Abstract
The Rho GTPases Rac (Ras-related C3 botulinum toxin substrate) and Cdc42 (cell division control protein 42 homolog) regulate cell functions governing cancer malignancy, including cell polarity, migration, and cell cycle progression. Accordingly, our recently developed Rac inhibitor EHop-016 (IC50, 1,100 nM) inhibits cancer cell migration and viability, and reduces tumor growth, metastasis, and angiogenesis in vivo. Herein, we describe MBQ-167, which inhibits Rac and Cdc42 with IC50s of 103 nM and 78 nM respectively, in metastatic breast cancer cells. Consequently, MBQ-167 significantly decreases Rac and Cdc42 downstream effector p21-activated kinase (PAK) signaling and the activity of signal transducer and activator of transcription (STAT3), without affecting Rho, MAPK, or Akt activities. MBQ-167 also inhibits breast cancer cell migration, viability, and mammosphere formation. Moreover, MBQ-167 affects cancer cells that have undergone epithelial to mesenchymal transition by a loss of cell polarity, and inhibition of cell surface actin-based extensions, to ultimately result in detachment from the substratum. Prolonged incubation (120 h) in MBQ-167 decreases metastatic cancer cell viability with a GI50 of ~130 nM, without affecting non-cancer mammary epithelial cells. The loss in cancer cell viability is due to MBQ-167-mediated G2/M cell cycle arrest and subsequent apoptosis, especially of the detached cells. In vivo, MBQ-167 inhibits mammary tumor growth and metastasis in immunocompromised mice by ~90%. In conclusion, MBQ-167 is 10X more potent than other currently available Rac/Cdc42 inhibitors, and has potential to be developed as an anticancer drug, as well as a dual inhibitory probe for the study of Rac and Cdc42.
Keywords: Rac, Cdc42, Rac/Cdc42 inhibitor, breast cancer, anoikis
Introduction
Rho GTPases regulate migration and invasion, cytoskeletal organization, transcriptional regulation, cell cycle progression, apoptosis, vesicle trafficking, and cell-to-cell and cell-to-extracellular matrix adhesions. The Rho GTPases Rac and Cdc42 are potent inducers of actin polymerization and extension of actin structures at the leading edge of motile cells. In addition, Cdc42 plays a critical role in cell polarity, and thus, promotes directed and persistent migration (1).
Studies have implicated hyperactive Rac and Cdc42 with increased cancer cell survival, proliferation, and invasion, as well in Ras and other oncogene-mediated transformation (2, 3). Furthermore, oncogenic cell surface receptors, such as tyrosine kinase, cytokine, and G protein coupled receptors, activate Rac and Cdc42 via regulation of their upstream effectors guanine nucleotide exchange factors (GEFs) (4, 5). Accordingly, Rac and Cdc42 proteins are generally not mutated in cancer but rather overexpressed or hyperactivated (6, 7). Even though ~9% of melanomas contain an activating Rac(P29S) mutation (8), and the hyperactive splice variant Rac1b is overexpressed in some cancers (7), a majority of the Rac and Cdc42 in human cancer is activated due to upregulated GEFs (9). Therefore, targeting the binding of GEFs to Rac and Cdc42 is a rational strategy to inhibit their activation.
Of the direct downstream effectors of Rac and Cdc42, p21-activated kinases (PAK) are overexpressed in a number of cancers and contribute to cancer transformation and progression by regulating key cellular functions, including cytoskeletal organization, cell migration, adhesion, growth, and development (10, 11). Therefore, a number of PAK inhibitors have been developed as anti-cancer therapeutics. However, these have been limited by specificity, bioavailability, and toxicity, and have yet to successfully complete clinical trials (12).
Both Rac and Cdc42 have also been used as drug targets, although to our knowledge, none of the available inhibitors have entered clinical studies. NSC23766 was the first Rac inhibitor shown to block the interaction of Rac with the GEFs Trio and Tiam1; however, its high effective concentrations (IC50>75 μM) limits its therapeutic use (13). Therefore, we developed a panel of putative Rac and Cdc42 inhibitors (14), which led to the identification of EHop-016 (15). EHop-016 blocks the interaction of the GEF Vav2 with Rac, and inhibits Rac activity at an IC50 of ~1.1 μM, which makes it ~100X more potent than NSC23766. EHop-016 also inhibits Cdc42 activity at concentrations of ≥10 μM, without affecting Rho activity (16).
We further reported that at 25 mg/kg body weight (BW), EHop-016 reduces mammary tumor growth, metastasis, and angiogenesis without apparent toxicity in nude mice. The pharmacokinetics analysis of EHop-016, after oral and intraperitoneal (i.p.) administration, demonstrated a bioavailability of ~30% with an average half-life of ~4.5h, indicating its potential as a cancer therapeutic in breast cancer (4, 14–18), and subsequently in other types of cancer (19–21).
Although other small molecule inhibitors, such as the NSC23766 derivative Aza-1 (inhibits both Rac and Cdc42) and CID2950007/ML141 (selective for Cdc42) are currently available, they are effective in the micromolar range (22–24). Our goal to develop a Rac/Cdc42 inhibitor with improved activities led to the identification of MBQ-167. Compared to EHop-016, MBQ-167 is a 10X more potent inhibitor of Rac and a 100X more potent inhibitor of Cdc42, which resulted in an enhanced inhibition of cancer malignancy.
Materials and Methods
Synthesis of MBQ-167
All reagents were purchased from Sigma-Aldrich Chemical Company. The synthesis of 3-Azido-9-ethyl-9H-carbazole 3 is as follows (Fig. 1).
Figure 1.

Design and synthesis of MBQ-167. Synthesis of MBQ-167. Reaction conditions: (i) conc. H2SO4, NaNO2, water 0–5 °C, 1 h; (ii) NaN3, 0 °C, 1 h, 76%; (iii) THF, 3, 50 °C, 1 h; (iv) NH4Cl (aq), 86%.
Step 1
To a solution of 2.10 g (10.0 mmol) 9-Ethyl-9H-carbazol-3-yl-amine 1 in 20 mL water, 2.0 mL (40.0 mmol) of concentrated sulfuric acid (H2SO4) was added. When all the amine was converted to the sulfate (green precipitate), 10 mL more of water was added and the suspension cooled to 0–5°C in an ice-water bath. A solution of 0.828 g (12.0 mmol) sodium nitrite (NaNO2) in 5 mL of water was added dropwise, and the mixture stirred for 1 h. Next, a solution of 0.780 g (12.0 mmol) of sodium azide (NaN3) in 5 mL of water was added dropwise and stirred continuously for 2–8 h. After completion of the reaction, the reaction mixture was warmed to 25°C, 30 mL of ethyl acetate and 20 mL of distilled water were added, and after vigorous mixing, the layers were separated. The organic layer was extracted with 10 mL brine, separated, dried on sodium sulfate, filtered and concentrated on a rotary evaporator. After silica gel chromatography using 3:1 hexanes/ethyl acetate as the eluent, 3-Azido-9-ethyl-9H-carbazole 3 was obtained as an off-white solid in a yield of 1.79 g (7.58 mmol = 76%). The product was identified with TLC and NMR spectroscopy. Rf = 0.82 (3:1, Hexane/Ethyl Acetate); 1HNMR (CDCl3, 400 MHz) δ 1.43 (t, J = 7.19 Hz, 3H), 4.37 (q, J = 7.19 Hz, 2H), 7.14 (dd, J = 2.17, 8.63 Hz, 1H), 7.24 (d, J = 7.13 Hz, 1H), 7.37 (d, J = 8.64 Hz, 1H), 7.41 (d, J = 8.24 Hz, 1H), 7.49 (t, J = 7.14 Hz, 1H), 7.75 (d, J = 2.19 Hz, 1H), 8.07 (d, J = 7.80 Hz, 1H); 13C (CDCl3, 100 MHz) δ 14.1, 38.0, 109.0, 109.8, 110.7, 117.5, 119.3, 120.9, 122.5, 124.2, 126.6, 131.4, 137.8 140.9.
Step 2
Synthesis of 1-(9-Ethyl-9H-carbazol-3-yl)-5-phenyl-1H-1,2,3-triazole 6 (MBQ-167)
In a 25 mL 3-neck round bottom flask containing phenylacetylene 0.11 g (1.1 mmol) under a nitrogen atmosphere, a solution of ethylmagnesium bromide in THF 1.1 mL (1.1 mmol) was added dropwise at 25°C. After the Grignard reagent was added, the mixture was heated at 50°C for 15 min and cooled to 25°C. A solution of 0.24 g (1.0 mmol) of azide 3 in THF (1.0 M) was added dropwise and heated to 50°C for 1 h. After quenching with 10% ammonium chloride, the products were extracted with ethyl acetate (3X). The organic layer was washed with 10 mL of brine, separated and dried on sodium sulfate, filtered and concentrated on a rotary evaporator to obtain crude material (0.33 g). The crude oil was purified via silica gel chromatography to obtain 0.29 g (0.86 mmol = 86%) of 1-(9-Ethyl-9H-carbazol-3-yl)-5-phenyl-1H-1,2,3-triazole MBQ-167 as a white solid. Purity (≥98%) was verified by TLC, NMR spectroscopy, and GC/MS: Rf = 0.26 (3:1, hexane/ethyl acetate); 1HNMR (CDCl3, 400 MHz) δ 1.47 (t, J = 7.22 Hz, 3H), 4.38 (q, J = 7.22 Hz, 2H), 7.26 –7.33 (m, 6H), 7.36 (dt, J = 1.76, 8.60 Hz, 1H), 7.41 (d, J = 8.84 Hz, 1H), 7.46 (d, J = 8.32 Hz, 1H), 7.53 (t, J = 7.32 Hz, 1H), 7.93 (s, 1H), 8.20 (d, J = 7.84 Hz, 1H), 8.15 (d, J = 1.80 Hz, 1H); 13C (CDCl3, 100 MHz) δ 13.8, 37.9, 108.7, 108.9, 117.9, 119.5, 120.8, 122.5, 123.0, 123.2, 126.6, 127.1, 128.3, 128.5, 128.8, 129.0, 133.1, 138.0, 139.8, 140.7. LRGC-MS m/z (rel%): [M]+338 (37), [M-C2H5]+ 310 (55), [M-C2H5N]+ 295 (100), [M-C2H5N2]+ 281 (34), [M-C9H9N3]+ 179 (34).
Cell Culture
MDA-MB-231, MCF-7 (ATCC), green fluorescent protein (GFP) tagged bone metastatic variant of MDA-MB-435 (GFP-HER2-BM) (characterized in (25), from Dr. Danny Welch, The University of Kansas Cancer Center), and MCF10A mammary epithelial cells (ATCC) were cultured and maintained as previously described (16). MDA-MB-231 and MCF-7 cell lines were obtained in 2000, the MCF-10A cell line was purchased in 2013, and the GFP-HER2-BM cell line was a gift from Dr. Danny Welch in 2008. The cell lines were authenticated by ATCC in 2015.
Rac and Cdc42 Activation Assays
For the IC50 curves
Rac1/2/3 and Cdc42 activation was determined as described (16), using a G-LISA kit (Cytoskeleton, Inc., Denver, CO). MDA-MB-231 cell lysates were prepared from 24 h MBQ-167 treatment by combining attached and detached cell populations (N=3). Four-parameter dose-response IC50 curves were fitted using the non-linear regression function of GraphPad Prism®.
Additionally, Rac, Cdc42, or Rac activation was determined, by pulldowns using the P21-binding domain (PBD) of PAK, or Rho binding domain of Rhotekin as described (2, 16). The GTP bound active Rac, Cdc42, or Rho was detected by Western blot (N=3).
Western blot analysis
Total cell lysates or pull-downs were Western blotted using routine procedures. The primary antibodies used were: Rac (Rac1,2,3), Cdc42, Bcl-xL, Bcl-2, Mcl-1, PAK1, PAK2, phospho (p) -PAK1(T423)/PAK2(T402), p-PAK1(S199/204)/PAK2(S192/197), p-PAK1(S144/204)/PAK2(S141), LIM kinase (LIMK1), p-LIMK1/2(Tyr507/Thr508), Cofilin, p-cofilin(S3), STAT3, p-STAT3(Y705), p-P-38 MAPK (T180/Y182), p-ERK (T202/Y204), p-Akt (S473), and Akt (Cell Signaling Technology, Inc.) and β–actin (Sigma).
Fluorescence Microscopy
MDA-MB-231 cells were treated with vehicle or MBQ-167 at 250 or 500 nM for 24 h. Cells were fixed, permeabilized, and stained with Rhodamine phalloidin to visualize F-actin, and with p-tyrosine or vinculin to visualize focal adhesions, as described (2). Fluorescence micrographs were acquired at 600X in an Olympus BX40 fluorescence microscope using a Spot digital camera.
Cell Migration Assays
Transwell assay
As described (2), quiescent MDA-MB-231 cells were treated with vehicle or MBQ-167 (250 nM) for 24 h. The attached and detached populations were separated and exactly 2×105 cells were placed on the top well of Transwell chambers with 5% FBS in the bottom well. The number of cells that migrated to the underside of the membrane following a 7 h incubation was quantified after staining fixed cells with propidium iodide (PI). For each treatment (N=3), cells in 20 microscopic fields were quantified.
Wound healing scratch assay
MDA-MB-231 cells plated on 6-well plates at equal cell density were incubated in 10% FBS until confluent. The media was changed to 2% FBS and a single scratch was made in the center of the monolayer culture with a pipet tip. MBQ-167 was added at 0, 250, or 500 nM immediately following wounding. Images were digitally acquired from an Olympus microscope (4× magnification) at 0, 8, 12, and 24 h and the scratch distance quantified in Adobe Photoshop. N=3 biological replicates (with 2 technical replicates each).
Mammosphere Formation Assay
As described (26), Equal numbers of MDA-MB-231 cells treated with vehicle or MBQ-167 were seeded in ultra-low attachment plates (Corning) at a density of 500 cells/well in serum-free mammary epithelium basal medium (Lonza). Mammospheres were counted after 4 days incubation in 0 or 250 nM MBQ-167 at 37°C, 5%CO2. Mammosphere-forming efficiency was calculated as the number of mammospheres divided by the number of cells seeded per well and expressed relative to vehicle controls.
Cell Viability Assays
As described (16), equal numbers of MDA-MB-231, GFP-HER2-BM, or MCF-10A cells were incubated in 0–1 μM MBQ-167 for 120 h. The CellTiter 96® Non-Radioactive Cell Proliferation Assay (Promega, Fitchburg, WI) was used according to the manufacturer’s instructions. This assay allows the quantification of the viability of both attached and detached cells in the same well. GI50 was determined as 100×(T−T0)/(C−T0) = 50 (T = the optical density of drug treatment after 120 h, T0 = the optical density at time zero, and C = the optical density of the untreated cells). Curves were fitted using the four-parameter logistic nonlinear regression models in GraphPad Prism software.
Cell Cycle Progression
MDA-MB-231 cells were incubated with 0 or 250 nM MBQ-167 for 48 h and all cells (detached and attached) were stained with PI, as in (27). Cell cycle stage was analyzed using a four-color flow cytometer (FACSCalibur, BD Biosciences, San Jose, CA). A total of 20,000 events were analyzed for each sample. List-mode files were collected using Cell Quest software 3.3 and analyzed using the Flow Jo software vX.0.7 (BD Biosciences, San Jose, CA).
Apoptosis Assay
Apoptosis was measured using a Caspase-Glo3/7 Luminescence Assay Kit as per manufacturer’s instructions (Promega, Corp., Madison, WI, USA), as in (17). Following treatment of equal numbers of cells with vehicle or MBQ-167 for 24 h, Caspase-3/7 Glo reagent was added and incubated at room temperature for 60 min. Caspase-3/7 activities were determined by quantifying luminescence.
Annexin V staining
Apoptotic cells were detected by fluorescence microscopy of Annexin V-Cy3-18 stained cells as per manufacturer’s instructions (Sigma–Aldrich, St Louis, MO, USA). Briefly, GFP-MDA-MB-231 cells grown on coverslips were treated with vehicle, or 250 or 500 nM MBQ-167 for 6 h and stained with Annexin V-Cy3-18 in binding buffer (10 mM HEPES/NaOH, pH 7.5, 0.14 M NaCl, 2.5 mM CaCl2) for 15 min at room temperature. Coverslips were washed in binding buffer and fixed with 3.7% paraformaldehyde prior to fluorescence microscopy. Images were digitally acquired from an Olympus inverted fluorescence microscope.
Animal protocol
All animal studies were conducted under approved protocol #A8180112 Institutional Animal Care and Use Committee, in accordance with the NIH Guideline for the Care and Use of Laboratory Animals. Female athymic nu/nu mice, 4 to 5wk old (Charles River Laboratories, Inc., Wilmington, MA) were maintained under pathogen-free conditions in HEPA-filtered cages.
Tumor establishment
GFP-HER2-BM cells (~5×105) in Matrigel (BD Biosciences, San Jose, CA) were injected at the fourth right mammary fat pad under isofluorane inhalation (1–3% in oxygen using an inhalation chamber at 2L/min) to produce orthotopic primary tumors, as described (17). After tumor establishment (1wk post-inoculation), animals were randomly divided into treatment groups (n=6).
Administration of MBQ-167
Mice were treated with vehicle (12.5% ethanol, 12.5% Cremophor (Sigma-Aldrich, St. Louis, MO), and 75% 1X PBS pH 7.4), or 1 or 10 mg/kg BW MBQ-167 by i.p. injection in a 100 μL volume 3X a wk. Treatments continued until sacrifice at day 65.
Whole body fluorescence image analysis
Mammary tumor growth was quantified as changes in the integrated density of GFP fluorescence, as in (28). Mice were imaged on day 1 of treatment administration, and once a week thereafter for 65 days, using the FluorVivo small animal in vivo imaging system (INDEC Systems, Inc., Santa Clara, CA). Tumor fluorescence intensities were analyzed using Image J software (National Institutes of Health, Bethesda, MD). Relative tumor growth was calculated as the integrated density of fluorescence of each tumor on each day of imaging relative to the integrated density of fluorescence of the same tumor on day 1 of treatment, as described (17). As in (29), Optimal Tumor growth was calculated as %T/C= (δT/δC) × 100 when δT>0, δT=average tumor size on day 65 of treated mice- average tumor size on day 01 of treated mice. δC= average tumor size on day 65 of control mice- average tumor size on day 01 of control mice. Tumor growth delay was calculated as the percentage by which the treated group tumor size is delayed in attaining a specified number of doublings (from day 1) compared with controls using: [(T−C)/C] × 100, where T and C are the median times in days for treated and control groups to double in tumor size.
Analysis of metastases
Following sacrifice, lungs, kidneys, livers, bones, and spleens were excised and immediately stored in liquid N2. Stored organs were thawed and analyzed by fluorescence microscopy, as described (17).
Liver enzyme assays
Frozen stored livers were thawed and homogenized to measure alkaline phosphatase (ALP) and alanine transaminase (ALT) activities using colorimetric assay kits from Abcam and Cayman Chemicals respectively, as per manufacturer’s instructions.
Statistical Analysis
Statistical analyses used Microsoft Excel and GraphPad Prism, and differences were considered statistically significant at P ≤ 0.05.
Results
We previously characterized the Rac inhibitor EHop-016 with an IC50 of 1.1μM in highly metastatic breast cancer cell lines, and determined that the carbazole fragment of EHop-016 was a key contributor to its activity. Additional observations from molecular docking studies suggested that EHop-016 binds to Rac in a bent U-shaped conformation (4, 16). This led to the synthesis of 9-Ethyl-3-(5-phenyl-[1,2,3]triazol-1-yl)-9H-carbazole (MBQ-167) (Fig. 1), which contains the essential carbazole group, and due to its ortho-substitution on a central triazole ring, is forced to adapt the desired bent shape. In silico modeling predicts that MBQ-167 binds deeper into the putative binding pocket of Rac with potential H bonding with Asn39, which is present in the switch region of both Rac and Cdc42 (Suppl. Fig. 1).
MBQ-167 affects cancer cell polarity
Human breast cancer cells were visualized by bright field microscopy following MBQ-167 treatment. At ≥100 nM, starting at six hours, MBQ-167 induced a loss of polarity in metastatic breast cancer cells. Treatment with 500 nM MBQ-167 for 24 h resulted in ~95% cell rounding and detachment from the substratum in metastatic MDA-MB-231 cells (Fig. 2A upper panel). Moreover, MBQ-167 induced this phenotype in multiple mesenchymal cancer cell types including GFP-HER2-BM, MDA-MB-468, and Hs578t human breast cancer cells (data not shown), as well as Mia-PaCa-2 pancreatic cancer cells, SKOV3 ovarian cancer cells, AGS and NCI-N87 gastric cancer cells, and SH-SY5Y neuroblastoma cells (Suppl. Fig. 2). On the other hand, non-cancer mammary epithelial MCF10A and epithelial breast cancer MCF-7 cells were resistant to MBQ-167 and remained polarized and attached to each other and the substratum (Fig. 2A, bottom panel).
Figure 2.

Breast cancer cell phenotype following MBQ-167 treatment. A, Upper panel, bright field images of MDA-MB-231 human metastatic breast cancer cells in response to MBQ-167. Lower panel, MCF-7 cells in response to MBQ-167. B,C, Focal adhesions and actin cytoskeleton following MBQ-167 treatment showing representative fluorescence micrographs of MDA-MB-231 cells. MDA-MB-231 cells were treated with 0, 250, or 500 nM MBQ-167 for 24h and fixed and stained in B, with phospho-tyrosine antibodies for focal adhesions (green) and Rhodamine phalloidin for F-actin (red) and in C, with anti vinculin (red). Arrows, invadopodia; arrowheads, focal adhesions.
To further investigate the effect of MBQ-167 on MDA-MB-231 cells, we performed immunofluorescence microscopy following 0–500 nM MBQ-167 to detect actin dynamics (by Rhodamine phalloidin) and focal adhesions (by anti-p-tyrosine and anti-vinculin). MBQ-167 rearranged the actin cytoskeleton and focal adhesions to result in loss of cell polarity and attachment to the extracellular matrix (ECM), with a marked reduction in both Rac-regulated lamellipodia/invadopodia and Cdc42-induced microspikes and filopodia (Fig. 2B,C). Moreover, in MBQ-167-treated cells, the focal adhesions were reduced from the cell edge and rearranged from the cytoskeleton to the center of the rounded detaching cells.
MBQ-167 inhibits Rac and Cdc42 activation in metastatic cancer cells
Since MBQ-167 disrupted the Rac and Cdc42-mediated cytoskeletal architecture, we investigated its potential to inhibit both Rac and Cdc42 activation in metastatic cancer cells. MDA-MB-231 and GFP-HER2-BM cells were treated for 24 h with vehicle or MBQ-167, and the detached and attached cells (~50% for each population), were recovered and lysed immediately. Equal amounts of protein were subjected to activation assays for Rac or Cdc42. Following treatment with 250 nM MBQ-167 for 24 h, the attached population of MDA-MB-231 cells demonstrated a ~25% decrease in Rac activation while the detached cells were more responsive with a ~75% decrease (Fig. 3A). At earlier times (6h), treatment with 250 or 500 nm MBQ-167, induced a 10–20% inhibition in Rac activity in the attached cell population, while the detached population demonstrated a ~40–50% inhibition (Suppl. Fig. 3). Similarly, Cdc42 activity was inhibited by 60% in the attached cells and 78% in the detached MDA-MB-231 cells following 250 nM MBQ-167 for 24 h (Fig. 3C). These results indicate that both Rac and Cdc42 activities are inhibited while the cells are still attached to the substratum but the more responsive cells gets detached first. Incubation of MDA-MB-231 cells with 500 nM MBQ-167 for 24 h resulted in ~90% detachment of cells and a parallel decrease in Rac and Cdc42 activities, demonstrating that a majority of the cells were responsive to MBQ-167. Similarly the GFP-HER2-BM highly metastatic breast cancer cell line responded to MBQ-167 by inhibition of Rac and Cdc42 activities significantly in the detached cell populations (Suppl. Figs. 4A,B). However, the non-metastatic more epithelial MCF-7 cell line, which did not respond to MBQ-167 by the cell detachment phenotype, was also insensitive to MBQ-167 treatment in Rac inhibition (Suppl. Table 2). This may be due to differences in the Rac and Cdc42 GEFs that are expressed and activated in metastatic breast cancer cell lines (MDA-MB-231) compared to the less metastatic more epithelial MCF-7 cell line.
Figure 3.

Inhibitory effect of MBQ-167 on Rac and Cdc42 activation. MDA-MB-231 human breast cancer cells were treated for 24 h with 250 nM MBQ-167. A–C, The attached (A) and detached (D) cell populations were recovered and equal amounts of proteins subjected to pulldown assays using the p21-binding domain of PAK to isolate the GTP bound Rac and Cdc42. Cell lysates were western blotted with antibodies to Rac or Cdc42. Results from positive bands in western blots were quantified using image J. A, Left, Representative western blot for Rac1/2/; right, quantification of Rac activation at 24 h following 0 or 250nM MBQ-167. B, Left, Representative western blot for Cdc42; right, quantification of Cdc42 activation following 24 h treatment with 0 or 250 nM MBQ-167. The integrated density for active Rac or Cdc42 (GTP) was divided by the total Rac or Cdc42 from the same cell lysates. Rac or Cdc42 activity for each MBQ-167 treatment was divided by the vehicle controls for each experiment to obtain Relative Rac or Cdc42 activity. N=3, * = P < 0.05, *** = P < 0.001. Error bars represent ± S.E.M. C,D, MDA-MB-231 cells with vehicle control (0.1% DMSO) or varying concentrations of MBQ-167 (0–1000 nM) were treated for 24 hrs. Total cell lysates using combined attached and detached treated populations were subjected to the G-LISA Rac1/2/3 or Cdc42 activation assay. IC50 curves for percentage Rac (C) or Cdc42 (D) activation are relative to vehicle from three biological replicates each with two technical replicates. Error bars represent ± S.D. Four-parameter dose-response curves generated using GraphPad Prism® are shown.
Next, as a measure of the specificity of MBQ-167 as a Rac/Cdc42 inhibitor, the IC50s for Rac and Cdc42 activation were also determined following 24 h in MBQ-167 using combined attached and detached populations. Results show that MBQ-167 inhibits Rac 1/2/3 activity in the MDA-MB-231 cells with an IC50 of 103 nM, and Cdc42 activity with an IC50 of 78 nM (Fig. 3D,E). Since the IC50 for Rac inhibition by EHop-016 is 1.1 μM and Cdc42 inhibition is ~8 μM (16), MBQ-167 is 10X more potent than EHop-016 for Rac inhibition and 100X more potent for Cdc42 inhibition.
To indirectly determine the specificity of MBQ-167 for inhibiting Rac activation by GEFs, Rac activity was determined from our previously characterized MDA-MB-435Br cells expressing a control vector or constitutively active (Rac1G12V) (2). However, MBQ-167 did not affect the Rac activity of this cell line expressing a Rac1(G12V) (data not shown), indicating that constitutive activation of Rac1 desensitizes the cells to inhibition by MBQ-167. We have also determined that MBQ-167 inhibits the interaction of Rac and Cdc42 with their GEFs by pulldown assays using a Rac1 or Cdc42 (G15A) mutant, as in (16) (data not shown). Moreover, as demonstrated from activation assays for Rac, Cdc42, and Rho from attached and detached cell populations, MBQ-167 did not affect the related GTPase Rho activation in both cell populations of MDA-MB-231 and GFP-HER2-BM metastatic cancer cells (Suppl. Table 1, Suppl. Fig. 4C).
MBQ-167 inhibits Rac and Cdc42 downstream effectors
To investigate the effect of MBQ-167 on Rac/Cdc42 signaling, we investigated its effect on the major Rac/Cdc42 downstream effector PAK. The phosphorylation status of several PAK residues was analyzed by western blotting, as a measure of its activity. At 250 nM, 24 h treatment with MBQ-167 inhibited PAK1 and PAK2 phosphorylation at the T423/T402 and S199/S192 residues in the detached population of MDA-MB-231 cells. Except for PAK1T423, phosphorylation of all of these residues was significantly decreased in the attached population as well. Even though PAK1T423 phosphorylation was not inhibited in the attached cells, the reduction in the homologous PAK2 phosphorylation sites indicates a preferential inhibition of PAK2 in the attached cells (Fig. 4A,B). Interestingly, MBQ-167 induced a dramatic increase in the phosphorylation of the PAK1S144 (Fig. 4A). However, overall PAK activity is inhibited by MBQ-167, because the activating phosphorylation (Y507/T508) of the direct PAK substrate LIM kinase (LIMK) and the inactivating phosphorylation (S3) of cofilin (actin depolymerization factor), a downstream effector of LIMK, were both decreased following MBQ-167 treatment. The decrease in cofilin phosphorylation was evident after 12 h following 250 nM MBQ-167 (Fig. 4C), indicating activation of cofilin, which account for the observed actin cytoskeletal restructuring (Fig. 2B). Moreover, in the MCF7 cell line, which did not respond to MBQ-167 by cell detachment or inhibition of Rac activation, MBQ-167 also did not affect PAK activity (Suppl. Fig. 5).
Figure 4.

The effect of MBQ-167 on signaling downstream of Rac and Cdc42. A, The effect of MBQ-167 on PAK1 and PAK2 phosphorylation as measured by western blotting for pPAK1 (T423)/pPAK2 (T402), pPAK1 (S199)/pPAK2 (S192), and pPAK1 (S144) levels in MDA-MB-231 cells after 24 hours of treatment in 0 or 250 nM MBQ-167. Data for separate attached (Att) and detached (Det) populations are shown. Left, representative western blots (N=3). Right, relative PAK activity following MBQ-167 treatment. Positive bands from all western blots were quantified using image J. The integrated density of p-PAK was divided by that of total PAK for the same cell lysate and used as a measure of PAK activity for each phospho PAK residue. Relative PAK activity was determined relative to vehicle controls for each experiment. N=3, * = P < 0.05, ** = P < 0.01, *** = P < 0.001. Error bars represent ± S.E.M. B, Effect of MBQ-167 on PAK downstream effectors LIMK and cofilin phosphorylation. MDA-MB-231 cells were incubated for 4, 12, or 24 h in vehicle or 250 nM MBQ-167, the attached (A) and detached (D) populations were separated, lysed, and equal protein used for western blotting. Representative western blot of total LIMK1 or p-LIMK1/2 (Y507/T508) following 24 h in 0 or 250 nM MBQ-167 (N=2) is shown. C, Representative western blot of total or p-cofilin (S3) of equal amounts of total protein lysates following 4, 12, or 24 h in 250nM MBQ-167 (N=3). Separated attached (A) and detached (D) populations are shown for 12 and 24h of MBQ-167 treatment. D, Effect of MBQ-167 on STAT3 phosphorylation and expression. Representative western blot is shown for pSTAT3 (Y705) and total STAT3 expression in GFP-HER2-BM cells after 24h treatment with vehicle or 100, 200, or 500 nM MBQ-167. Representative western blot (left) and quantification (right). N=3, * = P< 0.05, Error bars represent ± S.E.M. E, F, Effect of MBQ-167 on cell migration. E, The effect of MBQ-167 on MDA-MB-231 cellular migration as measured by a transwell assay. Images are representative of three independent experiments. The Graph below shows quantification of 20 microscopic fields per treatment per experiment of PI stained cells that migrated to the underside of the membrane through 8 micron diameter pores N=3,* = P < 0.05, Error bars represent ± S.E.M. F, The effect of MBQ-167 on cell migration in a scratch assay. MDA-MB-231 cells plated at equal density were subjected to a scratch in the center and treated with MBQ-167 at 0, 250, or 500 nM. Micrographs were digitally acquired at 0 and 24 h and the distance of the scratch quantified for each treatment and presented relative to the distance at time 0. Results are an average of two technical replicates and two biological replicates for each treatment ± S.D,* = P < 0.05. G, The effect of MBQ-167 on mammosphere forming efficiency in MDA-MB-231 cells. MDA-MB-231 cells treated with 0 or 250 nM MBQ-167 were subjected to mammosphere assays for 4 days. Cells were treated with MBQ-167 only once prior to placing on the mammosphere medium. Mammosphere forming efficiency was calculated as the percentage of the number of mammospheres divided by the number of cells seeded per well. N=3, *** = P < 0.001, Error bars represent ± S.D.
Rac activity has also been shown to directly stimulate the activity of the transcription factor, STAT3 (30). As seen in Figure 4D and Supplemental Figure 6A, MBQ-167 decreased STAT3 activity following 24 h exposure in both the attached and detached populations of MDA-MB-231 and GFP-HER2-BM cells. However, MBQ-167 did not affect mitogen activated protein kinase (MAPK) activities, either p38-MAPK or the p42/44 MAPK, as well as Akt activities as demonstrated by western blotting with phospho-specific antibodies (Supplemental Fig. 7).
MBQ-167 inhibits cell migration and mammosphere formation
Since Rac/Cdc42 and its downstream effector PAK directly regulate cell migration, we investigated the effect of MBQ-167 on MDA-MB-231 cell migration. The two cell populations (detached and attached) were recovered following 18 h in MBQ-167 and equal numbers of cells (vehicle-treated, and MBQ-167-treated attached and detached populations) were used for a Transwell assay for 6 h. This short incubation time is not sufficient for MDA-MB-231 cell division (doubling time of ~38 h) or inhibition of cell viability (Suppl. Fig. 8B,C). Therefore, the assay only measures the efficiency of cell migration.
MBQ-167 treatment reduced directed migration of the attached MDA-MB-231 cell population by ~60–70% at 250 and 500 nM. In the detached population, MBQ-167 (250 and 500 nM) inhibited cell migration by ~90% in a statistically significant manner (Fig. 4E). In the more metastatic GFP-HER2-BM cell line, 250 and 500 nM MBQ-167 inhibited cell migration by 80–90% in both attached and detached cells (Suppl. Fig. 6B). These results were confirmed in a wound healing assay where 250 and 500 nM MBQ-167 treatment for 24 h resulted in statistically significant ~80 and 90 % inhibition of wound closure, respectively (Fig. 4F).
STAT3 and Rac activities have been implicated in enhanced breast cancer stem cell-like properties and therapy resistance (31, 32). Therefore, we evaluated the capacity of MBQ-167 to target cancer stem cell populations using a mammosphere formation assay. Addition of MBQ-167 once for four days reduced the mammosphere forming efficiency of MDA-MB-231 cells by ~50% following 24 h treatment with MBQ-167 (Fig. 4F).
MBQ-167 inhibits cell survival
MBQ-167 induces a phenotype characterized by cell rounding, loss of lamellipodia, and eventual detachment from the surface substratum (Fig. 2). Therefore, we tested the potential of MBQ-167 to induce anoikis: apoptosis due to dissolution of integrin-mediated cell to ECM attachments (33). It should be emphasized that the metastatic cancer cells that detach in response to MBQ-167 following 24 h treatment are viable, as evidenced by trypan blue exclusion from live cells (Suppl. Fig. 8A). These detached cells also have the capacity for regrowth when replated without MBQ-167 (data not sown). As shown in Supplemental Figure 8B, MDA-MB-231 cells are 100% viable at concentrations ≤ 300 nM for 24 h. At 24 h MBQ-167 treatment, ~75% of MDA-MB-231, GFP-HER2-BM, and MCF-7 breast cancer cells, as well as the MCF-10 mammary epithelial cells are viable even at 5 μM MBQ-167 (Suppl. Fig. 8C,D). Prolonged treatment for 48, 96, and 120 h with MBQ-167 results in cell detachment from the substratum and loss of cell viability (Suppl. Fig. 8B).
Figure 5A shows a MTT assay following MBQ-167 treatment for 120 h for the metastatic cancer cells MDA-MB-231 and GFP-HER2-BM and the non-cancer mammary epithelial cells MCF10A, when we obtained ~100% cell death for all cell types at high concentrations (1000 nM) of MBQ-167. This assay includes both detached and attached cells in the case of the metastatic cancer cells. MBQ-167 at 120 h decreased the viability of MDA-MB-231 and GFP-HER2-BM cells with a GI50 of 110 nM and 150 nM respectively. However, the GI50 for the MCF10A epithelial cells at 350 nM MBQ-167 was ~3X higher (Fig. 5A). It should be noted that MBQ-167 inhibits Rac and Cdc42 activities with IC50s in the ~100 nM range at 24 h when the MDA-MB-231, GFP-HER2-BM, MCF-7, and MCF-10 cells are still viable (Suppl. Fig. 8B,C). Next, we determined whether the effect of MBQ-167 on cell viability is due to cell cycle arrest by flow cytometry. As shown in Figure 5B, MBQ-167 significantly arrested the cell cycle of MDA-MB-231 cells in the G2/M phase.
Figure 5. Effect of MBQ-167 on cell survival.
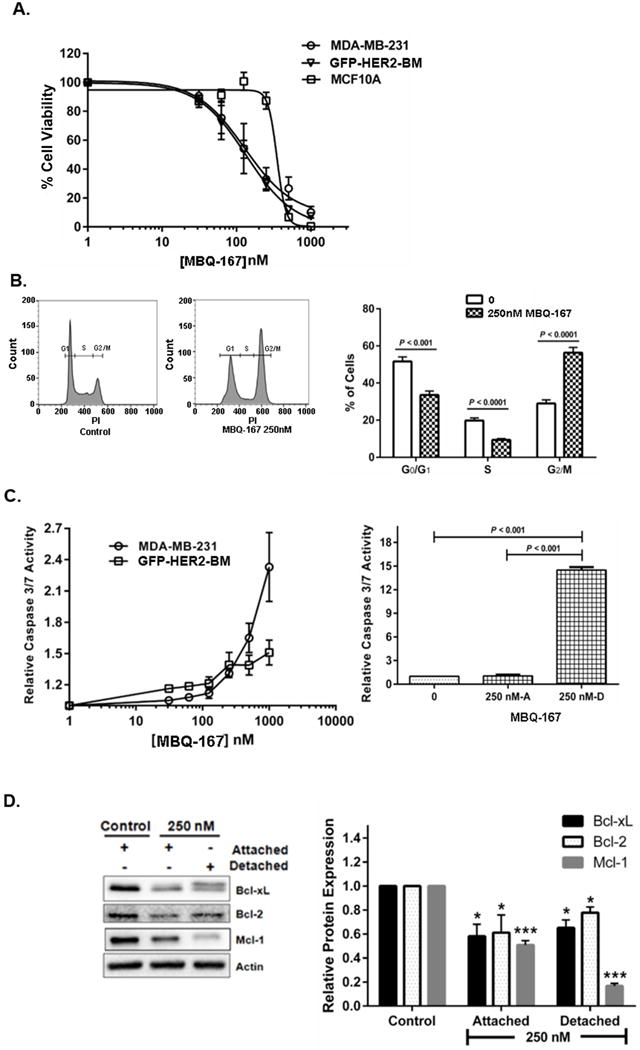
A, The effect of MBQ-167 on MDA-MB-231, GFP-HER2-BM, and MCF10A cell viability. Equal numbers of each respective cell line were treated with vehicle control (0.1% DMSO) or varying concentrations of MBQ-167 (0–1000 nM) for 120 h. GI50 curves for percentage cell viability are relative to vehicle from three biological replicates each with two technical replicates. Four-parameter dose-response curves generated using GraphPad Prism® are shown. N=3, Error bars represent ± S.E.M. B, The effect of MBQ-167 on cell cycle progression. Equal numbers of MDA-MB-231 cells in either vehicle control or treatment groups were treated for 24 h with 0 or 250 nM MBQ-167. Graphs represent the percentage of control versus 250 nM MBQ-167 treated cells stained with PI in G0/G1, S, or G2/M phases of the cell cycle. Left graph, representative flow cytometry analysis; right graph, quantification of cell cycle stage. N=3, Error bars ± S.E.M. C, The effect of MBQ-167 on caspase3,-7 activity. Left graph, caspase3/7 activity of MDA-MB-231 and GFP-HER2-BM cells (including attached and detached populations) following vehicle (0.1% DMSO) or varying concentrations of MBQ-167 (0–1000 nM) for 24 h. N=3, Error bars represent ± S.E.M. Right graph, the effect of MBQ-167 on caspase-3,-7 activity of MDA-MB-231 cells. Cells were treated with 250 nM MBQ-167 for 24 h and equal numbers of separated attached and detached cells were lysed and used for caspase 3/7 assays. Caspase-3,-7 activity relative to equal number of attached cells from control cells is shown. N=3, Error bars represent ± S.E.M. D, The effect of MBQ-167 on mitochondrial regulation of apoptosis. The effect of MBQ-167 on the expression of the pro-survival proteins Bcl-2, Bcl-xL, and Mcl-1 in MDA-MB-231 cells after 24h of treatment. Left panel, representative western blot; Right panel, quantification of the integrated density of positive bands using image J. N=3, * = P < 0.05, *** = P < 0.001, Error bars represent ± S.E.M.
To evaluate whether the cell cycle arrest was accompanied by an increase in apoptosis, we measured the activity of the effector caspases3/7 in whole cell populations (both attached and detached). A dose-dependent increase was observed for caspase-3/7 activity in both MDA-MB-231 and GFP-HER2-BM cell lines after 24 h in MBQ-167 (Fig. 5C, left panel). To determine whether MBQ-167 induces anoikis, the relative levels of caspase-3/7 activities were analyzed in the attached and detached MDA-MB-231 cell populations following 24 h at 250 nM MBQ-167. There was significant ~15-fold increase in caspase-3/7 activity in the detached population compared to the attached population of MDA-MB-231 cells (Fig. 5C, right panel). In Supplemental Figure 9, we validate the effect of MBQ-167 on apoptosis by showing increased Annexin V staining in MDA-MB-231 cells following 250 or 500 nM MBQ-167. In 500 nM MBQ-167, cells also demonstrated the classic blebbing associated with apoptosis. Finally, to explore the effect of MBQ-167 on mitochondrial apoptosis, we analyzed the expression of the pro-survival proteins Bcl-2, Bcl-xL, and Mcl-1 by western blotting. We found a significant decrease in the expression of pro-survival proteins after 24 h with 250 nM MBQ-167 (Fig. 5D).
MBQ-167 inhibits mammary tumor progression in nude mice
To test the effect of MBQ-167 on mammary tumor progression, nude mice were used to establish mammary fatpad tumors from GFP-HER2-BM cells. One week following mammary tumor establishment, the mice were treated 3X a week with 0, 1, or 10 mg/kg BW MBQ-167 by i.p. for 65 days. The vehicle treated mice demonstrated a linear increase in tumor growth, while MBQ-167-treated mice demonstrated a statistically significant reduction in tumor growth (Fig. 6A). At sacrifice, 1.0 mg/kg BW of MBQ-167 resulted in a ~80% reduction in tumor growth, and the 10 mg/kg BW MBQ-167 treatment resulted in ~95% reduction in tumor growth. Since EHop-016 only exerts ~40% reduction of tumor growth at 10 mg/kg BW (17), MBQ-167 is 10X more effective than EHop-016 (Fig. 6A).
Figure 6.

In-vivo efficacy of MBQ-167 compared with EHop-016. Mammary fatpad tumors were established in nude mice by inoculating 5 × 105 GFP-HER2-BM cells. Following one week, mice were treated with vehicle control or 1.0, or 10.0 mg/kg body weight (BW) MBQ-167 3X a week by i.p. injection. A, Left panel, representative excised tumors following 0, 1, 10 mg/kg BW MBQ-167. Right panel, average relative tumor growth from fluorescence in situ images up to 65 days following 0, 1.0, or 10 mg/kg BW MBQ-167 (3X a wk) (N=6). B, Representative fluorescence micrographs of lungs, spleens, and kidneys from vehicle or MBQ-167 treated mice following necropsy. C, Mouse weights from 1–65 days. D, E, Liver enzyme activities following MBQ-167 treatment. Following necropsy, livers were harvested, lysed and subjected to D, ALP activity or E, ALT activity assays. N=4, error bars represent ± S.E.M.
The optimal % change in tumor size, which takes into account the individual tumor growth for each treatment, showed that the tumors from mice treated with 1mg/kg BW MBQ-167 demonstrated a 58% growth change compared to controls (100%), while tumors from mice treated with 10 mg/kg BW MBQ-167 demonstrated only a 9% increase in tumor size (Suppl. Table 3). These data indicate that even though there was no tumor regression during the time of study, there was a drastic reduction in tumor growth in the 10 mg/kg BW MBQ-167 treated mice.
When the tumor growth delay was quantified, the control mice doubled in 8 days, and the MBQ-167 treated mice demonstrated similar doubling times for both treatments (10 and 11 days). However, at the second doubling (22), there was a delay in tumor growth of the MBQ-167 treated mice, where the tumors from control treated mice reached 22 in 14.5 days, while the tumors from 1 and 10mg/kg BW treated mice were similar to each other by reaching 22 in 30 days. By the 3rd doubling, there was also a disparity between the two MBQ-167 treatments, where the tumors from control mice reached 23 in 27 days, the 1 mg/kg BW MBQ-167 treated tumors took 57 days to reach the same size, and the 10mg/kg BW treated tumors never reached 23 in tumor growth. Similarly, only the control tumors reached 24 in 33 days, while the tumors from both MBQ-167 treated (1 and 10 mg/kg BW) mice never reached this size. This result demonstrates a significant inhibition in tumor growth initiated after 24 days of MBQ-167 treatment (Fig. 6A, Suppl. Table 4). This drastic reduction in tumor growth following MBQ-167 treatment resulted in no metastases to all of the organs tested (Fig. 6B).
The mice from this study were also examined once a week for potential toxicity. The mice treated with vehicle or MBQ-167 did not show any significant weight loss or phenotypic changes during the 65-day study (Fig. 6C). At necropsy, livers were harvested, lysed and subjected to liver enzyme assays as a test for potential toxic effects. Figure 6D shows that MBQ-167 does not affect ALP activity in the livers of MBQ-167 treated nude mice (1 and 10 mg/kg BW). However, the liver ALT levels were significantly increased by 10 mg/kg BW MBQ-167 treatment, indicating a potential metabolism of MBQ-167 at higher concentrations (Fig. 6E).
Discussion
Herein, we describe the biochemical characterization of MBQ-167, which inhibits Rac and Cdc42 activation. Rac and Cdc42 are pivotal signaling intermediates whose dysregulation has been implicated in oncogenic transformation, cancer progression, metastasis, and multiple diseases (1, 6, 34). Recent studies, including our own, have shown that targeting Rac and Cdc42 has potential for metastatic cancer therapy. However, the current small molecule inhibitors of Rac and Cdc42 are only effective at micromolar concentrations (4, 16, 17, 22).
The aim of this study was to develop a new, more potent inhibitor of Rac and Cdc42 with an IC50 < 1.0 μM. Accordingly, we report that the novel derivative MBQ-167 shows improved efficacy in metastatic breast cancer cells by inhibiting Rac activity with an IC50 of ~103 nM, and Cdc42 with an IC50 of 78 nM. This concentration dependent response of breast cancer cells to MBQ-167 demonstrates that MBQ-167 is specifically inhibiting the biochemical activation of the Rac and Cdc42. However, MBQ-167 did not inhibit the Rac activity of cells expressing dominant active Rac1(G12V) indicating that MBQ-167 is specifically inhibiting Rac1 activation. Moreover, our data show that MBQ-167 does not affect the activation of the related GTPase Rho. Therefore, we expect this new molecule to be useful as a tool for probing Rac and Cdc42 function in responsive cell types.
We also found that MBQ-167 was an effective inhibitor of the Rac and Cdc42 downstream effector PAK. Interestingly, MBQ-167 induced an increased autophosphorylation of S144 (PAK1), the activation of which is not essential, but contributes to the activity of the PAK kinase domain (35). This result may be due to a feedback mechanism compensating for Rac/Cdc42 inhibition. Nevertheless, phosphorylations in the PAK kinase domains, as well as the PAK effectors LIMK and cofilin, a potent regulator of actin filament dynamics during cell migration (36), were significantly inhibited by MBQ-167. Therefore, we conclude that overall, MBQ-167 inhibits PAK activity, contributing to a reduction in actin cytoskeletal extensions and cell migration. Since Rac and Cdc42 also regulate Wiskott Aldrich Syndrome protein (WASP)-family members that contribute to actin dynamics (37, 38), MBQ-167 may exert additional inhibitory effects on the cytoskeleton.
Furthermore, Cdc42 regulates cell polarity through the polarity protein portioning defective proteins (PAR6, 3), which stabilize microtubules during directed migration (39). Using a haploid derivative of the yeast strain BY4741, where the essential gene Cdc42 was knocked out conditionally via a tetracycline inducible promoter (40), we show that MBQ-167 exerts a similar phenotype to the cells with reduced Cdc42 expression. Supplemental Figure 10 demonstrates that Cdc42 knockdown abolishes cell polarity where the yeast buds (daughter cells) are not aligned symmetrically with the mother cells. A similar non-polar effect was also observed on yeast cell budding in the presence of MBQ-167. This mutant phenotype was more pronounced in the yeast cells with both Cdc42 knockdown and MBQ-167 treatment demonstrating that MBQ-167 may inhibit the highly conserved yeast Cdc42 to regulate cell polarity. As expected, MBQ-167 treatment also enhanced the growth inhibitory effects of Cdc42 knockdown.
The regulation of microtubule dynamics by Rac and Cdc42 activities are also critical for cell cycle progression, where Cdc42, and thus PAK, controls mitotic spindle formation and cell cycle progression in G2/M (41, 42). Therefore, the observed MBQ-167-mediated metastatic breast cancer cell cycle arrest in the G2/M phase may be a consequence of Rac/Cdc42/PAK inhibition by MBQ-167.
We also show that the decreased Rac/Cdc42/PAK activities, cell viability, loss of cell polarity, and detachment from the substratum in response to MBQ-167 is limited to cancer cells that have undergone epithelial to mesenchymal transition (EMT) but not to epithelial cancer or non-cancer cells. This selective response to MBQ-167 may be due to the differential expression and activities of Rac and Cdc42 GEFs in different breast cancer cell lines (43), where only a subset of the ~80 known Rac and Cdc42 GEFs are expected to be expressed and activated in the metastatic breast cancer cell lines that were investigated. Moreover, the currently available Rac/Cdc42 inhibitors also inhibit only a subset of Rac/Cdc42 GEFs. For instance NSC23766 inhibits only Tiam-1/and Trio activation of Rac, while EHop-016 is a specific inhibitor of the Vav/Rac interaction (13, 16). Therefore, MBQ-167 may inhibit only a subset of Rac/Cdc42 GEFs that are preferentially expressed/activated in the more metastatic mesenchymal-like cancer cells lines. We are currently investigating the specific GEFs inhibited by MBQ-167.
Additionally, the relative insensitivity of epithelial-like cells to MBQ-167 may be because the hemidesmosomes in epithelial cells are primarily regulated by α6β4-integrin mediated attachments to the intermediate filament cytoskeleton, which are not directly regulated by Rac and Cdc42. In contrast, the focal adhesions in mesenchymal cells, which are regulated by multiple integrin sub units to form attachments with the actin cytoskeleton, are under Rac/Cdc42/PAK regulation (42, 44). Therefore, the observed reduction and reorganization of focal adhesions in MBQ-167 treated cancer cells may reflect inhibition of the Rac/Cdc42/PAK regulated integrin-mediated focal adhesion assembly at the cell leading edge.
Focal adhesions are not only important for directed migration, disruptions in proper regulation of cell adhesion to the ECM can result in anoikis, apoptosis induced by inadequate or inappropriate cell–matrix interactions (45). In this context, Rac1 has been shown to confer anoikis resistance (46). Our data with caspase assays and reduction in pro-survival BCl2-homology proteins validate the hypothesis that MBQ-167 acts as an anticancer agent by inducing anoikis. Data show that only the detached breast cancer cells respond to MBQ-167 increased caspase3/7 activities, indicating that cell detachment precedes apoptosis signaling, as would be predicted during anoikis. Moreover, we show that MBQ-167 selectively decreases the viability of cancer cell lines that have undergone EMT, without affecting the non-cancer cell line MCF10A. This cell line specificity could be due to differences in the dependence on Rac/Cdc42/PAK signaling and the accompanying integrin engagement and focal adhesion assembly, in the more migratory mesenchymal cells compared to the epithelial cells. In addition, MBQ-167 may have similar effects in multiple other cancers, including a number of ovarian, gastric, pancreatic, and neuroblastoma cell lines that have undergone EMT. Since EMT is associated with more stem cell-like properties, therapy resistance, and disease recurrence (47), MBQ-167 has potential to reduce therapy resistance. Moreover, the fact that MBQ-167 is effective against the KRAS mutant MIA PaCa-2 cell line demonstrates its ability to target oncogenic RAS dependent cancers.
MBQ-167 also inhibits STAT3 phosphorylation, a Rac-regulated transcription factor shown to be active in several cancers (48). Since STAT3 activity increases the expression of several genes involved in cell cycle progression, its decrease in activity may contribute to the observed cell cycle arrest by MBQ-167 treatment. Importantly, STAT3 transcriptionally regulates all three of the pro-survival BCl-2 family genes analyzed in this study (49). Furthermore, several reports show that cancer stem cell like properties are dependent on STAT3 activity (50). Accordingly, MBQ-167 decreases the mammosphere forming efficiency of MDA-MB-231 cells by ~ 50%. These results suggest that MBQ-167 may be further effective as an anti-cancer therapeutic by targeting cancer stem cell like populations; however, more studies are needed to demonstrate that MBQ-167 specifically inhibits cancer stem cell activity.
Finally, we show that MBQ-167 reduces mammary fat pad tumor size starting as early as 3 wks following treatment, with a 91% reduction by 2 months at a non-toxic concentration of 10 mg/kg BW. The drastic reduction in mammary tumors also resulted in a 100% inhibition of metastases to all organs tested, probably because less cells were shed by the small tumors. As evidenced by the in vitro data, the reduced tumor size in response to MBQ-167 treatment is predicted to be due to inhibition of Rac/Cdc42/PAK signaling ultimately leading to a loss in cell viability, growth, and polarity causing the cells to detach from the tumor and undergo anoikis. Since we did not observe any metastases in mice treated with MBQ-167, any cells detached from the primary tumor probably go through anoikis, and do not survive in the circulation.
Taken together, we have shown that MBQ-167 is an effective Cdc42 and Rac inhibitor that significantly decreases downstream signaling and cancer promoting cell functions to ultimately reduce mammary tumor growth with 10X more potency than our first described Rac inhibitor EHop-016. However, the effects of MBQ-167 on the metastatic cancer cell phenotype, where the cells detach from the substratum to ultimately undergo apoptosis by anoikis mechanisms, may be due to additional effects of MBQ-167 on integrin signaling or alternate mechanisms, which will be explored in future investigations. Nevertheless, the dramatic effect of MBQ-167 on mouse mammary tumor growth warrants further development of MBQ-167 as an anticancer therapeutic.
Supplementary Material
Acknowledgments
We acknowledge the technical contributions of Cristina Del Valle, Gabriela Asencio Torres, Fabiola Pagan, and Sahily Gonzales-Crespo. We also acknowledge the kind receipt of yeast strains with conditional Cdc42 knockouts from Dr. Brian Rymond (University of Kentucky) and the GFP-HER2-BM cell line from Dr. Danny Welch (University of Kansas). Financial assistance was available from the following awards: Susan G Komen for the Cure and Puerto Rico Science, Technology, and Research Trust (PRSTRT) (to S. Dharmawardhane), UPR RCM NIH/NIMHHD grants G12MD007600, R25GM061838 (to T. Humphries-Bickley and L.D. Borrero-Garcia), NIH/NIGMS SC3GM116713 (to C.P. Vlaar), NIH/NIGMS 2P20GM103475 (to J. R. Rodriguez-Medina), and NIH/NIMHHD G12MD007583, and Title V PPOHA P031M10505 and Title V Cooperative P031S130068 from U.S. Department of Education (to Universidad Central del Caribe).
Financial support: Susan G Komen for the Cure and Puerto Rico Science, Technology, and Research Trust (PRSTRT) grants (to S. Dharmawardhane), NIH/NIGMS SC3GM116713 (to C.P. Vlaar), UPR RCM NIH/NIMHHD grants G12MD007600, R25GM061838 (to T. Humphries-Bickley and L.D. Borrero), NIH/NIGMS 2P20GM103475 (to J.R. Rodriguez-Medina), and NIH/NIMHHD G12MD007583, and Title V PPOHA P031M10505 and P031S130068 from U.S. Department of Education (to Universidad Central del Caribe).
Footnotes
Conflict of interest: Authors: E. Hernandez-O-Farrill, C. Vlaar, and S. Dharmawardhane have a conflict of interest due to Provisional patent application: 1,5-disubstituted 1,2,3-triazoles Compounds and Method of Using the Same, Ser. No.: 62/328,282, Filing Date: April 27, 2016
References
- 1.Ridley AJ. Rho GTPase signalling in cell migration. Curr Opin Cell Biol. 2015;36:103–12. doi: 10.1016/j.ceb.2015.08.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Baugher PJ, Krishnamoorthy L, Price JE, Dharmawardhane S. Rac1 and Rac3 isoform activation is involved in the invasive and metastatic phenotype of human breast cancer cells. Breast Cancer Res. 2005;7:R965–R74. doi: 10.1186/bcr1329. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Qiu RG, Chen J, Kirn D, McCormick F, Symons M. An essential role for Rac in Ras transformation. Nature. 1995;374:457–59. doi: 10.1038/374457a0. [DOI] [PubMed] [Google Scholar]
- 4.Dharmawardhane S, Hernandez E, Vlaar C. Development of EHop-016: a small molecule inhibitor of Rac. Enzymes. 2013;33(Pt A):117–46. doi: 10.1016/B978-0-12-416749-0.00006-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Wertheimer E, Gutierrez-Uzquiza A, Rosemblit C, Lopez-Haber C, Soledad SM, Kazanietz MG. Rac signaling in breast cancer: A tale of GEFs and GAPs. Cell Signal. 2011;24:353–62. doi: 10.1016/j.cellsig.2011.08.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Stengel K, Zheng Y. Cdc42 in oncogenic transformation, invasion, and tumorigenesis. Cell Signal. 2011;23:1415–23. doi: 10.1016/j.cellsig.2011.04.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Schnelzer A, Prechtel D, Knaus U, Dehne K, Gerhard M, Graeff H, et al. Rac1 in human breast cancer: overexpression, mutation analysis, and characterization of a new isoform, Rac1b. Oncogene. 2000;19:3013–20. doi: 10.1038/sj.onc.1203621. [DOI] [PubMed] [Google Scholar]
- 8.Krauthammer M, Kong Y, Ha BH, Evans P, Bacchiocchi A, McCusker JP, et al. Exome sequencing identifies recurrent somatic RAC1 mutations in melanoma. Nat Genet. 2012;44:1006–14. doi: 10.1038/ng.2359. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Vigil D, Cherfils J, Rossman KL, Der CJ. Ras superfamily GEFs and GAPs: validated and tractable targets for cancer therapy? Nat Rev Cancer. 2010;10:842–57. doi: 10.1038/nrc2960. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Rane CK. Minden A 21 activated kinases: structure, regulation, and functions. Small Gtpases. 2014;5:e28003. doi: 10.4161/sgtp.28003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.DZ Ye. Field J PAK signaling in cancer. Cell Logist. 2012;2:105–16. doi: 10.4161/cl.21882. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Crawford JJ, Hoeflich KP, Rudolph J. p21-Activated kinase inhibitors: a patent review. Expert Opin Ther Pat. 2012;22:293–310. doi: 10.1517/13543776.2012.668758. [DOI] [PubMed] [Google Scholar]
- 13.Gao Y, Dickerson JB, Guo F, Zheng J, Zheng Y. Rational design and characterization of a Rac GTPase-specific small molecule inhibitor. Proc Natl Acad Sci USA. 2004;101:7618–23. doi: 10.1073/pnas.0307512101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Hernandez E, De La Mota Peynado A, Dharmawardhane S, Vlaar CP. Novel inhibitors of Rac1 in metastatic breast cancer. PR Health Sci J. 2010;29:348–56. [PubMed] [Google Scholar]
- 15.Hernandez E, Vlaar C, Dharmawardhane S. Small-molecule inhibtors of Rac1 in metastatic breast cancer. US8884006B2. US Patents. 2014 Nov 11; US9278956B1, March 08, 2016.
- 16.Montalvo-Ortiz BL, Castillo-Pichardo L, Hernandez E, Humphries-Bickley T, De La Mota-Peynado A, Cubano LA, et al. Characterization of EHop-016, novel small molecule inhibitor of Rac GTPase. J Biol Chem. 2012;287:13228–238. doi: 10.1074/jbc.M111.334524. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Castillo-Pichardo L, Humphries-Bickley T, de la Parra C, Forestier-Roman I, Martinez-Ferrer M, Hernandez E, et al. The Rac Inhibitor EHop-016 Inhibits Mammary Tumor Growth and Metastasis in a Nude Mouse Model. Transl Oncol. 2014;7:546–55. doi: 10.1016/j.tranon.2014.07.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Humphries-Bickley T, Castillo-Pichardo L, Corujo-Carro F, Duconge J, Hernandez-O’Farrill E, Vlaar C, et al. Pharmacokinetics of Rac inhibitor EHop-016 in mice by ultra-performance liquid chromatography tandem mass spectrometry. J Chromatogr B Analyt Technol Biomed Life Sci. 2015;981–982C:19–26. doi: 10.1016/j.jchromb.2014.12.021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Martin H, Mali RS, Ma P, Chatterjee A, Ramdas B, Sims E, et al. Pak and Rac GTPases promote oncogenic KIT-induced neoplasms. J Clin Invest. 2013;123:4449–63. doi: 10.1172/JCI67509. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Maes H, Van ES, Krysko DV, Vandenabeele P, Nys K, Rillaerts K, et al. BNIP3 supports melanoma cell migration and vasculogenic mimicry by orchestrating the actin cytoskeleton. Cell Death Dis. 2014;5:e1127. doi: 10.1038/cddis.2014.94. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Manes TD, Pober JS. TCR-driven transendothelial migration of human effector memory CD4 T cells involves Vav, Rac, and myosin IIA. J Immunol. 2013;190:3079–30. doi: 10.4049/jimmunol.1201817. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Zins K, Lucas T, Reichl P, Abraham D, Aharinejad S. A Rac1/Cdc42 GTPase-specific small molecule inhibitor suppresses growth of primary human prostate cancer xenografts and prolongs survival in mice. PLoS One. 2013;8:e74924. doi: 10.1371/journal.pone.0074924. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Shutes A, Onesto C, Picard V, Leblond B, Schweighoffer F, Der CJ. Specificity and mechanism of action of EHT 1864, a novel small molecule inhibitor of Rac family small GTPases. J Biol Chem. 2007;282:35666–78. doi: 10.1074/jbc.M703571200. [DOI] [PubMed] [Google Scholar]
- 24.Oprea TI, Sklar LA, Agola JO, Guo Y, Silberberg M, Roxby J, et al. Novel Activities of Select NSAID R-Enantiomers against Rac1 and Cdc42 GTPases. PLoS One. 2015;10:e0142182. doi: 10.1371/journal.pone.0142182. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Phadke PA, Mercer RR, Harms JF, Jia Y, Frost AR, Jewell JL, et al. Kinetics of metastatic breast cancer cell trafficking in bone. Clin Cancer Res. 2006;12:1431–40. doi: 10.1158/1078-0432.CCR-05-1806. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.de la Parra C, Borrero-Garcia LD, Cruz-Collazo A, Schneider RJ, Dharmawardhane S. Equol, an isoflavone metabolite, regulates cancer cell viability and protein synthesis initiation via c-Myc and eIF4G. J Biol Chem. 2015;290:6047–57. doi: 10.1074/jbc.M114.617415. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Rivera RA, Castillo-Pichardo L, Gerena Y, Dharmawardhane S. Anti-Breast Cancer Potential of Quercetin via the Akt/AMPK/Mammalian Target of Rapamycin (mTOR) Signaling Cascade. PLoS One. 2016;11:e0157251. doi: 10.1371/journal.pone.0157251. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Bouvet M, Hoffman RM. Tumor imaging technologies in mouse models. Methods Mol Biol. 2015;1267:321–48. doi: 10.1007/978-1-4939-2297-0_16. [DOI] [PubMed] [Google Scholar]
- 29.Alley MC, Hollingshead MG, Dykes DJ, Waud WR. Human tumor xenograft models in NCI drug development. In: Teicher BA, Andrews PA, editors. Cancer drug discovery and development. Humana Press, Inc; Totowa, NJ: 2004. pp. 125–52. [Google Scholar]
- 30.Faruqi TR, Gomez D, Bustelo XR, Bar-Sagi D, Reich NC. Rac1 mediates STAT3 activation by autocrine IL-6. Proc Natl Acad Sci USA. 2001;98:9014–19. doi: 10.1073/pnas.161281298. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Sehl ME, Shimada M, Landeros A, Lange K, Wicha MS. Modeling of Cancer Stem Cell State Transitions Predicts Therapeutic Response. PLoS One. 2015;10:e0135797. doi: 10.1371/journal.pone.0135797. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Dokmanovic M, Hirsch DS, Shen Y, Wu WJ. Rac1 contributes to trastuzumab resistance of breast cancer cells: Rac1 as a potential therapeutic target for the treatment of trastuzumab-resistant breast cancer. Mol Cancer Ther. 2009;8:1557–69. doi: 10.1158/1535-7163.MCT-09-0140. [DOI] [PubMed] [Google Scholar]
- 33.Frisch SM, Ruoslahti E. Integrins and anoikis. Curr Opin Cell Biol. 1997;9:701–06. doi: 10.1016/s0955-0674(97)80124-x. [DOI] [PubMed] [Google Scholar]
- 34.Boettner B, Van Aelst L. The role of Rho GTPases in disease development. Gene. 2002;286:155–74. doi: 10.1016/s0378-1119(02)00426-2. [DOI] [PubMed] [Google Scholar]
- 35.Chong C, Tan L, Lim L, Manser E. The mechanism of PAK activation: auto-phosphorylation events in both regulatory and kinase domains control activity. J Biol Chem. 2001;276:17347–53. doi: 10.1074/jbc.M009316200. [DOI] [PubMed] [Google Scholar]
- 36.Edwards DC, Sanders LC, Bokoch GM, Gill GN. Activation of LIM-kinase by Pak1 couples Rac/Cdc42 GTPase signalling to actin cytoskeletal dynamics. Nat Cell Biol. 1999;1:253–59. doi: 10.1038/12963. [DOI] [PubMed] [Google Scholar]
- 37.Kurisu S, Suetsugu S, Yamazaki D, Yamaguchi H, Takenawa T. Rac-WAVE2 signaling is involved in the invasive and metastatic phenotypes of murine melanoma cells. Oncogene. 2005;24:1309–19. doi: 10.1038/sj.onc.1208177. [DOI] [PubMed] [Google Scholar]
- 38.Rohatgi R, Ma L, Miki H, Lopez M, Kirchhausen T, Takenawa T, et al. The interaction between N-WASP and the Arp2/3 complex links Cdc42-dependent signals to actin assembly. Cell. 1999;97:221–31. doi: 10.1016/s0092-8674(00)80732-1. [DOI] [PubMed] [Google Scholar]
- 39.Etienne-Manneville S, Hall A. Cell polarity: Par6, aPKC and cytoskeletal crosstalk. Curr Opin Cell Biol. 2003;15:67–72. doi: 10.1016/s0955-0674(02)00005-4. [DOI] [PubMed] [Google Scholar]
- 40.Mnaimneh S, Davirerwala AP, Haynes J, Moffat J, Peng WT, Zhang W, et al. Exploration of essential gene functions via titratable promoter alleles. Cell. 2004;118:31–44. doi: 10.1016/j.cell.2004.06.013. [DOI] [PubMed] [Google Scholar]
- 41.Chircop M. Rho GTPases as regulators of mitosis and cytokinesis in mammalian cells. Small Gtpases. 2014;5:e29770. doi: 10.4161/sgtp.29770. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Maroto B, Ye MB, von L K, Schnelzer A, Knaus UG. P21-activated kinase is required for mitotic progression and regulates Plk1. Oncogene. 2008;27:4900–08. doi: 10.1038/onc.2008.131. [DOI] [PubMed] [Google Scholar]
- 42.Burridge K, Chrzanowska-Wodnicka M, Zhong C. Focal adhesion assembly. Trends Cell Biol. 1997;7:342–47. doi: 10.1016/S0962-8924(97)01127-6. [DOI] [PubMed] [Google Scholar]
- 43.Sosa MS, Lopez-Haber C, Yang C, Wang H, Lemmon MA, Busillo JM, et al. Rac-GEF P-Rex1 as an essential mediator of ErbB signaling in breast cancer. Mol Cell. 2010;40:877–92. doi: 10.1016/j.molcel.2010.11.029. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Manser E, Huang HY, Loo TH, Chen XQ, Dong JM, Leung T, et al. Expression of constitutively active alpha-PAK reveals effects of the kinase on actin and focal complexes. Mol Cell Biol. 1997;17:1129–43. doi: 10.1128/mcb.17.3.1129. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Taddei ML, Giannoni E, Fiaschi T, Chiarugi P. Anoikis: an emerging hallmark in health and diseases. J Pathol. 2012;226:380–393. doi: 10.1002/path.3000. [DOI] [PubMed] [Google Scholar]
- 46.Coniglio SJ, Jou TS, Symons M. Rac1 protects epithelial cells against anoikis. J Biol Chem. 2001;276:28113–120. doi: 10.1074/jbc.M102299200. [DOI] [PubMed] [Google Scholar]
- 47.Smith BN, Bhowmick NA. Role of EMT in Metastasis and Therapy Resistance. J Clin Med. 2016;5:E17. doi: 10.3390/jcm5020017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Simon AR, Vikis HG, Stewart S, Fanburg BL, Cochran BH, Guan KL. Regulation of STAT3 by direct binding to the Rac1 GTPase. Science. 2000;290:144–47. doi: 10.1126/science.290.5489.144. [DOI] [PubMed] [Google Scholar]
- 49.Carpenter RL, Lo HW. STAT3 Target Genes Relevant to Human Cancers. Cancers (Basel) 2014;6:897–925. doi: 10.3390/cancers6020897. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Marotta LL, Almendro V, Marusyk A, Shipitsin M, Schemme J, Walker SR, et al. The JAK2/STAT3 signaling pathway is required for growth of CD44(+)CD24(−) stem cell-like breast cancer cells in human tumors. J Clin Invest. 2011;121:2723–35. doi: 10.1172/JCI44745. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.


