Abstract
Although miR-145 is the most frequently down-regulated miRNA in bladder cancer (BC), its exact stage-association and downstream effector have not been defined. Here, we found that miR-145 was upregulated in human BC patients with lymph node metastasis and in metastatic T24T cell line. Forced expression of miR-145 promoted anchorage-independent growth of T24T cells accompanied by the down-regulation of forkhead box class O1 (FOXO1). In contrast, in non-metastatic T24 cells, miR-145 overexpression inhibited cell growth with upregulation of FOXO1 and the knockdown of FOXO1 abolished the miR-145-mediated inhibition of cell growth. Mechanistic studies revealed that miR-145 directly bound to and attenuated 3′UTR activity of foxo1 mRNA in both T24 and T24T cells. Interestingly, miR-145 suppressed STAT3 phosphorylation at Tyr705 and increased foxo1 promoter transcriptional activity in T24 cells, but not in T24T cells, suggesting a role of STAT3 in the divergent responses to miR-145. Supporting this was our finding that STAT3 knockdown mimicked miR-145-mediated upregulation of FOXO1 in T24T cells and inhibition of anchorage-independent growth. Consistently, ectopic expression of miR-145 promoted tumor formation of xenograft T24T cells, whereas such promoting effect became inhibitory due to specific knockdown of STAT3. Together, our findings demonstrate the stage-specific association and function of miR-145 in BCs, and provide novel insights into the therapeutic targeting of miR-145.
Keywords: Bladder cancer, miR-145, STAT-3, FOXO1
INTRODUCTION
miRNAs, a class of small (21-23nt) non-protein-coding RNAs, are important regulators of target genes in eukaryotic cells and have significant roles in various diseases (1). In cancer cells, miRNAs function as crucial regulatory molecules, acting as oncogenes or tumor suppressors (2). Although miRNAs exert their functions through sequence-specific binding to mRNA of their target genes, it has been revealed that miRNAs also have cell-type-specific signatures on target mRNA expression and are stage-specific during cancer progression (3). miR-145 is previously found to be a main tumor suppressive miRNA, being down-regulated in various human cancers, including gastric cancer, breast cancer, prostate cancer, colon cancer, as well as in B-cell malignancies (4). Both in vitro and in vivo studies have shown that miR-145 can significantly inhibit proliferation, migration and invasion in cancer cells (4). However, it has recently been found that tumor-specific deletion of miR-145 in an autochthonous mouse model of lung adenocarcinoma did not affect tumor development, and that stromal expression of miR-145 promotes neoangiogenesis in lung cancers (5), thus arguing against the delivery of this miRNA as an agent in cancer therapeutics. Moreover, miR-145 displays dramatic up-regulation in hepatocellular carcinoma and colorectal cancers with lymph node metastasis in comparison to those without lymph node metastasis (6, 7), suggesting that miR-145 may promote lymph node metastasis of cancer, or it may even play an oncogene role in metastatic cancer.
Bladder cancer (BC) is the most common malignancy of urinary system, and is the number one cause of deaths in patients with urinary tract disease (8). The incidence of BC has steadily risen worldwide in recent decades. It is estimated that more than 74,000 Americans are newly diagnosed with BC and more than 16,000 die of this disease in 2015. BC is also the costliest cancer to treat on a per-case basis because of the need for the lifetime monitoring and treatment (9). Muscle-invasive BC (MIBC) represents 25–40% of all BC and accounts for virtually all the mortality from BC (10). Although current treatment methods that range from radical cystectomy to systemic chemotherapy are effective in some MIBC patients, the overall therapeutic efficacy is still far from satisfactory, indicating the need of new precise therapeutic strategies (11). Since half of the MIBC patients who have undergone radical cystectomy died of tumor metastasis, the high metastasis rate of MIBC has always been the major obstacle in clinical treatment (11). MIBC spreads from the bladder in a predictable stepwise manner to the pelvic lymph nodes and then to visceral organs (10). miR-145 is reported to be the most frequently down-regulated miRNA in BCs and has been shown to significantly inhibit proliferation, migration and invasion in BC cells (12). Nevertheless, the expression profile miR-145 in lymph node metastatic BC and its effects on metastatic BC cells have yet to be explored.
Signal transducer and activator of transcription 3 (STAT3) signaling is an important intrinsic pathway for cancer because it is frequently activated in malignant cells (13). The transcriptional activity of STAT3 is dependent on the phosphorylation at the tyrosine residue 705 (Tyr705) by upstream kinases and subsequent nuclear translocation after dimerization (13). Overexpression of STAT3 is associated with the increased risk of recurrence and decreased survival for patients with BC (14), and activation of STAT3 has also been demonstrated to be crucial for bladder cancer cell growth and survival (15). Moreover, MIBC tissues have been characterized by nuclear expression of activated STAT3 (15). In a STAT3 transgenic mouse model, MIBC developed directly from carcinoma in situ, and MIBC driven by active STAT3 was predominantly composed of stem cells (16). Furthermore, activation of STAT3 has been revealed to be a key mediator of MIBC metastasis (17, 18). In the present study, we addressed the stage-specific and cell-type-specific roles of miR-145 in BCs. We discovered that miR-145 was overexpressed in BC tissues with lymph node metastasis, and forced expression of miR-145 inhibited anchorage-independent growth of non-metastatic MIBC T24 cells, but promoted such growth of the metastatic BC T24T cells. Importantly, STAT3 overexpression and activation status contributed to the differential responses of MIBC cells to miR-145.
MATERIALS AND METHODS
Plasmids, antibodies, and other reagents
The miR-145 expression construct was kindly provided by Dr. Renato Baserga (Department of Cancer Biology, Thomas Jefferson University and Kimmel Cancer Center, Philadelphia, PA) (19). The shRNA sets for human FOXO1 and STAT3 were purchased from Open Biosystems (Thermo Fisher Scientific, Huntsville, AL), and the siRNA targeting jak2 mRNA coding region was purchased from Ribobio (Guangzhou, China). The FLAG-tagged human FOXO1 expression construct was kindly provided by Dr. Haojie Huang (Department of Biochemistry and Molecular Biology, Mayo Clinic College of Medicine, Rochester, MN) (20). Human FOXO1 promoter-driven luciferase reporter was cloned into the pGL3 luciferase assay vector, and was a gift from Dr. Jean-Baptiste Demoulin (De Duve Institute, Catholic University of Louvain, BE-1200 Brussels, Belgium) (21). A mutation of STAT3 binding site in the FOXO1 promoter was created using site-directed mutagenesis by the overlap extension PCR method with mutagenic primers 5′-ACAGAAACACTCGAGAAGCGCCATCCAATAATAGAGATCCAAA-3′ (sense), 5′-TTTGGATCTCTATTATTGGATGGCGCTTCTCGAGTGTTTCTGT-3′ (anti-sense), and flanking primers 5′-ATTGAGTAGAATTCCTCGCGGCCGCC-3′ (forward), 5′-TGGAAGATCTCTGCGCTGCTGCCTGTTGAAT-3′ (reverse). The 762bp-1176bp of FOXO1 3′UTR containing miR-145 target sites was cloned into psi-CHECKTM-2 dual luciferase reporter, and was obtained from Dr. Bruce A. White (Departments of Cell Biology and Molecular, Microbial and Structural Biology, University of Connecticut Health Center, Farmington, CT) (22). The mutation of miR-145 binding site in the FOXO1 3′UTR was created using site-directed mutagenesis by the overlap extension PCR method with mutagenic primers 5′-CATTTTGTATTCTAACGCCATTAGTACTAATTTTATACATGC-3′ (sense), 5′-GCATGTATAAAATTAGTACTAATGGCGTTAGAATACAAAATG-3′ (anti-sense), and flanking primers 5′-TCGCTCGAGTGGGACTTAAGAACA-3′ (forward), 5′-TATTGCGGCCAGCGGCCGCGAAGACTTGAT-3′ (reverse). The JAK2 expression vector was kindly provided by Dr. Zhizhuang J Zhao (Department of Pathology, University of Oklahoma Health Sciences Center, Oklahoma City, Oklahoma) (23). A mutation of miR-145 target sites in the JAK2 coding region was created using site-directed mutagenesis by the overlap extension PCR with mutagenic primers 5′-GCAACCATTATAATAACTCC AAACGGTGGAATTCAGTGG-3′ (sense), 5′-CCACTGAATTCCACCGTTTGGAGTT ATTATAATGGTTGC-3′ (anti-sense), and flanking primers 5′-CAGATTTATTCAGC AATTCAGCC-3′ (forward), 5′-AACTGTGTAGGATCCCGGTC-3′ (reverse). The antibodies against FOXO1, STAT1, p-STAT1, STAT3, p-STAT3, JAK1, JAK2, p-JAK2, and SHP-2 were commercially purchased from Cell Signaling Technology (Boston, MA). The antibodies against p27 and GAPDH were bought from Santa Cruz Biotechnology (Santa Cruz, CA).
Human BC tissues
72 pairs of BC samples and their paired adjacent normal bladder tissues were obtained from patients who underwent radical cystectomy at Department of Urology, Union Hospital, Tongji Medical College, Huazhong University of Science and Technology (Wuhan, China) between 2013 and 2015. All specimens were immediately snap-frozen in liquid nitrogen after surgical removal. Histological and pathological diagnoses were confirmed and the tissues were defined by a certified clinical pathologist according to the 2004 World Health Organization Consensus Classification and Staging System for bladder neoplasms. All specimens were obtained with appropriate informed consent from the patients and a supportive grant obtained from the Medical Ethics Committee of China.
Cell culture and transfection
Human invasive BC cell line T24 and the paired metastatic cell line T24T was kindly provided by Dr. Dan Theodorescu (Departments of Urology, University of Virginia, Charlottesville, VA) (24) in 2010 and used in our previous studies (25, 26). Both T24 and T24T cells were subjected to DNA tests and authenticated in our previous studies (27). The cell lines were regularly authenticated on the basis of viability, growth, morphology, recovery and chemical response as well, and were most recently confirmed 4–6 months before use by using a short tandem repeat method. Cells were cultured in DMEM/Ham’s F-12 (1:1 volume) mixed medium supplemented with 5% FBS, 1% penicillin/streptomycin and 2 mM L-glutamine. Transfections were carried out using PolyJetTM DNA In Vitro Transfection Reagent (SignaGen Laboratories, Gaithersburg, MD) according to the manufacturer’s instructions (26). The transfected cells were then respectively selected with hygromycin, G418 or puromycin (Life Technologies, Rockville, MD) for 4–6 weeks. Surviving cells were pooled as stable mass transfectants as described in our previous studies (28).
Anchorage-independent growth assay
Anchorage-independent growth ability was evaluated in soft agar as described in our previous studies (29). Briefly, 3 ml of 0.5% agar in basal modified Eagle’s medium supplemented with 10% FBS was layered onto each well of 6-well cell culture plates. Cells (3×104) suspended in 1 ml of normal medium were mixed with 2 ml of 0.5% agar in basal modified Eagle’s medium supplemented with 10% FBS, and 1 ml of mixture was added into each well over top of the 0.5% agar layer. After incubation at 37°C in 5% CO2 for 2 to 3 weeks, colonies with more than 32 cells were scored and presented as colonies per 104 cells.
Western blotting
Western blot was performed as described previously (30). Briefly, cells were plated into each well of 6-well plate and cultured in normal cell culture medium. The cells were then washed once with ice-cold phosphate-buffered saline and cell lysates were prepared with a lysis buffer (10 mM Tris-HCl (pH 7.4), 1% SDS, and 1 mM Na3VO4). An equal amount (80 μg) of total protein from each cell lysate was subjected to Western blot with the indicated antibody as described in previous studies (30). The specific antibody-bound bands were detected by using the alkaline phosphatase-linked secondary antibody and ECF Western blotting system (Amersham Biosciences, Piscataway, NJ). The images were acquired and the protein levels were quantified by Typhoon FLA 7000 imager (GE Healthcare, Pittsburgh, PA).
Reverse transcription-polymerase chain reaction (RT-PCR) and Quantitative RT-PCR
Total RNA was extracted with TRIzol reagent (Invitrogen Corp., Carlsbad, CA) and cDNAs were synthesized with the SuperScript® III First-Strand Synthesis System for RT-PCR (Invitrogen Corp., Carlsbad, CA). A pair of oligonucleotides (Forward: 5′-GATGATCTTGAGGCTGTTGTC-3′ and Reverse: 5′-CAGGGCTGCTTTTAACT CTG-3′) were used to detect human gapdh cDNA as loading control. The foxo1 cDNA fragments were amplified by human foxo1 specific PCR primers (Forward: 5′-AACCTGGCATTACAGTTGGCC-3′; Reverse: 5′-AAATGCAGGAGGCATGAC TACGT-3′). The endogenous jak2 mRNA level was evaluated by specific human jak2 PCR primers (Forward: 5′-GAGCCTATCGGCATGGAATA-3′; Reverse: 5′-ACT GCCATCCCAAGACATTC-3′). The PCR products were separated on 2% agarose gels and stained with ethidium bromide. The images were visualized and scanned with UV lights with FluorChem SP imaging system (Alpha Innotech Inc., San Leandro, CA) (31). For quantitative analysis of exogenous jak2 mRNA level, we designed the Forward primer targeting the jak2 mRNA (5′-CAGATGGATGCCCAGATGAG-3′), and the Reverse primer targeting the sequence transcribed from the backbone of the vector (5′-GCATTTAGGTGACACTATAG-3′). The Quantitative RT–PCR analysis was carried out using the SYBR Green PCR Kit (Qiagen, Santa Clarita, CA) and the 7900HT Fast Real-time PCR system (Applied Biosystems, Carlsbad, CA). For miRNA assay, total RNA was extracted using the miRNeasy Mini Kit (Qiagen, Valencia, CA). Analysis of miR-145 expression was performed by the miScript PCR system (Qiagen, Valencia, CA) and the 7900HT Fast Real-time PCR system. The ΔΔCT value was used to calculate the relative expression of miR-145, using U6 as endogenous control.
Luciferase reporter assay
As described in our previous studies (27, 32), dual-luciferase reporter assay was performed with the luciferase assay system (Promega Corp., Madison, WI). Each of the indicated luciferase reporter was transfected into human bladder cancer cells. Cells were then extracted with lysis buffer [25 mM Tris-phosphate (pH 7.8), 2 mM EDTA, 1% Triton X-100, and 10% glycerol]. The luciferase activity was determined with a microplate luminometer LB 96V (Berthold GmbH & Co. KG, Bad Wildbad, Germany). The firefly luciferase signal was normalized to the Renilla luciferase transfection control.
Animal experiments
All procedures for the mouse experiments were approved by the Animal Care Committee of Tongji Medical College. Eighteen female nude mice were divided into three groups (6/group). The stable T24T transfectants as indicated (1×106) were injected s.c. into 6-week-old nude mice in 100 μl PBS to produce tumor growth. Four weeks later, the animals were sacrificed and weighed simultaneously. No animal was dead before the end of the experiment. The tumors were excised, photographed and weighed. Tumor tissue sections obtained from the in vivo experiments were then made for immunohistochemical (IHC) staining. Briefly, tissues were excised and fixed overnight in 4% paraformaldehyde at 4°C. Fixed tissues were processed for paraffin embedding, and the serial 5 μm thick sections were then stained using specific antibodies against STAT3 and FOXO1 (Cell Signaling Technology), respectively. The resultant immunostaining images were captured using the AxioVision Rel.4.6 computerized image analysis system (Carl Zeiss, Oberkochen, Germany).
Statistical methods
Statistical analyses were performed using SPSS 19.0 software (SPSS Inc., Chicago, IL). Paired t-test was used to compare the difference between paired tissues using real-time PCR analysis. The Chi-square test was used to evaluate the difference in clinical pathological features. Student’s t-test was utilized to compare continuous variables, summarized as means ± SD, between different groups. P< 0.05 was considered significant statistically difference.
RESULTS
miR-145 was upregulated in BC tissues from patients with lymph node metastasis and in metastatic T24T cell line
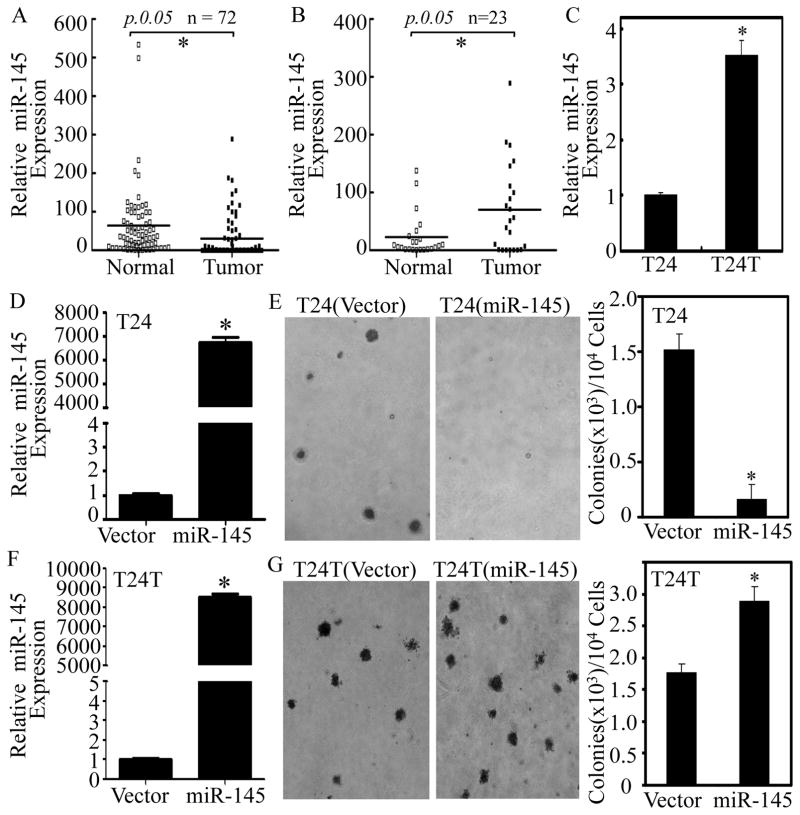
Although miR-145 has been reported to be the most frequently downregulated miRNA in BCs and has been shown to significantly inhibit proliferation and migration in BC cells (12), the expression profile miR-145 in human bladder tissues obtained from patients with lymph node metastasis and its biological function in metastatic BC development have yet to be explored. To determine the stage-specific expression of miR-145 in human BC, miR-145 expression was evaluated in 72 pairs of clinical BC tissues with different stages and their adjacent normal bladder tissues surgically removed from patients diagnosed with BC. In line with previous studies, a significant reduction of miR-145 expression was observed in 53/72 BC tissues compared with their adjacent normal bladder tissues (p<0.05, Figure 1A). Unexpectedly, miR-145 expression in BC patients with the lymph nodes metastasis was higher than in their adjacent normal bladder tissues (p<0.05, Table 1, & Figure 1B). Consistent with these findings, metastatic T24T cells showed increased miR-145 expression in comparison to parental non-metastatic T24 cells (p<0.05, Figure 1C). This is the first demonstration of miR-145 overexpression in BC patients with the lymph nodes metastasis, raising the interesting possibility that miR-145 contributes to BC metastasis.
Figure 1. Upregulation of miR-145 in human BC patients with lymph node metastasis and in T24T cells, and the differential responses to miR-145 on anchorage-independent growth between metastatic T24T cells and its parental non-metastatic T24 cells.
(A) Total RNA was extracted from human normal bladder (Normal) and the paired cancerous (Tumor) tissues surgically removed from 72 patients diagnosed with different stages of BC. RNA was then subjected to Quantitative real-time PCR for determining miR-145 expression. (B) The expression of miR-145 was compared in 23 pairs of normal bladder (Normal) and cancerous patients (Tumor) with lymph node metastasis. (C) miR-145 expression levels were evaluated by Quantitative real-time PCR in non-metastatic T24 and metastatic T24T BC cells. (D–G) miR-145 expression construct was stably transfected into T24 (D) and T24T (F) cells. The stable transfectants, T24(Vector) vs. T24(miR-145) (E) or T24T(Vector) vs. T24T(miR-145) (G) were subjected to determination of their anchorage-independent growth in soft agar assay. Colonies with more than 32 cells were scored and presented as colonies/104 cells. Results were presented as means ± SD from three independent experiments. Symbol “*” indicates a significant difference between the two groups as indicated (p< 0.05).
Table 1.
Correlation between miR-145 expression and clinicopathological factors
| Parameters | Group | Total | miR-145 expression | P value | |
|---|---|---|---|---|---|
|
| |||||
| Low | high | ||||
| Gender | Male | 56 | 40 | 16 | |
| Female | 16 | 13 | 3 | 0.533 | |
|
| |||||
| Age (years) | <55 | 8 | 6 | 2 | |
| ≥55 | 64 | 47 | 17 | 1.000 | |
|
| |||||
| Histological grade | Low | 27 | 20 | 7 | |
| High | 45 | 33 | 12 | 0.945 | |
|
| |||||
| Tumor stage | Ta-T1 | 29 | 21 | 8 | |
| T2-T4 | 43 | 32 | 11 | 0.850 | |
|
| |||||
| Tumor size | <3.0cm | 37 | 27 | 10 | 0.899 |
| ≥3.0cm | 35 | 26 | 9 | ||
|
| |||||
| Tumor multiplicity | Unifocal | 19 | 13 | 6 | |
| Multifocal | 53 | 40 | 13 | 0.550 | |
|
| |||||
| Lymph nodes metastasis | Absent | 49 | 44 | 5 | |
| Present | 23 | 9 | 14 | 0.000* | |
P<0.05
Ectopic expression of miR-145 exerted different effects between T24 and T24T cells on anchorage-independent growth
To test whether miR-145 promotes BC metastasis, we stably transfected miR-145 construct into non-metastatic T24 and metastatic T24T cells and established stable transfectants (Figures 1D and 1F). The stable ectopic expression of miR-145 blocked anchorage-independent growth in T24 cells, while forced expression of miR-145 promoted anchorage-independent growth in T24T cells (Figures 1E & 1G). The differential responses to miR-145 between metastatic T24T and its parental non-metastatic T24 BC cells, together with the findings of opposite expression of miR-145 in metastatic vs. non-metastatic BC patients, and in untransfected T24 cells vs. T24T cells, indicate that miR-145 exerts context-dependent effects in relation to the metastatic characteristics of BC.
miR-145 differentially regulated FOXO1, which mediated its opposite effects on anchorage-independent growth in T24 and T24T cells
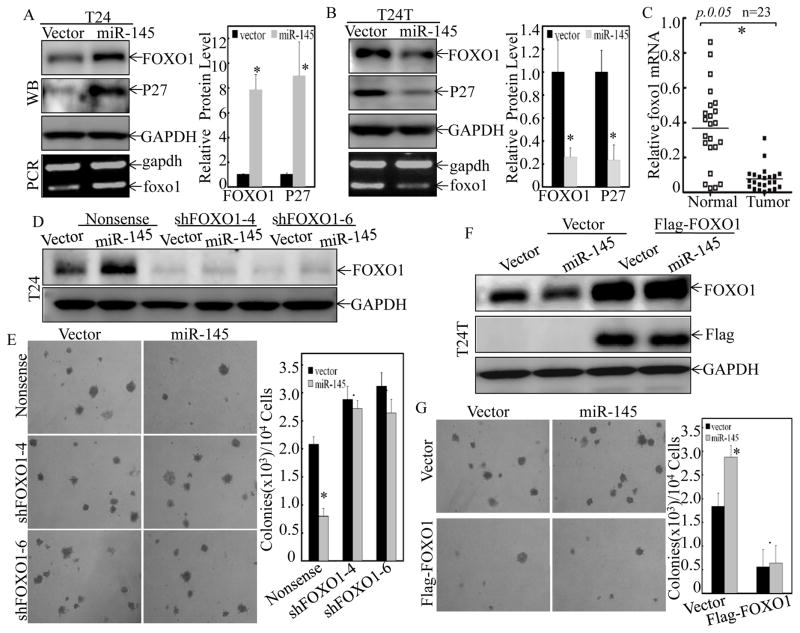
FOXO proteins have been demonstrated to play important roles in cancer cell growth through up-regulating negative regulators of the G1/S cell cycle transition, such as p27KIP1, p21WAF1 and p130, and repressing positive regulators, such as cyclin D1 and D2 (33). FOXO1 has also been proposed as a potential prognostic marker for BC progression (34). Thus, the potential effect of miR-145 on FOXO1 expression was evaluated between T24(Vector) vs. T24(miR-145), and T24T(Vector) vs. T24T(miR-145) cells. Consist with the biological activity of miR-145 overexpression in both cells, miR-145 overexpression resulted in upregulation of both mRNA and protein of FOXO1 in T24 cells (Figure 2A), while it impaired FOXO1 expression in T24T cells (Figure 2B). In addition, protein level of p27, the downstream effector of FOXO1, showed corresponding changes in T24 cells and T24T cells upon forced miR-145 expression (Figures 2A & 2B), suggesting potential association of FOXO1 expression with miR-145 effect on anchorage-independent in two cells. In line with these findings, in the lymph nodes metastatic BC tissues that the expression of miR-145 was high, the expression of foxo1 mRNA was downregulated (Figure 2C). To test this further, shRNA specific targeting FOXO1 was used to stably knock down foxo1 in T24 cells, and Flag-FOXO1 expression construct was used to stably transfected into T24T cells. As shown in Figures 2D & 2E, knockdown of FOXO1 reversed the growth inhibition of T24 cells by miR-145 in soft agar assay. On the other hand, overexpression of FOXO1 diminished the miR-145-induced anchorage-independent growth in T24T cells (Figures 2F & 2G). These results clearly demonstrate that FOXO1 mediates miR-145 regulation of anchorage-independent growth in both T24 and T24T cells, and that the differential effects on FOXO1 expression are a key factor for miR-145-caused divergent responses T24 versus T24T cells.
Figure 2. The differential effects of miR-145 on anchorage-independent growth between T24 and T24T cells were mediated by FOXO1.
(A & B), The cell extracts from T24(Vector) vs. T24(miR-145) (A) or T24T(Vector) vs. T24T(miR-145) (B) cells were subjected to Western Blotting for detection of FOXO1 and p27 protein levels as indicated. The foxo1 mRNA expression was also determined by RT-PCR as indicated. GAPDH protein or mRNA was used as loading control, respectively. (C) The expression of foxo1 mRNA was detected by Quantitative real-time PCR in 23 pairs of normal bladder (Normal) and cancerous patients (Tumor) with lymph node metastasis. (D & E), T24(Vector) and T24(miR-145) cells were stably transfected with either Nonsense shRNA or two FOXO1 shRNA constructs (shFOXO1-4, shFOXO1-6), respectively. The knockdown efficiency of FOXO1 was assessed by Western Blotting (D), and the stable transfectants were then used for determination of their anchorage-independent growth ability (E). (F & G), Flag-FOXO1 expression construct was used to stably transfected into T24T(Vector) and T24T(miR-145) cells, respectively. The overexpression efficiency was identified by Western blotting (F), and the stable transfectants were then subjected to determination of their anchorage-independent growth ability (G). Colonies with more than 32 cells were scored and presented as colonies/104 cells. Western Blotting experiments were repeated at least three times and the representative blots were shown in the figures. Results were presented as the means±SD of triplicates. Symbol “*” indicates a significant difference between Vector and miR-145 transfectants, while symbol “♣” indicates a significant difference in transfectants with either FOXO1 knockdown or overexpression in comparison to the scramble transfectant (p < 0.05).
miR-145 enhanced foxo1 promoter transcriptional activity in T24, but not in T24T cells, without affecting mRNA degradation
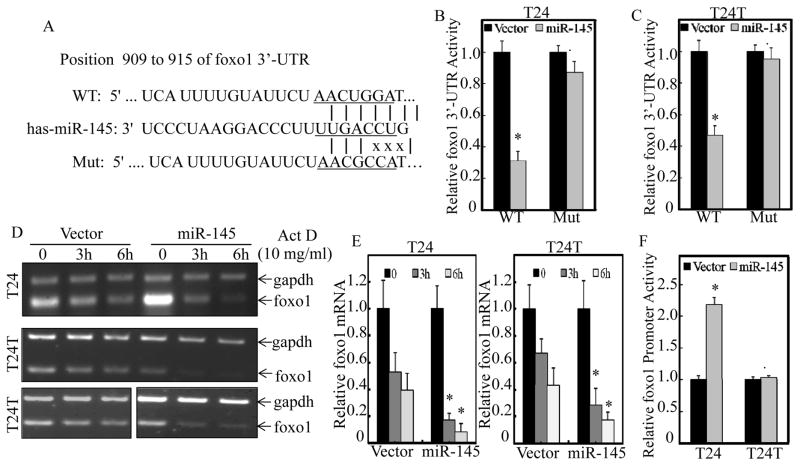
miRNAs often bind to the 3′-UTR of their target mRNAs and function as regulators via either modulation of mRNA stability or translational inhibition. As shown in Figure 3A, 3′-UTR of foxo1 mRNA has the predicted target site of miR-145. To determine whether miR-145 could regulate foxo1 expression via direct binding, the miRcode, miRWalk and TargetScan database were used. Both the wild type and miR-145 targeted site mutated FOXO1 3′-UTR luciferase reporters were constructed. The potential effect of miR-145 on foxo1 mRNA 3′-UTR activity was then tested by stable transfection of foxo1 mRNA 3′-UTR luciferase reporter into T24 and T24T cells. The results indicated that miR-145 ectopic expression had a comparable inhibition of foxo1 mRNA 3′-UTR activity between T24 and T24T cells (Figures 3B & 3C), and the point mutation of miR-145 bind site in foxo1 3′-UTR luciferase reporter abolished such miR-145 inhibition in both cells (Figures 3B & 3C). These results reveal that although 3′-UTR of foxo1 mRNA was directly targeted and inhibited by miR-145, the inhibition of foxo1 mRNA 3′-UTR activity is not involved in the differential regulatory effects of miR-145 on FOXO1 expression in T24 and T24T cells. In line with this notion, FOXO1 mRNA degradation rates were also comparable between T24 and T24T cells, although miR-145 overexpression accelerated its degradation in both cells (Figures 3D & 3E). It was important to note that miR145 overexpression led to marked upregulation of foxo1 mRNA in T24 cells, but not in T24T cells, suggesting that differential regulation of FOXO1 protein expression between T24 and T24T cells might be regulated on a transcriptional level. Therefore, we next compared the effect of miR-145 on foxo1 transcription between two cells by determining foxo1 promoter-driven luciferase reporter activity. The results showed that forced expression of miR-145 resulted in enhanced FOXO1 promoter activity in T24 cells, but not in T24T cells (Figure 3F). Taken together, these data indicate that in T24 cells, miR-145 promotes foxo1 mRNA degradation, whereas it also enhances foxo1 transcription in a more predominant manner and subsequently upregulating FOXO1 expression. On the other hand, failure of increasing transcription of foxo1 results in the downregulation of foxo1 mRNA and protein in T24T cells transfected with miR-145.
Figure 3. miR-145 inhibited foxo1 mRNA 3′UTR activity in both T24 and T24T cells, while it enhanced foxo1 promoter activity only in T24, but not in T24T cells.
(A) Schematic Sequence of the intact miR-145-binding site in WT (wide-type) foxo1 mRNA 3′-UTR and its mutation (Mut) of foxo1 3′UTR luciferase reporter. (B & C), WT foxo1 3′-UTR and its mutated luciferase reporters were stably transfected into T24(Vector) vs. T24(miR-145) (B) or T24T(Vector) vs. T24T(miR-145) (C) cells. Luciferase activity was evaluated by the Dual-Luciferase Reporter Assay System, and the results were presented as foxo1 3′-UTR luciferase activity relative to medium control (Relative foxo1 3′-UTR activity). Symbol “*” indicates a significant difference between Vector and miR-145 transfectants, while symbol “♣” indicates a significant difference in mutant reporter transfectant in comparison to WT reporter transfectant (p< 0.05). (D & E), For measuring mRNA decay rate, T24(Vector) vs.T24(miR-145) or T24T(Vector) vs. T24T(miR-145) cells were treated with actinomycin D (Act D) for 3 hours and 6 hours. The total RNA was isolated and then subjected to RT-PCR (D) and Quantitative real-time PCR (E) for determination of foxo1 mRNA levels. The gadph mRNA levels were used as loading control. (F) The indicated T24 and T24T cells were stably transfected with foxo1 promoter-driven luciferase reporter, and Dual-Luciferase Reporter Assay System was used to evaluate the Luciferase activity. Results are presented as means±SD of triplicates. Symbol “*” indicates a significant difference between Vector and miR-145 transfectants, while symbol “♣” indicates a significant difference in T24 in comparison to T24T cells (p< 0.05).
miR-145 enhanced foxo1 transcription by inhibiting STAT3 phosphorylation at Tyr705 in T24 cells, but not in T24T cells
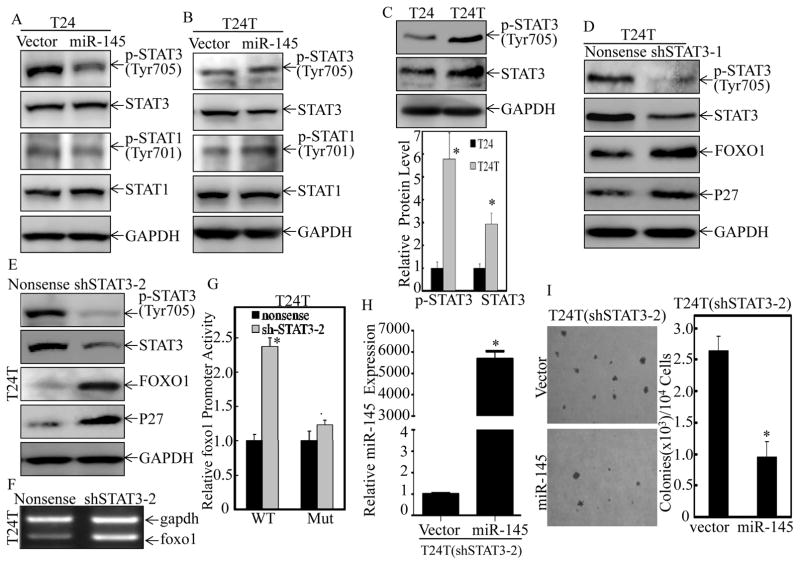
Although FOXO1 post-translational modifications and subcellular localization have been intensively investigated, the upstream regulators of its promoter transcription remains to be explored (35). To address the differential responses of foxo1 promoter to miR-145 between T24 and T24T cells, TFANSFAC® Transcription Factor Binding Sites Software (Biological Database, Wolfenbutel, Germany) was first employed. The results revealed that the promoter of human FOXO1 gene contained the putative DNA-binding sites of STATs transcription factors. Previous studies have showed the overexpressed total STAT3 protein and high nuclear expression of phosphorylated STAT3 at Tyr705 in muscle invasive BC cells J82 and UMUC3 (36). To test whether the STATs transcription factor was the mediator for the differential responses of foxo1 promoter to miR-145 in the two cells, both expression and activated levels of STAT1 and STAT3 were evaluated in T24 and T24T cells. Forced expression of miR-145 resulted in the dramatic decrease of STAT3 phsphorylation at Tyr705 with no effect on total protein level of STAT3 and both total protein level and phosphorylated level of STAT1 in T24 cells (Figure 4A). In contrast to the observations in T24 cells, miR-145 overexpression did not show any observable inhibition of either total protein level or phosphorylated level of STAT3 or STAT1 in T24T cells (Figure 4B). Further studies showed that T24T cells exhibited markedly elevated phosphorylation of STAT3 at Tyr705 with a slightly increase STAT3 protein expression in T24T cells in comparison to T24 cells (Figure 4C). These results demonstrate that STAT3 phosphorylation at Tyr705 is regulated by miR-145 in T24 cells, but not in T24T cells, further suggesting that the difference between STAT3 phosphorylation at Tyr705 might be associated with the differential responses to miR-145 between the two cells. To test this hypothesis, shRNAs specific targeting human STAT3 were co-transfected with foxo1 promoter-driven luciferase reporter into T24T cells. As shown in Figures 4D & 4E, knockdown of STAT3 in T24T cells could mimic the miR-145 upregulation of FOXO1 and p27 protein levels in T24 cells. The mRNA level and promoter activity of foxo1 were also upregulated in T24T cells upon STAT3 knockdown (Figures 4F & 4G), and the mutation of STAT3 binding site at foxo1 promoter effectively diminished the upregulation of foxo1 promoter activity caused by knockdown of STAT3 expression (Figure 4G). Moreover, knockdown STAT3 completely restored the response of T24T cells to miR-145 on inhibition of anchorage-independent growth (Figures 4H & 4I). These results strongly indicate that STAT3 expression and activation status contributed to the differential responses of human BC cells to miR-145 through regulation of foxo1 transcription.
Figure 4. Inhibition of STAT3 phosphorylation at Thr705 by miR-145 was crucial for its promotion of foxo1 transcription in BC cells.
(A & B), The cell extracts from T24(Vector) vs. T24(miR-145) (A) or T24T(Vector) vs. T24T(miR-145) (B) cells were subjected to Western Blotting for determination of protein levels of STAT3, p-STAT3 at Tyr705, STAT1 and p-STAT1 at Tyr701 protein as indicated. (C) STAT3 and p-STAT3 (Tyr705) level was evaluated in T24 and T24T cells by Western Blotting. (D & E), T24T(Nonsense) vs. T24T(shSTAT3-1) (D) and T24T(Nonsense) vs. T24T(shSTAT3-2) (E) were used for evaluation of STAT3, p-STAT3 (Tyr705), FOXO1 and P27 protein level by Western Blotting. GAPDH protein level was used as loading control. (F) The total RNA was isolated and subjected to RT-PCR for determination of foxo1 mRNA levels in T24T(Nonsense) and T24T(shSTAT3-2) cells. The gadph mRNA levels were used as loading control. (G) T24T(Nonsense) and T24T(shSTAT3-2) cells were stably transfected with foxo1 promoter-driven luciferase reporter or its STAT3 binding site mutant. Dual-Luciferase Reporter Assay System was performed to determine the Luciferase activity and the results were presented as foxo1 promoter activity relative to medium control (Relative foxo1 promoter activity). (H & I), miR-145 was stably transfected to T24T(shSTAT3-2) cells (H), and the stable transfectants were used for determination of their anchorage-independent growth ability (I). Colonies with more than 32 cells were scored and presented as colonies/104 cells. Western Blotting experiments were repeated at least three times and the representative blots were shown. Results were presented as the means±SD of triplicates. Symbol “*” indicates a significant difference in T24 vs. T24T, T24T(Nonsense) vs. T24T(shSTAT3), or T24T(shSTAT3/Vector) vs. T24T(shSTAT3/miR-145) transfectants, while symbol “♣” indicates a significant difference in mutant reporter transfectant in comparison to WT reporter transfectant (p<0.05).
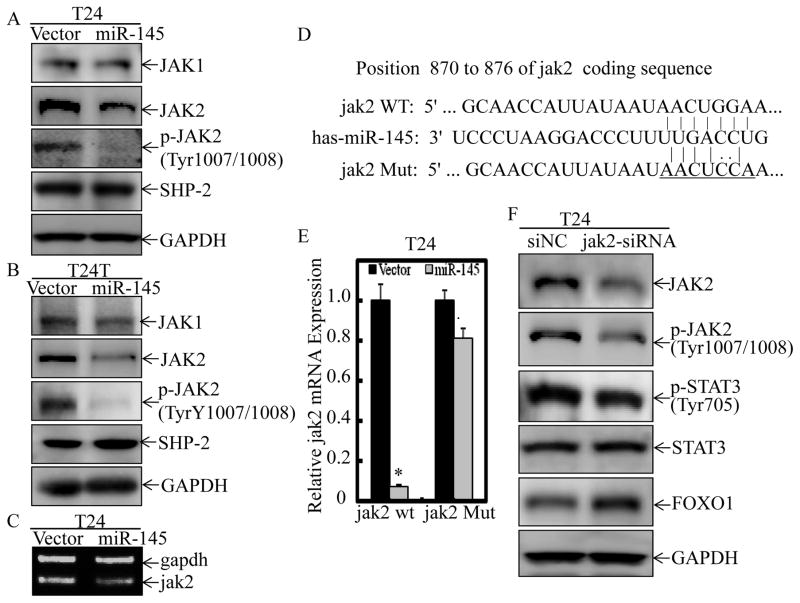
miR-145 inhibited STAT3 phosphorylation at Tyr705 through targeting JAK2 mRNA coding sequence
STAT proteins are phosphorylated, thus activated, by members of the tyrosine kinase Janus kinase (JAK) and dephosphorylated by Protein Tyrosine Phosphatase SHP-2 (37). It has been reported that muscle invasive BC tissues are characterized by nuclear expression of phosphorylated STAT3, and JAK2 is responsible for its phosphorylation at Tyr705 in muscle invasive BC cells (36). Besides, JAK-2/STAT-3 pathway is involved in inducing chemoresistance and stemness in muscle invasive BC cells (38). As shown in Figure 5A & 5B, forced expression of miR-145 in T24 cells and T24T cells resulted in a specific profound inhibition of JAK2 protein and its phosphorylation at Tyr1007/1008 respectively, whereas it did not show remarkably inhibition of either JAK1 or SHP-2 under same experimental conditions. Consistent with the result obtained from JAK2 protein levels, endogenous jak2 mRNA expression was also markedly decreased upon miR-145 overexpression in T24 cells (Figure 5C). The results obtained from analysis of jak2 mRNA indicated that coding region, but not 3′-UTR of JAK2 mRNA, has the predicted target site of miR-145 (Figure 5D). Therefore, we subsequently evaluated the expression of exogenous transcript from the JAK2 expression vector which lacks 3′-UTR. As expected, wild-type JAK2 was attenuated by miR-145 overexpression in comparison to Vector control transfectants in T24 cells (Figure 5E), whereas miR-145 had minimal effect on the JAK2 mRNA expression in which the two coding region of miR-145 targeted sites were mutated (Figures 5D & 5E). To further confirm the role of JAK2 in modulation of STAT3 phosphorylation at Tyr705, and FOXO1 expression, we transfected siRNA specific targeting JAK2 coding region into T24 cells and found that it could mimic the effects of miR-145 on inhibition of STAT3 phosphorylation at Tyr705 and upregulation of FOXO1 (Figure 5F). These results reveal that miR-145 can target JAK2 mRNA coding sequence to downregulate its expression, thus inhibits STAT3 phosphorylation at Tyr705 in T24 cells, but may inefficiently suppress this phosphorylation because of the constitutive activation of STAT3 in T24T cells.
Figure 5. miR-145 inhibited STAT3 phosphorylation at Tyr705 through targeting JAK2 mRNA coding sequence.
(A & B), The cell extracts from T24(Vector) vs. T24(miR-145) (A) and T24T(Vector) vs. T24T(miR-145) cells (B) were subjected to Western Blotting for detection of JAK1, JAK2, p-JAK2 at Tyr1007/1008 and SHP-2 protein levels as indicated. GAPDH protein was used as protein loading control. (C) The mRNA levels of jak2 in T24(Vector) and T24(miR-145) cells were determined by RT-PCR, and gapdh was used as loading control. (D) Schematic Sequence of the intact miR-145-binding site in WT (wide-type) jak2 mRNA coding sequence (wide-type, WT) and its mutation (Mut). (E) T24(Vector) and T24(miR-145) cells were stably transfected with JAK2 expression vector and its mutant, respectively. Exogenous jak2 mRNA expression was evaluated by Quantitative real-time PCR. (F) The siRNA specific targeting jak2 mRNA coding sequence (jak2-siRNA) and the control siRNA (siNC) were transfected into T24 cells for 72 hours. The cell extracts were then subjected to Western Blotting for detection of JAK2, p-JAK2 (Tyr1007/1008), STAT3, p-STAT3 (Tyr705) and FOXO1 protein levels as indicated. GAPDH protein was used as protein loading control. Western Blotting experiments were repeated at least three times and the representative blots were shown in the figures. Symbol “*” indicates a significant difference between T24(Vector) and T24(miR-145) transfectants, while symbol “♣” indicates a significant difference in mutant JAK2 transfectant in comparison to WT JAK2 transfectant (p<0.05).
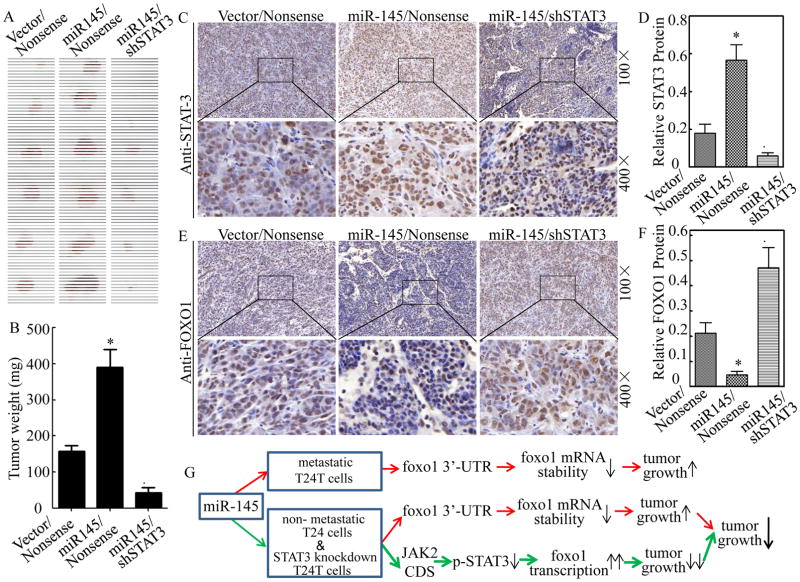
Knockdown of STAT3 impaired miR-145 promotion of xenograft tumor formation in nude mice injected with T24T transfectants in vivo
To determine the effect of miR-145 and its regulated STAT3 on tumorigenicity in vivo, T24T transfectants, including T24T(Vector/Nonsense), T24T(miR-145/Nonsense), and T24T(miR-145/shSTAT3), were injected subcutaneously into in the flanks of nude mice for evaluation of their tumor formation abilities. Four weeks after cell injection, the mice were sacrificed and the tumors were excised, photographed and weighed. Tumor tissue sections obtained from the mice were then made for determination of protein expression of STAT3 and FOXO1 using immunohistochemical (IHC) staining with specific antibodies. Consistent with in vitro results, miR-145 overexpression transfectant, T24T(Vector/miR-145), showed a considerable increase in tumor size and weight as compared with them from the mice injected with T24T(Vector/Nonsense) cells, while knockdown of STAT3 inhibited miR-145 promotion of tumor growth (Figures 6A & 6B). Histologic analysis of tumors showed that increased tumor growth by miR-145 ectopic expression was accompanied with FOXO1 downregulation, whereas the attenuation of tumor growth by knockdown of STAT3 expression was negatively associated with FOXO1 upregulation (Figures 6C–6F). These results clearly reveal the contribution of STAT3 activation to the differential responses of human BC cells to miR-145 regulation of tumor growth through modulation of foxo1 transcription as summarized in Figure 6G.
Figure 6. Knockdown of STAT3 impaired miR-145 promotion of xenograft tumor formation in nude mice injected with T24T transfectants in vivo.
(A) T24T stable transfectants as indicated were s.c. into in the flanks of nude mice for evaluation of their tumor formation abilities. Four weeks after cell injection, the mice were sacrificed and the tumors were excised, photographed and weighed (A & B). Tumor tissue sections obtained from the mice were then made for determination of protein expression of STAT3 and FOXO1 using immunohistochemical (IHC) staining as indicated (C & E). Protein expression levels were analyzed by calculating the integrated optical density per stained area (IOD/area) using Image-Pro Plus version 6.0 (D & F). Results were presented as the means±SD of 6 mice from each group. Symbol “*” indicates a significant difference between T24T(Vector/miR-145) and T24T(Vector/Nonsense) transfectants, while symbol “♣” indicates a significant difference in T24T(miR-145/shSTAT3) transfectant in comparison to T24T(miR-145/Nonsense) transfectant (p<0.05). (G) Proposed modeling for differential responses of various kinds of BC cells to miR-145.
DISSCUSSION
It has been reported that stromal expression of miR-145 could promote neoangiogenesis in lung cancer development (5). miR-145 has also been suggested as one of the predictive factors for the early prediction of lymph node metastasis in patients with hepatocellular carcinoma (6). Although previous studies report miR-145 is one of the most down-regulated miRNAs in both superficial Ta-T1 bladder tumors and invasive T2–T4 tumors with no significant difference between these two stages of human BC (39–42), the correlation of miR-145 expression with BC with lymph node metastatic status has never been explored. In the present study we discovered that the presence of lymph node metastasis was positively correlated with increased miR-145 expression in BC tissues. This was distinctly different from miR-145 downregulation in BC tissues in comparison to their paired normal bladder tissues obtained from patients with radical cystectomy and no correlation between miR-145 expression and BC invasion status by previous studies and current study. The results from our in vitro studies also indicated that metastatic T24T cells showed increased miR-145 expression in comparison to its parental non-metastatic T24 cells. We further discovered that ectopic expression of miR-145 promoted anchorage-independent growth of T24T cells in vitro and xenograft tumor formation in vivo. Our data revealed the stage-specific importance of highly expressed miR-145 in metastatic BC development. The important role of miR-145 in cancer metastasis is also supported by the results obtained from nude mouse model of orthotropic transplantation of colorectal cancer, in which miR-145 overexpression promoted lymph node metastasis (7). Previous studies have shown that miR-145 expression can be transcriptionally regulated by DNA methylation and p53 gene mutation (41), hypoxia-inducible factor (HIF)(43), Ras responsive element binding protein (RREB1) (44), and CCAAT/enhancer binding protein beta (C/EBP-β) (45). Whether some of these mechanisms are operative in metastatic BC deserves further investigation.
FOXO1 functions as transcription factor that regulates the expression of a series of cancer-related genes, such as p27KIP1, p21WAF1, p130, cyclin D1, cyclin D2, FasL and Bim. It plays important biological roles as tumor suppressor in regulating cell cycle arrest, apoptosis, DNA damage repair, and/or oxidative stress resistance (33). The miR-135a has been found to target 3′UTR of foxo1 mRNA and inhibit its expression to promote BC cell proliferation (46). miR-145 acts as negative regulator of lipolysis in white adipose tissue through directly targeting and repressing FOXO1 (47). However, the regulation of miR-145 targeting FOXO1 seems inconsistent in cancer cells, because both of them are regarded as tumor suppressors. Here, we found that miR-145 upregulated anchorage-independent growth of metastatic T24T BC cell through directly targeting FOXO1 mRNA 3′UTR, demonstrating the existence of miR-145/FOXO1 regulatory pathway in cancer and elucidating the growth-promoting role of miR-145 in metastatic BC. On the other hand, miR-145 regulation of foxo1 transcription has never been explored. The foxo1 promoter contains STATs binding sites, and it has been shown that STAT3 can bind to these sites in immune system T cells (48). We recently showed that phosphorylated STAT1 can bind to the foxo1 promoter to inhibit its transcription in BC cells (49). In the present study, we found that miR-145 suppressed STAT3 activation and increased foxo1 promoter transcriptional activity, which overcame its inhibition on foxo1 mRNA 3′UTR activation, and subsequently upregulated FOXO1 expression and suppressed anchorage-independent growth of non-metastatic T24 cells. Meanwhile, knockdown of the relative high expression of STAT3 restored the response of miR-145-induced upregulation of FOXO1 and inhibition of anchorage-independent growth of T24T cells. These results indicate that miR-145 differentially regulates FOXO1 expression depending on STAT3 expression and activation status. In the event that STAT3 is constitutively activated, miR-145 may inefficiently inhibit STAT3 phosphorylation at Tyr705 and therefore fail to upregulate foxo1 promoter transcriptional activity. Thus, STAT3 expression and activation status should be taken into consideration in future therapeutic approaches relevant to miR-145-associated treatment of BC, and probably some other cancers as well. Delivery of miR-145 combined with STAT3 knockdown might represent a much more effective strategy.
miRNAs usually form imperfect interactions with sequences in the 3′UTR of their targeted mRNAs, causing translational suppression and mRNA decay (2). Additiontally, recent findings suggest that miRNAs are able to target gene promoter, 5′UTR and coding region of mRNA (50–53). For example, miR-148 inhibits DNA methyltransferase 3b (Dnmt3b) gene expression through a region in its coding sequence (52). Dicer is also revealed to be targeted by the miRNA let-7 by means of sites within its coding region (53). JAK2 is a crucial upstream kinase of STAT3 phosphorylation at Tyr705. Here, we demonstrated that JAK2 was a novel target of miR-145, and the binding sites reside in JAK2 mRNA coding region rather than 3′UTR. Taken together, our results indicate that miR-145 regulates FOXO1 expression via two mechanisms: (i) directly targeting foxo1 mRNA 3′UTR to promote its mRNA degradation, and (ii) indirectly targeting JAK2/STAT3 axis to enhance foxo1 transcription. The final outcome of miR-145 upregulation of FOXO1 protein expression results from promoting mRNA transcription, which overcomes foxo1 mRNA degradation mediated by 3′UTR of foxo1 mRNA in T24 cells.
In conclusion, we demonstrate that miR-145 is upregulated in human BC tissues with lymph node metastasis and in metastatic BC cells. We also show that STAT3 expression and activation status contribute to the differential responses of human BC cells to miR-145 for its promotion of anchorage-independent growth through regulation of FOXO1 expression. In addition, we demonstrate that the coding region of JAK2 mRNA is a novel target of miR-145. Our findings underscore the stage-specific function and cell type-specific role of miR-145, and provide new insights into the therapeutic targeting of miR-145 for metastatic bladder cancer.
Acknowledgments
Financial support:
This work was partially supported by grants from NIH/NCI CA165980 (to C. Huang), CA177665 (to C. Huang) and CA112557 (to C. Huang), as well as NIH/NIEHS ES000260 (to C. Huang), and Key Project of Science and Technology Innovation Team of Zhejiang Province (2013TD10) (to C. Huang and H. Huang).
We thank Dr. Renato Baserga from Thomas Jefferson University and Kimmel Cancer Center for providing human miR-145 expression construct; Dr. Jean-Baptiste Demoulin from Catholic University of Louvain for generous gift of human foxo1 promoter-driven luciferase reporter; Dr. Haojie Huang from Mayo Clinic College of Medicine for providing FLAG-tagged human FOXO1 expression construct; Dr. Bruce A. White from University of Connecticut Health Center for the kind gift of human foxo1 3′-UTR luciferase reporter; and Dr. Zhizhuang J Zhao from University of Oklahoma Health Sciences Center for providing human JAK2 expression vector.
Abbreviations
- BC
bladder cancer
- RT-PCR
Reverse transcription-polymerase chain reaction
- FOXO1
forkhead box class O1
- MIBC
Muscle-invasive bladder cancer
- STAT3
Signal transducer and activator of transcription 3
- Tyr705
tyrosine residue 705
- JAK
tyrosine kinase Janus kinase
- IHC
immunohistochemical
- HIF
hypoxia-inducible factor
- RREB1
Ras responsive element binding protein
- C/EBP-β
CCAAT/enhancer binding protein beta
- Dnmt3b
DNA methyltransferase 3b
Footnotes
Disclosure statement: The authors disclose no potential conflicts of interest.
References
- 1.Das J, Podder S, Ghosh TC. Insights into the miRNA regulations in human disease genes. BMC Genomics. 2014;15:1010. doi: 10.1186/1471-2164-15-1010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Shenouda SK, Alahari SK. MicroRNA function in cancer: oncogene or a tumor suppressor? Cancer Metastasis Rev. 2009;28:369–78. doi: 10.1007/s10555-009-9188-5. [DOI] [PubMed] [Google Scholar]
- 3.Olson P, Lu J, Zhang H, Shai A, Chun MG, Wang Y, et al. MicroRNA dynamics in the stages of tumorigenesis correlate with hallmark capabilities of cancer. Genes Dev. 2009;23:2152–65. doi: 10.1101/gad.1820109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Cui SY, Wang R, Chen LB. MicroRNA-145: a potent tumour suppressor that regulates multiple cellular pathways. J Cell Mol Med. 2014;18:1913–26. doi: 10.1111/jcmm.12358. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Dimitrova N, Gocheva V, Bhutkar A, Resnick R, Jong RM, Miller KM, et al. Stromal expression of miR-143/145 promotes neoangiogenesis in lung cancer development. Cancer Discov. 2015 doi: 10.1158/2159-8290.CD-15-0854. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Zhang L, Xiang ZL, Zeng ZC, Fan J, Tang ZY, Zhao XM. A microRNA-based prediction model for lymph node metastasis in hepatocellular carcinoma. Oncotarget. 2015 doi: 10.18632/oncotarget.6534. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Yuan W, Sui C, Liu Q, Tang W, An H, Ma J. Up-regulation of microRNA-145 associates with lymph node metastasis in colorectal cancer. PLoS One. 2014;9:e102017. doi: 10.1371/journal.pone.0102017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Van Batavia J, Yamany T, Molotkov A, Dan H, Mansukhani M, Batourina E, et al. Bladder cancers arise from distinct urothelial sub-populations. Nat Cell Biol. 2014;16:982–91. 1–5. doi: 10.1038/ncb3038. [DOI] [PubMed] [Google Scholar]
- 9.Siegel RL, Miller KD, Jemal A. Cancer statistics, 2015. CA Cancer J Clin. 2015;65:5–29. doi: 10.3322/caac.21254. [DOI] [PubMed] [Google Scholar]
- 10.Youssef RF, Raj GV. Lymphadenectomy in management of invasive bladder cancer. Int J Surg Oncol. 2011;2011:758189. doi: 10.1155/2011/758189. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Knollman H, Godwin JL, Jain R, Wong YN, Plimack ER, Geynisman DM. Muscle-invasive urothelial bladder cancer: an update on systemic therapy. Ther Adv Urol. 2015;7:312–30. doi: 10.1177/1756287215607418. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Yoshino H, Seki N, Itesako T, Chiyomaru T, Nakagawa M, Enokida H. Aberrant expression of microRNAs in bladder cancer. Nat Rev Urol. 2013;10:396–404. doi: 10.1038/nrurol.2013.113. [DOI] [PubMed] [Google Scholar]
- 13.Yu H, Pardoll D, Jove R. STATs in cancer inflammation and immunity: a leading role for STAT3. Nat Rev Cancer. 2009;9:798–809. doi: 10.1038/nrc2734. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Mitra AP, Pagliarulo V, Yang D, Waldman FM, Datar RH, Skinner DG, et al. Generation of a concise gene panel for outcome prediction in urinary bladder cancer. J Clin Oncol. 2009;27:3929–37. doi: 10.1200/JCO.2008.18.5744. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Chen CL, Cen L, Kohout J, Hutzen B, Chan C, Hsieh FC, et al. Signal transducer and activator of transcription 3 activation is associated with bladder cancer cell growth and survival. Mol Cancer. 2008;7:78. doi: 10.1186/1476-4598-7-78. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Ho PL, Lay EJ, Jian W, Parra D, Chan KS. Stat3 activation in urothelial stem cells leads to direct progression to invasive bladder cancer. Cancer Res. 2012;72:3135–42. doi: 10.1158/0008-5472.CAN-11-3195. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Zhao D, Besser AH, Wander SA, Sun J, Zhou W, Wang B, et al. Cytoplasmic p27 promotes epithelial-mesenchymal transition and tumor metastasis via STAT3-mediated Twist1 upregulation. Oncogene. 2015;34:5447–59. doi: 10.1038/onc.2014.473. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Wei JH, Cao JZ, Zhang D, Liao B, Zhong WM, Lu J, et al. EIF5A2 predicts outcome in localised invasive bladder cancer and promotes bladder cancer cell aggressiveness in vitro and in vivo. Br J Cancer. 2014;110:1767–77. doi: 10.1038/bjc.2014.52. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.La Rocca G, Shi B, Sepp-Lorenzino L, Baserga R. Expression of micro-RNA-145 is regulated by a highly conserved genomic sequence 3’ to the pre-miR. J Cell Physiol. 2011;226:602–7. doi: 10.1002/jcp.22368. [DOI] [PubMed] [Google Scholar]
- 20.Lu H, Liu P, Pan Y, Huang H. Inhibition of cyclin-dependent kinase phosphorylation of FOXO1 and prostate cancer cell growth by a peptide derived from FOXO1. Neoplasia. 2011;13:854–63. doi: 10.1593/neo.11594. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Essaghir A, Dif N, Marbehant CY, Coffer PJ, Demoulin JB. The transcription of FOXO genes is stimulated by FOXO3 and repressed by growth factors. J Biol Chem. 2009;284:10334–42. doi: 10.1074/jbc.M808848200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Guttilla IK, White BA. Coordinate regulation of FOXO1 by miR-27a, miR-96, and miR-182 in breast cancer cells. J Biol Chem. 2009;284:23204–16. doi: 10.1074/jbc.M109.031427. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Zhao W, Gao R, Lee J, Xing S, Ho WT, Fu X, et al. Relevance of JAK2V617F positivity to hematological diseases--survey of samples from a clinical genetics laboratory. J Hematol Oncol. 2011;4:4. doi: 10.1186/1756-8722-4-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Gildea JJ, Golden WL, Harding MA, Theodorescu D. Genetic and phenotypic changes associated with the acquisition of tumorigenicity in human bladder cancer. Genes Chromosomes Cancer. 2000;27:252–63. doi: 10.1002/(sici)1098-2264(200003)27:3<252::aid-gcc5>3.0.co;2-9. [DOI] [PubMed] [Google Scholar]
- 25.Harding MA, Theodorescu D. RhoGDI signaling provides targets for cancer therapy. Eur J Cancer. 2010;46:1252–9. doi: 10.1016/j.ejca.2010.02.025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Jin H, Yu Y, Hu Y, Lu C, Li J, Gu J, et al. Divergent behaviors and underlying mechanisms of cell migration and invasion in non-metastatic T24 and its metastatic derivative T24T bladder cancer cell lines. Oncotarget. 2015;6:522–36. doi: 10.18632/oncotarget.2680. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Fang Y, Cao Z, Hou Q, Ma C, Yao C, Li J, et al. Cyclin d1 downregulation contributes to anticancer effect of isorhapontigenin on human bladder cancer cells. Mol Cancer Ther. 2013;12:1492–503. doi: 10.1158/1535-7163.MCT-12-0922. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Song L, Li J, Zhang D, Liu ZG, Ye J, Zhan Q, et al. IKKbeta programs to turn on the GADD45alpha-MKK4-JNK apoptotic cascade specifically via p50 NF-kappaB in arsenite response. J Cell Biol. 2006;175:607–17. doi: 10.1083/jcb.200602149. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Luo W, Liu J, Li J, Zhang D, Liu M, Addo JK, et al. Anti-cancer effects of JKA97 are associated with its induction of cell apoptosis via a Bax-dependent and p53-independent pathway. J Biol Chem. 2008;283:8624–33. doi: 10.1074/jbc.M707860200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Ouyang W, Ma Q, Li J, Zhang D, Liu ZG, Rustgi AK, et al. Cyclin D1 induction through IkappaB kinase beta/nuclear factor-kappaB pathway is responsible for arsenite-induced increased cell cycle G1-S phase transition in human keratinocytes. Cancer Res. 2005;65:9287–93. doi: 10.1158/0008-5472.CAN-05-0469. [DOI] [PubMed] [Google Scholar]
- 31.Zhang D, Li J, Costa M, Gao J, Huang C. JNK1 mediates degradation HIF-1alpha by a VHL-independent mechanism that involves the chaperones Hsp90/Hsp70. Cancer Res. 2010;70:813–23. doi: 10.1158/0008-5472.CAN-09-0448. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Che X, Liu J, Huang H, Mi X, Xia Q, Li J, et al. p27 suppresses cyclooxygenase-2 expression by inhibiting p38beta and p38delta-mediated CREB phosphorylation upon arsenite exposure. Biochim Biophys Acta. 2013;1833:2083–91. doi: 10.1016/j.bbamcr.2013.04.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Huang H, Tindall DJ. Dynamic FoxO transcription factors. J Cell Sci. 2007;120:2479–87. doi: 10.1242/jcs.001222. [DOI] [PubMed] [Google Scholar]
- 34.Kim TH, Jo SW, Lee YS, Kim YJ, Lee SC, Kim WJ, et al. Forkhead box O-class 1 and forkhead box G1 as prognostic markers for bladder cancer. J Korean Med Sci. 2009;24:468–73. doi: 10.3346/jkms.2009.24.3.468. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Zhang X, Rielland M, Yalcin S, Ghaffari S. Regulation and function of FoxO transcription factors in normal and cancer stem cells: what have we learned? Curr Drug Targets. 2011;12:1267–83. doi: 10.2174/138945011796150325. [DOI] [PubMed] [Google Scholar]
- 36.Sun Y, Cheng MK, Griffiths TR, Mellon JK, Kai B, Kriajevska M, et al. Inhibition of STAT signalling in bladder cancer by diindolylmethane: relevance to cell adhesion, migration and proliferation. Curr Cancer Drug Targets. 2013;13:57–68. [PubMed] [Google Scholar]
- 37.Zhang J, Zhang F, Niu R. Functions of Shp2 in cancer. J Cell Mol Med. 2015;19:2075–83. doi: 10.1111/jcmm.12618. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Ojha R, Singh SK, Bhattacharyya S. JAK-mediated autophagy regulates stemness and cell survival in cisplatin resistant bladder cancer cells. Biochim Biophys Acta. 2016;1860:2484–97. doi: 10.1016/j.bbagen.2016.07.021. [DOI] [PubMed] [Google Scholar]
- 39.Guancial EA, Bellmunt J, Yeh S, Rosenberg JE, Berman DM. The evolving understanding of microRNA in bladder cancer. Urol Oncol. 2014;32:41e31–40. doi: 10.1016/j.urolonc.2013.04.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Dip N, Reis ST, Srougi M, Dall’Oglio MF, Leite KR. Expression profile of microrna-145 in urothelial bladder cancer. Int Braz J Urol. 2013;39:95–101. doi: 10.1590/S1677-5538.IBJU.2013.01.12. discussion 2. [DOI] [PubMed] [Google Scholar]
- 41.Villadsen SB, Bramsen JB, Ostenfeld MS, Wiklund ED, Fristrup N, Gao S, et al. The miR-143/-145 cluster regulates plasminogen activator inhibitor-1 in bladder cancer. Br J Cancer. 2012;106:366–74. doi: 10.1038/bjc.2011.520. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Dyrskjot L, Ostenfeld MS, Bramsen JB, Silahtaroglu AN, Lamy P, Ramanathan R, et al. Genomic profiling of microRNAs in bladder cancer: miR-129 is associated with poor outcome and promotes cell death in vitro. Cancer Res. 2009;69:4851–60. doi: 10.1158/0008-5472.CAN-08-4043. [DOI] [PubMed] [Google Scholar]
- 43.Blick C, Ramachandran A, McCormick R, Wigfield S, Cranston D, Catto J, et al. Identification of a hypoxia-regulated miRNA signature in bladder cancer and a role for miR-145 in hypoxia-dependent apoptosis. Br J Cancer. 2015;113:634–44. doi: 10.1038/bjc.2015.203. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Kent OA, Fox-Talbot K, Halushka MK. RREB1 repressed miR-143/145 modulates KRAS signaling through downregulation of multiple targets. Oncogene. 2013;32:2576–85. doi: 10.1038/onc.2012.266. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Sachdeva M, Liu Q, Cao J, Lu Z, Mo YY. Negative regulation of miR-145 by C/EBP-beta through the Akt pathway in cancer cells. Nucleic Acids Res. 2012;40:6683–92. doi: 10.1093/nar/gks324. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Mao XP, Zhang LS, Huang B, Zhou SY, Liao J, Chen LW, et al. Mir-135a enhances cellular proliferation through post-transcriptionally regulating PHLPP2 and FOXO1 in human bladder cancer. J Transl Med. 2015;13:86. doi: 10.1186/s12967-015-0438-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Lin YY, Chou CF, Giovarelli M, Briata P, Gherzi R, Chen CY. KSRP and MicroRNA 145 are negative regulators of lipolysis in white adipose tissue. Mol Cell Biol. 2014;34:2339–49. doi: 10.1128/MCB.00042-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Oh HM, Yu CR, Golestaneh N, Amadi-Obi A, Lee YS, Eseonu A, et al. STAT3 protein promotes T-cell survival and inhibits interleukin-2 production through up-regulation of Class O Forkhead transcription factors. J Biol Chem. 2011;286:30888–97. doi: 10.1074/jbc.M111.253500. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Jiang G, Wu AD, Huang C, Gu J, Zhang L, Huang H, et al. Isorhapontigenin (ISO) inhibits invasive bladder cancer (BC) formation in vivo and human BC invasion in vitro by targeting STAT1/FOXO1 Axis. Cancer Prev Res (Phila) 2016 doi: 10.1158/1940-6207.CAPR-15-0338. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Orom UA, Nielsen FC, Lund AH. MicroRNA-10a binds the 5′UTR of ribosomal protein mRNAs and enhances their translation. Mol Cell. 2008;30:460–71. doi: 10.1016/j.molcel.2008.05.001. [DOI] [PubMed] [Google Scholar]
- 51.Kim DH, Saetrom P, Snove O, Jr, Rossi JJ. MicroRNA-directed transcriptional gene silencing in mammalian cells. Proc Natl Acad Sci U S A. 2008;105:16230–5. doi: 10.1073/pnas.0808830105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Duursma AM, Kedde M, Schrier M, le Sage C, Agami R. miR-148 targets human DNMT3b protein coding region. RNA. 2008;14:872–7. doi: 10.1261/rna.972008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Forman JJ, Legesse-Miller A, Coller HA. A search for conserved sequences in coding regions reveals that the let-7 microRNA targets Dicer within its coding sequence. Proc Natl Acad Sci U S A. 2008;105:14879–84. doi: 10.1073/pnas.0803230105. [DOI] [PMC free article] [PubMed] [Google Scholar]