Abstract
The activity of the E3 ligase, SMURF2, is antagonized by an intramolecular, autoinhibitory interaction between its C2 and Hect domains. Relief of SMURF2 autoinhibition is induced by TGFβ and is mediated by the inhibitory SMAD, SMAD7. In a proteomic screen for endomembrane interactants of the RING-domain E3 ligase, RNF11, we identified SMURF2, among a cohort of Hect E3 ligases previously implicated in TGFβ signaling. Reconstitution of the SMURF2·RNF11 complex in vitro unexpectedly revealed robust SMURF2 E3 ligase activity, with biochemical properties previously restricted to the SMURF2·SMAD7 complex. Using in vitro binding assays, we find that RNF11 can directly compete with SMAD7 for SMURF2 and that binding is mutually exclusive and dependent on a proline-rich domain. Moreover, we found that co-expression of RNF11 and SMURF2 dramatically reduced SMURF2 ubiquitylation in the cell. This effect is strictly dependent on complex formation and sorting determinants that regulate the association of RNF11 with membranes. RNF11 is overexpressed in certain tumors, and, importantly, we found that depletion of this protein down-regulated gene expression of several TGFβ-responsive genes, dampened cell proliferation, and dramatically reduced cell migration in response to TGFβ. Our data suggest for the first time that the choice of binding partners for SMURF2 can sustain or repress TGFβ signaling, and RNF11 may promote TGFβ-induced cell migration.
Keywords: cancer, E3 ubiquitin ligase, endosome, transforming growth factor beta (TGF-B), ubiquitylation (ubiquitination)
Introduction
Endocytosis of plasma membrane-associated growth factor receptors can either attenuate or propagate and sustain receptor-mediated key signaling pathways. For example, down-regulation of the epidermal growth factor receptor (EGFR)3 occurs through clathrin-dependent endocytosis. Here, cargo destined for degradation traffics first to the early endosome (also known as the sorting endosome) and is then sorted to the lysosome. Alternatively, activated EGF receptor internalized through clathrin-dependent endocytosis can be routed to the sorting endosome, where it continues to signal, or it can be recycled back to the plasma membrane directly or indirectly via recycling endosomes (1). Thus, clathrin-dependent endocytosis of the EGFR functions in both the transduction and the termination of growth factor signaling. In contrast, the regulation of TGFβ signaling is compartmentalized from its initial activation at the plasma membrane (2). At the plasma membrane, the receptor (TGFβR) is uniformly distributed between lipid-rich microdomains, termed lipid rafts, and non-raft regions. Following ligand engagement, the TGFβR·ligand complex can be endocytosed using two distinct mechanisms. In one pathway, ligand·receptor complexes internalized via clathrin-dependent endocytosis continue to actively signal on early endosomes through receptor-mediated phosphorylation of SMAD proteins. In a second pathway, internalization of ligand·receptor complexes residing in lipid rafts occurs via clathrin-independent/caveolin-dependent endocytosis, which is driven by ubiquitylation of the TGFβR by the SMURF2·SMAD7 E3 ligase complex and which targets the receptor for degradation by the lysosome or the proteasome (2). Thus, in contrast with EGFR signaling, there is physical segregation of the molecular machineries that mediate the propagation and termination of TGFβ signaling (2), because endosomes generated by caveolin-dependent endocytosis of the activated TGFβR and the SMURF2·SMAD7 complex are thought to bypass the sorting endosome en route to the lysosome.
Mechanistically, compartmentalized activation of SMURF2 ligase in response to TGFβ stimulation is achieved by regulated assembly of a stable complex with SMAD7 in the nucleus and subsequent recruitment to activated TGFβ receptors in lipid raft domains in the plasma membrane. This complex is stabilized by a protein-protein interaction between WW domains in SMURF2 and a PPXY motif in SMAD7, generating an active E3 ligase that is essential for the termination of TGFβ signaling (3, 4). The modularity of PPXY and WW domains prompts speculation that an additional layer of regulation, such as the sequestration of SMURF2, could be achieved by other proteins harboring proline-rich motifs that reside outside of the nucleus. Under these circumstances, the net effect of this type of regulation would be a prolonged or sustained level of responsiveness to TGFβ, because less SMURF2 would be available for recruitment to the nucleus for complex formation with SMAD7. Such a scenario could contribute to the deregulation of TGFβ signaling observed in disease (5).
A strong candidate for this novel type of regulation is RNF11 (RING finger protein 11), a gene identified in screens for cDNAs encoding RING finger proteins and genes highly expressed in breast cancer (6, 7). RNF11 encodes a protein of 154 amino acids with several potential interaction domains, including a PPXY motif, a RING domain, a ubiquitin interaction motif (UIM), and two acidic dileucine or DXXLL motifs. In growing cells, RNF11 displays a complex intracellular vesicular distribution, and it is thought to localize at steady state to the early endosome and recycling endosomes (8–11). Entry into these compartments is strictly regulated by multiple sorting determinants, which regulate its association with membranes and trafficking within the cell. First, acylation of RNF11 was shown to be essential for association with endosomes, because mutation of a putative myristoylation site prevented its trafficking to cellular membranes (8–10). Analysis of both acidic dileucine motifs also provided critical insight into how nascent RNF11 enters the endosomal compartment. A mutation (Lc) in the C-terminal DXXLL motif of RNF11 (amino acids 144–148) led to Golgi accumulation, suggesting a defect in export from this organelle, whereas a mutation (Ln) in the N-terminal DXXLL motif (amino acids 12–16) or the double mutation (Lc/Ln) caused the protein to be restricted to the plasma membrane (11). Based on these and other data, it has been proposed that newly synthesized RNF11 is directed to the plasma membrane from the sorting endosome, following its export from the Golgi, and reentry into the endosomal compartment from the plasma membrane is thought to occur through clathrin-dependent endocytosis (10,11). There is also accumulating evidence that ubiquitylation of RNF11 and/or its trafficking machinery could also regulate the subcellular localization of RNF11 (8–11). The intricate and strict controls that the cell has developed to establish the steady-state localization of RNF11 led us to test whether this might impact its function in the cell.
In this report, we show for the first time that RNF11 is able to block SMURF2-mediated repression of TGFβ signaling by sequestering SMURF2 in an endosomal compartment. We further show that a key element of this regulation entails the proper sorting of RNF11 to antagonize SMURF2 function. Moreover, we find for the first time that the SMURF2·RNF11 complex is a functional E3 ligase. Most importantly, whereas RNF11, like SMAD7, can stimulate the enzymatic activity of SMURF2, these proteins exhibit antagonistic effects on silencing of TGFβ activity in vivo, due to sequestration of SMURF2 to different compartments. Furthermore, depletion of RNF11 in the metastatic tumor cell line, 4T1, significantly mitigated TGFβ-responsive gene expression and cell migration. We propose that up-regulation of RNF11, which has been observed in multiple types of cancer, could inappropriately sustain TGFβ signaling by sequestering SMURF2 in an endosomal compartment, thereby preemptively antagonizing the function of SMURF2·SMAD7 complex in suppressing cellular TGFβ responsiveness, enhancing cell migration, and promoting the progression of these cancers.
Results
RNF11 associates with RING and Hect E3 ligases on membranes
Because all previous screens for RNF11-interacting proteins were yeast two-hybrid screens, and it was subsequently shown that RNF11 resides in an endosomal compartment, we initiated a proteomic screen in HEK293T cells to determine whether any known or novel interactions were localized to endomembranes. First, we generated a high-quality antibody against RNF11 and demonstrated its specificity (Fig. 1). As shown in Fig. 1A, our affinity-purified antibody detected a single band of ∼25 kDa in control but not RNF11-depleted extracts. Next, we used our antibody to analyze the subcellular localization of endogenous and C-terminally FLAG-tagged RNF11 and confirmed a prior observation that both proteins are restricted to punctate structures (Fig. 1B), some of which co-stain with the early endosomal marker EEA1 within the cytoplasm (8). No further analysis in this regard was performed, because RNF11-positive puncta have been extensively characterized and were definitively established to reside throughout the endosomal compartment (8–11). Cells transiently expressing C-terminally tagged RNF11 were then lysed in a detergent-free isosmotic buffer to preserve protein interactions associated with membranes, and the resulting crude cytosol (S800) was fractionated by low-speed centrifugation (20,000 × g) to enrich for endosomal proteins (Fig. 2A). Immunoblot analysis revealed enrichment of RNF11 in the crude endosomal fraction, as judged by the presence of markers for two different endosomal compartments (Fig. 2B). After solubilizing the resulting membranous pellet (P20K), we immunopurified RNF11 and associated proteins from this fraction and from crude cytosolic input and subjected the enriched proteins to mass spectrometric sequencing (Fig. 2A). Next, we “subtracted” common contaminants, collected from multiple, control immunoaffinity purification procedures using an identical strategy, from our list of peptides (12). Interestingly, a group of potential interacting proteins recovered from both cytosolic and membrane fractions was highly concordant, and inspection of gene functions linked to this group identified two RING E3 ligases and multiple Hect E3 ligases with an overlapping set of functional motifs (Table 1). Notably, the recovery of a substantial number of non-overlapping peptides indicates that these were robust interactions. Importantly, our screen provides compelling support for the notion that all RNF11·E3 ligase complexes identified here, including those previously identified in yeast two-hybrid screens, are stably associated with intracellular membranes.
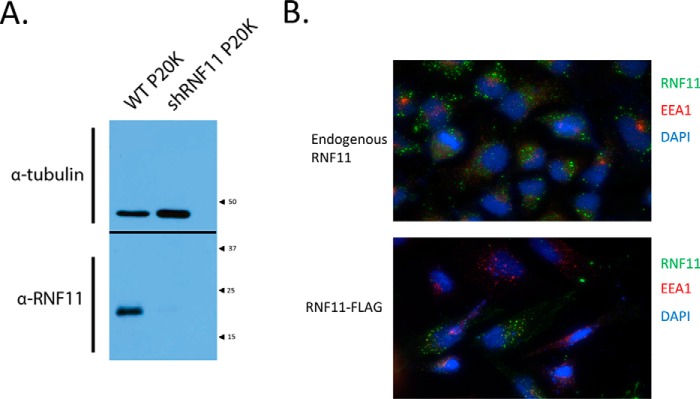
Figure 1.
Validation of specificity of anti-RNF11 polyclonal antibody. A, validation of affinity-purified RNF11 rabbit polyclonal antibody by knockdown in 293T cells. Shown is immunoblotting of 20 μg of solubilized P20K pellet. Inputs were resolved by SDS-PAGE and blotted. The blot was then cut in half at the 37 kDa marker. The top and bottom halves of the blot were probed with tubulin antibody and then developed together. Inputs show a decrease in RNF11 signal in cells stably expressing an shRNA specific for RNF11. α-Tubulin was used as a loading control. B, endogenous and C-terminally tagged RNF11 localize to the endosomal compartment. HeLa cells were transfected with a C-terminal FLAG fusion with RNF11 mammalian expression vector, fixed after 48 h, and visualized with FLAG antibody and the early endosomal marker EEA1 (bottom). Endogenous RNF11 immunostaining with the in-house RNF11 antibody and EEA1 antibody is shown in the top panel.
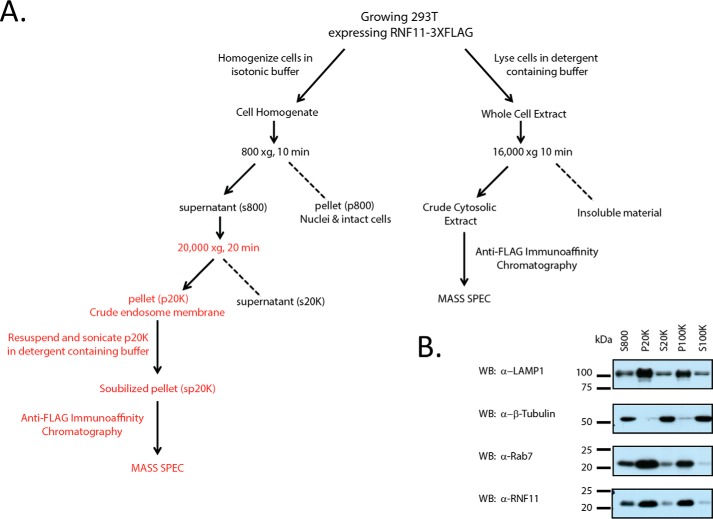
Figure 2.
RNF11 is enriched in the crude endosomal fraction. A, schema of the procedure used to prepare samples for mass spectrometry. B, RNF11 is enriched in a HEK293T membrane fraction obtained after subcellular fractionation. 25 μg of protein were loaded per lane. Immunoblots (WB) were probed with the in-house RNF11 antibody, anti-β-tubulin antibody, anti-Lamp1 antibody (lysosomal marker), and anti-Rab7 antibody (lysosomal marker). The HEK293T P20K and P100K fractions are defined under “Experimental procedures.”
Table 1.
RNF11 associates with RING and Hect E3 ligases and E2s on membranes
Depicted are the results of two proteomic screens, P20K pellet and crude cytosol (S800).
| Gene name | Accession number | Number of peptides (P20K pellet) | Number of peptides (cytosol) |
|---|---|---|---|
| ITCH | sp|q96j02 | 18 | 19 |
| NEDD4 | sp|p46934 | 0 | 18 |
| NEDD4-2 | sp|q96pu5 | 6 | 26 |
| SMURF1 | sp|b7zmb6 | 0 | 9 |
| SMURF2 | tr|q9hau4 | 9 | 47 |
| WWP1 | sp|q9h0m0 | 40 | 54 |
| WWP2 | sp|o00308 | 55 | 54 |
| LISTERIN | sp|094822 | 13 | 55 |
| hRul138 | sp|q86y13 | 3 | 0 |
| UBCH5C | sp|61077 | 0 | 3 |
| UBC7 | sp|p6107 | 4 | 0 |
| UBCH7 | sp|68036 | 0 | 8 |
| UBE2N | sp|p61088 | 16 | 23 |
| UBE2V1 | sp|q13404 | 0 | 6 |
Ubiquitylation of a subset of Hect E3 ligases is mediated by RNF11
A striking feature of the set of Hect E3 ligases identified in our RNF11 endosomal proteomic screen is the presence of all known members of the NEDD4 family (Table 1). This group of enzymes shares a common set of motifs, including an N-terminal C2 domain, several WW domains, and a catalytic C-terminal Hect domain (13). Interestingly, many of the Hect E3 ligases identified in our screen have been implicated in the regulation of TGFβ signal transduction. For example, SMURF1, SMURF2, NEDD4-2, and WWP1 participate in the suppression of TGFβ signaling by multiple mechanisms, which include polyubiquitylation of receptor SMADs (14) and receptor down-regulation (3, 15–17). In addition, ITCH/AIP4, which has been implicated in TGFβ signaling, was identified in our screen, although its role is less clear, and there is support for a role in both suppression and activation of the TGFβ pathway (18, 19).
To begin to understand mechanistically how multiple RNF11·Hect E3 ligase complexes regulate TGFβ signaling, we asked whether RNF11 impacts the ubiquitylation of Hect E3 ligases in vivo. We co-expressed untagged wild-type RNF11 with a subset of these enzymes and His-tagged ubiquitin. We focused on SMURF1 and SMURF2 because they are highly related (83% identity) and share multiple, common mechanisms of TGFβ inhibition. Importantly, one major regulatory difference between these two enzymes is that SMURF2 is uniquely subject to autoinhibition (20, 21). Thus, we speculated that RNF11 might differentially impact the ubiquitylation of these proteins in vivo.
Accordingly, we transiently co-expressed FLAG-tagged SMURF1 or SMURF2 with untagged wild-type RNF11 and His-tagged ubiquitin in HEK293T cells. Cell lysates were prepared under denaturing conditions and subjected to nickel chelation chromatography, and the resulting pool of His-tagged ubiquitin-conjugated proteins was analyzed through Western blotting detection of RNF11 and SMURF1/2 (Fig. 3A). We evaluated protein ubiquitylation within the entire cellular pool under denaturing conditions, an important distinction from all previous analyses, which were based on immunopurification strategies and are therefore complicated by DUB activity and co-purification of proteins that associate under native conditions (9, 11, 22, 23). Moreover, the status of protein ubiquitylation in our experiments was evaluated without manipulation of cellular levels of specific E2s, as had been done previously (22). Remarkably, we observed a dramatic reduction in cellular pools of ubiquitylated SMURF2 when this protein was co-expressed with RNF11 (Fig. 3A). This is likely to result from inhibition of autoubiquitylation, because the SMURF2 catalytic mutant exhibits no polyubiquitylation (see Fig. 6C). In striking contrast, ubiquitylation of SMURF1 was not affected by RNF11. Next, using the same assay, we compared SMURF2 ubiquitylation in the presence or absence of RNF11 by silencing expression of the latter with an shRNA. As shown in Fig. 3B, the level of SMURF2 ubiquitylation observed in a 293T cell line stably silenced for RNF11 is equivalent to that observed in control cells. In contrast, overexpression of RNF11 in this RNF11 knock-out cell line has the same effect on SMURF2 ubiquitylation as in control cells. This experiment suggests that SMURF2 ubiquitylation in vivo is subject to regulation by RNF11 when the expression of the latter vastly exceeds endogenous levels. We note that high levels of RNF11 have been reported in reported in late-stage breast and pancreatic cancers (7, 22).
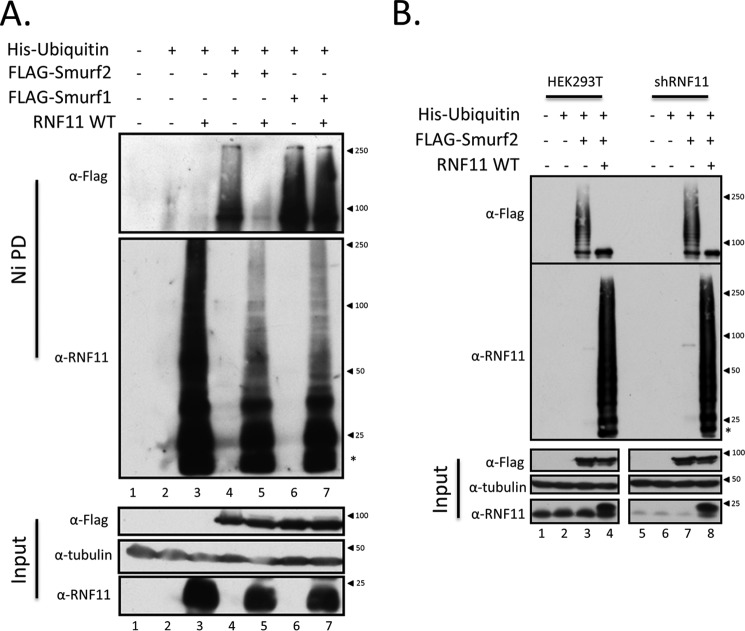
Figure 3.
RNF11 suppresses SMURF2 but not SMURF1 polyubiquitylation in the cell. A, RNF11 differentially affects polyubiquitylation of SMURF Hect E3 ligase family members. Untagged wild-type RNF11 was co-transfected with His-tagged ubiquitin alone or with N-terminally FLAG-tagged SMURF1 or SMURF2 for 48 h in HEK293T cells. Cell lysates were then prepared under denaturing conditions and subjected to nickel-agarose purification. Ubiquitylation of SMURFs and RNF11 was detected by probing of Western blots with anti-FLAG or RNF11 antibody. Untransfected cells (lane 1) and cells transfected with His-ubiquitin alone (lane 2) were included as negative controls. Predicted molecular masses were as follows: α-tubulin, 50 kDa; FLAG-SMURF2, 87 kDa; FLAG-SMURF1, 86 kDa; RNF11, 17.4 kDa. *, mobility of unmodified RNF11. B, SMURF2 polyubiquitylation in the cell is not impacted by RNF11 silencing. Untagged wild-type RNF11 was co-transfected with His-tagged ubiquitin alone or with N-terminally FLAG-tagged SMURF2 for 48 h in HEK293T or HEK293T stably expressing a RNF11 short hairpin. Cell lysates were then prepared under denaturing conditions and subjected to nickel-agarose purification. Ubiquitylation of SMURF2 and RNF11 was detected by probing of Western blots with anti-FLAG or RNF11 antibody. Untransfected cells (lane 1) and cells transfected with His-ubiquitin alone (lane 2) were included as negative controls.
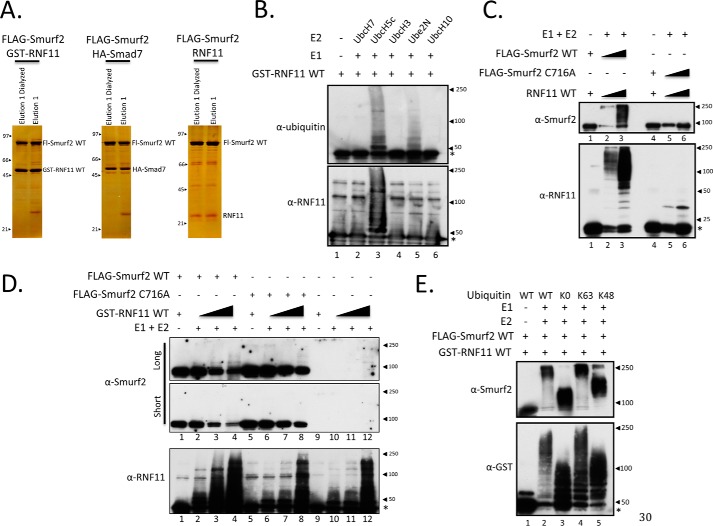
Figure 6.
Formation of the SMURF2·RNF11 complex activates SMURF2 E3 ligase activity toward RNF11. A, representative silver staining of FLAG-purified protein complexes assembled in vitro. Lysates expressing exogenous proteins were combined, incubated, purified with FLAG-agarose, and eluted with peptide, and an aliquot was assessed by silver staining. Shown are protein complexes purified from 5 mg of crude lysate. Purified complexes were subsequently assayed in C and E and Fig. 7. B, evaluation of RNF11 activity in vitro. GST-tagged RNF11 was purified from transformed BL21 bacteria with glutathione-agarose, eluted from the resin, dialyzed, and quantified by Coomassie stain. GST-RNF11 autoubiquitylation and E2 preference were assessed in solution in an in vitro ubiquitylation assay. The reaction was resolved by SDS-PAGE and immunoblotted. Lane 1 serves as a negative control and contains only the recombinant E3 and no E1 or E2. C, SMURF2 and RNF11 are both active when assembled as a complex in vitro. Protein complexes were reconstituted from Hi5 cells expressing FLAG-SMURF2 WT, FLAG-SMURF2 C716A, and RNF11 (untagged) as described under “Experimental procedures.” The purified protein complexes were then assessed for E3 ligase activity in solution by an in vitro ubiquitylation assay. The reaction was resolved by SDS-PAGE and immunoblotted. For each protein complex, there is a lane without E1 or E2 to detect unmodified protein. UBCH5C was used as the E2 in this experiment. D, SMURF2 E3 ligase activity is activated by association with RNF11. Two types of SMURF2·RNF11 complexes were assembled, each consisting of recombinant wild-type RNF11 produced in bacteria and wild-type or catalytically inactive (C716A) FLAG-tagged SMURF2 produced in insect cells. Each SMURF2·RNF11 complex was assembled under limiting conditions for SMURF2 while varying the amount of RNF11. The ubiquitylation reactions were resolved by SDS-PAGE, blotted, and probed for RNF11 or SMURF2. Two exposures are shown for SMURF2. E, evaluation of linkage specificity of SMURF2·RNF11 complex in vitro. The protein complex was reconstituted from Hi5 cells expressing FLAG-SMURF2 WT and bacterial lysate expressing GST-RNF11 as described under “Experimental procedures.” The activity of the assembled FLAG-SMURF2·GST-RNF11 complex was assessed in solution by an in vitro ubiquitylation assay using wild-type or indicated ubiquitin mutants. The reaction was resolved by SDS-PAGE and immunoblotted. K0, ubiquitin lacking lysine residues; K63, a form of ubiquitin that contains only lysine 63; K48, ubiquitin with only lysine 48. Lane 1 serves as a negative control and contains only the recombinant E3 and no E1 or E2. UBCH5C was used as the E2 in this experiment.
RNF11 localization, but not its catalytic activity, is required for suppression of SMURF2 ubiquitylation
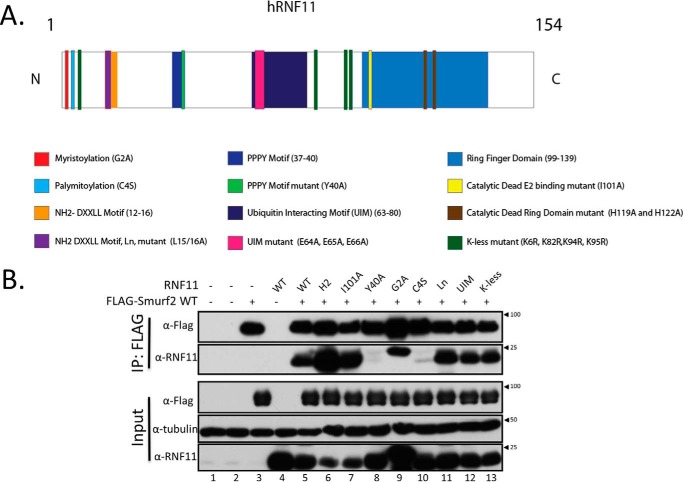
To understand how RNF11 regulates SMURF2 activity, we asked whether myristoylation, palmitoylation, and ubiquitylation of RNF11, which regulate its sorting, also contribute to the stabilization of this complex. Second, we asked whether the regulation of SMURF2 activity was restricted to the endosomal compartment and whether this was impacted by its association with RNF11. Assembly of the SMURF2·RNF11 complex is stabilized by robust protein-protein interactions involving the PPPY motif in RNF11 and two adjacent WW domains in SMURF2 (22). Therefore, because N-terminally tagged RNF11 delocalizes this protein (8–11), we expressed untagged versions of wild type and selected RNF11 mutants, together with SMURF2 (Fig. 4A). Cell lysates were prepared in RIPA buffer, which, importantly, is sufficient to solubilize membrane-associated proteins. Here, we found that in addition to the wild-type protein, SMURF2 can robustly complex with most of the members of our panel of RNF11 mutants, including myristoylation-deficient, lysine-less (K-less), and putative UIM mutants, as well as two different catalytically inactive versions of RNF11 (Fig. 4B). Therefore, stability and assembly of this complex do not require association with membranes and are not dependent on prior RNF11 myristoylation, ubiquitylation, or catalytic activity or a ubiquitin-dependent interaction in trans. In contrast, the C4S mutant, which disrupts the palmitoylation of RNF11, proved to be the exception, suggesting that acylation at this residue might also contribute to the stability of the complex. Furthermore, we confirmed that the PPXY motif of RNF11 is required for stable complex assembly with SMURF2, because the Y40A mutant did not interact (22).
Figure 4.
RNF11 palmitoylation and its PPXY motif of RNF11 are sufficient to stabilize the SMURF2·RNF11 complex. A, schematic of RNF11 protein-protein interaction domains and mutants used in this study. B, SMURF2·RNF11 complex is stabilized by the PPXY motif in RNF11. N-terminally FLAG-tagged wild-type SMURF2 was co-transfected alone or with untagged RNF11 wild-type and a panel of mutants for 48 h in HEK293T cells. Binding of FLAG-tagged SMURF2 to RNF11 wild type and mutants in lysates prepared with RIPA buffer was assessed by immunoblot detection of FLAG immunoprecipitates (IP). Untransfected cells (lane 1) and cells transfected with empty mammalian expression vector (lane 2) were included as negative controls.
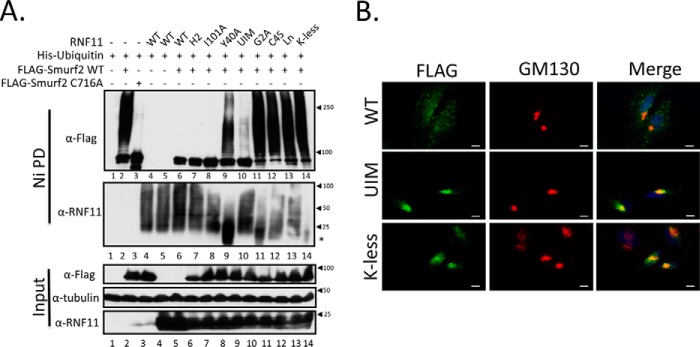
Next, we sought to determine whether the observed regulation of SMURF2 activity by RNF11 required stable interactions and whether it was restricted to a specific cellular compartment(s). As before, we evaluated ubiquitylation of both proteins after expression of untagged mutant and wild-type RNF11 and SMURF2 under denaturing conditions. Immunoblotting analysis showed that expression of wild-type RNF11, but not the PPXY motif mutant (Y40A), dramatically reduced total SMURF2 ubiquitylation (Fig. 5A). These data suggest for the first time that the association between RNF11 and SMURF2 in the cell results in the suppression of SMURF2 ubiquitylation in vivo and that this regulation requires stable complex assembly. We also tested our panel of RNF11 mutants that exhibit defects in acylation-dependent localization, catalytic activity, and sorting from the trans-Golgi network as well as the K-less version, which cannot be ubiquitylated, and the UIM mutant. Because RNF11 primarily localizes to sorting and recycling endosomes, we hypothesized that mutants that exhibited defective trafficking to these compartments would have no effect on SMURF2 ubiquitylation. Indeed, we found that RNF11 mutations that destabilized its association with membranes (G2A) or that prevented entry into the sorting endosomal compartment (C4S and Ln), either from the Golgi (C4S) (9) or the plasma membrane (Ln) (11), strongly compromised the ability of RNF11 to extinguish SMURF2 ubiquitylation (Fig. 5A). Further, we found that mutations that ablate the catalytic function of RNF11, by disrupting the integrity of either the RING domain (H119A/H122A) or the E2-binding site (I101A), also abolished SMURF2 ubiquitylation. Importantly, these mutants, unlike those just discussed, are able to traffic to the same compartments (through the same pathways) as the wild-type protein (8, 9). This suggests that RNF11 catalytic activity is not essential for suppression of SMURF2 ubiquitylation on intracellular membranes. Moreover, we observed that a K-less version of RNF11 and a putative UIM mutant, both of which significantly accumulate in the Golgi as compared with the wild-type protein, also failed to suppress SMURF2 ubiquitylation (Fig. 5, A and B). Finally, although the Y40A mutant of RNF11 localized correctly (11), it too failed to suppress SMURF2 ubiquitylation. Collectively, these data provide compelling evidence, for the first time, that RNF11 regulates SMURF2 ubiquitylation through a mechanism that requires 1) complex formation via interaction of the PPXY and WW domains within RNF11 and SMURF2, respectively, and 2) co-localization to endosomal compartments, where RNF11 normally resides.
Figure 5.
RNF11 suppresses SMURF2 polyubiquitylation in a compartment-specific manner. A, RNF11 antagonizes SMURF2 ubiquitylation in vivo. N-terminally FLAG-tagged wild-type and C716A mutant SMURF2 were co-transfected as indicated with His-tagged ubiquitin and with untagged wild-type RNF11 and a panel of mutants for 48 h in HEK93T cells. Cell lysates were prepared under denaturing conditions and subjected to nickel-affinity agarose purification. Ubiquitylation of SMURF2 and RNF11 was detected by immunoblotting with anti-FLAG or RNF11 antibody as indicated. Transfection with His-ubiquitin alone (lane 1) was included as a negative control. B, subcellular localization of K-less and UIM mutants of RNF11. HeLa cells were transfected with the indicated C-terminal FLAG fusion mammalian expression vectors, fixed after 48 h, and visualized with FLAG antibody and Golgi marker GM130. Scale bar, 10 μm.
Biochemical characterization of RNF11 and associated complexes
The SMURF2·RNF11 complex has not been extensively characterized at the biochemical level to our knowledge, and therefore, to provide further insight into the mechanism of activation of the SMURF2·RNF11 complex, we initiated a series of experiments to study this complex in vitro. First, we assessed whether recombinant RNF11 produced in bacteria was an active enzyme with a preference for specific E2s (Fig. 6B). We observed robust activity with one E2, UBE2D3/UBCH5C, and moderate autoubiquitylation with UBE2N (Fig. 6B). It is possible that this E2 preference may also be observed in the cell, because both UBCH5C and UBE2N were identified in our RNF11 proteomic screen (Table 1). Next, we evaluated the E3 ligase activity of both proteins present in the complex. We assembled SMURF2·RNF11 complexes consisting of recombinant wild-type RNF11 produced in bacteria and wild-type or catalytically inactive (C716A) SMURF2 produced in insect cells. Each SMURF2·RNF11 complex was assembled under limiting conditions for SMURF2 while simultaneously varying the amount of RNF11. Interestingly, we observed enhanced ubiquitylation of RNF11 by the wild-type SMURF2·RNF11 complex as compared with equimolar amounts of the SMURF2 (C716A)·RNF11 complex (Fig. 6D). Further, we observed that increasing amounts of RNF11 in the assembled complex resulted in the conversion of SMURF2 to a more slowly migrating form, which may reflect an increase in autoubiquitylation, a hallmark of enzymatic activation (4, 21). These data suggest that in vitro, RNF11 may serve both as both an activator and a substrate of SMURF2 in this complex. We also evaluated RNF11 ubiquitylation in vitro after co-expression and immunopurification of a complex of untagged RNF11 and FLAG-tagged wild-type or catalytically inactive (C716A) SMURF2 from insect cells (Fig. 6A). As shown in Fig. 6C, the wild-type SMURF2·RNF11 complex, but not the SMURF2 (C716A) complex, also exhibited robust RNF11 ubiquitylation. To our knowledge, there have been no prior reports of the assembly and functional testing of the SMURF2·RNF11 complex in vitro. Indeed, this result was unexpected because, until now, SMURF2 activity has only been demonstrated in complexes with SMAD7. Interestingly, this experiment further revealed RNF11 autoubiquitylation within the SMURF2 C716A·RNF11 complex, suggesting that both subunits of the complex could potentially recruit E2s in the context of the wild-type complex (compare lanes 2 and 3 versus lanes 5 and 6 of the RNF11 blot). Importantly, however, these data suggest that in the wild-type complex, most of the activity emanates from SMURF2 rather than RNF11 (Fig. 6C).
Finally, to further biochemically characterize this complex, we analyzed its ability to promote several different types of ubiquitin linkage by assembling a complex using FLAG-tagged SMURF2 produced in insect cells and bacterially produced GST-RNF11, which behaves similarly to untagged RNF11 in vitro. As shown in Fig. 6E, we found that the SMURF2·GST-RNF11 complex has robust enzymatic activity and can efficiently catalyze Lys-48- and Lys-63-type polyubiquitin linkages in vitro. Both types of ubiquitin linkages have been implicated in the activity of the SMURF2·SMAD7 complex toward SMURF2 and the TGFβ receptor (3). Together, our in vitro data establish that the SMURF2·RNF11 complex can indeed function as an E3 ligase and strongly suggest that it could also be active in the cell in some settings.
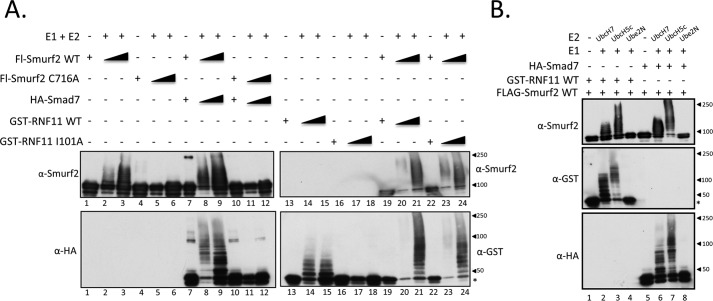
SMURF2 complexes with RNF11 and SMAD7 assemble biochemically related E3 ligases in vitro
To further biochemically characterize the RNF11/SMURF2 complex in vitro, we performed a side-by-side functional comparison with the well-characterized SMURF2·SMAD7 complex. SMURF2·RNF11 and SMURF2·SMAD7 complexes were assembled in vitro by incubating insect cell lysates expressing wild-type FLAG-SMURF2 or the C716A mutant of FLAG-SMURF2 together with those prepared from insect cells expressing recombinant SMAD7-HA or bacterially produced wild-type or I101A mutant GST-RNF11. After complex assembly, recombinant SMURF2 complexes were isolated by immunopurification and tested for E3 ligase activity in vitro. Strikingly, these experiments revealed notable biochemical similarities between SMURF2·SMAD7 and SMURF2·RNF11 complexes. First, as shown in Fig. 7A, robust ubiquitylation of SMURF2 and SMAD7 or RNF11 was observed, indicating that both complexes are functional E3 ligases (compare lanes 8 and 9 with lanes 20 and 21). Importantly, ubiquitylation of wild-type RNF11 and the I101A mutant of RNF11 was observed when complexed to SMURF2 (compare lanes 20 and 21 with lanes 23 and 24). Together, our biochemical characterization of the SMURF2·RNF11 complex presented here definitively establishes, for the first time, that RNF11 is a SMURF2 substrate and further suggests that it may also function to stimulate SMURF2 activity by a mechanism analogous to that used by SMAD7. We note that in contrast to a previous observation, we found robust ubiquitylation of SMAD7 when complexed to SMURF2 in vitro (4). Next, we compared E2 usage for both SMURF2 complexes (Fig. 7B). Remarkably, we observed no significant difference in SMURF2 activity toward itself or its partners (RNF11 and SMAD7) in reactions programmed with UBCH7 or UBCH5C. Given the observed biochemical similarities between the SMURF2·SMAD7 and SMURF2·RNF11 complexes, our data suggested that RNF11 should be able to functionally antagonize SMAD7 function if binding to SMURF2 was mutually exclusive.
Figure 7.
SMURF2·RNF11 and SMURF2·SMAD7 complexes are enzymatically similar. A, SMURF2·SMAD7 and SMURF2·RNF11 complexes are functional E3 ligases. SMURF2 protein complexes were reconstituted from Hi5 cells expressing FLAG-SMURF2 WT, FLAG-SMURF2 C716A, or HA-SMAD7 and bacterial lysates expressing GST-RNF11 WT or GST-RNF11 I101A as described under “Experimental procedures.” The activities of the assembled complexes were assessed in solution by an in vitro ubiquitylation assay. For each protein or protein complex, there is a lane without E1 or E2 to detect unmodified protein. UBCH5C was used as the E2 in this experiment. B, SMURF2·RNF11 and SMURF2·SMAD7 complexes show similar E2 preference in vitro. SMURF2 protein complexes were reconstituted from Hi5 cells expressing FLAG-SMURF2 WT and HA-SMAD7 as well as bacterial lysates expressing GST-RNF11 WT as described under “Experimental procedures.” The activities of the assembled complexes were assessed in solution by an in vitro ubiquitylation assay. Various E2s were used to determine preference. Lanes 1 and 5 serve as negative controls and contain only the recombinant E3 but no E1 or E2. Predicted molecular masses are as follows: RNF11 (untagged), 17.4 kDa; FLAG-SMURF2, 87 kDa; HA-SMAD7, 49.7 kDa; GST-RNF11, 44.4 kDa. *, mobility of unmodified RNF11.
SMURF2 forms mutually exclusive complexes with RNF11 or SMAD7
To directly test this notion, a fixed amount of preassembled recombinant SMAD7-SMURF2 complexes was incubated in vitro with increasing amounts of recombinant GST-RNF11 or RNF11 Y40A, and complexes were purified through the GST moiety and detected by immunoblotting. As shown in Fig. 8A, wild-type RNF11, but not the Y40A mutant, was able to compete directly with SMAD7 for SMURF2 binding. To better understand the molecular basis of this competition mechanistically, we asked whether SMAD7 or RNF11 binding to SMURF2 is mutually exclusive. Recombinant SMURF2, SMAD7, and RNF11 were incubated together, and the resulting complexes were subjected to two successive rounds of immunoprecipitation through SMAD7 and RNF11. As shown in Fig. 8B, this experiment conclusively demonstrated that the binding of SMURF2 to RNF11 or SMAD7 is mutually exclusive, and it is dependent on the integrity of their respective PY domains.
Figure 8.
SMURF2 binding to RNF11 or SMAD7 is mutually exclusive. A, RNF11 competes with SMAD7 for SMURF2 binding. Cell lysates from insect cells expressing FLAG-SMURF2 or HA-SMAD7 were first combined (1 mg of total protein each) and incubated for 1 h to preassemble complex. Three different amounts of bacterial lysates expressing GST, GST-RNF11 WT, or GST-RNF11 Y40A were then added (0.1, 1, and 5 mg of protein) to the reaction and incubated and supplemented with glutathione-agarose for another 2 h and resolved by SDS-PAGE. Titration of GST-RNF11 WT results in loss of SMAD7 interaction with SMURF2. B, RNF11 and SMAD7 form mutually exclusive complexes with SMURF2. Cell lysates from insect cells expressing FLAG-SMURF2 or HA-SMAD7 (1 mg of total protein each) were mixed with bacterial lysates expressing GST, GST-RNF11 WT, or GST-RNF11 Y40A (1 mg of total protein each) and incubated for 2 h, and the resulting complexes were immunoprecipitated with anti-FLAG-agarose. Elution from the FLAG immunoprecipitates was performed with FLAG peptide, and 5% of the eluate was resolved by Western blotting (top). The remainder of the eluate was then immunoprecipitated a second time with glutathione-agarose (middle) and anti-HA agarose (bottom).
RNF11 regulation of SMURF2 in the cell requires compartmentalization to membranes
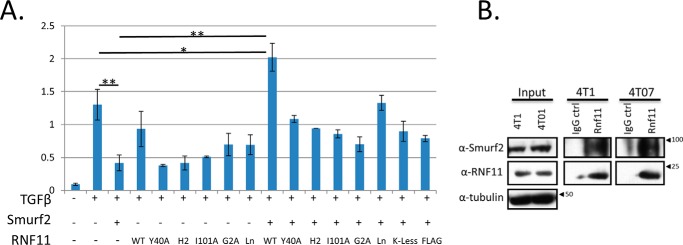
The foregoing studies suggested that direct competition with SMAD7 is one mechanism by which RNF11 could antagonize the function and formation of the SMAD7/SMURF2 complex. This would require, however, that RNF11 and SMAD7 co-localize to the same compartments in the cell. A second, distinct but complementary mechanism could be envisioned if RNF11 protein levels exceeded those of SMAD7 in unstimulated cells and RNF11 did not reside in the same compartment as SMAD7. In this setting, RNF11 could antagonize the function of SMURF2·SMAD7 complex through a sequestration mechanism. All previous subcellular localization studies of SMAD7 suggest that it does not reside in endosomal compartments wherein RNF11 localizes (2, 3). Therefore, we proceeded to test whether RNF11 can antagonize SMURF2-mediated repression of TGFβ responsiveness via a sequestration mechanism. Co-expression of N-terminally tagged RNF11 and SMURF2 has been previously reported to antagonize SMURF2 function in hepatoma cells transiently expressing the TGFβ reporter, p3TP Lux (22). However, the aberrant subcellular localization of the SMURF2·RNF11 complex assembled in this study (which resulted from delocalized RNF11) suggested a caveat regarding its physiological relevance (8–11). Therefore, we proceeded to test whether compartmentalization of the SMURF2·RNF11 complex was integral to its ability to suppress SMURF2-mediated repression of TGFβ signaling. Accordingly, we co-expressed SMURF2 together with untagged, wild-type RNF11 or the cohort of mutants characterized above, in Mv1lu cells stably carrying a TGFβ-responsive luciferase reporter (24). As previously reported, we found that exogenous expression of SMURF2 alone is sufficient to suppress TGFβ signaling as compared with controls (Fig. 9A). However, these studies also led to several novel conclusions. First, in contrast to a previous report (22), we found that ectopic RNF11 expression alone could not augment cellular responsiveness to TGFβ (Fig. 9A). Second, we observed that expression of wild-type RNF11 antagonized SMURF2-mediated repression and, strikingly, augmented TGFβ responsiveness well above levels seen in the SMURF2 control. Importantly, an equivalent level of expression using the same N-terminally tagged form of RNF11 used in the previous study (22) was significantly less effective in antagonizing SMURF2 function in this assay.
Figure 9.
RNF11 functionally antagonizes SMURF2 by sequestration in an endosomal compartment. A, SMURF2·RNF11 wild-type complex antagonizes SMURF2 repression of TGFβ-dependent transcriptional response. Mink lung epithelial cells stably expressing the TGFβ-responsive PAI-1 reporter fused to firefly luciferase were transfected with wild-type SMURF2, wild-type and mutant RNF11, and Renilla luciferase (to control for transfection efficiency) and serum-starved overnight, followed by treatment with recombinant TGFβ (400 pm for 16 h). The y axis represents relative light units as determined by the ratio of detected firefly luminescence to Renilla luminescence. Values represent means from at least two independent experiments, and error bars represent S.D. *, p < 0.02; **, p < 0.005. B, endogenous SMURF2·RNF11 complex is present in a mouse model for metastatic breast cancer. RNF11 immunoprecipitates using our in-house RNF11 antibody coupled to Protein A-Sepharose were prepared from 2 mg of crude cell lysates from 4T07 and 4T1 cells. IgG coupled to Protein A-Sepharose served as a control for nonspecific interactions in each case. The immunoprecipitates were resolved by SDS-PAGE, blotted, and then probed for endogenous RNF11 and SMURF2 (see “Experimental procedures”). Input blots were probed with anti-tubulin antibody.
A key question pertains to the location of RNF11-mediated antagonism of SMURF2 activity and whether this function of RNF11 activity is restricted to a cellular compartment. To examine this question in detail, we used our reporter assay to test the activity of untagged RNF11 mutants whose localization to membrane compartments was crippled. Under steady-state conditions, RNF11 is distributed principally between the early and recycling endosomal compartments (8–11). As shown in Fig. 9A, we found for the first time that myristoylation of RNF11, which is essential for stable association with the endosomal compartment but not for complex formation, played a critical role, because the G2A mutant displayed significantly less activity than the wild-type protein (∼3-fold reduction). This may also explain why N-terminally tagged RNF11 is not as effective as the wild-type protein in this assay, because it is possible that this epitope tag masks the myristoylation motif (MGNCLK) present in the endogenous protein (8, 9). Likewise, the K-less RNF11 mutant, which can stably bind to SMURF2 and exhibits a marked defect in entry into the sorting endosome from the Golgi, also failed to suppress SMURF2 activity in this assay (Figs. 4B and 9A). Interestingly, we also observed that the Ln mutant of RNF11, which can enter the sorting endosome upon Golgi exit but which is defective in reentry into this compartment from the plasma membrane, was the least compromised mutant in this assay (Fig. 9A). This result suggests that RNF11 could potentially sequester SMURF2 in the endosomal compartment or at the plasma membrane by direct recruitment. The former, however, is likely to be the principal mode of sequestration, because RNF11 is rapidly endocytosed upon reaching the plasma membrane (10).
We also asked whether the catalytic activity of RNF11 contributes to the mechanism through which it suppresses SMURF2 function. Interestingly, although both RING domain (H2) and E2 binding mutants of RNF11 (I101A) correctly localize to endosomes (8), both were defective in their ability to suppress SMURF2 function. Importantly, both of these mutants can stably bind SMURF2 through the robust interaction between their respective PPXY and WW domains (Fig. 4B). Thus, these data suggest that the mechanism through which SMURF2 function is antagonized by RNF11 may also require the integrity and/or function of its RING domain. The latter is likely, because although the I101A mutant is not a functional E3 ligase, its RING domain remains intact (8).
Our results therefore strongly suggest that the ability of RNF11 to stably bind and sequester SMURF2 in the endosomal compartment is critical for suppressing the function of SMURF2. That RNF11 suppresses SMURF2 function by sequestering it on membranes establishes a new mechanism and significantly extends and clarifies previous models (22).
Depletion of Rnf11 decreases TGFβ responsiveness in a tissue culture model for metastatic breast cancer
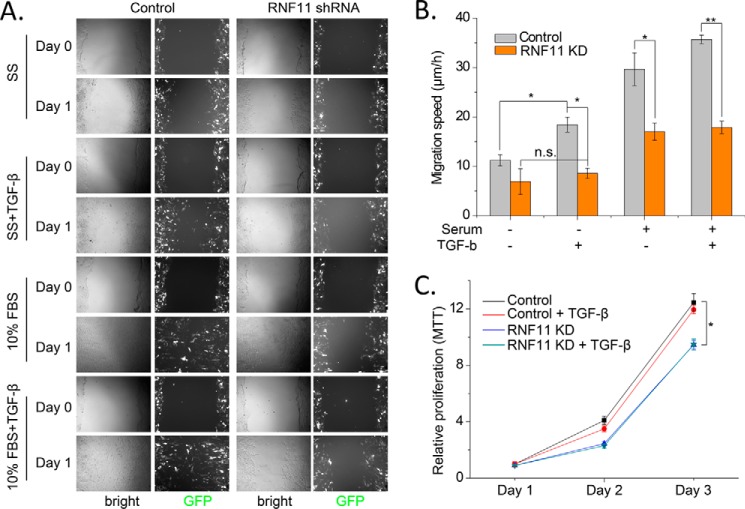
RNF11 has been reported to be overexpressed in multiple types of cancer, including breast tumors (7, 22). However, to date there has been no detailed investigations of endogenous SMURF2·RNF11 complex in vivo or of the impact of RNF11 overexpression in human cancers. Because RNF11 was shown to be highly expressed in metastatic human breast cancers, we investigated whether this complex could be detected in an established TGFβ-responsive mouse tissue culture model for metastatic breast cancer (25). The 4T1 and 4T07 mouse cell lines are single-cell subclones from the 410.4 tumor and were isolated from a spontaneously arising mammary tumor in a BALB/cfC3H mouse (25). Both 4T07 and 4T1 are highly tumorigenic but differ in their degree of metastatic behavior. For example, although 4T07 can be detected in the blood and lungs, it cannot metastasize to bone (25). 4T1, in contrast, can spontaneously metastasize to multiple sites, including lymph nodes, blood, lung, and bone. Importantly, 4T1 migration in vitro is enhanced by TGFβ in vitro and is required for the establishment of metastasis in vivo (26). We therefore hypothesized that if the RNF11·SMURF2 complex exists in 4T1 cells, a direct assessment of its role in cellular migration should be possible. As shown in Fig. 9B, we were able to detect a robust complex between the endogenous SMURF2 and RNF11 proteins in unstimulated, growing 4T07 and 4T1 cells, thus confirming that the two proteins interact under native conditions. Accordingly, we proceeded to test in a wound-healing assay whether Rnf11 knockdown impacted cell migration. Remarkably, we observed that whereas TGFβ increased migration in the control cells, as expected, the effect was abolished in Rnf11 shRNA-infected cells (Fig. 10, A and B). These data suggest for the first time that Rnf11 is an essential positive regulator of TGFβ-dependent cell migration. Next, we tested the impact of Rnf11 knockdown on the proliferation of 4T1 cells. Here we found that TGFβ had no effect on 4T1 proliferation, confirming a previous observation (25). On the other hand, we observed that Rnf11 knockdown moderately, but significantly, reduced cell proliferation in the absence of TGFβ (Fig. 10C). These data suggest that Rnf11 may impact both cell migration and proliferation, although the latter may be TGFβ-independent.
Figure 10.
Rnf11 regulates cell proliferation and TGFβ-dependent cell migration. A, Rnf11 knockdown reduces TGFβ-dependent cell migration in mouse 4T1 breast cancer cells. Adenoviruses expressing GFP together with control and RNF11 shRNAs were infected into 4T1 cells. Cell migration was evaluated by wound-healing assays under different conditions as indicated: serum starvation (SS) only, SS with TGFβ, 10% FBS only, and 10% FBS with TGF-β. Both bright field and green channels of the representative images are shown. B, quantification of cell migration speed as in A. C, RNF11 knockdown moderately reduces proliferation of mouse 4T1 cells. Cell proliferation was evaluated by MTT assays during 3 days. Scale bar, 400 μm. Data are shown as mean ± S.D. (error bars). *, p < 0.05; **, p < 0.01.
Rnf11 depletion differentially affects TGFβ-dependent gene expression
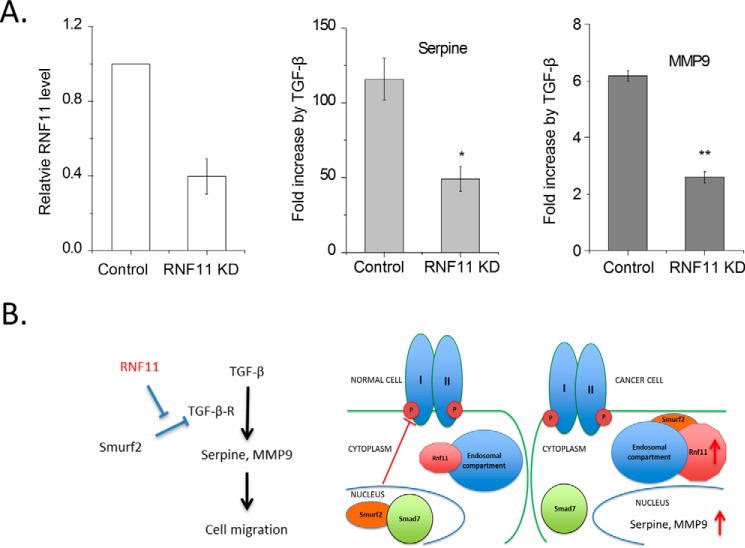
In this study, we have provided evidence suggesting that Rnf11 may function to enhance or sustain cellular TGFβ-dependent signaling through the sequestration of a key negative regulator, SMURF2. Our observation that Rnf11 promotes TGFβ-dependent migration in the 4T1 breast cancer cell line prompted an evaluation of the role of Rnf11 in modulating the expression of several genes implicated in this process. As shown in Fig. 11, we observed a reduction in TGFβ-induced expression of Serpine1 and Mmp9 upon Rnf11 knockdown. Importantly, both of these genes are widely reported to mediate cellular migration, tumor growth, and metastasis (27–29). These findings suggest that Rnf11 may play a role in the expression of genes that promote tumor growth and metastasis.
Figure 11.
A role for RNF11 in the regulation of TGFβ-dependent transcription in cancer cells. A, RNF11 knockdown mitigates TGFβ-dependent transcription. 4T1 cells were infected overnight with adenoviruses expressing GFP and a shRNA sequence designed to knock down RNF11 protein expression. Control cells were infected with a control virus expressing GFP and a scrambled shRNA sequence. The cells were subsequently serum-starved (1% FBS) overnight after 48 h of viral infection. At t = 70 h, half of the experimental and control wells were stimulated with 1 nm TGFβ for 2 h and subsequently harvested for mRNA expression analysis. Student's t test was performed: *, p < 0.05; **, p < 0.01. Error bars, S.E. B, model depicting RNF11 sequestration of SMURF2 in cancer cells. In normal cells, RNF11 is restricted to the endosomal compartment and does not complex to SMURF2, the majority of which resides in the nucleus. TGFβ stimulation, which induces SMAD7 protein expression, therefore favors the formation of the SMURF2·SMAD7 complex, which is then recruited to the activated TGFβR in the plasma membrane. In cancer cells, RNF11 expression is aberrantly high (depicted by a larger size in the schematic), which favors formation of the SMURF2·RNF11 complex in the endosomal compartment. TGFβ stimulation in this scenario does not favor formation of the SMURF2·SMAD7 complex and therefore could result in sustaining TGFβ signaling.
Discussion
A novel function for RNF11 in SMURF2 regulation
In a proteomic screen for endomembrane interactants of the RING-domain E3 ligase RNF11, we identified SMURF2, among a cohort of Hect E3 ligases previously implicated in TGFβ signaling. It is important to distinguish our experimental results from a prior study, which first reported an interaction between RNF11 and SMURF2 (22). In that report, the authors inadvertently expressed an N-terminally tagged form of RNF11, which cannot associate with cellular membranes and which is, therefore, delocalized with respect to the wild-type protein (8–11). Here, we have characterized the SMURF2·RNF11 complex in its native setting, the endosomal compartment, and this approach has enabled us to uncover a novel role for RNF11 in the regulation of SMURF2 activity and function, which has not been previously reported.
We find for the first time that SMURF2 ubiquitylation is essentially abolished by overexpression of RNF11. Further, we show that RNF11 regulation of SMURF2 ubiquitylation is achieved through the sequestration of SMURF2·RNF11 complex in the endosomal compartment. Interestingly, biochemical characterization of the SMURF2·RNF11 complex unexpectedly revealed robust SMURF2 E3 ligase activity principally directed toward RNF11. To date, the only known regulator of SMURF2 was the SMAD7 protein, which has no intrinsic enzymatic activity. Stimulation of SMURF2 ligase activity through association with SMAD7 in response to TGFβ stimulation is thought to relieve autoinhibition by competing with the C2 domain of SMURF2 for binding to its Hect domain (4, 21). The SMURF2·SMAD7 complex is further stabilized by interactions between WW and PPXY motifs in these proteins.
To our knowledge, there have been no other reports of an adaptor that can function to relieve the autoinhibition of SMURF2 other than SMAD7. In this report, we provide multiple lines of evidence that the RING finger protein, RNF11, can function through a mechanism analogous to SMAD7. First, we show that recombinant RNF11 and SMAD7 can each assemble functional E3 ligase complexes with SMURF2 that are indistinguishable with respect to E2 usage and formation of ubiquitin chain linkages. Second, we show that in vitro, RNF11 can compete with SMAD7 for SMURF2 and, therefore, that complex formation with either protein is mutually exclusive. Further, we show that the function of the SMURF2·RNF11 complex in the cell is strongly dependent on the sorting determinants that govern the subcellular localization of RNF11. Thus, we propose that RNF11, like SMAD7, can relocalize SMURF2 outside of the nucleus using a sequestration mechanism that requires complex assembly and localization motifs resident in the adaptor (Fig. 11B). Importantly, RNF11 sequestration of SMURF2, unlike that of SMAD7, is not TGFβ-dependent. Interestingly, although we found that SMURF2 ubiquitylation in the cell is effectively silenced by ectopic expression of RNF11, we are able to reconstitute an active E3 ligase in vitro composed of recombinant SMURF2 and RNF11, suggesting that the complex is inhibited by additional interactions that occur in vivo. Therefore, we posit (Fig. 11B) that the ability of RNF11 to interact with SMURF2 and localize the latter protein to the endosomal compartment confers a novel level of regulation on the enzymatic activity of SMURF2, thereby altering cellular TGFβ responsiveness.
A role for RNF11 in TGFβ pathway and tumorigenesis
We have also shown that RNF11 overexpression can antagonize SMURF2 suppression of TGFβ-dependent transcription. Importantly, we have shown that the ability of RNF11 to antagonize SMURF2 repression of TGFβ-mediated transcription is also dependent on RNF11-sorting determinants, its ability to recruit E2s, and its PPXY motif. Thus, we propose that under some conditions, RNF11 can function as an adaptor for SMURF2 to antagonize repression of the TGFβ pathway. Our findings may be particularly relevant to pathological situations. Indeed, aberrantly elevated RNF11 levels have been reported in late-stage breast and pancreatic cancers (7, 22). In this setting, our studies predict that the ability to down-regulate the TGFβ receptor in the stroma and/or the parenchyma would be severely compromised, and these cells would therefore exhibit aberrant and sustained TGFβ pathway activation. The latter has been implicated in the promotion of the epithelial-to-mesenchymal transition in late-stage breast cancers. In support of our model, we have now shown that ablation of RNF11 severely circumscribes the ability of metastatic breast cancer cells to migrate, in part by interfering with expression of key genes involved in cell migration. Further, disruption of RNF11 by insertional mutagenesis has recently been reported to impair the metastatic potential of murine melanoma B16F10 cells in vivo (30). These findings suggest that it may be therapeutically beneficial to inhibit RNF11 in multiple types of tumors, a possibility that we will explore in the future.
Our discovery that the E3 ligase activity of the SMURF2·RNF11 complex is functional and that both subunits exhibit activity raises additional interesting questions. Does this complex function as a bona fide Hect E3 ligase in the cell, beyond its ability to antagonize SMURF2-mediated repression of TGFβ responsiveness? If so, the identification of the substrates of this complex will be essential in the development of strategies to neutralize the effect of RNF11 overexpression in tumors. Finally, although the focus of this study has been on the SMURF2·RNF11 complex, it is important to note that RNF11 interacts with other NEDD4 family members as well. Future efforts will be directed toward understanding the physiological relevance of these complexes.
Experimental procedures
Plasmids
Bacterial expression vectors
pGEX6P1hRnf11 and hRnf11I101A were made by PCR amplification of the RNF11 open reading frame from pcDNA3.1 hRnf11 wild-type and I101A, respectively, with the hRnf11BamH1F and hRnf11XhoI R primers.
Mammalian expression vectors
pcDNA3.1 hRnf11 (wild type), H2, 101A, GSA, and C4S were a kind gift from Sanjita Beterbet (Emory University School of Medicine). pcDNA3.1 hRnf11 Y40A, UIM, and Ln mutant were created from pcDNA3.1 hRnf11 (wild type) with a kit (QuikChange II site-directed mutagenesis, Agilent) using the three primer pairs RNF11 Y40A and RNF11 Y40AS, E64-65S and E64-65AAS, and L15-16S and L15-16AS. pcDNA3.1 hRnf11 K-less was created in a stepwise fashion, by successively mutating individually each of the 4 lysines present in the RNF11 open reading frame. The pcB6His6 ubiquitin plasmid was a kind gift from Richard Baer (Columbia University School of Medicine). FLAG-SMURF1, FLAG-SMURF2, and FLAG-SMURF2 C716A were purchased from ADDGENE (plasmids 11752, 11746, and 11747).
Baculovirus expression vectors
pFastBac1 hRnf11 (untagged) was made by PCR amplification of the RNF11 open reading frame from pcDNA3.1 hRnf11 using the primers hRnf11BamHIF and hRnf11XhoIR and subcloned into pFastBac1. pFASTBACHTA hSMAD7-HA was made by PCR amplification of the hSMAD7-HA open reading frame from pCMV5-SMAD7-HA (Addgene 11733) using the primers EcoRI SMAD7F and SalI SMAD7-HA R. pFASTBAC HTA FLAG-SMURF2 and FLAG-SMURF2C716A were made by PCR amplification of the open reading frame from the mammalian expression vectors FLAG-SMURF2 and C716A using the primers EcoRI SMURF2F and NotI SMURF2R.
Antibodies and affinity resins
All antibodies used in this study with the exception of the RNF11 antibody were purchased from commercial sources. Our in-house RNF11 antibody was typically stored at a concentration of 1 μg/μl in 1× PBS, 10% glycerol. This antibody was used at 1:200 dilution for immunofluorescence and 1:1000 dilution for immunoblots. Rabbit polyclonal antibodies against Rab7 (sc-10767, 1:500) and GST (sc-33613, 1:1000) and the 12CA5 mouse monoclonal antibody (sc57592, 1:1000) were purchased from Santa Cruz Biotechnology, Inc. The goat polyclonal antibody against EEA1 (sc-6415, 1:200) was also purchased from Santa Cruz Biotechnology. Three antibodies were purchased from Sigma: mouse anti-α-tubulin T5168 (1:5000) and two anti M2 FLAG antibodies, F1804 (mouse polyclonal) and F7425 (rabbit polyclonal). The anti-GM130 antibody (catalog number 610822, 1:200) is a mouse monoclonal and was purchased from BD Transduction Laboratories. The anti-SMURF2 antibody (SAB2701310, 1:1000) was a rabbit polyclonal and was purchased from Sigma-Aldrich. Anti-ubiquitin is a rabbit polyclonal antibody purchased from R&D Systems (A-100, 1:500). Glutathione-agarose (G4510) and anti-FLAG M2 agarose (A2220) were purchased from Sigma. Ni-NTA-agarose was purchased from Qiagen Inc. Protein A- Sepharose (17-0780-01) and Protein G-Sepharose (17-0618-01) were purchased from GE Healthcare Life Sciences.
Primer sequences for cloning and mutagenesis (5′ to 3′)
Primers used were as follows: hRNF11_BamHI_F, AGTCGGATCCATGGGGAACTGCCTCAAAT; hRNF11_XhoI_R, AGTCCTCGAGTCAATTAGTCTCATAGGATGAAAGCA; RNF11 Y40A, GGAGCCGCCGCCGCCAGCTCAGGAACAAGTTC; RNF11 Y40AS, GAACTTGTTCCTGAGCTGGCGGCGGCGGCTCC; E64,65,66S, GCTAGCAACTCAGCTGACTGCAGCGGCACAAATTAGGATAGCTCAAA; E64,65,66 AS, TTTGAGCTATCCTAATTTGTGCCGCTGCAGTCAGCTGAGTTGCTAGC; L15,L16S, CCTCGGATGACATCTCCGCGGCTCACGAGTCTCAGTCCG; L15,16 AS, CGGACTGAGACTCGTGAGCCGCGGAGATGTCATCCGAGG; a17g (K6R), GGGAACTGCCTCAGATCCCCCACCTCG; a17g anti, CGAGGTGGGGGATCTGAGGCAGTTCCC; a245g (K82R), GTCTTATACAACATCTGCCTAGAGGAGTTTATGACCCTGG; a245g anti, CCAGGGTCATAAACTCCTCTAGGCAGATGTTGTATAAGAC; a281g (K94R), CCCTGGAAGAGATGGATCAGAAAGAAAGATCCGGGAG; a281g anti, CTCCCGGATCTTTCTTTCTGATCCATCTCTTCCAGGG; a284g (K95R), TGGAAGAGATGGATCAGAAAAAAGGATCCGGGAGTGT; a284g anti, ACACTCCCGGATCCTTTTTTCTGATCCATCTCTTCCA; EcoRI SMAD7 F, ATGCGAATTCATGTTCAGGACCAAACGATCTG; SalI SMAD7-HA R, ATGCGTCGACTTAGAGGCTAGCATAATCAGGAACA; EcoRI SMURF2 F, ATGCGAATTCATGGACTACAAGGACGACGATG; NotI SMURF2 R, ATGCGCGGCCGCTCATTCCACAGCAAATCCAC.
Primer sequences used for quantitative PCR (5′ to 3′)
Primers used were as follows: mGAPDH_F, CATGGCCTTCCGTGTTCCTA; mGAPDH_R, GCCTGCTTCACCACCTTCTT; mMMP9_F, CAGCCGACTTTTGTGGTCTTC; mMMP9_R, CGGTACAAGTATGCCTCTGCCA; mSerpine1_F, TCAACTACACTGAGTTCACCA; mSerpine1_R, GCACATCTTTCTCAAAGGGT; mRnF11_F, ATGACATCTCCCTGCTTCAC; mRnF11_R, AACTTGTTCCTGATATGGCG.
Cell lines and tissue culture
All cell lines were routinely cultured at 37 °C, 5% CO2, in Dulbecco's modified Eagle's medium supplemented with fetal calf serum (10%, v/v), penicillin/streptomycin (2.5%, v/v), and l-glutamine (2.5%, v/v). HEK293T, HeLa, and 3T3 L1 cells were purchased from ATCC. Mink lung epithelial cells (p800Luc) were a kind gift from D. B. Rifkin (New York University School of Medicine). These cells stably carry −799 to +71 of the human plasminogen activator inhibitor I gene fused to the firefly luciferase (24).
Production of RNF11 antibody
The RNF11 antibody was produced in rabbits using GST-RNF11 as the antigen. The antibody was purified by affinity chromatography by first depleting the sera of anti-GST antibodies by passage over a GST-coupled resin. The eluate from this step was then passed over a GST-RNF11-coupled resin and collected in fractions. Fractions containing the highest concentration of antibody as judged by Coomassie staining of a preparative SDS-polyacrylamide gel were pooled, dialyzed against 1× PBS (10% (v/v) glycerol), aliquotted, and then frozen at −80 °C for long-term storage.
Sample preparation for mass spectrometry
Fractionation protocol
Forty confluent 15-cm plates of pCDEF3 hRNF11 HA/3XFLAG-transfected 293T cells were harvested by scraping. The pellet washed twice with cold PBS (centrifuged for 5 min at 1000 rpm at 4 °C). Homogenization buffer (HB) was added to the pellet (containing protease and phosphatase inhibitors) at a ratio of 1 ml of cell pellet to 5 ml of HB. HB contained 250 mm sucrose, 20 mm Tris-HCl, pH 7.4, 1 mm EDTA, pH 8, 1.5 mm MgCl2, 10 mm KCl, 1 mm DTT). The cell pellet was homogenized using a 40-ml Dounce homogenizer on ice (or in a cold room). To check how many intact cells remained in the homogenate, a sample was removed after every 10 strokes and evaluated using a microscope. Dounce homogenization was performed until only 10% of the homogenate contained intact cells. This material was then centrifuged for 10 min at 800 × g at 4 °C. Pellet 1 (P800), which contained nuclei and intact cells, was discarded. Supernatant 1 (S800) resulting from this step was transferred to a fresh tube and centrifuged for 15 min at 20,000 × g at 4 °C. Pellet 2 (P20K) resulting from this step was washed by resuspending in HB and centrifuged again for 15 min at 20,000 × g at 4 °C. Pellet 2 (P20K) was resuspended in 1 ml of 0.1 HEMGN + 0.1% Nonidet P-40 (protease and phosphatase inhibitors and DTT). The resuspended pellet 2 was sonicated four times for 10 s (low setting) and then centrifuged for 15 min at 14,000 rpm at 4 °C. The protein concentration of the resulting supernatant (SP20K, sonication of 20,000 × g pellet) from this step was evaluated by Bradford and then snap-frozen at −80 °C.
FLAG purification of SP20K pellet for mass spectrometry
40 μl of 50% FLAG-agarose/ELB slurry was added to 2–3 mg of SP20K sample and incubated on a rocker for 3 h in the cold room. The resin was washed six times with at least 1 ml of ELB (50 mm Hepes, pH 7, 250 mm NaCl, 5 mm EDTA, pH 8, 0.1% Nonidet P-40, 1 mm DTT, 0.2 m 4-benzenesulfonyl fluoride hydrochloride, 2 g/ml leupeptin, 2 μg/ml aprotinin, 10 mm NaF, 50 mm β-glycerophosphate, glycerol at a final concentration of 10% (v/v)). The resin was then washed one time with TBS/D (Tris-buffered saline supplemented with DTT to 1 mm, no other additives included). 3× FLAG peptide was added to a final concentration of 500 μg/ml in TBS/D, the sample was eluted at room temperature for 30 min and spun at 1000 × g, supernatant was collected, and a second elution was performed. The FLAG elution was then TCA-precipitated and then resolved by SDS-PAGE. The gel was stained with Coomassie Blue, and samples of the excised gel were subjected to proteolytic digestion and mass spectrometric sequencing. Sequence analysis was performed at the Harvard microchemistry facility by microcapillary reverse-phase HPLC.
Preparation of cell extracts and purification of protein complexes
GST-tagged proteins were produced in BL21 bacteria, and all other proteins were expressed in baculovirus-infected Hi5 insect cells. Hi5 and BL21 cells were lysed with 0.1 HEMGN buffer (100 mm KCl, 25 mm HEPES, pH 7.6, 0.1 mm EDTA, 12.5 mm MgCl2, 10% glycerol, 0.1% Nonidet P-40, and protease inhibitors), and protein concentration was determined by a Bradford assay. Protein complexes were assembled by combining and incubating 5 mg of each crude lysate before purification. Affinity purification was performed with FLAG-M2 beads (elution buffer: 20 mm Tris, pH 7.5, 150 mm NaCl, 1 mm DTT, 500 μg/ml 3× FLAG peptide) for the following proteins/protein complexes: FLAG-SMURF2 WT; FLAG-SMURF2 C716A; FLAG-SMURF2 WT·GST-RNF11 WT; FLAG-SMURF2 WT·GST-RNF11 I101A; FLAG-SMURF2 C716A·GST-RNF11 WT; FLAG-SMURF2 WT·RNF11 (untagged); FLAG-SMURF2 C716A·RNF11 (untagged); FLAG-SMURF2 WT·HA-SMAD7; FLAG-SMURF2 C716A·HA-SMAD7. Affinity purification was performed with glutathione-agarose (elution buffer: 100 mm Tris, pH 8.0, 120 mm NaCl, 7 mg/ml glutathione, 1 mm DTT) for GST-RNF11 WT and GST-RNF11 I101A. All proteins and protein complexes were dialyzed in the following buffer: 50 mm Tris, pH 7.5, 5 mm MgCl2, 0.5 mm DTT, 100 mm NaCl, 10% glycerol.
In vitro binding assays and ubiquitylation assays
Cell lysates were combined, incubated for 1 h on ice, and then subjected to immunoprecipitation. For the sequential immunoprecipitation, FLAG-M2 beads were first added to combined lysates, washed four times with 0.1 HEMGN, and then eluted with FLAG peptide at a concentration of 500 μg/ml. The elution was then added to either glutathione-agarose or HA-agarose. Ubiquitylation assays were performed in 10-μl reactions at 30 ºC for 1 h using purified proteins (75 mm Tris, pH 7.5, 7.5 mm MgCl2, 3 mm ATP, 0.275 mm ubiquitin, 1 μm ubiquitin aldehyde, 0.055 μm E1, 0.55 μm E2, 400 nm to 1 μm E3). Recombinant human ubiquitin-activating enzyme (UBE1), recombinant E2s (UBCH7, UBCH5c, UBE2N, UBCH3, and UBCH10), ubiquitin aldehyde, and ubiquitin (WT, K0, K48 only, and K63 only; see the legend to Fig. 6) were obtained from R&D Systems.
Co-transfections and immunoprecipitation
HEK293 cells were transiently transfected with FLAG-tagged wild-type or inactive SMURF2, wild-type or mutant RNF11, or empty vector, as indicated. Cells were harvested and lysed in RIPA buffer (150 mm NaCl, 1% Nonidet P-40, 0.5% deoxycholate, 0.1% SDS) 48 h post-transfection. FLAG-M2 beads (Sigma) were added to lysate (1 mg of total protein/immunoprecipitation) and incubated at 4 ºC for 2 h. Beads were washed four times with RIPA buffer, treated with SDS loading buffer, separated by SDS-PAGE, and detected by Western blotting.
Immunofluorescence
Cells were fixed with 10% formalin solution (Sigma-Aldrich) for 15 min at room temperature, followed by permeabilization with 1× PBS and 0.1% Triton X-100 for 10 min and blocking in PBS, 0.1% Triton X-100, and 3% BSA for 1 h before incubation with primary antibodies (dilution 1:200) for 3 h. Images were acquired with a Zeiss Axiovert 200M microscope (×63 objective lens, numeric aperture 1.4, 1.63 Optovar), equipped with a cooled Retiga 2000R CCD (QImaging). Images represent the maximum projection of 12 deconvoluted planes using Metamorph software (Molecular Devices).
In vivo ubiquitylation assay
HEK293 cells were transiently transfected with pcB6-His ubiquitin, FLAG-tagged SMURF2, and wild-type or mutant RNF11. After 36 h, cells were treated with 10 mm MG132 (Peptides International) for 6 h. Cells were lysed, sonicated in Buffer A (100 mm NaH2PO4, 10 mm Tris-Cl, 6 m guanidine-HCl, 10 mm imidazole, pH 8.0), and incubated with Ni-NTA resin (Qiagen) for 3 h at room temperature. Ni-NTA beads were eluted in SDS loading buffer containing 0.2 m imidazole, separated by SDS-PAGE, and detected by Western blotting.
TGFβ-responsive luciferase reporter assay
Luciferase assays were performed using the Dual-Glo® luciferase assay system from Promega. Recombinant human TGFβ1 was purchased from R&D. Mink lung epithelial cells were stably transfected with an expression construct containing a truncated PAI-1 promoter fused to the firefly luciferase reporter gene. Cells were seeded at 5000 cells/well in a 96-well plate 24 h before transfection with plasmids of interest as well as pRL-TK co-reporter vector for constitutive expression of wild-type Renilla luciferase. Three wells were used per condition. Cells were treated with TGFβ1 24 h post-transfection at a concentration of 400 pm for 16 h. Luminescent signal was measured using a PerkinElmer Life Sciences EnVision® 2103 multilabel reader. The ratio of firefly luminescence to Renilla luminescence was then calculated to normalize for transfection efficiency. The statistical significance of differences was calculated with a two-tailed Student's t test.
Adenovirus infection for proliferation and migration assays and quantitative PCR analysis
The virus infection was performed similarly as described (31). Specifically, adenoviruses expressing either RNF11 shRNA or scrambled control sequence were purchased directly from Vector Biolabs (Ad(RGD)-GFP-U6-m-RNF11-shRNA (shADV-270666) and Ad(RGD)-GFP-U6-scrmb-shRNA). Medium containing adenoviruses and Polybrene was used to infect mouse 4T1 breast cancer cells overnight. To reach maximum infection efficiency, virus was infected twice.
Wound-healing assay and 3-(4,5-dimethylthiazol-2-yl)-2,5-diphenyltetrazolium bromide (MTT) proliferation analysis
The mouse 4T1 cells expressing control and RNF11 shRNAs were grown to 90–100% confluence and serum-starved for 24 h. Scratches were made using yellow tips, and wound healing was induced under different conditions, including serum and TGF-β. Wound widths were measured, and the average migration speed was calculated based on reduction of the wound width over 22–24 h. Both control and RNF11 shRNA-infected mouse 4T1 cells were plated in 96-well plates with a density of 2000 cells/well, and cell proliferation was assessed by an MTT assay. In brief, MTT reagent was incubated with cells for 2 h, and the formed purple precipitate was dissolved by DMSO. MTT absorbance at 570 nm was measured in 3 days, and TGF-β was added on day 1.
mRNA expression analysis
4T1 cells grown in 6-well dishes were rinsed twice with warm PBS and then removed from the plate with trypsin EDTA. The pellet was washed with PBS, repelleted, and then solubilized in 1 ml of TRIzol (Ambion)/well. Total RNA was isolated as per the manufacturer's instructions. The Verso cDNA synthesis kit (Thermo Scientific) was used to synthesize cDNA from 250 ng of total RNA. cDNA was diluted to 400 μl in 10 mm Tris, pH 8, and assayed by quantitative PCR using the SYBR Green method. In each case, gene expression was normalized to GAPDH (control gene). All gene expression experiments were repeated at least twice independently.
Author contributions
I. S. conceived and coordinated the study and wrote the paper. R. J. M., W. F., M. J. J., and I. S. designed, performed, and analyzed the experiments. M. T., B. S. C., and F. E. provided technical assistance. All authors analyzed the results and approved the final version of the manuscript.
Acknowledgments
We thank L. Anderson, S. Betarbet, and A. Levey for sharing wild type and mutant RNF11 expression plasmids, C. Giorgi for advice about subcellular fractionation, and Dr. B. D. Dynlacht and members of the Dynlacht laboratory for engaging scientific discussion and contributing constructive advice throughout the development of this project. We are grateful to our colleague Dr. M. Pagano for constructive advice during this project.
This work was supported by Center to Reduce Cancer Health Disparities (CRCHD) of NCI, National Institutes of Health, Grants R21CA153179, R21CA15317902-S1, and R21CA15317902-S2 (to I. S.) and bridging funds from the Department of Pathology at the NYU School of Medicine. The authors declare that they have no conflicts of interest with the contents of this article. The content is solely the responsibility of the authors and does not necessarily represent the official views of the National Institutes of Health.
- EGFR
- epidermal growth factor receptor
- TGFβR
- TGFβ receptor
- UIM
- ubiquitin interaction motif
- RIPA
- radioimmune precipitation assay
- K-less
- lysine-less
- Ni-NTA
- nickel-nitrilotriacetic acid
- HB
- homogenization buffer
- MTT
- 3-(4,5-dimethylthiazol-2-yl)-2,5-diphenyltetrazolium bromide.
References
- 1. Seto E. S., Bellen H. J., and Lloyd T. E. (2002) When cell biology meets development: endocytic regulation of signaling pathways. Genes Dev. 16, 1314–1336 [DOI] [PubMed] [Google Scholar]
- 2. Di Guglielmo G. M., Le Roy C., Goodfellow A. F., and Wrana J. L. (2003) Distinct endocytic pathways regulate TGF-β receptor signalling and turnover. Nat. Cell Biol. 5, 410–421 [DOI] [PubMed] [Google Scholar]
- 3. Kavsak P., Rasmussen R. K., Causing C. G., Bonni S., Zhu H., Thomsen G. H., and Wrana J. L. (2000) Smad7 binds to Smurf2 to form an E3 ubiquitin ligase that targets the TGF β receptor for degradation. Mol. Cell 6, 1365–1375 [DOI] [PubMed] [Google Scholar]
- 4. Ogunjimi A. A., Briant D. J., Pece-Barbara N., Le Roy C., Di Guglielmo G. M., Kavsak P., Rasmussen R. K., Seet B. T., Sicheri F., and Wrana J. L. (2005) Regulation of Smurf2 ubiquitin ligase activity by anchoring the E2 to the HECT domain. Mol. Cell 19, 297–308 [DOI] [PubMed] [Google Scholar]
- 5. Akhurst R. J., and Hata A. (2012) Targeting the TGFβ signalling pathway in disease. Nat. Rev. Drug Discov. 11, 790–811 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Seki N., Hattori A., Hayashi A., Kozuma S., Sasaki M., Suzuki Y., Sugano S., Muramatsu M. A., and Saito T. (1999) Cloning and expression profile of mouse and human genes, Rnf11/RNF11, encoding a novel RING-H2 finger protein. Biochim. Biophys. Acta 1489, 421–427 [DOI] [PubMed] [Google Scholar]
- 7. Kitching R., Wong M. J., Koehler D., Burger A. M., Landberg G., Gish G., and Seth A. (2003) The RING-H2 protein RNF11 is differentially expressed in breast tumours and interacts with HECT-type E3 ligases. Biochim. Biophys. Acta 1639, 104–112 [DOI] [PubMed] [Google Scholar]
- 8. Anderson L. R. (2007) RNF11: A Candidate Gene from the Park10 Locus, Ph. D. thesis, Emory University, Atlanta, GA [Google Scholar]
- 9. Santonico E., Belleudi F., Panni S., Torrisi M. R., Cesareni G., and Castagnoli L. (2010) Multiple modification and protein interaction signals drive the Ring finger protein 11 (RNF11) E3 ligase to the endosomal compartment. Oncogene 29, 5604–5618 [DOI] [PubMed] [Google Scholar]
- 10. Kostaras E., Sflomos G., Pedersen N. M., Stenmark H., Fotsis T., and Murphy C. (2013) SARA and RNF11 interact with each other and ESCRT-0 core proteins and regulate degradative EGFR trafficking. Oncogene 32, 5220–5232 [DOI] [PubMed] [Google Scholar]
- 11. Santonico E., Mattioni A., Panni S., Belleudi F., Mattei M., Torrisi M. R., Cesareni G., and Castagnoli L. (2015) RNF11 is a GGA protein cargo and acts as a molecular adaptor for GGA3 ubiquitination mediated by Itch. Oncogene 34, 3377–3390 [DOI] [PubMed] [Google Scholar]
- 12. Mellacheruvu D., Wright Z., Couzens A. L., Lambert J. P., St-Denis N. A., Li T., Miteva Y. V., Hauri S., Sardiu M. E., Low T. Y., Halim V. A., Bagshaw R. D., Hubner N. C., Al-Hakim A., Bouchard A., et al. (2013) The CRAPome: a contaminant repository for affinity purification-mass spectrometry data. Nat. Methods 10, 730–736 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Rotin D., and Kumar S. (2009) Physiological functions of the HECT family of ubiquitin ligases. Nat. Rev. Mol. Cell Biol. 10, 398–409 [DOI] [PubMed] [Google Scholar]
- 14. Gao S., Alarcón C., Sapkota G., Rahman S., Chen P. Y., Goerner N., Macias M. J., Erdjument-Bromage H., Tempst P., and Massagué J. (2009) Ubiquitin ligase Nedd4L targets activated Smad2/3 to limit TGF-β signaling. Mol. Cell 36, 457–468 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Kuratomi G., Komuro A., Goto K., Shinozaki M., Miyazawa K., Miyazono K., and Imamura T. (2005) NEDD4-2 (neural precursor cell expressed, developmentally down-regulated 4-2) negatively regulates TGF-β (transforming growth factor-β) signalling by inducing ubiquitin-mediated degradation of Smad2 and TGF-β type I receptor. Biochem. J. 386, 461–470 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Ebisawa T., Fukuchi M., Murakami G., Chiba T., Tanaka K., Imamura T., and Miyazono K. (2001) Smurf1 interacts with transforming growth factor-β type I receptor through Smad7 and induces receptor degradation. J. Biol. Chem. 276, 12477–12480 [DOI] [PubMed] [Google Scholar]
- 17. Komuro A., Imamura T., Saitoh M., Yoshida Y., Yamori T., Miyazono K., and Miyazawa K. (2004) Negative regulation of transforming growth factor-β (TGF-β) signaling by WW domain-containing protein 1 (WWP1). Oncogene 23, 6914–6923 [DOI] [PubMed] [Google Scholar]
- 18. Bai Y., Yang C., Hu K., Elly C., and Liu Y. C. (2004) Itch E3 ligase-mediated regulation of TGF-β signaling by modulating smad2 phosphorylation. Mol. Cell 15, 825–831 [DOI] [PubMed] [Google Scholar]
- 19. Lallemand F., Seo S. R., Ferrand N., Pessah M., L'Hoste S., Rawadi G., Roman-Roman S., Camonis J., and Atfi A. (2005) AIP4 restricts transforming growth factor-β signaling through a ubiquitination-independent mechanism. J. Biol. Chem. 280, 27645–27653 [DOI] [PubMed] [Google Scholar]
- 20. David D., Nair S. A., and Pillai M. R. (2013) Smurf E3 ubiquitin ligases at the cross roads of oncogenesis and tumor suppression. Biochim. Biophys. Acta 1835, 119–128 [DOI] [PubMed] [Google Scholar]
- 21. Wiesner S., Ogunjimi A. A., Wang H. R., Rotin D., Sicheri F., Wrana J. L., and Forman-Kay J. D. (2007) Autoinhibition of the HECT-type ubiquitin ligase Smurf2 through its C2 domain. Cell 130, 651–662 [DOI] [PubMed] [Google Scholar]
- 22. Subramaniam V., Li H., Wong M., Kitching R., Attisano L., Wrana J., Zubovits J., Burger A. M., and Seth A. (2003) The RING-H2 protein RNF11 is overexpressed in breast cancer and is a target of Smurf2 E3 ligase. Br. J. Cancer 89, 1538–1544 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Chen C., Zhou Z., Liu R., Li Y., Azmi P. B., and Seth A. K. (2008) The WW domain containing E3 ubiquitin protein ligase 1 upregulates ErbB2 and EGFR through RING finger protein 11. Oncogene 27, 6845–6855 [DOI] [PubMed] [Google Scholar]
- 24. Abe M., Harpel J. G., Metz C. N., Nunes I., Loskutoff D. J., and Rifkin D. B. (1994) An assay for transforming growth factor-β using cells transfected with a plasminogen activator inhibitor-1 promoter-luciferase construct. Anal. Biochem. 216, 276–284 [DOI] [PubMed] [Google Scholar]
- 25. Heppner G. H., Miller F. R., and Shekhar P. M. (2000) Nontransgenic models of breast cancer. Breast Cancer Res. 2, 331–334 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. McEarchern J. A., Kobie J. J., Mack V., Wu R. S., Meade-Tollin L., Arteaga C. L., Dumont N., Besselsen D., Seftor E., Hendrix M. J., Katsanis E., and Akporiaye E. T. (2001) Invasion and metastasis of a mammary tumor involves TGF-β signaling. Int. J. Cancer 91, 76–82 [DOI] [PubMed] [Google Scholar]
- 27. Krstic J., and Santibanez J. F. (2014) Transforming growth factor-β and matrix metalloproteinases: functional interactions in tumor stroma-infiltrating myeloid cells. ScientificWorldJournal 2014, 521754. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Mazzoccoli G., Pazienza V., Panza A., Valvano M. R., Benegiamo G., Vinciguerra M., Andriulli A., and Piepoli A. (2012) ARNTL2 and SERPINE1: potential biomarkers for tumor aggressiveness in colorectal cancer. J. Cancer Res. Clin. Oncol. 138, 501–511 [DOI] [PubMed] [Google Scholar]
- 29. Duffy M. J., McGowan P. M., Harbeck N., Thomssen C., and Schmitt M. (2014) uPA and PAI-1 as biomarkers in breast cancer: validated for clinical use in level-of-evidence-1 studies. Breast Cancer Res. 16, 428. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Wang L., Yang H. J., Gao S. S., Wang M., Shi Y., Cheng B. F., and Feng Z. W. (2015) Identification of a novel role of RING finger protein 11 promoting the metastasis of murine melanoma cells. Am. J. Transl. Res. 7, 1629–1635 [PMC free article] [PubMed] [Google Scholar]
- 31. Fu W., Wang L., Kim S., Li J., and Dynlacht B. D. (2016) Role for the IFT-A complex in selective transport to the primary cilium. Cell Rep. 17, 1505–1517 [DOI] [PMC free article] [PubMed] [Google Scholar]