Abstract
It is generally believed that the mammalian nucleotide excision repair pathway removes DNA helix-distorting bulky DNA lesions, while small non-bulky lesions are repaired by base excision repair (BER). However, recent work demonstrates that the oxidativly generated guanine oxidation products, spiroimininodihydantoin (Sp), 5-guanidinohydantoin (Gh), and certain intrastrand cross-linked lesions, are good substrates of NER and BER pathways that compete with one another in human cell extracts. The oxidation of guanine by peroxynitrite is known to generate 5-guanidino-4-nitroimidazole (NIm) which is structurally similar to Gh, except that the 4-nitro group in NIm is replaced by a keto group in Gh. However, unlike Gh, NIm is an excellent substrate of BER, but not of NER. These and other related results are reviewed and discussed in this article.
Keywords: Base excision repair, nucleotide excision repair, oxidative stress, DNA damage, reactive oxygen species
Graphical Abstract
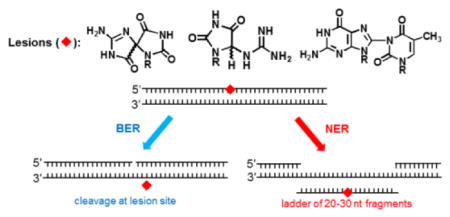
1. Introduction – oxidatively generated DNA damage is genotoxic
Environmental factors (infectious agents, asbestos, tobacco, UV light) are known to contribute to the development of chronic inflammation in human tissues [1, 2]. Reactive oxygen and nitrogen species (ROS and RNS, respectively) are overproduced at sites of chronic inflammation and induce persistent DNA damage that, if not properly repaired, can ultimately lead to the initiation and promotion of cell proliferation and cancer [3]. Epidemiological and clinical studies suggest that about 25% of all cancer cases worldwide are linked to chronic inflammation. Patients suffering from chronic inflammation are at a much higher risk of developing cancers [4]. A chronic imbalance between DNA damage and repair increases the risk of genomic instability, and it is therefore important to understand the mechanisms of the repair pathways that remove oxidatively generated DNA lesions from the genome. In this contribution, we consider some recent results that indicate that some of these DNA lesions can be excised by different overlapping repair pathways such as base excision, nucleotide incision, and nucleotide excision repair.
2. Mechanisms of DNA repair
2. 1. Base excision repair (BER)
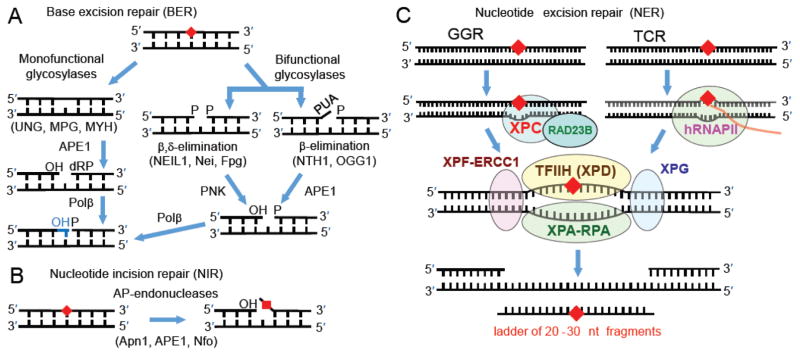
The repair of oxidatively generated lesions is critical for maintaining genomic stability during oxidative stress [5]. Existing paradigms suggest that base excision repair (BER) mechanisms are responsible for the removal of non-bulky oxidatively generated DNA lesions [6]. The mechanisms of BER are highly conserved from bacteria to humans [7, 8] and involve the distinct enzymatic reactions depicted in Figure 1A.
Fig. 1.
Excision of damaged nucleobases by (A) BER, (B) NIR (the lesion is not excised in this pathway), and (C) NER mechanisms.
BER proteins recognize damaged nucleobase by first binding to the damaged site and then cleaving the N-glycosyl bond to release the damaged base, thus forming an abasic site as shown in Figure 1A [9]. In the case of monofunctional glycosylases, the abasic site is cleaved by an apurinic (AP) human endonuclease (APE1) to form fragments with 3′-OH and 5′-deoxyribose phosphate (5′-dRP) ends [10]. Polymerase β (Pol β) subsequently adds a nucleotide to the 3′-OH end using the base in the complementary strand as the template. The 5′-dRP is concurrently removed by the AP lyase activity associated with Pol β (Figure 1A), and finally the nick is sealed by DNA ligase III/XRCC1 [11]. On the other hand, bifunctional glycosylases in addition possess AP lyase activity, which cleaves the abasic site in DNA resulting in the formation of a single-strand break containing either a phosphate (P) group (β, δ-elimination), or an α,βunsaturated aldehyde (PUA, (β-elimination) at the 3′-end [12, 13]. The 3′-PUA and 3′-phosphate groups are further removed by the diesterase activity of APE1 and the phosphatase activity of polynucleotide kinase (PNK) to form the same gapped product as in the case of the monofunctional glycosylase mechanism [14, 15]. This single nucleotide gap is filled by Pol β (Figure 1A), and the nick is sealed by DNA ligase III/XRCC1 as in the case of the monofunctional glycosylases [11].
2. 2. Nucleotide incision repair (NIR)
An alternative nucleotide incision repair pathway involving the AP endonucleases of E. coli Nfo, yeast Apn1 and human APE1, nick DNA on the 5-side of the damaged base, thus generating fragments with 3′-OH ends, as well as a damaged nucleotide on the 5′-end [16, 17] (Fig. 1B). It has been suggested that the NIR pathway can serve as a backup system for BER if the appropriate glycosylase is missing or inefficient. Once the AP endonuclease has produced the initial incision, the full regeneration and repair of the incised strand can occur as long as the other, subsequent repair factors are also available [18]
2. 3. Nucleotide excision repair (NER)
The mammalian global genomic nucleotide excision repair system (GG-NER) recognizes the distortions in the DNA double helix caused by the DNA lesions, rather than the lesions themselves [19] and the full repair of the DNA damage requires the sequential action of more than 30 proteins [20, 21]. In mammalian GG-NER, the recognition of DNA damage is achieved by the protein heterodimer XPC-RAD23B (Fig. 1C). The resulting XPC-RAD23B – damaged DNA complex recruits the ten-protein factor TFIIH, XPA, XPF and XPG that cooperate to excise the characteristic ~ 24 – 30 nucleotide (nt) dual incision products that contain the lesion and are the hallmarks of successful NER [22, 23]. By contrast, transcription coupled nucleotide excision repair (TC-NER) is initiated when human RNA polymerase II (hRNAPII) is stalled by DNA lesions. The stalled polymerase serves as the signal for recruiting the NER factor (TFIIH, etc.) and other NER factors that lead to the NER double incisions and, ultimately, the filling of the ~ 24–30 nt gap created by the dual incision and the removal of the damage-bearing 24–30 nt fragments. However, TC-NER removes DNA lesions only from the transcribed strand of active genes [24], and thus repairs a smaller, but critical fraction of cellular DNA damage.
3. Guanine is the major target of reactive oxygen and nitrogen species
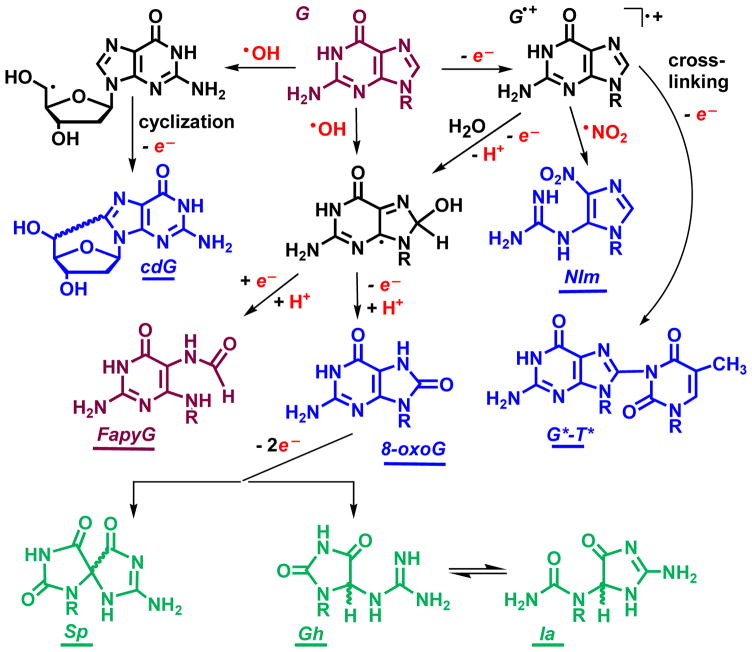
The primary target of oxidatively generated damage in DNA is guanine [25], the most easily oxidizable nucleic acid base in DNA [26]. The oxidation of guanine is typically initiated either by one-electron abstraction or by the addition of hydroxyl radicals (Fig. 2) [27].
Fig. 2.
Representative two-electron (blue) and four-electron (green) oxidation products of guanine produced by ROS and RNS. Abbreviations: cdG, 5′,8-cyclo-2′-deoxyguanosine; NIm, 5-guanidino-4-nitroimidazole; FapyG, 2,6-diamino-4-hydroxy-5-formamidopyrimidine; 8-oxoG, 8-oxo-7,8-dihydroguanine; G*–T*, intrastrand guanine(C8)-thymine(N3) cross-link; Sp, spiroiminodihydantoin; Gh, 5-guanidinohydantoin; Ia, iminoallantoin.
The radical intermediates formed are highly reactive and rapidly transform to stable end-products. The most abundant and best known oxidatively generated guanine lesion is 8-oxoG that is ubiquitous in cellular DNA [25, 28]. Other oxidation products include the diastereomeric 5,8′-cyclo-2′-deoxypurine lesions [29–31]. In gamma irradiated and aerated aqueous nucleoside solutions these lesions are formed at ~ 40 times lower concentrations than 8-oxoG; however, the levels of cdG and cdA were found to increase by factor of 10 – 20 as the oxygen concentration was diminished [32]. Alternatively radical intermediates can transform to 2,6-diamino-4-hydroxy-5-formamidopyrimidine (FapyG) [25, 27, 33]. Formation of FapyG has been detected upon exposure of DNA in aerated aqueous solutions to either hydroxyl radicals or one-electron oxidants [34, 35]. Furthermore, FapyG is produced in two-fold higher yield than 8-oxoG in gamma irradiated cellular DNA [36]. Radicals derived from the one-electron oxidation of guanine remain strong electrophiles and readily add to neighboring thymines to form guanine(C8)-thymine(N3) crosslinks (G*–T*) in air-equilibrated solutions (Fig. 2) [37]. Other forms of cross-linked DNA lesions include G[C8-5m]T and G[8–5]C lesions [29, 30, 38–42]. Guanine radicals readily combine with oxyl radicals to form stable end-products [43], while combination with nitrogen dioxide radicals leads to the formation of 5-guanidino-4-nitroimidazole (NIm) [44, 45]. NIm was found among a number of other products of guanine oxidation by peroxynitrite [46, 47] and serves as a marker of inflammation-related oxidation mechanisms [48].
It is well established that 8-oxoG is more easily oxidized than the parent guanine base [49]. Its further oxidation by diverse oxyl radicals (CO3•−, •NO2, SO4•−, RO•) [45, 50–53] and peroxynitrite [47], can lead to the formation of the diastereomeric spiroiminodihyadantoin (Sp) and 5-guanidinohydantoin (Gh) lesions [54–61]. The Sp and Gh lesions have been detected in mice with infection-induced colitis at concentration levels of about one percent, relative to the more abundant 8-oxoG levels [62]. Due to the presence of chiral carbon atoms, the Sp and Gh nucleobases exist as R and S diastereomers. Oligonucleotides containing single, site-specifically inserted S-Sp and R-Sp lesions can be isolated by anion-exchange HPLC [63]. In aqueous solutions, the Gh diastereomers are easily interconvertible, and can isomerize to iminoallantoin (Ia) (Fig. 2) [55]. In DNA, the isomerization of Gh to Ia occurs in basic solutions (pH > 8.2) [64]. The Sp and Gh lesions are products of four-electron oxidation mechanisms of guanine (in green color in Fig. 2).
4. BER of oxidatively modified guanine bases
A number of oxidatively modified guanine oxidation products are recognized by DNA glycosylases, which belong to the Nth (or Endonuclease III) and/or Fpg/Nei (or Endonuclease VIII) proteins [65]. The bifunctional human glycosylase NTH1 repairs oxidized pyrimidines and formamidopyrimidines by β-elimination mechanisms (Fig. 1A). NTH1 is a member of the helix-hairpin-helix (HhH) superfamily of DNA glycosylases that are highly conserved from bacteria to humans [66]. The 8-oxoG and FapyG lesions opposite cytosine in the complementary strand are recognized and removed by the human bifunctional glycosylase hOGG1 [67]. The latter also belongs to the HhH superfamily and removes the damaged bases also by the β-elimination mechanism. However, DNA replication past 8-oxoG occurs frequently in an error-prone manner by incorporating adenine across this lesion [68]. The DNA glycosylase MUTYH (also a member of the HhH superfamily) removes adenine paired with 8-oxoG or FapyG in the opposite strand [69].
The prokaryotic Fpg protein, referred also as MutM, which is known to repair 8oxoG and FapyG [70] belongs to the Fpg/Nei family of DNA glycosylases [65]. However, the eukaryotic Fpg homolog NEIL1 does not excise 8-oxoG, but recognizes and processes its oxidation products Sp and Gh [71, 72]. NEIL1 also recognizes oxidized pyrimidines and formamidopyrimidines [73–76] and removes these lesions by the β,δ-elimination mechanism (Fig. 1A). NEIL2 repairs the same lesions also by β,δ-elimination, but preferably in single-stranded rather than double-stranded DNA [77, 78]. NEIL3 excises the hydantoins Sp and Gh, as well as oxidized pyrimidines (with lower efficiencies) and cleaves the remaining AP site via β-elimination [79, 80].
5. NER of diastereomeric 5′,8-cyclopurines
The presently accepted DNA repair paradigms are based on the concept that small, non-bulky, oxidatively generated DNA lesions are repaired by base excision repair pathways, whereas bulky DNA lesions are recognized and removed by GG-NER or TC-NER mechanisms. While the mechanisms of lesion recognition are different in GG-NER and TC-NER, the resulting 24 – 30 nt dual incision products are common to both repair pathways.
A well known exception to the BER/NER paradigm, are the diastereomeric set of non-bulky, oxidatively generated 5′R and 5′S 5′,8-cyclo-2′-deoxyguanosine (cdG) and 5′,8-cyclo-2′-deoxyadenosine (cdA) lesions. The cdG and cdA DNA lesions are characterized by a covalent C-C bond between the purine C8 and the 2′-deoxyribose C5′ atoms within the same nucleoside. The N-glycosidic bond of the 5′,8-cyclopurine lesions is resistant to acid/base-catalyzed hydrolysis [81, 82], and is also resistant to base excision repair in mammalian cell extracts [83, 84], as well as to purified NEIL1, NEIL2, Fpg, OGG1, Endo III, and Endo VIII BER proteins [85]. However, the 5′,8-cyclopurines cause significant local distortions to the DNA double helix [86] and are therefore good substrates of mammalian [83, 84] and prokaryotic E. coli nucleotide excision repair systems [87]. The NER dual incision efficiencies of the diastereomeric cdA lesions are about four times greater in the case of the (5′R)- than the (5′S)-cdA lesion incorporated into plasmid DNA [83]. In linear 135-mer DNA duplexes, the 5′R cdG stereoisomer is incised ~ 1.5 more efficiently than the 5′S cdA lesion in 135-mer DNA duplexes in HeLa cell extracts [85]. More recently, the relative NER efficiencies of all four 5′R and 5′S diastereomeric 5′,8-cyclopurines were compared in human HeLa cell extracts [86] and the results are summarized in Table 1.
Table 1.
Relative efficiencies of formation of NER dual incision products in human HeLa cell extracts of the diastereomeric cdG and cdA lesions in 135-mer duplexes [86]. The NER dual incision efficiency of a bulky benzo[a]pyrene-derived DNA adduct ((+)-cis-B[a]P-N2-dG), known to be an excellent NER substrate [88], is shown as a reference.
| Lesion | Relative efficiency, % |
|---|---|
| 5′S–cdG | 39 ± 5 |
| 5′R–cdG | 72 ± 5 |
| 5′S–cdA | 30 ± 2 |
| 5′R–cdA | 51 ± 4 |
| (+)-cis-B[a]P-N2-dG | 100 |
In these experiments, The 5′R diastereomers of both the cdA or cdG lesions are recognized ~two-fold better than the respective 5′S lesions (Table 1), and the 5′R–cdG lesions is a somewhat better NER substrate than the 5′R cdA lesion. These results are qualitatively consistent with the observations of Kuraoka et al. [83] who reported a higher (5′R–cdA)/(5′S–cdA) NER efficiency in plasmid DNA, and Pande et al. [85] who obtained similar 5′S–cdG/5′S–cdA excision ratios of ~1.3–1.5 as in Table 1 in linear DNA. The quantitative differences between these published values and those reported here are most likely attributable to base sequence context effects since the effects of base sequence have been observed in the case of the benzo[a]pyrene diol epoxide-derived guanine adducts [89, 90].
Detailed molecular modeling and molecular dynamic simulation studies indicate that there are differences in the local perturbations and dynamics of the DNA backbone imposed by the cross-linked 5′,8 bonds and their absolute configurations. The absolute configurations of the 5′R cdA and 5′R cdG lesions give rise to a greater distortion and a more pronounced weakening of the local base stacking interactions in the case of the diastereomeric 5′R lesions [86]. These stereochemical effects are correlated with the observed higher NER dual incision efficiencies in the case of the 5′R cdA and 5′R cdG lesions than the stereoisomeric 5′S lesions, respectively.
6. Interplay between BER and NER pathways in the repair of oxidatively generated lesions
We have recently identified a set of non-bulky DNA lesions that are oxidatively generated by free radical mechanisms, and that are substrates of overlapping BER and NER mechanisms.
6.1. Intrastrand guanine(C8) – thymine(N3) crosslinked DNA lesions
Oxidatively generated DNA lesions with covalent bonds between neighboring nucleobases include interstrand and intrastrand crosslinked nucleotides. The interstrand cross-linked (ICL) lesions are characterized by covalent bonds between two nucleotides positioned on opposite DNA strands, whereas the coupling of two nucleotides on the same strand give rise to intrastrand cross-linked (IntraCL) lesions. ICL lesions generated by a variety of bifunctional agents such as cisplatin [91], are difficult to remove by DNA repair mechanisms and are therefore highly genotoxic [42, 92]. Well-known examples of IntraCL lesions are the UV radiation-induced cyclobutane pyrimidine dimers and the pyrimidine (6–4) pyrimidone photoproducts [23, 93] that are genotoxic if not removed by repair mechanisms.
Oxidatively generated IntraCL lesions with covalent linkages between the C8-atom of guanine and the methyl group of an adjacent thymine (G[8–5m]T) on its 3′-side [38], and the analogous G[8–5]C [39], and G[8–5m]C [94] intrastrand tandem lesions are formed in DNA solutions exposed to γ-radiation or under the action of Fenton reagents. These IntraCL lesions are known to be repaired by prokaryotic NER pathways [95–97] and evidence has been presented that these IntraCL lesions may be substrates of NER in mammalian tissues [98].
The more recently discovered guanine(C8)-thymine(N3) lesions (Fig. 2) identified in vitro [37, 47], have also been detected in human HeLa cells by isotope dilution LC-MS/MS methods [99]. There are two types of G*–T* lesions: in one case, the cross-linked guanine and thymine bases are either adjacent to one another (G*T*) or separated by one intervening cytosine (G*CT*) [37]. We have shown that both types of lesions are good substrates of NER in human HeLa cell extract experiments, with the G*T* lesion being removed less efficiently than the G*CT* lesion [100]. Thermal DNA melting studies reveal that both lesions significantly destabilize duplex DNA, and that the destabilization induced by the G*CT* cross-link is considerably greater [100]. Computational and modeling studies have also shown that both lesions dynamically distort and destabilize DNA duplexes by distorting normal Watson–Crick base-pairing and base-stacking interactions, and by causing the untwisting of base pairs accompanied by an opening of the minor groove [100]. These structural perturbations are much more pronounced in the G*CT* than in the G*T* cross-linked lesions, and are correlated with the differences in the NER activities of these two substrates in HeLa cell extracts.
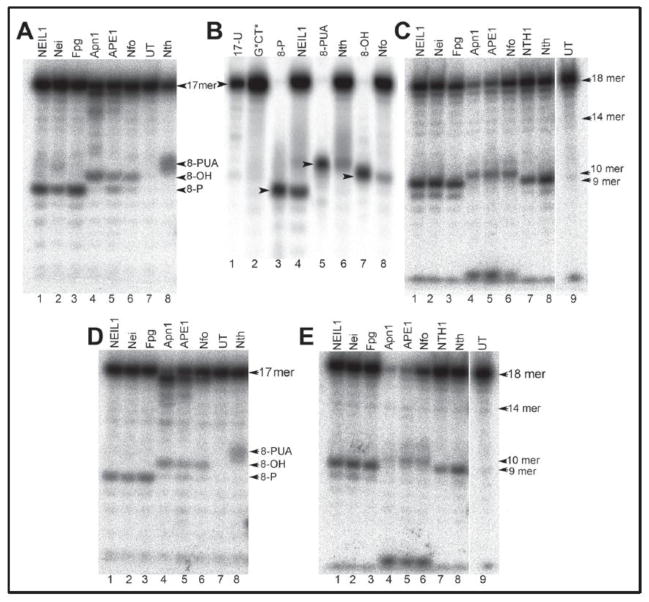
In contrast to 5′,8-cyclopurine lesions, the G*CT* and G*T* IntraCL lesions are incised in human HeLa cell extracts by BER as well as by NER mechanisms [101]. Detailed studies with purified repair proteins have shown that the bacterial, yeast, and human bifunctional DNA glycosylases (Nei, NEIL1, Nth, NTH1), as well as AP endonucleases (Nfo, Apn1 and APE1) cleave the strands adjacent to the G*CT* and G*T* IntraCL lesions (Fig. 3) embedded in site-specifically modified oligonucleotide duplexes.
Fig. 3.
Denaturing polyacrylamide gel electrophoresis analysis of the cleavage patterns generated by DNA glycosylases/AP lyases (BER) and NIR-AP endonucleases (NIR) in duplexes containing G*CT* and G*T* lesions. Panels A and D: duplexes constructed from either 5′-32P-labeled 17 mer G*CT* (A), or G*T* (D) strands hybridized with their natural complementary strands. Panel B: size marker 8-mer oligonucleotide standards with 5′-32P-labled 8-mer containing 3′-hydroxyl (3′-OH), or 3′-phosphoaldehyde (3′-PUA), or 3′-phosphate (3′-P) ends are shown in lanes 3, 5, and 7, respectively. These standards were derived from the parent 17-mer strands containing uracil. Panels C and E: duplexes constructed from cordycepin 3′-32P-endlabeled 18 mer G*CT* (C) or G*T* (E) strands hybridized with their complementary 17-mer complementary strands. UT indicates untreated oligonucleotide. (Reproduced from Talhaoui et al., J. Biol. Chem. 290 (2015) 4610-14617 [101]).
Analysis of the cleavage products by denaturing polyacrylamide gel electrophoresis (Fig. 3), in combination with MALDI-TOF/MS methods, showed that the DNA glycosylases/AP lyases excise the cross-linked guanine and cleave the resulting abasic sites via β- and β,δ-elimination mechanisms. In turn, the AP endonucleases of E. coli, Nfo, yeast Apn1 and human APE1 cleave the duplex DNA containing G*CT* and G*T* lesions on the 5′-side of the cross-linked guanine, and are thus capable of initiating the nucleotide incision repair pathway [101]. These experiments were performed with purified proteins in vitro and clearly show that the G*T* and G*CT* intrastrand crosslinks are efficiently incised by BER and NIR pathways at the sites of these lesions.
In HeLa cell extracts both G*CT* and G*T* lesions are also removed by NER mechanisms [100]. It is interesting to note, that the yields of NER products are higher (~ 14%) than of BER products (~ 3%) in the very same cell extract experiments in the case of the G*CT* lesion. In contrast, in the case of the G*T* lesions, the NER yield is ~ 5 times smaller, while the BER yield is ~ 2 times greater than in the case of G*CT* lesions in the same cell extracts.
6.2. Spiroiminodihyantoin and 5-guanidinohydantoin lesions are substrates of both NER and BER pathways
The Sp and Gh lesions in double-stranded DNA are good substrates of BER enzymes in vitro that include the bifunctional DNA glycosylases E. coli Fpg [102], Nei [103], mammalian NEIL1 and NEIL2 [78], NEIL3 [79, 104–106], human NEIL1 [71, 72], and human NEIL3 [80]. These hydantoins, are also repaired by the prokaryotic NER pathways initiated by UvrABC proteins in vitro [107].
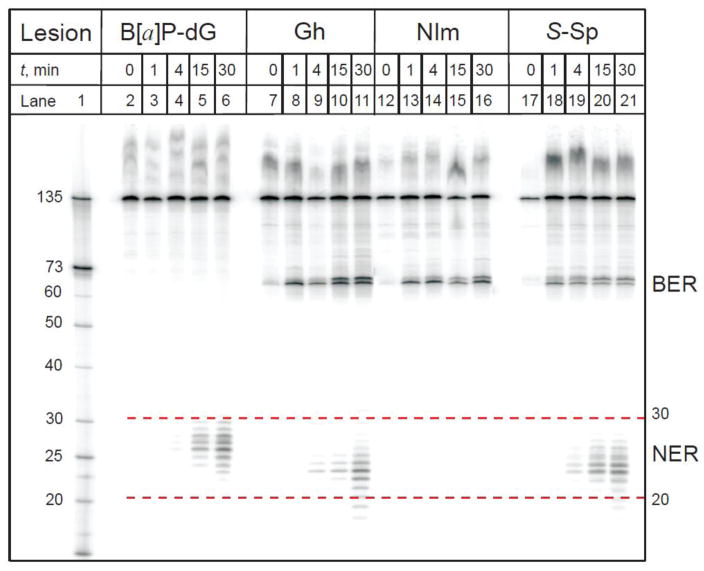
Recently, we found that the Sp and Gh lesions are repaired by competing BER and NER pathways in cell-free extracts derived from human fibroblasts and HeLa cells [108] as demonstrated by the HeLa cell extract experiments shown in Fig. 4.
Fig. 4.
A denaturing polyacrylamide gel showing the appearance of excision (BER) and dual incision (NER) products obtained by incubating, as a function of time, 135-mer duplexes with single Gh, S-Sp, or NIm lesions in HeLa cell extracts. The lesions were positioned at the 67th nucleotide counted from the 5′-end of the modified strand. The 10R (+)-cis-anti-B[a]P-N2-dG adducts, a bulky DNA lesion derived from the binding of a diol epoxide metabolite of benzo[a]pyrene, was used as positive control of NER activity in these cell extracts [88, 89]. Lane 1: oligonucleotide size markers. The apparent size range of the NER dual incision products is shown by the dotted lines (red). (Reproduced from Shafirovich et al., J. Biol. Chem. 291 (2016) 5309-5319 [108]).
The autoradiograph of the denaturing polyacrylamide gels clearly shows that incubation of the 135 nt oligonucleotide duplexes containing single S-Sp and Gh lesions in cell free extracts generates two group of products: (1) the characteristic ladders of NER dual incision products of ~20–30 nucleotides in lengths, and (2) the 67-mer BER products with 3′-P and 3′-OH ends (Fig. 1) arising from incisions at the sites of the lesions [108]. The BER products are not observed in the case of the bulky B[a]P-dG adducts, which are exclusively repaired by NER pathways [88, 109]. In contrast to the Sp and Gh DNA lesions, the NIm lesion yields only the BER product of incision, and NER products are absent in this case [46, 108]. Thus, the Sp and Gh lesions are good substrates of both NER and BER pathways, whereas the B[a]P-dG adducts are substrates of NER only, while the NIm lesions are uniquely substrates of BER only.
These conclusions are supported by experiments in cell extracts derived from human cells deficient in the XPC-RAD23B heterodimer, the initial NER sensor of structural distortions caused by DNA lesions, or XPA that plays a critical downstream role in the complex, multistep mammalian NER pathway. In such extracts, the NER pathway is non-functional as shown by the lack of dual incision products when DNA duplexes with B[a]P-dG adducts, Gh, or Sp are incubated in XPC- and XPA-deficient cell extracts. However, when the XPC-deficient cell extracts are complemented with purified XPC-RAD23B, the NER dual incision products reappear [108]. Additional evidence that the dual incision products derived from the Sp- and Gh-containing 135 bp duplexes is indeed due to NER activity, is the observation that the characteristic NER dual incision products disappear in the presence of low concentrations of a monoclonal antibody against XPA. Inhibition of XPA activity by anti-XPA antibodies has been widely used for validating the existence of NER pathways of repair [83, 110–112]. For example, the mouse monoclonal anti-XPA [5F12] antibody strongly inhibits NER of oxidatively generated 5′,8-cyclopurine lesions [83], and the 1,3-intrastrand d(GpTpG)-cisplatin cross-linked lesions [111]. Indeed, addition of anti-XPA [5F12] to the cell extracts containing 135 bp Sp-S duplexes selectively inhibits the appearance of the dual incision products, whereas the levels of the 67-mer incision fragments produced by the BER activity remain unaffected
Observation of parallel BER and NER pathways in the repair of the Sp and Gh modified 135 bp oligonucleotide duplexes in cell free extracts from cultured human cells can be explained by competitive binding of the Sp and Gh substrates by NER and BER proteins. Indeed, the addition of exogeneous bifunctional DNA glycosylase NEIL1 enhances the yield of 67 nt BER products, whereas the yield of dual incision products decreases [108]. These results are attributed to a competition between NEIL1 and the initial NER DNA lesion recognition factor XPC-RAD23B for binding to the same DNA lesion that increasingly favors NEIL1 as its concentration is increased.
An interesting question is why the NIm lesion is resistant to NER while the structurally similar Gh lesion is an excellent NER substrate. There are two important structural differences between these two lesions (Fig. 1): (i) the keto group in Gh is replaced by an –NO2 group in NIm, (ii) the 5-membered 5-nitroimidazole ring in NIm is planar, whereas in Gh the hydantoin ring is non-planar. Molecular modeling studies indicate that the NIm lesions adopt flexible and multiple ring-opened structures with the nitro and guanidino groups providing multiple hydrogen bonding possibilities [113]. By contrast, Gh can exist in the forms of two slowly interconverting R and S stereoisomers [55, 114]. On the other hand, the stereoisomeric Sp structures exist as stable, and identifiable R and S stereoisomers [60, 115]. All three lesions, NIm, Gh, and Sp, cause significant thermodynamic destabilization of DNA duplexes [63, 113–120], which is a common feature of DNA substrates that are recognized by the NER machinery. Thus, the Sp and Gh lesions are recognized by the prokaryotic UvrABC nuclease [107], as well as by the eukaryotic NER machinery in human cell extracts as demonstrated here. However, the NER-resistance of NIm appears to be correlated not with a destabilization, but with its conformational flexibility in double-stranded DNA [113].
7. Conclusion
The susceptibility of the oxygen free radical generated 5′,8-cyclodeoxypurine DNA lesions to repair by the nucleotide excision repair pathway, but not the base excision repair pathway, has been known for more than 15 years. Our recent work shows that other oxidatively generated DNA lesions such as spiroiminodihydantoin and guanidinohydantoin, both oxidation products of 8-oxoG, are substrates of overlapping and competing NER and BER pathways in human cell extracts. Under the same conditions, 5-guanidino-4-nitroimidazole (NIm), a lesion derived from the oxidation of guanine in DNA by peroxynitrate, is removed only by a BER pathway, but not the NER mechanism. The recent results reviewed in this article indicate that the range of oxidatively generated DNA lesions that are substrates of the nucleotide excision repair pathway may be much more extensive than previously thought. Understanding the relationships between susceptibility to NER and molecular structure of the lesion is challenging, because the thermodynamic destabilization of the DNA by the lesions is often, but not always correlated with NER activity.
Highlights.
Non-bulky guanine lesions are removed by Nucleotide Excision Repair mechanisms
DNA Intrastrand cross-linked G[C8-N3]T-thymine lesions are substrates of BER and NER
DNA hydantoin lesions are repaired by competitive BER and NER pathways
The DNA guanine lesion 5-guanidino-4-nitroimidazole is repaired by BER but not NER
Acknowledgments
This work was supported by the National Institute of Environmental Health Sciences Grants R01 ES 027059 to VS and R01 ES024050 to NEG. Components of this work were conducted in the Shared Instrumentation Facility at NYU that was constructed with support from a Research Facilities Improvement Grant (C06 RR-16572) from the National Center for Research Resources, National Institutes of Health. The acquisition of the MALDI-TOF mass spectrometer was supported by the National Science Foundation (CHE-0958457).
Abbreviations
- BER
base excision repair
- NER
nucleotide excision repair
- NIR
nucleotide incision repair
- ROS
reactive oxygen species
- RNS
reactive nitrogen species
- GG
global genomic
- TC
transcription coupled
- cdG
5′,8-cyclo-2′-deoxyguanosine
- cdA
5′,8-cyclo-2′-deoxyadenosine
- NIm
5-guanidino-4-nitroimidazole
- FapyG
2,6-diamino-4-hydroxy-5-formamidopyrimidine
- 8-oxoG
8-oxo-7,8-dihydroguanine
- G*–T*
intrastrand guanine(C8)-thymine(N3) cross-link
- Sp
spiroiminodihydantoin
- Gh
5-guanidinohydantoin
- Ia
iminoallantoin
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- 1.Coussens LM, Werb Z. Inflammation and cancer. Nature. 2002;420:860–867. doi: 10.1038/nature01322. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Grivennikov SI, Greten FR, Karin M. Immunity, inflammation, and cancer. Cell. 2010;140:883–899. doi: 10.1016/j.cell.2010.01.025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Kidane D, Chae WJ, Czochor J, Eckert KA, Glazer PM, Bothwell AL, Sweasy JB. Interplay between DNA repair and inflammation, and the link to cancer. Crit Rev Biochem Mol Biol. 2014;49:116–139. doi: 10.3109/10409238.2013.875514. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Hussain SP, Harris CC. Inflammation and cancer: an ancient link with novel potentials. Int J Cancer. 2007;121:2373–2380. doi: 10.1002/ijc.23173. [DOI] [PubMed] [Google Scholar]
- 5.Scott TL, Rangaswamy S, Wicker CA, Izumi T. Repair of oxidative DNA damage and cancer: recent progress in DNA base excision repair. Antioxid Redox Signal. 2014;20:708–726. doi: 10.1089/ars.2013.5529. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Wallace SS, Murphy DL, Sweasy JB. Base excision repair and cancer. Cancer Lett. 2012;327:73–89. doi: 10.1016/j.canlet.2011.12.038. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Lindahl T. An N-glycosidase from Escherichia coli that releases free uracil from DNA containing deaminated cytosine residues. Proc Natl Acad Sci U S A. 1974;71:3649–3653. doi: 10.1073/pnas.71.9.3649. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Mitra S, Hazra TK, Roy R, Ikeda S, Biswas T, Lock J, Boldogh I, Izumi T. Complexities of DNA base excision repair in mammalian cells. Mol Cell. 1997;7:305–312. [PubMed] [Google Scholar]
- 9.Porello SL, Leyes AE, David SS. Single-turnover and pre-steady-state kinetics of the reaction of the adenine glycosylase MutY with mismatch-containing DNA substrates. Biochemistry. 1998;37:14756–14764. doi: 10.1021/bi981594+. [DOI] [PubMed] [Google Scholar]
- 10.Doetsch PW, Cunningham RP. The enzymology of apurinic/apyrimidinic endonucleases. Mutat Res. 1990;236:173–201. doi: 10.1016/0921-8777(90)90004-o. [DOI] [PubMed] [Google Scholar]
- 11.Ide H, Kotera M. Human DNA glycosylases involved in the repair of oxidatively damaged DNA. Biol Pharm Bull. 2004;27:480–485. doi: 10.1248/bpb.27.480. [DOI] [PubMed] [Google Scholar]
- 12.Bailly V, Verly WG. Escherichia coli endonuclease III is not an endonuclease but a beta-elimination catalyst. Biochem J. 1987;242:565–572. doi: 10.1042/bj2420565. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Bailly V, Derydt M, Verly WG. Delta-elimination in the repair of AP (apurinic/apyrimidinic) sites in DNA. Biochem J. 1989;261:707–713. doi: 10.1042/bj2610707. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Chen DS, Herman T, Demple B. Two distinct human DNA diesterases that hydrolyze 3′-blocking deoxyribose fragments from oxidized DNA. Nucleic Acids Res. 1991;19:5907–5914. doi: 10.1093/nar/19.21.5907. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Wiederhold L, Leppard JB, Kedar P, Karimi-Busheri F, Rasouli-Nia A, Weinfeld M, Tomkinson AE, Izumi T, Prasad R, Wilson SH, Mitra S, Hazra TK. AP endonuclease-independent DNA base excision repair in human cells. Mol Cell. 2004;15:209–220. doi: 10.1016/j.molcel.2004.06.003. [DOI] [PubMed] [Google Scholar]
- 16.Ischenko AA, Saparbaev MK. Alternative nucleotide incision repair pathway for oxidative DNA damage. Nature. 2002;415:183–187. doi: 10.1038/415183a. [DOI] [PubMed] [Google Scholar]
- 17.Redrejo-Rodriguez M, Saint-Pierre C, Couve S, Mazouzi A, Ishchenko AA, Gasparutto D, Saparbaev M. New insights in the removal of the hydantoins, oxidation product of pyrimidines, via the base excision and nucleotide incision repair pathways. PLoS One. 2011;6:e21039. doi: 10.1371/journal.pone.0021039. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Prorok P, Alili D, Saint-Pierre C, Gasparutto D, Zharkov DO, Ishchenko AA, Tudek B, Saparbaev MK. Uracil in duplex DNA is a substrate for the nucleotide incision repair pathway in human cells. Proc Natl Acad Sci U S A. 2013;110:E3695–3703. doi: 10.1073/pnas.1305624110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Schärer OD. Achieving broad substrate specificity in damage recognition by binding accessible nondamaged DNA. Mol Cell. 2007;28:184–186. doi: 10.1016/j.molcel.2007.10.006. [DOI] [PubMed] [Google Scholar]
- 20.Marteijn JA, Lans H, Vermeulen W, Hoeijmakers JH. Understanding nucleotide excision repair and its roles in cancer and ageing. Nat Rev Mol Cell Biol. 2014;15:465–481. doi: 10.1038/nrm3822. [DOI] [PubMed] [Google Scholar]
- 21.Sancar A. Mechanisms of DNA Repair by Photolyase and Excision Nuclease (Nobel Lecture) Angew Chem Int Ed Engl. 2016 doi: 10.1002/anie.201601524. [DOI] [PubMed] [Google Scholar]
- 22.Naegeli H, Sugasawa K. The xeroderma pigmentosum pathway: decision tree analysis of DNA quality. DNA Repair (Amst) 2011;10:673–683. doi: 10.1016/j.dnarep.2011.04.019. [DOI] [PubMed] [Google Scholar]
- 23.Gillet LC, Scharer OD. Molecular mechanisms of mammalian global genome nucleotide excision repair. Chem Rev. 2006;106:253–276. doi: 10.1021/cr040483f. [DOI] [PubMed] [Google Scholar]
- 24.Hanawalt PC, Spivak G. Transcription-coupled DNA repair: two decades of progress and surprises. Nat Rev Mol Cell Biol. 2008;9:958–970. doi: 10.1038/nrm2549. [DOI] [PubMed] [Google Scholar]
- 25.Cadet J, Douki T, Ravanat JL. Oxidatively generated damage to the guanine moiety of DNA: mechanistic aspects and formation in cells. Acc Chem Res. 2008;41:1075–1083. doi: 10.1021/ar700245e. [DOI] [PubMed] [Google Scholar]
- 26.Steenken S, Jovanovic SV. How easily oxidizable is DNA? One-electron reduction potentials of adenosine and guanosine radicals in aqueous solution. J Am Chem Soc. 1997;119:617–618. [Google Scholar]
- 27.Steenken S. Purine bases, nucleosides, and nucleotides: aqueous solution redox chemistry and transformation reactions of their radical cations and e- and OH adducts. Chem Rev. 1989;89:503–520. [Google Scholar]
- 28.Nishimura S. 8-Hydroxyguanine: a base for discovery. DNA Repair (Amst) 2011;10:1078–1083. [PubMed] [Google Scholar]
- 29.Jaruga P, Dizdaroglu M. 8,5′-Cyclopurine-2′-deoxynucleosides in DNA: mechanisms of formation, measurement, repair and biological effects. DNA Repair (Amst) 2008;7:1413–1425. doi: 10.1016/j.dnarep.2008.06.005. [DOI] [PubMed] [Google Scholar]
- 30.Brooks PJ. The 8,5′-cyclopurine-2′-deoxynucleosides: candidate neurodegenerative DNA lesions in xeroderma pigmentosum, and unique probes of transcription and nucleotide excision repair. DNA Repair (Amst) 2008;7:1168–1179. doi: 10.1016/j.dnarep.2008.03.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Chatgilialoglu C, Ferreri C, Terzidis MA. Purine 5′,8-cyclonucleoside lesions: chemistry and biology. Chem Soc Rev. 2011;40:1368–1382. doi: 10.1039/c0cs00061b. [DOI] [PubMed] [Google Scholar]
- 32.Belmadoui N, Boussicault F, Guerra M, Ravanat JL, Chatgilialoglu C, Cadet J. Radiation-induced formation of purine 5′,8-cyclonucleosides in isolated and cellular DNA: high stereospecificity and modulating effect of oxygen. Org Biomol Chem. 2010;8:3211–3219. doi: 10.1039/c004531d. [DOI] [PubMed] [Google Scholar]
- 33.Dizdaroglu M, Kirkali G, Jaruga P. Formamidopyrimidines in DNA: mechanisms of formation, repair, and biological effects. Free Radic Biol Med. 2008;45:1610–1621. doi: 10.1016/j.freeradbiomed.2008.07.004. [DOI] [PubMed] [Google Scholar]
- 34.Douki T, Cadet J. Modification of DNA bases by photosensitized one-electron oxidation. Int J Radiat Biol. 1999;75:571–581. doi: 10.1080/095530099140212. [DOI] [PubMed] [Google Scholar]
- 35.Frelon S, Douki T, Ravanat JL, Pouget JP, Tornabene C, Cadet J. High-performance liquid chromatography--tandem mass spectrometry measurement of radiation-induced base damage to isolated and cellular DNA. Chem Res Toxicol. 2000;13:1002–1010. doi: 10.1021/tx000085h. [DOI] [PubMed] [Google Scholar]
- 36.Pouget JP, Frelon S, Ravanat JL, Testard I, Odin F, Cadet J. Formation of modified DNA bases in cells exposed either to gamma radiation or to high-LET particles. Radiat Res. 2002;157:589–595. doi: 10.1667/0033-7587(2002)157[0589:fomdbi]2.0.co;2. [DOI] [PubMed] [Google Scholar]
- 37.Crean C, Uvaydov Y, Geacintov NE, Shafirovich V. Oxidation of single-stranded oligonucleotides by carbonate radical anions: generating intrastrand cross-links between guanine and thymine bases separated by cytosines. Nucleic Acids Res. 2008;36:742–755. doi: 10.1093/nar/gkm1092. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Box HC, Budzinski EE, Dawidzik JD, Wallace JC, Evans MS, Gobey JS. Radiation-induced formation of a crosslink between base moieties of deoxyguanosine and thymidine in deoxygenated solutions of d(CpGpTpA) Radiat Res. 1996;145:641–643. [PubMed] [Google Scholar]
- 39.Box HC, Dawidzik JB, Budzinski EE. Free radical-induced double lesions in DNA. Free Radic Biol Med. 2001;31:856–868. doi: 10.1016/s0891-5849(01)00653-0. [DOI] [PubMed] [Google Scholar]
- 40.Wang Y. Bulky DNA lesions induced by reactive oxygen species. Chem Res Toxicol. 2008;21:276–281. doi: 10.1021/tx700411g. [DOI] [PubMed] [Google Scholar]
- 41.Cadet J, Douki T, Ravanat JL. Oxidatively generated base damage to cellular DNA. Free Radic Biol Med. 2010;49:9–21. doi: 10.1016/j.freeradbiomed.2010.03.025. [DOI] [PubMed] [Google Scholar]
- 42.Cadet J, Ravanat JL, TavernaPorro M, Menoni H, Angelov D. Oxidatively generated complex DNA damage: tandem and clustered lesions. Cancer Lett. 2012;327:5–15. doi: 10.1016/j.canlet.2012.04.005. [DOI] [PubMed] [Google Scholar]
- 43.Shafirovich V, Geacintov NE. Reactions of reactive nitrogen species and carbonate radical anions with DNA. In: Greenberg M, editor. Radical and radical ion reactivity in nucleic acid chemistry. John Willey&Sons, Inc; Hoboken, New Jersey: 2009. pp. 325–355. [Google Scholar]
- 44.Niles JC, Wishnok JS, Tannenbaum SR. A novel nitroimidazole compound formed during the reaction of peroxynitrite with 2′,3′,5′-tri-O-acetyl-guanosine. J Am Chem Soc. 2001;123:12147–12151. doi: 10.1021/ja004296k. [DOI] [PubMed] [Google Scholar]
- 45.Joffe A, Mock S, Yun BH, Kolbanovskiy A, Geacintov NE, Shafirovich V. Oxidative generation of guanine radicals by carbonate radicals and their reactions with nitrogen dioxide to form site specific 5-guanidino-4-nitroimidazole lesions in oligodeoxynucleotides. Chem Res Toxicol. 2003;16:966–973. doi: 10.1021/tx025578w. [DOI] [PubMed] [Google Scholar]
- 46.Gu F, Stillwell WG, Wishnok JS, Shallop AJ, Jones RA, Tannenbaum SR. Peroxynitrite-induced reactions of synthetic oligo 2′-deoxynucleotides and DNA containing guanine: Formation and stability of a 5-guanidino-4-nitroimidazole lesion. Biochemistry. 2002;41:7508–7518. doi: 10.1021/bi020148q. [DOI] [PubMed] [Google Scholar]
- 47.Yun BH, Geacintov NE, Shafirovich V. Generation of guanine-thymidine cross-links in DNA by peroxynitrite/carbon dioxide. Chem Res Toxicol. 2011;24:1144–1152. doi: 10.1021/tx200139c. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Lonkar P, Dedon PC. Reactive species and DNA damage in chronic inflammation: reconciling chemical mechanisms and biological fates. Int J Cancer. 2011;128:1999–2009. doi: 10.1002/ijc.25815. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Steenken S, Jovanovic SV, Bietti M, Bernhard K. The trap depth (in DNA) of 8-oxo-7,8-dihydro-2′deoxyguanosine as derived from electron-transfer equilibria in aqueous solution. J Am Chem Soc. 2000;122:2373–2374. [Google Scholar]
- 50.Joffe A, Geacintov NE, Shafirovich V. DNA lesions derived from the site-selective oxidation of guanine by carbonate radical anions. Chem Res Toxicol. 2003;16:1528–1538. doi: 10.1021/tx034142t. [DOI] [PubMed] [Google Scholar]
- 51.Misiaszek R, Crean C, Geacintov NE, Shafirovich V. Combination of nitrogen dioxide radicals with 8-oxo-7,8-dihydroguanine and guanine radicals in DNA: Oxidation and nitration end-products. J Am Chem Soc. 2005;127:2191–2200. doi: 10.1021/ja044390r. [DOI] [PubMed] [Google Scholar]
- 52.Shao J, Geacintov NE, Shafirovich V. Oxidation of 8-oxo-7,8-dihydro-2′-deoxyguanosine by oxyl radicals produced by photolysis of azo compounds. Chem Res Toxicol. 2010;23:933–938. doi: 10.1021/tx100022x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Rokhlenko Y, Geacintov NE, Shafirovich V. Lifetimes and reaction pathways of guanine radical cations and neutral guanine radicals in an oligonucleotide in aqueous solutions. J Am Chem Soc. 2012;134:4955–4962. doi: 10.1021/ja212186w. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Luo W, Muller JG, Rachlin EM, Burrows CJ. Characterization of spiroiminodihydantoin as a product of one-electron oxidation of 8-oxo-7,8-dihydroguanosine. Org Lett. 2000;2:613–616. doi: 10.1021/ol9913643. [DOI] [PubMed] [Google Scholar]
- 55.Luo W, Muller JG, Rachlin EM, Burrows CJ. Characterization of hydantoin products from one-electron oxidation of 8- oxo-7,8-dihydroguanosine in a nucleoside model. Chem Res Toxicol. 2001;14:927–938. doi: 10.1021/tx010072j. [DOI] [PubMed] [Google Scholar]
- 56.Niles JC, Wishnok JS, Tannenbaum SR. Spiroiminodihydantoin is the major product of the 8-oxo-7,8-dihydroguanosine reaction with peroxynitrite in the presence of thiols and guanosine photooxidation by methylene Blue. Org Lett. 2001;3:963–966. [PubMed] [Google Scholar]
- 57.Sugden KD, Campo CK, Martin BD. Direct oxidation of guanine and 7,8-dihydro-8-oxoguanine in DNA by a high-valent chromium complex: a possible mechanism for chromate genotoxicity. Chem Res Toxicol. 2001;14:1315–1322. doi: 10.1021/tx010088+. [DOI] [PubMed] [Google Scholar]
- 58.Burrows CJ, Muller JG, Kornyushyna O, Luo W, Duarte V, Leipold MD, David SS. Structure and potential mutagenicity of new hydantoin products from guanosine and 8-oxo-7,8-dihydroguanine oxidation by transition metals. Environ Health Perspect 110 Suppl. 2002;5:713–717. doi: 10.1289/ehp.02110s5713. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Fleming AM, Muller JG, Dlouhy AC, Burrows CJ. Structural context effects in the oxidation of 8-oxo-7,8-dihydro-2′-deoxyguanosine to hydantoin products: electrostatics, base stacking, and base pairing. J Am Chem Soc. 2012;134:15091–15102. doi: 10.1021/ja306077b. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Fleming AM, Orendt AM, He Y, Zhu J, Dukor RK, Burrows CJ. Reconciliation of chemical, enzymatic, spectroscopic and computational data to assign the absolute configuration of the DNA base lesion spiroiminodihydantoin. J Am Chem Soc. 2013;135:18191–18204. doi: 10.1021/ja409254z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Cui L, Ye W, Prestwich EG, Wishnok JS, Taghizadeh K, Dedon PC, Tannenbaum SR. Comparative analysis of four oxidized guanine lesions from reactions of DNA with peroxynitrite, singlet oxygen, and gamma-radiation. Chem Res Toxicol. 2013;26:195–202. doi: 10.1021/tx300294d. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Mangerich A, Knutson CG, Parry NM, Muthupalani S, Ye W, Prestwich E, Cui L, McFaline JL, Mobley M, Ge Z, Taghizadeh K, Wishnok JS, Wogan GN, Fox JG, Tannenbaum SR, Dedon PC. Infection-induced colitis in mice causes dynamic and tissue-specific changes in stress response and DNA damage leading to colon cancer. Proc Natl Acad Sci U S A. 2012;109:E1820–E1829. doi: 10.1073/pnas.1207829109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Kornyushyna O, Berges AM, Muller JG, Burrows CJ. In vitro nucleotide misinsertion opposite the oxidized guanosine lesions spiroiminodihydantoin and guanidinohydantoin and DNA synthesis past the lesions using Escherichia coli DNA polymerase I (Klenow fragment) Biochemistry. 2002;41:15304–15314. doi: 10.1021/bi0264925. [DOI] [PubMed] [Google Scholar]
- 64.Zhu J, Fleming AM, Orendt AM, Burrows CJ. pH-Dependent Equilibrium between 5-Guanidinohydantoin and Iminoallantoin Affects Nucleotide Insertion Opposite the DNA Lesion. J Org Chem. 2016;81:351–359. doi: 10.1021/acs.joc.5b02180. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Wallace SS. Base excision repair: a critical player in many games. DNA Repair (Amst) 2014;19:14–26. doi: 10.1016/j.dnarep.2014.03.030. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Wallace SS. DNA glycosylases search for and remove oxidized DNA bases. Environ Mol Mutagen. 2013;54:691–704. doi: 10.1002/em.21820. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.David SS, O’Shea VL, Kundu S. Base-excision repair of oxidative DNA damage. Nature. 2007;447:941–950. doi: 10.1038/nature05978. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Hsu GW, Ober M, Carell T, Beese LS. Error-prone replication of oxidatively damaged DNA by a high-fidelity DNA polymerase. Nature. 2004;431:217–221. doi: 10.1038/nature02908. [DOI] [PubMed] [Google Scholar]
- 69.Takao M, Zhang QM, Yonei S, Yasui A. Differential subcellular localization of human MutY homolog (hMYH) and the functional activity of adenine:8-oxoguanine DNA glycosylase. Nucleic Acids Res. 1999;27:3638–3644. doi: 10.1093/nar/27.18.3638. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Duclos S, Aller P, Jaruga P, Dizdaroglu M, Wallace SS, Doublie S. Structural and biochemical studies of a plant formamidopyrimidine-DNA glycosylase reveal why eukaryotic Fpg glycosylases do not excise 8-oxoguanine. DNA Repair (Amst) 2012;11:714–725. doi: 10.1016/j.dnarep.2012.06.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Krishnamurthy N, Zhao X, Burrows CJ, David SS. Superior removal of hydantoin lesions relative to other oxidized bases by the human DNA glycosylase hNEIL1. Biochemistry. 2008;47:7137–7146. doi: 10.1021/bi800160s. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Zhao X, Krishnamurthy N, Burrows CJ, David SS. Mutation versus repair: NEIL1 removal of hydantoin lesions in single-stranded, bulge, bubble, and duplex DNA contexts. Biochemistry. 2010;49:1658–1666. doi: 10.1021/bi901852q. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Bandaru V, Sunkara S, Wallace SS, Bond JP. A novel human DNA glycosylase that removes oxidative DNA damage and is homologous to Escherichia coli endonuclease VIII. DNA Repair (Amst) 2002;1:517–529. doi: 10.1016/s1568-7864(02)00036-8. [DOI] [PubMed] [Google Scholar]
- 74.Hazra TK, Kow YW, Hatahet Z, Imhoff B, Boldogh I, Mokkapati SK, Mitra S, Izumi T. Identification and characterization of a novel human DNA glycosylase for repair of cytosine-derived lesions. J Biol Chem. 2002;277:30417–30420. doi: 10.1074/jbc.C200355200. [DOI] [PubMed] [Google Scholar]
- 75.Takao M, Kanno S, Kobayashi K, Zhang QM, Yonei S, van der Horst GT, Yasui A. A back-up glycosylase in Nth1 knock-out mice is a functional Nei (endonuclease VIII) homologue. J Biol Chem. 2002;277:42205–42213. doi: 10.1074/jbc.M206884200. [DOI] [PubMed] [Google Scholar]
- 76.Jaruga P, Birincioglu M, Rosenquist TA, Dizdaroglu M. Mouse NEIL1 protein is specific for excision of 2,6-diamino-4-hydroxy-5-formamidopyrimidine and 4,6-diamino-5-formamidopyrimidine from oxidatively damaged DNA. Biochemistry. 2004;43:15909–15914. doi: 10.1021/bi048162l. [DOI] [PubMed] [Google Scholar]
- 77.Dou H, Mitra S, Hazra TK. Repair of oxidized bases in DNA bubble structures by human DNA glycosylases NEIL1 and NEIL2. J Biol Chem. 2003;278:49679–49684. doi: 10.1074/jbc.M308658200. [DOI] [PubMed] [Google Scholar]
- 78.Hailer MK, Slade PG, Martin BD, Rosenquist TA, Sugden KD. Recognition of the oxidized lesions spiroiminodihydantoin and guanidinohydantoin in DNA by the mammalian base excision repair glycosylases NEIL1 and NEIL2. DNA Repair (Amst) 2005;4:41–50. doi: 10.1016/j.dnarep.2004.07.006. [DOI] [PubMed] [Google Scholar]
- 79.Liu M, Bandaru V, Bond JP, Jaruga P, Zhao X, Christov PP, Burrows CJ, Rizzo CJ, Dizdaroglu M, Wallace SS. The mouse ortholog of NEIL3 is a functional DNA glycosylase in vitro and in vivo. Proc Natl Acad Sci U S A. 2010;107:4925–4930. doi: 10.1073/pnas.0908307107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Krokeide SZ, Laerdahl JK, Salah M, Luna L, Cederkvist FH, Fleming AM, Burrows CJ, Dalhus B, Bjoras M. Human NEIL3 is mainly a monofunctional DNA glycosylase removing spiroimindiohydantoin and guanidinohydantoin. DNA Repair (Amst) 2013;12:1159–1164. doi: 10.1016/j.dnarep.2013.04.026. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Theruvathu JA, Jaruga P, Dizdaroglu M, Brooks PJ. The oxidatively induced DNA lesions 8,5′-cyclo-2′-deoxyadenosine and 8-hydroxy-2′-deoxyadenosine are strongly resistant to acid-induced hydrolysis of the glycosidic bond. Mech Ageing Dev. 2007;128:494–502. doi: 10.1016/j.mad.2007.06.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Das RS, Samaraweera M, Morton M, Gascon JA, Basu AK. Stability of N-glycosidic bond of (5′S)-8,5′-cyclo-2′-deoxyguanosine. Chem Res Toxicol. 2012;25:2451–2461. doi: 10.1021/tx300302a. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.Kuraoka I, Bender C, Romieu A, Cadet J, Wood RD, Lindahl T. Removal of oxygen free-radical-induced 5′,8-purine cyclodeoxynucleosides from DNA by the nucleotide excision-repair pathway in human cells. Proc Natl Acad Sci U S A. 2000;97:3832–3837. doi: 10.1073/pnas.070471597. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84.Brooks PJ, Wise DS, Berry DA, Kosmoski JV, Smerdon MJ, Somers RL, Mackie H, Spoonde AY, Ackerman EJ, Coleman K, Tarone RE, Robbins JH. The oxidative DNA lesion 8,5′-(S)-cyclo-2′-deoxyadenosine is repaired by the nucleotide excision repair pathway and blocks gene expression in mammalian cells. J Biol Chem. 2000;275:22355–22362. doi: 10.1074/jbc.M002259200. [DOI] [PubMed] [Google Scholar]
- 85.Pande P, Das RS, Sheppard C, Kow YW, Basu AK. Repair efficiency of (5′S)-8,5′-cyclo-2′-deoxyguanosine and (5′S)-8,5′-cyclo-2′-deoxyadenosine depends on the complementary base. DNA Repair (Amst) 2012;11:926–931. doi: 10.1016/j.dnarep.2012.09.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86.Kropachev K, Ding S, Terzidis MA, Masi A, Liu Z, Cai Y, Kolbanovskiy M, Chatgilialoglu C, Broyde S, Geacintov NE, Shafirovich V. Structural basis for the recognition of diastereomeric 5′,8-cyclo-2′-deoxypurine lesions by the human nucleotide excision repair system. Nucleic Acids Res. 2014;42:5020–5032. doi: 10.1093/nar/gku162. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87.Jasti VP, Das RS, Hilton BA, Weerasooriya S, Zou Y, Basu AK. (5′S)-8,5′-cyclo-2′-deoxyguanosine is a strong block to replication, a potent pol V-dependent mutagenic lesion, and is inefficiently repaired in Escherichia coli. Biochemistry. 2011;50:3862–3865. doi: 10.1021/bi2004944. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88.Mocquet V, Kropachev K, Kolbanovskiy M, Kolbanovskiy A, Tapias A, Cai Y, Broyde S, Geacintov NE, Egly JM. The human DNA repair factor XPC-HR23B distinguishes stereoisomeric benzo[a]pyrenyl-DNA lesions. EMBO J. 2007;26:2923–2932. doi: 10.1038/sj.emboj.7601730. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 89.Kropachev K, Kolbanovskii M, Cai Y, Rodriguez F, Kolbanovskii A, Liu Y, Zhang L, Amin S, Patel D, Broyde S, Geacintov NE. The sequence dependence of human nucleotide excision repair efficiencies of benzo[a]pyrene-derived DNA lesions: insights into the structural factors that favor dual incisions. J Mol Biol. 2009;386:1193–1203. doi: 10.1016/j.jmb.2008.12.082. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 90.Cai Y, Patel DJ, Geacintov NE, Broyde S. Differential nucleotide excision repair susceptibility of bulky DNA adducts in different sequence contexts: hierarchies of recognition signals. J Mol Biol. 2009;385:30–44. doi: 10.1016/j.jmb.2008.09.087. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 91.Jung Y, Lippard SJ. Direct cellular responses to platinum-induced DNA damage. Chem Rev. 2007;107:1387–1407. doi: 10.1021/cr068207j. [DOI] [PubMed] [Google Scholar]
- 92.Clauson C, Scharer OD, Niedernhofer L. Advances in understanding the complex mechanisms of DNA interstrand cross-link repair. Cold Spring Harb Perspect Biol. 2013;5:a012732. doi: 10.1101/cshperspect.a012732. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 93.Friedberg EC. DNA damage and repair. Nature. 2003;421:436–440. doi: 10.1038/nature01408. [DOI] [PubMed] [Google Scholar]
- 94.Zhang Q, Wang Y. Independent generation of 5-(2′-deoxycytidinyl)methyl radical and the formation of a novel cross-link lesion between 5-methylcytosine and guanine. J Am Chem Soc. 2003;125:12795–12802. doi: 10.1021/ja034866r. [DOI] [PubMed] [Google Scholar]
- 95.Yang Z, Colis LC, Basu AK, Zou Y. Recognition and incision of g-radiation-induced cross-linked guanine-thymine tandem lesion G[8,5-Me]T by UvrABC nuclease. Chem Res Toxicol. 2005;18:1339–1346. doi: 10.1021/tx050147+. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 96.Gu C, Zhang Q, Yang Z, Wang Y, Zou Y, Wang Y. Recognition and incision of oxidative intrastrand cross-link lesions by UvrABC nuclease. Biochemistry. 2006;45:10739–10746. doi: 10.1021/bi060423z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 97.Raychaudhury P, Basu AK. Genetic requirement for mutagenesis of the G[8,5-Me]T cross-link in Escherichia coli: DNA polymerases IV and V compete for error-prone bypass. Biochemistry. 2011;50:2330–2338. doi: 10.1021/bi102064z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 98.Wang J, Cao H, You C, Yuan B, Bahde R, Gupta S, Nishigori C, Niedernhofer LJ, Brooks PJ, Wang Y. Endogenous formation and repair of oxidatively induced G[8–5 m]T intrastrand cross-link lesion. Nucleic Acids Res. 2012;40:7368–7374. doi: 10.1093/nar/gks357. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 99.Madugundu GS, Wagner JR, Cadet J, Kropachev K, Yun BH, Geacintov NE, Shafirovich V. Generation of guanine-thymine cross-links in human cells by one-electron oxidation mechanisms. Chem Res Toxicol. 2013;26:1031–1033. doi: 10.1021/tx400158g. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 100.Ding S, Kropachev K, Cai Y, Kolbanovskiy M, Durandina SA, Liu Z, Shafirovich V, Broyde S, Geacintov NE. Structural, energetic and dynamic properties of guanine(C8)-thymine(N3) cross-links in DNA provide insights on susceptibility to nucleotide excision repair. Nucleic Acids Res. 2012;40:2506–2517. doi: 10.1093/nar/gkr1087. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 101.Talhaoui I, Shafirovich V, Liu Z, Saint-Pierre C, Akishev Z, Matkarimov BT, Gasparutto D, Geacintov NE, Saparbaev M. Oxidatively Generated Guanine(C8)-Thymine(N3) Intrastrand Cross-links in Double-stranded DNA Are Repaired by Base Excision Repair Pathways. J Biol Chem. 2015;290:14610–14617. doi: 10.1074/jbc.M115.647487. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 102.Leipold MD, Muller JG, Burrows CJ, David SS. Removal of hydantoin products of 8-oxoguanine oxidation by the escherichia coli DNA repair enzyme, FPG. Biochemistry. 2000;39:14984–14992. doi: 10.1021/bi0017982. [DOI] [PubMed] [Google Scholar]
- 103.Hazra TK, Muller JG, Manuel RC, Burrows CJ, Lloyd RS, Mitra S. Repair of hydantoins, one electron oxidation product of 8-oxoguanine, by DNA glycosylases of Escherichia coli. Nucleic Acids Res. 2001;29:1967–1974. doi: 10.1093/nar/29.9.1967. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 104.Sejersted Y, Hildrestrand GA, Kunke D, Rolseth V, Krokeide SZ, Neurauter CG, Suganthan R, Atneosen-Asegg M, Fleming AM, Saugstad OD, Burrows CJ, Luna L, Bjoras M. Endonuclease VIII-like 3 (Neil3) DNA glycosylase promotes neurogenesis induced by hypoxia-ischemia. Proc Natl Acad Sci U S A. 2011;108:18802–18807. doi: 10.1073/pnas.1106880108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 105.Liu M, Imamura K, Averill AM, Wallace SS, Doublie S. Structural characterization of a mouse ortholog of human NEIL3 with a marked preference for single-stranded DNA. Structure. 2013;21:247–256. doi: 10.1016/j.str.2012.12.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 106.Rolseth V, Krokeide SZ, Kunke D, Neurauter CG, Suganthan R, Sejersted Y, Hildrestrand GA, Bjoras M, Luna L. Loss of Neil3, the major DNA glycosylase activity for removal of hydantoins in single stranded DNA, reduces cellular proliferation and sensitizes cells to genotoxic stress. Biochim Biophys Acta. 2013;1833:1157–1164. doi: 10.1016/j.bbamcr.2012.12.024. [DOI] [PubMed] [Google Scholar]
- 107.McKibbin PL, Fleming AM, Towheed MA, Van Houten B, Burrows CJ, David SS. Repair of hydantoin lesions and their amine adducts in DNA by base and nucleotide excision repair. J Am Chem Soc. 2013;135:13851–13861. doi: 10.1021/ja4059469. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 108.Shafirovich V, Kropachev K, Anderson T, Liu Z, Kolbanovskiy M, Martin BD, Sugden K, Shim Y, Chen X, Min JH, Geacintov NE. Base and Nucleotide Excision Repair of Oxidatively Generated Guanine Lesions in DNA. J Biol Chem. 2016;291:5309–5319. doi: 10.1074/jbc.M115.693218. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 109.Hess MT, Gunz D, Luneva N, Geacintov NE, Naegeli H. Base pair conformation-dependent excision of benzo[a]pyrene diol epoxide-guanine adducts by human nucleotide excision repair enzymes. Mol Cell Biol. 1997;17:7069–7076. doi: 10.1128/mcb.17.12.7069. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 110.Miura N, Miyamoto I, Asahina H, Satokata I, Tanaka K, Okada Y. Identification and characterization of xpac protein, the gene product of the human XPAC (xeroderma pigmentosum group A complementing) gene. J Biol Chem. 1991;266:19786–19789. [PubMed] [Google Scholar]
- 111.Saijo M, Matsuda T, Kuraoka I, Tanaka K. Inhibition of nucleotide excision repair by anti-XPA monoclonal antibodies which interfere with binding to RPA, ERCC1, and TFIIH. Biochem Biophys Res Commun. 2004;321:815–822. doi: 10.1016/j.bbrc.2004.07.030. [DOI] [PubMed] [Google Scholar]
- 112.Shen JC, Fox EJ, Ahn EH, Loeb LA. A rapid assay for measuring nucleotide excision repair by oligonucleotide retrieval. Sci Rep. 2014;4:4894. doi: 10.1038/srep04894. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 113.Jia L, Shafirovich V, Shapiro R, Geacintov NE, Broyde S. Flexible 5-guanidino-4-nitroimidazole DNA lesions: Structures and thermodynamics. Biochemistry. 2006;45:6644–6655. doi: 10.1021/bi0601757. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 114.Aller P, Ye Y, Wallace SS, Burrows CJ, Doublie S. Crystal structure of a replicative DNA polymerase bound to the oxidized guanine lesion guanidinohydantoin. Biochemistry. 2010;49:2502–2509. doi: 10.1021/bi902195p. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 115.Eckenroth BE, Fleming AM, Sweasy JB, Burrows CJ, Doublie S. Crystal Structure of DNA Polymerase beta with DNA Containing the Base Lesion Spiroiminodihydantoin in a Templating Position. Biochemistry. 2014;53:2075–2077. doi: 10.1021/bi500270e. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 116.Khutsishvili I, Zhang N, Marky LA, Crean C, Patel DJ, Geacintov NE, Shafirovich V. Thermodynamic profiles and nuclear magnetic resonance studies of oligonucleotide duplexes containing single diastereomeric spiroiminodihydantoin lesions. Biochemistry. 2013;52:1354–1363. doi: 10.1021/bi301566v. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 117.Jia L, Shafirovich V, Shapiro R, Geacintov NE, Broyde S. Structural and thermodynamic features of spiroiminodihydantoin damaged DNA duplexes. Biochemistry. 2005;44:13342–13353. doi: 10.1021/bi050790v. [DOI] [PubMed] [Google Scholar]
- 118.Yennie CJ, Delaney S. Thermodynamic consequences of the hyperoxidized guanine lesion guanidinohydantoin in duplex DNA. Chem Res Toxicol. 2012;25:1732–1739. doi: 10.1021/tx300190a. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 119.Jin Q, Fleming AM, Ding Y, Burrows CJ, White HS. Structural destabilization of DNA duplexes containing single-base lesions investigated by nanopore measurements. Biochemistry. 2013;52:7870–7877. doi: 10.1021/bi4009825. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 120.Neeley WL, Henderson PT, Essigmann JM. Efficient synthesis of DNA containing the guanine oxidation-nitration product 5-guanidino-4-nitroimidazole: generation by a postsynthetic substitution reaction. Org Lett. 2004;6:245–248. doi: 10.1021/ol036188j. [DOI] [PubMed] [Google Scholar]