Abstract
AU-rich element-binding proteins (ARE-BPs) offer post-transcriptional regulation of gene expression via physical interaction and recruitment of RNA decay machinery to the AU-rich elements within the 3′-UTR of the target transcripts. However, the role of ARE-BPs in lung cancer remains poorly understood. In this study, we have identified that K-homology splicing regulatory protein (KSRP), an ARE-BP, is robustly up-regulated in human lung cancer. Importantly, Kaplan-Meier survival analysis indicated that elevated KSRP expression was correlated with poor overall survival of lung cancer patients. Furthermore, cigarette smoke, a leading risk factor for lung cancer, was also identified to be an important contributor to increased KSRP expression. Remarkably, silencing of KSRP decreased cell proliferation, reversed anchorage-independent growth, and reduced migration/invasion, suggesting an oncogenic role for KSRP in lung cancer. Finally, we provide mechanistic evidence that KSRP promotes the down-regulation of Spry4 by a previously unidentified mechanism, i.e. post-transcriptional mRNA regulation.
Keywords: cell invasion, cell migration, mRNA decay, post-transcriptional regulation, RNA-binding protein, KSRP, NSCLC, Spry4
Introduction
Lung cancer, which primarily arises from lung epithelial cells, is the leading cause of cancer deaths for both men and women in the United States (1, 2). Importantly, the numbers of lung cancer deaths are more than breast, prostate, and colorectal cancers combined (1, 2). Despite advances in early detection and the development of molecular targeted therapies, which have shown some promise (3), a large number of mortalities still persists. Hence, identification of additional molecular targets is a research priority in lung cancer therapeutic research.
AU-rich element-binding proteins (ARE-BPs)2 offer post-transcriptional regulation of gene expression via physical interaction and recruitment of RNA decay machinery to the AU-rich elements within the 3′-untranslated region (UTR) of the target transcripts (4–6). The mRNAs with AU-rich elements (AREs) in their 3′-UTR constitute ∼10–15% of all the transcripts that span genes related to inflammation, transcription, cell proliferation, RNA metabolism, development, and cellular signaling, which play important roles in tumorigenesis (4–6). However, the role of ARE-BPs in lung cancer remains poorly understood. K-homology splicing regulatory protein (KSRP) belongs to the family of ARE-BPs, which promote rapid decay of select ARE-containing mRNAs (5, 6). Of note, KSRP has never been previously associated with lung cancer.
In this study, we observed a strong up-regulation in the expression of KSRP by using The Cancer Genome Atlas (TCGA) lung cancer data sets. To fully appreciate the functional role of KSRP in lung tumorigenesis, first we validated the expression of KSRP in lung cancer tissue microarrays (TMAs) and a panel of non-small-cell lung cancer (NSCLC) cell lines. We also identified cigarette smoke, a leading risk factor for lung cancer, as an important contributor for increased KSRP expression. Finally, by utilizing a battery of cell-based assays, we demonstrate a novel function for KSRP in the regulation of transformed cell growth through the down-regulation of Spry4, a tumor suppressor protein, by a previously unidentified mechanism, i.e. post-transcriptional regulation.
Results
Expression of KSRP in human lung cancer and NSCLC cell lines
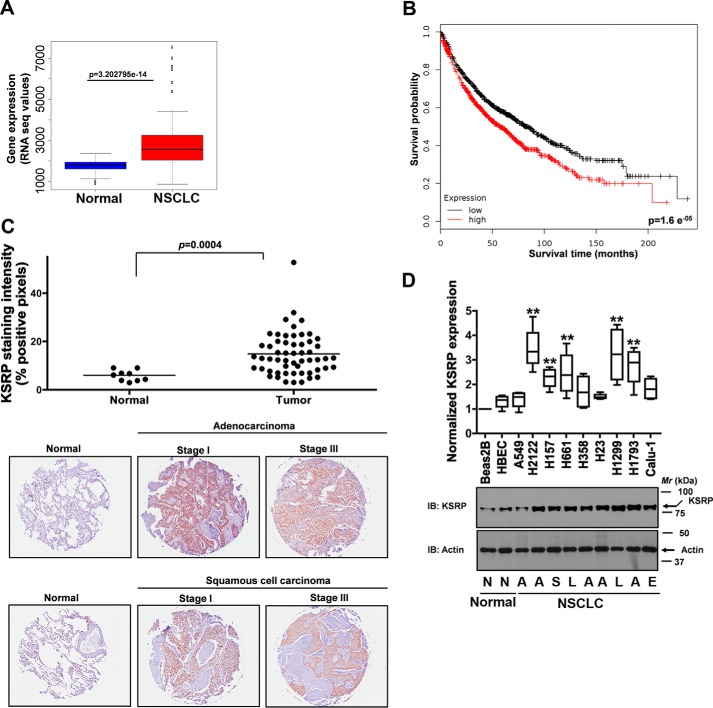
To determine the expression of KSRP in human lung tumors, we downloaded the lung cancer data sets from TCGA. The lung cancer data sets comprise 103 normal-tumor matched samples. The expression of KSRP is considered to be up-regulated if the Wald statistics value of the differential expression (normal versus tumor) is greater than 0 with a significant p value (<0.05), and the expression is considered to be down-regulated if the Wald statistics value is less than 0 with a significant p value (<0.05). A strong up-regulation of KSRP expression was observed in lung cancer data sets (Fig. 1A). Additionally, we performed Kaplan-Meier survival analysis by using the Kaplan-Meier plotter, an online tool (7). Our analyses indicated that higher KSRP expression was correlated with poor overall survival of lung cancer patients (Fig. 1B). To corroborate our in silico findings, we next looked for the expression of KSRP in human lung cancer specimens via staining lung cancer TMA slides (Biomax) with KSRP-specific antibodies followed by image analysis using a positive pixel count algorithm (Aperio). The TMA included 90 cases of lung carcinoma and 10 normal tissues in duplicate cores per case. Of note, the normal tissues were not matched samples. Consistent with our in silico findings, KSRP expression was significantly up-regulated in lung cancer tissues when compared with normal tissues (Fig. 1C). Representative immunohistochemistry images of normal lung, lung adenocarcinoma, and lung squamous carcinoma are shown in Fig. 1C. Finally, the expression of KSRP was also evaluated in non-transformed bronchial epithelial cell lines (Beas2B and human bronchial epithelial cell line (HBEC)) and NSCLC cell lines via immunoblotting (Fig. 1D). In agreement with our in silico analysis and TMA data, up-regulation in KSRP expression was also observed in 56% of NSCLC cell lines evaluated (Fig. 1D). Collectively, we show for the first time that KSRP expression is significantly up-regulated in lung cancer.
Figure 1.
KSRP is up-regulated in lung cancer. A, analysis of publicly available TCGA lung cancer data sets revealed a significant up-regulation in KSRP expression in lung cancer. B, online survival analysis software (Kaplan-Meier plotter) was used to assess the prognostic value of KSRP expression in lung cancer. C, KSRP staining of a TMA that includes normal and lung cancer tissue sections. The upper panel represents the staining intensity of KSRP, whereas representative images are displayed in the lower panel. p = 0.0004, t test. D, cell lysates of human non-transformed bronchial epithelial cells (Beas2B and HBEC) and a panel of NSCLC cell lines were probed for the expression of KSRP using anti-KSRP antibodies. N, normal; A, adenocarcinoma; S, squamous carcinoma; L, large cell carcinoma; E, epidermoid carcinoma. Error bars represent mean ± S.E. **, p < 0.01, ANOVA. RNA seq, RNA sequencing.
Effects of cigarette smoke on KSRP expression
Because cigarette smoking is a leading cause for lung cancer, we evaluated the effects of cigarette smoke (CS) on KSRP expression. For these studies, we first measured KSRP protein expression in the lung tissue samples from people who never smoked (non-smokers) and from people who smoked 10–25 cigarettes per day for at least 3 years (smokers). When compared with non-smokers, KSRP expression was significantly elevated in smokers (supplemental Fig. 1A). We also assessed the effects of CS exposure on KSRP expression in C57/BL6 mice (supplemental Fig. 1B). Consistent with the observations made in human tissues, a significant increase in KSRP expression was also observed in the lung tissue samples of mice exposed to CS for 6 months (supplemental Fig. 1B). To further confirm our observation that CS induced the expression of KSRP, we exposed Beas2B cells to cigarette smoke condensate (CSC). Exposure of Beas2B cells to CSC also induced KSRP expression (supplemental Fig. 1C). Taken together, these data suggest that CS is an important contributing factor for increased KSRP expression.
Effects of KSRP knockdown on cell proliferation
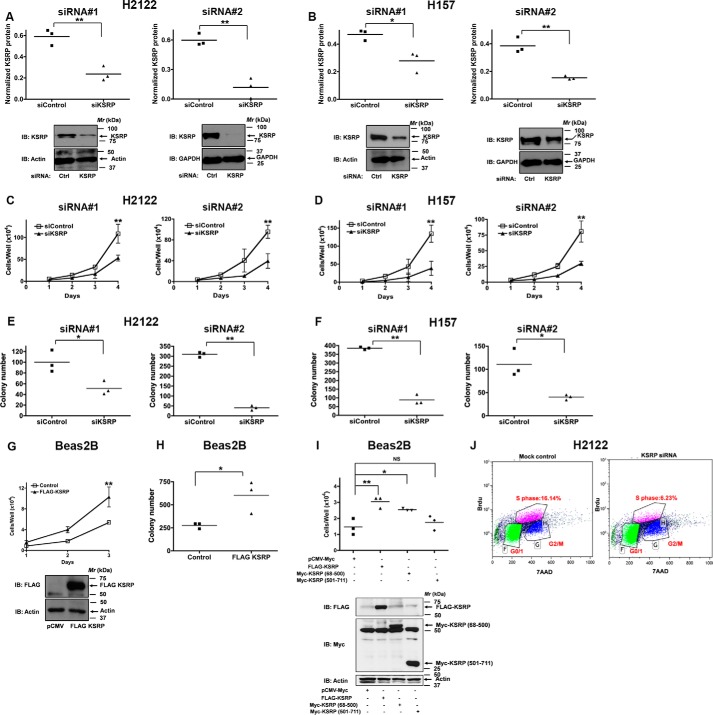
Having established an association of increased KSRP expression in lung cancer, we next sought to identify the role of KSRP in driving cell proliferation by using KSRP-specific small interfering RNAs (siRNAs) and short hairpin RNAs (shRNAs) in NSCLC cells (Fig. 2). Because KSRP is up-regulated in NSCLC, we chose to study the effects of KSRP knockdown on cell proliferation in several NSCLC cell lines, e.g. H2122 (derived from lung adenocarcinoma) and H157 (derived from lung squamous carcinoma). Transfection of H2122 and H157 with two different KSRP-specific siRNAs resulted in a significant reduction of KSRP expression when compared with cells transfected with scrambled siRNA controls (Fig. 2, A and B). Under similar knockdown conditions, a significant reduction in cell proliferation rates was observed in H2122 and H157 cells as determined by hemocytometer cell counting (Fig. 2, C and D) and clonogenic cell proliferation assays (Fig. 2, E and F). Additionally, we assessed cell proliferation via bromodeoxyuridine (BrdU) incorporation (Fig. 2J). The proportion of KSRP siRNA-treated H2122 cells in S phase (6%) was lower than the proportion of mock-transfected H2122 cells (16%), indicating that silencing KSRP expression suppressed the proliferation of H2122 cells (Fig. 2J). To further explore the role of KSRP in cell proliferation, we examined the effects of KSRP overexpression in Beas2B cells (Fig. 2, G and H). For these studies, a FLAG-tagged version of human KSRP (amino acids 68–711) was used in transient transfections of Beas2B cells (Fig. 2G), and the expression of FLAG-KSRP was ascertained by immunoblotting (Fig. 2G). Forced overexpression of KSRP in Beas2B cells resulted in an increased cell proliferation as determined by hemocytometer cell counting (Fig. 2G) and clonogenic cell proliferation assays (Fig. 2H). We next sought to identify the region of KSRP that promotes cell proliferation (Fig. 2I). Expression of the N terminus of KSRP harboring the K-homology motifs (Myc-KSRP(68–500)), but not the C terminus of KSRP (Myc-KSRP(501–711)), promoted Beas2B cell proliferation (Fig. 2I). These observations suggest that KSRP promotes cell proliferation via its N terminus, which encodes K-homology domains.
Figure 2.
KSRP regulates NSCLC cell proliferation. A and B, H2122 and H157 cells were treated with either control siRNAs or KSRP-specific siRNAs, and the extent of KSRP knockdown was determined by immunoblotting. *, p < 0.05; **, p < 0.01, t test. C and D, H2122 and H157 cells were treated with KSRP-specific siRNAs, and the effects of KSRP knockdown on cell proliferation were determined by hemocytometer cell counting. Error bars represent mean ± S.E. **, p < 0.01, ANOVA. E and F, H2122 and H157 cells were treated with KSRP-specific siRNAs, and the effects of KSRP knockdown on cell proliferation were determined by clonogenic assays. *, p < 0.05; **, p < 0.01, t test. G and H, Beas2B cells were transiently transfected with either control or FLAG-KSRP (amino acids 68–711) expression vectors, and the proliferation rates of the cells were determined by hemocytometer cell counting (error bars represent mean ± S.E; **, p < 0.01, ANOVA) and clonogenic assays (*, p < 0.05, t test). I, Beas2B cells were transiently transfected with control, FLAG-KSRP (amino acids 68–711), Myc-KSRP(68–500), and Myc-KSRP(501–711) expression vectors, and the proliferation rates of the cells were determined by hemocytometer cell counting. *, p < 0.05; **, p < 0.01, ANOVA. J, mock-transfected or KSRP siRNA-transfected H2122 cells were pulse-labeled with BrdU. The cell proliferation rates were later determined by staining with FITC-anti-BrdU antibodies followed by flow cytometry as described under “Experimental procedures.” Representative data of two independent, highly reproducible experiments are displayed. Ctrl, control; IB, immunoblotting; 7AAD, 7-aminoactinomycin D.
Effects of KSRP knockdown on anchorage-independent cell growth
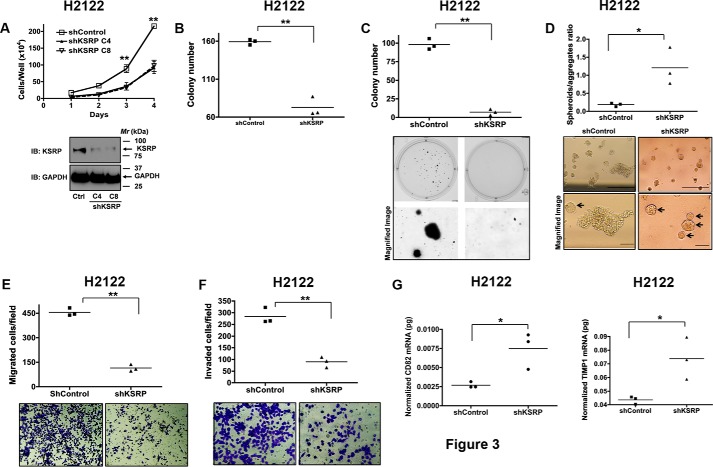
To determine the effects of KSRP knockdown on anchorage-independent cell growth, we developed multiple H2122 clones with stable expression of either control shRNA or KSRP shRNA to exclude the effects of clonal variation. Consistent with the effects of KSRP siRNAs on cell proliferation; stable knockdown of KSRP in H2122 cells also resulted in reduced cell proliferation as determined by hemocytometer cell counting (Fig. 3A) and clonogenic cell proliferation assays (Fig. 3B). The soft agar colony formation assay is a commonly used tool to measure anchorage-independent growth, a strict measure of transformed cell growth (8). To evaluate the effects of KSRP knockdown on anchorage-independent growth, H2122 clones stably expressing either control shRNA or KSRP shRNA were seeded into soft agar cultures. KSRP knockdown resulted in a remarkable reduction in the number of colonies formed when compared with H2122 clones stably expressing control shRNAs (Fig. 3C).
Figure 3.
KSRP regulates anchorage-independent growth, cell migration, and invasion. A and B, clones with stable expression of control shRNA or KSRP shRNA were generated as described under “Experimental procedures.” The cell proliferation rates of the stable clones were later determined by hemocytometer cell counting (A; error bars represent mean ± S.E.; **, p < 0.01, ANOVA) or clonogenic assays (B; **, p < 0.01, t test). C, anchorage-independent growth of H2122 clones with stable expression of either control shRNA or KSRP shRNA was assayed using soft agar assays as described under “Experimental procedures.” **, p < 0.01, t test. D, H2122 clones stably expressing either control shRNA or KSRP shRNA were embedded in Matrigel as single cells as described under “Experimental procedures.” The number of spheroids (arrows) and aggregates were counted and represented in the graph. Scale bar, 200 μm. *, p < 0.05, t test. E, migration of H2122 clones with stable expression of either control shRNA or KSRP shRNA was assayed in Transwell inserts as described under “Experimental procedures.” The top panel represents the number of cells migrated, whereas representative images are displayed in the bottom panel. **, p < 0.01, t test. F, invasion of H2122 clones with stable expression of either control shRNA or KSRP shRNA was assayed in Transwell inserts coated with Matrigel as described under “Experimental procedures.” The top panel represents the number of cells that invaded, whereas representative images are displayed in the bottom panel. **, p < 0.01, t test. G and H, total RNA from H2122 clones with stable expression of either control shRNA or KSRP shRNA was isolated, and the expression of CD82 (G) and TIMP1 (H) was later evaluated by qPCR as described under “Experimental procedures.” *, p < 0.05, t test. Ctrl, control; IB, immunoblotting.
Three-dimensional (3D) cultures allow phenotypic differentiation of non-malignant cells from malignant cells: non-malignant cells form highly organized spheroid-like structures, whereas malignant cells form unorganized, poorly proliferative structures. To examine whether KSRP plays a role in the phenotypic differentiation, H2122 clones stably expressing KSRP shRNAs were embedded in Matrigel matrices as single cells. As illustrated in Fig. 3D, KSRP knockdown induced spheroid formation similar to that of non-malignant cells (Fig. 3D), whereas control shRNA-treated cells formed poorly differentiated structures, a characteristic of malignant cells (Fig. 3D).
Effects of KSRP knockdown on migration and invasion
To evaluate the possible role of KSRP in NSCLC cell migration, we performed cell migration assays using Transwell (8-μm pore size) cell culture inserts. It was interesting to note that the stable knockdown of KSRP resulted in reduced cell migration when compared with H2122 clones stably expressing control shRNAs (Fig. 3E). We also evaluated the invasive abilities of H2122 clones stably expressing KSRP shRNAs by seeding the cells into Transwell cell culture inserts coated with Matrigel. The cells that invaded the Matrigel and migrated through the pores were later stained and counted and are represented in Fig. 3F. When compared with H2122 clones expressing control shRNAs, H2122 clones with stable knockdown of KSRP also displayed reduced invasive abilities (Fig. 3F). It was shown earlier that CD82-TIMP1 signaling suppresses tumor invasion via the inactivation of MMP9 (9). In accordance with the effects of KSRP knockdown on cell invasion, we also observed a significant increase in CD82 and TIMP1 expression in H2122 clones stably expressing KSRP shRNA (Fig. 3, G and H).
KSRP is a novel regulator of Spry4 tumor suppressor protein
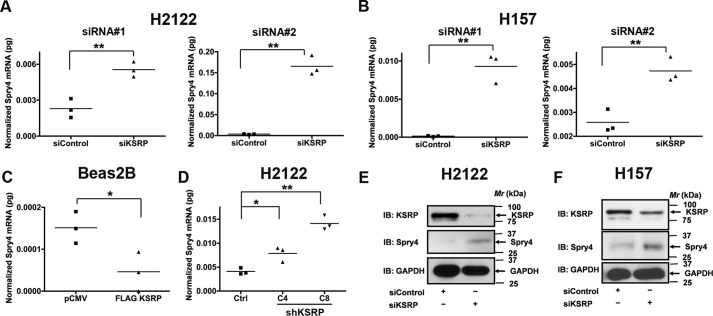
We chose to interrogate the role of KSRP in the regulation of Spry4 for the following reasons. 1) Spry4 has been previously shown to regulate NSCLC transformed cell growth, migration, and invasion (9). 2) Microarray data on KSRP-associated mRNAs showed enrichment of Spry4 (10). 3) In silico analysis of the 3′-UTR of Spry4 mRNA revealed eight class I (AUUUA) AREs. Because ARE-containing mRNAs are targeted by KSRP for degradation, we postulated that KSRP might represent a novel regulator of Spry4. To resolve this question, we first examined the mRNA expression of Spry4 in H2122 and H157 cells following siRNA-mediated KSRP knockdown (Fig. 4). Treatment of H2122 and H157 cells with KSRP siRNAs resulted in a robust increase in the expression of Spry4 transcripts (Fig. 4, A and B). Identical results were obtained in H2122 clones with stable knockdown of KSRP (Fig. 4D). Forced overexpression of KSRP in a non-transformed bronchial epithelial cell line, i.e. Beas2B, resulted in a significant reduction in the expression of Spry4 mRNA levels (Fig. 4C). Likewise, treatment of H2122 and H157 cells with KSRP siRNAs also resulted in a significant increase in Spry4 protein (Fig. 4, E and F).
Figure 4.
KSRP is a novel regulator of Spry4. A and B, total RNA from H2122 and H157 cells treated with KSRP-specific siRNAs was isolated, and the expression of Spry4 was later determined by qPCR as described under “Experimental procedures.” **, p < 0.01, t test. C, total RNA from Beas2B cells transiently transfected with either control or FLAG-KSRP expression vectors was isolated, and the expression of Spry4 was later determined by qPCR as described under “Experimental procedures.” *, p < 0.05, t test. D, total RNA from H2122 clones with stable expression of control shRNA or KSRP shRNA was isolated, and the expression of Spry4 was later determined by qPCR as described under “Experimental procedures.” *, p < 0.05; **, p < 0.01, ANOVA. E and F, proteins from H2122 and H157 cells treated with KSRP-specific siRNAs were isolated, and the expression of Spry4 was later determined by immunoblotting. Representative data of two independent, highly reproducible experiments are displayed. Ctrl, control; IB, immunoblotting.
KSRP binds to the 3′-UTR of Spry4 mRNA
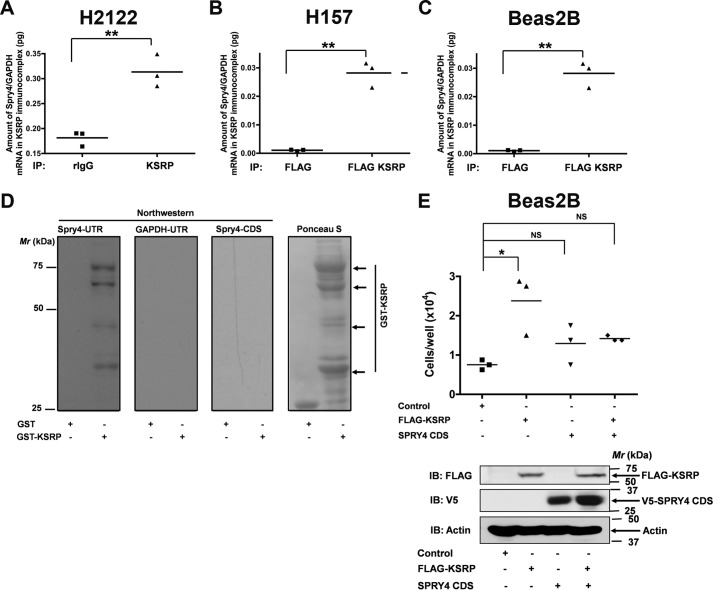
Because KSRP-mediated mRNA regulation involves the physical interaction of KSRP with the ARE-containing mRNAs, we evaluated whether KSRP was able to bind directly to Spry4 mRNA in a series of experiments (Fig. 5). To this end, we first performed RNA immunoprecipitations. In these experiments, KSRP was first immunoprecipitated from H2122 and H157 total cell lysates. Later, total RNA from the KSRP immunoprecipitate was isolated, and the amount of Spry4 and GAPDH transcripts in the RNA was evaluated by reverse transcription (RT)-qPCR (Fig. 5, A and B). This procedure has been successfully used in the past to demonstrate the interaction of KSRP with ARE-containing mRNAs (11). Increased enrichment of Spry4/GAPDH transcripts was detected in the KSRP immunocomplexes (Fig. 5, A and B). Similar results were obtained in an independent experiment wherein FLAG-KSRP was immunoprecipitated with anti-FLAG antibodies, indicating the validity of our assay (Fig. 5C).
Figure 5.
KSRP binds to Spry4 3′-UTR. A and B, RNA immunoprecipitation assay was performed on H2122 (A) and H157 (B) cell lysates with either control IgG or KSRP antibodies as described under “Experimental procedures.” The amounts of Spry4/GAPDH mRNA in the KSRP immunoprecipitates were later quantified using qPCR. *, p < 0.05; **, p < 0.01, t test. C, RNA immunoprecipitation assay was performed on the lysates of Beas2B cells expressing FLAG-KSRP with anti-FLAG antibodies. The amounts of Spry4/GAPDH mRNA in the KSRP immunoprecipitates were later quantified using qPCR. **, p < 0.01, t test. D, Northwestern analysis of KSRP and Spry4 UTR. Recombinant GST and GST-KSRP were separated by SDS-PAGE and transferred onto nitrocellulose membrane. Northwestern analysis was then performed either using DIG-labeled Spry4 UTR, GAPDH UTR, or Spry4 CDS probes as described under “Experimental procedures.” GAPDH UTR and Spry4 CDS probes were used as negative controls. Representative data of two independent, highly reproducible experiments are displayed. E, Beas2B cells were co-transfected with FLAG-KSRP (amino acids 68–711) and Spry4 CDS expression vectors. The proliferation rates of the cells were determined after 24 h by hemocytometer cell counting. KSRP-induced proliferative effects were attenuated upon co-expression of Spry4 CDS, suggesting that the proproliferative effects of KSRP were mediated through the down-regulation of Spry4. *, p < 0.05, ANOVA. IP, immunoprecipitation; IB, immunoblotting; rIgG, rabbit IgG; NS, not significant.
The interaction between KSRP and Spry4 was also investigated using Northwestern analysis. For these studies, recombinant GST or GST-KSRP proteins transferred onto nitrocellulose membranes were probed with in vitro transcribed digoxigenin (DIG)-labeled full-length Spry4 3′-UTR (bases 1160–4941). Spry4 UTR probe demonstrated binding to GST-KSRP but not to GST alone (Fig. 5D), whereas the DIG-labeled GAPDH 3′-UTR probe and Spry4 coding sequence (CDS) probe, tested as negative controls, did not bind to GST-KSRP (Fig. 5D). Because the GST-KSRP fusion protein was partially purified, we believe that the multiple bands were the result of endoproteolytic cleavage of GST-KSRP fusion protein in bacteria.
To determine whether KSRP promotes cell proliferation via the down-regulation of Spry4, we co-transfected human non-transformed bronchial epithelial cells (Beas2B) with KSRP and Spry4 CDS (Spry4 coding sequence that cannot be regulated by KSRP; Fig. 5D). Significantly, KSRP-induced proliferative effects were attenuated upon co-expression of Spry4 CDS (Fig. 5E), suggesting that the proproliferative effects of KSRP were mediated through the down-regulation of Spry4. These data provide a mechanistic link between KSRP and Spry4 in the context of the regulation of cell proliferation.
KSRP promotes post-transcriptional destabilization of Spry4 mRNA
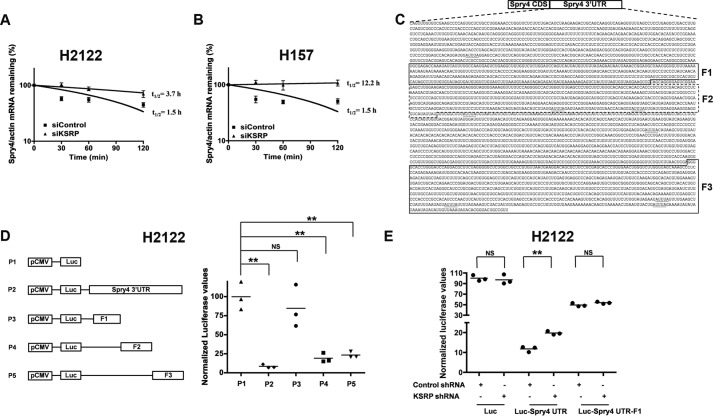
KSRP is an mRNA decay-promoting factor, which promotes rapid decay of ARE-containing mRNAs (11). We therefore aimed to determine the influence of KSRP knockdown on Spry4 mRNA stability. For these studies, H2122 and H157 cells were transiently transfected with KSRP siRNAs followed by the treatment with actinomycin D to block de novo mRNA synthesis. The cells were harvested at various times (0, 30, 60, and 120 min) after actinomycin D treatment. The half-lives of Spry4 mRNA were later determined by RT-PCR. In H2122 and H157 cells, Spry4 mRNA displayed significant mRNA decay (Fig. 6, A and B). Treatment of H2122 and H157 cells with KSRP siRNAs, on the contrary, stabilized Spry4 mRNA as demonstrated by an increase in the half-life of Spry4 mRNA (Fig. 6, A and B).
Figure 6.
KSRP promotes post-transcriptional destabilization of Spry4 transcripts. A and B, H2122 (A) and H157 (B) cells were transfected with either control or KSRP siRNAs followed by treatment with actinomycin D for the indicated periods of time. Spry4 and actin mRNA levels were quantified using qPCR, and the amounts of Spry4/actin mRNA remaining were plotted on a semilogarithmic scale. Error bars represent mean ± S.E. **, p < 0.01, ANOVA. C, schematic representation of the Spry4 mRNA with 3′-UTR displaying class I AREs. D, H2122 cells were transiently transfected with pcDNA-Luc or pcDNA-Luc with either full-length Spry4 3′-UTR (bases 1160–4961) or Spry4 3′-UTR truncations F1 (bases 2012–2650), F2 (bases 2651–3006), and F3 (bases 4072–4941). Later, the luciferase activities in the cell lysates were assayed using a luminometer, and normalized luciferase values are displayed in the graph. **, p < 0.01, ANOVA. E, H2122 cells were co-transfected with the indicated plasmids. Later, the luciferase activities in the cell lysates were assayed using a luminometer, and normalized luciferase values are displayed in the graph. **, p < 0.01, ANOVA. NS, not significant.
To evaluate the functional significance of the 3′-UTR of Spry4 mRNA (Fig. 6C), we developed a chimeric luciferase-Spry4 3′-UTR construct wherein the Spry4 3′-UTR was cloned downstream of firefly luciferase gene. If the Spry4 3′-UTR was functionally significant, it is anticipated that RNA-binding proteins, e.g. KSRP, would repress luciferase expression by targeting Spry4 3′-UTR. Indeed, the presence of Spry4 3′-UTR (Fig. 6D, P2) significantly reduced the expression of luciferase when compared with the luciferase reporter without Spry4 3′-UTR (Fig. 6D, P1). We next sought to identify the regulatory region within the Spry4 3′-UTR. For these experiments, we focused on the regions of Spry4 3′-UTR with clustered AREs. Dissection of the Spry4 3′-UTR revealed that bases 2651–4941 of the Spry4 3′-UTR (F2 and F3 regions) are functionally important (Fig. 6D, P4 and P5). It is noteworthy to mention that six class I (AUUUA) AREs were encoded within the F2 and F3 regions of Spry4 3′-UTR. These data indicate that Spry4 transcripts encode a functional 3′-UTR.
To ascertain whether the effects of KSRP on Spry4 3′-UTR were specific, we performed studies using a luciferase-Spry4 3′-UTR chimeric construct and KSRP shRNAs. If the effects of KSRP on Spry4 3′-UTR were specific, then upon KSRP knockdown the KSRP-mediated mRNA repression should be relaxed as detected by an increase in luciferase activities (Fig. 6E). Indeed, knockdown of KSRP increased the luciferase activities of Luc-Spry4 UTR but not the luciferase activities of Luc-Spry4 UTR-F1 (Fig. 6E). The small increase in the luciferase activities in the presence of KSRP shRNAs can be attributed to the efficacy of KSRP knockdown in these experiments.
Discussion
This is the first report demonstrating a novel mechanism of Spry4 regulation via a post-transcriptional mechanism mediated by KSRP. Spry4 is an important regulator of receptor tyrosine kinase signaling, which plays an essential role during growth, differentiation, and tumorigenesis (9, 12). Of note, Spry was shown to be an important negative regulator of protumorigenic fibroblast growth factor (FGF) and epidermal growth factor (EGF) signaling pathways (9, 12), underscoring the importance of this molecule and the newly identified mechanism in tumor suppression. Spry proteins are frequently down-regulated in cancers (9, 12); however, the mechanism of Spry4 down-regulation remains incompletely understood. Decreased mRNA expression can be attributed to reduced transcription and/or increased mRNA degradation. In the current study, we demonstrate that the loss of Spry4 in lung cancers is due to increased Spry4 mRNA degradation as a result of up-regulated KSRP expression.
KSRP is an ARE-BP that specifically binds and recruits RNA decay machinery to ARE-containing transcripts, subjecting them to degradation (13, 14). The importance of KSRP in several developmental processes has been highlighted (6, 11, 13–24, 26–40). However, the role of KSRP in lung cancer has not been previously demonstrated. In the current study, we show that KSRP is robustly up-regulated in lung cancer (Fig. 1). Subsequently, we also highlight the importance of KSRP in cell proliferation, migration/invasion, and transformed cell growth (Figs. 2–4). The observed differences in the cell proliferation rates of H2122, H157, and Beas2B might be due to the differences in plating efficiencies. Most importantly, we provide mechanistic evidence that KSRP promotes the down-regulation of Spry4 by a previously unidentified mechanism, i.e. post-transcriptional mRNA regulation (Figs. 4–6). Taken together, these studies form the preclinical framework to determine whether KSRP represents a suitable future molecular therapeutic target for treating lung cancer.
Although we have demonstrated that KSRP was up-regulated in lung cancer (Fig. 1), several questions still remain to be resolved. It was shown earlier that Dishevelled 3 (Dvl3), a critical regulator of Wnt signaling, regulates β-catenin transcript levels via the interaction with KSRP (11). In another study, Wnt7a/Fzd9 signaling was shown to regulate the expression of Spry4 in NSCLC (9). Based on these observations, it is tempting to speculate that Wnt7a/Fzd9 signaling-mediated regulation of Spry4 might be KSRP-dependent. However, how Wnt7a and/or its signaling intermediates, i.e. Dvl3, MEK5, ERK5, and peroxisome proliferator-activated receptor-γ, impair the mRNA decay-promoting abilities of KSRP remains obscure at the level of fine detail and is the subject of future study. Another unanswered research question includes deciphering the mechanism(s) that trigger KSRP expression in response to cigarette smoke.
We have identified a novel role for KSRP in the regulation of cell migration and invasion, which play an indispensable role for cancer metastasis. Spry4 is a demonstrated player that regulates cell migration and invasion in NSCLC (9). Therefore, the influence of KSRP on cell migration and invasion might be through the regulation of Spry4. The observations from the current study suggest that regulated expression of KSRP in the bronchial epithelium leads to the optimal expression of Spry4, which functions as a brake to cell proliferation, cell migration, and invasion. On the contrary, up-regulation of KSRP in NSCLC leads to the destabilization of Spry4 transcripts, resulting in increased cell proliferation, cell migration, and invasion, which contribute to the development of lung cancer.
Experimental procedures
Cell culture
Human non-transformed bronchial epithelial cell lines (Beas2B) and NSCLC cell lines (A549, H2122, H157, H661, H358, H23, H1299, H1793, and Calu1) were obtained from the tissue culture core facility of the University of Colorado Anschutz Medical Campus. Beas2B, H157, and H2122 were cultured in RPMI 1640 medium supplemented with 10% FBS in a humidified 5% CO2 incubator at 37 °C. HBEC, obtained from Dr. John Minna (University of Texas South Western), was cultured in bronchial epithelial cell growth medium (BEGMTM, Lonza). All the cell lines were cultured biweekly, and stocks of cell lines were passaged no more than 10 times for use in experiments.
For generating H2122 clones with stable expression of non-targeting shRNAs and KSRP shRNAs, H2122 cells were transfected with either pENTR/Lacz-control shRNA or pENTR/KSRP shRNA vectors followed by treatment with 100 or 200 μg/ml Zeocin (R25001, Invitrogen). Several Zeocin-resistant clones were subsequently isolated and screened for KSRP knockdown via immunoblotting.
Immunohistochemistry and tissue microarray
Lung cancer tissue microarrays with 90 cases of lung cancer and 10 normal tissues that were not from the same patient were obtained from Biomax (BC041114, US Biomax Inc., Rockville, MD). The TMAs were deparaffinized, rehydrated with a xylene and graded alcohol series, and subjected to antigen recovery using a citrate-based reagent (H-3300, Vector Laboratories, Burlingame, CA). After blocking nonspecific binding with normal horse serum, the TMAs were subjected to immunostaining with anti-KSRP antibody (HPA034739, Sigma) followed by incubation with ImmPRESS goat anti-rabbit horseradish peroxidase reagent (MP-7401, Vector Laboratories). Visualization was carried out using diaminobenzidine chromogen and then counterstaining with hematoxylin QS (H-3404, Vector Laboratories). Negative controls for immunostaining were performed by omitting the primary antibody and substituting with rabbit IgG. The stained slides were scanned using an Aperio digital pathology slide scanner (Leica Biosystems Inc., Buffalo Grove, IL) and scored using the positive pixel counting algorithm (Leica Biosystems Inc.), and the number of positive pixels were represented in a graph.
Cell proliferation studies
To measure the cell growth rates, 15,000 cells in complete growth medium were seeded per well in a 24-well culture plate. On subsequent days, cells were trypsinized from the wells with 100 μl of trypsin, diluted with 400 μl of growth medium, and counted using a hemocytometer.
Clonogenic assays were performed in triplicates by seeding 1000 cells/well in a 12-well culture plate followed by incubation at 37 °C in a 5% CO2 incubator. After 5–7 days, colonies were stained using a staining solution (0.5% crystal violet, 12% glutaraldehyde, 87.5% H2O) for 1 h at room temperature. After destaining in water and drying, colonies were quantified using a Bio-Rad Chemidoc imaging system and Quantity One software.
Cell proliferation was also determined by immunofluorescence staining of incorporated BrdU followed by flow cytometric analysis using a BD Pharmingen BrdU flow kit (FITC BrdU Flow kit, 559619) according to the manufacturer's recommendations. Briefly, H2122 cells were treated with KSRP siRNAs. After 48 h, the cells were pulse-labeled with 1 mm BrdU in PBS for 1 h. After 1 h, the cells were harvested by trypsin digestion followed by staining with FITC-labeled anti-BrdU antibodies and 7-aminoactinomycin D. Analysis was performed using a GalliosTM flow cytometer (Beckman Coulter Life Sciences). The S phase populations were compared and are presented in Fig. 2J.
Cell migration and invasion assays
For assessing cell migration, 15,000 cells in serum-free medium were seeded into Transwell inserts (353097, Corning) containing 8-μm permeable pores and allowed to migrate toward 10% FBS-containing medium. After 16 h, the Transwell inserts containing the cells were removed and washed in PBS three times. The migrated cells on the bottom of the insert were fixed with 5% glutaraldehyde solution followed by crystal violet (1%) staining. After washing the inserts three times with water, the inserts were allowed to air dry, and pictures were taken using an inverted microscope. Ten independent fields were counted for each Transwell, and the average numbers of cells/field were represented in graphs. For assessing cell invasion, 15,000 cells in serum-free medium were seeded into the Matrigel-coated Transwell inserts (BD Biosciences). After 16 h, the invaded cells were stained, counted, and represented in a graph similarly to the migration assay protocol.
Anchorage-independent growth
Soft agar assays were performed as described previously (8, 41). Briefly, 5000 cells were plated in triplicates in a 6-well plate in a volume of 1.5 ml of growth medium containing 0.3% Noble agar onto a base of 1.5 ml of growth medium containing 0.5% agar (8). The plates were incubated in a 37 °C CO2 incubator for 14 days. Later, colonies were stained for 16 h at 37 °C with nitroblue tetrazolium chloride (1 mg/ml), visualized under a microscope, counted, and represented in a graph.
3D cell culture
H2122 clones were grown in growth factor-reduced Matrigel (BD Biosciences) basement membrane according to Debnath et al. (42). Briefly, 5000 cells/well were grown in 4% Matrigel basement membrane with EGF on a 100% Matrigel layer. After 5–8 days, pictures of the colonies were taken using an inverted microscope equipped with a digital camera. Images were later analyzed by determining the number of spheroids and aggregates, and the spheroid/aggregate ratio was represented in figures.
RNA immunoprecipitation
Ribonucleoprotein complexes were immunoprecipitated from Beas2B, H2122, and H157 cells according to Bikkavilli and Malbon (11). Briefly, ribonucleoprotein complexes were immunoprecipitated from Beas2B cells expressing either empty vector or FLAG-KSRP using anti-FLAG antibodies, and complexes from H2122 and H157 cells were immunoprecipitated using anti-KSRP antibodies. Later, the RNA from the complexes was isolated using TRIzol reagent (Thermo Fisher). Real-time quantitative PCR amplification was later performed as described under “RNA isolation and real-time PCR.”
Northwestern assay
Northwestern assays were performed as described earlier (43). Briefly, DIG-labeled 3′-UTR probes of KSRP, GAPDH, and Spry4 CDS were synthesized in vitro using T7 RNA polymerase (Roche Applied Sciences) according to the manufacturer's recommendations in the presence of rNTPs, DIG-UTP, and pcDNA3.1 vectors harboring KSRP or GAPDH UTRs and pCDNA3.1 V5/His TOPO hSpry4 CDS as templates. For Northwestern analysis, recombinant proteins were resolved by SDS-PAGE and electrophoretically transferred to nitrocellulose membranes (“blots”). The blots were blocked in TBST (50 mm Tris, pH 7.4, 150 mm NaCl, and 0.1% Tween 20) containing 5% not-fat milk for 1 h at room temperature. Later, DIG-labeled probes (1 μg/ml in TBST with milk) were added to the blots and incubated at 4 °C with gentle rocking. After 16 h, the blots were washed thrice in TBST at 5-min intervals. The binding of RNA probes to KSRP was then revealed by probing the blots with anti-DIG alkaline phosphatase fragments diluted (1:1000) in TBST with 5% milk (11093274910, Roche Applied Science) followed by chemiluminescence detection of alkaline phosphatase using CDP-Star (Roche Applied Science).
Transfections and luciferase reporter assays
The reporter plasmids (pcDNA-Luc and pCDNA-Luc-Spry4 3′-UTR) and CMV-β-galactosidase control plasmids were transiently transfected into cells using Lipofectamine reagent (11668019, Invitrogen) according to the manufacturer's recommendations. After 48 h, the lysates were assayed for luciferase activities. The luciferase values were normalized to β-galactosidase values and represented in graphs.
KSRP mRNA stability
The half-life of KSRP mRNA was determined as described before (11). Briefly, H157 and H2122 cells treated with either control siRNA or KSRP siRNA were serum-starved for 16 h followed by actinomycin D treatment. Cells were then harvested at the indicated time points after the addition of actinomycin D (5 μg/ml). Total cellular RNA was isolated using TRIzol reagent, and KSRP mRNA levels were quantified by quantitative PCR as described earlier (11). Spry4/actin mRNA remaining (%) versus time after actinomycin D treatment was plotted on a semilogarithmic scale, and half-lives were calculated from the slope of fitted lines for each data set. An average of three independent half-life determinations was used for analysis.
Knockdown protocol
Double-stranded RNAs (siRNAs) targeting human KSRP (5′-GAUCAACCGGAGAGCAAGA-3′) and control siRNAs (5′-UCUGUGAUUUGAAAGACUAGCCAAG-3′) were procured from Invitrogen (22844075) and Qiagen (SI00300587). NSCLC cells were treated with 100 nm siRNAs using Lipofectamine 2000 reagent according to the manufacturer's protocol. Briefly, siRNAs were incubated with 5 μl of Lipofectamine 2000 for 20 min in 200 μl of Opti-MEM medium (Invitrogen), and then the mixture was added into 1 ml of growth medium in a 6-well plate in which cells were cultured to 80% confluence.
Immunoblotting analysis
Cell extracts were prepared in a lysis buffer (0.5% Triton X-100, 50 mm β-glycerophosphate, pH 7.20, 0.1 mm sodium vanadate, 2 mm MgCl2, 1 mm EDTA, 1 mm dithiothreitol, 2 μg/ml leupeptin, and 4 μg/ml aprotinin). The following antibodies were used for immunoblotting: anti-KSRP (HPA034739, Sigma-Aldrich), GAPDH (5174S, Cell Signaling Technology), actin (A3853, Sigma-Aldrich), anti-FLAG (F7425, Sigma-Aldrich), and anti-human Spry4 (AF5070, R&D Systems). Aliquots of various extracts were resolved by 10% SDS-PAGE and transferred to nitrocellulose membranes. The filters were blocked in Tris-buffered saline (10 mm Tris-Cl, pH 7.4, and 140 mm NaCl) containing 0.1% Tween 20 (TTBS) with 3% nonfat dry milk and then incubated with the same blocking solution containing the indicated antibodies at 0.5 μg/ml for 12–16 h. The filters were extensively washed in TTBS, and bound antibodies were visualized with horseradish peroxidase (HRP)-coupled secondary antibodies.
RNA isolation and real-time PCR
Total RNA from the cells was obtained using TRIzol reagent (10296-010, Invitrogen) according to the manufacturer's recommendations. For quantitative RT-PCR, 3 μg of total RNA was reverse transcribed using random primers, and real-time PCR was performed using the QuantiTect SYBR Green PCR kit (204050, Qiagen) and the Bio-Rad CFX real-time PCR detection system. Each cDNA sample was analyzed in triplicate, and the absolute amounts of transcripts were determined using an external standard. Briefly, a standard curve was generated using Ct values obtained from the real-time PCR using pGFP2-N2-mDvl2 plasmid. The Ct values of the target mRNA were later substituted in the equation from the standard curve to calculate the amount of the amplicon, and the calculated amounts of the amplicons (pg) were represented in graphs. The primers utilized in the PCR experiments were as follows: Spry4-F, 5′-CCAGGATGTCACCCACCATTG-3′; Spry4-R, 5′-TGTGCTGCTGCTGCTC-3′; actin-F, 5′-CCAGCTCACCATGGATGATG-3′; actin-R, 5′-ATGCCGGAGCCGTTGTC-3′; GAPDH-F,5′-CAAGGCTGTGGGCAAGGT-3′; GAPDH-R, 5′-GGAAGGCCATGCCAGTGA-3′; Dvl2-F, 5′-ACGACGATGCTGTACGAGTG-3′; Dvl2-R, 5′-ATTTCGAGGGAGGGTGAAGT-3′. Primers for CD82 were obtained from Integrated DNA Technologies, Inc. (345221533), and the primers for TIMP1 were obtained from SA Biosciences (PPH00771B-200).
The Cancer Genome Atlas analysis
For determining the expression of KSRP in lung cancer, lung cancer data sets were downloaded from the TCGA data portal (44, 45) and analyzed using the “R” program for statistical computing. For analysis, 103 matched normal-tumor lung cancer data sets were utilized, and the KSRP gene expression values (RNA sequence values) were represented in a graph.
Exposure of bronchial epithelial cells to CSC
Cultures of human bronchial epithelial cells, i.e. Beas2B, were grown to 80% confluence. Treatment with CSC (R100402, Murty Pharmaceuticals, Inc.) at a final concentration of 25 μg/ml was carried out in the culture medium at 37 °C for 4 h. The cell lysates were later probed for the expression of KSRP.
Exposure of mice to cigarette smoke
C57/BL6 mice were exposed to CS as described earlier (25). Briefly, 8-week-old control group mice were housed in a filtered air environment, whereas the experimental (CS exposure) group was subjected to CS exposure for 5 h/day, 5 days/week for 6 months using a whole-body smoke exposure system (TE-10, Teague Enterprises, Woodland, CA) using 2R4F cigarettes with 2.45 mg of nicotine/cigarette (Tobacco Research Institute, University of Kentucky, Lexington, KY).
Human tissues
We obtained deidentified human lungs not suitable for transplantation and donated for medical research from the National Disease Research Interchange (Philadelphia, PA) and the International Institute for the Advancement of Medicine (Edison, NJ). Smokers were individuals who smoked 10–25 cigarettes per day for at least 3 years, and non-smokers were individuals who had never smoked. The Committee for the Protection of Human Subjects at National Jewish Health and University of Illinois at Chicago approved this research.
Data analysis
Data were compiled from at least three independent, replicate experiments, each performed on separate cultures and on separate occasions. Comparisons of data among experimental groups were performed using Student's t test or one-way ANOVA as indicated in the figure legends. A p value <0.01 is denoted with a ** symbol, and a p value <0.05 is denoted with a * symbol.
Author contributions
RK. B., S. A. Z., M. V. S., L. W., P. Y. W., A. B., S. R., B. A. S., S. A., and R. A. W. designed, performed, and analyzed the experiments. C. D. C. analyzed the TMA slides shown in Fig. 1. N. M. R. and S. P. R. designed and analyzed cigarette smoke exposure in mice shown in supplemental Fig. 1B. B. K. provided the human tissue samples from non-smokers and smokers and analyzed the expression of KSRP shown in supplemental Fig. 1A. K. T. performed the analysis on TCGA data sets shown in Fig. 1A. RK. B., S. A. Z., S. A., and R. A. W. wrote the manuscript. All the authors reviewed the results and approved the final version of the manuscript.
Supplementary Material
Acknowledgments
We thank Drs. Roberto Gherzi (Istituto Nazionale per la Ricerca sul Cancro, Genova, Italy) and Diane Harris (UCLA Center for Human Nutrition) for the generous gift of FLAG-KSRP and pCDNA3.1 V5/His TOPO hSpry4 CDS plasmids, respectively. We also thank Ryan Deaton (Research Histology and Tissue Imaging Core, University of Illinois at Chicago) and Dr. Balaji Ganesh (Flow Cytometry Core, University of Illinois at Chicago) for assistance with TMA image analysis and BrdU incorporation analysis by flow cytometry, respectively.
This work was supported by National Institutes of Health Grants R01 CA138528 (to R. A. W.) and R01 HL118171 (to B. K.). The authors declare that they have no conflicts of interest with the contents of this article. The content is solely the responsibility of the authors and does not necessarily represent the official views of the National Institutes of Health.
This article contains supplemental Fig. 1.
- ARE-BP
- AU-rich element-binding protein
- KSRP
- K-homology splicing regulatory protein
- ARE
- AU-rich element
- TCGA
- The Cancer Genome Atlas
- TMA
- tissue microarray
- NSCLC
- non-small-cell lung cancer
- HBEC
- human bronchial epithelial cell line
- CS
- cigarette smoke
- CSC
- cigarette smoke condensate
- qPCR
- quantitative PCR
- DIG
- digoxigenin
- CDS
- coding sequence
- Luc
- luciferase
- Dvl
- Dishevelled
- ANOVA
- analysis of variance.
References
- 1. Jemal A., Siegel R., Ward E., Hao Y., Xu J., Murray T., and Thun M. J. (2008) Cancer statistics, 2008. CA 58, 71–96 [DOI] [PubMed] [Google Scholar]
- 2. Kohler B. A., Ward E., McCarthy B. J., Schymura M. J., Ries L. A., Eheman C., Jemal A., Anderson R. N., Ajani U. A., and Edwards B. K. (2011) Annual report to the nation on the status of cancer, 1975–2007, featuring tumors of the brain and other nervous system. J. Natl. Cancer Inst. 103, 714–736 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Sechler M., Borowicz S., Van Scoyk M., Avasarala S., Zerayesus S., Edwards M. G., Kumar Karuppusamy Rathinam M., Zhao X., Wu P. Y., Tang K., Bikkavilli RK, and Winn R. A. (2015) Novel role for γ-catenin in the regulation of cancer cell migration via the induction of hepatocyte growth factor activator inhibitor type 1 (HAI-1). J. Biol. Chem. 290, 15610–15620 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Barreau C., Paillard L., and Osborne H. B. (2005) AU-rich elements and associated factors: are there unifying principles? Nucleic Acids Res. 33, 7138–7150 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Khabar K. S. (2016) Hallmarks of cancer and AU-rich elements. Wiley Interdiscip. Rev. RNA 8, e1368. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Gherzi R., Lee K. Y., Briata P., Wegmüller D., Moroni C., Karin M., and Chen C. Y. (2004) A KH domain RNA binding protein, KSRP, promotes ARE-directed mRNA turnover by recruiting the degradation machinery. Mol. Cell 14, 571–583 [DOI] [PubMed] [Google Scholar]
- 7. Győrffy B., Surowiak P., Budczies J., and Lánczky A. (2013) Online survival analysis software to assess the prognostic value of biomarkers using transcriptomic data in non-small-cell lung cancer. PLoS One 8, e82241. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Borowicz S., Van Scoyk M., Avasarala S., Karuppusamy Rathinam M. K., Tauler J., Bikkavilli R. K., and Winn R. A. (2014) The soft agar colony formation assay. J. Vis. Exp. e51998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Tennis M. A., Van Scoyk M. M., Freeman S. V., Vandervest K. M., Nemenoff R. A., and Winn R. A. (2010) Sprouty-4 inhibits transformed cell growth, migration and invasion, and epithelial-mesenchymal transition, and is regulated by Wnt7A through PPARγ in non-small cell lung cancer. Mol. Cancer Res. 8, 833–843 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Winzen R., Thakur B. K., Dittrich-Breiholz O., Shah M., Redich N., Dhamija S., Kracht M., and Holtmann H. (2007) Functional analysis of KSRP interaction with the AU-rich element of interleukin-8 and identification of inflammatory mRNA targets. Mol. Cell. Biol. 27, 8388–8400 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Bikkavilli RK, and Malbon C. C. (2010) Dishevelled-KSRP complex regulates Wnt signaling through post-transcriptional stabilization of β-catenin mRNA. J. Cell Sci. 123, 1352–1362 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Masoumi-Moghaddam S., Amini A., and Morris D. L. (2014) The developing story of Sprouty and cancer. Cancer Metastasis Rev. 33, 695–720 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Briata P., Chen C. Y., Giovarelli M., Pasero M., Trabucchi M., Ramos A., and Gherzi R. (2011) KSRP, many functions for a single protein. Front. Biosci. 16, 1787–1796 [DOI] [PubMed] [Google Scholar]
- 14. Briata P., Chen C. Y., Ramos A., and Gherzi R. (2013) Functional and molecular insights into KSRP function in mRNA decay. Biochim. Biophys. Acta 1829, 689–694 [DOI] [PubMed] [Google Scholar]
- 15. Bird C. W., Gardiner A. S., Bolognani F., Tanner D. C., Chen C. Y., Lin W. J., Yoo S., Twiss J. L., and Perrone-Bizzozero N. (2013) KSRP modulation of GAP-43 mRNA stability restricts axonal outgrowth in embryonic hippocampal neurons. PLoS One 8, e79255. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Bollmann F., Art J., Henke J., Schrick K., Besche V., Bros M., Li H., Siuda D., Handler N., Bauer F., Erker T., Behnke F., Mönch B., Härdle L., Hoffmann M., et al. (2014) Resveratrol post-transcriptionally regulates pro-inflammatory gene expression via regulation of KSRP RNA binding activity. Nucleic Acids Res. 42, 12555–12569 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Briata P., Forcales S. V., Ponassi M., Corte G., Chen C. Y., Karin M., Puri P. L., and Gherzi R. (2005) p38-dependent phosphorylation of the mRNA decay-promoting factor KSRP controls the stability of select myogenic transcripts. Mol. Cell 20, 891–903 [DOI] [PubMed] [Google Scholar]
- 18. Briata P., Lin W. J., Giovarelli M., Pasero M., Chou C. F., Trabucchi M., Rosenfeld M. G., Chen C. Y., and Gherzi R. (2012) PI3K/AKT signaling determines a dynamic switch between distinct KSRP functions favoring skeletal myogenesis. Cell Death Differ. 19, 478–487 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Cammas A., Sanchez B. J., Lian X. J., Dormoy-Raclet V., van der Giessen K., López de Silanes I., Ma J., Wilusz C., Richardson J., Gorospe M., Millevoi S., Giovarelli M., Gherzi R., Di Marco S., and Gallouzi I. E. (2014) Destabilization of nucleophosmin mRNA by the HuR/KSRP complex is required for muscle fibre formation. Nat. Commun. 5, 4190. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Chou C. F., Lin Y. Y., Wang H. K., Zhu X., Giovarelli M., Briata P., Gherzi R., Garvey W. T., and Chen C. Y. (2014) KSRP ablation enhances brown fat gene program in white adipose tissue through reduced miR-150 expression. Diabetes 63, 2949–2961 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Chou C. F., Mulky A., Maitra S., Lin W. J., Gherzi R., Kappes J., and Chen C. Y. (2006) Tethering KSRP, a decay-promoting AU-rich element-binding protein, to mRNAs elicits mRNA decay. Mol. Cell. Biol. 26, 3695–3706 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Chou C. F., Zhu X., Lin Y. Y., Gamble K. L., Garvey W. T., and Chen C. Y. (2015) KSRP is critical in governing hepatic lipid metabolism through controlling Per2 expression. J. Lipid Res. 56, 227–240 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Gherzi R., Chen C. Y., Ramos A., and Briata P. (2014) KSRP controls pleiotropic cellular functions. Semin. Cell Dev. Biol. 34, 2–8 [DOI] [PubMed] [Google Scholar]
- 24. Gherzi R., Chen C. Y., Trabucchi M., Ramos A., and Briata P. (2010) The role of KSRP in mRNA decay and microRNA precursor maturation. Wiley Interdiscip. Rev. RNA 1, 230–239 [DOI] [PubMed] [Google Scholar]
- 25. Reddy N. M., Vegiraju S., Irving A., Paun B. C., Luzina I. G., Atamas S. P., Biswal S., Ana N. A., Mitzner W., and Reddy S. P. (2012) Targeted deletion of Jun/AP-1 in alveolar epithelial cells causes progressive emphysema and worsens cigarette smoke-induced lung inflammation. Am. J. Pathol. 180, 562–574 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Giovarelli M., Bucci G., Pasero M., Gherzi R., and Briata P. (2013) KSRP silencing favors neural differentiation of P19 teratocarcinoma cells. Biochim. Biophys. Acta 1829, 469–479 [DOI] [PubMed] [Google Scholar]
- 27. King P. H., and Chen C. Y. (2014) Role of KSRP in control of type I interferon and cytokine expression. J. Interferon Cytokine Res. 34, 267–274 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Linker K., Pautz A., Fechir M., Hubrich T., Greeve J., and Kleinert H. (2005) Involvement of KSRP in the post-transcriptional regulation of human iNOS expression-complex interplay of KSRP with TTP and HuR. Nucleic Acids Res. 33, 4813–4827 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Liu Y., and Liu Q. (2011) ATM signals miRNA biogenesis through KSRP. Mol. Cell 41, 367–368 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Michlewski G., and Cáceres J. F. (2010) Antagonistic role of hnRNP A1 and KSRP in the regulation of let-7a biogenesis. Nat. Struct. Mol. Biol. 17, 1011–1018 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Min H., Turck C. W., Nikolic J. M., and Black D. L. (1997) A new regulatory protein, KSRP, mediates exon inclusion through an intronic splicing enhancer. Genes Dev. 11, 1023–1036 [DOI] [PubMed] [Google Scholar]
- 32. Nechama M., Peng Y., Bell O., Briata P., Gherzi R., Schoenberg D. R., and Naveh-Many T. (2009) KSRP-PMR1-exosome association determines parathyroid hormone mRNA levels and stability in transfected cells. BMC Cell Biol. 10, 70. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Nicastro G., García-Mayoral M. F., Hollingworth D., Kelly G., Martin S. R., Briata P., Gherzi R., and Ramos A. (2012) Noncanonical G recognition mediates KSRP regulation of let-7 biogenesis. Nat. Struct. Mol. Biol. 19, 1282–1286 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Pasero M., Giovarelli M., Bucci G., Gherzi R., and Briata P. (2012) Bone morphogenetic protein/SMAD signaling orients cell fate decision by impairing KSRP-dependent microRNA maturation. Cell Rep. 2, 1159–1168 [DOI] [PubMed] [Google Scholar]
- 35. Repetto E., Briata P., Kuziner N., Harfe B. D., McManus M. T., Gherzi R., Rosenfeld M. G., and Trabucchi M. (2012) Let-7b/c enhance the stability of a tissue-specific mRNA during mammalian organogenesis as part of a feedback loop involving KSRP. PLoS Genet. 8, e1002823. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36. Ruggiero T., Trabucchi M., Ponassi M., Corte G., Chen C. Y., al-Haj L., Khabar K. S., Briata P., and Gherzi R. (2007) Identification of a set of KSRP target transcripts upregulated by PI3K-AKT signaling. BMC Mol. Biol. 8, 28. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37. Seok H., Cho J., Cheon M., and Park I. S. (2002) Biochemical characterization of apoptotic cleavage of KH-type splicing regulatory protein (KSRP)/far upstream element-binding protein 2 (FBP2). Protein Pept. Lett. 9, 511–519 [DOI] [PubMed] [Google Scholar]
- 38. Suswam E. A., Nabors L. B., Huang Y., Yang X., and King P. H. (2005) IL-1β induces stabilization of IL-8 mRNA in malignant breast cancer cells via the 3′ untranslated region: involvement of divergent RNA-binding factors HuR, KSRP and TIAR. Int. J. Cancer 113, 911–919 [DOI] [PubMed] [Google Scholar]
- 39. Trabucchi M., Briata P., Filipowicz W., Ramos A., Gherzi R., and Rosenfeld M. G. (2010) KSRP promotes the maturation of a group of miRNA precursors. Adv. Exp. Med. Biol. 700, 36–42 [PMC free article] [PubMed] [Google Scholar]
- 40. Trabucchi M., Briata P., Garcia-Mayoral M., Haase A. D., Filipowicz W., Ramos A., Gherzi R., and Rosenfeld M. G. (2009) The RNA-binding protein KSRP promotes the biogenesis of a subset of microRNAs. Nature 459, 1010–1014 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41. Winn R. A., Marek L., Han S. Y., Rodriguez K., Rodriguez N., Hammond M., Van Scoyk M., Acosta H., Mirus J., Barry N., Bren-Mattison Y., Van Raay T. J., Nemenoff R. A., and Heasley L. E. (2005) Restoration of Wnt-7a expression reverses non-small cell lung cancer cellular transformation through frizzled-9-mediated growth inhibition and promotion of cell differentiation. J. Biol. Chem. 280, 19625–19634 [DOI] [PubMed] [Google Scholar]
- 42. Debnath J., Muthuswamy S. K., and Brugge J. S. (2003) Morphogenesis and oncogenesis of MCF-10A mammary epithelial acini grown in three-dimensional basement membrane cultures. Methods 30, 256–268 [DOI] [PubMed] [Google Scholar]
- 43. Bikkavilli RK, and Malbon C. C. (2011) Arginine methylation of G3BP1 in response to Wnt3a regulates β-catenin mRNA. J. Cell Sci. 124, 2310–2320 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44. Cancer Genome Atlas Research Network, Weinstein J. N., Collisson E. A., Mills G. B., Shaw K. R., Ozenberger B. A., Ellrott K., Shmulevich I., Sander C., and Stuart J. M. (2013) The Cancer Genome Atlas Pan-Cancer analysis project. Nat. Genet. 45, 1113–1120 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45. Zhang K., and Wang H. (2015) Cancer Genome Atlas Pan-cancer Analysis Project. Zhongguo Fei Ai Za Zhi 18, 219–223 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.