clostridialesand family Clostridiaceae), Eubacterium(order Clostridialesand family Eubacteriaceae) and Blautia(order Clostridialesand family Lachnospiraceae). Bacteroidetes-associated transcripts were assigned to the order Bacteroidiawith the majority being further assigned to the genus Bacteroides(order Bacteroidalesand family Bacteroidaceae) and Alistipes(order Bacteroidalesand family Rikenellaceae). Comparing the classification of transcripts by taxonomic group with the microbial community composition determined from 16S rDNA sequencing, it appears that relative abundance of mRNA by taxonomic groups is not a function of the group's relative abundance within the gut microbial community.
Statistical analyses of the pooled samples using STAMP revealed differences in the contribution of mRNA at every taxonomic level among gut microbiota prior to and after treatment with any diet (Table S4). After 5-weeks on the SD diet, the relative abundance of mRNA transcripts associated with Firmicuteswas decreased and that of Proteobacteria-associated transcripts was increased compared to cecal mRNA at T0(Table S2). The gut microbial metatranscriptome of squirrels on LF and RD diets for 5-weeks was decreased in relative abundance of Firmicutes- and Proteobacteria-associated transcripts and increased relative proportion of Bacteroidetes- and Verrucomicrobia-associated transcripts as compared to HF squirrels. HF squirrels contained a greater relative abundance of Firmicutes-associated mRNA and lower relative abundance of Verrucomirobia-associated mRNA in their gut than all squirrels fed any other diet (Table S2).
The relative abundances of transcripts were compared among treatments using STAMP, and several KEGG functional categories of mRNAs were differentially abundant among gut microbial communities sampled prior to and after 5-weeks on any diet and among diet treatments and the control diet (Table S4). Within these functional categories, the majority of transcripts were attributable to genera within the Firmicutes (Figure 4). Transcripts associated with cell motility, specifically related to flagellar assembly genes (flgE and fliC; Table S4), were in higher relative abundance in RD compared to T0and HF and LF diet treatments. In the RD group, the majority of flgE transcripts derived from the phylum Firmicutesand class Clostridiaand fliC transcripts were linked to the phylum Firmicutes, classes Bacilliand Clostridia, as well as organisms classified to the phylum Proteobacteria.
Figure 4.
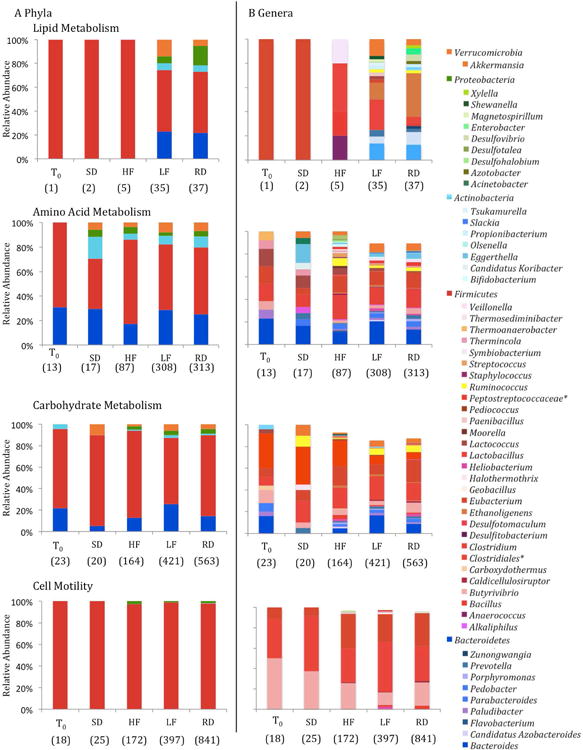
The relative abundance of KEGG level 2 functional transcripts attributable to dominant taxa at (A) phylum and (B) genus level. Taxa with less than 1% relative abundance were not included. The total number of transcripts for all taxa are listed below each bar. *Represents unclassified families or genera.
Additionally, LF and RD diet treatment resulted in lower abundances of carbohydrate metabolism-associated transcripts (KO level 2 subsystems; Table S4), in particular those involved in pyruvate metabolism (KO level 3 subsystem; Table S4) as compared to T0and the HF diet treatment. Of the transcripts that were attributable to pyruvate-metabolism and differed among treatments, the majority were functionally categorized to formate C-acetyltranserase (pflD; Table S4). The pflD-attributable transcripts were primarily associated with the phylum Firmicutesand genera Lactobacillusand Clostridium. The transcriptome of HF squirrels had greater relative abundance of transcripts associated with translation (KO level 2), specifically ribosome proteins (KO level 3 subsystem; Table S4) and the majority of those transcripts were attributable to the phylum Firmicutesand genera Clostridium, Lactobacillusand Eubacteria.
Short Chain Fatty Acid Concentrations and Percent Composition
Acetate was the most abundant SCFA, comprising >65% of all SCFAs measured, followed by butyrate (10.9-17.7%) and propionate (10.6-14.0%). The relative proportions of branched SCFAs (iso-butyrate, valerate, and iso-valerate) were lower than acetate, butyrate, and propionate (Table S3).
Neither cecal SCFA concentration, nor composition or pH differed across time, but differences in SCFA concentrations (but not pH) were apparent among cecal contents sampled from squirrels on different diets (Table S3). Although an inverse relationship between the pH of cecal contents and total SCFA concentrations was observed, the pH did not vary among groups sampled at T0and at 10-weeks post-hibernation or among diet groups (Table 1). Squirrels on the HF diet had a higher total SCFA concentration than those in the RD group (Table S3; p = 0.021). No other differences in SCFA concentrations among diet treatments were observed.
SCFA molar concentrations correlated with relative abundance of several taxa by treatment (Figure 5). The molar concentration of iso-butyrate and iso-valerate correlated to the genus Prevotellaat T0and to Coprococcusand an unclassified family and genus of the order Clostridiain LF squirrels (Figure 5). The genera Oscillospiraand Ruminococcuscorrelated with the concentrations of propionic acid and iso-valerate and iso-valerate, respectively, in SD squirrels. An unclassified genus of the family Lachnospiraceaepositively correlated with the concentration of acetate and negatively correlated with the concentration of valerate in HF squirrels, while the concentration of valerate was positively correlated with an unclassified genus of the family Ruminococcaceaein RD squirrels. Additional correlations were found within treatments between the concentration or relative proportion of specific SCFAs and minor taxa (not shown).
Figure 5.
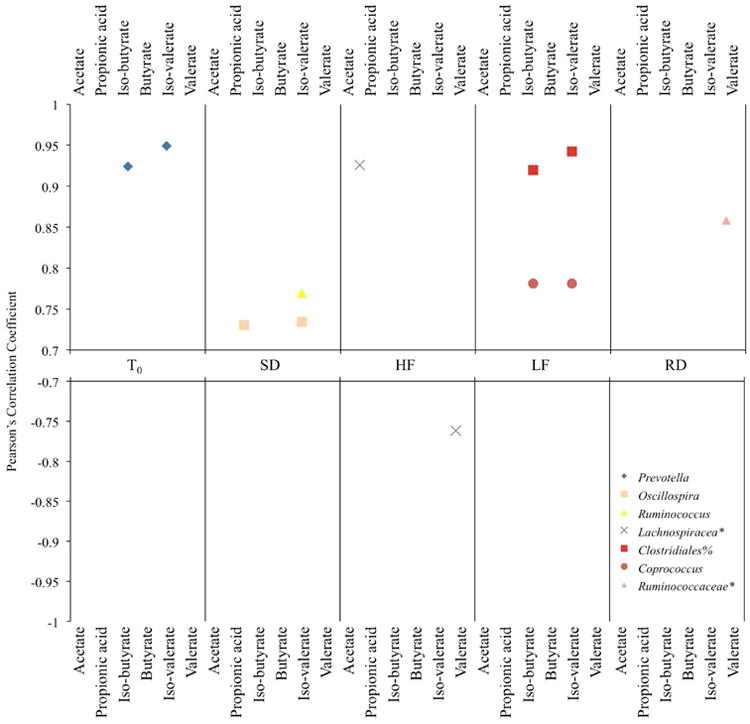
Pearson's correlations between bacterial genera and SCFA molar concentrations where * and % represents an unclassified genus or order, respectively.
Discussion
We set out to determine the influence of diet on fattening and the gut microbial community of arctic ground squirrels late in their active season. Body composition (measured across the active season), gut microbial community diversity and function, and cecal SCFA concentrations (characterized before and after treatment) were compared across time (T0vs. SD) and among diet groups (HF, LF, RD, and SD) to correlate changes in fattening and gut microbiota with changes in diet. Surprisingly, we found that during the pre-hibernation fattening period, arctic ground squirrels increased in adiposity at similar rates and to similar degrees irrespective of calories consumed or the macronutrient composition of the diet. Furthermore, we found that differences in diet did not influence microbial community structure or diversity, or relative SCFA concentrations in the cecum, but were associated with differences in the total SCFA concentration and gut microbial metatranscriptome. Additionally, several bacterial taxa correlated with specific SCFA concentrations and these correlations were unique across time and among diet groups. That arctic ground squirrels fatten at similar rates and to similar extents despite differences in calorie and macronutrient content of their diets suggests that pre-hibernation fat accumulation can be accomplished through decreased energy demand in the absence of changes in food intake.
The Effect of Diet on Pre-hibernation Fattening
Diet is a primary environmental factor contributing to obesity in non-hibernating mammals. At the most basic level, excess food intake leads to increased adiposity. Even without the consumption of excess calories, a diet high in fat also leads to weight gain in non-hibernators (Townsend et al., 2008). While excessive energy intake and HF diet leads to increased adiposity, restricting calorie intake or consuming a LF diet leads to decreased adiposity (Wadden et al., 1993; Vieira et al., 2009). Because of the strong influence of diet on adiposity in non-hibernators, it is important, in a comparative sense, to consider dietary influence on fattening in hibernating mammals and especially those species known to have an endogenously driven annual cycle of metabolism, body mass and body composition (Williams et al., 2014).
Free-living arctic ground squirrels exhibit extreme seasonal fluctuations in body fat, punctuated by a rapid (<3 weeks) increase from 5% to 45% body fat during the pre-hibernation fattening period (Buck and Barnes, 1999a). Increased energy uptake may be accomplished through either increased duration or intensity of foraging, selection of higher energy foods, or some combination of these factors. In our study of captive adult squirrels, diet manipulation in the latter half of the active season resulted in no differences in either the rate or degree of fat accumulation. LF and SD groups increased in fat mass as effectively as the HF treatment, despite squirrels receiving 75% to 50% less energy from fat, respectively. Additionally, squirrels maintained on RD showed no difference in accumulated fat mass as compared to the SD treatment squirrels. That the rate and degree of fat accumulation in arctic ground squirrels was not affected by macronutrient content supports the assertion by Sherriff et al. (2013) that the positive energy balance required for fattening is influenced to a greater degree by reduced energy utilization (via decreased metabolic rate, Tband activity) rather than increased food intake.
While the interpretation of our results corroborates the findings of Sheriff et al. (2013), discrepancies exist between our results and those found in studies of non-hibernating species. These differences may be explained by different life histories and physiologies of hibernators and non-hibernators. Alternatively, the lipid composition of the chow we used in our study could have reduced the obesogenic influence of the HF diet. Mice, humans and rats on a Westernized diet containing high saturated fatty acids become obese, gaining more fat than humans and rats consuming a high-fat diet rich in monounsaturated fatty acids (Harlan-Teklad TD96132; Posey et al., 2009; Fleisser et al., 2010; Turnbaugh et al., 2010; Daniel et al., 2014; Jakobsdottir et al., 2015; Piers et al., 2003; de Wit et al., 2012). The fat content in our HF diet was elevated as compared to the SD and LF diets through the addition of soybean oil, a source low in saturated fatty acids, rich in unsaturated fatty acids, and likely more similar to the predominantly plant-based diet naturally consumed by arctic ground squirrels (Frank et al., 2008). Consequently, our HF diet may not have triggered the high-fat, diet-inducible adipose accumulation seen in other studies and model systems. It is also possible that high individual variation in fattening rate among squirrels explains the lack of difference among diet treatments.
Gut Microbial Community Structure
Another important factor that could influence fattening is a diet-induced shift in the microbial community (composition or function) that promotes increased energy uptake. While we did not measure energy uptake in our squirrels and therefore cannot make inferences from this study about the role of gut microbiota on pre-hibernation fattening, we did investigate the effect of a high-fat diet on the arctic ground squirrel gut microbial community. The introduction of a high-fat diet is associated with changes in gut microbial community structure and bacterial diversity in mice and humans (Hildebrandt et al., 2009; Turnbaugh et al., 2010; Serino et al., 2012; Daniel et al., 2014), and some studies show an increase in the relative abundance of Firmicutesto Bacteroidetes(Ley et al., 2005; Ley et al., 2006; Turnbaugh and Gordon, 2009) regardless of host phenotype (obese vs. lean; Hildebrandt et al., 2009; de La Serre et al., 2010).
We found that increased dietary fat content had no significant effect on the gut microbial diversity or community structure of arctic ground squirrels. Although our findings are novel and could indicate a gut microbiota robust to changes in diet, it has been shown that the gut microbial community in mice is responsive to dietary lipid composition—mice on diets high in polyunsaturated fats host a microbiota that significantly differs from the microbiota of mice on a diet rich in saturated fatty acids (de Wit et al., 2012; Huang et al., 2013; Patterson et al., 2014). de Wit and colleagues (2012) found that the gut microbial community of mice maintained on HF diets (45% energy from fat) with increased polyunsaturated fat-to-saturated fat ratios (P/S) did not differ from those maintained on LF diets (10% energy from fat), while the gut microbial community of mice maintained on a high-fat, low P/S diet responded in a manner consistent with the studies mentioned above: i.e., reduced diversity, decreased Bacteroidetes, and increased Firmicutes. However, this response is not universal, and other studies have found that the gut microbial community in mice maintained on high P/S, low P/S and LF diets were all significantly different than one another (Patterson et al., 2014; Huang et al., 2013). Furthermore, Deol et al. (2015) found that mice on a diet enriched in soybean oil (40% energy from fat) exhibited greater weight gain and higher adiposity than mice on a standard diet (13.5% energy from fat) or a diet high in saturated fat (coconut oil; 40% energy from fat). Administration of the LF and RD diet also resulted in no change in arctic ground squirrel gut microbiota from standard diet, results that are consistent with some studies reported in mice (Henderson et al., 1998).
In addition to the impact of fatty acid type, our results may have been influenced significantly by the timing of our diet manipulations and the “vulnerability” of the arctic ground squirrel gut microbiota to change. During hibernation the gut microbiota of ground squirrels is reduced in diversity and density, and the relative abundances of bacterial taxa known to degrade host derived substrates increases (Carey et al., 2013; Stevenson et al., 2014b). These alterations due to hibernation persist immediately following the terminal arousal from hibernation (prior to feeding; Stevenson et al., 2014b), and subsequent shifts toward the active season microbial community are likely influenced by the reintroduction of dietary substrates. For example, in 13-lined ground squirrels that have completed at least one hibernation season, gut microbiotas of squirrels sampled after two weeks of feeding following hibernation cluster with mid-active season microbiotas, and there are few differences in their taxonomic compositions (Carey et al., 2013), suggesting that the “active-season” gut microbiota stabilizes quickly after reintroduction of food. While no studies have explicitly investigated the influence of diet on restructuring of the gut community after hibernation, it may be that had we begun the diet manipulation immediately after terminating hibernation, we would have observed differences in the gut microbiota and/or host fattening among diet treatments; this possibility warrants further investigation.
Because diet and other environmental factors are known to influence a host's gut microbiota, it is important to consider the squirrels' life history prior to capture and the effect of translocation to captivity from the wild. The early gut microbial community is shaped by maternal influences which have a profound impact on gut microbial diversity that persists into the adulthood (Benson et al., 2010; Dominguez-Bello et al., 2010). Once weaned, the gut microbiota transitions to an adult community and stabilizes within 2-3 weeks post-weaning (Palmer et al., 2007; Koenig et al., 2011; Schloss et al., 2012). We have observed a strong litter effect in arctic ground squirrels born in captivity to wild-caught mothers across their first active season (Stevenson et al., 2014a), indicating a strong maternal influence shaping their gut microbiota. Kohl and Dearing (2014) showed that wild-caught rodents maintain much of their gut microbial community when relocated to captivity and fed commercially available rodent chow. All squirrels in this study were either wild-caught within the same area or birthed from mothers trapped in that same area. While we cannot rule out the influence of maternal effects and early wild-diet exposure on the results of our study, and we do not know if (and to what degree) maternal influences persist beyond the first hibernation season, we did attempt to lessen the confounding effects of age class and prior dietary exposure: all squirrels had a) consumed a rodent chow diet for at least 2 months prior to entering hibernation and b) completed at least one hibernation cycle in captivity before entering the study (all were at least yearling adults).
While our results suggest that the gut microbial community of arctic ground squirrels may respond to fat composition of diets in a similar fashion to mice fed a high P/S ratio diet, it is also possible that our selected diets were not sufficiently different or extreme to elicit a response in the gut microbiota of a host that naturally experiences radical fluctuations in physiology and food availability annually. For example, after fasting for 7-9 months each year, arctic ground squirrels spend 3-5 months consuming a diet of tundra shrubs, seeds, mushrooms, and even scavenged animal material (Batzli and Sobaski, 1980; McLean, 1984; Gillis et al., 2005; Zazula et al., 2005) that varies in macronutrient content and availability across a short active season. Considering the effects of maternal influences, as well as captive rodents maintaining core members of the gut community from the wild, it may be that the macronutrient content of the diets used in this study fell well within the range that arctic ground squirrels naturally experience in the wild, and thus did not elicit a strong response in the gut microbial community.
Gut Microbial Function
Despite the lack of changes in gut microbial composition and diversity, diet treatment resulted in alterations in gut microbial function (gene expression and SCFA production) among diets but not across time. The gut microbial metatranscriptome and SCFA concentrations at the start of diet treatment (T0) did not differ from those observed at the end (SD), indicating that alterations in microbial function resulted from a shift in diet, rather than progression of time.
Analysis of the metatranscriptome provides a snapshot of the active functional profile of the microbiota; however, few studies have characterized the metatranscriptome of gut microbiota, fewer still have investigated the effect of host diet or obesity on metatranscriptome (effects of dietary milk: McNulty et al., 2011; Donovan et al., 2012; effects of obesity: Ridaura et al., 2013), and none have been conducted in a hibernating species. In our study, community metatranscriptomics analysis revealed changes in microbial function with diet, with notable differences in transcripts related to cell motility, carbohydrate metabolism, and translation (Table S3). In humans, surface proteins of the intestinal microbiota, such as flagellin, are more abundant in obese versus lean individuals (Ferrer et al., 2013), and are considered important because they enable microbes to interact with host cells (O'Connel Motherway et al., 2011; Kolmeder et al., 2012) and to better reach their food source (Vijay-Kumar et al., 2008). The increase in flagellar assembly genes flgE and fliC (Table S3) in the RD group compared to other diet treatments may have enabled fattening to the same degree and rate as other treatments by increasing dietary substrate accessibility and facilitating host-microbe communication.
The microbial metatranscriptome of HF squirrels was enriched in carbohydrate metabolism associated transcripts as compared to LF and RD squirrels. Many of the carbohydrate associated transcripts were related to the pyruvate metabolism enzyme pyruvate formate-lyase (pflD), which catalyzes the reversible conversion of pyruvate and coenzyme A to acetyl-CoA and formate in anaerobic fermentation, and replaces pyruvate dehydrogenase during anaerobic glucose metabolism in Escherichia, Clostridium, Streptococcusand a number of other prokaryotes (Abbe et al., 1982; Schomburg, 1996; Carere et al., 2008). Although we did not observe a change in relative abundance of microbes among diet groups, the majority of pflD transcripts were attributable to the genera Lactobacillusand Clostridium. Pyruvate is an important intermediate in many energy metabolic pathways, and genes involved in pyruvate metabolism are increased in mice fed a Westernized diet which includes both high saturated fat and high sugar content (Turnbaugh et al., 2008).
Transcripts related to ribosomal proteins were more abundant in HF squirrels than any other diet group (Table S4). Microbial genes encoding ribosomal proteins are among the most expressed in humans, and upregulation of transcripts relating to ribosomal synthesis are indicative of microbes responding to environmental changes by altering growth rate (Franzosa et al., 2014). Additionally, alterations in the metatranscriptome can be detected prior to alterations in microbial community structure and predict changes prior to their onset (Franzosa et al., 2014). The greater relative abundance of ribosome-associated transcripts in HF squirrels attributable to the bacteria Clostridium, Eubacteriumand Lactobacillusmay indicate that members of the gut microbial community had increased growth rates. Although we did not find increased relative abundance of specific taxa, had we allowed squirrels to continue on the HF diet and sampled the gut microbiota after longer exposure, we may have captured significant changes in the gut community reflecting the upregulation of ribosomal proteins by Clostridium, Eubacteriumand Lactobacillus.
The abundances of metabolism- and cell motility- associated microbial transcripts are lower at T0and for SD diet treatment than for all other treatment groups, and the decrease in abundance of microbial transcripts is concomitant with a decrease in diversity of microbial taxa to which transcripts were assigned at both a phylum and genus level. For many of the functional categories that differed among diet treatments (Table S2) the majority of transcripts were attributable to Firmicutes(Figure 4). The metabolism associated transcripts in LF or RD treatment were enriched in Bacteroidetesattributable mRNA, a profile that is consistent with the increased representation of Bacteroidetesin low-fat diets relative to the Westernized diet in mice and humans (Ley et al., 2006; Turnbaugh et al., 2009). However, the LF diet, which also contained a higher carbohydrate content compared to the HF diet, resulted in a decrease in the relative abundance of Eubacterium-attributable and Roseburia-attributable transcripts, taxa that have been previously shown to decrease in relative abundance within the gut community of mice consuming a low-carbohydrate diet (Duncan et al., 2007).
We found profound differences in the gene expression of microbial taxonomic groups across time and among diet groups (Figure 3, Table S2). The shift in transcriptome parallels the changes in gut microbial diversity associated with consumption of HF diet and fattening in mice and humans; the relative abundance of Firmicutes-associated transcripts increased and relative abundance of Bacteroidetes-associated transcripts decreased in squirrels fed a HF diet compared to a LF diet. The relative abundance of transcripts attributed to the genus Akkermansiawere lower at T0and under SD and HF diets. Of the transcripts associated with Akkermansia, all were further assigned to the species Akkermansia muciniphila, a known mucin-degrading bacteria that resides in the mucous layer of the healthy mammalian gut (Derrien et al., 2004). A.muciniphilais lower in relative abundance in the gut microbial community of obese mice and humans (Everard et al., 2013; Parks et al., 2013), and fewer A. muciniphilaassociated transcripts in squirrels could facilitate fattening.
Short chain fatty acid concentrations in the gut can also provide insight into the host-gut microbe relationship; however, we did not measure the rate of SCFA flux and therefore cannot infer their effect on host energy balance. The molar concentrations of specific SCFAs are correlated to the taxonomic families Ruminococcaceae, Prevotellaceae, and Lachnospiraceaeand numerous minor taxa, which may indicate they are key players in SCFA production in the squirrels. Because we did not observe changes in the relative abundance of these taxa among diet groups, we did not uncover connections between specific microbiota, relative SCFA concentrations, and diet. That we were unable to make these connections does not point to their absence, but rather indicates that these relationships are complex and may not be revealed within the snapshot of time that our sampling occurred or were obscured by high individual variation among squirrels.
Conclusion
In this study, we tested whether changes in diet would affect arctic ground squirrel fattening and shift the gut microbial community to one that facilitates fattening. We found that animals fatten at similar rates and to similar extents irrespective of calories consumed or the macronutrient composition of their diet. Furthermore, we found that diet did not alter gut microbial community composition and that despite changes in gut microbial function (determined by metatranscriptome analysis), SCFA production of the squirrels remained the same. While our results are derived from captive animals on controlled diets that are not representative of wild squirrels, they support the findings of Sheriff et al. (2012, 2013) in which free-living arctic ground squirrels gain fat by minimizing energy expenditure through physiological changes (i.e., reductions in metabolic rate and decreased body temperature) rather than increased food uptake. The Arctic is characterized by a brief but intense season of primary production and has likely selected for an arctic ground squirrel phenotype that readily and rapidly fattens in preparation for hibernation: failure to fatten sufficiently independent of changes in macronutrient composition of forage and caloric intake would most certainly compromise arctic ground squirrel overwinter survivorship.
Methods
Study Species
Free-living juvenile and adult female arctic ground squirrels (Urocitellus parryii) were trapped between 2010 and 2013 in the northern foothills of the Brooks Range, Alaska near the Atigun River (68° 38′ N; 149° 38′ W) and transferred to the University of Alaska Anchorage (UAA) animal holding facility. Squirrels were housed individually in hanging wire cages (46 × 30 × 30 cm) at 20°C with a 12:12 light:dark cycle and provided Mazuri rodent chow (Brentwood, MO, USA) and water ad libitumuntil hibernation. Prior to hibernation, squirrels were transferred to environmental chambers maintained at 2.0°C ± 1.0°C with a reduced food ration and no light. Once squirrels became torpid, as indicated by a Tb≤30°C (Buck et al., 2008), they were transferred to plastic tubs (Nalgene, Rochester, NY, USA; 36 × 56 × 20cm) containing pine shavings and cotton bedding and held at 0.0°C ± 1.0°C with neither food nor water and in constant darkness for the remainder of hibernation. At the completion of hibernation (determined by four consecutive days of activity at euthermic temperature [Tb>30°C]), squirrels were returned to hanging wire cages at 20°C on a 12:12 light:dark cycle, and provided water ad libitum. All animal procedures were approved by the UAA Institutional Animal Care and Use Committee (protocol numbers 418912, 418919, and 418923) and all animal permits issued by Alaska Department Fish & Game (14-071 and 15-094).
Diet Manipulation
All squirrels had been held in captivity for a minimum of one year by the time the diet manipulation began. All had consumed standard rodent chow for a minimum of two months prior to their first hibernation in captivity, and all had completed at least one hibernation cycle in captivity; thus, all squirrels began the diet treatment as either yearlings or adults (Table S5). For the first five weeks following hibernation, we fed squirrels a standardized rodent chow diet ad libitum(18%, 60% and 22% energy from fat, carbohydrates and protein, respectively; 3.0 kcal/g chow; Chow ID: TD.120766, Harlan Laboratories, Madison, WI, USA) and determined individual daily caloric intake by averaging the daily food consumption over the final three weeks post-hibernation (week 3, 4, and 5). Five-weeks after ending hibernation (T0), squirrels in the control group (n=8) were euthanized for sample collection and analysis (detailed below). The remaining squirrels were assigned to one of four experimental diet groups (n=8/group). Each group was maintained on their respective diet for the next five consecutive weeks: standardized (SD; maintained on the standardized diet), low fat (LF; 10%, 66% and 24% energy from fat, carbohydrates and protein, respectively; 2.8 kcal/g chow [TD.120765, Harlan Laboratories]), high fat (HF; 40%, 44% and 16% energy from fat, carbohydrates and protein, respectively; 4.2 kcal/g chow [TD.120767, Harlan Laboratories]) and restricted calorie (RD; 50% of the average energy intake of standardized diet prior to T0). Experimental diets were not provided ad libitum; rather, with the exception of squirrels in the RD group, each squirrel was provided a daily portion equal in calorie content to the daily average it consumed during the standardization period. Squirrels in the RD group received 50% of their individual average daily intake. Tap water was added to chow to create a soft mush; this deterred squirrels from caching or dropping food through the metal grating, and insured that the daily-allotted portion was entirely consumed. After five-weeks on the experimental diet (i.e., 10-weeks post-hibernation), squirrels were euthanized and samples collected for analyses.
Sample Collection
Procedures for terminal sampling, processing of ceca and collection of cecal content for pyrosequencing, flow cytometry, and short chain fatty acid analysis were completed according to Stevenson et al. (2014a and b). Briefly, animals were euthanized, ceca were excised and cecal content (both lumen and mucosal) was subsampled for pyrosequencing (1-2g) and short chain fatty acid (SCFA) analysis (1-2g). In addition, we collected subsamples for determination of pH (0.5g), and preserved subsamples for RNA extraction (0.5g) using Lifeguard™ Soil Preservation Solution (MoBio, Carlsbad, CA, USA). Subsamples were analyzed using flow cytometry upon terminal sampling and all other samples were stored at -80°C until analysis. After collection of cecal content, the remainder of the squirrel carcasses (including ceca and remaining cecal content) were weighed, sealed in plastic bags and stored at -20°C until analyzed for body composition. Precautions during sampling were taken to minimize the loss of mass through blood loss and evaporation.
Microbial Diversity Analysis
We extracted DNA from cecal contents using MoBio PowerSoil kits (MoBIO, Carlsbad, CA, USA) with modifications as described by Stevenson et al. (2014a). DNA concentrations were quantified and purity determined using a Nanodrop spectrophotometer (ND1000; Thermo-Scientific, Wilmington, DE, USA). The V4-V5 regions of the 16S ribosomal RNA (rRNA) gene were amplified from cecal DNA using universal bacterial primers 530F (5′-GTGCCAGCMGCNGCGG) and 1100R (5′-GGGTTNCGNTCGTTR) and sequenced on a Roche GS FLX Titanium 454 pyrosequencer following manufacture guidelines (Research and Testing Laboratories, Lubbock, TX, USA). Sequencing data were processed as described by Stevenson et al. (2014b). Briefly, we used QIIME 1.8.0 (Caporaso et al., 2010) to analyze raw 454-pyrosequenced reads. Sequences were de-multiplexed and processed to remove primers, short sequences (<200bp), sequences with ambiguous characters, and sequences with low-quality scores (> six ambiguous base calls and homopolymeric runs exceeding six base pairs). We pre-clustered and denoised sequences using Denoiser (Reeder and Knight, 2010) and clustered sequences into operational taxonomic units (OTUs) at 97% similarity using USEARCH 5.2.32 (Edgar et al., 2011). Rarefaction analysis of alpha diversity metrics (observed species, Phylogenetic Diversity-Whole tree and Chao1) as well as Good's Coverage estimates were calculated in QIIME.
Community Transcriptomics Analysis
RNA was extracted from cecal samples using the PowerMicrobiome RNA Isolation kit (MoBio, Carlsbad, CA) according to manufacture protocol. RNA quality was assessed with agarose gel electrophoresis and spectrophotometry (NanoDrop ND1000; Thermo-Scientific). Pooled extracts for each diet group were prepared by combining equal amounts of RNA (in nanograms) from each of five squirrels per diet treatment. Microbial rRNA was depleted and paired-end mRNA libraries were constructed with the NebNext Ultra Directional RNA Library Prep Kit (Illumina; Research and Testing Laboratory, Lubbock, TX). Strand specific paired-end sequencing was performed on the MiSeq (Illumina, San Diego, CA, USA) platform using a 2×300 kit (Research and Testing Laboratory, Lubbock, TX).
Raw sequence results were processed to detect and remove sequencing errors and contaminant sequences in read ends using AlienTrimmer (Criscuolo and Brisse, 2013), and paired using QIIME. Processed sequences were uploaded to the open source MetaGenome Rapid Annotation using Subsystem Technology (MG-RAST) server, version 3.6 (Meyer et al., 2008) for assignment of gene function (maximum e-value = 10-5, minimum identity = 60%, minimum alignment length = 15bp). The taxonomic affiliation of transcripts was assigned using MG-RAST against M5NR by the lowest common ancestor method (maximum e-value = 10-5; minimum identity = 60%; minimum alignment length = 15bp). Annotations are available under MG-RAST ID 4644360.3, 4644361.3, 4644362.3, 4644363.4, and 4644364.3 for the HF, LF, RD, SD, and T0samples, respectively.
Flow Cytometry
We determined cecal bacterial densities and viability using flow cytometry following the methods outlined in Ben-Amor et al. (2005) with modifications by Stevenson et al. (2014a). Briefly, cecal contents were suspended, pelleted via centrifugation and washed twice in anaerobic PBS. The resulting suspension was serially diluted and analyzed for bacterial viability by incubating with 5mM SYTCO BC and propidium iodide (PI). Viability controls contained unstained cells, live-stained (SYTCO BC only), or heat killed (PI only) samples. Microbial density was determined by incubation with 1000× SYTO BC plus beads (1 × 106beads/ml). Flow cytometry was performed using a FACScalibur (Becton Dickinson [BD], San Jose, California, USA) calibrated with Calibrite3 beads (BD). Data was analyzed using the CellQuest (BD) software.
Short Chain Fatty Acid Analysis
We prepared SCFA samples for each squirrel by extracting and acidifying supernatant from cecal content according to Stevenson et al. (2014a). Samples were analyzed at the United States Department of Agriculture Dairy Forage research Center (Madison, WI, USA). Extractions were passed through a glass pre-column containing 30mg of packed glass wool to remove particulates and analyzed using a Shimadzu GC 2010 Plus (Nakagyo-ku, Kyoto, Japan) equipped with a flame ionization detector (350°C) connected to a Phenomenex (Torrance, CA, USA) Zebron ZB-FFAP column (30 × 0.53mm id × 1.0μm; 110°C for 1min, ramped [6.0°C/min] to 160°C then 30.0°C/min to 220.0°C for 1min) with split injection (250.0°C). We created individual standard curves for each analyte (acetate, propionate, butyrate, valerate, iso-butyrate and iso-valerate) using a multi-point calibration (0, 15, 25, 45mM) with 2-ethylbutyrate as the internal standard.
Cecal pH Measurements
The pH of cecal content was determined using a pH meter (VWR SympHony Meter SB80PI, Buffalo Grove, IL, USA) as described by Thomas (1996) with modifications. Thawed cecal contents were diluted to 10-4by addition of contents to deionized water with continuous stirring. The pH of the suspension was measured after 10min.
Estimation of Body Composition
We estimated body composition across the active season using the deuterium (δ2H) dilution method every 10 days from the end of hibernation until terminal sampling (Speakman, 1997). Squirrels were weighed, anesthetized (Isoflorane, 3-5%) and blood was collected for analysis of background deuterium concentration (Ib) by nail clip at the distal edge of the quick and filling 1-2 heparinized Natelson capillary tubes (∼200μl/tube). The tubes were immediately flame sealed and stored at 4°C. A mass specific dose (injection volume [ml] = body mass [g]*0.000867; Iinj; Speakman, 1997) of 3% deuterium-enriched water was then administered by intraperitoneal injection, and the squirrel was allowed to recover from anesthesia for one hour to allow incorporation of deuterium into the body water pool (Lee et al., 2011). Following equilibration, the squirrel was anesthetized for collection of a second blood sample (enriched in deuterium; Ienr) as previously described. Blood samples were distilled by transferring blood from the capillary tube into a glass Pasteur pipette, flame sealing each end, and heating on a hot plate (90°C). Water condensed in the narrow end of the pipette and was collected for analysis (Lee et al., 2011).
Water samples were analyzed at the UAA Environment and Natural Resources Institute Stable Isotope Laboratory (ENRI-SIL) using a Thermo Finnigan TC/EA (Thermo Scientific, Bremen, Germany) in line with a Thermo Finnigan Delta Plus XP continuous-flow isotope ratio mass spectrometer (Thermo Scientific, Bremen, Germany). Samples were manually injected (1-1.2 μl per run) into the TC/EA in quintuplets. International reference standards (VSMOW & VSLAP; IAEA, Vienna, Austria) were used to calibrate the instrument and working standards (USGS45: δ2H= -10.3‰, and USGS46: δ2H= -235.8‰) were injected with each analytical run to correct for instrument drift.
Body composition was estimated according to the methods of Speakman (1997). The total body water pool (TBWiso) of each squirrel was calculated as the ratio of dilution of the injectate [(Iinj– Ib)/(Ienr– Ib)] multiplied by the volume of injectate administered (Vinj).
| (Equation 1) |
Lean mass (LMiso) of the squirrel was estimated by dividing TBWisoby the hydration constant (HC) determined via proximate chemical analysis (water mass divided by lean mass; see below). An equation was also derived using LMp, TBWiso, and body mass to predict lean mass via isotope dilution. The percentage of body fat was determined as the difference between whole body mass and lean mass and is inversely related to hydration of lean mass (Speakman, 1997).
Proximate Chemical Analysis
We determined final body composition of squirrels by proximate chemical analysis via Soxhlet extraction of lipids as described by Mason et al. (2006) with modifications. Frozen carcasses were thawed, sectioned, and passed three times through a meat grinder (once through a 7mm sieve plate and twice through a 4.5mm grinding plate; Cabela's ½ Horsepower Commercial Grade, Model #8, Sidney, NE, USA). We dried three subsamples of each homogenate (∼15g each) to a constant mass using a FreeZone 4.5 (Labconco, Kansas City, MO, USA) connected to a vacuum pump (Fisher Scientific Maxima C Plus Vacuum Pump, Waltham, MA, USA) and determined water content by subtraction (wet mass – dry mass).
Lipids from dried samples were extracted in petroleum ether using an automated Soxtherm (Model 416; Gerhardt, Germany) with the following program settings: extraction temperature 150°C; reduction interval 3.5min; reduction pulse 2s; hot extraction 45min; evaporation A 5×; rinsing time 80min; evaporation B 3×; evaporation C 10min. The lipid mass of homogenate was determined gravimetrically (varied between replicated samples by ±0.60% of wet mass), while lean mass was determined by subtracting lipid mass from initial homogenate mass. Whole body composition was determined by extrapolation.
Statistical Analysis
We calculated alpha diversity by rarefying the OTU table from 1 to 7000 sequences per sample at increasing intervals of 500 sequences per sample (total of 14 sample points per curve; 50 repetitions per rarefaction). A maximum value of 7000 sequences per sample was chosen to ensure all curves were continuous. We calculated three alpha diversity metrics: observed species (number of observed species at 97% species threshold), Phylogenetic Diversity-Whole tree (PD whole; the minimum total length of all the phylogenetic branches required to span a given set of taxa on a phylogenetic tree) and Chao1 (species richness estimate accounting for the number of observed species, along with the ratio of singleton to doubleton OTUs). For the Chao1 metric, a unique OTU table that included singletons was constructed. Statistical differences were determined for all three metrics using non-parametric t-tests with Monte Carlo permutations (n=999) on the 7000 sequences per sample rarified OTU table.
Beta diversity was determined by standardizing the OTU table to the minimum sequences/sample (1431) to decrease the weight of more abundant OTUs. We generated two-dimensional principal coordinates analysis (PCoA) plots from individual distance matrices produced from both unweighted (based on phylogenetic trees) and weighted (based on phylogenetic trees while taking into account relative abundance) UniFrac (Lozupone and Knight, 2005) distance, as well as Bray Curtis (relative abundance only) distance metrics. Analysis of similarities (ANOSIM) of Bray Curtis community structure dissimilarity was performed to determine if diets were significantly different. Differences among treatment groups were determined using pair-wise one-way permutational multivariate analysis of variance (group dispersion) using the PRIMER 6 software (Clark and Gorely, 2006).
Bacterial taxonomies were summarized at the phylum, class, order, family, and genus levels from the same standardized OTU table used in the beta diversity analysis. Taxonomies at each level were grouped by diet group and compared using one-way analysis of variance (ANOVA) followed by Tukey's HSD test in R (version 3.1.1). We determined statistical differences in bacterial cell density and viability, relative SCFA concentrations, cecal pH and percent body fat using ANOVA followed by a Tukey's HSD test (R version 3.1.1). Statistical differences in metatranscriptomes of pooled samples were identified using Fisher's exact test and were corrected using Storey's FDR approach using STAMP (Statistical Analysis between Metagenomic Profiles; Parks and Beiko, 2010). Pearson correlation analysis was used to test the correlation between SCFA concentrations and taxonomy (R version 3.1.1). Differences were considered significant at a pof <0.05 for all analyses.
The equation to predict lean mass from deuterium dilution was determined through multiple regression analysis (using R version 3.1.1) of TBWisoand body mass (BM) against lean mass (LMp). We compared estimates of lean mass, determined by the predictive equation or average HCapproach, to lean mass determined by proximate chemical analysis following methods by Bland and Altman (1986).
To test for differences in rate of fattening among diet groups, we fit data for percent change in body fat (determined by deuterium dilution) from T0to terminal sampling to mixed effects models using R (version 3.1.1), assuming a linear trend in change of mass over time. The mixed effects model including the average intercept for each diet group d(αd), the average slope over time for each diet group (βd), the random effects for individual (i) including differences in the intercept (ai,d) and slope (bi,d) from their group means, and the error (εt) including a first-order autoregressive coefficient (ϕ1) can be written as
| (Equation 2) |
where the ηi,d, are assumed to be independent, normally distributed errors
We included all possible interactions one-by-one and the best-fit model was determined by Akaike information criterion (AIC) values. Mixed effects models were compared using a likelihood ratios test.
Nucleotide sequence accession number
All sequencing data in the study, including 16S rRNA pyrosequencing data and metatranscriptomic data, are archived at NCBI Sequence Read Archive under Accession SRP070500.
Supplementary Material
Originality-Significance Statement.
This work is the first to examine the influence of diet on the functional and phenotypic diversity of the gut microbiota of a hibernating species during the pre-hibernation fattening period. Additionally, it is the first to characterize the effects of altered macronutrient content and calorie consumption on the metatranscriptome of a gut microbial community.
Acknowledgments
Funding was provided by the National Institute of General Medical Sciences of the National Institutes of Health under Award Numbers P20GM103395 and 1R15GM098938-01, and the National Science Foundation under Award DBI No 1263415, and by University of Alaska Anchorage Office of Undergraduate Research and Scholarship and The Alaska Heart Institute. We thank H.V. Carey for providing the SCFA extraction protocol, W.J. Radloff of the US Dairy Forage Research Center for technical assistance and use of instrumentation for SCFA analysis, M.D. Dillon and B. Briggs for assistance with the QIIME 1.6.0, AlienTrimmer and MG-RAST software and C. Cayabyab and S. Cain for assistance with animal care and sampling. The content is solely the responsibility of the authors and does not necessarily represent the official views of NIH or NSF.
Footnotes
The authors declare no conflicts of interest.
References
- Abbe K, Takahashi S, Yamada T. Involvement of oxygen-sensitive pyruvate formate-lyase in mixed-acid fermentation by Streptococcus mutansunder strictly anaerobic conditions. Journal of Bacteriology. 1982;152(1):175–182. doi: 10.1128/jb.152.1.175-182.1982. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bäckhed F, Manchester JK, Semenkovich CF, Gordon JI. Mechanisms underlying the resistance to diet-induced obesity in germ-free mice. PNAS. 2007;104(3):979–984. doi: 10.1073/pnas.0605374104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Barnes BM. Freeze avoidance in a mammal: body temperatures below 0±C in an arctic hibernator. Science. 1989;244:1593–95. doi: 10.1126/science.2740905. [DOI] [PubMed] [Google Scholar]
- Batzli GO, Sobaski ST. Distribution, abundance, and foraging patterns of ground squirrels near Atkasook, Alaska. Arct Alp Res. 1980;12:501–510. [Google Scholar]
- Ben-Amor K, Heilig H, Smidt H, Vaughan EE, Abee T, De Vos WM. Genetic diversity, injured, dead fecal bacterial assessed by fluorescence-activated cell sorting and 16S rRNA gene analysis. Appl Environ Microbiol. 2005;71(8):4679–4798. doi: 10.1128/AEM.71.8.4679-4689.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Benson AK, Kelly SA, Legge R, Ma F, Low SJ, Kim J, Zhang M, Oh PL, Nehrenberg D, Hua K, Kachman SD. Individuality in gut microbiota composition is a complex polygenic trait shaped by multiple environmental and host genetic factors. PNAS USA. 2010;107:18933–18938. doi: 10.1073/pnas.1007028107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bland JM, Altman DG. Statistics notes: measurement error proportional to the mean. Bmj. 1996;313(7049):106. doi: 10.1136/bmj.313.7049.106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Buck CL, Barnes BM. Annual cycle of body composition and hibernation in free-living arctic ground squirrels. J Mammal. 1999a;80:430–442. [Google Scholar]
- Buck CL, Barnes BM. Temperatures of hibernacula and changes in body composition of arctic ground squirrels over winter. J Mammal. 1999b;80(4):1264–1276. [Google Scholar]
- Buck CL, Barnes BM. Effects of ambient temperature on metabolic rate, respiratory quotient, and torpor in an arctic hibernator. Am J Physiol Regul Integr Comp Physiol. 2000;279:R255–R262. doi: 10.1152/ajpregu.2000.279.1.R255. [DOI] [PubMed] [Google Scholar]
- Buck CL, Barnes BM. Androgen in free-living arctic ground squirrels: seasonal changes and influence of staged male-male aggressive encounters. Horm Behav. 2003;43:318–326. doi: 10.1016/s0018-506x(02)00050-8. [DOI] [PubMed] [Google Scholar]
- Buck CL, Brenton A, Tøien Ø, Barnes BM. Overwinter body temperature patterns of free-living arctic ground squirrels (Spermophilis parryii) In: Lovegrove BG, McKenchie AE, editors. Hypometabolism in animals: Torpor, hibernation and cryobiology. University of KwaZulu-Natal, Pietermaritzburg; 2008. pp. 317–326. [Google Scholar]
- Carere CR, Kalia V, Sparling R, Cicek N, Levin DB. Pyruvate catabolism and hydrogen synthesis pathway genes of Clostridium thermocellum ATCC 27405. Indian J Microbiol. 2008;48(2):252–266. doi: 10.1007/s12088-008-0036-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Carey HV, Andrews MT, Martin SL. Mammalian hibernation: cellular and molecular responses to depressed metabolism and low temperature. Physiol Rev. 2003;83:1153–1181. doi: 10.1152/physrev.00008.2003. [DOI] [PubMed] [Google Scholar]
- Carey HV, Walters WA, Knight R. Seasonal restructuring of the ground squirrel gut microbiota over the annual hibernation cycle. Am J Physiol Regul Integ Comp Physiol. 2013;304(1):R33–R42. doi: 10.1152/ajpregu.00387.2012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Caporaso JG, Kuczynski J, Stombaugh J, Bittinger K, Bushman FD, Costello EK, Fierer N, Pena AG, Goodrich JK, Gordon JI, Huttley GA. QIIME allows analysis of high-throughput community sequencing data. Nat Methods. 2010;7(5):335–336. doi: 10.1038/nmeth.f.303. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Clarke KR, Gorley RN. Primer 6 User Manual. Plymouth Marine Laboratory; Plymouth: 2006. [Google Scholar]
- Criscuolo A, Brisse S. AlienTrimmer: a tool to quickly and accurately trim off multiple short contaminant sequences from high-throughput sequencing reads. Genomics. 2013;102(5-6):500–506. doi: 10.1016/j.ygeno.2013.07.011. [DOI] [PubMed] [Google Scholar]
- Daniel H, Gholami AM, Berry D, Desmarchelier C, Hahne H, Loh G, Mondot S, Lepage P, Rothballer M, Walker A, Böhm C. High-fat diet alters gut microbiota physiology in mice. ISME J. 2014;8(2):295–308. doi: 10.1038/ismej.2013.155. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dark J. Annual lipid cycles in hibernators: integration of physiology and behavior. Annu Rev Nutr. 2005;25:469–497. doi: 10.1146/annurev.nutr.25.050304.092514. [DOI] [PubMed] [Google Scholar]
- de La Serre CB, Ellis CL, Lee J, Hartman AL, Rutledge JC, Raybould HE. Propensity to high-fat diet-induced obesity in rats is accociated with changes in the gut microbiota and gut inflammation. Am J Physiol Gastrointest Liver Physiol. 2010;299(2):G440–448. doi: 10.1152/ajpgi.00098.2010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- de Wit N, Derrien M, Bosch-Vermeulen H, Oosterink E, Keshtkar S, Duval C, de Vogel-van den Bosch J, Kleerebezem M, Müller M, van der Meer R. Saturated fats stimulate obesity and hepatic steatosis and affects gut microbiota composition by an enhanced overflow of dietary fat to the distal intestine. Am J Physiol Gastrointest Liver Physiol. 2012;303(5):G589–599. doi: 10.1152/ajpgi.00488.2011. [DOI] [PubMed] [Google Scholar]
- Derrien M, Vaughan EE, Plugge CM, de Vos WM. Akkermansia muciniphilagen nov, sp nov, a human intestinal mucin-degrading bacterium. Int J Syst Evol Microbiol. 2004;54(5):1469–1476. doi: 10.1099/ijs.0.02873-0. [DOI] [PubMed] [Google Scholar]
- Deol P, Evans JR, Dhahbi J, Chellappa K, Han DS, Spindler S, Sladek FM. Soybean oil is more obesogenic and diabetogenic than coconut oil and fructose in mouse: potential role for the liver. PloS one. 2015;10(7):e0132672. doi: 10.1371/journal.pone.0132672. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dominguez-Bello MG, Costello EK, Contreras M, Magris M, Hidalgo G, Fierer N, Knight R. Delivery mode shapes the acquisition and structure of the initial microbiota across multiple body habitats in newborns. Proc Matl Acad Sci U S A. 2010;107(26):11971–11975. doi: 10.1073/pnas.1002601107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Donovan SM, Wang M, Li M, Friedberg I, Schwartz SL, Chapkin RS. Host-microbe interactions in the neonatal intestine: role of human milk oligosaccharides. Adv Nutr. 2012;3(3):450S–455S. doi: 10.3945/an.112.001859. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Duncan SH, Belenguer A, Holtrop G, Johnstone AM, Flint HJ, Lobley GE. Reduced dietary intake of carbohydrates by obese subjects results in decreased concentrations of butyrate and butyrate-producing bacteria in feces. Appl Environ Microbiol. 2007;73(4):1073–1078. doi: 10.1128/AEM.02340-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Duncan SH, Lobley GE, Holtrop G, Ince J, Johnstone AM, Louis P, Flint HJ. Human colonic microbiota associated with diet, obesity and weight loss. Int J Obes (Lond) 2008;32(11):1720–1724. doi: 10.1038/ijo.2008.155. [DOI] [PubMed] [Google Scholar]
- Edgar RC, Haas BJ, Clemente JC, Quince C, Knight R. UCHIME improves sensitivity and speed of chimera detection. Bioinformatics. 2011;27(16):2194–2200. doi: 10.1093/bioinformatics/btr381. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Everard A, Belzer C, Geurts L, Ouwerkerk JP, Druart C, Bindels LB, Guiot Y, Derrien M, Muccioli GG, Delzenne NM, De Vos WM. Cross-talk between Akkermansia muciniphilaand intestinal epithelium controls diet-induced obesity. Proc Natl Acad Sci U S A. 2013;110(22):9066–9071. doi: 10.1073/pnas.1219451110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ferrer M, Ruiz A, Lanza F, Haange S, Oberbach A, Till H, Bargiela R, Campoy C, Segura MT, Richter M, von Bergen M, Seifert J, Suarez A. Microbiota from the distal guts of lean and obese adolescents exhibit partial functional redundancy besides clear differences in community structure. Envir Micro. 2013;15(1):211–226. doi: 10.1111/j.1462-2920.2012.02845.x. [DOI] [PubMed] [Google Scholar]
- Fleissner CK, Huebel N, Abd El-Bary MM, Loh G, Klaus S, Blaut M. Absence of intestinal microbiota does not protect mice from diet-induced obesity. Br J Nutr. 2010;104(6):19–29. doi: 10.1017/S0007114510001303. [DOI] [PubMed] [Google Scholar]
- Frank CL, Karpovich S, Barnes BM. Dietary fatty acid composition and the hibernation patterns in free-ranging arctic ground squirrels. Physiological Biochem Zool. 2008;81(4):486–95. doi: 10.1086/589107. [DOI] [PubMed] [Google Scholar]
- Franzosa EA, Morgan XC, Segata N, Waldron L, Reyes J, Earl AM, Giannoukos G, Boylan MR, Ciulla D, Gevers D, Izard J. Relating the metatranscriptome and metagenome of teh human gut. PNAS USA. 2014;111(22):E2329–E2338. doi: 10.1073/pnas.1319284111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Geiser F. Reduction of metabolism during hibernation and daily torpor in mammals and birds: temperature effect or physiological inhibition. Annu Rev Physiol. 2004;66:239–274. doi: 10.1007/BF00692726. [DOI] [PubMed] [Google Scholar]
- Gillis EA, Morrison SF, Zazula GD, Hik DS. Evidence for selective caching by arctic ground squirrels living in alpine meadows in the Yukon. Arctic. 2005;58:354–360. [Google Scholar]
- Henderson AL, Cao W, Wang R, Lu M, Cerniglia CE. The effect of food restriction on the composition of intestinal microflora in rats. Exp Geront. 1998;33(3):239–247. doi: 10.1016/s0531-5565(97)00091-0. [DOI] [PubMed] [Google Scholar]
- Hildebrandt MA, Hoffmann C, Sherrill–Mix SA, Keilbaugh SA, Hamady M, Chen YY, Knight R, Ahima RS, Bushman F, Wu GD. High-fat diet determines the composition of the murine gut microbiome independently of obesity. Gasteroenterology. 2009;137(5):1716–1724. doi: 10.1053/j.gastro.2009.08.042. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hock RJ. Seasonal variation in physiologic functions of arctic ground squirrels and black bears. In: Lyman CP, Dawe AR, editors. Mammalian hibernation. Vol. 124. Harvard College; 1960. pp. 155–171. [Google Scholar]
- Huang EY, Leone VA, Devkota S, Wang Y, Brady MJ, Chang EB. Composition of dietary fat source shapes gut microbiota architecture and alters host inflammatory mediators in mouse adipose tissue. J Parent Ent Nutr. 2013 doi: 10.1177/0148607113486931. 0148607113486931. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jakobsdottir G, Xu J, Molin G, Ahrné S, Nyman M. High-fat diet reduces the formation of butyrate, but increases succinate, inflammation, liver fat and cholesterol in rats, while dietary fiber counteracts these effects. PLoS ONE. 2015;8(11):e80476. doi: 10.1371/journal.pone.0080476. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Koenig JE, Spor A, Scalfone N, Fricker AD, Stombaugh J, Knight R, Angenent LT, Ley RE. Succession of microbial consortia in the developing infant gut microbiome. 2011. PNAS USA. 2011;109(Supplemental 1):4578–4585. doi: 10.1073/pnas.1000081107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kolmeder CA, De Been M, Nikkilä J, Ritamo I, Mättö J, Valmu L, Salojärvi J, Palva A, Salonen A, de Vos WM. Comparative metaproteomics and diversity analysis of human intestinal microbiota testifies for its temporal stability and expression of core functions. PLoS ONE. 2012;7:e29913. doi: 10.1371/journal.pone.0029913. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Körtner G, Heldmaier G. Body weight cycles and energy balance in the alpine marmot (Marmota marmota) Physiol Zool. 1995;68(1):149–163. [Google Scholar]
- Kunz TH, Wrazen JA, Bernett CD. Changes in body mass and body composition in pre-hibernating little brown bats (Myotis lucifugus) Ecoscience. 1998;5:8–17. [Google Scholar]
- Lee TN, Fridinger RW, Barnes BM, Buck CL, O'Brien DM. Estimating lean mass over a wide range of body composition: a calibration of deuterium dilution in the arctic ground squirrel. Rapid Commun Mass Spectrom. 2011;25:3491–3496. doi: 10.1002/rcm.5253. [DOI] [PubMed] [Google Scholar]
- Ley RE, Bäckhed F, Turnbaugh PJ, Lozupone CA, Knight RD, Gordon JI. Obesity alters gut microbial ecology. Proc Natl Acad Sci. 2005;102(31):11070–11075. doi: 10.1073/pnas.0504978102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ley RE, Turnbaugh PJ, Klein S, Gordon JI. Human gut microbes associated with obesity. Nature. 2006;444(7122):1022–1023. doi: 10.1038/4441022a. [DOI] [PubMed] [Google Scholar]
- Ley RE. Obesity and the human microbiome. Curr Opin Gastroenterol. 2010;26(1):5–11. doi: 10.1097/MOG.0b013e328333d751. [DOI] [PubMed] [Google Scholar]
- Lozupone C, Knight R. UniFrac: a phylogenetic method for comparing microbial communities. Appl Environ Microbiol. 2005;71(12):8228–8235. doi: 10.1128/AEM.71.12.8228-8235.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lyman CP. Entering Hibernation. In: Lyman CP, Willis JS, Malan A, Wang LCH, editors. Hibernation and torpor in mammals and birds. Academic Press; New York: 1982. pp. 37–53. [Google Scholar]
- Mason DD, Barboza PS, Ward DH. Nutritional condition of Pacific Black Brant wintering at the extremes of their range. Condor. 2006;108:678. [Google Scholar]
- Mayer WV, Roche ET. Developmental patterns in the Barrow ground squirrel, Spermophilus undulates barrowensis. Growth. 1964;10:53–69. [PubMed] [Google Scholar]
- McLean IG. Seasonal patterns and sexual differences in the feeding ecology of arctic ground squirrels (Spermophilus parryii plesius) Can J Zool. 1984;63(6):1298–1301. [Google Scholar]
- McNulty NP, Yatsunenko T, Hsiao A, Faith JJ, Muegge BD, Goodman AL, Henrissat B, Oozeer R, Cools-Portier S, Gobert G, Chervaux C, Knights D, Lozupone CA, Knight R, Duncan AE, Bain JR, Muehlbauer MJ, Newgard CB, Heath AC, Gordon JI. The impact of a consortium of fermented milk strains on the gut microbiome of gnotobiotic mice and monozygotic twins. Sci Trans Med. 2011;3(106):106ra106–106ra106. doi: 10.1126/scitranslmed.3002701. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Meyer F, Paarmann D, D'Souza M, Olson R, Glass EM, Kubal M, Paczian T, Rodrigues A, Stevens R, Wilke A, Wilkening J, Edwards RA. The metagenomics RAST server – a public recourse for the automatic phylogenetic and functional analysis of metagenomes. BMC Bioinformatics. 2008;9:386. doi: 10.1186/1471-2105-9-386. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mrosovsky N. Lipid programs and life strategies in hibernators. Am Zool. 1976;16:685–697. [Google Scholar]
- Murphy EF, Cotter PD, Healy S, Marques TM, O'Sullivan O, Fouhy F, Clarke SF, O'Toole PW, Quigley EM, Stanton C, Ross PR, O-Doherty RM, Shanahan F. omposition and energy harvesting capacity of the gut microbiota: relationship to diet, obesity and time in mouse models. Gut. 2010;59:1635–1642. doi: 10.1136/gut.2010.215665. [DOI] [PubMed] [Google Scholar]
- O'Connell Motherway M, Zomer A, Leahy SC, Reunanen J, Bottacini F, Claesson MJ, O'Brien F, Flynn K, Casey PG, Munoz JAM, Kearney B. Functional genome analysis of Bifidobacterium breve UCC2003 reveals type IVb tight adherence (Tad) pili as an essential and conserved host-colonization factor. Proc Natl Acad Sci USA. 2011;108:11217–11222. doi: 10.1073/pnas.1105380108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Palmer C, Bik EM, DiGiulio DB, Relman DA, Brown PO. Development of the human infant intestinal microbiota. PLoS Biology. 2007;5:e177. doi: 10.1371/journal.pbio.0050177. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Parks DH, Beiko RG. Identifying biologically relevant differences between metagenomic communities. Bioinformatics. 2010;26(6):715–721. doi: 10.1093/bioinformatics/btq041. [DOI] [PubMed] [Google Scholar]
- Parks BW, Nam E, Org E, Kostem E, Norheim F, Hui ST, Pan C, Civelek M, Rau CD, Bennett BJ, Mehrabian M. Genetic control of obesity and gut microbiota composition in response to high-fat, high-sucrose diet in mice. Cell Metab. 2013;17(1):141–152. doi: 10.1016/j.cmet.2012.12.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Patterson E, O'Doherty RM, Murphy EF, Wall R, O'Sullivan O, Nilaweera K, Fitzgerald GF, Cotter PD, Ross RP, Stanton C. Impact of dietary fatty acids on metabolic activity and host intestinal microbiota composition in C57BL/6J mice. Br J Nutr. 2014;111(11):1905–1917. doi: 10.1017/S0007114514000117. [DOI] [PubMed] [Google Scholar]
- Piers LS, Walker KZ, Stoney RM, Soares MJ, O'Dea K. Substitution of saturated with monounsaturated fat in a 4-week diet affects body weight and composition of overweight and obese men. Br J Nutr. 2003;90(3):717–727. doi: 10.1079/bjn2003948. [DOI] [PubMed] [Google Scholar]
- Posey KA, Clegg DJ, Printz RL, Byun J, Morton GJ, Vivekanandan-Girl A, Pennathur S, Baskin DG, Heinecke JW, Woods SC, Schwartz MW, Niswender KD. Hypothalamic proinflammatory lipid accumulation, inflammaction, and insulin resistance in rats fed a high-fat diet. Am J Physiol Endocrinol Metab. 2009;6(5):E1003–1012. doi: 10.1152/ajpendo.90377.2008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Reeder J, Knight R. Rapidly denoising pyrosequencing amplicon reads by exploiting rank-abundance distributions. Nat Methods. 2010;7:668–669. doi: 10.1038/nmeth0910-668b. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Richter MM, Barnes BM, O'Reilly KM, Fenn AM, Buck CL. The influence of androgens on hibernation phenology of free-living male arctic ground squirrels. Hormones and Behavior. 2017 doi: 10.1016/j.yhbeh.2016.12.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Richter MM, Williams CT, Lee TN, Toien Ø, Florant GL, Barnes BM, Buck CL. Thermogenic capacity at sub-zero temperatures: how low can a hibernator go? Physiol Biochem Zool. 2015;88(1):81–89. doi: 10.1086/679591. [DOI] [PubMed] [Google Scholar]
- Ridaura VK, Faith JJ, Rey FE, Cheng J, Duncan AE, Kau AL, Griffin NW, Lombard V, Henrissat B, Bain JR, Muehlbauer MJ, Ilkayeva O, Semenkovich CF, Funai K, Hayashi DK, Lyle BJ, Martini MC, Ursell LK, Clemente JC, Van Treuren W, Walters WA, Knight R, Newgard CB, Heath AC, Gordon JI. Gut microbiota for twins discordant for obesity modulate metabolism in mice. Science. 2013;341(6150):1241214. doi: 10.1126/science.1241214. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schloss PD, Schubert AM, Zackular JP, Iverson KD, Young VB, Petrosino JF. Stabilization of thh murine gut microbiome following weaning. Gut Microbes. 2012;3(4):383–393. doi: 10.4161/gmic.21008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schomburg D, Stephan D. Enzyme Handbook. Vol. 11. Springer; Berlin Heidelberg: 1996. Formate C-acetyltransferase; pp. 871–875. [Google Scholar]
- Schwiertz A, Taras D, Schäfer K, Beijer S, Bos NA, Donus C, Hardt PD. Microbiota and SCFA in lean and overweight healthy subjects. Obesity. 2010;18(1):190–195. doi: 10.1038/oby.2009.167. [DOI] [PubMed] [Google Scholar]
- Serino M, Luche E, Gres S, Paylac A, Bergé M, Cenac C, Waget A, Klopp P, Iavovoni J, Klopp C, Mariette J, Bouchez O, Lluch J, Ouarné F, Monsan P, Valet P, Roques C, Amar J, Bouloumié A, Théodorou V, Burcelin R. Metabolic adaptation to a high-fat diet is associated with a change in the gut microbiota. Gut. 2012;61:543–553. doi: 10.1136/gutjnl-2011-301012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sheriff MJ, Williams CT, Kenagy GJ, Buck CL, Barnes BM. Thermoregulatory changes anticipate hibernation by 45 days: data from free-living arctic ground squirrels. J Comp Physiol B. 2012;182:841–847. doi: 10.1007/s00360-012-0661-z. [DOI] [PubMed] [Google Scholar]
- Sheriff MJ, Fridinger RW, Tøien Ø, Barnes BM, Buck CL. Metabolic Rate and Prehibernation Fattening in Free-Living Arctic Ground Squirrels. Physiol Biochem Zool. 2013;86(5):515–27. doi: 10.1086/673092. [DOI] [PubMed] [Google Scholar]
- Smith K, McCoy KD, Macpherson AJ. Use of axenic animals in studying the adaptation of mammals to their commensal intestinal microbiota. Sem Immunol. 2007;19(2):59–69. doi: 10.1016/j.smim.2006.10.002. [DOI] [PubMed] [Google Scholar]
- Sommer F, Ståhlman M, Ilkayeva O, Arnemo JM, Kindberg J, Josefsson J, Newgard CB, Fröbert O, Bäckhed F. The Gut Microbiota Modulates Energy Metabolism in the Hibernating Brown Bear Ursus arctos. Cell Rep. 2016;14(7):1655–1661. doi: 10.1016/j.celrep.2016.01.026. [DOI] [PubMed] [Google Scholar]
- Speakman JR. Doubly labeled water: Theory and practice. Chapman & Hall; London: 1997. [Google Scholar]
- Stevenson TJ, Buck CL, Duddleston KN. Temporal dynamics of the cecal gut microbiota of juvenile arctic ground squirrels: strong litter effect across the active season. Appl Environ Microbiol. 2014a;80(14):4260–4268. doi: 10.1128/AEM.00737-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stevenson TJ, Duddleston KN, Buck CL. Diversity, density and activity of the arctic ground squirrel cecal microbiota: effects of season and host physiological state. Appl Environ Microbiol. 2014b;80(18):5611–5622. doi: 10.1128/AEM.01537-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thomas GW. Soil pH and Soil Acidity. In: Sparks DL, editor. Methods of Soil Analysis: Chemical Methods Part 3. Soil Sci Soc of Am; Madison WI: 1996. [Google Scholar]
- Townsend KL, Lorenzi MM, Widmaier EP. High-fat diet-induced changes in body mass and hypothalamic gene expression in wild-type and leptin-deficient mice. Endocrine. 2008;33(2):176–188. doi: 10.1007/s12020-008-9070-1. [DOI] [PubMed] [Google Scholar]
- Turnbaugh PJ, Bäckhed F, Fulton L, Gordon JI. Diet-induced obesity is linked to marked but reversible alterations in the mouse distal gut microbiome. Cell host & microbe. 2008;3(4):213–223. doi: 10.1016/j.chom.2008.02.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Turnbaugh PJ, Gordon JI. The core gut microbiome, energy balance and obesity. J Physiol. 2009;587:4153–8. doi: 10.1113/jphysiol.2009.174136. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Turnbaugh PJ, Ridaura VK, Faith JJ, Rey FE, Knight R, Gordon JI. The effect of diet on the human gut microbiome: A metagenomic analysis in humanized gnotobiotic mice. Sci Trans Med. 2010;1(6):6ra14. doi: 10.1126/scitranslmed.3000322. [DOI] [PMC free article] [PubMed] [Google Scholar]
- van Breukelen F, Martin SL. Hibernation Continuum: Physiological and Molecular Aspects of Metabolic Plasticity in Mammals. Physiology. 2015;4:273–81. doi: 10.1152/physiol.00010.2015. [DOI] [PubMed] [Google Scholar]
- Vieira VJ, Valentine RJ, Wilund KR, Antao N, Baynard T, Woods JA. Effects of exercise and low-fat diet on adipose tissue inflammation and metabolic complications in obese mice. Am J Physiol Endocrinol Metab. 2009;296(5):E1164–E1171. doi: 10.1152/ajpendo.00054.2009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vijay-Kumar M, Aitken JD, Gewirtz AT. Toll like receptor-5: protecting the gut from enteric microbes. Semin Immunopathol. 2008;30(1):11–21. doi: 10.1007/s00281-007-0100-5. [DOI] [PubMed] [Google Scholar]
- Wadden TA. Treatment of obesity by moderate and severe caloric restriction: results of clinical research trials. Ann of Intern Med. 1993;119(7 Part 2):688–693. doi: 10.7326/0003-4819-119-7_part_2-199310011-00012. [DOI] [PubMed] [Google Scholar]
- Williams CT, Goropashnaya AV, Buck CL, Fedorov VB, Kohl F, Lee TN, Barnes BM. Hibernating above the permafrost: effects of ambient temperature and season on expression of metabolic genes in liver and brown adipose tissue of arctic ground squirrels. J Exp Biol. 2011;214:1300–1306. doi: 10.1242/jeb.052159. [DOI] [PubMed] [Google Scholar]
- Williams CT, Barnes BM, Kenagy GJ, Buck CL. Phenology of hibernation and reproduction in squirrels: integration of environmental cues with endogenous programming. J Zool. 2014 doi: 10.1111/jzo.12103. [DOI] [Google Scholar]
- Zazula GD, Mathewes RW, Harestad AS. Cache selection by arctic ground squirrels inhabiting boreal-steppe meadows of Southwest Yukon Territory, Canada. Arct Alp Res. 2005;38(4):631–638. [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.


