Abstract
Glycogen synthase kinase-3β (GSK3β) has diverse biological roles including effects on cellular differentiation, migration, and inflammation. GSK3β phosphorylates proteins to generate phosphodegrons necessary for recognition by Skp1/Cullin-1/F-box (SCF) E3 ubiquitin ligases leading to subsequent proteasomal degradation of these substrates. However, little is known regarding how GSK3β protein stability itself is regulated and how its stability may influence inflammation. Here we show that GSK3β is degraded by the ubiquitin-proteasome pathway in murine lung epithelial cells through lysine 183 as an acceptor site for K48 polyubiquitination. We have identified FBXO17 as an F-box protein subunit that recognizes and mediates GSK3β polyubiquitination. Both endogenous and ectopically expressed FBXO17 associate with GSK3β, and its overexpression leads to decreased protein levels of GSK3β. Silencing FBXO17 gene expression increased the half-life of GSK3β in cells. Furthermore, overexpression of FBXO17 inhibits agonist-induced release of keratinocyte-derived cytokine (KC) and interleukin-6 (IL-6) production by cells. Thus, the SCFFBXO17 E3 ubiquitin ligase complex negatively regulates inflammation by targeting GSK3β in lung epithelia.
Keywords: E3 ubiquitin ligase, inflammation, lung, lung injury, ubiquitin
Introduction
The ubiquitin-proteasome system (UPS)3 is the primary mechanism for protein degradation in eukaryotic cells (1). Ubiquitination of protein substrates involves the stepwise transfer of ubiquitin from an E1 ubiquitin-activating enzyme to an E2 ubiquitin-conjugating enzyme, and finally to an E3 ubiquitin ligase complex. In the final step of the reaction, the E3 ubiquitin ligase transfers ubiquitin chains to the substrate to facilitate degradation by the proteasome or sorting to the endosome-lysosome pathway (2). Two E1 enzymes, almost 40 E2 enzymes, and more than 1000 E3 enzymes have been identified in mammalian cells (3). The SCF superfamily represents the largest group of E3 ligases that mediate critical roles in cell biology, including tumorigenesis and inflammation. The SCF E3 ligase is a modular complex comprised of core proteins Rbx1, Skp1, Cul1, and an F-box protein (1, 2). The F-box protein subunit confers specificity for the target substrate. Approximately 70 human F-box proteins have been identified in the genome, but most have not been characterized (4–6). Proteins targeted by the ubiquitin-proteasome pathway contain primary sequences, post-translational modifications, or structural changes named “degrons” that allow recognition by ubiquitin ligases in the appropriate context (7, 8). Regulation of protein homeostasis is a growing area of interest in cancer biology and inflammatory diseases. F-box proteins in particular are attractive therapeutic targets because recognition of degrons serves as a key regulatory point for protein stability (1, 4, 9). A few F-box proteins have been implicated in the pathogenesis of acute respiratory distress syndrome (ARDS) (9–11). This devastating illness is characterized by an initial robust inflammatory response in the host that is linked to the activation of key regulatory kinases in effector cell populations in the setting of sepsis or severe pneumonia.
An important regulatory step for commitment of a protein substrate to proteasomal degradation is phosphorylation (8, 12). Glycogen synthase kinase 3β (GSK3β) is a serine-threonine kinase with over 50 known protein substrates (13, 14). Although basal constitutive kinase activity is high in cells, substrates of GSK3β usually require priming with phosphorylation by upstream kinases, leading to optimal kinetics of GSK3β-mediated phosphorylation (14, 15). Many substrates of SCF E3 ligases are phosphorylated by GSK3β, generating the phosphodegron required for E3 ligase recognition, ubiquitination, and proteasomal degradation (12). For example, GSK3β phosphorylates the ST2L receptor for IL-33 leading to degradation by SCFFBXL19 (11). TRAF proteins are phosphorylated by GSK3β prior to degradation by SCFFBXL2 (16), and the F-box protein FBXL2 itself is phosphorylated by the same kinase prior to ubiquitination and degradation by another SCF complex, SCFFBXO3(12, 16, 17).
In recent years there has been a resurgence in interest in GSK3β as its list of substrates grows in the literature (12, 18). GSK3β was initially discovered in the context of glucose metabolism, but it has now been established as a point of convergence for adaptive and innate immunity pathways (19, 20). GSK3β has been shown to phosphorylate serine 468 on the p65 subunit of NF-κB, inhibiting binding to promoters (21). TNFα-induced expression of IL-6 and CXCL1 and p65 recruitment to the promoter of select inflammatory genes also requires GSK3β activity in mouse embryonic fibroblast cells (22). GSK3β is required for TNFα-mediated NF-κB signaling, and suppression or deletion of GSK3β enhances sensitivity to TNFα-induced apoptosis (23, 24). GSK3β also negatively regulates NF-κB activity and mediates cross-tolerance to LPS in macrophages (25). Overall, the role of GSK3β in downstream inflammatory pathways varies depending on the cell type and stimuli used (13, 19).
A growing number of studies demonstrate a pathogenic role for GSK3β in end-organ damage in murine models of sepsis. Liver injury has been shown to be mediated by GSK3β activity and hepatocellular injury decreases with the use of GSK3β inhibitors (26, 27). More recently, murine models of ARDS demonstrate an active role of GSK3β in mediating acute lung inflammation and alveolar damage (28, 29). Despite critical roles in phosphorylating proteins to direct their fate to the UPS and studies supporting its role in inflammation and sepsis, very little is known about the regulation of GSK3β stability itself. Studies show that GSK3β is degraded by the proteasome in some cell types but the mechanism has not been well characterized (30). Regulation of GSK3β protein stability in lung biology has not been studied, and to date, no E3 ubiquitin ligase complex has been identified that targets GSK3β to the proteasome.
In this study we elucidate the mechanism of GSK3β polyubiquitination and degradation by the proteasome. We have identified a previously uncharacterized F-box protein subunit, FBXO17, which targets GSK3β for polyubiquitination by the SCF complex. We also show that FBXO17 attenuates inflammatory responses in lung epithelial cells by facilitating degradation of GSK3β protein. These studies describe a mechanism for regulation of GSK3β protein stability and potentially has broad implications in the regulation of immune responses in acute lung injury.
Results
GSK3β degradation occurs through the ubiquitin-proteasome pathway
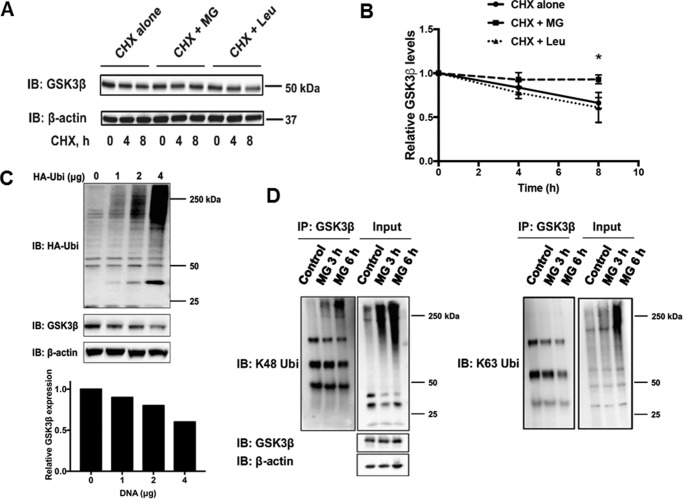
We examined the half-life of GSK3β in lung epithelial cells and determined whether degradation was primarily dependent on the UPS or endosome-lysosome pathway. Using cycloheximide (CHX) to inhibit protein synthesis, we treated cells with the proteasome inhibitor, MG132, or a lysosome inhibitor, leupeptin. The half-life of GSK3β exceeded ∼8 h. Protein levels were stabilized in the presence of MG132, but not leupeptin, supporting the supposition that GSK3β is processed primarily through the UPS (Fig. 1, A and B). To determine whether GSK3β is polyubiquitinated, control or HA-ubiquitin (HA-Ub) plasmids were transfected into mouse lung epithelial (MLE) cells. Endogenous GSK3β levels decreased in a dose-dependent manner in cells with increasing amounts of ectopically expressed HA-Ub plasmid (Fig. 1C). It has been demonstrated that lysine 48-linked ubiquitin chains are associated with targeting proteins to the UPS, whereas lysine 63 chains facilitate protein sorting to the endosome-lysosome pathway (31, 32). After treating MLE cells with MG132 to allow for accumulation of polyubiquitinated proteins, endogenous GSK3β was immunoprecipitated. GSK3β was shown to be polyubiquitinated by lysine 48-linked ubiquitin, but not lysine 63-linked ubiquitin chains (Fig. 1D). The data support the UPS as the dominant mechanism for GSK3β degradation in lung epithelia.
Figure 1.
GSK3β degradation occurs through the ubiquitin-proteasome pathway. A, MLE-12 cells were treated with CHX alone (40 μg/ml) or in combination with MG132 (20 μm) or leupeptin (20 μg/ml) for 0, 2, 4, and 8 h. Immunoblots of lysates for endogenous GSK3β and β-actin as a loading control were performed. B, shown are the relative densitometries of GSK3β protein over time for each immunoblot. The data represent mean ± S.E. of n = 4 independent experiments. *, p value <0.05 by a nonparametric test for trend. C, MLE-12 cells were transfected with plasmid encoding HA-tagged ubiquitin (HA-Ub) using 0, 1, 2, and 4 μg of DNA. Cells were cultured for 48 h. Immunoblots for GSK3β and β-actin as a loading control are shown. Bar graph depicts relative densitometry representative of the immunoblot shown. D, MLE-12 cells were treated with MG132 for 3 and 6 h prior to harvesting lysates. Immunoprecipitation (IP) of endogenous GSK3β was performed and samples were immunoblotted (IB) with antibodies against K48-ubiquitin or K63-ubiquitin.
Lysine 183 is an acceptor site for K48 polyubiquitination in GSK3β
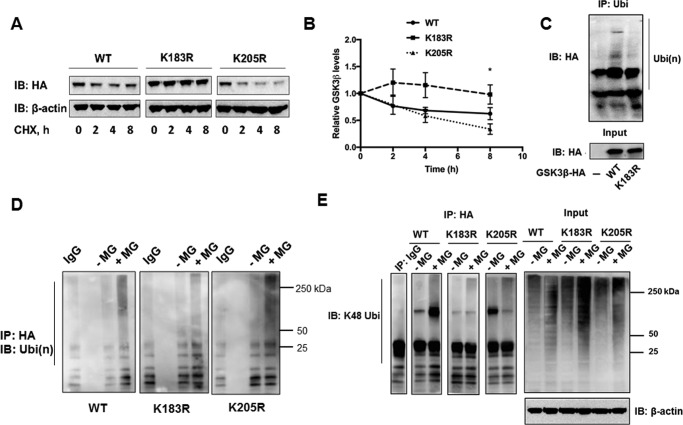
Ubiquitination most often occurs on lysine residues in target proteins. Human and mouse GSK3β share 99% homology and contain 23 lysine residues. Based on prediction algorithms (7, 33) 16 sites were screened and selected for site-directed mutagenesis. Plasmids expressing GSK3β with single lysine to arginine mutations were generated and transfected into MLE cells. CHX chase assays were performed with all mutations. Expression of a variant GSK3β plasmid harboring a K183R mutation in cells conferred stability of protein levels in CHX chase assays compared with expression of a wild-type or a K205R plasmid (Fig. 2, A and B). Cells transfected with plasmids expressing HA-tagged wild-type, K183R, or K205R variants were also subjected to co-immunoprecipitation and immunoblotting. First, after immunoprecipitation of ubiquitinated products and probing with HA antibody, polyubiquitination of mutant K183R-GSK3β was reduced compared with wild-type (Fig. 2C). In other experiments the antibodies were reversed using co-immunoprecipitation demonstrating similar findings (Fig. 2D). Last, in separate studies, immunoprecipitation with HA antibody followed by probing with the K48 antibody demonstrated that GSK3β was modified by K48 polyubiquitin chains. The intensity of this signal on immunoblots was reduced after analysis of expressed K183R-GSK3β plasmid (Fig. 2E). Of note, Lys-205 had been previously predicted as a ubiquitin acceptor site by the Phosphosite database (34).
Figure 2.
Lysine 183 is an acceptor site for K48 polyubiquitination in GSK3β. A, plasmids expressing HA-tagged wild-type, K183R, or K205R mutant GSK3β were transfected into MLE-12 cells. Cells were cultured for 48 h and then treated with CHX for 0, 2, 4, and 8 h. Lysates were prepared and immunoblotted for HA and β-actin as a loading control. B, the relative densitometries of GSK3β protein plotted over time for each immunoblot are shown. The data represent mean ± S.E. of n = 4 independent experiments. *, p value <0.05 by a nonparametric test for trend. C, MLE-12 cells were transfected with HA-tagged wild-type, or K183R GSK3β plasmids and cultured for 48 h. Cells were then treated with MG132 for 6 h and harvested. Immunoprecipitation (IP) was performed with ubiquitin antibody and samples were probed with HA antibody. D and E, cells were transfected with HA-tagged wild-type, K183R, or K205R mutant GSK3β plasmids and subjected to MG132 treatment prior to HA-antibody pulldown and ubiquitin (D) or K48 chain specific immunoblotting (E). Far right panel: input samples were immunoblotted (IB) with HA-antibody and β-actin as a loading control.
FBXO17 mediates GSK3β degradation
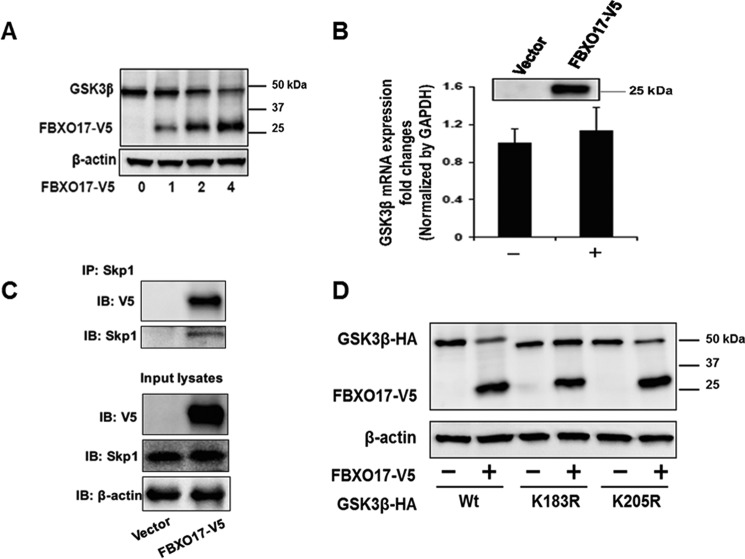
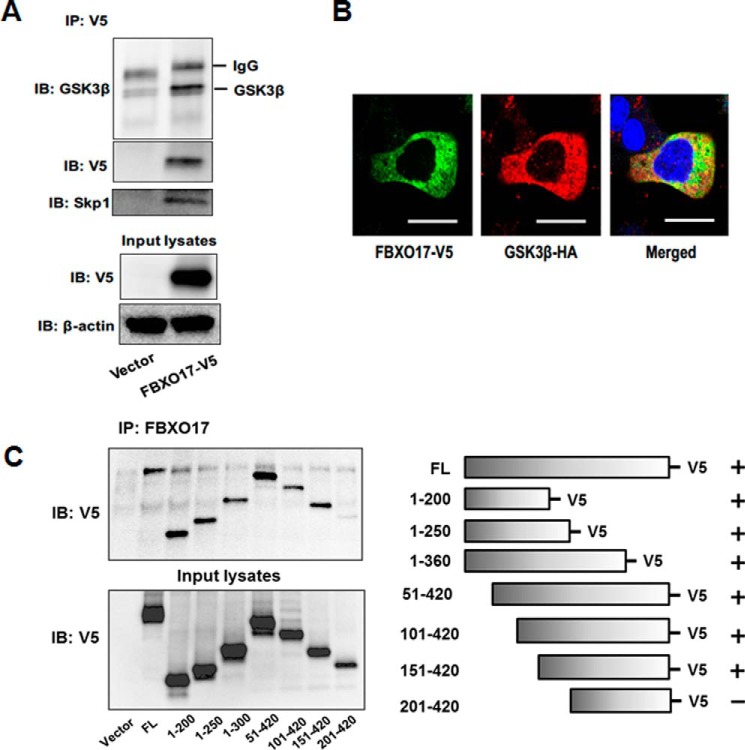
GSK3β is a phosphoenzyme and SCF E3 ligases target phosphoproteins for ubiquitination. Thus, with data supporting GSK3β degradation through the UPS, we sought to identify a potential F-box subunit of the SCF E3 ligase complex that could recognize and target GSK3β for polyubiquitination. Using a library of over 30 F-box protein expression plasmids, HEK293 cells were transfected with these plasmids and immunoblotted to analyze GSK3β protein levels. FBXO17 was identified in this screen as being associated with lower expression levels of GSK3β (data not shown). We then transfected a FBXO17-V5 plasmid in MLE cells and observed a trend for a decrease of GSK3β protein levels with no effect on mRNA transcript expression (Fig. 3, A and B). To confirm that this F-box protein was a subunit of the canonical SCF E3 ubiquitin ligase apparatus, we overexpressed FBXO17-V5 and performed co-immunoprecipitation. Here, Skp1 was shown to be associated with FBXO17 (Fig. 3C).
Figure 3.
FBXO17 targets GSK3β for degradation in lung epithelial cells. A, MLE-12 cells were transfected with 0, 1, 2, and 4 μg of FBXO17-V5 expression plasmids and cultured for 48 h. Endogenous GSK3β, FBXO17-V5, and β-actin protein levels were analyzed by immunoblotting (IB). B, MLE12 cells were transfected with 2 μg of FBXO17-V5 plasmid for 48 h. RNA was isolated and analyzed by RT-PCR using primers against GSK3β and GAPDH as an internal control. Inset, FBXO17-V5 protein expression was confirmed by immunoblotting using V5 antibody. C and D, FBXO17-V5 expression plasmid (2 μg) was transfected into MLE-12 cells and cells were cultured for 48 h. Immunoprecipitation (IP) of lysates was performed using Skp1 antibody. Samples were immunoblotted with Skp1, V5, and β-actin (loading control) antibodies. The data in each panel are representative of at least n = 3 independent experiments. D, MLE-12 cells were co-transfected with HA-tagged wild-type, K183R, or K205R mutant GSK3β plasmids with or without FBXO17-V5 plasmid. Samples were immunoblotted with HA, V5, and β-actin (loading control) antibodies.
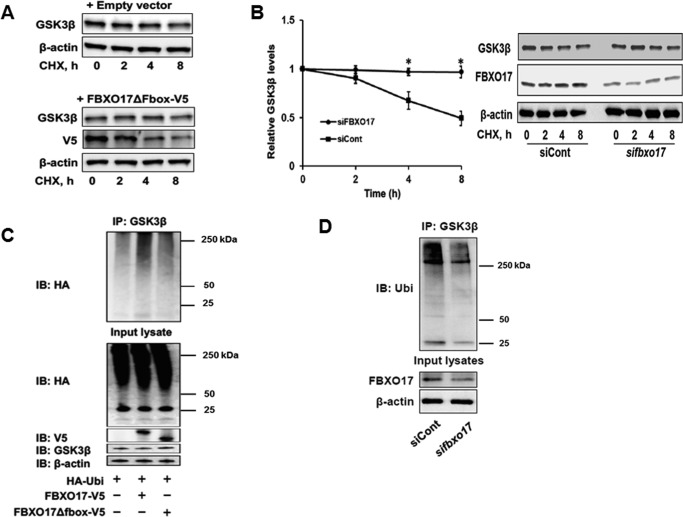
To further assess behavior of SCFFBXO17 to target GSK3β for degradation, plasmids expressing HA-tagged wild-type, K183R, or K205R variants were co-transfected with or without FBXO17-V5 plasmid into MLE cells. Expression of wild-type FBXO17-V5 decreased wild-type and K205R protein levels but not K183R protein (Fig. 3D). Additional studies were conducted expressing an empty vector or ΔFboxFBXO17-V5 in MLE cells in CHX chase studies. The latter construct lacks the ability to engage other components of the SCF apparatus. Here, unlike effects of FBXO17-V5, expression of ΔFboxFBXO17-V5 did not alter GSK3β lifespan nor did it induce kinase polyubiquitination (Fig. 4, A and C). When FBXO17 gene knockdown is performed, the GSK3β half-life increases and polyubiquitination of the kinase decreases (Fig. 4, B and D). These experiments suggest that FBXO17, as a component of the SCF complex, mediates K48 polyubiquitination of GSK3β at a distinct molecular site (Lys-183) for its proteasomal degradation.
Figure 4.
FBXO17 overexpression increases polyubiquitination and proteasomal degradation of GSK3β. A, MLE-12 cells were transfected with 2 μg of empty pcDNA 3.1 TOPO vector or ΔFboxFBXO17-V5-expressing plasmids and cultured for 48 h. Cells were then treated with cycloheximide (40 μg/ml) and lysates were collected at 0, 2, 4, and 8 h. Samples were immunoblotted with GSK3β, V5, and β-actin (loading control) antibodies. B, knockdown experiments were performed by co-transfecting GSK3β-V5 plasmids combined with FBXO17 siRNA (100 nm) or control scrambled RNA into BEAS-2B cells. Cells were cultured for 72 h and then treated with CHX (40 μg/ml). Samples were collected at 0, 2, 4, and 8 h. The relative densitometries of GSK3β protein plotted over time for each immunoblot are shown. The data represent mean ± S.E. of n = 3 independent experiments. *, p value <0.05 by a nonparametric test for trend. C, MLE-12 cells were all transfected with plasmids expressing HA-ubiquitin combined with empty vector, FBXO17-V5, or ΔFboxFBXO17-V5. Cells were cultured for 48 h and then treated with MG132 (20 μm) for 6 h prior to preparing lysates. Immunoprecipitation (IP) was done using GSK3β antibody. Samples were immunoblotted (IB) with antibodies against HA, V5, GSK3β, and β-actin as a loading control. D, BEAS-2B cells were transfected with 100 nm scrambled RNA (negative control) or FBXO17 siRNA. Cells were cultured for 72 h and then treated with MG132 (20 μm) for 6 h. Lysates were prepared and immunoprecipitation was done using a GSK3β antibody. Samples were immunoblotted with antibodies for ubiquitin, FBXO17, and β-actin (loading control). The data in each panel are representative of at least n = 3 independent experiments.
FBXO17 associates with GSK3β through a docking motif
Although our data supports the hypothesis that GSK3β is a substrate for FBXO17, GSK3β phosphorylates many substrates of the SCF E3 ligase complexes. Thus, we sought to clarify if FBXO17 specifically associates with GSK3β as a substrate for polyubiquitination. We examined this issue by expressing FBXO17-V5. Immunoprecipitation of GSK3β demonstrated an association between endogenous GSK3β and ectopically expressed FBXO17 (Fig. 5A). Colocalization of both proteins in the cytoplasm was demonstrated by immunofluorescence (Fig. 5B). To further characterize molecular interactions between FBXO17 and V5-tagged GSK3β, GSK3β truncation mutants were cloned and used in in vitro transcription and translation assays. When FBXO17 was immunoprecipitated, all mutants were found to associate with FBXO17 by immunoblotting with the exception of loss of binding of the 201–420 amino acid NH2-terminal mutant (Fig. 5C). These experiments demonstrate that amino acids 151–200 contain a putative binding domain required for association of GSK3β-V5 with FBXO17.
Figure 5.
FBXO17 associates with GSK3β through a docking motif. A, MLE cells were transfected with FBXO17-V5 plasmid and cultured for 48 h. Samples were collected and V5 antibody was used for immunoprecipitation (IP). Cells were collected and assayed for GSK3β, V5, Skp1, and β-actin (loading control) by immunoblotting (IB). B, MLE-12 cells were transfected with FBXO17-V5 and GSK3β-HA plasmids and cultured on glass bottom dishes for 48 h. Cells were fixed and immunostained with antibodies to V5 (green) and HA (red). Nuclei were stained with DAPI (blue). Colocalization is demonstrated by yellow on the merged image. Representative images from three independent experiments are shown. Scale bars = 10 μm. C, in vitro transcription and translation of wild-type and deletion mutants of V5-tagged GSK3β and FBXO17 (no tag) was performed. V5-GSK3β samples included full-length protein (FL) and deletion mutants expressing amino acids 1–200, 1–250, 1–360, 51–420, 101–420, 151–420, and 201–420. Samples were immunoprecipitated using the FBXO17 antibody and immunoblotted with V5 antibody. Data represent mean ± S.D. of duplicate measurements. The data in each panel are representative of at least n = 3 independent experiments.
Degradation of GSK3β by FBXO17 abrogates inflammatory cytokine production
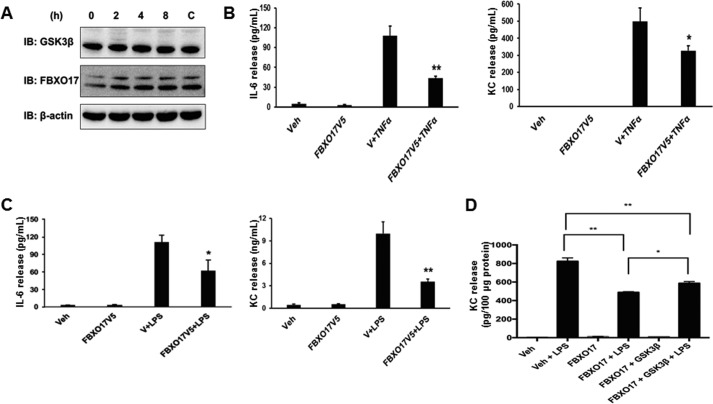
To determine whether FBXO17 has a role in inflammatory responses in lung epithelial cells, we first examined if endogenous levels of FBXO17 in MLE cells changed after stimulation with lipopolysaccharide (LPS). Kinetics studies of expression showed no significant changes in FBXO17 or GSK3β protein levels (Fig. 6A). Next, we overexpressed various plasmids in MLE cells and evaluated responses to LPS and tumor necrosis factor α (TNFα). We found that IL-6 and KC production was significantly reduced in response to LPS and TNFα when FBXO17 is ectopically expressed in cells (Fig. 6, B and C). Partial rescue of KC production in response to LPS was observed with overexpression of HA-GSK3β (Fig. 6D). FBXO17 expression did not modulate IL-10 levels with any stimuli used, and the F-box protein did not significantly alter IL-6 and KC production by MLE cells in response to lipoteichoic acid (LTA) or PamCys3 (data not shown). These results differ from a genome-wide study by Alper et al. (35) in which FBXO17 was identified as a possible proinflammatory gene in innate immune responses to LTA. In that study, knockdown of FBXO17 expression in RAW macrophages resulted in a blunted response to LTA stimulation, and its overexpression led to enhanced NF-κB promoter activity. However, our data suggest that FBXO17 has anti-inflammatory effects through negative regulation of GSK3β stability in lung epithelial cells.
Figure 6.
FBXO17 abrogates inflammatory cytokine production in lung epithelial cells. A, MLE-12 cells were stimulated with LPS (10 μg/ml) and lysates were collected at 0, 2, 4, and 8 h. Samples were immunoblotted (IB) with antibodies against GSK3β, FBXO17, and β-actin (loading control). A control (C) untreated sample is shown collected at 8 h. B, MLE-12 cells were transfected with FBXO17-V5 plasmid and cultured for 48 h. Cells were treated with TNFα (10 ng/ml) for 16 h. Samples were collected and analyzed by ELISA for IL-6 or KC. C, MLE-12 cells were transfected with FBXO17-V5 plasmid and cultured for 48 h. Cells were then treated with LPS (10 μg/ml) for 16 h. Samples were collected and analyzed by ELISA for IL-6 or KC. Data represent mean ± S.D. of duplicate measurements and were analyzed by Student's t test. The data in each panel are representative of at least n = 3 independent experiments with p values <0.05 (*) and <0.01 (**). D, MLE-12 cells were transfected with FBXO17-V5 with or without GSK3β-HA plasmid and cultured for 48 h. Cells were then treated with LPS (10 μg/ml) for 6 h. Samples were collected and analyzed by ELISA for KC. Data represent mean ± S.D. of duplicate measurements and were analyzed by Student's t test. The data in panel D are representative of at least n = 3 independent experiments with p values <0.01 (*) and <0.001 (**).
Discussion
This study elucidates a unique mechanism for attenuation of immune responses by regulation of GSK3β protein stability through a poorly characterized SCF E3 component. FBXO17, also known as FBG4, is an F-box protein and one of five members of the F-box-associated family thought to be involved in glycoprotein ER-associated degradation (36, 37). Glenn et al. (38) identified sulfated and galactose-terminated glycoproteins as likely binding substrates of FBXO17 by a glycan array, with no specific proteins identified as substrates for ubiquitination. This latter study also demonstrated FBXO17 expression in mouse brain and weaker expression in the lung, but not muscle, liver, pancreas, adipose tissue, or kidney, contrary to broader human tissue expression identified by others (37). Mouse and human FBXO17 share 85% homology. We confirmed that this F-box protein subunit is an authentic subunit associated with the core SCF E3 ubiquitin ligase machinery. Our identification of a specific protein substrate, GSK3β, targeted by the SCFFBXO17 E3 ligase complex has potentially important biologic implications for a wide array of fundamental processes. Given the many biological roles of GSK3β, this study raises additional questions regarding how altered stability of this kinase by FBXO17 influences downstream cellular pathways in different cell types. Moreover, as with other F box proteins (9, 16), small molecule FBXO17 targeting might be a strategy to modulate various processes by maintaining cellular levels of GSK3β.
There is growing interest in understanding how dysregulation of protein stability contributes to the pathogenesis of lung diseases (39–41). Findings from our group have identified several F-box protein subunits with anti-inflammatory effects. IL-33 stimulates inflammation in asthma and acute lung injury through the receptor ST2L. GSK3β phosphorylates ST2L and subsequently SCFFBXL19 targets the receptor for polyubiquitination, internalization, and degradation (11). FBXL19 overexpression decreases ST2L levels and reduces inflammation while improving animal survival in these studies. Another example is FBXL2, an F-box subunit that targets TRAF proteins in NF-κB signaling (17). FBXO3 was identified as an F-box subunit that targets FBXL2 for degradation after phosphorylation by GSK3β, thus promoting inflammation. Pharmacologic inhibition or depletion of FBXO3 leads to normal levels of FBXL2, decreased cytokine levels, and improved survival in murine models of sepsis (9, 16). These studies represent a subset of F-box protein subunits involved in modulating inflammatory processes. They also support prior studies that show GSK3β-mediated phosphorylation as a critical step for substrate recognition by SCF E3 ubiquitin ligases and commitment to the UPS pathway (12).
Lysine 183 appears to be the critical acceptor site for most, but not all K48-linked polyubiquitin chains covalently attached to GSK3β. However, other lysine residues may provide fine-tuning of regulation of kinase degradation by the UPS. In our studies, expression of the K183R mutant protein reduces responses to LPS compared with wild-type GSK3β, despite increased stability of mutant protein levels (data not shown). This may be attributed to reduced catalytic activity of the kinase possibly impacted by the K183R mutation given its close proximity to the activation loop (15, 42). Interestingly, a recent study demonstrated that GSK3β is ubiquitinated by K63, but not K48-linked ubiquitin chains on lysine 183 in response to poly(I:C)-mediated TLR3 stimulation of RAW macrophages (43). The K183R mutant was shown to function as a dominant-negative protein in this study. K63 polyubiquitination was required for TLR3 signaling, and the physiologic role of protein degradation by the proteasome was not suggested by this study. Another study showed that SCFFBXO7 mediates K63 polyubiquitination of GSK3β and negatively regulates its activity in HEK293T cells (44). In our experiments, we did not observe significant levels of K63-linked polyubiquitin chains when GSK3β was immunoprecipitated from MLE cells after MG132 treatment, indicating mechanistic variability of GSK3β processing by the UPS depending on the experimental context. Proteomic data suggest ubiquitination at lysine 205, but the K205R mutation was less stable than wild-type GSK3β (34). This finding may reflect the importance of lysine 205 for overall stability because it is located within the catalytic loop of GSK3β (14, 15).
GSK3β kinase activity is integral to Toll-like receptor and NFκB signaling in diverse cell types. Of note, ectopic expression of FBXO17 in MLE cells did not substantially abrogate cytokine release in response to TLR2 agonists suggesting a more restricted effect for the F-box protein in modulating TLR4 stimulation. Martin et al. (45) was the first group to show that GSK3β is required for TLR4-induced responses in RAW macrophages and in murine models of sepsis. In murine models of ARDS, Akt-mediated phosphorylation of GSK3β inhibits inflammation, and pharmacologic inhibition of GSK3β reduces inflammation and improves survival (29, 46). GSK3β also promotes neuroinflammation in glial cells from mice treated with LPS through STAT3 activation (47). A recent study by Peng and colleagues (48) demonstrated that FBXO17 reduces type I IFN signaling through recruitment of protein phosphatase 2A (PP2A) for dephosphorylation of interferon recruitment factor 3 (IRF3) in A549 and HEK293T cells. Intriguingly, these effects were not related to SCF E3 ligase function and were independent of the F-box domain and ubiquitin-proteasome pathway. These data support FBXO17 as a negative regulator of inflammation. Dexamethasone treatment of pancreatic islet cells has been associated with proteasomal degradation of GSK3β, indicating a relationship between suppression of inflammation and decreased stability of GSK3β (30). Our study suggests that GSK3β promotes inflammatory responses in lung epithelial cells, and FBXO17 is a negative regulator of these pathways.
In summary, this study is the first to identify an SCF E3 ligase that targets GSK3β for its proteasomal degradation. Physiologically, our data suggest that SCFFBXO17 is a negative regulator of GSK3β-mediated inflammatory responses in lung epithelia. It is likely that FBXO17 recognizes other substrates, and additional studies may implicate other targets of this F-box protein subunit. Drugs targeting the ubiquitin proteasome pathway are already being used in the clinical setting to treat cancer and rejection in organ transplantation (1, 3). Further studies of this pathway may reveal key therapeutic targets to exploit GSK3β degradation as a means for therapy in neoplasia and inflammatory disorders.
Experimental procedures
Antibodies and reagents
FBXO17 antibody was obtained from Protein Tech Group, Inc. (Rosemont, IL). Anti-mouse IgG and anti-rabbit IgG were obtained from Santa Cruz Biotechnology (Santa Cruz, CA). Antibodies against ubiquitin, K48 ubiquitin, K63 ubiquitin, Skp1, and Cullin 1 were purchased from Cell Signaling (Danvers, MA). β-Actin antibodies, lipoteichoic acid (Staphylococcus aureus), and CHX were obtained from Sigma. QuikChange II site-directed mutagenesis kits were purchased from Agilent Technologies (Santa Clara, CA). Pierce protein A/G-agarose beads and protease inhibitors were purchased from Thermo Scientific (Rockford, IL). MG132 and leupeptin was from Calbiochem (Darmstadt, Germany). Mouse V5 antibodies, miniprep, midiprep, and pcDNA3.1 directional cloning kits were purchased from Invitrogen and New England Biolabs (Ipswich, MA). In vitro transcription and translation (TnT) kits were from Promega (Madison, WI).
Cell culture
Mouse lung epithelial (MLE-12) cells and human bronchial epithelial cells (BEAS-2B) were obtained from ATCC (Manassas, VA). Ham's F-12 medium was purchased from Gibco (Life Technologies). Cells were supplemented with fetal bovine serum (FBS) from Gemini (Sacramento, CA). MLE cells were cultured in HITES medium supplemented with 10% FBS. Prior to CHX treatment, cells were starved for 1 h with 0% FBS, HITES medium. CHX treatment was carried out at a concentration of 40 μg/ml at varying time points in 0% FBS medium, with or without MG132 (20 μg/ml) or leupeptin (20 μg/ml).
Immunoprecipitation and immunoblotting
Cell lysates in 150 μl of lysis buffer (20 mm Tris-HCl (pH 7.4), 150 mm NaCl, 2 mm EGTA, 5 mm β-glycerophosphate, 1 mm MgCl2, 1% Triton X-100, 1 mm sodium orthovanadate, 10 μg/ml of protease inhibitors, 1 μg/ml of aprotinin, 1 μg/ml of leupeptin, and 1 μg/ml of pepstatin) were sonicated on ice for 12 s and centrifuged at 10,000 × g for 10 min at 4 °C in a microcentrifuge. For immunoprecipitation, equal amounts of cell lysates (1 mg) were incubated with 5 μg/ml of specific primary antibodies overnight at 4 °C followed by the addition of 40 μl of protein A/G-agarose for 4 h at 4 °C. For immunoblotting, equal amounts of supernatant (20 μg) were subjected to 10% SDS-PAGE gels, transferred to nitrocellulose membranes, blocked with 5% (w/v) nonfat milk in TBST (25 mm Tris-HCl, pH 7.4, 137 mm NaCl, and 0.1% Tween 20) for 1 h, and incubated with primary antibodies in 5% (w/v) BSA in TBST for 1–2 h. The membranes were washed at least three times with TBST at 10-min intervals followed by a 1-h incubation with mouse, rabbit, or goat horseradish peroxidase-conjugated secondary antibody (1:2,000). The membranes were developed with an enhanced chemiluminescence detection system according to manufacturer's instructions (49, 50).
RT-PCR
RNA was isolated from cells using RNeasy Mini Kits (Qiagen) per the protocol provided. Isolated RNAs were immediately converted to cDNA using High Capacity RNA-to-cDNA Kits (Life Technologies) after their concentrations were measured. Real-time PCR assays were performed using SYBR® Select Master Mix for CFX (2X) (Life Technologies) per the protocol provided and assays were performed with the C1000 Thermal Cycler (Bio-Rad).
Cloning and mutagenesis
Human GSK3β plasmid was purchased from Addgene (Cambridge, MA) for subsequent cloning into pcDNA3.1D/V5-His vector (Invitrogen). Site-directed mutagenesis of lysine to arginine residues was carried out via a PCR-based approach using the QuikChange II XL kit from Agilent Technologies (Santa Clara, CA) and appropriate primers. Primers used for lysine to arginine mutagenesis were designed with the following sequences: Lys-183 forward, 5′-AGGTTCTGCGGTCTAATATCCCGATGGCAGATTCCA-3′; reverse, 5′-TGGAATCTGCCATCGGGATATTAGACCGCAGAACCT-3′; K205R forward, 5′-GGACCAGCTGCCTTGCACTTCCAAAGTCACAG-3′ and reverse, 5′-CTGTGACTTTGGAAGTGCAAGGCAGCTGGTCC-3′. Human FBXO17 cDNA was cloned into a pcDNA3.1D/V5-His vector using the following primers: forward, 5′-GAGAGACCTTTATGAGAGAGCAAAAAGAGATGAG-3′ and reverse, 5′-CTCATCTCTTTTTGCTCTCTCATAAAGGTCTCTC-3′. F-box motif deletion was performed using a sequence overlapping extension cloning technique (New England Biolabs) for amino acids 15–60. Generated mutants were sequence-confirmed (Genewiz, South Plainfield, NJ).
Gene silencing
Small interfering RNAs (siRNA) for human FBXO17 were purchased from Santa Cruz (sc-97555). FBXO17 siRNA was transfected into BEAS-2B cells by electroporation. Concentrations used ranged from 10 to 100 nm. Cells were cultured for 72 h prior to lysate collection and immunoblotting.
Transfection
2.5 × 106 MLE cells were suspended in 100 μl of 20 mm HEPES in PBS and mixed with 2 μg of DNA in a cuvette. Cells were nucleofected on the T-013 protocol using an Amaxa Nucleofector II device (Basel, Switzerland). Immediately following nucleofection, 1 ml of 10% HITES medium was added to the cuvette and the cell solution was plated in 2 ml of 10% HITES culture medium in a 6-well plate. Cells were allowed to grow until 80% confluent prior to half-life experiments (48–72 h). Cells were then treated with 40 μg/ml of CHX at different time points for half-life studies and were harvested in lysis buffer using a rubber policeman for subsequent immunoblot analysis.
Immunofluorescence staining
MLE cells grown on glass bottom dishes were fixed with 3.7% formaldehyde for 20 min, and immunostained with anti-HA or anti-FBXO17 antibody. Cells were washed three times, and incubated with fluorescent-conjugated secondary antibodies. Images were captured by a Nikon ECLIPSE TE 300 inverted microscope.
In vitro transcription/translation and binding assays
The details of this assay were described previously and adapted from the manufacturer's protocol (9). Briefly, 1 μg of plasmid expressing full-length V5-GSK3β, truncated V5-GSK3β mutants, or FBXO17 was used in 25-μl reactions using the TnT Coupled Reticulocyte Lysate System (Promega). FBXO17 antibody was incubated with protein A/G-agarose resin for 1 h at room temperature and washed with PBS prior to adding reactions containing in vitro synthesized FBXO17. Reactions were incubated with beads for 1 h at room temperature and after washing with PBS, in vitro synthesized V5-GSK3β reactions (full-length and truncation mutants) were added. After incubating for 1 h at room temperature and washing, the proteins were eluted and processed for V5-GSK3β immunoblotting.
Enzyme-linked immunosorbent assay (ELISA)
IL-6 and IL-10 ELISA kits were purchased from Affymetrix/Ebioscience (San Diego, CA). KC ELISA Duoset kits were purchased from R & D Biosystems (Minneapolis, MN). After treating MLE cells with TNFα (10 ng/ml), LPS (10 μg/ml), or LTA (10 μg/ml) for 6 or 16 h in serum-free medium, supernatants were collected. The quantitative assays were performed following protocols provided by the manufacturers.
Statistical analysis
Descriptive statistics were reported with mean ± S.D. or S.E. as indicated. As our sample sizes of each experimental group were all less than 10, the sample sizes limited the ability to study a normal sample distribution. Thus, we employed appropriate non-parametric methods such as a Mann-Whitney U test and a Kruskal-Wallis equality-of-populations rank test to compare multiple groups within and between experiments. Also, a Wilcoxon-type test for trend was used where appropriate to check trends in data significance. Using these methods provided conservative analysis to determine statistical significance. All analyses were performed using one-way analysis of variance with Dunnett's post test using GraphPad Prism version 7.0 for Windows and Mac OS (GraphPad Software, La Jolla, CA).
Author contributions
T. S. performed most of the experiments, data analysis, and wrote the manuscript. J. W. and A. J. assisted with in vitro protein-binding assays and ELISA experiments. I. N. assisted with experiments. Y. Z. assisted with experiment design and data analysis. J. Z. and R. M. supervised the project and provided constructive review and edited the manuscript.
Acknowledgments
We thank Bill Chen and James Londino for helpful discussions.
This work was supported, in part, by the United States Department of Veterans Affairs, Veterans Health Administration, Office of Research and Development, Biomedical Laboratory Research and Development, a Merit Review Award from the United States Department of Veterans Affairs, the Flight Attendant Medical Research Institute, and National Institutes of Health R01 Grants HL096376, HL097376, HL098174, HL081784, 1UH2HL123502, P01HL114453 (to R. K. M.), R01 HL131665 (to Y. Z.), R01 GM115389 (to J. Z), an American Heart Association GIA award (to Y. Z), and American Lung Association Grant BRG RG350146 (to J. Z). The authors declare that they have no conflicts of interest with the contents of this article. The content is solely the responsibility of the authors and does not necessarily represent the official views of the National Institutes of Health or the Department of Veterans Affairs.
- UPS
- ubiquitin proteasome system
- Fbx
- F-box
- GSK3β
- glycogen synthase kinase-3β
- ARDS
- acute respiratory distress syndrome
- SCF
- Skp1-Cullin-F box
- Skp1
- S-phase kinase-associated protein 1
- Cul1
- Cullin 1
- Rbx1
- ring-box protein 1
- ST2L
- membrane-bound IL-33 receptor
- CHX
- cycloheximide
- MLE
- mouse lung epithelial
- V5
- histidine
- KC
- keratinocyte-derived cytokine or CXCL1
- TLR
- Toll-like receptor
- NFκB
- nuclear factor κB
- LTA
- lipoteichoic acid.
References
- 1. Skaar J. R., Pagan J. K., and Pagano M. (2014) SCF ubiquitin ligase-targeted therapies. Nat. Rev. Drug Discov. 13, 889–903 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2. Frescas D., and Pagano M. (2008) Deregulated proteolysis by the F-box proteins SKP2 and β-TrCP: tipping the scales of cancer. Nat. Rev. Cancer 8, 438–449 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Weathington N. M., and Mallampalli R. K. (2014) Emerging therapies targeting the ubiquitin proteasome system in cancer. J. Clin. Invest. 124, 6–12 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Wang Z., Liu P., Inuzuka H., and Wei W. (2014) Roles of F-box proteins in cancer. Nat. Rev. Cancer 14, 233–247 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Skaar J. R., Pagan J. K., and Pagano M. (2013) Mechanisms and function of substrate recruitment by F-box proteins. Nat. Rev. Mol. Cell Biol. 14, 369–381 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Nakayama K. I., and Nakayama K. (2006) Ubiquitin ligases: cell-cycle control and cancer. Nat. Rev. Cancer 6, 369–381 [DOI] [PubMed] [Google Scholar]
- 7. Guharoy M., Bhowmick P., Sallam M., and Tompa P. (2016) Tripartite degrons confer diversity and specificity on regulated protein degradation in the ubiquitin-proteasome system. Nat. Commun. 7, 10239. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Ravid T., and Hochstrasser M. (2008) Diversity of degradation signals in the ubiquitin-proteasome system. Nat. Rev. Mol. Cell Biol. 9, 679–690 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Mallampalli R. K., Coon T. A., Glasser J. R., Wang C., Dunn S. R., Weathington N. M., Zhao J., Zou C., Zhao Y., and Chen B. B. (2013) Targeting F box protein Fbxo3 to control cytokine-driven inflammation. J. Immunol. 191, 5247–5255 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Han S., and Mallampalli R. K. (2015) The acute respiratory distress syndrome: from mechanism to translation. J. Immunol. 194, 855–860 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Zhao J., Wei J., Mialki R. K., Mallampalli D. F., Chen B. B., Coon T., Zou C., Mallampalli R. K., and Zhao Y. (2012) F-box protein FBXL19-mediated ubiquitination and degradation of the receptor for IL-33 limits pulmonary inflammation. Nat. Immunol. 13, 651–658 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Xu C., Kim N.-G., and Gumbiner B. M. (2009) Regulation of protein stability by GSK3 mediated phosphorylation. Cell Cycle 8, 4032–4039 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Beurel E., Grieco S. F., and Jope R. S. (2015) Glycogen synthase kinase-3 (GSK3): regulation, actions, and diseases. Pharmacol. Ther. 148, 114–131 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Doble B. W., and Woodgett J. R. (2003) GSK-3: tricks of the trade for a multi-tasking kinase. J. Cell Sci. 116, 1175–1186 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. ter Haar E., Coll J. T., Austen D. A., Hsiao H. M., Swenson L., and Jain J. (2001) Structure of GSK3β reveals a primed phosphorylation mechanism. Nat. Struct. Biol. 8, 593–596 [DOI] [PubMed] [Google Scholar]
- 16. Chen B. B., Coon T. A., Glasser J. R., McVerry B. J., Zhao J., Zhao Y., Zou C., Ellis B., Sciurba F. C., Zhang Y., and Mallampalli R. K. (2013) A combinatorial F box protein directed pathway controls TRAF adaptor stability to regulate inflammation. Nat. Immunol. 14, 470–479 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Chen B. B., Glasser J. R., Coon T. A., and Mallampalli R. K. (2013) Skp-cullin-F box E3 ligase component FBXL2 ubiquitinates Aurora B to inhibit tumorigenesis. Cell Death Dis. 4, e759. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Cohen P., and Frame S. (2001) The renaissance of GSK3. Nat. Rev. Mol. Cell Biol. 2, 769–776 [DOI] [PubMed] [Google Scholar]
- 19. Beurel E., Michalek S. M., and Jope R. S. (2010) Innate and adaptive immune responses regulated by glycogen synthase kinase-3 (GSK3). Trends Immunol. 31, 24–31 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Wang H., Kumar A., Lamont R. J., and Scott D. A. (2014) GSK3β and the control of infectious bacterial diseases. Trends Microbiol. 22, 208–217 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Buss H., Dörrie A., Schmitz M. L., Frank R., Livingstone M., Resch K., and Kracht M. (2004) Phosphorylation of serine 468 by GSK-3β negatively regulates basal p65 NF-κB activity. J. Biol. Chem. 279, 49571–49574 [DOI] [PubMed] [Google Scholar]
- 22. Steinbrecher K. A., Wilson W. 3rd, Cogswell P. C., and Baldwin A. S. (2005) Glycogen synthase kinase 3β functions to specify gene-specific, NF-κB-dependent transcription. Mol. Cell Biol. 25, 8444–8455 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Takada Y., Fang X., Jamaluddin M. S., Boyd D. D., and Aggarwal B. B. (2004) Genetic deletion of glycogen synthase kinase-3β abrogates activation of IκBα kinase, JNK, Akt, and p44/p42 MAPK but potentiates apoptosis induced by tumor necrosis factor. J. Biol. Chem. 279, 39541–39554 [DOI] [PubMed] [Google Scholar]
- 24. Liao X., Zhang L., Thrasher J. B., Du J., and Li B. (2003) Glycogen synthase kinase-3β suppression eliminates tumor necrosis factor-related apoptosis-inducing ligand resistance in prostate cancer. Mol. Cancer Ther. 2, 1215–1222 [PubMed] [Google Scholar]
- 25. Park S. H., Park-Min K.-H., Chen J., Hu X., and Ivashkiv L. B. (2011) Tumor necrosis factor induces GSK3 kinase-mediated cross-tolerance to endotoxin in macrophages. Nat. Immunol. 12, 607–615 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Ren F., Duan Z., Cheng Q., Shen X., Gao F., Bai L., Liu J., Busuttil R. W., Kupiec-Weglinski J. W., and Zhai Y. (2011) Inhibition of glycogen synthase kinase 3β ameliorates liver ischemia reperfusion injury by way of an interleukin-10-mediated immune regulatory mechanism. Hepatology 54, 687–696 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Zhang H., Wang W., Fang H., Yang Y., Li X., He J., Jiang X., Wang W., Liu S., Hu J., Liu A., Dahmen U., and Dirsch O. (2014) GSK-3β inhibition attenuates CLP-induced liver injury by reducing inflammation and hepatic cell apoptosis. Mediators Inflamm. 2014, 629507. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Ren F., Zhang L., Zhang X., Shi H., Wen T., Bai L., Zheng S., Chen Y., Chen D., Li L., and Duan Z. (2016) Inhibition of glycogen synthase kinase 3β promotes autophagy to protect mice from acute liver failure mediated by peroxisome proliferator-activated receptor α. Cell Death Dis. 7, e2151. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Park D. W., Jiang S., Liu Y., Siegal G. P., Inoki K., Abraham E., and Zmijewski J. W. (2014) GSK3β-dependent inhibition of AMPK potentiates activation of neutrophils and macrophages and enhances severity of acute lung injury. Am. J. Physiol. Lung Cell Mol. Physiol. 307, L735–L745 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Failor K. L., Desyatnikov Y., Finger L. A., and Firestone G. L. (2007) Glucocorticoid-induced degradation of glycogen synthase kinase-3 protein is triggered by serum- and glucocorticoid-induced protein kinase and Akt signaling and controls β-catenin dynamics and tight junction formation in mammary epithelial tumor cells. Mol. Endocrinol. 21, 2403–2415 [DOI] [PubMed] [Google Scholar]
- 31. Reichard E. L., Chirico G. G., Dewey W. J., Nassif N. D., Bard K. E., Millas N. E., and Kraut D. A. (2016) Substrate ubiquitination controls the unfolding ability of the proteasome. J. Biol. Chem. 291, 18547–18561 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Nathan J. A., Kim H. T., Ting L., Gygi S. P., and Goldberg A. L. (2013) Why do cellular proteins linked to K63-polyubiquitin chains not associate with proteasomes? EMBO J. 32, 552–565 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Huang C.-H., Su M.-G., Kao H.-J., Jhong J.-H., Weng S.-L., and Lee T.-Y. (2016) UbiSite: incorporating two-layered machine learning method with substrate motifs to predict ubiquitin-conjugation site on lysines. BMC Syst. Biol. 10, 6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Hornbeck P. V., Kornhauser J. M., Tkachev S., Zhang B., Skrzypek E., Murray B., Latham V., and Sullivan M. (2012) PhosphoSitePlus: a comprehensive resource for investigating the structure and function of experimentally determined post-translational modifications in man and mouse. Nucleic Acids Res. 40, D261–D270 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. Alper S., Warg L. A., De Arras L., Flatley B. R., Davidson E. J., Adams J., Smith K., Wohlford-Lenane C. L., McCray P. B. Jr., Pedersen B. S., Schwartz D. A., and Yang I. V. (2016) Novel innate immune genes regulating the macrophage response to Gram-positive bacteria. Genetics 204, 327–336 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36. Winston J. T., Koepp D. M., Zhu C., Elledge S. J., and Harper J. W. (1999) A family of mammalian F-box proteins. Curr. Biol. 9, 1180–1182 [DOI] [PubMed] [Google Scholar]
- 37. Ilyin G. P., Sérandour A.-L., Pigeon C., Rialland M., Glaise D., and Guguen-Guillouzo C. (2002) A new subfamily of structurally related human F-box proteins. Gene 296, 11–20 [DOI] [PubMed] [Google Scholar]
- 38. Glenn K. A., Nelson R. F., Wen H. M., Mallinger A. J., and Paulson H. L. (2008) Diversity in tissue expression, substrate binding, and SCF complex formation for a lectin family of ubiquitin ligases. J. Biol. Chem. 283, 12717–12729 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39. Balch W. E., Sznajder J. I., Budinger S., Finley D., Laposky A. D., Cuervo A. M., Benjamin I. J., Barreiro E., Morimoto R. I., Postow L., Weissman A. M., Gail D., Banks-Schlegel S., Croxton T., and Gan W. (2014) Malfolded protein structure and proteostasis in lung diseases. Am. J. Respir. Crit. Care Med. 189, 96–103 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40. Bouchecareilh M., and Balch W. E. (2011) Proteostasis: a new therapeutic paradigm for pulmonary disease. Proc. Am. Thorac. Soc. 8, 189–195 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41. Weathington N. M., and Mallampalli R. K. (2013) New insights on the function of SCF ubiquitin E3 ligases in the lung. Cell Signal. 25, 1792–1798 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42. Dajani R., Fraser E., Roe S. M., Young N., Good V., Dale T. C., and Pearl L. H. (2001) Crystal structure of glycogen synthase kinase 3β. Cell 105, 721–732 [DOI] [PubMed] [Google Scholar]
- 43. Ko R., Park J. H., Ha H., Choi Y., and Lee S. Y. (2015) Glycogen synthase kinase 3β ubiquitination by TRAF6 regulates TLR3-mediated pro-inflammatory cytokine production. Nat. Commun. 6, 6765. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44. Teixeira F. R., Randle S. J., Patel S. P., Mevissen T. E., Zenkeviciute G., Koide T., Komander D., and Laman H. (2016) Gsk3β and Tomm20 are substrates of the SCFFbxo7/PARK15 ubiquitin ligase associated with Parkinson's disease. Biochem. J. 473, 3563–3580 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45. Martin M., Rehani K., Jope R. S., and Michalek S. M. (2005) Toll-like receptor-mediated cytokine production is differentially regulated by glycogen synthase kinase 3. Nat. Immunol. 6, 777–784 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46. Liu Z., Bone N., Jiang S., Park D. W., Tadie J.-M., Deshane J., Rodriguez C. A., Pittet J.-F., Abraham E., and Zmijewski J. W. (2015) AMP-activated protein kinase and glycogen synthase kinase 3β modulate the severity of sepsis-induced lung injury. Mol. Med. 21, 937–950 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47. Beurel E., and Jope R. S. (2009) Lipopolysaccharide-induced interleukin-6 production is controlled by glycogen synthase kinase-3 and STAT3 in the brain. J. Neuroinflammation 6, 9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48. Peng D., Wang Z., Huang A., Zhao Y., and Qin F. X. (2017) A novel function of F-box protein FBXO17 in negative regulation of type I IFN signaling by recruiting PP2A for IFN regulatory factor 3 deactivation. J. Immunol. 198, 808–819 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49. Mialki R. K., Zhao J., Wei J., Mallampalli D. F., and Zhao Y. (2013) Overexpression of USP14 protease reduces I-κB protein levels and increases cytokine release in lung epithelial cells. J. Biol. Chem. 288, 15437–15441 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50. Chen B. B., and Mallampalli R. K. (2009) Masking of a nuclear signal motif by monoubiquitination leads to mislocalization and degradation of the regulatory enzyme cytidylyltransferase. Mol. Cell Biol. 29, 3062–3075 [DOI] [PMC free article] [PubMed] [Google Scholar]