Abstract
Photoactivated adenylyl cyclase (PAC) and guanylyl cyclase rhodopsin increase the concentrations of intracellular cyclic nucleotides upon illumination, serving as promising second-generation tools in optogenetics. To broaden the arsenal of such tools, it is desirable to have light-activatable enzymes that can decrease cyclic nucleotide concentrations in cells. Here, we report on an unusual microbial rhodopsin that may be able to meet the demand. It is found in the choanoflagellate Salpingoeca rosetta and contains a C-terminal cyclic nucleotide phosphodiesterase (PDE) domain. We examined the enzymatic activity of the protein (named Rh-PDE) both in HEK293 membranes and whole cells. Although Rh-PDE was constitutively active in the dark, illumination increased its hydrolytic activity 1.4-fold toward cGMP and 1.6-fold toward cAMP, as measured in isolated crude membranes. Purified full-length Rh-PDE displayed maximal light absorption at 492 nm and formed the M intermediate with the deprotonated Schiff base upon illumination. The M state decayed to the parent spectral state in 7 s, producing long-lasting activation of the enzyme domain with increased activity. We discuss a possible mechanism of the Rh-PDE activation by light. Furthermore, Rh-PDE decreased cAMP concentration in HEK293 cells in a light-dependent manner and could do so repeatedly without losing activity. Thus, Rh-PDE may hold promise as a potential optogenetic tool for light control of intracellular cyclic nucleotides (e.g. to study cyclic nucleotide-associated signal transduction cascades).
Keywords: biophysics, cyclic nucleotide, enzyme mechanism, membrane enzyme, optogenetics, phosphodiesterases, photoreceptor, rhodopsin
Introduction
Rhodopsins, heptahelical membrane proteins containing retinal as chromophore, are subdivided into the animal and microbial types (1). Whereas animal rhodopsins serve almost exclusively as G-protein coupled receptors (2–5), the functions of microbial rhodopsins are diverse, including light-driven cation and anion pumps, light-gated cation and anion channels, positive and negative phototaxis sensors, photochromic sensors, and light-activated enzymes (1, 6–9). The first discovered light-activated enzyme was an algal histidine kinase (HK)3 (10), whereas fungal light-activated guanylyl cyclase (GC) was reported recently (11–13). These proteins are composed of a membrane-embedded rhodopsin domain and a C-terminal cytoplasmic enzyme domain, activated by light absorption by the all-trans-retinal chromophore. The molecular mechanisms of enzyme rhodopsin activation are understood much less than those of other microbial rhodopsins, mainly because of the difficulty of preparing functional full-length proteins. In fact, the light-dependent enzymatic activity has never been demonstrated for Rh-HK, and detailed spectroscopic study of Rh-GC photochemistry has been performed only for the transmembrane domain without the soluble enzyme domain (12).
Optogenetics has revolutionized life sciences, due to its ability to control various biological activities by light (14–18). Since the early stages of rhodopsin-based optogenetics, light-gated channels and light-driven ion pumps have been used to excite and silence neurons (19, 20). This is highly advantageous for applications in neuroscience when fast temporal resolution is required to change transmembrane potential. In contrast, for optogenetic control of a wider spectrum of biological functions, light-induced enzyme activation is more attractive, because intracellular signaling processes can be manipulated by light. In particular, optogenetic control of second messengers, such as Ca2+ and cyclic nucleotides, is in high demand.
Photoactivated adenylyl cyclase (PAC) with flavin adenine nucleotide (FAD) chromophore was discovered in Euglena gracilis in 2002 (21). It could be used as an optogenetic tool, which increases intracellular cAMP concentration under blue light (22–25). More recently, a microbial rhodopsin containing a GC domain was discovered in Blastocladiella emersonii (11) and applied to optogenetics (12, 13). In contrast to these light-activated proteins able to increase cAMP and cGMP concentrations, an efficient native light-dependent enzyme capable of lowering cyclic nucleotide concentration has not been known until now. It should be noted that engineered light-activated cyclic nucleotide phosphodiesterase (PDE) (26) and Gi-coupled animal opsin (27) are able to lower cyclic nucleotide concentration upon illumination. The former combines a GAF (cGMP-binding PDEs, adenylyl cyclase, and FhlA) domain and PDE domain, which allows control of the PDE activity by light (26). However, because of the photochromic nature of the GAF domain, its light-induced enzymatic activity has to be stopped by light of a different color. Gi-coupled animal opsin from mosquitos (Opn3) decreases cAMP concentration under illumination, because Gi inhibits the activity of adenylyl cyclase (AC) (27). On the other hand, enzymatic activity of mosquito Opn3 persists for several tens of minutes (27), being much longer than that of Rh-GC (12, 13), presumably because of the AC-related signaling network. In addition, GPCRs regulate other signaling cascades through Gβ,γ and arrestin, triggering complex intracellular events. Therefore, proteins composed of a light-sensor domain and a PDE domain, with fast enzymatic turnover induced by single light excitation, are highly desirable.
In this paper, we report on a novel type of microbial rhodopsin with a C-terminal PDE domain, found in the choanoflagellate Salpingoeca rosetta, a unicellular and colonial single flagellate eukaryote, considered to be one of the closest living relatives of metazoans. We successfully expressed and purified the full-length Rh-PDE from mammalian cells, and its enzymatic functions were studied in cells, in membranes, and in detergent, both biochemically and spectroscopically. We discuss a putative molecular mechanism of Rh-PDE activation based on the presented studies of the full-length protein and its applicability to optogenetics.
Results
A new microbial rhodopsin containing a PDE domain
The genome of the choanoflagellate S. rosetta (sequenced by the Broad Institute, NCBI accession number PRJNA37927) contains an unusual gene (NCBI Gene ID 16078606) encoding a rhodopsin fused to a C-terminal PDE domain (Rh-PDE). Fig. 1A shows a schematic drawing of the Rh-PDE architecture, which is composed of an N-terminal seven-transmembrane helical rhodopsin domain and a cytoplasmic PDE domain (see supplemental Fig. S1 for the complete amino acid sequence). It is possible that Rh-PDE exists as a dimer, as is typical for known PDEs (28–30). From the sequence analysis, we propose that Rh-PDE is a microbial rhodopsin that binds all-trans-retinal as a chromophore, where light absorption triggers protein structural changes to induce (or modulate) the enzymatic activity.
Figure 1.
Architecture and amino acid composition of Rh-PDE. A, Rh-PDE is composed of the seven-transmembrane helical rhodopsin domain and the cytoplasmic PDE domain. B, phylogenetic tree of selected microbial rhodopsins based on the sequences of the transmembrane domains. C, conservation of important residues of the Rh domain. Red and blue colors represent acidic and basic amino acids, respectively. Residue numbers in Rh-PDE and BR are shown. D, conservation of important residues of the PDE domain. Residue numbers in Rh-PDE and PDE10A are shown, and sequences are grouped according to the substrate specificity.
Fig. 1B shows a phylogenetic tree of selected microbial rhodopsins, based on the amino acid sequences of their transmembrane domains. Rh-PDE clusters with other enzyme rhodopsins of lower eukaryotes (e.g. histidine kinases and guanylate cyclases). Many common residues of microbial rhodopsins are conserved in Rh-PDE (Fig. 1C; see supplemental Fig. S2 for the complete alignment). Specifically, Asp-85, the primary counterion of the protonated Schiff base in light-driven proton pump bacteriorhodopsin (BR), is conserved as Glu-164 in Rh-PDE. The residues constituting the all-trans-retinal binding pocket of BR (Trp-86, Thr-89, Trp-182, and Tyr-185) (1) are also conserved as Trp-165, Thr-168, Trp-264, and Phe-267, respectively, in Rh-PDE. The PDE domain contains many conserved residues typical for the active centers of soluble PDEs (Fig. 1D), but it is not possible to predict the substrate specificity of Rh-PDE (cAMP, cGMP, or both) from the sequence.
Enzymatic activity of Rh-PDE in mammalian cells
To investigate enzyme rhodopsins of lower eukaryotes, it is important to find a suitable expression system. Previous studies on Rh-GC used yeast expression, where the full-length protein could not be obtained (12). In this study, we first measured enzymatic activity of Rh-PDE using HEK293 cells, from which the full-length protein was subsequently purified and characterized in vitro. To measure the in-cell enzymatic activity of Rh-PDE, we used the GloSensor assay, which is based on a cyclic nucleotide-dependent luciferase. In this assay, the Rh-PDE-expressing cells were first incubated in the serum-containing culture medium after the addition of all-trans-retinal in the dark at room temperature (∼27 °C), and the luminescence intensity was increased by sodium nitroprusside or forskolin, which activate GC or AC, resulting in elevation of the cytoplasmic cGMP or cAMP level, respectively.
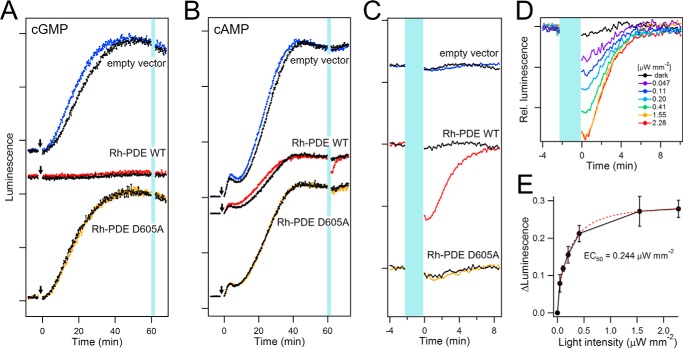
Fig. 2A shows the activity of Rh-PDE toward cGMP. In the control experiments, the luminescence intensity gradually increased after the addition of sodium nitroprusside and reached a plateau after 40 min for HEK293 cells expressing GloSensor only (empty vector). As expected, there was no effect of light on the level of cGMP in this system (Fig. 2A, top). In contrast, such a gradual increase of luminescence after addition of sodium nitroprusside has not been observed upon expression of Rh-PDE (Fig. 2A, middle), which suggests constitutive activity of Rh-PDE toward cGMP, both in the dark and upon illumination. Consistent with this idea, light-independent gradual increase of luminescence occurred for the D605A mutant (Fig. 2A, bottom), with inactivating replacement in the active center of the PDE domain.
Figure 2.
Enzymatic activity of Rh-PDE in HEK293 cells. A, luminescence signals representing cGMP levels, as observed in HEK293 cells with the empty vector (top), Rh-PDE (middle), and the PDE-inactive D605A mutant (bottom). HEK293 cells were preincubated in the culture medium with all-trans-retinal in the dark. The arrows and vertical light-blue line indicate sodium nitroprusside treatments and irradiation at 510 nm with 2.28 μW mm−2 intensity, respectively, and the luminescence signals of non-irradiated cells are shown as a control (black lines). B, luminescence signals representing cAMP level, as observed in HEK293 cells with the empty vector (top), Rh-PDE (middle), and the PDE-inactive D605A mutant (bottom). HEK293 cells were preincubated in the culture medium with all-trans-retinal in the dark. The arrows and vertical light-blue line indicate forskolin treatments and irradiation at 510 nm with 2.28 μW mm−2 intensity, respectively, and the luminescence signals of non-irradiated cells are shown as a control (black lines). Only HEK293 cells expressing Rh-PDE (red line in the middle panel) exhibited a decrease of the luminescence signal, indicating that cAMP concentration is lowered by light. C, changes of luminescence signals upon 2-min 510-nm irradiation (light-blue line) of HEK293 cells with the empty vector (top), Rh-PDE (middle), the PDE-inactive D605A mutant (bottom), expanded from B. Colored and black lines represent the data with and without irradiation, respectively. D, intensity dependence of the light-induced cAMP concentration decrease by Rh-PDE (cells were irradiated for 2 min with 510-nm light with 0–2.28 μW mm−2 intensities). E, peak amplitudes of luminescence decrease plotted against the light intensity (n ≧ 5 cells). Error bars, S.D. The half-maximal light intensity (EC50) was determined to be 0.244 μW mm−2 by a single exponential decay fit (red dashed line).
Fig. 2, B and C, shows the activity of Rh-PDE toward cAMP. The luminescence intensity gradually increased after the addition of forskolin and reached a plateau after 40 min, similar to the cGMP assay (Fig. 2B). At that time point, illumination at 510 nm with 2.28 μW mm−2 intensity caused a decrease of the luminescence in cells expressing Rh-PDE (red line) but not in those with the empty vector (blue line) (Fig. 2, B and C). 28 ± 2.3% (S.D.) reduction in the luminescence intensity was reproducibly observed under these experimental conditions (n = 6). Thus, it can be concluded that the decrease of cAMP concentration must originate from the light-induced Rh-PDE activity, which is further supported by the lack of signal for the catalytically inactive D605A mutant (Fig. 2 (B and C), yellow line). It should be noted that after the light was switched off, the light-induced luminescence decrease persisted for a significant amount of time (∼10 s) and then decayed with τ1/e of 195 s (Fig. 2C, red line). This kinetics involves at least two components, deactivation of Rh-PDE and production of cAMP by endogenous adenylyl cyclases. Based on the exponential fitting, we estimate the lifetime of the light-activated state of Rh-PDE as about 71 s (supplemental Fig. S3). Another notable feature is that the stationary luminescence intensity in the dark was smaller for Rh-PDE (Fig. 2B, middle) than for the empty vector and the D605A mutant (Fig. 2B, top and bottom), suggesting constitutive dark activity toward cAMP, although smaller than toward cGMP. Fig. 2D shows that the light-induced reduction of luminescence increased with the applied light intensity, and saturated with an EC50 of 0.244 μW/mm2 (Fig. 2E). One could expect that the intensity increase was a sigmoidal function if any two-photon processes were involved (e.g. both protein molecules in a dimer need to be activated). Nearly linear increase at lower light intensities in Fig. 2E suggests that Rh-PDE activation is a single-photon event, and activation of one molecule is sufficient even if Rh-PDE is likely to form a dimer. In summary, the in-cell assays demonstrated that Rh-PDE shows strong constitutive activity toward cGMP, whereas the activity toward cAMP is light-dependent.
Suitability of Rh-PDE for optogenetic applications
The observed light-dependent decrease of intracellular cAMP concentration suggests that Rh-PDE may be useful as an optogenetic tool. Together with PAC and Gs-coupled opsins capable of increasing intracellular cAMP concentration, Rh-PDE increases the diversity of the optogenetics toolkit. Unlike the engineered light-activated PDE (26), Rh-PDE is switched off by its photocycle, without the need for an additional illumination. On the other hand, Gi-opsin, which can decrease intracellular cAMP concentration (27), also switches off in the dark, warranting a comparison of the properties of Rh-PDE and Gi-opsin.
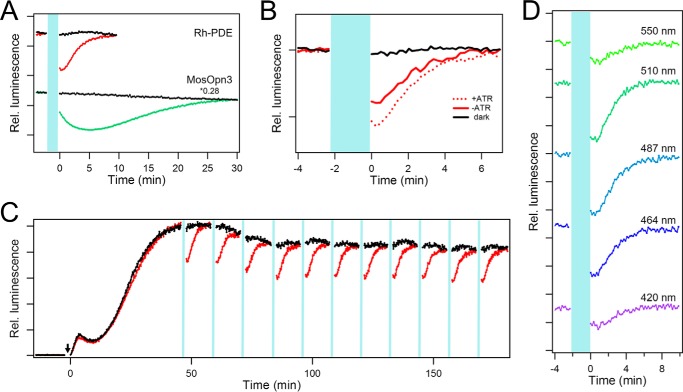
Thus, we expressed mosquito Opn3 in HEK cells with the addition of all-trans-retinal, using the same protocol as for Rh-PDE. Although the native chromophore of mosquito Opn3 is 11-cis-retinal, it is known that this opsin binds 13-cis-retinal, which can be easily formed from all-trans-retinal, and that the regenerated pigment shows an absorption maximum at 500 nm (27). Upon application of the 510-nm light, the cells expressing mosquito Opn3 show a strong decrease in the intracellular cAMP concentration, which continues to decrease after the light is switched off, reaching a minimum in ∼5 min, and then recovers with τ1/e of 16 min (Fig. 3A, green line). These data reproduce the data reported previously (27), and it is obvious that the luminescence signal is smaller and faster (both in the rise and the decay) in the case of Rh-PDE under the same illumination conditions (Fig. 3A, red line). In the case of mosquito Opn3, intermolecular protein-protein interactions between opsin and Gi and between Gi and AC should amplify the signal. Thus, as an optogenetic tool, Rh-PDE exhibits a less sensitive, but about 5 times faster, response than mosquito Opn3. In addition, Rh-PDE only acts on cyclic nucleotides, whereas additional intracellular effects of mosquito Opn3 are unclear.
Figure 3.
Rh-PDE as a potential optogenetic tool. A, comparison of in-cell hydrolytic activities of Rh-PDE (top) and mosquito Opn3 (bottom) toward cAMP. The top panel is reproduced from Fig. 2C. Mosquito Opn3 was expressed in HEK293 cells similar to Rh-PDE. During the time period shown in light blue, 510-nm light with 2.28 μW mm−2 intensity was applied to the cells. B, effect of the addition of all-trans-retinal on the Rh-PDE activity. C, repeatability of the in-cell Rh-PDE activation. 2-min irradiation at 510 nm with 2.28 μW mm−2 intensity was repeated every 15 min, and other conditions were as in Fig. 2C. D, irradiation wavelength dependence of the in-cell activity of Rh-PDE toward cAMP. All of the data were obtained from experiments at the same light intensity (0.4 μW mm−2).
Another important parameter for optogenetic applications is chromophore availability. We compared the activity of Rh-PDE toward cAMP with and without the addition of all-trans-retinal in the cell culture medium after transfection (Fig. 3B). Although the luminescence decrease is smaller without the addition of all-trans-retinal, it still retains robust activity at 70% of the amplitude upon its addition, which is explained by the fact that all-trans-retinal is commonly present in cells. Although mosquito Opn3 11-cis-retinal chromophore is not abundant in cells, Koyanagi et al. (27) found that mosquito Opn3 can bind 13-cis-retinal, a ubiquitously present retinal isomer. Because 13-cis- and all-trans-retinals exist in photoequilibrium, mosquito Opn3 is usable in general tissues, although its activity without the addition of 11-cis-retinal was reduced > 2-fold (27). In contrast, Rh-PDE binds naturally abundant all-trans-retinal, and the active state is rapidly reverted to the resting state as a result of the photocycle. This comes along with the high repeatability, another important property for optogenetics. Fig. 3C shows that activation of Rh-PDE can be repeated for longer than 2 h, showing the suitability of Rh-PDE as an optogenetic tool. To define the usable spectral range of Rh-PDE, we systematically changed the irradiation wavelength at the same light intensity (Fig. 3D) and found that the luminescence is maximally decreased by the blue-green light (480–510 nm).
HPLC analysis of the enzymatic activity of Rh-PDE
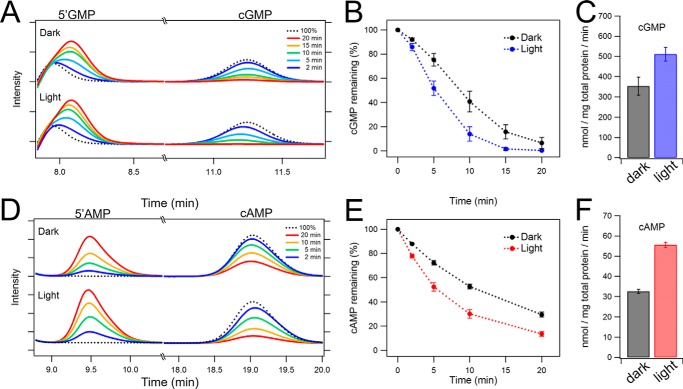
We further characterized the enzymatic activity of Rh-PDE by HPLC analysis using crude membranes of HEK293 cells. In these experiments, cGMP or cAMP was mixed with the crude cell membranes expressing Rh-PDE, and time- and light-dependent hydrolysis of cyclic nucleotides into 5′-GMP or 5′-AMP was monitored by HPLC. Fig. 4A shows the HPLC profile for cGMP, where Rh-PDE decreases the amount of cGMP and concomitantly increases that of 5′-GMP. Importantly, a 10-min incubation under illumination results in an 85% decrease of cGMP concentration, compared with only 60% in the dark (Fig. 4B). This indicates that Rh-PDE has light-dependent enzymatic activity toward cGMP, although its dark constitutive activity is significant. The hydrolysis rates were calculated from the slopes of the curves (0–10 min) in Fig. 4B as 353 nmol/mg total protein/min in the dark and 511 nmol/mg total protein/min under illumination, giving 142% activation by light.
Figure 4.
HPLC analysis of the enzymatic activity of Rh-PDE. A, HEK293 cell membranes expressing Rh-PDE were incubated with 100 μm cGMP for 2 min (blue), 5 min (light blue), 10 min (green), 15 min (orange), and 20 min (red) in the dark (top) and under the illumination (bottom), and the reaction products were analyzed by HPLC. Dotted black line, profile for 100 μm cGMP without Rh-PDE. B, time course of the decrease of cGMP concentration in the dark (black) and under illumination (blue) (n = 3). C, cGMP hydrolysis activity in the dark and under the illumination calculated from B. D, HEK293 cell membranes expressing Rh-PDE were incubated with 100 μm cAMP for 2 min (purple), 5 min (light blue), 10 min (green), and 20 min (red) in the dark (top) and under illumination (bottom), and the reaction products were analyzed by HPLC. Dotted black line, profile for 100 μm cAMP without Rh-PDE. E, time course of the decrease of cAMP concentration in the dark (black) and under illumination (red) (n = 3). F, cAMP hydrolysis activity in the dark and under illumination calculated from E. Error bars, S.D.
Fig. 4D shows the HPLC profiles for cAMP in the dark (top) and under illumination (bottom). Rh-PDE decreases cAMP concentration in both cases, but the activity is smaller compared with that for cGMP. It should be noted that a 10-fold higher amount of the membranes was used for the cAMP hydrolysis assay (see “Experimental procedures”). A clear light-dependent activation can be seen for the cAMP hydrolysis, as was observed in the cGMP assay above. The hydrolysis rates were calculated as 32.6 nmol/mg total protein/min in the dark and 53.6 nmol/mg total protein/min under illumination, giving ∼164% stimulation by light (Fig. 4, E and F). The activity toward cGMP is ∼10-fold higher than toward cAMP (Fig. 4), consistent with the in-cell results.
Hydrolysis of cGMP and cAMP was entirely abolished in the D605A mutant, with inactivating replacement in the active center of the PDE domain (data not shown). Although crude cell membranes contain many other proteins, the comparison between the wild-type and the D605A mutant Rh-PDE indicates that the observed hydrolysis of both nucleotides originates exclusively from Rh-PDE, consistent with the in-cell results (Fig. 2).
Molecular properties of purified full-length Rh-PDE
The in-cell and in-membrane studies described above showed that the enzymatic activity of Rh-PDE is stimulated by light toward both cAMP and cGMP. To gain insight into the molecular mechanism of photoactivation, it is necessary to purify and characterize the full-length protein. Although expression levels of rhodopsins are usually much smaller in HEK293T cells than in yeast and Escherichia coli, previously, we have been able to conduct infrared spectroscopic studies on monkey red- and green-sensitive pigments based on HEK293 cell expression (31, 32). We thus applied the sample preparation procedure as used for monkey red- and green-sensitive pigments to Rh-PDE, using HEK293T cell expression, solubilization by dodecyl-β-d-maltopyranoside (DDM), and purification by monoclonal antibody column chromatography, with all-trans-retinal being added into the culture medium. As a result, we were able to obtain purified full-length protein (Fig. 5A, inset) with yields comparable with those of monkey color pigments (0.4 mg of Rh-PDE versus 0.3 mg of monkey color pigments from 300 culture plates of HEK293 cells with a 10-cm diameter (31, 32)).
Figure 5.
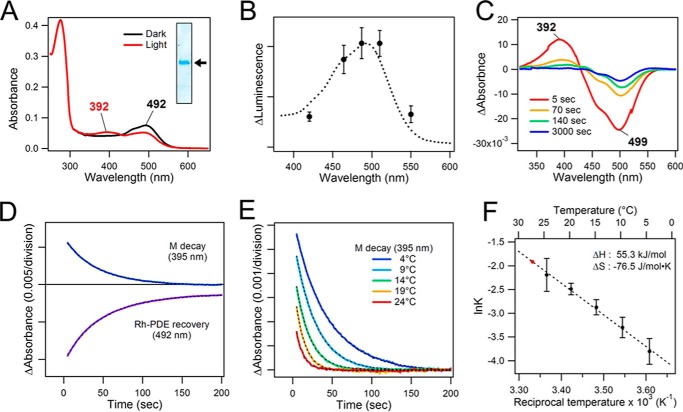
Molecular properties of purified full-length Rh-PDE. A, absorption spectra of full-length Rh-PDE in the dark (black) and after irradiation with 510-nm light (red) at 4 °C. Inset, Coomassie Brilliant Blue-stained purified Rh-PDE on 12% SDS-PAGE. The arrow corresponds to about 70 kDa. B, absorption spectrum of purified full-length Rh-PDE (dotted line; reproduced from the black line in A) superimposed with the action spectrum of the light-induced enzymatic activity toward cAMP in HEK293T cells (the peak amplitudes in Fig. 3D are plotted versus irradiation wavelength with 0.4 μW mm−2 intensity). C, difference absorption spectra of the photoactivation. D, M decay and the parent state recovery kinetics at 4 °C. E, temperature dependence of the M decay. F, Arrhenius plot for the M-decay kinetics. From the linear fit (dotted line), the decay time constant at 27 °C was estimated to be 6.78 s (red point). Error bars, S.D.
We characterized the photoreactions of the purified full-length Rh-PDE using UV-visible spectroscopy. Fig. 5A shows the absorption spectrum of Rh-PDE in the dark, with the λmax located at 492 nm (black). This absorption spectrum shows good correlation with the action spectrum obtained from the GloSensor assays (Fig. 5B) using the wavelength dependence of the light-induced Rh-PDE activity toward cAMP (Fig. 3D). The spectrum possesses a shoulder at about 460 nm, which resembles that of sensory rhodopsin II (SRII) (33). The absorption at 280 nm is 4.8 times higher than that at 492 nm, which is significantly higher than the value of ∼2 known for most microbial rhodopsins. Among the possible reasons is the additional large extramembrane domain, incomplete chromophore regeneration or bleaching, and/or co-purified proteins other than Rh-GC (see “Experimental procedures” for details). Upon the irradiation, the absorption peak at 492 nm decreases, and a new absorption band appears in the 350–420 nm region. It should be noted that the measurements shown in Fig. 5A were performed at 4 °C because the decay of the blue-shifted photointermediate was too fast to record its spectrum at room temperature. Fig. 5C shows the difference spectra of the photoactivation and its relaxation, demonstrating bleaching of the dark state at 499 nm and concomitant appearance of the intermediate state with the maximum at 392 nm. This peak wavelength suggests the light-induced deprotonation of the retinal Schiff base, in a state known as the M intermediate, which decays to the original spectral state.
Fig. 5D represents the M decay and the parent state recovery kinetics at 4 °C, with τ1/e of about 45 s (43.1 s at 395 nm and 48.7 s at 492 nm). It should be noted that the initial absorption at 492 nm does not fully recover after the complete decay of the M intermediate, indicating some irreversible processes. We infer that Rh-PDE in detergent tends to form aggregates (and/or partially bleaches) upon continuous illumination, which does not occur in membranes, judging from the perfect repeatability of the in-cell Rh-PDE activation (Fig. 3C). To estimate the M decay kinetics under the conditions of the enzyme activity experiments (27 °C), we measured its temperature dependence. As expected, the M decay becomes faster at higher temperatures (Fig. 5E), and the temperature dependence follows the Arrhenius behavior well (Fig. 5F), with an activation energy of 57.8 kJ/mol, and activation enthalpy and entropy of 55.3 kJ/mol and −76.5 J/mol·K, respectively. We estimated the M decay time constant at 27 °C to be 6.78 s, which suggests that the enzymatic activity persists well after its decay.
We co-expressed the 1D4-tagged Rh-PDE and its eGFP-tagged version in HEK293 cells followed by solubilization and purification by 1D4-agarose resin. The eluate contained not only the 1D4-tagged protein but also the eGFP-tagged version, judging from the fluorescence spectrum and SDS-PAGE (supplemental Fig. S4). This result indicates that Rh-PDE forms stable oligomers that survive solubilization.
Discussion
We report on a novel type of microbial rhodopsin from the choanoflagellate S. rosetta containing a C-terminal PDE domain. Animal rhodopsins and sensory microbial rhodopsins have their respective transducers (G proteins, Htr membrane transducers, and others) as separate proteins (1–8). In contrast, enzyme rhodopsins such as Rh-HK, Rh-GC, and Rh-PDE contain covalently bound enzymatic domains, whose activation mechanism is unknown and intriguing. Above, we presented data showing that Rh-PDE exhibits light-dependent hydrolysis activity both toward cGMP and cAMP while retaining some levels of constitutive activity in the dark. The data in Fig. 4 show that Rh-PDE has higher activity toward cGMP than toward cAMP, both in the dark and in the light-activated form. It is known that substrate specificity differs among PDE families, as shown in Fig. 1D (28). The observed dark activity was not anticipated, because the light-sensitive rhodopsin domain was expected to contribute to the tight regulation of the enzymatic activity. In fact, the Rh-GC exhibits a very tight light-dependent cyclase activity, in which the enzyme has only 1% basal activity in the dark (12). Whether or not the high constitutive activity of Rh-PDE is derived from the background activity of the PDE domain, from a specific conformation of the rhodopsin part, or from instability of the oligomer in the heterologous expression system will have to be addressed in future studies. At this point, we believe that it is logical to suggest that the membrane and cellular environments differ between S. rosetta and HEK293 cells, preventing hydrolytic activity in the dark in the former. For example, it is possible that the PDE domain is post-translationally modified (phosphorylated), or other regulatory protein(s) associate with it and keep Rh-PDE inactive in S. rosetta in the dark.
During the in-cell cyclic nucleotide concentration measurements by the GloSensor assay, a light-dependent cAMP decrease was observed (Fig. 2), consistent with the results obtained with crude membranes shown in Fig. 4. In contrast, we did not observe any light-dependent change of the intracellular cGMP level in the GloSensor assay in Fig. 2A but observed an enhancement of the Rh-PDE cGMP activity in crude membranes by light. This can be explained, taking into account that the cGMP hydrolysis rate by Rh-PDE is about 10-fold faster than that for cAMP (Fig. 4, C and F). Thus, the dark activity toward cGMP is high enough to completely degrade cellular cGMP, preventing observation of the effects of light. In addition, the concentration of endogenous cGMP is about 10-fold lower than that of cAMP, which is another plausible reason for our observations.
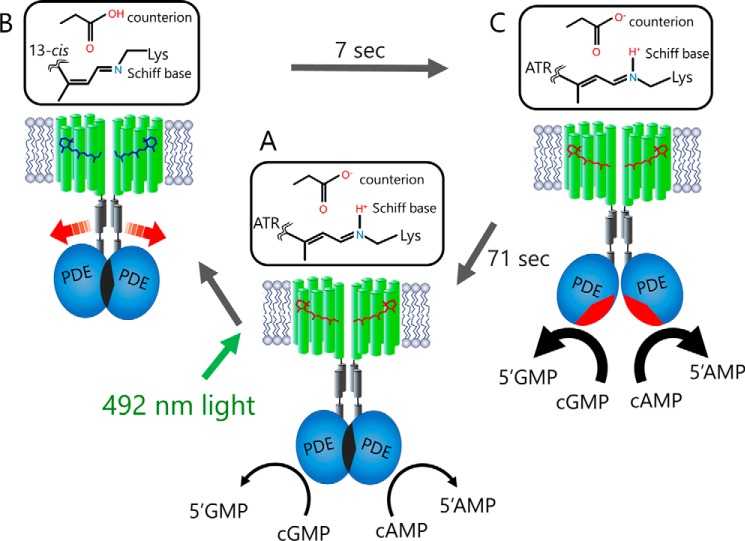
Based on the presented results, we propose the activation mechanism of Rh-PDE shown in Fig. 6. We suggest that Rh-PDE exists in a putative dimeric form, consistent with the reported PDE structures (21, 22). The crystal structure of the full-length PDE-2A, where the elongated C-terminal helix of the GAF domain directly connects to the catalytic region of the PDE domain (30), may offer some insights into the architecture of Rh-PDE. In Rh-PDE, the linker region between the end of transmembrane helix 7 of the rhodopsin domain and the beginning of the catalytic region contains 50–60 residues (supplemental Fig. S1), which corresponds to 7–8 nm in length, assuming a single helix. On the other hand, it is well known that the HAMP (histidine kinases, adenylyl cyclases, methyl-accepting chemotaxis proteins, and phosphatases) domain, which connects transmembrane and cytosolic signaling domains in many sensory proteins, forms a coiled-coil bundle of two amphipathic helices (34, 35). Indeed, two such helical amphipathic sequences can be seen in the linker region of Rh-PDE (supplemental Fig. S1). Therefore, we suggest that transmembrane helix 7 of the Rh domain elongates and forms the HAMP-like domain, which connects to the catalytic region of the PDE domain (Fig. 6). Dimer formation of the PDE domain makes the catalytic region inaccessible to the cytoplasmic bulk, so that the PDE activity remains shut off in the dark (a condition which may not be fully reproduced in experiments with HEK293 cells).
Figure 6.
Schematic representation of the Rh-PDE activation mechanism. The model shows a putative Rh-PDE dimer. A, in the dark, Rh-PDE exhibits constitutive activity toward cGMP and cAMP, suggesting that the catalytic region is somewhat exposed to the cytoplasm in mammalian cells. B, upon absorption of blue-green light by the Rh domain, photoisomerization of the retinal chromophore from all-trans-retinal (ATR) to the 13-cis form produces the M intermediate with deprotonated Schiff base (Rh domain switch on), but the PDE domain has not changed yet (PDE domain switch off). C, next, the M intermediate decays to the parent spectral state in 7 s (Rh domain switch off), accompanied by the structural changes in the PDE domain (PDE domain switch on). The resulting increased enzymatic activity is turned off within ∼70 s (PDE domain switch off).
The resting state of Rh-PDE shows the λmax at 492 nm, which coincides well with the action spectrum of the light-induced enzymatic activity (Fig. 5B). The absorption in the visible region indicates that the chromophore Schiff base is protonated in Rh-PDE. In light-driven proton pump BR, the protonated Schiff base, Arg-82, Asp-85, and Asp-212 constitute a quadrupole inside the protein, where Asp-85 acts as a primary counterion of the Schiff base (1). Similarly, the Schiff base, Arg-161, Glu-164, and Asp-292 probably form a quadrupole in Rh-PDE (Fig. 1C). The absorption spectrum is considerably blue-shifted compared with other microbial rhodopsins and is close to those of SRII and channelrhodopsin-2 (ChR2). The position corresponding to Ala-215 in BR is Thr in SRII, whose side chain OH group contributes to the blue spectral shift (36, 37). ChR2 has O-H bearing Ser at the homologous position as well, but Rh-PDE has Ala (Fig. 1C), so that the mechanism of its blue-shifted absorption is not clear.
After the photoexcitation, the M intermediate with deprotonated retinal Schiff base forms in the Rh-PDE photocycle (Fig. 5, A and C), similar to many other microbial and animal rhodopsins. In many cases, proton transfer from the Schiff base induces large structural changes, leading to the respective functional processes (1). In Rh-PDE, the M formation probably triggers protein structural changes at the cytoplasmic surface of the rhodopsin domain, which results in the light-induced increase in PDE activity. The observed increase in the in-cell enzymatic activity toward cAMP for ∼10 s after the light was switched off (Fig. 2C) coincides with the time scale of the M decay (7 s; Fig. 5F), suggesting that the enzymatic activity is not yet induced in the PDE domain in the M intermediate. In other words, structural changes of the Rh domain in the M intermediate lead to those of the PDE domain at the later times after photoactivation, presumably altering the structure of helix 7 and the HAMP-like domain (Fig. 6). Linear light intensity dependence of the activity toward cAMP (Fig. 2E) suggests that activation of just one Rh domain in a putative dimer is sufficient to trigger an increase in the enzymatic activity. An alternative (although less plausible) explanation for the linear intensity dependence is that Rh-PDE is monomeric in the mammalian membranes, which could also explain its significant dark activity, although this contradicts our results with eGFP-tagged Rh-PDE, which suggest oligomerization (supplemental Fig. S4).
The recovery of the original spectral state occurs parallel to the M decay (Fig. 5D), suggesting that the photocycle completes in ∼7 s at 27 °C (∼45 s at 4 °C). This fact appears to contradict the long-lived enzymatic activity (71 s) of PDE in cells (Fig. 2C and supplemental Fig. S3). Because the light absorption originates from the retinal chromophore in the membrane core of the Rh domain, it may be not sensitive to the conformation of the PDE domain, which probably switches on upon the transition from the M intermediate to the initial spectral state in the Rh domain. Structural changes of the Rh domain, particularly in helix 7 (and ensuing changes in the HAMP-like domain) possibly dissociate the PDE dimer, leading to the two catalytic domains being exposed to the aqueous phase (Fig. 6). The photocycles of microbial rhodopsins are generally too short to keep sustained enzymatic activity, so that a special mechanism is needed to achieve this. The presence of the two domains, Rh and PDE, connected through the HAMP-like linker could allow for 10 times longer enzymatic activity than the photocycle of the Rh domain alone.
The molecular mechanism of the light-activated sensory function has been extensively studied for SRII, which forms a 2:2 complex with two transmembrane helical Htr transducer (38, 39). The linker region of the transducer contains a HAMP-like domain. The M-intermediate structure of the receptor-transducer complex (40) and numerous EPR studies (41, 42) revealed significant outward tilting of the cytoplasmic half of helix F in SRII, which causes rotational motion of the second transmembrane helix of the transducer. Outward motion of helix F on the cytoplasmic side is common for activation of microbial and animal rhodopsins and also causes motion of helix G. Presently, there is no structural information on Rh-PDE, and further studies will be needed to reveal the activation mechanism of the PDE domain by light absorption in the Rh domain.
Light control of second messengers is in high demand in optogenetics, and PAC (22–25) and Rh-GC (12, 13) are promising tools for increasing intracellular concentration of cyclic nucleotides by light. In addition to the engineered light-activated PDE (26) and mosquito Opn3 (27), Rh-PDE may become a promising optogenetics tool, but all of them have their specific drawbacks. The enzymatic activity of the engineered light-activated PDE has to be stopped by additional irradiation of a different color due to the photochromic nature of the GAF domain. Although the chromophore of mosquito Opn3 is 11-cis-retinal, not abundant in mammalian cells, it can be used for optogenetics because it can bind 13-cis-retinal. Mosquito Opn3 is more sensitive and has a longer activation time than Rh-PDE, because of the amplification processes. In addition, mosquito Opn3 regulates other signaling cascades through Gβ,γ and arrestin, triggering complex intracellular events. In contrast, Rh-PDE regulates only catalysis of cyclic nucleotides hydrolysis. Whereas Rh-PDE lowers cAMP concentration by light, its constitutive activity would be problematic for optogenetic applications, because Rh-PDE maintains very low intracellular cGMP levels. This requires further efforts to make it suitable for optogenetic applications, and additional engineering of the active center of the PDE domain is possibly needed.
Experimental procedures
Molecular biology
A full-length gene encoding Rh-PDE (NCBI Gene ID 16078606) was synthesized after a human codon optimization (GenScript) and cloned into peGFP-N1 vector between HindIII and BamHI sites. We amplified the Rh-PDE gene by PCR with the forward primer encoding the first 20 bp and the reverse primer encoding the last 15 bp of the Rh-PDE gene followed by the 1D4 epitope tag (TETSQVAPA) sequence. The amplified fragment was inserted into pCDNA3.1 vector (Invitrogen) between HindIII and XbaI sites. The plasmid for D605A Rh-PDE was generated by QuikChange site-directed mutagenesis kit (Agilent Technologies). The MosOpn3 gene, which lacks 99 C-terminal amino acids (27), was synthesized after a human codon optimization (GenScript) and fused to the cDNA encoding a Venus fluorescent protein and cloned into pCDNA3.1 vector between HindIII and XbaI sites. All constructs were verified by the DNA sequencing. Expression of Rh-PDE with the 1D4 epitope tag was confirmed by Western blotting.
Assay of the enzymatic activity of Rh-PDE in mammalian cells
HEK293 cells were purchased from the JCRB Cell Bank and cultured in Eagle's minimal essential medium with l-glutamine and phenol red (Wako) containing 10% (v/v) FBS and penicillin/streptomycin. The cells were co-transfected with the PDE or MosOpn3 plasmid and the pGloSensor-42F cGMP or pGloSensor-22F cAMP vector (Promega) by using Lipofectamine 2000 (Invitrogen). Changes in the intracellular cGMP or cAMP concentration of the HEK293 cells were measured by using the GloSensor assay (Promega). The transfected cells were incubated with or without 0.5 μm all-trans-retinal (Toronto Research Chemicals). Before measurements, the culture medium was replaced with a CO2-independent medium containing 10% (v/v) FBS and 2% (v/v) GloSensor cAMP or cGMP stock solution (Promega). Cells were then incubated for 2 h at room temperature in the dark. Intracellular cGMP or cAMP level was observed by monitoring luminescence by using a microplate reader (Corona Electric) at 27 °C. The cells were treated with 100 μm sodium nitroprusside (Wako), a direct activator of endogenous guanylyl cyclase, to elevate the intracellular cGMP level. Alternatively, the cells were treated with 3.5 μm forskolin (Wako), a direct activator of adenylyl cyclase, to elevate intracellular cAMP level. The cells were illuminated with a xenon lamp (LAX-103, Asahi Spectra Co., Ltd., Tokyo, Japan) through an interference filter (420, 464, 487, 510, and 550 nm). The light intensity was adjusted to desired values by a variable ND filter mounted in the LAX-103. The light intensity was measured by a power meter LP1 (Sanwa Electric Instruments Co., Ltd., Tokyo, Japan).
HPLC analysis for in vitro assay of the enzymatic activity of Rh-PDE
HEK293T cells were transfected with plasmid pCDNA3.1-Rh-PDE by the calcium phosphate method. DMEM/F-12 medium contained 0.5 μm all-trans-retinal and penicillin and streptomycin. The cells were harvested after 24 h and washed in buffer A (140 mm NaCl, 3 mm MgCl2, 50 mm HEPES-NaOH, pH 6.5). The cells were resuspended with buffer A and homogenized using a Potter-Elehjem grinder (Wheaton) and a syringe with a 27-gauge needle. The syringe was filled and drained five times while stirring the homogenate. The protein amount was determined by a BCA protein assay (Thermo Fisher Scientific). Samples were kept in the dark before measurement for at least 2 h. Catalytic activity was measured at room temperature in 100 μl of buffer A with 1.6–1.8 μg (in the case of cGMP) or 16–18 μg (in the case of cAMP) of total protein in a 1.5-ml sample tube. The sample was illuminated with a xenon lamp (MAX-303, Asahi Spectra Co.) through a Y52 filter (7 mW mm−2). Reaction was initiated by adding cyclic nucleotides (final concentration 100 μm). Aliquots were taken out at different time points, and the reactions were immediately terminated by adding 100 μl of 0.1 n HCl and frozen in liquid nitrogen. After thawing, the samples were centrifuged to remove the membranes and denatured proteins. Nucleotides (20 μl of aliquot) were separated by HPLC (Shimadzu Corp., Kyoto, Japan) with a C18 reversed-phase column (Waters) and 100 mm potassium phosphate (pH 5.9), 4 mm tetrabutylammonium iodide, and 10% (v/v) methanol as eluent (12, 26). Nucleotides were monitored at 254 nm. Data were evaluated with LabSolutions (Shimadzu). Peak areas were integrated and assigned to the educt cyclic nucleotide based on retention times of corresponding standard compound.
Purification of Rh-PDE
HEK293T cells expressing Rh-PDE, described above, were harvested after 24 h and homogenized by a Dounce tissue grinder (Wheaton). Membrane proteins were then extracted with 2% (w/v) DDM in buffer A, followed by centrifugation (6,500 × g, 1 h, 4 °C) to remove unsolubilized fractions. The extract was mixed with 1D4-agarose resin and incubated at room temperature for 15–30 min. After washing the resin with buffer A containing 0.02% (w/v) DDM (buffer B), proteins were eluted with buffer B containing 0.11–0.18 mg/ml 1D4 peptide. Protein fractions were concentrated with Amicon Ultra 30K centrifugal devices (Millipore).
Spectroscopy
The samples were illuminated with a 1-kW tungsten-halogen projector lamp (Master HILUX-HR, Rikagaku) through a Y52 filter (>500 nm), and the absorption spectra of Rh-PDE were recorded at various temperatures by using a JASCO V-650 spectrometer.
Estimation of chromophore regeneration
The extinction coefficient of the full-length Rh-PDE protein was determined as 106,340 m−1 cm−1 based on the protein sequence by using the ProtParam tool of Expasy (43). The absorption change, representing the bleaching of a purified Rh-PDE by hydroxylamine (HA), was measured with a UV-visible spectrometer (Shimadzu) equipped with an integrating sphere after the addition of HA to a final concentration of 500 mm. The molecular extinction coefficient of the chromophore in Rh-PDE (ϵ) was determined as 33,710 m−1 cm−1 from the ratio between the absorbance of Rh-PDE at 492 nm and retinal oxime (ϵ = 33,600 m−1 cm−1 at 360 nm (44)) produced by the reaction between retinal Schiff base and HA. From the values above and the absorptions at 280 and 492 nm in Fig. 5A (black spectrum), the extent of chromophore regeneration was estimated.
Phylogenetic analysis of rhodopsin genes
The amino acid sequences of transmembrane domains of selected rhodopsins (corresponding to residues 9–234 of bacteriorhodopsin) were aligned using ClustalO (45), and the resulting phylogenetic trees were visualized with TreeView (46).
Author contributions
S. P. T., L. S. B., and H. K. contributed to the study design. S. P. T. designed the construct. K. Y. conducted all measurements. S. P. T., L. S. B., and H. K. wrote the paper. All authors discussed and commented on the manuscript.
Supplementary Material
Acknowledgments
We thank Dr. Shoko Hososhima, Mizuna Tanaka, and Shunta Nakamura for helpful discussions.
This work was supported by grants from the Japanese Ministry of Education, Culture, Sports, Science, and Technology, 25104009 and 15H02391 (to H. K.), by PRESTO, Japan Science and Technology Agency Grant JPMJPR1688 (to S. P. T.), and by Natural Sciences and Engineering Research Council of Canada (NSERC) Grant RGPIN-2013-250202 (to L. S. B.). The authors declare that they have no conflicts of interest with the contents of this article.
This article contains supplemental Figs. S1–S4.
- HK
- histidine kinase
- GC
- guanylyl cyclase
- W
- watts
- AC
- adenylyl cyclase
- PAC
- photoactivated adenylyl cyclase
- PDE
- phosphodiesterase
- Rh
- rhodopsin
- BR
- bacteriorhodopsin
- DDM
- dodecyl-β-d-maltopyranoside
- SRII
- sensory rhodopsin II
- eGFP
- enhanced GFP
- ChR2
- channelrhodopsin-2
- HA
- hydroxylamine.
References
- 1. Ernst O. P., Lodowski D. T., Elstner M., Hegemann P., Brown L. S., and Kandori H. (2014) Microbial and animal rhodopsins: structures, functions, and molecular mechanisms. Chem. Rev. 114, 126–163 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2. Palczewski K. (2006) G Protein-coupled receptor rhodopsin. Annu. Rev. Biochem. 75, 743–767 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Shichida Y., and Morizumi T. (2007) Mechanism of G-protein activation by rhodopsin. Photochem. Photobiol. 83, 70–75 [DOI] [PubMed] [Google Scholar]
- 4. Hofmann K. P., Scheerer P., Hildebrand P. W., Choe H. W., Park J. H., Heck M., and Ernst O. P. (2009) A G protein-coupled receptor at work: the rhodopsin model. Trends Biochem. Sci. 34, 540–552 [DOI] [PubMed] [Google Scholar]
- 5. Koyanagi M., and Terakita A. (2014) Diversity of animal opsin-based pigments and their optogenetic potential. Biochim. Biophys. Acta 1837, 710–716 [DOI] [PubMed] [Google Scholar]
- 6. Jung K. H., Trivedi V. D., and Spudich J. L. (2003) Demonstration of a sensory rhodopsin in eubacteria. Mol. Microbiol. 47, 1513–1522 [DOI] [PubMed] [Google Scholar]
- 7. Grote M., Engelhard M., and Hegemann P. (2014) Of ion pumps, sensors and channels: perspectives on microbial rhodopsins between science and history. Biochim. Biophys. Acta 1837, 533–545 [DOI] [PubMed] [Google Scholar]
- 8. Brown L. S. (2014) Eubacterial rhodopsins: unique photosensors and diverse ion pumps. Biochim. Biophys. Acta 1837, 553–561 [DOI] [PubMed] [Google Scholar]
- 9. Inoue K., Kato Y., and Kandori H. (2015) Light-driven ion-translocating rhodopsins in marine bacteria. Trends Microbiol. 23, 91–98 [DOI] [PubMed] [Google Scholar]
- 10. Luck M., Mathes T., Bruun S., Fudim R., Hagedorn R., Tran Nguyen T. M., Kateriya S., Kennis J. T. M., Hildebrandt P., and Hegemann P. (2012) A photochromic histidine kinase rhodopsin (HKR1) that is bimodally switched by ultraviolet and blue light. J. Biol. Chem. 287, 40083–40090 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Avelar G. M., Schumacher R. I., Zaini P. A., Leonard G., Richards T. A., and Gomes S. L. (2014) A rhodopsin-guanylyl cyclase gene fusion functions in visual perception in a fungus. Curr. Biol. 24, 1234–1240 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Scheib U., Stehfest K., Gee C. E., Körschen H. G., Fudim R., Oertner T. G., and Hegemann P. (2015) The rhodopsin-guanylyl cyclase of the aquatic fungus Blastocladiella emersonii enables fast optical control of cGMP signaling. Sci. Signal. 8, rs8. [DOI] [PubMed] [Google Scholar]
- 13. Gao S., Nagpal J., Schneider M. W., Kozjak-Pavlovic V., Nagel G., and Gottschalk A. (2015) Optogenetic manipulation of cGMP in cells and animals by the tightly light-regulated guanylyl-cyclase opsin CyclOp. Nat. Commun. 6, 8046. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Deisseroth K. (2011) Optogenetics. Nat. Methods 8, 26–29 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Yizhar O., Fenno L. E., Davidson T. J., Mogri M., and Deisseroth K. (2011) Optogenetics in neural systems. Neuron 71, 9–34 [DOI] [PubMed] [Google Scholar]
- 16. Miesenböck G. (2011) Optogenetic control of cells and circuits. Annu. Rev. Cell Dev. Biol. 27, 731–758 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Zhang F., Vierock J., Yizhar O., Fenno L. E., Tsunoda S., Kianianmomeni A., Prigge M., Berndt A., Cushman J., Polle J., Magnuson J., Hegemann P., and Deisseroth K. (2011) The microbial opsin family of optogenetic tools. Cell 147, 1446–1457 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Dugué G. P., Akemann W., and Knöpfel T. (2012) A comprehensive concept of optogenetics. Prog. Brain Res. 196, 1–28 [DOI] [PubMed] [Google Scholar]
- 19. Boyden E. S., Zhang F., Bamberg E., Nagel G., and Deisseroth K. (2005) Millisecond-timescale, genetically targeted optical control of neural activity. Nat. Neurosci. 8, 1263–1268 [DOI] [PubMed] [Google Scholar]
- 20. Zhang F., Wang L.-P., Brauner M., Liewald J. F., Kay K., Watzke N., Wood P. G., Bamberg E., Nagel G., Gottschalk A., and Deisseroth K. (2007) Multimodal fast optical interrogation of neural circuitry. Nature 446, 633–639 [DOI] [PubMed] [Google Scholar]
- 21. Iseki M., Matsunaga S., Murakami A., Ohno K., Shiga K., Yoshida K., Sugai M., Takahashi T., Hori T., and Watanabe M. (2002) A blue-light-activated adenylyl cyclase mediates photoavoidance in Euglena gracilis. Nature 415, 1047–1051 [DOI] [PubMed] [Google Scholar]
- 22. Schröder-Lang S., Schwärzel M., Seifert R., Strünker T., Kateriya S., Looser J., Watanabe M., Kaupp U. B., Hegemann P., and Nagel G. (2007) Fast manipulation of cellular cAMP level by light in vivo. Nat. Methods 4, 39–42 [DOI] [PubMed] [Google Scholar]
- 23. Ryu M. H., Moskvin O. V., Siltberg-Liberles J., and Gomelsky M. (2010) Natural and engineered photoactivated nucleotidyl cyclases for optogenetic applications. J. Biol. Chem. 285, 41501–41508 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Stierl M., Stumpf P., Udwari D., Gueta R., Hagedorn R., Losi A., Gärtner W., Petereit L., Efetova M., Schwarzel M., Oertner T. G., Nagel G., and Hegemann P. (2011) Light modulation of cellular cAMP by a small bacterial photoactivated adenylyl cyclase, bPAC, of the soil bacterium Beggiatoa. J. Biol. Chem. 286, 1181–1188 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Costa W. S., Liewald J., and Gottschalk A. (2014) Photoactivated adenylyl cyclases as optogenetic modulators of neuronal activity. Methods Mol. Biol. 1148, 161–175 [DOI] [PubMed] [Google Scholar]
- 26. Gasser C., Taiber S., Yeh C.-M., Wittig C. H., Hegemann P., Ryu S., Wunder F., and Möglich A. (2014) Engineering of a red-light-activated human cAMP/cGMP-specific phosphodiesterase. Proc. Natl. Acad. Sci. U.S.A. 111, 8803–8808 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Koyanagi M., Takada E., Nagata T., Tsukamoto H., and Terakita A. (2013) Homologs of vertebrate Opn3 potentially serve as a light sensor in nonphotoreceptive tissue. Proc. Natl. Acad. Sci. U.S.A. 110, 4998–5003 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Bender A. T., and Beavo J. A. (2006) Cyclic nucleotide phosphodiesterases: molecular regulation to clinical use. Pharmacol. Rev. 58, 488–520 [DOI] [PubMed] [Google Scholar]
- 29. Francis S. H., Blount M. A., and Corbin J. D. (2011) Mammalian cyclic nucleotide phosphodiesterases: molecular mechanisms and physiological functions. Physiol. Rev. 91, 651–690 [DOI] [PubMed] [Google Scholar]
- 30. Pandit J., Forman M. D., Fennell K. F., Dillman K. S., and Menniti F. S. (2009) Mechanism for the allosteric regulation of phosphodiesterase 2A deduced from the X-ray structure of a near full-length construct. Proc. Natl. Acad. Sci. U.S.A. 106, 18225–18230 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Katayama K., Furutani Y., Imai H., and Kandori H. (2010) An FTIR study of monkey green- and red-sensitive visual pigments. Angew. Chem. Int. Ed. Engl. 49, 891–894 [DOI] [PubMed] [Google Scholar]
- 32. Katayama K., Furutani Y., Imai H., and Kandori H. (2012) Protein-bound water molecules in primate red- and green-sensitive visual pigments. Biochemistry 51, 1126–1133 [DOI] [PubMed] [Google Scholar]
- 33. Shimono K., Ikeura Y., Sudo Y., Iwamoto M., and Kamo N. (2001) Environment around the chromophore in pharaonis phoborhodopsin: mutation analysis of the retinal binding site. Biochim. Biophys. Acta 1515, 92–100 [DOI] [PubMed] [Google Scholar]
- 34. Bordignon E., Klare J. P., Doebber M., Wegener A. A., Martell S., Engelhard M., and Steinhoff H. J. (2005) Structural analysis of a HAMP domain: the linker region of the phototransducer in complex with sensory rhodopsin II. J. Biol. Chem. 280, 38767–38775 [DOI] [PubMed] [Google Scholar]
- 35. Hulko M., Berndt F., Gruber M., Linder J. U., Truffault V., Schultz A., Martin J., Schultz J. E., Lupas A. N., and Coles M. (2006) The HAMP domain structure implies helix rotation in transmembrane signaling. Cell 126, 929–940 [DOI] [PubMed] [Google Scholar]
- 36. Shimono K., Iwamoto M., Sumi M., and Kamo N. (2000) Effects of three characteristic amino acid residues of pharaonis phoborhodopsin on the absorption maximum. Photochem. Photobiol. 72, 141–145 [DOI] [PubMed] [Google Scholar]
- 37. Shimono K., Hayashi T., Ikeura Y., Sudo Y., Iwamoto M., and Kamo N. (2003) Importance of the broad regional interaction for spectral tuning in Natronobacterium pharaonis phoborhodopsin (sensory rhodopsin II). J. Biol. Chem. 278, 23882–23889 [DOI] [PubMed] [Google Scholar]
- 38. Gordeliy V. I., Labahn J., Moukhametzianov R., Efremov R., Granzin J., Schlesinger R., Büldt G., Savopol T., Scheidig A. J., Klare J. P., and Engelhard M. (2002) Molecular basis of transmembrane signalling by sensory rhodopsin II-transducer complex. Nature 419, 484–487 [DOI] [PubMed] [Google Scholar]
- 39. Klare J. P., Bordignon E., Engelhard M., and Steinhoff H.-J. (2004) Sensory rhodopsin II and bacteriorhodopsin: light activated helix F movement. Photochem. Photobiol. Sci. 3, 543–547 [DOI] [PubMed] [Google Scholar]
- 40. Moukhametzianov R., Klare J. P., Efremov R., Baeken C., Göppner A., Labahn J., Engelhard M., Büldt G., and Gordeliy V. I. (2006) Development of the signal in sensory rhodopsin and its transfer to the cognate transducer. Nature 440, 115–119 [DOI] [PubMed] [Google Scholar]
- 41. Wegener A. A., Chizhov I., Engelhard M., and Steinhoff H. J. (2000) Time-resolved detection of transient movement of helix F in spin-labelled pharaonis sensory rhodopsin II. J. Mol. Biol. 301, 881–891 [DOI] [PubMed] [Google Scholar]
- 42. Wegener A. A., Klare J. P., Engelhard M., and Steinhoff H. J. (2001) Structural insights into the early steps of receptor-transducer signal transfer in archaeal phototaxis. EMBO J. 20, 5312–5319 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43. Gasteiger E., Gattiker A., Hoogland C., Ivanyi I., Appel R. D., and Bairoch A. (2003) ExPASy: the proteomics server for in-depth protein knowledge and analysis. Nucleic Acids Res. 31, 3784–3788 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44. Scharf B., Pevec B., Hess B., and Engelhard M. (1992) Biochemical and photochemical properties of the photophobic receptors from Halobacterium halobium and Natronobacterium pharaonis. Eur. J. Biochem. 206, 359–366 [DOI] [PubMed] [Google Scholar]
- 45. Sievers F., Wilm A., Dineen D., Gibson T. J., Karplus K., Li W., Lopez R., McWilliam H., Remmert M., Söding J., Thompson J. D., and Higgins D. G. (2011) Fast, scalable generation of high-quality protein multiple sequence alignments using Clustal Omega. Mol. Syst. Biol. 7, 539. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46. Page R. D. M. (2002) Visualizing phylogenetic trees using TreeView. Curr. Protoc. Bioinformatics 10.1002/0471250953.bi0602s01 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.