Abstract
Tropomyosin receptor kinase C (TrkC) is involved in cell survival, apoptosis, differentiation, and tumorigenesis. TrkC diverse functions might be attributed to the hypothetical non-coding RNAs embedded within the gene. Using bioinformatics approaches, a novel microRNA named TrkC-miR2 was predicted within the TrkC gene capable of regulating the Wnt pathway. For experimental verification of this microRNA, the predicted TrkC-premir2 sequence was overexpressed in SW480 cells, which led to the detection of two mature TrkC-miR2 isomiRs, and their endogenous forms were detected in human cell lines as well. Later, an independent promoter was deduced for TrkC-miR2 after the treatment of HCT116 cells with 5-azacytidine, which resulted in differential expression of TrkC-miR2 and TrkC host gene. RT-quantitative PCR and luciferase assays indicated that the APC2 gene is targeted by TrkC-miR2, and Wnt signaling is up-regulated. Also, Wnt inhibition by using small molecules along with TrkC-miR2 overexpression and TOP/FOP flash assays confirmed the positive effect of TrkC-miR2 on the Wnt pathway. Consistently, TrkC-miR2 overexpression promoted SW480 cell survival, which was detected by flow cytometry, MTT (3-(4,5-dimethylthiazol-2-yl)-2,5-diphenyltetrazolium bromide) assays, and crystal violate analysis. RT-qPCR analysis revealed that TrkC-miR2 is significantly up-regulated (∼70 times) in colorectal tumor tissues compared with their normal pairs. Moreover, the TrkC-miR2 expression level discriminated grades of tumor malignancies, which was consistent with its endogenous levels in HCT116, HT29, and SW480 colorectal cancer cell lines. Finally, an opposite expression pattern was observed for TrkC-miR2 and the APC2 gene in colorectal cancer specimens. In conclusion, here we introduce TrkC-miR2 as a novel regulator of Wnt signaling, which might be a candidate oncogenic colorectal cancer biomarker.
Keywords: annexin, biomarker, cancer, cell cycle, colorectal cancer, TrkC-miR2, Survival, Wnt signaling, Colorectal cancer, Biomarker
Introduction
TrkC2 (tropomyosin receptor kinase C (N_000015.9)) or neurotrophin-tyrosine kinase receptor type 3 (NTRK3) is a member of the Trk family of neurotrophin receptors (1). Expression of Trk family receptors in some cell types either promotes cell proliferation or differentiation (2). TrkC is implicated in regulation of growth and survival of many human cancer tissues, acting as an oncogene or a tumor suppressor gene, and also is inactivated by epigenetic mechanisms in colorectal cancer (CRC) (3–5).
MicroRNAs (miRNAs) are highly conserved, are 18–27 nucleotides long, and are endogenously made non-coding RNAs in many organisms (6). The complicated secondary structure of miRNA precursor (pri-miRNA) is quickly processed into 1 or more (∼70) nucleotides long hairpin-structured pre-miRNA molecule(s), which is further processed into its mature form located either at the 5′ or 3′ side of the stem loop (7, 8). Mature miRNA in mammalian cells often pairs imperfectly to its target transcripts, resulting in either mRNA degradation or translation inhibition (9). Although ∼55,000 miRNA genes are estimated to be encoded within the human genome (10), ∼2500 human miRNAs are now registered in miRBase database. Therefore, several bioinformatics tools have been developed for prediction of novel miRNAs. This software is designed based on conservation of predicted miRNA sequence, its precursor secondary structure, stability information, and similarity of the predicted miRNA to the known miRNAs (11, 12).
Colorectal cancer is the third most common cancer worldwide with an estimated one million new cases and a half-million deaths each year (13). Irregular Wnt signaling pathway, which occurs through mutations mainly of APC, is a primary progression event in 90% of CRCs (14, 15). Considering the invasive nature and the cost of colonoscopic screening of CRC and also the limitation of low sensitivity of fecal occult blood test, there is a pressing need for new non-invasive biomarkers with high sensitivity and specificity to improve the diagnosis of CRC (13, 16). Recently, the discovery of miRNAs that play important roles in oncogenesis and also in Wnt signaling regulation has opened new opportunities for the early diagnosis of CRC (13, 17, 18).
After our successful bioinformatics prediction and experimental verification of hsa-miR-11181 (19) as the first miRNA located in TrkC gene, we predicted and verified TrkC-miR2, which is located in the vicinity of hsa-miR-11181. Functional analysis of the novel TrkC-miR2 confirmed its regulatory effect on Wnt signaling pathway and suggested its potential as a CRC biomarker.
Results
Computational prediction of a novel miRNA in TrkC gene
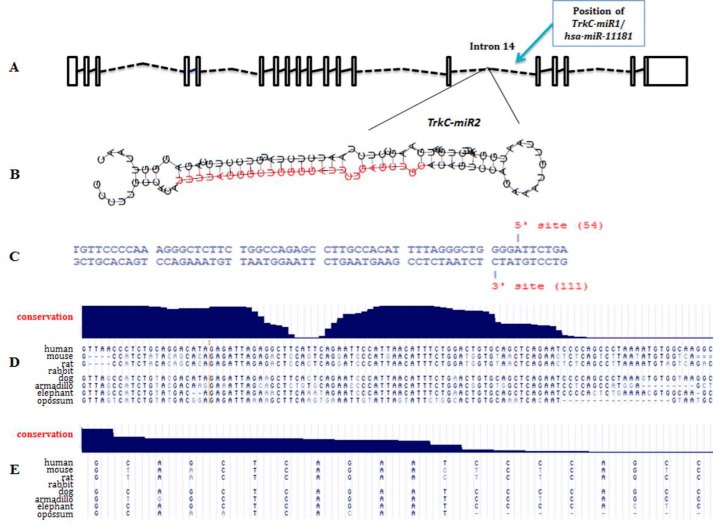
Among 150 predicted stem loops within the introns of TrkC gene, one of them located in 14th intron of the gene (hg17, chr15: 86307074–86307177) and at the vicinity of TrkC-miR1 (hsa-miR-11181) locus (Fig. 1A) had the most characteristics of a real miRNA precursor. SSC profiler predicted a bona fide mature miRNA sequence within this precursor (Fig. 1B), and Microprocessor SVM tool recognized Drosha and Dicer recognition sites in it (Fig. 1C). UCSC software also plotted a saddle-like pattern of conservation for the stem loop, which is a prominent characteristic of real miRNAs (Fig. 1D). A miRNA mature form was also conserved between mammals including dog and cat with a subtle variation of sequence compared with rodents (Fig. 1E). The predicted mature miRNA and its precursor were named TrkC-miR2 and TrkC-premir2, respectively. Moreover, miRNA Spotter, MiRmat, Pmirp, and Mature Bayes software predicted TrkC-miR2–5p as bona fide miRNA. No identical or similar miRNA for TrkC-miR2 has been reported in miRBase database. Alibaba2.1 Prediction Server online bioinformatics tool predicted an independent promoter for TrkC-miR2 at ∼800 bp upstream of the corresponding stem-loop, which may interact with some transcription factors including NF-κB.
Figure 1.
Bioinformatics prediction of TrkC-miR2 encoded within the 14th intron of human TrkC gene. A, TrkC gene introns (broken lines) and exons (rectangles) adapted from Ensemble are shown. The position of the stem loop encoding TrkC-miR2 is shown at in the 17-kb vicinity of the newly discovered hsa-miR-11181. B, predicted stem loop encoding TrkC-miR2. The red sequence was predicted by SSC profiler as the TrkC-miR2 mature form. C, Drosha enzyme cutting sites, predicted by Microprocessor SVM tool, are shown on the sequence of hairpin structure. Blat search by the UCSC genome browser shows high conservation of TrkC-premir2 (D) and TrkC-miR2–5p (E) between several organisms.
Detection of exogenous and endogenous TrkC-miR2 and its sequence determination
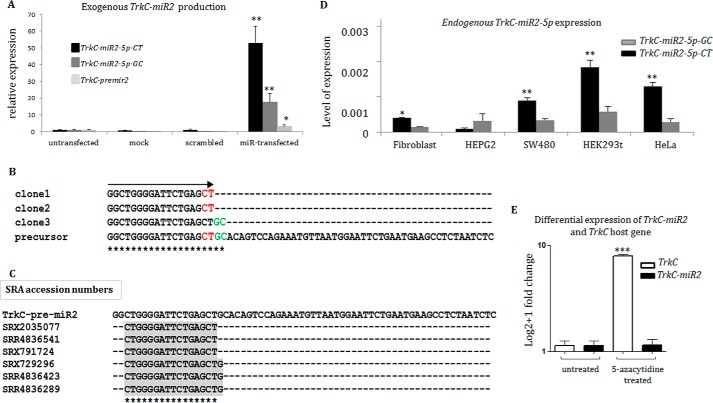
After the overexpression of a recombinant plasmid vector containing sequences of predicted TrkC-miR2 precursor in SW480 cell line, mature predicted miRNA was specifically amplified (Fig. 2A). When, the amplification products were cloned in the TA vector and sequenced, TrkC-miR2–5p sequence was represented in multiple sequencing results showing two nucleotides variation in its 3′ end, which resulted in TrkC-miR2–5p-CT and TrkC-miR2–5p-GC introduction (Fig. 2B). The minimum size of these sequences was submitted to the EMBL-EBI database, accessible by EBI accession numbers HG969187, HG969188, and HG969189 for TrkC-miR2–5p-CT, TrkC-miR2–5p-GC, and TrkC-premir2, respectively. In several RNA-sequencing data (NCBI-SRA) analysis attempts, multiple reads were detected showing TrkC-miR2–5p-CT expression, whereas no read was found for TrkC-miR2–5p-GC. Instead, several reads were detected in which the miRNA sequence was one nucleotide smaller than TrkC-miR2-5p-GC, meaning one nucleotide longer than TrkC-miR2–5p-CT. This may suggest a novel TrkC-miR2 isomiR (Fig. 2C). Endogenous TrkC-miR2 expression was also detected in HEPG2, SW480, HEK293t, HeLa, and fibroblast human cell lines through RT-qPCR (Fig. 2D). The highest expression level of TrkC-miR2 was detected in HEK293t cells, and the TrkC-miR2–5p-CT level was higher than TrkC-miR2–5p-GC isomiR in most of these cell lines (Fig. 2D).
Figure 2.
TrkC-miR2–5p sequence characterization and evidence for an independent promoter. A, overexpression of TrkC-premir2 in SW480 cells and detection of predicted TrkC-miR2–5p mature forms (-CT and -GC isomiRs) by RT-qPCR. Note that TrkC-miR2–5p-CT isomiR is dominantly produced in the transfected cells. B, sequencing results of three TA vector clones containing TrkC-miR2–5p sequences created in A. Clones 1 and 2 show the sequence of TrkC-miR2–5p-CT isomiR, and clone 3 shows the sequence of TrkC-miR2–5p-GC isomiR. TrkC-miR2 sequences are aligned with the sequence of their precursor. The significance of asterisks in panels C and B is the complete matching of aligned sequences at each position. C, shown is the presence of TrkC-miR2 in the SRA data. Three reads for detected TrkC-miR2-5p-CT and TrkC-miR2-5p-CTG isomiRs are shown. However, no read for TrkC-miR2-5p-GC was detected in the SRA data set. D, shown is the expression level of TrkC-miR2–5p-CT and -GC isomiRs in several human cell lines. TrkC-miR2–5p-CT relative expression was higher than TrkC-miR2–5p-GC isomiR in most of these cell lines. E, treatment of HCT-116 cells with epidrug resulted in TrkC expression elevation (100 times). However, it did not have such an effect on TrkC-miR2–5p expression level. Error bars indicate S.D. of duplicate experiments. U48 RNA and GAPDH were used as internal controls for the amplifications. *, p < 0.05; **, p < 0.01; ***, p < 0.001.
Evidence for an independent promoter for TrkC-miR2
The HCT-116 cell line was treated with 5-azacytidin epidrug, and the expression levels of TrkC-miR2 and TrkC were measured and compared with the levels of these genes in untreated HCT-116 cells. RT-qPCR results indicated that the TrkC expression level has been elevated up to 100 times in the epidrug-treated cells compared with the untreated ones. However, TrkC-miR2-5p expression level was not affected by epidrug treatment (Fig. 2E).
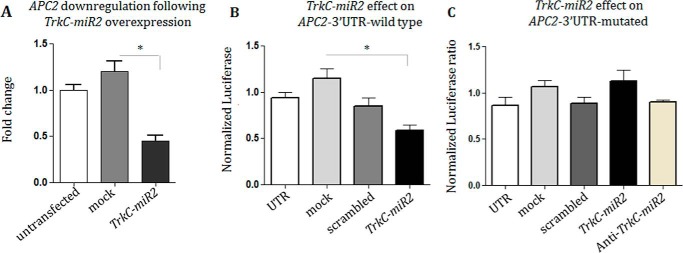
Direct interaction of TrkC-miR2 with APC2–3′-UTR
∼700 target genes were predicted for TrkC-miR2-5p using Dianna lab software. APC2 (ENSG00000115266) gene was the highest scored target gene in which 23 highly conserved TrkC-miR2-5p-specific MREs were predicted in its 3′-UTR. A DNA fragment containing the TrkC-miR2 precursor was PCR-amplified from the human genome and cloned in fusion with GFP/ORF in the pEGFP-C1 expression vector. Then, RT-qPCR indicated significant down-regulation (60%) of the APC2 expression level in the SW480 cells transfected with TrkC-miR2 precursor (Fig. 3A). When 3′-UTR sequence of APC2 was cloned downstream of Renilla luciferase ORF and co-expressed with TrkC-miR2 in HEK293t cells, a dual luciferase assay supported direct interaction with APC2 transcript, showing 55% reduction in luciferase count (Fig. 3B). When, mutated APC2, 3′-UTR (supplemental Fig. 1) was used in the same experiment, and TrkC-miR2 overexpression did not significantly change the luciferase expression level (Fig. 3C). Co-expression of TrkC-miR2 with cassettes containing APC (supplemental Fig. 2A), AxinI (supplemental Fig. 2B), and AxinII (supplemental Fig. 2C) 3′-UTRs sequences downstream of Renilla luciferase ORF showed no significant alteration of Renilla luciferase activity compared with mock or scrambled controls.
Figure 3.
TrkC-miR2 direct interaction with its predicted target gene. A, RT-qPCR result shows APC2 gene down-regulation after TrkC-premir2 overexpression in the SW480 cell line compared with the related controls. B, dual luciferase assay supported TrkC-miR2 direct interaction with 3′-UTR sequence of APC2 target gene. C, lack of interaction between the overexpressed TrkC-miR2 and mutated APC2,3′-UTR detected by dual luciferase assay and supported direct interaction between this miRNA and the wild-type sequence of APC2,3′-UTR sequence shown in B. *, p < 0.05.
TrkC-miR2 as a novel regulator of Wnt signaling in SW480 cells
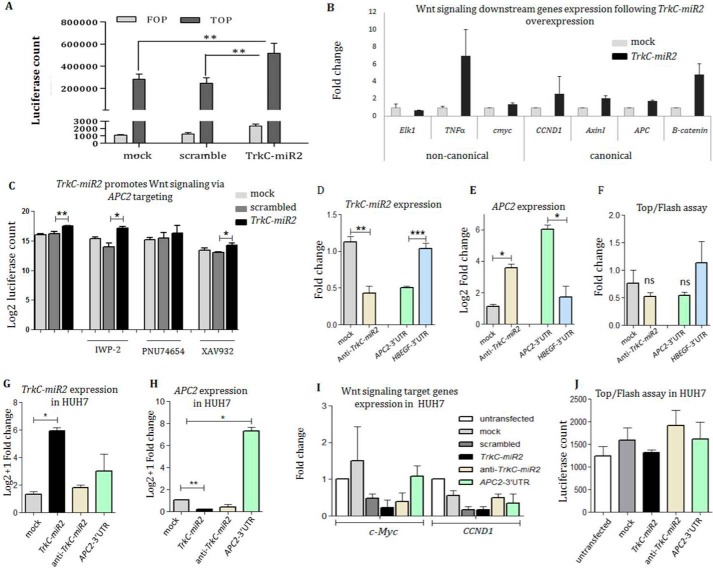
TrkC-miR2 overexpression effect on the Wnt signaling pathway was investigated through TOP/FOP flash assay. To this aim, Wnt signaling positive SW480 cells was transiently co-transfected using pGL4-TOP and TrkC-miR2-overexpressing vectors, and results were compared with the situation that pGL4-TOP was co-transfected with scrambled or mock recombinant vectors. Luciferase activity in the SW480-TOP cells overexpressing TrkC-miR2 was significantly increased (∼2-fold, p < 0.05) compared with the mock and scrambled negative vectors. For the reporter assay, relative luciferase activities of SW480-TOP cells were strongly higher than those of SW480-FOP cells (Fig. 4A).
Figure 4.
TrkC-miR2 expression alteration effect on the Wnt signaling pathway. A, Wnt signaling up-regulation after TrkC-miR2 overexpression in the SW480 cells. Luciferase activity was significantly increased in the cells overexpressing TrkC-premir2 compared with the controls. B, increased expression level of the genes involved in both canonical and non-canonical Wnt signaling pathway after TrkC-miR2 overexpression. C, Wnt inhibitor small molecules and TrkC-miR2 co-treatment effect on the Wnt signaling is shown through Top/flash assay. Although Wnt signaling is down-regulated after the XAV932 and IWP-2 small molecules application, this down-regulation has been compensated by TrkC-miR2 overexpression compared with the controls. When the cells were treated with the PNU74654 small molecule, significant Wnt signaling alteration was not detected with or without TrkC-miR2 overexpression. Error bars indicate S.D. of three experiments. D, successful down-regulation of TrkC-miR2 using anti-TrkC-miR2 sequence in SW480 cells (compared with the mock control). Also, APC2-3′-UTR overexpression as a scavenger, resulted in down-regulation of TrkC-miR2 (compared with the non-scavenger HBEGF-3′-UTR sequence), detected by RT-qPCR. Expression data were normalized against U48 as an internal control. E, RT-qPCR results showed significant up-regulation of APC2 expression after the down-regulation of TrkC-miR2 (using anti-TrkC-miR2) or after the scavenging of TrkC-miR2 (by APC2-3′-UTR overexpression) in SW480 cells. Expression data were normalized against GAPDH as an internal control. F, top/Fop flash assay after transfection of anti-TrkC-miR2 in SW480 cells showing non-significant Wnt signaling reduction compared with the mock transfected control. Also, overexpression of APC2-3′-UTR as a TrkC-miR2 scavenger again resulted in non-significant Wnt signaling attenuation compared with the off-target HBEGF-3′-UTR overexpression. ns, not significant. G, RT-qPCR analysis of TrkC-miR2 expression in HUH7 cell lines after TrkC-premiR2, anti-TrkC-miR2, and APC2-3′-UTR overexpression compared with mock control. TrkC-miR2 level was significantly increased in the cells overexpressing TrkC-premiR2; however, anti-TrkC-miR2 and APC2-3′-UTR scavenger constructs were not capable of significant reduction in TrkC-miR2 level. H, shows APC2 expression alteration after TrkC-premiR2, anti-TrkC-miR2, and APC2-3′-UTR overexpression. APC2-3′-UTR-specific primers were used for detection of APC2 expression alteration. Only successful overexpression of TrkC-miR2 (G) has resulted in significant reduction of APC2 transcripts. I, RT-qPCR against c-Myc and CCND1 genes (as the Wnt signaling target genes) after the overexpression of interested constructs. Data indicate no significant Wnt signaling alteration after the TrkC-miR2 expression alteration in Huh7 cells. J, shown is the Top/flash assay in the HUH7 cell line before and after TrkC-miR2 expression alteration. Similar to I, no significant Wnt signaling alteration was detected. *, p < 0.05; **, p < 0.01; ***, p < 0.001.
The effect of TrkC-miR2 overexpression on the Wnt signaling pathway was further investigated through RT-qPCR analysis against downstream genes of the pathway. The expression level of the genes involved in canonical and non-canonical Wnt signaling pathways (c-MYC, CCND1, AxinI, APC1, B-catenin, and TNFα) was significantly elevated after TrkC-miR2 overexpression in SW480 cell line (Fig. 4B).
SW480 cells were also treated with XAV932 (increases AxinI expression), PNU74654 (inhibits β-catenin and TCF interaction), and IWP-2 (inhibits LRP) small molecules in order to manipulate Wnt signaling pathway at different steps. Then, these cells were co-transfected with pGL4-TOP and TrkC-miR2-overexpressing vectors, and results were compared with the situation that pGL4-TOP was co-transfected with scrambled or mock constructs. Luciferase activity in SW480-TOP cells, which were treated with XAV932 and IWP-2, was significantly increased when TrkC-miR2 was overexpressed compared with the mock and scrambled negative vectors (p < 0.05). However, when β-catenin and TCF interaction was inhibited by using PNU74654, overexpression of TrkC-miR2 did not significantly change the Wnt signaling (Fig. 4C).
Using anti-TrkC-miR2, this miRNA was significantly down-regulated in SW480 cells (Fig. 4D), which was followed by the elevation of APC2 gene expression (Fig. 4E) and non-significant reduction of the Wnt activity (Fig. 4F). Scavenging of TrkC-miR2 through overexpression of APC2-3′-UTR (Fig. 4D) also resulted in elevation of APC2 gene expression (Fig. 4E) and also non-significant reduction of Wnt activity (Fig. 4F). Mock and HBEGF-3′-UTR constructs were applied as controls.
Although the overexpression of TrkC-premiR2 in HUH7 cells (Fig. 4G) resulted in significant down-regulation of APC2 gene expression (Fig. 4H), it was not effective on the Wnt signaling activity (Fig. 4, I and J).
Up-regulated TrkC-miR2 in CRC-originated cell lines and tissue samples
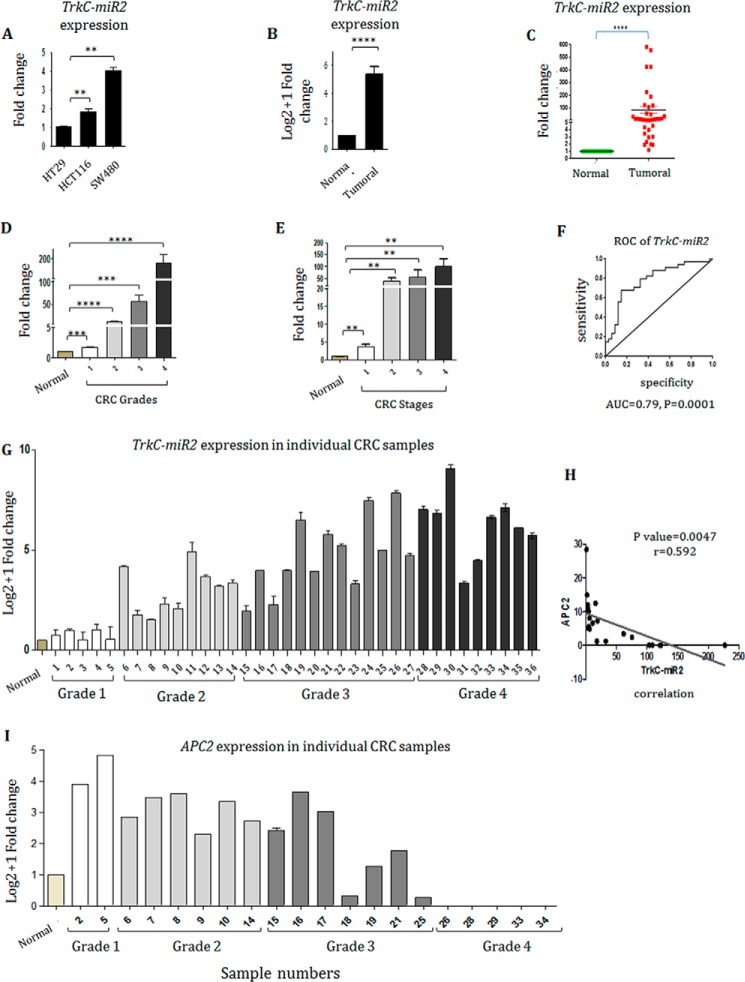
Endogenous expression of TrkC-miR2–5p-CT was detected through RT-qPCR in low (HT29), intermediate (HCT-116), and high (SW480) grades of human colon cancer cell lines. The highest expression level of TrkC-miR2–5p-CT was detected in SW480 cell line; however, the minimum expression level of it was detected in the HT29 cell line (Fig. 5A).
Figure 5.
Implication of TrkC-miR2 in colorectal cancer. A, TrkC-miR2-5p-CT expression level in HCT-116 and SW480 cells was 2- and 4-fold higher than HT29 cell line (p < 0. 005), respectively. B, TrkC-miR2 expression status in 36 CRC tumor (T) tissues and normal (N) pairs. C, Mann-Whitney analysis indicated that TrkC-miR2 has been significantly increased in CRCs (∼70-fold) compared with the paired adjacent non-CRC tissue samples (p < 0.0001). D, increased expression of TrkC-miR2 detected in the more advanced grades of CRCs compared with the normal ones (p < 0.001). E, TrkC-miR2 expression in different stages of CRC samples. Error bars indicate S.E. of tetraplicate experiments. F, ROC curve analysis of TrkC-miR2 expression in CRC patients. The up-regulated TrkC-miR2 expression yielded an area under the curve (AUC) value of 0.79 (95% confidence interval: 0.6714–0.8944) with 75% sensitivity and 75% specificity, supported TrkC-miR2 expression as a diagnostic value for discriminating CRC from healthy controls. G, TrkC-miR2 expression analysis in individual samples, distributed in high and low grade samples. Samples are shown by the numbers. H, a significant negative correlation was calculated between TrkC-miR2 and APC2 expression with a correlation coefficient (r) of −0.592 and a significant p value of 0.0046. I, APC2 expression analysis in individual high and low grade samples. Error bars indicate S.D. of duplicated experiments. *, p < 0.05; **, p < 0.01; ***, p < 0.001; ****, p < 0.0001.
TrkC-miR2
5p-CT was detected in 36 colorectal cancer sample pairs, whereas TrkC-miR2-5p-GC expression was not detected. TrkC-miR2–5p-CT was ∼70-fold up-regulated in tumor tissues compared with the paired adjacent non-tumor samples (p < 0.0001) (Fig. 5B), which was supported with Mann-Whitney analysis (Fig. 5C). CRC tumor samples were distributed in all four grades (1–4). These data showed significant differences between different grades of malignancy (Fig. 5D). Although TrkC-miR2 expression was not significantly altered between the tumors at stages of 2–4 (Mann-Whitney test, p > 0.05), a non-significant steady/gradual increase of TrkC-miR2 expression was detected during the increase of CRC stages (p < 0.005) (Fig. 5E).
To investigate the suitability of TrkC-miR2-5p-CT for discrimination of tumor versus non-tumor states of CRC samples, sensitivity and specificity were calculated using ROC (receiver operating characteristic) curve analysis. An area under the curve = 0.79 for TrkC-miR2-5p-CT (p value = 0.0001; Fig. 5F) was calculated, which is a score greater than the cutoff (0.7) needed for a considerable biomarker. The expression levels of TrkC-miR2 and APC2 were compared in >20 individuals tumor samples showing a significant negative correlation between them with a correlation coefficient (r) of −0.592 (p = 0.0046) (Fig. 5, G–I).
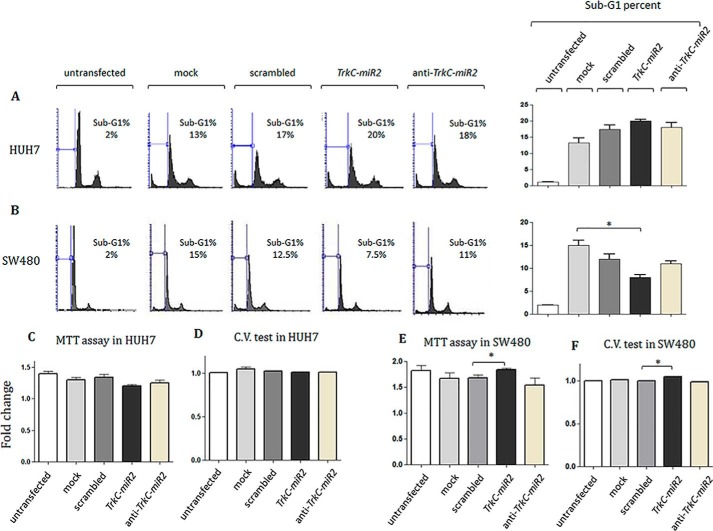
Survival effect of TrkC-premir2 overexpression in SW480 cell line
SW480 and HUH7 cell lines were transfected using TrkC-premir2-overexpressing vector, and cell cycle distribution of the cells was examined (Fig. 6). No significant sub-G1 population alteration was detected in HUH7 cells overexpressing TrkC-premir2 compared with the control cells (Fig. 6A). However, overexpression of TrkC-premir2 in SW480 cells rendered a significant reduction in sub-G1 cell population compared with the negative control. Knockdown of this miRNA did not significantly alter the subG1 population rate (Fig. 6B). Consistently, an MTT assay (Fig. 6C) and crystal violet analysis (Fig. 6D) indicated no significant proliferation rate alteration in the transfected HUH7 cells overexpressing TrkC-premir2 compared with the related control cells. Furthermore, overexpression of TrkC-premir2 in SW480 cells resulted in significant elevation of survival rate detected by both MTT (Fig. 6E) and crystal violet assays (Fig. 6F) compared with the negative control.
Figure 6.
Cell cycle and survival analysis for the cells overexpressing TrkC-premir2. A and B, shown is flow cytometry propidium iodide staining 36 h after transfection of two different cell lines in which TrkC-premir2 has been overexpressed or down-regulated. Although no significant alteration was detected for HUH7 cell line (A), t test analysis indicated a significant reduction in sub-G1 cell cycle distribution of SW480 (B)-transfected cells after TrkC-miR2 overexpression. However, knockdown of TrkC-miR2 in both cell types did not significantly change the sub-G1 percentages of the cells. C and D, MTT and crystal violet assay against HUH7 cells under TrkC-miR2 expression alteration. No significant cell survival alteration rate was calculated in these situations. E and F, MTT and crystal violet assay against SW480 cells under TrkC-miR2 expression alteration. A significant cell survival alteration rate was calculated when TrkC-miR2 was overexpressed. *, p < 0.05
Discussion
miRNAs are known to be involved in many processes including cancer and differentiation (20). Because their discovery through forward genetics is relatively inefficient (21), bioinformatics tools are established for prediction of novel miRNAs (22). TrkC receptor is known to be involved in neurotrophin signaling, which is related to cell death, survival, cancer, and differentiation, similar to Wnt signaling (3, 5, 23–25). On the other hand TrkC is a potential tumor suppressor gene commonly inactivated by epigenetic mechanisms in CRC (3, 5). Despite its widespread involvement in cell signaling, molecular mechanisms that regulate Wnt signaling pathway are poorly understood (26). Hence, discovery of common regulatory factors for the Wnt signaling pathway may provide the possibility of cell fate manipulation in the cases of diseases like CRC and tissue regeneration. Our previous attempts resulted in discovery of hsa-miR-6165 (27) and hsa-miR-11181 (19), which are located in NGFR and TrkC genes, respectively. Herein, we gathered bioinformatics and supportive experimental evidences showing the presence of a second novel miRNA located in the TrkC gene that has the potential of being considered as a regulator of Wnt signaling pathway and also has the potential to be used as a valuable biomarker for diagnosis of CRC.
Detection of TrkC-miR2 that is driven by an independent promoter
One of the bona fide predicted stem loops in TrkC intron 14th (Fig. 1A), named TrkC-premir2, showed the most criteria for producing novel TrkC-miR2 (Fig. 1B). A Drosha processing site was predicted in TrkC-premir2 (Fig. 1C), and like most of known miRNAs precursors (12), a saddle-like conservation plot was predicted for it (Fig. 1D). TrkC-miR2 is conserved in mammals like dog and cat (Fig. 1E), and no identical sequence has been reported for TrkC-miR2 in the miRBase database.
Similar to the approaches used by others (19, 27, 28), exogenous production of mature TrkC-miR2 was detected when a DNA fragment corresponding to TrkC-premir2 sequence was overexpressed in SW480 cells (Fig. 2A). Then, production of mature TrkC-miR2 was confirmed by sequencing of specifically amplified RT-qPCR products (Fig. 2B). Interestingly, similar to many other reported miRNAs (29–31), TrkC-miR2–5p-CT and TrkC-miR2–5p-GC isomiRs were processed from the 5′ arm of its precursor. Next generation sequencing (NGS) data analysis revealed the existence of several reads for TrkC-miR2–5p-CT, which once again supported the identity of this miRNA. Also, there was no SRA (Sequence Read Archive) read for TrkC-miR2–5p-GC consistent with the lower expression level we have detected for this isomiR compared with the level of TrkC-miR2–5p-CT. Interestingly several SRA reads were found containing one nucleotide longer than TrkC-miR2–5p-CT, which could be considered as another TrkC-miR2 isomiR (Fig. 2C). Endogenous production of these isomiRs was confirmed in several cell lines in which TrkC-miR2–5p-CT expression level was higher than TrkC-miR2–5p-GC isomiR (Fig. 2D).
Differential expression of TrkC and TrkC-miR2 in different tumor samples (data not shown) suggested an independent promoter for this miRNA in TrkC gene sequences. Further evidence for the presence of an independent promoter came from HCT-116 cells treated with 5-azacytidine epidrug (3, 5). By default, TrkC gene is expressed in a very low level as a result of promoter methylation in HCT-116 cells (5). TrkC gene expression level was increased 100 times in the HCT-116 cells treated with 5-azacytidine, whereas TrkC-miR2–5p expression was not altered before and after 5-azacytidine treatment (Fig. 2E). Overall, accumulative evidence supported the presence of two or maybe three isomiRs for the novel TrkC-miR2, which is driven by an independent promoter.
Association between TrkC-miR2 and Wnt signaling pathway
APC2 as a major Wnt signaling pathway component was predicted as the highest scored target gene for TrkC-miR2–5p. Therefore, TrkC-premir2 was overexpressed in SW480 cells, and RT-qPCR results indicated that the APC2 transcription level has been down-regulated in these cells (Fig. 3A). Furthermore, a dual luciferase assay supported direct interaction of TrkC-miR2 with the wild-type sequence of APC2, 3′-UTR (Fig. 3B), whereas it did not have a similar effect on the mutated APC2, 3′-UTR sequence (Fig. 3C). Meanwhile, TrkC-premir2 overexpression did not show such an effect on the other tested Wnt signaling pathway components (supplemental Fig. 2). These results suggested TrkC-miR2 as a novel regulator that affects the Wnt signaling pathway inhibitory complex only through APC2 gene expression, and further analysis in physiological condition was needed to confirm its function.
During Wnt signaling, Wnt ligands interact with Frizzled and LRP co-receptors leading to inactivation of the tumor suppressive genes APC, GSK-3β, and Axin and finally release β-catenin oncogenic protein (32). After nuclear translocation of β-catenin, which complexes with TCF/LEF transcription factors, Wnt-responsive genes such as CyclinD1 (CCND1) is up-regulated, and cell cycle is motivated (33).
APC2 as a negative regulator of Wnt signaling pathway (26) was shown to be targeted by TrkC-miR2 (Fig. 3). Also, overexpression of TrkC-miR2 resulted in Wnt signaling up-regulation (Fig. 4, A and B). Therefore, it was interesting to know if the effect of this novel miRNA is confined to the APC2 transcripts. To do so, Wnt signaling was blocked via small molecules at three points of this pathway, and then activity of the pathway was measured through TOP/FOPflash assay system after TrkC-miR2 overexpression. When TrkC-miR2 was overexpressed in SW480 cells that were treated with IWP-2 small molecule (inhibitor of LRP receptor) (34), Wnt signaling was up-regulated compared with the cells that were treated with this small molecule as well as mock and scrambled controls (Fig. 4C). That means TrkC-miR2 effect is downstream to the LRP receptor in the Wnt signaling pathway. When TrkC-miR2 was overexpressed in the SW480 cells that were treated with PNU74654 (inhibits β-catenin and TCF interaction; Refs. 35 and 36), Wnt signaling was not significantly affected compared with the cells that were co-treated with this small molecule as well as control vectors (Fig. 4C). This result once again emphasizes that TrkC-miR2 works upstream to the β-catenin where the APC2 protein does its function. When TrkC-miR2 was overexpressed in the SW480 cells that were treated with XAV932 (up-regulates AxinI expression and down-regulates Wnt signaling; Refs. 37 and 38), the result was an elevation of Wnt signaling consistent with its down-regulation effect on APC2 expression (Fig. 4C). This experiment again introduces TrkC-miR2 as a positive regulator of Wnt signaling pathway, potentially via targeting of APC2. Finally, using antisense against the APC2 gene expression, the APC2 gene was successfully down-regulated, and then TrkC-miR2 overexpression no longer had a significant effect on the Wnt signaling activity (supplemental Fig. 4).
Although TrkC-miR2 was down-regulated and APC2 was up-regulated after the application of anti-TrkC-miR2 or APC2-3′-UTR scavenger constructs in SW480 cells, the expected Wnt signaling attenuation was not significant (Fig. 4, D–F). This might be justified by the suggestion of a saturation status for Wnt signaling inhibitory complex in which APC2 protein is a component along with APC, GSK3, and Axin proteins.
The expression level of TrkC-miR2 in HUH7 cells was much lower (1/100) than its level in SW480 cells detected by RT-qPCR (supplemental Fig. 3A). Therefore, it may justify that down-regulation of TrkC-miR2 in HUH7 cells (Fig. 4G) was not as efficient as in SW480 cells (Fig. 4D).
Successful overexpression of TrkC-miR2 in HUH7 cells (Fig. 4G) was also followed by down-regulation of the APC2 target gene (Fig. 4H), but it was not capable of affecting Wnt signaling (Fig. 4, I and J), unlike in SW480 cells (Fig. 4, A and B). This might be justified by lower Wnt activity in HUH7 cells. RT-qPCR indicated a 6000× higher APC2 expression level in HUH7 cells compared with SW480 cells (supplemental Fig. 3B). That means, although TrkC-miR2 overexpression has resulted in APC2 down-regulation, its level still has been too high to allow Wnt signaling up-regulation. Consistently, Top/flash assay counts (relative light units) compared with SW480 and HUH7 cells indicate that Wnt activity in HUH7 cells is much lower than in SW480 cells (Fig. 4, A and J). A similar low count for Wnt activity in HUH7 cells has been reported elsewhere (39). Overall, higher APC2 gene expression levels along with lower TrkC-miR2 levels in HUH7 cells compared with SW480 may justify the differential effect of TrkC-miR2 overexpression in these cell lines.
Differential expression of TrkC-miR2 in CRCs and marginal non-tumor tissue samples
Wnt signaling is generally activated in CRCs (14, 15, 40), and this pathway was activated after TrkC-miR2 overexpression followed by APC2 reduction in SW480 cell lines (Figs. 3 and 4). Therefore, TrkC-miR2 expression level was investigated in HT29, HCT-116, and SW480 cell lines, which originated from grades 1–3 of CRC tumors, respectively (41, 42). Results indicated that the TrkC-miR2 expression level is up-regulated as the grade of the cell lines increase (Fig. 5A), suggesting a TrkC-miR2-positive expression relationship with grades of malignancy. TrkC-miR2 expression level was also analyzed in CRC tumor samples compared with their non-tumor pairs. RT-qPCR results indicated an up-regulation of TrkC-miR2 in CRC tissues (Fig. 5, B–D). Pathological tumor node metastasis (PTNM) staging and histopathological analysis indicated that TrkC-miR2 expression was positively associated with different stages and grades of malignancy (Fig. 5, D and E). In other words, TrkC-miR2 was expressed more in tumors with higher malignancy grade and advanced stage. Such an effect has been reported for some other tumor biomarkers (43). Furthermore, ROC curve analysis (44) evaluated the sensitivity and specificity of TrkC-miR2 expression level for discrimination of CRC specimens (Fig. 5F). Analysis of TrkC-miR2 and APC2 target gene expression in the individual tumor samples suggested a negative correlation between them (Fig. 5, G and I). Then the Pearson correlation coefficient test confirmed a significant negative correlation between TrkC-miR2 and APC2 expression (Fig. 5H). Overall, accumulative evidence suggested that TrkC-miR2 might be a bona fide biomarker for CRC diagnosis. Of course, analysis on a greater number of CRC samples is necessary to draw a confident conclusion.
Ectopic expression of TrkC-miR2 induces cell survival
Flow cytometry analysis, MTT assay, and crystal violet staining of the cells overexpressing TrkC-miR2 indicated significant survival rate elevation of SW480 cells (Fig. 6). This effect was consistent with up-regulation of Wnt signaling (Fig. 4A) in SW480 cells overexpressing TrkC-miR2. The result is also in accordance with previously reported survival effect of TrkC (45, 46), which highlighted the effective cellular functionality of TrkC-miR2 in parallel with TrkC host gene function. It has been reported that Wnt signaling pathway is active in the HUH7 cell line (47, 48). However, our analysis indicated that the Wnt pathway is not as strong as in SW480 cells (Fig. 4, A and J), probably due to the much higher expression level of APC2 gene in HUH7 cells (supplemental Fig. 3B). That means a significant reduction of APC2 transcript levels by TrkC-miR2 overexpression in HUH7 cells (Fig. 4H) has not been critical for reduction of Wnt activity (Fig. 4, I and J). Accordingly, TrkC-miR2 overexpression has not been able to affect the cell cycle status in HUH7 cells (Fig. 6A). Therefore, differential cell cycle effects of TrkC-miR2 in HUH7 and SW480 cells could be attributed to different physiological conditions, and activity of the Wnt signaling pathway in transcript or protein levels, cell content, and genetics/epigenetics background (3, 5, 49) existed within the studied cell lines.
In conclusion, we here introduced TrkC-miR2 as a functional miRNA mapped onto the 14th intron of TrkC gene together with accumulated evidence for its identity and oncogenic functionality against the components of Wnt signaling pathway, especially against APC2 gene expression. The present evidence revealed a significant up-regulation of TrkC-miR2 in CRC tumors and suggest it as a potential biomarker for CRC progression.
Experimental procedures
Bioinformatics prediction of miRNA and its candidate target genes
SSC profiler and miPRED bioinformatics tools were used to predict bona fide hairpin structures within TrkC gene. Drosha processing sites were predicted by using CID-miRNA software along with Microprocessor SVM program. TrkC-miR2 and its precursor sequence conservation status were examined by using Mireval along with blat search against human genome and other organisms in UCSC database. MatureBayes, Pmirp, MiRNA Spotter, MiRmat, and MirZ online tools also predicted TrkC-miR2. The miRBase database was used to search for similar sequences of TrkC-miR2 and its precursor in different species. RNAFOLD algorithm was used for prediction of RNA secondary structure. Potential target genes of TrkC-miR2 were analyzed by RNAHybrid and DIANA-microT tools. Alibaba 2.1, P-Match, Tfsitescan, and Promoter2.0 Prediction Server bioinformatics tools were used for prediction of potential promoter sequences upstream of the putative stem-loop. To find the pathway in which TrkC-miR2 is involved, DAVID (david.abcc.ncifcrf.gov/), Diana-mirpath, and geneset2 miRNA online tools were applied. RNA-sequencing data were searched by using NCBI-SRA to examine the existence of novel discovered miRNAs in small RNA-sequencing data.
Cell lines
HCT116 and HT-29 cell lines were cultured in RPMI 1640 medium (Invitrogen), and SW480, HEK293t and HUH7 cell lines were cultured in DMEM-HG (Invitrogen). These media were supplemented with 100 units/ml penicillin, 100 μg/ml streptomycin (Sigma), and 10% fetal bovine serum (Invitrogen) and followed by incubation at 37 °C with 5% CO2. HCT116 (ID# C570) and SW480 (ID# C506) cell lines were obtained from Pasteur Institute/Iran, and HEK293t (ID# IBRC C10683) and HT-29 (ID# IBRC C10097) were purchased from the National Center for Genetic and biological reserves in Iran.
Tissue samples
36 CRC tissue samples were obtained from Imam Khomeini and Dr. Shariati Hospitals, Tehran, Iran, and stored at −80 °C until used.
DNA constructs
To clone the region corresponding to TrkC-miR2 sense and antisense sequences, ∼802 bp of human TrkC-intron-14 were PCR-amplified using Int-F and Int-R primers (Table 1) and cloned into pEGFP-C1 expression vector (Clontech) downstream of the GFP sequence both in sense and antisense directions. Human genomic DNA was extracted from white blood cells using standard protocol (50). A previously described hairpin structure sequence (51) was cloned into pEGFP-C1 vector as the scrambled control. TOP/FOP flash was also constructed into the pGL4 vector. All recombinant vectors were sequenced for verification of the correct insert.
Table 1.
Primer and oligo sequences that were used in the study
| Primer name | Primer sequence, 5′ to 3′ | Amplicon size (base pairs) |
|---|---|---|
| TrkC-real time | Forward, CCTGTGTCCTGTTGGTGGTTCTC | 195 |
| Reverse, GAGTCATGCCAATGACCACAGTGTC | ||
| TrkC-premir2 | GATTCTGAGCTGCACAGTCCAG | |
| TrkC-miR2-5p | GGCTGGGGATTCTGAGCT | |
| U48 | Forward, TGACCCCAGGTAACTCTGAGTGTGT | |
| Anchored oligo (dT) | GCGTCGACTAGTACAACTCAAGGTTCTTCCAGTCACGACG (T)18V | |
| Universal-outer | GCGTCGACTAGTACAACTCAAG | |
| Universal-inner | AACTCAAGGTTCTTCCAGTCACG | |
| B-cat-real time | Forward, AGAACAGAGCCAATGGCTTG | 130 |
| Reverse, CCTGGCCATATCCACCAGAG | ||
| c-myc-real time | Forward, CTCCTACGTTGCGGTCACAC | 142 |
| Reverse, CGGGTCGCAGATGAAACTCT | ||
| GAPDH | Forward, GCCACATCGCTCAGACAC | 115 |
| Reverse, GGCAACAATATCCACTTTACCAG | ||
| TrkC-Intron | Int-F: CTGGCGGCCGCTGAACAAGGGAGATGGCTCAGTGG | 802 |
| Int-R: TAGACGCGTGGCTTTGCTGTCACCGCTGAGG | ||
| APC2-3′-UTR | Forward, GGGCGAAGCCTGTAATCACTGC | 2515 |
| Reverse, GAGTCGGACAGCTGACGGTG | ||
| Axin1-3′-UTR | Forward, AAGGTGGACTGATAGGCTGGT | 715 |
| Reverse, AGAAGACACACCACAGCCAGG | ||
| APC1-3′-UTR | Forward, TGGAACCCAAAGTCCTAAGC | 2140 |
| Reverse, CTGGGAAAACAACAGAAGTAG | ||
| APC2-real time | Forward, TCCCAGCTCCCTGCCTCTGT | 129 |
| Reverse, AGCCAGCCAGACCCAAGTTCT | ||
| APC1-real time | Forward, TATTACGGAATGTGTCCAGCTTG | 133 |
| Reverse, CCACATGCATTACTGACTATTGTC | ||
| Axin1-real time | Forward, ATGCAGGAGAGCGTGCAGGTC | 237 |
| Reverse, TGACGATGGATCGCCGTCCTC |
Dual luciferase assay
Wild-type sequence of APC23′-UTRs was cloned in psiCHECK vector downstream of luciferase gene for dual luciferase assay analysis according to its manufacture's protocol (Promega kit). Also, predicted MREs in this sequence were mutated, and amplified products were cloned in psiCHECK vector as negative controls (supplemental Fig. 1).
TOP/FOP flash assay
A TOP/FOP flash assay was used for Wnt signaling pathway analysis, and to this aim the TCF/LEF-responsive luciferase construct was made under the control of minimal TK promoter and tandem repeats of the TCF/LEF transcriptional response element (TRE). The cells were plated in 48-well plates and transfected with 400 ng of miRNA-encoding vector and with 200 ng of luciferase-encoding vector. The cells were harvested 48 h post-transfection, and luciferase activity was measured by using the Dual-Glo luciferase assay kit (Promega).
RNA preparation
Total RNA was isolated by using TRIzol (Invitrogen) according to the manufacturer's protocol and treated with RNase-free DNAaseI (Fermentas) and qualified on the agarose gel.
Primer designing
RT-qPCR was applied for detection of TrkC-miR2 and its precursor and expression analysis of miRNA host and targets genes. Related primers were designed using NCBI Primer-blast (www.ncbi.nlm.nih.gov), IDT oligo analyzer, and MWG online PCR primer design tools. Primer and oligo sequences that were used in the study are listed in Table 1.
CDNA synthesis and RT-qPCR for detection of TrkC-miR2 and its precursor
CDNAs were made from polyadenylated RNAs according to the protocol (27), and each cDNA sample was amplified using specific primers in a real-time PCR system (Applied Biosystems) using the following conditions for 45 cycles: stage 1, 95 °C for 5 s; stage 2, 60 °C for 20 s; stage 3, 72 °C for 30s. RT-qPCR was performed according to Minimum Information for Publication of Quantitative Real-Time PCR Experiments (MIQE) guidelines using SYBRPremix Ex-TaqTM II PCR Mastermix (Takara, Japan) in experimental duplicates. Expression data were analyzed using endogenous U48 and GAPDH as the reference genes and were normalized using the 2−ΔCt and 2−ΔΔCt methods (52).
Overexpression of TrkC-premir2 in cell lines
The pEGFP-C1 expression vector containing TrkC-miR2 precursor (0.8 μg DNA) was engulfed in Lipofectamine 2000 (Invitrogen) and used for transfection of HEK293t, SW480, and HUH7 cell lines. 24 h later GFP microscopy (by Nikon eclipse Te2000-s) ensured successful transfection.
Azacytidine treatment
Azacytidine dissolved in water (Sigma) was used for treatment of HCT116 cells with a final concentration of 10 μm for 48 h. Then total RNA was extracted from these cells and used for RT-qPCR analysis.
Small molecules treatment
SW480 cells were seeded in 48-well plates, and 30 h later IWP-2, PNU74654, and XAV932 small molecules were applied in concentrations of 5, 6, and 5 μm for 10 h, respectively. Then, after 8 h of starvation, interested genetic constructs were transfected to the cells, and media were refreshed after 6 h. Then again small molecules were applied to resume their inhibitory effects. 48 h after transfection, the cells were lysed, and TOP/FOP flash assays were performed.
Cell cycle analysis
Cells were transfected with overexpression cassettes of TrkC-premir2 and anti-TrkC-miR2 and were harvested 36 h after transfection and stained with propidium iodide. All of the samples were analyzed with a FACS Calibur flow cytometer using Cell Quest software (BD Biosciences).
MTT assay
HUH7 and SW480 cells (8000 cells/well) were plated in a 96-well plate in tetraplicate. After 24 h they were transfected by interested constructs engulfed with Lipofectamine 2000. 20 μl of 5 mg/ml MTT (Sigma) was added to each well 36 h post-transfection followed by further incubation at 37 °C for 4 h, after which the culture medium was removed, and 100 μl of DMSO (Sigma) was added to each well to dissolve the formazan crystals. A490 was measured with an ELISA Microplate Reader (Biotek) as a function of cell viability
Statistical analysis
RT-qPCR data were analyzed Using DataAssist software V3.0 (53). Other statistical analysis was performed with GraphPad Prism 5.04 (GraphPad, San Diego, CA). For flow cytometry studies, data showing the percent of cell population within the negative group and test group were compared by using the Repeated Measures analysis of variance test followed by the Bonferroni test using GraphPad. RT-qPCR data resulting from 36 CRC tissue samples were analyzed using an unpaired non-parametric Mann-Whitney test by treating tumor and non-tumor samples as two independent groups using GraphPad Prism and SPSS software.
Author contributions
S. D. and B. M. S. conceived and designed the experiments. S. D., A Y., H. N., and M. J. performed the experiments. S. D. and B. M. S. analyzed the data. B. M. S., S. J. M., and M. N. contributed the reagents, materials, and analysis tools. S. D. and B. M. S. wrote the paper.
Supplementary Material
Acknowledgments
We thank Dr. Saman Hosseinkhani, Dr. Masood Soleimani, and Ali Fasihi for kind advice.
This work was supported by financial aid from Tarbiat Modares University and Iran National Science Foundation Grant 91001522. The authors declare that they have no conflicts of interest with the contents of this article.
This article contains supplemental Fig. S1–S4.
- TrkC
- tropomyosin receptor kinase C
- CRC
- colorectal cancer
- miRNA
- microRNA
- qPCR
- quantitative PCR
- MTT
- 3-(4,5-dimethylthiazol-2-yl)-2,5-diphenyltetrazolium bromide
- SRA
- Sequence Read Archive
- ROC
- receiver operating characteristic
- PTNM
- pathological tumor node metastasis
- TCF
- T-cell factor
- LEF
- lymphoid enhancer factor
- oligo
- oligonucleotide
- MRE
- miRNA recognition element.
References
- 1. Reichardt L. F. (2006) Neurotrophin-regulated signalling pathways. Philos. Trans. R. Soc. Lond. B. Biol. Sci. 361, 1545–1564 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2. McGregor L. M., McCune B. K., Graff J. R., McDowell P. R., Romans K. E., Yancopoulos G. D., Ball D. W., Baylin S. B., and Nelkin B. D. (1999) Roles of trk family neurotrophin receptors in medullary thyroid carcinoma development and progression. Proc. Natl. Acad. Sci. U.S.A. 96, 4540–4545 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Genevois A.-L., Ichim G., Coissieux M.-M., Lambert M.-P., Lavial F., Goldschneider D., Jarrosson-Wuilleme L., Lepinasse F., Gouysse G., Herceg Z., Scoazec J. Y., Tauszig-Delamasure S., and Mehlen P. (2013) Dependence receptor TrkC is a putative colon cancer tumor suppressor. Proc. Natl. Acad. Sci. U.S.A. 110, 3017–3022 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Jin W., Kim G. M., Kim M. S., Lim M. H., Yun C., Jeong J., Nam J.-S., and Kim S.-J. (2010) TrkC plays an essential role in breast tumor growth and metastasis. Carcinogenesis 31, 1939–1947 [DOI] [PubMed] [Google Scholar]
- 5. Luo Y., Kaz A. M., Kanngurn S., Welsch P., Morris S. M., Wang J., Lutterbaugh J. D., Markowitz S. D., and Grady W. M. (2013) NTRK3 is a potential tumor suppressor gene commonly inactivated by epigenetic mechanisms in colorectal cancer. PLoS Genet. 9, e1003552. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Aranha M. M., Santos D. M., Solá S., Steer C. J., and Rodrigues C. (2011) miR-34a regulates mouse neural stem cell differentiation. PloS ONE 6, e21396. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Krol J., Loedige I., and Filipowicz W. (2010) The widespread regulation of microRNA biogenesis, function and decay. Nat. Rev. Genet. 11, 597–610 [DOI] [PubMed] [Google Scholar]
- 8. Wang Z. (2010) MicroRNA: a matter of life or death. World J. Biol. Chem. 1, 41–54 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Wang Y., and Lee C. G. (2009) MicroRNA and cancer: focus on apoptosis. J. Cell. Mol. Med. 13, 12–23 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Miranda K. C., Huynh T., Tay Y., Ang Y.-S., Tam W.-L., Thomson A. M., Lim B., and Rigoutsos I. (2006) A pattern-based method for the identification of MicroRNA binding sites and their corresponding heteroduplexes. Cell 126, 1203–1217 [DOI] [PubMed] [Google Scholar]
- 11. Berezikov E., Cuppen E., and Plasterk R. H. (2006) Approaches to microRNA discovery. Nat. Genet. 38, S2–S7 [DOI] [PubMed] [Google Scholar]
- 12. Berezikov E., Guryev V., van de Belt J., Wienholds E., Plasterk R. H., and Cuppen E. (2005) Phylogenetic shadowing and computational identification of human microRNA genes. Cell 120, 21–24 [DOI] [PubMed] [Google Scholar]
- 13. Ng E. K., Chong W. W., Jin H., Lam E. K., Shin V. Y., Yu J., Poon T. C., Ng S. S., and Sung J. J. (2009) Differential expression of microRNAs in plasma of colorectal cancer patients: a potential marker for colorectal cancer screening. Gut 58, 1375–1381 [DOI] [PubMed] [Google Scholar]
- 14. Suzuki H., Watkins D. N., Jair K.-W., Schuebel K. E., Markowitz S. D., Chen W. D., Pretlow T. P., Yang B., Akiyama Y., Van Engeland M., Toyota M., Tokino T., Hinoda Y., Imai K., Herman J. G., and Baylin S. B. (2004) Epigenetic inactivation of SFRP genes allows constitutive WNT signaling in colorectal cancer. Nat. Genet. 36, 417–422 [DOI] [PubMed] [Google Scholar]
- 15. Bienz M., and Clevers H. (2000) Linking colorectal cancer to Wnt signaling. Cell 103, 311–320 [DOI] [PubMed] [Google Scholar]
- 16. Yang L., Belaguli N., and Berger D. H. (2009) MicroRNA and colorectal cancer. World J. Surg. 33, 638–646 [DOI] [PubMed] [Google Scholar]
- 17. Slaby O., Svoboda M., Fabian P., Smerdova T., Knoflickova D., Bednarikova M., Nenutil R., and Vyzula R. (2007) Altered expression of miR-21, miR-31, miR-143, and miR-145 is related to clinicopathologic features of colorectal cancer. Oncology 72, 397–402 [DOI] [PubMed] [Google Scholar]
- 18. Bandrés E., Cubedo E., Agirre X., Malumbres R., Zárate R., Ramirez N., Abajo A., Navarro A., Moreno I., Monzó M., and García-Foncillas J. (2006) Identification by real-time PCR of 13 mature microRNAs differentially expressed in colorectal cancer and non-tumoral tissues. Mol. Cancer 5, 29. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Dokanehiifard S., Soltani B. M., Parsi S., Hosseini F., Javan M., and Mowla S. J. (2015) Experimental verification of a conserved intronic microRNA located in the human TrkC gene with a cell type-dependent apoptotic function. Cell. Mol. Life Sci. 72, 2613–2625 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Dalmay T. (2008) MicroRNAs and cancer. J. Intern. Med. 263, 366–375 [DOI] [PubMed] [Google Scholar]
- 21. Abbott A. L., Alvarez-Saavedra E., Miska E. A., Lau N. C., Bartel D. P., Horvitz H. R., and Ambros V. (2005) The let-7 MicroRNA family members mir-48, mir-84, and mir-241 function together to regulate developmental timing in Caenorhabditis elegans. Dev. Cell 9, 403–414 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Gomes C. P., Cho J.-H., Hood L., Franco O. L., Pereira R. W., and Wang K. (2013) A review of computational tools in microRNA discovery. Front. Genet. 4, 81. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Hapner S. J., Boeshore K. L., Large T. H., and Lefcort F. (1998) Neural differentiation promoted by truncated trkC receptors in collaboration with p75 NTR. Dev. Biol. 201, 90–100 [DOI] [PubMed] [Google Scholar]
- 24. Verdi J. M., Birren S. J., Ibáñez C. F., Persson H., Kaplan D. R., Benedetti M., Chao M. V., and Anderson D. J. (1994) p75 LNGFR regulates Trk signal transduction and NGF-induced neuronal differentiation in MAH cells. Neuron 12, 733–745 [DOI] [PubMed] [Google Scholar]
- 25. Nishita M., Hashimoto M. K., Ogata S., Laurent M. N., Ueno N., Shibuya H., and Cho K. W. (2000) Interaction between Wnt and TGF-β signalling pathways during formation of Spemann's organizer. Nature 403, 781–785 [DOI] [PubMed] [Google Scholar]
- 26. Kunttas-Tatli E., Zhou M.-N., Zimmerman S., Molinar O., Zhouzheng F., Carter K., Kapur M., Cheatle A., Decal R., and McCartney B. M. (2012) Destruction complex function in the Wnt signaling pathway of Drosophila requires multiple interactions between adenomatous polyposis coli 2 and Armadillo. Genetics 190, 1059–1075 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Parsi S., Soltani B. M., Hosseini E., Tousi S. E., and Mowla S. J. (2012) Experimental verification of a predicted intronic microRNA in human NGFR gene with a potential pro-apoptotic function. PloS ONE 7, e35561. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Li B., Duan H., Li J., Deng X. W., Yin W., and Xia X. (2013) Global identification of miRNAs and targets in Populus euphratica under salt stress. Plant Mol. Biol. 81, 525–539 [DOI] [PubMed] [Google Scholar]
- 29. Neilsen C. T., Goodall G. J., and Bracken C. P. (2012) IsomiRs: the overlooked repertoire in the dynamic microRNAome. Trends Genet. 28, 544–549 [DOI] [PubMed] [Google Scholar]
- 30. McGahon M. K., Yarham J. M., Daly A., Guduric-Fuchs J., Ferguson L. J., Simpson D. A., and Collins A. (2013) Distinctive profile of isomiR expression and novel microRNAs in rat heart left ventricle. PloS ONE 8, e65809. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Kalkman H. O. (2009) Altered growth factor signaling pathways as the basis of aberrant stem cell maturation in schizophrenia. Pharmacol. Ther. 121, 115–122 [DOI] [PubMed] [Google Scholar]
- 32. Nelson W. J., and Nusse R. (2004) Convergence of Wnt, β-catenin, and cadherin pathways. Science 303, 1483–1487 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Mosimann C., Hausmann G., and Basler K. (2006) Parafibromin/hyrax activates Wnt/Wg target gene transcription by direct association with β-catenin/armadillo. Cell 125, 327–341 [DOI] [PubMed] [Google Scholar]
- 34. Chen B., Dodge M. E., Tang W., Lu J., Ma Z., Fan C.-W., Wei S., Hao W., Kilgore J., Williams N. S., Roth M. G., Amatruda J. F., Chen C., and Lum L. (2009) Small molecule-mediated disruption of Wnt-dependent signaling in tissue regeneration and cancer. Nat. Chem. Biol. 5, 100–107 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. Demilly A., Steinmetz P., Gazave E., Marchand L., and Vervoort M. (2013) Involvement of the Wnt/β-catenin pathway in neurectoderm architecture in Platynereis dumerilii. Nat. Commun. 4, 1915. [DOI] [PubMed] [Google Scholar]
- 36. Kahn M. (2014) Can we safely target the WNT pathway? Nat. Rev. Drug Discov. 13, 513–532 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37. Bao R., Christova T., Song S., Angers S., Yan X., and Attisano L. (2012) Inhibition of tankyrases induces Axin stabilization and blocks Wnt signalling in breast cancer cells. PloS ONE 7, e48670. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38. Cammarata P. R., Neelam S., and Brooks M. M. (2015) Inhibition of hypoxia inducible factor-1α down-regulates the expression of epithelial to mesenchymal transition early marker proteins without undermining cell survival in hypoxic lens epithelial cells. Mol. Vis. 21, 1024–1035 [PMC free article] [PubMed] [Google Scholar]
- 39. Liu J., Ding X., Tang J., Cao Y., Hu P., Zhou F., Shan X., Cai X., Chen Q., Ling N., Zhang B., Bi Y., Chen K., Ren H., and Huang A. (2011) Enhancement of canonical Wnt/β-catenin signaling activity by HCV core protein promotes cell growth of hepatocellular carcinoma cells. PLoS ONE 6, e27496. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40. Morin P. J., Sparks A. B., Korinek V., Barker N., Clevers H., Vogelstein B., and Kinzler K. W. (1997) Activation of β-catenin-Tcf signaling in colon cancer by mutations in β-catenin or APC. Science 275, 1787–1790 [DOI] [PubMed] [Google Scholar]
- 41. Kondoh N., Schweinfest C. W., Henderson K. W., and Papas T. S. (1992) Differential expression of S19 ribosomal protein, laminin-binding protein, and human lymphocyte antigen class I messenger RNAs associated with colon carcinoma progression and differentiation. Cancer Res. 52, 791–796 [PubMed] [Google Scholar]
- 42. McInroy L., and Määttä A. (2011) Plectin regulates invasiveness of SW480 colon carcinoma cells and is targeted to podosome-like adhesions in an isoform-specific manner. Exp. Cell Res. 317, 2468–2478 [DOI] [PubMed] [Google Scholar]
- 43. Link A., Balaguer F., Shen Y., Nagasaka T., Lozano J. J., Boland C. R., and Goel A. (2010) Fecal MicroRNAs as novel biomarkers for colon cancer screening. Cancer Epidemiol. Biomarkers Prev. 19, 1766–1774 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44. Hartwell L., Mankoff D., Paulovich A., Ramsey S., and Swisher E. (2006) Cancer biomarkers: a systems approach. Nat. Biotechnol. 24, 905–908 [DOI] [PubMed] [Google Scholar]
- 45. Kumar S., Kahn M. A., Dinh L., and de Vellis J. (1998) NT-3-mediated TrkC receptor activation promotes proliferation and cell survival of rodent progenitor oligodendrocyte cells in vitro and in vivo. J. Neurosci. Res. 54, 754–765 [DOI] [PubMed] [Google Scholar]
- 46. Minichiello L., and Klein R. (1996) TrkB and TrkC neurotrophin receptors cooperate in promoting survival of hippocampal and cerebellar granule neurons. Genes Dev. 10, 2849–2858 [DOI] [PubMed] [Google Scholar]
- 47. Xu N., Shen C., Luo Y., Xia L., Xue F., Xia Q., and Zhang J. (2012) Upregulated miR-130a increases drug resistance by regulating RUNX3 and Wnt signaling in cisplatin-treated HCC cell. Biochem. Biophys. Res. Commun. 425, 468–472 [DOI] [PubMed] [Google Scholar]
- 48. Zhang Y., Wei W., Cheng N., Wang K., Li B., Jiang X., and Sun S. (2012) Hepatitis C virus-induced up-regulation of microRNA-155 promotes hepatocarcinogenesis by activating Wnt signaling. Hepatology 56, 1631–1640 [DOI] [PubMed] [Google Scholar]
- 49. Zhu S., Wu H., Wu F., Nie D., Sheng S., and Mo Y.-Y. (2008) MicroRNA-21 targets tumor suppressor genes in invasion and metastasis. Cell Res. 18, 350–359 [DOI] [PubMed] [Google Scholar]
- 50. Sambrook J., Fritsch E., and Maniatis T. (1989) Molecular Cloning: A Laboratory Manual, Cold Spring Harbor Laboratory Press, Cold Spring Harbor, New York [Google Scholar]
- 51. Xu N., Papagiannakopoulos T., Pan G., Thomson J. A., and Kosik K. S. (2009) MicroRNA-145 regulates OCT4, SOX2, and KLF4 and represses pluripotency in human embryonic stem cells. Cell 137, 647–658 [DOI] [PubMed] [Google Scholar]
- 52. Livak K. J., and Schmittgen T. D. (2001) Analysis of relative gene expression data using real-time quantitative PCR and the 2−ΔΔCt method. Methods 25, 402–408 [DOI] [PubMed] [Google Scholar]
- 53. Mestdagh P., Van Vlierberghe P., De Weer A., Muth D., Westermann F., Speleman F., and Vandesompele J. (2009) A novel and universal method for microRNA RT-qPCR data normalization. Genome Biol. 10, R64. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.