Abstract
Hyaluronan (HA) is an extremely large polysaccharide (glycosaminoglycan) involved in many cellular functions. HA catabolism is thought to involve the initial cleavage of extracellular high-molecular-weight (HMW) HA into intermediate-size HA by an extracellular or cell-surface hyaluronidase, internalization of intermediate-size HA, and complete degradation into monosaccharides in lysosomes. Despite considerable research, the identity of the hyaluronidase responsible for the initial HA cleavage in the extracellular space remains elusive. HYAL1 and HYAL2 have properties more consistent with lysosomal hyaluronidases, whereas CEMIP/KIAA1199, a recently identified HA-binding molecule that has HA-degrading activity, requires the participation of the clathrin-coated pit pathway of live cells for HA degradation. Here we show that transmembrane protein 2 (TMEM2), a mammalian homolog of a protein playing a role in zebrafish endocardial cushion development, is a cell-surface hyaluronidase. Live immunostaining and surface biotinylation assays confirmed that mouse TMEM2 is expressed on the cell surface in a type II transmembrane topology. TMEM2 degraded HMW-HA into ∼5-kDa fragments but did not cleave chondroitin sulfate or dermatan sulfate, indicating its specificity to HA. The hyaluronidase activity of TMEM2 was Ca2+-dependent; the enzyme's pH optimum is around 6–7, and unlike CEMIP/KIAA1199, TMEM2 does not require the participation of live cells for its hyaluronidase activity. Moreover, TMEM2-expressing cells could eliminate HA immobilized on a glass surface in a contact-dependent manner. Together, these data suggest that TMEM2 is the long-sought-after hyaluronidase that cleaves extracellular HMW-HA into intermediate-size fragments before internalization and degradation in the lysosome.
Keywords: cell surface, glycosaminoglycan, hyaluronan, hyaluronidase, membrane function, CEMIP/KIAA1199, TMEM2
Introduction
Hyaluronic acid (HA)2 is a glycosaminoglycan composed of repeating disaccharide units of glucuronic acid and N-acetylglucosamine. It is a linear polymer of extremely large molecular mass, often exceeding 106 Da (1). The sheer size of HA suggests that cells should have very efficient mechanisms for its metabolism. In fact, one-third of the total body HA, which is estimated to be 15 g in a human with a 70-kg body weight, is thought to be turned over daily (2). In skin, the metabolic half-life of HA is 1 to 1.5 days (3). It is believed that high-molecular-weight (HMW) HA (106–107 Da) is first degraded extracellularly into intermediate-size fragments of 10–100 kDa. These are then internalized and degraded to monosaccharides by the combined actions of lysosomal hyaluronidase and exoglucosidases (4). Considering the accumulating evidence for the role of HA degradation in tumor invasion and metastasis (5), identifying the molecule(s) that degrade HA on the cell surface is an important biological issue.
The HYAL family molecules have been implicated as the major players in HA catabolism. HYAL1 and HYAL2 are expressed widely and postulated to be the key hyaluronidases involved in HA catabolism in somatic tissues. However, some data on HYAL1 and HYAL2 are inconsistent with the notion that these molecules are the physiological hyaluronidases that cleave HMW-HA on the cell surface or in the extracellular space. Both HYAL1 and HYAL2 favor acidic pH for their activity. HYAL1 is enzymatically active only below pH 5.5 (6), and the pH optimum for HYAL2 is pH 4 (7). Such low pH optima suggest that they are more likely to be lysosomal enzymes. In fact, there is evidence that both HYAL1 and HYAL2 are primarily present in lysosomes (7, 8). Although HYAL2 is GPI-anchored and a part of HYAL2 molecules are fractionated in the membrane fraction (9), this does not entirely substantiate its physiological role as a cell-surface hyaluronidase, as its HA-degrading activity is very low, at least 50 times lower than that of HYAL1 (9, 10). Other HYAL family molecules, PH-20/SPAM1 and HYAL5, can function at neutral pH (11, 12). Although there are reports that PH-20/SPAM1 is expressed in some cancers (13–15), their expression in normal tissues is largely restricted in the testis, and it is unlikely that PH-20/SPAM1 and HYAL5 act as principal extracellular hyaluronidase in a broad range of normal somatic tissues.
Other than the HYAL family molecules, the cell migration-inducing and hyaluronan-binding protein (CEMIP), also known as KIAA1199, has HA-binding and -degrading activities (16). CEMIP is a putative secretory protein containing an N-terminal signal sequence, and therefore it is considered to be a strong candidate as a hyaluronidase that acts in the extracellular space. CEMIP-mediated HA degradation, however, requires the participation of the clathrin-coated pit pathway (16), suggesting that CEMIP may not simply act as an extracellular hyaluronidase. Also noteworthy is that neither conditioned media of CEMIP-expressing cells nor recombinant CEMIP show HA-degrading activity. In addition, CEMIP is detected mainly inside the cells (16, 17).
Against this backdrop, we sought to identify a novel hyaluronidase that is present and functions on the cell surface. Here we report that transmembrane protein 2 (TMEM2), a type II transmembrane protein with sequence similarities to CEMIP, is such a cell-surface hyaluronidase. TMEM2 depolymerizes HMW-HA to HA fragments of ∼5 kDa in a Ca2+-dependent manner with a pH optimum between pH 6 and 7. Surface biotinylation and live cell immunostaining confirm that TMEM2 is indeed expressed on the cell surface, and TMEM2-expressing cells can degrade substrate-bound HA in a contact-dependent manner. Finally, absolute quantification of transcript copy numbers reveals that Tmem2 mRNA is expressed ubiquitously in adult mouse tissues and during development at far higher levels than Cemip mRNA. Together, these data suggest that TMEM2 is a novel hyaluronidase that cleaves extracellular HMW-HA into intermediate-sized fragments prior to internalization and complete degradation of these HA fragments in the lysosome.
Results
TMEM2 is expressed on the cell surface with a type II membrane topology
In search of a novel hyaluronidase that is present on the cell surface, we searched the database for mammalian proteins that have similarity to CEMIP. This led to the identification of TMEM2 (HUGO Gene Nomenclature ID: 11869), a member of the TMEM family, as a candidate. The TMEM family is a heterogeneous collection of more than 300 different human open reading frames that are grouped based only on the presence of at least one putative transmembrane domain (18). The functions of many of TMEM proteins are unknown. In the case of TMEM2, however, the phenotype of zebrafish mutants of the TMEM2 homolog suggests its role in cardiac morphogenesis (19, 20).
The open reading frame of human TMEM2 (NCBI RefSeq: NM_013390) predicts a type II transmembrane protein containing PbH1 repeats, a structural feature present in CEMIP (16) and in members of the bacterial polysaccharide lyase family (21). Domain structures of TMEM2 in comparison with CEMIP are shown in Fig. 1A. The deduced amino acid sequence of TMEM2 consists of (from its N terminus) an 82-residue cytoplasmic domain, a single transmembrane domain, and 1278-residue extracellular domain. The extracellular domain contains one G8 domain (22), one GG domain (23), and three PbH1 repeats. In comparison, CEMIP contains one G8, two GG, and four PbH1 repeats, and the overall amino acid identity between CEMIP and the extracellular domain of TMEM2 is 48%.
Figure 1.
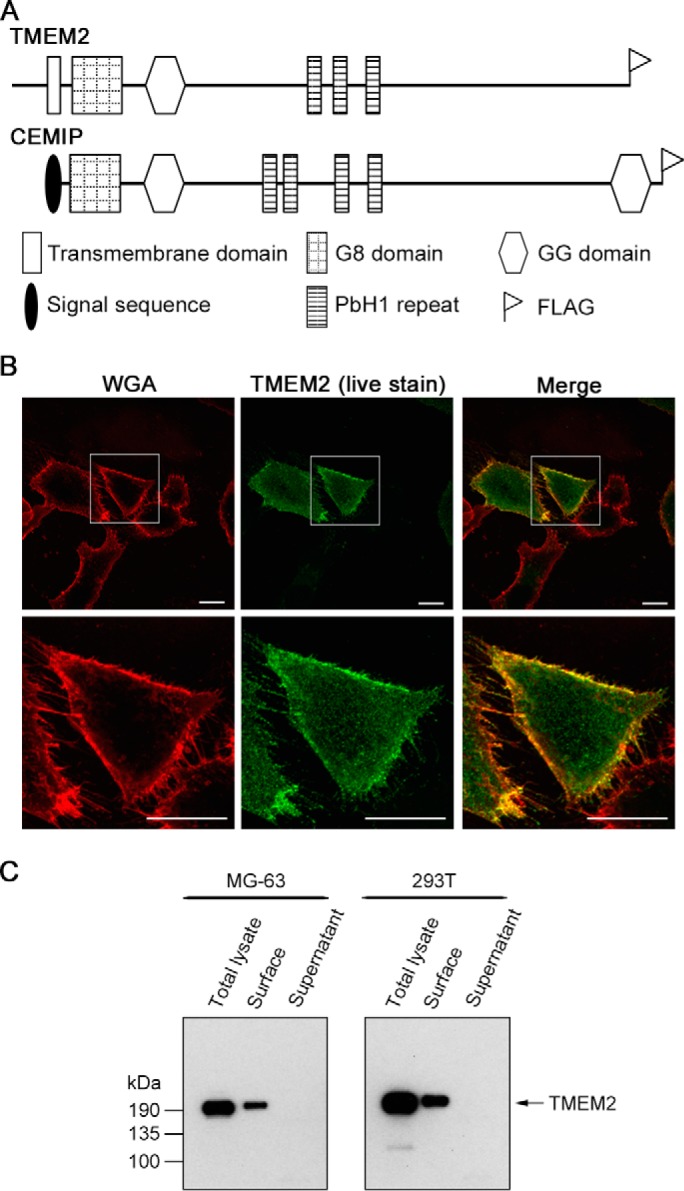
TMEM2 is expressed as a type II transmembrane protein. A, domain structure of TMEM2 and CEMIP. The location of the FLAG epitope tag added to recombinant proteins is also indicated. B, live immunostaining of MG-63 cells transiently transfected with TMEM2WT. Cells were stained live with anti-FLAG antibody to detect surface-expressed TMEM2. Co-staining with Alexa Fluor 594-conjugated wheat germ agglutinin reagent (WGA) was used to visualize cell contours. Scale bars, 10 μm. The experiment was repeated three times with similar results. C, analysis of cell surface expression by surface biotinylation assays. Cells were transfected with TMEM2WT. Forty eight h after transfection, culture supernatants were harvested (Supernatant). Cells were then incubated with membrane-impermeable sulfo-NHS-SS-biotin and solubilized with RIPA buffer (Total lysate). Biotinylated proteins were isolated from total lysates with streptavidin-agarose (Surface). Equivalent amounts of these fractions were analyzed by immunoblotting with anti-FLAG M2 antibody. The experiment was repeated two times with similar results.
We examined whether TMEM2 is indeed expressed as a type II transmembrane protein. Mouse TMEM2 cDNA tagged with a FLAG epitope at its C terminus was transiently transfected to MG-63 cells, and the expression of TMEM2 was probed by live immunostaining with anti-FLAG antibody. As shown in Fig. 1B, live staining with anti-FLAG antibody detected TMEM2 immunoreactivity on the cell surface, demonstrating not only the presence of the protein on the cell surface but also that the C terminus is outside the cell. To further investigate cell-surface expression of TMEM2, we performed a surface biotinylation assay on TMEM2-transfected MG-63 and 293T cells. This analysis confirms cell-surface localization of TMEM2 in both MG-63 and 293T cells (Fig. 1C, surface). No TMEM2 was detected in culture supernatants (Fig. 1C, supernatant). Together, these results demonstrate that TMEM2 is a type II transmembrane protein, as predicted from its deduced sequence.
TMEM2 depolymerizes HA into fragments of ∼5 kDa
To determine whether TMEM2 has the ability to depolymerize HA, we used a cell-based HA degradation assay, in which fluorescein-labeled HA (FA-HA) of various sizes was added to culture media of 293T cells transiently transfected with the FLAG-tagged, full-length TMEM2 cDNA (TMEM2WT). Parental 293T cells show no HA-depolymerizing activity in this assay (16) (see also the elution patterns with mock-transfected cells in Fig. 2A). With TMEM2-transfected cells, HMW-HA with an average molecular size of 1500 kDa (FA-HA1500) was depolymerized into fragments of ∼5 kDa (Fig. 2A, HA1500). Smaller HA (FA-HA200 and FA-HA20) were also depolymerized into ∼5-kDa fragments (Fig. 2A, HA200 and HA20), whereas HA fragments with an average size of 5 kDa (Fig. 2A, HA5) were not further degraded, suggesting that ∼5 kDa (∼100 sugar residues long) is the smallest size on which TMEM2 can act.
Figure 2.
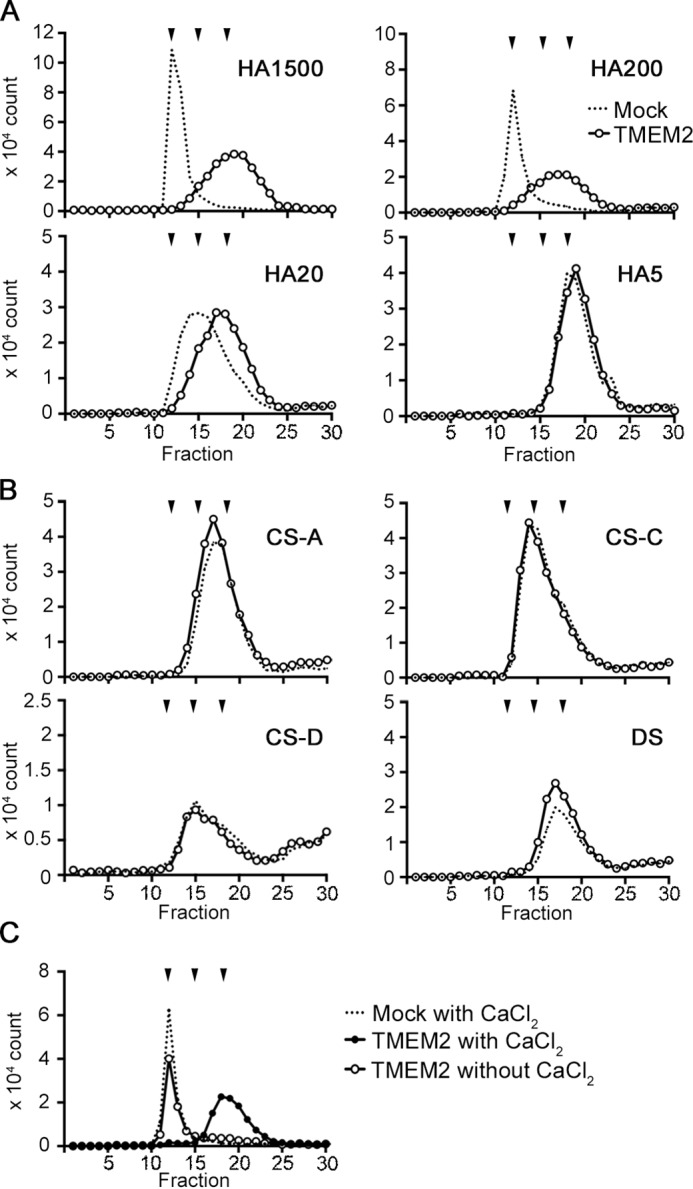
HA-specific degrading activity of TMEM2 and its Ca2+ dependence. A and B, 293T cells were transiently transfected with TMEM2WT (TMEM2) or without cDNA (Mock) and cultured with 0.1 μg/ml of FA-labeled GAGs to examine GAG-degrading activity of TMEM2. Degradation of GAGs was analyzed by gel filtration on a Sephacryl S-300HR column. A, degradation of HA of different average sizes (FA-HA1500, FA-HA200, FA-HA20, and FA-HA5). The experiment was repeated three times with similar results. B, degradation of chondroitin sulfate A (FA-CSA), B (FA-CSC), D (FA-CSD), and dermatan sulfate (DS). The experiment was repeated two times with similar results. C, effect of Ca2+ on the HA-degrading activity of TMEM2. HA degradation assay was performed with membrane fraction of TMEM2WT-transfected 293T cells and FA-HA1500 in the presence and absence of 1 mm CaCl2. A–C, downward arrowheads above the chromatograms represent, from left to right, elution peaks of FA-HA1500/FA-HA200 (both are eluted at V0), FA-HA20, and FA-HA5, respectively. The experiment was repeated three times with similar results.
It has been reported that the HYAL family hyaluronidases, such as HYAL1 and HYAL2, have broad substrate specificity and depolymerize not only HA but chondroitin sulfate and dermatan sulfate (11). To examine the substrate specificity of TMEM2, we tested chondroitin sulfate (CS)-A, CS-C, CS-D, and dermatan sulfate (DS) in the same cell-based assay. None of these GAGs were depolymerized by TMEM2 (Fig. 2B), indicating that the GAG depolymerizing activity of TMEM2 is specific for HA.
HA-depolymerizing activity of TMEM2 is Ca2+-dependent
To gain insight into the enzymatic mechanism of TMEM2, we first examined the Ca2+ dependence of HA-depolymerizing activity. Although there has been no report that the HYAL family hyaluronidases require Ca2+ for their activity, some of the bacterial polysaccharide lyases, such as pectate lyases, with which TMEM2 shares the PbH1 repeats, are Ca2+-dependent (24–26). To examine the requirement for Ca2+, the HA degradation assay was performed with the membrane fraction of TMEM2-transfected 293T cells as an enzyme source. As shown in Fig. 2C, FA-HA1500 was depolymerized into ∼5-kDa fragments in the presence of 1 mm CaCl2 but not in the absence of CaCl2, demonstrating that TMEM2 requires Ca2+ for its HA-depolymerizing activity.
Identification of amino acid residues critical for the enzymatic activity of TMEM2
Although a detailed elucidation of its exact enzymatic mechanism would ultimately require the three-dimensional structure of TMEM2 complexed with HA fragments, we sought to gain initial insight into this issue by examining the effect of mutagenesis of certain amino acid residues that are considered to be functionally important based on sequence information. One such residue is Arg265 (Fig. 3A), which positionally corresponds to the site of the deafness mutation (Arg187) in the human CEMIP gene (16, 27). Because this mutation in CEMIP has been shown to reduce its HA-depolymerizing activity (16), we surmised that Arg265 may also be important for the HA-depolymerizing activity of TMEM2. Moreover, an interesting feature in the region surrounding Arg265 is the DQD275 sequence just downstream of Arg265. DXD motifs have been shown to serve as functionally critical Ca2+-coordination sites in Erwinia pectate lyases, which, like TMEM2 and CEMIP, contain the PbH1 repeats (24, 28). Also, many glycosyltransferases possess DXD motifs that are involved in the coordination with divalent cations and/or a ribose (29). Thus we performed a series of mutagenesis experiments focused on Arg265, Asp273, and Asp275. In addition, we generated mutants at other acidic residues in the region (Glu260, Asp262, Glu281, Glu283, and Asp286). Among these residues, Arg265, Asp273, and Asp286 are completely conserved between TMEM2 and CEMIP (Fig. 3A).
Figure 3.
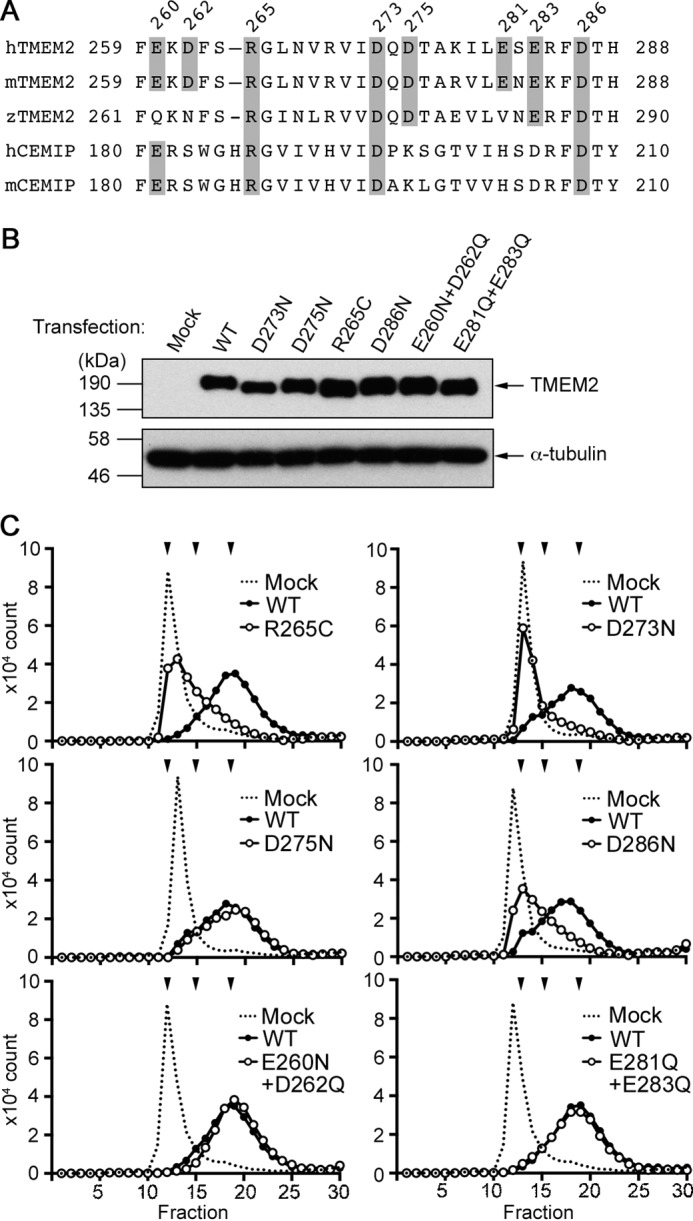
Identification of amino acid residues critical for the hyaluronidase activity of TMEM2. A, sequence alignment of human, mouse, and zebrafish TMEM2 and human and mouse CEMIP in the region surrounding the deafness mutation site (Arg187) of CEMIP. Shaded residues in TMEM2 were mutagenized as follows: R265C, D273N, D275N, D286N, E260N/D262Q, and E281Q/E283Q. B, analysis of the expression level of TMEM2 point mutants in 293T cells. Total cell lysates of transfected 293T cells were analyzed by immunoblotting with anti-FLAG M2 (upper panel) and anti-α-tubulin antibodies (lower panel; loading controls). C, HA-degrading activity of TMEM2 mutants. HA degradation assays with 293T cells were performed with TMEM2 mutants as indicated in the panels. The experiment with D273N and D275N was performed as a single experiment with a single mock sample, and gel filtration chromatography was run consecutively; therefore, the trace of the mock sample was reused in these two panels. The same is true for the experiment with E260N/D262Q and E281Q/E283Q. Downward arrowheads above the chromatograms represent, from left to right, elution peaks of FA-HA1500/FA-HA200 (both are eluted at V0), FA-HA20, and FA-HA5, respectively. Each experiment was repeated three times with similar results.
We generated single- and double-point mutants in which residues have been mutagenized as follows: R265C, D273N, D275N, D286N, E260N/D262Q, and E281Q/E283Q. We first confirmed that these mutagenized TMEM2 proteins were expressed at similar levels upon transfection into 293T cells (Fig. 3B). HA degradation assays with 293T cells were performed with these mutants (Fig. 3C). D275N, E260N/D262Q, and E281Q/E283Q mutants are as enzymatically active as wild-type TMEM2. In contrast, the activities of R265C, D273N, and D286N mutants were greatly reduced in comparison with wild-type TMEM2. Although the elucidation of the role of these residues requires the crystal structure of TMEM2, these results demonstrate that this region is important for the hyaluronidase activity of TMEM2.
pH optimum of TMEM2
The presence of TMEM on the cell surface (see Fig. 1) and its activity under standard cell culture conditions (see Fig. 2) strongly suggest that TMEM2 acts primarily on the cell surface. We sought additional insight into the cellular site of action of TMEM2 by examining the pH optimum for its HA-degrading activity. To prevent the contamination in this analysis by endogenous hyaluronidases residing in intracellular compartments, we produced an N-terminally truncated, soluble TMEM2 consisting only of its extracellular domain. Truncated TMEM2 was secreted into culture supernatants of 293T cells as a soluble protein migrating at ∼180 kDa (Fig. 4A). HA degradation assays were performed in a range of pH 4–8 with concentrated culture supernatants as an enzyme source. Results from these experiments indicate that the soluble TMEM2 is active in the range of pH 5–8 with the highest activity around pH 6 (Fig. 4, B and C), consistent with the notion that TMEM2 is functional in the extracellular environment. Below pH 5, TMEM2 quickly loses activity, becoming totally inactive at pH 4. This suggests that TMEM2 is unlikely to be enzymatically active in lysosomes. Additionally, the retention of enzymatic activity of this soluble form demonstrates that, unlike CEMIP, TMEM2 can exert the hyaluronidase activity independent of the participation of cellular endocytic pathways.
Figure 4.
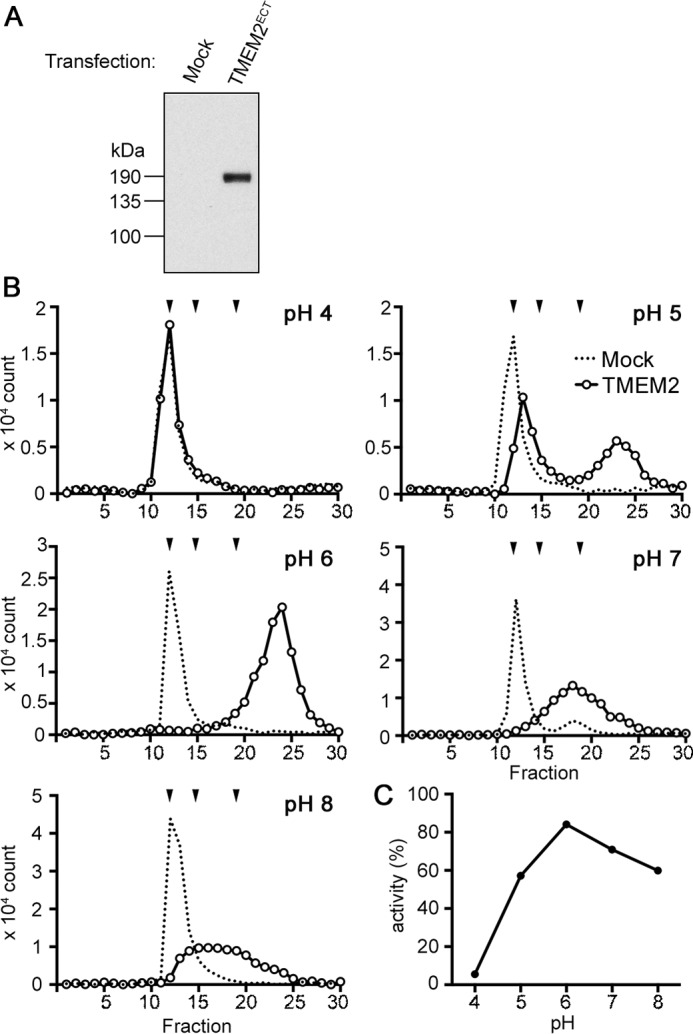
Effect of pH on the hyaluronidase activity of TMEM2. A, analysis of the expression of soluble TMEM2. 293T cells were transfected with an N-terminally truncated TMEM2 cDNA (TMEM2ECT) or without cDNA (Mock), and culture supernatants were immunoblotted with rabbit polyclonal anti-TMEM2 antibody. B, HA-degrading activity of soluble TMEM2 at different pH conditions. FA-HA1500 (0.06 μg/ml) was incubated for 16 h with concentrated culture supernatants from TMEM2ECT-transfected or mock-transfected 293T cells in reaction buffers adjusted to pH 4–8 as indicated in the figure. Degradation of HA was analyzed by gel filtration on Sephacryl S-300 HR. Downward arrowheads above the chromatograms represent, from left to right, elution peaks of FA-HA1500/FA-HA200 (both are eluted at V0), FA-HA20, and FA-HA5, respectively. The experiment was repeated two times with similar results. C, pH profile of the hyaluronidase activity of soluble TMEM2. Enzyme activity is expressed as a percentage of fluorescence counts that represent depolymerized HA relative to the total fluorescence input.
TMEM2-expressing cells degrade substrate-bound HA in a contact-dependent manner
Cell-surface localization of TMEM2 and the lack of release into culture supernatants (see Fig. 1) suggest that TMEM2 degrades matrix-bound HA upon cell contact or adhesion. To examine this mode of action, we devised an assay in which TMEM2-transfected 293T cells are cultured on a substrate of FA-HA1500 immobilized to amino-silanized glass (30). In this assay, the spatial nature of HA degradation is revealed as “dark holes” in a fluorescent background. Cultures with mock-transfected cells revealed no holes in HA substrates (Fig. 5, mock). In contrast, TMEM2-transfected cells created conspicuous holes, corresponding to the locations of cell clusters adhering to the substrate (Fig. 5, Wild-type TMEM2). As an additional control, we examined cells transfected with the D273N mutant, which has greatly reduced hyaluronidase activity (see Fig. 3C). D273N-expressing cells created less significant holes than cells transfected with wild-type TMEM2 (Fig. 5, D273N). Together, these results indicate that cells expressing TMEM2 can degrade matrix-bound HA at sites of cell-substratum contacts.
Figure 5.
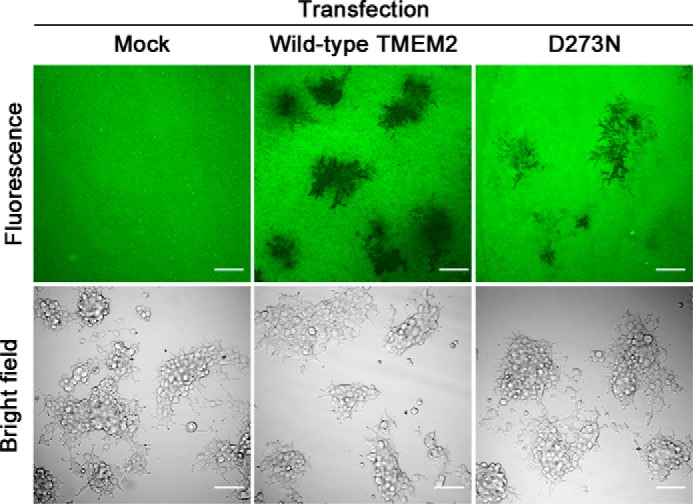
TMEM2-expressing cells degrade substrate-bound HA in a contact-dependent manner. 293T cells transfected with TMEM2WT (WT), D273N (D273N), or without cDNA (Mock) were plated on a substrate of FA-HA1500 immobilized to amino-silanized glass. After incubating for 60 h, cultures were examined on a confocal microscope with both fluorescence imaging (top panels) and bright field imaging (bottom panels). Note that TMEM2-expressing cells eliminate substrate-bound HA corresponding to the location of cell clusters. Scale bar, 100 μm. The experiment was repeated four times with similar results.
Absolute quantification of transcript copy numbers reveals that Tmem2 is expressed much more highly than Cemip in normal mouse tissues
To gain insight regarding the physiological sites of TMEM2 action, we analyzed Tmem2 mRNA expression in various mouse tissues. In doing so, we also sought to compare the expression levels of Tmem2 and Cemip, because both are presumed to act on the same substrate. However, standard expression profiling techniques, such as qPCR and microarray analysis, do not allow comparison of expression levels between different genes (“inter-gene” comparison). Therefore, to enable inter-gene comparison between Tmem2 and Cemip, we performed absolute quantification of mRNA copy numbers using the standard curve method (31). As shown in Fig. 6A, Tmem2 is widely expressed in essentially all organs examined, with copy numbers greater than 2 × 105 copies per μg of total RNA. (In comparison, Hyal2 mRNA is expressed at 4.1 × 105, 7.3 × 105, and 1.4 × 105 copies per μg of total RNA in the heart, liver, and lung, respectively.) An especially high level of Tmem2 expression was found in the lung (1.6 × 106 copies per μg of total RNA). Also noteworthy is that the expression level of Tmem2 is much higher than that of Cemip in the majority of organs, except for the brain, spleen, and synovium where they are expressed at similar levels. Essentially no Cemip transcripts were detected in the lung, liver, salivary gland, colon, kidney, and ureter. Developmental expression was examined in whole mouse embryos at embryonic day (E) E11, E15, and E17 (Fig. 6B). A high level of Tmem2 expression was found at embryonic day E11, after which expression declines. Little Cemip expression was detected at any time point examined. Together, these data demonstrate that in essentially all adult tissues and during embryonic development, Tmem2 is much more abundantly expressed than Cemip.
Figure 6.
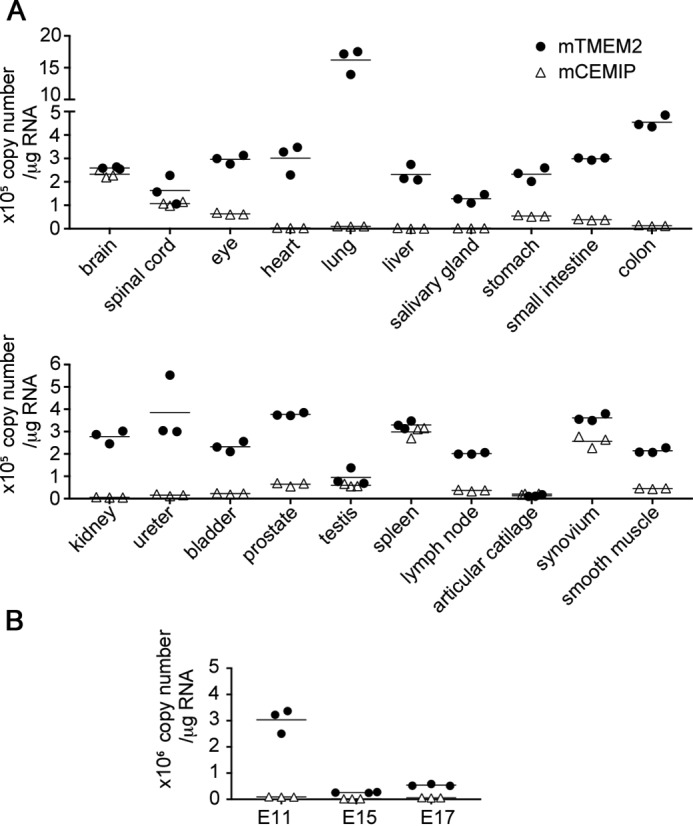
Expression profiles of Tmem2 and Cemip in normal mouse tissues (A) and during embryonic development (B) determined by absolute quantification by RT-qPCR. Copy numbers of Tmem2 and Cemip transcripts were determined by TaqMan gene expression assay with standard curves generated from reference plasmids. All samples were analyzed in biological triplicates. Filled circles, Tmem2; open triangles, Cemip.
Discussion
Catabolism of HA involves multiple steps mediated by enzymes present in different cellular compartments. The widely accepted model (11) proposes that HMW-HA in the extracellular matrix is first cleaved into intermediate-size HA by extracellular/cell-surface hyaluronidase. Subsequently, intermediate-size HA is internalized and further degraded by lysosomal hyaluronidase. Finally, these HA fragments are degraded into monosaccharides by lysosomal exoglucosidases. Considering the extremely large molecular size of HA, it is reasonable that the size of HA needs to be reduced significantly before cells can internalize it for further degradation. One of the key questions regarding this catabolic cascade has been the molecular identity of the hyaluronidase(s) that cleaves HMW-HA on the cell surface and/or in the extracellular space. Although the HYAL family molecules, including HYAL1 and HYAL2, have been considered to play an important role in the catabolism of HA, the biochemical and biological properties of these molecules are not consistent with those expected of an enzyme that cleaves HMW-HA on the cell surface and/or in the extracellular space. Their acidic pH optima and localization in lysosomes suggest that they function primarily as lysosomal enzymes (7, 8). Interestingly, Harada and Takahashi (32) reported that HYAL2, a GPI-anchored molecule that is localized intracellularly when transfected alone into 293T cells, is translocated to the cell surface upon co-transfection of CD44, suggesting the possibility that it may act as a cell-surface hyaluronidase in the presence of CD44. Still, in view of a number of conflicting data regarding its pH optimum (9, 32–34) and subcellular localization (9, 33–36), the significance of HYAL2 as a cell-surface hyaluronidase is far from clear. Two other HYAL family molecules, PH-20/SPAM1 and HYAL5, are active in near neutral pH and can degrade HA in the extracellular environment (12, 37). However, because their expression is largely restricted to the testis (12, 37), it is unlikely that they function as principal cell surface/extracellular hyaluronidase in a broad range of somatic tissues and cell types.
In contrast, the molecular and biochemical properties of TMEM2 are consistent with those expected for a hyaluronidase that functions on the cell surface. Cell-surface localization of TMEM2, as well as its membrane topology as a type II transmembrane protein, is confirmed by two independent approaches, namely surface biotinylation assay and live immunocytochemistry (Fig. 1). Elimination of substrate-bound HA in the area of cell-substrate contact (Fig. 5) shows that TMEM2 is not only present on the cell surface but is also functional in this location. The pH optimum of TMEM2 (Fig. 4) is also consistent with the notion that TMEM2 acts in the extracellular environment. The steep loss of activity below pH 5 suggests that TMEM2 is unlikely to function in lysosomes, although this does not entirely rule out the possibility that TMEM2 is enzymatically active in early endosomes where pH ranges from 6.0 to 6.5.
Along with TMEM2, CEMIP is another potential hyaluronidase that can function on or in the vicinity of the cell surface. Nevertheless, CEMIP has several features that are inconsistent with the notion that it is a hyaluronidase that functions in the extracellular space. For example, although CEMIP is a secreted protein with an N-terminal signal sequence (38), culture supernatants of CEMIP-transfected cells show little HA-degrading activity (16), and CEMIP is detected mainly intracellularly (16, 17). Concerning the lack of activity in culture supernatant, Yoshida et al. (16) reported that CEMIP-mediated HA degradation requires the participation of the clathrin-coated pit pathway, suggesting that CEMIP may function in intracellular compartments rather than in the extracellular space. Thus despite structural similarities, the properties of TMEM2 and CEMIP are distinct, and the primary site of hyaluronidase activity of CEMIP may not be cell surface or extracellular space.
This study has also revealed a stark difference in the expression levels of Tmem2 and Cemip mRNAs in various normal tissues and organs in mice. Our absolute quantification analysis demonstrates that at the mRNA level, Tmem2 is much more highly expressed than Cemip in essentially all normal adult tissues and organs (see Fig. 6). The extremely low copy number of Cemip transcripts in several tissues, such as the heart, lung, liver, salivary gland, colon, and kidney, suggests that only a very small amount of CEMIP protein, if any, is present in these tissues. Although the copy number of transcripts does not necessarily reflect the level of enzymatic activity, it is not unreasonable to suppose that CEMIP plays only a minor physiological role as a hyaluronidase in these tissues. It has been reported that the expression of CEMIP is induced by histamine and in inflammatory tissues (16) and that CEMIP is strongly up-regulated in human cancers, especially in those with poor prognosis (17, 39–41). These observations, together with low expression in normal tissues, may indicate that CEMIP is expressed primarily under pathological conditions.
At present, nothing is known about the physiological role of TMEM2 in mammals, but its broad expression pattern suggests that it plays a functional role in various developmental and physiological contexts. The observations that mutations in the zebrafish tmem2 (frozen ventricle and wickham) manifest as lethal defects in heart development suggest that the embryonic heart is one of the important sites in which TMEM2 plays a critical role in mammals. These zebrafish mutants exhibit abnormal cardiac looping accompanied by diminished constriction of the atrioventricular canal. Detailed analyses of the zebrafish heart phenotype indicate that Tmem2 plays a critical role in endocardial and myocardial morphogenesis by facilitating the migration of myocardial and endocardial cells while restricting endocardial cushion formation (19, 20). These observations resonate with the fact that HA, being the main component of cardiac jelly, plays a critical role in endocardial cushion development; mutant mice of hyaluronan synthase 2 (Has2) and the HA-binding proteoglycan versican (Vcan) exhibit defects in endocardial cushion development (42–44). In fact, Smith et al. (19) noticed in zebrafish wickham mutants that HA levels are increased in the developing heart, which is consistent with the possibility that zebrafish Tmem2 is a physiological hyaluronidase. Because these zebrafish studies were conducted and interpreted while unaware of the function of TMEM2 as a hyaluronidase, it will be interesting to revisit the data to see if some or all of these phenotypes can be explained by the premise that TMEM2 functions as a hyaluronidase in this developmental context. This would provide insight into whether TMEM2 functions solely as a hyaluronidase or has an additional function, possibly as a signaling receptor.
In conclusion, this study provides evidence that TMEM2 is a novel cell-surface hyaluronidase. The biochemical and cell biological properties of TMEM2 suggest that it is the long sought-after hyaluronidase that cleaves extracellular HMW-HA into intermediate-size fragments on the cell surface. Identification of TMEM2 as a hyaluronidase should promote our understanding of the catabolism of HA and its functional significance in development and disease.
Experimental procedures
Cell culture
293T and MG-63 cells were obtained from American Type Culture Collection (Manassas, VA) and cultured in DMEM and minimum Eagle's medium, respectively. Both media were supplemented with 10% fetal bovine serum and 100 units/ml penicillin and 100 μg streptomycin.
Construction of expression vectors
Mouse TMEM2 cDNA (BC076570) was amplified by PCR and subcloned into a pcDNA3 (Thermo Fisher Scientific, Waltham, MA)-based vector that had been modified to fuse a FLAG tag to the C terminus of TMEM2 (designated as TMEM2WT). Point mutants of TMEM2 were generated from TMEM2WT by using the QuikChange site-directed mutagenesis kit (Agilent Technologies, Santa Clara, CA). Six constructs with single and two double point mutations were generated, namely R265C (i.e. Arg265 was mutagenized to Cys, and so on), D273N, D275N, D286N, E260N/D262Q, and E281Q/E283Q. All these mutagenized TMEM2 constructs were C-terminally FLAG-tagged as is the case with TMEM2WT. All constructs were confirmed by sequencing.
HA degradation assay with cells in culture
293T cells in a 12-well plate (6 × 105 per well) were transiently transfected with FLAG-tagged wild-type and mutagenized TMEM2 constructs (see above) using TransIT-X2 (Mirus Bio, Madison, WI). After 8 h, fluoresceinamine (FA)-labeled GAGs (0.1 μg/ml or 0.5 μg/ml) were added to cells. FA-labeled HA of the average molecular size of 1500, 200, 20, and 5 kDa (FA-HA1500, FA-HA200, FA-HA20, and FA-HA5), FA-labeled chondroitin sulfate A, C, and D (FA-CSA, FA-CSC and FA-CSD), and FA-labeled dermatan sulfate (FA-DS) were purchased from PG Research (Tokyo, Japan). After a 72-h incubation with GAGs, the medium was harvested, and the size of GAGs was analyzed on a Sephacryl S-300 HR (GE Healthcare) column (1.6 × 18 cm) equilibrated with in PBS containing 0.2% NaN3. Fractions of 1.0 ml were collected at 0.7 ml/min, and fluorescence signals (excitation, 485 nm; emission, 535 nm) were measured by a Beckman DTX880 multimode plate reader.
HA degradation assay with the membrane fraction
293T cells (3 × 106 cells in a 6-cm dish) were transfected with TMEM2WT. After a 48-h incubation, cells were harvested with PBS, and homogenized with Dounce homogenizer (Wheaton) in 50 mm HEPES buffer (pH 7.0). The suspensions of disrupted cells were first centrifuged at 500 × g for 5 min at 4 °C, and then the supernatant was centrifuged at 10,000 × g for 10 min. The pellets (“P2 fraction”) were resuspended in 0.15 ml of 50 mm HEPES buffer (pH 7.0). For reaction, 0.05 ml of the resuspended P2 fraction was mixed with 0.55 ml of FA-HA1500 (final HA concentration of 0.1 μg/ml) in 50 mm HEPES buffer (pH 7.0) with or without 1 mm CaCl2 and incubated at 37 °C for 12 h, followed by gel filtration on Sephacryl S-300 HR and fluorescence measurement, as described above.
HA degradation assay with recombinant soluble TMEM2
To examine the effect of pH on enzymatic activity, recombinant soluble TMEM2 was produced. The cDNA of the extracellular domain of mouse TMEM2 (residues Ser104 to Leu1383) was amplified by PCR and subcloned into pSecTag2A (Thermo Fisher Scientific), which contains a mouse Igκ signal sequence. The Myc and His tags present in pSecTag2A were not added to the recombinant protein, as their presence was found to have detrimental effects on the expression level. The resultant construct (designated as TMEM2ECT) was transfected into 5 × 106 293T cells in a 10-cm dish. After a 72-h incubation, culture supernatants were collected and concentrated using Vivaspin concentrators (molecular mass cutoff, 5000 Da; Sigma). For reaction, 0.03 ml of the concentrates were mixed with 0.77 ml of FA-HA1500 (final HA concentration of 0.06 μg/ml) in either 50 mm acetate buffer (pH 4.0), 50 mm acetate buffer (pH 5.0), 50 mm MES buffer (pH 6.0), 50 mm HEPES buffer (pH 7.0), or 50 mm HEPES buffer (pH 8.0), all containing 1 mm CaCl2 and protease inhibitor mixture. Reaction mixtures were incubated for 16 h at 37 °C. Degradation of HA was analyzed on a Sephacryl S-300 HR column as described above. Enzyme activity was expressed as a percentage of fluorescence counts that represent depolymerized HA relative to the total fluorescence input.
Live immunolabeling
MG-63 cells cultured on glass coverslips (Glaswarenfabrik Karl Hecht GmbH, Sondheim v. d. Rhön, Germany) coated with type I collagen (Rat tail; Corning) were transfected with TMEM2WT. Forty-eight h after transfection, cells were stained live by incubating with anti-FLAG M2 antibody (1:2000) for 30 min at 4 °C to label TMEM2 present on the cell surface. After washing, cells were fixed with 4% paraformaldehyde in PBS and incubated with Alexa Fluor 488-conjugated donkey anti-mouse IgG (1:1000; catalog no. 715545151, lot 121992, Jackson ImmunoResearch, West Grove, PA) for 1 h at room temperature. Then to visualize cell contour, cells were incubated with Alexa Fluor 594-conjugated wheat germ agglutinin reagent (W11262, lot 1837939, Thermo Fisher Scientific) in PBS (1:200) for 10 min at room temperature. Coverslips were mounted using Prolong Gold antifade reagent (Thermo Fisher Scientific). Images were taken with Zeiss LSM710 laser-scanning microscope.
Surface biotinylation
MG-63 and 293T cells (1 × 106 in a 6-cm dish) were transfected with TMEM2WT. Forty eight h after transfection, cell-surface proteins were biotinylated by incubation with 1 mg/ml of membrane-impermeable EZ-link sulfo-NHS-SS-biotin (Thermo Fisher Scientific) in PBS for 30 min at room temperature. Non-reacted biotin was quenched by 50 mm Tris (pH 8.0). Cells were washed with PBS three times and then lysed with 500 μl of RIPA buffer. Cell lysates were incubated with 25 μl of streptavidin-agarose (Novagen) for 30 min at room temperature. After washing precipitates with RIPA buffer, biotinylated cell surface proteins were eluted by heating in SDS-sample buffer at 95 °C for 10 min. Equivalent amounts of whole-cell lysates and biotinylated surface fractions were separated by SDS-PAGE. TMEM2 was detected by immunoblotting with mouse monoclonal anti-FLAG antibody (M2, Sigma).
HA immobilization on glass bottom dishes and in situ HA degradation assay
To prepare glass substrates to which HA is immobilized, we employed the method described by Werb et al. (30). Glass bottom dishes (35 mm in diameter; MatTek Corp., Ashland, MA) were washed sequentially with 20% HNO3, water, 0.1 n NaOH, and water. Dried dishes were exposed to 2% γ-aminopropyltriethoxysilane in acetone for 5 min at room temperature, followed by rinses with water and PBS. The dishes were treated with 0.25% glutaraldehyde in PBS for 30 min at room temperature, washed with PBS, and incubated with 0.5 mg/ml FA-HA1500 in PBS for 2 h at room temperature. After rinsing with PBS, dishes were immediately used for HA degradation assays, in which 293T cells transfected with TMEM2WT or D273N were seeded at a density of 0.2 × 105 cells per dish and cultured for 60 h in a CO2 incubator. Degradation of substrate-bound HA by cells was examined by live imaging on a Zeiss LSM710 confocal microscope.
Absolute quantification of mRNA copy numbers
Mouse total RNA of the adult brain, eye, heart, kidney, liver, lung, salivary grand, spinal cord, spleen, stomach, testis, smooth muscle, and whole embryos at 11, 15, and 17 embryonic days was purchased from Clontech (mouse total RNA master panel, 636644). Total RNA from the small intestine, colon, kidney, ureter, bladder, prostate, lymph node, articular cartilage and synovium was isolated using RNeasy mini kit (Qiagen). cDNA was synthesized from 1 μg of total RNA using the SuperScript VILO Master Mix (Thermo Fisher Scientific). For qRT-PCR, TaqMan gene expression assay was carried out using LightCycler 96 system (Roche Applied Science) with TaqMan primer/probe sets for mouse TMEM2 (Mm00459599_m1) and CEMIP (Mm00472921_m1) obtained from Applied Biosystems (Foster City, CA). For absolute quantification, standard curves were generated with Ct values obtained from qPCR of various copy numbers of reference plasmids containing the coding regions of mouse TMEM2 and CEMIP (pcDNA3-TMEM2 and pcDNA3-CEMIP). Biological triplicates of qPCR assay were analyzed, and copy number of mRNA was calculated based on the standard curves.
Immunoblotting
For immunoblotting analysis of wild-type and point mutants of TMEM2, transfected cells were lysed in 4 m urea, 4% SDS in 100 mm Tris buffer (pH 8) containing protease inhibitor mixture (Sigma). For soluble TMEM2, culture supernatants of TMEM2ECT-transfected cells were collected and supplemented with protease inhibitor mixture. In these samples, protein concentration was determined by the BCA method (Thermo Fisher Scientific). Samples containing equal amount of proteins were separated by 8–16% SDS-PAGE under a reducing condition and transferred to Immobilon-P membranes (Millipore). After blocking, membranes were probed with anti-FLAG M2 antibody (1:2500, F3165, lot 60K9181, Sigma) for the detection of full-length TMEM2, rabbit polyclonal anti-TMEM2 antibody (1:2500, SAB2105088, lot QC12843; Sigma) for the detection of soluble TMEM2, and anti-α-tubulin (1:5000, T6074, lot 046K4770, Sigma; as loading controls). After incubation with anti-mouse IgG HRP (1:2000, catalog no. 1721011; Bio-Rad) or anti-rabbit IgG HRP antibody (1:2000, catalog no. 1706515; Bio-Rad), protein bands were visualized using SuperSignal West Pico chemiluminescence substrate (Thermo Fisher Scientific).
Author contributions
Y. Y. and C. O. conceived and coordinated the research. H. Y., F. I., and Y. Y. designed the experiments. H. Y. conducted most of the experiments and analyzed the results. Y. T. and T. I. performed absolute quantification of mRNA. Y. Y. and H. Y. wrote the paper.
Acknowledgment
We thank Dr. Bill Stallcup for reading the manuscript.
Note added in proof
In the version of this article that was published as a Paper in Press on February 28, 2017, the wrong traces were inadvertently used in the HA1500 panel in Fig. 2A. This error has now been corrected and does not affect the results or the conclusions of this work.
This work was supported in part by National Institutes of Health Grant R01 AR062692. The authors declare that they have no conflicts of interest with the contents of this article. The content is solely the responsibility of the authors and does not necessarily represent the official views of the National Institutes of Health.
This article was selected as one of our Editors' Picks.
- HA
- hyaluronan
- HMW
- high molecular weight
- FA-HA
- fluorescein-labeled hyaluronan
- GAG
- glycosaminoglycan
- CS
- chondroitin sulfate
- E
- embryonic day
- CEMIP
- cell migration-inducing and hyaluronan-binding protein
- DS
- dermatan sulfate
- GPI
- glycosylphosphatidylinositol
- qPCR
- quantitative PCR.
References
- 1. Laurent T. C., and Fraser J. R. (1992) Hyaluronan. FASEB J. 6, 2397–2404 [PubMed] [Google Scholar]
- 2. Fraser J. R., and Laurent T. C. (1989) Turnover and metabolism of hyaluronan. Ciba Found. Symp. 143, 41–53 [PubMed] [Google Scholar]
- 3. Stern R. (2003) Devising a pathway for hyaluronan catabolism: are we there yet? Glycobiology 13, 105R–115R [DOI] [PubMed] [Google Scholar]
- 4. Stern R., Kogan G., Jedrzejas M. J., and Soltés L. (2007) The many ways to cleave hyaluronan. Biotechnol. Adv. 25, 537–557 [DOI] [PubMed] [Google Scholar]
- 5. Chanmee T., Ontong P., and Itano N. (2016) Hyaluronan: A modulator of the tumor microenvironment. Cancer Lett. 375, 20–30 [DOI] [PubMed] [Google Scholar]
- 6. Afify A. M., Stern M., Guntenhöner M., and Stern R. (1993) Purification and characterization of human serum hyaluronidase. Arch. Biochem. Biophys. 305, 434–441 [DOI] [PubMed] [Google Scholar]
- 7. Lepperdinger G., Strobl B., and Kreil G. (1998) HYAL2, a human gene expressed in many cells, encodes a lysosomal hyaluronidase with a novel type of specificity. J. Biol. Chem. 273, 22466–22470 [DOI] [PubMed] [Google Scholar]
- 8. Triggs-Raine B., Salo T. J., Zhang H., Wicklow B. A., and Natowicz M. R. (1999) Mutations in HYAL1, a member of a tandemly distributed multigene family encoding disparate hyaluronidase activities, cause a newly described lysosomal disorder, mucopolysaccharidosis IX. Proc. Natl. Acad. Sci. U.S.A. 96, 6296–6300 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Rai S. K., Duh F. M., Vigdorovich V., Danilkovitch-Miagkova A., Lerman M. I., and Miller A. D. (2001) Candidate tumor suppressor HYAL2 is a glycosylphosphatidylinositol (GPI)-anchored cell-surface receptor for jaagsiekte sheep retrovirus, the envelope protein of which mediates oncogenic transformation. Proc. Natl. Acad. Sci. U.S.A. 98, 4443–4448 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Vigdorovich V., Strong R. K., and Miller A. D. (2005) Expression and characterization of a soluble, active form of the jaagsiekte sheep retrovirus receptor, Hyal2. J. Virol. 79, 79–86 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Stern R., and Jedrzejas M. J. (2006) Hyaluronidases: their genomics, structures, and mechanisms of action. Chem. Rev. 106, 818–839 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Kim E., Baba D., Kimura M., Yamashita M., Kashiwabara S., and Baba T. (2005) Identification of a hyaluronidase, Hyal5, involved in penetration of mouse sperm through cumulus mass. Proc. Natl. Acad. Sci. U.S.A. 102, 18028–18033 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Bouga H., Tsouros I., Bounias D., Kyriakopoulou D., Stavropoulos M. S., Papageorgakopoulou N., Theocharis D. A., and Vynios D. H. (2010) Involvement of hyaluronidases in colorectal cancer. BMC Cancer 10, 499. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Godin D. A., Fitzpatrick P. C., Scandurro A. B., Belafsky P. C., Woodworth B. A., Amedee R. G., Beech D. J., and Beckman B. S. (2000) PH20: a novel tumor marker for laryngeal cancer. Arch. Otolaryngol. Head Neck Surg. 126, 402–404 [DOI] [PubMed] [Google Scholar]
- 15. Beech D. J., Madan A. K., and Deng N. (2002) Expression of PH-20 in normal and neoplastic breast tissue. J. Surg. Res. 103, 203–207 [DOI] [PubMed] [Google Scholar]
- 16. Yoshida H., Nagaoka A., Kusaka-Kikushima A., Tobiishi M., Kawabata K., Sayo T., Sakai S., Sugiyama Y., Enomoto H., Okada Y., and Inoue S. (2013) KIAA1199, a deafness gene of unknown function, is a new hyaluronan binding protein involved in hyaluronan depolymerization. Proc. Natl. Acad. Sci. U.S.A. 110, 5612–5617 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Evensen N. A., Kuscu C., Nguyen H. L., Zarrabi K., Dufour A., Kadam P., Hu Y. J., Pulkoski-Gross A., Bahou W. F., Zucker S., and Cao J. (2013) Unraveling the role of KIAA1199, a novel endoplasmic reticulum protein, in cancer cell migration. J. Natl. Cancer Inst. 105, 1402–1416 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Wrzesiński T., Szelag M., Cieślikowski W. A., Ida A., Giles R., Zodro E., Szumska J., Poźniak J., Kwias Z., Bluyssen H. A., and Wesoly J. (2015) Expression of pre-selected TMEMs with predicted ER localization as potential classifiers of ccRCC tumors. BMC Cancer 15, 518. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Smith K. A., Lagendijk A. K., Courtney A. D., Chen H., Paterson S., Hogan B. M., Wicking C., and Bakkers J. (2011) Transmembrane protein 2 (Tmem2) is required to regionally restrict atrioventricular canal boundary and endocardial cushion development. Development 138, 4193–4198 [DOI] [PubMed] [Google Scholar]
- 20. Totong R., Schell T., Lescroart F., Ryckebüsch L., Lin Y. F., Zygmunt T., Herwig L., Krudewig A., Gershoony D., Belting H. G., Affolter M., Torres-Vázquez J., and Yelon D. (2011) The novel transmembrane protein Tmem2 is essential for coordination of myocardial and endocardial morphogenesis. Development 138, 4199–4205 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Jenkins J., Shevchik V. E., Hugouvieux-Cotte-Pattat N., and Pickersgill R. W. (2004) The crystal structure of pectate lyase Pel9A from Erwinia chrysanthemi. J. Biol. Chem. 279, 9139–9145 [DOI] [PubMed] [Google Scholar]
- 22. He Q. Y., Liu X. H., Li Q., Studholme D. J., Li X. W., and Liang S. P. (2006) G8: a novel domain associated with polycystic kidney disease and non-syndromic hearing loss. Bioinformatics 22, 2189–2191 [DOI] [PubMed] [Google Scholar]
- 23. Guo J., Cheng H., Zhao S., and Yu L. (2006) GG: a domain involved in phage LTF apparatus and implicated in human MEB and non-syndromic hearing loss diseases. FEBS Lett. 580, 581–584 [DOI] [PubMed] [Google Scholar]
- 24. Scavetta R. D., Herron S. R., Hotchkiss A. T., Kita N., Keen N. T., Benen J. A., Kester H. C., Visser J., and Jurnak F. (1999) Structure of a plant cell wall fragment complexed to pectate lyase C. Plant Cell 11, 1081–1092 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Lietzke S. E., Yoder M. D., Keen N. T., and Jurnak F. (1994) The three-dimensional structure of pectate lyase E, a plant virulence factor from Erwinia chrysanthemi. Plant Physiol. 106, 849–862 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Pickersgill R., Jenkins J., Harris G., Nasser W., and Robert-Baudouy J. (1994) The structure of Bacillus subtilis pectate lyase in complex with calcium. Nat. Struct. Biol. 1, 717–723 [DOI] [PubMed] [Google Scholar]
- 27. Abe S., Usami S., and Nakamura Y. (2003) Mutations in the gene encoding KIAA1199 protein, an inner-ear protein expressed in Deiters' cells and the fibrocytes, as the cause of nonsyndromic hearing loss. J. Hum. Genet. 48, 564–570 [DOI] [PubMed] [Google Scholar]
- 28. Yoder M. D., and Jurnak F. (1995) The refined three-dimensional structure of pectate lyase C from Erwinia chrysanthemi at 2.2 angstrom resolution (implications for an enzymatic mechanism). Plant Physiol. 107, 349–364 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Lairson L. L., Henrissat B., Davies G. J., and Withers S. G. (2008) Glycosyltransferases: structures, functions, and mechanisms. Annu. Rev. Biochem. 77, 521–555 [DOI] [PubMed] [Google Scholar]
- 30. Werb Z., Tremble P. M., Behrendtsen O., Crowley E., and Damsky C. H. (1989) Signal transduction through the fibronectin receptor induces collagenase and stromelysin gene expression. J. Cell Biol. 109, 877–889 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Yu Y., Lee C., Kim J., and Hwang S. (2005) Group-specific primer and probe sets to detect methanogenic communities using quantitative real-time polymerase chain reaction. Biotechnol. Bioeng. 89, 670–679 [DOI] [PubMed] [Google Scholar]
- 32. Harada H., and Takahashi M. (2007) CD44-dependent intracellular and extracellular catabolism of hyaluronic acid by hyaluronidase-1 and -2. J. Biol. Chem. 282, 5597–5607 [DOI] [PubMed] [Google Scholar]
- 33. Chow G., Knudson C. B., and Knudson W. (2006) Expression and cellular localization of human hyaluronidase-2 in articular chondrocytes and cultured cell lines. Osteoarthritis Cartilage 14, 849–858 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Duterme C., Mertens-Strijthagen J., Tammi M., and Flamion B. (2009) Two novel functions of hyaluronidase-2 (Hyal2) are formation of the glycocalyx and control of CD44-ERM interactions. J. Biol. Chem. 284, 33495–33508 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. Hida D., Danielson B. T., Knudson C. B., and Knudson W. (2015) CD44 knock-down in bovine and human chondrocytes results in release of bound HYAL2. Matrix Biol. 48, 42–54 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36. Miller A. D., Vigdorovich V., Strong R. K., Fernandes R. J., and Lerman M. I. (2006) Hyal2, where are you? Osteoarthritis Cartilage 14, 1315–1317 [DOI] [PubMed] [Google Scholar]
- 37. Cherr G. N., Yudin A. I., and Overstreet J. W. (2001) The dual functions of GPI-anchored PH-20: hyaluronidase and intracellular signaling. Matrix Biol. 20, 515–525 [DOI] [PubMed] [Google Scholar]
- 38. Yoshida H., Nagaoka A., Nakamura S., Tobiishi M., Sugiyama Y., and Inoue S. (2014) N-terminal signal sequence is required for cellular trafficking and hyaluronan-depolymerization of KIAA1199. FEBS Lett. 588, 111–116 [DOI] [PubMed] [Google Scholar]
- 39. Matsuzaki S., Tanaka F., Mimori K., Tahara K., Inoue H., and Mori M. (2009) Clinicopathologic significance of KIAA1199 overexpression in human gastric cancer. Ann. Surg. Oncol. 16, 2042–2051 [DOI] [PubMed] [Google Scholar]
- 40. Fink S. P., Myeroff L. L., Kariv R., Platzer P., Xin B., Mikkola D., Lawrence E., Morris N., Nosrati A., Willson J. K., Willis J., Veigl M., Barnholtz-Sloan J. S., Wang Z., and Markowitz S. D. (2015) Induction of KIAA1199/CEMIP is associated with colon cancer phenotype and poor patient survival. Oncotarget 6, 30500–30515 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41. Kao J., Salari K., Bocanegra M., Choi Y. L., Girard L., Gandhi J., Kwei K. A., Hernandez-Boussard T., Wang P., Gazdar A. F., Minna J. D., and Pollack J. R. (2009) Molecular profiling of breast cancer cell lines defines relevant tumor models and provides a resource for cancer gene discovery. PLoS ONE 4, e6146. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42. Camenisch T. D., Spicer A. P., Brehm-Gibson T., Biesterfeldt J., Augustine M. L., Calabro A. Jr., Kubalak S., Klewer S. E., and McDonald J. A. (2000) Disruption of hyaluronan synthase-2 abrogates normal cardiac morphogenesis and hyaluronan-mediated transformation of epithelium to mesenchyme. J. Clin. Invest. 106, 349–360 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43. Mjaatvedt C. H., Yamamura H., Capehart A. A., Turner D., and Markwald R. R. (1998) The Cspg2 gene, disrupted in the hdf mutant, is required for right cardiac chamber and endocardial cushion formation. Dev. Biol. 202, 56–66 [DOI] [PubMed] [Google Scholar]
- 44. Hatano S., Kimata K., Hiraiwa N., Kusakabe M., Isogai Z., Adachi E., Shinomura T., and Watanabe H. (2012) Versican/PG-M is essential for ventricular septal formation subsequent to cardiac atrioventricular cushion development. Glycobiology 22, 1268–1277 [DOI] [PubMed] [Google Scholar]


