Abstract
Type I interferons (IFN-1) are cytokines that affect the expression of thousands of genes, resulting in profound cellular changes. IFN-1 activates the cell by dimerizing its two-receptor chains, IFNAR1 and IFNAR2, which are expressed on all nucleated cells. Despite a similar mode of binding, the different IFN-1s activate a spectrum of activities. The causes for differential activation may stem from differences in IFN-1-binding affinity, duration of binding, number of surface receptors, induction of feedbacks, and cell type-specific variations. All together these will alter the signal that is transmitted from the extracellular domain inward. The intracellular domain binds, directly or indirectly, different effector proteins that transmit signals. The composition of effector molecules deviates between different cell types and tissues, inserting an additional level of complexity to the system. Moreover, IFN-1s do not act on their own, and clearly there is much cross-talk between the activated effector molecules by IFN-1 and other cytokines. The outcome generated by all of these factors (processing step) is an observed phenotype, which can be the transformation of the cell to an antiviral state, differentiation of the cell to a specific immune cell, senescence, apoptosis, and many more. IFN-1 activities can be divided into robust and tunable. Antiviral activity, which is stimulated by minute amounts of IFN-1 and is common to all cells, is termed robust. The other activities, which we term tunable, are cell type-specific and often require more stringent modes of activation. In this review, I summarize the current knowledge on the mode of activation and processing that is initiated by IFN-1, in perspective of the resulting phenotypes.
Keywords: cell signaling, interferon, protein-protein interaction, receptor structure-function, STAT transcription factor
Introduction
From their discovery in 1957, type I interferons (IFN-1s)2 have been known for their antiviral activity, and are found in all nucleated cell types (1). IFNs are members of the cytokine family mediating diverse biological and cellular responses such as resistance to viral infections, regulation of cell survival, promotion of antitumor activities, and immune response modulation (2). Human type I interferons include 13 similar IFNαs with 80% homology, and single IFNω, IFNκ, IFNϵ, and IFNβ with lower homology (30–50%). In addition to type I, there are type II (with a single member: IFNγ) and type III (IFNλ) interferons. Common to all interferons is the activation of antiviral activity. However, both the ligand and the receptor components differ between type I, II, and III interferons. Interestingly, the different interferons share many of the same signaling cascade components. In this review, I focus on the activity of IFN-1. IFN-1s are found in all vertebrates, they are intron-less, and they have undergone relatively rapid gene duplication and evolution (3). This results in species specificity, i.e. human IFN-1s are not active in mouse in physiological concentrations and vice versa (4). All IFN-1s bind the same surface receptor, composed of two proteins, IFNAR1 and IFNAR2, found on the surface of all nucleated cells (Fig. 1A). With the exception of ΙFNα1 and IFNϵ, binding to IFNAR2 is much tighter then to IFNAR1, with the weak binding to IFNAR1 being evolutionary conserved (5). IFN-1s were between the first cytokines that were heterologously expressed, making them a preferred drug candidate against various diseases, including multiple sclerosis (IFNβ), hepatitis C (IFNα2), and various malignancies (6). In recent years, detrimental functions of IFN-1s in immunologically relevant scenarios have also been revealed (7). These multitudinous activities of IFN-1s are mediated through the induction or repression of thousands of genes (8). Although IFN-1s activate their receptors on all nucleated cells, the effects were often found to be cell type-specific. In addition, there is an important role to the IFN-1 subtype used, the duration of activation, and concentration (reviewed in Ref. 9). Following ternary complex assembly, the Janus family kinases (JAKs) Tyk2 and Jak1, which are associated with the membrane-proximal part of the cytoplasmic domains of IFNAR1 and IFNAR2, respectively, are activated by reciprocal trans-phosphorylation (10), followed by receptor phosphorylation, which in turn recruits and activates downstream signaling, and signal transducer and activator of transcription (STAT) proteins are the best studied examples of this (Fig. 1A). Here, I aim to provide a current view on the molecular basis for various IFN-1 activities, with emphasis toward the less common pathways.
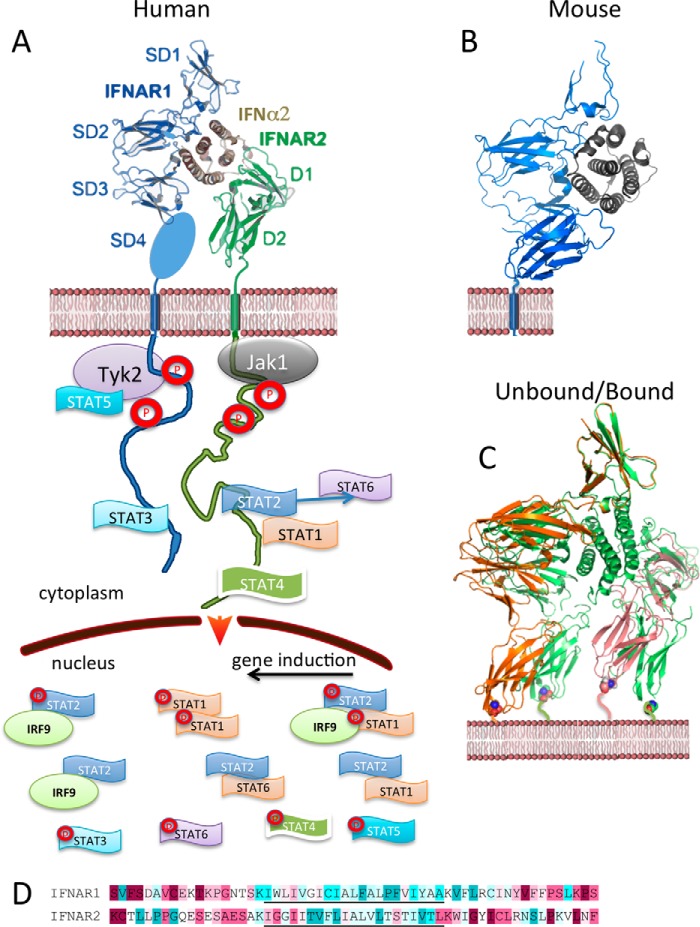
Figure 1.
Structure of IFN-1 ligand·receptor complex. A, ternary structure of the IFNα2 mutant YNS bound to IFNAR1 (domains 1–3, with domain 4 missing) and IFNAR2 (Protein Data Bank (PDB) 3SE3). IFN-1-induced receptor dimerization drives the cross-phosphorylation of associated JAKs and STATs. Although STAT1 and STAT2 are common to all cells, other STAT phosphorylations may be cell type-specific. Activated STATs are imported to the nucleus, where they serve as transcription factors. In addition to phosphorylated STATs, U-STAT1 and U-STAT2 were also observed in the nucleus. B, mouse IFNβ-IFNAR1 structure (PDB 3WCY). C, comparing the IFN-1 ternary complex structure (PDB 3SE3 in green) with their unbound counterparts (PDB IDs: IFNAR1, 3S98; IFNAR2, 1N6U). The alignment was done on SD1. D, sequence conservation of cytokine receptor TMDs (underlined) and their surroundings. Sequence conservation was determined using ConSurf. Highest to lowest conservation is colored from magenta to cyan.
Structural studies of IFN-1/receptor interactions
The ECD of human IFNAR1 spans amino acids (aa) 28–436 and is composed of four fibronectin type III subdomains of ∼100 aa each. This is followed by a 21-aa-long helical TMD, which in turn is connected to the 100-aa-long, mostly natively unstructured ICD. IFNAR2 has a similar architecture, although with a shorter ECD composed of two fibronectin type III subdomains and a more extended ICD of 251 aa. Structures of unbound IFNAR1, IFNAR2, IFNα2, IFNβ, the binary complexes IFN·IFNAR1 and IFN·IFNAR2, and the ternary complexes containing IFNAR1·IFNα2/IFNω·IFNAR2 were solved to high resolution (Fig. 1) (11–14). The ligand-docking modes of IFNα2 and IFNω in the two independently solved ternary complex structures appears to be shared by other type I IFN-1s, including IFNβ. This was verified by mutations, single particle electron microscopy, and blocking-antibody experiments (11, 15, 16). Comparing the determined structures of human and mice complexes shows differences in the details of binding, but with the same general ligand-receptor architecture (Fig. 1, A and B) (13). For a detailed review on the IFN-1 structures, see Ref. 17. Comparison of the unbound receptor subunits with the bound forms revealed a large movement in the receptor orientation upon binding. In particular, an outward movement of IFNAR1 was observed (Fig. 1C), which was verified by FRET measurements (18). As the IFNAR1 domain SD4 is not directly involved in ligand binding (19), the observed conformational movement in SD4 suggests a transfer of signal from the IFN-binding site to the membrane-proximal domain of IFNAR1. However, extensive mutagenesis, including deletions and insertions to the TMD of IFNAR1 and its immediate surroundings, did not shown any change in ligand binding, magnitude of signaling, or biological phenotype (20). This suggests a lack of flow of structural information between the ECD and ICD of IFNAR1. This is not surprising, taking into account the natively unstructured nature of the ICD of IFNAR1. Moreover, the TMDs of IFNAR1 and IFNAR2 are between the least conserved regions of the receptor sequences (Fig. 1D) (20), suggesting lack of importance of the specific sequence of this region. For IFN-1, it is now established that in the absence of ligand, the receptors do not dimerize, and thus signaling is initiated by IFN-1-mediated receptor dimerization and not by a structural signal transmitted from the ECD to the ICD (21, 22).
The varying binding affinities of IFN-1s and their functional implications
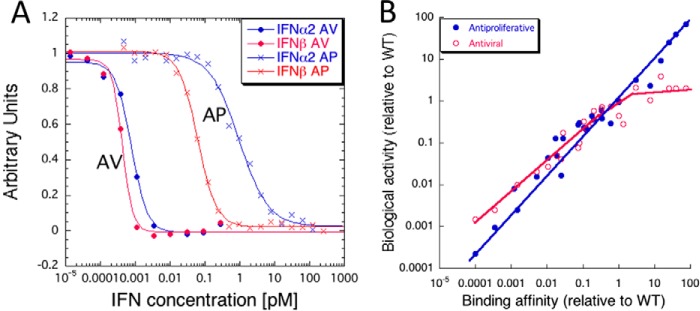
The main difference between IFN-1 subtypes is in their binding affinity toward the receptor subunits. The weakest binder to IFNAR2 is IFNα1, and the tightest is IFNβ (200 nm versus 0.2 nm affinity) (23). All IFNαs bind IFNAR1 with a similar low affinity of 1–5 μm, whereas IFNβ binds significantly tighter, with 100 nm affinity (23, 24). Despite some allosteric cross-talk (25), the binding affinity of IFNs to the ECD of either one of the receptor subunits is not influenced by the presence of the second receptor ECD (26). Reproducing the entire range of binding affinities of natural IFN-1s by mutant IFNα2 proteins showed that indeed, the variation in binding affinity accounts for many of the differences in biological potencies of natural IFNs (5, 11, 23), validating the receptor-centric approach toward understanding IFN-1 signaling. Antiviral and antiproliferative activities of IFN-1s are hallmarks of their differential responses. The EC50 for antiviral activity requires ∼1000-fold lower IFN concentration than required for activating antiproliferative activity (pm versus nm). Comparing the potency of IFNβ with that of IFNα2 shows only a minor difference of EC50 for antiviral activity, whereas a 50-fold lower concentration of the former is required to elicit antiproliferative activity (Fig. 2A). Fig. 2B shows the relation between antiviral and antiproliferative potencies of many IFNα2 mutants and their receptor-binding affinities. Both activities scale with affinity; however, antiproliferative activity directly scales over a 5 orders of magnitude affinity change, whereas antiviral activity is already optimized at the affinity of IFNα2.
Figure 2.
Affinity-activity relation of IFN-1 signaling. A, antiviral (AV) and antiproliferative (AP) response of WISH cells to IFN-1 treatment. Each dot represents the mean values of six independent antiviral and antiproliferative experiments (5). For clarity, the fraction of antiviral response (y axis) is shown as relative light transmission, while the fraction of antiproliferative response is shown as absorbance upon crystal violet staining (thus antiviral cell survival increases with IFN-1, whereas cell numbers decrease with increased antiproliferative response). B, relative to WT IFNα2, biological potency (EC50, antiviral, antiproliferative) is plotted against interferon receptor-binding affinity of IFNα2 mutants as a measure using surface plasmon resonance.
The good relation between binding and activity allowed for the generation of IFN-1s with higher affinities than naturally observed. In particular, the H57Y/E58N/Q61S triple mutation (YNS) was found to increase affinity toward IFNAR1 by 60-fold. Interestingly, these three positions are evolutionary conserved, suggesting that weak binding to IFNAR1 (which is found for all IFNs except IFNβ) is of benefit (27). Combining YNS with the tail of IFNα8 on the template of IFNα2 raises the affinity to IFNAR2 by an additional 15-fold (28), making it the tightest binding and most potent human IFN-1 available. The antiproliferative and antiviral potencies of the tighter binding mutants resemble IFNβ, strongly suggesting that increased binding affinity is the main difference between IFNβ and IFNα2 (Fig. 2B). The superior activity of YNSα8-tail fused to a 600-proline-alanine-serine tail (for prolonged serum half-life) in comparison with IFNα2 and IFNβ was clearly shown in an engineered mouse, harboring the human receptor ECDs (IFNAR1 and IFNAR2) fused to mouse TMDs and ICDs (called HyBNAR), for treating experimental autoimmune encephalomyelitis (29).
A recently engineered IFN variant, sIFN-I, was shown to bind IFNAR1 ∼5-fold tighter and IFNAR2 10-fold weaker than IFNα2, and was more potent as an antitumorigenic agent in mouse in comparison with IFNα2. As its activity on cell lines was not much increased, its enhanced in vivo activity may be due to its extended plasma lifetime (30).
Tight binding is not always an advantage, as is shown by a number of human diseases that are enhanced by elevated IFN-1 signaling. Examples include lupus, tuberculosis, AIDS, psoriatic skin inflammation, and cognitive decline (7, 31–35). A number of approaches have been taken to circumvent increased IFN-1 signaling, particularly in lupus. One strategy is to use neutralizing antibodies against IFN-1s. This resulted in only partial reduction of IFN-1 signaling due to the very low amount of IFN-1 required for signaling. A second approach was to use neutralizing antibodies against IFNAR1. This approach almost completely blocked IFN signaling (36, 37). A novel finding is that sphingosine 1-phosphate receptor 1 (S1PR1) agonists are effective in treating infectious and multiple autoimmune pathologies inhibiting IFN-1 responses specifically in plasmacytoid dendritic cells through acceleration of IFNAR1 turnover. This results in lower surface receptor numbers and thus reduced signaling (38). This finding is interesting, as system-wide blocking of IFN-1 activities as done by anti-IFNAR1 antibodies may result in susceptibility to infectious disease and cancer. Another approach is to block receptor signaling by engineering an antagonist based on IFNα2 that binds tightly to one receptor, but does not bind the second receptor (39–41). The antagonist, termed IFN-1ant, completely blocked IFN-1 antiproliferative activity, but left some of the antiviral activity intact. IFN-1ant was tested for efficacy in rhesus macaques, showing that it indeed significantly reduced IFN-1 signaling (34), which increased the severity of simian immunodeficiency virus infection.
The relation between IFN surface receptor levels, signaling, and disease
IFN-1 can activate diverse responses in different cells, tissues, and individuals, drawing attention to the variability of surface receptor expression levels. It was suggested by Wagner et al. (42) that IFNAR expression regulates the antiproliferative effects of interferons on cancer cells and solid tumors. These findings were confirmed by a multitude of studies showing that IFN-1s have a role in oncogene-induced senescence (43, 44), that IFNAR levels are predictive toward its antitumor effect (45), and that varying IFNAR levels directly affects the antitumorigenic potency of IFN-1s (46, 47). Indeed, enhanced tumor development was observed in mice lacking functional IFN-1 receptors (48).
Interestingly, in cells with abundant receptors, IFNα2 matched or even surpassed IFNβ activity, including antiproliferative potency, resulting in closure of the gap between these two IFN-1s at high receptor levels (49). Gradually reducing interferon receptor levels by increasing concentrations of siRNA resulted in a decreased signal, with the antiproliferative activity and STAT activation being linearly related to receptor numbers. Conversely, antiviral activity was not affected until receptor levels were less than 50% of the original one. Genes associated with immunomodulation, such as CXCL10 and CXCL11, were expressed in line with residual antiproliferative activity, whereas expression levels of genes associated with antiviral activity (such as MX1 and PKR) were affected in line with levels of antiviral activity (28). Although in the same cell line surface receptor levels were predictive toward levels of IFN-1 activation, this was not the case when comparing different cell lines with one another (28).
The most obvious way surface receptor levels can affect signaling is by increasing the magnitude of the signal. However, biophysical binding measurements on artificial membranes have also shown a direct link between receptor levels and the affinity of IFN-1 to form the ternary complex (19). Measuring binding in the presence of both receptor chains at very low surface receptor concentrations results in a similar affinity to that measured toward the individual receptor. However, at increased receptor concentrations, the observed binding affinity of IFN-1 was significantly increased due to increased avidity (17, 24, 50). This can explain why at high receptor concentrations the antiproliferative activity of IFNα2 equals that of IFNβ (49), and suggests a mechanism for cells to control IFN-1 activity by varying their receptor concentration.
STAT1- and STAT2-mediated signaling
The canonical pathway of IFN-1 signaling involves the phosphorylation of JAKs and STATs (51). Dimerization of IFNAR1 and IFNAR2 promotes interactions between Jak1 and Tyk2, which activate kinase activity via cross-phosphorylation, generating docking sites for other effector proteins, particularly STATs (Fig. 1A). Although STAT proteins are supposedly recruited to Tyr(P) via their SH2 domains, STAT2 was found to bind IFNAR2 via a constitutive, phosphorylation-independent binding site (52). STAT1 in turn was shown to be recruited via STAT2 (21), which was essential for STAT1 phosphorylation in U6A cells (52, 53). Conversely, in other cell lines, STAT1 activation by IFN-1 was also observed in STAT2-null cells (54), suggesting it to be independent on STAT2. The reason for these opposite findings is not clear.
The hallmark of IFN-1 signaling is the formation of a pSTAT1·pSTAT2 heterodimer, which in complex with IRF9 forms the transcription factor ISGF3 that promotes transcription of key interferon-stimulated genes (ISGs) (Fig. 1A). Next to pSTAT1·pSTAT2 heterodimers, homodimeric pSTAT1 is also being generated upon IFNAR activation, which is responsible for regulation of candidate IFN-γ activated sites (interferon-γ-activated sequence (GAS) elements). Although STAT1 is ubiquitously found to participate in cytokine signaling, STAT2 is specific for type I and type III interferons. However, although both type I and III IFNs act through the formation of the ISGF3 transcription factor, type III IFNs only activate a subset of genes found to be activated by type I IFNs. In the canonical pathway of IFN-1-mediated signaling, phosphorylation of STAT1 on Tyr-701 and STAT2 on Tyr-690 leads to heterodimerization in a parallel conformation. Apart from the canonical complex, STAT2·IRF9 in the absence of STAT1 was shown to be active. This complex continually shuttles in and out of the nucleus (55), even in its unphosphorylated form (U-STAT2). Moreover, it interacts with many of the known ISGs and was suggested to play a role in regulating ISG expression and execution of IFN-1-dependent biological activities, both independent and dependent on phosphorylation (56–60). In STAT1 KO cells overexpressing STAT2, ISG expression seems to correlate with STAT2 phosphorylation and the presence of a STAT2·IRF9 complex. The STAT2·IRF9-induced ISG expression persists for a longer time when compared with the transient nature of ISGF3 expression. Among genes activated by the STAT2·IRF9 complex are many known ISGs involved in antiviral response, leading to an antiviral response in cells lacking STAT1 (57, 61). The STAT2·IRF9 complex recognizes the core ISRE sequence at lower binding affinity when compared with ISGF3, explaining its partial gene activation and delayed activity (62). The STAT2·IRF9 response was absent in STAT1-Y701F homozygote mouse, where STAT1-Y701F suppressed the formation of the STAT2·IRF9 complex (63), maybe through the formation of U-STAT1·U-STAT2 complexes.
mTORC2 is a complex of the proteins Rictor and Sin1 with mTOR. It was found that KO of either Rictor or Sin1 significantly reduces pSTAT2, and thereby IFN-1 gene induction (64), in line with strongly reduced gene induction upon STAT2 knockdown (65). Also, in both cases, IFN-1-induced antiproliferative activity was absent. Although STAT2 is not directly involved in IFNγ signaling, the formation of U-STAT2 complex with STAT1 was shown to inhibit pSTAT1 homodimer nuclear translocation in response to IFNγ and thereby has an indirect effect on IFNγ signaling (66).
Phosphorylation of STAT1 and STAT2 is transient, diminishing within hours of the addition of IFN-1. However, ISG gene induction persists for a long time in the presence of IFN-1, and even in its absence (67). IFN-1s stimulate the overproduction of U-STAT1 and U-STAT2, which persists long after pSTAT is not observed. It was shown that expression of U-STAT1 and U-STAT2 to high levels resulted in ISG gene activation in the absence of IFN-1, with U-STAT1 also being found in the nuclei (68). In line with these findings, it was suggested that increased levels of U-ISGF3 drive a persistent response of IFN-1, in terms of both gene activation and the induction of the antiproliferative response, which requires the prolonged, continuous presence of IFN-1 (69). It should be noted that the slow induction of the antiproliferative response is a result of the presence of the caspase8 inhibitor, cFLIP. Knockdown of cFLIP significantly speeds up induction of antiproliferation, suggesting that this response can also proceed in the absence of high levels of U-STAT1 (65).
Non-canonical IFN-1-mediated signaling
In addition to STAT1 and STAT2, IFN-1 also strongly activates (phosphorylates) STATs 3–6 (Fig. 1A) (11, 70). STAT3 is activated by many cytokines, modulating the transcription of genes involved in cell differentiation, proliferation, apoptosis, angiogenesis, metastasis, and immune responses. STAT3 was shown to negatively regulate IFN-1-mediated responses. As a result, STAT3 knockdown cells display enhanced gene expression and antiviral activity in response to IFN-1s (71, 72). Mutually repressive roles for STAT3 and IFN-1 signaling following viral infection were identified (73). The effect of pSTAT3 on ISGF3-dependent transcription depends on the Sin3a complex as a cell-specific repressor of STAT3 activity, exerting opposite regulation on STAT3- and ISGF3-dependent transcription and thus acting as a cell-specific modulator of IFN-1 action (74).
STAT4 binds IFNAR2 and is activated by IFN-1s (75). STAT4 promotes IFNγ production during viral infection, and conversely, pSTAT1 appears to negatively regulate IFNγ production (76). IFN-1-activated STAT4 was required for peak expansion of antigen-specific CD8 T cells, which together with low STAT1 levels result in resistance to IFN-1-mediated antiproliferation (77).
STAT5 interacts constitutively with IFNAR-associated Tyk2. During IFN-1 stimulation, its tyrosine-phosphorylated form acts as a docking site for the SH2 domain of CrkL. CrkL and STAT5 then form a complex that translocates to the nucleus and promotes the production of a subset of ISGs (78, 79). STAT5 is phosphorylated on serines 725/730 in an IFN-1-dependent manner. In mouse fibroblasts, disrupting the STAT5a and STAT5b genes hinders IFN-1-dependent gene transcription via GAS elements (79), suggesting STAT5 to be an integral part of IFN-1-induced signaling. Moreover, in primary human B and CD4 T cells, ISGF3 is not the main activator of gene expression upon IFNβ treatment, but STAT5 activation of specific pro-survival genes has a primary role. The differences in the activation of STAT1 and STAT5 in different leukocyte subsets result in the induction of pro- and antiapoptotic genes, respectively, explaining some of the varied effects of IFN-1s (80).
Activation of STAT6 results in the formation of a STAT2·STAT6 complex in response to IFN-1 stimulation as detected in B cells and is suggested to take part in the IFN-1-mediated antiproliferative activity (70). Similarly, it was recently suggested that STAT6, in a STAT2-dependent manner, exerts antiproliferative activity also in Daudi cells upon IFN-1 induction (81).
In addition to STATs, other signaling factors have a role in IFN-mediated activities. These include isoforms of the protein kinase C and the multifunctional adaptor protein CrkL, activation of the MAPK p38 via the small G-protein Rac1, and activation of the AKT/mTOR/ULK1 pathway via PI3K and the ERK/MAP kinase pathway (2, 82–86). Recently, ULK1 was identified as an important regulator of IFN activity that is independent of STAT activation (83). It is not clear whether ULK1 is directly or indirectly activated by IFNAR, and it is suggested that ULK1 acts through p38 to affect ISG signaling and IFN-induced biological activity.
Feedback mechanisms
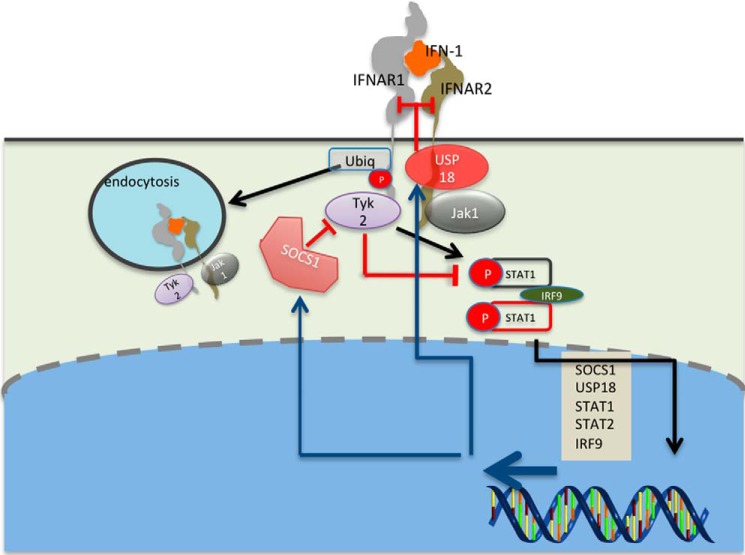
As mentioned above, elevated, prolonged IFN-1 responses are detrimental, causing increased levels of inflammation (6, 36, 87). Therefore, multiple layer feedback mechanisms on the receptor and on its activation and signaling exist (Fig. 3). Endocytosis of IFN-1 receptors was recorded minutes after IFN stimulation. This is initiated upon the formation of the ternary complex via ubiquitination of IFNAR1, which exposes an endocytic motif masked by Tyk2 in the inactive state (88, 89). At high levels of surface IFNAR1, a Tyk2-independent pathway also promotes IFNAR1 ubiquitination and degradation (90). The level of endocytosis is directly related to the strength of the signal (i.e. to the concentration of the ternary complex): low IFN-1 concentration and/or low-affinity IFN-1 weakly affects surface receptor levels, whereas at high concentrations of tightly binding IFN-1s, the receptor levels are reduced by over 60% (50). In the extreme case of the K152R IFNω mutant, IFNAR2 levels were reduced by >90% (11), whereas for the IFN-1ant mutant that binds tightly to IFNAR2, but does not bind IFNAR1, no endocytosis was observed (40). IFNAR1 levels can stay suppressed for days upon induction with IFNβ, but not IFNα2, whereas IFNAR2 levels return to normal (50). Receptor endocytosis does not immediately abrogate signaling, as it was shown that the receptors keep on signaling in the early endosome, with the VPS35 subunit binding to IFNAR2, resulting in its recycling to the plasma membrane, whereas IFNAR1 is sorted to the lysosome for degradation (91, 92).
Figure 3.
Feedback mechanisms regulating IFN-1 signaling. Endocytosis is already observed minutes after interferon receptor activation by IFN-1. Endocytosis is activated by Tyk2 phosphorylation of Ser-535 on IFNAR1, facilitating its polyubiquitination. This in turn exposes a masked linear endocytic motif enabling the recruitment of the AP2 complex and ensuing internalization of the type I IFN receptor. STAT phosphorylation peaks ∼1 h after initiation of IFN-1 induction, and is then suppressed by SOCS1 binding to Tyk2. SOCS1 expression is rapidly up-regulated by IFN-1, enhancing the negative feedback. A third layer of feedback is mediated by USP18, which binds IFNAR2 and obstructs complex formation. USP18 expression is regulated by IFN-1, enhancing the negative feedback with increased expression levels. Obstruction of any one of these feedback mechanisms was shown to cause disease due to enhanced IFN-1 signaling (6).
A second feedback mechanism is the IFN-1-stimulated USP18 protein (Fig. 3). The ISG15-specific protease activity of USP18 is not required for IFNAR desensitization. Rather, binding to IFNAR2 and STAT2 was shown to be responsible for its activity as a negative feedback regulator (93, 94). USP18 reduces the cell surface-binding affinity of IFN-1s by potently interfering with the recruitment of IFNAR1 to the ternary complex (22, 95, 96). Thus, the responsiveness to IFNα is reduced after the first wave of gene induction, whereas IFNβ, due to its 30-fold higher binding affinity to IFNAR1, is still able to efficiently recruit IFNAR1 and form a signaling ternary complex even at high USP18 concentrations. This may also explain the evolutionary conservation of weak binding affinity of all IFNαs toward IFNAR1, which permits the USP18 feedback to function. As USP18 levels remain high for many hours after secession of an IFN-1 signal, for a prolonged time, it prevents reactivation of the receptor using IFNα, but not IFNβ. The modulation of ligand-binding affinity by USP18 makes it a prime candidate to explain differential activation by IFNβ versus IFNα, as was further verified by using high-affinity IFNα mutants (22, 95, 96). Lack of USP18 results in a persistent, strong IFN-1 signal, as shown for mouse brain that developed destructive interferonopathy (97). On the other hand, reduced USP18 can increase antiviral immunity (98).
The third layer of negative regulation is SOCS1 (suppressor of cytokine signaling) (Fig. 3), which is a potent inhibitor of JAKs and binds to pTyk2 (99). Tyk2 inhibition by SOCS1 results in a reduced IFN-1 response due to decreased Tyk2-mediated activity, and also through the negative impact on IFNAR1 surface expression, which is stabilized by Tyk2. The SOCS1-SH2 domain is only functionally relevant if the correct phosphorylated target is present, and thus the SOCS rely on an active signal, adding a further intrinsic level of regulation to the system (100). SOCS1 expression is rapidly up-regulated, even with low amounts of IFNα2; however, its production is maintained over time only with a continuous, strong IFN-1 signal generated by IFNβ or high amounts of IFNα2 (40).
IFN-1 induces robust and tunable activities
The most basic and easily measured modes of IFN-1 activities are robust and tunable. Robust activities are those that require only minute amounts of IFN-1, independent of its binding affinity. These activities are common to all cells, and are activated even with low surface receptor levels. The most redundant robust activity is the antiviral activity of IFN-1s, which serves as the first line of defense for viral and other pathogen attacks. Robust activities are driven by ISRE promoter elements (40). On the other hand, some of the IFN-1-induced activities are cell type-specific, requiring 1000-fold higher IFN-1 concentration, are more strongly activated by the high-affinity binding IFNβ (or IFNα2 variants engineered for tight binding), are induced after longer times of IFN-1 treatment, and require higher surface receptor numbers. We define these activities as tunable. Antiproliferative and immunomodulatory activities are good examples of those. An analysis of the promoter regions of genes involved in tunable activities showed no enrichment in ISRE sequences (40). Robust responses are saturated by IFNα, with only tunable activities taking advantage of the higher affinity of IFNβ (23, 50). USP18 seems to be a perfect candidate to explain the molecular mechanism supporting the deviation between robust and tunable activities, as it inhibits binding of lower affinity IFN-1s over time (22). A still open question is whether disease states resulting from high persistent interferon levels are due to up-regulation of robust or tunable genes. Robust genes are annotated by Gene Ontology biological process terms (using DAVID Bioinformatics Resources 6.8) as related to response to viruses, biotic stimulus, and MHC1, whereas tunable genes are annotated to cytokine and chemokine activities, taxis, inflammation, and antiproliferative activities (50). The distinction is clinically relevant, as blocking the induction of robust genes requires total shutdown of the system, whereas blocking tunable genes requires only partial shutdown, leaving at least partial antiviral activity intact. For the latter, an antagonist such as IFN-1ant or its derivatives would be perfect (41), whereas for the former, a receptor blocking antibody such as anifrolumab (which progressed to phase III clinical trials) would be preferable (37). As mentioned above, tunable activities are cell type-specific. It is interesting to note that that in cells resistant to IFN-1-induced antiproliferative activity, very little expression of tunable genes was observed (40). Although most of these genes are not related to antiproliferative activity, it suggests that the same program controls them.
Conclusions and perspectives
60 years have passed since the discovery of IFN-1 by Isaacs and Lindenmann (1). At the beginning, IFN-1 was a great hope, and was compared with the discovery of antibiotics, but to combat viruses. Although IFN-1 was initially recognized as an antiviral agent, it became clear with time that cellular activation by IFN-1 is much broader than just fighting viruses. This acknowledgment resulted in a revival of interferon research and to our realization that there is much we yet do not understand. In addition to being an antiviral agent, it connects innate to acquired immunity, drives immune cell differentiation, is involved in monitoring cellular health, and has the ability to drive senescence and apoptosis. All these different activities are governed by binding of IFN-1 to its cell surface receptors, with no evidence to refute the receptor-centric view. One could compare this with turning on the same switch, resulting in multiple different outcomes. In this review, I tried to provide the current knowledge of how this is possible. How can binding generate a multitude of signals? Although we now have a quantitative understanding of differences in ligand-receptor binding, much is to be learned on how the different modes of binding are processed to molecular differences in signaling. IFN-1 activates six different STATs, in addition to other signaling molecules. How the outcome of this complex processing stage is dictated is not well-understood and provides a challenge for the future. It should be clear that many of the outcomes are cell type-specific. In this context, one of the complications of IFN-1 research is that many of the findings were not repeated on multiple cellular backgrounds, making it difficult to assess the generality of these findings. This is particularly true for tunable activities, which generate much of the interest in this field. Better understanding of IFN-1 signaling will enable us to engineer IFN-1 variants to optimally drive specific activities, making it again a drug of choice.
This research was funded by the United States-Israel Binational Science Foundation (BSF) Grant 2011093 and the I-CORE Program of the Planning and Budgeting Committee and the Israel Science Foundation Grant 1775/12. The author declares that he has no conflicts of interest with the contents of this article.
- IFN-1
- type I interferon
- IFNAR
- IFN-α/β receptor
- ECD
- extracellular domain
- ICD
- intracellular domain
- TMD
- transmembrane domain
- mTOR
- mechanistic target of rapamycin
- ISG
- interferon-stimulated gene
- ISRE
- interferon-stimulated response element
- SOCS
- suppressor of cytokine signaling
- aa
- amino acid(s)
- p
- phosphorylated
- U
- unphosphorylated
- GAS
- interferon-γ-activated sequence.
References
- 1. Isaacs A., and Lindenmann J. (1957) Virus interference. I. The interferon. Proc. R Soc. Lond. B Biol. Sci. 147, 258–267 [PubMed] [Google Scholar]
- 2. Platanias L. C. (2005) Mechanisms of type-I- and type-II-interferon-mediated signalling. Nat. Rev. Immunol. 5, 375–386 [DOI] [PubMed] [Google Scholar]
- 3. Krause C. D., and Pestka S. (2015) Cut, copy, move, delete: the study of human interferon genes reveal multiple mechanisms underlying their evolution in amniotes. Cytokine 76, 480–495 [DOI] [PubMed] [Google Scholar]
- 4. Harari D., Abramovich R., Zozulya A., Smith P., Pouly S., Köster M., Hauser H., and Schreiber G. (2014) Bridging the species divide: transgenic mice humanized for type-I interferon response. PLoS ONE 9, e84259. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Jaitin D. A., Roisman L. C., Jaks E., Gavutis M., Piehler J., Van der Heyden J., Uze G., and Schreiber G. (2006) Inquiring into the differential action of interferons (IFNs): an IFN-α2 mutant with enhanced affinity to IFNAR1 is functionally similar to IFN-β. Mol. Cell. Biol. 26, 1888–1897 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Trinchieri G. (2010) Type I interferon: friend or foe? J. Exp. Med. 207, 2053–2063 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. McNab F., Mayer-Barber K., Sher A., Wack A., and O'Garra A. (2015) Type I interferons in infectious disease. Nat. Rev. Immunol. 15, 87–103 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Samarajiwa S. A., Forster S., Auchettl K., and Hertzog P. J. (2009) INTERFEROME: the database of interferon regulated genes. Nucleic Acids Res. 37, D852–7 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Schreiber G., and Piehler J. (2015) The molecular basis for functional plasticity in type I interferon signaling. Trends Immunol. 36, 139–149 [DOI] [PubMed] [Google Scholar]
- 10. Cohen B., Novick D., Barak S., and Rubinstein M. (1995) Ligand-induced association of the type I interferon receptor components. Mol. Cell. Biol. 15, 4208–4214 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Thomas C., Moraga I., Levin D., Krutzik P. O., Podoplelova Y., Trejo A., Lee C., Yarden G., Vleck S. E., Glenn J. S., Nolan G. P., Piehler J., Schreiber G., and Garcia K. C. (2011) Structural linkage between ligand discrimination and receptor activation by type I interferons. Cell 146, 621–632 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Chill J. H., Quadt S. R., Levy R., Schreiber G., and Anglister J. (2003) The human type I interferon receptor: NMR structure reveals the molecular basis of ligand binding. Structure 11, 791–802 [DOI] [PubMed] [Google Scholar]
- 13. de Weerd N. A., Vivian J. P., Nguyen T. K., Mangan N. E., Gould J. A., Braniff S. J., Zaker-Tabrizi L., Fung K. Y., Forster S. C., Beddoe T., Reid H. H., Rossjohn J., and Hertzog P. J. (2013) Structural basis of a unique interferon-β signaling axis mediated via the receptor IFNAR1. Nat. Immunol. 14, 901–907 [DOI] [PubMed] [Google Scholar]
- 14. Senda T., Shimazu T., Matsuda S., Kawano G., Shimizu H., Nakamura K. T., and Mitsui Y. (1992) Three-dimensional crystal structure of recombinant murine interferon-β. EMBO J. 11, 3193–3201 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Li Z., Strunk J. J., Lamken P., Piehler J., and Walz T. (2008) The EM structure of a type I interferon-receptor complex reveals a novel mechanism for cytokine signaling. J. Mol. Biol. 377, 715–724 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Slutzki M., Jaitin D. A., Yehezkel T. B., and Schreiber G. (2006) Variations in the unstructured C-terminal tail of interferons contribute to differential receptor binding and biological activity. J. Mol. Biol. 360, 1019–1030 [DOI] [PubMed] [Google Scholar]
- 17. Piehler J., Thomas C., Garcia K. C., and Schreiber G. (2012) Structural and dynamic determinants of type I interferon receptor assembly and their functional interpretation. Immunol. Rev. 250, 317–334 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Strunk J. J., Gregor I., Becker Y., Li Z., Gavutis M., Jaks E., Lamken P., Walz T., Enderlein J., and Piehler J. (2008) Ligand binding induces a conformational change in ifnar1 that is propagated to its membrane-proximal domain. J. Mol. Biol. 377, 725–739 [DOI] [PubMed] [Google Scholar]
- 19. Lamken P., Gavutis M., Peters I., Van der Heyden J., Uzé G., and Piehler J. (2005) Functional cartography of the ectodomain of the type I interferon receptor subunit ifnar1. J. Mol. Biol. 350, 476–488 [DOI] [PubMed] [Google Scholar]
- 20. Sharma N., Longjam G., and Schreiber G. (2016) Type I interferon signaling is decoupled from specific receptor orientation through lenient requirements of the transmembrane domain. J. Biol. Chem. 291, 3371–3384 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Löchte S., Waichman S., Beutel O., You C., and Piehler J. (2014) Live cell micropatterning reveals the dynamics of signaling complexes at the plasma membrane. J. Cell Biol. 207, 407–418 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Wilmes S., Beutel O., Li Z., Francois-Newton V., Richter C. P., Janning D., Kroll C., Hanhart P., Hötte K., You C., Uzé G., Pellegrini S., and Piehler J. (2015) Receptor dimerization dynamics as a regulatory valve for plasticity of type I interferon signaling. J. Cell Biol. 209, 579–593 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Lavoie T. B., Kalie E., Crisafulli-Cabatu S., Abramovich R., DiGioia G., Moolchan K., Pestka S., and Schreiber G. (2011) Binding and activity of all human α interferon subtypes. Cytokine 56, 282–289 [DOI] [PubMed] [Google Scholar]
- 24. Lamken P., Lata S., Gavutis M., and Piehler J. (2004) Ligand-induced assembling of the type I interferon receptor on supported lipid bilayers. J. Mol. Biol. 341, 303–318 [DOI] [PubMed] [Google Scholar]
- 25. Akabayov S. R., Biron Z., Lamken P., Piehler J., and Anglister J. (2010) NMR mapping of the IFNAR1-EC binding site on IFNα2 reveals allosteric changes in the IFNAR2-EC binding site. Biochemistry 49, 687–695 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Waichman S., Bhagawati M., Podoplelova Y., Reichel A., Brunk A., Paterok D., and Piehler J. (2010) Functional immobilization and patterning of proteins by an enzymatic transfer reaction. Anal. Chem. 82, 1478–1485 [DOI] [PubMed] [Google Scholar]
- 27. Kalie E., Jaitin D. A., Abramovich R., and Schreiber G. (2007) An interferon α2 mutant optimized by phage display for IFNAR1 binding confers specifically enhanced antitumor activities. J. Biol. Chem. 282, 11602–11611 [DOI] [PubMed] [Google Scholar]
- 28. Levin D., Harari D., and Schreiber G. (2011) Stochastic receptor expression determines cell fate upon interferon treatment. Mol. Cell. Biol. 31, 3252–3266 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Harari D., Kuhn N., Abramovich R., Sasson K., Zozulya A. L., Smith P., Schlapschy M., Aharoni R., Köster M., Eilam R., Skerra A., and Schreiber G. (2014) Enhanced in vivo efficacy of a type I interferon superagonist with extended plasma half-life in a mouse model of multiple sclerosis. J. Biol. Chem. 289, 29014–29029 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Zhang K.-J., Yin X.-F., Yang Y.-Q., Li L. L., Xu Y.-N., Chen L.-Y., Liu X.-J., Yuan S.-J., Fang X.-L., Xiao J., Wu S., Xu H.-N., Chu L., Katlinski K. V., et al. (2017) A potent in vivo antitumor efficacy of novel recombinant type I interferon. Clin. Cancer Res. 10.1158/1078-0432.CCR-16-1386 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Baruch K., Deczkowska A., David E., Castellano J. M., Miller O., Kertser A., Berkutzki T., Barnett-Itzhaki Z., Bezalel D., Wyss-Coray T., Amit I., and Schwartz M. (2014) Aging-induced type I interferon response at the choroid plexus negatively affects brain function. Science 346, 89–93 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Mayer-Barber K. D., Andrade B. B., Oland S. D., Amaral E. P., Barber D. L., Gonzales J., Derrick S. C., Shi R., Kumar N. P., Wei W., Yuan X., Zhang G., Cai Y., Babu S., Catalfamo M., et al. (2014) Host-directed therapy of tuberculosis based on interleukin-1 and type I interferon crosstalk. Nature 511, 99–103 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Sandling J. K., Garnier S., Sigurdsson S., Wang C., Nordmark G., Gunnarsson I., Svenungsson E., Padyukov L., Sturfelt G., Jönsen A., Bengtsson A. A., Truedsson L., Eriksson C., Rantapää-Dahlqvist S., Mälarstig A., et al. (2011) A candidate gene study of the type I interferon pathway implicates IKBKE and IL8 as risk loci for SLE. Eur. J. Hum. Genet. 19, 479–484 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Sandler N. G., Bosinger S. E., Estes J. D., Zhu R. T., Tharp G. K., Boritz E., Levin D., Wijeyesinghe S., Makamdop K. N., del Prete G. Q., Hill B. J., Timmer J. K., Reiss E., Yarden G., Darko S., et al. (2014) Type I interferon responses in rhesus macaques prevent SIV infection and slow disease progression. Nature 511, 601–605 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. Gui J., Gober M., Yang X., Katlinski K. V., Marshall C. M., Sharma M., Werth V. P., Baker D. P., Rui H., Seykora J. T., and Fuchs S. Y. (2016) Therapeutic elimination of the type 1 interferon receptor for treating psoriatic skin inflammation. J. Invest. Dermatol. 136, 1990–2002 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36. Crow M. K., Olferiev M., and Kirou K. A. (2015) Targeting of type I interferon in systemic autoimmune diseases. Transl. Res. 165, 296–305 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37. Oon S., Wilson N. J., and Wicks I. (2016) Targeted therapeutics in SLE: emerging strategies to modulate the interferon pathway. Clin. Transl. Immunology 5, e79. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38. Teijaro J. R., Studer S., Leaf N., Kiosses W. B., Nguyen N., Matsuki K., Negishi H., Taniguchi T., Oldstone M. B. A., and Rosen H. (2016) S1PR1-mediated IFNAR1 degradation modulates plasmacytoid dendritic cell interferon-α autoamplification. Proc. Natl. Acad. Sci. U.S.A. 113, 1351–1356 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39. Pan M., Kalie E., Scaglione B. J., Raveche E. S., Schreiber G., and Langer J. A. (2008) Mutation of the IFNAR-1 receptor binding site of human IFN-α2 generates type I IFN competitive antagonists. Biochemistry 47, 12018–12027 [DOI] [PubMed] [Google Scholar]
- 40. Levin D., Schneider W. M., Hoffmann H. H., Yarden G., Busetto A. G., Manor O., Sharma N., Rice C. M., and Schreiber G. (2014) Multifaceted activities of type I interferon are revealed by a receptor antagonist. Sci. Signal 7, ra50. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41. Urin V., Levin D., Sharma N., Harari D., and Schreiber G. (2015) Fine tuning of a type 1 interferon antagonist. PLoS ONE 10, e0130797. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42. Wagner T. C., Velichko S., Chesney S. K., Biroc S., Harde D., Vogel D., and Croze E. (2004) Interferon receptor expression regulates the antiproliferative effects of interferons on cancer cells and solid tumors. Int. J. Cancer 111, 32–42 [DOI] [PubMed] [Google Scholar]
- 43. Katlinskaya Y. V., Katlinski K. V., Yu Q., Ortiz A., Beiting D. P., Brice A., Davar D., Sanders C., Kirkwood J. M., Rui H., Xu X., Koumenis C., Diehl J. A., and Fuchs S. Y. (2016) Suppression of type I interferon signaling overcomes oncogene-induced senescence and mediates melanoma development and progression. Cell Rep. 15, 171–180 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44. Chiantore M. V., Vannucchi S., Accardi R., Tommasino M., Percario Z. A., Vaccari G., Affabris E., Fiorucci G., and Romeo G. (2012) Interferon-β induces cellular senescence in cutaneous human papilloma virus-transformed human keratinocytes by affecting p53 transactivating activity. PLoS ONE 7, e36909. [DOI] [PMC free article] [PubMed] [Google Scholar] [Retracted]
- 45. Booy S., van Eijck C. H. J., Dogan F., van Koetsveld P. M., and Hofland L. J. (2014) Influence of type-I interferon receptor expression level on the response to type-I interferons in human pancreatic cancer cells. J. Cell. Mol. Med. 18, 492–502 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46. Bhattacharya S., HuangFu W. C., Dong G., Qian J., Baker D. P., Karar J., Koumenis C., Diehl J. A., and Fuchs S. Y. (2013) Anti-tumorigenic effects of Type 1 interferon are subdued by integrated stress responses. Oncogene 32, 4214–4221 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47. Fuchs S. Y. (2013) Hope and fear for interferon: the receptor-centric outlook on the future of interferon therapy. J. Interferon Cytokine Res. 33, 211–225 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48. Picaud S., Bardot B., De Maeyer E., and Seif I. (2002) Enhanced tumor development in mice lacking a functional type I interferon receptor. J. Interferon Cytokine Res. 22, 457–462 [DOI] [PubMed] [Google Scholar]
- 49. Moraga I., Harari D., Schreiber G., Uzé G., and Pellegrini S. (2009) Receptor density is key to the α2/β interferon differential activities. Mol. Cell. Biol. 29, 4778–4787 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50. Kalie E., Jaitin D. A., Podoplelova Y., Piehler J., and Schreiber G. (2008) The stability of the ternary interferon-receptor complex rather than the affinity to the individual subunits dictates differential biological activities. J. Biol. Chem. 283, 32925–32936 [DOI] [PubMed] [Google Scholar]
- 51. Stark G. R., and Darnell J. E. Jr. (2012) The JAK-STAT pathway at twenty. Immunity 36, 503–514 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52. Li X., Leung S., Kerr I. M., and Stark G. R. (1997) Functional subdomains of STAT2 required for preassociation with the α interferon receptor and for signaling. Mol. Cell. Biol. 17, 2048–2056 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53. Leung S., Qureshi S. A., Kerr I. M., Darnell J. E. Jr., and Stark G. R. (1995) Role of STAT2 in the α interferon signaling pathway. Mol. Cell. Biol. 15, 1312–1317 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54. Romero-Weaver A. L., Wang H.-W., Steen H. C., Scarzello A. J., Hall V. L., Sheikh F., Donnelly R. P., and Gamero A. M. (2010) Resistance to IFN-α-induced apoptosis is linked to a loss of STAT2. Mol. Cancer Res. 8, 80–92 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55. Veals S. A., Schindler C., Leonard D., Fu X. Y., Aebersold R., Darnell J. E. Jr., and Levy D. E. (1992) Subunit of an α-interferon-responsive transcription factor is related to interferon regulatory factor and Myb families of DNA-binding proteins. Mol. Cell. Biol. 12, 3315–3324 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56. Testoni B., Völlenkle C., Guerrieri F., Gerbal-Chaloin S., Blandino G., and Levrero M. (2011) Chromatin dynamics of gene activation and repression in response to interferon α (IFNα) reveal new roles for phosphorylated and unphosphorylated forms of the transcription factor STAT2. J. Biol. Chem. 286, 20217–20227 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57. Blaszczyk K., Nowicka H., Kostyrko K., Antonczyk A., Wesoly J., and Bluyssen H. A. R. (2016) The unique role of STAT2 in constitutive and IFN-induced transcription and antiviral responses. Cytokine Growth Factor Rev. 29, 71–81 [DOI] [PubMed] [Google Scholar]
- 58. Blaszczyk K., Olejnik A., Nowicka H., Ozgyin L., Chen Y.-L., Chmielewski S., Kostyrko K., Wesoly J., Balint B. L., Lee C.-K., and Bluyssen H. A. R. (2015) STAT2/IRF9 directs a prolonged ISGF3-like transcriptional response and antiviral activity in the absence of STAT1. Biochem. J. 466, 511–524 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59. George C. X., Das S., and Samuel C. E. (2008) Organization of the mouse RNA-specific adenosine deaminase Adar1 gene 5′-region and demonstration of STAT1-independent, STAT2-dependent transcriptional activation by interferon. Virology 380, 338–343 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60. Perry S. T., Buck M. D., Lada S. M., Schindler C., and Shresta S. (2011) STAT2 mediates innate immunity to Dengue virus in the absence of STAT1 via the type I interferon receptor. PLoS Pathog. 7, e1001297. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61. Kraus T. A., Lau J. F., Parisien J. P., and Horvath C. M. (2003) A hybrid IRF9-STAT2 protein recapitulates interferon-stimulated gene expression and antiviral response. J. Biol. Chem. 278, 13033–13038 [DOI] [PubMed] [Google Scholar]
- 62. Bluyssen H. A. R., and Levy D. E. (1997) Stat2 is a transcriptional activator that requires sequence-specific contacts provided by Stat1 and p48 for stable interaction with DNA. J. Biol. Chem. 272, 4600–4605 [DOI] [PubMed] [Google Scholar]
- 63. Majoros A., Platanitis E., Szappanos D., Cheon H., Vogl C., Shukla P., Stark G. R., Sexl V., Schreiber R., Schindler C., Müller M., and Decker T. (2016) Response to interferons and antibacterial innate immunity in the absence of tyrosine-phosphorylated STAT1. EMBO Rep. 17, 367–382 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64. Kaur S., Kroczynska B., Sharma B., Sassano A., Arslan A. D., Majchrzak-Kita B., Stein B. L., McMahon B., Altman J. K., Su B., Calogero R. A., Fish E. N., and Platanias L. C. (2014) Critical roles for Rictor/Sin1 complexes in interferon-dependent gene transcription and generation of antiproliferative responses. J. Biol. Chem. 289, 6581–6591 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65. Apelbaum A., Yarden G., Warszawski S., Harari D., and Schreiber G. (2013) Type I interferons induce apoptosis by balancing cFLIP and Caspase-8 independent of death ligands. Mol. Cell. Biol. 33, 800–814 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66. Ho J., Pelzel C., Begitt A., Mee M., Elsheikha H. M., Scott D. J., and Vinkemeier U. (2016) STAT2 is a pervasive cytokine regulator due to its inhibition of STAT1 in multiple signaling pathways. PLoS Biol. 14, e2000117. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67. Jaitin D. A., and Schreiber G. (2007) Upregulation of a small subset of genes drives type I interferon-induced antiviral memory. J. Interferon Cytokine Res. 27, 653–664 [DOI] [PubMed] [Google Scholar]
- 68. Cheon H., and Stark G. R. (2009) Unphosphorylated STAT1 prolongs the expression of interferon-induced immune regulatory genes. Proc. Natl. Acad. Sci. U.S.A. 106, 9373–9378 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69. Cheon H., Holvey-Bates E. G., Schoggins J. W., Forster S., Hertzog P., Imanaka N., Rice C. M., Jackson M. W., Junk D. J., and Stark G. R. (2013) IFNβ-dependent increases in STAT1, STAT2, and IRF9 mediate resistance to viruses and DNA damage. EMBO J. 32, 2751–2763 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70. Gupta S., Jiang M., and Pernis A. B. (1999) IFN-α activates Stat6 and leads to the formation of Stat2:Stat6 complexes in B cells. J. Immunol. 163, 3834–3841 [PubMed] [Google Scholar]
- 71. Wang W. B., Levy D. E., and Lee C. K. (2011) STAT3 negatively regulates type I IFN-mediated antiviral response. J. Immunol. 187, 2578–2585 [DOI] [PubMed] [Google Scholar]
- 72. Zhao L.-J., He S.-F., Wang W., Ren H., and Qi Z.-T. (2016) Interferon α antagonizes STAT3 and SOCS3 signaling triggered by hepatitis C virus. Cytokine 80, 48–55 [DOI] [PubMed] [Google Scholar]
- 73. Ray J. P., Marshall H. D., Laidlaw B. J., Staron M. M., Kaech S. M., and Craft J. (2014) Transcription factor STAT3 and type I interferons are corepressive insulators for differentiation of follicular helper and T helper 1 cells. Immunity 40, 367–377 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74. Icardi L., Mori R., Gesellchen V., Eyckerman S., De Cauwer L., Verhelst J., Vercauteren K., Saelens X., Meuleman P., Leroux-Roels G., De Bosscher K., Boutros M., and Tavernier J. (2012) The Sin3a repressor complex is a master regulator of STAT transcriptional activity. Proc. Natl. Acad. Sci. U.S.A. 109, 12058–12063 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75. Tyler D. R., Persky M. E., Matthews L. A., Chan S., and Farrar J. D. (2007) Pre-assembly of STAT4 with the human IFN-α/β receptor-2 subunit is mediated by the STAT4 N-domain. Mol. Immunol. 44, 1864–1872 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76. Nguyen K. B., Watford W. T., Salomon R., Hofmann S. R., Pien G. C., Morinobu A., Gadina M., O'Shea J. J., and Biron C. A. (2002) Critical role for STAT4 activation by type 1 interferons in the interferon-γ response to viral infection. Science 297, 2063–2066 [DOI] [PubMed] [Google Scholar]
- 77. Gil M. P., Ploquin M. J. Y., Watford W. T., Lee S.-H., Kim K., Wang X., Kanno Y., O'Shea J. J., and Biron C. A. (2012) Regulating type 1 IFN effects in CD8 T cells during viral infections: changing STAT4 and STAT1 expression for function. Blood 120, 3718–3728 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78. Fish E. N., Uddin S., Korkmaz M., Majchrzak B., Druker B. J., and Platanias L. C. (1999) Activation of a CrkL-Stat5 signaling complex by type I interferons. J. Biol. Chem. 274, 571–573 [DOI] [PubMed] [Google Scholar]
- 79. Uddin S., Lekmine F., Sassano A., Rui H., Fish E. N., and Platanias L. C. (2003) Role of Stat5 in Type I interferon-signaling and transcriptional regulation. Biochem. Biophys. Res. Commun. 308, 325–330 [DOI] [PubMed] [Google Scholar]
- 80. van Boxel-Dezaire A. H., Zula J. A., Xu Y., Ransohoff R. M., Jacobberger J. W., and Stark G. R. (2010) Major differences in the responses of primary human leukocyte subsets to IFN-β. J. Immunol. 185, 5888–5899 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81. Hsu Y. A., Huang C. C., Kung Y. J., Lin H. J., Chang C. Y., Lee K. R., and Wan L. (2016) The anti-proliferative effects of type I IFN involve STAT6-mediated regulation of SP1 and BCL6. Cancer Lett. 375, 303–312 [DOI] [PubMed] [Google Scholar]
- 82. Katsoulidis E., Li Y., Mears H., and Platanias L. C. (2005) The p38 mitogen-activated protein kinase pathway in interferon signal transduction. J. Interferon Cytokine Res. 25, 749–756 [DOI] [PubMed] [Google Scholar]
- 83. Saleiro D., Mehrotra S., Kroczynska B., Beauchamp E. M., Lisowski P., Majchrzak-Kita B., Bhagat T. D., Stein B. L., McMahon B., Altman J. K., Kosciuczuk E. M., Baker D. P., Jie C., Jafari N., Thompson C. B., et al. (2015) Central role of ULK1 in type I interferon signaling. Cell Rep. 11, 605–617 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84. Thyrell L., Hjortsberg L., Arulampalam V., Panaretakis T., Uhles S., Dagnell M., Zhivotovsky B., Leibiger I., Grandér D., and Pokrovskaja K. (2004) Interferon α-induced apoptosis in tumor cells is mediated through the phosphoinositide 3-kinase/mammalian target of rapamycin signaling pathway. J. Biol. Chem. 279, 24152–24162 [DOI] [PubMed] [Google Scholar]
- 85. Hjortsberg L., Lindvall C., Corcoran M., Arulampalam V., Chan D., Thyrell L., Nordenskjold M., Grandér D., and Pokrovskaja K. (2007) Phosphoinositide 3-kinase regulates a subset of interferon-α-stimulated genes. Exp. Cell Res. 313, 404–414 [DOI] [PubMed] [Google Scholar]
- 86. de Weerd N. A., and Nguyen T. (2012) The interferons and their receptors: distribution and regulation. Immunol. Cell Biol. 90, 483–491 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87. Bhattacharya S., Katlinski K. V., Reichert M., Takano S., Brice A., Zhao B., Yu Q., Zheng H., Carbone C. J., Katlinskaya Y. V., Leu N. A., McCorkell K. A., Srinivasan S., Girondo M., Rui H., et al. (2014) Triggering ubiquitination of IFNAR1 protects tissues from inflammatory injury. EMBO Mol. Med. 6, 384–397 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88. Kumar K. G., Barriere H., Carbone C. J., Liu J., Swaminathan G., Xu P., Li Y., Baker D. P., Peng J., Lukacs G. L., and Fuchs S. Y. (2007) Site-specific ubiquitination exposes a linear motif to promote interferon-α receptor endocytosis. J. Cell Biol. 179, 935–950 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 89. Kumar K. G., Varghese B., Banerjee A., Baker D. P., Constantinescu S. N., Pellegrini S., and Fuchs S. Y. (2008) Basal ubiquitin-independent internalization of interferon α receptor is prevented by Tyk2-mediated masking of a linear endocytic motif. J. Biol. Chem. 283, 18566–18572 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 90. Liu J., Plotnikov A., Banerjee A., Suresh Kumar K. G., Ragimbeau J., Marijanovic Z., Baker D. P., Pellegrini S., and Fuchs S. Y. (2008) Ligand-independent pathway that controls stability of interferon α receptor. Biochem. Biophys. Res. Commun. 367, 388–393 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 91. Chmiest D., Sharma N., Zanin N., Viaris de Lesegno C., Shafaq-Zadah M., Sibut V., Dingli F., Hupé P., Wilmes S., Piehler J., Loew D., Johannes L., Schreiber G., and Lamaze C. (2016) Spatiotemporal control of interferon-induced JAK/STAT signaling and gene transcription by the retromer complex. Nat. Commun. 7, 13476. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 92. Marchetti M., Monier M. N., Fradagrada A., Mitchell K., Baychelier F., Eid P., Johannes L., and Lamaze C. (2006) Stat-mediated signaling induced by type I and type II interferons (IFNs) is differentially controlled through lipid microdomain association and clathrin-dependent endocytosis of IFN receptors. Mol. Biol. Cell 17, 2896–2909 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 93. Malakhova O. A., Kim K. I., Luo J. K., Zou W., Kumar K. G., Fuchs S. Y., Shuai K., and Zhang D. E. (2006) UBP43 is a novel regulator of interferon signaling independent of its ISG15 isopeptidase activity. EMBO J. 25, 2358–2367 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 94. Arimoto K. I., Löchte S., Stoner S. A., Burkart C., Zhang Y., Miyauchi S., Wilmes S., Fan J. B., Heinisch J. J., Li Z., Yan M., Pellegrini S., Colland F., Piehler J., and Zhang D. E. (2017) STAT2 is an essential adaptor in USP18-mediated suppression of type I interferon signaling. Nat. Struct. Mol. Biol. 24, 279–289 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 95. François-Newton V., Magno de Freitas Almeida G., Payelle-Brogard B., Monneron D., Pichard-Garcia L., Piehler J., Pellegrini S., and Uzé G. (2011) USP18-based negative feedback control is induced by type I and type III interferons and specifically inactivates interferon α response. PLoS ONE 6, e22200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 96. Francois-Newton V., Livingstone M., Payelle-Brogard B., Uzé G., and Pellegrini S. (2012) USP18 establishes the transcriptional and anti-proliferative interferon α/β differential. Biochem. J. 446, 509–516 [DOI] [PubMed] [Google Scholar]
- 97. Goldmann T., Zeller N., Raasch J., Kierdorf K., Frenzel K., Ketscher L., Basters A., Staszewski O., Brendecke S. M., Spiess A., Tay T. L., Kreutz C., Timmer J., Mancini G. M. S., Blank T., et al. (2015) USP18 lack in microglia causes destructive interferonopathy of the mouse brain. EMBO J. 34, 1612–1629 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 98. Speer S. D., Li Z., Buta S., Payelle-Brogard B., Qian L., Vigant F., Rubino E., Gardner T. J., Wedeking T., Hermann M., Duehr J., Sanal O., Tezcan I., Mansouri N., Tabarsi P., et al. (2016) ISG15 deficiency and increased viral resistance in humans but not mice. Nat. Commun. 7, 11496. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 99. Piganis R. A., De Weerd N. A., Gould J. A., Schindler C. W., Mansell A., Nicholson S. E., and Hertzog P. J. (2011) Suppressor of cytokine signaling (SOCS) 1 inhibits type I interferon (IFN) signaling via the interferon α receptor (IFNAR1)-associated tyrosine kinase Tyk2. J. Biol. Chem. 286, 33811–33818 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 100. Linossi E. M., and Nicholson S. E. (2015) Kinase inhibition, competitive binding and proteasomal degradation: resolving the molecular function of the suppressor of cytokine signaling (SOCS) proteins. Immunol. Rev. 266, 123–133 [DOI] [PubMed] [Google Scholar]