Abstract
Ceramide and more complex sphingolipids constitute a diverse group of lipids that serve important roles as structural entities of biological membranes and as regulators of cellular growth, differentiation, and development. Thus, ceramides are vital players in numerous diseases including metabolic and cardiovascular diseases, as well as neurological disorders. Here we show that acyl-coenzyme A-binding protein (ACBP) potently facilitates very-long acyl chain ceramide synthesis. ACBP increases the activity of ceramide synthase 2 (CerS2) by more than 2-fold and CerS3 activity by 7-fold. ACBP binds very-long-chain acyl-CoA esters, which is required for its ability to stimulate CerS activity. We also show that high-speed liver cytosol from wild-type mice activates CerS3 activity, whereas cytosol from ACBP knock-out mice does not. Consistently, CerS2 and CerS3 activities are significantly reduced in the testes of ACBP−/− mice, concomitant with a significant reduction in long- and very-long-chain ceramide levels. Importantly, we show that ACBP interacts with CerS2 and CerS3. Our data uncover a novel mode of regulation of very-long acyl chain ceramide synthesis by ACBP, which we anticipate is of crucial importance in understanding the regulation of ceramide metabolism in pathogenesis.
Keywords: ceramide, ceramide synthase, fatty acid metabolism, lipid binding protein, sphingolipid, ACBP, Acyl-CoA
Introduction
Sphingolipids (SLs),4 and in particular ceramides, mediate a number of cellular effects (1, 2) including insulin resistance and are among the most deleterious lipids that perturb pancreatic β-cell function, vascular reactivity, and mitochondrial metabolism, which underlie the development of diabetes, obesity, and cardiovascular disease (3–5). Importantly, inhibition of ceramide synthesis ameliorates many metabolic disorders in rodents (5). The ceramide N-acyl chain length significantly impacts the function of ceramides (6). For instance, ablation of ceramide synthase (CerS) 2, which catalyzes the formation of very-long (C22–24) acyl chain ceramides, results in hepatic insulin resistance in mice (7). Despite increased appreciation of the importance of ceramides, little information is available to allow delineation of the mechanism(s) of regulation of ceramide synthesis. Six CerS isoforms exist in mammals, each synthesizing ceramides with a specific acyl chain length (8). Thus, we only have limited knowledge about the structure and function of the CerS isoforms; however, it is known that the active site resides in a sequence of 150 residues within the Tram-Lag-CLN8 domain (9) and that CerS can be regulated by phosphorylation, by homo- or heterodimerization, and by interaction with fatty acid elongase, ELOVL1 (10–12). The CerS are highly homologous proteins and reside in the endoplasmic reticulum where they have access to their two substrates, sphinganine and acyl-CoA esters.
A number of attempts have been made to model flux through the SL biosynthetic pathway (13, 14). In contrast, experimental analysis of flux is limited to studies showing that the availability of fatty acids and serine can affect ceramide synthesis. This indicates that SL flux is coupled to other metabolic routes including synthesis and degradation of storage lipids like triacylglycerols (15, 16), fatty acid elongation, and fatty acid availability (12). Interestingly, acyl-CoA synthetase 5 (ACSL5) has recently been functionally coupled to ceramide synthesis via interaction with ceramide synthases (17). Acyl-CoA esters are generated in the cytosol via the action of acyl-CoA synthetases (18) and subsequently sequestered by high-affinity acyl-CoA-binding domain proteins (ACBDs), which determine the metabolic routes and regulatory properties of acyl-CoA esters. The ACBDs comprise a highly conserved multigene family of intracellular lipid-binding proteins (19). Expression analyses confirm that ACBD1, also commonly known as ACBP, is by far the most abundantly expressed paralog (19). Recent genetic studies show that loss of ACBP in Saccharomyces cerevisiae and Caenorhabditis elegans abrogates fatty acid chain elongation and SL synthesis, resulting in compromised plasma membrane integrity and membrane function (20).5
We now show for the first time that ACBP potently activates CerS2 and CerS3 activity via direct interaction, and we show that ACBP ablation has deleterious effects on SL levels in mouse testis. We suggest that this novel interaction acts as a critical mode of regulation of very-long-chain ceramide synthesis and as a putative site of interconnection between pathways of glycerolipid and SL synthesis. Hence, we suggest that the interaction between ACBP and CerS is important for directional channeling of acyl moieties in various cellular states.
Results
ACBP stimulates CerS activity
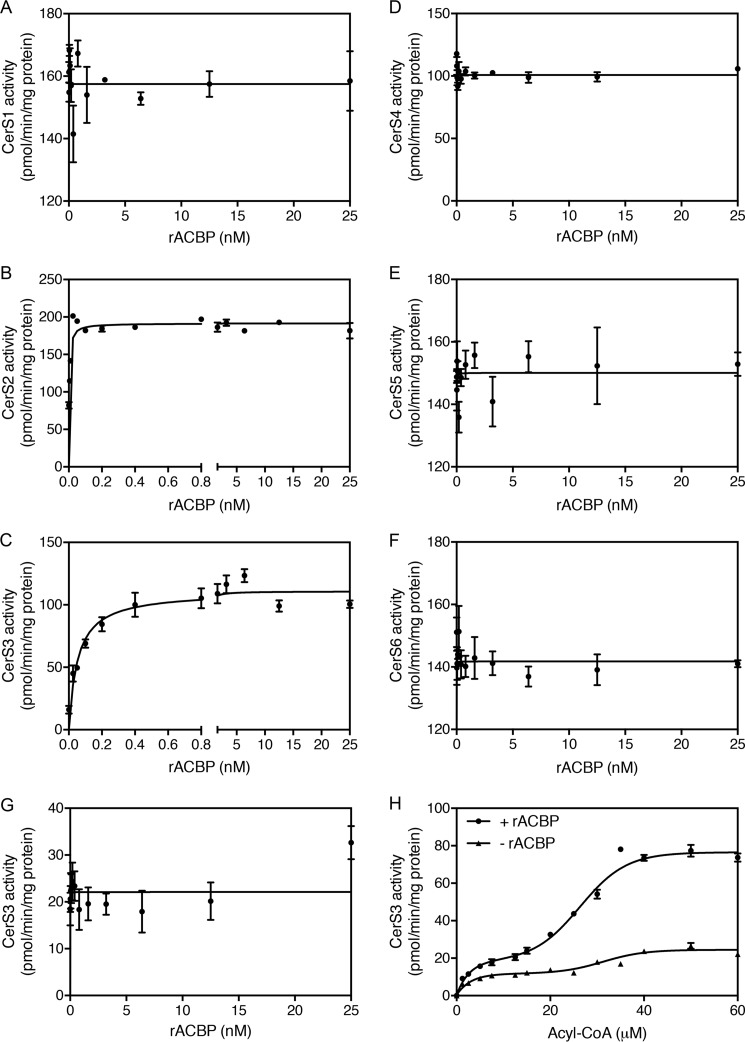
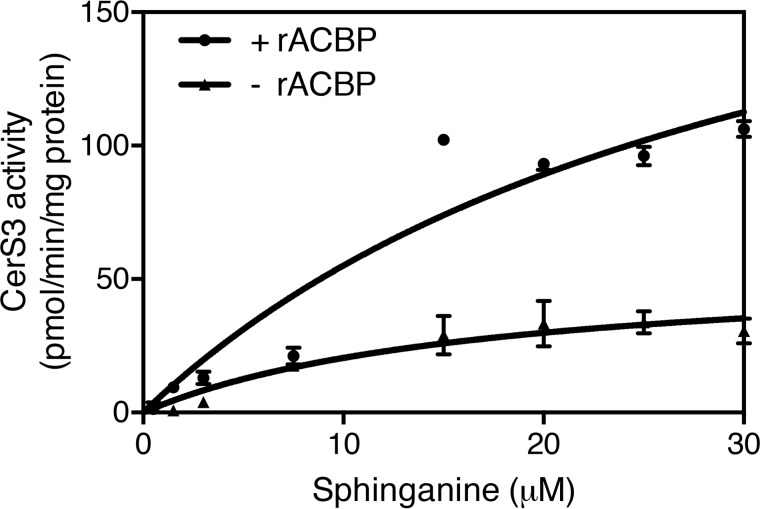
Ceramide synthases catalyze the N-acylation of sphinganine using acyl-CoA esters as substrate. By virtue of its ability to bind acyl-CoA esters, we speculated that CerS activity is determined by the availability of acyl-CoA esters. We therefore assayed CerS activity in homogenates of HEK293T cells overexpressing each of the six CerS isoforms in the presence or absence of ACBP. These studies showed that recombinant ACBP potently stimulates both CerS2 and CerS3 activity. In the absence of ACBP, CerS2 activity was 82 ± 7 pmol/mg/min, which increased more than 2-fold in the presence of as little as 0.25 nm ACBP (Fig. 1B). In the absence of ACBP, CerS3 activity was 16.0 ± 5.4 pmol/mg/min, which increased nearly 7-fold upon addition of ACBP (Fig. 1C). ACBP had no effect on the activity of the other CerS isoforms (Fig. 1) and did not affect the activity of a catalytic inactive form of CerS3 (Fig. 1). CerS activity increased in a biphasic manner (Fig. 1) upon increasing C26:0-CoA levels while keeping ACBP levels constant, indicating that CerS3 has both low- and high-affinity binding components. Furthermore, the Vmax of CerS3 toward sphinganine was increased from 55 to 235 pmol/mg/min in the presence of ACBP (Fig. 2).
Figure 1.
ACBP stimulates CerS2 and CerS3 activity in vitro. Homogenates were prepared from HEK293T cells overexpressing Cers1 (A), CerS2 (B), CerS3 (C), CerS4 (D), CerS5 (E), or CerS6 (F) or a catalytic inactive form of CerS3 (W15R) (G). CerS activity was determined using C18:0-CoA (A), C24:1-CoA (B), C26:0-CoA (C, G, and H), C20:0-CoA (D), and C16:0-CoA (E and F), respectively, in the presence of increasing amounts of ACBP. H, CerS3 activity was assayed in the presence of increasing amounts of C26:0-CoA in the presence or absence of 1 nm recombinant ACBP. The results are mean values ± S.D., n = 3 for A–G, and n = 6 for H.
Figure 2.
ACBP increases CerS3 activity. Homogenates were prepared from HEK293T cells overexpressing CerS3, and CerS3 activity was determined using C26:0-CoA in the presence of increasing amounts of sphinganine. The results are shown as means ± S.D., n = 3.
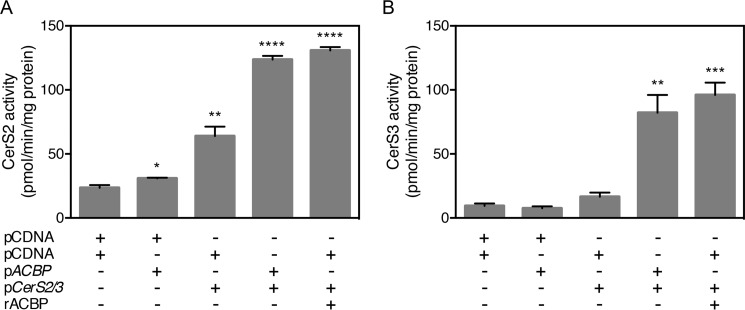
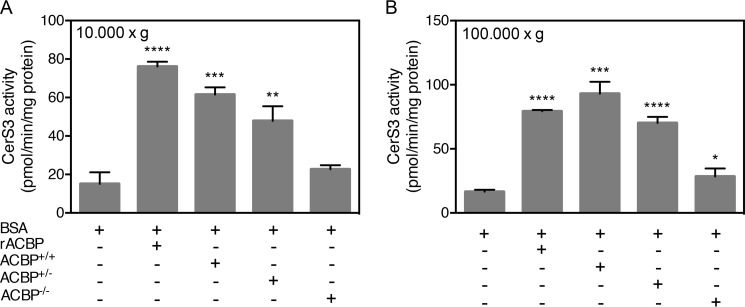
To further substantiate the finding that ACBP stimulates CerS activity, HEK293T cells were co-transfected with ACBP and CerS2 or CerS3. The activities of both CerS2 and CerS3 significantly increased upon co-transfection to levels that were comparable with CerS activity upon addition of purified recombinant ACBP to cell lysates (Fig. 3). Moreover, because ACBP is a highly abundant cytosolic protein, we hypothesized that addition of cytosol would stimulate CerS3 activity. Indeed, we demonstrated that both low speed (10,000 × g) and high-speed (100,000 × g) supernatants from wild-type mouse liver tissue stimulate CerS3 activity in vitro (Fig. 4). Importantly no stimulation of CerS3 activity was observed when using cytosol from an ACBP−/− mouse, confirming that ACBP is the main factor in liver cytosol capable of activating CerS. This was further supported by the dose-dependent stimulation of CERS activity using cytosol from heterozygous ACBP+/− mouse liver resulting in ∼50% activity relative to wild-type cytosol (Fig. 4). Together, these observations demonstrate that the activity of both CerS2 and CerS3 is highly regulated by substrate availability mediated by ACBP.
Figure 3.
Co-expression of ACBP in HEK293T cells increases CerS2 and CerS3 activity. CerS2-HA (A) or CerS3-HA (B) were co-expressed in HEK293T cells with Flag-tagged ACBP, and CerS2 or CerS3 activity was assayed. CerS2 or CerS3 activity was also assayed in cell lysates from cells transfected with CerS2 or CerS3 after the addition of purified, recombinant ACBP. The data shown represent means ± S.D., n = 3. Statistical analyses were performed using unpaired t test. *, p < 0.05; **, p < 0.01; ***, p < 0.001; ****, p < 0.0001.
Figure 4.
CerS3 activity is activated by high-speed cytosol. Homogenates were prepared from HEK293T cells overexpressing CerS3, and CerS3 activity was determined using C26:0-CoA after addition of purified recombinant ACBP or liver 10,000 × g cytosol (A) or liver 100,000 × g cytosol (B) prepared from ACBP+/+, ACBP+/−, or ACBP−/− mice. The data shown represent means ± S.D., n = 3. The statistical analyses were performed using unpaired t test. *, p < 0.05; **, p < 0.01; ***, p < 0.001; ****, p < 0.0001.
ACBP ablation alters CerS activity and the sphingolipidome of mouse testis
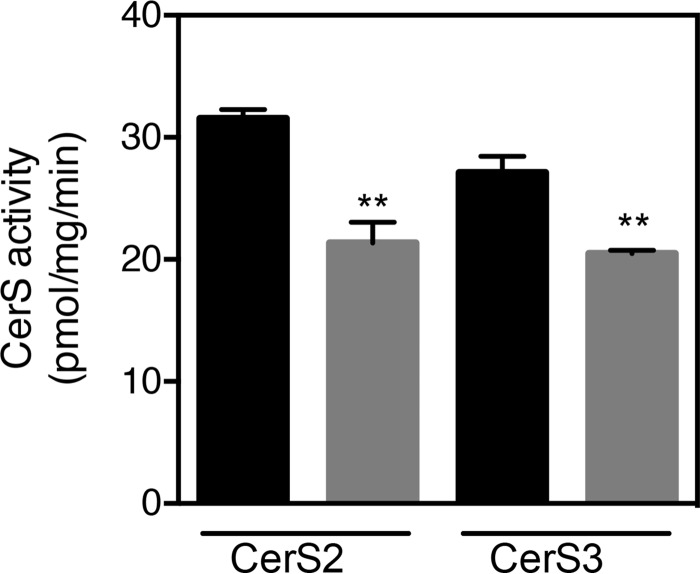
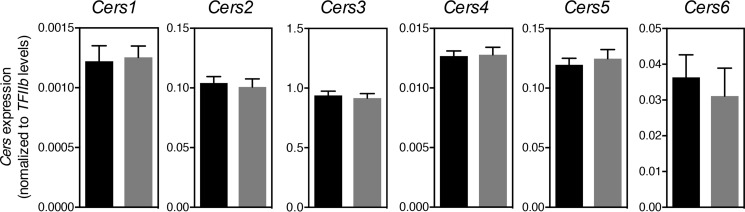
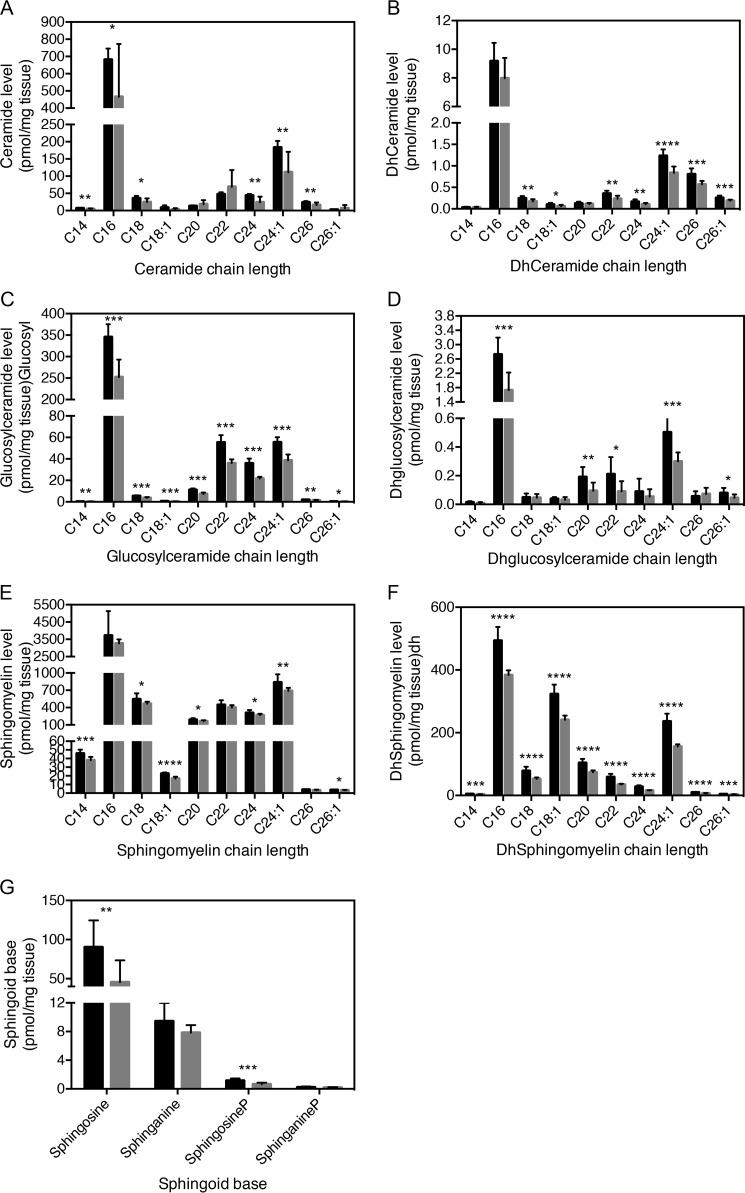
Because ACBP potently stimulates CerS2 and CerS3 activity in vitro, we examined CerS2 and CerS3 activities in testis from ACBP depleted (ACBP−/−) mice. Consistently, we observed significantly reduced CerS2 and CerS3 activity in testis from ACBP−/− mice (Fig. 5), which was not due to altered Cers mRNA expression (Fig. 6). This prompted us to examine the sphingolipidome in ACBPv mice. Thus, we analyzed the SL profile of testis from ACBP+/+ and ACBP−/− mice by ESI-MS/MS and found significant changes in ceramide, dihydroceramide, glucosylceramide, dihydroglucosylceramide, sphingomyelin, and dihydrosphingomyelin levels (Fig. 7). CerS2 utilizes primarily C22-C24-CoAs and C18-CoA to a very minor extent (21), whereas CerS3 mainly uses C26:0-CoA. Reduction in CerS2 and CerS3 activities might cause changes in levels of SLs containing other acyl chain lengths as we observe, because of altered dimer formation, which can significantly affect ceramide formation (10).
Figure 5.
Loss of ACBP in mice reduces CerS activity in vivo. CerS2 and CerS3 activities were determined in homogenates from testes from either ACBP+/+ mice (black bars) or ACBP−/− mice (gray bars) using C22:0-CoA and C26:0-CoA as substrates, respectively. The values are mean values ± S.D., n = 3. Statistical analyses were performed using unpaired t test. **, p < 0.01.
Figure 6.
Cers1–6 expression is unaffected in testes of ACBP−/− mice. Total RNA was isolated from testis from ACBP+/+ (black bars) or ACBP−/− mice (gray bars), and expression levels of Cers1–6 were determined by quantitative PCR. The data shown are mean values ± S.D., n = 9. Statistical analyses were performed using unpaired t test.
Figure 7.
Ablation of ACBP affects the testis sphingolipidome. LC-ESI-MS/MS analysis of ceramide (A), dihydroceramide (B), glucosylceramide (C), dihydroglucosylceramide (D), sphingomyelin (E), dihydrosphingomyelin (F), and sphingoid bases (G) from testis from ACBP+/+ mice (black bars) or ACBP−/− mice (gray bars). The values represent means ± S.D., n = 3. Statistical analyses were performed using unpaired t test. *, p < 0.05; **, p < 0.01; ***, p < 0.001; ****, p < 0.0001.
Activation of CerS3 requires ligand binding to ACBP
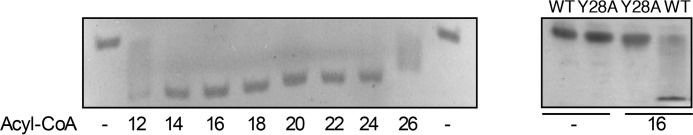
ACBP binds saturated and unsaturated C14-C22 acyl-CoA esters with an affinity of 1–15 nm in a one-to-one binding mode, whereas free CoA binds with a much lower affinity (KD of 2 μm) (22–24). The present finding that ACBP activates CerS2 and CerS3 suggests that ACBP can also bind very-long-chain acyl-CoA esters. Using gel shift assays under non-denaturing conditions, we observed that C12:0-C26:0-CoAs alter the mobility of ACBP (Fig. 8A), arguing that ACBP binds these acyl-CoA esters, although C12 and C26-CoA is bound with lower affinity than the other acyl-CoA esters.
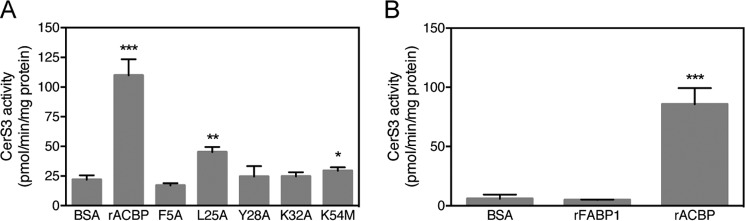
Figure 8.
ACBP binds long-chain and very-long-chain acyl-CoA esters. Left panel, recombinant rat ACBP was incubated with the indicated saturated acyl-CoA (1:1 molar ratio); apo- and holo-ACBP were subsequently separated by native gel electrophoresis; and protein bands were visualized by Coomassie staining. Right panel, recombinant rat ACBP (WT) or ACBP Y28A were incubated with C16:0-CoA and subsequently separated by native gel electrophoresis, and protein bands were visualized by Coomassie staining.
To test whether ligand binding to ACBP is required for CerS3 activation, we examined the effect of a panel of ACBP mutants, which do bind acyl-CoA (Ref. 23 and Fig. 8B), on CerS3 activity. CerS3 activity was not stimulated by ACBP in which Tyr28, Lys32 or Lys54 were mutated to alanine, which all interact with the 3′-phosphate on CoA and are required for ligand binding (Fig. 9). Moreover, altering the interaction between ACBP and the ω-end of the acyl-chain or the adenine ring by altering Leu-25 or Phe-5 to alanine, respectively, also abolishes the activating properties of ACBP (Fig. 9). Liver-type fatty-acid-binding protein-1 (FABP1), which also binds acyl-CoA esters and is highly abundant in liver (25), did not activate CerS3 (Fig. 9), which further substantiates that activation of ceramide synthase activity by ACBP is specific.
Figure 9.
CerS3 activity is activated by ACBP and depends on ligand binding to ACBP. Homogenates were prepared from HEK293T cells overexpressing CerS3 and CerS3 activity was determined using C26:0-CoA after addition of purified recombinant ACBP or a mutated form of ACBP (A), which binds ligand with very low affinity, or purified recombinant liver FABP1 (B). The values are shown as means ± S.D., n = 3. Statistical analyses were performed using unpaired t test. *, p < 0.05; **, p < 0.01; ***, p < 0.001; ****, p < 0.0001.
CerS interacts with ACBP
Our results clearly demonstrate that CerS2 and CerS3 are both activated by ACBP, suggesting that ACBP specifically target acyl-CoA esters to CerS2 or CerS3 via direct interaction. We were unable to detect such interactions by gel-shift assays or by co-immunoprecipitation with or without cross-linking (not shown), likely because of the transient nature of this putative interaction. We therefore used a proximity ligation assay (PLA) (26) in which we co-expressed CerS3-HA and ACBP-FLAG in HEK293T cells and were able to detect an interaction between CerS3 and ACBP (Fig. 10). Consistent with the intracellular localization of CerS3, the CerS3-ACBP interaction localized to the perinuclear/ER (Fig. 10, A and B). We demonstrate that ACBP also interacts with other members of the CerS family (Fig. 10B), even though their activity is not modulated by ACBP (Fig. 1) and also interacts with CerS2 and CerS3 even though they are catalytic inactive (Fig. 10C). Neither deletion of the HOX domain nor the C terminus of CerS2 affected the interaction with ACBP (results not shown).
Figure 10.
CerS and ACBP interact in situ. FLAG-tagged ACBP was co-expressed in HEK293T cells together with HA-tagged CerS2 or CerS3 (A), the indicated HA-tagged CerS (B), and HA-tagged wild-type and mutated (both W15R) forms of CerS2 or CerS3 (C). PLA using primary antibodies against HA and FLAG tags and secondary antibodies carrying PLA probes were applied to visualize the ACBP-CerS interaction. DAPI was used to detect nuclei. Scale bars, 10 or 20 μm (ACBP/CerS2 in B). Controls were cells transfected with empty plasmids alone or in combination with either ACBP or CerS3 plasmids. Other controls received either only one primary antibody or none. D, Western blots show expression of FLAG-tagged ACBP and HA-tagged CerS in HEK293T cells transfected with the indicated plasmid. GAPDH was used as loading control. The arrows mark the CerS. 2m and 3m indicate cells expressing CerS2 W15R and CerS3 W15R, respectively.
Discussion
In the present study we show for the first time that CerS interact with and are regulated by ACBP, suggesting a novel mode of regulating sphingolipid synthesis. Despite the importance of ceramides and other sphingolipids as structural entities and as signaling molecules, the molecular mechanisms governing their synthesis remain unclear. In mammals, regulation of de novo ceramide synthesis may involve a complex balance between the six CerS isoforms because CerS activity can be modulated by reversible dimerization (10). Thus, different CerS isoforms physically interact, and the co-expression of one particular isoform can modulate the activity of another. Although the mechanisms driving dimerization remains unresolved, it is likely to be dynamic because pharmacologic stimulation of ceramide synthesis leads to an increase in CerS dimerization (10, 27). Accordingly, the recently reported transcriptional and post-transcriptional regulation of CerS expression (28) may contribute to their dimerization and activity. Moreover, sphingosine-1-phosphate specifically inhibits CerS2 activity in vitro via a regulatory site, indicating an important regulatory interplay between sphingolipid metabolites (21).
The fact that CerS2 and CerS3 interact with and are regulated by ACBP suggests that these proteins might form a functional complex that drives ceramide synthesis. It is also interesting to note that ELOVL1 interacts with CerS2, suggesting that synthesis of C24-CoA by ELOVL1 is coordinated with the utilization of C24-CoA by CerS2 for ceramide production (12). We therefore speculate that CerS2, ELOVL1 and ACBP form a functional platform, which promotes coordinated and directional synthesis of very-long-chain acyl-CoA esters and ceramides. As a consequence, this interaction might determine the flux through the de novo ceramide synthesis pathway. To this end, generation of acyl-CoA esters by ACSL5 has recently been functionally coupled to ceramide synthesis (17), suggesting that ACSLs also could be part in such platforms. Notably, serine palmitoyl transferase, which catalyzes the first and rate-limiting step in sphingolipid synthesis, is also regulated by specific protein interactions by reversibly forming a SPOTS (SPT, ORM1/2, TSC3, SAC1) complex with a family of conserved Orm proteins, the serine palmitoyltransferase accessory subunit Tsc3, and the phosphoinositide phosphatase Sac1 (29, 30). However, even though CerS4–6 also interact with ACBP, their (and CerS1) kinetics are not affected by increasing levels of ACBP. Similarly, CerS2 also interacts with ELOVL1–7, although CerS2 only affects ELOVL1 activity (12). As the acyl chain length increases, the solubility of acyl-CoA esters in aqueous solutions decreases rapidly. Thus, whereas CerS1 and CerS4–6 all are characterized by having a preference for long-chain acyl-CoA esters, CerS2 and CerS3 both prefer very-long-chain acyl-CoA (31). Thus, we speculate that ACBP interacts with all the CerS to maintain a critical level of available acyl-CoA substrates, which is particularly important for CerS2 and CerS3.
Further kinetic analyses of CerS3 activation by ACBP revealed a biphasic saturation profile suggesting the presence of both low- and high-affinity binding sites. Such a kinetic profile has been observed for a number of other enzymes, e.g. naproxen demethylation by CYP2C9 (32, 33), CYP3A4-mediated metabolism of naphthalene (32), and 7-ethoxycoumarin O-deethylation and aminopyrine N-demethylation by NADPH-P450 reductase (34).
Even though the level of FABP1, which also binds acyl-CoA esters, is ∼10-fold higher than ACBP in liver, which also contains other acyl-CoA-binding domain containing proteins (19), we show that ACBP is the primary factor in liver cytosol that stimulates CerS activity in vitro. This and the fact that ACBP mutants, incapable of binding ligand, cannot stimulate ceramide synthase activity, underline the suggestion that ACBP specifically channels acyl-CoA esters for activation of CerS2 and CerS3. Accordingly, ablation of ACBP in mice testis, in which both CerS2 and CerS3 are highly expressed (21), results in global alterations in ceramide and sphingomyelin levels. This, however, does not affect reproduction of ACBP-null mice (35). We previously reported that loss of ACBP in mice impairs the epidermal barrier function resulting in heat and water loss (36). Because synthesis of ceramides containing ultra-very-long-chain acyl chains generated by CerS3 in keratinocytes are required for the integrity of the epidermal barrier in both mice and humans (37–39), we speculate that the impaired epidermal barrier in ACBP knock-out mice might be due to reduced levels of ceramides produced by CerS3 in the absence of ACBP.
In conclusion, we have identified a novel mode of regulating CerS2 and CerS3 activity in vivo via an interaction with ACBP. The present work and the genetic mice models that we have available provide an excellent framework to further delineate how ceramide synthases are regulated in vivo by acyl-CoA-binding proteins and how this interaction affects mammalian physiology. By analogy to other lipid binding proteins involved in intra- and intercompartmental trafficking of sphingolipids like FAPP2 and CERT (40), we anticipate that ACBP constitutes a new regulatory lipid binding protein, which plays a central role in the sphingolipid network and is therefore important in our understanding of human diseases triggered by altered sphingolipid metabolism.
Experimental procedures
Materials
d-Erythro-[4,5-3H]sphinganine was synthesized as described (41). Fatty acyl-CoAs, sphinganine, lipid standards for TLC, and internal standards for LC-ESI-MS/MS were from Avanti Polar Lipids (Alabaster, AL). Silica gel 60 TLC plates were from Merck. All solvents were of analytical grade and were from Biolab (Jerusalem, Israel). Defatted bovine serum albumin and a protease inhibitor mixture were from Sigma-Aldrich.
Mice
Acbp knock-out mice have been described previously (35, 36). The mice were housed under standard conditions (36). Animal experiments and breeding of transgenic mice were approved by the Danish Animal Experiment Inspectorate. Tissues were dissected and processed according to previously described procedures (36).
Cell culture
HEK293T cells were cultured in Dulbecco's modified Eagle's medium (Gibco) supplemented with 10% fetal calf serum, 100 IU/ml penicillin, and 100 μg/ml streptomycin. HEK293T cells were transfected with human CerS with an HA tag at the C terminus (CerS-HA) and/or murine ACBP (ACBD1) with a Flag tag at the C terminus using a jetPEIaTM, DNA in vitro transfection reagent (Polypus transfection) (8 μg of DNA/10-cm culture dish). 48 h after transfection, the cells were collected by trypsinization and stored at −80 °C.
Expression and purification of recombinant rat ACBP
Protein expression and purification of rat ACBP was performed as described (42). Selected residues in rat ACBP were replaced with alanine by site-directed mutagenesis (43).
CerS assays and SL measurements
CerS was assayed in homogenates as described using [4,5-3H]sphinganine, 15 μm sphinganine, 20 μm defatted bovine serum albumin, and 50 μm fatty acyl-CoA (Avanti Polar Lipids, Alabaster, AL) (44). For CerS1 and CerS4 assays 150 μg of protein was used; for CerS2, CerS5, and CerS6 100 μg of protein was used; and for CerS3 assays 250 μg of protein was used. When the effects of high-speed cytosols on CerS activity were examined, 6 μg of cytosol protein was added for each μl assay. SL levels were measured by ESI-MS/MS using a PE-Sciex API 3000 triple quadrupole mass spectrometer and an ABI 4000 quadrupole-linear ion trap mass spectrometer (45, 46).
Proximity ligation assay
The interaction between ACBP and CerS was examined by PLA in HEK293T cells essentially as described previously (47). Briefly, 0.48 μg of DNA was mixed with 1 μg of polyethylenimine, diluted with 20 μl of DMEM, and incubated for 10 min at room temperature. The cells were grown to 90–95% confluence in a 10-cm Petri dish, detached by incubation with 500 μl of trypsin, and neutralized with 5.5 ml DMEM, mixed with the transfection mixture and seeded into 16 chambered cover glass (C37000; Thermo). The cells were incubated as described above for 48 h. DMEM was aspirated, the cells were washed with PBS, the chambers were removed, and the cells were fixed with 4% paraformaldehyde in H2O for 10 min at room temperature. The cells were washed three times for 5 min each in PBS in a 10-cm Petri dish with gentle agitation. The cells were permeabilized with 0.5% Triton X-100 in PBS for 10 min, followed by three washes in 0.05% Tween 20 in TBS for 5 min each in a 10-cm Petri dish with gentle agitation. The cells were blocked with 1 drop of Duolink® in situ blocking solution (DUO82007; Sigma-Aldrich) for 1 h at 37 °C in a humidity chamber. The cells were incubated with 40 μl of primary antibodies: anti-DDK (TA50011; Origene) and anti-HA (H6908; Sigma-Aldrich), 1 μg/ml diluted in Duolink® antibody diluent (DUO82008; Sigma-Aldrich) for 1 h at 37 °C in a humidity chamber. The cells were washed for 2 min followed by two 10-min washes in Duolink® in situ wash buffer A (DUO82047; Sigma-Aldrich) in a 10-cm Petri dish with gentle agitation and incubated with 40 μl of Duolink® in situ PLA probes, anti-rabbit (DUO82002; Sigma-Aldrich) or anti-mouse (DUO82004; Sigma-Aldrich) diluted 5× in Duolink® antibody diluent (DUO82008; Sigma-Aldrich) for 1 h at 37 °C in a humidity chamber. The cells were washed for 2 min followed by two 10-min washes in Duolink® in situ wash buffer A in a 10-cm Petri dish with gentle agitation. PLA probes were ligated by incubation with 40 μl of Duolink® in situ ligase diluted 1:40 in Duolink® in situ ligation (DUO82009; Sigma-Aldrich) for 30 min at 37 °C in a humidity chamber followed by two washes for 2 min in Duolink® in situ wash buffer A in a 10-cm Petri dish with gentle agitation. PLA probes were amplified by incubation with 40 μl of Duolink® in situ amplification Red diluted 1:40 in 1× Duolink® in situ ligation (DUO82011; Sigma-Aldrich) for 100 min at 37 °C in a humidity chamber and washed twice for 10 min in Duolink® in situ wash buffer B (DUO82048; Sigma-Aldrich) in a 10-cm Petri dish with agitation (30 rpm). The cover slides were dipped in 10× diluted Duolink® in situ wash buffer B (DUO82048; Sigma-Aldrich), the silicone membrane around the cells was removed, and the cover slides were mounted on microscopy slides with Duolink® in situ mounting medium with DAPI (DUO82040; Sigma-Aldrich). Florescent signals were visualized at ambient temperature on a Leica TCS SP8X confocal microscope with a hybrid detector using a HCX PL APO 100×/1.40 oil objective using Leica Application Suite software. The images were processed using ImageJ (48).
RNA isolation and real-time PCR
Isolation of total RNA and determination of mRNA levels by real-time PCR were carried out as described (36). TFIIB expression was used for normalization. The following real-time primers were used: TfIIb forward, 5′-GTT CTG CTC CAA CCT TTG CCT-3′; TfIIb reverse, 5′-TGT GTA GCT GCC ATC TGC ACT T-3′; CerS1 forward, 5′-GCC ACC ACA CAC ATC TTT CGG-3′; CerS1 reverse, 5′-GGA GCA GGT AAG CGC AGT AG-3′; CerS2 forward, 5′-AGA GTG GGC TCT CTG GAC G-3′; CerS2 reverse, 5′-CCA GGG TTT ATC CAC AGT GAC-3′; CerS3 forward, 5′-CCT GGC TGC TAT TAG TCT GAT G-3′; CerS3 reverse, 5′-CTG CTT CCA TCC AGC ATA GG-3′; CerS4 forward, 5′-CTG TGG TAC TGT TGT TGC ATG AC-3′; CerS4 reverse, 5′-GCG CGT GTA GAA GAA GAC TAA G-3′; CerS5 forward, 5′-TGG CCA ATT ATG CCA GAC GTG AG-3′; CerS5 reverse, 5′-GGT AGG GCC CAA TAA TCT CCC AGC-3′; CerS6 forward, 5′-GCA TTC AAC GCT GGT TTC GAC-3′; and CerS6 reverse, 5′-TTC AAG AAC CGG ACT CCG TAG-3′.
Western blotting
Transfected HEK293T cells were harvested in SDS lysis buffer and protein extracts were prepared as described in Ref. 36. The protein concentration was determined using a Pierce BCA protein assay kit, followed by SDS-PAGE protein separation, blotting, and ECL as described in Ref. 36. The following antibodies were used: anti-HA (H6908; Sigma), anti-DDK (TA50011; Origene), anti-GAPDH (sc-25778; Santa Cruz), and horseradish peroxidase-conjugated anti-rabbit and anti-mouse antibodies (W4011 and W4021; Promega).
Statistics
Statistical analysis was performed using the Student's t test, and differences among groups were considered significant for p < 0.05 (*), p < 0.01 (**), p < 0.001 (***), and p < 0.0001 (****).
Author contributions
N. S. F. performed CerS assays, H. E. performed the proximity ligation assays and Western blotting, D. N. managed all mice work, G. V. analyzed the enzyme kinetic analyses, S. L. K. and A. H. M. carried out the ESI-MS/MS analyses of sphingolipids, N. J. F. expressed and purified all recombinant ACBP proteins, and N. J. F. and A. H. F. managed and funded the studies and wrote the manuscript
Acknowledgment
We thank Dr. Elad L. Laviad (Weizmann Institute of Science) for help with the initial part of this study.
This work was supported by Israel Science Foundation Grants 0888/11, by Danish Council for Independent Research-Natural Science Grant 23459, and by the Novo Nordisk Foundation. The authors declare that they have no conflicts of interest with the contents of this article.
N. J. Færgeman, unpublished results.
- SL
- sphingolipid
- ACBP
- acyl-coenzyme A-binding protein
- ACBD
- acyl-CoA-binding domain proteins
- CerS
- ceramide synthase
- ELOVL
- elongase
- ESI
- electrospray ionization
- PLA
- proximity ligation assay
- ACSL
- acyl-CoA synthetase
- FABP
- fatty-acid-binding protein-1.
References
- 1. Hannun Y. A., and Obeid L. M. (2008) Principles of bioactive lipid signalling: lessons from sphingolipids. Nat. Rev. Mol. Cell Biol. 9, 139–150 [DOI] [PubMed] [Google Scholar]
- 2. Maceyka M., and Spiegel S. (2014) Sphingolipid metabolites in inflammatory disease. Nature 510, 58–67 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Astudillo L., Sabourdy F., Therville N., Bode H., Ségui B., Andrieu-Abadie N., Hornemann T., and Levade T. (2015) Human genetic disorders of sphingolipid biosynthesis. J. Inherit Metab. Dis. 38, 65–76 [DOI] [PubMed] [Google Scholar]
- 4. Hla T., and Kolesnick R. (2014) C16:0-ceramide signals insulin resistance. Cell Metab. 20, 703–705 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Chaurasia B., and Summers S. A. (2015) Ceramides: lipotoxic inducers of metabolic disorders. Trends Endocrinol. Metab. 26, 538–550 [DOI] [PubMed] [Google Scholar]
- 6. Raichur S., Wang S. T., Chan P. W., Li Y., Ching J., Chaurasia B., Dogra S., Öhman M. K., Takeda K., Sugii S., Pewzner-Jung Y., Futerman A. H., and Summers S. A. (2014) CerS2 haploinsufficiency inhibits β-oxidation and confers susceptibility to diet-induced steatohepatitis and insulin resistance. Cell Metab. 20, 687–695 [DOI] [PubMed] [Google Scholar]
- 7. Park J. W., Park W. J., Kuperman Y., Boura-Halfon S., Pewzner-Jung Y., and Futerman A. H. (2013) Ablation of very long acyl chain sphingolipids causes hepatic insulin resistance in mice due to altered detergent-resistant membranes. Hepatology 57, 525–532 [DOI] [PubMed] [Google Scholar]
- 8. Tidhar R., and Futerman A. H. (2013) The complexity of sphingolipid biosynthesis in the endoplasmic reticulum. Biochim. Biophys. Acta 1833, 2511–2518 [DOI] [PubMed] [Google Scholar]
- 9. Tidhar R., Ben-Dor S., Wang E., Kelly S., Merrill A. H. Jr., and Futerman A. H. (2012) Acyl chain specificity of ceramide synthases is determined within a region of 150 residues in the Tram-Lag-CLN8 (TLC) domain. J. Biol. Chem. 287, 3197–3206 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Laviad E. L., Kelly S., Merrill A. H. Jr., and Futerman A. H. (2012) Modulation of ceramide synthase activity via dimerization. J. Biol. Chem. 287, 21025–21033 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Sassa T., Hirayama T., and Kihara A. (2016) Enzyme activities of the ceramide synthases CERS2–6 are regulated by phosphorylation in the C-terminal region. J. Biol. Chem. 291, 7477–7487 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Ohno Y., Suto S., Yamanaka M., Mizutani Y., Mitsutake S., Igarashi Y., Sassa T., and Kihara A. (2010) ELOVL1 production of C24 acyl-CoAs is linked to C24 sphingolipid synthesis. Proc. Natl. Acad. Sci. U.S.A. 107, 18439–18444 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Alvarez-Vasquez F., Sims K. J., Cowart L. A., Okamoto Y., Voit E. O., and Hannun Y. A. (2005) Simulation and validation of modelled sphingolipid metabolism in Saccharomyces cerevisiae. Nature 433, 425–430 [DOI] [PubMed] [Google Scholar]
- 14. Wronowska W., Charzynska A., Nienaltowski K., and Gambin A. (2015) Computational modeling of sphingolipid metabolism. BMC Syst. Biol. 9, 47. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Rajakumari S., Rajasekharan R., and Daum G. (2010) Triacylglycerol lipolysis is linked to sphingolipid and phospholipid metabolism of the yeast Saccharomyces cerevisiae. Biochim. Biophys. Acta 1801, 1314–1322 [DOI] [PubMed] [Google Scholar]
- 16. Radner F. P., Grond S., Haemmerle G., Lass A., and Zechner R. (2011) Fat in the skin: triacylglycerol metabolism in keratinocytes and its role in the development of neutral lipid storage disease. Dermatoendocrinol. 3, 77–83 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Senkal C. E., Salama M. F., Snider A. J., Allopenna J. J., Rana N. A., Koller A., Hannun Y. A., and Obeid L. M. (2017) Ceramide is metabolized to acylceramide and stored in lipid droplets. Cell Metab. 25, 686–697 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Grevengoed T. J., Klett E. L., and Coleman R. A. (2014) Acyl-CoA metabolism and partitioning. Annu. Rev. Nutr. 34, 1–30 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Neess D., Bek S., Engelsby H., Gallego S. F., and Færgeman N. J. (2015) Long-chain acyl-CoA esters in metabolism and signaling: role of acyl-CoA binding proteins. Prog. Lipid Res. 59, 1–25 [DOI] [PubMed] [Google Scholar]
- 20. Gaigg B., Neergaard T. B., Schneiter R., Hansen J. K., Faergeman N. J., Jensen N. A., Andersen J. R., Friis J., Sandhoff R., Schrøder H. D., and Knudsen J. (2001) Depletion of acyl-coenzyme A-binding protein affects sphingolipid synthesis and causes vesicle accumulation and membrane defects in Saccharomyces cerevisiae. Mol. Biol. Cell 12, 1147–1160 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Laviad E. L., Albee L., Pankova-Kholmyansky I., Epstein S., Park H., Merrill A. H. Jr., and Futerman A. H. (2008) Characterization of ceramide synthase 2: tissue distribution, substrate specificity, and inhibition by sphingosine 1-phosphate. J. Biol. Chem. 283, 5677–5684 [DOI] [PubMed] [Google Scholar]
- 22. Faergeman N. J., Sigurskjold B. W., Kragelund B. B., Andersen K. V., and Knudsen J. (1996) Thermodynamics of ligand binding to acyl-coenzyme A binding protein studied by titration calorimetry. Biochemistry 35, 14118–14126 [DOI] [PubMed] [Google Scholar]
- 23. Kragelund B. B., Poulsen K., Andersen K. V., Baldursson T., Krøll J. B., Neergård T. B., Jepsen J., Roepstorff P., Kristiansen K., Poulsen F. M., and Knudsen J. (1999) Conserved residues and their role in the structure, function, and stability of acyl-coenzyme A binding protein. Biochemistry 38, 2386–2394 [DOI] [PubMed] [Google Scholar]
- 24. Fulceri R., Knudsen J., Giunti R., Volpe P., Nori A., and Benedetti A. (1997) Fatty acyl-CoA-acyl-CoA-binding protein complexes activate the Ca2+ release channel of skeletal muscle sarcoplasmic reticulum. Biochem. J. 325, 423–428 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Rolf B., Oudenampsen-Krüger E., Börchers T., Faergeman N. J., Knudsen J., Lezius A., and Spener F. (1995) Analysis of the ligand binding properties of recombinant bovine liver-type fatty acid binding protein. Biochim. Biophys. Acta 1259, 245–253 [DOI] [PubMed] [Google Scholar]
- 26. Söderberg O., Leuchowius K. J., Gullberg M., Jarvius M., Weibrecht I., Larsson L. G., and Landegren U. (2008) Characterizing proteins and their interactions in cells and tissues using the in situ proximity ligation assay. Methods 45, 227–232 [DOI] [PubMed] [Google Scholar]
- 27. Jensen S. A., Calvert A. E., Volpert G., Kouri F. M., Hurley L. A., Luciano J. P., Wu Y., Chalastanis A., Futerman A. H., and Stegh A. H. (2014) Bcl2L13 is a ceramide synthase inhibitor in glioblastoma. Proc. Natl. Acad. Sci. U.S.A. 111, 5682–5687 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Wegner M. S., Wanger R. A., Oertel S., Brachtendorf S., Hartmann D., Schiffmann S., Marschalek R., Schreiber Y., Ferreirós N., Geisslinger G., and Grösch S. (2014) Ceramide synthases CerS4 and CerS5 are upregulated by 17β-estradiol and GPER1 via AP-1 in human breast cancer cells. Biochem. Pharmacol. 92, 577–589 [DOI] [PubMed] [Google Scholar]
- 29. Breslow D. K., Collins S. R., Bodenmiller B., Aebersold R., Simons K., Shevchenko A., Ejsing C. S., and Weissman J. S. (2010) Orm family proteins mediate sphingolipid homeostasis. Nature 463, 1048–1053 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Han S., Lone M. A., Schneiter R., and Chang A. (2010) Orm1 and Orm2 are conserved endoplasmic reticulum membrane proteins regulating lipid homeostasis and protein quality control. Proc. Natl. Acad. Sci. U.S.A. 107, 5851–5856 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Levy M., and Futerman A. H. (2010) Mammalian ceramide synthases. IUBMB Life 62, 347–356 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Korzekwa K. R., Krishnamachary N., Shou M., Ogai A., Parise R. A., Rettie A. E., Gonzalez F. J., and Tracy T. S. (1998) Evaluation of atypical cytochrome P450 kinetics with two-substrate models: evidence that multiple substrates can simultaneously bind to cytochrome P450 active sites. Biochemistry 37, 4137–4147 [DOI] [PubMed] [Google Scholar]
- 33. Hutzler J. M., Hauer M. J., and Tracy T. S. (2001) Dapsone activation of CYP2C9-mediated metabolism: evidence for activation of multiple substrates and a two-site model. Drug Metab. Dispos. 29, 1029–1034 [PubMed] [Google Scholar]
- 34. Inouye K., Mizokawa T., Saito A., Tonomura B., and Ohkawa H. (2000) Biphasic kinetic behavior of rat cytochrome P-4501A1-dependent monooxygenation in recombinant yeast microsomes. Biochim. Biophys. Acta 1481, 265–272 [DOI] [PubMed] [Google Scholar]
- 35. Neess D., Bloksgaard M., Bek S., Marcher A. B., Elle I. C., Helledie T., Due M., Pagmantidis V., Finsen B., Wilbertz J., Kruhøffer M., Færgeman N., and Mandrup S. (2011) Disruption of the acyl-CoA-binding protein gene delays hepatic adaptation to metabolic changes at weaning. J. Biol. Chem. 286, 3460–3472 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36. Neess D., Bek S., Bloksgaard M., Marcher A. B., Færgeman N. J., and Mandrup S. (2013) Delayed hepatic adaptation to weaning in ACBP−/− mice is caused by disruption of the epidermal barrier. Cell Rep. 5, 1403–1412 [DOI] [PubMed] [Google Scholar]
- 37. Jennemann R., Rabionet M., Gorgas K., Epstein S., Dalpke A., Rothermel U., Bayerle A., van der Hoeven F., Imgrund S., Kirsch J., Nickel W., Willecke K., Riezman H., Gröne H. J., and Sandhoff R. (2012) Loss of ceramide synthase 3 causes lethal skin barrier disruption. Hum. Mol. Genet. 21, 586–608 [DOI] [PubMed] [Google Scholar]
- 38. Eckl K. M., Tidhar R., Thiele H., Oji V., Hausser I., Brodesser S., Preil M. L., Onal-Akan A., Stock F., Müller D., Becker K., Casper R., Nürnberg G., Altmüller J., Nürnberg P., et al. (2013) Impaired epidermal ceramide synthesis causes autosomal recessive congenital ichthyosis and reveals the importance of ceramide acyl chain length. J. Invest. Dermatol. 133, 2202–2211 [DOI] [PubMed] [Google Scholar]
- 39. Radner F. P., Marrakchi S., Kirchmeier P., Kim G. J., Ribierre F., Kamoun B., Abid L., Leipoldt M., Turki H., Schempp W., Heilig R., Lathrop M., and Fischer J. (2013) Mutations in CERS3 cause autosomal recessive congenital ichthyosis in humans. PLoS Genet. 9, e1003536. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40. Yamaji T., and Hanada K. (2015) Sphingolipid metabolism and interorganellar transport: localization of sphingolipid enzymes and lipid transfer proteins. Traffic 16, 101–122 [DOI] [PubMed] [Google Scholar]
- 41. Hirschberg K., Rodger J., and Futerman A. H. (1993) The long-chain sphingoid base of sphingolipids is acylated at the cytosolic surface of the endoplasmic reticulum in rat liver. Biochem. J. 290, 751–757 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42. Wadum M. C., Villadsen J. K., Feddersen S., Møller R. S., Neergaard T. B., Kragelund B. B., Højrup P., Faergeman N. J., and Knudsen J. (2002) Fluorescently labelled bovine acyl-CoA-binding protein acting as an acyl-CoA sensor: interaction with CoA and acyl-CoA esters and its use in measuring free acyl-CoA esters and non-esterified fatty acids. Biochem. J. 365, 165–172 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43. Fisher C. L., and Pei G. K. (1997) Modification of a PCR-based site-directed mutagenesis method. BioTechniques 23, 570–571, 574 [DOI] [PubMed] [Google Scholar]
- 44. Lahiri S., Lee H., Mesicek J., Fuks Z., Haimovitz-Friedman A., Kolesnick R. N., and Futerman A. H. (2007) Kinetic characterization of mammalian ceramide synthases: determination of Km values towards sphinganine. FEBS Lett. 581, 5289–5294 [DOI] [PubMed] [Google Scholar]
- 45. Sullards M. C., Liu Y., Chen Y., and Merrill A. H. Jr. (2011) Analysis of mammalian sphingolipids by liquid chromatography tandem mass spectrometry (LC-MS/MS) and tissue imaging mass spectrometry (TIMS). Biochim. Biophys. Acta 1811, 838–853 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46. Shaner R. L., Allegood J. C., Park H., Wang E., Kelly S., Haynes C. A., Sullards M. C., and Merrill A. H. Jr. (2009) Quantitative analysis of sphingolipids for lipidomics using triple quadrupole and quadrupole linear ion trap mass spectrometers. J. Lipid Res. 50, 1692–1707 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47. Thymiakou E., and Episkopou V. (2011) Detection of signaling effector-complexes downstream of bmp4 using PLA, a proximity ligation assay. J. Vis. Exp. pii, 2631. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48. Schneider C. A., Rasband W. S., and Eliceiri K. W. (2012) NIH Image to ImageJ: 25 years of image analysis. Nat. Methods 9, 671–675 [DOI] [PMC free article] [PubMed] [Google Scholar]